Abstract
Acute erythroid leukemia (AEL) is a high risk leukemia of poorly understood genetic basis, with controversy regarding diagnosis in the spectrum of myelodysplasia and myeloid leukemia. We compared genomic features of 159 childhood and adult AEL cases to non-AEL myeloid disorders, and defined 5 age-related subgroups with distinct transcriptional profiles: adult, TP53-mutated; NPM1-mutated; KMT2A-mutated/rearranged; adult, DDX41-mutated; and pediatric, NUP98-rearranged. Genomic features influenced outcome, with NPM1 mutations and HOXB9 over-expression associated with favorable prognosis, and TP53, FLT3 or RB1 alterations associated with poor survival. Targetable signaling mutations were present in 45% of cases, and included recurrent mutations of ALK and NTRK1, the latter of which drive erythroid leukemogenesis sensitive to TRK inhibition. This genomic landscape of AEL provides the framework for accurate diagnosis and risk stratification of this disease, and the rationale for testing targeted therapies in this high-risk leukemia.
Editorial summary:
Analysis of genomic and clinical features of acute erythroid leukemia in comparison to other myeloid disorders argues for its distinct classification, defines subgroups and suggests therapeutic vulnerabilities.
INTRODUCTION
The last decade has seen major advances in the identification of clinically relevant markers for diagnosis, prognostication and disease monitoring in acute myeloid leukemia (AML)1,2, however the genetic basis of several AML subtypes, including acute erythroid leukemia (AEL), remains poorly characterized.3–5 AEL is characterized by proliferation of erythroid and myeloid blast cells in the bone marrow and is associated with a poor prognosis4,6,7. Since its first description by Di Guglielmo8, AEL has been diagnosed and subclassified by morphology alone, using variable criteria of questionable clinical significance without consideration of underlying biological features. In the initial French-American-British (FAB)9 classification and the subsequent World Health Organization (WHO) 2008 classification of myeloid neoplasms, two AEL subtypes were defined based on erythroid percentage and non-erythroid blast proportion10. “M6a” cases had at least 50% erythroid cells and at least 20% blasts of non-erythroid cells in bone marrow. “M6b” cases were characterized by at least 80% of bone marrow cells consisting of erythroid precursors without an increase in myeloblasts. Both entities were regarded as subtypes of AML, and consequently, the use of intensive cytarabine-based anti-leukemic treatment regimens. However, erythroleukemia classification was significantly changed in the revision of the WHO classification of hematopoietic malignancies11. M6a was merged into a hybrid subtype of myelodysplasia and AML (specifically, “myelodysplastic syndrome (MDS) or AML, not otherwise specified (NOS) (non-erythroid subtype)” based on the percentage of blasts in the bone marrow rather than biological or genetic features. M6b remained as a subtype of “AML, NOS, acute erythroid leukemia (pure erythroid type)” but with the new criteria of at least 30% of proerythroblasts due to its association with poor prognosis12.
There are few data regarding the genomic basis of AEL13,14, and none to guide appropriate classification. For example, only three cases with AEL were included in The Cancer Genome Atlas (TCGA) study of AML.1 As patients with AEL are more likely to receive aggressive induction chemotherapy rather than the less intensive treatments used for MDS, diagnosis and sub-classification has important implications for therapy. In view of these diagnostic uncertainties13–15 and the high risk nature of this leukemia16 we performed a comprehensive genomic analysis of both childhood and adult AEL and compared the mutational landscape to that of MDS and non-erythroid AML. Moreover, we examined associations between genomic characteristics and outcome, and developed experimental models of AEL to examine leukemogenic potential of genomic alterations and to test the efficacy of targeted therapeutic approaches.
RESULTS
Clinical characteristics
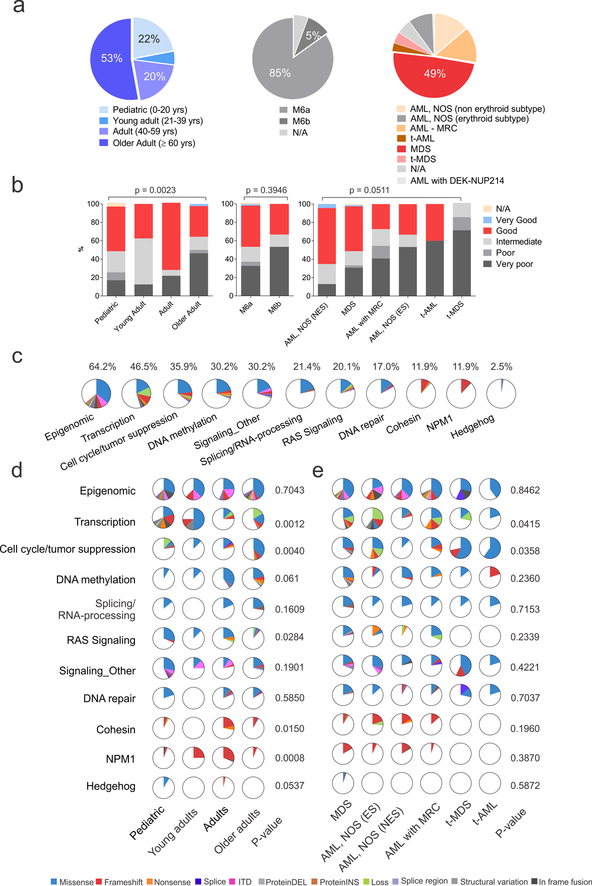
The cohort of 159 cases included 35 pediatric cases (0–20 years, 22%), 8 young adults (21–39 years, 5%), 32 adults (40–59 years, 20%), and 84 older adults (≥ 60 years, 53%) (Supplementary Tables 1–2 and Fig. 1a). Eighty-five percent of cases were centrally confirmed as M6a and 5% as M6b according to FAB/WHO 2008 criteria. M6a cases were re-classified under WHO 2016 criteria as MDS (49.1%), AML, NOS (non-erythroid subtype, 13.8%) or AML with myelodysplasia-related changes (AML-MRC, 13.8%). Very poor cytogenetic risk group as defined by the Revised International Prognostic Scoring System (IPSS-R)17 was more frequent in patients age 60 and older (46.4%) compared to those under age 20 (17.1%; P=0.0023; Fig. 1b).
Figure 1. Demographic, clinical and genomic patient’s characteristics.
(a) Distribution of patients according to age (left), WHO 2008 criteria (middle) and revised WHO 2016 criteria. (b) Revised International Prognostic Scoring System (IPSS-R)-based cytogenetic risk according to age (left panel), WHO 2008 (middle) and WHO 2016 (right panel) criteria (n= 159 biologically independent samples). Two-sided P values are from exact-Chi-square test (age and WHO 2008) and Chi-square test (WHO 2016) (c) Pie charts showing the distribution of the recurrently mutated pathways in the whole AEL cohort. (d) Pie charts showing the distribution of the recurrently mutated pathways according to age (n= 159 biologically independent samples) and WHO 2016 criteria (e) (n= 149 biologically independent samples). The similarity of somatic alteration prevalence in different leukemia subtypes was evaluated by two-sided Chi-Square test. See also Supplementary Tables 8 for numbers and P values for each pathway and gene. Abbreviations: yrs, years; N/A, information not available; ns, not significant; AML, NOS acute myeloid leukemia, not otherwise specified; NES, non erythroid subtype; ES, erythroid subtype; t-AML, therapy-related AML; t-MDS, therapy-related MDS.
Mutation burden and recurrently mutated genes in AEL
By genome and transcriptome sequencing analysis we identified a mean of 14.5 non-silent mutations (range 1–39) per case (Supplementary Tables 3–6 and Supplementary Fig. 1a). The mutational burden was independent of age and WHO 2008/2016 subtype, but was higher in cases of very poor cytogenetic risk (18.60, range 1–33) compared to good risk (12.29, range 1–30, P=2.81×10−6) and intermediate risk (11.65, range 1–28, P=0.0001; Supplementary Fig. 1b). Multimodal analysis of genomic alterations using the Genomic Random INterval (GRIN)18 model, identified 289 altered driver genes, 80 of which retained significance after multiple test correction (Supplementary Table 7). Fourteen genes are not included in the Catalogue of Somatic Mutations in Cancer (COSMIC v81; Supplementary Table 7) or described as targets of mutation in AML and MDS19. These included CCDC30 (n=4), MAD1L1 (n=3), UBTF (n=3) and DDX25 (n=2). Mutations in FAM186A, MYH4, UBTF, or PTPN1 were confirmed to be somatic in 9 cases with available germline material. Eleven pathways were recurrently mutated: epigenetic regulation (n=102, 64.2%), transcriptional regulation (n=74, 46.5%), cell cycle/tumor suppression including TP53 mutations (n=57, 35.9%), DNA methylation (N=48, 30.2%), splicing/RNA processing (n=34, 21.4%), Ras signaling (n=32, 20.1%), non-Ras signaling (n=48, 30.2%), DNA repair (n=27, 17.0%), cohesin (n=19, 12.0%), NPM1 (n=19, 12.0%) and Sonic Hedgehog signaling (n=4, 2.5%) (Supplementary Figs. 1c and 2–3; Supplementary Table 8 and Supplementary Note).
The frequency of alteration of these pathways varied according to age at diagnosis and subtype of myeloid malignancy. Genes encoding transcriptional regulators (e.g. WT1 and UBTF) were more frequently mutated in children (P=0.0012) and in AML-MRC (P=0.0415). Mutations in Ras signaling genes were also most common in patients under 20 years of age (P=0.0284), whereas older patients were more likely to harbor mutations in DNMT3A, TET2 and ASXL1, genes implicated in clonal hematopoiesis of undetermined potential (CHIP)20–22. Cohesin and NPM1 genes were most commonly mutated in patients 20–59 years of age than in patients less than 20 or over 60 years of age(P=0.0150 and 0.0008, respectively) (Fig. 1d). The majority of sequence mutations in TP53 and genes encoding for epigenetic modifiers (e.g. KMT2D, KMT2C, DNMT3A, IDH1/2, and TET2) had clonal mutant allele fractions, consistent with the notion that they are acquired early during tumorigenesis, but as they occurred with other recurrent mutations, are insufficient for full leukemic transformation. For example, clonal mutations in splicing factors or cohesin genes were frequently associated with mutations in epigenetic modifiers. Subclonal mutations were most frequently identified in transcription factors and signaling genes, suggesting that these are secondary events. Detailed results describing the type and frequency of mutations in pediatric and adult AEL cases are reported in the Supplementary Note and Supplementary Tables 9–11.
TP53 alterations in AEL
Mutations in TP53 genes and tumor suppression genes were present in 57 (35.9%) cases, and were more frequent in patients age 60 and older (P=1.60×10−5) (Fig. 1e) (Supplementary Tables 8–9). All but one of the TP53-mutated cases exhibited alterations of both alleles, as two clonal sequence mutations (28.0% of mutated cases), a mutation and DNA copy-neutral loss of heterozygosity (29.0%), or mutation and deletion of the other allele (39%) (Supplementary Fig. 2e). Sequence mutations were predominantly missense and occurring in the DNA binding domain (Supplementary Fig. 2f) and they were almost exclusively in adult patients with only two mutated pediatric case. Notably, in two cases we identified cryptic complex structural variations (in one case only identifiable by whole genome sequencing) involving the first intron of TP53, and predicted to prevent translation (Supplementary Fig. 2g–i). These cases had the lowest expression of TP53 of all cases (Supplementary Fig. 2j). Thus, biallelic alterations of TP53 through sequence or structural alterations are a hallmark of a subset of AEL in adults.
