Abstract
DNA double-strand breaks (DSBs) are especially toxic DNA lesions that, if left unrepaired, can lead to wide-ranging genomic instability. Of the pathways available to repair DSBs, the most accurate is homologous recombination (HR), where a homologous sequence is used as a donor template to restore genetic information at the break site. While much of the biochemical aspects of HR repair have been characterized, how the repair machinery locates and discriminates between potential homologous donor templates throughout the genome remains elusive. We use Drosophila melanogaster to investigate whether there is a preference between intrachromosomal and interhomolog donor sequences in mitotically dividing cells. Our results demonstrate that, although interhomolog HR is possible and frequent if another donor template is not available, intrachromosomal donor templates are highly preferred. This is true even if the interhomolog donor template is less diverged than the intrachromosomal donor template. Thus, despite the stringent requirements for homology, the chromosomal location of the donor template plays a more significant role in donor template choice.
Keywords: DSB repair, homologous recombination, Drosophila melanogaster, interhomolog recombination, intrachromosomal recombination
Genomic integrity is crucial for proper maintenance and dissemination of genetic information. However, cells frequently encounter DNA lesions that challenge the integrity of the genome. Of these lesions, double-strand breaks (DSBs) are especially toxic. They arise from both endogenous (e.g., reactive metabolites, programmed DSBs in meiotic and immune cells) and exogenous sources (e.g., ultraviolet or ionizing radiation, chemotherapeutic drugs) (reviewed in So et al. 2017). If left unrepaired, DSBs can lead to loss of genetic information, chromosomal rearrangements and apoptosis (Khanna and Jackson 2001; Hakem 2008). Consequently, inability to repair DSBs has been implicated in numerous genetic pathologies, including many cancers (reviewed in Jackson and Bartek 2009; Negrini et al. 2010; Janssen and Medema 2013).
Organisms have evolved multiple mechanisms to repair DSBs. These repair mechanisms have been broadly categorized into two pathways: non-homologous end joining (NHEJ) and homologous recombination (HR). NHEJ is active throughout the cell cycle and has been suggested to play a major role in DSB repair in mammalian cells (reviewed in Burma et al. 2006). In this repair pathway, the two broken ends of a DSB are directly ligated. Depending on the nature of the DSB, end processing including insertions and deletions (indels) may be required, resulting in potentially mutagenic repair outcomes. By contrast, HR uses a homologous sequence as a donor template to restore sequences at the broken ends of a DSB. Homologous donor templates may be located elsewhere on the chromosome or, in late S/G2 phases of the cell cycle, on the sister chromatid (collectively referred to as intrachromosomal HR) or on the homologous chromosome (interhomolog HR) in diploid organisms. HR most commonly occurs in S/G2 phases of the cell cycle in eukaryotic cells and can accurately repair DSBs with minimal or no loss of genetic information (reviewed in Shrivastav et al. 2008).
The molecular machinery, genetic components, and events required for HR repair have been well characterized (reviewed in San Filippo et al. 2008; Mazón et al. 2010; Heyer et al. 2010; Jasin and Rothstein 2013; Mimitou and Symington 2011; Bell and Kowalczykowski 2016). HR is initiated by resection of the 5′ end of a DSB, producing a single-stranded DNA (ssDNA) overhang. The 3′ overhang invades a homologous sequence that is used as a donor template for DNA repair synthesis. In mitotic cells, HR proceeds via synthesis-dependent strand annealing (SDSA), and the newly synthesized DNA strand dissociates from its homologous donor template. This strand then anneals to the other broken end of the DSB, restoring the intervening sequence through gene conversion, where the sequence that received the DSB is converted to the donor template sequence. While the biochemistry of HR repair is well characterized, the mechanism by which the HR repair machinery is able to locate and discriminate between potential homologous sequences across the genome (“homology search”) remains unclear.
An important factor in the homology search is the degree of homology between the DSB end and the homologous donor template. Several studies in eukaryotic systems have demonstrated that recombination between diverged sequences is highly suppressed; as the degree of homology between substrates decreases, the frequency of HR also decreases. In yeast, one to three mismatches in a stretch of homologous sequence is enough to suppress HR repair (Datta et al. 1996; Chen and Jinks-Robertson 1999). Suppression of repair between diverged sequences is also observed in homology-dependent template switching during break-induced repair (Anand et al. 2014). In mouse embryonic stem cells, a modest 1.2% sequence divergence results in sixfold reduction in HR (Elliott et al. 1998), a pattern consistent in human cells, though less pronounced (LaRocque and Jasin 2010). Recombination between diverged sequences is also markedly reduced in Drosophila melanogaster, with as little as 0.4% divergence significantly suppressing HR in a mismatch repair-dependent manner (Nassif and Engels 1993; Do et al. 2014; Do and LaRocque 2015).
