Abstract
Our current knowledge of mycobacterial infections in humans has progressively increased over the past few decades. The infection of Mycobacterium tuberculosis causes tuberculosis (TB) disease, which has reasoned for excessive morbidity and mortality worldwide, and has become a foremost issue of health problem globally. Mycobacterium leprae, another member of the family Mycobacteriaceae, is responsible for causing a chronic disease known as leprosy that mainly affects mucosa of the upper respiratory tract, skin, peripheral nerves, and eyes. Ample amount of existing data suggests that pathogenic mycobacteria have skilled in utilizing different mechanisms to escape or offset the host immune responses. They hijack the machinery of immune cells through the modulation of microRNAs (miRs), which regulate gene expression and immune responses of the host. Evidence shows that miRs have now gained considerable attention in the research, owing to their involvement in a broad range of inflammatory processes that are further implicated in the pathogenesis of several diseases. However, the knowledge of functions of miRs during mycobacterial infections remains limited. This review summarises recent findings of differential expression of miRs, which are used to good advantage by mycobacteria in offsetting host immune responses generated against them.
Keywords: Mycobacterium tuberculosis, Tuberculosis, microRNAs, Macrophages, Immune response, Apoptosis
INTRODUCTION
Mycobacterium tuberculosis, thrives in its only ecological niche provided by humans, The time period of co-existence of M. tuberculosis within humans dates back to about 3 million years ago, and has contributed in making M. tuberculosis a smart and competent human pathogen (1). M. tuberculosis mainly affects the lungs and causes pulmonary tuberculosis (TB), however it can even disseminate to other parts of the body to cause extra-pulmonary TB. In 2017, TB accounted for nearly 1.3 million deaths worldwide, among HIV-negative people, and an additional 300,000 deaths were reported in HIV-positive people from TB (2). Despite of being potentially curable, the treatment drugs for TB are costly and toxic, and its easy transmission to other individuals makes it challenging to conquer. Another mycobacterium, Mycobacterium leprae or Hansen's bacillus spirally, causes leprosy, which mainly affects the skin and peripheral nerves, and gives rise to disfigured, pale-colored skin sores, loss of sensation in the fore and hind limb and muscle weakness (3). The conventional treatment protocol of leprosy includes multidrug therapy (MDT); however, the morbidity rates due to nerve damage even after the conclusion of MDT in leprosy are significantly high. In 2017 alone, 211,009 new cases of leprosy were reported, which indicated an active transmission of the disease, even after implication of intensive treatment regimens (4). Further there are few opportunistic mycobacteria like Mycobacterium avium, which resides ubiquitously in soil and water sources, and can cause non-tuberculous mycobacterial (NTM) pulmonary disease in immuno-deficient patients (like HIV). However, the prevalence of M. avium pulmonary disease in non-HIV populations, which are suffering from chronic obstructive pulmonary disease (COPD) like bronchiectasis or cystic fibrosis, has also increased over the past few decades (5). Similarly, Mycobacterium marinum a water-borne mycobacteria that usually infects fish and amphibians, can also cause opportunistic infections in humans that initially develops as injuries to soft-tissues and skin, in the regions that were previously exposed to aquatic equipment like fish lines, fishhooks or aquarium. The disease condition is occupation specific and relatively rare however, there is no good treatment regime for the disease (6).
Mycobacteria have different routes of infection to the host, however once they reach inside the host body, immune system begins to marshal forces against them, and in order to survive within this competing environment, mycobacteria have devised ways to regulate host immune machinery. One of the well-known mechanisms to regulate the expression of various genes and gene products that participate in signaling pathways is through microRNAs (miRs), which are a class of small non-coding RNAs that play a critical role in post-transcriptional regulation of gene expression (7). In the last decade, miR-mediated regulation of host cells, upon mycobacterial infection has been extensively explored, which has resulted in the identification of several key genes and pathways targeted by them. In this review, pathogenesis and severity of mycobacterial diseases has been discussed, which is followed by a detailed summary of emerging roles of miRs in manipulating host defence mechanism against mycobacterial infection, through regulation of apoptosis and modulation of immune responses of immune pathways (8,9,10).
MYCOBACTERIAL INFECTION AND DISEASE
M. tuberculosis is an airborne pathogen and can be easily transmitted from person to person with lung TB cough, sneeze or spit, and inhalation of only a few viable bacilli can make a healthy individual infected with M. tuberculosis (11). Following the lung exposure, the alveolar macrophages (AMs) and dendritic cells (DCs) in the air spaces readily internalise M. tuberculosis but do not kill the mycobacteria. At this stage, the growth of mycobacteria occurs logarithmically, and the phagocytes start to produce cytokines and chemokines, which subsequently recruit blood polymorphonuclear leukocytes and additional mononuclear leukocytes at the site of infection (11,12). When the foci of infected cells gets surrounded by mononuclear leukocytes, a macrophage-rich cell mass known as granuloma is formed, and within these granulomas, macrophages play an essential role in controlling the growth of intracellular M. tuberculosis and limiting its dissemination to other sites (12). Gradually, the granulomas decrease in size and cellularity (13). However, if the granulomatous response is not effective enough, the bacilli will continue to replicate, and lead to pulmonary cavitation, a condition of high drug resistance and mycobacterial burden. Further, a rupture in nearby bronchi can pave the way for bacilli to reach other parts of the lungs through the airways and even to the extra-pulmonary sites through haematogenous routes (14). Finally, the outcome of whether active TB disease will supervene or not will depend upon a balance between the efficiency of immune system in outgrowing and clearing M. tuberculosis and the extent of destruction caused during primary cavitation to the lung parenchyma, and the meshwork of fibrillar collagen supporting normal lung structure (14). If the response hence generated is optimal, it will lead to complete bacillary control and development of latency. The ability of M. tuberculosis to establish latency (a state of immune unresponsiveness) should be credited to its site of residence within the humans, its remarkable ability to avoid phagolysosomal degradation in macrophages (15), as well as to regulate various immune responses mediated by them, such as inhibition of production of pro-inflammatory cytokines like TNF-α and IL-1, which ensures its persistence within these infected cells (15,16).
M. leprae a slow multiplying pathogen has an incubation period of up to 5 years. Its symptoms can take 1–20 or even more years to develop, however a prompt treatment of leprosy is required once the symptoms begin to surface, as if left untreated it can lead to progressive and permanent damage to the affected site (17). The most acceptable route for transmission of leprosy is through nasal droplet, spit, or by building frequent and close contacts with the affected individuals (18). However, there is little evidence to the mechanism of how the symptoms of leprosy are produced, owing to the fact that M. leprae is an obligate intracellular parasite and therefore it cannot be cultured in the laboratory or outside the host. So far, it has been reported that M. leprae uses numerous molecular mechanisms to cause nerve damage as it binds to Schwann cells through a laminin-binding protein present on its surface, which may lead to nerve injury through demyelination (19). As part of the human immune responses, macrophages have been identified as key players in the pathogenesis of leprosy, as they can engulf M. leprae by phagocytosis (20). Further, during an inflammatory response, bone marrow derived macrophages are recruited at the site of infection where they enter the tissues in large numbers and engage in defence mechanisms against pathogens. Keratinocytes are another type of cells that play an important role in innate immune responses against M. leprae (21) by generating cytokines and chemokines like TNF-α and IL-1β, that are critically required for recruiting neutrophils, DCs, and T cells at the site of infection (21).
M. avium has a highly hydrophobic cell wall, which along with providing it resistance from antibiotics and disinfectants, facilitates its aerosolization, and confers it an additional ability to adhere to surfaces (22). They are oligotrophic, therefore can thrive in environments with very less nutrients (23). Besides, these NTM, have a key ability to form biofilms in the environment and hypothetically in vivo (24), which can allow these bacilli to persist for very lengthy periods in the environment. Although the exact mode of transmission of these non-conventional mycobacteria in humans has not been defined and even the knowledge of host immune responses against M. avium is relatively miniscule (25). Despite of it, in few studies M. avium replication has been reported to retard when it was cultured in activated macrophages. The macrophage activation resulted in generation of ROS, nitric oxide (NO), and pro-inflammatory cytokines like TNF-α which effectively inhibited the growth of M. avium (26).
The immune responses generated against mycobacteria are majorly determined by host-pathogen interactions (27) and the changes in concentrations of immune-regulatory molecules upon such interactions, play a crucial role in triggering various downstream signaling pathways, so as to effectively evade off the invading pathogen. However, these pathogenic mycobacteria can alter host immune responses to suit their own benefit, by modulating host miRs, which bind to complementary target mRNAs, involved in a complex set of immune interactions and cause their translational repression or degradation (28).
THE BIOGENESIS OF miRs
The biogenesis of miRs generally starts with transcription of primary microRNAs (pri-miRs) by miR genes in the nucleus. These pri-miRs, which are at times even more than 1,000 nucleotide in length, have a double-stranded hairpin like structure consisting of 60–120 nucleotides, along with 5′ and 3′ overhangs (29). These features of pri-miR structure facilitate in its recoginition by RNA binding protein DiGeorge Syndrome Critical Region 8 (DGCR8), while the other motifs within the pri-miR structure are recognised by a double-strand-specific ribonuclease, Drosha (30). The DGCR8 and Drosha, together make a microprocessor complex, which cleaves the pri-miR duplex at the base of its hairpin structure (31). The hairpin structure hence formed bears a 2-nucleotide 3′ overhang and is translocated from the nucleus into the cytoplasm by a Ran-GTP dependent exportin 5 (XPO5) protein, which functions as a dsRNA binding protein (32). After entering into the cytoplasm the pre-miRs bind to a DICER (RNase III endonuclease), which cleaves the loop region of pre-miR hairpin and releases a duplex miR consisting of nearly 22 nucleotides (7). Out of the two strands of the duplex miR, one strand gets degraded and the remaining strand known as the ‘guide strand’ gets loaded onto an RNA-induced silencing complex (RISC), which comprises of a family of argonaute proteins. The miRISC complex hence formed with the association of miR and RISC assists the miR in finding sequence complementarity within the target mRNA. Upon binding to the target mRNAs, miRs can either degrade or translationally suppress the expression of the respective genes (8).
Today, it is well accepted that a single miR can target hundreds of different mRNAs, and conversely several different miRs can also target the same mRNA, resulting in enhanced translational inhibition (33). Further, a single biological system can be influenced by many miRs and a single miR can influence many biological systems (34), including those of protective immune responses, both innate and acquired, that play a pivotal role in controlling and eliminating the invading pathogens. One of the hallmark strategies used by mycobacteria to evade the immune attack, is through the deregulation of host miRs, wherein it hijacks the host phagocytic cell machinery and reprograms it. This recently explored area has unveiled a significant modulation of host apoptotic and immune response pathways, through the modulation of miRs, which catalyses the ability of mycobacteria to lush within the host (35,36).
