Abstract
CRISPR-Cas9 is an RNA guided endonuclease that has revolutionized the ability to edit genome and introduce desired manipulations in the target genomic sequence. It is a flexible methodology and is capable of targeting multiple loci simultaneously. Owing to the fact that cancer is an amalgamation of several genetic mutations, application of CRISPR-Cas9 technology is considered as a novel strategy to combat cancer. Genetic and epigenetic modulations in cancer leads to development of resistance to conventional therapy options. Given the abundance of transcriptomic and genomic alterations in cancer, developing a strategy to decipher these alterations is critical. CRISPR-Cas9 system has proven to be a promising tool in generating cellular and animal models to mimic the mutations and understand their role in tumorigenesis. CRISPR-Cas9 is an upheaval in the field of cancer immunotherapy. Furthermore, CRISPR-Cas9 plays an important role in the development of whole genome libraries for cancer patients. This approach will help understand the diversity in genome variation among the patients and also, will provide multiple variables to scientists to investigate and improvise cancer therapy. This review will focus on the discovery of CRISPR-Cas9 system, mechanisms behind CRISPR technique and its current status as a potential tool for investigating the genomic mutations associated with all cancer types.
Keywords: CRISPR-Cas9, Cancer, Genome editing
1. Introduction
Since the development of advanced genome sequencing, researchers have made an excellent improvement in the field of genetic mapping. However, the major challenge faced by the scientists is to understand the multiple roles of signaling molecules [1]. Homologous recombination in the early 2000 gained colossal attention for its ability to alter genome. However, disadvantages like high cost and high man-power limited its applicability. Recently, the three most frequently used genome engineering tools are transcription activator-like effector nucleases (TALENs), zinc finger nucleases (ZFNs) and clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 (CRISPR-Cas9) systems. ZFNs contain a cleavage region of Fok1 restriction endonucleases and a common Cys2-His2-DNA binding domain [1]. TALENs contains 33-35 evolutionary conserved sequence of amino acids that can be shuffled to target a specific nucleotide. These genome editing techniques generate double-strand DNA break and have been utilized to understand pathophysiology of disease, normal gene function and development of novel therapeutic techniques [2–4].The major difference between ZFN or TALEN as compared with CRISPR-Cas9 is that ZFN and TALEN are protein based DNA editing techniques, whereas CRISPR is a small RNA-mediated sequence specific cleavage technique. CRISPR-Cas9’s ability to specifically target the gene of interest using guide RNA (gRNA) makes this methodology highly programmable and nucleotide specific.
CRISPR-Cas9 is a bacterial adaptive defense mechanism against invading pathogens. In the past years, this technique in prokaryotes has been repurposed as a modern tool for epigenetic modulations, transcriptional modifications, genome editing and imaging. Double-strand DNA break created by the Cas9 enzyme in CRISPR technique is repaired by non-homologous end joining (NHEJ) repair mechanism, thus inducing small deletions or insertions, which further elucidate the function of that gene. Another way to repair double-strand break by Cas9 is using homology directed repair (HDR) using the template DNA. This technology enables researchers to specifically target a pool of genes using single guide RNA (sgRNA). sgRNA can modulate target genes, thus permitting the identification of a specific gene function in disease progression, and understanding disease-specific mutations. Most importantly, CRISPR can be utilized to target multiple genes at once thus making this technique highly efficient [1]. This endonuclease technology enhances our ability to decode mutations associated with disease progression especially in advanced tumor development where multiple genes are involved [52].
Cancer is characterized by a multitude of mutations and the resulting genetic aberrations leads to a heterogeneous population of cells in tumors. Approximately, 140 genes with detrimental mutations have been reported in tumorigenic progression [5]. These mutations then results in the uncontrolled activation of several signaling pathways and play a significant role in cancer metastasis, proliferation, inhibition of tumor suppressor genes, and resistance to current therapeutical options. Cancer treatment demands a novel and strategical approach to understand the role of tumor specific genes in the process of carcinogenesis. Over the past decade, several oncogenic mutations have been mimicked by CRISPR-Cas9 system invitro and invivo. Consequently, it has potentiated our ability to understand the specific role of several oncogenes. There is a severe demand for such novel tools that can recapitulate the tumor characteristics and enhance our ability to specifically target mutated genes with minimal toxic effects to the surrounding normal cells. Here in this review, we have compiled the CRISPR-Cas9 technique in a stepwise hierarchy from its discovery to its application as a promising tool in field of cancer that holds a tremendous capability of engineering normal and cancer genomes.
1.1. Discovery of CRISPR-Cas9
CRISPR-Cas9 system was initially discovered in 1987 when Ishino et al., identified a novel protein sequence while performing the sequence analysis of IAP gene expressed in Escherichia coli [6]. During the sequencing, they observed 29 nucleotides separated by 32 non-repeating spacer nucleotide units, which is now coined as CRISPR sequence [6, 7]. Until 30 years ago, it was tough to assume the biological relation behind these unique repeating sequences in prokaryotes due to insufficient DNA sequence data. However, in 1993 advancement in genome analysis techniques lead to the identification of the CRISPRs in archaea (84%) and bacteria (45%) [8, 9]. Complete genomic sequencing revealed that these repeating sequences were specific to the prokaryotic genome and were absent in the genome of eukaryotes and viruses [7]. These short non-repeating spacer nucleotide sequence was unique for each strain demonstrating the importance of these sequence in preserving function(s) of all strains [10]. In early 2000, Monjica et al., linked the expression of these sequences with the immune system in prokaryotic organisms. However, this massive discovery remained unrecognized until 2005 when three groups autonomously discovered the role of CRISPR loci in adaptive immunity. Analogous to this phenomenon, the genes earlier proposed in hyper-thermophilic archaea to encode for DNA repair mechanisms were identified to be precisely associated with CRISPR and thus identified as CRISPR-associated (Cas) genes [11, 12]. Traditionally used Cas9 is derived from S. pyogenes (SpCas9). However, diverse Cas enzymes belonging to Cpf1, class 2 CRISPR-Cas9 system have been discovered ever since the potential of CRISPR technology has been identified [13]. Genomic analysis indicated that similar to eukaryotic RNA interference (RNAi) system, CRISPR and Cas9 genes work together to establish adaptive immunity against invading plasmids and viruses in prokaryotes [8, 14, 15]. It was observed that the non-repeating sequences were acquired from the invading phage DNA and conjugative plasmids [16]. Moreover, manipulation or removal of these non-repeating sequences sensitized phage resistant strain, thus confirming their role in adaptive immunity [17]. It has been observed that CRISPR captures a piece of invading DNA to initiate the memory which later helps the prokaryotes to fight against pathogens [8]. Comprehensive genome studies ascertained particular characteristics common to CRISPR loci found in prokaryotes a) Located in intergenic regions i.e a stretch of DNA located within genes and is mostly non-coding, b) Contain repeated sequences that are common in all prokaryotes, c) Possess interspersed sequences that are non-conserved. In beginning of the century, genomic analysis enabled scientists to identify that certain genes are regularly expressed around the CRISPR region and were termed as Cas genes. Cas-3 and Cas-4 subtypes of Cas genes are involved in transcriptional regulation, DNA metabolism, DNA repair-recombination and chromosomal segregation [11]. At the same time, another group identified that Cas genes are genes encoding for helicase, DNA polymerase, and RecB like nucleases. In further studies, it was observed that these foreign sequences were integrated into the CRISPR locus. These repeated sequences of foreign DNA along with the CRISPR spacers transcribe as a long transcript that later gets processed into small CRISPR RNAs (crRNAs) [18, 19]. Transcription of CRISPR repeats and spacers either form a secondary hairpin RNA structure or an unstructured RNA [20]. In 2011, trans-activating CRISPR RNAs (tracrRNA’s) were discovered in Streptococcus pyogenes by utilizing the latest RNA sequencing tools [21]. crRNA and tracrRNA facilitate the immunological actions in prokaryotes [1]. It was observed that prokaryotes like S. pyogenes possess tracrRNA in a duplex form with pre-crRNA (hairpin RNA structure), where it forms a complete base pairing with pre-crRNA with only one mismatch. In order for the crRNA to mature and mediate immunity, it must undergo a two-step process a) cleavage of tracrRNA from pre-crRNA, b) maturation of pre-crRNA to crRNA. It is important to mention that none of the Cas proteins have RNAse III like motif, necessary for the cleavage of mature crRNA from tracrRNA, thus RNAse III from the organism is recruited to the complex for cleavage. Deltcheva et al., identified that only csn1 facilitates the maturation of crRNA by aiding proper base pairing between tracrRNA and crRNA. Later, csn1 was named as Cas9 [7, 21]. Pre-crRNA, tracrRNA, Cas1, Cas2 and Cas9 belong to a larger subunit called CRISPR ASsociated Complex for Antiviral Defense (CASCADE). When prokaryotes such as S. pyogenes encounter a foreign plasmid or phage DNA, they incorporate protospacer gene (gene obtained from the pathogen) followed by PAM (Protospacer Adjacent Motif, 2-6 base pairs) from the invading pathogen in the CRISPR loci. Cas9 enzyme in prokaryotes do not target the protospacer gene unless it is followed by a PAM sequence. PAM is characterized as an identification sequence for Cas9 endonuclease mediated cleavage. In prokaryotes, this sequence is obtained from the invading pathogen. When a prokaryote encounters a new pathogen, a new protospacer sequence is incorporated near the AT-rich region, a region between the Cas gene and CRISPR loci. The incorporation of this new spacer sequence is performed by Cas1 and Cas2 metal-dependent endoribonucleases [10, 19]. Once the mechanism of CRISPR-Cas9 was clearly understood in prokaryotes, Jinek and colleagues devised a unique technique for genome editing where they deciphered the role of tracrRNA and magnesium. It has been shown that, in spite of the presence of Cas9 enzyme, crRNA, PAM, absence of tracrRNA and magnesium in the complex makes the cleavage of target gene unsuccessful. Subsequently, a single guide RNA was designed which eliminates the use of multiple components, thus making the system re-programmable. The sgRNA encodes the tracrRNA-crRNA complex along with magnesium. sgRNA guided Cas9 endonuclease facilitates the cleavage of target gene that is followed by PAM at 3’ end. Hence, this RNA programmed Cas9 machinery has enabled the scientists to target and cleave any sequence of interest [22]. The developmental cascade of CRISPR-Cas9 technology has been schematically represented in Figure 1.
