Abstract
Genetic and nongenetic factors are involved in the individual ability to physiologically acclimatize to high-altitude hypoxia through processes that include increased heart rate and ventilation. High-altitude acclimatization is thought to have a genetic component, yet it is unclear if other factors, such as epigenetic gene regulation, are involved in acclimatization to high-altitude hypoxia in nonacclimatized individuals. We collected saliva samples from a group of healthy adults of European ancestry (n = 21) in Kathmandu (1,400 m; baseline) and three altitudes during a trek to the Everest Base Camp: Namche (3,440 m; day 3), Pheriche (4,240 m; day 7), and Gorak Shep (5,160 m; day 10). We used quantitative bisulfite pyrosequencing to determine changes in DNA methylation, a well-studied epigenetic marker, in LINE-1, EPAS1, EPO, PPARa, and RXRa. We found significantly lower DNA methylation between baseline (1,400 m) and high altitudes in LINE-1, EPO (at 4,240 m only), and RXRa. We found increased methylation in EPAS1 (at 4,240 m only) and PPARa. We also found positive associations between EPO methylation and systolic blood pressure and RXRa methylation and hemoglobin. Our results show that incremental exposure to hypoxia can affect the epigenome. Changes to the epigenome, in turn, could underlie the process of altitude acclimatization.
Keywords: altitude, epigenetics, DNA methylation, hypoxia, incremental ascent
Introduction
More than 140 million people worldwide permanently live at high altitudes, and 40 million more visit altitudes above 2,500 m annually (Ward et al., 2000; Moore, 2001). Atmospheric oxygen partial pressure decreases with increasing altitude, and most individuals experience physiological changes in low-oxygen environments, including increased ventilation, increased red blood cell production, and increased heart rate (HR) (Houston and Riley, 1947). A combination of molecular- to the organismal-level changes occurs during high-altitude acclimatization (Sarkar et al., 2003).
The acute response to hypoxia (seconds to hours) involves changes in homeostatic regulation, whereas chronic acclimatization (hours to years) is characterized by gene expression changes in the carotid body, endothelial cells, and other tissues (Kourembanas et al., 1990; Wiener et al., 1996; Huey and Powell, 2000). One of main responders to decreasing levels of oxygen is the hypoxia-inducible factor 1 (HIF) pathway. HIF-1 consists of two subunits, oxygen-regulated HIF-1α, and constitutively expressed HIF-1β (Ivan et al., 2001). In normoxic conditions, HIF-1α is hydroxylated by HIF prolyl hydroxylase (EGLN) and destined for degradation by ubiquitination via the von Hippel–Lindau ubiquitin ligase (Ohh et al., 2000; Epstein et al., 2001). HIF-1α hydroxylation is decreased in hypoxic conditions allowing it to accumulate and dimerize with HIF-1β forming an active HIF-1 transcription factor in the nucleus (Semenza, 2007). HIF is involved in promoting angiogenesis, regulating erythropoiesis, stimulating glycolysis, and inhibiting fatty acid oxidation (Formenti et al., 2010; Haase, 2013; Huang et al., 2014).
Previous studies have shown that mRNA levels of genes involved in the HIF pathway change upon hypoxic exposure, including HIF1A and ARNT in rats and mice (Wiener et al., 1996), mRNA levels of the platelet-derived growth factor (PDGF-B) in human endothelial cells (Kourembanas et al., 1990), and dopamine D2 receptor (D2R) in rat carotid body (Huey and Powell, 2000). Given their ability to change upon exposure to environmental factors, epigenetic mechanisms have been hypothesized to play a role the hypoxic response (Brown and Rupert, 2014). Epigenetics refers to mitotically and, in some cases, meiotically heritable changes to gene expression that do not involve changes to DNA sequence and may be reversible (Wolffe and Guschin, 2000; Feil and Fraga, 2011). The most widely studied and best understood epigenetic modification is DNA methylation, an addition of a methyl group to the nucleotide cytosine in a cytosine-guanine dinucleotide (CpG) (Mohn and Schubeler, 2009; Lam et al., 2012). DNA methylation is most commonly associated with gene repression when located in promoter regions of genes (Klose and Bird, 2006).
Epigenetic modifications are known for their plasticity and ability to change based on the environmental conditions (Bollati and Baccarelli, 2010). Previous studies found associations between DNA methylation and pharmaceuticals, exercise, stress, and other exposures (Dolinoy, 2007; Dolinoy and Jirtle, 2008; Senut et al., 2012; Faulk et al., 2014; Non et al., 2016). Decreased oxygen levels are associated with increased production of reactive oxygen species (ROS) that are genotoxic and can affect DNA methylation and the posttranslational modifications to histone proteins (James et al., 2004; Niu et al., 2015). Moreover, epigenetic changes have been observed in cancer cells that are often hypoxic due to the lower oxygen availability of solid tumors (Shahrzad et al., 2007; Baxter et al., 2014). Here, we focused on DNA methylation and exposure to high-altitude hypoxia.
Epigenetic regulation has been studied in the context of high-altitude adaptation in Andeans and Ethiopians (Alkorta-Aranburu et al., 2012; Childebayeva et al., 2019). Despite this, epigenetic changes associated with acclimatization to high-altitude hypoxia are not well understood (Julian et al., 2014). To determine if short-term exposure to hypoxia affects the epigenome, we recruited individuals trekking to Everest Base Camp in the Nepal Himalaya. We collected saliva samples and various physiological measurements at four different altitudes: Kathmandu [1,400m; baseline (BL)], Namche (3,440m; day 3), Pheriche (4,240m; day 7), and Gorak Shep (5,160m; day 10).
We determined the DNA methylation status of the repetitive element LINE-1 and the hypoxia-associated genes EPAS1, EPO, PPARa, and RXRa. We chose LINE-1 as the marker of global methylation as it has been shown to have different methylation profiles at high compared to low altitude in multigenerational Andeans of Quechua ancestry (Childebayeva et al., 2019). We examined methylation at EPAS1 as polymorphisms near this locus are associated with hemoglobin levels in Tibetans (Beall et al., 2010), EPO as it is involves in red blood cell production (Eckardt et al., 1992; Dame et al., 1998; Beall et al., 2010), and PPARa and RXRa as these hypoxia-associated genes are involved in lipid metabolism regulation (Keller et al., 1993; Chinetti et al., 2000) and PPARA is associated with adaptation in high-altitude populations in the Himalaya (Keller et al., 1993; Chinetti et al., 2000; Simonson et al., 2010; Horscroft et al., 2017). RXRa and PPARa form a heterodimer that is necessary for PPARa functioning (Chan and Wells, 2009). The aforementioned HIF pathway genes have been chosen due to previous evidence that methylation levels at these genes are associated hypoxia (Rossler et al., 2004; Lachance et al., 2014; Cortese et al., 2016).
Materials and Methods
Ethics and Participant Recruitment
This study abided by the Canadian Government Tri-Council policy on research ethics with human participants (TCPS2) and the Declaration of Helsinki, except for registration in a database. Ethical approval was received in advance through Mount Royal University Human Research Ethics Board (protocol 100012 and 101361), the Syracuse University Institutional Review Board (protocol 18-006), and the University of Michigan Institutional Review Board (HUM00141118) and harmonized with the Nepal Health Research Council (protocol 109-2017).
This study took place in May 2018 as part of a research expedition in the Khumbu Valley, Everest region of Nepal. We recruited 21 healthy, nonpregnant, nonlactating, nonsmokers between 19 and 52 years of age from a larger research expedition to Everest Base Camp in the Nepal Himalaya. All participants were recruited in Kathmandu via verbal communication and provided written and informed consent prior to voluntary participation in the study. Even though these participants were recruited as a part of a larger research expedition, the research questions and data collection reported here were planned a priori in advance. Participants either were all altitude naive or had an extended period since the last altitude experience (> 1year). All participants were of self-reported European descent to control for population effects on epigenetics. Participant characteristics can be found in Table 1.
Table 1.
