Abstract
Obesity is understood to be an inflammatory condition characterized in part by changes in resident immune cell populations in adipose tissue. However, much of this knowledge has been obtained through experimental animal models. Epigenetic mechanisms, such as DNA methylation may be useful tools for characterizing the changes in immune cell populations in human subjects. In this study, we introduce a simple and intuitive method for assessing cellular infiltration by blood into other heterogeneous, admixed tissues such as adipose tissue, and apply this approach in a large human cohort study. Associations between higher leukocyte infiltration, measured by evaluating a distance measure between the methylation signatures of leukocytes and adipose tissue, and increasing body mass index (BMI) or android fat mass (AFM) were identified and validated in independent replication samples for CD4 (pBMI=0.009, pAFM=0.020), monocytes (pBMI=0.001, pAFM=4.3×10−4) and dendritic cells (pBMI=0.571, pAFM=0.012). Patterns of depletion with increasing adiposity were observed for plasma B (pBMI=0.430, pAFM=0.004) and immature B (pBMI=0.022, pAFM=0.042) cells. CD4, dendritic, monocytes, immature B, and plasma B cells may be important agents in the inflammatory process. Finally, the method used to assess leukocyte infiltration in this study is straightforwardly extended to other cell types and tissues in which infiltration might be of interest.
Keywords: epigenetics, DNA methylation, adipose tissue, methylation signatures, leukocyte infiltration, cellular admixture
INTRODUCTION
Estimating the distribution of leukocyte populations in complex, heterogeneous tissue has been an active area of methodological development in the molecular study of human inflammatory diseases. Epigenetic patterns, and more specifically, DNA methylation (DNAm), have been shown to impact gene expression and cellular differentiation and function. Prior studies have identified several differentially methylated DNA regions (DMRs) for most major types of leukocytes. Building on this work, methods for deconvoluting cellular mixtures have been developed (Houseman et al., 2012), including a reference-free approach (Houseman, Molitor, & Marsit, 2014) which permits admixture estimation of different cell-types using all available methylation markers. Deconvolution estimates may then be used to regress out the effects of the leukocyte subpopulations in the primary tissue of interest. However, the adjustment process requires a large number of degrees of freedom which may present challenges with respect to power in studies with small sample sizes and artificially separate portions of signals which are inherently joint in nature. Motivated by a biological interest in the infiltrative effects of leukocytes in obesity, rather than seeking to directly deconvolve certain cellular populations in a heterogenous tissue sample, we instead explore the effect of particular cell-types conditional on their presence in adipose tissue vis-à-vis degree of similarity in methylation patterns between leukocyte DMRs and methylation levels in adipose tissue samples.
Obesity, now itself considered to be an inflammatory condition (Das, 2001) is often characterized by chronic, low-grade inflammation that is sustained on both a systemic and tissue level (Oishi & Manabe, 2016). Obesity-induced inflammation in adipose tissue has been shown to promote insulin resistance (McNelis & Olefsky, 2014), and is thought to be mediated mostly by adipose tissue macrophage accumulation in fat (Weisberg et al., 2003; H. Xu et al., 2003). Macrophage populations are understood to dynamically shift during obesity, where anti-inflammatory M2 macrophages commonly present in lean adipose tissue are replaced with pro-inflammatory M1 macrophages with increasing adiposity (Castoldi, Naffah de Souza, Câmara, & Moraes-Vieira, 2016; Hill, Reid Bolus, & Hasty, 2014; Lumeng, Bodzin, & Saltiel, 2007; Odegaard et al., 2007). However, reports from a study by Xu et al. (X. Xu et al., 2013) demonstrated no difference in transcriptional levels of inflammatory cytokines in M1 versus M2 macrophages in obese mice, suggesting that increased inflammatory cytokines in obese adipose tissue could not be explained solely by the accumulation of pro-inflammatory macrophages. Further characterization of the heterogeneous composition of adipose tissue beyond the M1-versus-M2 paradigm, particularly the role of infiltrating immune cells, is necessary to understand adipose inflammation (Carvalheira, Qiu, & Chawla, 2013; Grant & Dixit, 2015; Schipper, Prakken, Kalkhoven, & Boes, 2012; Wensveen, Valentić, Šestan, Turk Wensveen, & Polić, 2015).
Much of what is known about inflammation in obesity has been obtained from experimental mouse models, but little is known about how these observations might directly translate to humans. Interestingly, in recent work by our group, pathway analyses of the top BMI-associated CpG loci in female adipose tissue also populated a large number of terms associated with hematopoietic and immune-related biological processes (Chu et al., 2018). Furthermore, in another study we demonstrated that high epigenome-wide correlation between adipose tissue and blood is associated with higher BMI (Huang et al., 2016). At the same time, the relationship between specific leukocyte infiltration and obesity is still unclear with respect to the manner of association, and also with respect to the specific blood cell types that might jointly characterize obesity-induced inflammation. Epigenetic signatures could potentially be used as biomarkers for identifying leukocyte infiltration and risk of insulin resistance. Thus, our primary aim is to use adipose DNA methylation data to identify which leukocyte cell types are prominent in human adipose inflammation in obesity and their roles.
MATERIALS AND METHODS
An overview of the leukocyte infiltration analyses in the discovery, replication, and pooled stages can be seen in Figure 1.
Figure 1. Overview of the leukocyte infiltration and methylation Euclidean Distance (mED) analyses.
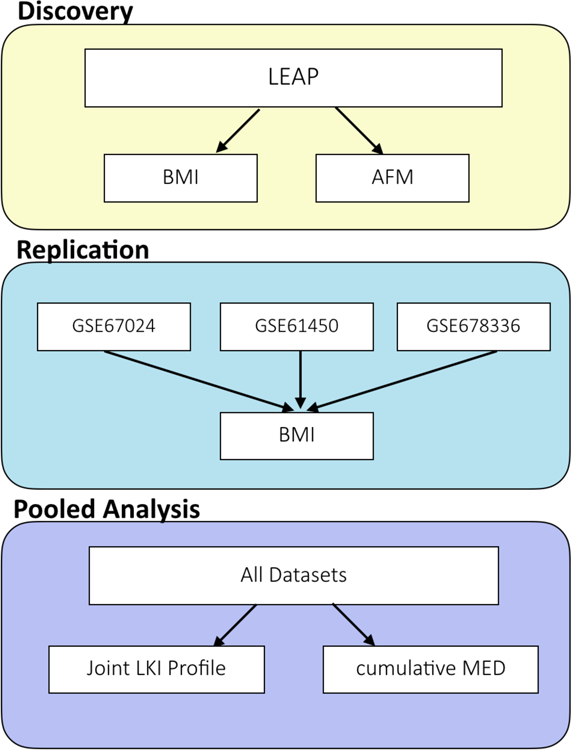
The study design consisted of three general stages: 1) discovery in the LEAP cohort, 2) replication in three independent datasets from the Gene Expression Omnibus database with measures of DNAm in adipose tissue and BMI, and 3) a pooled analysis of joint infiltration effects and cumulative mED effects.