HOST APOPTOTIC PATHWAY
Apoptosis is a genetically determined programmed cell death pathway, which is essential for maintaining the normal cell turnover in the body. It also plays a critical role in immune system, as in this pathway an intact plasma membrane bearing the cytoplasmic contents of dead cells is maintained around the infected cells, which is later ingested by professional phagocytes (37). This result in elimination of a favourable intracellular environment required for replication of mycobacteria, and thus forcing it to re-habilitate a naive host cell (38). Therefore, the intracellular pathogens like mycobacteria replicate within the host phagocytes by inhibiting apoptotic pathway and inducing necrotic cell death pathway, wherein a cell injury results in pre-mature death of cells (39), which is used by bacteria to egress form the cells.
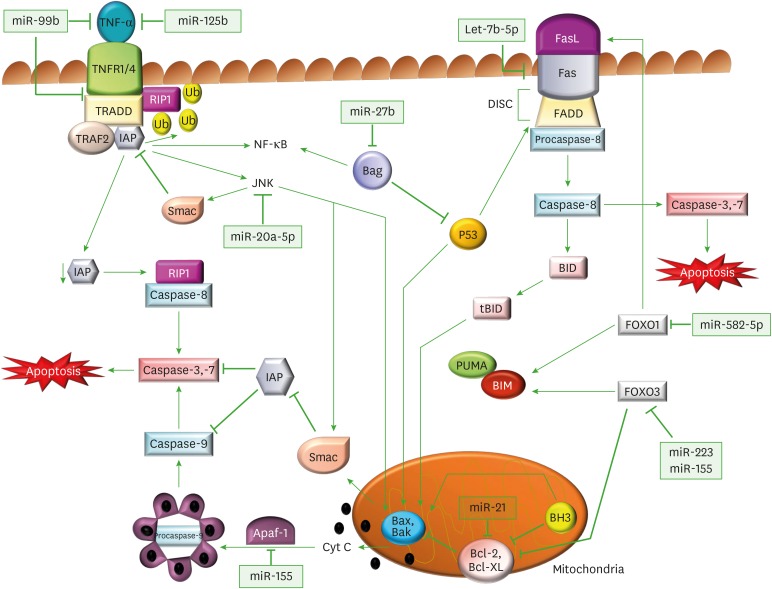
The apoptotic pathway can be activated via an extrinsic/death-receptor-mediated pathway and an intrinsic/mitochondrial pathway. The extrinsic apoptotic pathway is initiated by the activation of death receptors, which are expressed on some cells like monocytes, macrophages, activated T cells and some other immune and non-immune cells (40). Cell surface receptors like TNF receptors (TNFRs), make an important family of transmembrane death receptors that undergo protein cross-linking upon ligand interaction, to trigger a downstream signalling pathway which ultimately leads to apoptosis (Fig. 1) (41). TNF-α, is a soluble pro-inflammatory cytokine, and is primarily produced by activated macrophages in response to infection and inflammation. It elicits its biological response by interacting with a transmembrane TNFR1 that undergoes trimerization upon ligand interaction, and allows the recruitment of TNF receptor-associated death domain protein (TRADD) upon the assembled receptor complex. TRADD, which is an adaptor molecule, acts as a bridge between TNFR1 and pro-cysteine aspartase (caspase)-8 activating molecule receptor interacting protein kinase-1 (RIP1). Procaspase-8 upon activation gets converted into caspase 8, which in turn activates executioner caspases 3 and 7 (nucleases) that degrade the DNA and play a critical role in inducing apoptosis (42,43). However, TNFα-TNFR1 interaction can also follow an antagonising cell survival pathway, where TNFR1 can allow binding of TNF-receptor-associated factor (TRAF)-2 and inhibitory apoptosis protein (IAP) to the receptor complex. The protein complex so formed can mediate anti-apoptotic signals from TNF receptors due to ubiquitin ligase activity of IAP (44,45). IAPs ubiquitinate and degrade RIP1, and therefore prevent interaction of RIP1 with caspase-8, thus result in inhibition of apoptotic pathway. However, in the absence of IAPs, RIP1 cannot be ubiquitinated (46), due to which it is able to form a cytosolic complex with caspase-8 and induce apoptosis (47,48). Similarly, Fas ligand (FasL), a specialised member of TNF cytokine family, binds with its Fas receptor (FasR), and transduces an apoptotic signal upon its trimerisation. The clustered FasRs facilitate the recruitment of Fas-associated protein with death domain (FADD) that forms a death-inducing signaling complex with procaspase-8. This ultimately results in nuclear condensation and fragmentation of genomic DNA (Fig. 1) (49). In most of the cells, caspase-8 also catalyses the cleavage of the pro-apoptotic BH3-only protein BH3 interacting-domain death agonist (BID) into its truncated BID form. These BH-3 only members of the B-cell lymphoma 2 (Bcl-2) family are exclusively involved in downregulating anti-apoptotic members of the family (Bcl-2, Bcl-extra-large [Bcl-xl]), and in upregulating pro-apoptotic Bcl2-homologous antagonist killer (Bak) and Bcl-2-associated x protein (Bax) proteins, which act as antagonist of anti-apoptotic proteins (like IAPs) (50).
Figure 1. Regulation of apoptotic signaling pathways through miRs by M. tuberculosis. Diagrammatic representation of miRs and their target genes, which alter the canonical apoptotic pathways of immune cells, and assist in M. tuberculosis survival. In the extrinsic apoptotic pathway, TNFα binds to its receptors (TNFR1/4) and triggers its trimerization, which recruits TRADD, an adaptor molecule that allows binding of TRAF-2 and IAP to the receptor complex. RIP1, a kinase then binds TNFR1 through TRADD association. IAP proteins are responsible for RIP1 ubiquitination, and in their absence, RIP1 cannot be ubiquitinated. Non-ubiquitinated RIP1 can form a cytosolic complex with the caspase-8, leading to induction of apoptosis. The binding of TRAF-2 to the receptor complex stimulates the JNK pathway, which is a proapoptotic pathway. JNK can promote the release of Smac, from mitochondria, which inhibits TRAF2/IAP complex formation, and relieves the inhibition of caspase-8, thereby triggering caspase activation. JNK also participates in intrinsic apoptotic pathway, as after activation, JNK translocates to the mitochondria and phosphorylates BH3-only family of Bcl-2 proteins, which antagonize the anti-apoptotic activity of Bcl-2 or Bcl-Xl proteins. Further, JNK stimulates the release cyt C from mitochondrial inner membrane through BID-Bax dependent mechanism and promotes apoptosomes formation from released cyt C, caspase-9, and Apaf-1. The apoptosomes initiates caspase-9-dependent caspase cascade. In FasR pathway, a FasL binds to FasR and induces the recruitment of FADD followed by activation of procaspase-8, which further activates caspase-3 (the executioner) causing apoptosis. Caspase-8 also cleaves BID, a BH3 domain-containing protein of the BCL-2, which triggers intrinsic apoptosis and amplifies the signal from the extrinsic pathway. The expression of another member of BH3 only protein is increased by FOXO1 and FOXO3, which initiates the Bax/Bak-dependent apoptotic pathway.
The binding of TRAF-2 to the receptor complex can stimulate another pathway, the JNK pathway, which is a pro-apoptotic pathway. The JNK activation stimulates BID-Bax dependent mitochondrial apoptotic pathway, wherein the activated multidomain, proapoptotic proteins Bax and Bak, oligomerise on mitochondrial outer membrane to form supramolecular openings, which causes membrane permeabilization and release of cytochome C (cyt C) from intermembrane space into the cytosol. The released cyt C promotes the formation of apoptosomes consisting of cyt C, caspase-9, and apoptotic protease activating factor 1 (Apaf-1), in the cytosol, which finally activates caspase-9 dependent caspase cascade (Fig. 1) (51,52). Further, JNK can also translocate to the mitochondria and trigger apoptosis by promoting Bcl-2 associated death promoter (BAD)-mediated neutralization and multiple site phosphorylation of Bcl-2 family of anti-apoptotic proteins like Bcl-xl, to suppress their anti-apoptotic activity and initiate apoptosis (53,54). The BH3-only family of anti-apoptotic proteins are located on outer mitochondrial membrane, where they block the release of cyt C. Additionally, JNK can also promote the release of a pro-apoptotic mitochondrial protein second mitochondria-derived activator of caspase (Smac), which inhibits the activity of TRAF2/IAP1 complex, and relieves the inhibition of caspase-8. Therefore, JNK can initiate caspase activation through multiple pathways to ultimately bring out apoptosis (55).
Forkhead box protein O1 (FOXO1), a protein encoded by the FOXO1 gene, belonging to FOXO family of transcription factors, also plays a crucial role in cell death, by transactivating Bim, a pro-apoptotic protein, which is also a member of BH3-only subset of Bcl-2 family proteins. Bim induces Bax-Bak oligomerization and the release of cyt C from mitochondrial outer membrane, and thus FOXO1 plays a key role in promoting apoptosis (Fig. 1) (56). Similarly, the activation of another FOXO protein FOXO-3 also enhances the concentration of Bim protein as it supresses the action of Bim neutralizing Bcl-2 proteins. Therefore, FOXO3 enhances apoptosis by overcoming the protective effects of anti-apoptotic Bcl-xl and IAPs in cell survival (57). Additionally, FOXO1 and FOXO3 can also transcriptionally upregulate FasL and ROS respectively, both of which contribute to apoptotic cell death (56,57).
miRs AS REGULATORS OF APOPTOSIS
In order to regulate this complex and intertwined mechanism of apoptosis, mycobacteria especially, M. tuberculosis has been reported to regulate the expression of an array of miRs in macrophages (Tables 1 and 2).