Chromothripsis affects hematopoietic regulators
A notable finding was the presence of chromothripsis, a massive shattering and reassembly of chromosomes23,24, in 13.1% of AEL cases (18/137 cases with available DNA copy number data (Supplementary Tables 12–13 and Supplementary Figs. 4–5). Chromothripsis was more frequent than in MDS (3/301, 1.2%; two-sided P = 1.68×10−7, Fisher’s exact test)25 and de novo non-erythroid AML (0/197; two-sided P = 5.36×10−8, Fisher’s exact test)1. It was observed only in adults TP53-mutated cases and was associated with very poor cytogenetic risk (two-sided P = 1.99×10−9, Fisher’s exact test). The most frequent chromothriptic chromosome was 19 (n=6), which has not previously been reported as a target of chromothripsis in cancer. A minimal common deleted segment on chromosome 19 was seen in all cases (Supplementary Fig. 5); this segment harbors the erythroid transcription factor Kruppel-like factor-1 (KLF1)26 and the hematopoietic regulator nuclear factor I X (NFIX)27, suggesting that the chromosomal regions targeted by chromothripsis may have direct roles in leukemic transformation.
Comparative genomic analysis of AEL, AML and MDS
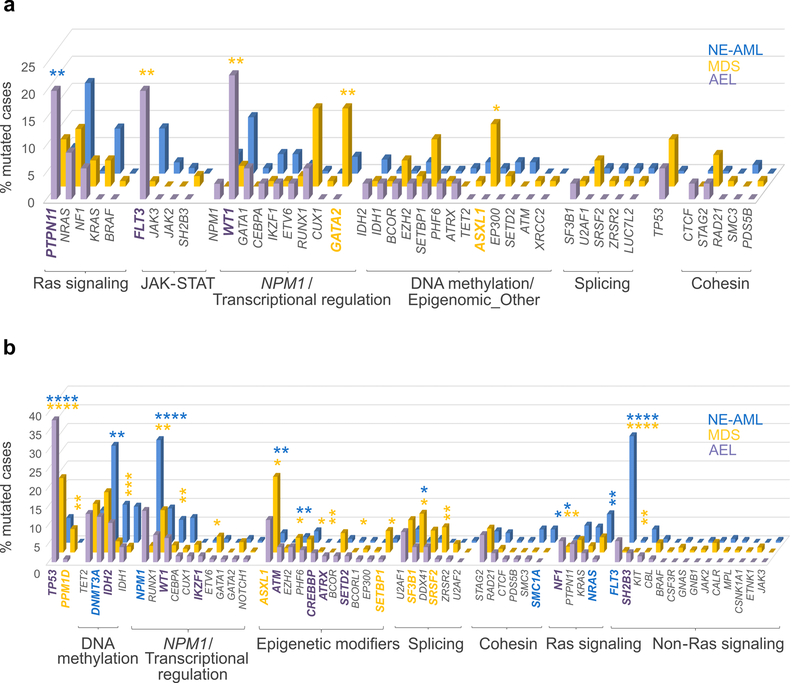
To examine the appropriateness of reclassification of many AEL cases as MDS or non erythroid AML from the WHO 2008 to 2016 criteria, we compared the genomic features of AEL to independent cohorts of pediatric and adult MDS (Supplementary Table 14) and non-erythroid AML (Supplementary Tables 15–16). Compared to childhood MDS, childhood AEL was characterized by higher frequency of mutations in FLT3 and WT1 (20.0% v 0.96%, P=0.0003 and 22.86% v 3.9%, P=0.002, respectively) and a lower frequency of mutations in GATA2 and ASXL1 (0 v 14.4%, P=0.012 and 0 v 11.5%, P=0.037, respectively). Compared to non-erythroid childhood AML, childhood AEL cases showed a higher frequency of mutations in the Ras pathway gene PTPN11 (20% v 4.7%, respectively, P=0.005), in epigenetic regulators such as KMT2A (8.6% v 0.5%, P=0.012), KMT2D, KMT2C and PHF8 (5.7 % v 0, P=0.023), in the transcriptional regulator UBTF (8.6% v 0.5%, P=0.012) and in the RNA-processing gene ELL (5.7 % v 0, P=0.023) (Fig. 2a and Supplementary Table 17).
Figure 2. Mutation rates in AEL (WHO 2008), non-erythroid AML and MDS patients.
(a) Comparison of mutation rates in pediatric (0–20 years) AEL (n=35 biologically independent samples), MDS (n=104 biologically independent samples) and non erythroid AML (n=192 biologically independent samples) patients. (b) Comparison of mutation rates in adult AEL (n=124 biologically independent samples), MDS (n=1410 biologically independent samples) and non-erythroid AML (n=197 biologically independent samples) patients. Mutated genes are grouped according to their function and the order within each group is based on the mutation frequency in AEL patients (from largest to smallest). Only cases for which sequencing data were available for all three cohorts are reported in the figure. Data are from non-silent SNV, indel or internal tandem duplication (ITD) sequence mutations. Frequency of mutations in the different leukemia subtypes were compared by two-sided Fisher’s exact tests (see Supplementary Tables 17–18 for numbers for each group and P values for each gene). P values are from Fisher’s exact test. *, P-value ≤ 0.05; **, P-value ≤ 0.01; ***, P-value ≤ 0.001; ****, P-value ≤ 0.0001. P values in purple are from AEL v MDS; those in blue are from: AEL v non erythroid AML (NE-AML). Genes whose mutation frequency is statistically different among the different myeloid entities are in bold and their color depends on the subtype with the higher frequency (purple: AEL, yellow: MDS and blue: NE-AML).
The mutational spectrum of adult AEL was intermediate between MDS and AML. For example, we observed much lower frequency of canonical genes mutated in AML such as FLT3 and NPM1 in AEL when compared to non-erythroid AML (P=2.13×10−7 and P=0.004, respectively) but they were more common than in MDS (P=2.18×10−5 and P=4.19×10−9, respectively). Conversely, MDS-associated mutations such as SF3B1 and ASXL1 were less frequent in AEL compared to MDS (P=0.018 and P=0.017, respectively), but more common than in non-erythroid AML (P=0.034 and P=0.002). Moreover, compared to MDS, adult AEL cases had a higher frequency of mutations in in the epigenetic regulators ATM (P=0.023), CREBBP (P=0.021), ATRX1 (P=0.006) and SETD2 (P=0.035), in the Ras signaling gene NF1 (P=0.007) and in the transcription factors WT1 (P=0.005) and IKZF1 (P=0.035). In contrast, infrequently mutated genes in adult AEL were PPM1D (P=0.008) and SRSF2 (P=0.006). Interestingly, disease-associated Ras mutations were identified, with NRAS rarely mutated in AEL compared to non-erythroid AML (0.8% v 7.6%, P=0.007) and NF1 with an opposite trend (5.6% v 1.0%, P=0.030) (Fig. 2b, Supplementary Note and Supplementary Tables 18–19). TP53 mutations are a hallmark of AEL and they were present in 37.9% of adult cases (39.5% of all adult AEL cases if cases with structural variations of TP53 were also included) but were less common in MDS (19.9%, P=1.02×10−5) and non-erythroid AML (6.6 %, P=4.15×10−12). Thus, mutational prevalence varies significantly between the three major subtypes of myeloid neoplasms (AEL, non-erythroid AML and MDS) in both children and adults, suggesting the recent reclassification of many AEL case as MDS or AML is unfounded from a mutational perspective.
Gene fusions targeting erythroid development and signaling
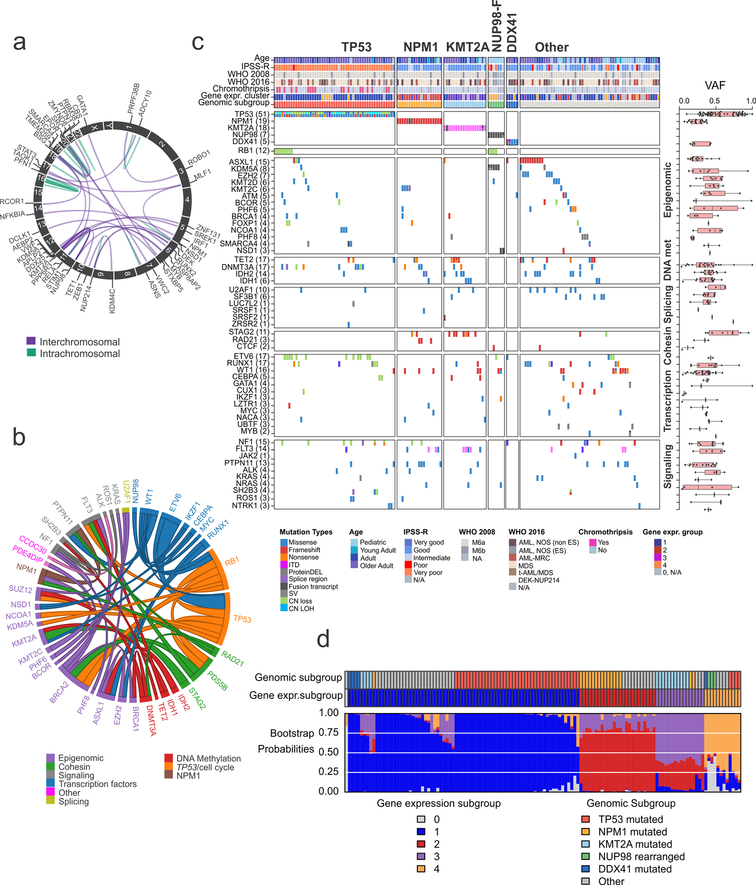
Chromosomal rearrangements resulting in expression of an in-frame fusion of two genes were found in 26.0% of AEL cases and were more frequent in cases with very poor cytogenetic risk. The only recurrent fusions were those involving NUP98 in pediatric cases (20.0%). ZMYND8-RELA, identified in a single case, has been previously reported28, indicating it is also recurrent in pediatric AEL. Additional gene fusions involved the erythroid transcription factor GATA1 (MYB-GATA1)29 or most commonly affected epigenetic regulators (KMT2A-rearrangements, ZEB1-KDM4C and SMARCA4-CBS) or signaling pathways (APLP2-EPOR, DEK-NUP214, ASNS-PTPN1, SRC-VWC2, RUNX2-STAT3 and PRKAR2B-PIK3C) (Supplementary Table 5, Fig. 3a and Supplementary Fig. 7).
Figure 3. Genomic classification of AEL.
(a) Schematic chimeric inter- (purple) and intra- (green) chromosomal in frame fusions in AEL. Gene labels are shown for interchromosomal fusions. (b) Pairwise gene associations with a Fisher’s exact test P value < 0.05. Different functional annotation categories are in different colors. The size of the ribbon is proportional to the number of cases (n= 159 biologically independent samples). (c) Heatmap showing the 5 genomic AEL subgroups (TP53, TP53-mutated AEL; NPM1, NPM1-mutated AEL; KMT2A, KMT2A-mutated or rearranged AEL; NUP98-F, AEL with NUP98 fusions; DDX41, DDX41-mutated AEL) and “other”, lacking a recurrent exclusive mutated gene and/or fusion gene (n= 159 biologically independent samples). (d) Color map of genomic subgroups, expression subgroup, and expression subgroup bootstrap probabilities. Each column of the color map corresponds to one patient (n= 130 biologically independent samples). The top row indicates the genomic subgroup of each patient according to the color legend at the bottom right. The middle row indicates the expression subgroup according to the color legend at the bottom left. The third row provides the expression subgroup bootstrap assignment probabilities according to the color legend at the bottom left. The primary assignment bootstrap probability was greater than 50% in 128 of 130 subjects; the mean primary assignment bootstrap probability across all subjects was 82.6%. Abbreviations: gene expr. group, gene expression group; mut: mutated; CN LOH: copy neutral loss of heterozygosity.