Figure 1:
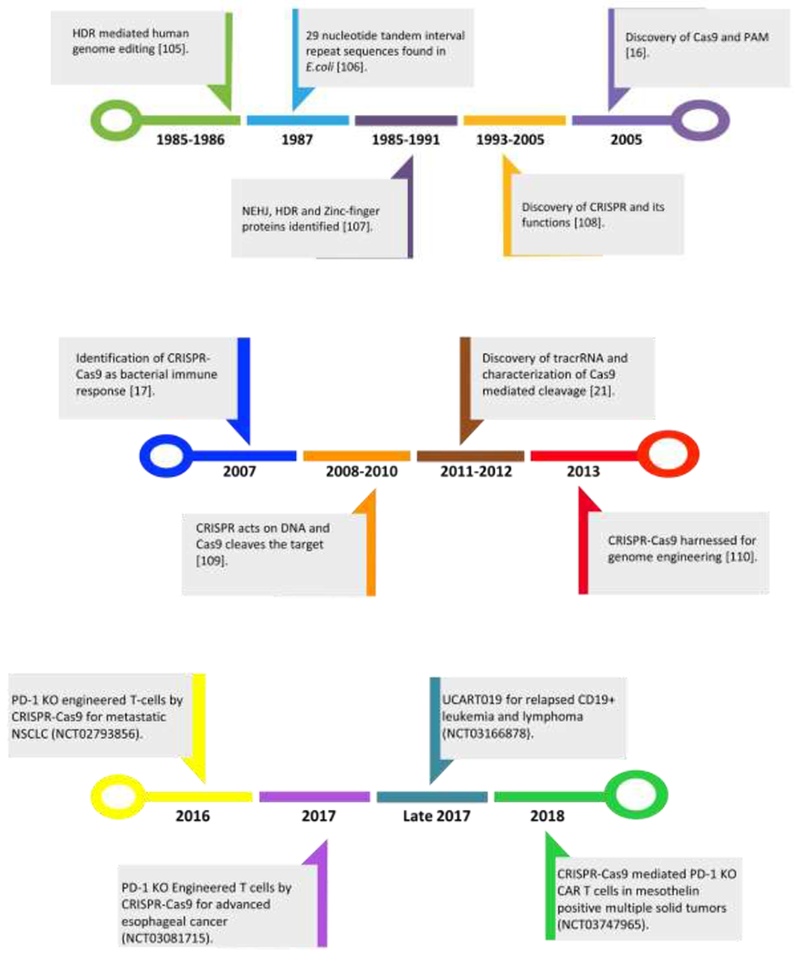
Schematic representation of various stages in the discovery of CRISPR-Cas9 technology
1.2. CRISPR as a genetic element
CRISPR-Cas9 genome editing technology is derived from the concept of adaptive immune response in prokaryotes [23]. It holds several advantages over the conventional genome editing technologies including a) simplicity in target designing, b) regulation and c) multiple gene targeting ability [24]. CRISPR consists of two major components: sgRNA and Cas protein. The sgRNA consists of a sequence also called as scaffold. sgRNA encompasses Cas enzyme binding site and a spacer sequence, called the target sequence which is specific to the gene of interest. The spacer sequence contains ~20 nucleotides and it is followed by PAM [7, 25]. PAM sequences vary based upon the type of Cas9 enzyme [26]. Several types of Cas enzymes have been identified and the flexibility in manipulating their properties have led to a multiplexed approach for advanced genome engineering [27]. CRISPR-Cas9 system can target any gene of interest provided the targeting complex consists of a target gene sequence along with Cas9 or Cpf1 that has endonuclease activity [28]. Cas9 enzyme binds to the PAM in a sequence specific manner. Once the sgRNA and Cas9 enzymes are incorporated in the system, Cas9 binds to the scaffold with its positively charged grooves forming a ribonucleotide complex. Once the complex forms, Cas9 undergoes a conformational change rendering it in an active state. In its active conformation, Cas9-sgRNA complex binds to the target gene in a highly specific manner [26]. The complex will not mediate the effects until there is high sequence specificity between sgRNA and the target locus. The template strand binds to the gRNA, whereas the non-template strand binds to HNH/RuvC groove of Cas9 [29, 30]. Once nucleotide specificity is achieved, the sgRNA will bind to the target locus in a 3’-5’ direction. However, mismatches at the 3’ end will abolish the whole mechanism. On the other hand, minimal mismatches at the 5’ end within the close proximity of PAM sequence can still lead to effective cleavage [30, 31]. This cleavage results in the formation of a double-strand break in the targeted sequence which therefore enhances the activation of two repair mechanism 1) Non-homologus end joining (NHEJ) repair and 2) Homology directed repair (HDR). NHEJ repair is an error prone mechanism that repairs DNA double-strand break by performing insertion of indels, causing amino acid deletion, mutation, insertions or frame shift mutations in the open reading frame of target locus [32, 33]. HDR is a natural repair mechanism initiated in microorganisms and humans following a DNA double-strand break [34]. This repair mechanism can be utilized to anneal the DNA double-strand breaks following CRISPR mediated cleavage [35, 36]. sgRNA mediated cleavage of DNA can be annealed by providing donor template to the targeted cells. The donor DNA contains desired insertions or modifications. These insertions are flanked with segments of DNA homologous to the cleaved DNA. Thus, the homology directed repair mechanism of the cells utilize the donor template and create an insertion of the desired gene in the targeted DNA segment [37, 38].
