Abstract
The strain denominated TRQ65 was isolated from wheat (Triticum turgidum subsp. durum) commercial fields in the Yaqui Valley, Mexico. Here, we report its draft genome sequence, which presented ~ 4.5 million bp and 45.5% G + C content. Based on the cutoff values on species delimitation established for average nucleotide identity (> 95 to 96%), genome-to-genome distance calculator (> 70%), and the reference sequence alignment-based phylogeny builder method, TRQ65 was strongly affiliated to Bacillus paralicheniformis. The rapid annotation using subsystem technology server revealed that TRQ65 contains genes related to osmotic, and oxidative stress response, as well as auxin biosynthesis (plant growth promotion traits). In addition, antiSMASH and BAGEL revealed the presence of genes involved in lipopeptides and antibiotic biosynthesis. The function of those annotated genes was validated at a metabolic level, observing that strain TRQ65 was able to tolerate saline (91.0%), and water (155.0%) stress conditions, besides producing 28.8 ± 0.9 µg/mL indoles. In addition, strain TRQ65 showed growth inhibition (1.6 ± 0.4 cm inhibition zone) against the causal agent of wheat spot blotch, Bipolaris sorokiniana. Finally, plant–microbe interactions assays confirm the ability of strain TRQ65 to regulate wheat growth, showing a significant increment in shoot height (26%), root length (40%), shoot dry weight (48%), stem diameter (55%), and biovolume index (246%). These findings provide insights for future agricultural studies of this strain.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-1972-5) contains supplementary material, which is available to authorized users.
Keywords: Plant growth-promoting rhizobacteria, Biocontrol agent, Average nucleotide identity, Genome to genome distance calculator, Biofertilizer
Introduction
The genus Bacillus was first isolated and described as a rod-shaped, heat-resistant, and endospore-forming Gram-positive bacterium (Cohn 1872). The species of this genus are widely distributed due to their ability to form endospores, which provide them resistance to several habitats, such as: environments under optimal or extreme conditions (Tejera-Hernández et al. 2011). Soil is considered the main reservoir of Bacillus, due to the great metabolic diversity of this genus associated with metabolizing a large source of organic compounds (McSpadden Gardener 2004).
In agriculture, the Bacillus species are the most extensively studied bacteria for (1) controlling/inducing plant systemic resistance against phytopathogens, by consumption of leached exudates, production of siderophores, activity of lytic enzymes (chitinases, glucanases, proteases), production of antibiotics, and biosynthesis of cyclic lipopeptides (Villarreal-Delgado et al. 2017; Tiwari et al. 2019), and (2) promoting plant growth and development, through the production or regulation of phytohormones, solubilization of phosphates, activity of 1-aminocyclopropane-1-carboxylate (ACC) deaminase, production of siderophores, and biological nitrogen fixation (Berendsen et al. 2012; Trabelsi and Mhamdi 2013; Barra et al. 2015; García-Meléndez et al. 2017; Valenzuela-Ruiz et al. 2018; Robles-Montoya et al. 2019). Thus, Bacillus has been used as an active ingredient for the formulation of (1) biofungicides, v.gr. Ballad Plus (Bacillus pumilus QST2808 against Erysiphe, and Puccinia), Serenade ASO (B. subtilis QST713 against Pythium, Rhizoctonia, and Fusarium), Fungifree AB (B. subtilis 83 against Colletotrichum, and Leveillula), EcoGuard-GN (Bacillus licheniformis SB3086 against Colletotrichum, and Sclerotinia), and DiPel WG (Bacillus thuringiensis against Cydia, and Otiorhychus) (Villarreal-Delgado et al. 2017; Villa-Rodríguez et al. 2019), and (2) biofertilizers, v.gr. BIOXTERRA BS (Bacillus subtilis), BIOXTERRA BT (Bacillus thuringiensis), Bio-P (Bacillus subtilis, and Azotobacter chroococcum), and Hydroguard (Bacillus amyloliquefaciens), which colonize and protect the rhizosphere improving root biomass and vigor of plants (Botanicare 2012; Valenzuela-Aragon et al. 2018; Kashyap et al. 2019; BioAgro Chemical 2019a, b; AGSOL 2019).