Table 1. Regulation of host miRs during M. tuberculosis infections.
| miRNAs | Species | Targets | Biological outcome | Reference |
|---|---|---|---|---|
| miR-20a-5p | M. tuberculosis | JNK-2 | Inhibition of apoptosis | (61) |
| miR-21 | M. tuberculosis MPT64 | Bcl-2 | Inhibits apoptosome formation | (63) |
| miR-125b | M. tuberculosis | TNF | Destabilises TNF-α miR | (59) |
| miR99b | M. tuberculosis | TNF and TNF-α receptor | Lowers production of TNF-α and TNF-α receptor | (58) |
| miR-582-5p | M. tuberculosis | FOXO1 | Inhibits activation of multiple pro-apoptotic genes | (64) |
| miR-155 | M. tuberculosis | FOXO3 | Inhibition of apoptosis | (66) |
| miR-223 | M. tuberculosis | FOXO3 | Inhibition of apoptosis | (65) |
| miR-27b | M. tuberculosis | Bag2 | p53 dependent apoptosis | (62) |
| hsa-let-7b-5p | M. tuberculosis | Fas protein | M. tuberculosis survival in THP-1 cells | (60) |
| miR-155 | M. tuberculosis | Atg3 | Reduced autolysosome fusion and decrease autophagosome number | (92) |
| miR-142-3p | M. tuberculosis | N-WASP | Deregulation of actin dynamics during phagocytosis | (90) |
| miR-99b | M. tuberculosis strain H37Rv | TNF-α | TNF-α, IL-6, IL-12, and IL-1 | (58) |
| miR-223 | M. tuberculosis | p65 phosphorylation, CXCL2, CCL3 and IL-6 | NF-κB, TNF-α and IL-6 and IL-12p40, chemotaxis | (107,108) |
| Let-7f | M. tuberculosis | A20 | Reduced production of IL-1β, TNF-α, and NO | (106) |
| miR-26a and miR-132 | M. tuberculosis | P300 | Inhibition of IFN-γ signalling cascade | (115) |
| miR-29 | M. tuberculosis | IFN-γ | Regulates Th1 response | (114) |
| miR-125b | M. tuberculosis lipomannan | TNF-α | Regulates Th1 response | (59) |
| miR-144* | M. tuberculosis | TNF-α, IFN-γ | Cytokine signalling and cell proliferation | (116) |
| miR-1178 | M. tuberculosis | TLR-4 | IFN-γ, IL-6, IL-1 and TNF-α | (117) |
| miR-106b-5p | M. tuberculosis | CtsS mRNA | Reduced T cell activation and HLA-DR expression | (124) |
Table 2. miR-mediated modulation of host immune response during M. bovis and other mycobacterial infections.
| miRs | Species | Target | Biological outcome | Reference |
|---|---|---|---|---|
| miR-203 | M. bovis BCG | MyD88 | Inhibits production of NF-κB, TNF-α, and IL-6 | (101) |
| miR-149 | M. bovis BCG | MyD88 | Reduced production of NF-κB, TNF-α, and IL-6 | (103) |
| miR-124 | M. bovis BCG | TLR6, MyD88, TRAF-6, TNF-α | Blocks inflammatory response | (96) |
| miR-31 and miR-150 | M. bovis BCG | MyD88 | Interferes with TLR2 signalling | (101,102) |
| miR-142-3p | M. bovis BCG | IRAK-1 | NF- κB, TNF-α, and IL-6 | (91) |
| miR-146a | M. bovis BCG | IRAK-1 and TRAF-6 | Reduces activation of NF-κB and MAPK signalling, PTGS2, suppresses NO production | (97) |
| miR-146a | M. bovis BCG | IFN-γ | Autoimmune disease involving STAT1 pathway | (113) |
| miR-21 | M. bovis BCG | IL-12p35 mRNA | Inhibits IL-12 production | (105) |
| miR-155 | M. marium | CEBP | Inhibits production NO synthase | (69) |
| Let-7a and miR-29a | M. avium | Caspase 3 and 7 | Inhibition of apoptosis | (10) |
| miR-142-3p | M. smegmatis | N-WASP | Deregulation of actin dynamics during phagocytosis | (91) |
| miR-125b | M. smegmatis | TNF | Inhibition of TNF-α, IL-6, IL-12, and IL-1 production | (58) |
| miR-21 | M. leprae | CAMP, DEFB4 | Inhibition of vitamin D dependent anti-microbial pathway | (118) |
miR-99b: M. tuberculosis modulates host microbicidal responses and suppresses the Ag presentation and production of host protective cytokines by infected cells, by inhibiting the synthesis of TNF and its receptor through upregulation of miR-99b expression in infected macrophages and DCs, which targets TNF mRNA (58), and hence causes dysregulation of host protective adaptive immune responses.
miR-125b: In another study, TNF biosynthesis was found to be inhibited by miR-125b, as it binds to the 3′-untranslated region (UTR) region of TNF mRNA and obstructs its transcription. When M. tuberculosis infection was compared with non-virulent Mycobacterium smegmatis infection higher hsa-miR-125b expression was found in human alveolar macrophages infected with virulent M. tuberculosis (58), which lowered TNF production and assisted M. tuberculosis to potentially increase its virulence and subvert host immune defence (58).
Let-7b-5p: M. tuberculosis can significantly up regulate miR-hsa-let-7b-5p expression in infected THP-1 human macrophages, which directly targets 3′-UTR of Fas protein; it therefore facilitates intracellular survival of M. tuberculosis by inhibiting apoptosis. The results were further confirmed by inhibiting hsa-let-7b-5p, which correspondingly augmented apoptosis of THP-1 cells and enabled enhanced M. tuberculosis clearance (60).
miR-20a-5p: It was found to target JNK-2 in human macrophages, and therefore functions as a negative regulator of apoptosis (61). The overexpression of miR-20a-5p in response to mycobacterial infection resulted in marked survival of M. tuberculosis within the macrophages. A knockdown study of miR-20a-5p confirmed a more efficient M. tuberculosis clearance and its reduced survival in macrophages (61).
miR-27b: In a separate study M. tuberculosis H37Ra, was reported to induce the expression of miR-27b, which directly targets anti-apoptotic protein Bcl-2 associated anthogene (Bag)-2 in macrophages. Bag-2 acts as a negative regulator of p53 apoptosis signalling pathway, wherein the transcription factor p53 transcriptionally activates pro-apoptotic proteins of BH3-only family. Therefore, inhibition of anti-apoptotic Bag-2 protein by miR-27b in H37Ra, facilitates bacterial elimination by removing inhibition on p53 signalling pathway (62), and this upregulation of miR-27b in an attenuated strain of mycobacteria, which affects the fate of apoptotic proteins, can be a possible reason for inverse correlation between mycobacterial virulence and effectuation of apoptosis.
miR-21: M. tuberculosis can also secrete certain proteins like MPT64, which induces overexpression of miR-21 that causes an upregulation of anti-apoptotic proteins like Bcl-2, which are localized to the outer membrane of mitochondria. miR-21 can even inhibit the action of pro-apoptotic proteins like Bax and Bak, which act on the mitochondrial membrane to increase permeabilization and release of cyt C and ROS, so as to trigger apoptotic cascade. Therefore, upon cumulating both actions, miR-21 was found to promote macrophage survival by inhibiting the action of pro-apoptotic proteins and upregulating the action of anti-apoptotic proteins. Later, it was reported that MPT64 inhibits apoptosis through NF-κB-miR21-Bcl-2 pathway in RAW264.7 cells (63).
miR-582-5p: In a study, differentially expressed miRs in patients with active TB were investigated, miR-582-5p was found to directly target FOXO1, which inhibited apoptosis in M. tuberculosis-infected macrophages. This apoptotic miR was found to be upregulated in monocytes obtained from active TB patients (64).
miR-223: It is also an anti-apoptotic miR as it suppresses macrophage apoptotic pathway by targeting FOXO3. The expression of miR-223 was reported to be highly upregulated in macrophages obtained from active TB patients (65).
miR-155: Furthermore, in another study, serum miR-155 levels were found to be negatively associated with FOXO3 and TB-suppressing activity of cells (66), therefore miR-155 was reported to inhibit apoptosis in monocytes of active TB patients by downregulating the expression of FOXO3 (67). In another study, miR-155 was found to be the most highly upregulated miR in M. tuberculosis-infected murine RAW264.7 cells and bone marrow-derived macrophages where it inhibited the expression of cyclooxygenase-2 and IL-6, which are modulators of the innate immune response (68). In a different study done on M. marium infection, miR-155 was reported to bind at 3′-UTR of CCAAT/enhancer binding protein β (CEBPB), a transcriptional activator of inducible nitric oxide synthase (iNOS), which resulted in its inhibition and favoured mycobacterium survival (69).
Let-7a and miR-29a: The miRs let-7e and miR-29a were reported to be upregulated by M. avium in human monocyte derived macrophages, which accounted for decreased expression of caspase-3 and caspase-7 (10).
Collectively, these studies highlight the regulation of apoptosis by miRs and their exploitation by mycobacteria to persist within the host. These findings about diverse roles of miRs are consistent with the theory that, a single miR can target multiple genes at the same time and have varied consequences. It also emphasizes the regulatory role of miRs at different steps in distinct pathways.
REGULATION OF SIGNALING PATHWAYS DURING MYCOBACTERIAL INFECTION
Other than apoptosis, which is an important host defence mechanism, phagocytes can also clear foreign particles through a process known as phagocytosis. In this process, the foreign particle is engulfed by a cell into a phagosome, which fuses with a lysosome, and finally, the acidic environment of the phagolysosome so formed, degrades the harboured foreign particle (70). After degradation, the mycobacterial Ags are processed and transported to the cell surface where they are made available for priming of T cell responses, and to generate adaptive immune response against the phagocytosed pathogen. However, in an active TB state, M. tuberculosis resides and replicates within these phagolysosomal compartments of macrophages, due to its ability to combat the acidic environment generated by lysosomal enzymes and to prevent the development of phagolysosome into acid-rich compartments (71). Furthermore, viable and virulent M. tuberculosis can even bud from the phagosomes with no subsequent fusion with lysosomes in specialised vesicles.
Cdc42/N-Wiskott Aldrich syndrome (WASP)/actin-related proteins (Arp)2/3 and TLR signaling pathway
Phagocytosis is a multi-step process, which involves local reorganisation of actin cytoskeleton to internalise the foreign particles such as pathogens (72). The actin assembly is initiated through ligation of one of the best-studied opsonic phagocytic receptors, complement receptor (CR) 3 (or αMβ2) and Fcγ receptor (FcγR), which is followed by membrane extension and fusion, ultimately leading to particle internalization (73). FcγR enables the engulfment of IgG-coated targets through the formation of phagocytic cup, consisting of extensive disarranged actin-rich network around the foreign particle (74). This process requires the activity of Rho family GTPases (Cdc42) (75), and phosphatidylinositol 4,5-bisphosphate, which phosphorylates WASP family of proteins (76) at its 291 tyrosine residue, and converts it from an auto-inhibited state to an activated state (77). This enhances the ability of WASP to promote Arp 2/3-mediated actin polymerisation (78), which is present in almost every actin cytoskeleton-containing eukaryotic cell.