AEL comprises distinct genomic and gene expression groups
Both genomic alterations and gene expression profiles classified AEL cases into prognostically significant groups, independent of WHO subtype and with close but incomplete overlap in membership between the groups in each classifier. Based on patterns of mutation association and exclusivity, we defined five distinct genomic subtypes (Fig. 3c, Supplementary Table 20 and Supplementary Fig. 8): mostly adult, TP53-deregulated (n=51, 32%); NPM1-mutated (n=19, 12%); KMT2A-mutated/rearranged (n=18, 11%); pediatric, NUP98-rearranged (n=7, 4%); and adult, DDX41-mutated (n=5, 3%). Thirty-seven percent of cases lacked an identifiable exclusive recurrent founding genetic alteration, but as a group exhibited a higher frequency of mutations in ASXL1 (16.6% v 5.1%, P=0.02), and in the splicing factor gene SF3B1 (8.3% v 1.0%, P=0.03) compared to the other groups. Mutation patterns were subtype-dependent with mutations in tumor suppression genes more frequent in NUP98-fusion-positve AEL and TP53-mutated AEL (P=3.84×10−29) and mutation in cohesin genes more frequent in NPM1 and KMT2A-mutated/rearranged subgroups (P=7.39×10−5). Mutations in genes with a role in DNA methylation never co-occurred with NUP98 fusions nor DDX41 mutations (P=0.028) (Supplementary Table 21).
Genomic subtype-defining lesions were grouped by gene expression analysis in four distinct AEL subgroups (Fig. 3d). Group 1 was characterized by mutations in TP53 (54.9%) and overexpression of LTF, DLK1 and MECOM. Group 2 and group 3 were characterized by mutations in NPM1 (56.0%) and KMT2A (68.8%) genes, respectively, and by overexpression of PRDM16 and HOX genes (HOXB5, HOXB6, HOXB8 and HOXB9). Group 4 included cases with NUP98 fusions (25%) or additional cases with TP53 mutations (33.3%) and was characterized by overexpression of TMEM246, PLOD2, FREM1, MECOM and low expression of DEFA1B and DEF4A, compared to the other groups (Supplementary Table 22 and Supplementary Data).
Genomic determinants of outcome
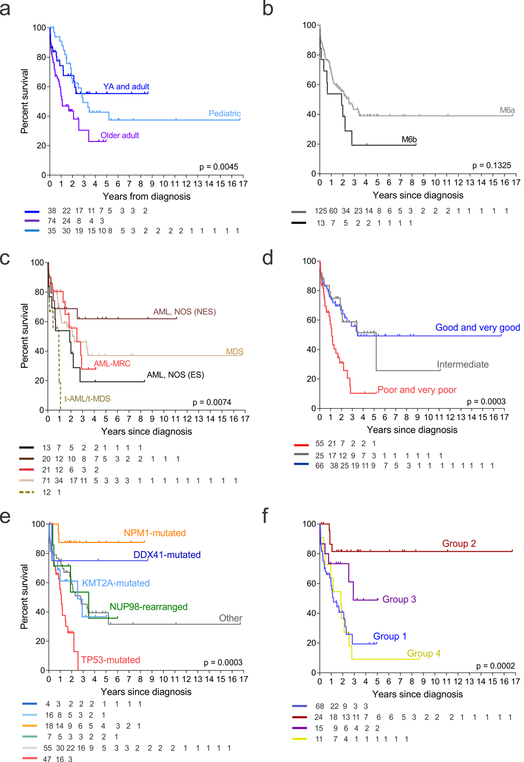
AEL is often associated with poor prognosis7, however except for cytogenetic risk factors the biological reasons for this have been unclear. Age, IPSS-R cytogenetic risk groups, therapy-related leukemia, genomic subgroups, gene expression classes, genetic pathways (tumor suppression) and individual genetic lesions (TP53, NPM1, FLT3, RB1) were associated with outcome in univariate analysis (Fig. 4, Supplementary Tables 24–25 and Supplementary Fig. 9). Among the genomic subgroups, TP53-mutationswere associated with very poor prognosis (median survival of 13 months, with no patients surviving at 5 years) while NPM1-mutated cases had excellent outcome with a 5-year survival of 87.5 % (95% confidential interval, CI: 60.5–100). Subgroups defined by gene expression also had marked variation in outcome, with group 4 (NUP98-fusions or a subset of TP53-mutated cases) having the worst outcome with an estimated 5-year survival of 9.1% (0.0–21.1). Conversely, group 2 (NPM1-mutated, HOXB-overexpressing) showed a better outcome with a 5-year survival of 81.6% (55.7–100). These associations were also observed in a subset of patients treated most intensively, with chemotherapy-based regimens (n= 81), with IPSS-R cytogenetic risk groups, genomic and gene expression subgroups, genetic pathways (tumor suppression) and individual genetic lesions (TP53 and NPM1) being associated with outcome in univariate analysis (Supplementary Table 26).Chromothripsis or the number or mutation burden (as measured by number of driver genes per individual patients was not independently associated with outcome in this cohort (Supplementary Table 25, and Supplementary Fig. 9). In a multivariate analysis incorporating genetic, clinical, and diagnostic variables, the gene expression classes but not WHO subgroups were the most powerful predictors of outcome (Supplementary Table 25).
Figure 4. Association with clinical outcome.
Kaplan–Meier survival curves with overall survival distributions according to age (a), WHO 2008 (b) and 2016 classification criteria (c), IPSS-R cytogenetic risk (d), genomic subgroups (e) and gene expression groups (f) (n= 147 independent individuals). At risk numbers for each analysis are provided in the figures. Outcome associations were analyzed with the log-rank test. Abbreviations: AML, NOS acute myeloid leukemia, not otherwise specified; NES, non erythroid subtype; ES, erythroid subtype; t-AML, therapy-related AML; t-MDS, therapy-related MDS.
Functional modeling of gene fusions
NUP98-KDM5A, as previously described30, but not other fusions (MYB-GATA1, ASNS-PTPN1, PFN1-SCHIP1, CDC37-IL27RA, NPM1-MLF1 or RUNX2-STAT3) was sufficient to confer self-renewal in vitro (Supplementary Fig. 10) and promoted the development of a serially transplantable myeloid (MPO and B220 positive) leukemia in mice. These results support our genomic observations that NUP98-KDM5A-positive cases, in contrast to cases with non NUP98-fusions, harbor few additional mutations, and this lesion is likely sufficient for leukemogenesis (Supplementary Note, Supplementary Tables 29–30 and Supplementary Fig. 10). In contrast, the other fusions were observed together with alterations in other recurrently mutated genes, suggesting cooperativity between these events. Although NUP98-KDM5A fusion was recurrent in AEL, the leukemia established in mice was myeloid, suggesting that additional alterations (e.g. RB1 deletions) or a different cell of origin are responsible for the leukemia phenotype.
Targetable signaling mutations in AEL
A mutation in at least one signaling pathway amenable to inhibition by tyrosine kinase/JAK2/Ras inhibitors was identified in 44.7% of cases (Supplementary Fig. 3a, b) and in 15.09% (24/159) co-occurred with TP53 mutations (Supplementary Table 24), known to confer refractoriness to conventional chemotherapy2. Mutations in tyrosine kinase-Ras pathway genes (NF1, PTPN11, KRAS and NRAS) occurred in 20.1% of all cases with multiple signaling mutations in 12.6% (20/159) of cases (Supplementary Fig. 3c). In these cases the mutations had lower variant allele frequencies than in cases with only one mutation, indicating presence in subclones (Supplementary Fig. 3d). Mutations in the JAK-STAT pathway genes (FLT3, JAK2, EPOR and SH2B3) occurred in 12.6% of all cases and in 20% (4/20) of cases they co-occurred with TP53 mutations.
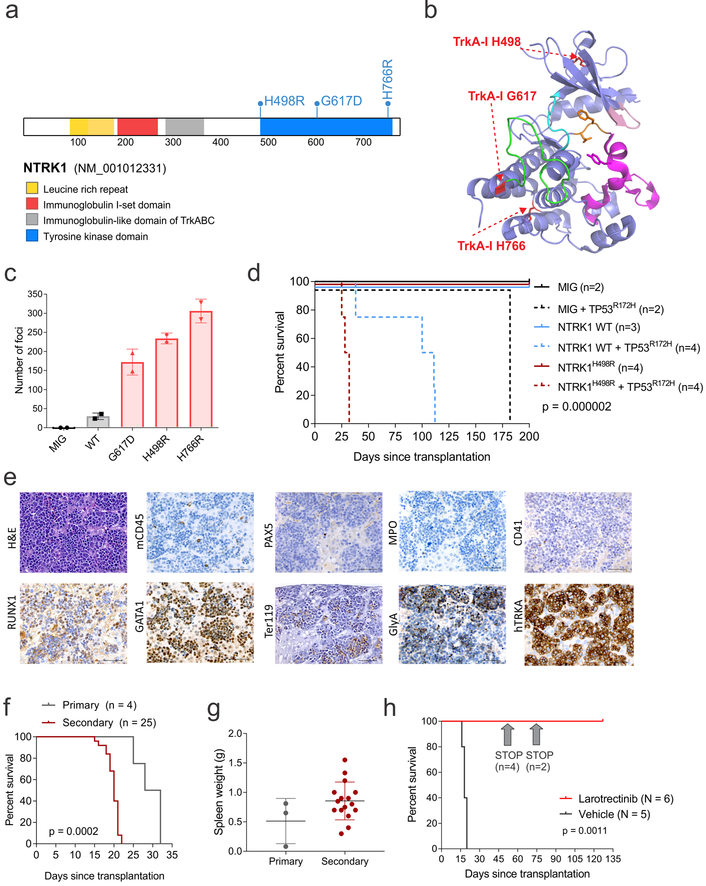
Additional mutations affected the phosphatidylinositol 3’ – kinase (PI3K) - AKT signaling pathway (7.5%) and a variety of different kinases (e.g. ALK, NTRK1, ABL class genes, PDGFRB) in 15.7% of all cases (Supplementary Fig. 3). NTRK1 mutations were observed in 3 cases, and while expressed, were not associated with higher NTRK1 expression compared to cases with wild-type NTRK1 (Supplementary Figure 11a), supporting the notion that NTRK1 is expressed in erythroid lineages as previously shown31,32. NTRK1-fusions or mutant alleles have been described to occur and play a role in solid tumors and hematological malignancies33–36, but they have not been reported in AEL, so we sought to examine their role in leukemogenesis and targetability. The mutations affected three residues in the tyrosine kinase domain (H498R, G617D and H766R) that are not involved in the binding of TRK inhibitors such as entrectinib (Fig. 5a, b) and do not alter cellular localization (Supplementary Fig. 11b). In focus formation assays, all three NTRK1 mutants promoted morphological transformation and loss of contact inhibition of NIH/3T3 cells (Fig. 5c), which was abrogated by entrectinib (Supplementary Fig. 11c, d).
Figure 5. NTRK1 mutations in AEL.
(a) NTRK1 mutations in AEL. (b) Structural modeling of NTRK1 mutations (in red) (PDB 4F0I43). The structure is represented similar to that of ref.44 with the DFG-motif in orange, the activation segment in magenta, the kinase insert domain in green, the hinge in cyan, and the G-loop in pink. (c) Focus formation assay in NIH/3T3 cells. Number of foci are from 2 week culture and two replicates. Mean and S.D. are shown. (d) Kaplan–Meier survival curves from mice transplanted with wild-type (WT, plain lines) or TP53R172H (dotted lines) HSPCs expressing NTRK1, NTRK1H498R or empty vector (MIG). (e) Hematoxylin and eosin staining and IHC of liver from a representative primary tumor with NTRK1H498R/TP53R172H induced erythroid leukemia. Scale bars represent 50 μm. This experiment was performed in an independent mouse obtaining similar results. (f) Kaplan–Meier survival curves of primary and secondary recipient mice. Outcome associations were analyzed with the log-rank test. (g) Spleen weight from primary (n = 3) and secondary (n=16) recipient mice with NTRK1H498R/TP53R172H induced leukemia. The mean expression is shown by the horizontal line in the scatter dot plot and the error bars represent the S.D. (h) Kaplan–Meier survival curves in mice treated with larotrectinib (n=6) or vehicle (n=5). In the drug-treated group larotrectinib was stopped after 49 (n=4 mice) and 69 (n=2 mice) days. Outcome associations between treated (n=6) and untreated (n=5) mice were analyzed with the log-rank test.