Participant characteristics and DNA methylation.
| Kathmandu (1,400m) | Namche (3,400m) | Pheriche (4,370m) | Gorak Shep (5,160m) | |
|---|---|---|---|---|
| LINE-1 | 64.55 (3.05) | 62.83 (3.41)** | 63.40 (2.12)* | 63.70 (3.63). |
| EPAS1 | 6.48 (1.19) | 6.55 (1.09) | 6.95 (1.38)* | 6.90 (1.35) |
| EPO | 72.78 (4.88) | 71.79 (4.14) | 69.57 (4.13)* | 71.22 (3.98) |
| PPARa | 13.68 (4.16) | 14.92 (4.51) | 15.49 (3.79)** | 16.12 (4.09)*** |
| RXRa | 40.01 (11.75) | 35.13 (12.82)* | 32.44 (9.05)*** | 33.57 (12.98)** |
| Hemoglobin (g/dL) | 132.20 (26.67) | 146.32 (18.13)** | 149.42 (16.84)** | 149.63 (26.06) ** |
| Body mass index | 22.69 (2.52) | 22.58 (2.46) | 22.45 (2.36)** | 22.25 (2.34)*** |
| Systolic blood pressure (mmHg) | 119.62 (12.29) | 124.62 (10.60). | 120.62 (10.91) | 126.67 (17.98)* |
| Diastolic blood pressure (mmHg) | 83.00 (6.61) | 87.95 (7.61)** | 87.19 (8.45)** | 86.38 (9.01)* |
| Peripheral oxygen saturation | 96.86 (1.11) | 92.67 (3.26)*** | 89.29 (2.72)*** | 81.48 (4.80)*** |
| % Female | 45.45 (24.65, 66.26) | |||
| Age (year) | 24.41 (8.20) | |||
Data are means (SD) of average measurement per individual, 95% confidence interval (CI) for proportions in brackets. Age is presented as mean (SD).
Significance symbols denote the difference between Kathmandu baseline and each altitude.
.p<0.10. *p<0.05. **p<0.01. ***p<0.001.
Ascent Profile and Data Collection
Over the course of 10 days, a team of researchers and study participants trekked from 2,800 to 5,160 m. The ascent profile included three nontrekking rest days at 3,440 m (day 3), 3,820 m (day 5), and 4,240 m (day 7; Figure 1). In the morning between 6:00 and 8:00 local time at 1,400m (Kathmandu; day 0), 3,440 m (Namche; day 3), 4,240 m (Pheriche; day 7), and 5,160 m (Gorak Shep; day 10), saliva samples for DNA and physiological measures were taken following one night of sleep at each altitude.
Figure 1.
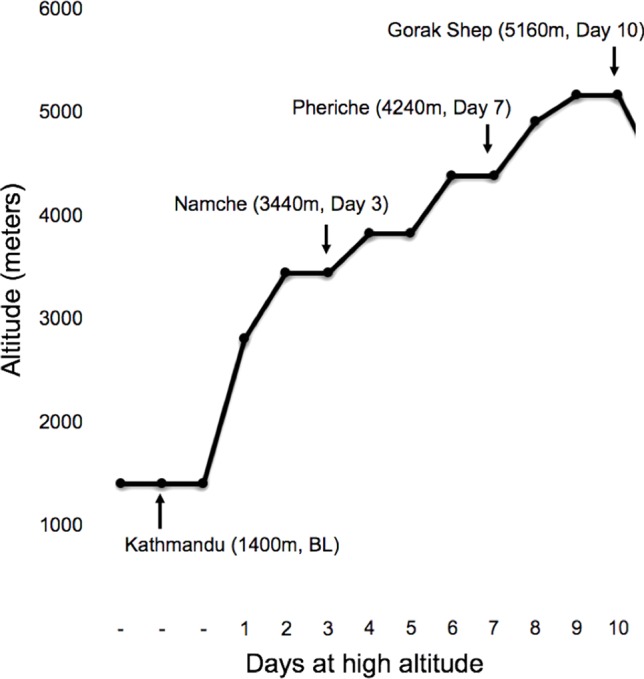
Trekking profile with the sampling locations marked by arrows.
With respect to physiological measures, body weight was measured using a portable digital scale (model HBF-516B; Omron, San Ramon, CA, United States). All physiological measures were obtained at rest in a seated position following >2-min rest with eyes closed and white noise played through headphones to limit distraction. Blood pressure was assessed using an automated sphygmomanometer. Peripheral oxygen saturation and HR (min−1) were measured using a portable finger pulse oximeter (Masimo SET Rad-5, Danderyd, Sweden). Hemoglobin concentration [(Hb); Hemocue Hemoglobin System, Hb201+, Angelholm, Sweden] was assessed via finger capillary blood sample using sterile lancets and universal precautions. Self-reported acute mountain sickness scores were obtained using the standard Lake Louise Questionnaire (Roach Rc et al., 1993). All phenotypic measures were performed at the same time of day for each participant. Physiological data can be found in Table 1 and Supplementary Table 1.
DNA Methylation
Saliva samples were collected, and DNA was extracted following a well-established protocol (Quinque et al., 2006). Quantitative pyrosequencing was performed to assess DNA methylation levels of LINE1, EPAS1, EPO, PPARa, and RXRa. Five hundred nanograms of DNA from each sample was bisulfite converted using the EZ-96 DNA Methylation™ Kit (Zymo Research, Irvine, CA, USA). Bisulfite-converted DNA was amplified using primers for LINE1 and each targeted gene and HotstarTaq plus Master Mix (Qiagen, Valencia, CA, USA). Primer sequences and the locations of the amplicons can be found in Supplementary Table 2. Each sample was pyrosequenced in duplicate using the Pyromark Q96 pyrosequencer (Qiagen). Quality control of the data was assessed using quality control measures built into the pyrosequencing software. All measurements outside of 2 standard deviations from the mean of all samples for each CpG position were excluded. Moreover, measurements with the coefficient of variance between replicates of more than 10 were excluded from further analyses. Duplicate measurements were averaged, as was DNA methylation at CpG sites within each gene. Statistical modeling was performed on these average DNA methylation values for each subject at each gene. Statistical analyses were performed using the samples collected from the 21 individuals at four altitudes for each LINE1, EPAS1, EPO, and PPARa and 19 for RXRa. No template controls and 0% methylated DNA and 100% methylated DNA controls (Qiagen) were included in all experiments.
Statistical Analysis
We used R version 3.5.1 (R Core Team, 2018). Packages lme4 (Bates et al., 2015), lmerTest (Kuznetsova et al., 2017), ggplot2 (Wickham, 2009), and directlabels (Hocking, 2018) were employed in our statistical analysis and plotting. Linear mixed-effects modeling was used to account for replicate measurements at each altitude. The following linear mixed-effects models were tested. Study participants were modeled as random effects to account for repeated measurements. We included age and sex in the models, since both are known to affect DNA methylation (Liu et al., 2010; Hernandez et al., 2011; Lam et al., 2012; Horvath, 2013).
Yi (% methylation) ∼ B00 + B01(X) + B02(Sex) + B03(Age) + (1|ID) + ei, where X = low altitude (1,440m, BL) vs. high altitude [3,440m (day 3), vs. 4,240m (day 7), vs. 5,160m (day 10) combined], ID = sample ID.
Yi (% methylation) ∼ B00 + B01(X) + B02(Sex) + B03(Age) + (1|ID) + ei, where X = altitude [1,400 (BL) vs. 3,440m (day 3), vs. 4,240m (day 7), vs. 5,160m (day 10)], ID = sample ID.
Yi (% methylation) ∼ B00 + B01(X) + B02(Altitude) + B03(Sex) + B04(Age) + (1|ID) + ei, where X = phenotype, altitude = 1,400m (BL), 3,440m (day 3), 4,240m (day 7), 5,160m (day 10), ID = sample ID.