1.3. Dual functional modulations mediated by CRISPR-Cas9
CRISPR-Cas9 can mediate both activation and repression of the target gene. Cas9 can bind to the target DNA without causing any cleavage [39]. The nuclease sites (HNH and RuvC) can be rendered inactive by performing point mutations at D10A and H840A in spCas9. This results in the production of dead Cas9 (dCas9). dCas9 retains the ability to bind to the gene sequence but loses the ability to cleave the gene sequence at any locus due to mutations in its nuclease site. dCas9 can be tagged with transcriptional activators and repressors to form dCas9 fusion proteins. These fusion proteins (dCas9 + transcriptional activators/ repressors) can be directed to bind to the transcriptional start site [40, 41] .Once the fusion proteins bind to the start site, they mediate the activation or suppression of the target gene similar to the activators and repressors [37, 42]. Furthermore, Cas9 enzymes can be engineered by combining with epigenetic modifiers such as p300 and TET1. The resultant modified Cas9 enzymes can be used to perform high-grade epigenetic engineering for a particular chromatin. This helps researchers to understand the effect of a single epigenetic marker. For example, methylation or acetylation of a particular chromatin. However, it is important to note that these epigenetic modifications are inheritable and can be expressed in the future progeny of the cells [43].
2. CRISPR and cancer
The specificity of CRISPR-Cas9 technology in genome editing underscores its potential to treat various diseases like cancer, cystic fibrosis, AIDS, hematologic and neuronal disorders [44–48]. A multitude of uncontrolled genetic and epigenetic mutations characterizes the progression of cancer. Certain oncogenic mutations are evidences of genomic instability in cancer. Recent advances in genome editing has enabled our understanding about the genetic profile in healthy and diseased state [49, 50]. CRISPR-Cas9 technology provides better understanding about the functions of genetic anomalies through invitro, invivo and exvivo experimental mutation models.
Genetically engineered pre-clinical tumor models facilitate the understanding of complex tumor microenvironment and designing of novel anti-cancer agents. In the current scenario, genetically editing each gene is laborious and exorbitant. Henceforth, an affordable and precise method to meet the above-mentioned constraints is warranted. CRISPR-Cas9 mediated genetically modified tumor models has become an indispensable tool in oncology [51]. It holds the potential to modify a wide array of genes, which are aberrantly regulated in human cancers (Table: 1). Several studies have reported the ease of developing genetically modified mice models by CRISPR-Cas9 [51–55]. A multitude of genes have been simultaneously edited by manipulating the embryo in a RNA guided manner to generate an efficient manipulated mice model [52]. In another study, Cre dependent Cas9 mouse models were created by viral/non-viral delivery of guide RNAs in neurons, immune cells and endothelial cells (Figure 2). Introduction of Cas9 by Cre recombinase in the lung generated a lung adenocarcinoma model. Adeno associated virus vector carrying sgRNA was injected in the mice which generated loss of function mutations in p53, LKB1 and HDR mediated mutation in Kras (G12D), leading to the formation of tumors in lungs [53]. A recent study has reported the specific targeting of oncogenes by CRISPR-Cas9. Tumor promoting genes like TMEM135-CCDC67 and MAN2A1-FER fusion genes were found to be aberrantly regulated in prostate cancer and hepatocellular carcinoma respectively [54, 55]. In this study, catalytic domain of Cas9 was mutated at D10A which converts Cas9 into nickase. Consequently, adenovirus mediated introduction of nickase Cas9 into cancer cells caused single – strand breaks in the target DNA and sgRNA targeted the breakpoint sequences of the mutated fusion genes. Another adenovirus was used to deliver the suicide gene herpes simplex virus (HSV1) thymidine kinase (TK) into the chromosal breakpoints of the fusion genes TMEM135-CCDC67 and MAN2A1-FER in nickase Cas9 mediated manner. Subsequently, HSV1-TK phosphorylate the prodrug ganciclovir (synthetic nucleoside homolog) to ganciclovir monophosphate and subsequently to ganciclovir triphosphate, which disrupts the target DNA synthesis by elongation termination. Xenograft transplantation of TMEM135-CCDC67+ and MAN2A1-FER+ human prostate and liver cancer cells resulted in tumor formation in immune-deficient mice. In due course, tumor bearing mice were injected with adenovirus carrying HSV1-TK and treated with ganciclovir. HSV1-TK mediated activation of ganciclovir resulted in suppression of tumor growth by 30%. Furthermore, it also inhibited the metastatic efficiency of the tumors. These findings suggest that mutated Cas9 intervened suicide gene insertion into cancer cells may be a futuristic and novel genome editing strategy for cancer treatment [56]. Trp53 has been shown to be mutated in several tumor types and therefore serves as a potential candidate for mutagenesis. It has been demonstrated that, Trp53 was inactivated in mouse embryonic fibroblasts (MEFs) with lentiviral and retroviral delivery of Cas9 and sgRNA. The subsequent population obtained after Trp53 loss showed a significant increase in the Trp53 mutant population. Moreover, it has been reported that, Trp53 specific sgRNA had very high specificity to Trp53 thus showed minimal off target effects [57]. Apart from the mutations that result in uncontrolled tumor progression, cancer cells also exhibit characteristics of acquiring resistance to chemotherapeutic drugs. For example, estrogen receptor targeting drugs such as tamoxifen and fulvestrant are highly successful in treatment of breast cancer. However, it has been reported that, breast cancer cells acquire resistance with prolonged treatment with tamoxifen and fulvestrant. This phenomenon can be corroborated to mutations in amino acids that are located on the helix12 region of the estrogen-binding site. Harrod et al., performed a knock-in experiment utilizing the CRISPR-Cas9 technique, where they inserted a single tyrosine 537 in wild type and estrogen responsive MCF7 cells. They observed that Y537S mutation promoted estrogen receptor activity in MCF7 cells thus confirming the role of ER mutation in acquired tamoxifen resistant in breast cancer [58]. Shalem et al., generated a sgRNA lentiviral library to identify the genes whose inactivation results in the acquired resistant to BRAF inhibitor vemurafenib. Moreover, in their later studies, they utilized the CRISPR-Cas9 lentiviral technique to design a genomic library to identify the genes responsible for the acquired resistance in vemurafenib resistant melanoma cells [59, 60].
Table 1:
CRISPR-Cas9 targeted genes in various cancer models
| Cancer Type | CRISPR-Cas9 targeted genes | Reference |
|---|---|---|
| Bladder cancer | p21, E-cadherin, hBAX | [104] |
| Tripple negative breast cancer | SHCBP1 | [105] |
| Non-small cell lung cancer | Cd74-Ros1, Eml4-Alk, Ki5b-Ret | [106] |
| Non-small cell lung cancer | Met | [107] |
| Melanoma | Id1, Id3 | [108] |
| Breast Cancer | Her2 | [109] |
| Cervical cancer | HPV E6, E7 | [110] |
| Lymphoma | MCl1, TP53 | [111] |
| PDAC | p57 | [112] |
| Glioblastoma | Pten, Apc, Nf1 | [113] |
| Medulloblastoma | Ptch1 | [113] |
Figure 2:
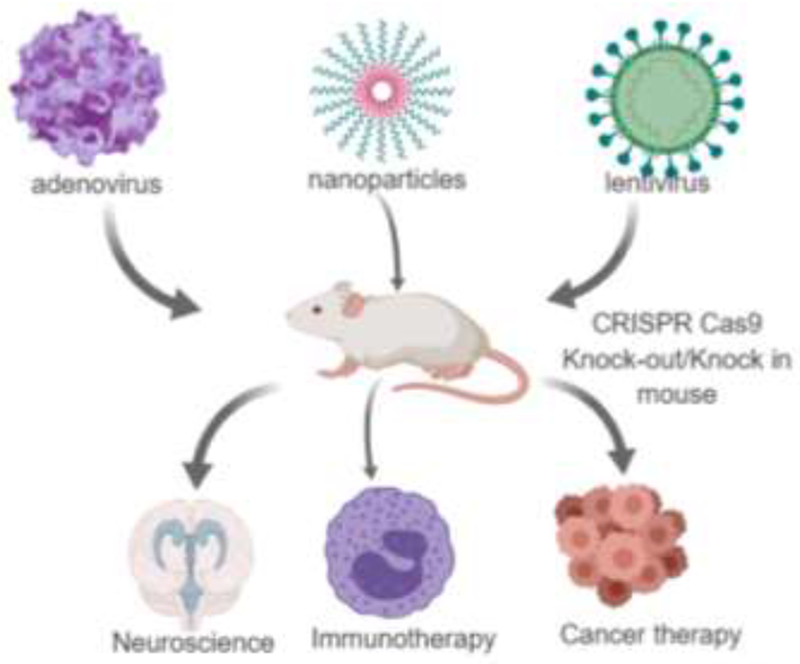
CRISPR-Cas9 as a tool for in-vivo mouse modelling
Within the last decade, miRNAs have gained tremendous importance for its role in upregulation of oncogenic pathways. miRNA gene is responsible for the transcription of pre-miRNA which is then processed by Drosha and Dicer to produce mature miRNA. Several studies have shown that miRNA expression can be modified by targeting the 5’ region of pre-miRNA using CRISPR-Cas9 technology. CRISPR-Cas9 has been widely used to modulate miRNA expression in various cancer models such as hepatocellular carcinoma, renal cell carcinoma, pancreatic cancer, ovarian cancer, and chronic myeloid leukemia [61]. The precision and accuracy of the CRISPR-Cas9 technique has also been tested in invivo mouse model. However, this strategy requires adequate precision to develop successful experimental models. Insertion of a specific recombinase site similar to LoxP is useful in modifying somatic cells in adults. Notably, for the first time, Tyler Jack’s laboratory employed the CRISPR-Cas9 machinery to developed Pten and Trp53 knockout mice, which resulted in the formation of liver tumors. It has been shown that, tumor formation was a result of Pten phenocopying its mutations from CRISPR deletion through Cre-LoxP technology [62]. Sanchez-Rivera et al., demonstrated that, direct intra-tracheal delivery of Cre-LoxP, Cas9 and specific sgRNAs combined the over-expression of mutant Kras and suppression of Trp53, resulting in the production of tumor with different tumor histopathology [63].