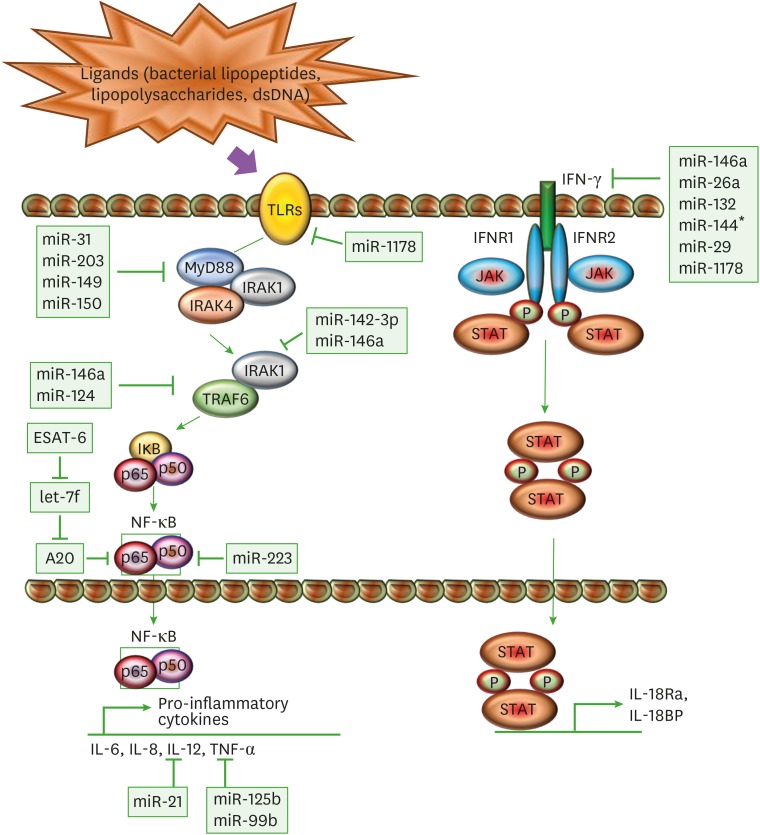
After internalization of mycobacteria by alveolar macrophages or other innate cells through phagocytosis, their Ags get exposed to a variety of intracellular receptors including TLRs, which are a class of protein receptors that are known to play significant roles in the innate immune system, by recognising pathogen-associated molecular patterns (PAMPs) derived from various microbes. The different set of TLRs like, TLR2 in association with TLR1/TLR6, with TLR4, or with TLR9 recognise distinct mycobacterial components (79). TLR2 is involved in generating responses in innate immune cells such as macrophages and DCs in response to a variety of mycobacterial cell wall Ags including bacterial lipoproteins and lipopolysaccharides (80). TLR4 is activated by heat shock proteins, whereas TLR9 recognizes unmethylated CpG motifs of mycobacterial DNA (79,81). The ligation of ligands to TLRs leads to the formation of the Myddosome, that consists of interactions between the death domains of an adaptor protein myeloid differentiation primary response 88 (MyD88), with those of the protein kinases IL-1R-associated kinase (IRAK) 4 and IRAK1 (Fig. 2). This interaction induces auto-phosphorylation of IRAK4, which then phosphorylates and activates IRAK1 (82). Once the different components of the Myddosome are phosphorylated, they promote the activation of the ubiquitin E3 ligases (like TRAF6) that provide ubiquitin-binding sites for TAB2 and TAB3. Subsequently, TGF-β-activated kinase (TAK)1 gets activated, which then phosphorylates IκB kinase (IKK) α and IKKβ, and gives rise to a canonical signaling-competent IKK complex. This further phosphorylates components of the transcription factor NF-κB such as IκBα, p105 (NF-κB1) and relA (p65) (83). The phosphorylation of p105 leads to its cleavage into p50 and activation of MAPK signaling cascade. TAK1-mediated phosphorylation and activation results in activation of other MAPKs p38α/β and JNK1/2 pathways as well that cumulatively activate the various transcription factors to drive the production of pro-inflammatory cytokines such as IL-12 and TNFα (Fig. 2) (84), which acts as a key regulator of host immune responses to mycobacterial infections (85). TNF mediates several mechanisms (1,85,86), to cause macrophage-mediated induction of chemokine and cytokine expression (58), and activation of apoptosis (87,88,89) to efficiently kill the bacilli. Therefore mycobacteria (mainly M. tuberculosis) uses miRs as weapons to regulate several key steps of these signalling pathways.
Figure 2. Immunomodulation of macrophages through miRs. A schematic diagram represents different miRs and their target genes, which allows M. tuberculosis to mitigate the immune response through modulation of TLR and IFN-γ pathway. The ligation of mycobacterial ligands to TLRs leads to the interaction of MyD88 with IRAK4 and IRAK1. IRAK4 auto-phosphorylates and activates IRAK1. This promotes the activation of TRAF6, which further activates TAK1. TAK1 gives rise to canonical IKK complex by phosphorylating IKKα and IKKβ, which phosphorylates components of the transcription factor NF-κB such as IκBα, p50, and p65. TAK1-mediated activation of NF-κB transcription factors drives the production of pro-inflammatory cytokines such as IL-12 and TNF-α. In another pathway, IFN-γ acts as a principal mediator of macrophage activation, which modulates pro-inflammatory cytokine production and induces production of anti-inflammatory molecules. Upon ligand binding, oligomerization and transphosphorylation of IFN-γ receptors (IFNR1 and IFNR2) activate JAK1 and JAK2, which creates a docking site for STAT1. STAT1 homodimerizes upon phosphorylation (P) in an antiparallel configuration, forming a complex γ-activated factor, which translocates to the nucleus and binds to the γ-activated site, located at the promoters of primary response genes, increasing their transcription, like IL-1Ra and IL-18BP. SOCS proteins negatively regulate the IFN-γ pathway by inhibiting JAKs and STAT1 phosphorylation.
miR-142-3p: M. tuberculosis and non-pathogenic M. smegmatis infection enhances the expression of miR-142-3p, which targets N-WASP, and deregulates actin dynamics during phagocytosis. Although the expression of miR-142-3p gets upregulated in murine macrophages in both the infections, but in primary human macrophages, high levels of miR-142-3p were only reported upon infection with M. tuberculosis. This disparity between the results in murine and human macrophages was attributed to the fact that evolution of M. tuberculosis has taken place through millions of years of human interactions, with a propensity to reside in macrophages. Therefore, where M. tuberculosis could downregulate N-WASP in human macrophages by upregulation of miR-142-3p, M. smegmatis had no effect on miR-142-3p expression (90). This outcome also indicates that, different mycobacterial species have different effects on host immune system due to the dissimilar miR regulation by them. It was also reported that M. bovis BCG-mediates downregulation of miR-142-3p which targets IRAK-1 and plays a critical role in the regulation of innate immune response through transfer signaling from TLR (Table 2). Decreased expression of miR-142-3p results in a subsequent increase in production of NF-κB, TNF-α and IL-6 in the macrophages (91).
miR-155: It was reported that live and virulent M. tuberculosis infection can stimulate active translation of miR-155, which was presumed to pair with Atg3, an E2 conjugating enzyme involved in LC3-lipidation and autophagosome formation. The expression level of miR-155 in a study was found to be inversely related with the level of Atg3 and number of autophagosomes. The role of miR-155 in assisting the survival of M. tuberculosis was confirmed by silencing miR-155, which rescued autophagosome number in DC and enhanced autolysosome fusion (92). During mycobacterial infection, miR-155 was also reported to directly target a Ras homolog in the brain, which plays a critical role in autophagy pathway, thus this miR-155 mediated inhibition contributes to enhanced survival of intracellular mycobacteria (93). It acts as a positive regulator of TLR signaling (94) and this protective role of miR-155 against mycobacterial infection was also confirmed through miR-155 knockout in mice, which were found to be more susceptible to M. tuberculosis infection, and died relatively early, as compared to wild-type mice (95).
miR-124: In peripheral blood leukocytes obtained from active pulmonary TB patients, the overexpression of miR-124 was observed, which targeted the mRNAs of TLR6, MyD88, TRAF-6 and TNF-α, therefore it was found to block the inflammatory responses (96). The elevated expression of miR-124 was also found in murine alveolar macrophages in response to Mycobacterium bovis bacillus Calmette–Guérin (BCG; a non-virulent vaccine strain of M. tuberculosis) in vitro and in vivo infection model (96).
miR-146a: M. tuberculosis induces the expression of miR-146a, which mechanistically targets IRAK-1 and TRAF-6 and significantly enhances intracellular growth of bacilli by reducing activation of NF-κB and MAPK signaling, required for expression of iNOS and therefore NO production (97). Nitric oxide has been found to kill M. tuberculosis tubercule with a molar potency comparable to chemotherapy and reduced production of iNOS provides a survival advantage to M. tuberculosis (98). Additionally, in the same study, it was also reported that overexpression of miR-146a can strongly reduce the production of prostaglandin synthetase 2 (PTGS2). PTGS2 is responsible for increasing iNOS levels and it plays a decisive role in the early containment of M. tuberculosis infection (99). miR-146a can also repress mycobacteria-mediated inflammatory response in RAW264.7 macrophages by inhibiting the induction of pro-inflammatory cytokines TNF-α, IL-1β, IL-6 and chemokine monocyte chemoattractant protein-1(MCP-1) (100).
MyD88 targeting miRs: M. bovis BCG induces overexpression of certain miRs like miR-203 (101), miR-31, miR-150 (102) and miR-149 (103) which directly target MyD88. The downregulation of MyD88 results in suppression of inflammatory cytokines like NF-κB, TNF-α and IL-6, as MyD88 enhances the expression of genes of these inflammatory cytokines and it also plays a critical role in activation of protein kinases like MAPK (Table 2) (101,104).
miR-21: The expression of another miR, miR-21was also found to be enhanced in M. bovis BCG infected macrophages and DCs. miR-21, which directly targets IL-12p35 mRNA and inhibits IL-12 production, results in impaired T cells responses in both in vitro and in vivo models (Table 2) (105).
miR-let-7f: M. tuberculosis secretes an effector Ag known as early secreted antigenic target 6 (ESAT-6), which downregulates miR-Let-7f in macrophages that targets A20, an inhibitor of NF-κB signaling. It was reported that there was an enhanced production of IL-1β, TNF-α and NO in A20-deficient macrophages, which resulted in reduced bacilli survival (106).
miR-223: Another miR, miR-223 was reported to be involved in negative regulation of NF-κB signaling, by suppressing phosphorylation of p65 and its nuclear translocation. M. tuberculosis significantly enhances the expression of miR-223, in monocytes and monocyte-derived macrophages of TB patients (107). In another study done in myeloid cells, it was reported that miR-223 targets chemoattractants like Chemokine C-X-C motif ligand 2 (CXCL2), Chemokine Ligand (CCL) 3, and IL-6. Conclusively, overexpression of miR-223 causes negative regulation of NF-κB signaling, which subsequently results in reduced production of IL-1β, IL-6, TNF-α and IL-12p40 (108).
miR-27b: M. tuberculosis H37Ra in murine macrophages, restrains the activity of NF-κB and the production of pro-inflammatory factors, through upregulation of miR-27b in macrophages, which involves TLR2/MyD88/NF-κB signaling pathway. Since miR-27b reduced the production of pro-inflammatory cytokines in macrophages, it was suggested to be involved in avoiding excessive inflammation during M. tuberculosis infection (62).
miR-99b: In another finding, miR-99b was found to be highly upregulated in M. tuberculosis H37Rv infected DCs and macrophages, and the expression level of miR-99b was inversely related with the concentration levels of pro-inflammatory cytokines such as IL-6, IL-12, and IL-1β. Additionally, miR-99b was also reported to target TNF-α receptor gene, as inhibition of miR-99b enhanced their production (58).