Study Sample
Study subjects were recruited from the Longitudinal Effects on Aging Perinatal (LEAP) Project, a nested sub-study of the New England Family Study (NEFS), which has been described in detail elsewhere (Agha et al., 2015; Loucks et al., 2016; Slopen et al., 2015). Briefly, the LEAP study included Providence-born participants recruited from the NEFS and assessed between 2010–2011. Among 796 participants who met inclusion criteria including: not deceased or incarcerated, available assessments taken at age 7 years, and located within 100 miles of a clinical site during 2010–2011, 400 participants consented for further participation and were included for the present sub-study. Among these, 316 subjects had adequate adipose tissue biopsies, 16 had inadequate specimens, and 68 refused. Adipose tissue methylation analyses were conducted on a final, representative set of 143 randomly sampled subjects out of 316 participants due to budgetary constraints. The study protocol was approved by the institutional review boards at Brown University and Memorial Hospital of Rhode Island (#0908000028).
Collection of Covariates and Methylation Data
Body mass index was calculated as weight per height squared (kg/m2). Body weight and height were obtained by trained personnel using a calibrated stadiometer and weighing scale. Dual-energy x-ray absorbtiometry (DXA) scans were also conducted using the Lunar Prodigy Advance scanner (GE Healthcare, Madison, WI) to obtain direct measures of android fat mass (AFM) (i.e., centrally located fat). To monitor the reproducibility of the DXA assessments, weekly assessments were performed using models simulating varying levels of body fat.
Subcutaneous adipose tissue samples were obtained from the upper outer quadrant of the buttock using a 16-gauge needle, and buffy coat was obtained from centrifuged whole blood samples. DNA was extracted from the adipose tissue samples and buffy coat using the Qiagen DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA) and the Zymo Genomic DNA Clean & Concentrator Kit, and sodium bisulfite conversion was performed using the EZ-96 DNA Methylation-Direct and EZ DNA Methylation-Direct kits (Zymo Research, Orange, CA using manufacturer protocols. Adipose tissue samples were then analyzed on the Infinium HumanMethylation450 BeadChip array (Illumina, San Diego, CA) at the Core Genomics Facility in the UCSF Institute for Human Genetics (San Francisco, CA) using manufacturer protocols.
Pre-processing of the methylation data is described in Huang et al (Huang et al., 2016).
Briefly, the ‘methylumi’ package in R was employed to make background and dye-bias corrections, and correction for Type 1 and Type 2 probe bias was conducted via the Beta-Mixture Quantile Dilation approach (Teschendorff et al., 2013); batch effect adjustments were made using linear mixed models. Methylation levels were recorded on the beta scale, but were converted to the M-scale prior to analysis (Du et al., 2010).
Additional measured demographic and risk factors included included age, race (white or non-white), sex, maternal smoking (cigarettes per day), and socioeconomic index at age 7.
External Replication Datasets
Three external datasets with both methylation and BMI data were obtained from the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/), a publicly available repository of genomic data to validate infiltration cell-type candidates identified in our primary analyses. From the GEO, we selected GSE67024 (Arner et al., 2015) which contained 29 samples of abdominal subcutaneous fat cells from 15 obese and 14 never-obese women, GSE61450 (Bonder et al., 2014) which contained 71 samples of abdominal subcutaneous fat in subjects who had a BMI of 35 or greater, and GSE68336 (Pietiläinen et al., 2016) which contained 70 samples of abdominal subcutaneous adipose tissue from 35 monozygotic weight-discordant twin pairs, otherwise completely matched for age, race, sex, and family background. BMI was assessed as a continuous measure in the GSE67024 and GSE61450 datasets, but was categorized as a binary covariate (High (mean 31, s.d. 5.18) or Low (mean 25, s.d. 4.52)) in the GSE68336 twin study. All methylation data in the replication datasets were also measured on the Infinium HumanMethylation450K BeadChip array. For analyses of the GEO datasets, no additional normalization steps were employed to the already pre-processed beta values.
Statistical Analyses
mED Construction
To assess cellular infiltration in adipose tissue, we first constructed a metric for comparing epigenetic signatures between leukocytes and adipose tissue. Previously reported differentially methylated region (DMR) reference panels were obtained for 12 leukocyte cell-types of interest: activated natural killer (aNK) (Wiencke et al., 2016), CD4, CD8, dendritic, granulocytes, immature B, memory B, monocytes, naïve B, NK, plasma B, and T-regulatory (Treg) cells (Kim et al., 2016). For each cell type, methylation levels at 50 uniquely identifying CpG sites were obtained to create a reference methylation signature.
The leukocyte reference panels were then compared to the methylation levels at the corresponding CpG sites in participant adipose tissue by calculating the methylation Euclidean distance (mED) between the expression profiles. More specifically, for each cell type , we calculate the mED for subject as:
where, for cell type j, and denote the methylation value of CpG site k in 1) adipose tissue for subject i and 2) the reference panel, respectively. The mED constructed here can be thought of as representing a similarity score between adipose tissue and the reference leukocyte methylation profiles, where smaller values denote greater similarity between the tissues, suggesting higher infiltration by that cell-type. Prior to calculating mED, all CpG sites were scaled by the adipose tissue standard deviation of DNAm per site to ensure equal weighting of all CpG sites within a given leukocyte reference panel.
Cell-Type Specific Infiltration Analysis: Discovery and Replication
Calculated adipose mEDs for each leukocyte type were individually regressed on BMI and AFM in marginal and covariate adjusted models using least-squares regression. Adjustment covariates for the analyses in LEAP included age, sex, race, and smoking (cigarettes/day). Given expected high correlation between leukocytes, an initial, lenient significance threshold was applied for the discovery phase: cell-types that were significant (p<0.05) in either the BMI or AFM analyses were selected as candidate infiltrating leukocytes for replication. Six AFM scores were missing and were therefore excluded from the analysis.