Several pioneering studies have reported the role of CRISPR-Cas9 in enhancing anti-tumor immunotherapy by targeting immunological molecules like program cell death protein 1 (PD-1) and cytotoxic T- lymphocyte antigen – 4 (CTLA-4) [64]. It has been reported that, CRISPR-Cas9 technology has been strongly implicated in engineering immune cell receptors like chimeric antigen receptor (CAR) in T- lymphocytes, which is collectively termed as CAR – T cells [65]. Patient - derived T-cells are engineered and expanded to express a highly specific CAR. Subsequently, CARs facilitate T-cells to recognize and lodge an immune response against antigen expressing cancer cells [66]. Recently, FDA has approved two CAR-T cell therapy for treatment of non-Hodgkin lymphoma and diffuse B-cell lymphoma [67]. This mode of anti-tumor immunotherapy has shown relapse free long-term patient survival and a complete remission of cancer was achieved in more than 80% of patients [68, 69]. However, CAR – T cell therapy has become ineffective due to the shared expression of CAR in normal and malignant T-cells, which results in internal self-killing of CAR-T cells. Secondly, generating enormous amount of T-cells pose a technical and financial challenge. Moreover, CAR-T cell therapy pose a huge risk of graft vs host disease which can lead to the deterioration of the patient’s body system. Current studies suggest that development of second-generation CAR-T cells by genomic editing CRISPR-Cas9 technology will overcome the limitations in CAR-T cell therapy [64, 67, 70, 71]. CD7 has been reported to be an attractive immunotherapeutic target for treatment of T-cell malignancies. However, normal T-cells also express CD7, which makes CD7 targeted CAR- T cell therapy ineffective and pave way to induce fratricide in T-cells. In a ground-breaking study by Cooper et al., authors have enrolled the genomic engineering CRISPR-Cas9 system to generate CD7 and T-cell receptor alpha chain (TRAC) deleted CAR-T targeting CD7 (UCART7) cells. In other words, CD7 expression was eliminated from healthy T-cells by the CRISPR-Cas9 system. It has been evidently demonstrated that, injection of UCART7 cells kills T-cell acute lymphoblastic leukemia (T-ALL) cells and patient - derived T-ALL, and no adverse events like xenogenic graft vs host disease was reported [70]. The immune functions of T-cells are attenuated due to the expression of various inhibitory or immune checkpoint signaling molecules like PD-1, CTLA-4, lymphocyte activated gene -3 (LAG-3) and domain containing protein-3 (TIM-3). Therefore, specifically knocking out these signaling molecules reverses T-cell exhaustion and promotes anti-tumor immune response. The multiplex genome editing capacity of the guide RNAs in the CRISPR-Cas9 system has facilitated the knockdown of a multitude of T-cell inhibitory molecules on a one-shot protocol. Fas receptor, a member of the tumor necrosis factor – α (TNF- α) family of death receptors plays a crucial role in functioning of T-cells. Binding of Fas ligand to Fas receptor results in T-cell apoptosis, thereby deteriorating T-cell mediated killing of cancer cells. Ren et al., proved that one-shot CRISPR-Cas9 machinery was able to generate CD3− HLA-1− Fas− CAR T-cells that are resistant to apoptosis. Furthermore, dual inhibitory resistant universal CAR-T cells were generated using CRISPR system. These cells are characterized by TCR−, HLA-1−, PD-1−, CTLA-4− genotype [72]. PD-1 is a cell surface receptor and an immune checkpoint protein, which boosts immune-evasion in cancer cells. Binding of PD-L1 surface proteins on tumor cells to PD-1 receptor in T-cells results in diminished activity of T-cell mediated cytotoxic activity against tumor cells. CRISPR-Cas9 genome modulation system was used to engineer patient derived T-lymphocytes to knockdown PD-1 in T-cells. Subsequently, these specifically edited T-lymphocytes was expanded and re-injected in the patient to stimulate T-cell mediated immune response against cancer cells. In 2016, the first clinical trial in tumor immunotherapy using the CRISPR-Cas9 technology to engineer PD-1 receptors in T-cells was performed [73].
3. Industrial prospects of CRISPR technology
The precise applicability of the CRISPR-Cas9 system has attracted several large-scale and small-scale pharmaceutical industries to formulate the clinical applications of CRISPR-Cas9 machinery. Biotech companies CRISPR Therapeutics, Intellia Therapeutics and Editas Medicine are currently the pioneers in the CRISPR-Cas9 market [64]. CRISPR Therapeutics has collaborated with Vertex Pharmaceuticals and submitted its clinical trial application for evaluating the effects of CRISPR-Cas9 system in treatment of β-thalassemia. The therapy is known as CTX001 and has revolutionized the CRISPR-Cas9 market. The company is also involved in designing a CRISPR-Cas9 based treatment option for sickle cell disease. CTX001 employs an ex-vivo approach where the genomes are engineered by CRISPR-Cas9 system to cleave and inactivate the target gene BCL11A. CRISPR edited genome is re-injected into the patients [74, 75]. Editas Medicine employs CRISPR-Cas9 and CRISPR/Cpf-1 system for genome editing. EDIT-101 is the company’s leading research program, which aims at engineering CEP290 gene in retinal tissue for treatment of Leber Congenital Amaurosis type 10, an inherited eye disorder that causes blindness in children. Editas has partnered with Allergan for the EDIT-101 program. Moreover, Editas is also commissioning CRISPR-Cas9 system for T-cell mediated immunotherapy in cancer in collaboration with Juno Therapeutics. Juno therapeutics has sanctioned around $ 47 million for research support to develop CRISPR based therapies for cancer [76]. Intellia Therapeutics has employed CRISPR-Cas9 system to counter-act genetic anomalies, hematologic disorders and autoimmune diseases. The company has developed a CRISPR based treatment in alliance with Regeneron Pharmaceuticals for the treatment of transthyretin amyloidosis. Novartis is also a part of the company’s collaboration program for the development of CRISPR based CAR- T-cell tumor immunotherapy [77]. Apart from biotech firms, Microsoft giant Bill Gates has acknowledged CRISPR-Cas9 system as a revolutionary tool in the field of health sciences. The billionaire realized the potential of CRISPR genome editing technology and is one of the early investors in Editas Medicine for developing CRISPR based treatment. Researchers from Gates foundation are evaluating the effects of CRISPR-Cas9 mechanism in improving the livestock, crops and malaria [78]. According to an authenticated report from Forbes, $120 million has been invested in Editas Medicine, where technology giants Bill Gates and Google contribute a major part to the investment [78, 79]. Development of CRISPR-Cas9 machinery is financially backed up by several tech giants and industries, which shows a promising futuristic approach for treatment of human diseases and inherited anomalies.