Results
Hypoxic Exposure Is Associated With Changes in DNA Methylation
We found LINE1 methylation to be negatively associated with altitude when comparing low altitude 1,400 m (BL) to high altitude [3,440 m (day 3) + 4,240 m (day 7) + 5,160 m (day 10)] (β = −1.62 (high), p = 0.005) (Table 2, Figure 2, and Supplementary Figure 1A). Methylation levels of LINE1 were also significantly lower at 3,440 m (day 3) and 4,240 m (day 7) compared to 1,400 m (BL) (Table 2, Figure 2 and Supplementary Figure 1B).
Table 2.
Associations between DNA methylation and altitude.
| β | p | ||
|---|---|---|---|
| LINE-1 | |||
| Low vs. high* | −1.62 (High) | 0.005 ** | |
| 3,440m (day 3)** | −2.04 | 0.003 ** | |
| 4,240m (day 7)** | −1.47 | 0.037 * | |
| 5,160m (day 10)** | −1.32 | 0.055. | |
| EPAS1 | |||
| Low vs. high* | 0.36 (High) | 0.096. | |
| 3,440m (day 3)** | 0.27 | 0.310 | |
| 4,240m (day 7)** | 0.58 | 0.033* | |
| 5,160m (day 10)** | 0.22 | 0.393 | |
| EPO | |||
| Low vs. high* | −1.34 (High) | 0.171 | |
| 3,440m (day 3)** | −0.61 | 0.603 | |
| 4,240m (day 7)** | −2.71 | 0.023 * | |
| 5,160m (day 10)** | −0.69 | 0.553 | |
| PPARa | |||
| Low vs. high* | 1.97 (High) | 0.002 ** | |
| 3,440m (day 3)** | 1.10 | 0.125 | |
| 4,240m (day 7)** | 2.05 | 0.005 ** | |
| 5,160m (day 10)** | 2.76 | < 0.001*** | |
| RXRA | |||
| Low vs. high* | −7.14 (High) | < 0.001*** | |
| 3,440m (day 3)** | −5.13 | 0.039 * | |
| 4,240m (day 7)** | −8.70 | < 0.001*** | |
| 5,160m (day 10)** | −7.58 | 0.003 ** |
Low vs. high refers to 1,400m (BL) vs. 3,440, 4,240, and 5,160m combined. Otherwise, results indicate the difference between 1,400m (BL) and the altitude listed. All mixed-effects models were adjusted for age and sex.
.p<0.10. *p<0.05. **p<0.01. ***p<0.001.
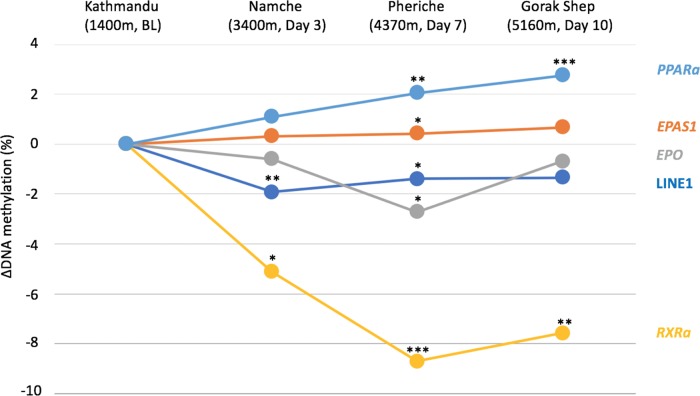
Figure 2.
DNA methylation difference between Kathmandu and each altitude is plotted for each gene over time. The baseline is Kathmandu, which is 0. Significance levels are shown by *p<0.05, **p<0.005, ***p<0.001.
The association between EPAS1 methylation and high vs. low altitude approached significance (β = 0.36 (high), p = 0.096, Supplementary Figure 1B), and only the comparison between 1,400 m (BL) and 4,240m (day 7) was significant at p<0.05 (β = 0.58, p = 0.033, Table 2).
EPO methylation was not significantly different between high and low altitude [β = −1.34 (high), p = 0.171] (Table 2, Figure 2 and Supplementary Figure 1C), and only the comparison between 1,400 and 4,240 m (day 7) was significant (β = −2.71, p = 0.023, Table 2, Figure 2).
PPARa methylation was positively associated with high altitude [β = 1.97 (high), p = 0.002, Figure 2 and Supplementary Figure 1D], and the comparisons between 1,400 m (BL) vs. 4,240 m (day 7) and 5,160 m (day 10) were significant (Table 2).
We observed decreased methylation of RXRa associated with high altitude [β = −7.14 (high), p<0.001, Figure 2 and Supplementary Figure 1E]. Moreover, RXRa methylation levels at 3,440 m (day 3), 4,240 m (day 7), and 5,160 m (day 10) were significantly lower than at 1,400 m (BL) (Table 2).
Associations Between DNA Methylation and Phenotypic Data
Systolic blood pressure was positively associated with EPO methylation (β = 0.63, p = 0.022, Supplementary Table 3). This relationship was seen for baseline, day 3, and day 10 but not day 7 (Supplementary Figure 2B). We found a significant association between increased RXRa DNA methylation and increased hemoglobin levels (β = 0.54, p = 0.038, Supplementary Table 3). This general relationship was observed for each altitude, except the low-altitude baseline (Supplementary Figure 2A). We also identified associations approaching significance between PPARa methylation and hemoglobin, RXRa methylation and systolic blood pressure, body mass index, and EPO, and mean arterial pressure and EPO methylation (Supplementary Table 3). We did not find any significant associations between hemoglobin levels and EPAS1 or EPO methylation (data not shown).
Discussion
Oxygen homeostasis is an essential component of basic physiological homeostasis. In the mitochondria, oxygen is used to produce ATP in the process of oxidative phosphorylation (Semenza, 2007). HIF downregulates oxygen consumption by mitochondria and stimulates the glycolytic pathway enzymes (Semenza, 2010). In addition, the HIF pathway is involved in the regulation of fatty acid metabolism (Huang et al., 2014; Liu et al., 2014). Fatty acid oxidation is inhibited in hypoxic conditions as a result of the switch to glycolysis. This leads to accumulation of lipid droplets that have been shown to play a role in protection against ROS (Whitmer et al., 1978; Bensaad et al., 2014). Moreover, the HIF pathway is involved in increasing hemoglobin levels in response to decreased oxygen levels (Franke et al., 2013). In this study, we determined DNA methylation levels of four HIF pathway genes involved in oxygen homeostasis and metabolism, EPAS1, EPO, PPARa, and RXRa, and the marker of global methylation, LINE-1, to better understand how the epigenome responds to changes in ambient oxygen availability.
This is the first study to report changes in DNA methylation associated with an incremental ascent to high altitude in a cohort of European ancestry. Previous studies have shown that DNA methylation is affected by chronic exposure to hypoxia (Watson et al., 2010; Yuen et al., 2013; Brown and Rupert, 2014; Childebayeva et al., 2019). However, the effects of short-term hypoxic exposure on the epigenome have not been studied in the context of acclimatization to high altitude in nonacclimatized individuals.
We found decreased LINE-1 methylation, increased PPARa, and decreased RXRa methylation at high compared to low altitude. We also identified increased EPAS1 methylation at 4,240 m (seven days of ascent) and decreased EPO methylation at 4,240 m compared to 1,400m. We also found positive associations between RXRa methylation and hemoglobin and between EPO methylation and systolic blood pressure. These findings show that short-term exposure to high-altitude hypoxia can influence the epigenome, which may in turn influence gene expression and phenotype and thus contribute to high-altitude acclimatization.
LINE-1 is a repetitive element, and its methylation level is associated with the global genomic methylation level (Ogino et al., 2008a; Iwagami et al., 2012). The methylation status of LINE-1 has been used as a proxy for the status of the methylome upon exposure to toxicants and in cancer (Chalitchagorn et al., 2004; Kile et al., 2012). Decreased LINE-1 methylation has been shown in cancer and has been associated with genomic instability (Ogino et al., 2008b; Pattamadilok et al., 2008). We found lower LINE-1 methylation levels associated with high-altitude exposure in our cohort. This could be explained by the effect of ROS on the genome, since ROS production is higher in hypoxic conditions (Wongpaiboonwattana et al., 2013; Kloypan et al., 2015). This finding likely reflects the effects of hypoxia as a stressor on the genome.
