Abstract
Integrative and conjugative elements (ICEs) are mobile genetic elements that transfer from cell to cell by conjugation (like plasmids) and integrate into the chromosomes of bacterial hosts (like lysogenic phages or transposons). ICEs are prevalent in bacterial chromosomes and play a major role in bacterial evolution by promoting horizontal gene transfer. Exclusion prevents redundant transfer of conjugative elements into host cells that already contain a copy of the element. Exclusion has been characterized mostly for conjugative elements of Gram-negative bacteria. Here, we report the identification and characterization of an exclusion mechanism in ICEBs1 from the Gram-positive bacterium Bacillus subtilis. We found that cells containing ICEBs1 inhibit the activity of the ICEBs1-encoded conjugation machinery in other cells. This inhibition (exclusion) was specific to the cognate conjugation machinery and the ICEBs1 gene yddJ was both necessary and sufficient to mediate exclusion by recipient cells. Through a mutagenesis and enrichment screen, we identified exclusion-resistant mutations in the ICEBs1 gene conG. Using genes from a heterologous but related ICE, we found that exclusion specificity was determined by ConG and YddJ. Finally, we found that under conditions that support conjugation, exclusion provides a selective advantage to the element and its host cells.
Abbreviated Summary
Type IV secretion systems encoded by plasmids and integrative and conjugative elements (ICEs) mediate transfer of DNA from donor to recipient cells, enabling bacteria to acquire new traits, including antibiotic resistances. Exclusion systems in conjugative elements inhibit the activity of the cognate conjugation machinery. We identified and characterized an exclusion system in ICEBs1 of Bacillus subtilis: the exclusion protein YddJ targets ConG (VirB6 homolog) in its cognate conjugation machinery inhibiting conjugative DNA transfer and providing a selective advantage to the element and its host cells.
INTRODUCTION
Integrative and conjugative elements (ICEs), also known as conjugative transposons, are self-transmissible genetic elements whose life cycle combines characteristics of lysogenic phages or transposons and conjugative plasmids. ICEs integrate into the host chromosome and are passively replicated with the rest of the genome (like some lysogenic phages and transposons), and under specific conditions, or stochastically, excise and transfer to other cells via self-encoded conjugation machinery (like conjugative plasmids) (Johnson & Grossman, 2015). ICEs are remarkably prevalent and thought to outnumber conjugative plasmids across major bacterial clades (Guglielmini et al., 2011). ICEs contribute to bacterial evolution by enabling the acquisition of various phenotypes by horizontal gene transfer. These phenotypes are conferred to host cells by so-called ‘cargo genes’ that are within an ICE but that are not required for the ICE life cycle. Some of the phenotypes conferred by ICEs include antibiotic resistances, pathogenicity, symbiosis, and metabolic capabilities (reviewed in Burrus & Waldor, 2004; Johnson & Grossman, 2015). In addition, many ICEs can mobilize other genetic elements, typically plasmids, that do not encode their own transfer machinery (e.g., Naglich & Andrews, 1988; Valentine et al., 1988; Hochhut et al., 2000; Lee et al, 2012).
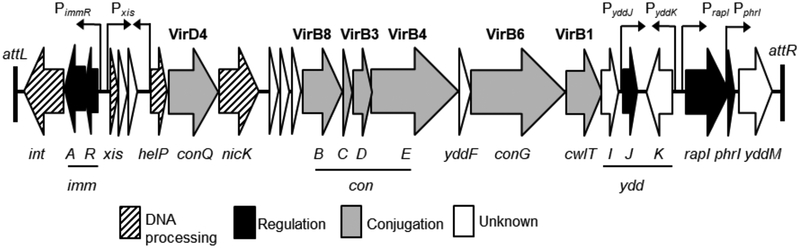
ICEBs1 (Fig. 1) is approximately 20 kb and found in the chromosome of various isolates of the Gram-positive bacterium Bacillus subtilis (Burrus et al., 2002; Auchtung et al., 2005; Earl et al., 2007). ICEBs1 integrates into a specific site in trnS-leu2 (encoding a leucyl-tRNA) in the host chromosome. Integration does not disrupt the tRNA gene. When stably integrated, most of the ICEBs1 genes are repressed. Multiple mechanisms control induction (de-repression) and transfer of ICEBs1. The ICEBs1-encoded RapI-PhrI signaling system controls induction by sensing cues about growth phase, the concentration of potential recipients, and whether or not the neighboring cells already contain a copy of ICEBs1 (Auchtung et al., 2005). RecA also controls induction of ICEBs1 in response to DNA damage, independently of the RapI-PhrI signaling system (Auchtung et al., 2005). Both the RapI and RecA pathways cause the ICEBs1 repressor ImmR to be cleaved by the protease and anti-repressor ImmA (Bose et al., 2008).
Fig.1. Genetic map of ICEBs1.
Organization of ICEBs1 open reading frames, indicated by horizontal arrows pointing in the direction of transcription, with the name of the gene indicated below. The color and patterns of each arrow indicates the gene’s function as DNA processing (diagonal stripes), regulation (black), conjugation (gray), and unknown (white). Conjugation genes encoding proteins homologous/analogous to the VirB/D type IV secretion system are indicated by the corresponding protein names in bold above the arrows. The positions of the promoters for immR, xis, yddJ, yddK, rapI, phrI, and an uncharacterized small antisense RNA are indicated by vertical arrows with the arrow head pointing in the direction of transcription. Black boxes indicate the 60 bp repeats marking the ends of the element (Auchtung et al., 2016).
The rapI-phrI signaling system and ImmR both help to limit acquisition of ICEBs1 by a cells that already contain a copy of the element. PhrI-mediated inhibition of RapI activity inhibits SOS-independent de-repression of ICEBs1 gene expression (Auchtung et al., 2005), thereby inhibiting the earliest step in activation of the element in potential donor cells. ImmR mediates a phage-like immunity that inhibits integration of additional copies of ICEBs1 that enter a host cell that already contains the element (Auchtung et al., 2007). Thus, if ICEBs1 is activated in some cells and actually does transfer to a recipient that already contains a copy, then integration of the newly acquired element is inhibited by ImmR-mediated immunity (Auchtung et al., 2007). Similar to repressor-mediated immunity in some lysogenic phages, ImmR-mediated immunity is bypassed by overproduction of the ICEBs1 integrase Int (Auchtung et al., 2007). Thus, these two mechanisms that inhibit acquisition of a second copy of ICEBs1 work at the first and last steps in acquisition.
The conjugation machinery encoded by ICEBs1 is a type IV secretion system (T4SS) that resembles other well characterized T4SSs encoded by conjugative elements found in Gram-positive bacteria, such as pCW3, pIP501, and pCF10 (Goessweiner-Mohr et al., 2014; Guglielmini, et al., 2014; Leonetti et al., 2015). The T4SS of ICEBs1 is classified as a member of the MPFFA class of T4SSs, which are found in Firmicutes and Actinobacteria and do not contain dedicated adhesion proteins (Guglielmini et al., 2014). T4SSs involved in conjugation transfer ssDNA that is attached to a relaxase protein from a donor cell to a recipient cell when the two cells are in direct contact (Bhatty et al., 2013; Goessweiner-Mohr et al., 2014; Grohmann et al., 2018). All T4SSs appear to share a conserved set of proteins (Bhatty et al., 2013; Guglielmini, et al., 2014). Using nomenclature from the Agrobacterium tumefaciens Ti plasmid, six of these proteins and the ICEBs1 counterparts are, respectively: VirB1/CwlT (cell wall hydrolase), VirD4/ConQ (the coupling protein), VirB4/ConE (an ATPase), and VirB3/ConD, VirB6/ConG, and VirB8/ConB (three membrane channel components) (Fig. 1).
Exclusion is a mechanism encoded by conjugative elements to prevent redundant transfer; i.e., the entry of an identical or highly similar element into a cell that already contains a copy of the element (Garcillan-Barcia & de la Cruz, 2008). Exclusion systems (also called entry exclusion) have been described for many of the major conjugative plasmid groups. They prevent transfer of DNA by the cognate conjugation machinery from a cell that has the plasmid to another cell that also has the plasmid. The general mechanism for exclusion in Gram-negative bacteria involves an exclusion protein present in the inner membrane of the “recipient” cell that already contains the element. The exclusion protein recognizes a component of the cognate conjugation machinery in a would-be donor, and through means yet unknown, prevents successful transfer of DNA. To date, the exclusion protein and its target in the conjugation machinery have been identified for three different conjugative systems, all from elements found in Gram-negative bacteria: 1) the F/R100 plasmids (Anthony et al., 1999; Audette et al., 2007); 2) the SXT/R391 ICEs (Marrero et al., 2005); and 3) the R64/R621a plasmids (Sakuma et al., 2013).
Selective advantages for elements with exclusion have been proposed and are mostly theoretical. For example, mathematical modeling of competition between incompatible conjugative plasmids with and without exclusion predicts an exclusion-competent plasmid can penetrate a cell population containing an exclusion-less plasmid, potentially expelling it from the population (van der Hoeven, 1985). Experimental evidence for the advantage of exclusion has come from studies of the F plasmid in E. coli, where it has been shown that exclusion prevents lethal zygosis, a phenomenon where F- recipient cells are killed in the presence of excess Hfr or F+ donors that lack the exclusion functions (Skurray et al., 1973, 1974, 1976; Ou, 1980).
In this study, we describe an exclusion mechanism in ICEBs1 that specifically inhibits its cognate conjugation machinery. We identified the ICEBs1 gene yddJ as necessary and sufficient for exclusion in the recipient cell. Through a mutagenesis and enrichment screen, we identified exclusion-resistant mutations in the ICEBs1 gene conG, an essential component of the conjugation machinery. Using homologs of conG and yddJ from a heterologous ICE, we found that conG and yddJ determine specificity of exclusion. We provide evidence that exclusion protects ICEBs1 and its host cell from cell death caused by redundant transfer, providing experimental evidence that there is selective pressure to maintain exclusion, at least for some conjugative elements.
RESULTS
Rationale
Cells containing the mobile element ICEBs1 have multiple element-encoded mechanisms for limiting acquisition of a second copy of ICEBs1. One mechanism involves RapI-PhrI mediated cell-cell signaling (Auchtung et al., 2005). A second mechanism is element-encoded immunity, analogous to phage immunity, and is mediated by the ICEBs1 repressor ImmR (Auchtung et al., 2007). In the course of analyzing ImmR-mediated immunity, it seemed that there was at least one additional mechanism by which cells that contain ICEBs1 limit acquisition of another copy of the element (Auchtung et al., 2007). To uncover and analyze this third mechanism, we used conditions and assays that bypassed the effects of cell-cell signaling and repressor-mediated immunity. Cell-cell signaling was bypassed by inducing ICEBs1 by over-production of the activator RapI (Auchtung et al., 2005). ImmR-mediated immunity was bypassed by assaying transfer of a mobilizable plasmid (Lee et al., 2012) that does not need to integrate into the chromosome of the transconjugant and is not affected by ICEBs1-mediated immunity. Results described below demonstrate that ICEBs1 has a mechanism of exclusion that functions to inhibit transfer of DNA through the ICEBs1-encoded conjugation machinery.