IFN-γ-mediated signaling
The production of lymphokine IFN-γ in response to infection is a hallmark of innate and adaptive immune response. It is a principal mediator of macrophage activation and therefore provides resistance to intracellular pathogens (109). IFN-γ modulates pro-inflammatory cytokine production and induces the production of some anti-inflammatory molecules such as IL-1 receptor antagonist (IL-1Ra) and IL-18 binding protein (IL-18BP) (Fig. 2). Moreover, IFN-γ also triggers apoptosis by interfering with the signal transduction of suppressors of cytokine signaling (SOCS) which negatively regulates several pathways, particularly the Janus kinase/Signal transducer and activator of transcription (JAK/STAT) pathway, and since this signaling pathway is itself responsible for JAK-STAT induction, it forms a classical negative feedback circuit (110). Together, it has several anti-inflammatory characteristics, which assist in the regulation of cytokine network (111). Corresponding to other signaling pathways discussed so far, mycobacteria can also regulate IFN-γ signaling pathway by modulating expressions of various miRs by either decreasing or increasing their expression. Further, the significance of IFN-γ was understood from the knockdown studies of IFN-γ performed in xenograft models. Upon infection of these models with M. tuberculosis, it was found that they developed granulomas but were incapable to restrict the growth of the bacilli, due to their inability to produce reactive nitrogen intermediates. IFN-γ knockdown mice also exhibited heightened tissue necrosis and ultimately succumbed to TB (112).
miR-146a: The expression of miR-146a was found to be decreased in alveolar macrophages of pulmonary TB patients (99). Subsequently, in another report, mice deficient in miR-146a was reported to develop fatal IFN-γ-dependent autoimmune disease, which was speculated to involve STAT1 signaling pathway. Additionally, it was found that miR-146a also negatively regulated TNF-α (Table 2) (113).
miR-29: It is upregulated in response to M. tuberculosis infection and it assists in controlling innate and adaptive immune responses against M. tuberculosis by targeting IFN-γ. It was reported that in transgenic models endogenous blockage of miR-29 resulted in increased resistance against M. tuberculosis infection (114).
miR-26a and miR-132: M. tuberculosis also causes overexpression of miR-26a and miR-132 in macrophages, that target transcriptional co-activator of IFN-γ, p300. Therefore, overexpression of these miRs blocks the macrophage response to IFN-γ, by blocking IFN-γ downstream signalling (115).
miR-144*: Similarly, a significant overexpression of miR-144* was found in T cells of active TB patients which inhibited T cell proliferation and reduced production of TNF-α and IFN-γ. Therefore miR-144* was suggested to modulate cytokine production and T cell proliferation (116).
miR-1178: Recently overexpression of miR-1178 was delineated in human macrophages, which was linearly correlated to M. tuberculosis infection in a concentration- and time-dependent manner. The overexpression of miR-1178 was found to significantly decrease the accumulation of pro-inflammatory cytokines like IFN-γ, IL-6, IL-1β, and TNF-α in the M. tuberculosis-infected macrophages, which decreased the production of inflammatory cytokine against M. tuberculosis and enhanced its intracellular growth. Additionally, it was also observed that miR-1178 directly targeted TLR4 and therefore negatively regulated its expression in M. tuberculosis-infected macrophages (117).
miR-21: In leprosy, miR-21 was found to be upregulated in monocytes derived from lepromatous leprosy skin lesion samples. The overexpression of miR-21 was directly related to the reduced expression of vitamin D-dependent antimicrobial peptides, for example- human cathelicidin antimicrobial peptide (CAMP) and defensin beta 4 (DEFB4) (118). The tissues that are exposed to environmental microbes like skin and the squamous epithelia of lungs secrete CAMP in significant amounts, which mediates chemoattraction and bactericidal activities (119). Whereas, DEFB4 a defensin made by neutrophils, and it encodes for a cytotoxic and microbicidal peptide (120). Thus, miR-21 dependent inhibition of CAMP and DEFB4 has unveiled the role of miR-21 in inhibiting antibacterial activity and adaptive immune responses generated against M. leprae.
T CELL ACTIVATION THROUGH Ag PRESENTATION DURING MYCOBACTERIAL INFECTION
MHC is highly essential for acquired immune system as it aids in recognition of foreign molecules. MHC encodes for the HLA class II genes, which are highly polymorphic genes and are responsible for Ag presentation to CD4+ T cells. This results in the generation of an immune response against the presented Ag. CD4 T cells after Ag presentation are positively selected on the basis of class II alleles and they become predetermined to generate a specific immune response which is also known as MHC restriction of T cells (121). M. tuberculosis secretes certain proteins that are involved in the generation of immune response among which ESAT-6 is a major secretory and most reactive Ag of M. tuberculosis. It is involved in T cell activation and macrophage inhibition (122).
miR-106b-5p: In one of the recent findings, miR-106b-5p, which directly targets cathepsin S (CtsS) mRNA by binding to the 3′-UTR, was reported to be specifically upregulated by M. tuberculosis. CtsS belongs to an important group of degradative lysosomal enzymes, which includes cysteine cathepsin proteases that expose intracellular pathogens to degradative enzymes and are also involved in Ag processing and presentation (123,124). Furthermore, it was reported that downregulation of CtsS by M. tuberculosis also reduces HLA-DR class II surface expression. Therefore, upregulation of miR-106b-5p by M. tuberculosis not only obstructs bacterial clearance but also inhibits Ag presentation and T cell activation, through the involvement of CtsS and MHC class II molecules (125). However, there are not many findings about the role of different miRs in the regulation of T cell differentiation, but it opens possibilities for future research, which can assist in the generation of vaccines by providing valuable underlying information about the mechanism behind TB regulated miRs and their role in Ag presentation and T cell differentiation.
CONCLUSIONS AND FUTURE PERSPECTIVES
M. tuberculosis causes one of the world's most deadly communicable disease TB, which is difficult to eradicate, due to the capability of M. tuberculosis to camouflage within the host immune cells and remain undetected till the disease is well established. M. leprae is well known to cause another potentially curable yet severe infectious disease, leprosy. Similarly, the disease prevalence of M. avium and M. marinum is also on a rise. Today, several studies have confirmed the regulation of multiple immunological responses by these pathogens, such as the nature of the cell receptor recognition, signaling, inflammation and Ag presentation pathways during different stages of infection and disease. The recent researches have bridged the gap between these known mechanisms exploited by mycobacteria and their genetic regulation, by highlighting the role of miRs in host-pathogen interactions and transcriptional regulation of target genes. However, the gap between the cause and cure still remains wide. Therefore, further work should be extrapolated in the direction to provide critical information for the development of novel preventive and therapeutic cure by utilising miRs as biomarkers for these diseases. The intervention of nanotechnology in medicine and exploitation of nanoparticle-mediated delivery of miRs in host-directed therapies have been proposed for various hallmark diseases. Similarly, detection of target regulatory mRNAs and miRs in mycobacterial diseases can allow exogenous manipulation of miRs and offer an opportunity to restrain the multiplication or enhance the clearance of mycobacteria from the host cells. Furthermore, the findings done so far have been based upon discerning the effect of observed/reported altered expression of miR(s), in response to M. tuberculosis infection as compared to normal cells/tissue. However, research on deciphering discrete host macrophage immunoregulatory pathways altered by a set of miRs affecting the same pathway is yet to be done. Therefore, apart from individual studies on knockdown and overexpression of genes that alter miR profiles, researchers need to pool together different findings into one, to understand the stepwise in-vivo alteration and the extent of manipulation of miRs and gene response done by M. tuberculosis. This also opens new avenues for research on miR mediated mechanisms, through which mycobacteria interacts, grows, persists and reactivates within the host cells.
ACKNOWLEDGEMENTS
This work was supported by the Ramalingaswami Re-entry Fellowship awarded to K.K.N. The authors acknowledge the Department of Biotechnology, New Delhi, India for providing this fellowship.
Abbreviations
- AM
alveolar macrophages
- Apaf-1
apoptotic protease activating factor 1
- Arp
actin-related proteins
- BAD
bcl-2-associated death promoter
- Bag
bcl-2 associated anthogene
- Bak
bcl2-homologous antagonist killer
- Bax
bcl-2-asscoiated x protein
- BCG
bacillus Calmette–Guérin
- Bcl-2
B-cell lymphoma 2
- Bcl-xl
B-cell lymphoma-extra large
- BID
BH3 interacting-domain death agonist
- CAMP
cathelicidin antimicrobial peptide
- caspase
cysteine aspartases
- CEBPB
CCAAT/enhancer binding protein β
- CtsS
cathepsin S
- cyt C
cytochrome C
- DC
dendritic cell
- DEFB4
defensin beta 4
- DGCR8
DiGeorge Syndrome Critical Region 8
- ESAT-6
early secreted antigenic target 6
- FADD
Fas-associated protein with death domain
- FasL
Fas ligand
- FasR
Fas receptor
- FcγR
Fcγ receptor
- FOXO
forkhead box protein O
- IAP
inhibitor of apoptosis
- IKK
IκB kinase
- IL-1Ra
IL-1 receptor antagonist
- IL-18BP
IL-18 binding protein
- iNOS
inducible nitric oxide synthase
- IRAK
IL-1R-associated kinase
- MDT
multidrug therapy
- miR
microRNA
- MyD88
myeloid differentiation primary response 88
- NO
nitric oxide
- NTM
non-tuberculous mycobacterial
- PTGS2
prostaglandin synthetase 2
- RIP1
receptor interacting protein kinase-1
- RISC
RNA induced silencing complex
- Smac
second mitochondria-derived activator of caspases
- SOCS
suppressors of cytokine signalling
- TAK
TGF-β-activated kinase
- TB
tuberculosis
- TNFR
TNF receptor
- TRADD
TNF receptor type 1-associated death domain
- TRAF
TNF-receptor-associated factor
- UTR
untranslated region
- WASP
Wiskott Aldrich syndrome
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
- Conceptualization: Nyati KK.