EPAS1 is involved in activation of oxygen-regulated genes, plays a role in vascular remodeling (Peng et al., 2000), and is an important regulator of EPO, which controls erythropoiesis (Rankin et al., 2007). Importantly, EPAS1 contributes to high-altitude adaptation in Tibetans and shows altered methylation in Andeans (Beall et al., 2010; Childebayeva et al., 2019). We found increased methylation of EPAS1 associated with high altitude at 4,240m (day 7), potentially suggesting decreased EPAS1 expression. In comparison, high-altitude adapted Andeans show decreased methylation in hypoxic conditions compared to normoxia (Childebayeva et al., 2019). It is possible that the increase in EPAS1 methylation corresponds to the increase in EPAS1 hydroxymethylation. Hydroxymethylation of a CpG site is an intermediate stage in the demethylation pathway catalyzed by the ten-eleven translocation (TET) family enzymes (Tahiliani et al., 2009). The bisulfite conversion method we used does not differentiate between methylated and hydroxymethylated cytosines (Nestor et al., 2010). It is possible that EPAS1 is in the process of demethylation, which would be expected in hypoxic conditions, although we are seeing a general increase in DNA methylation in our participants.
EPO plays a major role in increased erythropoiesis under HIF control (Koury and Bondurant, 1990; Yoon et al., 2006; Risso et al., 2007). EPO levels rise quickly upon hypoxic exposure (Eckardt et al., 1989). In previous studies of high-altitude acclimatization, EPO has been shown to peak after 1 to 3 days at altitude, followed by a decline in an altitude-dependent manner (Abbrecht and Littell, 1972; Ge et al., 2002). EPO has a conserved HIF-1 binding site (HBS) CGTG in its 3′ UTR containing a CpG site (Wang and Semenza, 1993; Wang and Semenza, 1996; Wenger et al., 1998). Decreased methylation status of the HBS has been associated with activation of EPO in hypoxic conditions (Wenger et al., 1998). We found lower methylation upstream of the HBS site at all higher altitudes compared with 1,400m, and this was statistically significant at 4,240m. These findings are concordant with previously observed increased EPO expression levels at high altitude (Robach et al., 2004) and suggest that DNA methylation may play a role in this process.
We found a significant positive association between EPO methylation and systolic blood pressure suggesting that lower levels of erythropoietin may be correlated with higher blood pressure, since higher DNA methylation around this locus just upstream of an HBS has been linked to decreased expression of EPO based on previous research (Wenger et al., 1998). Other studies have shown a positive relationship between erythropoietin and human recombinant erythropoietin and blood pressure at rest and exercise in humans and in hypertensive and normotensive rats (Berglund and Ekblom, 1991; Raine and Roger, 1991; Muntzel et al., 1993). More research is necessary to establish if decreased EPO methylation of the locus targeted here is truly associated with higher levels of erythropoietin and if there is a link between EPO methylation and systolic blood pressure, especially since we did not find an association with hemoglobin. Of note, we observed a negative relationship between EPO methylation and systolic blood pressure at 4,240m (day 7), indicating a potential positive link between EPO expression and blood pressure at this altitude. This is the only altitude where we observed a significant change in methylation compared to the baseline. Further investigation is necessary to determine why the positive relationship between EPO expression and systolic blood pressure was identified only at 4,240 m of altitude (day 7).
PPARa is a transcription factor involved in controlling fatty acid metabolism and oxidation (Keller et al., 1993). PPARs activate the gene for acyl coenzyme A oxidase, which is the rate-limiting enzyme of the peroxisomal β-oxidation pathway (Dreyer et al., 1992; Keller et al., 1993). In addition to its role in fatty acid metabolism, PPARa is also associated with conditions such as obesity and diabetes, as well as various cardiovascular conditions including hypertension and atherosclerosis (Belanger et al., 2002). PPARa promotes fatty acid oxidation and may be involved in the switch from fatty acid oxidation to glucose oxidation via regulation of uncoupling protein 3 (Teruel et al., 2000; Gilde et al., 2003).
HIF transcription factors are known regulators of metabolism (Formenti et al., 2010). Previous studies in cell cultures have shown that PPARa is downregulated by HIF-1 in hypoxic conditions, which may be an adaptive response to hypoxia-induced inflammatory stimuli and metabolic changes (Narravula and Colgan, 2001). Another study of hypoxia exposure during an incremental ascent to the Everest Base Camp has found lower capacity for fatty acid oxidation in skeletal muscle and lower PPARa expression at altitude in the Himalayan Sherpa compared to lowlanders (Horscroft et al., 2017).
We found increased PPARa methylation associated with increasing altitude. The region we targeted is in the promoter region of PPARa suggesting that increased methylation here would be associated with a decrease in expression of PPARa, which is consistent with previous findings of decreased PPARa expression in hypoxic conditions (Narravula and Colgan, 2001). Decreased expression of PPARa is associated with diminished breakdown of fatty acids (Yoon, 2009). Lower levels of fatty acid oxidation are hypothesized to occur in hypoxic conditions due to the switch to anaerobic glycolysis (Ge et al., 2012).
RXRa is a transcription factor involved in fat metabolism and intracellular receptor signaling. RXRa binds to PPARa forming an active transcriptional complex able to bind to target genes known as proliferator-responsive elements (Dreyer et al., 1993). Several studies have shown that the activity of the PPARa/RXRa complex is reduced in hypoxic conditions to enable suppression of fatty acid metabolism (Huss et al., 2001; Belanger et al., 2007). The RXRa pathway was altered by hypobaric hypoxia exposure in the rat brain (Sethy et al., 2011). RXRs play a protective role in H9c2 cardiomyocytes from hypoxia/reoxygenation–induced oxidative injury in rats (Shan et al., 2014). We found decreased methylation of the CpG island located in the promoter region of RXRa, which may be associated with increased expression of RXRa.
Interestingly, we observed opposite trends in PPARa and RXRa methylation change. For example, individuals with increased PPARa methylation at high altitude have decreased RXRa methylation (Supplementary Figures 1D, E). Interestingly, individuals with decreased PPARa methylation (IDs 7 and 17) at high altitude have increased RXRa methylation (IDs 7 and 17), further highlighting the interactive nature of PPARa/RXRa. Since we see opposite change in PPARa and RXRa methylation levels, it is unclear whether it indicates increased or decreased activity of the PPARa/RXRa complex.
We found a significant positive association between RXRa methylation and hemoglobin levels. RXRa is a member of the retinoic acid receptor family and is necessary for normal hematopoiesis during development (Melnick and Licht, 1999; Oren et al., 2003). Retinoic acid signaling, specifically retinoic acid receptor α, is also involved in adult hematopoiesis (Canete et al., 2017). RXRa has been shown to play a role in hematopoietic signaling in mice (Ricote et al., 2006). However, the role of RXRa in this process is not well understood in adult humans. Our data suggest that there is a relationship between RXRa and hemoglobin levels. Previous studies have shown that PPARa is associated with hemoglobin levels, which could explain why we see a significant association with RXRa as well, since PPARa and RXRa are known to interact (Simonson et al., 2010; Scheinfeldt et al., 2012).
One of the limitations of our study is the use of saliva as a source of DNA. Saliva comprised white blood cells and epithelial buccal cells. Due to differences in DNA methylation signatures between tissues, any changes in methylation associated with altitude exposure may be confounded by cell type composition differences between altitudes (Lokk et al., 2014). We did not quantify saliva cell types at each altitude and thus were unable to control for this limitation. Furthermore, we only assessed the methylation levels of saliva. Therefore, it is unclear how other tissues responded to hypoxic exposure. However, there are known correlations between saliva and blood. Studies of genome-wide DNA methylation have reported 88.5% to 96.7% Pearson correlation between blood and saliva CpG sites within an individual (Thompson et al., 2013; Smith et al., 2015a). Saliva buccal epithelial cell methylation is also similar to the methylation patters of the brain due to the same ectodermal developmental origin and thus may serve as a proxy for DNA methylation changes in the brain (Smith et al., 2015b). Methylation levels of certain CpG sites are known to be tissue-specific, while methylation levels of other CpG sites correlate between tissue types (Varley et al., 2013). To our knowledge, the genes and loci we chose to study have not been shown to be tissue-specific in terms of methylation levels.