The presence of ICEBs1 in recipient cells inhibits acquisition of a plasmid mobilized by the ICEBs1 conjugation machinery
We used a plasmid mobilization assay to monitor the efficiency of conjugation through the ICEBs1-encoded type IV secretion system. The conjugation machinery encoded by ICEBs1 can mobilize at least three different rolling circle replicating (RCR) plasmids that do not encode their own conjugation machinery (Lee et al., 2012). We measured the ability of recipient cells with or without ICEBs1 (strain CAL88 or CAL89, respectively) to obtain the RCR plasmid pC194 from donor cells via the ICEBs1-encoded conjugation machinery. Donor cells containing ICE and pC194 (ICEBs1 Δ(rapI-phrI)342::kan, Pxyl-rapI, pC194, StrS; MA116) were grown in defined minimal medium to mid-exponential phase in presence of chloramphenicol to select for pC194. Pxyl-rapI expression was induced with xylose, thereby causing induction of ICEBs1, and these donor cells were mixed with recipients and placed on filters to allow efficient mating (Materials and Methods). The transfer efficiency of pC194 into recipients was calculated as the number of transconjugants (determined by CmR StrR CFUs) per initial number of donors at the time of cell mixing, converted to percent transconjugants per donor (Materials and Methods).
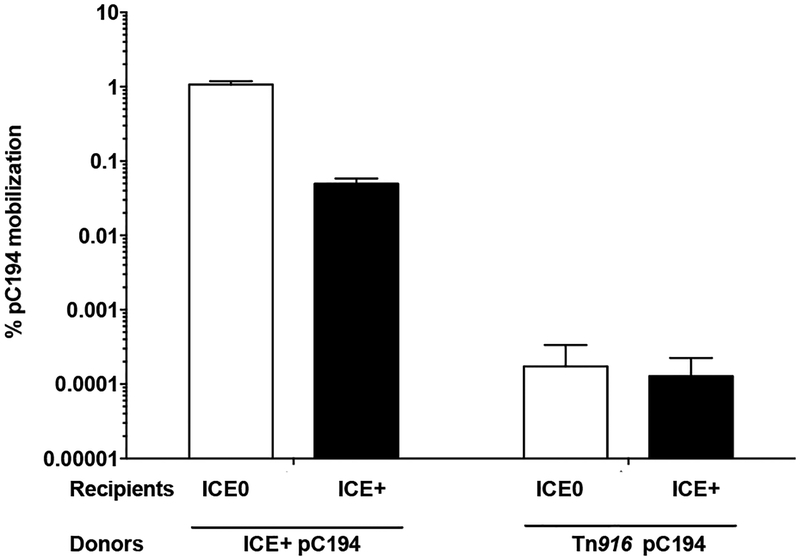
The transfer efficiency of pC194 into recipient cells that did not contain ICEBs1 was ~1% transconjugants (containing pC194) per donor (Fig. 2), similar to previous findings (Lee et al., 2012). In contrast, the efficiency of acquisition of pC194 by recipients that contained ICEBs1 was ~0.05% transconjugants per donor (Fig. 2), a reduction of ~20-fold. Transfer of the plasmid from donor to recipient by transformation with free or released plasmid was unlikely because recipient cells were defective in competence due to a comK null mutation. These results indicate that ICEBs1 likely has an exclusion mechanism that inhibits acquisition of plasmid DNA through the ICEBs1-encoded conjugation machinery.
Fig. 2. ICEBs1 in recipient cells inhibits acquisition of pC194 mobilized by ICEBs1, but not by Tn916.
The percent mobilization of pC194 (CmR) by indicated donors and recipients. Left two bars: donors with ICEBs1 (MA116; ICEBs1 Δ(rapI-phrI)342::kan, Pxyl-rapI; pC194(cat), StrS). Right two bars: donors with Tn916 (MA1100; ICEBs10 Tn916; pC194(cat), SpcS). White bars: mobilization into recipients without ICEBs1 (CAL89; ICEBs10 str-84 comK::spc). Black bars: mobilization into recipients with ICEBs1 (CAL88; ICEBs1 str-84 comK::spc). Mobilization was calculated as the percent number of transconjugants (CmR StrR cells for ICEBs1 donors, and CmR SpcR cells for Tn916 donors) per number of initial donors. Data presented are averages from three independent experiments, with error bars depicting standard deviations.
ICEBs1-mediated exclusion is specific for the ICEBs1 conjugation machinery
We found that the inhibition of acquisition of pC194 by ICEBs1 in recipients was specific to the ICEBs1 conjugation machinery in donors. We used Tn916, an ICE that can also mobilize pC194 (Naglich & Andrews, 1988) and that does not encode an exclusion function (Norgren & Scott, 1991) to test specificity of ICEBs1-mediated exclusion. Donor cells containing Tn916 and pC194 (ICEBs10 Tn916; pC194, SpcS; MA1100) were grown in defined minimal medium with chloramphenicol to mid-exponential phase, induced with 2.5 μg/mL tetracycline for 2 hours (to boost activation of Tn916), mixed with recipients with or without ICEBs1, filtered and incubated for 3 hours and then harvested, as above.
The efficiency of transfer of pC194 by the Tn916-encoded conjugation machinery was ~1×10−4 % transconjugants per donor (Fig. 2), similar to previously reported results (Naglich & Andrews, 1988). The presence of ICEBs1 in recipient cells did not reduce the efficiency of transfer of pC194 by the Tn916-encoded conjugation machinery (Fig. 2). Although transfer of pC194 by Tn916 was low, our limit of detection was ~10−6 %, so we would have easily detected inhibition by ICEBs1-containing recipients. Based on these results, we conclude that 1) pC194 is not the target of the inhibition by ICEBs1 in the recipient, and 2) ICEBs1 has an exclusion mechanism that inhibits acquisition of DNA that is transferred through the ICEBs1, but not the Tn916, conjugation machinery.
In recipient cells, the ICEBs1 gene yddJ is both necessary and sufficient for exclusion
We expected the exclusion gene(s) to be transcriptionally active in recipient cells containing ICEBs1 that is integrated in the chromosome. In the integrated state, most ICEBs1 genes are repressed and only a few genes are expressed, including genes at the left and right ends of the element (Fig. 1). We tested the ability of various recipient cells containing deletions within ICEBs1 to exclude pC194 mobilized by ICEBs1 from donor cells. Because the functions of all three genes (immR, immA, int) at the left end of ICEBs1 are known, we focused on genes at the right end (Fig. 1A). Preliminary analyses of deletion-insertion mutations in yddM, rapI-phrI, and yddI indicated that none of these genes was required for exclusion. In contrast, deletion-insertion mutations that removed yddJ, alone or in combination with other genes, caused a defect in exclusion. Based on these preliminary analyses, we tested directly the effects of yddJ.
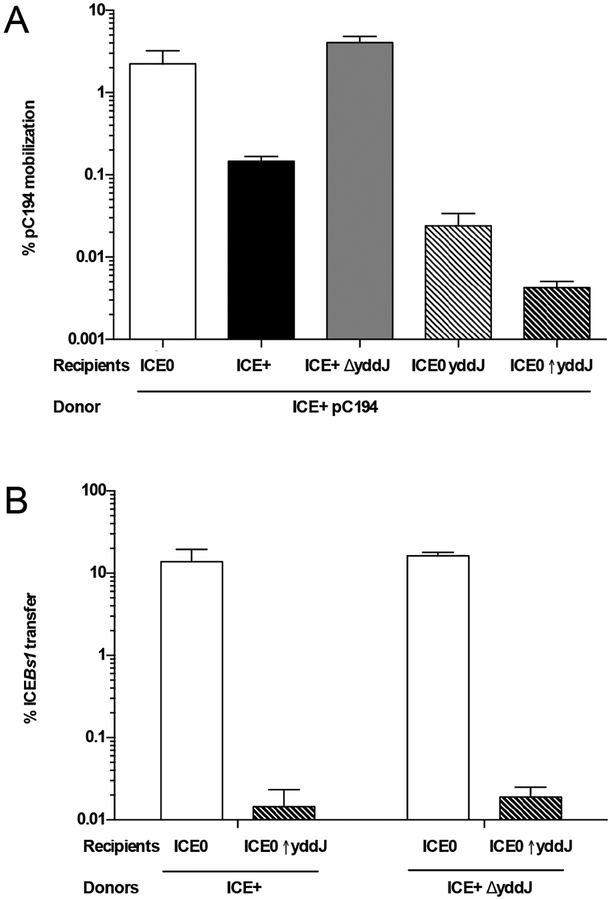
We found that yddJ in recipient cells was necessary for exclusion. We constructed a deletion of yddJ and compared the ability of recipients without ICEBs1 (ICEBs10 str-84; CAL89), with ICEBs1 containing yddJ+ (ICEBs1 yddJ+ str-84; CAL88) or not containing yddJ (ICEBs1 ΔyddJ str-84; MA665) to acquire pC194 by mobilization through the ICEBs1 conjugation machinery in donor cells. As expected, cells with ICEBs1 (yddJ+) had reduced acquisition of pC194 (~0.2% transconjugants per donor) (Fig. 3A). In contrast, cells with ICEBs1 ΔyddJ acquired pC194 at ~3% transconjugants per donor, a frequency similar to that of cells with no ICEBs1 (Fig. 3A). These results indicate that yddJ in recipients is necessary for exclusion.
Fig. 3. In recipient cells, ICEBs1 gene yddJ is necessary and sufficient for exclusion.
A. yddJ is necessary and sufficient in the recipient cell for exclusion of pC194 mobilized by ICEBs1. The percent mobilization of pC194 by ICEBs1 (MA116) donors into various recipients: without ICEBs1 (CAL89), white bars; with ICEBs1 (CAL88), black bars; with ICEBs1 with yddJ deleted (MA665; ICEBs1 ΔyddJ str-84), gray bars; without ICEBs1 and yddJ expressed from its own promoter (MA996; ICEBs10 lacA::PyddJ-yddJ str-84), light dashed bars; without ICEBs1 and yddJ over-expressed from the Pspank(hy) promoter (MA982; ICEBs10 lacA::Pspank(hy)-yddJ str-84), dark dashed bars. Mobilization was calculated as the percent number of transconjugants (CmR StrR cells) per number of initial donors. Data presented are averages from three independent experiments, with error bars depicting standard deviations.
B. yddJ is sufficient in the recipient cell and not required in the donor cell for exclusion of ICEBs1. Left two bars: percent transfer of ICEBs1 with yddJ (MMB970; ICEBs1 Δ(rapI-phrI)342::kan, Pxyl-rapI). Right two bars: percent transfer of ICEBs1 without yddJ (MA11; ICEBs1 ΔyddJ Δ(rapI-phrI)342::kan, Pxyl-rapI). White bars: transfer into recipients without ICEBs1 (CAL89). Black bars: transfer into recipients without ICEBs1 and overexpressing yddJ (MA982). Transfer was calculated as the percent number of transconjugants (KanR StrR cells) per number of initial donors. Data presented are averages from three independent experiments, with error bars depicting standard deviations.
