Abstract
The PIWI-interacting RNA (piRNA) pathway protects genome integrity in part through establishing repressive heterochromatin at transposon loci. Silencing requires piRNA-guided targeting of nuclear PIWI proteins to nascent transposon transcripts, yet the subsequent molecular events are not understood. Here, we identify SFiNX (Silencing Factor interacting Nuclear eXport variant), an interdependent protein complex required for Piwi-mediated co-transcriptional silencing in Drosophila. SFiNX consists of Nxf2–Nxt1, a gonad-specific variant of the heterodimeric mRNA export receptor Nxf1–Nxt1, and the Piwi-associated protein Panoramix. SFiNX mutant flies are sterile and exhibit transposon de-repression because piRNA-loaded Piwi is unable to establish heterochromatin. Within SFiNX, Panoramix recruits heterochromatin effectors, while the RNA binding protein Nxf2 licenses co-transcriptional silencing. Our data reveal how Nxf2 might have evolved from an RNA transport receptor into a co-transcriptional silencing factor. Thus, NXF variants, which are abundant in metazoans, can have diverse molecular functions and might have been co-opted for host genome defense more broadly.
INTRODUCTION
Eukaryotic cells establish heterochromatin at genomic repeats and transposon insertions to suppress transcription and recombination 1,2. One strategy to confer sequence specificity to this process is via repressor proteins (e.g. KRAB-type zinc finger repressors in tetrapods) that bind defined DNA motifs and recruit heterochromatin inducing factors 3. A second strategy for sequence-specific heterochromatin formation builds on nuclear small RNAs 4–6. These ~20–30nt long regulatory RNAs guide Argonaute proteins to complementary sequences in nascent target transcripts, which are attached to chromatin via transcribing RNA polymerases 7. Binding of nuclear Argonautes to nascent transcripts leads to transcriptional repression and heterochromatin formation. As Argonaute recruitment requires the nascent target RNA, this process is defined as ‘co-transcriptional silencing’. Besides impacting chromatin and transcription, nuclear small RNA pathways have also been linked to the co-transcriptional processes of splicing, RNA quality control and turnover 8–10. Together, this hints at complex molecular connections between nuclear Argonautes, the nascent target RNA, and chromatin.
The principal nuclear Argonaute pathway in animals is the PIWI-interacting small RNA (piRNA) pathway 11,12. It acts predominantly in gonads and safeguards the integrity of the germline genome. In Drosophila melanogaster, a single nuclear Argonaute protein (Piwi) orchestrates co-transcriptional silencing and heterochromatin formation at hundreds of transposon insertions throughout the genome 13–16. Although transposons contain strong promoters and enhancers, binding of Piwi to nascent transposon transcripts effectively suppresses their transcription, resulting in up to several hundred-fold reductions in steady state RNA levels. How Piwi orchestrates co-transcriptional silencing at the molecular level is poorly understood. Genetic studies identified the piRNA pathway-specific proteins Maelstrom, Asterix (Gtsf1), and Panoramix (Silencio) as essential co-factors for Piwi-mediated silencing 14,17–21. In the absence of any of these proteins, Piwi is abundantly expressed, localizes to the nucleus, is loaded with transposon-targeting piRNAs, but is incapable of target silencing. Among these three Piwi co-factors, only Panoramix is capable of inducing co-transcriptional silencing and heterochromatin formation if targeted to a nascent RNA through aptamer-based tethering. Silencing via tethered Panoramix is independent of Piwi but requires the H3K9 methyltransferase SetDB1 (Eggless), the H3K4 demethylase Lsd1 (Su(var)3–3), and the H3K9me2/3 reader protein HP1a (Su(var)205) 20,21. This places Panoramix downstream of Piwi and upstream of the cellular heterochromatin machinery. How Panoramix, which does not resemble any known protein, is connected to the nascent RNA, or to chromatin effectors and chromatin itself, is unknown.
Here, we show that Panoramix functions in the context of an interdependent protein complex with Nxf2–Nxt1, a variant of the highly conserved Nxf1–Nxt1 (Tap–p15) heterodimer. From budding yeast to humans, Nxf1–Nxt1 is the principal nuclear mRNA export receptor that mediates the translocation of export-competent mRNAs through nuclear pore complexes into the cytoplasm 22–24. Nxf2 is one of the three nuclear RNA export factor (NXF) variants in Drosophila melanogaster, but no mRNA export function could be attributed to it 25,26. Using genetics, biochemistry and structural biology, we reveal an unexpected, RNA-export independent role for Nxf2 in piRNA guided co-transcriptional silencing of transposons. Our findings provide a functional link between the genome-transposon conflict and NXF variants, which have largely unknown function in animals.
RESULTS
The Nxf2–Nxt1 heterodimer interacts with Panoramix
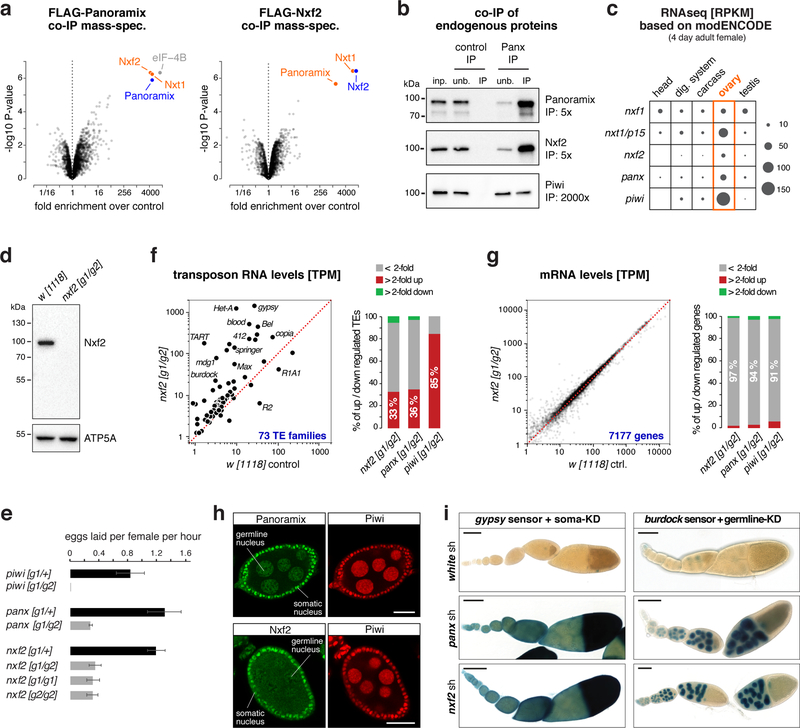
To elucidate the molecular function of Panoramix, we determined its protein interactors in cultured ovarian somatic cells (OSCs), which express a nuclear Piwi/piRNA pathway 27,28. We isolated Panoramix via immunoprecipitation (IP) from nuclear lysate of a clonal OSC line expressing FLAG-tagged Panoramix and identified co-eluting proteins by quantitative mass spectrometry. The most prominent interactors were nuclear RNA export factor 2 (Nxf2), the mRNA export co-factor Nxt1, and eIF-4B (Fig. 1a left; Table S1). Among those, Nxf2 and Nxt1 were also identified in genetic transposon de-repression screens 19,29,30. We confirmed the interaction between Panoramix, Nxf2 and Nxt1 using reciprocal co-IP mass-spectrometry with FLAG-tagged Nxf2 as bait (Fig. 1a right; Table S1). In both experiments, peptide levels for bait and interactors were in a similar range, suggesting that Panoramix, Nxf2, and Nxt1 form a stable protein complex (see below; Supplementary Fig. 1a). In comparison, the previously identified Panoramix-interactor Piwi 20,21 was only ~2-fold enriched and Piwi peptide levels in the IP eluates were ~20-fold lower than the other interactors (Supplementary Fig. 1a, Table S1), indicating a transient or sub-stoichiometric association between Piwi and Panoramix or Nxf2. A co-IP experiment using a monoclonal antibody against Panoramix confirmed these findings for the endogenous Panoramix, Nxf2 and Piwi proteins (Fig. 1b).
Figure 1. The Nxf2–Nxt1 heterodimer interacts with Panoramix.
a, Volcano plots showing fold enrichments of proteins determined by quantitative mass spectrometry in FLAG-Panoramix (left) or FLAG-Nxf2 (right) co-immunoprecipitates versus control (n=3 biological replicates). Experimental OSC lines stably expressed tagged Panoramix or Nxf2, wildtype OSCs served as control.
b, Western blot analysis of co-immunoprecipitation of endogenous Panoramix from nuclear OSC lysate. Relative amount loaded in immunoprecipitation (IP) compared to input (inp.) is indicated (unb. refers to unbound fraction; negative control: mouse IgGs).
c, Expression levels of indicated genes in indicated tissues based on modENCODE mRNA-seq data 31.
d, Western blot analysis of ovarian lysates with anti-Nxf2 antibody (ATP5A: loading control).
e, Average egg-laying rates of females with indicated genotype (n=3 biological replicates; error bars indicate s.d.). None of the eggs laid by panoramix or nxf2 mutant females developed into larvae.
f, g Left: Steady state ovarian RNA levels (transcripts per million) of transposons (f) and genes (g) in nxf2 mutants compared to control. Right: Number of differentially expressed genes or transposons in indicated genotypes.
h, Confocal images of wildtype egg chambers (scale bar: 20 μm) co-stained for Panoramix (top) or Nxf2 (bottom) with Piwi (for antibody specificity see Sup. Fig. 1f).
i, X-Gal stained ovarioles (scale bar: 0.1mm) from flies expressing gypsy-lacZ (soma sensor) or burdock-lacZ (germline sensor) plus indicated knockdown (KD) constructs.
Source data: panel a: Supplementary Table 1; panel f, g: Supplementary Table 2; uncropped blot images: Supplementary Data Set 1.
Nxf2 belongs to the NXF protein family, which in Drosophila is composed of the principal mRNA export receptor Nxf1, and the NXF variants Nxf2, Nxf3, and Nxf4 24,25. Like piwi and panoramix, nxf2 is expressed predominantly in ovaries (Fig. 1c) 31. To follow up on the connection between Panoramix and Nxf2, we generated nxf2 null mutant flies (Fig. 1d; Supplementary Fig. 1b). In contrast to nxf1 and nxt1 mutants, which are lethal 32,33, nxf2 mutants were viable and developed gonads (Supplementary Fig. 1c). However, as panoramix mutants, nxf2 mutant females were sterile; compared to control flies, they laid fewer eggs (Fig. 1e) and none of these developed into a larva. To investigate whether the sterility of nxf2 mutants is linked to defects in transposon silencing, we sequenced total ovarian RNA from nxf2 mutants and control flies. Similar to panoramix or piwi mutants, nxf2 mutants expressed elevated levels of several transposon families (30%; TPM>5) while only very few endogenous mRNAs exhibited changes in their levels (Fig. 1f, g; Supplementary Fig. 1d, e; Table S2). Among the de-silenced transposons were germline-specific (e.g. HeT-A, burdock) and soma-specific (e.g. gypsy, mdg1) elements, indicating that Nxf2 is required for transposon silencing in both ovarian tissues. In support of this, Nxf2 was expressed, like Panoramix, in ovarian somatic and germline cells (Fig. 1h; Supplementary Fig. 1f) and RNAi mediated depletion of Nxf2 specifically in the ovarian soma or germline resulted in de-repression of cell-type specific transposon reporters (Fig. 1i). Taken together, Nxf2 interacts with Panoramix and is required for transposon silencing.
Nxf2 is required for Piwi-mediated heterochromatin formation
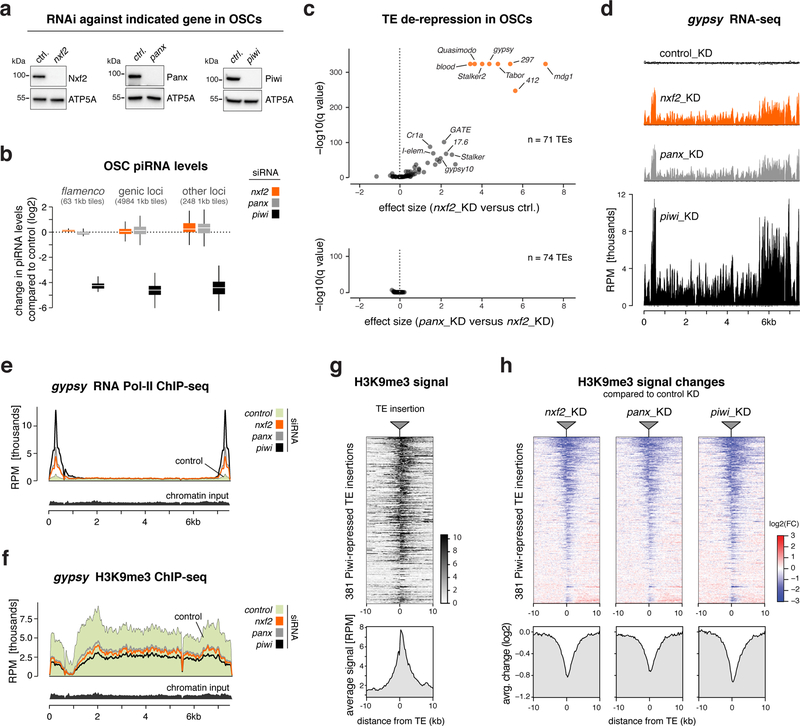
Given its interaction with Panoramix, we hypothesized that Nxf2 is required for Piwi to induce co-transcriptional silencing. If so, loss of Nxf2 should not affect piRNA biogenesis, but would result in piRNA-loaded Piwi being unable to specify heterochromatin at target loci 20,21. Indeed, Piwi levels and localization were unchanged in nxf2 mutants (Supplementary Fig. 2a), indicative of intact piRNA biogenesis and loading. Furthermore, sequencing of small RNAs from Nxf2-depleted OSCs, revealed that Nxf2, similar to Panoramix, is not required for piRNA production (Fig. 2a, b). We obtained similar results when we depleted Nxf2 specifically in the ovarian germline via transgenic RNAi (Supplementary Fig. 2b, c). Thus, irrespective of tissue and genomic origin, loss of Nxf2 does not impact piRNA biogenesis.