- Data curation: Nyati KK.
- Formal analysis: Sharma P.
- Funding acquisition: Nyati KK.
- Investigation: Agarwal RG.
- Methodology: Agarwal RG.
- Project administration: Nyati KK.
- Resources: Sharma P.
- Supervision: Nyati KK.
- Validation: Nyati KK.
- Visualization: Agarwal RG.
- Writing - original draft: Agarwal RG.
- Writing - review & editing: Sharma P, Nyati KK.
References
- 1.Gutierrez MC, Brisse S, Brosch R, Fabre M, Omaïs B, Marmiesse M, Supply P, Vincent V. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis . PLoS Pathog. 2005;1:e5. doi: 10.1371/journal.ppat.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization (WHO) Global tuberculosis report 2018 [Internet] [accessed on 1 April 2019]. Available at https://www.who.int/tb/publications/global_report/en/
- 3.Fischer M. Leprosy - an overview of clinical features, diagnosis, and treatment. J Dtsch Dermatol Ges. 2017;15:801–827. doi: 10.1111/ddg.13301. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization (WHO) WHO fact sheets: leprosy [Internet] [accessed on 12 August 2019]. Available at https://www.who.int/news-room/fact-sheets/detail/leprosy.
- 5.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 6.Parent LJ, Salam MM, Appelbaum PC, Dossett JH. Disseminated Mycobacterium marinum infection and bacteremia in a child with severe combined immunodeficiency. Clin Infect Dis. 1995;21:1325–1327. doi: 10.1093/clinids/21.5.1325. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe Y, Kanai A. Systems biology reveals microRNA-mediated gene regulation. Front Genet. 2011;2:29. doi: 10.3389/fgene.2011.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabir N, Hussain T, Shah SZ, Peramo A, Zhao D, Zhou X. miRNAs in Tuberculosis: New avenues for diagnosis and host-directed therapy. Front Microbiol. 2018;9:602. doi: 10.3389/fmicb.2018.00602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin Y, Wang Q, Zhou Y, Duan Y, Gao Q. Inhibition of IFN-γ-induced nitric oxide dependent antimycobacterial activity by miR-155 and C/EBPβ. Int J Mol Sci. 2016;17:535. doi: 10.3390/ijms17040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharbati J, Lewin A, Kutz-Lohroff B, Kamal E, Einspanier R, Sharbati S. Integrated microRNA-mRNA-analysis of human monocyte derived macrophages upon Mycobacterium avium subsp. hominissuis infection. PLoS One. 2011;6:e20258. doi: 10.1371/journal.pone.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guirado E, Schlesinger LS, Kaplan G. Macrophages in tuberculosis: friend or foe. Semin Immunopathol. 2013;35:563–583. doi: 10.1007/s00281-013-0388-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russell DG, Cardona PJ, Kim MJ, Allain S, Altare F. Foamy macrophages and the progression of the human tuberculosis granuloma. Nat Immunol. 2009;10:943–948. doi: 10.1038/ni.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mack U, Migliori GB, Sester M, Rieder HL, Ehlers S, Goletti D, Bossink A, Magdorf K, Hölscher C, Kampmann B, et al. LTBI: latent tuberculosis infection or lasting immune responses to M. tuberculosis? A TBNET consensus statement. Eur Respir J. 2009;33:956–973. doi: 10.1183/09031936.00120908. [DOI] [PubMed] [Google Scholar]
- 14.Lee JY. Diagnosis and treatment of extrapulmonary tuberculosis. Tuberc Respir Dis (Seoul) 2015;78:47–55. doi: 10.4046/trd.2015.78.2.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flynn JL, Goldstein MM, Chan J, Triebold KJ, Pfeffer K, Lowenstein CJ, Schreiber R, Mak TW, Bloom BR. Tumor necrosis factor-alpha is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity. 1995;2:561–572. doi: 10.1016/1074-7613(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 16.Roach DR, Bean AG, Demangel C, France MP, Briscoe H, Britton WJ. TNF regulates chemokine induction essential for cell recruitment, granuloma formation, and clearance of mycobacterial infection. J Immunol. 2002;168:4620–4627. doi: 10.4049/jimmunol.168.9.4620. [DOI] [PubMed] [Google Scholar]
- 17.Geluk A. Correlates of immune exacerbations in leprosy. Semin Immunol. 2018;39:111–118. doi: 10.1016/j.smim.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Chavarro-Portillo B, Soto CY, Guerrero MI. Mycobacterium leprae's evolution and environmental adaptation. Acta Trop. 2019;197:105041. doi: 10.1016/j.actatropica.2019.105041. [DOI] [PubMed] [Google Scholar]
- 19.Jin SH, An SK, Lee SB. The formation of lipid droplets favors intracellular Mycobacterium leprae survival in SW-10, non-myelinating Schwann cells. PLoS Negl Trop Dis. 2017;11:e0005687. doi: 10.1371/journal.pntd.0005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inkeles MS, Teles RM, Pouldar D, Andrade PR, Madigan CA, Lopez D, Ambrose M, Noursadeghi M, Sarno EN, Rea TH, et al. Cell-type deconvolution with immune pathways identifies gene networks of host defense and immunopathology in leprosy. JCI Insight. 2016;1:e88843. doi: 10.1172/jci.insight.88843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyrio EC, Campos-Souza IC, Corrêa LC, Lechuga GC, Verícimo M, Castro HC, Bourguignon SC, Côrte-Real S, Ratcliffe N, Declercq W, et al. Interaction of Mycobacterium leprae with the HaCaT human keratinocyte cell line: new frontiers in the cellular immunology of leprosy. Exp Dermatol. 2015;24:536–542. doi: 10.1111/exd.12714. [DOI] [PubMed] [Google Scholar]
- 22.Falkinham JO., 3rd Surrounded by mycobacteria: nontuberculous mycobacteria in the human environment. J Appl Microbiol. 2009;107:356–367. doi: 10.1111/j.1365-2672.2009.04161.x. [DOI] [PubMed] [Google Scholar]
- 23.Falkinham JO. Impact of human activities on the ecology of nontuberculous mycobacteria. Future Microbiol. 2010;5:951–960. doi: 10.2217/fmb.10.53. [DOI] [PubMed] [Google Scholar]
- 24.Williams MM, Yakrus MA, Arduino MJ, Cooksey RC, Crane CB, Banerjee SN, Hilborn ED, Donlan RM. Structural analysis of biofilm formation by rapidly and slowly growing nontuberculous mycobacteria. Appl Environ Microbiol. 2009;75:2091–2098. doi: 10.1128/AEM.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orme IM, Ordway DJ. Host response to nontuberculous mycobacterial infections of current clinical importance. Infect Immun. 2014;82:3516–3522. doi: 10.1128/IAI.01606-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bermudez LE, Young LS. Tumor necrosis factor, alone or in combination with IL-2, but not IFN-gamma, is associated with macrophage killing of Mycobacterium avium complex. J Immunol. 1988;140:3006–3013. [PubMed] [Google Scholar]
- 27.Tezera LB, Bielecka MK, Chancellor A, Reichmann MT, Shammari BA, Brace P, Batty A, Tocheva A, Jogai S, Marshall BG, et al. Dissection of the host-pathogen interaction in human tuberculosis using a bioengineered 3-dimensional model. eLife. 2017;6:e21283. doi: 10.7554/eLife.21283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 29.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 30.O'Brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326:1275–1279. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- 32.Song MS, Rossi JJ. Molecular mechanisms of Dicer: endonuclease and enzymatic activity. Biochem J. 2017;474:1603–1618. doi: 10.1042/BCJ20160759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 34.Williams AE. Functional aspects of animal microRNAs. Cell Mol Life Sci. 2008;65:545–562. doi: 10.1007/s00018-007-7355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bettencourt P, Pires D, Anes E. Immunomodulating microRNAs of mycobacterial infections. Tuberculosis (Edinb) 2016;97:1–7. doi: 10.1016/j.tube.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 36.Singh PK, Singh AV, Chauhan DS. Current understanding on micro RNAs and its regulation in response to Mycobacterial infections. J Biomed Sci. 2013;20:14. doi: 10.1186/1423-0127-20-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krysko DV, Vanden Berghe T, D'Herde K, Vandenabeele P. Apoptosis and necrosis: detection, discrimination and phagocytosis. Methods. 2008;44:205–221. doi: 10.1016/j.ymeth.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 38.Fratazzi C, Arbeit RD, Carini C, Balcewicz-Sablinska MK, Keane J, Kornfeld H, Remold HG. Macrophage apoptosis in mycobacterial infections. J Leukoc Biol. 1999;66:763–764. doi: 10.1002/jlb.66.5.763. [DOI] [PubMed] [Google Scholar]
- 39.Leppo J. Imaging cell injury and death. Curr Cardiol Rep. 2003;5:40–44. doi: 10.1007/s11886-003-0036-x. [DOI] [PubMed] [Google Scholar]
- 40.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Behar SM, Martin CJ, Booty MG, Nishimura T, Zhao X, Gan HX, Divangahi M, Remold HG. Apoptosis is an innate defense function of macrophages against Mycobacterium tuberculosis . Mucosal Immunol. 2011;4:279–287. doi: 10.1038/mi.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stutz F, Neville M, Rosbash M. Identification of a novel nuclear pore-associated protein as a functional target of the HIV-1 Rev protein in yeast. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- 43.Wong WW, Gentle IE, Nachbur U, Anderton H, Vaux DL, Silke J. RIPK1 is not essential for TNFR1-induced activation of NF-κB. Cell Death Differ. 2010;17:482–487. doi: 10.1038/cdd.2009.178. [DOI] [PubMed] [Google Scholar]
- 44.Park YC, Ye H, Hsia C, Segal D, Rich RL, Liou HC, Myszka DG, Wu H. A novel mechanism of TRAF signaling revealed by structural and functional analyses of the TRADD-TRAF2 interaction. Cell. 2000;101:777–787. doi: 10.1016/s0092-8674(00)80889-2. [DOI] [PubMed] [Google Scholar]
- 45.Vucic D, Dixit VM, Wertz IE. Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol. 2011;12:439–452. doi: 10.1038/nrm3143. [DOI] [PubMed] [Google Scholar]