To limit potential false discoveries via study design, independent datasets were obtained to protect against false positives through replication. Methods identical to those used in the primary analyses were used to calculate mEDs and assess infiltration in the external replication datasets, GSE67024 and GSE61450. However, conditional logistic regression was applied in GSE68336 to accommodate both the binary categorization of BMI, and the matched study design. In GSE61450, available adjustment covariates included sex and age, and in GSE67024, only age. As the analysis in GSE68336 was applied in monozygotic twin-pairs, no additional adjustments were made.
Pooled Analysis of Joint Infiltration Effects and Infiltration Profile Selection
There is evidence that inflammatory immune infiltration involves populations of multiple leukocytes, which may vary by the adiposity of the individual (Carvalheira et al., 2013; Castoldi et al., 2016). To capture this variability, and to further assess the use of DNAm sites as biomarkers for infiltration, we conducted lasso regression to select predictive profiles of leukocyte infiltration after pooling the primary data from LEAP with the two replication study datasets with continuous BMI measures (GSE67024 and GSE61450) to conduct an analysis of joint effects. Prior to combining all three datasets, mEDs were standardized by cell-type, within each cohort, to ensure comparability across the datasets when pooled.
For the joint multicellular analyses, BMI was regressed on all 12 leukocyte types, adjusting for age, race (white or non-white), gender, and cohort (LEAP, GSE67024, GSE61450). To select predictive joint infiltration profiles, we employed lasso regression with 10-fold cross validation in the pooled data sample using the ‘glmnet’ and ‘selectiveInference’ packages in R (Lee, Sun, Sun, & Taylor, 2016; Taylor & Tibshirani, 2017). The most parsimonious model demonstrating a comparable error to the model which minimized cross-validation error was selected, and the active set of covariates selected for inclusion were identified. To account for the inferential bias induced by the model-search process of cross-validated lasso, post-selection confidence intervals and p-values were obtained for each coefficient.
Cumulative mED Risk Score and BMI
To determine the extent to which cumulative effect of mEDs across different cell-types might associate with BMI, we next assessed the mEDs across 1) the full panel of 12 leukocytes, 2) the top validated leukocytes in the marginal analyses, and 3) the significant leukocytes in the joint multicellular analyses, both within each cohort and within the pooled sample. For each subject, cumulative mEDs (cmEDs) were obtained by first weighting each cell-type by the pooled covariate-adjusted associations with BMI, then summing mED values across the relevant cell-types for each of the three analyses as follows:
where S corresponds to the different cmED constructions denoted in 1)-3) above. Finally, BMI was regressed on each calculated cmED type in each cohort and in the pooled sample, with respective covariate adjustments as described in the preceding analyses.
RESULTS
Study Sample
Participants of this study had a mean age of 47 (range 44–50) years; 66% were white, and 52% were females. Between men and women, the distributions of age, race, BMI at age 7, maternal smoking, and adult BMI, our primary outcome of interest, were highly similar (Table 1). However, for AFM, the proportion of women decreased at higher levels.
Table 1.
Characteristics of the LEAP study sample.
| Characteristics | Total | Males | Females |
|---|---|---|---|
| (n=143) | (n=69) | (n=74) | |
| Age in Years | |||
| Mean (SD) | 46.9 (1.7) | 47.1 (1.6) | 46.8 (1.8) |
| Race %(no.) | |||
| White | 66.4 (95) | 63.8 (44) | 68.9 (51) |
| Non-White | 33.6 (48) | 36.2 (25) | 31.1 (23) |
| Age 7 BMI | |||
| Mean (SD) | 16.3 (2.5) | 16.5 (2.8) | 16.1 (2.1) |
| Maternal Cig/Day | |||
| Median (IQR) | 5 (0–20) | 4.5 (0–20) | 6 (0–20) |
| Range | 0–50 | 0–40 | 0–50 |
| SEI7 | |||
| Median (IQR) | 37.5 (22.3–57.8) | 38 (26–58) | 37 (21–57) |
| Range | 2–96 | 5–94 | 2–96 |
| Adult BMI | |||
| Mean (SD) | 31.5 (7.4) | 31.7 (5.8) | 31.3 (8.7) |
| Range | 19.4–65.5 | 21.2–46.1 | 19.4–65.5 |
| Adult AFM | |||
| Median (IQR) | 17.1 (12.8–22.3) | 18.2 (13.8–25.2) | 15.5 (11.5–20) |
| Range | 3.4–39.5 | 3.4–39.5 | 3.9–33.1 |
BMI: body mass index; AFM: android fat mass; SEI7: socioeconomic index (age 7); SD: standard deviation; IQR: interquartile range
Cell-Type Specific Infiltration Associated with Adiposity
To assess leukocyte infiltration into adipose tissue, we constructed a novel and intuitive metric to compare the methylation patterns in adipose tissue with differentially methylated region (DMR) signature panels for leukocytes reported in prior literature (Kim et al., 2016; Wiencke et al., 2016) by calculating methylation Euclidean distances (mED). In the LEAP cohort, we identified the mEDs of aNK (p=4.9×10−4), monocytes (p=0.001), NK (p=0.001), CD4 (p=0.009) and granulocytes (p=0.039) to be significantly negatively associated with BMI (Table 2). Negative associations are consistent with our hypothesis of increased infiltration with increased adiposity – in other words, as the distance scores decrease, adiposity increases, thus suggesting higher infiltration in subjects with higher levels of adiposity. Immature B (p=0.022) cells were identified as positively associated with BMI, indicating depletion of these leukocytes in the cellular composition of adipose tissue in subjects with higher BMI. In the analyses of android fat mass, we identified the mEDs of CD4 (p=0.020), aNK (p=0.001), NK (p=1.6×10−4), and dendritic (p=0.002) cells to be significantly negatively associated with AFM. Plasma B (p=0.004), immature B (p=0.020), and naïve B (p=0.030) cells were identified as positively associated with AFM.
Table 2.