Figure 2. Nxf2 is required for Piwi-mediated heterochromatin formation.
a, Western blots showing knockdown efficiencies of indicated proteins in OSCs (ATP5A: loading control).
b, Box plot showing fold change (compared to control knockdown) of piRNA levels in OSCs depleted of indicated genes (box plots indicate median, first and third quartiles (box), whiskers show 1.5× interquartile range; outliers were omitted).
c, Volcano plots showing changes (as effect size) in transposon levels (RNA-seq; n=3 biological replicates; upper plot: nxf2 knockdown versus control, lower plot: panoramix versus nxf2 knockdowns).
d, Density profiles showing normalized reads from OSC RNA-seq experiments mapping to gypsy (knockdowns indicated).
e, f, Density profiles showing normalized reads from OSC RNA Pol II ChIP-seq (e), or H3K9me3 ChIP-seq (f) experiments mapping to gypsy (knockdowns indicated).
g, Heatmap (top) and metaplot (bottom) showing H3K9me3 levels within the 20kb region flanking all 381 Piwi-repressed euchromatic transposon insertions in OSCs (sorted for average H3K9me3 signal).
h, Heatmaps (top) and metaplots (bottom) showing control-normalized log2-fold changes in H3K9me3 levels at genomic regions surrounding transposon insertions defined in Fig. 2g (sorting as in g).
Source data: panel c: Supplementary Table 3; uncropped blot images: Supplementary Data Set 1.
To ask whether Nxf2 is required for piRNA-guided co-transcriptional silencing, we turned to OSCs. Here, loss of Piwi results in up to hundred-fold elevated RNA levels of a subset of LTR-retrotransposons due to increased transcription accompanied by loss of the heterochromatic H3K9me3 mark (Supplementary Fig. 2d) 14. In Nxf2-depleted OSCs, all piRNA pathway-repressed transposons were de-silenced despite normal piRNA levels (Fig. 2c top, 2d; Table S3). The extent of de-repression was indistinguishable to that seen in Panoramix-depleted cells (Fig. 2c bottom, 2d). Consistent with a role of Nxf2 in co-transcriptional silencing, ChIP-seq experiments revealed that loss of Nxf2 resulted in increased RNA Polymerase II occupancy and reduced H3K9me3 levels for piRNA-targeted transposons like gypsy, or mdg1 (Fig. 2e, f; Supplementary Fig. 2e). In contrast, transposons not under piRNA control in OSCs (e.g. burdock, F-element) showed no such changes (Supplementary Fig. 2f, g).
To assess these effects at the level of individual genomic loci, we examined the euchromatic insertion sites of Piwi-repressed transposons in the OSC genome. At these stand-alone transposon insertions, H3K9me3 marks spread into flanking genomic regions, to which sequencing reads could be mapped unambiguously (Fig. 2g) 14. Focusing on the ~380 Piwi-silenced transposon insertions revealed that Nxf2 loss phenocopies the decrease in H3K9me3 levels seen in cells depleted of Panoramix or Piwi (Fig. 2h). Piwi-independent H3K9me3 domains instead were unaffected in Nxf2-depleted cells (Supplementary Fig. 2h).
Several dozen piRNA-repressed transposons are inserted in the vicinity of endogenous gene loci. In the absence of Piwi or Panoramix, loss of repressive heterochromatin at these transposon insertions results in elevated transcription of these genes (Supplementary Fig. 2i; Table S4) 14. A highly similar set of genes was differentially expressed in Nxf2- or Panoramix-depleted OSCs (Supplementary Fig. 2i; Table S4). The rare changes in gene expression caused by Nxf2 loss can, therefore, be attributed to impaired Piwi-mediated heterochromatin formation at transposon loci. We confirmed all findings based on RNAi-mediated depletion of Nxf2 in OSCs with an independent siRNA targeting nxf2 (Supplementary Fig. 2j, k; Table S4). Our combined results support a model where Nxf2—rather than acting as a cellular RNA transport receptor—is required for co-transcriptional silencing and heterochromatin formation downstream of Piwi.
Targeting Nxf2 to nascent RNA induces co-transcriptional silencing
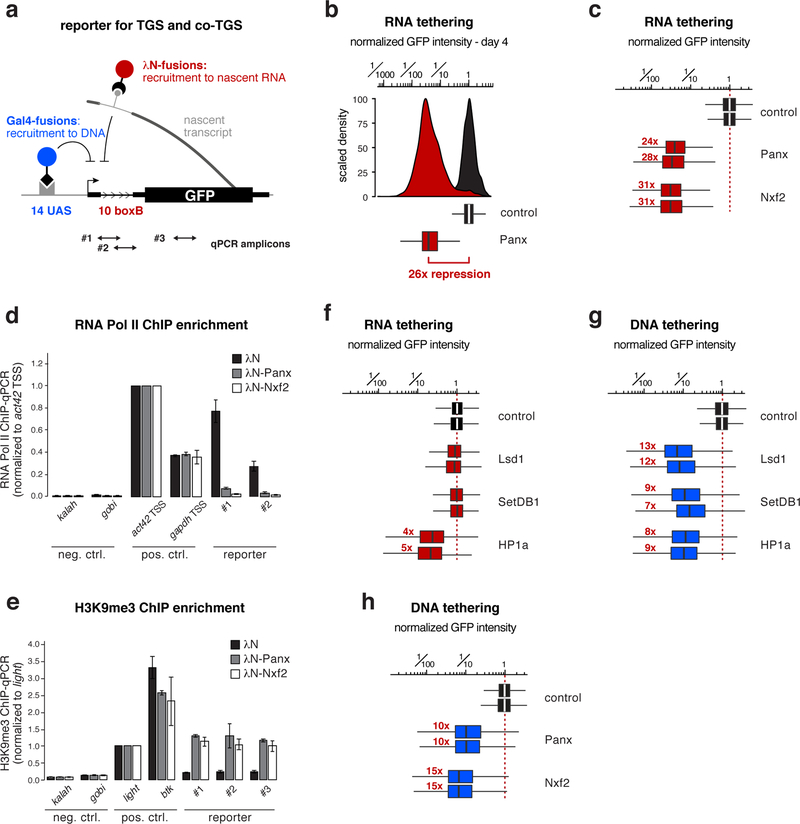
To test Nxf2’s involvement in heterochromatin-mediated silencing more directly, we converted a reporter system in fly ovaries that assays co-transcriptional silencing independently of piRNAs 20,21 into a quantitative cell culture assay. We generated a clonal OSC line harboring a single-copy transgene that expresses GFP under control of a strong enhancer (Supplementary Fig. 3a). To mimic piRNA-guided target silencing, any factor of interest can be recruited to the nascent reporter RNA as a λN-fusion protein, through boxB sites located in the intron of the reporter construct (Fig. 3a). The same reporter cell line also allows recruitment of factors of interest to the reporter DNA (via the Gal4-UAS system) in order to assay transcriptional silencing independent of targeting the nascent RNA (Fig. 3a).
Figure 3. Targeting Nxf2 to nascent RNA induces co-transcriptional silencing.
a, Cartoon of the GFP silencing reporter, which allows recruitment of proteins to the nascent RNA via intronic boxB sites and to DNA via UAS sites (positions of qPCR primers used in d, e are indicated).
b, Histogram (top) and corresponding box plot (bottom) showing the λN-control normalized GFP intensity in transfected OSCs (4 days after transfection of the tagged fusion proteins; box plots indicate median, first and third quartiles (box), whiskers show 1.5× interquartile range; outliers were omitted; n=2500 cells; red number indicates fold-repression value based on median, normalized to control).
c, Box plots showing GFP intensity in OSCs 4 days after transfection with plasmids expressing indicated λN-fusion proteins (red numbers indicate fold-repression value based on median, normalized to control for two biological replicates; box plot definition: Fig. 3b; n=2500 cells).
d, e, Normalized RNA Pol II occupancy (d) and H3K9me3 levels (e) at indicated loci determined by chromatin immunoprecipitation (ChIP) followed by quantitative PCR using OSCs expressing indicated λN-fusions (n=2 biological replicates; error bars: s.d.).
f, As in c.
g, h, As in c, but for indicated Gal4 DNA binding domain fusions.
Source data: panel d, e: available online.
While expression of λN alone had no impact on GFP reporter levels, transient expression of λN-Panoramix resulted in > 25-fold reporter repression for four to five days (Fig. 3b; Supplementary Fig. 3b). Expression of λN-Nxf2 led to similarly potent repression (Fig. 3c; Supplementary Fig. 3c). In both cases, silencing correlated with reduced RNA Pol-II occupancy and establishment of an H3K9me3 domain at the reporter locus (Fig. 3d, e). Experimental tethering of either Panoramix or Nxf2 to a nascent RNA, therefore, induces co-transcriptional silencing accompanied by heterochromatin formation. The efficiency of this silencing process is remarkable considering that the boxB sites reside in an intron of the reporter construct, and thus one would expect that the target RNA is only transiently present at the encoding DNA locus. Indeed, the heterochromatin promoting factors SetDB1, Lsd1, or HP1a, which act downstream of Piwi, were not capable of inducing comparable co-transcriptional silencing when targeted to the nascent reporter RNA (Fig. 3f; Supplementary Fig. 3d). The same factors silenced the reporter as efficiently as Panoramix or Nxf2 when targeted directly to the reporter DNA (Fig. 3g, h; Supplementary Fig. 3e, f). Therefore, we propose that co-transcriptional silencing, i.e. silencing via targeting nascent RNA, by Panoramix and Nxf2 requires more than merely recruiting the so-far identified heterochromatin effectors to the nascent RNA.
Panoramix and Nxf2–Nxt1 form the interdependent SFiNX complex
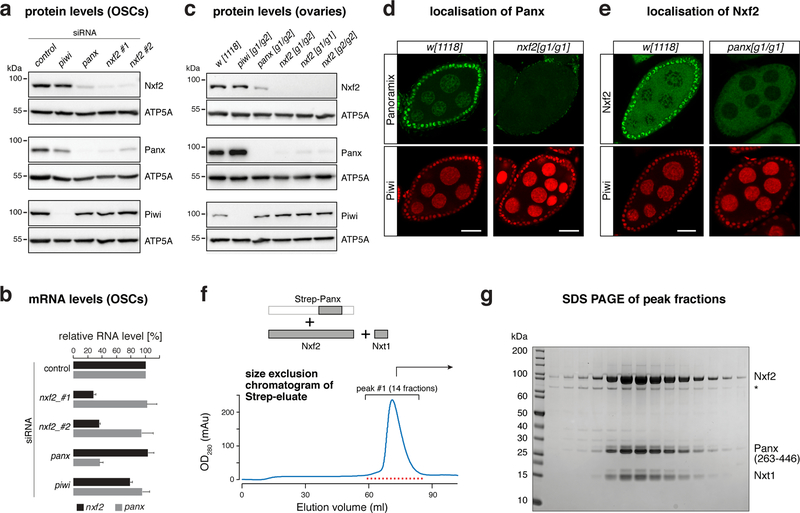
Nxf2 and Panoramix interact, are both capable of inducing co-transcriptional silencing, and loss of either protein results in highly similar phenotypes (Fig. 1–3). These findings suggested a close molecular connection between Nxf2 and Panoramix. In support of this, we found a reciprocal dependency between both proteins: Depletion of Nxf2 or Panoramix in OSCs resulted in substantial reductions of the respective other protein, while the corresponding mRNA levels were unchanged (Fig. 4a, b). Similarly, in nxf2 mutant ovaries, Panoramix protein was hardly detectable by western blotting or immunofluorescence analysis (Fig. 4c, d), despite unchanged panoramix mRNA levels (Supplementary Fig. 4a). Conversely, Nxf2 protein levels were reduced in panoramix mutant ovaries (Fig. 4c), and the remaining Nxf2 protein was excluded from the nucleus (Fig. 4e), suggesting that Nxf2’s nuclear localization depends on Panoramix. Consistent with this, Panoramix harbors a Lysine-rich sequence stretch (residues 196–262), which is required for its nuclear localization (Supplementary Fig. 4b). Based on these results, we hypothesized that Panoramix and Nxf2 stabilize each other via forming a nuclear protein complex.
Figure 4. Panoramix and Nxf2–Nxt1 form the interdependent SFiNX complex.
a, Western blots showing levels of indicated proteins in lysates from OSCs with indicated gene knockdowns (ATP5A: loading control).
b, Bar graph showing fold change of RNA-seq levels of indicated genes compared to control knockdown in OSCs (n=3 biological replicates; error bars: s.d.).
c, Western blots showing levels of indicated proteins in ovary lysates from flies with indicated genotypes (ATP5A: loading control).
d, Confocal images of egg chambers (scale bar: 20 μm) from w[1118] control and nxf2 mutant flies stained for Panoramix (upper panels) and Piwi (lower panels).
e, As in d, but for panoramix mutants and Nxf2 protein.
f, Top: schematics of the trimeric complex expressed in insect cells (grey boxes indicate co-expressed proteins/fragments). Bottom: size exclusion chromatogram of the affinity-purified Strep-Panoramix eluate (mAU = milli-absorbance unit).
g, SDS-PAGE of the 14 peak fractions indicated in f (* indicates HSP70 as determined by mass spectrometry).