To examine leukemogenic potential of NTRK1 mutations, wild-type or mutated NTRK1 were expressed in lineage negative hematopoietic stem and progenitor cells (lin- HSPCs) from wild-type or TP53R172H (equivalent to human TP53R175H, present in 2 NTRK1-mutant cases) C57BL/6 mice37. While the expression of TP53R172H alone resulted in disseminated T-cell lymphoma, NTRK1H498R/G617D/H766R or wild type NTRK1, together with TP53R172H, promoted the development of an aggressive and transplantable erythroid leukemia, with significantly shorter latency observed in mice inoculated with lin- HSPCs harboring NTRK1/TP53 co-mutated mice compared to TP53-mutant, NTRK1 wild type HSPCs (Fig. 5d–g and Supplementary Fig. 11). To further investigate the mechanisms of leukemogenesis in the NTRK1/TP53 models of leukemia, we examined leukemic gene expression profiles by RNA-seq (Supplementary Tables 31–32) and mutational burdens by whole exome sequencing (Supplementary Table 33) in both primary and secondary recipient mice (total = 16 samples for RNA-seq and 16 samples for exome sequencing) and in untransduced lin- HSPCs. NTRK1/TP53 co-mutated tumors had a distinct gene expression profile characterized by up-regulation of genes overlapping significantly with CBFA2T3 target genes (Supplementary Fig. 11f and Supplementary Tables 31–32). However, these results can be due to a relative frequency of HSPCs within the heterogeneous populations that are compared and not linked to the molecular mechanism of transformation by NTRK1/TP53. Compared to NTRK1 WT/ TP53 mutated tumors, the genes perturbed by NTRK1/TP53 co-mutations overlapped significant with KLF1, EWSR1-FLI38 and TP53 target genes and showed enrichment of genes up-regulated in HSPCs from adult bone marrow and fetal liver (Supplementary Tables 32).
NTRK1/TP53 co-mutated tumors harbored few additional somatic mutations (N=8 in NTRK1G617D/TP53R172H; n=4 in NTRK1H766R/TP53R172H; and n=6 in NTRK1H498R/TP53R172H), none of which involved known cancer genes or targets of mutation in AEL. In contrast, the NTRK1 wild-type/TP53 mutated tumor harbored 54 non-silent mutations, several of which involved orthologs of genes mutated in human AEL (Brca1 and Ubtf1) (Supplementary Table 33). These findings and the significantly shorter latency of NTRK1-mutated tumors indicates NTRK1 is an oncogenic driver, but additional mutations are required to drive leukemogenesis in cells expressing WT NTRK1. Both tumors from wild-type NTRK1/mutated TP53 and NTRK1/TP53 co-mutated mice were characterized by the development of multiple chromosomal aneuploidies (Supplementary Fig. 11i), with recurrent amplifications of chromosomes 3, 8, 11 and 15 (Supplementary Fig. 11j).
NTRK1/TP53 co-mutated tumors were highly sensitive in vivo to larotrectinib, an oral, potent and selective inhibitor of TRK39,40. Leukemia remained undetectable for at least three months after cessation of treatment (Fig. 5h and Supplementary Fig. 11k).
DISCUSSION
Here, we describe the genomic landscape of childhood and adult acute erythroid leukemia, that has been subjected to variable classification schema, and was excluded from the erythroid/myeloid category of “AML, NOS” in the revised WHO 2016 classification.11 This reclassification has resulted in great uncertainty regarding diagnosis, risk classification and assignment of therapy of appropriate intensity13,14. We show that although many of the most common targets of mutation in AEL are observed in MDS and AML, the frequency and constellations of lesions are distinct in both childhood and adult AEL. Moreover, mutational spectra are highly age-dependent, with mutations of genes driving clonal hematopoiesis (e.g. DNMT3A, TET2, ASXL1 and TP53) commonly observed in older adults compared to children. This suggests that in adults, AEL may arise from acquisition of mutations on the background of clonal hematopoiesis. In contrast, in children we observed recurrent NUP98-fusions which harbor few additional alterations, suggesting that this lesion is sufficient for leukemogenesis.
In this study we have shown that AEL comprises multiple molecular subgroups in both children and adults. The mutational spectrum is intermediate between MDS and AML: mutations common in non erythroid AML are less frequent in AEL, but they are more common than in MDS. Conversely, MDS-associated mutations are less frequent in AEL, but more common than in non erythroid AML.
Genomic alterations and gene expression profiles closely correlated and classified cases. Moreover, they were the strongest predictors of outcome suggesting that they should be incorporated in a revision of the diagnostic and prognostic criteria. Low HOXB9 expression, TP53 mutations, RB1 copy number loss and FLT3 mutations were associated with poor outcome whereas NPM1 conferred good prognosis.
The importance of comprehensive genomic analysis incorporating detection of structural variants and gene expression is highlighted by the heterogeneity and complexity of the genomic alterations identified in this study, such as chromothripsis and cryptic TP53 structural variations, suggesting that targeted-sequencing approaches will underestimate and potentially misclassify leukemia cases. Here we showed that not only the multiple types of alteration, but the uniform biallelic alteration of TP53 that has not been characterized in prior reports, is a feature of adult AEL and a key driver of AEL leukemogenesis as supported by the potent interaction with NTRK1 in our in vivo mouse model.
AML therapy has not changed significantly in over thirty years and is poorly tolerated by older patients, who comprise the majority of cases. However in recent years, considerable progress has been made in understanding disease pathogenesis in AML with the identification of multiple somatically acquired driver mutations which affect prognosis and suggest targets for novel therapies. Clinical trials based on genetics are commonly utilized and provide prognostic and clinical management regardless of WHO classification or morphologic subtype of leukemia for most of the AML patients with recurrent genetic alterations (e.g. FLT3 mutations)41. However, except for few cases, AEL was often underrepresented in the genomic screening studies of AML and/or MDS and clear guidelines on how treat these patients are lacking. Thus the identification and validation of specific biomarkers as provided in this study will guide future studies that tailor therapy in specific subtypes. We developed a highly penetrant erythroid leukemia mouse model of NTRK1 and TP53 mutations which was exquisitely sensitive in vivo to inhibition with larotrectinib, an oral, potent and selective inhibitor of NTRK. The efficacy of selective NTRK inhibitors on NTRK-driven AML and ALL has been also recently reported34,36. All together these data demonstrate the sensitivity of NTRK1 mutant patient’s leukemic cells to NTRK inhibitors and enhance the relevance of these inhibitors for future clinical trials.
Moreover, since TP53 mutations and/or adverse cytogenetics were recently found to have very high response rates to hypomethylating agents (HMA) in AML42 and AEL7, HMA could represent a new therapeutic approach to improve outcome for this disease where TP53 mutations represent the most common initiating event for AEL.
While our data show that each erythroleukemia consists of multiple subgroups with distinct constellations of genomic alterations, they do not directly address the relative importance of genomic alteration versus the hematopoietic (or erythroid) progenitor in which the lesions are acquired as determinants of erythroid lineage. As many recurrently mutated genes are also observed in other myeloid leukemias, cell of origin is likely important, however the remarkably aggressive leukemia induced by concomitant NTRK1 and TP53 alterations indicates that mutation co-occurrence is an important determinant of disease lineage.
These data indicate that genomic analysis provides a more accurate foundation for the diagnosis and therapy of AEL by identifying different recurrent genetic alterations in subgroups with different clinical outcomes and potential targets for novel therapies. Toward this direction our study provides insights into genetic alterations that predominate in AEL compared to AML and MDS and define distinct subtypes with different genomic background and therapeutic targeting.
ONLINE METHODS
Study cohort
Acute erythroid leukemia
The diagnosis of AEL was centrally reviewed and confirmed for 159 pediatric and adult cases according to FAB and WHO 200810, and revised WHO 2016 criteria11. Unless specified we subclassified AEL according to WHO 2008 criteria. After central review, 10% of cases (Supplementary Table 1) could not be classified under WHO 2008 or 2016 criteria due to lack of archived slides. Cytogenetic risk groups were defined according to the Revised International Prognostic Scoring System (IPSS-R) for MDS17. The full patient characteristics are described in Supplementary Tables 1 and 2 and Fig. 1a, b. Patients with confirmed AEL were divided into the following four age groups: pediatric cases (n=35; age 0 to 20 years), young adults (n=8; age 21 to 39 years), adults (n=32; age 40 to 59 years), and older adults (n=84; age ≥ 60 years). Patients were from the MLL Munich Leukemia Laboratory, Germany (n=100); the Children’s Oncology Group (COG; see URLs) (n=12); the Centre for Cancer Biology, University of South Australia, Australia and SA Pathology (n=10); the Australian Leukaemia and Lymphoma Group (ALLG) Tissue Bank (n=9); the Japanese Paediatric Leukaemia/Lymphoma Study Group (JPLSG), Japan (n=9); St. Jude Children’s Research Hospital, USA (n=7); the Associazione Italiana di Ematologia e Oncologia Pediatrica (AIEOP), Italy (n=4); the Australian Centre for Blood Diseases, Alfred Hospital and Monash University, Australia (n=4); and the University of California at San Francisco, USA (n=1). Patients were enrolled onto clinical trial protocols of the following groups or centers: AML 2002/01 protocol from the AIEOP45, JPLSG AML-05 trial46, German-Austrian AML Study Group (AMLSG) 07–04 study (ClinicalTrials.gov identifier: )47, Children’s Cancer Group (CCG)- 2961 study ()48, European LeukemiaNet (ELN)49 and/or institutional protocols. AEL samples from COG were from patients treated on recent COG AML trials who achieved an initial remission to induction chemotherapy. These trials randomized type or timing of induction therapy (CCG-2961) and the addition of Gemtuzumab ozogamicin to backbone therapy in a single arm pilot (AAML03P1) or randomized fashion (AAML0531)50. All patients or their legal guardians gave written informed consent for sample collection and research. The study was approved by the St. Jude Children’s Research Hospital Institutional Review Board.
Non-erythroid AML
Pediatric and adult samples with non-erythroid AML were from Therapeutically Applicable Research to Generate Effective Treatments (TARGET) (n=192) and from The Cancer Genome Atlas (TCGA) study, respectively (n=197)51. TARGET samples were from COG AML trials as described above and publicly available data can be found at The National Cancer Institute Portal (see URLs) and in reference52. TCGA patient’s characteristics were described in details in reference51 and publicly available data can be found at the Genomic Data Commons Data Portal (see URLs).
Myelodysplastic syndrome
1514 cases (n=104, age 0 to 20 years; n=1410, age ≥ 21 years) with myelodysplastic syndrome (MDS) were from the Center for International Blood and Marrow Transplant Research (CIBMTR) repository and their characteristics have been reported in Lindsley RC et al53. Patients were not included if the percentage of blasts in the bone marrow or blood was 20% or more or if they had received a diagnosis of chronic myelomonocytic leukemia or overlap myelodysplastic–myeloproliferative neoplasms.
Cell lines
TF-154 and HEL 92.1.755 (referred to as HEL from here on) cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were thawed and cultured according to ATCC’s instructions. Immunophenotypic and cytogenetic analyses were performed prior DNA and RNA extraction.