4. Pitfalls of CRISPR technology
Despite of extensive and profound applications of the CRISPR-Cas9 technology in genome re-organization and treatment of various diseases, its applicability is hindered by certain limitations. The efficiency and specificity of the CRISPR-Cas9 system is limited by numerous factors like Cas9 activity, selection of target site, design of sgRNA, Cas9 delivery and off-target effects [80]. The precision and specificity of CRISPR can be modulated at the sgRNA level and Cas9 level [13]. Even though the sgRNA is designed utilizing high specificity for the target locus, there are multiple genes in the human genome that might share a homology with the target gene of interest. Hence, these off target effects must be minimized while designing the guide RNA [81]. Cas9 cleaves the target DNA sequences by binding to the 20 nucleotides of the target sequences [36]. It has been demonstrated that sgRNA encoding +85 nucleotide tracrRNA enhanced Cas9 activity and incorporated increased genetic aberrations invivo. On the other hand, mutations in proximal region of PAM abrogated the Cas9 activity [82]. An equilibrated Cas9 activity is essential for a successful sgRNA guided Cas9 genome editing. As mentioned earlier, sgRNA-Cas9 complex aids in knockdown of the target gene. However, it has been evidently reported that, a disproportion in Cas9 activity leads to off-target effects. Researchers have modified the conventional spCas9 to Cas9 nickase. The modified nickase encodes a D10A mutation and as a result, it has retained only one nuclease domain of spCas9 enzyme out of two (HNH/RuvC) [83, 84]. Cas9 nickase causes only a single strand nick rather than a double-stranded break. Owing to this phenomenon, Cas9 nickase mediated genome editing requires at least two Cas9 nickase enzymes, which can target the same sequence at different locations, thus reducing the possibility of off target effects [85]. Slaymaker et al., designed a Cas9 mutant with 32 substitutions and tested its ability in HEK (human embryonic kidney) cells to reduce off target effects. They discovered that, mutated SpCas9 (K855A) reduced off target effects as compared to wild-type SpCas9 [86]. Similarly, the off target nicks produced by Cas9 enzyme were studied by Kleinstiver et al., and Joung’s Lab where they hypothesized that, Cas9 with mutations in the phosphate backbone binding region can reduce the binding and cleaving of non-template DNA strand by Cas9 independent of sgRNA. This mutated Cas9 was termed as SpCas9-HF1 containing the mutations in the following amino acid sequence: N497A, R661A, Q695A, and Q926A [83]. It has also been shown that, RNA guided Cas9 endonuclease activity is high, irrespective of the binding of noncomplementary sequences in the target nucleotides, which leads to off-target mutations in human cells. Therefore, a tight control in proportions of sgRNA-Cas9 complexes proves to be a crucial factor to improve specificity of target gene mutations [87]. Cas9 mediated silencing of target genes is facilitated by transfection of plasmids encoding sgRNA and Cas9 into target cells [88]. The unmethylated CpG nucleotides in the bacterial DNA can activate an immune response and diminish the CRIPSR-Cas9 genome editing process [89]. Moreover, lipid based transfection reagents like lipofectamine that are used to transfect sgRNA-Cas9 encoded plasmid can cause cytotoxic effects to the target cells. Alternative non-cytotoxic transfection methods like electroporation, nucleofection, microinjection, sgRNA-Cas9 chemical conjugation, Cas9 – peptide conjugation and cationic lipid mediated delivery of Cas9 have been adopted to transfect sgRNA-Cas9 plasmids in mice, human and zebrafish [80, 90–92]. sgRNA-Cas9 complexes delivered by viral vectors display greater efficiency and less cytotoxicity in transfected cells [80]. In addition, viral vectors like integrase defective lenti-viral vectors (IDLVs), adenoviral vectors (ADVs) and recombinant adeno associated viral vectors (rAAVs) possess huge capacity to encode sgRNA-Cas9 complexes for efficient delivery into target cells. Furthermore, the compatibility of viral vectors to enter into a large number of cell types makes it a widely used system to deliver CRISPR machinery in mammalian cells [93–95].
In a revolutionary study by Charlesworth et al., it has been discovered that, humans possess pre-existing humoral and cell mediated adaptive immunity to Cas9 endonuclease enzyme derived from Streptococcus pyogenes and Staphylococcus aureus, thus limiting the applicability of CRISPR-Cas9 technology in management of various diseases [75]. This claim is underpinned by another study led by Simhadri et al., In this study, they have shown that, American population possess pre-existing antibodies to Cas9 enzyme, which raises a major concern in terms of safety and efficacy. However, the levels of currently detected anti-Cas9 antibodies are low to elicit an immune response, but high titers of anti-Cas9 antibodies can exert a deadly immune response, which can hinder the clinical applications of CRISPR-Cas9 technology. The immunological action against Cas9 enzyme is currently under extensive investigation in pre-clinical animal models [96].
Apart from the technical challenges, ethical and legal issues are major pressing concerns that prevent the applicability of CRISPR-Cas9 system to a vast extent. As mentioned earlier, specific knockout of targeted genes by CRISPR-Cas9 technology can be achieved by altering the genome in germline. Scientists have employed CRISPR-Cas9 mechanism to engineer human embryos to replace defective DNA [36, 97–101]. Embryonic genome editing has raised several ethical and legal concerns in using CRISPR-Cas9 system for the betterment of living beings in the environment. Scientists speculate that, embryonic genome editing may lead to irreversible effects in future generations and ecological imbalance due to the inheritance of unintended alterations [102]. Off-target mutagenesis by CRISPR-Cas9 system has resulted in some deleterious effects such as cell death and transformation in humans. Although, inherent mutations caused by CRISPR-Cas9 genome editing in the embryos can generate novel invivo models and treat various diseases, it can also lead to an imbalance in the ecological system. Uncontrolled inherent mutations can generate organisms with genetic traits of modified sequences and disrupt the ecological system. In a controversial study by Huang et at., researchers applied CRISPR-Cas9 system to modify beta-globulin gene in tripronuclear zygotes (non-viable human zygotes) for treatment of β-thalassemia [101]. However, this study was rejected by Nature and Science journals on ethical grounds, but it created an uproar among the public. Even though, CRISPR-Cas9 technology is in its primitive stage and major aspects have to be unearthed, strict policies underscoring the safety assessment and regulatory norms should be employed for further application of CRISPR technology.