In addition to homology, the location of the donor template relative to the DSB is also important. Numerous studies have found a pronounced usage of sister chromatid donor templates in both yeast and mammalian cells (Kadyk and Hartwell 1992; Liang et al. 1998; Johnson and Jasin 2000), though evidence exists for recombination between sequences on homologous, heterologous, or ectopic chromosomes (Lichten and Haber 1989; Malkova et al. 1996; Donoho et al. 1998; Richardson et al. 1998). Furthermore, intrachromosomal HR repair frequency in yeast varies according to the position of the donor template relative to the DSB: as the genomic distance between the DSB site and the homologous sequence increases, the frequency of HR decreases (Lichten and Haber 1989; Lee et al. 2016). This pattern is consistent in Drosophila, where HR repair of P-element-induced DSBs occurs more frequently between proximal sequences located on the same chromosome as the DSB (Engels et al. 1994), despite the observation that homologs are able to effectively serve as donor templates for HR repair (Rong and Golic 2003; Brunner et al. 2019). Taken together, these studies suggest that proximity plays a role in the efficiency and prevalence of HR repair and that the sister chromatid, being more proximal, is utilized more often as a donor template.
However, these studies lack in the ability to simultaneously track repair events from several homologous donor templates located throughout the genome. If given a choice, does the HR repair machinery preferentially distinguish between intrachromosomal donor templates and those located elsewhere in the genome? In the present study, we investigated this in mitotically dividing tissues of Drosophila, a strong model to test proximity effects given that homologous chromosomes have been shown to pair throughout the cell cycle (Metz 1916). We show that in the absence of an intrachromosomal donor template, interhomolog recombination is possible and occurs as frequently as intrachromosomal recombination. Yet, provided a choice between intrachromosomal and interhomolog donor templates, intrachromosomal donor templates are highly preferred. In accordance with previous studies, the degree of homology between the DSB and donor templates significantly affects the choice of donor template. Our findings suggest a sophisticated repair mechanism, whereby the HR repair machinery can distinguish between several donor templates while also discriminating against diverged sequences, regardless of genomic location.
Methods
DNA manipulations and cloning
All DNA construct development followed manufacturer’s protocol, unless otherwise stated. The DR-whiteΔ9 construct (containing Sce.white, yellow transgene, iwhiteΔ9 donor template, and attB sequence) was created by a two-step cloning process. First, the iwhiteΔ9 donor template was created by identifying a spontaneous HR repair event from the original DR-white assay (Do et al. 2014) that resulted in orange-colored eyes. The repair event contained a 9-bp deletion at the original SacI site in the white cDNA, resulting in a 3-amino acid deletion and reduced w+ gene product (thus orange eyes). This repair event was used as a template for PCR amplification of iwhiteΔ9 fragment using CloneAmp HiFi PCR Pre-mix (Clontech) with primers 5′ GCTCCACCGCGGTGGCGGCCGCTTGGCCAAGAGGATCAGGAGCTA (forward, contains underlined NotI sequence) and 5′ CTTGATATCGAATTCCTGCAGTTGCAGATCGGCGGCGGAGAAGTTAA (reverse, contains underlined PstI sequence). The iwhiteΔ9 PCR fragment with flanking restriction sites was cloned into NotI/PstI–digested pBlueScript.KS−.attB vector (pBSKS–.attB; described previously in Do et al. 2014) using In-Fusion Cloning (Clontech) to create pBSKS−.iwhiteΔ9.attB. Next, a Sce.white and yellow+ fragment from the original DR-white construct (Do et al. 2014) was amplified with primers 5′ GAGCTCCACCGCGGTGGCGGCCGCCAAGTTTGTACAAAAAAGCAG (forward, contains underlined NotI sequence) and 5′ GCTCCTGATCCTCTTGGCCAAGCGGCCGCCAACTTTATTATACAAAGTTGTTT (reverse, contains underlined NotI sequence) using HiFi PCR Pre-mix. The Sce.white_y+ PCR fragment with flanking restriction sites was cloned into NotI–digested pBSKS−.iwhiteΔ9.attB vector using In-Fusion Cloning to create DR-whiteΔ9. Similarly, for the Sce.white DSB recipient construct (containing Sce.white, yellow transgene, and attB sequence), the Sce.white_y+ PCR fragment with flanking NotI restriction sites was cloned into NotI–digested pBSKS−.attB vector using In-Fusion Cloning.