Genomic studies
Genomic profiling of patient samples and the TF-1 and HEL cell lines was performed by whole genome (n=6), exome (n=142 patients and two cell lines), transcriptome (n=139 tumor patients and two cell lines) or targeted sequencing (n=12 patients) and single nucleotide polymorphism (SNP) microarray genotyping to identify DNA copy number alterations (n=137 patients and 2 cell lines) (Supplementary Table 1).
Whole genome sequencing
Five pediatric cases (paired tumor/germline samples) and one older adult (unpaired tumor sample) had whole genome sequencing (WGS) (Supplementary Table 1). Sequencing libraries were constructed for WGS cases from genomic DNA and sequenced using combinatorial probe anchor ligation by Complete Genomics Inc. (CGI)56. Reads were mapped to the GRCh37 reference human genome assembly by the CGI Cancer Sequencing service using software version 2.1 of the CGI cancer analysis pipeline (see URLs). Methods and publicly data are provided in details at the Therapeutically Applicable Research to Generate Effective Treatment (TARGET) data portal (see URLs).
Whole exome sequencing
Whole exome sequencing was performed in 122 unpaired and 20 paired tumor/germline samples with AEL (Supplementary Table 1). Germline samples were from flow sorted CD45high/CD3+ positive cells (n=2), mesenchymal cells (n=7), and remission (n=15); four cases had whole exome sequencing of both remission samples and mesenchymal cells. DNA was extracted using the AllPrep DNA/RNA Mini Kit (Qiagen). Library construction utilized DNA tagmentation (fragmentation and adapter attachment) performed using the reagent provided in the Illumina Nextera rapid exome kit, and was performed using the Caliper Biosciences Sciclone G3 (Perkin Elmer). First-round PCR (10 cycles) was performed using Illumina Nextera kit reagents, and clean-up steps using AMPure XP beads (Beckman Coulter Genomics/Agencourt). Target capture utilized Illumina Nextera Rapid Capture Expanded Exome and supplied hybridization and associated reagents. The pre-hybridization pool size was 12 samples, and second round PCR (10 cycles) performed with Nextera kit reagents. Library quality control and sequencing was performed as previously described.57
Whole exome sequencing analysis
Whole exome sequencing mapping, coverage and quality assessment, single nucleotide variation (SNV) and insertion/deletion (indel) detection and annotation for mutations have been described previously58. The reference human genome assembly NCBI Build 37 was used to map all samples. The mapping statistics and coverage for each sample are summarized in Supplementary Table 34.
More specifically, putative SNVs and indel variants were detected by SNPdetector59. Further evaluation of SNVs and indels was performed by manual review of the BAM files using Bambino60. Non-silent coding variations present in tumors, but absent in normal tissue, were considered somatic mutations. To remove additional germline variations from the data set generated by sequencing tumors without matching germline samples, new non-silent mutations were compared to the Exome Variant Server (EVS) (National Heart, Lung, and Blood Institute Exome Sequencing Project; see URLs) and to a database of germline variations identified in the Pediatric Cancer Genome Project61. Novel variants that passed this germline filter were annotated with dbSNP v141 and manually reviewed. Those at a site of known somatic sequence mutations in the Catalogue of Somatic Mutations in Cancer (COSMIC) database (see URLs)19 or at a gene included in COSMIC cancer gene census were grouped as putative somatic mutations, and others were considered variants of unknown origin. The analysis pipeline is shown in Supplementary Fig. 12a, b.
RNA-sequencing
RNA was extracted by AllPrep DNA/RNA Mini Kit (Qiagen). Transcriptome sequencing (RNA-seq) was performed using the TruSeq library preparation on the Illumina HiSeq 2000 platform as previously described62. Briefly, all sequencing was paired end and performed using either total RNA and stranded RNA sequencing (n=104), polyadenylated-selected non-stranded sequencing (n=31) or both (n=6 including the two AEL cell lines) (Supplementary Table 1).
RNA-sequencing analysis
Paired-end reads from RNA-seq were aligned using STAR63,64 to the following four databases: (i) human NCBI Build 37 reference sequence, (ii) RefSeq, (iii) a sequence file that represents all possible combinations of non-sequential pairs in RefSeq exons, and (iv) AceView flat file (University of California Santa Cruz, UCSC), representing transcripts constructed from human expressed sequence tags (ESTs). The mapping results were aligned to human reference genome coordinates, and final BAM files were constructed by selecting the best alignment, as previously described35. The mapping statistics and coverage for each RNA-seq sample are summarized in Supplementary Table 35. Structural variation detection in RNA-seq was carried out using CICERO, an algorithm that uses de novo assembly to identify structural variations in RNA-seq data35. Gene expression was quantitated using HTSeq and normalized using Trimmed Mean of M values (TMM) method in edgeR65. In order to simultaneously analyze polyA enriched and total stranded RNAseq samples, we utilized 6 cases that have been sequenced by both protocols to calculate an averaged scale factor for each gene, and used this scale factor to correct all polyA enriched samples to remove platform specific bias. For samples without WES but with RNA-seq available, we double checked all the cancer gene SNVs identified from WES in this cohort, to see if there is evidence in these RNA-seq only samples. The analysis pipeline is shown in Supplementary Fig. 12c. Statistical testing of differences in gene expression between subgroups were performed using limma66.
Bootstrap Evaluation of Hierarchical Clustering
A rigorous bootstrap procedure was applied to the RNA-seq data (n = 139) to define gene expression subgroups in an unsupervised manner. One hundred-thirty cases were evaluable for this analysis (Supplementary Table 1). In order to avoid variations due to technical artifacts 9 samples (SJAEL018159 to SJAEL018168_D1, Supplementary Table 1) were excluded since sequencing data were not generated at St. Jude Children’s Research Hospital.
The bootstrap procedure developed by Pawlikowska et al67 was used to quantify the reproducibility of 504 hierarchical cluster analysis methods defined by all 504 combinations of 28 considered values for the number of selected genes (m= 1,2,3,…,10,20,30,…,100, 200, 300, …, 1000 genes), 9 considered values for the number of subgroups to be defined (k=2, 3,…,10), and 2 considered dendrogram linkage methods (complete or average). Each cluster analysis method was applied to the observed RNA-seq data (log2 counts per million, CPM, mapped reads) and each of 500 bootstrap data sets obtained by resampling subjects with replacement. The reproducibility of each cluster analysis method was computed by summarizing the consistency of its subgroup assignments across the 500 bootstrap data sets.
We then selected specific cluster analysis results for further analysis. Among each set of cluster analysis methods that sought to identify a given number of clusters k, we selected the method that with the best bootstrap reproducibility among those that discovered k subgroups, assigned at least 10 subjects to each discovered subgroup, and used at least 2k features. Based on these criteria, we selected one set of results yielding two subgroups, one set of results yielding three subgroups, and one set of results yielding four subgroups for further consideration.
In this study, we then used the total covariance about the densest interval (total CADI) metric to select features for subgroup discovery analysis. For one pair of genes, the CADI is computed as
where xi and yi represent gene expression values of two genes for individual i and x* and y* are robust center estimates defined as the mean of observations in the narrowest interval containing at least half the data values (the densest interval)68. For each gene, its CADI with every other gene was computed using the formula above and its total CADI was computed as the sum of its CADI with all other genes.
Targeted-next generation sequencing
Seven AEL cases from COG AML0531 (Supplementary Table 1) were analyzed by targeted next-generation sequencing (NGS) of 400 candidate genes. The procedure has been described in details in an independent manuscript52. One sample (SJAEL047111) was analyzed by an integrated DNA/RNA target capture NGS assay, which is able to detect different classes of genomic events, including base substitutions, indels, copy number aberrations (CNAs), and chromosomal rearrangements as previously described.69
Sequencing Validation
For validation of SNV, indels or internal duplications we performed a comparison between the data generated at St. Jude Children’s Research Hospital (Memphis, USA) by whole exome and RNA-seq with those previously generated from the same samples but from independent DNA/RNA extractions at MLL Munich Leukemia Laboratory (Munich, Germany) by Sanger or targeted sequencing approaches. Sequence variations were analyzed by either Sanger sequencing using BigDye Term v1.1 cycle sequencing chemistry (Applied Biosystems), or by the 454 GS platform (454 Life Sciences)70 or were studied using a combination of a microdroplet-based assay (RainDance, Billerica, MA) and the MiSeq sequencing instrument (Illumina, San Diego, CA).71 The partial tandem duplication (PTD) in the KMT2A gene was confirmed by quantitative PCR,72 the internal tandem duplication (ITD) in the FLT3 gene was analyzed by fragment length analysis (Supplementary Table 39).73
For validation of gene fusions primers were designed upstream and downstream of each fusion’s breakpoint and used for reverse transcription and polymerase chain reaction (PCR) using Phusion enzyme (New England Biosciences) from patient samples (Supplementary Table 36). The PCR products were purified by Wizard SV Gel and PCR Clean-up system (Preprotech) and sequence was verified by Sanger sequencing. The sequenced amplicon was aligned to a reference fusion sequence generated from National Center for Biotechnology Information (NCBI) using the contigs obtained from RNA-sequencing. The results were analyzed using CLC Main Workbench (Qiagen).
Single nucleotide polymorphism microarrays
Samples (122 unpaired and 15 paired tumor/germline samples) were genotyped using Affymetrix SNP 6.0 microarrays according to the manufacturer’s instructions. CEL files were generated using GeneChip Command Console Software. SNP calls were generated using Genotyping Console (Affymetrix) and the Birdseed v2 algorithm with default parameters. Array normalization and copy number inference were performed according to a previously published workflow74. Normalized data were viewed in dChip75 and regions with abnormal copy number identified computationally by circular binary segmentation (CBS)76 and analyzed as previously described.74 Evidence of chromothripsis was defined as the presence of at least ten changes in segmental copy number between two or three copy number states on an individual chromosome.24 Inferred copy number log2 data from chromosomes with chromothripsis were exported from dChip and visualized in USCS Genome Browser.
Fluorescence in situ hybridization
Cell pellets from various primary leukemia samples that had been fixed in Carnoy’s fixative were used for fluorescence in situ hybridization (FISH) analysis. Probes were prepared from appropriate BAC clones or contigs of fosmid clones. Purified DNAs were labeled by nick translation with either Alexa Fluor 488 dUTP or Alexa Fluor 594 dUTP. All hybridizations were performed as two step, three probe experiments. The first hybridization consisted of 5´ and 3´ probes from the target gene to show disruption due to chromosomal rearrangement. Images were made from this first hybridization and microscope coordinates were recorded. Those cases having a positive result on the first hybridization were subjected to a second hybridization using a probe for the suspected fusion partner. The same cells that had been imaged previously after the first hybridization were imaged again after the second hybridization. This second experiment confirmed that when there is a gene disruption found in the first hybridization it either is or is not accompanied by fusion in the second hybridization. This type of approach can distinguish between gene fusion events vs gene disruption events not accompanied by fusion.
Genomic Random INterval (GRIN) model
GRIN was used to perform an integrative analysis of multimodality genomic data (DNA copy number alterations, mutations, structural rearrangements and expression levels from RNA-sequencing) to systematically identify genes in which perturbation of more than one genomic modality is likely to signify a role as a putative tumor suppressor. GRIN computes a P-value for the number of subjects with a copy number gain, copy number loss, sequence mutation, or structural rearrangement, as previously described.57
To evaluate the association of each type of lesion with expression for each gene, the Wilcoxon rank-sum test77 was used to compare the RNA-seq log2-counts per million reads (CPM) expression values of subjects with the specific type of lesion (mutation, fusion, copy number gain, copy number loss) with those of subjects with no detected lesion. For each type of lesion, the q-value78 with the P-value moment-based estimator78 was used to characterize the false discovery rate of performing multiple tests across many genes.