We also found that yddJ alone, of all the ICEBs1 genes, was sufficient to cause exclusion. We expressed yddJ, from its own promoter, in cells that were missing ICEBs1 (ICEBs10 lacA::{PyddJ-yddJ} str-84; MA996). This strain was used as a recipient and compared to recipients without ICEBs1, and therefore no yddJ (ICEBs10 str-84; CAL89). The mobilization of pC194 from donor cells into recipients without yddJ was ~2%. In contrast, the mobilization of pC194 was reduced ~100-fold to ~0.03% into recipients expressing yddJ (Fig. 3A). We also expressed yddJ from the strong promoter Pspank(hy) in cells that were missing ICEBs1 (ICEBs10 lacA::{Pspank(hy)-yddJ} str-84; MA982). The mobilization of pC194 from donor cells into these recipients was reduced ~500-fold to ~0.004% (Fig. 3A). Based on these results, we conclude that yddJ is sufficient to cause exclusion in the absence of all other ICEBs1 genes, and that exogenous expression (from an exogenous locus, using its own or an exogenous strong promoter) causes more exclusion than normal levels of expression of yddJ from its endogenous locus within ICEBs1. In most of the experiments described below, we used strains over-expressing yddJ to cause exclusion.
Assays described above used mobilization of pC194 as a read-out for conjugation, largely to bypass the effects of repressor-mediated immunity that inhibits stable acquisition of ICEBs1 (Auchtung et al., 2007). Since recipients over-expressing yddJ do not have ICEBs1, there is no immR (repressor)-mediated immunity. Therefore, we could determine if yddJ also inhibited acquisition of ICEBs1 itself.
We found that yddJ in recipients inhibited acquisition of ICEBs1 from donor cells. We monitored transfer of ICEBs1 from donor cells (ICEBs1 Δ(rapI-phrI)342::kan, Pxyl-rapI, StrS; MMB970) into recipient cells without or with over-expression of yddJ, as above. The transfer efficiency of ICEBs1 was calculated as the number of transconjugants (KanR StrR CFUs) per initial number of donors at the time of cell mixing, converted to percent transconjugants per donor. We found the transfer of ICEBs1 into a yddJ over-expressing recipient was ~0.01%, a reduction of ~1000-fold compared to ~14% into a recipient without yddJ (Fig. 3B). These results indicate that yddJ is sufficient for exclusion of ICEBs1. In experiments below, we use transfer of ICEBs1 into recipients with or without over-expression of yddJ as an assay for exclusion.
yddJ in donor cells is not required for transfer or exclusion
We wondered if yddJ in donor cells was needed for yddJ-mediated exclusion coming from recipients. In some exclusion systems, the gene mediating exclusion in the recipient is also needed in the donor. For example, in plasmid R27, the genes eexA and eexB are needed in both the recipient and donor cells for exclusion (Gunton et al., 2008).
We found that loss of yddJ in donor cells had virtually no effect on transfer efficiency or exclusion. In the experiment above, we also monitored transfer of ICEBs1 from donor cells that contained a deletion of yddJ in ICEBs1 (ICEBs1 ΔyddJ Δ(rapI-phrI)342::kan, Pxyl-rapI; MA11) into recipient cells with or without over-expression of yddJ. Deletion of yddJ in donors did not affect the ability of ICEBs1 to transfer into ICEBs10 recipient cells, indicating yddJ is not required for transfer. The ΔyddJ donor was as sensitive as the yddJ+ donor to exclusion by the presence of yddJ in recipient cells (Fig. 3B). Based on these results, we conclude that yddJ is needed only in recipients to mediate exclusion.
YddJ is a putative lipoprotein with a cystatin-like fold
yddJ is predicted to encode a 126 aa lipoprotein (Zhou et al., 2008). Based on what is known about B. subtilis lipoproteins, the N-terminal 18 aa should serve as a signal peptide that is cleaved at the cell surface. The cysteine at amino acid position 19 would serve as the site of lipid modification, resulting in a mature 108 aa form that is tethered to the cell membrane by the lipid anchor (Simonen & Palva, 1993).
Previous findings indicate that YddJ is a lipoprotein. In proteomic studies with glucose-starved B. subtilis cells (Otto et al., 2010), YddJ was detected in the enriched membrane fraction, but not in the biotinylation enrichment (which purifies membrane proteins containing an extracellular cysteine) or membrane-shaved (which purifies integral membrane proteins) fractions. These findings are consistent with the predicted lack of exposed, unmodified cysteines in mature YddJ available for biotinylation, and the fact that YddJ does not have any predicted transmembrane domains.
We searched for conserved motifs and structural similarity between YddJ and other proteins. Analysis using Phyre2 (Kelley et al., 2015) indicated that the residues 5–49 of the processed, mature YddJ (36% of the protein) modeled with 95.1% confidence to the crystal structure of a protein from S. aureus from the DUF4467 protein family. KEGG’s SSDB Motif Search (Kanehisa et al., 2017) also identified DUF4467 as matching residues 24–118 of YddJ with an E-value of 4×10−31. DUF4467 is a large family of Gram-positive lipoproteins with a cystatin-like fold (Finn et al., 2017). Cystatins are a superfamily of proteins which act as inhibitors of C1 and C13 cysteine peptidase families (Brown & Dziegielewska, 1997). ICEBs1 contains a cell wall hydrolase, CwlT, with a peptidase domain belonging to the C1 family (Fukushima et al., 2008; Dewitt & Grossman, 2014). This similarity indicated that YddJ in the recipient cell might target the peptidase domain of CwlT in the donor cell.
CwlT is not the target of YddJ-mediated exclusion
CwlT is a bifunctional cell wall hydrolase, containing an N-terminal muramidase domain and a C-terminal peptidase domain (Fukushima et al., 2008; Dewitt & Grossman, 2014). Muramidase activity is virtually required for transfer, and peptidase activity is important, but partially dispensable; mutating the active site or deleting the peptidase domain results in a mutant ICEBs1 that has an approximately 1,000-fold decrease in transfer efficiency (Dewitt & Grossman, 2014).
If the peptidase domain and peptidase activity of CwlT was the target of YddJ-mediated exclusion, then CwlT mutants that lack peptidase activity or the peptidase domain should be insensitive to exclusion. We found the contrary. We monitored transfer of ICEBs1 from donor cells that contained wild type cwlT (MMB970) or a deletion of the peptidase domain of cwlT in ICEBs1 (MA980) into recipient cells with or without over-expression of yddJ. ICEBs1 donors with the mutant cwlT were excluded by yddJ over-expressing recipients ~2,000-fold (0.02% transconjugants/donor into recipient cells without yddJ, compared to 2×10−5 % transconjugants/donor into recipient cells with yddJ), nearly the same extent (~3000-fold) as ICEBs1 containing wild type cwlT (~18% transconjugants/donor into recipient cells without yddJ, compared to 6×10−3 % transconjugants/donor into recipient cells with yddJ), revealing that the peptidase domain of CwlT is not the target of YddJ.
Isolation of exclusion-resistant mutations in ICEBs1
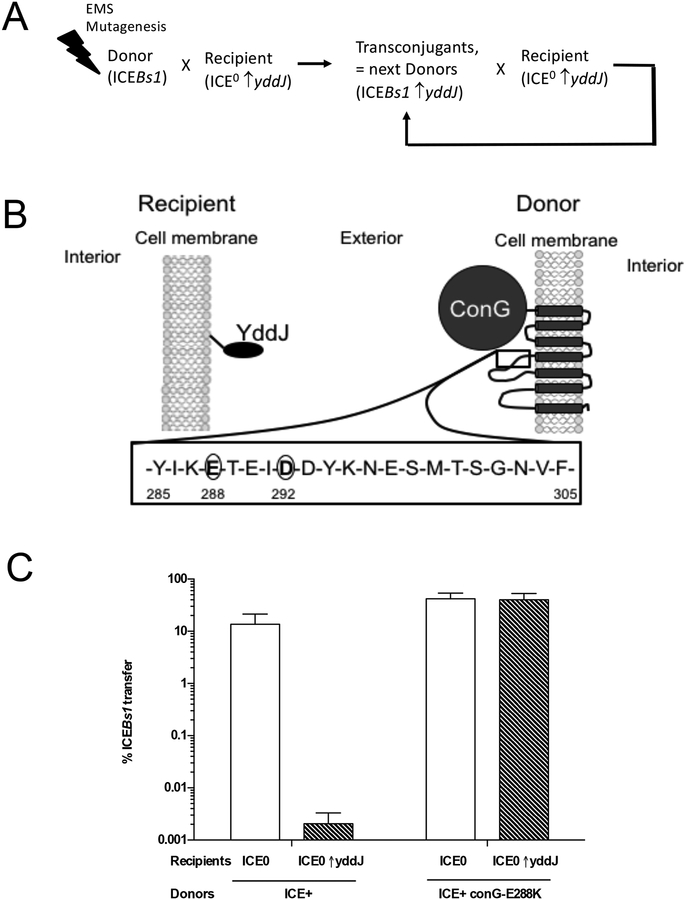
We hypothesized that YddJ on recipient bacteria interacted with some part of the ICEBs1-encoded conjugation machinery in the donor. If so, then mutations in the target gene(s) encoding the ICEBs1 conjugation machinery might make donors resistant to YddJ-mediated exclusion. To identify potential target(s) of yddJ-mediated exclusion in ICEBs1, we performed a mutagenesis and enrichment screen to identify exclusion-resistant, transfer-competent mutants of ICEBs1. We randomly mutagenized a pool of ICEBs1 donor cells with ethyl methanesulfonate to generate point mutations throughout the chromosome. We used this pool of mutagenized cells as donors, and selected for transfer of ICEBs1 into recipients that were over-expressing yddJ. We selected for and pooled transconjugants and then used the transconjugant pool as donors in successive rounds of mating with recipients that over-expressed yddJ (Fig. 4A). We expected that any exclusion-resistant ICEBs1 mutants that were fully functional for conjugation would transfer up to 1000-fold more efficiently than wild type (exclusion-sensitive) ICEBs1. After several rounds of conjugation, such an exclusion-resistant mutant should be enriched in the transconjugant population. We mutagenized 16 independent cultures of donor cells and enriched for mutants with increased transfer efficiency from each separate culture. We purified transconjugants and sequenced ICEBs1 to identify exclusion-resistant mutations.
Fig. 4. Isolation of exclusion-resistant conG mutations in ICEBs1.
A. Schematic of the mutagenesis and enrichment screen for exclusion-resistant mutations in ICEBs1 (described in the text and Methods).
B. Schematic of YddJ and ConG predicted topologies. YddJ is a putative lipoprotein. Results from proteomic fractionation studies indicated that YddJ is associated with the cell membrane but that it is not a transmembrane protein (Otto et al., 2010). ConG is predicted to have seven transmembrane regions. Residues 285–305 of the extracellular loop between the third and fourth transmembrane regions are shown with the residues (288 and 292) identified in the screen for exclusion-resistance circled.
C. ICEBs1 conG-E288K donors are resistant to yddJ-mediated exclusion. Left two bars: percent transfer of ICEBs1 (MA1049; ICEBs1 Δ(rapI-phrI)342::kan, Pxyl-rapI). Right two bars: percent transfer of exclusion-resistant ICEBs1 (MA1089; ICEBs1 conG-E288K Δ(rapI-phrI)342::kan, Pxyl-rapI). White bars: recipients without ICEBs1 (CAL89). Dashed bars: recipients without ICEBs1 and overexpressing yddJ (MA982). Transfer was calculated as the percent number of transconjugants (KanR StrR cells) per number of initial donors. Data presented are averages from three independent experiments, with error bars depicting standard deviations.