For iwhite interhomolog donor construct (containing yellow transgene, iwhite donor template, and attB sequence), the y+ transgene was amplified from the original DR-white construct with primers 5′ GAGCTCCACCGCGGTGGCGGCCGCCAACTTTTCTATACAAAGTTG (forward, contains underlined NotI sequence) and 5′ GCTCCTGATCCTCTTGGCCAAGCGGCCGCCAACTTTATTATACAAAGTTGTTT (reverse, contains underlined NotI sequence) using HiFi PCR Pre-mix. The y+ PCR fragment with flanking NotI restriction sites was cloned into NotI-digested pBSKS-.iwhite.attB vector using In-Fusion Cloning to create iwhite interhomolog donor construct. The iwhiteΔ9 interhomolog donor construct (containing yellow transgene, iwhiteΔ9 donor template, and attB sequence) was similarly created by inserting the y+ PCR fragment with flanking NotI restriction sites into NotI–digested pBSKS−.iwhiteΔ9.attB vector using In-Fusion Cloning.
Drosophila stocks and genetics
Drosophila were maintained on standard NutriFly Bloomington Formulation medium (Genesee Scientific, San Diego, CA) at 25° with 12-hour light/dark cycles.
The whiteΔ allele was created using CRISPR/Cas9 tools. Two sequences homologous to the start (5′ GTGTGAAAAATCCCGGCAAT) and stop (5′ ACATATATCCGAAATAACTGCC) codon regions of the white gene were cloned adjacent to guide RNA (gRNA) scaffolds in the pCFD4 Drosophila expression vector as described in (Port et al. 2014) and CRISPR Fly Design (http://www.crisprflydesign.org). Briefly, a pCFD4 fragment including the two described sequences were amplified from pCFD4 (Addgene) with primers 5′ TATATAGGAAAGATATCCGGGTGAACTTCGTGTGAAAAATCCCGGCAATGTTTTAGAGCTAGAAATAGCAAG (forward) and 5′ ATTTTAACTTGCTATTTCTAGCTCTAAAACACATATATCCGAAATAACTGCCGACGTTAAATTGAAAATAGGTC (reverse) using HiFi PCR Pre-mix. The amplified PCR fragment was inserted into BbsI-linearized pCFD4 vector using In-Fusion Cloning. Positive clones, identified by restriction digest, were sequenced (5′ GACACAGCGCGTACGTCCTTCG) to confirm accurate incorporation of gRNA sequences. pCFD4 vector with incorporated gRNAs were injected into NIG-FLY CAS-0001 embryos (genotype y2 cho2 v1; attP40{nos-Cas9}/CyO) (BestGene). G0 males were isolated and crossed into balancer lines to remove the Cas9 transgene. G1 progeny were phenotypically screened for white eyes, crossed out to balancer lines, and their DNA extracted using the previously described DNA Preparation Protocol (Gloor et al. 1993). Briefly, genomic DNA was isolated using Squishing Buffer (50 µL; 10 mM Tris-Cl pH 8.2, 25 mM NaCl) and Proteinase K (10 µg), incubated at 37° for 30 min, followed by 95° inactivation for five minutes. PCR was performed using SapphireAmp Fast PCR Master Mix (Clontech) with primers 5′ GACAGCGAAAGAGCAACTACG (forward) and 5′ ACCAGGTTCTTTCGATTACCTC (reverse) flanking the white coding exons. Successful mutant lines were sequenced (5′ GACAGCGAAAGAGCAACTACG) to confirm deletion of the endogenous white sequence.
For recombination assays, purified DR-whiteΔ9, Sce.white, iwhite, and iwhiteΔ9 constructs were injected and integrated at the 51C1 locus of FlyC31 line M{3xP3-RFP’}ZH-51C using the PhiC3 integrase system (Bischof et al. 2007) (BestGene). Stable transformants were selected based on y+ expression and locus integration confirmed by PCR using primers 5′ CTGCAACTGCAGGAATTCG (forward) and 5′ GTCGTCCAGGCCTCGTTAAT (reverse). One line of each was selected based on fertility and health for downstream applications.
The heat-inducible I-SceI transgene is located on Chromosome 2 and linked to the dominant marker Sco (Sco–) (Rong and Golic 2003). For interhomolog recombination assays, this line was used to establish a recombinant line of I-SceI transgene and iwhite or iwhiteΔ9 constructs using standard genetic techniques. Briefly, crosses were set up for recombination to occur between the I-SceI transgene and the iwhite or iwhiteΔ9 constructs on Chromosome 2 within the female germline. Potential recombinant events in the next generation expressing y+ transgene and exhibiting Sco– phenotype were isolated. To confirm recombination, genomic DNA of potential recombinants was isolated using Squishing Buffer and PCR performed using SapphireAmp Fast PCR Master Mix with I-SceI-specific primers 5′ CCAGCTGATCGAACTGAACA (forward) and 5′ CGCAGACCCTTAACCAGGTA (reverse).