Strain TRQ65 was isolated from wheat (Triticum turgidum subsp. durum) rhizosphere of commercial fields, in the Yaqui Valley, Mexico (27.3692°, 110.3886°). This strain is preserved in Colección de Microorganismos Edáficos y Endófitos Nativos (México) (COLMENA, http://www.itson.edu.mx/COLMENA) (de los Santos Villalobos et al. 2018). According to the strong association of TRQ65 with wheat plants in the field, we inferred a synergistic interaction between them, which needs to be studied to propose its potential use in the biocontrol of phytopathogens that affect wheat production and/or to regulate the growth of this crop. Previously, Valenzuela-Aragon et al. (2018) and Villa-Rodríguez et al. (2019)—based on the 16S rRNA sequencing—affiliated the strain TRQ65 to the genus Bacillus; however, Diaz-Rodriguez et al. (2019) affiliated this strain to Bacillus licheniformis. Since it is one of the largest bacterial genera [comprising 377 named species and 7 subspecies, including synonyms (Parte 2018), with a great genomic and metabolic diversity], its taxonomic affiliation is complex on the basis of traditional phenotypic (Fan et al. 2017) and molecular methods (sequencing of the 16S rRNA gene) (Rooney et al. 2009). Thus, the genome of TRQ65 was sequenced to (1) clarify its taxonomic affiliation and (2) explore its genomic and metabolic background associated with biological control of phytopathogens and wheat growth promotion.
High-quality genomic DNA was extracted from a fresh culture of strain TRQ65, which was grown in Nutrient Broth [24 h at 32 °C, using an orbital shaker at 121 rpm, obtaining 1 × 106 colony forming units (CFU)/mL], and following the protocol described by Valenzuela-Aragon et al. (2018). Then, the bacterial DNA was sequenced by Illumina MiSeq platform, obtaining a total of 5,079,308 total reads [2 × 300 base pairs (bp)]. The quality of the obtained reads was analyzed by FastQC version 0.11.5 (Andrews 2010). Trimmomatic version 0.32 (Bolger et al. 2014) was used to remove adapter sequences and low-quality bases, and only 8.42% was dropped. Subsequently, de novo assembly was generated by SPAdes version 3.10.1 (Bankevich et al. 2012), using the “–careful” parameter for error correction in reads. The draft genome of TRQ65 presented 4,475,481 bp; 45.5% G + C content; 676,421 bp N50; 3 L50; and 32 contigs (> 200 bp). The assembled contigs were ordered by Mauve contig Mover version 2.4.0 (Darling et al. 2004), using the reference genome of Bacillus paralicheniformis KJ-16T [KY694465]. In addition, the presence of plasmids in the TRQ65 genome was analyzed by PLACNETw (https://castillo.dicom.unican.es/upload/) (Vielva et al. 2017); however, no plasmids were observed for strain TRQ65, and to our understanding the presence of plasmids has not been reported for this species.
The 16S rRNA gene sequence of TRQ65 was used to confirm the authenticity of the studied genome according to Chun et al. (2018). In addition, the gene sequence was submitted to NCBI and EzBioCloud database to determine the more closely related strains (based on the cutoff values on species delimitation established for the 16S rRNA gene > 98.7%) (Yoon et al. 2017a; Chun et al. 2018). Thus, the highest similarity values (100%) for the 16S rRNA gene sequence of TRQ65 corresponded to Bacillus paralicheniformis KJ-16T [KY694465], Bacillus haynesii NRRL B-41327T [MRBL01000076], and Bacillus licheniformis ATCC 14580T [AE017333], followed by Bacillus glycinifermentans GO-13 T [LECW01000063], 99.92%, and Bacillus sonorensis NBRC 101234T [AYTN01000016], 99.84% (Table 1). This finding supports the previous taxonomic affiliation of strain TRQ65 to the genus Bacillus (Valenzuela-Aragon et al. 2018; Villa-Rodríguez et al. 2019; Diaz-Rodriguez et al. 2019). To affiliate that strain at a species level, its genome was compared to its more closely related strains (Table 1), by using (1) the average nucleotide identity (ANI), by the OrthoANI algorithm (Yoon et al. 2017b) and (2) the Genome to Genome Distance Calculator (GGDC) version 2.1, by BLAST (Meier-Kolthoff et al. 2013). Those bio-informatics tools have been proposed as a strong approach to clarify the taxonomic affiliation of prokaryotes, which has been used to discover a novel Bacillus species, B. cabrialesii TE3T (de los Santos-Villalobos et al. 2019). Thus, based on the profound taxonomic affiliation provided by those tools, at a species level [ANI > 95-96% (Varghese et al. 2015), and GGDC > 70% (Yoon et al. 2017b)], TRQ65 was strongly affiliated to Bacillus paralicheniformis (Table 1).
Table 1.