Association between specific leukocyte infiltration as measured by mED and measures of adiposity in the LEAP study sample.
| BMI* | AFM* | |||||||
|---|---|---|---|---|---|---|---|---|
| Cell Type | Effect | 95% CI | LRTP | R2 | Effect | 95% CI | LRTP | R2 |
| ActivatedNK | −1.047 | (−1.635, −0.458) | 4.9E-04 | 0.080 | −0.801 | (−1.448, −0.154) | 0.014 | 0.036 |
| Monocyte | −0.961 | (−1.536, −0.387) | 0.001 | 0.073 | −1.091 | (−1.698, −0.484) | 4.3E-04 | 0.115 |
| NK | −0.942 | (−1.526, −0.359) | 0.001 | 0.068 | −0.500 | (−1.145, 0.145) | 0.122 | 0.017 |
| CD4 | −0.834 | (−1.464, −0.203) | 0.009 | 0.047 | −0.753 | (−1.396, −0.11) | 0.020 | 0.054 |
| Immature B | 0.828 | (0.108, 1.548) | 0.022 | 0.032 | 0.763 | (0.015, 1.512) | 0.042 | 0.023 |
| Granulocyte | −0.299 | (−0.588, −0.011) | 0.039 | 0.029 | −0.065 | (−0.366, 0.236) | 0.665 | 0.001 |
| Treg | −0.255 | (−0.757, 0.247) | 0.310 | 0.007 | 0.378 | (−0.171, 0.927) | 0.169 | 0.014 |
| Memory B | −0.247 | (−0.859, 0.365) | 0.420 | 0.004 | 0.627 | (−0.074, 1.329) | 0.075 | 0.031 |
| Plasma B | 0.235 | (−0.36, 0.83) | 0.430 | 0.004 | 0.957 | (0.31, 1.604) | 0.004 | 0.047 |
| Naïve B | −0.184 | (−0.763, 0.395) | 0.526 | 0.003 | 0.717 | (0.061, 1.372) | 0.030 | 0.036 |
| CD8 | −0.182 | (−0.813, 0.448) | 0.562 | 0.003 | 0.204 | (−0.452, 0.859) | 0.534 | 3.3E-07 |
| Dendritic | −0.168 | (−0.76, 0.425) | 0.571 | 0.003 | −0.767 | (−1.373, −0.162) | 0.012 | 0.066 |
Adjusted for age, sex, race, current smoking (cig/day); BMI: body mass index; AFM: android fat mass (kg); CI: confidence interval; LRTP: likelihood ratio test p-value; R2: marginal R-squared value
External Replication of Infiltrating Cell-Types
Replication analyses of the primary LEAP study findings were conducted in three independent datasets with BMI and DNAm data downloaded from the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database: GSE67024, GSE61450, and GSE68336. GSE67024 (Arner et al., 2015) included 29 samples of female abdominal subcutaneous fat cells, GSE61450 (Bonder et al., 2014) included 71 samples of abdominal subcutaneous fat from subjects with BMI≤35, and GSE678336 (Pietiläinen et al., 2016) included 70 samples of abdominal subcutaneous adipose tissue from 35 monozygotic weight-discordant twin pairs. For more specific descriptions of the replication datasets, see Materials and Methods.
In the GSE67024 dataset, the mEDs of CD4 (p=0.002), monocytes (p=6.9×10−5), aNK (p=0.002), and dendritic cells (p=3×10−8) were identified as significantly negatively associated with BMI (Table 3). In GSE61450, the mEDs of CD4 (p=0.031), monocytes (p=0.021), NK (p=0.030), and dendritic cells (p=0.007) were significantly negatively associated with BMI, and only immature B cells (p=0.032) were identified as having a significant positive association. In GSE68336, we conducted two analyses: first in all twin-pairs, then in BMI-discordant twin-pairs. The twin-pair analyses revealed one negative mED association among dendritic cells (pall = 3.2×10−4; pdiscordant = 1.5×10−4 and one positive association among Treg cells (pall = 0.002; pdiscordant = 0.019) with BMI category. The discordant twin-pair analyses additionally identified monocytes (pdiscordant = 0.009) as having a negative mED association with BMI category. Overall, CD4, monocytes, and dendritic cell mED associations with BMI in the LEAP analyses were validated by at least two of the replication analyses with respect to both direction of effect and significance (Table 3). Comparisons of available clinical characteristics between the primary LEAP dataset and the replication studies are available in Table S1.
Table 3.
Replication of cell-specific infiltration association
| GSE67024a | GSE61450b | GSE68336-A* | GSE68336-DC* | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cell Type | Effect | LRTP | R2 | Effect | LRTP | R2 | Effect | LRTP | Effect | LRTP |
| Activated NK | −2.830 | 0.002 | 0.168 | 0.189 | 0.836 | 8.9E-05 | −0.111 | 0.606 | −0.150 | 0.528 |
| Monocyte | −2.984 | 6.9E-05 | 0.003 | −1.470 | 0.021 | 0.074 | −0.216 | 0.124 | −0.560 | 0.009 |
| NK | −0.377 | 0.710 | 0.011 | −1.916 | 0.030 | 0.027 | 0.374 | 0.077 | 0.486 | 0.059 |
| CD4 | −3.226 | 0.002 | 0.050 | −2.121 | 0.031 | 0.033 | −0.337 | 0.116 | −0.454 | 0.113 |
| Immature B | 0.073 | 0.950 | 0.014 | 1.468 | 0.032 | 0.064 | 0.356 | 0.064 | 0.403 | 0.060 |
| Granulocyte | −0.978 | 0.150 | 0.073 | 0.345 | 0.125 | 0.04 | 0.046 | 0.448 | 0.058 | 0.393 |
| Treg | 1.653 | 0.221 | 0.380 | 0.179 | 0.847 | 1.95E-07 | 0.923 | 0.002 | 0.658 | 0.019 |
| Memory B | −0.755 | 0.415 | 0.002 | −0.885 | 0.452 | 0.010 | −0.069 | 0.699 | −0.101 | 0.629 |
| Plasma B | −1.607 | 0.096 | 0.031 | 0.797 | 0.199 | 0.009 | 0.301 | 0.108 | 0.542 | 0.037 |
| Naïve B | −0.863 | 0.583 | 1.7E-04 | 0.036 | 0.972 | 2.7E-04 | 0.134 | 0.499 | 0.430 | 0.096 |
| CD8 | −1.633 | 0.134 | 0.335 | −0.331 | 0.669 | 0.014 | 0.297 | 0.168 | 0.271 | 0.247 |
| Dendritic | −3.474 | 3E-08 | 0.029 | −1.578 | 0.007 | 0.107 | −0.721 | 3.2E-04 | −1.089 | 1.5E-04 |
Adjustment covariates for each dataset were as follows:
age
age and sex
represents results from matched conditional logistic regression in a monozygotic twin study, with body mass index (BMI) characterized as high or low. LRTP: likelihood ratio test p-value; R2: marginal R-squared value; A: All Twin Pairs; DC: Discordant Twin Pairs.