DSB repair assays
DSB repair assays were performed based on protocol described previously (Do et al. 2014). To induce DSBs, females containing the DSB recipient chromosome (either Sce.white, DR-white or DR-whiteΔ9) were crossed to males containing the heat-inducible I-SceI transgene (Rong and Golic 2003). For intrachromosomal repair assays, females carrying DR-white or DR-whiteΔ9 were crossed to males containing the I-SceI transgene alone. For interhomolog repair assay, females carrying the Sce.white construct were crossed to males containing I-SceI transgene and either iwhite or iwhiteΔ9. For intrachromosomal/interhomolog choice assay, females carrying DR-white were crossed to males containing I-SceI and iwhiteΔ9, while females carrying DR-whiteΔ9 were crossed to males containing I-SceI and iwhite. After three days, flies were removed and zero- to three-day old embryos and larvae were heat-shocked in a 38° water bath for one hour. Single F1 males containing both the DSB recipient chromosome (Sce.white, DR-white or DR-whiteΔ9) and the heat-inducible I-SceI transgene (along with the corresponding iwhiteΔ9 or iwhite sequence) were crossed to four–five y w tester females in vials. For each experiment/condition, vials containing ≥ 20 F2 progeny from 63-80 individual male germlines were scored. HR repair frequency was determined in F2 progeny containing Sce.white, DR-white, or DR-whiteΔ9.
Molecular analyses of DSB repair events using TIDE
Genomic DNA was extracted from whole fly samples using a genomic DNA prep protocol adapted from Sullivan, Ashburner, and Hawley (2000). Single, adult flies were homogenized in Buffer A (50 μL; 100 mM Tris-Cl pH 7.5, 100 mM EDTA, 100 mM sodium chloride, 0.5% SDS) and incubated at 65° for 30 min. Buffer B (100 μL; 1.4 M potassium acetate, 4.3 M lithium chloride) was added and the mixture incubated on ice for 30 min. Samples were centrifuged at 13,200 rpm for 15 min at 4°. Supernatant was transferred to tubes containing 100 μL of isopropanol and centrifuged at 13,200 rpm for 10 min at room temperature. The DNA pellet was washed with 250 μL of cold 70% ethanol, air-dried and resuspended in 20 μL H2O. PCR reactions were performed on 100 ng of purified genomic DNA using SapphireAmp Fast PCR Master Mix. The target I-SceI recognition site in Sce.white was amplified using primers 5′ GTGGATCAGGTAATCCAGG (forward) and 5′ CTTAAGCCATCGTCAGTTGC (reverse) under the following conditions: three minutes at 94°; 30 s at 94°, 30 s touchdown at 66° (-0.5°/cycle) and 30 s at 72° (16x); 30 s at 94°, 30 s at 58°, 30 s at 72° (20x); five minutes at 72° (1x); and held at 12°. PCR products were purified using the Wizard SV Gel and PCR Clean-Up System (Promega) and sequenced (5′ GAGCCCACCTCCGGACTGGAC). Sequences (Supplemental Files S2-26) were analyzed using the TIDE (Tracking across Indels by DEcomposition) algorithm, a computational protocol previously described (Brinkman et al. 2014) and customized for the DR-white assay (Janssen et al. 2016). The algorithm was further modified to include indels of up to 35 nucleotides surrounding the I-SceI DSB site in the Sce.white sequence (Supplemental File S1). HR products were identified as conversions of the I-SceI sequence to either the wild-type white+ sequence (a 23-bp deletion) or the modified iwhiteΔ9+ sequence (a 32-bp deletion). Other insertions and deletions of up to 35 nucleotides were categorized as NHEJ with processing products. An output of 0 nucleotide indels was classified as the control I-SceI recognition sequence where neither HR nor NHEJ with processing repair events occurred.
Data Availability
Strains and plasmids are available upon request. Supplemental Table S1, the R script algorithm (Supplemental File S1), and sequence files (Supplemental Files S2-S26) used for TIDE analyses are available on FigShare. https://doi.org/10.25387/g3.9271007.
