Abstract
Objective
Progression to insulin therapy in clinically diagnosed type 2 diabetes is highly variable. GAD65 autoantibodies (GADA) are associated with faster progression, but their predictive value is limited. We aimed to determine if a Type 1 Diabetes Genetic Risk Score (T1DGRS) could predict rapid progression to insulin treatment over and above GADA testing.
Research Design and Methods
We examined the relationship between T1DGRS, GADA (negative or positive) and rapid insulin requirement (within 5 years) using Kaplan-Meier survival analysis and Cox regression in 8,608 participants with clinical type 2 diabetes (onset >35 years, treated without insulin for ≥6 months). T1DGRS was analyzed both continuously (as standardized scores) and categorized based on previously reported centiles of a type 1 diabetes population (<5th (low), 5th-50th (medium), >50th (high)).
Results
In GADA positive participants (3.3%), those with higher T1DGRS progressed to insulin more quickly: Probability of insulin requirement at five years [95% CI]: 47.9%[35.0%,62.78%] (high T1DGRS) vs 27.6%[20.5%,36.5%] (medium T1DGRS) vs 17.6%[11.2%,27.2%] (low T1DGRS), p=0.001. In contrast T1DGRS did not predict rapid insulin requirement in GADA negative participants (p=0.4). In Cox regression analysis with adjustment for age of diagnosis, BMI and cohort, T1DGRS was independently associated with time to insulin only in the presence of GADA: hazard ratio per SD increase 1.48 (1.15,1.90), p=0.002.
Conclusions
A Type 1 Diabetes Genetic Risk Score alters the clinical implications of a positive GADA test in patients with clinical type 2 diabetes, and is independent of and additive to clinical features.
Type 2 diabetes is a progressive disease due to a gradual reduction in the capacity of the pancreatic islet cells (beta cells) to produce insulin (1). The clinical course of this progression is highly variable with some patients progressing very rapidly to requiring insulin treatment, whilst others can be successfully treated with lifestyle changes or oral agents for many years (1; 2). Being able to identify patients likely to rapidly progress may have clinical utility in prioritization monitoring and treatment escalation, and in choice of therapy.
It has previously been shown that many patients with clinical features of type 2 diabetes have positive GAD65 autoantibodies (GADA) and that the presence of this autoantibody is associated with faster progression to insulin (3; 4). This is often termed Latent Autoimmune Diabetes in Adults (LADA) (5; 6). However the predictive value of GADA testing is limited in a clinical type 2 diabetes population, with many GADA positive patients not requiring insulin treatment for many years (4; 7). Previous research has suggested that genetic variants in the HLA region associated with type 1 diabetes are associated with more rapid progression to insulin in patients with clinically defined type 2 diabetes and positive GADA (8).
We have recently developed a Type 1 Diabetes Genetic Risk Score (T1D GRS), which provides an inexpensive ($70 in our local clinical laboratory, <$20 where DNA has been previously extracted), integrated assessment of a person’s genetic susceptibility to type 1 diabetes (9). The score is composed of 30 type 1 diabetes risk variants weighted for effect size, and aids discrimination of type 1 diabetes from type 2 diabetes. The T1D GRS has advantages over HLA typing alone, as it includes more genetic information, is cheaper than conventional HLA typing, and represents a continuous scale of likelihood of type 1 diabetes susceptibility. In young-onset adults (diagnosed between 20-40 years) it can predict insulin dependence and is independent of and additive to islet autoantibodies and clinical features (9). It is not known if the T1D GRS will improve the prediction of insulin requirement by GADA in clinically defined type 2 diabetes.
We aimed to determine if the T1D GRS could predict rapid progression to insulin (within 5 years of diagnosis) over and above GADA testing in patients with a clinical diagnosis of type 2 diabetes treated without insulin at diagnosis.
Research design and methods
We examined the relationship between GADA, T1D GRS and progression to insulin therapy using survival analysis in 8,608 participants with clinical type 2 diabetes initially treated without insulin therapy.
Study population
Included participants had a clinical diagnosis of type 2 diabetes after the age of 35 years, and were treated without insulin for the first 6 months from diagnosis and were of white European origin.
Participants were identified in the following cohorts: Genetics of Diabetes Audit and Research Tayside Study (GoDARTS) (10), Hoorn Diabetes Care System (DCS) (11), Diabetes Alliance for Research in England (DARE) (12), Predicting Response to Incretin Based Agents in Type 2 Diabetes (PRIBA) (13), and MRC MASTERMIND Progressors (14). These cohorts were studies of participants with a clinical diagnosis of type 2 diabetes recruited from primary and secondary care, and are population based with the exception of PRIBA and MRC MASTERMIND Progressor which account for <10% of participants. Summaries of the cohort recruitment and data collection methods are shown in Supplementary Table 1, a flow diagram of sample selection is shown in Supplementary Figure 1.
Participants known to have had GADA testing performed either in clinical practice or prior to diagnosis (through review of electronic laboratory records) were excluded due to the risk of the result influencing the clinician’s treatment decision.
In the GoDarts cohort, participants diagnosed with diabetes before 1st January 1994 were excluded; due to insufficient prescribing information we were unable to define time to insulin prior to this date. In the DARE cohort, only the participants recruited in the Exeter Centre with saved serum were included.
