Abstract
Experimental, observational and clinical trials support a critical role of folate one-carbon metabolism (FOCM) in colorectal cancer (CRC) development. In this report, we focus on understanding the relationship between common genetic variants and metabolites of FOCM. We conducted a genome-wide association study of FOCM biomarkers among 1788 unaffected (without CRC) individuals of European ancestry from the Colon Cancer Family Registry. Twelve metabolites, including 5-methyltetrahydrofolate, vitamin B2 (flavin mononucleotide and riboflavin), vitamin B6 (4-Pyridoxic acid, pyridoxal and pyridoxamine), total homocysteine, methionine, S-adenosylmethionine, S-adenosylhomocysteine, cystathionine, and creatinine were measured from plasma using liquid chromatography-mass spectrometry (LC-MS) or LC-MS/MS. For each individual biomarker, we estimated genotype array-specific associations followed by a fixed-effect meta-analysis. We identified the variant rs35976024 (at 2p11.2 and intronic of ATOH8) associated with total homocysteine (P=4.9×10−08). We found a group of six highly correlated variants on chromosome 15q14 associated with cystathionine (all P<5×10−08), with the most significant variant rs28391580 (P=2.8×10−08). Two variants (rs139435405 and rs149119426) on chromosome 14q13 showed significant (P<5×10−8) associations with S-adenosylhomocysteine. These three biomarkers with significant associations are closely involved in homocysteine metabolism. Furthermore, when assessing the principal components (PCs) derived from seven individual biomarkers, we identified the variant rs12665366 (at 6p25.3 and intronic of EXOC2) associated with the first PC (P=2.3×10−08). Our data suggest that common genetic variants may play an important role in FOCM, particularly in homocysteine metabolism.
Keywords: folate, one-carbon metabolism, genome-wide association analysis, colorectal cancer, epidemiology
Introduction
Folate one-carbon metabolism (FOCM) is a metabolic process in which folate activates and transfers one carbon units to support a wide range of biological processes including nucleotide synthesis and methylation reactions (Ducker & Rabinowitz, 2017). Due to the essential role of FOCM in maintaining genome function and integrity, it is biologically plausible that folate and associated one-carbon metabolites may play an important role in cancer development, in particular for colorectal cancer (CRC). Indeed, the role between these biological reactions and carcinogenesis has been defined in vitro, in vivo (Giovannucci, 2002; Kim, 2003; Mason & Choi, 2000), and even in silico (Nijhout, Reed, Budu, & Ulrich, 2004; Reed, Nijhout, Sparks, & Ulrich, 2004; Ulrich et al., 2008). Epidemiological studies have also shown dietary folates are associated with decreased colorectal neoplasia risk (Giovannucci, 2002; Kennedy et al., 2011). Although supporting evidence from both experimental and observational studies makes folate and one-carbon metabolic cycle a good target to probe as a biomarker in CRC development, the exact role that folate plays is complicated with evidence of a dual role in colorectal carcinogenesis-protection prior to development of neoplastic lesions but promotion of growth after tumor development) (Kim, 2003). Furthermore, Mason et al. documented a significant trend toward increasing CRC incidence in the US and Canada coinciding with the fortification of grain products with folic acid (Mason et al., 2007). Moreover, some recent human studies, including randomized clinical trials, suggest risks for colon, breast or prostate cancers may be increased in those taking high doses of folic acid from supplements (Charles, Ness, Campbell, Davey Smith, & Hall, 2004; Cole et al., 2007; Figueiredo et al., 2009; Hirsch et al., 2009; Stolzenberg-Solomon et al., 2006) in the context of a folic acid fortified diet. Notably, a randomized clinical trial of colorectal adenoma suggested the risk of recurrent advanced colorectal adenoma or multiple adenomas increased with excessive levels of folate (Rees et al., 2017). Several other trials reported no such effect (Logan, Grainge, Shepherd, Armitage, & Muir, 2008; Song et al., 2012; Wu et al., 2009), although they had shorter follow-up periods.
The ability to combine genomics and metabolism data is a powerful tool to quantify and identify potential mechanisms within the biological system in question (Fiehn, 2002; Nicholson, Lindon, & Holmes, 1999). It has been extensively used in biomarker discovery to facilitate disease diagnosis (Madsen, Lundstedt, & Trygg, 2010) and mechanistic dissection of disease pathophysiology (Li et al., 2008). Targeted metabolomics is commonly considered to facilitate the accurate measure of selected endogenous metabolites in the biological samples. With the emergence of liquid chromatography mass spectrometry (LCMS)-based metabolomics, it is possible to profile and even quantify the analytes found in a particular pathway. In this study, we combined genome-wide genotype data with targeted metabolomics to profile the baseline relationships in the FOCM pathway among individuals disease-free of CRC. The goal of this study is to improve our understanding of the genetic contribution to the FOCM pathway in order to better elucidate its role in colorectal carcinogenesis.
Materials and Methods
Study participants.
The study population consisted of disease-free controls (i.e. individuals without CRC) enrolled in the Colon Cancer Family Registry (CCFR) (Newcomb et al., 2007). The CCFR is an international consortium of six study centers, including Sinai Health System (Ontario), Fred Hutchinson Cancer Research Center (FHCRC), Mayo Clinic, University of Hawaii (UHI, not included in this study), University of Southern California/Cedars-Sinai Medical Center (USC/CSMC) consortium, and the University of Melbourne (Australia). Details of the study design have been reported previously (Newcomb et al., 2007). Briefly, CCFR Phase I (1998–2002) focused on recruitment of CRC cases identified from population-based cancer registries and/or clinical centers. Disease-free controls were either age-and sex-matched population-based (Australia, FHCRC, Ontario), CRC case spouses (Mayo) or same-generation (sibling or cousin) family-based (USC/CSMC) consortium participants. CCFR Phase II (2002–2007) focused on recruitment of either CRC cases diagnosed under 50 years of age or clinically identified multi-case families. CCFR Phase II controls were age-and sex-matched from the general population (FHCRC) or case spouses (Mayo) and had no personal history of CRC. The study population had previously participated in a previous genome-wide association study (GWAS) of CRC (Schmit et al., 2018; Schumacher et al., 2015). All controls self-reported as non-Hispanic White. From the available controls with genotyping data, a total of 1,788 disease-free controls were included in the measurement of plasma FOCM and contributed to the analysis. Participants provided written informed consent, and the Institutional Review Boards at each center approved the study.
Genotyping and imputation.
Details of sample collection, genotyping, quality control (QC) and imputation have been reported elsewhere (Schmit et al., 2018). In brief, genotype data was generated from germline DNA on the Affymetrix Axiom, Illumina 1M/1M-Duo, Omni1 and OncoArray. Standard QC filters were applied to the high-density genotype array data at both the individual participant and SNP levels. Quality-controlled genotype data was imputed to the 1,000 Genomes Project (1KGP) Phase 1 multiethnic reference panel (March 2012 release, N=1,092) using SHAPE-IT/IMPUTE2 (Affymetrix Axiom; Illumina 1M/1M-Duo, and Omni1) (Eyre et al., 2012; Howie, Donnelly, & Marchini, 2009) or the 1KGP Phase 3 reference panel (Illumina OncoArray) (Amos et al., 2017). Imputation quality (info score> 0.3) and minor allele frequency filters (MAF ≥ 1%) were imposed on variants prior to the analysis phase. Approximately 44.4% (N=794) of the participants were run on Illumina 1M/1M-Duo while 12.1% (N=217) were on Illumina OncoArray; Affymetrix Axiom and Illumina Omni1 contributed similar proportions (20.5% (N=366) and 23.0% (N=411), respectively).
Biomarker measurement.
Blood samples were collected from participants at study entry to quantify circulating FOCM biomarkers (N=12), including folate (5-methyltetrahydrofolate), vitamin B2 (flavin mononucleotide and riboflavin), vitamin B6 (4-Pyridoxic acid, pyridoxal and pyridoxamine), total homocysteine (tHCY), methionine (MET), S-adenosylmethionine (SAM), S-adenosylhomocysteine (SAH), cystathionine (CYSTA), and creatinine. Detailed information regarding biomarker measurement, including QC is included in Supplementary Materials. We used internal standardization with controls to remove any batch effect.
Statistical analysis.
SNPs with allele frequency < 5% were excluded from the analysis. We only considered a SNP if it was identified in at least three arrays. Principal components (PCs) were calculated with EIGENSTRAT 6.1.4 (version) (Price et al., 2006) and used for ancestral adjustment in analyses. The percentage of samples returning below detection limit (BDL) measurements varied across the biomarkers (0.1%-47%). For biomarkers with a BDL <10%, including creatinine, MET, tHCY, SAH, SAM and CYSTA, we excluded samples with levels falling below the detection limit. We performed a sensitivity analysis replacing all the values falling below the detection limit with the minimal detectable values and results remained similar for biomarkers with a BDL <10% (data not shown). Biomarkers with BDL <10% were analyzed as continuous to achieve optimal power. Biomarkers with a BDL >10%, including FMN, 5-MTHF, pyridoxamine, pyridoxal, 4-pyridoxic acid and riboflavin, were dichotomized as high vs. low using median values as cutoffs. Due to skewed distributions, all the biomarkers were log-transformed except creatinine. Because the ratio of SAM to SAH is considered to reflect methylation potential, including histone methylation (Mentch et al., 2015), we also evaluated the ratio of SAM to SAH in this study. Furthermore, three additional ratios that are involved in FOCM, including the ratios of SAH to tHCY, tHCY to MET, and tHCY to CYSTA (Pacana et al., 2015; Stabler et al., 2013; Yi et al., 2000) were also assessed. The four ratios were dichotomized by median values.