From 16 independently mutagenized cultures, we identified three different point mutations in conG of ICEBs1 that caused an exclusion-resistant phenotype without causing a defect in transfer efficiency (Fig. 4B). A glutamate-to-lysine mutation at residue 288 (conG-E288K) was isolated from 14 of the independent cultures. Two other mutations, an aspartate-to-tyrosine and aspartate-to-glycine mutation at residue 292 (conG-D292Y; conG-D292G) were each isolated once. We compared transfer and exclusion of wild type and conG-E288K ICEBs1 donors and confirmed that the conG-E288K mutation fully abolished exclusion by yddJ without affecting transfer efficiency (Fig. 4C). In preliminary testing, we found that ICEBs1 donors containing conG-D292Y or conG-D292G mutations behaved similarly to conG-E288K.
ConG is essential for ICEBs1 conjugation and is predicted to have seven transmembrane domains that form part of the mating channel (Babic et al., 2011; Leonetti et al., 2015). The exclusion-resistant point mutations were in a region predicted to form part of an extracellular loop between the third and fourth transmembrane domains (Fig. 4B). The existence of exclusion-resistant mutations in conG strongly indicates that ConG in the donor is the target of YddJ in the recipient.
ConG and YddJ determine exclusion specificity
We postulated that YddJ specifically recognizes and targets its cognate ConG, as was indicated by the inability of YddJ to inhibit Tn916-mediated transfer, despite the presence of a conG homolog in Tn916. However, Tn916 does not have a homolog of yddJ, nor does it have exclusion. We identified a homolog of yddJ in ICEBat1, a putative ICE found in Bacillus atrophaeus.
Proteins from ICEBat1 compared to those from ICEBs1.
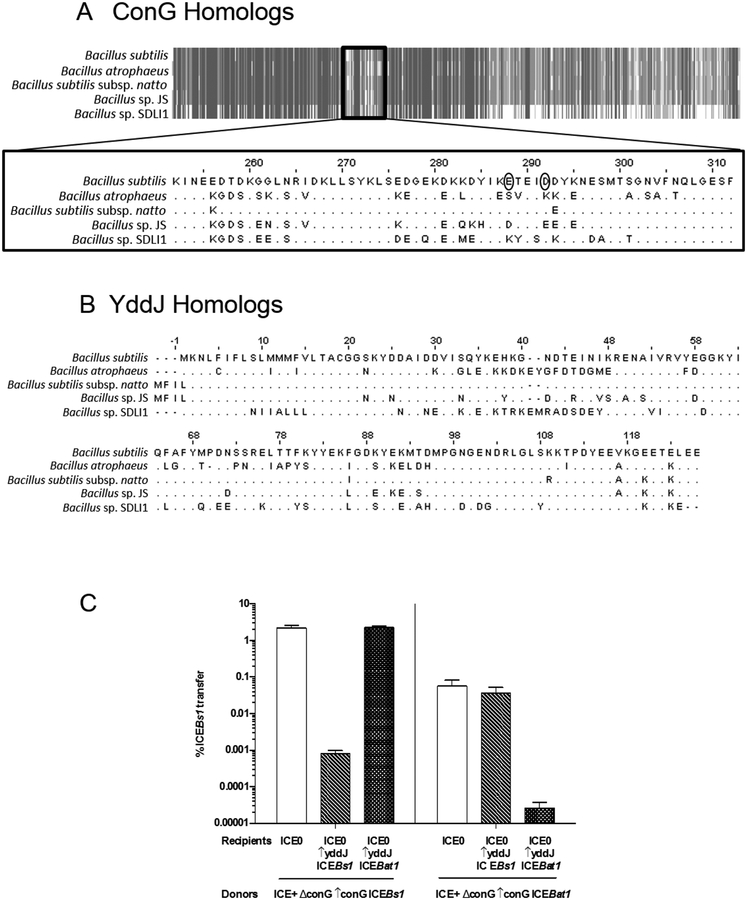
ICEBat1 is similar to ICEBs1. It contains homologs for all ICEBs1 genes needed for regulation and conjugation, including conG. Most conjugation proteins are virtually identical (>95%); however, ConG is less so (only 87% identical), and the differences are concentrated in two regions (Fig. 5A): the loop region between the predicted third and fourth transmembrane domains and the predicted extracellular C-terminal region. Notably, the loop region between the third and fourth transmembrane domains is the location of the mutations in ConGICEBs1 that confer resistance to YddJ-mediated exclusion.
Fig. 5. ICEBs1 and ICEBat1 homology and exclusion specificity.
Alignments of ConG (A) and YddJ (B) homologs from ICEBs1-like elements from five Bacillus species, including B. subtilis (ICEBs1) and B. atrophaeus (ICEBat1), were generated using Jalview (www.jalview.org). A. Schematic alignment of ConG homologs. Gray indicates regions that are identical, while white indicates regions that are dissimilar. Alignment of the dissimilar internal region that includes the residues mutated in exclusion-resistant ConG from ICEBs1 (circled) is shown in detail. B. Alignment of YddJ homologs.
C. The percent transfer of ICEBs1 strains with conG from either ICEBs1 (left panel; donor strain KPD225) or ICEBat1 (right panel; donor strain KPD224). White bars: recipients without ICEBs1 (CAL89). Dashed bars: recipients with yddJ from ICEBs1 over-expressed (MA982). Hatched bars: recipients with yddJ from ICEBat1 over-expressed (KPD219; ICEBs10 lacA::Pspank(hy)-yddJICEBat1 str-84). Transfer was calculated as the percent number of transconjugants (KanR StrR cells) per number of initial donors. Data presented are averages from three independent experiments, with error bars depicting standard deviations.
We also found that YddJ is more divergent (only 67% identical) between these two ICEs than most of the other ICE proteins (Fig. 5B). There are four regions of sequence divergence with at least two consecutive non-identical residues: 1) residues 30–48 on ICEBs1 YddJ and 30–50 on ICEBat1 YddJ, 2) residues 57–58 on ICEBs1 YddJ and 59–60 on ICEBat1 YddJ, 3) residues 65–81 on ICEBs1 YddJ and 67–82 on ICEBat1 YddJ, and 4) 86–95 on ICEBs1 YddJ and 87–96 on ICEBat1 YddJ. These differences are consistent with the notion that ConG and YddJ determine exclusion specificity.
ConG from ICEBat1 functions in the context of the ICEBs1 conjugation machinery.
B. atrophaeus is difficult to manipulate genetically and little is known about the function of ICEBat1. Therefore, to test function and specificity, we introduced conG and yddJ from ICEBat1 into B. subtilis.
We first determined that ConG from ICEBat1 was able to function with the ICEBs1-encoded conjugation machinery. We constructed donor strains with ICEBs1 containing conG from either ICEBs1 or ICEBat1. In both cases, conG was deleted from its native locus within ICEBs1, and a copy of conGICEBs1 (ICEBs1 ΔconG Δ(rapI-phrI)342::kan, thrC::{Pspank(hy)-conGICEBs1 mls}; KPD225) or conGICEBat1 (ICEBs1 ΔconG Δ(rapI-phrI)342::kan, thrC::{Pspank(hy)-conGICEBat1 mls}; KPD224) was expressed ectopically from Pspank(hy) located at thrC. Both strains contained Pxyl-rapI for xylose-inducible activation of ICE.
We found that both conGICEBs1 and conGICEBat1 complemented the transfer defect caused by loss of conG in ICEBs1. As determined previously, conGICEBs1 fully restored transfer to wild type levels (Babic et al., 2011; Leonetti et al., 2015). conGICEBat1 largely restored conjugation, but to levels ~10-fold lower than those with conGICEBs1 (Fig. 5C). Based on these results, we conclude that conGICEBat1 is largely functional in the context of the ICEBs1-encoded conjugation machinery.
YddJ inhibited transfer only when the conjugation machinery contained the cognate ConG.
We monitored ICE transfer from donors expressing either conGICEBs1 or conGICEBat1 into recipient strains that expressed yddJ from either ICEBs1 (MA982) or ICEBat1 (KPD219). Donor cells expressing conGICEBs1 as part of the conjugation machinery were sensitive to exclusion by recipient cells expressing yddJICEBs1 (Fig. 5C), recapitulating results presented above. However, they were not sensitive to exclusion by recipients expressing yddJICEBat1 (Fig. 5C). The failure of YddJICEBat1 to inhibit the conjugation machinery containing ConG from ICEBs1 is intriguing, but could simply reflect a non-functional yddJICEBat1, due perhaps to lack of expression, misfolding, or a defective gene. Alternatively, if YddJICEBat1 is functional, then it could indicate that the ConG-YddJ pair determines specificity of exclusion.
We found that YddJICEBat1 was indeed functional and was able to inhibit the conjugation machinery that contained ConG from ICEBat1. Donor cells expressing conGICEBat1 as part of the conjugation machinery were able to transfer ICEBs1 at a frequency of 6×10−2 % into cells without yddJ. Transfer was reduced ~2,000-fold into recipients that expressed yddJICEBat1, but there was no significant reduction into recipient cells that expressed yddJICEBs1 (Fig. 5C).
To summarize these results: 1) YddJICEBat1 is functional and capable of inhibiting transfer from the ICEBs1 conjugation machinery that contains ConGICEBat1, but does not inhibit transfer from the ICEBs1 conjugation machinery containing ConGICEBs1; 2) YddJICEBs1 is capable of inhibiting transfer from the ICEBs1 conjugation machinery that contains the ConGICEBs1, but does not inhibit transfer from the ICEBs1 conjugation machinery that contains ConGICEBat1. Together, these results demonstrate that the specificity for exclusion resides with ConG and YddJ, and that YddJ-mediated exclusion is specific for its cognate element, even between highly related ICEs.
Exclusion is beneficial to ICEBs1 and its host cells by preventing loss of viability due to redundant transfer
We tested whether exclusion conferred any benefit to ICEBs1 and/or its host cells by measuring the viability of cells containing ICEBs1 with and without exclusion. Cells containing ICEBs1 (with or without a functional exclusion system) were grown in defined minimal medium, ICEBs1 was activated by over-expression of rapI, and ~8×108 cells were filtered and then incubated as in a mating experiment (Materials and Methods). Cells were then recovered from the filters and the number of viable cells (CFUs) was determined and compared to that of the initial input onto the filter.