Assessment of diabetes progression (time to insulin)
For GoDarts and DCS cohorts, time to insulin was defined from electronic prescription records. For Exeter Cohorts (DARE, PRIBA and MRC MASTERMIND Progressors), insulin treatment, date of commencing insulin and date of diagnosis were self-reported at a single visit.
Laboratory Measurement
The Academic Department of Blood Sciences at the Royal Devon and Exeter Hospital measured GADA for all five cohorts at a median diabetes duration of 6.1 years, using the same assay from biobanked samples stored at -80C. GADA was performed using the RSR Limited ELISA assay (RSR Ltd, Cardiff, UK) on the Dynex DS2 ELISA Robot (Dynex Technologics, Worthing, UK). The cut-off for positivity was ≥11 units/ml, based on the 97.5th centile of 1,500 controls without diabetes (15). The lowest reportable value (lowest calibrant) was 5.0 units/ml. The laboratory participates in the International Autoantibody Standardization Programme.
The HbA1c value at latest follow up (closest available result, median 10.6 years diabetes duration) was obtained from electronic healthcare records or measured on a research sample by the Academic Department of Blood Sciences at the Royal Devon and Exeter Hospital.
Assessment of T1D GRS
The development of the T1D GRS has been described previously (9). In brief, T1D GRS consists of 30 common type 1 diabetes genetic variants (single nucleotide polymorphisms (SNPs)) from HLA and non-HLA loci; each variant is weighted by their effect size on type 1 diabetes risk from previously published literature, with weights for DR3/DR4-DQ8 assigned based on imputed haplotypes. The combined score represents an individual’s genetic susceptibility to type 1 diabetes. Variants used to derive the score are shown in Supplementary Table 2. For ease of clinical interpretation the score is presented in this article as the centile position of the distribution in the Wellcome Trust Case Control Consortium type 1 diabetes population (16).
In the Exeter cohorts, genotyping was performed using the KASP genotyping assay by LGC Genomics (Hoddesdon, UK) as previously described (9). Genotyping in the GoDarts cohort was performed using custom genotyping arrays (including Immunochip, Cardio-Metabochip (Metabochip) and Human Exome array) from Illumina as previously described (17). Genotyping in the DCS cohort was performed with Illumina’s HumanCoreExome Array and imputed using IMPUTE2 (18) into the 1000 Genomes March 2012 reference panel. All SNPs had an INFO > 0.8.
T1D GRS calculation was not performed if genotyping results were missing for either of the two alleles with the greatest weighting (DR3/DR4-DQ8 or HLA_DRB1_15) or if more than two of any other SNPs were missing.
Statistical analysis
We assessed the relationship between time to insulin treatment and each of GADA and T1D GRS using survival analysis. For this analysis, T1D GRS was categorized based on centiles of a type 1 diabetes population (Wellcome Trust Case Control Consortium (16)): <5th centile (< 0.234 (low)), 5th-50th centile (>= 0.234 & <= 0.280 (medium)), >50th centile (> 0.280 (high)) as previously reported (9; 19). GADA was dichotomized into negative or positive based on the cut-off for positivity. Participants were then classified into six risk groups from these categories 1) GADA negative, low T1D GRS 2) GADA negative, medium T1D GRS 3) GADA negative, high T1D GRS 4) GADA positive, low T1D GRS 5) GADA positive, medium T1D GRS 6) GADA positive, high T1D GRS.
Time to insulin data was censored at five years (or the latest available time point not on insulin, if earlier). Survival distributions for time to insulin, stratified by risk groups, were estimated using the Kaplan-Meier product limit estimator (20). The proportional hazard assumption was checked visually and failed. Differences in time to insulin between risk groups were therefore compared using the Wilcoxon (Breslow) test. Positive predicted values were obtained from the product limit estimator which makes allowances for censored observations.
To assess whether clinical characteristics were different across risk groups we performed Wilcoxon test for trend (21) on the continuous variables and Pearson chi-squared test for categorical variables.
To assess whether GADA, T1D GRS (as a continuous covariate), age of diagnosis and BMI (closest available to diagnosis, median 3 years diabetes duration) are independent predictors of rapid progression to insulin we performed multivariate Cox proportional hazards regression analysis (22). When T1D GRS was used as a continuous covariate, the proportional hazard assumption was satisfied. T1D GRS and GADA were added in as separate variables and as an interaction term. The log-linearity assumption was checked by examining Martingale-based residual plots and was considered valid. Study of origin was included as a strata variable to control for effects of cohort differences.
As a 10 SNP T1D GRS combining the 10 alleles with the greatest weightings ordered by published odds ratios (Supplementary Table 3) has also been proposed for clinical practice, we repeated survival analysis using T1D GRS defined by this 10 SNP score using the same centile cut-offs for categorization (9). We also estimated survival distributions for risk groups based on imputed HLA DR3/DR4 genotypes, individually and grouped by number of copies of at risk alleles.
Median follow-up time was calculated using the reverse Kaplan-Meier method (23). All analysis was performed in Stata/SE 15.1 (StataCorp, College Station, TX).
Results
We identified 8,608 participants with a clinical diagnosis of type 2 diabetes meeting all of our inclusion criteria, Table 1 shows the characteristics for these participants. 79.9% (n = 6,879) had been followed for at least five years; median follow up time, calculated as the median time to censoring (insulin treatment or latest follow up), was 10.5 (95% CI 10.3, 10.6) years. 7.8% (n = 533) of those participants with over five years follow up had progressed to insulin <= 5 years. 3.3% (n = 280) of participants were GADA positive (measured at a median 6.1 years diabetes duration). The distribution of participants by low, medium and high TID GRS category was 53.2% (n = 4,580), 40.7% (n = 3,504) and 6.1% (n = 524) respectively. Characteristics of the participants stratified by cohort are shown in Supplementary Table 4.