Results
Homologous recombination is suppressed in DR-whiteΔ9
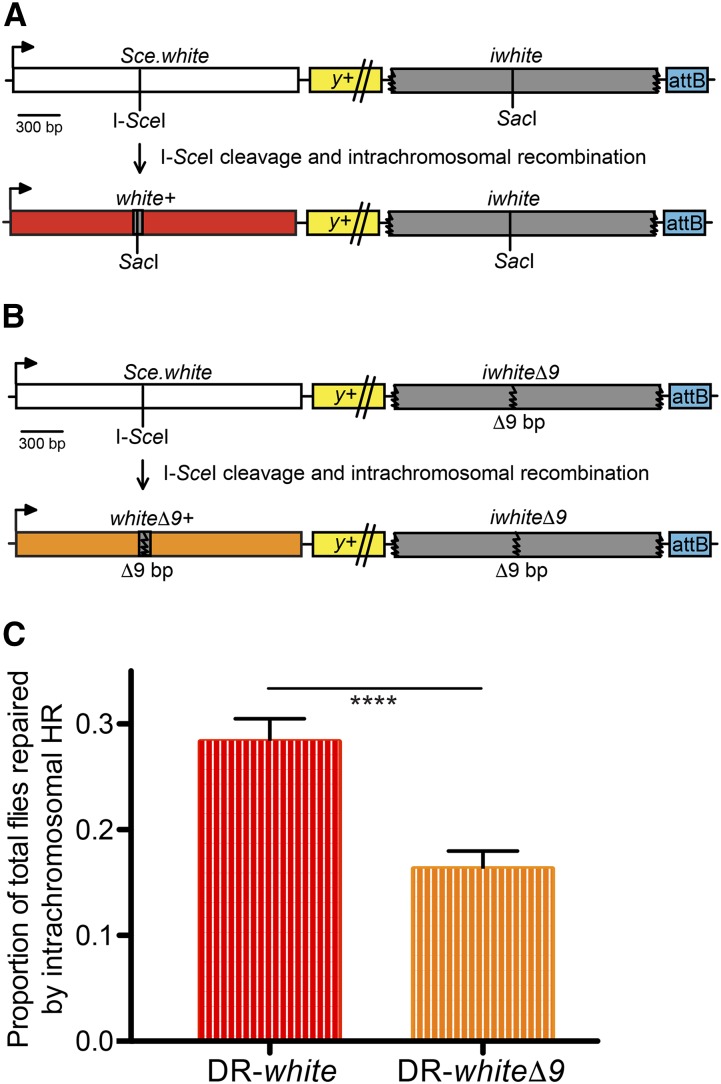
The previously characterized DR-white assay effectively measures intrachromosomal HR repair frequency in the Drosophila male premeiotic germline (Figure 1A)(Do et al. 2014; Do and LaRocque 2015; Janssen et al. 2016; Delabaere et al. 2017; Ertl et al. 2017). Briefly, flies containing the DR-white reporter assay and the I-SceI transgene are heat shocked to induce I-SceI expression in all cells, resulting in DSB formation followed by repair. Premeiotic germline events are isolated by crossing individual males to tester females and scoring F2 progeny for eye color (red eyes indicate gene conversion to wild-type white sequence).
Figure 1.
Sequence divergence significantly affects the frequency of homologous recombination repair. (A) The DR-white assay measures the frequency of intrachromosomal HR repair. An I-SceI recognition sequence is inserted into the wild-type SacI recognition site of white cDNA, resulting in a defective white sequence (Sce.white; white box). The downstream white sequence is defective because of 5′ and 3′ truncations (iwhite; gray box). Integration of DR-white is targeted using the attB sequence and followed with the yellow (y+) transgene (yellow box; not to scale). HR repair of an I-SceI-induced DSB results in gene conversion of the Sce.white sequence to white+ (red box), resulting in red-eyed progeny. (B) The DR-whiteΔ9 assay replaces the downstream iwhite sequence with iwhiteΔ9, which contains a 9-bp deletion including the wild-type SacI site in addition to 5′ and 3′ truncations (gray box). HR repair results in gene conversion of the Sce.white sequence to whiteΔ9+ (orange box), resulting in orange-eyed progeny. (C) F2 progeny for 38 individual male germlines of DR-white and 40 of DR-whiteΔ9 were scored for eye color. The average intrachromosomal HR frequency out of total flies scored are shown. Error bars are S.E.M.; ****P < 10−4 (two-tailed unpaired Student’s t-test).
Discriminating between interhomolog and intrachromosomal HR requires a distinguishing allele on the homolog that can reliably detect a choice between intrachromosomal and interhomolog repair. A novel iwhiteΔ9 sequence, containing a 9-bp deletion at the wild-type SacI site, was identified that can serve as an HR donor template and results in a distinguishing phenotype of orange eyes. However, the sequence divergence between the DSB recipient and the new iwhiteΔ9 donor template could potentially confound measures of interhomolog HR repair frequency. Thus, two modifications were made to the existing DR-white assay to establish if homology differences would affect HR repair frequency in our assay.
First, to ensure that the endogenous white gene on Chromosome X could not serve as a donor template for repair and distort measures of HR frequency, the white coding region was deleted via CRISPR-Cas9 methods. The whiteΔ allele contains a complete deletion of the white coding exons and intervening introns and a 7-bp insertion at the end-joining repair junction (5′ CTTGTTA). The novel whiteΔ mutant allows experimental control over the donor templates available for repair. Thus, all genetic crosses described herein were carried out in a whiteΔ mutant background.