5. Future prospects of CRISPR-Cas9 technology in cancer treatment
CRISPR-Cas9’s potential of specific genome editing has dawned a new era in the development of mankind. Recent advancements in technology has decoded the effects of CRISPR-Cas9 system in improving agriculture, livestock and treatment of various human diseases and genetic disorders. Markedly, we envision a new era in management of several diseases including cancer. This novel gene engineering machinery has opened a contemporary approach in understanding the role of several signaling molecules in the process of carcinogenesis. Consequently, it will aid us in administering unique strategies to design novel cancer therapeutics. The flexibility in programming CRISPR-Cas9 technique has highlighted its emergence as a modeling tool for cancer biologists [103]. CRISPR-Cas9 based technology has the potential to restructure the concepts in cancer biology by providing improvised approach for personalized therapy, gene therapy, immunotherapy and genetic disorder treatment. In future, we prospect this technology will provide an essential genome array across almost all cancer cell lines. Complete genome information along with the genetic and epigenetic data that already exists will be an exciting approach for the discovery of novel targets. CRISPR has emerged from a niche technique to a mainstream technology within a decade. Albeit of the rapid progression of the CRISPR-Cas9 system, it still possess several challenges that require keen attention for its improvised application. Off-target effects or unintended mutations, immune response, toxicity of transfection reagents, genomic editing in embryos, which can lead to inheritable mutations, ethical and legal concerns are some of the key limitations encountered in the application of CRISPR-Cas9 system. Several studies have been published to counteract the limitations and improve the feasibility of CRISPR-Cas9 applications. Despite the adverse effects of CRISPR technology, several biotech firms and technology giants like Bill Gates and Google have foreseen the future of CRISPR technology as a revolutionary tool for mankind to defend against various disorders. As a result, non-pharmaceutical investors and companies have bankrolled several millions of dollars for the development and application of CRISPR technology. Bill Gates has quoted “If the world is to continue the remarkable progress of the past few decades, it is vital that scientists, subject to safety and ethics guidelines, be encouraged to continue taking advantage of such promising tools as CRISPR”. Henceforth, progression in approaches to neutralize the limitations and adherence to ethical and legal norms will pave way for a well-developed application of the CRISPR-Cas9 technology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: Authors disclose no conflict of interest
References
- [1].Zhang F, Wen Y, Guo X, CRISPR/Cas9 for genome editing: progress, implications and challenges, Hum Mol Genet 23(R1) (2014) R40–6. [DOI] [PubMed] [Google Scholar]
- [2].Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD, Genome editing with engineered zinc finger nucleases, Nat Rev Genet 11 (9) (2010) 636–46. [DOI] [PubMed] [Google Scholar]
- [3].Hsu PD, Lander ES, Zhang F, Development and applications of CRISPR-Cas9 for genome engineering, Cell 157(6) (2014) 1262–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Strong A, Musunuru K, Genome editing in cardiovascular diseases, Nat Rev Cardiol 14(1) (2017) 11–20. [DOI] [PubMed] [Google Scholar]
- [5].Maresso KC, Tsai KY, Brown PH, Szabo E, Lippman S, Hawk ET, Molecular cancer prevention: Current status and future directions, CA Cancer J Clin 65(5) (2015) 345–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ishino S, Mizukami T, Yamaguchi K, Katsumata R, Araki K, Nucleotide sequence of the meso-diaminopimelate D-dehydrogenase gene from Corynebacterium glutamicum, Nucleic Acids Res 15(9) (1987) 3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lau V, Davie JR, The discovery and development of the CRISPR system in applications in genome manipulation, Biochem Cell Biol 95(2) (2017) 203–210. [DOI] [PubMed] [Google Scholar]
- [8].Ishino Y, Krupovic M, Forterre P, History of CRISPR-Cas from Encounter with a Mysterious Repeated Sequence to Genome Editing Technology, J Bacteriol 200(7) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mojica FJ, Juez G, Rodriguez-Valera F, Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites, Mol Microbiol 9(3) (1993) 613–21. [DOI] [PubMed] [Google Scholar]
- [10].Rath D, Amlinger L, Rath A, Lundgren M, The CRISPR-Cas immune system: biology, mechanisms and applications, Biochimie 117 (2015) 119–28. [DOI] [PubMed] [Google Scholar]
- [11].Jansen R, Embden JD, Gaastra W, Schouls LM, Identification of genes that are associated with DNA repeats in prokaryotes, Mol Microbiol 43(6) (2002) 1565–75. [DOI] [PubMed] [Google Scholar]
- [12].Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E, Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements, J Mol Evol 60(2) (2005) 174–82. [DOI] [PubMed] [Google Scholar]
- [13].Gao L, Cox DBT, Yan WX, Manteiga JC, Schneider MW, Yamano T, Nishimasu H, Nureki O, Crosetto N, Zhang F, Engineered Cpf1 variants with altered PAM specificities, Nat Biotechnol 35(8) (2017) 789–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV, A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis, Nucleic Acids Res 30(2) (2002) 482–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV, A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action, Biol Direct 1 (2006) 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bolotin A, Quinquis B, Sorokin A, Ehrlich SD, Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin, Microbiology 151 (Pt 8) (2005) 2551–61. [DOI] [PubMed] [Google Scholar]
- [17].Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P, CRISPR provides acquired resistance against viruses in prokaryotes, Science 315(5819) (2007) 1709–12. [DOI] [PubMed] [Google Scholar]
- [18].Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J, Small CRISPR RNAs guide antiviral defense in prokaryotes, Science 321(5891) (2008) 960–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Marraffini LA, Sontheimer EJ, CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea, Nat Rev Genet 11(3) (2010) 181–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kunin V, Sorek R, Hugenholtz P, Evolutionary conservation of sequence and secondary structures in CRISPR repeats, Genome Biol 8(4) (2007) R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E, CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III, Nature 471(7340) (2011) 602–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E, A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity, Science 337(6096) (2012) 816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cong L, Zhang F, Genome engineering using CRISPR-Cas9 system, Methods Mol Biol 1239 (2015) 197–217. [DOI] [PubMed] [Google Scholar]
- [24].Guernet A, Grumolato L, CRISPR/Cas9 editing of the genome for cancer modeling, Methods 121–122 (2017) 130–137. [DOI] [PubMed] [Google Scholar]
- [25].Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales AP, Li Z, Peterson RT, Yeh JR, Aryee MJ, Joung JK, Engineered CRISPR-Cas9 nucleases with altered PAM specificities, Nature 523(7561) (2015) 481–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Farboud B, Severson AF, Meyer BJ, Strategies for Efficient Genome Editing Using CRISPR-Cas9, Genetics (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, Lander ES, Voytas DF, Ting AY, Zhang F, RNA targeting with CRISPR-Cas13, Nature 550(7675) (2017) 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sanchez-Rivera FJ, Jacks T, Applications of the CRISPR-Cas9 system in cancer biology, Nat Rev Cancer 15(7) (2015) 387–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O, Crystal structure of Cas9 in complex with guide RNA and target DNA, Cell 156(5) (2014) 935–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Anders C, Niewoehner O, Duerst A, Jinek M, Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease, Nature 513(7519) (2014) 569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jiang F, Doudna JA, CRISPR-Cas9 Structures and Mechanisms, Annu Rev Biophys 46 (2017) 505–529. [DOI] [PubMed] [Google Scholar]
- [32].Ma Y, Zhang L, Huang X, Genome modification by CRISPR/Cas9, FEBS J 281 (23) (2014) 5186–93. [DOI] [PubMed] [Google Scholar]
- [33].Pellagatti A, Dolatshad H, Valletta S, Boultwood J, Application of CRISPR/Cas9 genome editing to the study and treatment of disease, Arch Toxicol 89(7) (2015) 1023–34. [DOI] [PubMed] [Google Scholar]