Table 1.
Participant characteristics. Median (IQR) or % (n = 8 608).
| Characteristic | Value |
|---|---|
| Sex (% Male) | 56.4% |
| Age at diagnosis (years) | 60 (52, 68) |
| BMI (kg/m2)* | 30.4 (27.2, 34.7) |
| Duration of diabetes (years) at latest follow up | 10.6 (6.0, 14.3) |
| Duration of diabetes (years) at GADA | 6.1 (3.3, 10.0) |
| Insulin treated within 5 years (%)† | 7.8% |
| HbA1c (%)‡ | 7.0 (6.4, 8.0) |
| HbA1c (mmol/mol)‡ | 53 (46, 64) |
| GADA positive (%) | 3.3% |
| T1D GRS centile§ | 4.2 (0.6, 16.1) |
Closest to diagnosis (median 3 years diabetes duration).
Percentage of participants observed for at least five years.
At latest follow up.
Centile of participants with type 1 diabetes from the Wellcome Trust Case Control Consortium.
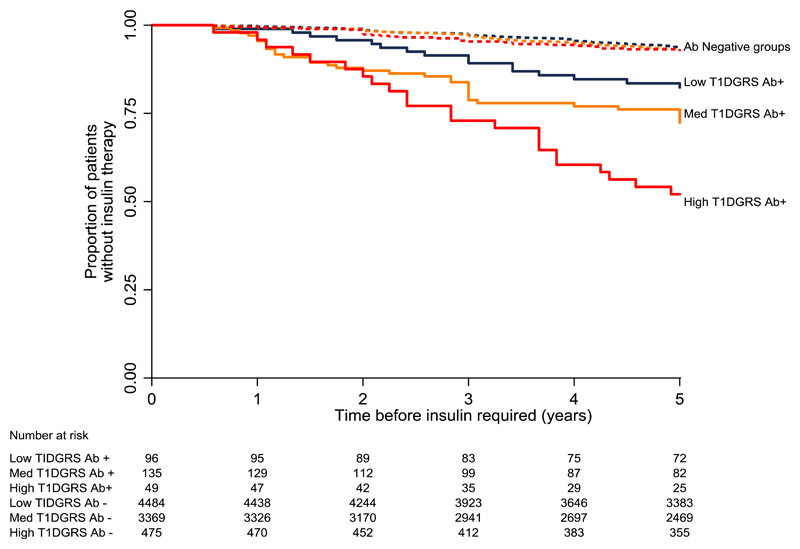
High T1D GRS is associated with markedly higher rates of rapid insulin requirement in participants with positive GADA, but is not associated in those who are GADA negative
T1D GRS was strongly predictive of rapid insulin requirement in participants with positive GADA (Figure 1). In GADA positive participants, those with higher T1D GRS progressed to insulin more quickly (p=0.001): probability of requiring insulin at five years post diagnosis (positive predictive value) [95% CI]: 47.9% [35.0%, 62.78%] (high T1D GRS) vs 27.6% [20.5%, 36.5%] (medium T1D GRS) vs 17.6% [11.2%, 27.2%] (low T1D GRS).
Figure 1.
Kaplan-Meier plot of probability of requiring insulin therapy during 5-year follow-up by risk group of T1D GRS. Solid lines represent GADA positive groups, dashed lines represent GADA negative groups. Blue = low T1D GRS (<5th centile of a type 1 diabetes population (< 0.234)), orange = medium T1D GRS (5th-50th centile of a type 1 diabetes population (>= 0.234 & <= 0.280)), red =high T1D GRS (>50th centile of a type 1 diabetes population (> 0.280)).
T1D GRS was not associated with rapid insulin requirement in GADA negative participants. For the GADA negative participants, the probability of requiring insulin at five years post diagnosis was similar across all risk groups (p=0.4): 7.4% [5.3%, 10.3%] (high T1D GRS) vs 7.3% [6.5%, 8.3%] (medium T1D GRS) vs 6.7% [5.9%, 7.5%] (low T1D GRS).
Differences in T1D GRS were associated with higher HbA1c in GADA positive participants but no differences in other clinical features
The characteristics of the GADA positive and negative participants split by T1D GRS category are shown in Table 2. In GADA positive participants, HbA1c increased (p = 0.04) and BMI decreased (p = 0.01) with higher T1D GRS category. In GADA negative participants, clinical characteristics were similar across all categories of T1D GRS.
Table 2.