- 46.Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J, Gillard JW, Jaquith JB, Morris SJ, Barker PA. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 47.Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 48.de Almagro MC, Vucic D. The inhibitor of apoptosis (IAP) proteins are critical regulators of signaling pathways and targets for anti-cancer therapy. Exp Oncol. 2012;34:200–211. [PubMed] [Google Scholar]
- 49.Waring P, Müllbacher A. Cell death induced by the Fas/Fas ligand pathway and its role in pathology. Immunol Cell Biol. 1999;77:312–317. doi: 10.1046/j.1440-1711.1999.00837.x. [DOI] [PubMed] [Google Scholar]
- 50.Adrain C, Creagh EM, Martin SJ. Apoptosis-associated release of Smac/DIABLO from mitochondria requires active caspases and is blocked by Bcl-2. EMBO J. 2001;20:6627–6636. doi: 10.1093/emboj/20.23.6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chinnaiyan AM. The apoptosome: heart and soul of the cell death machine. Neoplasia. 1999;1:5–15. doi: 10.1038/sj.neo.7900003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hill MM, Adrain C, Duriez PJ, Creagh EM, Martin SJ. Analysis of the composition, assembly kinetics and activity of native Apaf-1 apoptosomes. EMBO J. 2004;23:2134–2145. doi: 10.1038/sj.emboj.7600210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chauhan D, Li G, Hideshima T, Podar K, Mitsiades C, Mitsiades N, Munshi N, Kharbanda S, Anderson KC. JNK-dependent release of mitochondrial protein, Smac, during apoptosis in multiple myeloma (MM) cells. J Biol Chem. 2003;278:17593–17596. doi: 10.1074/jbc.C300076200. [DOI] [PubMed] [Google Scholar]
- 54.Dhanasekaran DN, Reddy EP. JNK signaling in apoptosis. Oncogene. 2008;27:6245–6251. doi: 10.1038/onc.2008.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deng Y, Ren X, Yang L, Lin Y, Wu X. A JNK-dependent pathway is required for TNFα-induced apoptosis. Cell. 2003;115:61–70. doi: 10.1016/s0092-8674(03)00757-8. [DOI] [PubMed] [Google Scholar]
- 56.Farhan M, Wang H, Gaur U, Little PJ, Xu J, Zheng W. FOXO signaling pathways as therapeutic targets in cancer. Int J Biol Sci. 2017;13:815–827. doi: 10.7150/ijbs.20052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hagenbuchner J, Ausserlechner MJ. Mitochondria and FOXO3: breath or die. Front Physiol. 2013;4:147. doi: 10.3389/fphys.2013.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh Y, Kaul V, Mehra A, Chatterjee S, Tousif S, Dwivedi VP, Suar M, Van Kaer L, Bishai WR, Das G. Mycobacterium tuberculosis controls microRNA-99b (miR-99b) expression in infected murine dendritic cells to modulate host immunity. J Biol Chem. 2013;288:5056–5061. doi: 10.1074/jbc.C112.439778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rajaram MV, Ni B, Morris JD, Brooks MN, Carlson TK, Bakthavachalu B, Schoenberg DR, Torrelles JB, Schlesinger LS. Mycobacterium tuberculosis lipomannan blocks TNF biosynthesis by regulating macrophage MAPK-activated protein kinase 2 (MK2) and microRNA miR-125b. Proc Natl Acad Sci U S A. 2011;108:17408–17413. doi: 10.1073/pnas.1112660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tripathi A, Srivastava V, Singh BN. hsa-let-7b-5p facilitates Mycobacterium tuberculosis survival in THP-1 human macrophages by Fas downregulation. FEMS Microbiol Lett. 2018;365:365. doi: 10.1093/femsle/fny040. [DOI] [PubMed] [Google Scholar]
- 61.Zhang G, Liu X, Wang W, Cai Y, Li S, Chen Q, Liao M, Zhang M, Zeng G, Zhou B, et al. Down-regulation of miR-20a-5p triggers cell apoptosis to facilitate mycobacterial clearance through targeting JNK2 in human macrophages. Cell Cycle. 2016;15:2527–2538. doi: 10.1080/15384101.2016.1215386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liang S, Song Z, Wu Y, Gao Y, Gao M, Liu F, Wang F, Zhang Y. MicroRNA-27b modulates inflammatory response and apoptosis during Mycobacterium tuberculosis infection. J Immunol. 2018;200:3506–3518. doi: 10.4049/jimmunol.1701448. [DOI] [PubMed] [Google Scholar]
- 63.Wang Q, Liu S, Tang Y, Liu Q, Yao Y. MPT64 protein from Mycobacterium tuberculosis inhibits apoptosis of macrophages through NF-kB-miRNA21-Bcl-2 pathway. PLoS One. 2014;9:e100949. doi: 10.1371/journal.pone.0100949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Y, Jiang J, Wang X, Zhai F, Cheng X. miR-582-5p is upregulated in patients with active tuberculosis and inhibits apoptosis of monocytes by targeting FOXO1. PLoS One. 2013;8:e78381. doi: 10.1371/journal.pone.0078381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xi X, Zhang C, Han W, Zhao H, Zhang H, Jiao J. MicroRNA-223 is upregulated in active tuberculosis patients and inhibits apoptosis of macrophages by targeting FOXO3. Genet Test Mol Biomarkers. 2015;19:650–656. doi: 10.1089/gtmb.2015.0090. [DOI] [PubMed] [Google Scholar]
- 66.Zhang C, Xi X, Wang Q, Jiao J, Zhang L, Zhao H, Lai Z. The association between serum miR-155 and natural killer cells from tuberculosis patients. Int J Clin Exp Med. 2015;8:9168–9172. [PMC free article] [PubMed] [Google Scholar]
- 67.Huang J, Jiao J, Xu W, Zhao H, Zhang C, Shi Y, Xiao Z. MiR-155 is upregulated in patients with active tuberculosis and inhibits apoptosis of monocytes by targeting FOXO3. Mol Med Rep. 2015;12:7102–7108. doi: 10.3892/mmr.2015.4250. [DOI] [PubMed] [Google Scholar]
- 68.Kumar R, Halder P, Sahu SK, Kumar M, Kumari M, Jana K, Ghosh Z, Sharma P, Kundu M, Basu J. Identification of a novel role of ESAT-6-dependent miR-155 induction during infection of macrophages with Mycobacterium tuberculosis. Cell Microbiol. 2012;14:1620–1631. doi: 10.1111/j.1462-5822.2012.01827.x. [DOI] [PubMed] [Google Scholar]
- 69.Tsai MC, Chakravarty S, Zhu G, Xu J, Tanaka K, Koch C, Tufariello J, Flynn J, Chan J. Characterization of the tuberculous granuloma in murine and human lungs: cellular composition and relative tissue oxygen tension. Cell Microbiol. 2006;8:218–232. doi: 10.1111/j.1462-5822.2005.00612.x. [DOI] [PubMed] [Google Scholar]
- 70.Weiss G, Schaible UE. Macrophage defense mechanisms against intracellular bacteria. Immunol Rev. 2015;264:182–203. doi: 10.1111/imr.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Botelho RJ, Grinstein S. Phagocytosis. Curr Biol. 2011;21:R533–R538. doi: 10.1016/j.cub.2011.05.053. [DOI] [PubMed] [Google Scholar]
- 72.Hoppe AD, Swanson JA. Cdc42, Rac1, and Rac2 display distinct patterns of activation during phagocytosis. Mol Biol Cell. 2004;15:3509–3519. doi: 10.1091/mbc.E03-11-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jongstra-Bilen J, Harrison R, Grinstein S. Fcγ-receptors induce Mac-1 (CD11b/CD18) mobilization and accumulation in the phagocytic cup for optimal phagocytosis. J Biol Chem. 2003;278:45720–45729. doi: 10.1074/jbc.M303704200. [DOI] [PubMed] [Google Scholar]
- 74.Miki H, Miura K, Takenawa T. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO J. 1996;15:5326–5335. [PMC free article] [PubMed] [Google Scholar]
- 75.Miki H, Takenawa T. Direct binding of the verprolin-homology domain in N-WASP to actin is essential for cytoskeletal reorganization. Biochem Biophys Res Commun. 1998;243:73–78. doi: 10.1006/bbrc.1997.8064. [DOI] [PubMed] [Google Scholar]
- 76.Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 77.Ma L, Rohatgi R, Kirschner MW. The Arp2/3 complex mediates actin polymerization induced by the small GTP-binding protein Cdc42. Proc Natl Acad Sci U S A. 1998;95:15362–15367. doi: 10.1073/pnas.95.26.15362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Doherty TM, Arditi M. TB, or not TB: that is the question -- does TLR signaling hold the answer? J Clin Invest. 2004;114:1699–1703. doi: 10.1172/JCI23867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jo EK, Yang CS, Choi CH, Harding CV. Intracellular signalling cascades regulating innate immune responses to Mycobacteria: branching out from Toll-like receptors. Cell Microbiol. 2007;9:1087–1098. doi: 10.1111/j.1462-5822.2007.00914.x. [DOI] [PubMed] [Google Scholar]
- 80.Kleinnijenhuis J, Oosting M, Joosten LA, Netea MG, Van Crevel R. Innate immune recognition of Mycobacterium tuberculosis . Clin Dev Immunol. 2011;2011:405310. doi: 10.1155/2011/405310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Basu J, Shin DM, Jo EK. Mycobacterial signaling through Toll-like receptors. Front Cell Infect Microbiol. 2012;2:145. doi: 10.3389/fcimb.2012.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Deguine J, Barton GM. MyD88: a central player in innate immune signaling. F1000Prime Rep. 2014;6:97. doi: 10.12703/P6-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang J, Clark K, Lawrence T, Peggie MW, Cohen P. An unexpected twist to the activation of IKKβ: TAK1 primes IKKβ for activation by autophosphorylation. Biochem J. 2014;461:531–537. doi: 10.1042/BJ20140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Clark K. Protein kinase networks that limit TLR signalling. Biochem Soc Trans. 2014;42:11–24. doi: 10.1042/BST20130124. [DOI] [PubMed] [Google Scholar]
- 85.Harris J, Hope JC, Keane J. Tumor necrosis factor blockers influence macrophage responses to Mycobacterium tuberculosis . J Infect Dis. 2008;198:1842–1850. doi: 10.1086/593174. [DOI] [PubMed] [Google Scholar]