Second, the DR-white assay was modified to create the DR-whiteΔ9 assay with similar features: Sce.white, yellow transgene, and an attB targeting sequence (Figure 1B)(Do et al. 2014). Unique to previous studies, DR-whiteΔ9 exchanges the downstream iwhite donor template for the iwhiteΔ9 donor template. In a whiteΔ mutant background, the downstream iwhiteΔ9 provides the only homologous donor template for repair. Repair of I-SceI-induced DSBs via intrachromosomal HR in DR-whiteΔ9 results in gene conversion of Sce.white to a whiteΔ9+ sequence producing orange eyes in the F2 generation (Figure 1B).
To determine whether the deletion in iwhiteΔ9 affects HR repair frequency, we compared the frequency of intrachromosomal HR in both DR-white and DR-whiteΔ9. Recombination in DR-whiteΔ9 decreased roughly 40% as compared to the original DR-white (16.3 ± 1.7% and 28.3 ± 2.2%, respectively; P < 10−4, two-tailed unpaired Student’s t-test; Figure 1C). While unsurprising, this finding revealed that the two repair donor templates, iwhite and iwhiteΔ9, are not equivalent in terms of repair frequency. Therefore, each construct was tested separately to mitigate any confounding effects of sequence divergence.
Interhomolog recombination is frequent in the male premeiotic germline
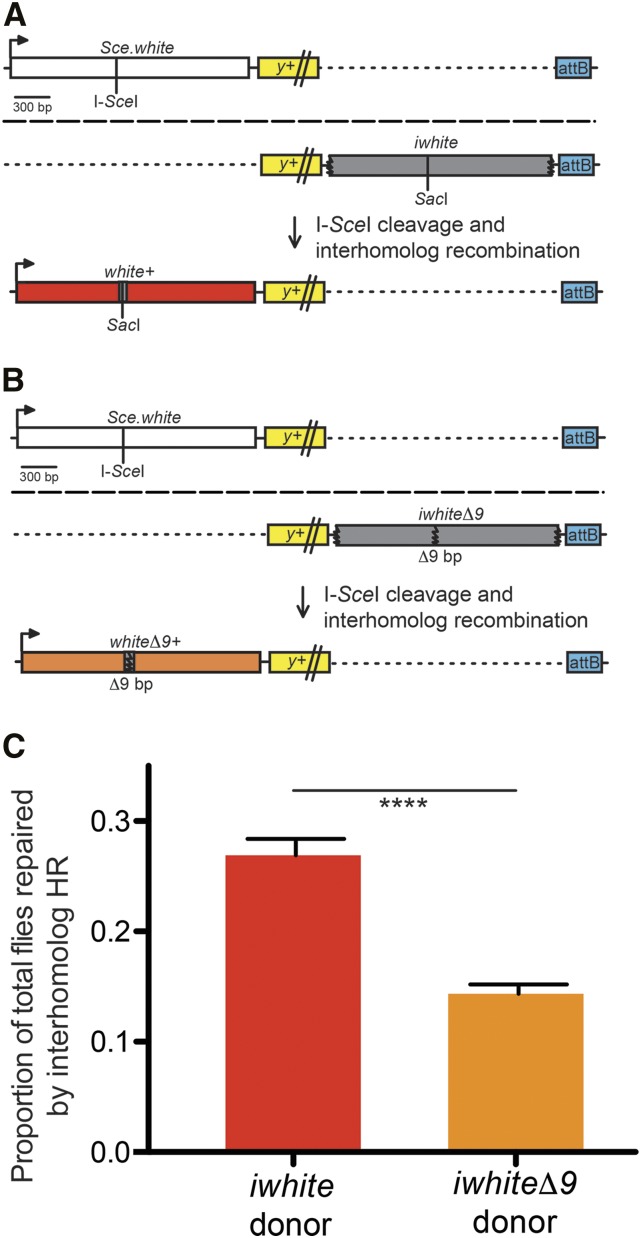
To confirm that interhomolog recombination is possible and detectable in our system, the interhomolog repair construct Sce.white, without a downstream intrachromosomal donor template, was utilized. Females carrying Sce.white were crossed with males carrying the iwhite or iwhiteΔ9 sequence on the allelic locus (Figures 2A and 2B). Gene conversion of the I-SceI-induced DSB to either white+ (red eyes) or whiteΔ9+ (orange eyes) indicates interhomolog HR repair, as the only viable donor template was located on the homolog.
Figure 2.