Source data: uncropped blot and gel images: Supplementary Data Set 1.
To test for a Panoramix–Nxf2 complex, we co-expressed both proteins in insect cells together with Nxt1, which we identified as Nxf2 and Panoramix interactor (Fig. 1), and which functions as a general NXF cofactor 24. While all three proteins were abundantly produced, Panoramix was degraded. Therefore, we expressed instead of full-length Panoramix a 25 kDa fragment that is necessary and sufficient for binding Nxf2 (see below). A single affinity purification step of the Strep-tagged Panoramix fragment, followed by size exclusion chromatography, resulted in a defined protein peak containing all three factors as a monomeric complex (Fig. 4f, g; Supplementary Fig. 4c, d). Panoramix, Nxf2, and Nxt1 therefore form a stable protein complex, which we named SFiNX (Silencing Factor interacting Nuclear eXport factor variant).
Panoramix-mediated silencing via nascent RNA requires Nxf2
Due to the interdependency between Panoramix and Nxf2, our experiments so far interrogated the function of the SFiNX complex rather than that of the individual proteins. To disentangle the molecular roles of Panoramix and Nxf2, we set out to generate interaction-deficient point-mutant variants. Panoramix consists of an N-terminal disordered half and a C-terminal half with predicted secondary structure elements (Fig. 5a). Using GFP-tagged full-length Nxf2 as bait, we mapped the interaction site within Panoramix to the first part of the structured domain (Fig. 5a; Supplementary Fig. 5a, b). Within there, we identified two regions that upon deletion impacted the Nxf2–Panoramix interaction (Fig. 5a; Supplementary Fig. 5c). Panoramix Δ308–386 failed to bind Nxf2, while Panoramix Δ387–446 interacted less efficiently with Nxf2. The 308–386 peptide harbors a predicted amphipathic alpha-helix (Supplementary Fig. 5d, e). Mutating four hydrophobic residues predicted to line one side of this helix abrogated the Nxf2 interaction (Fig. 5b; Supplementary Fig. 5e). Notably, both Panoramix variants that are defective in Nxf2 binding (Panoramix[Δ308–386] and Panoramix[helix mutant]) accumulated to lower levels compared to the wildtype protein (Supplementary Fig. 5f). Panoramix Δ387–446 on the other hand accumulated to higher levels than wildtype Panoramix (Supplementary Fig. 5f). Considering the co-dependency between Panoramix and Nxf2 in vivo, we hypothesized that the 387–446 peptide within Panoramix harbors a destabilizing element that induces protein degradation if not protected by Nxf2. Consistent with this, fusing the 387–446 Panoramix peptide to GFP led to a ~100-fold reduction in GFP levels (Supplementary Fig. 5g). Mutation of four hydrophobic residues within this “degron” led to increased Panoramix levels (Supplementary Fig. 5d, f). When combining both sets of point mutations, the resulting Panoramix[helix+degron mutant] accumulated to high levels and was unable to interact with Nxf2 (Fig. 5b; Supplementary Fig. 5f).
Figure 5. Panoramix-mediated silencing via nascent RNA requires Nxf2.
a, Panoramix protein cartoon showing predicted secondary structure elements. The summary below states the involvement of the [helix] and [degron] sites in Nxf2 interaction and Panoramix’ protein stability.
b, Western blot analysis of GFP-Nxf2 immunoprecipitation experiments from S2 cells transiently co-transfected with indicated FLAG-Panoramix plasmids (relative amount loaded in IP-lanes: 3x).
c, Schematics of the domain architecture of Nxf1 and Nxf2 (RRM: RNA recognition motif; LRR: leucine rich repeat; NTF2l: nuclear transport factor 2-like domain; UBA: ubiquitin associated domain).
d, Cartoon view of Nxf2’s UBA domain (pink) in complex with the Panoramix helix (cyan).
e, Detailed views of interfaces between Nxf2’s UBA domain (pink) and the Panoramix helix (cyan).
f, Sequence alignment of the Nxf2 and Nxf1 UBA domains (red squares: identical residues; blue triangles: Nxf2 residues involved in Panoramix interaction; red triangles: interaction residues that differ in Nxf1).
g, Western blot analysis of immunoprecipitation experiments (bait: stabilized GFP-Panoramix) using S2 cells transiently co-transfected with indicated FLAG-Nxf2 plasmids (relative amount loaded in IP lanes: 3x).
h, Shown is the transposon repression rescue potential of indicated constructs transfected into OSCs depleted of endogenous Panoramix (left) or Nxf2 (right). mdg1 levels determined via qRT-PCR (n=2 biological replicates; error bars: s.d.).
i, j, Box plots showing GFP intensity in OSCs 4 days after transfection with plasmids expressing indicated λN-tagged (red) or Gal4-tagged (blue) fusion proteins (red numbers indicate fold-repression values, normalized to control for two biological replicates; box plots indicate median, first and third quartiles (box), whiskers show 1.5× interquartile range; outliers were omitted; n=2500 cells).
Source data: panel h: available online; uncropped blot images: Supplementary Data Set 1.
We then turned to Nxf2, whose domain organization resembles that of the mRNA export receptor Nxf1 34, except that it has a duplicated putative RNA binding unit consisting of RNA recognition motif (RRM) and a Leucine-rich repeat (LRR) domain (Fig. 5c). Using stabilized Panoramix[degron-mutant] as bait, we determined that Nxf2’s NTF2-like and UBA domains together are sufficient to bind Panoramix, and that the UBA domain is required for the Panoramix interaction (Supplementary Fig. 5h). Consistent with this, the 30 amino-acid amphipathic Panoramix helix, that is required for the Nxf2 interaction (Fig. 5b), bound the Nxf2 UBA domain in vitro (Supplementary Fig. 5i). Building on this, we determined the structure of Nxf2’s UBA domain in complex with the Panoramix helix at 2.0 Å resolution (Supplementary Fig. 6a, b; Table 1). This revealed that the UBA domain of Drosophila Nxf2 is highly similar to that of human NXF1 (PDB ID: 1OAI; 35. Both domains consist of a three-helix bundle (α1-α3) with a short fourth helix (α4) at the C terminus (Fig. 5d). α1, α3 and α4 of the Nxf2 UBA domain form a hydrophobic core that interacts with the hydrophobic face of the amphipathic Panoramix helix involving the Panoramix residues L323, A324, V325, A328, V331, and L332. In addition, the N terminus of the Panoramix helix is stabilized by two flanking hydrogen bonds and salt bridges (Fig. 5e). The experimentally determined Panoramix[helix mutant] variant that cannot bind Nxf2 (Fig. 5b) supported this structure: three out of the six hydrophobic residues contributing to the interaction were mutated in the Panoramix[helix mutant] variant. Within Nxf2, ten hydrophobic residues contribute to the Panoramix interaction (Fig. 5f). Out of these, two residues (V800 and I827) are highly different in Drosophila Nxf1 (hydrophilic residues Q632 and E657), which does not interact with Panoramix (Supplementary Fig. 6c, d). When we mutated V800 and I827 in Nxf2, together with two flanking residues, into the corresponding Nxf1 amino acids, the resulting Nxf2[UBA mutant] was unable to bind Panoramix (Fig. 5g; Supplementary Fig. 6c).
Table 1.
Data collection and refinement statistics
| UBA-linker-Panx helix (PDB 6OPF) | NTF2l domain–Nxt1 (PDB 6MRK) | |
|---|---|---|
| Data collection | ||
| Space group | P21 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 47.89, 72.89, 59.68 | 64.54, 74.09, 155.81 |
| α, β, γ (°) | 90, 109.9, 90 | 90, 90, 90 |
| Resolution (Å) | 45.03–2.00 (2.11–2.00)a | 77.9–2.80(2.95–2.80) |
| Rmerge | 0.082(0.492) | 0.102(0.412) |
| I/σ(I) | 10.1(2.7) | 11.6(3.8) |
| CC1/2 | 0.998(0.946) | 0.995(0.904) |
| Completeness (%) | 95.7(94.6) | 99.9(99.9) |
| Redundancy | 5.4(5.3) | 5.4(5.3) |
| Refinement | ||
| Resolution (Å) | 45.03–2.00 (2.11–2.00) | 77.9–2.80(2.95–2.80) |
| No. reflections | 24,962 (3,660) | 19,083 (2,732) |
| Rwork / Rfree | 22.0/24.6 | 21.2/26.3 |
| No. atoms | 2802 | 5118 |
| Protein | 2706 | 5101 |
| Water | 96 | 17 |
| B factors | ||
| Protein | 34.4 | 43.8 |
| Water | 37.7 | 32.1 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.007 | 0.008 |
| Bond angles (°) | 0.809 | 1.565 |
Building on the interaction-deficient Nxf2 and Panoramix variants, we determined whether the individual proteins are capable of supporting Piwi-mediated silencing. We performed genetic rescue experiments in OSCs and asked whether expression of siRNA resistant Panoramix[helix+degron-mutant] or Nxf2[UBA-mutant] variants could restore silencing of the mdg1 transposon in OSCs depleted for endogenous Panoramix or Nxf2. While the respective wild-type proteins supported mdg1 silencing, neither of the interaction-deficient mutants (expressed with NLS sequence to assure nuclear localization; Supplementary Fig. 6e, f) displayed rescue activity (Fig. 5h). Therefore, both Nxf2 and Panoramix contribute essential activities to the silencing process beyond reciprocal protein stabilization. To investigate the function of Panoramix and Nxf2 as silencing factors more directly, we turned to the transcriptional silencing reporter assay in OSCs (Fig. 3a). Nxf2 with point mutated UBA domain was inert in inducing reporter silencing, irrespective of whether it was targeted to nascent RNA or to DNA directly (Fig. 5i; Supplementary Fig. 6g). Instead, the Panoramix[helix+degron mutant] variant, which is defective in Nxf2 binding, showed clear, though in comparison to the wildtype protein weak co-transcriptional silencing activity (Fig. 5j; Supplementary Fig. 6h). Remarkably, when recruited directly to the reporter DNA, Panoramix[helix+degron mutant] was as potent in inducing silencing as wildtype Panoramix (Fig. 5j; Supplementary Fig. 6h). Taken together, our data indicate that Panoramix, and not Nxf2, connect SFiNX to the silencing machinery. Nxf2 instead is required for Panoramix to achieve potent silencing via the nascent RNA. To understand the function of Nxf2 within SFiNX, we reasoned that a specific molecular feature intrinsic to Nxf1 was exploited by the evolutionary exaptation of an RNA transporter into co-transcriptional silencing. At the same time, Nxf2 must have lost other Nxf1 characteristics in order to avoid getting channeled into mRNA export. We therefore set out to systematically compare Nxf2 to the well-studied Nxf1 protein.
The Nxf2–Nxt1 heterodimer lost nucleoporin binding activity
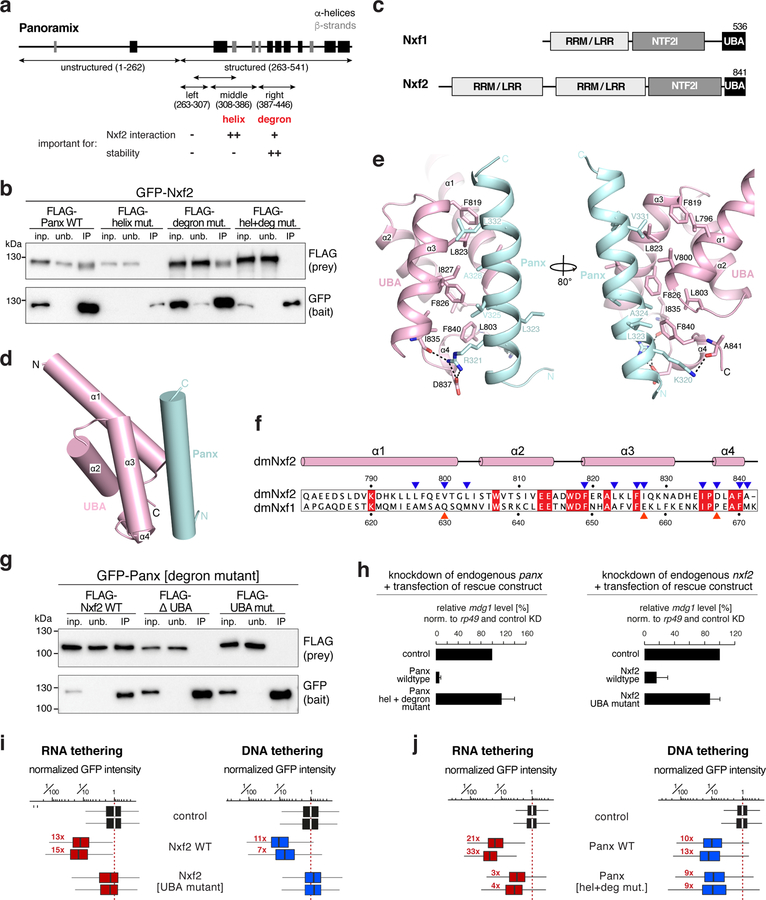
A central molecular feature of Nxf1 is its ability to shuttle through the selective phenylalanine-glycine (FG) repeat meshwork of the inner nuclear pore complex (NPC) 36. Two nucleoporin FG-binding pockets, one residing in the UBA domain and one in the NTF2-like domain, confer NPC shuttling ability to Nxf1 37. We examined both sites in Nxf2 at the structural level. The putative FG-binding pocket within Nxf2’s UBA domain lies on the opposite side of the Panoramix binding surface, making it per se accessible (Fig. 6a). In our UBA–Panoramix-helix structure, a salt bridge between E814 and K829 might restrict access to the hydrophobic core of the putative FG-binding pocket, rendering it probably non-functional for nucleoporin binding (Fig. 6b). Previous work has shown that modifications of the human NXF1 UBA domain in the region occupied by the Panoramix binding surface inhibit FxFG peptide binding 37,38. Further work will be required to resolve whether Panoramix binding modulates the functionality of this putative FG-binding pocket. To inspect the second putative FG-binding pocket, we determined the 2.8 Å resolution crystal structure of Nxf2’s NTF2-like domain bound to Nxt1 (Supplementary Fig. 7; Table 1). Based on this, Nxf2 interacts with Nxt1 in a manner very similar to human NXF1 (Fig. 6c) 39. But in contrast to the human protein, Nxf2’s putative FG-binding pocket within the NTF2-like domain is again concealed: It is occupied by the bulky side chains of its own Phe735 and Tyr690 and in addition, Arg747 may inhibit access by hydrogen-bonding to Tyr690 (Fig. 6d). Our combined structural evidence therefore suggests that the affinity of Nxf2–Nxt1 for FG-nucleoporins may be reduced relative to Nxf1–Nxt1. Because mutations in either of the two FG-binding pockets reduce nucleoporin binding for human NXF1 37, it might be that Drosophila Nxf2 lost nucleoporin binding ability. This hypothesis is supported by the observation that GFP-tagged Nxf2 did not accumulate at nuclear pores where GFP-tagged Nxf1 is highly enriched (Fig. 6e). Although further work will be needed to establish binding affinities between Nxf2 and the various FG-peptide motifs present in the NPC, our data suggest that Nxf2 has lost nucleoporin binding activity, an evolutionary change consistent with repurposing this NXF variant for co-transcriptional silencing.