Pooled Analysis of Joint Leukocyte Infiltration Profiles
In the pooled multivariable analysis including all 12 cell types in the model, we identified the mEDs of immature B (p=0.002), plasma B (p=0.002), and memory B (p=0.009) cells as being associated with BMI (Table 4). Applying lasso regression in the pooled data selected immature B, plasma B, memory B, monocytes, aNK, CD4, dendritic, CD8, granulocytes, and NK cells as predictive cell-types for BMI, in addition to selecting age, race, gender, and cohort for inclusion in the final model. Among the leukocytes, only the mEDs of plasma B (p=0.003), immature B (p=0.012) cells were identified as significant mED predictors of BMI via post-selection inference accounting for the model selection process of lasso regression (Table 4). Significant lasso model predictors were consistent with the top results of the joint pooled analyses. Memory B cells were identified as significant in the pooled multivariable analysis, but were shown not to be significant after post-selection inference. In general, that the joint multivariable and post-selection lasso models selected identical sets of significant jointly-associated leukocytes provides reassuring evidence for the robustness of these findings.
Table 4.
Pooled multivariable analysis and lasso regression of joint infiltration by all leukocytes.
| Joint Multicellular | LASSO | |||||
|---|---|---|---|---|---|---|
| Covariates* | Effect | 95% CI | P | Effect | 95% POSI | P |
| Immature B | 1.815 | (0.682, 2.948) | 0.002 | 1.808 | (0.593, 2.942) | 0.003 |
| Plasma B | 2.276 | (0.822, 3.731) | 0.002 | 2.267 | (0.339, 4.653) | 0.012 |
| Memory B | −2.081 | (−3.622, −0.539) | 0.009 | −2.072 | (−3.668, 0.807) | 0.069 |
| Monocyte | −1.271 | (−2.615, 0.073) | 0.065 | −1.266 | (−2.64, 0.439) | 0.064 |
| Activated NK | −1.216 | (−2.669, 0.237) | 0.102 | −1.211 | (−2.704, 0.776) | 0.102 |
| CD4 | −1.002 | (−2.212, 0.207) | 0.106 | −0.998 | (−2.213, 0.683) | 0.107 |
| Dendritic | −1.053 | (−2.331, 0.225) | 0.108 | −1.049 | (−3.014, 0.709) | 0.104 |
| CD8 | 0.814 | (−0.515, 2.143) | 0.231 | 0.811 | (−1.537, 2.984) | 0.227 |
| Granulocyte | 0.654 | (−0.598, 1.906) | 0.307 | 0.651 | (−1.888, 1.888) | 0.315 |
| NK | −0.572 | (−2.02, 0.876) | 0.439 | −0.570 | (−2.17, 3.146) | 0.443 |
| Naïve B | −0.358 | (−1.837, 1.12) | 0.635 | −0.357 | (−1.694, 5.878) | 0.657 |
| Treg | 0.215 | (−1.23, 1.66) | 0.771 | 0.214 | (−10.105, 1.386) | 0.788 |
| Age | −0.147 | (−0.297, 0.004) | 0.058 | −0.965 | (−1.958, 0.284) | 0.058 |
| Race | ||||||
| Not White | 0.450 | (−2.156, 3.056) | 0.735 | 0.180 | (−5.808, 1.077) | 0.740 |
| Gender | ||||||
| Female | −2.586 | (−4.795, −0.377) | 0.023 | −1.248 | (−2.315, −0.028) | 0.023 |
| Cohort | ||||||
| GSE67024 | 2.875 | (−0.347, 6.097) | 0.082 | 0.934 | (−0.452, 2.708) | 0.080 |
| GSE61450 | 14.117 | (11.78, 16.454) | 1.9E-25 | 6.433 | (5.383, 8.842) | 0 |
CI: confidence interval; POSI: post-selection interval
Italicized cell types indicate cell types that were selected in the active set by LASSO.
Cumulative mEDs and BMI
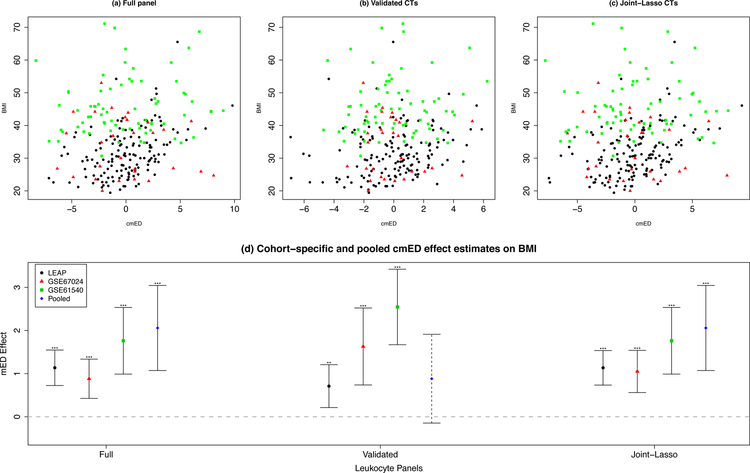
Given that multiple leukocyte populations likely act together during inflammatory dysregulation of adipose tissue, we constructed an intuitive measure of the cumulative effect of different leukocyte types called the cumulative methylation Euclidean distance (cmED). Within each cohort, each cell-type mED score was weighted by the pooled covariate-adjusted estimates of association with BMI. Then, these weighted scores were summed across all cell-types to create a risk score for infiltration by leukocytes across a given set of interest. In the pooled sample, the cmEDs across 1) the full panel of all 12 cell-types (p=5.9×10−5), 2) the top validated leukocytes from the marginal analyses (p=0.094) and 3) the leukocytes selected by lasso in the pooled analyses of joint leukocyte effects (p=0.001) were all positively associated with BMI (Table 5 and Figure 2). The covariate-adjusted associations between the composite measure of cmED and BMI for these panels in individual cohorts are also reported in Table 5.
Table 5. Cumulative infiltration by multiple cell-types and BMI.
Cumulative mED (cmED) scores, weighted by cell-specific effect estimates obtained from the pooled data, are associated with BMI across all cohorts, and after pooling.
| Full Panela | Validated CTsb | Joint-Lasso CTsc | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect | SE | P | Effect | SE | P | Effect | SE | P | ||||
| LEAP | 1.136 | 0.210 | 2.7E-07 | 0.175 | 0.710 | 0.254 | 0.006 | 0.051 | 1.135 | 0.204 | 1.4E-07 | 0.177 |
| GSE67024 | 0.881 | 0.232 | 3.2E-04 | 0.128 | 1.630 | 0.455 | 0.001 | 0.142 | 1.051 | 0.250 | 7.9E-05 | 0.159 |
| GSE61450 | 1.761 | 0.394 | 1.4E-04 | 0.427 | 2.545 | 0.446 | 5.3E-06 | 0.540 | 1.761 | 0.394 | 1.4E-04 | 0.427 |
| Pooled* | 2.058 | 0.503 | 5.9E-05 | 0.423 | 0.882 | 0.525 | 0.094 | 0.394 | 1.752 | 0.508 | 0.001 | 0.412 |
Panels for each cmED analysis included:
all twelve cell-types
CD4, monocytes, and dendritic cell-types
immature B, plasma B, Memory B, monocytes, aNK, CD4, dendritic, CD8, granulocytes, and NK cell-types.