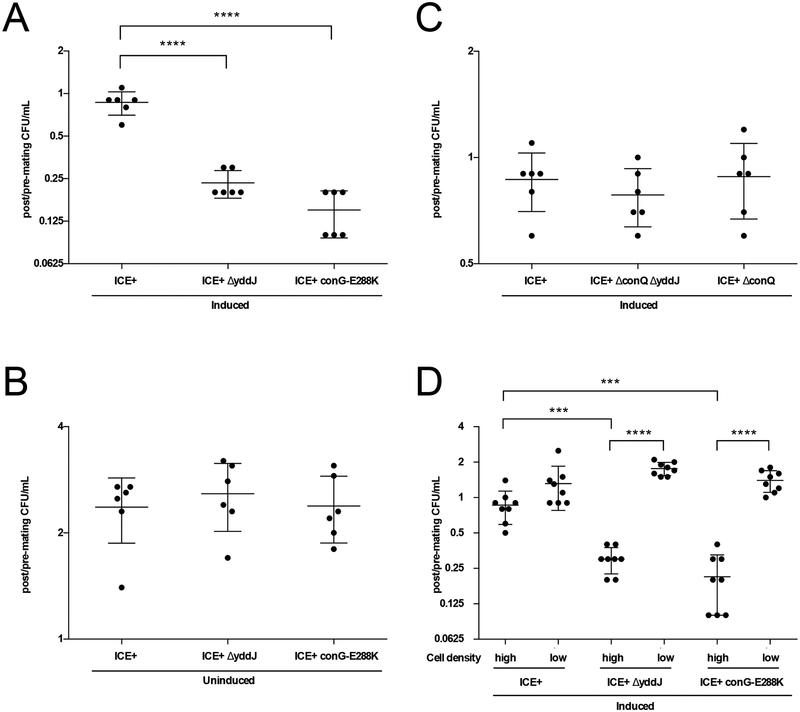
We found that cells containing ICEBs1 with a defective exclusion system had decreased viability relative to cells with a functional exclusion system. This decrease in viability was dependent on conditions that favor conjugation, including activation of ICEBs1, the presence of a functional conjugation system, and a sufficient cell density (see below). In all cases, the number of exclusion-competent cells (ICEBs1 yddJ+; MA1049) recovered from the filter was ~80–90% of the initial input. In contrast, in the absence of a functional exclusion system (ICEBs1 ΔyddJ; MA1050, and ICEBs1 conG-E288K; MA1089) the number of cells recovered was <25% of the input (Fig. 6A). These results indicate that something about exclusion was likely preventing cell death under the conditions tested.
Fig. 6. Exclusion is beneficial to ICEBs1 and its host cells by preventing loss of viability due to redundant transfer.
Data depicted in A-C. are from experiments where cells (monocultures) were grown in minimal medium, induced (as indicated) with 1% xylose for 2 hours, and placed at high cell density on filters for 3 hours. Cells were sampled before and after plating on filters to determine CFU/ml pre- and post-mating conditions. The y-axis shows the number of viable cells recovered post-mating per number of input cells pre-mating on a log2 scale. Each dot represents a value from an independent experiment (n=6). The middle bars represent averages and the shorter bars depict standard deviations. P-values were calculated by an ordinary one-way ANOVA with Dunnett’s correction for multiple comparisons (**** indicates p-value <0.0001) using GraphPad Prism version 6.
A and B. Cells containing exclusion-defective ICEBs1 exhibit a loss of viability under conditions that favor conjugation. Cell recovery is shown for cells containing wildtype (MA1049; ICEBs1, Pxyl-rapI) or exclusion-deficient ICEBs1 (MA1050; ICEBs1 ΔyddJ, Pxyl-rapI, and MA1089; ICEBs1 conG-E288K, Pxyl-rapI). A. Cell viability following activation (induction) of ICEBs1. B. Cell viability with NO activation (uninduced) of ICEBs1.
C. Loss of viability from defective exclusion depends on a functional conjugation machinery. Cell recovery is shown for induced cells containing wildtype (MA1049) and transfer-deficient ICEBs1 with exclusion (MA1070; ICEBs1 ΔconQ, Pxyl-rapI) and without exclusion (MA1069; ICEBs1 ΔconQ ΔyddJ, Pxyl-rapI).
D. Loss of viability depends on high cell density. Cells containing wildtype (MA1049) and exclusion-deficient ICEBs1 (MA1050 and MA1089) were grown in minimal medium, induced with 1% xylose for 2 hours, and placed at high and low cell density on filters for 3 hours. CFU/ml were determined before and after incubation on the filter. The y-axis depicts the number of viable cells recovered from the filter per number of input cells pre-filtration on a log2 scale. Each dot represents a value from an independent experiment (n=8). The middle bars represent averages and the shorter bars depict standard deviations. P-values were calculated by an unpaired two-tailed t-test with Welch’s correction (*** indicates p-value<0.0005; **** indicates p-value <0.0001) using GraphPad Prism version 6.
The cell death observed in the absence of a functional exclusion system could be due to cell autonomous effects of yddJ, or possibly excessive conjugation, or both. To test this, we used three different conditions that would limit or eliminate conjugation and monitored the effects of a functional exclusion system on cell death. We found that cell death in the absence of exclusion was dependent on conditions that favor conjugative transfer. The three conditions tested were:
No activation of ICEBs1. In experiments in which ICEBs1 was not activated (no expression of rapI), there was no detectable effect of the presence or absence of a functional exclusion system on cell viability (Fig. 6B).
An ICEBs1 mutant that is incapable of transfer. We introduced a conQ null mutation (ΔconQ) into ICEBs1 with yddJ (MA1070) or without yddJ (MA1069). conQ encodes the coupling protein that ‘brings’ the DNA substrate for transfer to the conjugation machinery and is essential for transfer (Lee et al., 2012). In the ΔconQ mutant, the absence of yddJ (no exclusion) did not cause a decrease in viability compared to ΔconQ, yddJ+ (Fig. 6C).
Conditions of low cell density. Conjugative transfer between cells is dependent on cell-cell contact. At low cell densities, there will be few mating pairs formed and low transfer frequencies. We measured the viability of cells with activated ICEBs1 at high and low cell density. ICEBs1-containing cells were grown and activated by over-expression of rapI as described above. After 2 hours of activation, ~8×108 or ~3×106 cells were filtered and incubated for 3 hours. Viable cells were recovered and quantified as described above.
We found that cells containing defective exclusion systems had increased viability under low cell density (conditions that prevent conjugation), compared to high cell density (conditions that promote conjugation). Cells containing exclusion-competent ICEBs1 (ICEBs1 yddJ+; MA1049) were recovered >100% (indicative of cell growth) of initial input at low cell density, and ~90% of initial input at high cell density. In contrast, cells containing exclusion-deficient ICEBs1 (ICEBs1 ΔyddJ; MA1050, and ICEBs1 conG-E288K; MA1089) were recovered >100% of initial input at low cell density, compared to <30% of initial input at high cell density (Fig. 6D). Together, these results indicate that exclusion confers a benefit to ICEBs1 and its host cell by preventing redundant transfer, thus protecting host cell viability under conditions of high transfer.
DISCUSSION
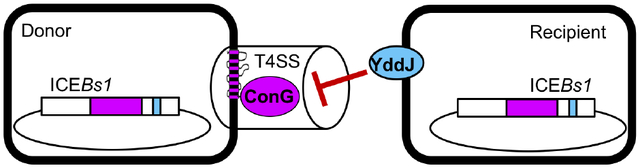
Experiments described here demonstrate that ICEBs1 encodes an exclusion mechanism that is beneficial to cells with the element. The ICEBs1 gene yddJ causes exclusion in recipient cells by inhibiting transfer from the ICEBs1 conjugation machinery in donor cells. The target of YddJ-mediated exclusion is the essential conjugation protein ConG, a conserved protein with seven predicted transmembrane domains. Exclusion protects ICEBs1 and its host cells against cell death caused by redundant transfer. Together with previous findings, we conclude that ICEBs1 has three distinct mechanisms to inhibit host cells from acquiring a second copy of the element. 1) RapI-dependent, SOS-independent activation under conditions of high population density, the earliest step in the ICEBs1 life cycle, is inhibited by PhrI-mediated cell signaling if a host cell is surrounded by other cells containing ICEBs1 (Auchtung et al., 2005). 2) Exclusion (described here), inhibits transfer of DNA through the ICEBs1 conjugation machinery. This inhibition appears to be independent of the DNA substrate to be transferred. 3) If ICEBs1 is transferred to a cell that already has a copy, then repressor-mediated immunity inhibits integration and stable acquisition of that copy of the element (Auchtung et al., 2007). We postulate that these mechanisms all contribute to the stability of ICEBs1 in a host genome and that introduction of a second copy will lead to recombination between and instability of the linked elements.
Selective advantage of exclusion
To date, experimental evidence demonstrating the selective advantages of exclusion has come from the characterization of lethal zygosis in the F plasmid (Skurray et al., 1973, 1974, 1976; Ou, 1980). Recipients lacking the F plasmid (F-) are killed when mixed with an excess of Hfr or F+ exclusion-null donors. F+ exclusion-null cells are also susceptible to lethal zygosis when used as recipients in matings with excess Hfr donors. These results indicate that exclusion is important for protecting the recipient cell from excess transfer, whether it is an established host or an unoccupied cell, and that unidirectional transfer from donor to recipient is sufficient for killing.
Our findings are similar to the lethal zygosis studies above, in that under conditions that support conjugative transfer, exclusion protects the element and host cell by preventing excessive transfer and cell death. We observe killing in the absence of exclusion with donors that are induced to transfer ICE amongst themselves. These experiments do not distinguish whether it is mutual or unidirectional transfer between donor cells that results in killing, though we speculate that the former is likely. The mechanism of killing in lethal zygosis was proposed to be excessive recipient membrane damage. Experiments in which Hfr donors were mixed with F- recipients whose cell walls were labeled with radioactive DAP resulted in a release of the recipient’s cell wall material into the medium (Ou, 1980). This may also be the case in the context of ICEBs1 and B. subtilis, where assembling the type IV secretion system and transferring to/from donor cells at high frequencies results in extensive degradation of the thick cell wall.
Under what natural scenarios would a conjugative element and its host cell encounter such conditions of extreme transfer that exclusion protects against? One possibility is when an element transfers rapidly through chains of recipient cells, as has been demonstrated for ICEBs1 and B. subtilis (Babic et al., 2011), or complex bacterial communities such as biofilms (Lécuyer et al., 2018). It is not hard to imagine that exclusion in such contexts would be the main mechanism to prevent excessive transfer and maintain host cell viability. In addition to its protective role under conditions of high transfer, it is also likely that exclusion contributes to the efficient propagation of an element through a cell population by preventing futile transfer into a cell already occupied by a copy of the element.
Diverse proteins mediating exclusion
YddJ is a member of the DUF4467 family consisting of ~250 lipoproteins found in various Gram-positive bacteria (multiple species of Bacillus, Staphylococcus, Streptococcus, Listeria, etc.) (Finn et al., 2017). YddJ is one of the first DUF4467 members with an established function, that of an exclusion protein. We suspect that other YddJ-like proteins associated with conjugative elements are exclusion proteins as well. It is also possible that there are YddJ-like proteins that, like cystatins, target cysteine peptidases. This was the scenario we initially hypothesized for ICEBs1 exclusion; our initial hypothesis was that YddJ inhibited the peptidase domain of the cell wall hydrolase CwlT from ICEBs1. Such peptidases inhibitors, if they exist, may regulate hydrolases from conjugative elements and modulate their transfer. It is also possible that such peptidase inhibitors have a role in regulating cell wall metabolism.
Genes that mediate exclusion have been identified in many conjugative elements, mostly from the major incompatibility groups of conjugative plasmids (Garcillan-Barcia & de la Cruz, 2008). Exclusion proteins are generally diverse with no consistently conserved domains. Any similarity is typically between proteins from the same plasmid groups. Exclusion proteins do have a few general features in common. They tend to be relatively small transmembrane proteins or lipoproteins. Most are not required in donor cells, either for conjugation or exclusion, with the noted exception of R27 (Gunton et al., 2008), and are sufficient for exclusion in the recipient cells.
A conserved target for exclusion proteins?