Figure 6. Nxf2 lost nucleoporin binding.
a, Superposition of the overall structure of dmNxf2 UBA domain (pink) complexed with Panoramix helix (cyan) and the crystal structure of hsNXF1 UBA domain (PDB ID: 1OAI) (in silver) 35.
b, Left: Ribbon (top) and electrostatic surface (bottom) representation showing the FG-repeat binding pocket in the crystal structure of hsNXF1 UBA domain (gray) in complex with a nucleoporin FxFG-repeat peptide (magenta; PDB ID: 1OAI) 35. Right: Ribbon (top) and electrostatic surface (bottom) representation showing the closed putative FG-repeat binding pocket in the crystal structure of dmNxf2’s UBA domain (pink).
c, Superposition of the overall structure of dmNxf2’s NTF2-like domain complexed with dmNxt1 (purple and yellow) and the crystal structure (silver) of hsNXF1–NXT1 (PDB ID: 1JKG) 39 (dashed curves: invisible loops in the structure).
d, Left: Ribbon (top) and electrostatic surface (bottom) representation showing the specific recognition of nucleoporin FG-repeat (magenta) in the hsNXF1 (light blue)–NXT1 (gray) structure (PDB ID: 1JN5) by a hydrophobic FG-repeat binding pocket 39. Right: Ribbon (top) and electrostatic surface (bottom) representation showing the blocked putative FG-repeat binding pocket in the crystal structure of Nxf2’s NTF2-like domain (purple) complexed with Nxt1 (yellow). The hydrogen bond is shown as black dashed line.
e, Confocal images showing individual nurse cell nuclei from flies expressing GFP-Nxf1 (upper row) or GFP-Nxf2 (lower row) (scale bar: 5 μm). Nuclear pore complexes (NPCs) are stained with wheat germ agglutinin (WGA).
A key role for Nxf2’s RNA binding unit within SFiNX
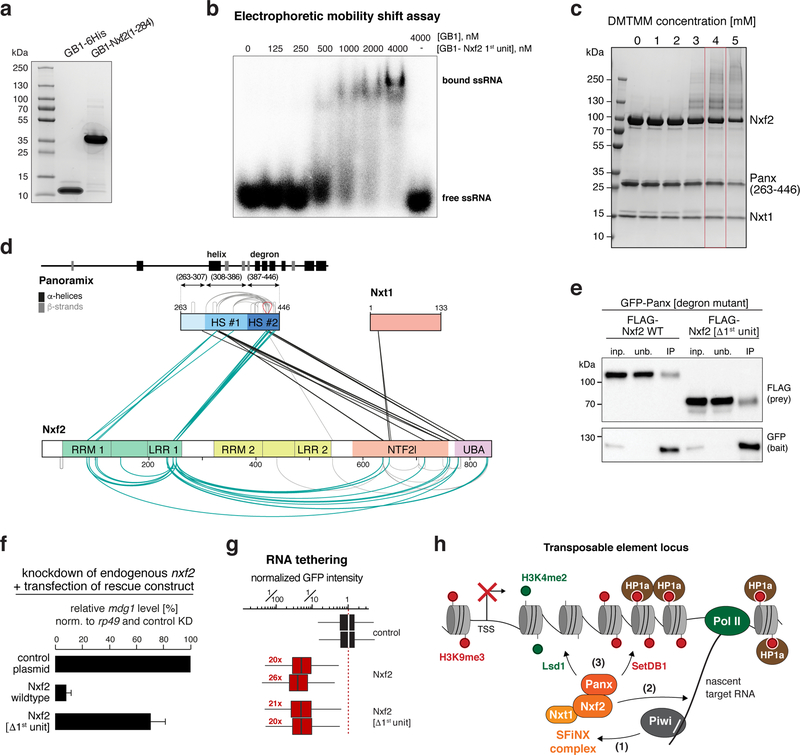
Nxf1’s second core feature is its ability to bind mRNA cargo via the RRM-LRR domains 23,40. This RNA binding activity is non-sequence-specific 41 and requires recruitment of Nxf1 to cargo RNA through adaptor proteins that mark the completion of the different mRNA processing steps 42. Nxf2 harbors a tandem RRM-LRR fold (Fig. 5c). Based on electrophoretic mobility shift assays, Nxf2’s N-terminal RRM-LRR domain (1st unit) is capable of binding single-stranded RNA in vitro (Fig. 7a, b; Supplementary Fig. 8a). The RNA binding activity of Nxf2’s 1st unit was abrogated upon mutating three positively charged amino acids, whose equivalent residues in human NXF1 contact the constitutive transport element (CTE) of simian type D retroviral transcripts (Supplementary Fig. 8b–d) 43,44. We did not succeed in obtaining Nxf2’s second RRM-LRR unit (2nd unit) as a soluble recombinant protein, preventing a statement whether it provides additional RNA binding activity.
Figure 7. A key role for Nxf2’s RNA binding unit within SFiNX.
a, SDS-PAGE showing the recombinant first RNA-binding unit of Nxf2 (1st unit) and GB1 control peptide used for the EMSA experiment (b).
b, Phosphorimage showing Electrophoretic Mobility Shift Assay (EMSA) with labelled single stranded RNA (ssRNA) and increasing amount of indicated recombinant protein.
c, SDS-PAGE showing chemically crosslinked recombinant SFiNX complex. The condition boxed in red was chosen for mass spectrometric analysis.
d, Schematic summary of crosslinks identified between Panoramix, Nxf2, and Nxt1 in the SFiNX complex. Protein domains in Nxf2 as in Fig. 5b. Within Panoramix, hotspot (HS) #1 includes the amphipathic helix, HS #2 the degron site. Straight lines indicate intermolecular crosslinks, half-circles indicate intramolecular crosslinks.
e, Western blot analysis of immunoprecipitation experiments (bait: stabilized GFP-Panoramix) using S2 cells transiently co-transfected with indicated FLAG-Nxf2 plasmids (relative amount loaded in IP lanes: 3x).
f, Bar graph showing the transposon repression rescue potential of indicated constructs transfected into OSCs depleted of endogenous Nxf2. mdg1 levels determined via qRT-PCR (n=2 biological replicates; error bars: s.d.).
g, Box plots showing GFP intensity in OSCs 4 days after transfection with plasmids expressing indicated λN-tagged fusion proteins (red numbers: fold-repression values, normalized to control for two biological replicates are indicated; box plots indicate median, first and third quartiles (box), whiskers show 1.5× interquartile range; outliers were omitted; n=2500 cells).
h, Schematic model for Piwi-mediated co-transcriptional silencing. Upon target binding Piwi interacts with the adaptor complex, SFiNX (1). Within SFiNX, Nxf2 links to the nascent target RNA (2). This allows Panoramix to recruit chromatin effectors (e.g. Lsd1 to demethylate H3K4me2, SetDB1 to establish H3K9me3) (3).
Source data: panel d: Supplementary Table 6; panel f: available online; uncropped blot and gel images: Supplementary Data Set 1.
Nxf2’s ability to bind RNA raised the question of how it avoids binding to random nuclear RNAs, which would bear the danger of ectopic silencing and heterochromatin formation. Inspired by the Nxf1 literature, we hypothesized that Nxf2’s RNA binding activity is regulated. It has been suggested that prior to mRNA cargo binding, Nxf1 is in a closed conformation with its RNA binding unit folding back onto the NTF2-like domain 45. Upon adaptor-mediated recruitment of Nxf1 to an export-competent mRNA 42, this intramolecular inhibition is released, RNA cargo is bound, and the complex shuttles through the NPC. To probe for a putative intramolecular interaction within SFiNX, we took advantage of the recombinant Panoramix–Nxf2–Nxt1 complex (Fig. 4). We used chemical crosslinking coupled to mass spectrometry and established an interaction map of residues that are in physical proximity within the complex (Fig. 7c, d; Table S5). One set of identified crosslinks (black in Fig. 7d) was in agreement with our structural and biochemical data: First, the two crosslinks involving Nxt1 map to the NTF2-like domain. Second, two crosslinking hotspots are apparent within Panoramix. Hotspot #1 corresponds to the amphipathic helix, hotspot #2 to the degron site. Both hotspots exhibit several crosslinks to the C-terminus of Nxf2, indicating that the Panoramix–Nxf2 interaction involves besides the helix–UBA interaction (Fig. 5), additional interactions, probably with the NTF2-like domain. Strikingly, nearly all other identified protein crosslinks involve Nxf2’s first RRM-LRR unit (1st unit). We identified multiple intramolecular crosslinks between the 1st unit and the NTF2-like and UBA domains, as well as intermolecular crosslinks to Panoramix, mostly to the degron site. As the 1st unit is dispensable for Panoramix binding (Fig. 7e), the identified intra-molecular interactions could indicate a regulatory interaction within Nxf2, similar to what has been proposed for Nxf1 45.
To determine the importance of Nxf2’s 1st RNA-binding unit for SFiNX function, we performed genetic rescue experiments. In flies and OSCs, expression of Nxf2[Δ1st unit] instead of the wildtype protein was not able to support transposon silencing (Fig. 7f; Supplementary Fig. 8e, f), although Nxf2[Δ1st unit] localized to the nucleus (Supplementary Fig. 8e) and interacted with Panoramix (Fig. 7e). Together with the finding that the 1st unit is able to bind RNA, this suggested a model where Nxf2 anchors SFiNX to the nascent target RNA via its 1st RRM-LRR unit. If this were true, SFiNX lacking Nxf2’s 1st unit should remain silencing competent if recruited to the target RNA via the λN-boxB tethering system. Indeed, expression of λN-tagged Nxf2[Δ1st unit] induced co-transcriptional silencing as efficiently as λN-tagged wildtype Nxf2 (Fig. 7g; Supplementary Fig. 8g). Taken together, our findings suggest a model where in the non-target-engaged state, Nxf2’s RNA binding unit folds back onto the SFiNX complex. Upon recruitment to a target transcript, Nxf2 interacts with RNA, thereby anchoring SFiNX via the nascent target transcript to chromatin and allowing Panoramix to recruit effectors to establish heterochromatin (Fig. 7h).
DISCUSSION
The discovery of SFiNX, a nuclear protein complex consisting of Panoramix, Nxf2, and Nxt1, provides a key molecular connection between Piwi, the nascent target RNA, and the cellular heterochromatin machinery. In the absence of SFiNX, piRNA-loaded Piwi is incapable of inducing co-transcriptional silencing (Fig. 2). Conversely, experimental recruitment of SFiNX, but not its individual components, to a nascent RNA results in potent silencing and local heterochromatin formation independently of Piwi (Fig. 3). Our data indicate that within SFiNX, Panoramix, and not Nxf2, links to the downstream cellular heterochromatin effectors (Fig. 5). Based on genetic experiments, the histone methyl-transferase SetDB1, the histone-demethylase Lsd1, and the heterochromatin binding protein HP1a are required for piRNA-guided co-transcriptional silencing 20,21. We did not find any of these factors enriched in our SFiNX co-IP mass-spectrometry experiments. Interestingly, the SUMO E3 ligase Su(var)2–10, which was recently linked to piRNA-guided co-transcriptional silencing 46, is enriched more than five-fold in Nxf2 and Panoramix IP experiments (Table S1). Two additional factors required for piRNA-guided transcriptional silencing—Maelstrom and Asterix (Gtsf1) 14,17–21—are also enriched in both IPs. It is currently unclear how these proteins relate to SFiNX, yet genetic experiments place them upstream or in parallel to SFiNX 20,21.