Lastly, we did not collect gene expression data from our participants and thus have not been able to directly link changes in DNA methylation to gene expression.
Overall, we found that short-term exposure to high-altitude hypoxia can affect the epigenome. We observed changes in LINE-1 and hypoxia-pathway associated genes EPAS1, EPO, PPARa, and RXRa. We also found significant associations between DNA methylation of EPO and RXRa and systolic blood pressure and hemoglobin, respectively. Our findings contribute to the growing literature on the role of epigenetics in acclimatization to high altitude. Future studies of the genome-wide effects of hypoxia on epigenetics are necessary to better understand the extent of DNA methylation change upon high-altitude exposure.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Ethics Statement
This study abided by the Canadian Government Tri-Council policy on research ethics with human participants (TCPS2) and the Declaration of Helsinki, except for registration in a data base. Ethical approval was received in advance through Mount Royal University Human Research Ethics Board (Protocol 100012 and 101361), the Syracuse University Institutional Review Board (Protocol 18-006), the University of Michigan Institutional Review Board (HUM00141118) and harmonized with the Nepal Health Research Council (Protocol 109-2017). All participants were recruited via verbal communication and provided written and informed consent prior to voluntary participation in the study.
Author Contributions
TB, AC, AB, TD, DD, and JG conceived and designed the research. AC, TH, and JW performed experiments. AC and TH analyzed data. TB, AC, and TH wrote the manuscript with contributions from all authors.
Funding
This work was supported by the University of Michigan (to AC) and the Michigan Lifestage Environmental Exposures and Disease (M-LEEaD) National Institute of Environmental Health Sciences Core Center (P30 ES017885 to DD). AC was supported by the Marshall Weinberg Award and the National Geographic Early Career Award (EC-50834R-18). TD was supported by a Natural Sciences and Engineering Research Council of Canada Discovery grant (RGPIN-2016-04915).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge our study participants from Syracuse University in Syracuse, NY, USA; Mount Royal University in Calgary, AB, Canada; and the MidSweden University in Sweden. This work would have not been possible without our Sherpa guides in Nepal: Nima Sherpa, Tsering Sherpa, Tashi Jangbu, and Fura Tashi Chuserwa, and their porter team. We would like to thank Mackenzie Bronson for her help with sample collection in Gorak Shep and Drs. Mark White and Kerby Shedden for statistical support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.01062/full#supplementary-material
References
- Abbrecht P. H., Littell J. K. (1972). Plasma erythropoietin in men and mice during acclimatization to different altitudes. J. Appl. Physiol. 32, 54–58. 10.1152/jappl.1972.32.1.54 [DOI] [PubMed] [Google Scholar]
- Alkorta-Aranburu G., Beall C. M., Witonsky D. B., Gebremedhin A., Pritchard J. K., Di Rienzo A. (2012). The genetic architecture of adaptations to high altitude in Ethiopia. PLoS Genet. 8, e1003110. 10.1371/journal.pgen.1003110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D., Machler M., Bolker B. M., Walker S. C. (2015). Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Software 67, 1–48. 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Baxter E., Windloch K., Gannon F., Lee J. S. (2014). Epigenetic regulation in cancer progression. Cell Biosci. 4, 45. 10.1186/2045-3701-4-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall C. M., Cavalleri G. L., Deng L., Elston R. C., Gao Y., Knight J., et al. (2010). Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl. Acad. Sci. U S A 107, 11459–11464. 10.1073/pnas.1002443107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger A. J., Lu H., Date T., Liu L. X., Vincent K. A., Akita G. Y., et al. (2002). Hypoxia up-regulates expression of peroxisome proliferator-activated receptor gamma angiopoietin-related gene (PGAR) in cardiomyocytes: role of hypoxia inducible factor 1alpha. J. Mol. Cell Cardiol. 34, 765–774. 10.1006/jmcc.2002.2021 [DOI] [PubMed] [Google Scholar]
- Belanger A. J., Luo Z., Vincent K. A., Akita G. Y., Cheng S. H., Gregory R. J., et al. (2007). Hypoxia-inducible factor 1 mediates hypoxia-induced cardiomyocyte lipid accumulation by reducing the DNA binding activity of peroxisome proliferator-activated receptor alpha/retinoid X receptor. Biochem. Biophys. Res. Commun. 364, 567–572. 10.1016/j.bbrc.2007.10.062 [DOI] [PubMed] [Google Scholar]
- Bensaad K., Favaro E., Lewis C. A., Peck B., Lord S., Collins J. M., et al. (2014). Fatty acid uptake and lipid storage induced by HIF-1alpha contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 9, 349–365. 10.1016/j.celrep.2014.08.056 [DOI] [PubMed] [Google Scholar]
- Berglund B., Ekblom B. (1991). Effect of recombinant human erythropoietin treatment on blood pressure and some haematological parameters in healthy men. J. Intern. Med. 229, 125–130. 10.1111/j.1365-2796.1991.tb00319.x [DOI] [PubMed] [Google Scholar]
- Bollati V., Baccarelli A. (2010). Environmental epigenetics. Heredity (Edinb) 105, 105–112. 10.1038/hdy.2010.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. J., Rupert J. L. (2014). Hypoxia and environmental epigenetics. High Alt. Med. Biol. 15, 323–330. 10.1089/ham.2014.1016 [DOI] [PubMed] [Google Scholar]
- Canete A., Cano E., Munoz-Chapuli R., Carmona R. (2017). Role of vitamin a/retinoic acid in regulation of embryonic and adult hematopoiesis. Nutrients 9, 1–18. 10.3390/nu9020159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalitchagorn K., Shuangshoti S., Hourpai N., Kongruttanachok N., Tangkijvanich P., Thong-Ngam D., et al. (2004). Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene 23, 8841–8846. 10.1038/sj.onc.1208137 [DOI] [PubMed] [Google Scholar]
- Chan L. S., Wells R. A. (2009). Cross-Talk between PPARs and the Partners of RXR: A Molecular Perspective. PPAR Res. 2009, 925309. 10.1155/2009/925309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childebayeva A., Jones T. R., Goodrich J. M., Leon-Velarde F., Rivera-Chira M., Kiyamu M., et al. (2019). LINE-1 and EPAS1 DNA methylation associations with high-altitude exposure. Epigenetics 14, 1–15. 10.1080/15592294.2018.1561117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinetti G., Fruchart J. C., Staels B. (2000). Peroxisome proliferator-activated receptors (PPARs): nuclear receptors at the crossroads between lipid metabolism and inflammation. Inflamm. Res. 49, 497–505. 10.1007/s000110050622 [DOI] [PubMed] [Google Scholar]
- Cortese R., Zhang C., Bao R., Andrade J., Khalyfa A., Mokhlesi B., et al. (2016). DNA methylation profiling of blood monocytes in patients with obesity hypoventilation syndrome: effect of positive airway pressure treatment. Chest 150, 91–101. 10.1016/j.chest.2016.02.648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame C., Fahnenstich H., Freitag P., Hofmann D., Abdul-Nour T., Bartmann P., et al. (1998). Erythropoietin mRNA expression in human fetal and neonatal tissue. Blood 92, 3218–3225. 10.1182/blood.V92.9.3218 [DOI] [PubMed] [Google Scholar]