The target of the exclusion system of ICEBs1 is ConG, an essential component of the conjugation machinery. ConG is predicted to have seven transmembrane domains and a large extracellular C-terminal region. orf15 of Tn916 encodes a similar protein (Burrus et al., 2002), although some isolates of Tn916 appear to encode a truncated form of Orf15 that is missing the large extracellular C-terminal region (Browne et al., 2015). ConG is thought to be part of the channel through which substrate is transferred (Auchtung et al., 2016) and is homologous to VirB6, a five transmembrane domain inner membrane protein predicted to form the channel of the Ti plasmid of A. tumefaciens (Jakubowski et al., 2004).
All type IV secretion systems have a polytopic protein homologous or analogous to VirB6 that performs a similar function (Bhatty et al., 2013), and these proteins have been identified as the donor targets for the exclusion systems of the F/R100 plasmids, SXT/R391 ICEs, and R64/R621a plasmids (isolated from Gram-negative bacteria and characterized in E.coli). The targets for both F/R100 and SXT/R391 exclusion are TraG proteins, VirB6 homologs (Beaber et al., 2002; Lawley et al., 2003) predicted to have 3–5 transmembrane domains (Audette et al., 2007; Marrero et al., 2007). The target for R64/R621a exclusion is TraY, proposed to be a VirB6 analog (Guglielmini et al., 2014) predicted to have seven transmembrane domains (Komano et al., 2000).
The targets of exclusion in the donor likely share the function of forming the channel of the secretion system through which the substrate travels. The fact that diverse exclusion proteins with no obvious sequence similarity converge upon targeting this function indicates that this is a conserved strategy for exclusion.
Even though all type IV secretion systems probably have a ConG/VirB6 homolog or analog, they do not all have exclusion systems. For example, Tn916, an ICE whose conjugation machinery is closely related to that of ICEBs1, does not have an exclusion system (Norgren & Scott, 1991). This could be due to some fundamental difference in the components of the T4SS, for example, differences in sequence or in key contacts made between those components. Alternatively, we believe that most conjugation systems likely have the potential for exclusion, and the presence or absence of exclusion depends on some significant selective advantage to having an exclusion system. Variables that might contribute to selective pressures could include the efficiency of activation of an element, the efficiency of transfer, and the potential cost to the host and/or element to having multiple copies of an element in a single cell.
MATERIALS and METHODS
Media and growth conditions
Cells were grown at 37°C with shaking in S750 defined minimum medium (Jaacks et al., 1989) supplemented with auxotrophic requirements (40 μg/ml tryptophan, 40 μg/ml phenylalanine, 200 μg/ml threonine, as needed), or LB medium as indicated. Antibiotics were used at the following concentrations for growth on LB agar plates: chloramphenicol (5 μg/ml), kanamycin (5 μg/ml), streptomycin (100 μg/ml), spectinomycin (100 μg/ml), tetracycline (12.5 μg/ml), and a combination of erythromycin (0.5 μg/ml) and lincomycin (12.5 μg/ml) to select for macrolide-lincosamide-streptogramin (MLS) resistance. Isopropyl-β-D-thiogalactopyranoside (IPTG, Sigma) was used at a final concentration of 1 mM to induce expression from the Pspank(hy) promoter. When 1mM IPTG was added to cells without ICEBs1 and with no genes under the control of the Pspank(HY) promoter, no deleterious effects on growth were observed. Tetracycline was used at a final concentration of 2.5 μg/ml to stimulate Tn916 gene expression and excision in donor cells.
Strains and alleles
The B. subtilis strains used are listed in Table 1. Standard techniques were used for cloning and strain construction (Harwood & Cutting, 1990). Some alleles related to ICEBs1 were previously described and are summarized below. Donor strains contained a derivative of ICEBs1 that contains a deletion of rapI-phrI and a kanamycin-resistance cassette inserted, Δ(rapI-phrI)342::kan (Auchtung et al., 2005). rapI was over-expressed from Pxyl-rapI to induce ICEBs1 gene expression and excision in donor cells. Alleles were integrated into amyE with various antibiotic resistances, and included: amyE::{Pxyl-rapI spc} (Berkmen et al., 2010), amyE::{Pxyl-rapI cat}, and amyE::{Pxyl-rapI mls} (Johnson & Grossman, 2014). ICEBs10 indicates that the strain is cured of ICEBs1. Recipients were typically streptomycin-resistant (str-84) (Auchtung et al., 2005; Lee et al., 2007), and streptomycin was used as a counter-selective marker in mating experiments unless otherwise indicated. The donor strain MA980 contains the peptidase deletion of cwlT, cwlTΔ(207–327), described previously (Dewitt & Grossman, 2014). Donor strains containing a deletion of conG, ΔconG(5–805), were derived from MMB1283 (Leonetti et al., 2015). Donor strains containing a deletion of conQ, ΔconQ848, were derived from CAL848 (Lee et al., 2012).
Table 1.
B. subtilis strainsa
| Strain | Relevant genotype (reference) |
|---|---|
| BOSE986 | ICEBs10 amyE::{Pxyl-rapI spc} |
| CAL88 | str-84 comK::spc (Auchtung et al., 2005) |
| CAL89 | ICEBs10 str-84 comK::spc (Auchtung et al., 2005) |
| KPD35 | ICEBs10 amyE::{Pxyl-rapI spc} lacA::{Pspank(hy)- yddJ mls} str-84 comK::tet |
| KPD36 | ICEBs10 amyE::{Pxyl-rapI cat} lacA::{Pspank(hy)- yddJ mls} str-84 comK::tet |
| KPD38 | ICEBs1 Δ(rapI-phrI)342::kan, amyE::{Pxyl-rapI mls} comK::cat |
| KPD80 | ICEBs1 conG-E288K Δ(rapI-phrI)342::kan amyE::{Pxyl-rapI cat} lacA::{Pspank(hy)- yddJ mls} str-84 comK::tet |
| KPD219 | ICEBs10 lacA::{Pspank(hy)- yddJICEBat1 mls} str-84 comK::spc |
| KPD224 | ICEBs1 ΔconG(5–805) Δ(rapI-phrI)342::kan amyE::{Pxyl-rapI cat} thrC::{Pspank(hy)-conGICEBat1 mls} |
| KPD225 | ICEBs1 ΔconG(5–805) Δ(rapI-phrI)342::kan amyE::{Pxyl-rapI cat} thrC::{Pspank(hy)-conG mls} |
| MMB970 | ICEBs1 Δ(rapI-phrI)342::kan amyE::{Pxyl-rapI spc} (Dewitt & Grossman, 2014) |
| MA11 | ICEBs1 ΔyddJ Δ(rapI-phrI)342::kan amyE::{Pxyl-rapI spc} |
| MA116 | ICEBs1 Δ(rapI-phrI)342::kan amyE::{Pxyl-rapI spc}; pC194 (cat) |
| MA980 | ICEBs1 cwlTΔ(207–327) Δ(rapI-phrI)342::kan amyE::{Pxyl-rapI spc} (Dewitt & Grossman, 2014) |
| MA982 | ICEBs10 lacA::{Pspank(hy)- yddJ mls} str-84 comK::spc |
| MA996 | ICEBs10 lacA::{PyddJ- yddJ mls} str-84 comK::spc |
| MA997 | ICEBs1 ΔyddJ str-84 comK::spc |
| MA1027 | ICEBs10 amyE::{Pxyl-rapI spc} comC::mls |
| MA1049 | ICEBs1 Δ(rapI-phrI)342::kan amyE::{Pxyl-rapI spc} comC::mls |
| MA1050 | ICEBs1 ΔyddJ Δ(rapI-phrI)342::kan amyE::{Pxyl-rapI spc} comC::mls |
| MA1069 | ICEBs1 ΔconQ848 ΔyddJ Δ(rapI-phrI)342::kan amyE::{Pxyl-rapI spc} comC::mls |
| MA1070 | ICEBs1 ΔconQ848 Δ(rapI-phrI)342::kan amyE::{Pxyl-rapI spc} comC::mls |
| MA1089 | ICEBs1 conG-E288K Δ(rapI-phrI)342::kan amyE::{Pxyl-rapI spc} comC::mls |
| MA1100 | ICEBs10 Tn916 (tet) comC::mls; pC194 (cat) |
all strains derived from JH642 and contain pheA1, trpC2 mutations (Perego et al., 1988)
Construction of comK and comC null mutations.
Null mutations in comK and comC were used to prevent transformation. The comK::spc allele has been described (Auchtung et al., 2005). The comK::tet allele replaced the comK open reading frame with the tet cassette from pDG1513 (Guerout-Fleury et al., 1995). The comK::cat allele was derived from CAL419 and has been described (Lee et al., 2007). The comC::mls allele replaced from 324 bp upstream to 26 bp downstream of the comC open reading frame with the mls cassette from pHP13 (Lee et al., 2012).
Construction of pC194-containing ICEBs1 and Tn916 donor strains for mobilization assays.
pC194-containing ICEBs1 strain MA116 was derived by transforming pC194 into the ICEBs1 donor strain MMB970 (ICEBs1 Δ(rapI-phrI)342::kan, Pxyl-rapI) and selecting for chloramphenicol resistance. pC194-containing Tn916 strain MA1100 was derived by first transforming JMA222 (ICEBs10) (Auchtung et al., 2005) with chromosomal DNA from BS49 (Browne et al., 2015; Haraldsen et al., 2003) and selecting for tetracycline resistance to introduce Tn916. The comC::mls allele was introduced by transformation. pC194 was introduced by mobilization (conjugation) from strain MA116 and selecting for chloramphenicol and tetracycline resistance.
Deletion of yddJ.
We constructed a deletion of yddJ that extends from the first base pair downstream from the yddI open reading frame (61 base pairs upstream from the yddJ start codon to the second bp downstream from the yddJ open reading frame. This deletion was constructed by amplifying two ~0.5 kb fragments containing DNA flanking the deletion endpoints by PCR and cloning them into pCAL1422 (a plasmid that contains E. coli lacZ) via isothermal assembly (Gibson et al., 2009), essentially as previously described for other alleles (Thomas et al., 2013; Wright et al., 2015). The resulting plasmid, pTD113, was integrated into the chromosome via single-crossover recombination. Transformants were screened for loss of lacZ, indicating loss of the integrated plasmid, and PCR was used to identify a clone that contained the ΔyddJ allele.
Construction of PyddJ-yddJ and Pspank(hy)-yddJ at lacA.
We expressed yddJ from its own promoter by inserting the region spanning 600 bp upstream to the end of the yddJ open reading frame into lacA to generate PyddJ-yddJ, present in strain MA996. We also fused yddJ to the LacI-repressible IPTG-inducible promoter Pspank(hy) to test yddJ function. Constructs included Pspank(hy)-yddJ (from ICEBs1), present in strain MA982, and Pspank(hy)-yddJICEBat1, (yddJ from ICEBat1) present in strain KPD219. yddJ from ICEBs1 was PCR amplified from genomic DNA from strain AG174. yddJICEBat1 was amplified by PCR from genomic DNA from B. atrophaeus strain 11A1 (from the Bacillus Genetic Stock Center; www.bgsc.org). For the PyddJ-yddJ construct, the PCR fragment was inserted by isothermal assembly between the PacI and SacI sites of pCJ80, a cloning vector that contains Pspank(hy), lacI, an mls cassette, and flanking homology for insertion by double-crossover into the chromosome at lacA (Wright & Grossman, 2016). For the Pspank(hy)-yddJ constructs, the PCR fragments were inserted by isothermal assembly between the SphI and SacI sites of pCJ80. The alleles were integrated by double cross-over into the chromosome by transformation and selecting for MLS resistance, generating the alleles lacA::{Pspank(hy)-yddJ mls} or lacA::{Pspank(hy)-yddJICEBat1 mls}.