SE: standard error;: marginal R-squared value; CTs: cell-types
includes a cohort effect in the model.
Figure 2. Cumulative mEDs and BMI.
The association between BMI and cmEDs employing a) the full panel of 12 leukocytes, b) the validated leukocytes, and c) the joint-lasso leukocytes. The cohort-specific and pooled cMED effect estimates can be seen in panel d).
DISCUSSION
This study demonstrates a novel, but simple and intuitive, approach for utilizing methylation data to identify and estimate the effects of leukocyte infiltration in adipose tissue. Much of the literature regarding inflammatory processes in adipose tissue does not include cell mixture deconvolution adjustment to separate signals from different kinds of cell types. However, our interest lies not in cell mixture estimation per se, but in the slightly more challenging task of observing the infiltration of complex, admixed tissue types, such as adipose tissue, by blood – which is itself a mixture. To accommodate this goal, we borrow information from established DMR reference panels to estimate infiltration effects of different cell types, both marginally and jointly. Importantly, this method can accommodate both pre-established reference libraries, or study-specific derived reference libraries, and is straightforward to apply. Although our study investigated adipose tissue as the target tissue of interest, this approach can easily be employed in other tissue types.
Among our top findings in the LEAP cohort, CD4 and monocyte mED associations with BMI were replicated in at least two of our replication studies. It is worth noting that dendritic cell mED, which was identified as associated with AFM but not with BMI in our primary LEAP analyses, was associated with BMI in all three replications. Evidence for infiltration by CD4 (Fabbrini et al., 2013; Rocha et al., 2008; Strissel et al., 2010; Winer et al., 2009), monocytes (Castoldi et al., 2016; Kanda, 2006; Masoodi, Kuda, Rossmeisl, Flachs, & Kopecky, 2015), and dendritic (Chen et al., 2014; Rajan & Longhi, 2016; Stefanovic-Racic et al., 2012) cells with increased adiposity has also been previously reported in mice and human studies (Mathis & Shoelson, 2011; Norata et al., 2015), providing biological plausibility for our findings. More importantly, the directions of association between methylation profile similarity as measured by mEDs and BMI among these cell-types were consistent both across all of our analyses (cohort-specific and pooled) and with findings described in literature.
In our pooled multicellular analysis, plasma B and immature B cells were associated with patterns of depletion in adipose tissue with higher adiposity: increasing profile dissimilarity was associated with higher BMI. The role of B cells, and specific subtypes, in inflammatory obesogenic processes has been less well studied. Main findings with regard to B cell involvement suggest an early accumulation in adipose tissue prior to the recruitment of T cells during inflammation (Duffaut, Galitzky, Lafontan, & Bouloumié, 2009; McLaughlin et al., 2014), but some recent studies have also identified IL-10 producing populations of B regulatory cells that may confer protective effects against insulin resistance (Nishimura et al., 2013; Wu, Parekh, Hsiao, Kitamura, & Van Kaer, 2014).
The associations between mED and BMI in immature B and plasma B cells were corroborated by our lasso variable selection model with respect to significance and directions of effect. Neither of these findings were among the top validated BMI-associated hits, but the full active set selected by the lasso regression procedure included the replicated LEAP findings of CD4, monocytes, and dendritic cells. In general, that all directions of effect for mED associations with BMI, across the validated cell-type and the pooled analyses, are largely consistent with each other and with prior research findings is compelling.
In the cmED analyses, associations with BMI were consistently positively associated and significant across both cohort-specific and pooled analyses, for all cell-type panels. This is supportive of the notion that profile-driven summary mED metrics derived from leukocyte methylation libraries may provide additional utility in the analysis of leukocyte infiltration. Total proportion of variability explained in BMI by the full panel cmEDs was not appreciably increased as compared to the joint panels in the pooled sample. In general, the full panel demonstrated higher performance than both the joint and validated cell type panels. This suggests that the interplay between the infiltrating activity by the different cell types is better summarized by an inclusive cmED panel (such as the full or joint panels). Taken together, these findings lend credibility to the evidence for significant associations between infiltration patterns, as measured by mED, and adiposity.
Although adjustment for multiple comparisons were not explicitly conducted, we reasoned that despite testing 12 different cell-types, moderate to high correlation observed between some of the leukocytes would suggest a much lower number of independent tests in practice (Supplementary Figures S1 and S2). These concerns are somewhat mitigated through study design, as we replicated our findings using three independent data sets. Other limitations of this study include the dependency of mEDs on the selection of the leukocyte DMR reference panels. Although the panels in our construction had good representation from broad, major classes of immune cell types, articulating small but important differences in adipose infiltration by highly correlated cell-types remains challenging. The selection of the reference panel is one of the most important considerations in this approach, as incomplete panels might result in missed associations, and impact the estimation of other cellular effects. In this study, we also used reference panels derived from healthy individuals, but it is possible that leukocyte signatures in the context of higher BMI might differ from those of healthy subjects. Finally, our use of blood DMR sites to identify cell-specific leukocyte invasion in adipose tissue does not preclude the possibility that the sites in our reference panels might also experience similar methylation in resident adipose cells, which could lead to signal obfuscation at those sites. Thus, experimental validation will be necessary as proof-of-concept for our approach.
CONCLUSION
Our findings support the belief that obesity-related inflammation arises as a result of excessive and inappropriate activation of the immune system. Much of the literature on inflammation in obesity is derived from experimental animal models, with, to our knowledge, few large-sample interrogations in humans. We applied our mED method to human tissue data obtained from a large cohort and conducted both replication and pooled association studies. That our findings are consistent with prior work on inflammation in obesity in animal models and smaller human studies is reassuring with respect to our approach and analytic results, but nonetheless will require experimental validation and replication, respectively.
Finally, we present a simple and intuitive approach for assessing cellular infiltration by blood into other heterogeneous, admixed tissues such as adipose tissue. Our approach is easy to apply and straightforwardly extended to other tissues in which leukocyte infiltration might be of interest. This method can also be extended to other kinds of cellular infiltration as well, provided reference libraries are available or can be derived.