As there is the potential for differential coverage and imputation quality by array, we first estimated genotype array-specific associations followed by a fixed-effect meta-analysis of the four arrays. Associations between imputed genetic dosage and plasma biomarkers were performed for each individual biomarker. Linear (for biomarkers treated as continuous) or logistic (for biomarkers treated as binary) regression models, adjusting for age, sex, centers and PCs of ancestry were used to obtain array-specific association estimates (regression coefficients and standard errors). All the biomarkers except creatinine were further adjusted for estimated Glomerular filtration rate (GFR) which was inferred from creatinine levels, age, sex and race using the Chronic Kidney Disease Epidemiology Collaboration equation (Levey et al., 2009) because GFR is associated with circulating one-carbon metabolite levels, particularly for tHCY (Francis, Eggers, Hostetter, & Briggs, 2004). An array-wide meta-analysis using fixed-effect models with inverse variance weighting was implemented in METAL (Willer, Li, & Abecasis, 2010).
Because the biomarkers were moderately correlated in this study (Supplementary Figure 1), we further evaluated biomarkers (those with BDL <15%, including creatinine, tHCY, MET, SAH, SAM, FMN and CYSTA) using principal component analysis (PCA) to derive patterns that best explained the maximal variation in these one-carbon metabolites. Since biomarker levels varied by study centers (Table 1 and Supplementary Table 2), for the PCA we first regressed each individual biomarker on variables known to be correlated with the biomarker levels (i.e. age, sex, current alcohol consumption status, current cigarette smoking status and body mass index [BMI]) in each study center and extracted the residuals from the multiple linear regression models. PCA was then performed on the residuals pooled from all study centers and principal components of the biomarkers were derived. We then evaluated the association between imputed genetic dosage and the biomarker principal components using similar analysis as we did for individual biomarker. We conducted analysis for the top four biomarker components accounting for approximately 70% of the variance in biomarker levels (20.3%, 18.5%, 15.6% and 14.7% respectively), respectively.
Table 1.
Study population characteristics and circulating biomarker levels according to arrays used for genotyping.
| GWAS Array |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Affymetrix Axiom (N=366) |
Illumina 1M/1M Duo (N=794) |
Illumina Omni1 (N=411) |
Illumina OncoArray (N=217) |
||||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD |
P
for differences |
|
| Age, yrs | 52.3 | 11.5 | 59.1 | 11.0 | 55.1 | 10.9 | 50.0 | 10.6 | <0.001 |
| BMI, kg/m2 | 26.8 | 5.3 | 26.6 | 5.0 | 26.8 | 4.9 | 27.2 | 5.4 | 0.50 |
| N | % | N | % | N | % | N | % | ||
| Female | 200 | 54.6 | 410 | 51.6 | 228 | 55.5 | 101 | 46.5 | 0.14 |
| Current alcohol intake status, yes | 223 | 60.9 | 517 | 65.1 | 207 | 50.4 | 104 | 47.9 | <0.001 |
| Current smoking status, yes | 49 | 13.4 | 84 | 10.6 | 58 | 14.1 | 20 | 9.2 | 0.13 |
| A history of adenomas | 83 | 22.7 | 91 | 11.5 | 115 | 28.0 | 15 | 6.9 | <0.001 |
| Center | <0.001 | ||||||||
| Ontario | 90 | 24.6 | 384 | 48.4 | 104 | 25.3 | 0 | ||
| USC/CSMC consortium | 79 | 21.6 | 0 | 0 | 114 | 27.7 | 0 | ||
| Australia | 93 | 25.4 | 173 | 21.8 | 10 | 2.4 | 0 | ||
| Mayo | 37 | 10.1 | 0 | 0 | 98 | 23.8 | 0 | ||
| Seattle | 67 | 18.3 | 237 | 29.8 | 85 | 20.7 | 217 | 100 | |
| Median | IQR | Median | IQR | Median | IQR | Median | IQR | ||
| Creatinine, mg/dl | 2.4 | 1.3 | 2.3 | 1.0 | 2.3 | 1.3 | 2.0 | 0.8 | <0.001 |
| SAH, ng/ml | 9.4 | 12.7 | 11.2 | 15.9 | 11.4 | 12.9 | 5.1 | 7.3 | <0.001 |
| tHCY, umol/l | 13.2 | 11.2 | 11.5 | 8.2 | 11.6 | 8.8 | 8.7 | 8.5 | <0.001 |
| METH, umol/l | 15.4 | 16.5 | 31.4 | 24.7 | 14.9 | 16.9 | 22.8 | 20.6 | <0.001 |
| CYSTA, ng/ml | 35.8 | 60.4 | 47.0 | 66.3 | 51.2 | 65.8 | 86.0 | 96.9 | <0.001 |
| SAM, ng/ml | 90.5 | 219.4 | 142.5 | 208.3 | 56.7 | 170.8 | 98.8 | 195.5 | <0.001 |
| Riboflavin, nmol/l | 1.1 | 11.7 | 0.6 | 18.0 | 1.9 | 16.2 | 15.3 | 43.3 | <0.001 |
| FMN, ng/ml | 2.7 | 12.0 | 11.4 | 30.0 | 3.0 | 15.7 | 6.2 | 11.5 | <0.001 |
| Pyridoxic acid, ng/ml | 2.5 | 13.9 | 2.1 | 24.4 | 7.2 | 23.1 | 7.3 | 23.2 | 0.0003 |
| Pyridoxal, ng/ml | 2.5 | 38.0 | 4.1 | 50.7 | 0.6 | 5.5 | 2.9 | 10.3 | <0.001 |
| Pyridoxamine, ng/ml | 1.9 | 2.1 | 2.1 | 1.9 | 2.1 | 2.5 | 1.5 | 2.0 | 0.001 |
| 5-MTHF, ng/ml | 6.3 | 16.2 | 9.0 | 19.0 | 10.0 | 17.1 | 21.3 | 24.4 | <0.001 |
| Ratio of SAM to SAH | 5.2 | 20.4 | 12.7 | 33.8 | 2.5 | 12.4 | 13.9 | 30.8 | <0.001 |
| Ratio of SAH to HCY | 0.005 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.005 | 0.01 | <0.001 |
| Ratio of tHCY to METH | 0.7 | 1.2 | 0.3 | 0.5 | 0.6 | 0.8 | 0.3 | 0.3 | <0.001 |
| Ratio of tHCY to CYSTA | 54.8 | 139.9 | 30.8 | 68.3 | 35.9 | 90.5 | 15.0 | 28.9 | <0.001 |
Abbreviation: SD, standard deviation. IQR, interquartile range. SAH, S-adenosylhomocysteine. SAM, S-adenosylmethionine. tHCY, total Homocysteine.
CYSTA, Cystathionine. FMN, Flavin-mononucleotide. 5-MTHF, 5-methyltetrahydrofolate.
Finally, as there are candidate genes known to play important roles in FOCM, we selected 24 genes (Supplementary Table 1) involved in FOCM according to prior studies (Cheng et al., 2015; Figueiredo, Levine, Crott, Baurley, & Haile, 2013; Levine et al., 2010; Ose et al., 2018). We used the ANNOVAR software to characterize the SNPs with gene-based annotation (K. Wang, Li, & Hakonarson, 2010) and included coding exonic, intronic and non-coding variants (Supplementary Table 1). A total of 1,316 SNPs associated with the 24 FOCM-related genes were identified and included in the analysis. For all the SNPs within a single gene, P values for SNPs were adjusted for multiple testing using Pact which takes into account the correlation among SNPs within a gene (Conneely & Boehnke, 2007). For a test within a single gene, an α-level of 0.05 after implementing Pact was used to determine statistical significance; across all 33 genes tested, a Bonferroni corrected α-level of 0.002 (0.05/33) was considered as the threshold.
For the identified genome-wide significant variants, we used ANNOVAR (K. Wang et al., 2010), HaploReg v4.1 (Ward & Kellis, 2012) and RegulomeDB (Boyle et al., 2012) to annotate the functional aspects of the variants. All statistical analysis was performed using R software (version 3.3.3).
Results
A total of 1,788 participants who had both imputed genetic data and biomarker measurements contributed to the analysis. We compared participants’ characteristics and plasma biomarker levels across the four arrays (Table 1 and Supplementary Table 2). The distributions of sex, BMI and current smoking status were similar across arrays while age, current alcohol consumption status, a history of adenomas and study center were significantly different by array. For instance, participants genotyped using Illumina 1M/1M-Duo array were older than those genotyped on the other three arrays and more likely to be current alcohol drinkers than those genotyped on Omni1 or OncoArray (Supplementary Table 2). This difference reflects study center differences – for example the Affymetrix Axiom included participants from all 5 participating study centers in this study while the OncoArray data for this investigation had participants from the Seattle center only. Plasma levels of all the biomarkers as well as the ratios of individual metabolites were significantly different across arrays, although the levels were similar between Illumina 1M/1M-Duo and Omni1. Small to moderate correlations were observed among seven one-carbon metabolites (Supplementary Figure 1). The strongest correlation was observed between SAH and MET (Spearman r = 0.31; P < 0.05). Plasma tHCY was significantly inversely correlated with MET (Spearman r = −0.25) and one of the vitamin B2 co-factors (i.e. FMN, Spearman r = −0.2), and positively correlated with SAM (Spearman r = 0.19).