Construction of Pspank(hy)-conG at thrC.
conG was amplified by PCR from B. subtilis or B. atrophaeus genomic DNA and fused to Pspank(hy) essentially as described (Leonetti et al., 2015). Constructs included Pspank(hy)-conGICEBs1, present in strain KPD225, and Pspank(hy)-conGICEBat1, present in strain KPD224. These alleles were used to complement the ΔconG(5–805) deletion in ICEBs1.
Construction of isogenic strains used for determining effects of exclusion on survival.
Transfer-competent wild type and exclusion-deficient mutants of ICEBs1 (containing the Δ(rapI-phrI)342::kan allele) were transferred by conjugation into MA1027 (ICEBs10 amyE::{Pxyl-rapI spc} comC::mls). The corresponding transfer-deficient mutants of ICEBs1 were transformed into BOSE986 (ICEBs10 amyE::{Pxyl-rapI spc}), then transformed with DNA from MA1012 (ICEBs10 comC::mls) selecting for mls, thereby making the cells defective in competence. MA1049 (ICEBs1), MA1050 (ICEBs1 ΔyddJ), and MA1089 (ICEBs1 conG-E288K) were derived by transferring ICE from MMB970, MA11, and KPD80 into MA1027, and selecting for kanamycin and MLS resistance. MA1069 (ICEBs1 ΔyddJ ΔconQ) and MA1070 (ICEBs1 ΔconQ) were derived by transforming chromosomal DNA from CAL848 with and without ΔyddJ into BOSE986, selecting for kanamycin resistance, and introducing comC::mls by transforming chromosomal DNA from MA1012.
Mating assays
Mating assays for ICEBs1 were performed essentially as described (Auchtung et al., 2005; Lee et al., 2007). Briefly, donor and recipient cells were grown in S750 defined minimal medium containing 0.1 % glutamate and 1% arabinose until they reached mid-exponential growth phase. At an OD600 of 0.2, 1% xylose was added to donors to induce expression of Pxyl-rapI, causing induction of ICEBs1. For recipients containing Pspank(hy)-yddJ, 1 mM IPTG was added as indicated. After 2 hours of growth in the presence of xylose, equal numbers (~4×108 cells each) of donor and recipient cells were mixed and collected by vacuum filtration onto a nitrocellulose filter. Filters were incubated at 37°C for 3 hours on 1.5% agar plates containing 1x Spizizen’s salts (2 g/l (NH4)SO4, 14 g/l K2HPO4, 6 g/l KH2PO4, 1 g/l Na3 citrate-2H2O, 0.2 g/l MgSO4-7H20) (Harwood & Cutting, 1990). Cells were resuspended from the filters, diluted and plated on LB agar plates containing the appropriate antibiotics to select for transconjugants. Plates were incubated at 37°C overnight to allow for colony growth. The number of donor cells (CFU/ml) was determined at the time of cell mixing (after growth in xylose for 2 hours). Mating efficiency was calculated as the percent transconjugants CFU/ml per initial donor CFU/ml. The number of initial donors, rather than the number of viable donors post-mating, was used in these calculations for two reasons: 1) there is limited growth of cells on filters during mating; and importantly, 2) there is some loss of viability of donor cells on the filters during mating. This loss of viability leads to an overestimate of mating efficiencies per initial donor.
Mutagenesis and enrichment screens
For the initial round of the screen, 16 independent cultures of ICEBs1 donor cells (strain KPD38) were grown in LB medium to mid-exponential phase, and mutagenized with 1.2% ethyl methylsulfonate for 40 minutes, resulting in ~50% killing and a ~100-fold increase in the frequency of streptomycin resistant mutants, essentially as described (Grossman et al., 1992).Cells were pelleted, washed twice with LB, then resuspended to an OD600 of 0.125 and allowed to continue growing. At an OD600 of 0.5, rapI expression was induced by adding 1% xylose, and cells were grown for 30 minutes to an OD600 of ~1. Recipient cells (KPD36: ICEBs10, Pxyl-rapI cat, Pspank(hy)-yddJ, comK::tet) were grown in LB to mid-exponential phase. At an OD600 of 0.1, yddJ expression in recipients was induced with 1mM IPTG, and cells were grown to an OD600 of ~1.0. Equal numbers of donor and recipient cells were mixed and collected by vacuum filtration onto a nitrocellulose filter. Filters were incubated at 37°C for 3 hours on 1.5% agar plates containing 1x Spizizen’s salts. Cells were resuspended from the filters and plated on LB agar containing tetracycline and kanamycin to select for transconjugants.
For the second round of the screen, transconjugants from the previous round (now donors) were scraped off of the selection plates (above), resuspended in LB containing kanamycin and tetracycline, and diluted to an OD600 of 0.125. These donor cells were grown to an OD600 of 0.5 and induced as described above. Recipient cells (KPD35: ICEBs10, Pxyl-rapI spc, Pspank(hy)- yddJ, str-84 comK::tet) for the second round were grown and induced as described above. As before, donor and recipient cells were mixed, filtered, and incubated at 37°C for 3 hours on 1.5% agar plates containing 1x Spizizen’s salts. Cells were resuspended from the filters and plated on LB agar containing spectinomycin and kanamycin to select for transconjugants.
Rounds three and four of the screen repeated the process described for round two with alternating recipient strains: In round three, KPD36 was used as the recipient strain and transconjugants were selected with chloramphenicol and kanamycin. In round four, KPD35 was used as the recipient strain and transconjugants were selected with spectinomycin and kanamycin. After four rounds of the screen, exclusion-resistant mutants were sufficiently enriched in the transconjugant population that exclusion by yddJ was no longer observed. At this point, transconjugants were restreaked to purity, genomic DNA was isolated from two colonies from each of the 16 independent parallel enrichments, and ICEBs1 was sequenced to identify the mutations that were likely causing the exclusion-resistant phenotype. Mating survival assays
Cells were grown in S750 defined minimal medium containing 0.1% glutamate and 1% arabinose until they reached mid-exponential growth phase. At an OD600 of 0.2, 1% xylose was added to induce expression of Pxyl-rapI, causing induction of ICEBs1. After 2 hours of growth in the presence of xylose, ~8×108 cells were mixed with 5 ml of medium and filtered onto a nitrocellulose filter and incubated at 37°C for 3 hours on 1.5% agar plates containing 1x Spizizen’s salts, as if for a mating experiment. Cells were resuspended from the filters, diluted and plated on LB agar to determine post-mating viable cell counts. The number of viable cells was also determined prior to filtering the cells.
For survival assays testing the effect of high and low cell density, cells were prepared as described above, with the exception that after 2 hours of induction with xylose, either ~8×108 or ~3×106 cells were mixed with 5 ml or 10 ml of medium, respectively, before sampling and filtering.
ACKNOWLEDGMENTS
We thank Jennifer Thornton and Chris Johnson for initial experiments demonstrating exclusion, Chris Johnson and Joshua Jones for initial investigations on ICEBat1, and Janet Smith, Luciane Schons Fonseca, M. Michael Harden, Mary Anderson, and Michael Laub for comments on the manuscript. Research reported here is based upon work supported, in part, by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01 GM050895 and R35 GM122538 to ADG. MA and KPD were also supported, in part, by the NIGMS pre-doctoral training grant T32 GM007287. Any opinions, findings, and conclusions or recommendations expressed in this report are those of the authors and do not necessarily reflect the views of the National Institutes of Health.
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Footnotes
DATA AVAILABILTY STATEMENT. The data that support the findings of this study are available from the corresponding author upon reasonable request.
CONFLICTS OF INTEREST. None. We have no conflicts of interests.