Participant characteristics by risk group. Median (IQR) or %. p values given for continuous variables are Wilcoxon-type test for trend, for categorical variables Pearson chi-squared.
| <5th T1D GRS centile for type 1 diabetes‡ (low) | 5th–50th T1D GRS centile for type 1 diabetes ‡ (medium) | >50th T1D GRS centile for type 1 diabetes ‡ (high) | p-value | |
|---|---|---|---|---|
| GADA negative | ||||
| n (% of GADA negative) | 4,484 (54%) | 3,369 (40%) | 475 (6%) | |
| Sex (% Male) | 56.2% | 57.0% | 56.2% | >0.1 |
| Age at diagnosis (years) | 60 (52, 68) | 60 (52, 68) | 60 (51, 68) | >0.1 |
| BMI (kg/m2)* | 30.5 (27.3, 34.9) | 30.4 (27.1, 34.6) | 30.7 (27.4, 34.3) | >0.1 |
| Duration of diabetes (years) at latest follow up | 10.6 (6.1, 14.4) | 10.5 (5.8, 14.1) | 10.6 (6.0, 14.4) | >0.1 |
| Duration of diabetes (years) at GADA | 6.3 (3.3, 10.0) | 6.0 (3.3, 10.0) | 6.3 (3.6, 10.2) | >0.1 |
| HbA1c (%)† | 7.0 (6.4, 8.0) | 7.0 (6.4, 8.0) | 6.9 (6.4, 8.0) | >0.1 |
| HbA1c (mmol/mol) † | 53 (46, 64) | 53 (46, 64) | 52 (46, 64) | >0.1 |
| Insulin treated within 5 years (%) (where observed ≥ five years) | 6.7% | 7.4% | 8.1% | >0.1 |
| GADA (units/mL) | 4.9 (4.9, 5.0) | 4.9 (4.9, 5.0) | 4.9 (4.9, 5.0) | >0.1 |
| GADA positive | ||||
| n (% of GADA positive) | 96 (34%) | 135 (48%) | 49 (18%) | |
| Sex (% Male) | 51.0% | 57.0% | 49.0% | >0.1 |
| Age at diagnosis (years) | 61 (50, 69) | 59 (51, 67) | 54 (49, 63) | 0.06 |
| BMI (kg/m2)* | 29.6 (26.7, 34.1) | 28.7 (25.6, 32.5) | 27.7 (25.4, 30.4) | 0.01 |
| Duration of diabetes (years) at latest follow up | 11.1 (9.0., 13.8) | 10.4 (6.7, 14.9) | 11.8 (9.1, 15.0) | >0.1 |
| Duration of diabetes (years) at GADA | 5.2 (3.1, 9.5) | 5.6 (3.0, 10.1) | 8.9 (4.9, 11.1) | 0.01 |
| HbA1c (%)† | 7.3 (6.6, 9.1) | 7.8 (6.7, 9.0) | 8.1 (7.1, 9.1) | 0.04 |
| HbA1c (mmol/mol) † | 56 (49, 76) | 62 (50, 75) | 66 (55, 77) | 0.04 |
| Insulin treated within 5 years (%) (where observed ≥ five years) | 18.4% | 27.8% | 40.5% | 0.03 |
| GADA (units/mL) | 77.6 (24.3, 1191.9) | 111.4 (28.8, 1354.9) | 175.9 (38.6, 1218.2) | >0.1 |
Closest to diagnosis (median 3 years diabetes duration).
At latest follow up.
Centile of participants with type 1 diabetes from the Wellcome Trust Case Control Consortium.
When comparing the characteristics of GADA positive and negative participants (Table 2), GADA positive participants had a higher T1D GRS (median 0.251 vs 0.231, p < 0.001) and a lower BMI (median 28.73 vs 30.48, p < 0.001) but similar age of diagnosis (median 59 vs 60 years, p = 0.052).
T1D GRS and GADA are predictors of rapid insulin requirement and are independent of age and BMI
Table 3 shows the Cox proportional hazards regression model for time to insulin (censored at 5 years) controlled for effects of cohort differences. As expected, the presence of GADA was a significant predictor of time to insulin (Hazard Ratio (HR) 3.43 [2.50, 4.71], p <0.001). T1D GRS was independently associated with time to insulin, but only in the presence of GADA (HR per 1 standard deviation (SD) increase in T1D GRS 1.48 [1.15, 1.90], p = 0.002). These associations were independent of age at diagnosis and BMI.
Table 3.
Hazard ratios from Cox proportional regression model (adjusted for cohort) for time to insulin censored at 5 years (30 SNP T1D GRS).
| Variable | Hazard Ratio (95% CI) | p value |
|---|---|---|
| GADA negative | 1 | |
| GADA positive | 3.43 (2.50, 4.71) | <0.001 |
| GADA negative:T1D GRS (per 1 SD increase in T1D GRS) | 1.02 (0.94, 1.12) | >0.1 |
| GADA positive:T1D GRS (per 1 SD increase in T1D GRS) | 1.48 (1.15, 1.90) | 0.002 |
| Age at diagnosis (per 1 year) | 0.97 (0.96, 0.97) | <0.001 |
| BMI (per kg/m2 unit)* | 1.00 (0.98, 1.01) | >0.1 |
Closest to diagnosis
A 10 SNP T1D GRS, and HLA type alone are predictive of future insulin requirement
The association between the 10 SNP T1D GRS and rapid insulin requirement was consistent with our findings using the full 30 SNP T1D GRS. The 10 SNP T1D GRS was associated with rapid insulin requirement in the GADA positive risk groups (p < 0.001) but was not associated in the GADA negative groups (p=0.4) (Supplementary Figure 2). In Cox proportional hazards regression model (Supplementary Table 5), the 10 SNP T1D GRS was independently associated with future insulin treatment in GADA positive participants (HR per 1 SD increase in T1D GRS 1.34 [1.05, 1.71], p = 0.02). Kaplan-Meier plots for HLA DR3/DR4 genotype risk groups, individually and grouped by number of at risk alleles, are shown in Supplementary Figures 3 and 4.