The involvement of Nxf2, a nuclear RNA export variant, in co-transcriptional silencing came as a surprise to us. Based on our biochemical and structural data, we propose that two molecular features of the ancestral NXF protein, the principal mRNA export receptor Nxf1, facilitated the evolutionary exaptation of Nxf2 into piRNA-guided silencing (Fig. 7). First, Nxf2 retained its ability to bind RNA, thereby providing SFiNX a molecular link to the nascent target RNA. Second, our crosslinking-mass spectrometry data implies that similar to Nxf1, the RNA binding activity of Nxf2 might be gated. We suggest that this could provide a critical regulatory switch to ensure that SFiNX only associates with transcripts that are specified as targets via the Piwi-piRNA complex. A central open question is, how SFiNX is recruited to piRNA complementary target RNAs, and whether the RRM-LRR units are involved in this recruitment. In the case of Nxf1, various proteins (e.g. SR-proteins, THO-complex, UAP56, Aly) that are recruited during co-transcriptional mRNA maturation, are required to restrict Nxf1 deposition onto export-competent mRNAs only 42,47,48. Whether any of these factors is also required for loading Nxf2 onto RNA, potentially by interacting with the RRM-LRR units, is currently unclear. It is, however, likely that target-engaged Piwi plays a central role in the deposition of SFiNX onto the target RNA. Despite considerable experimental efforts we were not able to establish a direct molecular link between Piwi and either Nxf2 or Panoramix. As piRNA-independent recruitment of Piwi to a nascent RNA is incapable of target silencing, the putative Piwi–SFiNX interaction most likely occurs only once Piwi is bound to a target RNA via a complementary piRNA 20,21,49. In light of this, it is possible that Nxf2 interacts preferentially with certain RNA structural features, for example the piRNA-target RNA duplex. Further biochemical experiments, as well as structural insight into the full SFiNX complex will be required to shed light onto the molecular logic of this intriguing silencing complex.
Although no direct ortholog of Nxf2 is identifiable in vertebrates, our finding that an NXF variant is involved in transposon silencing in Drosophila likely points to a more general scheme. The Nxf1 ancestor diversified through independent evolutionary radiations into numerous NXF variants in different animal lineages (Supplementary Fig. 8h; Table S6) 50. In flies, the three NXF variants exhibit gonad-specific expression with Nxf2 and Nxf3 being expressed predominantly in ovaries, and Nxf4 being testis-specific 51. Besides Nxf2, Drosophila Nxf3 is also an essential piRNA pathway component as it is required for the nuclear export of un-processed piRNA cluster transcripts 52. In mice and humans, several NXF variants are preferentially expressed in testes, and Nxf2 mutant mice are male sterile 53. Considering this, we speculate that also in vertebrates the host-transposon conflict has been a driver of NXF protein evolution through duplication and exaptation events. Our study highlights that some of these variants might have evolved novel molecular functions, not directly related to RNA export biology.
MATERIALS & METHODS
Fly strains
All fly strains used in this study are listed in Table S7 and are available from the VDRC (http://stockcenter.vdrc.at/control/main). Flies were kept at 25 °C. For each experiment, flies were aged for 5–6 days and kept on apple juice agar plates with yeast paste to ensure consistent ovarian morphology. Two independent nxf2 frameshift mutant alleles were generated by injecting the pDCC6 plasmid with nxf2 targeting gRNAs (Table S8) into w[1118] flies 54. Sequences for the two frameshift alleles are indicated in Supplementary Fig. 1c. N-terminal 3xFLAG_V5_GFP-tagging of endogenous nxf2 and panoramix loci was done by co-injecting a repair template and the gRNA containing plasmid (Addgene 45956) into act-Cas9 flies (BL-58492). Oligonucleotides used for gRNA cloning are listed in Table S8.
Germline and soma specific gene knockdowns were performed by crossing short hairpin (shRNA) transgene strains with the maternal triple driver (MTD)-GAL4 line or the traffic jam-GAL4 driver line, respectively. Oligonucleotides used for shRNA cloning are listed in Table S8. gypsy and burdock-LacZ sensor strains are described in 29.
Rescue strains with Nxf2 or its variant were generated by injecting respective rescue transgenes into nxf2 mutant flies containing attp landing sites on the same chromosome (attP154). The rescue transgenes contained the panoramix regulatory control region (chr2R: 21,308,437–21,313,490) and the panoramix coding sequence was replaced by the nxf2 rescue construct.
X-gal staining of ovaries
Ovaries were dissected into ice cold PBS, fixed in 0.5% glutaraldehyde (in PBS) for 15 min at room temperature, and then washed twice with PBS. Next, samples were incubated in staining solution (10 mM PBS, 1 mM MgCl2, 150 mM NaCl, 3 mM potassium ferricyanide, 3 mM potassium ferrocyanide, 0.1% Triton X-100, 0.1% X-gal (5-bromo-4-chloro-3-indolyl-β -d-galactoside)) overnight (gypsy-sensor) or for 2h (burdock-sensor) at room temperature.
Generation of Nxf2 and Panoramix antibodies
Purified His-tagged Nxf2 (1–326) protein was used to generate the mouse anti-Nxf2 antibody used for western blot. The mouse anti-Nxf2 and anti-Panoramix antibodies used for immunofluorescence were raised against the SFiNX complex consisting of his-tagged Nxf2 (541–841), Strep-tagged-Panoramix (263–446) and Flag-tagged Nxt1 (full length). All antibodies were generated at the MFPL Monoclonal Antibody Facility.
OSC cell culture
OSCs were obtained from the Siomi lab and cultured as described 27,28. Plasmid and siRNA transfections were performed using Cell Line Nucleofector kit V (Amaxa Biosystems) with the program T-029, using 8 million cells per transfection. siRNAs used in this study are listed in Table S9.
Stable OSC reporter line generation
The reporter construct (traffic jam enhancer driven GFP_P2A-Blasticidin-resistance harboring 10 intronic boxB sites and 14 upstream UAS sites; Addgene 128010) was integrated into chromosomal location chr2L:9,094,918, which is devoid of genes and major chromatin marks, using CRISPR-Cas9. In brief, 600bp long homology arms flanking the integration site were amplified from OSC genomic DNA. Oligonucleotides targeting the locus (Table S8) were cloned into the guide RNA expression plasmid (Addgene 49330). Two independent gRNA containing plasmids were mixed 1:1 and 200 ng of this mix were co-transfected with 1200 ng of the integration plasmid into OSCs. After two days, the cells were plated with different dilutions and on the following day Blasticidin containing media was added (1:1000) for a 4-day long selection. Afterwards, the cells were grown in normal medium for about 2 weeks until individual clones could be isolated.
Droplet PCR
To assess the copy number of the reporter construct integrated in the OSC genome, the QX200™ Droplet Digital™ PCR System (BIORAD) was used according to the manufacturer’s instructions. In brief, genomic OSC DNA was digested with EcoRI and HindIII restriction enzymes. The PCR reaction was set up with 10ng digested genomic DNA (primer sequences in Table S8) and the QX200™ ddPCR™ EvaGreen Supermix. The PCR mix and the QX200 Droplet Generation Oil for EvaGreen were added into a DG8™ cartridge and droplets were generated with QX200 Droplet generator. Thermal cycling and droplet reading were performed with the instructor’s standard protocol which gave the concentration of the amplicon in copies/reaction volume. Based on 2 house-keeping control genes, the copy number in the genome was determined for the integrated reporter.
Stable OSC line generation with extra genomic copy
The pAcm vector 28 was modified to create the integration constructs. Downstream of the act5C promoter, a 3xFLAG-HA tag was added followed by the open reading frames encoding full length Panoramix or Nxf2. The selection cassette consisted of an independent transcription unit driving mCherry_P2A_Puromycin-resistance via the traffic jam enhancer from Drosophila yakuba in combination with the Drosophila synthetic core promoter (DSCP) 55. The two transgenes were integrated into the chromosomal location chr2L:9,103,945, which is devoid of genes. 1200 bp long homology arms flanking the integration sequence were used for the integration. Stable integration was generated as described above for the reporter cell line, except that Puromycin was used for the selection (1:2000).
Tethering reporter assay
All tethering constructs are based on λN-entry or Gal4-entry vectors (Addgene 128011–128014). Various full-length CDS or CDS variants were inserted into the entry vectors to generate N-terminally tagged fusion proteins. Unless having full length genes, the SV40 NLS sequence (PKKKRKV) was included to ensure nuclear localization of the variants. A separate expression cassette driving mCherry via the traffic jam enhancer was used to select positively transfected cells.
OSCs harboring stably integrated GFP reporter were transfected with 4µg of λN/Gal4 fusion construct. As a negative control, λN/Gal4 empty vector was used. Two days after transfection, cells were harvested for WB analysis and four days after transfection cells were harvested for flow cytometry analysis using a FACS BD LSR Fortessa (BD Biosciences). Transfected cells were gated based on mCherry expression and the GFP intensity was determined in that population (per experiment 2500 cells). Data analysis was performed using FACS Diva and FlowJo. The gating strategy is described in Supplementary Data Set 2.
OSC rescue assay
OSCs were co-transfected with siRNAs targeting panoramix or nxf2 and a plasmid containing the act5c driven siRNA-resistant rescue construct. A second transfection was performed after two days and cells were collected after four days for WB analysis and RNA isolation for RT-qPCR. siRNAs used for the rescue experiments are listed in Table S9.
RT-qPCR
OSCs or 5–10 pairs of ovaries were collected into TRIzol reagent and RNA was isolated according to the manufacturer’s instructions. Total RNA was digested with RQ1 RNase-Free DNase (Promega) and cDNA was prepared using random hexamer oligonucleotides and Superscript II (Invitrogen). Primers used for qPCR analysis are listed in Table S8. Source data for qPCR in Fig. 3, 5 and 7 are available online.
Immunofluorescence staining of OSCs
2 days following transfection, cells were plated on concavalin A coated coverslips. After 4 hours, cells were fixed with formaldehyde solution (4% formaldehyde in PBS) for 15 min at room temperature. Fixed cells were washed twice with PBS for 5 min, permeabilized with PBX (0.1 % Triton X-100 in PBS) for 10 min and washed again with PBS for 5 min. Blocking was done in BBS (1 % BSA in PBS) for 30 min and the primary antibody was diluted in BBS and incubated ON at 4°C. Following three washing steps with PBS, the fluorophore-conjugated secondary antibody was diluted in BBS and cells were incubated with it for 1 hour at room temperature in the dark. The stained cells were washed three times with PBS, the second wash containing DAPI. The mounted samples were imaged with a Zeiss LSM-780 confocal microscope and the images were processed using FIJI/ImageJ. Antibodies are listed in Table S10.
Immunofluorescence staining of ovaries
After dissecting ovaries into ice cold PBS (max 30 min), ovaries were fixed with 4% formaldehyde and 0.3 % Triton X-100 in PBS for 20 min at room temperature. Fixed ovaries were washed 3x with PBX (0.3 % Triton X-100 in PBS) for 10 min and blocked in BBX (1 % BSA and 0.3 % Triton X-100 in PBS) for 30 min. Primary antibody was diluted in BBX and ovaries were incubated with it 24 hours at 4 °C. Following three washing steps with PBX, the fluorophore-conjugated secondary antibody was diluted in BBX and ovaries were incubated with it for ON at 4°C in the dark. The stained ovaries were washed three times with PBX, the second wash containing DAPI. The mounted samples were imaged with a Zeiss LSM-780 confocal microscope and the images were processed using FIJI/ImageJ. Antibodies are listed in Table S10.
Single molecule RNA Fluorescence In Situ Hybridization (FISH)
mdg1 RNA FISH on ovaries was performed as described 56 using CAL Fluor Red 590-labeled Stellaris oligo probes (Table S11). After the RNA FISH protocol, egg chambers were blocked with SBX (1% BSA; 0.1% Triton X-100; 2xSSC) for 30 min and then incubated with primary anti-GFP antibody (Abcam) for 24h at 4°C. After 3x washing (10 min with SBX), samples were incubated with fluorescent secondary antibody for 12h at 4°C. Stacks of soma nuclei were imaged on a Zeiss LSM780 confocal microscope and a maximum intensity projection of 3 slices was generated.
Small RNA-seq
Total RNA was isolated with TRIzol reagent according to the manufacturer’s instructions, and 2S rRNA was depleted as described 57. Small RNA libraries were generated as described 58. In brief, using radio-labelled oligonucleotides as size-markers, 18 to 29nt long RNAs were purified by PAGE. The 3′ linker (containing four random nucleotides) was ligated with T4 RNA ligase 2, truncated K227Q (NEB) overnight at 16°C. Following PAGE purification, the 5′ linker (containing four random nucleotides) was ligated to the small RNAs using T4 RNA ligase (NEB) overnight at 16°C. After PAGE purification, the linker-ligated RNAs were reverse transcribed and PCR amplified. Sequencing was performed with HiSeq2500 (Illumina) in single-read 50 mode.
Small RNA-seq analysis
Sequencing reads were trimmed by removal of the adaptor sequences and the four random nucleotides flanking the small RNA. These reads were pre-mapped to the Drosophila melanogaster rRNA precursor, the mitochondrial genome and unmapped reads were mapped to the Drosophila melanogaster genome (dm6), all using Bowtie 59 (release 1.2.2) with 0 mismatch allowed. Genome mapping reads were intersected with Flybase genome annotations (r6.18) using Bedtools 60 (2.27.1). Reads mapping to rRNA, tRNA, snRNA, snoRNA loci and the mitochondrial genome were removed from the analysis. The quantification of small RNAs was carried out as described in 61. with the following modifications: as a minimal count per 1kb tile cutoff, a value which includes 80% of all reads in the control libraries was used (98 for OSC-KD, 29 for GLKD). Tiles with a mappability below 20% were excluded from the analysis. Annotation groups were based on RefSeq assembly release 6. Tiles overlapping with genes and piRNA clusters were annotated as genic and respective cluster, tiles without annotation were grouped as ‘other’. All sequenced libraries with their GEO Accession number are listed in Table S12.