- Dolinoy D. C. (2007). Epigenetic gene regulation: early environmental exposures. Pharmacogenomics 8, 5–10. 10.2217/14622416.8.1.5 [DOI] [PubMed] [Google Scholar]
- Dolinoy D. C., Jirtle R. L. (2008). Environmental epigenomics in human health and disease. Environ. Mol. Mutagen. 49, 4–8. 10.1002/em.20366 [DOI] [PubMed] [Google Scholar]
- Dreyer C., Keller H., Mahfoudi A., Laudet V., Krey G., Wahli W. (1993). Positive regulation of the peroxisomal beta-oxidation pathway by fatty acids through activation of peroxisome proliferator-activated receptors (PPAR). Biol. Cell. 77, 67–76. 10.1016/S0248-4900(05)80176-5 [DOI] [PubMed] [Google Scholar]
- Dreyer C., Krey G., Keller H., Givel F., Helftenbein G., Wahli W. (1992). Control of the peroxisomal beta-oxidation pathway by a novel family of nuclear hormone receptors. Cell 68, 879–887. 10.1016/0092-8674(92)90031-7 [DOI] [PubMed] [Google Scholar]
- Eckardt K. U., Boutellier U., Kurtz A., Schopen M., Koller E. A., Bauer C. (1989). Rate of erythropoietin formation in humans in response to acute hypobaric hypoxia. J. Appl. Physiol. 66 (1985), 1785–1788. 10.1152/jappl.1989.66.4.1785 [DOI] [PubMed]
- Eckardt K. U., Ratcliffe P. J., Tan C. C., Bauer C., Kurtz A. (1992). Age-dependent expression of the erythropoietin gene in rat liver and kidneys. J. Clin. Invest. 66 (89), 753–760. 10.1172/JCI115652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein A. C., Gleadle J. M., Mcneill L. A., Hewitson K. S., O’rourke J., Mole D. R., et al. (2001). C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54. 10.1016/S0092-8674(01)00507-4 [DOI] [PubMed] [Google Scholar]
- Faulk C., Barks A., Sanchez B. N., Zhang Z., Anderson O. S., Peterson K. E., et al. (2014). Perinatal lead (Pb) exposure results in sex-specific effects on food intake, fat, weight, and insulin response across the murine life-course. PLoS One 9, e104273. 10.1371/journal.pone.0104273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R., Fraga M. F. (2011). Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet. 13, 97–109. 10.1038/nrg3142 [DOI] [PubMed] [Google Scholar]
- Formenti F., Constantin-Teodosiu D., Emmanuel Y., Cheeseman J., Dorrington K. L., Edwards L. M., et al. (2010). Regulation of human metabolism by hypoxia-inducible factor. Proc. Natl. Acad. Sci. U S A 107, 12722–12727. 10.1073/pnas.1002339107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke K., Gassmann M., Wielockx B. (2013). Erythrocytosis: the HIF pathway in control. Blood 122, 1122–1128. 10.1182/blood-2013-01-478065 [DOI] [PubMed] [Google Scholar]
- Ge R. L., Simonson T. S., Cooksey R. C., Tanna U., Qin G., Huff C. D., et al. (2012). Metabolic insight into mechanisms of high-altitude adaptation in Tibetans. Mol. Genet Metab. 106, 244–247. 10.1016/j.ymgme.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge R. L., Witkowski S., Zhang Y., Alfrey C., Sivieri M., Karlsen T., et al. (2002). Determinants of erythropoietin release in response to short-term hypobaric hypoxia. J. Appl. Physiol. 92 (1985), 2361–2367. 10.1152/japplphysiol.00684.2001 [DOI] [PubMed]
- Gilde A. J., Van Der Lee K. A., Willemsen P. H., Chinetti G., Van Der Leij F. R., Van Der Vusse G. J., et al. (2003). Peroxisome proliferator-activated receptor (PPAR) alpha and PPARbeta/delta, but not PPARgamma, modulate the expression of genes involved in cardiac lipid metabolism. Circ. Res. 92, 518–524. 10.1161/01.RES.0000060700.55247.7C [DOI] [PubMed] [Google Scholar]
- Haase V. H. (2013). Regulation of erythropoiesis by hypoxia-inducible factors. Blood Rev. 27, 41–53. 10.1016/j.blre.2012.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez D. G., Nalls M. A., Gibbs J. R., Arepalli S., Van Der Brug M., Chong S., et al. (2011). Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum. Mol. Genet. 20, 1164–1172. 10.1093/hmg/ddq561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocking T. D. (2018). “Package ‘directlabels’”. (R Core Team).
- Horscroft J. A., Kotwica A. O., Laner V., West J. A., Hennis P. J., Levett D. Z. H., et al. (2017). Metabolic basis to Sherpa altitude adaptation. Proc. Natl. Acad. Sci. U. S. A. 114, 6382–6387. 10.1073/pnas.1700527114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. (2013). DNA methylation age of human tissues and cell types. Genome Biol. 14, R115. 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston C. S., Riley R. L. (1947). Respiratory and circulatory changes during acclimatization to high altitude. Am. J. Physiol. 149, 565–588. 10.1152/ajplegacy.1947.149.3.565 [DOI] [PubMed] [Google Scholar]
- Huang Li, Li X., Zhang L., Sun L., He X., Zhong X., et al. (2014). HIF-1-mediated suppression of acyl-CoA dehydrogenases and fatty acid oxidation is critical for cancer progression. Cell Rep. 8, 1930–1942. 10.1016/j.celrep.2014.08.028 [DOI] [PubMed] [Google Scholar]
- Huey K. A., Powell F. L. (2000). Time-dependent changes in dopamine D(2)-receptor mRNA in the arterial chemoreflex pathway with chronic hypoxia. Brain Res. Mol. Brain Res. 75, 264–270. 10.1016/S0169-328X(99)00321-6 [DOI] [PubMed] [Google Scholar]
- Huss J. M., Levy F. H., Kelly D. P. (2001). Hypoxia inhibits the peroxisome proliferator-activated receptor alpha/retinoid X receptor gene regulatory pathway in cardiac myocytes: a mechanism for O2-dependent modulation of mitochondrial fatty acid oxidation. J. Biol. Chem. 276, 27605–27612. 10.1074/jbc.M100277200 [DOI] [PubMed] [Google Scholar]
- Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., et al. (2001). HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468. 10.1126/science.1059817 [DOI] [PubMed] [Google Scholar]
- Iwagami S., Baba Y., Watanabe M., Shigaki H., Miyake K., Ida S., et al. (2012). Pyrosequencing assay to measure LINE-1 methylation level in esophageal squamous cell carcinoma. Ann. Surg Oncol. 19, 2726–2732. 10.1245/s10434-011-2176-3 [DOI] [PubMed] [Google Scholar]
- James S. J., Cutler P., Melnyk S., Jernigan S., Janak L., Gaylor D. W., et al. (2004). Metabolic biomarkers of increased oxidative stress and impaired methylation capacity in children with autism. Am. J. Clin. Nutr. 80, 1611–1617. 10.1093/ajcn/80.6.1611 [DOI] [PubMed] [Google Scholar]
- Julian C., Subudhi A., Evero O., Pedersen B., Dvorkin D., Lovering A., et al. (2014). Epigenetic modification of gene expression during human acclimatization to hypobaric hypoxia (885.4). FASEB J. 28, 885.884. 10.1096/fasebj.28.1_supplement.885.4 [DOI] [Google Scholar]
- Keller H., Dreyer C., Medin J., Mahfoudi A., Ozato K., Wahli W. (1993). Fatty acids and retinoids control lipid metabolism through activation of peroxisome proliferator-activated receptor-retinoid X receptor heterodimers. Proc. Natl. Acad. Sci. U S A 90, 2160–2164. 10.1073/pnas.90.6.2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile M. L., Baccarelli A., Hoffman E., Tarantini L., Quamruzzaman Q., Rahman M., et al. (2012). Prenatal arsenic exposure and DNA methylation in maternal and umbilical cord blood leukocytes. Environ. Health Perspect. 120, 1061–1066. 10.1289/ehp.1104173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R. J., Bird A. P. (2006). Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 31, 89–97. 10.1016/j.tibs.2005.12.008 [DOI] [PubMed] [Google Scholar]