Several genome-wide significant associations were identified with plasma tHCY, CYSTA and SAH in the meta-analysis of the four arrays. Variant rs35976024, located at 2p11.2 and intronic of ATOH8, was associated with tHCY levels: the A allele was linked to an approximately 10% decrease in tHCY levels (P = 4.89E-08; Table 2, Supplementary Table 3 and Figure 1). The inverse association was found in participants tested in all four arrays with similar effect estimates (Pheterogeneity = 0.91). Six variants, located at Chr15, were found significantly associated with CYSTA levels. Variant rs28391580 was the most significant (P = 2.82E-08; Table 2 and Figure 1) and was highly correlated with the other 5 SNPs (r2: 0.86 – 1.0; the highest correlation with rs28416399: r2 = 1.0). rs28391580 maps to 15q14 with the nearest gene TMCO5A (approximately 200kb away; Supplementary Table 3 and Supplementary Figure 2). In the meta-analysis, the effect estimates of all the 6 SNPs for plasma CYSTA were very similar (the effect allele was associated with approximately 20% decreased CYSTA levels). Two variants (moderately correlated in this study, r2 = 0.57), located at chr14, were identified as significantly associated with SAH levels in the meta-analysis of three of the arrays except the OncoArray (SNPs excluded due to poor quality). The stronger association was seen with variant rs139435405, which maps to 14q13 with two nearest genes being PTCSC3 and MBIP (approximately 5kb and 112kb away, respectively; Supplementary Table 3 and Supplementary Figure 2). The effect alleles of both SNPs (i.e. T and C for variants rs139435405 and rs149119426, respectively) was associated with about 26% increased SAH levels. Regional association plots for the genome-wide significant variants in Supplementary Figure 2 depict the meta-analysis of GWAS results in the context of their surrounding linkage disequilibrium (LD) structures and nearby genes. When assessing the four ratios (i.e. SAM to SAH, SAH to tHCY, tHCY to MET and tHCY to CYSTA) which were treated as high vs. low, we did not find genome-wide significant variants that were associated with any of the four ratios (Supplementary Figure 3).
Table 2.
Genome-wide significant variants† for circulating total HCY, CYSTA and SAH levels.
| Meta-analysis | Affymetrix Axiom |
Illumina 1M/1M Duo |
Illumina Omni1 |
Illumina OncoArray |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rsID | Chr | Position (BP) |
Gene | Allele Eff/Ref |
Eff allele freq |
Beta | S.E. | P | Beta | S.E. | P | Beta | S.E. | P | Beta | S.E. | P | Beta | S.E. | P | Phet |
| tHCY | |||||||||||||||||||||
| rs35976024 | 2 | 86003971 | ATOH8 | A/G | 0.32 | −0.10 | 0.02 | 4.89E-08 | −0.10 | 0.05 | 0.06 | −0.11 | 0.02 | 2.11E-06 | −0.11 | 0.05 | 0.04 | −0.04 | 0.09 | 0.62 | 0.91 |
| CYSTA | |||||||||||||||||||||
| rs28391580 | 15 | 38033956 | T/C | 0.27 | −0.24 | 0.04 | 2.82E-08 | −0.24 | 0.10 | 0.02 | −0.22 | 0.06 | 1.62E-04 | −0.34 | 0.11 | 0.001 | −0.18 | 0.11 | 0.11 | 0.75 | |
| rs28416399 | 15 | 38028526 | G/A | 0.27 | −0.24 | 0.04 | 2.97E-08 | −0.24 | 0.10 | 0.02 | −0.22 | 0.06 | 1.75E-04 | −0.34 | 0.11 | 0.001 | −0.18 | 0.11 | 0.11 | 0.74 | |
| rs765683 | 15 | 38018791 | T/C | 0.30 | −0.23 | 0.04 | 3.20E-08 | −0.27 | 0.10 | 0.01 | −0.18 | 0.06 | 1.21E-03 | −0.35 | 0.10 | 0.001 | −0.21 | 0.11 | 0.05 | 0.53 | |
| rs4924190 | 15 | 38017922 | T/C | 0.25 | −0.24 | 0.04 | 3.30E-08 | −0.23 | 0.10 | 0.03 | −0.22 | 0.06 | 2.69E-04 | −0.39 | 0.11 | 3.26E-04 | −0.17 | 0.11 | 0.12 | 0.52 | |
| rs11856650 | 15 | 38025137 | A/G | 0.27 | −0.23 | 0.04 | 3.75E-08 | −0.24 | 0.10 | 0.02 | −0.22 | 0.06 | 2.24E-04 | −0.34 | 0.11 | 0.001 | −0.18 | 0.11 | 0.11 | 0.74 | |
| rs8023982 | 15 | 38021714 | A/G | 0.27 | −0.23 | 0.04 | 4.08E-08 | −0.24 | 0.10 | 0.02 | −0.22 | 0.06 | 2.23E-04 | −0.33 | 0.11 | 0.001 | −0.18 | 0.11 | 0.11 | 0.76 | |
| SAH | |||||||||||||||||||||
| rs139435405 | 14 | 36655451 | C/T | 0.75 | 0.23 | 0.04 | 1.95E-08 | 0.27 | 0.09 | 0.002 | 0.19 | 0.05 | 5.43E-04 | 0.27 | 0.08 | 0.001 | N/A | N/A | N/A | 0.62 | |
| rs149119426 | 14 | 36656744 | T/C | 0.63 | 0.23 | 0.04 | 4.26E-08 | 0.20 | 0.09 | 0.02 | 0.20 | 0.06 | 3.73E-04 | 0.30 | 0.08 | 2.02E-04 | N/A | N/A | N/A | 0.61 | |
| Biomarker PC1 | |||||||||||||||||||||
| rs12665366 | 6 | 541907 | EXOC2 | G/A | 0.12 | −0.35 | 0.06 | 2.33E-08 | −0.44 | 0.15 | 0.003 | −0.26 | 0.08 | 0.001 | −0.43 | 0.16 | 0.005 | −0.80 | 0.26 | 0.002 | 0.18 |
Abbreviation: Chr, chromosome. S.E., standard error. PC, principal component. tHCY, total Homocysteine. CYSTA, Cystathionine. SAH, S-adenosylhomocysteine. Phet. Pheterogeneity.
Variants identified in at least three arrays.
Figure 1.
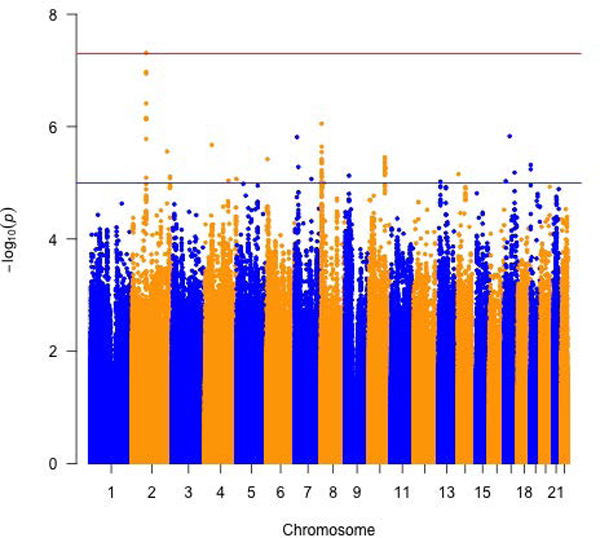
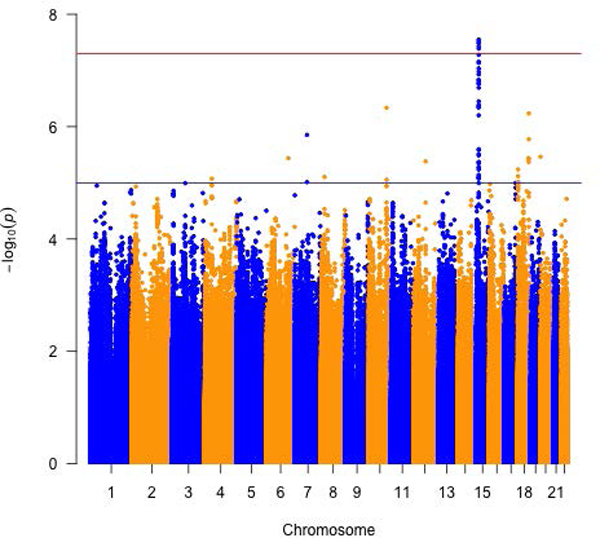
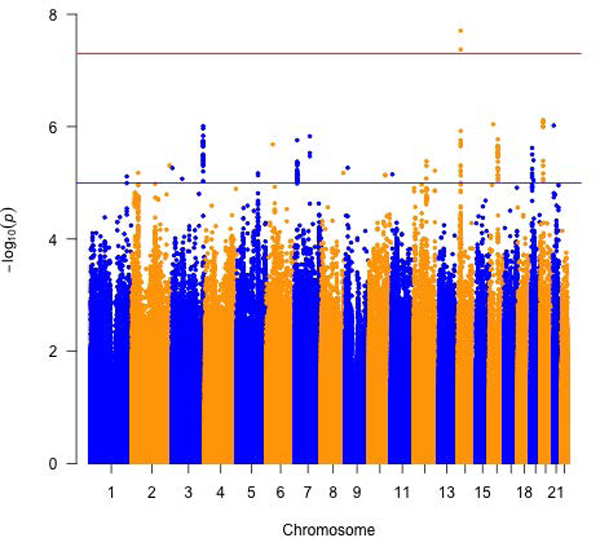
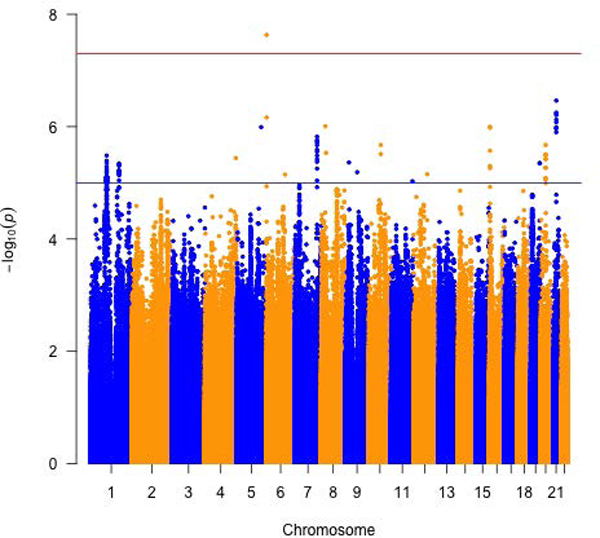
Meta-analysis of three or more arrays for circulating folate one-carbon metabolites. Manhattan plots for a) total Homocysteine, b) Cystathionine and c) S-adenosylhomocysteine and d) biomarker PC1.