Conclusions
In this large study of participants with a clinical diagnosis of type 2 diabetes, we have found that type 1 genetic susceptibility alters the clinical implications of a positive GADA when predicting rapid time to insulin. GADA positive participants with high T1D GRS were more likely to require insulin within 5 years of diagnosis, with 48% progressing to insulin in this time in contrast to only 18% in participants with low T1D GRS. The T1D GRS was independent of and additive to participant’s age of diagnosis and BMI. However, T1D GRS was not associated with rapid insulin requirement in participants who were GADA negative.
To our knowledge this is the first study to assess the association between an integrated assessment of type 1 genetic risk and GADA in patients with type 2 diabetes or LADA. A key strength of this study is use of large, predominantly population-based, cohorts of participants diagnosed with type 2 diabetes and to date, is the largest cohort with measured GADA in a western population. This means our results are likely to reflect true associations in patients seen in clinical practice. An additional key strength is the use of a single laboratory and assay for measuring GADA across cohorts, with a very robustly defined threshold for positive GADA based on a large predominantly adult control population. We have demonstrated that our results are independent of and additive to participants’ clinical features.
A limitation of our study is that time to insulin has been self-reported in the Exeter cohorts at a single visit, in contrast to other cohorts where electronic healthcare records were available. Insulin commencement was also based on clinical decision making rather than a trial protocol. Both these aspects may introduce imprecision but since both clinicians and participants were unaware of results, systematic bias would be unlikely. An additional limitation of cross-sectional study design is that GADA was measured at a median 6.1 years diabetes duration, which could result in a lower prevalence than if measurement was undertaken at diagnosis. However, in adult populations the difference is likely to be small, with GADA positivity being stable over the first 6 years in UKPDS study participants (adult onset type 2 diabetes) (24) and a modest reduction in prevalence (72% to 63%) observed after 8 years in adult onset type 1 diabetes (25). The results of this study can only be applied to white European populations and we do not have measurement of other islet autoantibodies in this cohort - the interaction between genetic risk and other islet autoantibodies would be an area of interest for future research (26).
Our findings are consistent with previous research in a population of participants diagnosed with diabetes between the ages of 20 to 40 years, where the same T1D GRS was predictive of insulin dependent diabetes (9), and other work which has shown this risk score to be additive to islet autoantibodies in predicting future type 1 diabetes (27). It is also consistent with previous research showing patients defined as LADA who have HLA type associated with type 1 diabetes susceptibility, have more rapid progression to insulin (8), and with research showing a combination of positive islet cell autoantibodies and high risk HLA is associated with low C-peptide in a cohort diagnosed as type 2 diabetes in contrast to either of these features alone (28). While the relationship between integrated genetic risk of type 1 diabetes and progression of type 2 diabetes or LADA has not been previously assessed, it has previously been shown that a type 2 diabetes genetic risk score covering 61 established type 2 diabetes risk variants is not associated with time to insulin (17) and that a 69 SNP type 2 diabetes genetic risk score has very limited utility in discriminating patients with type 1 from type 2 diabetes (9).
The prevalence of positive GADA in our cohorts was lower than in much of the previous literature, with previous multicenter studies reporting widely varying prevalence of positive GADA in type 2 diabetes populations ranging from 4% to 14% (29; 30). In addition to diabetes duration, differences in the prevalence of GADA positivity between our and other studies may be explained by our use of an assay with higher specificity than used in many other studies (29–33), our lack of an upper age limit (with lower GADA prevalence seen at older ages (4; 33; 34)), and our use of predominantly population cohorts not selected from secondary care where treatment with insulin is more frequent. We have used a robustly defined high specificity (97.5%) threshold to define positive GADA in line with current clinical laboratory practice, using a large control population. Detectable GADA are commonly found in healthy adult non-diabetic populations and therefore a threshold based on a control population is recommended to robustly define GADA positivity (31–33). An additional potential reason for low autoantibody prevalence is that we have excluded a small number of cohort participants who had GADA tested in clinical practice, which may have influenced treatment choice. However only 47 participants were excluded of whom only 13 were GADA positive, so the effect on overall prevalence is small.
Our findings have clear implications for clinical practice. The T1D GRS represents a novel clinical test that can be used to enhance the prognostic value of GADA testing. For predicting future insulin requirement in patients with apparent type 2 diabetes who are GADA positive, T1D GRS may be clinically useful and can be used as an additional test in the screening process. However, in patients with type 2 diabetes who are GADA negative, there is no benefit gained from genetic testing. This is unsurprising as the prevalence of underlying autoimmunity in patients with a clinical phenotype of type 2 diabetes who are GADA negative is likely to be extremely low, therefore most GADA negative participants with high T1D GRS will have non-autoimmune diabetes. The use of this two-step testing approach may facilitate a precision medicine approach to patients with apparent type 2 diabetes; patients who are likely to progress rapidly are identified for targeted management which may include increased monitoring, early therapy intensification and/or interventions aimed at slowing progression (35; 36).
The costs of analyzing the T1D GRS are relatively modest and may fall further as genetic testing is rapidly becoming less expensive (37). This may allow introduction of the T1D GRS into clinical practice. While the test cost could potentially be reduced further by using 10 SNPs or imputing HLA type alone, the majority of test costs are attributable to DNA extraction, sample handling and test interpretation, with cost for genotyping additional SNPs as low as 10 cents per SNP. Savings would therefore be modest and, while this study does not have sufficient statistical power to directly compare different risk scores in antibody positive participants, this may come at a cost of reduced test accuracy. The use of a risk score approach has an additional advantage over using HLA alone, as it provides genetic information expressed as a simple to use continuous variable.