REFERENCES
- Anthony KG, Klimke WA, Manchak J, & Frost LS (1999). Comparison of proteins involved in pilus synthesis and mating pair stabilization from the related plasmids F and R100–1: Insights into the mechanism of conjugation. J Bacteriol, 181(17), 5149–5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchtung JM, Aleksanyan N, Bulku A, & Berkmen MB (2016). Biology of ICEBs1, an integrative and conjugative element in Bacillus subtilis. Plasmid, 86, 14–25. 10.1016/j.plasmid.2016.07.001 [DOI] [PubMed] [Google Scholar]
- Auchtung JM, Lee C. a, Garrison KL, & Grossman AD (2007). Identification and characterization of the immunity repressor (ImmR) that controls the mobile genetic element ICEBs1 of Bacillus subtilis. Mol Microbiol, 64(6), 1515–1528. 10.1111/j.1365-2958.2007.05748.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchtung JM, Lee C. a, Monson RE, Lehman AP, & Grossman AD (2005). Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. PNAS, 102(35), 12554–12559. 10.1073/pnas.0505835102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audette GF, Manchak J, Beatty P, Klimke W. a, & Frost LS (2007). Entry exclusion in F-like plasmids requires intact TraG in the donor that recognizes its cognate TraS in the recipient. Microbiology, 153(Pt 2), 442–451. 10.1099/mic.0.2006/001917-0 [DOI] [PubMed] [Google Scholar]
- Babic A, Berkmen MB, Lee CA, & Grossman AD (2011). Efficient Gene Transfer in Bacterial Cell Chains. MBio, 2(2), e00027–11. 10.1128/mBio.00027-11.Editor [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaber JW, Hochhut B, & Waldor MK (2002). Genomic and Functional Analyses of SXT, an Integrating Antibiotic Resistance Gene Transfer Element Derived from Vibrio cholerae Genomic and Functional Analyses of SXT, an Integrating Antibiotic Resistance Gene Transfer Element Derived from Vibrio choler. J Bacteriol, 184(15), 4259–4269. 10.1128/JB.184.15.4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkmen MB, Lee CA, Loveday E, & Grossman AD (2010). Polar Positioning of a Conjugation Protein from the Integrative and Conjugative Element ICEBs1 of Bacillus subtilis. J Bacteriol, 192(1), 38–45. 10.1128/JB.00860-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatty M, Laverde Gomez JA, & Christie PJ (2013). The Expanding Bacterial Type IV Lexicon. Res Microbiol, 164(6). 10.1037/a0018493.Understanding [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose B, Auchtung JM, Lee CA, & Grossman AD (2008). A conserved anti-repressor controls horizontal gene transfer by proteolysis. Mol Microbiol, 70(3), 570–582. 10.1111/j.1365-2958.2008.06414.x.A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WM, & Dziegielewska KM (1997). Friends and relations of the cystatin superfamily--new members and their evolution. Protein Science : A Publication of the Protein Society, 6(1), 5–12. 10.1002/pro.5560060102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne HP, Anvar SY, Frank J, Lawley TD, Roberts AP, & Smits WK (2015). Complete genome sequence of BS49 and draft genome sequence of BS34A, Bacillus subtilis strains carrying Tn916. FEMS Microbiol Lett, 362(3), 1–4. 10.1093/femsle/fnu050 [DOI] [PubMed] [Google Scholar]
- Burrus V, Pavlovic G, Decaris B, & Guédon G (2002). The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid, 48(2), 77–97. 10.1016/S0147-619X(02)00102-6 [DOI] [PubMed] [Google Scholar]
- Burrus V, & Waldor MK (2004). Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol, 155(5), 376–386. 10.1016/j.resmic.2004.01.012 [DOI] [PubMed] [Google Scholar]
- Dewitt T, & Grossman AD (2014). The Bifunctional Cell Wall Hydrolase CwlT Is Needed for Conjugation of the Integrative and Conjugative Element ICEBs1 in Bacillus subtilis and B. anthracis. J Bacteriol, 196(8), 1588–1596. 10.1128/JB.00012-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl AM, Losick R, & Kolter R (2007). Bacillus subtilis Genome Diversity. J Bacteriol, 189(3), 1163–1170. 10.1128/JB.01343-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, … Mitchell AL (2017). InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res, 45(Database issue), D190–D199. 10.1093/nar/gkw1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima T, Kitajima T, Yamaguchi H, Ouyang Q, Furuhata K, Yamamoto H, … Sekiguchi J (2008). Identification and characterization of novel cell wall hydrolase CwlT: a two-domain autolysin exhibiting n-acetylmuramidase and DL-endopeptidase activities. J Biol Chem, 283(17), 11117–11125. 10.1074/jbc.M706626200 [DOI] [PubMed] [Google Scholar]
- Garcillan-Barcia MP, & de la Cruz F (2008). Why is entry exclusion an essential feature of conjugative plasmids? Plasmid, 60(1), 1–18. 10.1016/j.plasmid.2008.03.002 [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R, Venter JC Iii, H. CA, Smith HO, & America N (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods, 6(5), 12–16. 10.1038/NMETH.1318 [DOI] [PubMed] [Google Scholar]
- Goessweiner-Mohr N, Arends K, Keller W, & Grohmann E (2014). Conjugation in Gram-Positive Bacteria. Plasmids: Biology and Impact in Biotechnology and Discovery, 237–256. 10.1128/microbiolspec.PLAS-0004-2013 [DOI] [Google Scholar]
- Grohmann E, Christie PJ, Waksman G, & Backert S (2018). Type IV secretion in Gram-negative and Gram-positive bacteria. Mol Microbiol, 107(4), 455–471. 10.1111/mmi.13896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman AD, Lewis T, & Devivo R (1992). Suppressors of a spo0A missense mutation and their effects on sporulation in Bacillus subtilis. Biochimie, 74, 679–688. [DOI] [PubMed] [Google Scholar]
- Guerout-Fleury A-M, Shazand K, Frandsen N, & Stragier P (1995). Antibiotic-resistance cassettes for Bacillus subtilis. Gene, 167, 335–336. [DOI] [PubMed] [Google Scholar]
- Guglielmini J, Néron B, Abby SS, Garcillán-Barcia MP, La Cruz DF, & Rocha EPC (2014). Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res, 42(9), 5715–5727. 10.1093/nar/gku194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmini J, Quintais L, Garcillán-Barcia MP, de la Cruz F, & Rocha EPC (2011). The repertoire of ice in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet, 7(8). 10.1371/journal.pgen.1002222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunton JE, Ussher JER, Rooker MM, Wetsch NM, Alonso G, & Taylor DE (2008). Entry exclusion in the IncHI1 plasmid R27 is mediated by EexA and EexB. Plasmid, 59(2), 86–101. 10.1016/j.plasmid.2007.11.004 [DOI] [PubMed] [Google Scholar]
- Haraldsen JD, & Sonenshein AL (2003). Efficient sporulation in Clostridium difficile requires disruption of the s K gene. Mol Microbiol, 48(3), 811–821. [DOI] [PubMed] [Google Scholar]
- Harwood CR, & Cutting SM (1990). Molecular Biological Methods for Bacillus. Chichester: John Wiley & Sons. [Google Scholar]
- Hochhut B, Marrero J, & Waldor MK (2000). Mobilization of Plasmids and Chromosomal DNA Mediated by the SXT Element, a Constin Found in Vibrio cholerae O139. J Bacteriol, 182(7), 2043–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaacks KJ, Healy J, Losick R, & Grossman AD (1989). Identification and Characterization of Genes Controlled by the Sporulation-Regulatory Gene spo0H in Bacillus subtilis. J Bacteriol, 171(8), 4121–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski SJ, Krishnamoorthy V, Cascales E, & Christie PJ (2004). Agrobacterium tumefaciens VirB6 Domains Direct the Ordered Export of a DNA Substrate Through a Type IV Secretion System. J Mol Biol, 341(4), 961–977. 10.1037/a0017530.A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CM, & Grossman AD (2014). Identification of host genes that affect acquisition of an integrative and conjugative element in Bacillus subtilis. Mol Microbiol, 93(6), 1284–1301. 10.1111/mmi.12736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CM, & Grossman AD (2015). Integrative and Conjugative Elements (ICEs): What They Do and How They Work. Annu Rev Genet, 49, 577–601. 10.1146/annurev-genet-112414-055018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, & Morishima K (2017). KEGG : new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res, 45, 353–361. 10.1093/nar/gkw1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, & Sternberg MJE (2015). The Phyre2 web portal for protein modelling, prediction and analysis. Nat Protoc, 10(6), 845–858. 10.1038/nprot.2015.053.The [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komano T, Yoshida T, Narahara K, & Furuya N (2000). The transfer region of Incl1 plasmid R64: Similarities between R64 tra and Legionella icm/dot genes. Mol Microbiol, 35(6), 1348–1359. 10.1046/j.1365-2958.2000.01769.x [DOI] [PubMed] [Google Scholar]
- Lawley TD, Klimke WA, Gubbins MJ, & Frost LS (2003). F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett, 224(1), 1–15. 10.1016/S0378-1097(03)00430-0 [DOI] [PubMed] [Google Scholar]
- Lécuyer F, Bourassa J, Gélinas M, Charron-Lamoureux V, Burrus V, Beauregard PB (2018). Biofilm Formation Drives Transfer of the Conjugative Element ICEBs1 in Bacillus subtilis. mSphere, 3(5), e00473–18. 10.1128/mSphere.00473-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CA, & Grossman AD (2007). Identification of the origin of transfer (oriT) and DNA relaxase required for conjugation of the integrative and conjugative element ICEBs1 of Bacillus subtilis. J Bacteriol, 189(20), 7254–7261. 10.1128/JB.00932-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CA, Thomas J, & Grossman AD (2012). The Bacillus subtilis conjugative transposon ICEBs1 mobilizes plasmids lacking dedicated mobilization functions. J Bacteriol, 194(12), 3165–3172. 10.1128/JB.00301-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonetti CT, Hamada MA, Laurer SJ, Broulidakis MP, Swerdlow KJ, Lee CA, … Berkmen MB (2015). Critical components of the conjugation machinery of the integrative and conjugative element ICEBs1 of Bacillus subtilis. J Bacteriol, 197(15), 2558–2567. 10.1128/JB.00142-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrero J, & Waldor MK (2005). Interactions between inner membrane proteins in donor and recipient cells limit conjugal DNA transfer. Dev Cell, 8(6), 963–970. 10.1016/j.devcel.2005.05.004 [DOI] [PubMed] [Google Scholar]
- Marrero J, & Waldor MK (2007). Determinants of entry exclusion within Eex and TraG are cytoplasmic. J Bacteriol, 189(17), 6469–6473. 10.1128/JB.00522-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naglich JG, & Andrews RE (1988). Tn916-Dependent Conjugal Transfer of pC194 and pUB110 from Bacillus subtilis into Bacillus thuringiensis subsp. israelensis. Plasmid, 20, 113–126. [DOI] [PubMed] [Google Scholar]
- Norgren M, & Scott JR (1991). The presence of conjugative transposon Tn916 in the recipient strain does not impede transfer of a second copy of the element. J Bacteriol, 173(1), 319–324. 10.1128/jb.173.1.319-324.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto A, Bernhardt J, Meyer H, Schaffer M, Herbst FA, Siebourg J, … Becher D (2010). Systems-wide temporal proteomic profiling in glucose-starved Bacillus subtilis. Nat Commun, 1(9). 10.1038/ncomms1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou JT (1980). Role of surface exclusion genes in lethal zygosis in Escherichia coli K12 mating. Mol Gen Genet, 178(3), 573–581. 10.1007/BF00337863 [DOI] [PubMed] [Google Scholar]
- Perego M, Spiegelman GB, & Hoch JA (1988). Structure of the gene for the transition state regulator, abrB : regulator synthesis is controlled by the spoOA sporulation gene in Bacillus subtilis. Mol Microbiol, 2(6), 689–699. [DOI] [PubMed] [Google Scholar]
- Sakuma T, Tazumi S, Furuya N, & Komano T (2013). ExcA proteins of IncI1 plasmid R64 and IncIγ plasmid R621a recognize different segments of their cognate TraY proteins in entry exclusion. Plasmid, 69(2), 138–145. 10.1016/j.plasmid.2012.11.004 [DOI] [PubMed] [Google Scholar]
- Simonen M, & Palva I (1993). Protein Secretion in Bacillus Species. Microbiol Rev, 57(1), 109–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurray RA, & Reeves P (1973). Characterization of lethal zygosis associated with conjugation in Escherichia coli K-12. J Bacteriol, 113(1), 58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurray RA, & Reeves P (1974). F Factor-Mediated Immunity to Lethal Zygosis in Escherichia coli K-12 F Factor-Mediated Immunity to Lethal Zygosis in Escherichia coli K-12. J Bacteriol, 117(1), 100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurray RA, Willetts N, & Reeves P (1976). Effect of tra mutations on F factor-specified immunity to lethal zygosis. Mol Gen Genet, 146(2), 161–165. 10.1007/BF00268084 [DOI] [PubMed] [Google Scholar]
- Thomas J, Lee CA, & Grossman AD (2013). A Conserved Helicase Processivity Factor Is Needed for Conjugation and Replication of an Integrative and Conjugative Element. PLoS Genet, 9(1). 10.1371/journal.pgen.1003198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine PJ, Shoemaker NB, & Salyers AA (1988). Mobilization of Bacteroides Plasmids by Bacteroides Conjugal Elements. J Bacteriol, 170(3), 1319–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoeven N (1985). Evolution of bacterial surface exclusion against incompatible plasmids. J Theor Biol, 117(3), 431–452. 10.1016/S0022-5193(85)80153-3 [DOI] [PubMed] [Google Scholar]
- Wright LD, & Grossman AD (2016). Autonomous Replication of the Conjugative Transposon Tn 916. J Bacteriol, 198(24), 3355–3366. 10.1128/JB.00639-16.Editor [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright LD, Johnson CM, & Grossman AD (2015). Identification of a Single Strand Origin of Replication in the Integrative and Conjugative Element ICE Bs1 of Bacillus subtilis. PLoS Genet, 11(10), e1005556 10.1371/journal.pgen.1005556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Boekhorst J, Francke C, & Siezen RJ (2008). LocateP : Genome-scale subcellular-location predictor for bacterial proteins. BMC Bioinformatics, 9(173), 1–17. 10.1186/1471-2105-9-173 [DOI] [PMC free article] [PubMed] [Google Scholar]