Interhomolog HR repair is possible and as frequent as intrachromosomal HR repair. (A) The interhomolog repair assay targets the Sce.white construct (white box) to Chromosome 2 and an iwhite donor template on the same allelic locus (gray box; homologs are separated by long dashed line). HR repair of an I-SceI induced DSB from the homolog results in gene conversion of the Sce.white sequence to white+ (red box), resulting in red-eyed progeny. Graphics are aligned with the y+ transgene and attB sequence in order to compare with other figures. Dotted lines are for alignment purposes only. (B) The same assay is performed using the iwhiteΔ9 donor template (gray box), instead of iwhite, on the allelic Chromosome 2. HR repair results in gene conversion of the Sce.white sequence to whiteΔ9+ (orange box), resulting in orange-eyed progeny. (C) F2 progeny for 65 individual male germlines with iwhite interhomolog donor template and 80 with iwhiteΔ9 interhomolog donor template were scored. The average interhomolog HR frequency out of total flies scored are shown. Error bars are S.E.M.; ****P < 10−10 (two-tailed unpaired Student’s t-test).
When using the iwhite donor template (Figure 2C), interhomolog recombination occurred at a frequency of 26.6 ± 1.5%, comparable to the 28.3% frequency of intrachromsomal HR repair in the DR-white assay (P > 0.05, two-tailed unpaired Student’s t-test). A homology-dependent effect on interhomolog recombination frequency was also observed. Interhomolog recombination decreased significantly by about 45% when using the iwhiteΔ9 donor template (14.1 ± 0.8%; Figure 2C; P < 10−10, two-tailed unpaired Student’s t-test), similar to the 40% decrease observed in the DR-whiteΔ9 assay (P > 0.05, two-tailed unpaired Student’s t-test). Thus, interhomolog recombination in Drosophila occurs as frequently as intrachromsomal HR repair if no other donor template is available, and the efficiency of interhomolog repair is significantly affected by sequence divergence.
Intrachromosomal donor templates are highly preferred in the premeiotic male germline
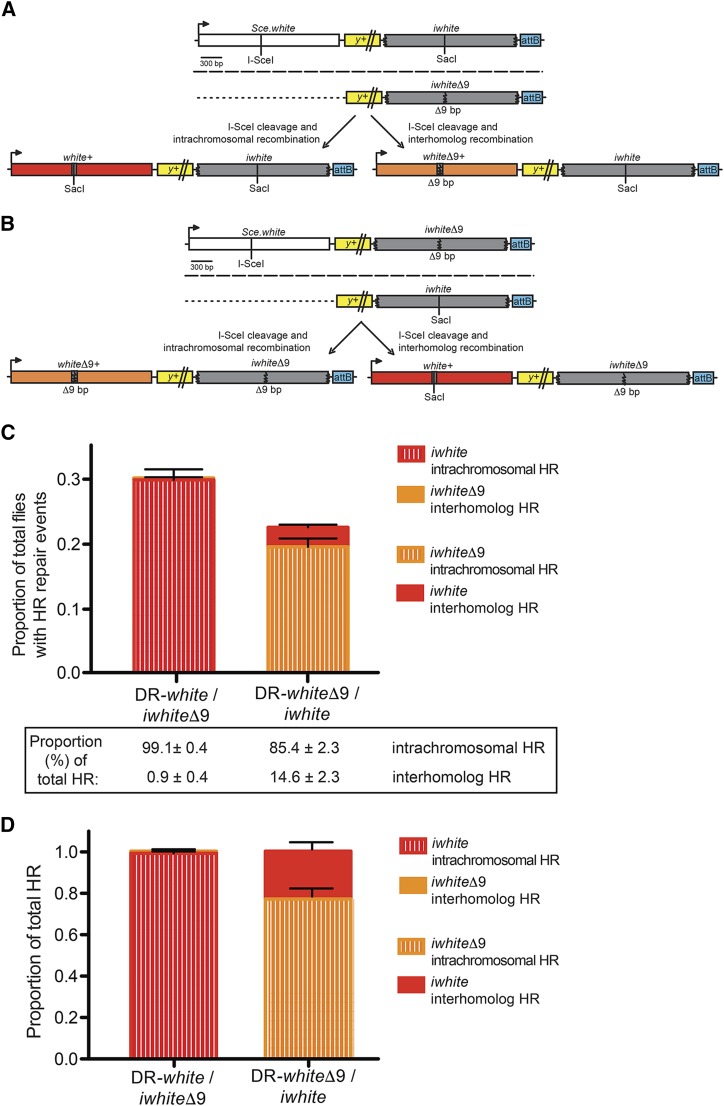
Having established that interhomolog HR repair is possible and frequent in Drosophila, we investigated the relative contribution of intrachromosomal and interhomolog HR repair when donor templates are simultaneously available on both homologous chromosomes. Females carrying either DR-white or DR-whiteΔ9 were crossed with males carrying the iwhiteΔ9 or iwhite interhomolog donor templates, respectively (Figure 3A and 3B).