16S rRNA similarity, ortho average nucleotide identity (ANI), and Genome to Genome Distance Calculator (GGDC) values by the genome comparison of TRQ65 vs. its more closely related species (16S rRNA > 98.7%)
| Strain TRQ65 compared to: | 16S rRNA similarity | Ortho ANI | GGDC |
|---|---|---|---|
| % | % | % | |
| Bacillus paralicheniformis KJ-16T [KY694465] | 100.00 | 99.07 | 92.40 |
| Bacillus haynesii NRRL B-41327T [MRBL01000076] | 100.00 | 95.13 | 61.40 |
| Bacillus licheniformis ATCC 14580T [AE017333] | 100.00 | 94.56 | 57.60 |
| Bacillus swezeyi NRRL B-41294T [MRBK01000096] | 99.67 | 83.27 | 26.10 |
| Bacillus sonorensis NBRC 101234T [AYTN01000016] | 99.84 | 81.55 | 24.70 |
| Bacillus glycinifermentans GO-13T [LECW01000063] | 99.92 | 80.84 | 23.70 |
| Bacillus subtilis subsp. inaquosorum KCTC 13429T [AMXN01000021] | 98.93 | 73.11 | 19.00 |
| Bacillus subtilis subsp. spizizenii NRRL B-23049T [CP002905] | 99.01 | 72.93 | 19.10 |
| Bacillus nakamurai NRRL B-41091T (LSAZ01000028) | 98.76 | 72.84 | 18.70 |
| Bacillus tequilensis KCTC 13622T [AYTO01000043] | 98.93 | 72.79 | 18.80 |
| Bacillus atrophaeus JCM9070T [AB021181] | 98.76 | 72.74 | 18.60 |
| Bacillus velezensis CR-502T [AY603658] | 98.70 | 72.73 | 19.50 |
| Bacillus subtilis subsp. subtilis NCIB 3610T [ABQL01000001] | 98.85 | 72.70 | 19.00 |
| Bacillus halotolerans ATCC 25096T [LPVF01000003] | 98.85 | 72.45 | 18.50 |
| Bacillus siamensis KCTC 13613T [AJVF01000043] | 98.76 | 72.42 | 19.00 |
| Bacillus subtilis subsp. stercoris D7XPN1T [JHCA01000027] | 98.85 | 72.39 | 18.80 |
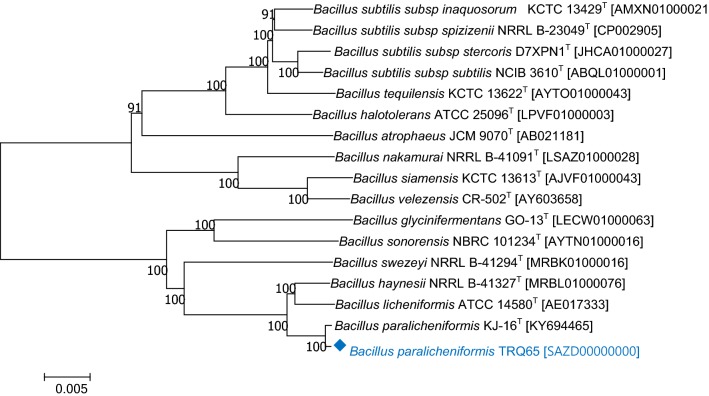
In addition, a phylogenetic tree was constructed to further support the authenticity of the genome data, as well as to determine the genetic relationship between strain TRQ65 and its more closely related species. Thus, the genome sequences were aligned using the reference sequence alignment-based phylogeny (REALPHY) builder method version 1.12 (Bertels et al. 2014), followed by the generation of the genome-based phylogenetic tree by MEGA version 7.0 (Kumar et al. 2016). The neighbor-joining method was used with a bootstrap support of 1000 replications, which confirmed that the taxonomic affiliation of TRQ65 is Bacillus paralicheniformis (Fig. 1).
Fig. 1.