- [34].Liang F, Han M, Romanienko PJ, Jasin M, Homology-directed repair is a major double-strand break repair pathway in mammalian cells, Proc Natl Acad Sci U S A 95(9) (1998) 5172–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mali P, Esvelt KM, Church GM, Cas9 as a versatile tool for engineering biology, Nat Methods 10(10) (2013) 957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F, Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity, Cell 154(6) (2013) 1380–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM, RNA-guided human genome engineering via Cas9, Science 339(6121) (2013) 823–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J, Saturation editing of genomic regions by multiplex homology-directed repair, Nature 513(7516) (2014) 120–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F, Genome engineering using the CRISPR-Cas9 system, Nat Protoc 8(11) (2013) 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM, CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering, Nat Biotechnol 31(9) (2013) 833–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R, Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system, Cell Res 23(10) (2013) 1163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, Lim WA, Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds, Cell 160(1–2) (2015) 339–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA, Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers, Nat Biotechnol 33(5) (2015) 510–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhan T, Rindtorff N, Betge J, Ebert MP, Boutros M, CRISPR/Cas9 for cancer research and therapy, Semin Cancer Biol (2018). [DOI] [PubMed] [Google Scholar]
- [45].Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent CK, Nieuwenhuis EE, Beekman JM, Clevers H, Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients, Cell Stem Cell 13(6) (2013) 653–8. [DOI] [PubMed] [Google Scholar]
- [46].Huang Z, Tomitaka A, Raymond A, Nair M, Current application of CRISPR/Cas9 gene-editing technique to eradication of HIV/AIDS, Gene Ther 24(7) (2017) 377–384. [DOI] [PubMed] [Google Scholar]
- [47].Mettananda S, Fisher CA, Hay D, Badat M, Quek L, Clark K, Hublitz P, Downes D, Kerry J, Gosden M, Telenius J, Sloane-Stanley JA, Faustino P, Coelho A, Doondeea J, Usukhbayar B, Sopp P, Sharpe JA, Hughes JR, Vyas P, Gibbons RJ, Higgs DR, Editing an alpha-globin enhancer in primary human hematopoietic stem cells as a treatment for beta-thalassemia, Nat Commun 8(1) (2017) 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Rohn TT, Kim N, Isho NF, Mack JM, The Potential of CRISPR/Cas9 Gene Editing as a Treatment Strategy for Alzheimer’s Disease, J Alzheimers Dis Parkinsonism 8(3) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, Leiserson MDM, Miller CA, Welch JS, Walter MJ, Wendl MC, Ley TJ, Wilson RK, Raphael BJ, Ding L, Mutational landscape and significance across 12 major cancer types, Nature 502(7471) (2013) 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr., Kinzler KW, Cancer genome landscapes, Science 339(6127) (2013) 1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Markossian S, Flamant F, CRISPR/Cas9: a breakthrough in generating mouse models for endocrinologists, J Mol Endocrinol 57(2) (2016) R81–92. [DOI] [PubMed] [Google Scholar]
- [52].Yang H, Wang H, Jaenisch R, Generating genetically modified mice using CRISPR/Cas-mediated genome engineering, Nat Protoc 9(8) (2014) 1956–68. [DOI] [PubMed] [Google Scholar]
- [53].Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F, CRISPR-Cas9 knockin mice for genome editing and cancer modeling, Cell 159(2) (2014) 440–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Yu YP, Ding Y, Chen Z, Liu S, Michalopoulos A, Chen R, Gulzar ZG, Yang B, Cieply KM, Luvison A, Ren BG, Brooks JD, Jarrard D, Nelson JB, Michalopoulos GK, Tseng GC, Luo JH, Novel fusion transcripts associate with progressive prostate cancer, Am J Pathol 184(10) (2014) 2840–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Chen ZH, Yu YP, Tao J, Liu S, Tseng G, Nalesnik M, Hamilton R, Bhargava R, Nelson JB, Pennathur A, Monga SP, Luketich JD, Michalopoulos GK, Luo JH, MAN2A1-FER Fusion Gene Is Expressed by Human Liver and Other Tumor Types and Has Oncogenic Activity in Mice, Gastroenterology 153(4) (2017) 1120–1132 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Chen ZH, Yu YP, Zuo ZH, Nelson JB, Michalopoulos GK, Monga S, Liu S, Tseng G, Luo JH, Targeting genomic rearrangements in tumor cells through Cas9-mediated insertion of a suicide gene, Nat Biotechnol 35(6) (2017) 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Malina A, Mills JR, Cencic R, Yan Y, Fraser J, Schippers LM, Paquet M, Dostie J, Pelletier J, Repurposing CRISPR/Cas9 for in situ functional assays, Genes Dev 27(23) (2013) 2602–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Harrod A, Fulton J, Nguyen VTM, Periyasamy M, Ramos-Garcia L, Lai CF, Metodieva G, de Giorgio A, Williams RL, Santos DB, Gomez PJ, Lin ML, Metodiev MV, Stebbing J, Castellano L, Magnani L, Coombes RC, Buluwela L, Ali S, Genomic modelling of the ESR1 Y537S mutation for evaluating function and new therapeutic approaches for metastatic breast cancer, Oncogene 36(16) (2017) 2286–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F, Genome-scale CRISPR-Cas9 knockout screening in human cells, Science 343(6166) (2014) 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F, Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex, Nature 517(7536) (2015) 583–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Aquino-Jarquin G, Emerging Role of CRISPR/Cas9 Technology for MicroRNAs Editing in Cancer Research, Cancer Res 77(24) (2017) 6812–6817. [DOI] [PubMed] [Google Scholar]
- [62].Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, Cai W, Yang G, Bronson R, Crowley DG, Zhang F, Anderson DG, Sharp PA, Jacks T, CRISPR-mediated direct mutation of cancer genes in the mouse liver, Nature 514(7522) (2014) 380–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Sanchez-Rivera FJ, Papagiannakopoulos T, Romero R, Tammela T, Bauer MR, Bhutkar A, Joshi NS, Subbaraj L, Bronson RT, Xue W, Jacks T, Rapid modelling of cooperating genetic events in cancer through somatic genome editing, Nature 516(7531) (2014) 428–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Mollanoori H, Shahraki H, Rahmati Y, Teimourian S, CRISPR/Cas9 and CAR-T cell, collaboration of two revolutionary technologies in cancer immunotherapy, an instruction for successful cancer treatment, Hum Immunol 79(12) (2018) 876–882. [DOI] [PubMed] [Google Scholar]
- [65].Wu HY, Cao CY, The application of CRISPR-Cas9 genome editing tool in cancer immunotherapy, Brief Funct Genomics (2018). [DOI] [PubMed] [Google Scholar]
- [66].Wang Z, Wu Z, Liu Y, Han W, New development in CAR-T cell therapy, J Hematol Oncol 10(1) (2017) 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Jung IY, Lee J, Unleashing the Therapeutic Potential of CAR-T Cell Therapy Using Gene-Editing Technologies, Mol Cells 41(8) (2018) 717–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kochenderfer JN, Dudley ME, Kassim SH, Somerville RP, Carpenter RO, Stetler-Stevenson M, Yang JC, Phan GQ, Hughes MS, Sherry RM, Raffeld M, Feldman S, Lu L, Li YF, Ngo LT, Goy A, Feldman T, Spaner DE, Wang ML, Chen CC, Kranick SM, Nath A, Nathan DA, Morton KE, Toomey MA, Rosenberg SA, Chemotherapy-refractory diffuse large B-cell lymphoma and indolent B-cell malignancies can be effectively treated with autologous T cells expressing an anti-CD19 chimeric antigen receptor, J Clin Oncol 33(6) (2015) 540–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Park JH, Geyer MB, Brentjens RJ, CD19-targeted CAR T-cell therapeutics for hematologic malignancies: interpreting clinical outcomes to date, Blood 127(26) (2016) 3312–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Cooper ML, Choi J, Staser K, Ritchey JK, Devenport JM, Eckardt K, Rettig MP, Wang B, Eissenberg LG, Ghobadi A, Gehrs LN, Prior JL, Achilefu S, Miller CA, Fronick CC, O’Neal J, Gao F, Weinstock DM, Gutierrez A, Fulton RS, DiPersio JF, An “off-the-shelf” fratricide-resistant CAR-T for the treatment of T cell hematologic malignancies, Leukemia 32(9) (2018) 1970–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJ, Hamieh M, Cunanan KM, Odak A, Gonen M, Sadelain M, Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection, Nature 543(7643) (2017) 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Ren J, Zhang X, Liu X, Fang C, Jiang S, June CH, Zhao Y, A versatile system for rapid multiplex genome-edited CAR T cell generation, Oncotarget 8(10) (2017) 17002–17011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Cyranoski D, Chinese scientists to pioneer first human CRISPR trial, Nature 535(7613) (2016) 476–7. [DOI] [PubMed] [Google Scholar]
- [74].C. OFFORD, US Companies Launch CRISPR Clinical Trial, 2018. https://www.the-scientist.com/news-opinion/us-companies-launch-crispr-clinical-trial-64746. (Accessed 12.11.2018 2018).
- [75].Carsten Trevor Charlesworth PSD, Dever Daniel P, Dejene Beruh, Gomez-Ospina Natalia, Mantri Sruthi, Pavel-Dinu Mara, Camarena Joab, Weinberg Kenneth I, Porteus Matthew H, Identification of Pre-Existing Adaptive Immunity to Cas9 Proteins in Humans, bioRxiv (2018). [Google Scholar]
- [76].Herper M, Bill Gates And 13 Other Investors Pour $120 Million Into Revolutionary Gene-Editing Startup, 2015. https://www.forbes.com/sites/matthewherper/2015/08/10/bill-gates-and-13-other-investors-pour-120-million-into-revolutionary-gene-editing-startup/#2ae8471e6369.2018).