RNA-seq with rRNA depletion
We modified the protocol published in 62. Total RNA was isolated with TRIzol reagent, which was further purified by RNAeasy columns with on-column DNase I digest (Qiagen), all according to the manufacturer’s instructions. Depletion of rRNA from the purified total RNA was done by using a mix of antisense oligonucleotides matching Drosophila melanogaster rRNAs (listed in Table S13) and the Hybridase Thermostable RNase H (Epicentre) which specifically degrades RNA in RNA-DNA hybrids. The oligonucleotides were added to the RNA in RNase H Buffer (20 mM Tris-HCl pH=8, 100 mM NaCl) and annealed with a temperature gradient from 95 °C to 45 °C. The hybrids were digested at 45 °C for 1 hour. Next, DNA was digested with TURBO DNase (Invitrogen) and RNA was purified using RNA Clean & Concentrator-5 (Zymo) according to the manufacturer’s instructions. Libraries were prepared using a NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB) according to the protocol and sequenced on a HiSeq2500 (Illumina) in single-read 50 mode.
RNA-seq with polyA selection
Total RNA was isolated with TRIzol reagent. Poly(A)+ RNA enrichment was performed with Dynabeads Oligo(dT)25 (Thermo Fisher), with two consecutive purifications according to the manufacturer’s instructions. Next, cDNA was prepared using NEBNext Ultra II RNA First and Second Strand Synthesis Module. The cDNA was purified with AmpureXP beads and library was prepared with NEBNext Ultra II DNA Library Prep Kit Illumina (NEB) according to the protocol and sequenced on a HiSeq2500 (Illumina) in single-read 50 mode.
RNA-seq analysis
Sequencing reads were trimmed by removal of the adaptor sequences. Reads were mapped to the Drosophila melanogaster rRNA precursor and the mitochondrial genome using Bowtie 59 (release 1.2.2) with 0 mismatches allowed. Remaining reads were mapped to the Drosophila melanogaster genome (dm6) using STAR 63 (v.2.5.2b; settings: --outSAMmode NoQS --readFilesCommand cat --alignEndsType Local --twopassMode Basic --outReadsUnmapped Fastx --outMultimapperOrder Random --outSAMtype SAM --outFilterMultimapNmax 1000 --winAnchorMultimapNmax 2000 --outFilterMismatchNmax 0 --seedSearchStartLmax 30 --alignSoftClipAtReferenceEnds No --outFilterType BySJout --alignSJoverhangMin 15 --alignSJDBoverhangMin 1 ). Genome mapping reads were intersected with Flybase genome annotations (r6.18) using Bedtools 60 (2.27.1). Reads mapping to rRNA, tRNA, mitoRNA were excluded from further analysis.
Differential gene expression analysis
Genome matching reads were randomized in order and quantified using Salmon 64 (v.0.10.2; settings: --dumpEqWeights --seqBias --gcBias --useVBOpt --numBootstraps 100 -l SF --incompatPrior 0.0 --validateMappings). Salmon results were further processed using wasabi (https://github.com/COMBINE-lab/wasabi commitID=478c133). DGE analysis was performed pairwise between libraries using sleuth 65 (v0.30.0; settings: extra_bootstrap_summary = TRUE transform_fun_tpm = function(x) log2(x + 0.5), read_bootstrap_tpm = TRUE, gene_mode = TRUE) and running the wald-test function. The sleuth model is a measurement error in the response model. It attempts to segregate the variation due to the inference procedure by Salmon from the variation due to the covariates -- the biological and technical factors of the experiment. For the Wald test, the effect-size represents the estimate of the selected coefficient. It is analogous to, but not equivalent to, the fold-change. The transformed values are on the log2 scale, thus the estimated coefficient is also on the log2 scale. This value takes into account the estimated ‘inferential variance’ estimated from the Salmon bootstraps. For TEs and mRNAs, we required a minimum of TPM >5 in any of the analyzed libraries.
ChIP-seq
Chromatin immunoprecipitation (ChIP) was carried out according to 66, with minor modifications. In brief, OSCs were crosslinked with 1% formaldehyde, quenched with glycine, washed with PBS, collected by centrifugation and pellets were flash-frozen in liquid nitrogen. Chromatin was prepared using Lysis Buffer 1, 2 and 3 from 66 and sonication was performed with a Covaris E220 Ultrasonicator for 20 min. For immunoprecipitation, anti H3K9me3 and RNA Pol II antibodies (Table S10), were coupled to Protein G and Protein A Dynabeads, respectively. Sheared chromatin was incubated with the bead-coupled antibodies for 4 hours at 4 °C, beads were washed, and elution plus de-crosslinking was performed at 65 °C overnight. Following RNase A and proteinase K treatment, DNA was purified with ChIP DNA Clean & Concentrator Kit (Zymo). ChIP-qPCR was performed to test the efficiency of the ChIP and libraries were prepared with NEBNext Ultra DNA Library Prep Kit Illumina (NEB) according to the protocol and sequenced on a HiSeq2500 (Illumina) in single-read 50 mode.
ChIP-seq analysis
Sequencing reads were trimmed by removal of the adaptor sequences and filtered for a minimal length of 18 nucleotides. Reads were mapped to the Drosophila melanogaster rRNA precursor, the mitochondrial genome and the genome (dm6) using Bowtie 59 (release 1.2.2), all with 0 (genome wide analysis) or 3 (TE-consensus analysis) mismatches allowed. BigWig files were generated using Homer 67 and UCSC BigWig tools 68. Heatmaps and meta profiles were generated with Deeptools within Galaxy using BigWig files. The genomic coordinates of euchromatic TE insertions were determined in 14 and the same Piwi-regulated TEs were used as in 20. To calculate log2 fold change values relative to control knockdown, bigwigCompare was used with a pseudo-count of 1. To determine Piwi-dependent H3K9me3 regions, the quantification of ChIP-seq reads was carried out as described 61 with the following modifications: As a minimal count per tile cutoff a value of 150 reads was used and tiles with a mappability below 20% were excluded from the analysis. Piwi-dependent regions were classified by a log2 fold change > 2 when comparing control knockdown with Piwi knockdown. For TE consensus analysis, genome mapping reads longer than 23 nucleotides were mapped to TE consensus sequences using STAR 63 (v.2.5.2b; settings: --outSAMmode NoQS --readFilesCommand cat --alignEndsType Local --twopassMode Basic --outReadsUnmapped Fastx --outMultimapperOrder Random --outSAMtype SAM --outFilterMultimapNmax 1000 --winAnchorMultimapNmax 2000 --outFilterMismatchNmax 3 --seedSearchStartLmax 30 --outFilterType BySJout --alignSJoverhangMin 15 --alignSJDBoverhangMin 1). Multiple mappings were only allowed within one transposon and read-counts were divided equally to the mapping positions. For plotting, read-counts were normalized to 10 million sequenced reads, converted to bedgraph tracks using Bedtools (2.27.1) 60 and plotted in RStudio. All sequenced libraries with their GEO Accession numbers are listed in Table S12.
Protein co-immunoprecipitation from nuclear OSC cell lysates
OSCs were collected after trypsinization by centrifugation, washed with PBS and centrifuged again. The cell pellet was resuspended in LB1 (10 mM Tris-HCl pH=7.5, 2 mM MgCl2, 3 mM CaCl2, freshly supplemented with Complete Protease Inhibitor Cocktail (Roche)), incubated at 4°C for 10 min followed by a centrifugation step. The pellet was resuspended in LB2 (10 mM Tris-HCl pH=7.5, 2 mM MgCl2, 3 mM CaCl2, 0,5 % IGEPAL CA-630, 10 % glycerol, freshly supplemented with Complete Protease Inhibitor Cocktail (Roche)), incubated at 4°C for 10 min followed by a centrifugation step. The isolated nuclei were lysed in LB3 (50 mM Tris-HCl pH=8, 150 mM NaCl, 2 mM MgCl2, 0,5 % Triton X-100, 0,25 % IGEPAL CA-630, 10 % glycerol, freshly supplemented with Complete Protease Inhibitor Cocktail (Roche)), incubated at 4°C for 20 min followed by a centrifugation step. Nuclear lysate was used for immunoprecipitation with Flag M2 Magnetic Beads (Sigma) for 2h at 4°C. The beads were washed 3× 10 min with LB3 and were either used for mass spectrometry analysis or the proteins were eluted in 1× SDS buffer with 5 min incubation at 95°C for western blotting.
Protein co-immunoprecipitation from S2 cell lysates
S2 cells were transfected using Cell Line Nucleofector kit V (Amaxa Biosystems) with the program G-030, using 8 million cells per transfection. S2 cells were co-transfected with FLAG-tagged and GFP-tagged protein encoding plasmids. After two days, cells were collected by centrifugation, washed with PBS and collected again. The cell pellet was resuspended in LB (30 mM Tris-HCl pH=7.5, 150 mM NaCl, 2 mM MgCl2, 0,5 % Triton X-100, 10 % glycerol, freshly supplemented with Complete Protease Inhibitor Cocktail (Roche)), incubated at 4°C for 20 min followed by a centrifugation step. The total cell lysate was used for immunoprecipitation with GFP-Trap magnetic beads (ChromoTek) for 2h at 4°C. The beads were washed 3× 10 min with LB and the proteins were eluted in 1× SDS buffer with 5 min incubation at 95°C.
Western blot
Proteins were separated by SDS–polyacrylamide gel electrophoresis (PAGE) and transferred to a 0.2 μm nitrocellulose membrane (Bio-Rad). The membrane was blocked with 5% milk in PBX (0.05 % Triton X-100 in PBS) and were incubated with primary antibody ON at 4°C. After three washes with PBX, the membrane was incubated with HRP-conjugated secondary antibody for 1h, followed by three PBX washes. The membrane was incubated with Clarity Western ECL Blotting Substrate (Bio-Rad) and imaged with a ChemiDoc MP imaging system (Bio-Rad). Antibodies are listed in Table S10.
Mass spectrometry analysis
Co-immunoprecipitated proteins coupled to magnetic beads were digested with LysC on the beads, eluted with glycine followed by trypsin digestion. Peptides were analyzed using an UltiMate 3000 RSLCnano System (Thermo Fisher Scientific) coupled to a Q Exactive HF mass spectrometer (Thermo Fisher Scientific), equipped with a Proxeon nanospray source (Thermo Fisher Scientific). Peptides were loaded onto a trap column (Thermo Fisher Scientific, PepMap C18, 5 mm × 300 μm ID, 5 μm particles, 100 Å pore size) at a flow rate of 25 μL/min using 0.1% TFA as mobile phase. After 10 min, the trap column was switched in line with the analytical column (Thermo Fisher Scientific, PepMap C18, 500 mm × 75 μm ID, 2 μm, 100 Å). Peptides were eluted using a flow rate of 230 nl/min and a binary 3h gradient. The gradient starts with the mobile phases: 98% A (water/formic acid, 99.9/0.1, v/v) and 2% B (water/acetonitrile/formic acid, 19.92/80/0.08, v/v/v), increases to 35%B over the next 180 min, followed by a gradient in 5 min to 90%B, stays there for 5 min and decreases in 2 min back to the gradient 98%A and 2%B for equilibration at 30°C.
The Q Exactive HF mass spectrometer was operated in data-dependent mode, using a full scan (m/z range 380–1500, nominal resolution of 60,000, target value 1E6) followed by MS/MS scans of the 10 most abundant ions. MS/MS spectra were acquired using normalized collision energy of 27, isolation width of 1.4 m/z, resolution of 30.000 and the target value was set to 1E5. Precursor ions selected for fragmentation (exclude charge state 1, 7, 8, >8) were put on a dynamic exclusion list for 60 s. Additionally, the minimum AGC target was set to 5E3 and intensity threshold was calculated to be 4.8E4. The peptide match feature was set to preferred and the exclude isotopes feature was enabled.
For peptide identification, the RAW-files were loaded into Proteome Discoverer (version 2.1.0.81, Thermo Scientific). All hereby created MS/MS spectra were searched using MSAmanda v2.1.5.9849, Engine version v2.0.0.9849 69. For the first step search the RAW-files were searched against Drosophila melanogaster reference translations retrieved from Flybase (dmel_all-translation-r6.13; 21,983 sequences; 20,112,742 residues), using the following search parameters: The peptide mass tolerance was set to ±5 ppm and the fragment mass tolerance to 15ppm. The maximal number of missed cleavages was set to 2. The result was filtered to 1 % FDR on protein level using Percolator algorithm integrated in Thermo Proteome Discoverer. A sub-database was generated for further processing. Peptide areas were quantified using an in-house developed tool APQuant: http://ms.imp.ac.at/index.php?action=peakjuggler 70.