Identification of potential oncogenic driver genes
To identify potential driver genes, the list of mutated genes obtained from sequencing and SNP array analyses was compared with the cancer gene census from the COSMIC database19 (see URLs) according to COSMIC v81 release (May 2017). Genes that were recurrently mutated (≥ 3 different cases) but not included in COSMIC were further searched in cBioPortal for Cancer Genomics (see URLs)79. A gene was considered a putative candidate oncogenic driver if previously reported in published literature or in the cancer gene census (COSMIC v81); if previously unreported but mutated in this study in ≥ 3 different cases (when considering the whole cohort, independently on age); and/or implicated in functional classes relevant for tumorigenesis (epigenetic regulation, cohesin, transcriptional regulation, tumor suppression, signaling pathways, splicing/RNA processing and DNA repair). The list of putative candidate driver genes was thereafter filtered for statistically significant alterations (P value < 0.05) by GRIN. Functional annotation classes were defined based on literature and The Gene Ontology Annotation (GOA) resource (see URLs)80. Epigenomic classes were defined according to Huether R. et al.81
Co-occurrence matrix
For pairwise gene association analysis we included those genes with alterations (mutations, fusion, focal and/or relevant copy number gain and loss) in at least 3 AEL cases. The association between genes was tested by Fisher’s exact test. The final plot was prepared using ggplot2–2.2.1 package under R-3.3.2.
Structural modeling
The residue positions for each of the three point mutants of TrkA (H498R, G617D and H766R) were identified in the apo crystal structure of the TrkA kinase domain (Protein Data Bank ID code 4F0I43). A cartoon representation (Fig. 5b) in a view analogous to that provided in reference44 was generated with the program PyMol82 and showed that the 3 mutation positions are structurally distant from each other and also removed from canonically functional features. The residue positions were numbered according to the TrkA-I isoform of this study, which differ by 5 residues when compared to the TrkA-II isoform used in numbering PDB entry 4F0I. In the numbering of entry PDB 4F0I, the H498, G617, and H766 residues are numbered as H503, G622, and H771. The TrkA-I and TrkA-II isoforms are fully identical within the kinase domain, but differ ahead of the kinase domain by a 5-residue insertion.
Retroviral cloning
Gene fusions
NUP98-KDM5A, NPM1-MLF1, MYB-GATA1, ASNS-PTPN1, PFN1-SHIP, RUNX2-STAT3 and CDC37-IL27RA fusion transcripts were amplified by reverse transcription and PCR using Phusion® High-Fidelity DNA Polymerase (New England Biolabs Inc) and primers listed in Supplementary Table 36 from leukemic cell cDNA. Amplification products were purified by Wizard SV Gel and PCR Clean-up system (Promega) and verified by Sanger sequencing. Purified PCR products from NUP98-KDM5A, ASNS-PTPN1, NPM1-MLF1, PFN1-SCHIP and CDC37-IL27RA were cloned into pCR-Blunt II-TOPO (Life Technologies) and sub-cloned into the Murine Stem Cell Virus- Internal Ribosome Entry Site- Green Fluorescent Protein (MSCV-IRES-GFP) retroviral vector. Purified PCR products from RUNX2-STAT3 and MYB-GATA1 were cloned into the gateway entry clone pDONR™221 Vector (Life Technologies) and then transferred into a Gateway-compatible MSCV-IRES-GFP vector using the LR Clonase enzyme (Life Technologies). Constructs were verified by Sanger sequencing.
NTRK1 mutations
A gateway compatible entry clone containing the NTRK1 cDNA was obtained from Genecopoeia (NTRK1 pDONR, GC-Z5973). The NTRK1 H498R, G617D and H766R mutations were generated by site directed mutagenesis using the Quikchange II XL Kit (Agilent Technologies, Inc.) and primers in Supplementary Table 36. Mutated cDNAs were then transferred into a Gateway-compatible MSCV-IRES-GFP vector using the LR Clonase enzyme (Life Technologies). Constructs were verified by Sanger sequencing. C-terminal 6X Histidine (His) tagged wild-type and mutated NTRK1 isoforms were generated by mutagenesis.
Virus Production
293T cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM)/10% fetal bovine serum (FBS) and L-glutamine, 200 units/mL penicillin, and 200u g/mL streptomycin (Invitrogen). To produce murine retrovirus, 293T cells were co-transfected with pEcopac helper packaging plasmid using FuGENE HD (Promega) and MSCV-IRES-GFP as an empty vector or containing the fusion gene or mutant of interest. Viral supernatants were harvested 48 hours post-transfection.
Functional modeling
Fusion transcripts and mutated genes were cloned into a murine stem cell virus (MSCV)-internal ribosome entry site (IRES)-green fluorescence protein (GFP) vector (MIG). Transformation capability was assessed in NIH/3T3 cells in focus formation assays and/or in mouse lin- HSPCs for clonogenic assays and transplantation of irradiated mice. Mice were housed in the American Association of Accredited Laboratory Animal Care (AAALAC)-accredited facility at St Jude and were treated on Institutional Animal Care and Use Committee (IACUC)-approved protocols in accordance with NIH guidelines.
Immunofluorescence
NIH/3T3 mouse fibroblast cells expressing wild-type or mutated C-terminus 6X His-tagged NTRK1 (H498R, G617D or H766R) proteins (0.5 million per sample) were seeded overnight to poly-D lysine-coated Millicell EZ slides (Millipore), fixed for 5 minutes at room temperature with 4% paraformaldehyde, permeabilized in 0.1%/PBS Triton-X 100 (Sigma-Aldrich) for 10 minutes and washed three times. Sites of non-specific antibody binding were blocked by incubating cells for 60 minutes in donkey serum (Sigma-Aldrich) 1x/PBS. Cells were stained at 37°C for 1 hour with a rabbit polyclonal anti-6X His tag® (ab9108, Abcam), washed three times in PBS, and then incubated for 45 minutes to a secondary antibody conjugated to Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 (Thermo Fisher Scientific, A-21428). Slides were washed three times and incubated for 10 minutes with DAPI (Sigma-Aldrich) for nucleic acid staining. Slides were mounted with Golden ProLong® Diamond Antifade Mountant (Life Technologies) and fluorescent images were captured using a Zeiss LSM 780 confocal microscope (Zeiss) and analyzed by ZEN 2012 software (Zeiss) as previously described83.
Focus formation assay
NIH/3T3 mouse fibroblast cells were transduced with retroviral supernatants for wild-type NTRK1, mutated NTRK1 (H498R, G617D or H766R) or empty vector using polybrene 10 mg/ml for 48 hours priors to sorting for GFP-positive cells (BD FACSAria II, BD Biosciences, Franklin Lakes, NJ). Transduction efficacy was similar for all constructs. Five-hundred thousand sorted GFP-positive cells were cultured in 6-well plate for 2 weeks in Dulbecco’s Modified Eagle’s Medium/10% FBS and L-glutamine, 200 units/mL penicillin, and 200μg/mL streptomycin. After 14 days, cells were fixed in 4% paraformaldehyde, stained with 0.05% crystal violet and foci were counted. Each experimental condition was performed in duplicate. For the drug treatment study with entrectinib, one million GFP-positive sorted cells were cultured in tissue culture dishes (100 mm) and processed as described above.
Colony forming unit assays
Bone marrow cells were harvested from 8-week old wild-type C57BL/6 mice and lin- HSPCs were isolated using the EasySep Mouse Hematopoietic Progenitor Cell Enrichment Kit (Stem Cell Technologies, Vancouver, Canada) according to the manufacturer’s recommendation. Isolated lin- HSPCs were cultured for 48 hours in Iscove’s Modified Dulbecco’s Medium/20% fetal bovine serum supplemented with penicillin-streptomycin, L-glutamine, recombinant mouse IL-3 (10 ng/ml), IL-6 (20 ng/ml), IL-7 (10 ng/ml), FLT-3 ligand (40 ng/ml) and stem cell factor (SCF; 50 ng/ml) (Peprotech). Cells were infected on RetroNectin-coated plates for 48 hours (Takara Bio Inc.) with MSCV-IRES-GFP retrovirus expressing the fusion gene of interest (NUP98-KDM5A, MYB-GATA1, ASNS-PTPN1, PFN1-SCHIP1, CDC37-IL27RA, NPM1-MLF1, RUNX2-STAT3) or empty vector control. Transduced GFP-positive cells were obtained by fluorescence-activated cell sorting. For clonogenic assays, 10,000 cells were plated in triplicate in Methocult M3231 (Stem Cell Technologies) with the appropriate factors (SCF, 100 ng/ml; FLT-3 ligand, 10 ng/ml; IL-7, 20 ng/ml) for mouse lymphoid progenitor cells, in M3534 (Stem Cell Technologies) with the addition of GM-CSF (10 ng/ml) for mouse myeloid progenitor cells and in M3436 (Stem Cell Technologies) for mouse erythroid progenitor cells. Colonies were scored 7–10 days later. For re-plating, 10,000 cells were cultured in identical conditions, with colonies counted on day 7–10. Colony identity was confirmed by morphological analysis through cytospin and Wright-Giemsa staining and by flow analysis for a panel of multi-lineage markers (B220-BV605, Gr-1-PerCP-Cy5.5, Ter119-V500, CD3-APC, Mac1-Alexa700, CD41-PE and CD19-APC-Cy7).
Genetically engineered mouse models of NUP98-KDM5A-positive leukemia
Fetal liver cells were harvested from embryos at embryonic day 14 from wild-type pregnant C57BL/6 female mice and lin- negative HSPCs were isolated using the EasySep Mouse Hematopoietic Progenitor Cell Enrichment Kit (Stem Cell Technologies) as described above. Cells were infected on RetroNectin-coated plates for 48 hours (Takara Bio Inc.) with MSCV-IRES-GFP retrovirus expressing NUP98-KDM5A or other chimeric fusions (MYB-GATA1, ASNS-PTPN1 and NPM1-MLF1) with/without the MSCV-luciferase-IRES-mCherry retrovirus. Transduced GFP-positive or GFP-mCherry-double positive cells were obtained by fluorescence-activated cell sorting. Recipient 8-week-old wild-type C57Bl/6 female mice were sublethally irradiated (550 Rad) 24 hours prior to transplantation with 0.2–0.5 × 106 GFP-positive or GFP-mCherry double-positive cells via tail-vein injection. To monitor the development of leukemia in transplanted mice that underwent transplantation, cohorts of each transplanted construct were followed over time by biweekly measurement of bioluminescence with Xenogen IVIS-200 (PerkinElmer) or by retro orbital bleeding and analysis of GFP-positive cells at flow cytometry. Region of interest (ROI) measurements and total fluxes (photons/second, p/s) were recorded and analyzed by the Living Imaging v.4.4 software (Caliper Life Sciences). Animals that became moribund were euthanized, and blood, bone marrow, spleen and additional tissues were analyzed for evidence of leukemia, using morphology, flow cytometry, and histopathologic analysis. Post-mortem flow analysis for a panel of multi-lineage markers (B220-BV605, Gr-1-PerCP-Cy5.5, Ter119-V500, CD3-APC, Mac1-Alexa700, CD41-PE and CD19-APC-Cy7) was performed on the GFP-positive or GFP-mCherry double positive population to determine the lineage of disease in engrafted samples. For secondary transplantations, 8-week-old wild-type C57BL/6 female mice were sublethally irradiated (550 Rad) 24 hours prior to transplantation with 0.5 × 106 GFP-mCherry double positive spleen cells from mice that underwent primary transplantation and that developed leukemia. The same monitoring criteria applied to mice that underwent primary transplantation were used for secondary transplanted recipients (bioluminescence, morphology, flow cytometry and histopathology).