Supplementary Material
ACKNOWLEDGEMENTS
FUNDING
This work was supported by the National Institute on Aging at the National Institutes of Health [grant numbers RC2AG036666, R01AG048825]; the Kansas IDeA Network of Biomedical Research Excellence Bioinformatics Core which is supported in part by the National Institute of General Medical Science [award P20GM103428]; and the Ministry of Science and Technology [grant number 105-2118-M-001-014-MY3].
LIST OF ABBREVIATIONS
- AFM
android fat mass
- DNAm
DNA methylation
- mED
methylation Euclidean distance
- cmED
cumulative methylation Euclidean distance
- DMR
differentially methylated regions
- NK
natural killer
- Treg
T-regulatory
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
DATA AVAILABILITY STATEMENT
The data underlying the results presented in the study are available via data request to the Longitudinal Effects on Aging Perinatal Project (https://sites.google.com/a/brown.edu/nefs/for-researchers/nefs-substudies/leap). The data underlying the replication analyses can be downloaded from the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/).
REFERENCES
- Agha G, Houseman EA, Kelsey KT, Eaton CB, Buka SL, & Loucks EB (2015). Adiposity is associated with DNA methylation profile in adipose tissue. International Journal of Epidemiology, 44(4), 1277–1287. doi: 10.1093/ije/dyu236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arner P, Sinha I, Thorell A, Rydén M, Dahlman-Wright K, & Dahlman I (2015). The epigenetic signature of subcutaneous fat cells is linked to altered expression of genes implicated in lipid metabolism in obese women. Clinical Epigenetics, 7(1), 1–13. doi: 10.1186/s13148-015-0126-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonder M, Kasela S, Kals M, Tamm R, Lokk K, Barragan I, … Milani L (2014). Genetic and epigenetic regulation of gene expression in fetal and adult human livers. BMC Genomics, 15(1), 860–813. doi: 10.1186/1471-2164-15-860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalheira JBC, Qiu Y, & Chawla A (2013). Blood spotlight on leukocytes and obesity. Blood, 122(19), 3263–3267. doi: 10.1182/blood-2013-04-459446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi A, Naffah de Souza C, Câmara NOS, & Moraes-Vieira PM (2016). The Macrophage Switch in Obesity Development. Frontiers in Immunology, 6(6), 637. doi: 10.3389/fimmu.2015.00637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Tian J, Tian X, Tang X, Rui K, Tong J, … Wang S (2014). Adipose Tissue Dendritic Cells Enhances Inflammation by Prompting the Generation of Th17 Cells. PLoS ONE, 9(3), e92450–92458. doi: 10.1371/journal.pone.0092450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu SH, Loucks EB, Kelsey KT, Gilman SE, Agha G, Eaton CB, … Huang Y-T (2018). Sex-specific epigenetic mediators between early life social disadvantage and adulthood BMI. Epigenomics, 16(3), 321. doi: 10.2217/epi-2017-0146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das UN (2001). Is obesity an inflammatory condition? Nutrition (Burbank, Los Angeles County, Calif.), 17(11–12), 953–966. [DOI] [PubMed] [Google Scholar]
- Du P, Zhang X, Huang C-C, Jafari N, Kibbe WA, Hou L, & Lin SM (2010). Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics, 11(1), 587. doi: 10.1186/1471-2105-11-587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffaut C, Galitzky J, Lafontan M, & Bouloumié A (2009). Unexpected trafficking of immune cells within the adipose tissue during the onset of obesity. Biochemical and Biophysical Research Communications, 384(4), 482–485. doi: 10.1016/j.bbrc.2009.05.002 [DOI] [PubMed] [Google Scholar]
- Fabbrini E, Cella M, Mccartney SA, Fuchs A, Abumrad NA, Pietka TA, … Klein S (2013). Association between specific adipose tissue CD4+ T-cell populations and insulin resistance in obese individuals. Gastroenterology, 145(2), 366–374.e361–363. doi: 10.1053/j.gastro.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant RW, & Dixit VD (2015). Adipose tissue as an immunological organ. Obesity, 23(3), 512–518. doi: 10.1002/oby.21003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, & Friedman J (2009). The Elements of Statistical Learning (2 ed.): Springer Science & Business Media. [Google Scholar]
- Hill AA, Reid Bolus W, & Hasty AH (2014). A decade of progress in adipose tissue macrophage biology. Immunological reviews, 262(1), 134–152. doi: 10.1111/imr.12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, … Kelsey KT (2012). DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics, 13(1), 86. doi: 10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Molitor J, & Marsit CJ (2014). Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics, 30(10), 1431–1439. doi: 10.1093/bioinformatics/btu029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y-T, Chu S, Loucks EB, Lin C-L, Eaton CB, Buka SL, & Kelsey KT (2016). Epigenome-wide profiling of DNA methylation in paired samples of adipose tissue and blood. Epigenetics, 11(3), 227–236. doi: 10.1080/15592294.2016.1146853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda H (2006). MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. Journal of Clinical Investigation, 116(6), 1494–1505. doi: 10.1172/JCI26498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Eliot M, Koestler DC, Houseman EA, Wetmur JG, Wiencke JK, & Kelsey KT (2016). Enlarged leukocyte referent libraries can explain additional variance in blood-based epigenome-wide association studies. Epigenomics, 8(9), 1185–1192. doi: 10.2217/epi-2016-0037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JD, Sun DL, Sun Y, & Taylor JE (2016). Exact post-selection inference, with application to the lasso. The Annals of Statistics, 44(3), 907–927. doi: 10.1214/15-AOS1371 [DOI] [Google Scholar]
- Loucks EB, Huang Y-T, Agha G, Chu S, Eaton CB, Gilman SE, … Kelsey KT (2016). Epigenetic Mediators Between Childhood Socioeconomic Disadvantage and Mid-Life Body Mass Index: The New England Family Study. Psychosomatic Medicine, 78(9), 1053–1065. doi: 10.1097/PSY.0000000000000411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumeng CN, Bodzin JL, & Saltiel AR (2007). Obesity induces a phenotypic switch in adipose tissue macrophage polarization. Journal of Clinical Investigation, 117(1), 175–184. doi: 10.1172/JCI29881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoodi M, Kuda O, Rossmeisl M, Flachs P, & Kopecky J (2015). Lipid signaling in adipose tissue: Connecting inflammation & metabolism. BBA - Molecular and Cell Biology of Lipids, 1851(4), 503–518. doi: 10.1016/j.bbalip.2014.09.023 [DOI] [PubMed] [Google Scholar]