Protein expression and purification for crystallization
The UBA domain of Drosophila melanogaster Nxf2 (residues 781–841) and Panoramix helix (residues 311–340) were covalently linked through a KLGSHM linker in one expression cassette. In addition, the NTF2-like domain of Drosophila melanogaster Nxf2 (residues 573–777, NTF2l) and full-length Nxt1 (residues 1–133) were cloned into a modified RSFduet-1 vector (Novagen) with an N-terminal His6-SUMO tag on the NTF2-like domain and no tag on Nxt1. Proteins were expressed in E. coli strain BL21(DE3) RIL (Stratagene). The cells were grown at 37°C until OD600 reached 0.8, then the media was cooled to 16°C and IPTG (isopropyl β-D-1-thiogalactopyranoside) was added to a final concentration of 0.35 mM to induce protein expression overnight at 16°C. The cells were harvested by centrifugation at 4°C and disrupted by sonication in Binding buffer (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 20 mM imidazole) supplemented with 1 mM PMSF (phenylmethylsulfonyl fluoride) and 3 mM β-mercaptoethanol. After centrifugation, the supernatants were loaded onto 5 ml HisTrap Fastflow column (GE Healthcare). After extensive washing with Binding buffer, the complex was eluted with Binding buffer supplemented with 500 mM imidazole. The His6-SUMO tag was removed by Ulp1 protease digestion during dialysis against Binding buffer and separated by reloading onto HisTrap column. The flow-through fraction was further purified by HiTrap Q FF column and Superdex 75 16/60 column (GE Healthcare). The pooled fractions were concentrated to 35 mg/ml (UBA-linker-helix) and 10 mg/ml in crystallization buffer (20 mM Tris-HCl pH 7.5, 300 mM NaCl, 1 mM DTT).
Crystallization, data collection and structure determination
As the complex of the UBA domain with the Panoramix (Panx) helix was not very stable and the yield was very low by co-expression, we covalently linked the UBA domain and the Panoramix helix in one cassette by linkers of different length (KL(GS)nHM, n=1, 2, 4, 6). Only the construct with the six-residue KLGSHM linker produced crystals. Crystals of the UBA-linker-Panx helix were grown in 0.095 M sodium citrate pH 5.6, 19% (v/v) Isopropanol, 19% (w/v) PEG 4000, 5% (v/v) glycerol. Crystals of the NTF2-like domain (NTF2l)–Nxt1 complex were grown from a solution containing 0.1 M MES pH 6.5, 1.6 M magnesium sulfate using the hanging drop vapor diffusion method at 20°C. For data collection, the crystals were flash frozen (100 K) in liquid nitrogen.
The data sets for UBA-linker-Panx helix and NTF2l–Nxt1 complexes were collected at 0.9791 Å on beamline 24ID-C and at 0.9792 Å on 24ID-E NE-CAT (Advanced Photo Source, Argonne National Laboratory), respectively. The diffraction data of both, the UBA-linker-Panoramix helix and the NTF2l–Nxt1 complex were processed with the iMosfilm 71 and the structures were solved by molecular replacement (MR) in PHENIX 72 using the structure of the UBA domain of human NXF1 in complex with FxFG peptide (PDB ID: 1OAI) 35 and the structure of the human NXF1(NTF2l)–NXT1 complex (PDB ID: 1JKG) 39 as search templates. The automatic model building was carried out using the program PHENIX AutoBuild 72. The resulting model was refined by PHENIX refinement 72 and Refmac5 73, and completed manually using COOT 74. The Ramachandran plot showed 97.2% favored and 2.8% allowed for UBA-linker-Panx helix structure and 92% favored and 7.8% allowed and 0.2% outliers for NTF2l–Nxt1 structure. The quality of the refined models was validated with MolProbity 75, and the overall MolProbity scores are 1.72 (UBA-linker-Panx helix) and 2.57 (NTF2l-Nxt1). Table 1 summarizes the statistics of the diffraction and refinement data. All the molecular graphics were generated with the PyMOL program (https://pymol.org/2/).
Recombinant protein expression in insect cells
To generate a Panoramix (263–446) / Nxf2 (full length) / Nxt1 (full length) co-expression plasmid, the individual open reading frames were cloned into a modified version of the pACEBac1 vector (Geneva Biotech), in which the expression cassette is flanked by BsaI restriction enzyme sites. Panoramix was cloned with an N-terminal Twin-Strep-tag, Nxf2 with an N-terminal His6-tag and Nxt1 with an N-terminal FLAG-tag. All constructs contained the intact polyhedrin leader sequence which harbors a mutated ATG (ATT) upstream of the actual start codon. A low-level of non-canonical initiation from an upstream ATT site results in low level of slightly larger versions of the proteins. The three expression cassettes were then combined into a single destination vector via Golden Gate cloning 76. The resulting plasmid was transposed into the EmBacY bacmid backbone 77 and transfected into Spodoptera frugiperda Sf9 cells to generate a single baculovirus expressing all three genes. The resulting virus was used to infect Trichoplusia ni High5 cells at a density of 1 × 106 cells/ml and expression was performed at 21°C. The cells were harvested 4 days after growth arrest, approximately 96–120 hours after infection, collected by centrifugation and the cell pellets were flash-frozen in liquid nitrogen.
Affinity purification with Twin-Strep-tag
High5 cells were lysed in lysis buffer (LB) (50 mM Tris-HCl pH=8, 150 mM NaCl, 0,05 % TX100, 1mM DDT) freshly supplemented with Complete Protease Inhibitor Cocktail (Roche) and with Benzonase (~10U/ml) for 30 min at 4°C and the lysate was cleared by centrifugation. For purification, a StrepTactin Superflow HC resin (IBA GmbH) was used with the AKTA Purifier FPLC system and the column was equilibrated with 2 column volumes of LB before sample loading. The bound protein complex was eluted with LB supplemented with 5 mM desthiobiotin and analyzed by SDS-PAGE and InstantBlue (Expedeon) staining.
Size exclusion chromatography (SEC)
After affinity purification, the complex containing fractions were pooled and further purified by SEC using a HiLoad 16/60 Superdex 200 prep grade column (GE Healthcare) with the AKTA Purifier FPLC system in SEC buffer (SB) (50 mM Tris-HCl pH=8, 150 mM NaCl, 1mM DDT). The column was equilibrated with 2 column volumes of SB before sample loading. The purified complex was analyzed by SDS-PAGE and Coomassie staining. The calibration curve was generated with the Biorad Gelfiltration Standard.
Size exclusion chromatography with in-line multi-angle light scattering (SEC-MALS)
SEC-MALS experiments were performed by using an Äkta-MALS system. Proteins (500 μl) at a concentration of 1.5 mg/ml were loaded on Superdex 75 10/300 GL column (GE Healthcare) and eluted with HEPES buffer (20 mM HEPES, pH=7.5, 200 mM NaCl) at a flow rate of 0.2 ml/min. The light scattering was monitored by a miniDAWN TREOS system (Wyatt Technologies) and concentration was measured by an Optilab T-rEX differential refractometer (Wyatt Technologies). Molecular masses of proteins were analyzed using the Astra program (Wyatt Technologies).
Crosslinking-mass spectrometry (XL-MS)
Protein crosslinking and digestion:
The purified complex was crosslinked with 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methyl-morpholinium chloride (DMTMM; final concentration 4mM). To identify the optimal crosslinker concentration, the complex was titrated with different concentrations and crosslinking yield was checked with SDS-PAGE. The complex was reacted with the crosslinker for 30 minutes at room temperature. To stop the reaction, Tris-HCl pH=8 was added to a final concentration of 100 mM. Sodium deoxycholate (SDC) was added to a final concentration of 1.5%. Samples were reduced with dithiothreitol (DTT, 10 mM, 30 min, 60°C), alkylated with iodoacetamide (IAA, 15 mM, 30 min at room temperature in the dark) and diluted to 1% SDC. Proteins were digested for 3h using trypsin (protein/enzyme 30:1, 37°C). With the addition of 1% trifluoro acetic acid (TFA), SDC was precipitated and the digest was stopped. The supernatant was decanted and stored until measurement.
Size-exclusion-chromatography (SEC) enrichment:
The digested samples were enriched for crosslinks (XLs) prior to LC-MS/MS analysis using SEC. Therefore, approx. 15 µg of the digest was separated on a TSKgel SuperSW2000 column (300 mm × 4.5 mm × 4 μm, Tosoh Bioscience). The three high mass fractions were collected and measured on the mass spectrometer.
LC-MS/MS:
Digested peptides were separated using a Dionex UltiMate 3000 HPLC RSLC nanosystem prior to MS analysis. The HPLC was interfaced with the mass spectrometer via a Nanospray Flex™ ion source. For sample concentrating, washing and desalting, the peptides were trapped on an Acclaim PepMap C-18 precolumn (0.3×5mm, Thermo Fisher Scientific), using a flowrate of 25 µl/min and 100% buffer A (99.9% H2O, 0.1% TFA). The separation was performed on an Acclaim PepMap C-18 column (50 cm x 75 µm, 2 µm particles, 100 Ä pore size, Thermo Fisher Scientific) applying a flowrate of 230 nl/min. For separation, a solvent gradient ranging from 2–35% buffer B (80% ACN, 19.92% H2O, 0.08% TFA) was applied. The applied gradient varied from 60–90 min, depending on the sample complexity.
Mass Spectrum Acquisition:
MS1 spectra were recorded at a resolution of 120000 ranging from 350–1600 m/z (AGC 1e6, 60 ms max. injection time). The top 10 most intense ions from MS1 were selected for fragmentation. MS2 spectra were recorded at 30,000 resolution (AGC 5e4, max. injection time 150 ms, isolation width 1.0 m/z). DMTMM crosslinks were fragmented with higher energy C-trap dissociation (HCD) using a stepped collision energy of 30–33-35%. Once a precursor was selected for MS2, it was excluded from fragmentation for 30sec.
Data Analysis:
Raw files were analyzed with pLink 78 (Version 2.3.3) using the settings as described above. Used crosslinker: DMTMM (−18.0116 Da, reactivity towards lysine, protein N-terminus, serine, threonine and tyrosine or aspartate, glutamate and the protein C-terminus, respectively); MS1 accuracy: 10 ppm; MS2 accuracy: 20 ppm; used enzyme: trypsin; max. missed cleavages: 4; minimum peptide length: 5; max. modifications: 4; static modifications: carbamidomethylation (cysteine, +57.021 Da); dynamic modifications: oxidation (methionine, +15.995 Da). For the database search a database containing the three crosslinked proteins was used and the false discovery rate (FDR) was set to 1%. To reduce the number of false positives, XLs were manually validated. For XL visualization, xiNET was used 79.
Recombinant protein expression in bacterial cells
The open reading frame encoding the Nxf2(1–284) fragment or its point mutant variant was cloned into pET21a with an N-terminal GB1 solubility enhancing tag. Protein expression was in BL21DE3 cells, which were grown at 37°C until OD600=0.6–0.8 and then induced with 0.1 mM IPTG for 18 hours at 18°C. The cell pellet was resuspended in lysis buffer (50mM NaPO4 pH=8, 150 mM NaCl, 0.1 % Triton X-100, 10 mM imidazole, 10% glycerol, 5 mM 2-mercaptoethanol, freshly supplemented with 1 mM PMSF and Complete Protease Inhibitor Cocktail (Roche). Lysozyme (10 mg/ml) was added and the cell suspension was incubated for 30 min at 4°C and then sonicated for 15 min at 40% power output with 30% duty cycle (3 sec ON / 7 sec OFF). The sonicated suspension was centrifuged for 20 min at 19,000 g at 4°C. The supernatant was loaded onto pre-equilibrated TALON Metal Affinity Resin (Takara) and incubated for 2 hours. The resin was washed with 20 x bed volume wash buffer (50 mM Tris-HCl pH=7.25, 500 mM NaCl, 20 mM imidazole, 0.1 % Triton X-100, 10 % glycerol, 5 mM 2-mercaptoethanol). The proteins were eluted with 5 x bed volume elution buffer (50 mM Tris-HCl pH=8 300 mM NaCl, 300 mM imidazole, 0.1 % Triton X-100, 10 % Glycerol) and dialyzed against PBS supplemented with 10 % glycerol and 5 mM 2-mercaptoethanol. Post dialysis the protein was aliquoted, flash-frozen in liquid nitrogen and stored at − 80°C for further analysis.
Electrophoretic mobility shift assay (EMSA)
10 nM [32P] 5’-labelled single-stranded 35 nt RNA (sequence: (CUCAUCUUGGUCGUACGCGGAAUAGUUUAAACUGU) was incubated with various concentrations of recombinant protein in 10 µl total volume with EMSA binding buffer (10 mM Tris-HCl pH=7.9, 2 mM MgCl2, 0.1 mM EDTA, 4% glycerol, 50 mM KCl, 1 mM DTT, 10 µg/ml BSA) for 20 min at 4°C. 2 µl of EMSA loading buffer (50% glycerol, 0.075% bromophenol blue) was added to the samples which were analyzed by 4.8 % PAGE gel in 0.5 x TBE. The radioactive bands were visualized with a Phosphorimager.
Alpha helix characterization
To determine the physicochemical properties of the predicted alpha-helix within Panoramix, the HeliQuest web server 80 was used. Using an 18 amino acid sliding window (corresponding to a complete helical wheel), the tool predicts hydrophobic surfaces and shows the sequence in a helical wheel representation.
Phylogenetic tree, orthologue identification and multiple sequence alignment
For phylogenetic reconstruction, Nxf family members from a set of species was extracted, aligned using mafft (v7.407) and the obtained protein sequence alignment was converted to a codon alignment using pal2nal (v14). From this alignment a maximum-likelihood tree was inferred with iqtree (v1.6.7) using best-fitting codon model selection by Modelfinder and 1000 ultrafast bootstrap replicates; visualization using iTol (v4.2.3).