When the seven metabolites (i.e. creatinine, MET, SAH, SAM, tHCY, CYSTA and FMN) were analyzed as principal components, the first PC correlated positively with all seven biomarkers. The strongest correlation was with tHCY (eigenvector = 0.63), followed by creatinine, SAM and SAH (eigenvector = 0.49, 0.38 and 0.32, respectively) (Supplementary Table 4). The G allele of variant rs12665366, located at 6p25.3 and intronic of EXOC2, was inversely associated with PC1 (P = 2.33E-08; Table 2 and Figure 1). No significant heterogeneity in array-specific associations was observed except the effect estimate from the OncoArray was stronger (Pheterogeneity = 0.18).
We further evaluated the 1,316 SNPs in 24 genes that are known to play important roles in FOCM according to prior studies (Cheng et al., 2015; Levine et al., 2010; Ose et al., 2018). Out of the 1,316 SNPs, 39 were mapped to coding exonic variants and the rest (1,277) were intronic or non-coding variants (Supplementary Table 1). After taking into account multiple comparisons, we observed several SNPs within a single gene that were significantly (Pact < 0.05) associated with a specific one-carbon metabolite (Table 3a and 3b). For instance, six SNPs of MTHFR, including one of the well-studied SNP (rs1801131, located at 1p36.3 and exonic of MTHFR), were associated with plasma tHCY levels. The A allele of variant rs1801131 was associated with approximately 7% increased tHCY levels in the meta-analysis and similar positive associations were observed across the four arrays.
Table 3.
Candidate gene SNPs involved in folate one-carbon metabolism and circulating folate one-carbon metabolites† in meta analysis (Table 3a) and by each array (Table 3b). Table 3a.
| Table 3a. | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Meta-analysis |
||||||||||
| rsID | Chr | Position (BP) | Gene | Location | Allele Eff/Ref |
Eff allele freq |
Beta | S.E. | P | Pact |
| Creatinine | ||||||||||
| rs12485165 | 22 | 31020341 | TCN2 | Intronic | T/G | 0.13 | 0.16 | 0.04 | 3.61E-04 | 0.01 |
| rs150167372 | 22 | 31020114 | TCN2 | Intronic | C/T | 0.88 | 0.16 | 0.05 | 4.92E-04 | 0.01 |
| rs9621049 | 22 | 31013419 | TCN2 | Exonic | T/C | 0.12 | 0.16 | 0.05 | 5.01E-04 | 0.01 |
| rs12168392 | 22 | 31022301 | TCN2 | Intronic | A/G | 0.12 | 0.16 | 0.05 | 5.72E-04 | 0.01 |
| rs4820886 | 22 | 31016539 | TCN2 | Intronic | G/T | 0.88 | 0.16 | 0.05 | 6.11E-04 | 0.01 |
| rs1004474 | 18 | 660383 | TYMS | Intronic | G/A | 0.54 | −0.08 | 0.03 | 4.84E-03 | 0.04 |
| HCY | ||||||||||
| rs1801131 | 1 | 11854476 | MTHFR | Exonic | G/T | 0.69 | 0.07 | 0.02 | 7.89E-04 | 0.01 |
| rs7526128 | 1 | 11857240 | MTHFR | Intronic | T/C | 0.61 | −0.06 | 0.02 | 2.36E-03 | 0.03 |
| rs1476413 | 1 | 11852300 | MTHFR | Intronic | T/C | 0.26 | 0.06 | 0.02 | 2.54E-03 | 0.03 |
| rs6541005 | 1 | 11857525 | MTHFR | Intronic | A/T | 0.43 | −0.06 | 0.02 | 2.68E-03 | 0.03 |
| rs3818762 | 1 | 11851003 | MTHFR | Intronic | C/G | 0.27 | 0.06 | 0.02 | 4.08E-03 | 0.04 |
| rs17367504 | 1 | 11862778 | MTHFR | Intronic | A/G | 0.85 | 0.07 | 0.03 | 5.14E-03 | 0.04 |
| SAM | ||||||||||
| rs4911253 | 20 | 31352585 | DNMT3B | Intronic | A/G | 0.56 | −0.17 | 0.06 | 3.00E-03 | 0.02 |
| rs2424905 | 20 | 31352927 | DNMT3B | Intronic | T/C | 0.57 | −0.16 | 0.06 | 4.00E-03 | 0.03 |
| rs6087995 | 20 | 31358253 | DNMT3B | Intronic | A/C | 0.57 | −0.16 | 0.06 | 5.00E-03 | 0.04 |
| rs72765189 | 9 | 140525434 | EHMT1 | Intronic | A/G | 0.11 | 0.28 | 0.09 | 2.19E-03 | 0.02 |
| rs190756635 | 9 | 140530183 | EHMT1 | Intronic | G/A | 0.89 | 0.28 | 0.09 | 2.66E-03 | 0.03 |
| rs111275198 | 9 | 140530274 | EHMT1 | Intronic | A/G | 0.89 | 0.27 | 0.09 | 2.80E-03 | 0.03 |
| rs57705093 | 5 | 162944172 | MAT2B | Intronic | T/A | 0.73 | −0.23 | 0.06 | 3.25E-04 | 0.005 |
| 5-MethyTHF | ||||||||||
| rs2235523 | 1 | 14096457 | PRDM2 | Intronic | A/T | 0.37 | 0.29 | 0.09 | 1.22E-03 | 0.04 |
| rs3891167 | 18 | 658423 | TYMS | Intronic | G/A | 0.75 | −0.33 | 0.11 | 2.09E-03 | 0.02 |
| METH | ||||||||||
| rs34973186 | 11 | 59598262 | GIF | Intronic | T/C | 0.07 | 0.17 | 0.05 | 1.92E-03 | 0.01 |
| Pyridoxal acid | ||||||||||
| rs3788202 | 21 | 46957218 | SLC19A1 | Intronic | G/A | 0.26 | 0.28 | 0.09 | 2.65E-03 | 0.03 |
| rs12801088 | 11 | 4116335 | RRM1 | Intronic | A/C | 0.08 | −0.42 | 0.16 | 7.07E-03 | 0.04 |
| Flavin mononucleotide | ||||||||||
| rs34973186 | 11 | 59598262 | GIF | Intronic | T/C | 0.07 | −0.29 | 0.11 | 1.10E-02 | 0.03 |
| Pyridoxamine | ||||||||||
| rs2244171‡ | 21 | 44496594 | CBS | Intronic | T/G | 0.08 | 0.58 | 0.22 | 8.08E-03 | N/A |
| rs3784621 | 15 | 48633092 | DUT | Intronic | C/T | 0.82 | −0.42 | 0.15 | 4.11E-03 | 0.01 |
| rs10851465 | 15 | 48629884 | DUT | Intronic | T/C | 0.18 | −0.41 | 0.15 | 4.96E-03 | 0.01 |
| rs11637235 | 15 | 48633153 | DUT | Intronic | T/C | 0.78 | −0.36 | 0.14 | 8.48E-03 | 0.02 |
| rs3784619 | 15 | 48625938 | DUT | Intronic | A/G | 0.85 | −0.33 | 0.15 | 2.80E-02 | 0.04 |
| rs12592155 | 15 | 48623500 | DUT | Intronic | A/C | 0.15 | 0.33 | 0.15 | 2.85E-02 | 0.03 |
| rs12592157 | 15 | 48623524 | DUT | Intronic | C/G | 0.85 | 0.33 | 0.15 | 2.92E-02 | 0.03 |
| Table 3b. | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Affymetrix Axiom | Illumina 1M/1M Duo | Illumina Omni1 | Illumina OncoArray | |||||||||
| rsID | Beta | S.E. | P | Beta | S.E. | P | Beta | S.E. | P | Beta | S.E. | P |
| Creatinine | ||||||||||||
| rs12485165 | 0.18 | 0.13 | 0.18 | 0.10 | 0.06 | 0.10 | 0.22 | 0.10 | 0.03 | 0.22 | 0.10 | 0.02 |
| rs150167372 | 0.17 | 0.14 | 0.23 | 0.13 | 0.07 | 0.05 | 0.20 | 0.10 | 0.05 | 0.20 | 0.10 | 0.05 |