While using the T1D GRS in a two stage approach may have clinical utility, approaches that go beyond single tests and thresholds to integrate multiple clinical features and biomarkers are likely to have the greatest use for clinical practice. The T1D GRS is additive to other predictive features such as age of diagnosis and BMI, and dichotomizing the test to use thresholds will lose predictive value. While the negative predictive value of a low T1D GRS in participants with GADA is high (<5th centile 92%), positive predictive values are modest, with the majority of high T1D GRS participants not requiring insulin by 5 years. Therefore approaches that combine different predictive features on a continuous basis, using prediction models (clinical calculators), may have the greatest utility in accurately predicting future insulin requirement in this group, and are an important area for future research (38). Additional areas for future research include the association between T1D GRS and progression where multiple islet autoantibodies have been tested, and assessment in a prospective setting where islet autoantibodies have been measured at diabetes diagnosis.
In conclusion, a Type 1 Diabetes Genetic Risk Score alters the clinical implications of a positive GADA test in patients with clinical type 2 diabetes, and is independent of and additive to clinical features. This therefore represents a novel test for identifying patients with rapid progression in this population.
Supplementary Material
Acknowledgments
The authors thank participants who took part in these studies and the research teams who undertook cohort recruitment. We thank Rachel Nice of the Blood Sciences Department, Royal Devon and Exeter Hospital for assistance with conducting the study.
GoDarts: We are grateful to all the participants in this study, the general practitioners, the Scottish School of Primary Care for their help in recruiting the participants, and to the whole team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. The study complies with the Declaration of Helsinki. We acknowledge the support of the Health Informatics Centre, University of Dundee for managing and supplying the anonymized data and NHS Tayside, the original data owner.
Funding
The Wellcome Trust United Kingdom Type 2 Diabetes Case Control Collection (GoDARTS) was funded by The Wellcome Trust (084727/Z/08/Z, 085475/Z/08/Z, 085475/B/08/Z) and as part of the EU IMI-SUMMIT program. GADA assessment in GoDARTS and DCS was funded by EU Innovative Medicines Initiative 115317 (DIRECT), resources of which are composed of financial contributions from the European Union's Seventh Framework Programme (FP7/2007-2013), and European Federation of Pharmaceutical Industries and Associations (EFPIA) companies in kind contribution. The DCS cohort was partially funded by the Netherlands Organization for Health Research and Development (Priority Medicines Elderly Programme 113102006). The Diabetes Alliance for Research in England (DARE) study was funded by the Wellcome Trust and supported by the Exeter NIHR Clinical Research Facility. The MASTERMIND study was funded by the UK Medical Research Council (MR/N00633X/) and supported by the NIHR Exeter Clinical Research Facility. The PRIBA study was funded by the National Institute for Health Research (U.K.) (DRF-2010-03-72) and supported by the NIHR Exeter Clinical Research Facility.
B.M.S and A.T.H. are supported by the NIHR Exeter Clinical Research Facility. T.J.M. is a National Institute for Health Research Senior Clinical Senior Lecturer. E.R.P. is a Wellcome Trust New Investigator (102820/Z/13/Z). A.T.H. is a Wellcome Trust Senior Investigator and NIHR Senior Investigator. R.A.O is supported by a Diabetes UK Harry Keen Fellowship (16/0005529). A.G.J. is supported by an NIHR Clinician Scientist award (CS-2015-15-018).
The views given in this article do not necessarily represent those of the National Institute for Health Research, the National Health Service or the Department of Health.
Footnotes
Duality of Interest
No potential conflicts of interest relevant to this article were reported.
Author Contributions
A.L.G, T.J.M, B.M.S and A.G.J conceived the idea and designed the study. A.L.G, T.J.M, F.R, L.A.D, A.T.H, R.A.O, C.N.A.P, A.A.V.D.H, F.C, P.J.M.E, M.N.W, R.C.S, L.M.H, E.R.P and A.G.J researched the data. A.L.G analyzed the data with assistance from B.M.S, T.J.M, A.T.H, and A.G.J. A.L.G drafted the manuscript with assistance from B.M.S and A.G.J. All authors critically revised the manuscript and approved the final version. A.G.J is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation
Parts of this study were presented in abstract form at the Diabetes UK Professional Conference, London, UK March 2018.