- Kloypan C., Srisa-Art M., Mutirangura A., Boonla C. (2015). LINE-1 hypomethylation induced by reactive oxygen species is mediated via depletion of S-adenosylmethionine. Cell Biochem. Funct. 33, 375–385. 10.1002/cbf.3124 [DOI] [PubMed] [Google Scholar]
- Kourembanas S., Hannan R. L., Faller D. V. (1990). Oxygen tension regulates the expression of the platelet-derived growth factor-B chain gene in human endothelial cells. J. Clin. Invest. 86, 670–674. 10.1172/JCI114759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koury M. J., Bondurant M. C. (1990). Erythropoietin retards DNA breakdown and prevents programmed death in erythroid progenitor cells. Science 248, 378–381. 10.1126/science.2326648 [DOI] [PubMed] [Google Scholar]
- Kuznetsova A., Brockhoff P. B., Christensen R. H. B. (2017). lmerTest Package: Tests in Linear Mixed Effects Models. J. Stat. Software 82, 1–26. 10.18637/jss.v082.i13 [DOI] [Google Scholar]
- Lachance G., Uniacke J., Audas T. E., Holterman C. E., Franovic A., Payette J., et al. (2014). DNMT3a epigenetic program regulates the HIF-2alpha oxygen-sensing pathway and the cellular response to hypoxia. Proc. Natl. Acad. Sci. U S A 111, 7783–7788. 10.1073/pnas.1322909111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam L. L., Emberly E., Fraser H. B., Neumann S. M., Chen E., Miller G. E., et al. (2012). Factors underlying variable DNA methylation in a human community cohort. Proc. Natl. Acad. Sci. U S A 109 Suppl 2, 17253–17260. 10.1073/pnas.1121249109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Morgan M., Hutchison K., Calhoun V. D. (2010). A study of the influence of sex on genome wide methylation. PLoS One 5, e10028. 10.1371/journal.pone.0010028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Ma Z., Zhao C., Wang Y., Wu G., Xiao J., et al. (2014). HIF-1alpha and HIF-2alpha are critically involved in hypoxia-induced lipid accumulation in hepatocytes through reducing PGC-1alpha-mediated fatty acid beta-oxidation. Toxicol. Lett. 226, 117–123. 10.1016/j.toxlet.2014.01.033 [DOI] [PubMed] [Google Scholar]
- Lokk K., Modhukur V., Rajashekar B., Martens K., Magi R., Kolde R., et al. (2014). DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome Biol. 15, r54. 10.1186/gb-2014-15-4-r54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick A., Licht J. D. (1999). Deconstructing a disease: RARalpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood 93, 3167–3215. 10.1182/blood.V93.10.3167.410k44_3167_3215 [DOI] [PubMed] [Google Scholar]
- Mohn F., Schubeler D. (2009). Genetics and epigenetics: stability and plasticity during cellular differentiation. Trends Genet. 25, 129–136. 10.1016/j.tig.2008.12.005 [DOI] [PubMed] [Google Scholar]
- Moore L. G. (2001). Human genetic adaptation to high altitude. High Alt. Med. Biol. 2, 257–279. 10.1089/152702901750265341 [DOI] [PubMed] [Google Scholar]
- Muntzel M., Hannedouche T., Lacour B., Drueke T. B. (1993). Erythropoietin increases blood pressure in normotensive and hypertensive rats. Nephron 65, 601–604. 10.1159/000187571 [DOI] [PubMed] [Google Scholar]
- Narravula S., Colgan S. P. (2001). Hypoxia-inducible factor 1-mediated inhibition of peroxisome proliferator-activated receptor alpha expression during hypoxia. J. Immunol. 166, 7543–7548. 10.4049/jimmunol.166.12.7543 [DOI] [PubMed] [Google Scholar]
- Nestor C., Ruzov A., Meehan R., Dunican D. (2010). Enzymatic approaches and bisulfite sequencing cannot distinguish between 5-methylcytosine and 5-hydroxymethylcytosine in DNA. Biotechniques 48, 317–319. 10.2144/000113403 [DOI] [PubMed] [Google Scholar]
- Niu Y., Desmarais T. L., Tong Z., Yao Y., Costa M. (2015). Oxidative stress alters global histone modification and DNA methylation. Free Radic. Biol. Med. 82, 22–28. 10.1016/j.freeradbiomed.2015.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Non A. L., Hollister B. M., Humphreys K. L., Childebayeva A., Esteves K., Zeanah C. H., et al. (2016). DNA methylation at stress-related genes is associated with exposure to early life institutionalization. Am. J. Phys. Anthropol. 161, 84–93. 10.1002/ajpa.23010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S., Kawasaki T., Nosho K., Ohnishi M., Suemoto Y., Kirkner G. J., et al. (2008. a). LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int. J. Cancer 122, 2767–2773. 10.1002/ijc.23470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S., Nosho K., Kirkner G. J., Kawasaki T., Chan A. T., Schernhammer E. S., et al. (2008. b). A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J. Natl. Cancer Inst. 100, 1734–1738. 10.1093/jnci/djn359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohh M., Park C. W., Ivan M., Hoffman M. A., Kim T. Y., Huang L. E., et al. (2000). Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat. Cell Biol. 2, 423–427. 10.1038/35017054 [DOI] [PubMed] [Google Scholar]
- Oren T., Sher J. A., Evans T. (2003). Hematopoiesis and retinoids: development and disease. Leuk. Lymphoma 44, 1881–1891. 10.1080/1042819031000116661 [DOI] [PubMed] [Google Scholar]
- Pattamadilok J., Huapai N., Rattanatanyong P., Vasurattana A., Triratanachat S., Tresukosol D., et al. (2008). LINE-1 hypomethylation level as a potential prognostic factor for epithelial ovarian cancer. Int. J. Gynecol. Cancer 18, 711–717. 10.1111/j.1525-1438.2007.01117.x [DOI] [PubMed] [Google Scholar]
- Peng J., Zhang L., Drysdale L., Fong G. H. (2000). The transcription factor EPAS-1/hypoxia-inducible factor 2alpha plays an important role in vascular remodeling. Proc. Natl. Acad. Sci. U S A 97, 8386–8391. 10.1073/pnas.140087397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinque D., Kittler R., Kayser M., Stoneking M., Nasidze I. (2006). Evaluation of saliva as a source of human DNA for population and association studies. Anal. Biochem. 353, 272–277. 10.1016/j.ab.2006.03.021 [DOI] [PubMed] [Google Scholar]
- Raine A. E., Roger S. D. (1991). Effects of erythropoietin on blood pressure. Am. J. Kidney Dis. 18, 76–83. [PubMed] [Google Scholar]
- Rankin E. B., Biju M. P., Liu Q., Unger T. L., Rha J., Johnson R. S., et al. (2007). Hypoxia-inducible factor-2 (HIF-2) regulates hepatic erythropoietin in vivo. J. Clin. Invest. 117, 1068–1077. 10.1172/JCI30117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricote M., Snyder C. S., Leung H. Y., Chen J., Chien K. R., Glass C. K. (2006). Normal hematopoiesis after conditional targeting of RXRalpha in murine hematopoietic stem/progenitor cells. J. Leukoc. Biol. 80, 850–861. 10.1189/jlb.0206097 [DOI] [PubMed] [Google Scholar]
- Risso A., Turello M., Biffoni F., Antonutto G. (2007). Red blood cell senescence and neocytolysis in humans after high altitude acclimatization. Blood Cells Mol. Dis. 38, 83–92. 10.1016/j.bcmd.2006.10.161 [DOI] [PubMed] [Google Scholar]
- Roach Rc, Bartsch P. P.H., Hackett O. O. (1993). The Lake Louise Acute Moutain Sickness Scoring System. Burlington, VT: Queen City Press. [Google Scholar]