| rs9621049 | 0.17 | 0.14 | 0.21 | 0.12 | 0.06 | 0.05 | 0.20 | 0.10 | 0.05 | 0.20 | 0.10 | 0.05 |
| rs12168392 | 0.17 | 0.14 | 0.20 | 0.12 | 0.06 | 0.06 | 0.19 | 0.10 | 0.05 | 0.20 | 0.10 | 0.05 |
| rs4820886 | 0.17 | 0.14 | 0.21 | 0.12 | 0.06 | 0.05 | 0.19 | 0.10 | 0.06 | 0.20 | 0.10 | 0.05 |
| rs1004474 | −0.12 | 0.08 | 0.13 | −0.09 | 0.04 | 0.03 | −0.005 | 0.07 | 0.94 | −0.09 | 0.06 | 0.13 |
| HCY | ||||||||||||
| rs1801131 | 0.14 | 0.05 | 0.01 | 0.06 | 0.02 | 0.02 | 0.05 | 0.06 | 0.38 | 0.07 | 0.09 | 0.41 |
| rs7526128 | −0.12 | 0.05 | 0.01 | −0.04 | 0.02 | 0.06 | −0.05 | 0.05 | 0.28 | −0.07 | 0.08 | 0.41 |
| rs1476413 | 0.14 | 0.06 | 0.01 | 0.05 | 0.03 | 0.04 | 0.03 | 0.06 | 0.65 | 0.08 | 0.09 | 0.37 |
| rs6541005 | −0.14 | 0.05 | 0.005 | −0.04 | 0.02 | 0.09 | −0.05 | 0.05 | 0.35 | −0.09 | 0.09 | 0.30 |
| rs3818762 | 0.14 | 0.05 | 0.01 | 0.05 | 0.03 | 0.05 | 0.04 | 0.06 | 0.51 | 0.04 | 0.09 | 0.62 |
| rs17367504 | 0.06 | 0.07 | 0.44 | 0.08 | 0.03 | 0.01 | 0.09 | 0.07 | 0.19 | −0.01 | 0.11 | 0.95 |
| SAM | ||||||||||||
| rs4911253 | −0.15 | 0.13 | 0.26 | −0.23 | 0.08 | 0.002 | −0.16 | 0.14 | 0.24 | 0.12 | 0.18 | 0.49 |
| rs2424905 | −0.14 | 0.13 | 0.27 | −0.23 | 0.08 | 0.003 | −0.15 | 0.14 | 0.27 | 0.15 | 0.18 | 0.39 |
| rs6087995 | −0.16 | 0.13 | 0.22 | −0.22 | 0.08 | 0.004 | −0.13 | 0.14 | 0.33 | 0.11 | 0.18 | 0.55 |
| rs72765189 | 0.32 | 0.21 | 0.11 | 0.34 | 0.13 | 0.01 | 0.29 | 0.24 | 0.20 | −0.01 | 0.27 | 0.97 |
| rs190756635 | 0.33 | 0.21 | 0.10 | 0.35 | 0.13 | 0.01 | 0.18 | 0.22 | 0.40 | 0.02 | 0.27 | 0.94 |
| rs111275198 | 0.32 | 0.21 | 0.12 | 0.33 | 0.13 | 0.01 | 0.23 | 0.22 | 0.28 | 0.01 | 0.27 | 0.96 |
| rs57705093 | 0.03 | 0.15 | 0.82 | −0.38 | 0.09 | 8.3E-06 | −0.14 | 0.15 | 0.34 | −0.02 | 0.19 | 0.92 |
| 5-MethyTHF | ||||||||||||
| rs2235523 | 0.31 | 0.21 | 0.13 | 0.20 | 0.13 | 0.13 | 0.33 | 0.20 | 0.09 | 0.59 | 0.27 | 0.02 |
| rs3891167 | −0.65 | 0.25 | 0.01 | −0.15 | 0.16 | 0.33 | −0.49 | 0.25 | 0.05 | −0.32 | 0.28 | 0.26 |
| METH | ||||||||||||
| rs34973186 | 0.07 | 0.13 | 0.57 | 0.20 | 0.07 | 0.01 | 0.07 | 0.14 | 0.62 | 0.23 | 0.13 | 0.06 |
| Pyridoxal acid | ||||||||||||
| rs3788202 | 0.63 | 0.21 | 0.003 | 0.21 | 0.14 | 0.13 | 0.30 | 0.20 | 0.13 | 0.01 | 0.25 | 0.98 |
| rs12801088 | −0.29 | 0.35 | 0.42 | −0.27 | 0.22 | 0.22 | −0.84 | 0.36 | 0.02 | −0.60 | 0.45 | 0.17 |
| Flavin mononucleotide | ||||||||||||
| rs34973186 | −0.11 | 0.20 | 0.57 | −0.36 | 0.19 | 0.06 | −0.34 | 0.27 | 0.20 | −0.42 | 0.28 | 0.13 |
| Pyridoxamine | ||||||||||||
| rs2244171‡ | −0.01 | 0.42 | 0.97 | 0.99 | 0.36 | 0.004 | 0.75 | 0.61 | 0.21 | 0.54 | 0.47 | 0.26 |
| rs3784621 | −0.21 | 0.37 | 0.58 | −0.51 | 0.22 | 0.02 | −0.08 | 0.37 | 0.83 | −0.58 | 0.29 | 0.05 |
| rs10851465 | −0.15 | 0.37 | 0.69 | −0.52 | 0.22 | 0.02 | −0.06 | 0.38 | 0.88 | −0.59 | 0.29 | 0.05 |
| rs11637235 | −0.14 | 0.35 | 0.68 | −0.35 | 0.20 | 0.08 | −0.33 | 0.35 | 0.34 | −0.54 | 0.28 | 0.06 |
| rs3784619 | −0.12 | 0.38 | 0.75 | −0.43 | 0.23 | 0.05 | −0.06 | 0.40 | 0.88 | −0.45 | 0.30 | 0.14 |
| rs12592155 | 0.13 | 0.38 | 0.73 | 0.43 | 0.23 | 0.05 | 0.06 | 0.40 | 0.89 | 0.43 | 0.30 | 0.15 |
| rs12592157 | 0.13 | 0.38 | 0.73 | 0.43 | 0.23 | 0.05 | 0.05 | 0.40 | 0.90 | 0.43 | 0.30 | 0.15 |
Abbreviation: SNP, Single nucleotide polymorphism. tHCY, total Homocysteine. SAM, S-adenosylmethionine. FMN, Flavin-mononucleotide. 5-MTHF, 5-methyltetrahydrofolate. MET: Methionine.
Only SNPs with Pact < 0.05 presented.
1 single SNP was included for CBS.
Finally, since several SNPs have been found significantly associated with circulating HCY, vitamin B12 and vitamin B6 levels in prior GWA studies (Hazra et al., 2009; Hazra et al., 2008; Tanaka et al., 2009), we assessed those variants in this study. We were only able to assess prior identified SNPs for plasma tHCY because vitamin B12 was not evaluated in our study and different metabolites of vitamin B6 were assessed in our study compared to previous ones (Hazra et al., 2009; Tanaka et al., 2009). However, we did not observe significant associations between previously identified SNPs and plasma tHCY levels (Supplementary Table 5).
Discussion
Genetic factors have long been hypothesized to influence circulating levels of folate and associated metabolites (Hustad et al., 2007; Nilsson, Read, Berg, & Johansson, 2009; Siva et al., 2007; Thuesen et al., 2010), however, only a few studies conducted genome-wide assessment of the genetic determinants of biomarkers involved in FOCM (Hazra et al., 2009; Hazra et al., 2008; Tanaka et al., 2009). Furthermore, none of the prior studies evaluated genetic determinants for metabolites other than B vitamins involved in FOCM, including MET, CYSTA and SAM. Circulating concentrations of several those metabolites, such as MET, have been found to be associated with CRC risk, individually (Myte et al., 2016) or together with folate levels (Nitter et al., 2014). Thus, we performed a GWAS to identify common genetic variants that influence plasma levels of 12 FOCM biomarkers among 1,788 unaffected (free of cancer) participants. We identified variant rs35976024 (located at 2p11.2 and intronic of ATOH8), 6 variants on chromosome 15q14 and 2 variants on chromosome 14q13 which demonstrated significant (P < 5 × 10−8) associations with tHCY, CYSTA and SAH, respectively. Additionally, when assessing principal components derived from 7 individual metabolites, we found that variant rs12665366 (located at 6p25.3 and intronic of EXOC2) was significantly associated with the first principal component, a component characterized with a strong positive correlation with plasma tHCY and moderate correlation with SAM, SAH, CYSTA and creatinine.