- [77].I. therapeutics, 2018. https://www.intelliatx.com/pipeline-2/.
- [78].Loria K, Bill Gates says it would be a ‘tragedy’ to pass up a controversial, revolutionary gene-editing technology, 2018. https://www.businessinsider.com/bill-gates-pushing-genetic-editing-with-crispr-2018-4.
- [79].Barry F, Google ventures part of $120m investment in CRISPR medicine, 2015. https://www.biopharma-reporter.com/Article/2015/08/11/Google-Ventures-part-of-120m-investment-in-CRISPR-medicine.
- [80].Peng R, Lin G, Li J, Potential pitfalls of CRISPR/Cas9-mediated genome editing, FEBS J 283(7) (2016) 1218–31. [DOI] [PubMed] [Google Scholar]
- [81].Rees HA, Komor AC, Yeh WH, Caetano-Lopes J, Warman M, Edge ASB, Liu DR, Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery, Nat Commun 8 (2017) 15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F, DNA targeting specificity of RNA-guided Cas9 nucleases, Nat Biotechnol 31(9) (2013) 827–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK, High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects, Nature 529(7587) (2016) 490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Trevino AE, Zhang F, Genome editing using Cas9 nickases, Methods Enzymol 546 (2014) 161–74. [DOI] [PubMed] [Google Scholar]
- [85].Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, E PRI, Lin S, Kiani S, Guzman CD, Wiegand DJ, Ter-Ovanesyan D, Braff JL, Davidsohn N, Housden BE, Perrimon N, Weiss R, Aach J, Collins JJ, G.M. Church, Highly efficient Cas9-mediated transcriptional programming, Nat Methods 12(4) (2015) 326–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F, Rationally engineered Cas9 nucleases with improved specificity, Science 351(6268) (2016) 84–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD, High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells, Nat Biotechnol 31(9) (2013) 822–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Mans R, van Rossum HM, Wijsman M, Backx A, Kuijpers NG, van den Broek M, Daran-Lapujade P, Pronk JT, van Maris AJ, Daran JM, CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae, FEMS Yeast Res 15(2) (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Wagner H, Toll meets bacterial CpG-DNA, Immunity 14(5) (2001) 499–502. [DOI] [PubMed] [Google Scholar]
- [90].Kim S, Kim D, Cho SW, Kim J, Kim JS, Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins, Genome Res 24(6) (2014) 1012–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Ramakrishna S, Kwaku Dad AB, Beloor J, Gopalappa R, Lee SK, Kim H, Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA, Genome Res 24(6) (2014) 1020–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen ZY, Liu DR, Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo, Nat Biotechnol 33(1) (2015) 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Wang X, Wang Y, Wu X, Wang J, Wang Y, Qiu Z, Chang T, Huang H, Lin RJ, Yee JK, Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors, Nat Biotechnol 33(2) (2015) 175–8. [DOI] [PubMed] [Google Scholar]
- [94].Holkers M, Maggio I, Henriques SF, Janssen JM, Cathomen T, Goncalves MA, Adenoviral vector DNA for accurate genome editing with engineered nucleases, Nat Methods 11(10) (2014) 1051–7. [DOI] [PubMed] [Google Scholar]
- [95].Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M, Zhang F, In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9, Nat Biotechnol 33(1) (2015) 102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Simhadri VL, McGill J, McMahon S, Wang J, Jiang H, Sauna ZE, Prevalence of Pre-existing Antibodies to CRISPR-Associated Nuclease Cas9 in the USA Population, Mol Ther Methods Clin Dev 10 (2018) 105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Friedland AE, Tzur YB, Esvelt KM, Colaiacovo MP, Church GM, Calarco JA, Heritable genome editing in C. elegans via a CRISPR-Cas9 system, Nat Methods 10(8) (2013) 741–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R, One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering, Cell 153(4) (2013) 910–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK, Efficient genome editing in zebrafish using a CRISPR-Cas system, Nat Biotechnol 31(3) (2013) 227–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Cyranoski D, Reardon S, Embryo editing sparks epic debate, Nature 520(7549) (2015) 593–4. [DOI] [PubMed] [Google Scholar]
- [101].Liang P, Xu Y, Zhang X, Ding C, Huang R, Zhang Z, Lv J, Xie X, Chen Y, Li Y, Sun Y, Bai Y, Songyang Z, Ma W, Zhou C, Huang J, CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes, Protein Cell 6(5) (2015) 363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Cribbs AP, Perera SMW, Science and Bioethics of CRISPR-Cas9 Gene Editing: An Analysis Towards Separating Facts and Fiction, Yale J Biol Med 90(4) (2017) 625–634. [PMC free article] [PubMed] [Google Scholar]
- [103].Torres-Ruiz R, Rodriguez-Perales S, CRISPR-Cas9: A Revolutionary Tool for Cancer Modelling, Int J Mol Sci 16(9) (2015) 22151–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Liu Y, Zeng Y, Liu L, Zhuang C, Fu X, Huang W, Cai Z, Synthesizing AND gate genetic circuits based on CRISPR-Cas9 for identification of bladder cancer cells, Nat Commun 5 (2014) 5393. [DOI] [PubMed] [Google Scholar]
- [105].Feng W, Li HC, Xu K, Chen YF, Pan LY, Mei Y, Cai H, Jiang YM, Chen T, Feng DX, SHCBP1 is over-expressed in breast cancer and is important in the proliferation and apoptosis of the human malignant breast cancer cell line, Gene 587(1) (2016) 91–7. [DOI] [PubMed] [Google Scholar]
- [106].Choi PS, Meyerson M, Targeted genomic rearrangements using CRISPR/Cas technology, Nat Commun 5 (2014) 3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Togashi Y, Mizuuchi H, Tomida S, Terashima M, Hayashi H, Nishio K, Mitsudomi T, MET gene exon 14 deletion created using the CRISPR/Cas9 system enhances cellular growth and sensitivity to a MET inhibitor, Lung Cancer 90(3) (2015) 590–7. [DOI] [PubMed] [Google Scholar]
- [108].Krachulec JM, Sedlmeier G, Thiele W, Sleeman JP, Footprintless disruption of prosurvival genes in aneuploid cancer cells using CRISPR/Cas9 technology, Biochem Cell Biol 94(3) (2016) 289–96. [DOI] [PubMed] [Google Scholar]
- [109].Wang H, Sun W, CRISPR-mediated targeting of HER2 inhibits cell proliferation through a dominant negative mutation, Cancer Lett 385 (2017) 137–143. [DOI] [PubMed] [Google Scholar]
- [110].Zhen S, Hua L, Takahashi Y, Narita S, Liu YH, Li Y, In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9, Biochem Biophys Res Commun 450(4) (2014) 1422–6. [DOI] [PubMed] [Google Scholar]
- [111].Aubrey BJ, Kelly GL, Kueh AJ, Brennan MS, O’Connor L, Milla L, Wilcox S, Tai L, Strasser A, Herold MJ, An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo, Cell Rep 10(8) (2015) 1422–32. [DOI] [PubMed] [Google Scholar]
- [112].Mazur PK, Herner A, Mello SS, Wirth M, Hausmann S, Sanchez-Rivera FJ, Lofgren SM, Kuschma T, Hahn SA, Vangala D, Trajkovic-Arsic M, Gupta A, Heid I, Noel PB, Braren R, Erkan M, Kleeff J, Sipos B, Sayles LC, Heikenwalder M, Hessmann E, Ellenrieder V, Esposito I, Jacks T, Bradner JE, Khatri P, Sweet-Cordero EA, Attardi LD, Schmid RM, Schneider G, Sage J, Siveke JT, Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma, Nat Med 21(10) (2015) 1163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Zuckermann M, Hovestadt V, Knobbe-Thomsen CB, Zapatka M, Northcott PA, Schramm K, Belic J, Jones DT, Tschida B, Moriarity B, Largaespada D, Roussel MF, Korshunov A, Reifenberger G, Pfister SM, Lichter P, Kawauchi D, Gronych J, Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling, Nat Commun 6 (2015) 7391. [DOI] [PMC free article] [PubMed] [Google Scholar]