For Nxf protein family collection, proteins showing significant sequence similarity to the NTF2-like domain of Drosophila melanogaster Sbr (Nxf1) were collected from the NCBI non-redundant protein database (NCBI nr) using blastp (query NP_524660.1:372–531; species filter: Arthropoda and selected other organisms). Hits were retained if they also showed reciprocal best blast hits to one of the 4 known Drosophila melanogaster Nxf family members in blastp searches against the Drosophila melanogaster proteome (PTHR10662 members: Nxf3/ FBgn0263232, nxf2/ FBgn0036640, nxf4/ FBgn0051501, sbr/ FBgn0003321). The obtained Nxf protein family set was supplemented with Drosophila melanogaster nxf4 protein and its orthologs identified in reciprocal-best-blast-procedure.
For UBA protein domain alignments, a subset of representative Nxf2 protein sequences was selected using an 80% identity cutoff over the C-terminal region (corresponding to NP_524111.3: 726–841). The species selection was also used in alignments of the Nxf1 ortholog groups.
Panoramix ortholog identification was performed using reciprocal psi-blast searches against NCBI nr, using the region of highest conservation among Drosophilid Panoramix proteins as a query (NP_611576.1:292–510). Multiple sequence alignments were visualized in Jalview (v2.10.4), and the secondary structure was predicted with JPRED. Sequence accessions from the alignments are listed in Table S6.
Supplementary Material
ACKNOWLEDGMENTS
We thank K. Meixner for experimental support, P. Duchek and J. Gokcezade for generating CRISPR edited and transgenic flies, the VBCF NGS unit for deep sequencing, VBCF Protein Technologies Facility for protein expression, VBCF VDRC unit for fly stocks, the MFPL monoclonal facility for antibodies, and TRiP & Bloomington stock centers for flies. We thank A. Koehler and G. Riddihough from Life Science Editors (lifescienceeditors.com) for comments on the manuscript. We thank the Brennecke lab, particularly P. Andersen, for support and feedback. The Brennecke lab is supported by the Austrian Academy of Sciences, the European Community (ERC-2015-CoG - 682181), and the Austrian Science Fund (F 4303 and W1207). J. Batki was supported by the Boehringer Ingelheim Fonds. X-ray diffraction studies were conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by NIGMS grant P30 GM124165 and U.S. Department of Energy grant DE-AC02–06CH11357. The Pilatus 6M detector on 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). MSKCC core facilities are supported by P30 CA008748. This work was supported by funds from the Maloris Foundation (DJP) and MSKCC core grant (P30 CA008748).
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Data availability statement
All sequencing data used for this study (Table S12) have been deposited at NCBI GEO (GSE120617). The mass spectrometry data have been deposited to the ProteomeXchange Consortium via PRIDE (PXD011201) 81. Coordinate and structure factors of the UBA-linker-helix and the dmNxf2–Nxt1 complex are available from Protein Data Bank (PDB; accession codes 6OPF and 6MRK). Source data for Figure 1a are available in Supplementary Table 1, Figure 1f, g and Supplementary Figure 1d, e in Supplementary Table 2, Figure 2c and Supplementary Figure 2d in Supplementary Table 3, Supplementary Figure 2i, k in Supplementary Table 4 and Figure 7d in Supplementary Table 6. Source data for Figures 3d, e, 5h, and 7f are available online.
Code availability statement
All custom code is based on the publicly available code used in 61 with modifications indicated in the Methods section.
Reporting Summary statement:
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
REFERENCES
- 1.Fedoroff NV Presidential address. Transposable elements, epigenetics, and genome evolution. Science 338, 758–67 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Slotkin RK & Martienssen R Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8, 272–85 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Yang P, Wang Y & Macfarlan TS The Role of KRAB-ZFPs in Transposable Element Repression sand Mammalian Evolution. Trends Genet 33, 871–881 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grewal SI RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev 20, 134–41 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castel SE & Martienssen RA RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 14, 100–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holoch D & Moazed D RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet 16, 71–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimada Y, Mohn F & Buhler M The RNA-induced transcriptional silencing complex targets chromatin exclusively via interacting with nascent transcripts. Genes Dev 30, 2571–2580 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dumesic PA et al. Stalled spliceosomes are a signal for RNAi-mediated genome defense. Cell 152, 957–68 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reyes-Turcu FE, Zhang K, Zofall M, Chen E & Grewal SI Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat Struct Mol Biol 18, 1132–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teixeira FK et al. piRNA-mediated regulation of transposon alternative splicing in the soma and germ line. Nature 552, 268–272 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czech B et al. piRNA-Guided Genome Defense: From Biogenesis to Silencing. Annu Rev Genet 52, 131–157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ozata DM, Gainetdinov I, Zoch A, O’Carroll D & Zamore PD PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet (2018). [DOI] [PubMed]
- 13.Wang SH & Elgin SC Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc Natl Acad Sci U S A 108, 21164–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sienski G, Donertas D & Brennecke J Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151, 964–80 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rozhkov NV, Hammell M & Hannon GJ Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev 27, 400–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Thomas A et al. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev 27, 390–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donertas D, Sienski G & Brennecke J Drosophila Gtsf1 is an essential component of the Piwi-mediated transcriptional silencing complex. Genes Dev 27, 1693–705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohtani H et al. DmGTSF1 is necessary for Piwi-piRISC-mediated transcriptional transposon silencing in the Drosophila ovary. Genes Dev 27, 1656–61 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muerdter F et al. A Genome-wide RNAi Screen Draws a Genetic Framework for Transposon Control and Primary piRNA Biogenesis in Drosophila. Mol Cell 50, 736–48 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sienski G et al. Silencio/CG9754 connects the Piwi-piRNA complex to the cellular heterochromatin machinery. Genes Dev 29, 2258–71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Y et al. Panoramix enforces piRNA-dependent cotranscriptional silencing. Science 350, 339–42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez-Navarro S & Hurt E Linking gene regulation to mRNA production and export. Curr Opin Cell Biol 23, 302–9 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Stewart M Polyadenylation and nuclear export of mRNAs. J Biol Chem 294, 2977–2987 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herold A et al. TAP (NXF1) belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol Cell Biol 20, 8996–9008 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herold A, Klymenko T & Izaurralde E NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA 7, 1768–80 (2001). [PMC free article] [PubMed] [Google Scholar]
- 26.Herold A, Teixeira L & Izaurralde E Genome-wide analysis of nuclear mRNA export pathways in Drosophila. EMBO J 22, 2472–83 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niki Y, Yamaguchi T & Mahowald AP Establishment of stable cell lines of Drosophila germ-line stem cells. Proc Natl Acad Sci U S A 103, 16325–30 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saito K et al. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461, 1296–9 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Handler D et al. The Genetic Makeup of the Drosophila piRNA Pathway. Mol Cell 50, 762–77 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Czech B, Preall JB, McGinn J & Hannon GJ A Transcriptome-wide RNAi Screen in the Drosophila Ovary Reveals Factors of the Germline piRNA Pathway. Mol Cell 50, 749–61 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown JB et al. Diversity and dynamics of the Drosophila transcriptome. Nature 512, 393–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilkie GS et al. Small bristles, the Drosophila ortholog of NXF-1, is essential for mRNA export throughout development. RNA 7, 1781–92 (2001). [PMC free article] [PubMed] [Google Scholar]
- 33.Caporilli S, Yu Y, Jiang J & White-Cooper H The RNA export factor, Nxt1, is required for tissue specific transcriptional regulation. PLoS Genet 9, e1003526 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valkov E, Dean JC, Jani D, Kuhlmann SI & Stewart M Structural basis for the assembly and disassembly of mRNA nuclear export complexes. Biochim Biophys Acta 1819, 578–92 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Grant RP, Neuhaus D & Stewart M Structural basis for the interaction between the Tap/NXF1 UBA domain and FG nucleoporins at 1A resolution. J Mol Biol 326, 849–58 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Kohler A & Hurt E Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8, 761–73 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Braun IC, Herold A, Rode M & Izaurralde E Nuclear export of mRNA by TAP/NXF1 requires two nucleoporin-binding sites but not p15. Mol Cell Biol 22, 5405–18 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grant RP, Hurt E, Neuhaus D & Stewart M Structure of the C-terminal FG-nucleoporin binding domain of Tap/NXF1. Nat Struct Biol 9, 247–51 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Fribourg S, Braun IC, Izaurralde E & Conti E Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol Cell 8, 645–56 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Liker E, Fernandez E, Izaurralde E & Conti E The structure of the mRNA export factor TAP reveals a cis arrangement of a non-canonical RNP domain and an LRR domain. EMBO J 19, 5587–98 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aibara S et al. Structural characterization of the principal mRNA-export factor Mex67-Mtr2 from Chaetomium thermophilum. Acta Crystallogr F Struct Biol Commun 71, 876–88 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tutucci E & Stutz F Keeping mRNPs in check during assembly and nuclear export. Nat Rev Mol Cell Biol 12, 377–84 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Teplova M, Wohlbold L, Khin NW, Izaurralde E & Patel DJ Structure-function studies of nucleocytoplasmic transport of retroviral genomic RNA by mRNA export factor TAP. Nat Struct Mol Biol 18, 990–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aibara S, Katahira J, Valkov E & Stewart M The principal mRNA nuclear export factor NXF1:NXT1 forms a symmetric binding platform that facilitates export of retroviral CTE-RNA. Nucleic Acids Res 43, 1883–93 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Viphakone N et al. TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export. Nat Commun 3, 1006 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ninova M, Chen YA, Godneeva B Rogers AK, Luo Y, Aravin AA, Fejes Tóth K The SUMO ligase Su(var)2–10 links piRNA-guided target recognition to chromatin silencing. BioRxiv [Preprint]. (2019) Available from: 10.1101/533091 [DOI] [PMC free article] [PubMed]
- 47.Heath CG, Viphakone N & Wilson SA The role of TREX in gene expression and disease. Biochem J 473, 2911–35 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muller-McNicoll M & Neugebauer KM How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat Rev Genet 14, 275–87 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Post C, Clark JP, Sytnikova YA, Chirn GW & Lau NC The capacity of target silencing by Drosophila PIWI and piRNAs. RNA 20, 1977–86 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stutz F & Izaurralde E The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends Cell Biol 13, 319–27 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Gramates LS et al. FlyBase at 25: looking to the future. Nucleic Acids Res 45, D663–D671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.ElMaghraby MF, Andersen PR, Pühringer F, Meixner K, Lendl T, Tirian L, Brennecke J A heterochromatin-specific RNA export pathway facilitates piRNA production. BioRxiv [Preprint]. (2019) Available from: 10.1101/596171. [DOI] [PubMed]
- 53.Pan J et al. Inactivation of Nxf2 causes defects in male meiosis and age-dependent depletion of spermatogonia. Dev Biol 330, 167–74 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gokcezade J, Sienski G & Duchek P Efficient CRISPR/Cas9 Plasmids for Rapid and Versatile Genome Editing in Drosophila. G3 (Bethesda) (2014). [DOI] [PMC free article] [PubMed]
- 55.Pfeiffer BD et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci U S A 105, 9715–20 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mohn F, Sienski G, Handler D & Brennecke J The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell 157, 1364–79 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Hayashi R et al. Genetic and mechanistic diversity of piRNA 3’-end formation. Nature 539, 588–592 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jayaprakash AD, Jabado O, Brown BD & Sachidanandam R Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res 39, e141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langmead B, Trapnell C, Pop M & Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–2 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andersen PR, Tirian L, Vunjak M & Brennecke J A heterochromatin-dependent transcription machinery drives piRNA expression. Nature 549, 54–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morlan JD, Qu K & Sinicropi DV Selective depletion of rRNA enables whole transcriptome profiling of archival fixed tissue. PLoS One 7, e42882 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patro R, Duggal G, Love MI, Irizarry RA & Kingsford C Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pimentel H, Bray NL, Puente S, Melsted P & Pachter L Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods 14, 687–690 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Lee TI, Johnstone SE & Young RA Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc 1, 729–48 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–89 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kent WJ, Zweig AS, Barber G, Hinrichs AS & Karolchik D BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26, 2204–7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dorfer V et al. MS Amanda, a universal identification algorithm optimized for high accuracy tandem mass spectra. J Proteome Res 13, 3679–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Doblmann J et al. apQuant: Accurate Label-Free Quantification by Quality Filtering. J Proteome Res (2018). [DOI] [PubMed]
- 71.Battye TG, Kontogiannis L, Johnson O, Powell HR & Leslie AG iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67, 271–81 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adams PD et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58, 1948–54 (2002). [DOI] [PubMed] [Google Scholar]
- 73.Murshudov GN, Vagin AA & Dodson EJ Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53, 240–55 (1997). [DOI] [PubMed] [Google Scholar]
- 74.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Engler C, Kandzia R & Marillonnet S A one pot, one step, precision cloning method with high throughput capability. PLoS One 3, e3647 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trowitzsch S, Bieniossek C, Nie Y, Garzoni F & Berger I New baculovirus expression tools for recombinant protein complex production. J Struct Biol 172, 45–54 (2010). [DOI] [PubMed] [Google Scholar]
- 78.Fan SB et al. Using pLink to Analyze Cross-Linked Peptides. Curr Protoc Bioinformatics 49, 8 21 1–19 (2015). [DOI] [PubMed] [Google Scholar]
- 79.Combe CW, Fischer L & Rappsilber J xiNET: cross-link network maps with residue resolution. Mol Cell Proteomics 14, 1137–47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gautier R, Douguet D, Antonny B & Drin G HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinformatics 24, 2101–2 (2008). [DOI] [PubMed] [Google Scholar]
- 81.Vizcaino JA et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44, D447–56 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.