The locus (rs35976024) which was found to be significantly associated with plasma tHCY is located at chromosome 2 (2p11.2) and intronic of ATOH8. The variant rs35976024 overlaps with an enhancer (i.e. H3K4me1) detected in colon tissue and is likely to affect transcription factor binding site and result in the binding motif changes (Supplementary Table 3). ATOH8 basic-helix-loop-helix transcription factor involved in embryogenesis (B. Wang, Balakrishnan-Renuka, Napirei, Theiss, & Brand-Saberi, 2015) and the development of multiple tissues (Fang et al., 2014; Guttsches et al., 2015; Inoue et al., 2001; Lynn, Sanchez, Gomis, German, & Gasa, 2008). It has been reported to be associated with tumor progression in several types of cancer, including CRC (Ye et al., 2017). ATOH8 expression measured by immunohistochemistry in CRC tumor tissue was significantly higher than that in tumor-adjacent normal tissue and was associated with a worse overall survival (Ye et al., 2017). Copy number amplification of ATOH8 was found in glioblastoma tissue (Freire et al., 2008). On the other hand, ATOH8 mRNA expression was found substantially lower in tumor tissue than in tumor-adjacent normal tissue in other types of cancer, including hepatocellular carcinoma (Song et al., 2015) and Nasopharyngeal carcinoma (Z. Wang et al., 2016). In addition, the decreased expression of ATOH8 in hepatocellular carcinoma was associated with a significant reduction in disease-free survival (Song et al., 2015). However, despite the apparent role of the ATOH8 gene in tumor progression, whether it is involved in FOCM remains largely unknown. Variants identified for plasma CYSTA or SAH are intergenic. A group (N=6) of highly correlated SNPs at chromosome 15 (15q14) were found to be associated with circulating CYSTA levels. Although it is likely that this specific region may influence CYSTA levels, the functional analysis of this region is largely unknown. These 6 variants appeared not to influence regulatory elements, such as enhancer or promoter elements, although some of them may lead to binding motif changes (Supplementary Table 3). The nearest gene to the 6 variants is TMCO5A which is approximately 200kb upstream of them. For plasma SAH, although the strongest SNP (rs139435405) is also intergenic, it is located approximately 5kb downstream of PTCSC3 which is a thyroid-specific long non-coding RNA. PTCSC3 is substantially down-regulated in papillary thyroid carcinoma (PTC) and appears to act as a tumor suppressor gene (Fan et al., 2013; Jendrzejewski et al., 2012). Despite no apparent association between folate status and thyroid cancer, the C677T polymorphism in MTHFR gene (rs1801133), a known SNP that affects folate metabolism, has been reported to be associated with thyroid cancer risk (Vu-Phan & Koenig, 2014). However, this variant rs139435405 does not appear to alter regulatory elements (Supplementary Table 3).
Given the correlation between metabolites involved in FOCM, we assessed 7 markers (i.e. those with BDL <15%, including creatinine, tHCY, MET, SAH, SAM, FMN and CYSTA) using PCA to derive patterns that best capture the maximal variation in these metabolites. We identified a SNP rs12665366, located at chromosome 6 (6p25.3) and intronic of EXOC2, that was significantly associated with the first PC. However, no evidence has shown that the variant rs12665366 may substantially affect regulatory elements (e.g. enhancer or promoter; Supplementary Table 3). The first PC accounts approximately 20.3% variation of the 7 metabolites and was positively correlated most strongly with tHCY. A SNP rs1540771, located at the same 6p25.3 region and lying between IRF4 and EXOC2, was identified in prior GWAS to be significantly associated with the presence of freckles in Europeans (Sulem et al., 2007), but it is not correlated with rs12665366 (r2 < 0.01; ~76kb away from it). Furthermore, another intronic variant in EXOC2 (rs9328342) which was approximately 192kb away from, but again not correlated with rs12665366 (r2 < 0.01), has been reported to be associated with serum 25-hydroxyvitamin D levels (Saternus et al., 2015). Therefore, if the association of rs12665366 with biomarkers of FOCM, particularly tHCY, is replicated in future studies, it suggests that 6p25.3 region may be biologically-relevant to metabolism of multiple nutrients.
Of the 12 one-carbon metabolites assessed individually or using principal component analysis, we identified genome-wide significant variants that are associated with plasma tHCY, CYSTA or SAH. Results from our study suggest that genetic factors may play an important role in HCY metabolism. HCY is metabolized through two major pathways: re-methylation and transsulfuration (Selhub, 1999). In re-methylation, HCY acquires a methyl group from 5-MTHF and vitamin B12 folate to form MET. Alternatively, it acquires the methyl group from betaine to form MET, independent of vitamin B12. In transsulfuration, HCY condenses with serine to irreversibly form CYSTA, which requires pyridoxal-5’-phosphate (the active form of vitamin B6). A substantial proportion of MET is activated to form SAM, the universal methyl group carrier. SAH which is generated by demethylation of SAM is then hydrolyzed to form HCY, which becomes available for a new cycle of methyl group transfer. In addition, changes in MET levels can lead to changes in SAM/SAH ratio (Mentch et al., 2015) that has a profound impact on methylation reactions. HCY which is not re-methylated to MET or transsulfurated to CYSTA is quickly exported to circulation. Thus, our findings generally support that genetic variants may influence pathways related to HCY metabolism. Furthermore, our results are consistent with findings of relatively high heritability of circulating levels of tHCY in previous studies. Heritability calculated from utilizing monozygotic (MZ) and dizygotic (DZ) twins is estimated to be more than 50% for circulating folate (Nilsson et al., 2009) and approximately 60% for circulating tHCY (Nilsson et al., 2009; Siva et al., 2007). Previous studies focusing on key genes involved in FOCM found a variant in MTHFR (i.e. C677T, rs1801133) to be associated with circulating folate and tHCY levels (Hustad et al., 2007; Thuesen et al., 2010). Thus, if replicated in future studies, our results provide new insights in the genetic influence on folate-mediated metabolism, particularly HCY metabolism. This is interesting given that high tHCY levels have been reported to be associated with CRC risk (Miller et al., 2013).
A major strength of this study is the genome-wide assessment of genetic determinants not only for B vitamins (i.e. folate, vitamin B2, vitamin B6) but also other components involved in FOCM, while previous studies focused mostly on B vitamins. Another major strength is the use of a single laboratory for the measurement of all the biomarkers despite participants in this study being recruited from multiple sites. Furthermore, we (Louie and Asante) have developed a validated and more sensitive and precise assay to quantify blood B vitamins. This validated multi-analyte LC-MS method can simultaneously measure the endogenous plasma levels of metabolites involved in FOCM (Asante et al., 2018). However, our study also has several limitations. A major limitation is the lack of validation of the identified GWAS SNPs in our studies. However, to our best knowledge, there seem no existing studies which conducted GWA analysis on biomarkers involved in one-carbon metabolism in addition to B vitamins. Another limitation was that participants were genotyped on 4 different arrays with a very small number of overlapping SNPs (N=26,797). However, we conducted a rigorous QC on genotyped SNPs and thus were able to generate accurately imputed SNP data. Furthermore, although this is a large study for the measurement of biomarkers, the sample size is small for a GWAS, possibly contributing to the lack of variants identified to have associations with plasma B vitamins. Other limitations include lack of racial/ethnic diversity (i.e. European ancestry only) in the study population and lack of information on dietary folic and methionine intake. FOCM-related dietary intake may modify the association of FOCM-related genetic variants with colon cancer risk (Liu et al., 2013); however, we were not able to perform the G-E interaction tests in this study (e.g. assess whether dietary folic and/or methionine intake may modify the SNP -FOCM biomarker association). This may be warranted further investigation in future larger-scale studies with information on FOCM-related dietary intake.
In conclusion, in this genome-wide association analysis of biomarkers involved in FOCM, some of which were assessed for the first time to our knowledge, we identified several genetic variants or regions which are associated with circulating tHCY, SAH and CYSTA. As these biomarkers are specifically involved in the pathway of HCY metabolism, if replicated in future studies, results from our study provides support that common gene variants may play an important role in FOCM, particularly HCY.
Supplementary Material
Acknowledgments
Financial support: This work has been funded by the following NIH grants: R01CA140561, U01CA122839, and P01CA196569. The Colon Cancer Family Registry (CCFR) Illumina GWAS was supported in part by NCI/NIH grants U01 CA122839, R01 CA143237 and U01 CA167551. The CORECT Affymetrix Axiom and OncoArray GWAS was supported in part by NCI/NIH grant U19 CA148107. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the CCFR, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government, any cancer registry, or the CCFR.
Abbreviations:
- GWAS
Genome-wide Association Study
- FOCM
Folate One-carbon Metabolism
- MET
Methionine
- tHCY
total Homocysteine
- SAH
S-adenosylhomocysteine
- SAM
S-adenosylmethionine
- CYSTA
Cystathionine
- FMN
Flavin-mononucleotide
- 5-MTHF
5-methyltetrahydrofolate
- CRC
colorectal cancer
- CCFR
Colon Cancer Family Registry
- BDL
Below Detection Limit
- LD
Linkage Disequilibrium
- PTC
Papillary Thyroid Carcinoma
- PCA
Principal Component Analysis
- LC-MS
Liquid chromatography–mass spectrometry
Footnotes
Conflicts of interest: The authors declare no potential conflicts of interest.
Data Availability
The genetic data in this study are openly available in a public repository and the circulating biomarker data are available upon reasonable request from the authors.