Phylogenetic relation between TRQ65 and closely related species: Bacillus paralicheniformis KJ-16T [KY694465]; Bacillus haynesii NRRL B-41327T [MRBL01000076]; Bacillus licheniformis ATCC 14580T [AE017333]; Bacillus glycinifermentans GO-13T [LECW01000063]; Bacillus sonorensis NBRC 101234T [AYTN01000016]; Bacillus swezeyi NRRL B-41294T [MRBK01000096]; Bacillus subtilis subsp. spizizenii NRRL B-23049T [CP002905]; Bacillus subtilis subsp. inaquosorum KCTC 13429T [AMXN01000021]; Bacillus tequilensis KCTC 13622T [AYTO01000043]; Bacillus subtilis subsp. subtilis NCIB 3610T [ABQL01000001]; Bacillus halotolerans ATCC 25096T [LPVF01000003]; Bacillus subtilis subsp. stercoris D7XPN1T [JHCA01000027]; Bacillus atrophaeus JCM 9070T [AB021181]; Bacillus nakamurai NRRL B-41091T [LSAZ01000028]; Bacillus siamensis KCTC 13613T [AJVF01000043]; Bacillus velezensis CR-502T [AY603658], constructed by the builder method in REALPHY 1.12 (Bertels et al. 2014) and MEGA 7 (Kumar et al. 2016) using the neighbor-joining algorithm (based on 1000 bootstrap replications). Scale bar (0.005) represents the number of nucleotide substitutions per site
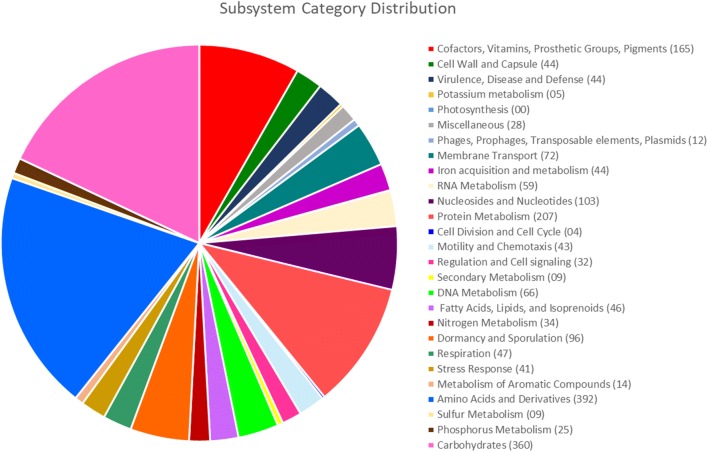
The genome annotation of the studied strain was created through Rapid Annotation Using Subsystem Technology (RAST) server version 2.0 (http://rast.nmpdr.org) (Aziz et al. 2008; Overbeek et al. 2013), by the RASTtk pipeline. Strain TRQ65 showed a total of 91 RNAs, and 4811 predicted coding DNA sequences (CDS)—distributed into 361 subsystems. The most abundant subsystem was amino acids and derivatives (392 genes), followed by carbohydrates (360 genes), protein metabolism (207 genes), cofactors, vitamins, prosthetic groups, and pigments (165 genes), nucleosides and nucleotides (103), and dormancy and sporulation (96) (Fig. 2).
Fig. 2.
Subsystem category distribution of coding DNA sequences (CDS) from strain TRQ65, generated through RASTtk pipeline. CDS: 4811, CDS in subsystems: 1419, and subsystems: 361
The genome of strain TRQ65 revealed the presence of genes involved in (1) the tolerance to abiotic factors in agrosystems (oxidative and water stress conditions), (2) the biological control of phytopathogens (lipopeptides and antibiotic biosynthesis), and (3) the promotion of plant growth (auxin biosynthesis) (Table S1). Putative annotated genes of strain TRQ65 were validated through a metabolic characterization according to Valenzuela-Aragon et al. (2018). The percentage of the abiotic stress tolerance by TRQ65 was calculated by subtracting the bacterial growth (cm) under abiotic stress condition minus the bacterial growth (cm) under optimal condition, and dividing by the bacterial growth (cm) under optimal condition. TRQ65 showed the ability to grow—compared to control conditions—on Petri dishes containing nutrient agar under saline (sodium chloride 5%, 6.8 dS m−1, for 3 days at 28 °C) stress, 91.0 ± 5.3%, and water (polyethylene glycol 6000 10%, − 0.84 mPa, for 3 days at 28 °C) stress, 155.0 ± 3.7%. Similar findings have been reported by Palacio-Rodríguez et al. (2017), Obeidat (2017), and Rajabi Agereh et al. (2019), associating the tolerance of abiotic stress conditions from bacterial strains to genes involved in glycerol, ferric, iron, and zinc uptake, as well as fumarate and nitrate regulation. Those and other promising genes were found in the TRQ65 genome (Table S1).