Genetically engineered mouse models of NTRK1/TP53 mutated leukemia
Bone marrow cells were harvested from 8-week old wild-type C57BL/6 mice or TP53R172H knockin mice37 and lineage negative HSPCs were isolated using the EasySep Mouse Hematopoietic Progenitor Cell Enrichment Kit (Stem Cell Technologies) as above described. Cells were infected on RetroNectin-coated plates for 48 hours (Takara Bio Inc.) with MSCV-IRES-GFP retrovirus expressing wild-type NTRK1 or mutated NTRK1 (H498R, G617D or H766R) and transduced GFP-positive cells were obtained by fluorescence-activated cell sorting. Recipient 8-week-old wild-type C57Bl/6 female mice were sublethally irradiated (550 Rad) 24 hours prior to transplantation with 0.2 × 106 GFP-positive cells via tail-vein injection. To monitor the development of leukemia in transplanted mice that underwent transplantation, cohorts of each transplanted construct were followed over time by analysis of GFP-positive cells in retro orbital bleeding samples. Animals that became moribund were killed, and blood, bone marrow, and spleen samples were analyzed for evidence of leukemia, using morphology, flow cytometry, and histopathologic analysis. Post-mortem flow analysis for a panel of multi-lineage markers (B220-BV605, Gr-1-PerCP-Cy5.5, Ter119-V500, CD3-APC, Mac1-Alexa700, CD41-PE and CD19-APC-Cy7) was performed on the GFP-positive population to determine the lineage of disease in engrafted samples. A second more specific lineage panel included CD34-BV421, Ter119-BUV396, Mac1-BV605, Gr1-BV711, CD117-APC-eFluor780, CD71-PE-Cy7 and CD44-APC. For secondary transplantations, 8-week-old wild-type C57BL/6 female mice were sublethally irradiated (550 Rad) 24 hours prior to transplantation with 0.2 × 106 GFP-positive spleen cells from mice that underwent primary transplantation and that developed leukemia. The same monitoring criteria applied to mice that underwent primary transplantation were used for secondary transplanted recipients (bioluminescence, morphology, flow cytometry and histopathology) as above described.
Immunohistochemistry
Immunohistochemistry (IHC) was performed on formalin-fixed paraffin-embedded tissues sectioned at 4μm. All assay steps for CD34, murine-specific CD45, GATA1, Glycophorin A (GlyA), and myeloperoxidase (MPO) and human TrkA, including deparaffinization, rehydration, and epitope retrieval, were performed on the Ventana Discovery Ultra autostainer with Ventana Reaction Buffer (Ventana, #950–300,) rinses between steps. Antibody binding of CD34 and CD45 was by the OmniMap Rat Detection kit (Roche, #760–4457), and binding of GATA1, GlyA, MPO and hTrkA was detected using the OmniMap Rabbit Detection kit (Roche, #760–4311) for 16 minutes, followed by ChromoMap DAB (Roche, #760–159) for 10 minutes. All assay steps for B220, Pax5 and RUNX1/AML1 were performed on the Bond Max with Bond wash buffer (# AR9590, Leica) rinses between steps. Slides were incubated with the primary antibody; slides for B220 were followed by the secondary antibody (rabbit anti-rat, Vector Labs #BA-4001) at 1:400 for 10 minutes. Antibody binding for these 3 antibodies was detected using the anti-rabbit Bond Polymer Refine Detection kit (Leica, #DS9800). All assay steps for CD41 and CD43 were performed on the Biocare intelliPATH with Biocare TBS wash buffer (#TWB954M, Biocare) rinses between steps. Slides were incubated with the primary antibody followed by the secondary antibody (for CD41 goat anti-rabbit, Vector Labs #BA-1000, and for CD43 rabbit anti-rat, Vector Labs #BA-4001) at 1:200 for 30 minutes, streptavidin conjugated to horse radish peroxidase (ThermoShandon, #TS-125-HR, 10 minutes) and substrate containing the chromagen DAB (ThermoShandon, #TA-125-HDX, 5 minutes). All assay steps for Ter119 were performed on the Biocare intelliPATH with Biocare TBS wash buffer (cat # TWB954M, Biocare) rinses between steps. Slides were incubated with the primary antibody followed by the secondary antibody (rabbit anti-rat, Vector Labs #BA-4001) at 1:200 for 30 minutes, streptavidin conjugated to horse radish peroxidase (ThermoShandon, #TS-125-HR, 10 minutes) and substrate containing the chromagen DAB (ThermoShandon, #TA-125-HDX, 5 minutes). Antibodies are listed in Supplementary Table 37.
Karyotype analysis
To evaluate chromosome ploidy and karyotypic analysis of NTRK1/TP53R172H mutated tumors, frozen mouse leukemic cells from spleen or liver were thawed in the Cytogenetic Shared Resource Laboratory (St. Jude Children’s Research Hospital) and processed by a direct harvest using routine cytogenetic methods after a five hour colcemid exposure time. The slides were allowed to air dry for optimal banding of the chromosomes with trypsin and Wright’s stain. At least twenty cells were analyzed for each tumor.
Spectral karyotype analysis
For spectral karyotype (SKY) analysis frozen murine leukemia cells were thawed in a 37°C water bath, added to 7 mls of media and colcemid for a five hour exposure time followed by a direct harvest using routine cytogenetic methods. A commercially prepared SKY probe from Applied Spectral Imaging (Carlsbad, CA) was used as the probe for this analysis. Applied Spectral Imaging protocols were followed for the hybridization and detection steps. A total of 21 metaphase cells were analyzed.
In vivo treatment with larotrectinib
For in vivo drug treatment studies, following engraftment and within two weeks since transplantation mice were randomized to receive larotrectinib (LOXO-101) (200 mg/kg) or vehicle (Labrafac) as described in reference39.
Statistical analysis
Associations between categorical values were examined using Fisher’s exact test or Chi-square as appropriate. These included associations between gene/pathway-level alterations and age groups or leukemia subtypes (AEL, non-erythroid AML or MDS). Data are reported as P values without correction for multiple comparisons. P values for the gene co-occurrence matrix Fisher exact test. The odds ratio (OR) was calculated as the ratio of odds of a mutation in gene A among those with mutation in gene B to the odds of a mutation in gene A among those without a mutation in gene B. OR>1 indicates a positive association between two genes. Conversely, OR<1 indicates a negative association. For clinical correlation analysis, the primary outcome was overall survival, which was defined as the time from initiation of therapy to death due to any cause or was censored at the date of last follow-up. The Kaplan–Meier method was used to estimate the survival function and overall survival distributions were compared with log-rank tests. The Cox proportional hazards model was used to identify independent risk factors for overall survival. A stepwise regression model with variables which are significantly associated with survival in univariate analysis was carried out84–86. In our analyses, we specified that a variable has to be significant at the 0.25 level before it can be entered into the model, and also specified that a variable in the model has to be significant at the 0.15 level for it to remain in the model.
For in vivo mouse studies, the ANOVA test was also used to determine significance in spleen weights. Kaplan-Meier analysis and the Mantel-Cox log rank test were used for survival data of mouse models. Analyses were performed using Prism v7.0 (GraphPad), R (see URLs)87, and SAS (v9.1.2, SAS Institute, Cary, NC).
LIFE SCIENCES REPORTING SUMMARY
Further information on research design is available in the Life Sciences Reporting Summary linked to this article.
STATEMENT OF DATA AVAILABILITY
Genomic data including both sequencing and copy number data have been deposited in the European Genome-phenome Archive (EGA), accession EGAS00001002537 (https://www.ebi.ac.uk/ega/studies/EGAS00001002537). This includes the following data sets: whole exome sequencing data (EGAD00001003413), RNA-Seq data (EGAD00001003412) and SNP6 Affymetrix copy number data (EGAD00010001443). Moreover, genomic data including non-silent SNV, indels, ITD sequence mutations, in-frame gene fusions, structural variations, copy number aberrations and gene expression data can be explored interactively at the St. Jude PeCan Data Portal88 (https://pecan.stjude.cloud/proteinpaint/study/ael).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the American Lebanese Syrian Associated Charities of St Jude Children’s Research Hospital; by a Stand Up to Cancer Innovative Research Grant and a St Baldrick’s Foundation Robert J. Arceci Innovation Award (to C.G.M.); by a Leukemia and Lymphoma Society Specialized Center of Research grant (to C.G.M.); by the Henry Schueler 41&9 Foundation (to C.G.M.); by a Lady Tata Memorial Trust Award (to I.I.), by St Jude Children’s Research Hospital Hematological Malignancies Program Garwood Fellowship (to I.I.); by Italian Scientists and Scholars in North America Foundation (ISSNAF) to I.I.; by a Leukemia and Lymphoma Society Translational Research Program (to C.G.M.); by a National Cancer Institute Outstanding Investigator Award R35 CA197695 (to C.G.M.); by the R25CA23944 from the National Cancer Institute (to St. Jude Pediatric Oncology Education program); by a St. Jude Summer Plus Fellowship, Rhodes College (to S.M.M.); by NIH Cancer Center Support Grant P30 CA21765 (to C.G.M.); by Fondazione Cariparo (to G.B.) and AIRC (to G.B.); by AIRC 5×1.000 “Immunity in Cancer Spreading and Metastasis” (to F.L.); by National Medical Research Council, Singapore (NMRC/CSA/0053/2013) (to A.E.J.Y.); and by Cancer Science Institute of Singapore (to A.E.J.Y.). This work was also supported by a Project Grant (516726 to B.T.K.) and Program Grants (1016647 and 1113577 to B.T.K.), a Fellowship (1063008 to B.T.K.), and an Independent Research Institutes Infrastructure Support Scheme Grant (361646) from the Australian National Health and Medical Research Council (to B.T.K.); the Leukaemia Foundation of Australia (to C.L.C. and B.T.K.); the Sylvia & Charles Viertel Foundation (to B.T.K.); the Australian Cancer Research Fund (to B.T.K.); Cancer Council of South Australia Beat Cancer Project (1145385, to A.L.B., H.S.S., and C.N.H) and a Victorian State Government Operational Infrastructure Support Grant (to B.T.K.). We thank the ALLG Tissue Bank at the Princess Alexandra Hospital, Brisbane (now the Cancer Collaborative Biobank), for providing samples. The ALLG Tissue Bank received funding support from the Leukaemia Foundation (to P.M.), the National Health and Medical Research Council (to P.M.), and Queensland Health and Pathology Queensland for the ALLG Tissue Bank (to P.M.). The South Australian Cancer Research Biobank was supported by the Cancer Council SA Beat Cancer Project, University of Adelaide, University of South Australia, South Australian Health and Medical Research Institute, SA Health, Health Service Gifts and Charitable Board of the Central Adelaide Local Health Network, Medvet Laboratories Pty Ltd and the Government of South Australia (to L.B.T., R.D.A., H.S., and I.L.). We thank the staff of the Biorepository, the Hartwell Centre for Bioinformatics and Biotechnology, the Flow Cytometry and Cell Sorting Core Facility, the Cell and Tissue Imaging Facility, the Animal Resources Center and the Small Animal Imaging Center, the Compound Management Center and the Department of Chemical Biology & Therapeutics of St. Jude Children’s Research Hospital. We thank Loxo Oncology, Inc. for providing larotrectinib and support in dosing.