References
- Amos CI, Dennis J, Wang Z, Byun J, Schumacher FR, Gayther SA, … Easton DF (2017). The OncoArray Consortium: A Network for Understanding the Genetic Architecture of Common Cancers. Cancer Epidemiol Biomarkers Prev, 26(1), 126–135. doi: 10.1158/1055-9965.epi-16-0106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asante I, Pei H, Zhou E, Liu S, Chui D, Yoo E, & Louie SG (2018). Simultaneous quantitation of folates, flavins and B6 metabolites in human plasma by LC-MS/MS assay: Applications in colorectal cancer. J Pharm Biomed Anal, 158, 66–73. doi: 10.1016/j.jpba.2018.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, … Snyder M (2012). Annotation of functional variation in personal genomes using RegulomeDB. Genome Res, 22(9), 1790–1797. doi: 10.1101/gr.137323.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles D, Ness AR, Campbell D, Davey Smith G, & Hall MH (2004). Taking folate in pregnancy and risk of maternal breast cancer. BMJ, 329(7479), 1375–1376. doi: 10.1136/bmj.329.7479.1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng TY, Makar KW, Neuhouser ML, Miller JW, Song X, Brown EC, … Ulrich CM (2015). Folate-mediated one-carbon metabolism genes and interactions with nutritional factors on colorectal cancer risk: Women’s Health Initiative Observational Study. Cancer, 121(20), 3684–3691. doi: 10.1002/cncr.29465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole BF, Baron JA, Sandler RS, Haile RW, Ahnen DJ, Bresalier RS, … Greenberg ER (2007). Folic acid for the prevention of colorectal adenomas: a randomized clinical trial. JAMA, 297(21), 2351–2359. doi: 10.1001/jama.297.21.2351 [DOI] [PubMed] [Google Scholar]
- Conneely KN, & Boehnke M (2007). So many correlated tests, so little time! Rapid adjustment of P values for multiple correlated tests. Am J Hum Genet, 81(6), 1158–1168. doi: 10.1086/522036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducker GS, & Rabinowitz JD (2017). One-Carbon Metabolism in Health and Disease. Cell Metab, 25(1), 27–42. doi: 10.1016/j.cmet.2016.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre S, Bowes J, Diogo D, Lee A, Barton A, Martin P, … Worthington J (2012). High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat Genet, 44(12), 1336–1340. doi: 10.1038/ng.2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Li X, Jiang W, Huang Y, Li J, & Wang Z (2013). A long non-coding RNA, PTCSC3, as a tumor suppressor and a target of miRNAs in thyroid cancer cells. Exp Ther Med, 5(4), 1143–1146. doi: 10.3892/etm.2013.933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F, Wasserman SM, Torres-Vazquez J, Weinstein B, Cao F, Li Z, … Pei X (2014). The role of Hath6, a newly identified shear-stress-responsive transcription factor, in endothelial cell differentiation and function. J Cell Sci, 127(Pt 7), 1428–1440. doi: 10.1242/jcs.136358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O (2002). Metabolomics–the link between genotypes and phenotypes. Plant molecular biology, 48(1–2), 155–171. [PubMed] [Google Scholar]
- Figueiredo JC, Grau MV, Haile RW, Sandler RS, Summers RW, Bresalier RS, … Baron JA (2009). Folic acid and risk of prostate cancer: results from a randomized clinical trial. J Natl Cancer Inst, 101(6), 432–435. doi: 10.1093/jnci/djp019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo JC, Levine AJ, Crott JW, Baurley J, & Haile RW (2013). Folate-genetics and colorectal neoplasia: what we know and need to know next. Mol Nutr Food Res, 57(4), 607–627. doi: 10.1002/mnfr.201200278 [DOI] [PubMed] [Google Scholar]
- Francis ME, Eggers PW, Hostetter TH, & Briggs JP (2004). Association between serum homocysteine and markers of impaired kidney function in adults in the United States. Kidney Int, 66(1), 303–312. doi: 10.1111/j.1523-1755.2004.00732.x [DOI] [PubMed] [Google Scholar]
- Freire P, Vilela M, Deus H, Kim YW, Koul D, Colman H, … Almeida JS (2008). Exploratory analysis of the copy number alterations in glioblastoma multiforme. PLoS One, 3(12), e4076. doi: 10.1371/journal.pone.0004076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannucci E (2002). Epidemiologic studies of folate and colorectal neoplasia: a review. J Nutr, 132(8 Suppl), 2350S–2355S. doi: 10.1093/jn/132.8.2350S [DOI] [PubMed] [Google Scholar]
- Guttsches AK, Balakrishnan-Renuka A, Kley RA, Tegenthoff M, Brand-Saberi B, & Vorgerd M (2015). ATOH8: a novel marker in human muscle fiber regeneration. Histochem Cell Biol, 143(5), 443–452. doi: 10.1007/s00418-014-1299-6 [DOI] [PubMed] [Google Scholar]
- Hazra A, Kraft P, Lazarus R, Chen C, Chanock SJ, Jacques P, … Hunter DJ (2009). Genome-wide significant predictors of metabolites in the one-carbon metabolism pathway. Hum Mol Genet, 18(23), 4677–4687. doi: 10.1093/hmg/ddp428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra A, Kraft P, Selhub J, Giovannucci EL, Thomas G, Hoover RN, … Hunter DJ (2008). Common variants of FUT2 are associated with plasma vitamin B12 levels. Nat Genet, 40(10), 1160–1162. doi: 10.1038/ng.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S, Sanchez H, Albala C, de la Maza MP, Barrera G, Leiva L, & Bunout D (2009). Colon cancer in Chile before and after the start of the flour fortification program with folic acid. Eur J Gastroenterol Hepatol, 21(4), 436–439. doi: 10.1097/MEG.0b013e328306ccdb [DOI] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, & Marchini J (2009). A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet, 5(6), e1000529. doi: 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hustad S, Midttun O, Schneede J, Vollset SE, Grotmol T, & Ueland PM (2007). The methylenetetrahydrofolate reductase 677C-->T polymorphism as a modulator of a B vitamin network with major effects on homocysteine metabolism. Am J Hum Genet, 80(5), 846–855. doi: 10.1086/513520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue C, Bae SK, Takatsuka K, Inoue T, Bessho Y, & Kageyama R (2001). Math6, a bHLH gene expressed in the developing nervous system, regulates neuronal versus glial differentiation. Genes Cells, 6(11), 977–986. [DOI] [PubMed] [Google Scholar]
- Jendrzejewski J, He H, Radomska HS, Li W, Tomsic J, Liyanarachchi S, … de la Chapelle A (2012). The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc Natl Acad Sci U S A, 109(22), 8646–8651. doi: 10.1073/pnas.1205654109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy DA, Stern SJ, Moretti M, Matok I, Sarkar M, Nickel C, & Koren G (2011). Folate intake and the risk of colorectal cancer: a systematic review and meta-analysis. Cancer Epidemiol, 35(1), 2–10. doi: 10.1016/j.canep.2010.11.004 [DOI] [PubMed] [Google Scholar]
- Kim YI (2003). Role of folate in colon cancer development and progression. J Nutr, 133(11 Suppl 1), 3731S–3739S. doi: 10.1093/jn/133.11.3731S [DOI] [PubMed] [Google Scholar]
- Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF 3rd, Feldman HI, … Coresh J (2009). A new equation to estimate glomerular filtration rate. Ann Intern Med, 150(9), 604–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Figueiredo JC, Lee W, Conti DV, Kennedy K, Duggan DJ, … Haile RW (2010). A candidate gene study of folate-associated one carbon metabolism genes and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev, 19(7), 1812–1821. doi: 10.1158/1055-9965.epi-10-0151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, … Zhang M (2008). Symbiotic gut microbes modulate human metabolic phenotypes. Proceedings of the National Academy of Sciences, 105(6), 2117–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AY, Scherer D, Poole E, Potter JD, Curtin K, Makar K, … Ulrich CM (2013). Gene-diet-interactions in folate-mediated one-carbon metabolism modify colon cancer risk. Mol Nutr Food Res, 57(4), 721–734. doi: 10.1002/mnfr.201200180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan RF, Grainge MJ, Shepherd VC, Armitage NC, & Muir KR (2008). Aspirin and folic acid for the prevention of recurrent colorectal adenomas. Gastroenterology, 134(1), 29–38. doi: 10.1053/j.gastro.2007.10.014 [DOI] [PubMed] [Google Scholar]
- Lynn FC, Sanchez L, Gomis R, German MS, & Gasa R (2008). Identification of the bHLH factor Math6 as a novel component of the embryonic pancreas transcriptional network. PLoS One, 3(6), e2430. doi: 10.1371/journal.pone.0002430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen R, Lundstedt T, & Trygg J (2010). Chemometrics in metabolomics—a review in human disease diagnosis. Analytica Chimica Acta, 659(1), 23–33. [DOI] [PubMed] [Google Scholar]
- Mason JB, & Choi SW (2000). The mechanisms by which folate depletion enhances colorectal carcinogenesis: a unified scheme. Nestle Nutr Workshop Ser Clin Perform Programme, 4, 87–99; discussion 99–101. [DOI] [PubMed] [Google Scholar]
- Mason JB, Dickstein A, Jacques PF, Haggarty P, Selhub J, Dallal G, & Rosenberg IH (2007). A temporal association between folic acid fortification and an increase in colorectal cancer rates may be illuminating important biological principles: a hypothesis. Cancer Epidemiol Biomarkers Prev, 16(7), 1325–1329. doi: 10.1158/1055-9965.epi-07-0329 [DOI] [PubMed] [Google Scholar]