- Mathis D, & Shoelson SE (2011). Immunometabolism: an emerging frontier. Nature Reviews Genetics, 11(2), 81–83. doi: 10.1038/nri2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin T, Liu LF, Lamendola C, Shen L, Morton J, Rivas H, … Engleman E (2014). T-Cell Profile in Adipose Tissue Is Associated With Insulin Resistance and Systemic Inflammation in Humans. Arteriosclerosis, Thrombosis, and Vascular Biology, 34(12), 2637–2643. doi: 10.1161/ATVBAHA.114.304636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNelis JC, & Olefsky JM (2014). Macrophages, Immunity, and Metabolic Disease. Immunity, 41(1), 36–48. doi: 10.1016/j.immuni.2014.05.010 [DOI] [PubMed] [Google Scholar]
- Nishimura S, Manabe I, Takaki S, Nagasaki M, Otsu M, Yamashita H, … Nagai R (2013). Adipose Natural Regulatory B Cells Negatively Control Adipose Tissue Inflammation. Cell metabolism, 18(5), 759–766. doi: 10.1016/j.cmet.2013.09.017 [DOI] [PubMed] [Google Scholar]
- Norata GD, Caligiuri G, Chavakis T, Matarese G, Netea MG, Nicoletti A, … Marelli-Berg FM (2015). The Cellular and Molecular Basis of Translational Immunometabolism. Immunity, 43(3), 421–434. doi: 10.1016/j.immuni.2015.08.023 [DOI] [PubMed] [Google Scholar]
- Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, … Chawla A (2007). Macrophage-specific PPARγ controls alternative activation and improves insulin resistance. Nature, 447(7148), 1116–1120. doi: 10.1038/nature05894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi Y, & Manabe I (2016). Integrated regulation of the cellular metabolism and function of immune cells in adipose tissue. Clinical and Experimental Pharmacology and Physiology, 43(3), 294–303. doi: 10.1111/1440-1681.12539 [DOI] [PubMed] [Google Scholar]
- Pietiläinen KH, Ismail K, Järvinen E, Heinonen S, Tummers M, Bollepalli S, … Ollikainen M (2016). DNA methylation and gene expression patterns in adipose tissue differ significantly within young adult monozygotic BMI-discordant twin pairs. International Journal of Obesity, 40(4), 654–661. doi: 10.1038/ijo.2015.221 [DOI] [PubMed] [Google Scholar]
- Rajan SS, & Longhi MP (2016). Dendritic cells and adipose tissue. Immunology, 149(4), 353–361. doi: 10.1111/imm.12653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha VZ, Folco EJ, Sukhova G, Shimizu K, Gotsman I, Vernon AH, & Libby P (2008). Interferon-gamma, a Th1 cytokine, regulates fat inflammation: a role for adaptive immunity in obesity. Circulation Research, 103(5), 467–476. doi: 10.1161/CIRCRESAHA.108.177105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schipper HS, Prakken B, Kalkhoven E, & Boes M (2012). Adipose tissue-resident immune cells: key players in immunometabolism. Trends in Endocrinology and Metabolism, 23(8), 407–415. doi: 10.1016/j.tem.2012.05.011 [DOI] [PubMed] [Google Scholar]
- Slopen N, Loucks EB, Appleton AA, Kawachi I, Kubzansky LD, Non AL, … Gilman SE (2015). Early origins of inflammation: An examination of prenatal and childhood social adversity in a prospective cohort study. Psychoneuroendocrinology, 51, 403–413. doi: 10.1016/j.psyneuen.2014.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic-Racic M, Yang X, Turner MS, Mantell BS, Stolz DB, Sumpter TL, … O'Doherty RM (2012). Dendritic cells promote macrophage infiltration and comprise a substantial proportion of obesity-associated increases in CD11c+ cells in adipose tissue and liver. Diabetes, 61(9), 2330–2339. doi:10.2337/db11-1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strissel KJ, DeFuria J, Shaul ME, Bennett G, Greenberg AS, & Obin MS (2010). T-cell recruitment and Th1 polarization in adipose tissue during diet-induced obesity in C57BL/6 mice. Obesity, 18(10), 1918–1925. doi: 10.1038/oby.2010.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J, & Tibshirani R (2017). Post-selection inference for ℓ1-penalized likelihood models. Canadian Journal of Statistics, 19(4), 1212. doi: 10.1002/cjs.11313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, & Beck S (2013). A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics, 29(2), 189–196. doi: 10.1093/bioinformatics/bts680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, & Ferrante AW (2003). Obesity is associated with macrophage accumulation in adipose tissue. Journal of Clinical Investigation, 112(12), 1796–1808. doi: 10.1172/JCI19246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensveen FM, Valentić S, Šestan M, Turk Wensveen T, & Polić B (2015). The “Big Bang” in obese fat: Events initiating obesity-induced adipose tissue inflammation. European Journal of Immunology, 45(9), 2446–2456. doi: 10.1002/eji.201545502 [DOI] [PubMed] [Google Scholar]
- Wiencke JK, Butler R, Hsuang G, Eliot M, Kim S, Sepulveda MA, … Kelsey KT (2016). The DNA methylation profile of activated human natural killer cells. Epigenetics, 11(5), 363–380. doi: 10.1080/15592294.2016.1163454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer S, Chan Y, Paltser G, Truong D, Tsui H, Bahrami J, … Dosch H-M (2009). Normalization of obesity-associated insulin resistance through immunotherapy. Nature Reviews Genetics, 15(8), 921–929. doi: 10.1038/nm.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Parekh VV, Hsiao J, Kitamura D, & Van Kaer L (2014). Spleen supports a pool of innate-like B cells in white adipose tissue that protects against obesity-associated insulin resistance. Proceedings of the National Academy of Sciences, 111(43), E4638–E4647. doi: 10.1073/pnas.1324052111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, … Chen H (2003). Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. Journal of Clinical Investigation, 112(12), 1821–1830. doi: 10.1172/JCI19451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Grijalva A, Skowronski A, van Eijk M, Serlie MJ, & Ferrante AW Jr. (2013). Obesity Activates a Program of Lysosomal-Dependent Lipid Metabolism in Adipose Tissue Macrophages Independently of Classic Activation. Cell metabolism, 18(6), 816–830. doi: 10.1016/j.cmet.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [dataset] Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. (2013).NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Research, 41(D1), Pages D991–D995, 10.1093/nar/gks1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.