On the other hand, antiSMASH version 5.0 (https://antismash.secondarymetabolites.org) and BAGEL version 4.0 (http://bagel4.molgenrug.nl/) were used to identify putative genes in the TRQ65 genome involved in the biological control of phytopathogens. Thus, eight genes associated with lipopeptide biosynthesis, bacitracin, bacillibactin, butirosin, lichenysin, haloduracin alpha, haloduracin beta, were identified by antiSMASH; and nine genes associated with lipopeptide biosynthesis, lichenicidin, haloduracin alpha, bottromycin, enterocin, and sonorensin, were identified by BAGEL (Table S1). These lipopeptides have been reported as having antitumor, immunosuppressant, surfactant, cytotoxic, and antimicrobial properties (Raaijmakers et al. 2010). To validate the functionality of those putative genes in the TRQ65 genome, the antagonistic ability of this strain was evaluated in vitro against Bipolaris sorokiniana, the causal agent of wheat spot blotch (Villa-Rodriguez et al. 2016). For this, a three replicate quantitative assay was performed according to Villa-Rodríguez et al. (2019), a volume of 10 µL of Bipolaris sorokiniana TPQ3 conidia suspension (1 × 105 conidia/mL) was placed in the center of Petri dishes containing potato dextrose agar, and 10 µL of Bacillus paralicheniformis TRQ65 cell suspension (1 × 106 CFU/mL) was inoculated in two equidistant points, at about 2 cm distance of the studied phytopathogen. After an incubation for 5 days at 28 °C, the inhibition halo of Bipolaris sorokiniana TPQ3 by strain TRQ65 was quantified. Bacillus paralicheniformis TRQ65 showed an inhibition zone of 1.6 ± 0.4 cm against Bipolaris sorokiniana TPQ3, which confirms the function of putative genes associated with the biological control of phytopathogens found in the genome of TRQ65 by antiSMASH and BAGEL (Table S1).
Regarding the ability of strain TRQ65 to promote the growth and development of plants, this strain was able to biosynthesize 28.8 ± 0.9 µg/mL indoles through the Salkowski method (Rahman et al. 2010). This finding confirms the functionality of the identified putative genes in the TRQ65 genome associated with the biosynthesis of that phytohormone (Figure S1). In addition, to validate the ability of strain TRQ65 to regulate the growth of plants, an axenic in vivo plant–bacterium interaction assay was performed in a growth chamber, under controlled conditions. Thus, wheat variety CIRNO C2008 seeds were washed three times in sterile distilled water, followed by soaking in 70% (v/v) ethanol for 1 min, washed with 3% (v/v) sodium hypochlorite for 10 min, and five additional washes with sterile distilled water. The strain was grown in nutrient broth for 24 h at 28 °C and 120 rpm; then, it was centrifuged at 3500 rpm for 10 min. The pellet was re-suspended in sterile distilled water up to the desired cell concentration. Then, plants (germinated under axenic conditions) were inoculated at day 0 and day 15 with 1 × 108 CFU of TRQ65. The control treatment (uninoculated plants) was only sprayed with sterile distilled water. Two biological replicates (n = 15 wheat plants) of each treatment were carried out, grown under a sterilized (5 times at 121 °C and 15 psi, for 1 h) soil–perlite (70:30) mixture. The assay was carried out in a growth chamber BJPX-A450-BIOBASE, under axenic conditions and 70% humidity, and a photoperiod 12 h light/dark (25 °C during the light, and 15 °C during dark) for 30 days. The analysis of plant biometrics was done according to Thilagar et al. (2016). The percentage of wheat growth promotion by TRQ65 was calculated by subtracting the value (cm or g) of inoculated plants minus the value (cm or g) of un-inoculated plants, and dividing by the value (cm or g) of uninoculated plants. The inoculation of strain TRQ65 to wheat plants showed a significant (Tukey–Kramer test, p = 0.05) increment (compared to uninoculated plants) of shoot height (26.14%), root length (36.43%), stem diameter (53.33%), stem circumference (54.34%), shoot dry weight (100%), and biovolume index (146.05%) (Table 2). These findings strongly validate the ability of strain TRQ65 to promote wheat growth, through metabolites produced by those putative genes found in its genome (Table S1) and/or novel genes that need to be studied.
Table 2.