COMPETING FINANCIAL INTERESTS
M.M.: Employment by MLL Munich Leukemia Laboratory; R.C.L.: research funding from MedImmune and Jazz, and consulting fees from Takeda; B.L.E.: research funding from Celgene and Deerfield, and consulting fees from GRAIL T.H.: Equity ownership of MLL Munich Leukemia Laboratory; C.G.M.: research funding from Loxo Oncology for TRK inhibitors in acute lymphoblastic leukemia. Correspondence and requests for materials should be addressed to Charles Mullighan; charles.mullighan@stjude.org
Footnotes
URLs
Children’s Oncology Group (COG): https://childrensoncologygroup.org/;
Therapeutically Applicable Research To Generate Effective Treatments (TARGET) Data Portal: https://ocg.cancer.gov/programs/target;
Genomic Data Commons Data Portal: https://portal.gdc.cancer.gov/;
Complete Genomics Inc. (CGI) cancer analysis pipeline: http://www.completegenomics.com/customer-support/documentation/;
Exome Variant Server: http://evs.gs.washington.edu/EVS/;
Catalogue of Somatic Mutations in Cancer (COSMIC): http://cancer.sanger.ac.uk/census/;
cBioPortal for Cancer Genomics: http://cbioportal.org
The Gene Ontology Annotation (GOA): http://www.ebi.ac.uk/GOA;
R Project for Statistical Computing: www.r-project.org;
European Genome-phenome Archive (EGA): https://www.ebi.ac.uk/ega/studies/EGAS00001002537;
Enrichr: http://amp.pharm.mssm.edu/Enrichr/;
St. Jude PeCan Data Portal: https://pecan.stjude.cloud/proteinpaint/study/ael;
UCSC browser session of chromothripsis: https://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=ilaria.iacobucci%40stjude.org&hgS_otherUserSessionName=Chromothripsis_AEL_hg18.
REFERENCES
- 1.Cancer Genome Atlas Research, N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368, 2059–74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papaemmanuil E et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med 374, 2209–21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cervera N et al. Molecular characterization of acute erythroid leukemia (M6-AML) using targeted next-generation sequencing. Leukemia 30, 966–70 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Grossmann V et al. Acute erythroid leukemia (AEL) can be separated into distinct prognostic subsets based on cytogenetic and molecular genetic characteristics. Leukemia 27, 1940–3 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Ping N et al. Exome sequencing identifies highly recurrent somatic GATA2 and CEBPA mutations in acute erythroid leukemia. Leukemia 31, 195–202 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Liu W et al. Pure erythroid leukemia: a reassessment of the entity using the 2008 World Health Organization classification. Mod Pathol 24, 375–83 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Almeida AM et al. Clinical Outcomes of 217 Patients with Acute Erythroleukemia According to Treatment Type and Line: A Retrospective Multinational Study. Int J Mol Sci 18(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dameshek W & Baldini M The Di Guglielmo syndrome. Blood 13, 192–4 (1958). [PubMed] [Google Scholar]
- 9.Bennett JM et al. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French-American-British Cooperative Group. Ann Intern Med 103, 620–5 (1985). [DOI] [PubMed] [Google Scholar]
- 10.Vardiman JW et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood 114, 937–51 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Arber DA et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127, 2391–405 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Kowal-Vern A et al. Diagnosis and characterization of acute erythroleukemia subsets by determining the percentages of myeloblasts and proerythroblasts in 69 cases. Am J Hematol 65, 5–13 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Lichtman MA The disappearance of acute erythroid leukemia: An act of legerdemain at the World Health Organization. Blood Cells Mol Dis 61, 54–7 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Arber DA Revisiting erythroleukemia. Curr Opin Hematol 24, 146–151 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Wang SA et al. Acute erythroid leukemia with <20% bone marrow blasts is clinically and biologically similar to myelodysplastic syndrome with excess blasts. Mod Pathol 29, 1221–31 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Hasserjian RP et al. Acute erythroid leukemia: a reassessment using criteria refined in the 2008 WHO classification. Blood 115, 1985–92 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenberg PL et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood 120, 2454–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pounds S et al. A genomic random interval model for statistical analysis of genomic lesion data. Bioinformatics 29, 2088–95 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forbes SA et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res 45, D777–D783 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaiswal S et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 371, 2488–98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie M et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 20, 1472–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genovese G et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 371, 2477–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephens PJ et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144, 27–40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rausch T et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 148, 59–71 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abaigar M et al. Chromothripsis Is a Recurrent Genomic Abnormality in High-Risk Myelodysplastic Syndromes. PLoS One 11, e0164370 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang CT et al. Activation of KLF1 Enhances the Differentiation and Maturation of Red Blood Cells from Human Pluripotent Stem Cells. Stem Cells 35, 886–897 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holmfeldt P et al. Nfix is a novel regulator of murine hematopoietic stem and progenitor cell survival. Blood 122, 2987–96 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panagopoulos I et al. Fusion of ZMYND8 and RELA genes in acute erythroid leukemia. PLoS One 8, e63663 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quelen C et al. Identification of a transforming MYB-GATA1 fusion gene in acute basophilic leukemia: a new entity in male infants. Blood 117, 5719–22 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Wang GG et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature 459, 847–51 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majeti R et al. Dysregulated gene expression networks in human acute myelogenous leukemia stem cells. Proc Natl Acad Sci U S A 106, 3396–401 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersson A, Eden P, Olofsson T & Fioretos T Gene expression signatures in childhood acute leukemias are largely unique and distinct from those of normal tissues and other malignancies. BMC Med Genomics 3, 6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gatalica Z, Xiu J, Swensen J & Vranic S Molecular characterization of cancers with NTRK gene fusions. Mod Pathol (2018). [DOI] [PubMed] [Google Scholar]
- 34.Taylor J et al. Oncogenic TRK fusions are amenable to inhibition in hematologic malignancies. J Clin Invest 128, 3819–3825 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts KG et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med 371, 1005–15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts KG et al. ETV6-NTRK3 induces aggressive acute lymphoblastic leukemia highly sensitive to selective TRK inhibition. Blood 132, 861–865 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lang GA et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 119, 861–72 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Grunewald TG et al. Chimeric EWSR1-FLI1 regulates the Ewing sarcoma susceptibility gene EGR2 via a GGAA microsatellite. Nat Genet 47, 1073–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doebele RC et al. An Oncogenic NTRK Fusion in a Patient with Soft-Tissue Sarcoma with Response to the Tropomyosin-Related Kinase Inhibitor LOXO-101. Cancer Discov 5, 1049–57 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khotskaya YB et al. Targeting TRK family proteins in cancer. Pharmacol Ther 173, 58–66 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Dohner H et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 129, 424–447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welch JS et al. TP53 and Decitabine in Acute Myeloid Leukemia and Myelodysplastic Syndromes. N Engl J Med 375, 2023–2036 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bertrand T et al. The crystal structures of TrkA and TrkB suggest key regions for achieving selective inhibition. J Mol Biol 423, 439–53 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Bertrand T Crystal Structures of Neurotrophin Receptors Kinase Domain. Vitam Horm 104, 1–18 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Pession A et al. Results of the AIEOP AML 2002/01 multicenter prospective trial for the treatment of children with acute myeloid leukemia. Blood 122, 170–8 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Tomizawa D et al. Excess treatment reduction including anthracyclines results in higher incidence of relapse in core binding factor acute myeloid leukemia in children. Leukemia 27, 2413–6 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Schlenk RF et al. All-trans retinoic acid as adjunct to intensive treatment in younger adult patients with acute myeloid leukemia: results of the randomized AMLSG 07–04 study. Ann Hematol 95, 1931–1942 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lange BJ et al. Outcomes in CCG-2961, a children’s oncology group phase 3 trial for untreated pediatric acute myeloid leukemia: a report from the children’s oncology group. Blood 111, 1044–53 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buchner T et al. Acute Myeloid Leukemia (AML): different treatment strategies versus a common standard arm--combined prospective analysis by the German AML Intergroup. J Clin Oncol 30, 3604–10 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Farrar JE et al. Genomic Profiling of Pediatric Acute Myeloid Leukemia Reveals a Changing Mutational Landscape from Disease Diagnosis to Relapse. Cancer Res 76, 2197–205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cancer Genome Atlas Research, N. et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368, 2059–74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolouri H et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med 24, 103–112 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindsley RC et al. Prognostic Mutations in Myelodysplastic Syndrome after Stem-Cell Transplantation. N Engl J Med 376, 536–547 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kitamura T et al. Establishment and characterization of a unique human cell line that proliferates dependently on GM-CSF, IL-3, or erythropoietin. J Cell Physiol 140, 323–34 (1989). [DOI] [PubMed] [Google Scholar]
- 55.Martin P & Papayannopoulou T HEL cells: a new human erythroleukemia cell line with spontaneous and induced globin expression. Science 216, 1233–5 (1982). [DOI] [PubMed] [Google Scholar]
- 56.Drmanac R et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327, 78–81 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Liu Y et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet 49, 1211–1218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang J et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481, 157–63 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang J et al. SNPdetector: a software tool for sensitive and accurate SNP detection. PLoS Comput Biol 1, e53 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edmonson MN et al. Bambino: a variant detector and alignment viewer for next-generation sequencing data in the SAM/BAM format. Bioinformatics 27, 865–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Downing JR et al. The Pediatric Cancer Genome Project. Nat Genet 44, 619–22 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roberts KG et al. High Frequency and Poor Outcome of Philadelphia Chromosome-Like Acute Lymphoblastic Leukemia in Adults. J Clin Oncol 35, 394–401 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H & Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–60 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–40 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ritchie ME et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pawlikowska I et al. Dunn Index Bootstrap (DIBS): A procedure to empirically select a cluster analysis method that identifies biologically and clinically relevant molecular disease subgroups. BMC Bioinformatics 16, P2 (2015). [Google Scholar]
- 68.Rousseeuw PJ Least Median of Squares Regression. Journal of the American Statistical Association 79, 871–880 (1984). [Google Scholar]
- 69.He J et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood 127, 3004–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kohlmann A et al. The Interlaboratory RObustness of Next-generation sequencing (IRON) study: a deep sequencing investigation of TET2, CBL and KRAS mutations by an international consortium involving 10 laboratories. Leukemia 25, 1840–8 (2011). [DOI] [PubMed] [Google Scholar]
- 71.Delic S et al. Application of an NGS-based 28-gene panel in myeloproliferative neoplasms reveals distinct mutation patterns in essential thrombocythaemia, primary myelofibrosis and polycythaemia vera. Br J Haematol 175, 419–426 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Weisser M et al. Risk assessment by monitoring expression levels of partial tandem duplications in the MLL gene in acute myeloid leukemia during therapy. Haematologica 90, 881–9 (2005). [PubMed] [Google Scholar]
- 73.Schnittger S et al. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood 100, 59–66 (2002). [DOI] [PubMed] [Google Scholar]
- 74.Mullighan CG et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446, 758–64 (2007). [DOI] [PubMed] [Google Scholar]
- 75.Lin M et al. dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics 20, 1233–40 (2004). [DOI] [PubMed] [Google Scholar]
- 76.Venkatraman ES & Olshen AB A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics 23, 657–63 (2007). [DOI] [PubMed] [Google Scholar]
- 77.Wilcoxon F Individual comparisons by ranking methods. Biometrics 1, 80–83 (1945). [Google Scholar]
- 78.Storey JD & Tibshirani R Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100, 9440–5 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gao J et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huntley RP et al. The GOA database: gene Ontology annotation updates for 2015. Nucleic Acids Res 43, D1057–63 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huether R et al. The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nat Commun 5, 3630 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.3r1. (2010). [Google Scholar]
- 83.Iacobucci I et al. Truncating Erythropoietin Receptor Rearrangements in Acute Lymphoblastic Leukemia. Cancer Cell 29, 186–200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mullighan CG et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med 360, 470–80 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mantel N Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep 50, 163–70 (1966). [PubMed] [Google Scholar]
- 86.Fine JP & Gray RJ A Proportional Hazards Model for the Subdistribution of a Competing Risk. J Am Stat Assoc 94, 496–509 (1999). [Google Scholar]
- 87.An D R Development Core Team. R: A language and environment for statistical computing in R Foundation for Statistical Computing (Vienna, Austria, 2009). [Google Scholar]
- 88.Zhou X et al. Exploring genomic alteration in pediatric cancer using ProteinPaint. Nat Genet 48, 4–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.