References
- 1.U.K. Prospective Diabetes Study Group: U.K. Prospective Diabetes Study 16: Overview of 6 Years' therapy of Type II Diabetes: A Progressive Disease. Diabetes. 1995;44:1249. [PubMed] [Google Scholar]
- 2.Fonseca VA. Defining and Characterizing the Progression of Type 2 Diabetes. Diabetes care. 2009;32:S151. doi: 10.2337/dc09-S301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groop LC, Bottazzo GF, Doniach D. Islet Cell Antibodies Identify Latent Type I Diabetes in Patients Aged 35–75 Years at Diagnosis. Diabetes. 1986;35:237. doi: 10.2337/diab.35.2.237. [DOI] [PubMed] [Google Scholar]
- 4.Turner R, Stratton I, Horton V, Manley S, Zimmet P, Mackay IR, Shattock M, Bottazzo GF, Holman R. UKPDS 25: autoantibodies to islet-cell cytoplasm and glutamic acid decarboxylase for prediction of insulin requirement in type 2 diabetes. The Lancet. 350:1288–1293. doi: 10.1016/s0140-6736(97)03062-6. [DOI] [PubMed] [Google Scholar]
- 5.Tuomi T, Groop LC, Zimmet PZ, Rowley MJ, Knowles W, Mackay IR. Antibodies to Glutamic Acid Decarboxylase Reveal Latent Autoimmune Diabetes Mellitus in Adults With a Non—Insulin-Dependent Onset of Disease. Diabetes. 1993;42:359. doi: 10.2337/diab.42.2.359. [DOI] [PubMed] [Google Scholar]
- 6.Pozzilli P, Di Mario U. Autoimmune Diabetes Not Requiring Insulin at Diagnosis (Latent Autoimmune Diabetes of the Adult) Diabetes care. 2001;24:1460. doi: 10.2337/diacare.24.8.1460. [DOI] [PubMed] [Google Scholar]
- 7.Liu L, Li X, Xiang Y, Huang G, Lin J, Yang L, Zhao Y, Yang Z, Hou C, Li Y, Liu J, et al. Latent autoimmune diabetes in adults with low-titer GAD antibodies: similar disease progression with type 2 diabetes: a nationwide, multicenter prospective study (LADA China Study 3) Diabetes care. 2015;38:16–21. doi: 10.2337/dc14-1770. [DOI] [PubMed] [Google Scholar]
- 8.Maioli M, Pes GM, Delitala G, Puddu L, Falorni A, Tolu F, Lampis R, Orru V, Secchi G, Cicalo AM, Floris R, et al. Number of autoantibodies and HLA genotype, more than high titers of glutamic acid decarboxylase autoantibodies, predict insulin dependence in latent autoimmune diabetes of adults. European journal of endocrinology. 2010;163:541–549. doi: 10.1530/EJE-10-0427. [DOI] [PubMed] [Google Scholar]
- 9.Oram RA, Patel K, Hill A, Shields B, McDonald TJ, Jones A, Hattersley AT, Weedon MN. A Type 1 diabetes genetic risk score can aid discrimination between Type 1 and Type 2 diabetes in young adults. Diabetes care. 2016;39:337–344. doi: 10.2337/dc15-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hebert HL, Shepherd B, Milburn K, Veluchamy A, Meng W, Carr F, Donnelly LA, Tavendale R, Leese G, Colhoun HM, Dow E, et al. Cohort Profile: Genetics of Diabetes Audit and Research in Tayside Scotland (GoDARTS) International journal of epidemiology. 2017 doi: 10.1093/ije/dyx140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Heijden AA, Rauh SP, Dekker JM, Beulens JW, Elders P, t Hart LM, Rutters F, van Leeuwen N, Nijpels G. The Hoorn Diabetes Care System (DCS) cohort. A prospective cohort of persons with type 2 diabetes treated in primary care in the Netherlands. BMJ open. 2017;7:e015599. doi: 10.1136/bmjopen-2016-015599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diabetes Alliance for Research in England (DARE) [Accessed 23 November 2017]; [article online], Available from http://www.diabetesgenes.org/content/diabetes-alliance-research-england-dare-previously-known-exeter-research-alliance-extra-stud.
- 13.Jones AG, McDonald TJ, Shields BM, Hill AV, Hyde CJ, Knight BA, Hattersley AT, for the PSG Markers of beta cell failure predict poor glycemic response to GLP-1 receptor agonist therapy in type 2 diabetes. Diabetes care. 2016;39:250–257. doi: 10.2337/dc15-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.RetroMASTER - Retrospective Cohort MRC ABPI STratification and Extreme Response Mechanism in Diabetes. [article online], Available from https://www.clinicaltrials.gov/ct2/show/NCT02109978.
- 15.McDonald TJ, Colclough K, Brown R, Shields B, Shepherd M, Bingley P, Williams A, Hattersley AT, Ellard S. Islet autoantibodies can discriminate maturity-onset diabetes of the young (MODY) from Type 1 diabetes. Diabetic medicine : a journal of the British Diabetic Association. 2011;28:1028–1033. doi: 10.1111/j.1464-5491.2011.03287.x. [DOI] [PubMed] [Google Scholar]
- 16.The Wellcome Trust Case Control C. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou K, Donnelly LA, Morris AD, Franks PW, Jennison C, Palmer CNA, Pearson ER. Clinical and Genetic Determinants of Progression of Type 2 Diabetes: A DIRECT Study. Diabetes care. 2014;37:718. doi: 10.2337/dc13-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS genetics. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel KA, Oram RA, Flanagan SE, De Franco E, Colclough K, shepherd M, Ellard S, Weedon MN, Hattersley AT. Type 1 Diabetes Genetic Risk Score: a novel tool to discriminate monogenic and type 1 diabetes. Diabetes. 2016;65:2094–2099. doi: 10.2337/db15-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan EL, Meier P. Nonparametric Estimation from Incomplete Observations. Journal of the American Statistical Association. 1958;53:457–481. [Google Scholar]