- 86.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Beg AA, Baltimore D. An essential role for NF-κB in preventing TNF-α-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 88.Van Antwerp DJ, Martin SJ, Kafri T, Green DR, Verma IM. Suppression of TNF-α-induced apoptosis by NF-κB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 89.Keane J, Balcewicz-Sablinska MK, Remold HG, Chupp GL, Meek BB, Fenton MJ, Kornfeld H. Infection by Mycobacterium tuberculosis promotes human alveolar macrophage apoptosis. Infect Immun. 1997;65:298–304. doi: 10.1128/iai.65.1.298-304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bettencourt P, Marion S, Pires D, Santos LF, Lastrucci C, Carmo N, Blake J, Benes V, Griffiths G, Neyrolles O, et al. Actin-binding protein regulation by microRNAs as a novel microbial strategy to modulate phagocytosis by host cells: the case of N-Wasp and miR-142-3p. Front Cell Infect Microbiol. 2013;3:19. doi: 10.3389/fcimb.2013.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu G, Zhang Z, Wei J, Zhang Y, Zhang Y, Guo L, Liu X. microR-142-3p down-regulates IRAK-1 in response to Mycobacterium bovis BCG infection in macrophages. Tuberculosis (Edinb) 2013;93:606–611. doi: 10.1016/j.tube.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 92.Wang J, Yang K, Zhou L, Minhaowu, Wu Y, Zhu M, Lai X, Chen T, Feng L, Li M, et al. MicroRNA-155 promotes autophagy to eliminate intracellular mycobacteria by targeting Rheb. PLoS Pathog. 2013;9:e1003697. doi: 10.1371/journal.ppat.1003697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Etna MP, Sinigaglia A, Grassi A, Giacomini E, Romagnoli A, Pardini M, Severa M, Cruciani M, Rizzo F, Anastasiadou E, et al. Mycobacterium tuberculosis-induced miR-155 subverts autophagy by targeting ATG3 in human dendritic cells. PLoS Pathog. 2018;14:e1006790. doi: 10.1371/journal.ppat.1006790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang J, Zhu X, Xiong X, Ge P, Liu H, Ren N, Khan FA, Zhou X, Zhang L, Yuan X, et al. Identification of potential urine proteins and microRNA biomarkers for the diagnosis of pulmonary tuberculosis patients. Emerg Microbes Infect. 2018;7:63. doi: 10.1038/s41426-018-0066-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Iwai H, Funatogawa K, Matsumura K, Kato-Miyazawa M, Kirikae F, Kiga K, Sasakawa C, Miyoshi-Akiyama T, Kirikae T. MicroRNA-155 knockout mice are susceptible to Mycobacterium tuberculosis infection. Tuberculosis (Edinb) 2015;95:246–250. doi: 10.1016/j.tube.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 96.Ma C, Li Y, Li M, Deng G, Wu X, Zeng J, Hao X, Wang X, Liu J, Cho WC, et al. microRNA-124 negatively regulates TLR signaling in alveolar macrophages in response to mycobacterial infection. Mol Immunol. 2014;62:150–158. doi: 10.1016/j.molimm.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 97.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nathan C. Inducible nitric oxide synthase in the tuberculous human lung. Am J Respir Crit Care Med. 2002;166:130–131. doi: 10.1164/rccm.2205016. [DOI] [PubMed] [Google Scholar]
- 99.Rangel Moreno J, Estrada García I, De La Luz García Hernández M, Aguilar Leon D, Marquez R, Hernández Pando R. The role of prostaglandin E2 in the immunopathogenesis of experimental pulmonary tuberculosis. Immunology. 2002;106:257–266. doi: 10.1046/j.1365-2567.2002.01403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu Z, Zhou G, Deng X, Yu Q, Hu Y, Sun H, Wang Z, Chen H, Jia C, Wang D. Analysis of miRNA expression profiling in human macrophages responding to Mycobacterium infection: induction of the immune regulator miR-146a. J Infect. 2014;68:553–561. doi: 10.1016/j.jinf.2013.12.017. [DOI] [PubMed] [Google Scholar]
- 101.Wei J, Huang X, Zhang Z, Jia W, Zhao Z, Zhang Y, Liu X, Xu G. MyD88 as a target of microRNA-203 in regulation of lipopolysaccharide or Bacille Calmette-Guerin induced inflammatory response of macrophage RAW264.7 cells. Mol Immunol. 2013;55:303–309. doi: 10.1016/j.molimm.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 102.Ghorpade DS, Holla S, Kaveri SV, Bayry J, Patil SA, Balaji KN. Sonic hedgehog-dependent induction of microRNA 31 and microRNA 150 regulates Mycobacterium bovis BCG-driven toll-like receptor 2 signaling. Mol Cell Biol. 2013;33:543–556. doi: 10.1128/MCB.01108-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xu G, Zhang Z, Xing Y, Wei J, Ge Z, Liu X, Zhang Y, Huang X. MicroRNA-149 negatively regulates TLR-triggered inflammatory response in macrophages by targeting MyD88. J Cell Biochem. 2014;115:919–927. doi: 10.1002/jcb.24734. [DOI] [PubMed] [Google Scholar]
- 104.von Bernuth H, Picard C, Jin Z, Pankla R, Xiao H, Ku CL, Chrabieh M, Mustapha IB, Ghandil P, Camcioglu Y, et al. Pyogenic bacterial infections in humans with MyD88 deficiency. Science. 2008;321:691–696. doi: 10.1126/science.1158298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wu Z, Lu H, Sheng J, Li L. Inductive microRNA-21 impairs anti-mycobacterial responses by targeting IL-12 and Bcl-2. FEBS Lett. 2012;586:2459–2467. doi: 10.1016/j.febslet.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 106.Kumar M, Sahu SK, Kumar R, Subuddhi A, Maji RK, Jana K, Gupta P, Raffetseder J, Lerm M, Ghosh Z, et al. MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-κB pathway. Cell Host Microbe. 2015;17:345–356. doi: 10.1016/j.chom.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 107.Liu Y, Wang R, Jiang J, Yang B, Cao Z, Cheng X. miR-223 is upregulated in monocytes from patients with tuberculosis and regulates function of monocyte-derived macrophages. Mol Immunol. 2015;67:475–481. doi: 10.1016/j.molimm.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 108.Dorhoi A, Iannaccone M, Farinacci M, Faé KC, Schreiber J, Moura-Alves P, Nouailles G, Mollenkopf HJ, Oberbeck-Müller D, Jörg S, et al. MicroRNA-223 controls susceptibility to tuberculosis by regulating lung neutrophil recruitment. J Clin Invest. 2013;123:4836–4848. doi: 10.1172/JCI67604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Keane J, Shurtleff B, Kornfeld H. TNF-dependent BALB/c murine macrophage apoptosis following Mycobacterium tuberculosis infection inhibits bacillary growth in an IFN-γ independent manner. Tuberculosis (Edinb) 2002;82:55–61. doi: 10.1054/tube.2002.0322. [DOI] [PubMed] [Google Scholar]
- 110.Larsen L, Röpke C. Suppressors of cytokine signalling: SOCS. APMIS. 2002;110:833–844. doi: 10.1034/j.1600-0463.2002.1101201.x. [DOI] [PubMed] [Google Scholar]
- 111.Mühl H, Pfeilschifter J. Anti-inflammatory properties of pro-inflammatory interferon-γ. Int Immunopharmacol. 2003;3:1247–1255. doi: 10.1016/S1567-5769(03)00131-0. [DOI] [PubMed] [Google Scholar]
- 112.Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, Bloom BR. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J Exp Med. 1993;178:2249–2254. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lu YJ, Shen N, Wang X. Genetic associations between miR-146a/499 polymorphisms and tuberculosis: a meta-analysis. Int J Clin Exp Med. 2016;9:6445–6452. [Google Scholar]
- 114.Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, Hua M, Li N, Yao H, Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat Immunol. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 115.Ni B, Rajaram MV, Lafuse WP, Landes MB, Schlesinger LS. Mycobacterium tuberculosis decreases human macrophage IFN-γ responsiveness through miR-132 and miR-26a. J Immunol. 2014;193:4537–4547. doi: 10.4049/jimmunol.1400124. [DOI] [PubMed] [Google Scholar]
- 116.Liu Y, Wang X, Jiang J, Cao Z, Yang B, Cheng X. Modulation of T cell cytokine production by miR-144* with elevated expression in patients with pulmonary tuberculosis. Mol Immunol. 2011;48:1084–1090. doi: 10.1016/j.molimm.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 117.Shi G, Mao G, Xie K, Wu D, Wang W. MiR-1178 regulates mycobacterial survival and inflammatory responses in Mycobacterium tuberculosis-infected macrophages partly via TLR4. J Cell Biochem. 2018;119:7449–7457. doi: 10.1002/jcb.27054. [DOI] [PubMed] [Google Scholar]
- 118.Liu PT, Wheelwright M, Teles R, Komisopoulou E, Edfeldt K, Ferguson B, Mehta MD, Vazirnia A, Rea TH, Sarno EN, et al. MicroRNA-21 targets the vitamin D-dependent antimicrobial pathway in leprosy. Nat Med. 2012;18:267–273. doi: 10.1038/nm.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Knapp GS, McDonough KA. Cyclic AMP signaling in mycobacteria. Microbiol Spectr. 2014;2 doi: 10.1128/microbiolspec.MGM2-0011-2013. [DOI] [PubMed] [Google Scholar]
- 120.Rivas-Santiago B, Schwander SK, Sarabia C, Diamond G, Klein-Patel ME, Hernandez-Pando R, Ellner JJ, Sada E. Human β-defensin 2 is expressed and associated with Mycobacterium tuberculosis during infection of human alveolar epithelial cells. Infect Immun. 2005;73:4505–4511. doi: 10.1128/IAI.73.8.4505-4511.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mangalam AK, Taneja V, David CS. HLA class II molecules influence susceptibility versus protection in inflammatory diseases by determining the cytokine profile. J Immunol. 2013;190:513–518. doi: 10.4049/jimmunol.1201891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Smart M, Behrens M, David L, Conway C, Taneja V. Immune response to immunodominant Mycobacterium tuberculosis antigen ESAT-6 derived peptide is HLA-haplotype dependent. Jacobs J Allergy Immunol. 2014;1:002. [PMC free article] [PubMed] [Google Scholar]
- 123.Costantino CM, Ploegh HL, Hafler DA. Cathepsin S regulates class II MHC processing in human CD4+ HLA-DR+ T cells. J Immunol. 2009;183:945–952. doi: 10.4049/jimmunol.0900921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Driessen C, Bryant RA, Lennon-Duménil AM, Villadangos JA, Bryant PW, Shi GP, Chapman HA, Ploegh HL. Cathepsin S controls the trafficking and maturation of MHC class II molecules in dendritic cells. J Cell Biol. 1999;147:775–790. doi: 10.1083/jcb.147.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pires D, Bernard EM, Pombo JP, Carmo N, Fialho C, Gutierrez MG, Bettencourt P, Anes E. Mycobacterium tuberculosis modulates miR-106b-5p to control cathepsin S expression resulting in higher pathogen survival and poor T-cell activation. Front Immunol. 2017;8:1819. doi: 10.3389/fimmu.2017.01819. [DOI] [PMC free article] [PubMed] [Google Scholar]