- Robach P., Fulla Y., Westerterp K. R., Richalet J. P. (2004). Comparative response of EPO and soluble transferrin receptor at high altitude. Med. Sci. Sports Exerc. 36, 1493–1498. 10.1249/01.MSS.0000139889.56481.E0 [DOI] [PubMed] [Google Scholar]
- Rossler J., Stolze I., Frede S., Freitag P., Schweigerer L., Havers W., et al. (2004). Hypoxia-induced erythropoietin expression in human neuroblastoma requires a methylation free HIF-1 binding site. J. Cell Biochem. 93, 153–161. 10.1002/jcb.20133 [DOI] [PubMed] [Google Scholar]
- Sarkar S., Banerjee P. K., Selvamurthy W. (2003). High altitude hypoxia: an intricate interplay of oxygen responsive macroevents and micromolecules. Mol. Cell Biochem. 253, 287–305. 10.1023/A:1026080320034 [DOI] [PubMed] [Google Scholar]
- Scheinfeldt L. B., Soi S., Thompson S., Ranciaro A., Woldemeskel D., Beggs W., et al. (2012). Genetic adaptation to high altitude in the Ethiopian highlands. Genome Biol. 13, R1. 10.1186/gb-2012-13-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza G. L. (2007). Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem. J. 405, 1–9. 10.1042/BJ20070389 [DOI] [PubMed] [Google Scholar]
- Semenza G. L. (2010). Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene 29, 625–634. 10.1038/onc.2009.441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senut M. C., Cingolani P., Sen A., Kruger A., Shaik A., Hirsch H., et al. (2012). Epigenetics of early-life lead exposure and effects on brain development. Epigenomics 4, 665–674. 10.2217/epi.12.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethy N. K., Singh M., Kumar R., Ilavazhagan G., Bhargava K. (2011). Upregulation of transcription factor NRF2-mediated oxidative stress response pathway in rat brain under short-term chronic hypobaric hypoxia. Funct. Integr. Genomics 11, 119–137. 10.1007/s10142-010-0195-y [DOI] [PubMed] [Google Scholar]
- Shahrzad S., Bertrand K., Minhas K., Coomber B. L. (2007). Induction of DNA hypomethylation by tumor hypoxia. Epigenetics 2, 119–125. 10.4161/epi.2.2.4613 [DOI] [PubMed] [Google Scholar]
- Shan P. R., Xu W. W., Huang Z. Q., Pu J., Huang W. J. (2014). Protective role of retinoid X receptor in H9c2 cardiomyocytes from hypoxia/reoxygenation injury in rats. World J. Emerg. Med. 5, 122–127. 10.5847/wjem.j.issn.1920-8642.2014.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonson T. S., Yang Y., Huff C. D., Yun H., Qin G., Witherspoon D. J., et al. (2010). Genetic evidence for high-altitude adaptation in Tibet. Science 329, 72–75. 10.1126/science.1189406 [DOI] [PubMed] [Google Scholar]
- Smith A. K., Kilaru V., Klengel T., Mercer K. B., Bradley B., Conneely K. N., et al. (2015. a). DNA extracted from saliva for methylation studies of psychiatric traits: evidence tissue specificity and relatedness to brain. Am. J. Med. Genet. Neuropsychiatr. Genet. 168, 36–44. 10.1002/ajmg.b.32278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. K., Kilaru V., Klengel T., Mercer K. B., Bradley B., Conneely K. N. et al. (2015. b). DNA extracted from saliva for methylation studies of psychiatric traits: evidence tissue specificity and relatedness to brain. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 168B, 36–44. 10.1002/ajmg.b.32278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M., Koh K. P., Shen Y., Pastor W. A., Bandukwala H., Brudno Y., et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935. 10.1126/science.1170116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teruel T., Smith S. A., Peterson J., Clapham J. C. (2000). Synergistic activation of UCP-3 expression in cultured fetal rat brown adipocytes by PPARalpha and PPARgamma ligands. Biochem. Biophys. Res. Commun. 273, 560–564. 10.1006/bbrc.2000.2982 [DOI] [PubMed] [Google Scholar]
- Thompson T. M., Sharfi D., Lee M., Yrigollen C. M., Naumova O. Y., Grigorenko E. L. (2013). Comparison of whole-genome DNA methylation patterns in whole blood, saliva, and lymphoblastoid cell lines. Behav. Genet. 43, 168–176. 10.1007/s10519-012-9579-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley K. E., Gertz J., Bowling K. M., Parker S. L., Reddy T. E., Pauli-Behn F., et al. (2013). Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 23, 555–567. 10.1101/gr.147942.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. L., Semenza G. L. (1993). General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc. Natl. Acad. Sci. U S A 90, 4304–4308. 10.1073/pnas.90.9.4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. L., Semenza G. L. (1996). Molecular basis of hypoxia-induced erythropoietin expression. Curr. Opin. Hematol. 3, 156–162. 10.1097/00062752-199603020-00009 [DOI] [PubMed] [Google Scholar]
- Ward M., Milledge J. S., West J. B. (2000). High altitude medicine and physiology. London: Arnold. [Google Scholar]
- Watson J. A., Watson C. J., Mccann A., Baugh J. (2010). Epigenetics, the epicenter of the hypoxic response. Epigenetics 5, 293–296. 10.4161/epi.5.4.11684 [DOI] [PubMed] [Google Scholar]
- Wenger R. H., Kvietikova I., Rolfs A., Camenisch G., Gassmann M. (1998). Oxygen-regulated erythropoietin gene expression is dependent on a CpG methylation-free hypoxia-inducible factor-1 DNA-binding site. Eur. J. Biochem. 253, 771–777. 10.1046/j.1432-1327.1998.2530771.x [DOI] [PubMed] [Google Scholar]
- Whitmer J. T., Idell-Wenger J. A., Rovetto M. J., Neely J. R. (1978). Control of fatty acid metabolism in ischemic and hypoxic hearts. J. Biol. Chem. 253, 4305–4309. [PubMed] [Google Scholar]
- Wickham H. (2009). ggplot2: Elegant Graphics for Data Analysis. Elegant Graphics for Data Analysis. New York, NY: Springer. 1–212. 10.1007/978-0-387-98141-3_1 [DOI]
- Wiener C. M., Booth G., Semenza G. L. (1996). In vivo expression of mRNAs encoding hypoxia-inducible factor 1. Biochem. Biophys. Res. Commun. 225, 485–488. 10.1006/bbrc.1996.1199 [DOI] [PubMed] [Google Scholar]
- Wolffe A. P., Guschin D. (2000). Review: chromatin structural features and targets that regulate transcription. J. Struct. Biol. 129, 102–122. 10.1006/jsbi.2000.4217 [DOI] [PubMed] [Google Scholar]
- Wongpaiboonwattana W., Tosukhowong P., Dissayabutra T., Mutirangura A., Boonla C. (2013). Oxidative stress induces hypomethylation of LINE-1 and hypermethylation of the RUNX3 promoter in a bladder cancer cell line. Asian Pac. J. Cancer Prev. 14, 3773–3778. 10.7314/APJCP.2013.14.6.3773 [DOI] [PubMed] [Google Scholar]
- Yoon D., Pastore Y. D., Divoky V., Liu E., Mlodnicka A. E., Rainey K., et al. (2006). Hypoxia-inducible factor-1 deficiency results in dysregulated erythropoiesis signaling and iron homeostasis in mouse development. J. Biol. Chem. 281, 25703–25711. 10.1074/jbc.M602329200 [DOI] [PubMed] [Google Scholar]
- Yoon M. (2009). The role of PPARalpha in lipid metabolism and obesity: focusing on the effects of estrogen on PPARalpha actions. Pharmacol. Res. 60, 151–159. 10.1016/j.phrs.2009.02.004 [DOI] [PubMed] [Google Scholar]
- Yuen R. K., Chen B., Blair J. D., Robinson W. P., Nelson D. M. (2013). Hypoxia alters the epigenetic profile in cultured human placental trophoblasts. Epigenetics 8, 192–202. 10.4161/epi.23400 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.