- 21.Cuzick J. A wilcoxon-type test for trend. Statistics in medicine. 1985;4:87–90. doi: 10.1002/sim.4780040112. [DOI] [PubMed] [Google Scholar]
- 22.Cox D. Regression models and life tables. Journal of the Royal Statistical Society, Series B (Methodological) 1972;34:187–220. [Google Scholar]
- 23.Schemper M, Smith TL. A note on quantifying follow-up in studies of failure time. Controlled Clinical Trials. 1996;17:343–346. doi: 10.1016/0197-2456(96)00075-x. [DOI] [PubMed] [Google Scholar]
- 24.Desai M, Cull CA, Horton VA, Christie MR, Bonifacio E, Lampasona V, Bingley PJ, Levy JC, Mackay IR, Zimmet P, Holman RR, et al. GAD autoantibodies and epitope reactivities persist after diagnosis in latent autoimmune diabetes in adults but do not predict disease progression: UKPDS 77. Diabetologia. 2007;50:2052–2060. doi: 10.1007/s00125-007-0745-6. [DOI] [PubMed] [Google Scholar]
- 25.Schölin A, Björklund L, Borg H, Arnqvist H, Björk E, Blohmé G, Bolinder J, Eriksson JW, Gudbjörnsdottir S, Nyström L, Östman J, et al. Islet antibodies and remaining β-cell function 8 years after diagnosis of diabetes in young adults: a prospective follow-up of the nationwide Diabetes Incidence Study in Sweden. Journal of Internal Medicine. 2004;255:384–391. doi: 10.1046/j.1365-2796.2003.01273.x. [DOI] [PubMed] [Google Scholar]
- 26.Bottazzo GF, Bosi E, Cull CA, Bonifacio E, Locatelli M, Zimmet P, Mackay IR, Holman RR. IA-2 antibody prevalence and risk assessment of early insulin requirement in subjects presenting with type 2 diabetes (UKPDS 71) Diabetologia. 2005;48:703–708. doi: 10.1007/s00125-005-1691-9. [DOI] [PubMed] [Google Scholar]
- 27.Redondo MJ, Geyer S, Steck AS, Sharp S, Wentworth J, Weedon M, Antinozzi P, Pugliese A, Oram R. A type 1 diabetes genetic risk score predicts progression of islet autoimmunity and development of type 1 diabetes in individuals at risk (Abstract) 53rd EASD Annual Meeting of the European Association for the Study of Diabetes: 51. Diabetologia. 2017;60:1–608. [Google Scholar]
- 28.Groop L, Miettinen A, Groop P-H, Meri S, Koskimies S, Bottazzo GF. Organ-Specific Autoimmunity and HLA-DR Antigens as Markers for β-Cell Destruction in Patients With Type II Diabetes. Diabetes. 1988;37:99–103. doi: 10.2337/diab.37.1.99. [DOI] [PubMed] [Google Scholar]
- 29.Buzzetti R, Zampetti S, Maddaloni E. Adult-onset autoimmune diabetes: current knowledge and implications for management. Nature Reviews Endocrinology. 2017;13:674. doi: 10.1038/nrendo.2017.99. [DOI] [PubMed] [Google Scholar]
- 30.Laugesen E, Østergaard JA, Leslie RDG, the Danish Diabetes Academy W, Workshop S Latent autoimmune diabetes of the adult: current knowledge and uncertainty. Diabetic Medicine. 2015;32:843–852. doi: 10.1111/dme.12700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonifacio E, Yu L, Williams AK, Eisenbarth GS, Bingley PJ, Marcovina SM, Adler K, Ziegler AG, Mueller PW, Schatz DA, Krischer JP, et al. Harmonization of Glutamic Acid Decarboxylase and Islet Antigen-2 Autoantibody Assays for National Institute of Diabetes and Digestive and Kidney Diseases Consortia. The Journal of clinical endocrinology and metabolism. 2010;95:3360–3367. doi: 10.1210/jc.2010-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bingley PJ. Clinical applications of diabetes antibody testing. The Journal of clinical endocrinology and metabolism. 2010;95:25–33. doi: 10.1210/jc.2009-1365. [DOI] [PubMed] [Google Scholar]
- 33.Tuomi T, Carlsson A, Li H, Isomaa B, Miettinen A, Nilsson A, Nissén M, Ehrnström BO, Forsén B, Snickars B, Lahti K, et al. Clinical and genetic characteristics of type 2 diabetes with and without GAD antibodies. Diabetes. 1999;48:150. doi: 10.2337/diabetes.48.1.150. [DOI] [PubMed] [Google Scholar]
- 34.Tuomi T, Santoro N, Caprio S, Cai M, Weng J, Groop L. The many faces of diabetes: a disease with increasing heterogeneity. The Lancet. 2014;383:1084–1094. doi: 10.1016/S0140-6736(13)62219-9. [DOI] [PubMed] [Google Scholar]
- 35.Florez JC. Precision Medicine in Diabetes: Is It Time? Diabetes care. 2016;39:1085–1088. doi: 10.2337/dc16-0586. [DOI] [PubMed] [Google Scholar]
- 36.Leslie RD, Palmer J, Schloot NC, Lernmark A. Diabetes at the crossroads: relevance of disease classification to pathophysiology and treatment. Diabetologia. 2016;59:13–20. doi: 10.1007/s00125-015-3789-z. [DOI] [PubMed] [Google Scholar]
- 37.Christensen KD, Dukhovny D, Siebert U, Green RC. Assessing the Costs and Cost-Effectiveness of Genomic Sequencing. Journal of Personalized Medicine. 2015;5:470–486. doi: 10.3390/jpm5040470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hattersley AT, Patel KA. Precision diabetes: learning from monogenic diabetes. Diabetologia. 2017;60:769–777. doi: 10.1007/s00125-017-4226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.