- Mentch SJ, Mehrmohamadi M, Huang L, Liu X, Gupta D, Mattocks D, … Locasale JW (2015). Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metab, 22(5), 861–873. doi: 10.1016/j.cmet.2015.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JW, Beresford SA, Neuhouser ML, Cheng TY, Song X, Brown EC, … Ulrich CM (2013). Homocysteine, cysteine, and risk of incident colorectal cancer in the Women’s Health Initiative observational cohort. Am J Clin Nutr, 97(4), 827–834. doi: 10.3945/ajcn.112.049932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myte R, Gylling B, Schneede J, Ueland PM, Haggstrom J, Hultdin J, … Van Guelpen B. (2016). Components of One-carbon Metabolism Other than Folate and Colorectal Cancer Risk. Epidemiology, 27(6), 787–796. doi: 10.1097/ede.0000000000000529 [DOI] [PubMed] [Google Scholar]
- Newcomb PA, Baron J, Cotterchio M, Gallinger S, Grove J, Haile R, … Seminara D (2007). Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev, 16(11), 2331–2343. doi: 10.1158/1055-9965.epi-07-0648 [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Lindon JC, & Holmes E (1999). ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica, 29(11), 1181–1189. [DOI] [PubMed] [Google Scholar]
- Nijhout HF, Reed MC, Budu P, & Ulrich CM (2004). A mathematical model of the folate cycle: new insights into folate homeostasis. J Biol Chem, 279(53), 55008–55016. doi: 10.1074/jbc.M410818200 [DOI] [PubMed] [Google Scholar]
- Nilsson SE, Read S, Berg S, & Johansson B (2009). Heritabilities for fifteen routine biochemical values: findings in 215 Swedish twin pairs 82 years of age or older. Scand J Clin Lab Invest, 69(5), 562–569. doi: 10.1080/00365510902814646 [DOI] [PubMed] [Google Scholar]
- Nitter M, Norgard B, de Vogel S, Eussen SJ, Meyer K, Ulvik A, … Riboli E (2014). Plasma methionine, choline, betaine, and dimethylglycine in relation to colorectal cancer risk in the European Prospective Investigation into Cancer and Nutrition (EPIC). Ann Oncol, 25(8), 1609–1615. doi: 10.1093/annonc/mdu185 [DOI] [PubMed] [Google Scholar]
- Ose J, Botma A, Balavarca Y, Buck K, Scherer D, Habermann N, … Ulrich CM (2018). Pathway analysis of genetic variants in folate-mediated one-carbon metabolism-related genes and survival in a prospectively followed cohort of colorectal cancer patients. Cancer Med. doi: 10.1002/cam4.1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacana T, Cazanave S, Verdianelli A, Patel V, Min HK, Mirshahi F, … Sanyal AJ (2015). Dysregulated Hepatic Methionine Metabolism Drives Homocysteine Elevation in Diet-Induced Nonalcoholic Fatty Liver Disease. PLoS One, 10(8), e0136822. doi: 10.1371/journal.pone.0136822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, & Reich D (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet, 38(8), 904–909. doi: 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- Reed MC, Nijhout HF, Sparks R, & Ulrich CM (2004). A mathematical model of the methionine cycle. J Theor Biol, 226(1), 33–43. [DOI] [PubMed] [Google Scholar]
- Rees JR, Morris CB, Peacock JL, Ueland PM, Barry EL, McKeown-Eyssen GE, … Baron JA (2017). Unmetabolized Folic Acid, Tetrahydrofolate, and Colorectal Adenoma Risk. Cancer Prev Res (Phila), 10(8), 451–458. doi: 10.1158/1940-6207.capr-16-0278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saternus R, Pilz S, Graber S, Kleber M, Marz W, Vogt T, & Reichrath J (2015). A closer look at evolution: Variants (SNPs) of genes involved in skin pigmentation, including EXOC2, TYR, TYRP1, and DCT, are associated with 25(OH)D serum concentration. Endocrinology, 156(1), 39–47. doi: 10.1210/en.2014-1238 [DOI] [PubMed] [Google Scholar]
- Schmit SL, Edlund CK, Schumacher FR, Gong J, Harrison TA, Huyghe JR, … Gruber SB (2018). Novel Common Genetic Susceptibility Loci for Colorectal Cancer. J Natl Cancer Inst. doi: 10.1093/jnci/djy099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher FR, Schmit SL, Jiao S, Edlund CK, Wang H, Zhang B, … Peters U (2015). Genome-wide association study of colorectal cancer identifies six new susceptibility loci. Nat Commun, 6, 7138. doi: 10.1038/ncomms8138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selhub J (1999). Homocysteine metabolism. Annu Rev Nutr, 19, 217–246. doi: 10.1146/annurev.nutr.19.1.217 [DOI] [PubMed] [Google Scholar]
- Siva A, De Lange M, Clayton D, Monteith S, Spector T, & Brown MJ (2007). The heritability of plasma homocysteine, and the influence of genetic variation in the homocysteine methylation pathway. QJM, 100(8), 495–499. doi: 10.1093/qjmed/hcm054 [DOI] [PubMed] [Google Scholar]
- Song Y, Manson JE, Lee IM, Cook NR, Paul L, Selhub J, … Zhang SM (2012). Effect of combined folic acid, vitamin B(6), and vitamin B(12) on colorectal adenoma. J Natl Cancer Inst, 104(20), 1562–1575. doi: 10.1093/jnci/djs370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Pan G, Chen L, Ma S, Zeng T, Man Chan TH, … Guan XY (2015). Loss of ATOH8 Increases Stem Cell Features of Hepatocellular Carcinoma Cells. Gastroenterology, 149(4), 1068–1081 e1065. doi: 10.1053/j.gastro.2015.06.010 [DOI] [PubMed] [Google Scholar]
- Stabler SP, Korson M, Jethva R, Allen RH, Kraus JP, Spector EB, … Mudd SH (2013). Metabolic profiling of total homocysteine and related compounds in hyperhomocysteinemia: utility and limitations in diagnosing the cause of puzzling thrombophilia in a family. JIMD Rep, 11, 149–163. doi: 10.1007/8904_2013_235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolzenberg-Solomon RZ, Chang SC, Leitzmann MF, Johnson KA, Johnson C, Buys SS, … Ziegler RG (2006). Folate intake, alcohol use, and postmenopausal breast cancer risk in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial. Am J Clin Nutr, 83(4), 895–904. doi: 10.1093/ajcn/83.4.895 [DOI] [PubMed] [Google Scholar]
- Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Magnusson KP, … Stefansson K (2007). Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet, 39(12), 1443–1452. doi: 10.1038/ng.2007.13 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Scheet P, Giusti B, Bandinelli S, Piras MG, Usala G, … Ferrucci L (2009). Genome-wide association study of vitamin B6, vitamin B12, folate, and homocysteine blood concentrations. Am J Hum Genet, 84(4), 477–482. doi: 10.1016/j.ajhg.2009.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuesen BH, Husemoen LL, Ovesen L, Jorgensen T, Fenger M, & Linneberg A (2010). Lifestyle and genetic determinants of folate and vitamin B12 levels in a general adult population. Br J Nutr, 103(8), 1195–1204. doi: 10.1017/s0007114509992947 [DOI] [PubMed] [Google Scholar]
- Ulrich CM, Neuhouser M, Liu AY, Boynton A, Gregory JF 3rd, Shane B, … Nijhout HF (2008). Mathematical modeling of folate metabolism: predicted effects of genetic polymorphisms on mechanisms and biomarkers relevant to carcinogenesis. Cancer Epidemiol Biomarkers Prev, 17(7), 1822–1831. doi: 10.1158/1055-9965.epi-07-2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu-Phan D, & Koenig RJ (2014). Genetics and epigenetics of sporadic thyroid cancer. Mol Cell Endocrinol, 386(1–2), 55–66. doi: 10.1016/j.mce.2013.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Balakrishnan-Renuka A, Napirei M, Theiss C, & Brand-Saberi B (2015). Spatiotemporal expression of Math6 during mouse embryonic development. Histochem Cell Biol, 143(6), 575–582. doi: 10.1007/s00418-014-1305-z [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, & Hakonarson H (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res, 38(16), e164. doi: 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Xie J, Yan M, Wang J, Wang X, Zhang J, … Liu, Q. (2016). Downregulation of ATOH8 induced by EBV-encoded LMP1 contributes to the malignant phenotype of nasopharyngeal carcinoma. Oncotarget, 7(18), 26765–26779. doi: 10.18632/oncotarget.8503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LD, & Kellis M (2012). HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res, 40(Database issue), D930–934. doi: 10.1093/nar/gkr917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, & Abecasis GR (2010). METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26(17), 2190–2191. doi: 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Platz EA, Willett WC, Fuchs CS, Selhub J, Rosner BA, … Giovannucci E (2009). A randomized trial on folic acid supplementation and risk of recurrent colorectal adenoma. Am J Clin Nutr, 90(6), 1623–1631. doi: 10.3945/ajcn.2009.28319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye M, He Y, Lin H, Yang S, Zhou Y, Zhou L, … Cai ZZ (2017). High expression of atonal homolog 8 predicts a poor clinical outcome in patients with colorectal cancer and contributes to tumor progression. Oncol Rep, 37(5), 2955–2963. doi: 10.3892/or.2017.5554 [DOI] [PubMed] [Google Scholar]
- Yi P, Melnyk S, Pogribna M, Pogribny IP, Hine RJ, & James SJ (2000). Increase in plasma homocysteine associated with parallel increases in plasma S-adenosylhomocysteine and lymphocyte DNA hypomethylation. J Biol Chem, 275(38), 29318–29323. doi: 10.1074/jbc.M002725200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