Wheat growth promotion by the inoculation of Bacillus paralicheniformis TRQ65 (growth chamber assay)
| Variable | Un-inoculated | Bacillus paralicheniformis TRQ65 |
|---|---|---|
| Shoot height (cm) | 18.59 ± 5.41a | 23.45 ± 3.26b |
| Root length (cm) | 7.41 ± 2.24a | 10.11 ± 2.53b |
| Stem diameter (cm) | 0.15 ± 0.04a | 0.23 ± 0.04b |
| Stem circumference (cm) | 0.46 ± 0.13a | 0.71 ± 0.13b |
| Shoot dry weight (g) | 0.05 ± 0.01a | 0.10 ± 0.03b |
| Root dry weight (g) | 0.09 ± 0.02a | 0.10 ± 0.01a |
| Biovolume index | 68.79 ± 29.96a | 169.26 ± 47.90b |
Means (2 × n = 15) with the same letter are not significantly different, according to Tukey–Kramer test (p = 0.05)
In conclusion, the obtained genomic findings—and the phenotypic traits previously reported by Valenzuela-Aragon et al. (2018), Villa-Rodríguez et al. (2019), and Diaz-Rodriguez et al. (2019)—strongly confirm that strain TRQ65 belongs to Bacillus paralicheniformis. In addition, its genome contains genes involved in tolerance of abiotic stress conditions, biological control of phytopathogens, and plant growth promotion. Therefore, the genomic, metabolic, and ecological background observed in Bacillus paralicheniformis TRQ65 suggests this strain as a promising plant growth-promoting bacterium, where further analysis regarding other functional genes are required for its industrial usage as a microbial inoculant to produce wheat and other economic crops.
Accession numbers The assembled contigs were deposited in the DDBJ/ENA/GenBank and published with the accession number SAZD00000000. The version described in this paper is the first version of the genome sequence deposited.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This study was supported by the CONACyT Project 257246 “Interacción trigo x microorganismos promotores del crecimiento vegetal: identificando genes con potencial agro-biotecnológico”, and the Instituto Tecnológico de Sonora (ITSON) Project PROFAPI 2019-0094 “Identificación de genes asociados al control biológico y promoción del crecimiento vegetal en el genoma de Bacillus sp. TRQ65”. In addition, we thank Abraham Chaparro Encinas for his support in the bacterial DNA extraction. Valeria Valenzuela-Ruiz and Rosa Robles Montoya were supported by CONACYT fellowship number 712969 and 627262, respectively.
Author contributions
VV: conceptualization; data curation; formal analysis; investigation; methodology; visualization; writing—original draft; writing—review and editing. RIRM: conceptualization; data curation; formal analysis; investigation; methodology; visualization; writing—original draft. FP: conceptualization; formal analysis; investigation; methodology; visualization; writing—original draft; writing—review and editing. GS: visualization; writing—original draft; writing—review and editing. MCO: visualization; writing—original draft; writing—review and editing. RRR: visualization; writing—original draft; writing—review and editing. SS: conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization; writing—original draft; writing—review and editing.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Agriculture Solution AGSOL What is Bio-P™?. https://www.agsolcanada.com/individual-product-info/bio-p. Accessed 30 June 2019
- Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed 8 may 2019
- Aziz RK, Bartels D, Best A, et al. The RAST server: rapid annotations using subsystems technology. BMC Genom. 2008;9:1–15. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barra F, Roscetto E, Soriano AA, Vollaro A, Postiglione I, Pierantoni GM. Photodynamic and antibiotic therapy in combination to fight biofilms and resistant surface bacterial infections. Int J Mol Sci. 2015;16:20417–20430. doi: 10.3390/ijms160920417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen R, Pieterse CM, Bakker PA. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Bertels F, Silander OK, Pachkov M, Rainey PB, Van Nimwegen E. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol Biol Evol. 2014;31:1077–1088. doi: 10.1093/molbev/msu088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BioAgro Chemical, BIO X TERRA BS (2019a) http://bioagrochemical.com.mx/images/ft_bioxterra_bs.pdf. Accessed 30 June 2019
- BioAgro Chemical, BIO X TERRA BT (2019b) http://bioagrochemical.com.mx/images/ft_bioxterra_bt.pdf. Accessed 30 June 2019
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botanicare, Hydroguard® (2012) https://www.botanicare.com/products/hydroguard/. Accessed 30 June 2019
- Chun J, Oren A, Ventosa A, et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol. 2018;68:461–466. doi: 10.1099/ijsem.0.002516. [DOI] [PubMed] [Google Scholar]
- Cohn F (1872) Untersuchungen Über Bakterien. Beitrage zur Biologie Pflanz 1:127–1224. https://doi.org/BHL/BeitraegezurBiologiederPflanzen
- Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14(7):1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos Villalobos S, Parra Cota F, Herrea Sepulveda A, Valenzuela Aragon B, Estrada Mora JC. Colmena: colección de microorganismos edáficos y endófitos nativos, para contribuir a la seguridad alimentaria nacional. Rev Mexicana Cienc Agric. 2018;9(1):191–202. doi: 10.2931/remexca.v9i1.858. [DOI] [Google Scholar]
- de los Santos-Villalobos S, Robles RI, Parra Cota FI, Larsen J, Lozano P, Tiedje MJ. Bacillus cabrialesii sp. nov., an endophytic plant growth promoting bacterium isolated from wheat (Triticum turgidum subsp. durum) in the Yaqui Valley, Mexico. Int J Syst Evol Microbiol. 2019 doi: 10.1099/ijsem.0.003711. [DOI] [PubMed] [Google Scholar]
- Diaz-Rodriguez A, Parra-Cota FI, Santoyo G, de los Santos-Villalobos S. Chlorothalonil tolerance of indole producing bacteria associated to wheat (Triticum turgidum L.) rhizosphere in the Yaqui Valley, Mexico. Ecotoxicology. 2019;28:569–577. doi: 10.1007/s10646-019-02053-x. [DOI] [PubMed] [Google Scholar]
- Fan B, Blom J, Klenk HP, Borriss R. Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis form an “operational group B. amyloliquefaciens” within the B. subtilis species complex. Front Microbiol. 2017;8:22. doi: 10.3389/fmicb.2017.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García Meléndez M, Zárate Camargo G, Meza Contreras JJ, Herrera Sepúlveda A, Parra Cota FI, de los Santos Villalobos S. Abiotic stress tolerance of microorganisms associated with oregano (Origanum vulgare L.) in the Yaqui Valley. Sonora. Open Agric J. 2017;2(1):260–265. doi: 10.1515/opag-2017-0029. [DOI] [Google Scholar]
- Kashyap BK, Solanki MK, Pandey AK, Prabha S, Kumar P, Kumari B. Bacillus as plant growth promoting rhizobacteria (PGPR): a promising green agriculture technology. In: Ansari R, Mahmood I, editors. Plant health under biotic stress. 1. Singapore: Springer; 2019. pp. 219–236. [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSpadden Gardener BB. Ecology of Bacillus and Paenibacillus spp. in agricultural systems. Phytopathology. 2004;94(11):1252–1258. doi: 10.1094/phyto.2004.94.11.1252. [DOI] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013;14:1–14. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeidat M. Isolation and characterization of extremely halotolerant Bacillus species from Dead Sea black mud and determination of their antimicrobial and hydrolytic activities. Afr J Microbiol Res. 2017;11(32):1303–1314. doi: 10.5897/ajmr2017.8608. [DOI] [Google Scholar]
- Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2013;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacio-Rodríguez R, Coria-Arellano JL, López-Bucio J, Sánchez-Salas J, Muro-Pérez G, Castañeda-Gaytán G, Sáenz-Mata J. Halophilic rhizobacteria from Distichlis spicata promote growth and improve salt tolerance in heterologous plant hosts. Symbiosis. 2017;73(3):179–189. doi: 10.1007/s13199-017-0481-8. [DOI] [Google Scholar]
- Parte AC. LPSN—list of prokaryotic names with standing in nomenclature (bacterio.net) 20 years on. Int J Syst Evol Microbiol. 2018;68:1825–1829. doi: 10.1099/ijsem.0.002786. [DOI] [PubMed] [Google Scholar]
- Raaijmakers JM, De Bruijn I, Nybroe O, Ongena M. Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol Rev. 2010;34(6):1037–1062. doi: 10.1111/j.1574-6976.2010.00221.x. [DOI] [PubMed] [Google Scholar]
- Rahman A, Sitepu IR, Tang SY, Hashidoko Y. Salkowski’s reagent test as a primary screening index for functionalities of rhizobacteria isolated from wild dipterocarp saplings growing naturally on medium-strongly acidic tropical peat soil. Biosci Biotechnol Biochem. 2010;74(11):2202–2208. doi: 10.1271/bbb.100360. [DOI] [PubMed] [Google Scholar]
- Rajabi Agereh S, Kiani F, Khavazi K, Rouhipour H, Khormali F. An environmentally friendly soil improvement technology for sand and dust storms control. Environ Health Eng Manag. 2019;6(1):63–71. doi: 10.15171/EHEM.2019.07. [DOI] [Google Scholar]
- Robles-Montoya RI, Parra Cota FI, de los Santos Villalobos S. Draft genome sequence of Bacillus megaterium TRQ8, a plant growth‐promoting bacterium isolated from wheat (Triticum turgidum subsp. durum) rhizosphere in the Yaqui Valley, Mexico. 3 Biotech. 2019;9:201. doi: 10.1007/s13205-019-1726-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney AP, Price NP, Ehrhardt C, Swezey JL, Bannan JD. Phylogeny and molecular taxonomy of the Bacillus subtilis species complex and description of Bacillus subtilis subsp. inaquosorum subsp. nov. Int J Syst Evol Microbiol. 2009;59(10):2420–2436. doi: 10.1099/ijs.0.009126-0. [DOI] [PubMed] [Google Scholar]
- Tejera-Hernández B, Rojas-Badía MM, Heydrich-Pérez M (2011) Potencialidades del género Bacillus en la promoción del crecimiento vegetal y el control de hongos fitopatógenos. Rev CENIC Cienc Biol 42(3):131–138. http://www.redalyc.org/articulo.oa?id=181222321004
- Thilagar G, Bagyaraj DJ, Podile AR, Vaikuntapu PR. Bacillus sonorensis, a novel plant growth promoting rhizobacterium in improving growth, nutrition and yield of chilly (Capsicum annuum L.) Proc Natl Acad Sci. 2016;88:813–818. doi: 10.1007/s40011-016-0822-z. [DOI] [Google Scholar]
- Tiwari S, Prasad V, Lata C. Bacillus: Plant growth promoting bacteria for sustainable agriculture and environment. In: Shankar Singh J, Singh DP, editors. New and future developments in microbial biotechnology and bioengineering. Lucknow: Elsevier; 2019. pp. 43–55. [Google Scholar]
- Trabelsi D, Mhamdi R. Microbial inoculants and their impact on soil microbial communities: a review. Biomed Res Int. 2013;2013:1–11. doi: 10.1155/2013/863240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela-Aragon B, Parra-Cota F, Santoyo G, Arellano G, de los Santos-Villalobos S. Plant-assisted selection: a promising alternative for in vivo identification of wheat (Triticum turgidum L. subsp. Durum) growth promoting bacteria. Plant Soil. 2018;435:367–384. doi: 10.1007/s11104-018-03901-1. [DOI] [Google Scholar]
- Valenzuela-Ruiz V, Ayala-Zepeda M, Arellano-Wattenbarger GL, Parra-Cota FI, García-Pereyra J, Aviña-Martínez GN, de los Santos-Villalobos S. Microbial culture collections and their potential contribution to current and future food security. Rev Lat Rec Nat. 2018;14(1):18–25. [Google Scholar]
- Varghese NJ, Mukherjee S, Ivanova N, Konstantinidis KT, Mavrommatis K, Kyrpides NC, Pati A. Microbial species delineation using whole genome sequences. Nucleic Acids Res. 2015;43(14):6761–6771. doi: 10.1093/nar/gkv657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielva L, de Toro M, Lanza VF, de la Cruz F. PLACNETw: a web-based tool for plasmid reconstruction from bacterial genomes. Bioinformatics. 2017;33(23):3796–3798. doi: 10.1093/bioinformatics/btx462. [DOI] [PubMed] [Google Scholar]
- Villa-Rodriguez E, Lugo-Enriquez C, de los Santos-Villalobos S. First report of Cochliobolus sativus causing spot blotch on durum wheat (Triticum durum) in the Yaqui Valley, Mexico. Plant Dis. 2016;100:2329. doi: 10.1094/PDIS-05-16-0634-PDN. [DOI] [Google Scholar]
- Villa-Rodríguez E, Parra-Cota F, Castro-Longoria E, López-Cervantes J, de los Santos-Villalobos S. Bacillus subtilis TE3: a promising biological control agent against Bipolaris sorokiniana, the causal agent of spot blotch in wheat (Triticum turgidum L. subsp. durum) Biol Control. 2019;132:135–143. doi: 10.1016/j.biocontrol.2019.02.012. [DOI] [Google Scholar]
- Villarreal-Delgado MF, Villa-Rodríguez ED, Cira-Chávez LA, Estrada-Alvarado MI, Parra-Cota FI, De los Santos-Villalobos S. The genus Bacillus as a biological control agent and its implications in the agricultural biosecurity. Rev Mex Fitopatol. 2017;36(1):95–130. doi: 10.18781/R.MEX.FIT.1706-5. [DOI] [Google Scholar]
- Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol. 2017;67(5):1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SH, Ha SM, Lim JM, Kwon SJ, Chun J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek. 2017;110(10):1281–1286. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.