Figure 3.
Intrachromosomal HR repair is preferred over interhomolog HR repair in mitotically dividing cells. (A) The intrachromosomal/interhomolog choice repair assay. Females carrying DR-white are crossed with males carrying the iwhiteΔ9 homolog donor template. HR repair of an I-SceI induced DSB from the intrachromosomal iwhite donor template results in gene conversion of the Sce.white sequence to white+ (red box), resulting in red-eyed progeny. HR repair from the interhomolog iwhiteΔ9 donor template results in gene conversion of the Sce.white sequence to whiteΔ9+ (orange box), resulting in orange-eyed progeny. Homologs are separated by long dashed line; dotted lines are used for direct comparison with other figures. (B) The same intrachromosomal/interhomolog choice repair assay is performed with flies carrying DR-whiteΔ9 and an iwhite homolog donor template. Intrachromosomal HR repair results in gene conversion of the Sce.white sequence to whiteΔ9+ (orange box), while interhomolog HR repair results in gene conversion to white+ (red box). (C) F2 progeny of 79 individual male germlines of DR-white/iwhiteΔ9 and 63 individual male germlines of DR-whiteΔ9/iwhite were scored. Average HR frequency out of total flies scored is shown. Error bars are S.E.M. Values given below are proportion of intrachromosomal or interhomolog HR out of total HR events ± SEM (D) Proportion of intrachromosomal (striped bars) or interhomolog (solid bars) HR out of total HR repair events in either DR-white/iwhiteΔ9 or DR-whiteΔ9/iwhite whole adult flies using TIDE analyses. Error bars are S.E.M.
When the iwhiteΔ9 sequence is used as the interhomolog donor template (Figure 3A), the frequency of interhomolog HR repair is only 0.3 ± 0.1% (Figure 3C). In contrast, when the iwhite sequence is used as the intrachromosomal donor template, the frequency of intrachromosomal HR repair is 29.4 ± 1.6%, accounting for the majority of all HR repair events (99.1 ± 0.4%, Figure 3C). Similarly, when iwhite is used as the interhomolog donor template (Figure 3B), the frequency of interhomolog HR repair is only 3.0 ± 0.4% (Figure 3C). When the more diverged iwhiteΔ9 sequence is used as the intrachromosomal donor template, the frequency of intrachromosomal HR repair is decreased (19.1 ± 1.3%) compared to intrachromosomal repair using the less diverged iwhite donor sequence. Although total HR frequency declined by about 25% (P < 0.001, two-tailed unpaired Student’s t-test), intrachromosomal recombination still occurred an impressive 85.4 ± 2.3% relative to total HR frequency (Figure 3C). Thus, despite the effects of sequence divergence on recombination frequency, the data strongly support a preference for intrachromosomal over interhomolog donor templates in the Drosophila male premeiotic germline.
Intrachromosomal donor templates are highly preferred in non-germline tissues
The significant preference for intrachromosomal HR repair in the male premeiotic germline prompted us to investigate the HR repair preferences in other Drosophila tissues, including mitotically dividing somatic cells. As described above, heat-shock-induced DSBs were generated at the I-SceI recognition site of Sce.white (Figure 3A and 3B). Cells in F1 larvae that contained repair events developed into adult tissues and were analyzed molecularly as a population of events. To quantify the proportion of intrachromosomal and interhomolog HR repair events across all tissues, DNA sequences from individual male and female F1 adults were analyzed with the established TIDE (Tracking across Indels by DEcomposition) algorithm (Janssen et al. 2016)(Supplemental File S1). The TIDE algorithm calculates the relative proportion of NHEJ with processing and HR repair events by comparing sequence deviations (Supplemental Files S2-S24) relative to the original I-SceI recognition sequence in Sce.white (Supplemental Files S25-S26).
Of the TIDE-detectable repair events, NHEJ with processing ranged from 15.2 to 93.4% and total HR events were lower on average (ranging from 6.6 to 84.8%; Supplemental Table S1). Within the HR events, with an iwhite intrachromosomal donor template, the proportion of intrachromosomal HR repair events relative to total HR frequency was 98.9 ± 0.9% (Figure 3D, Supplemental Table S1). Notably, the proportion of intrachromosomal HR repair events was similar to that previously observed in the male premeiotic germline (99.1 ± 0.4%, P > 0.05, two-tailed unpaired Student’s t-test). Expectedly, with an iwhiteΔ9 intrachromosomal donor template, the proportion of intrachromosomal HR repair in all tissues decreased (76.9 ± 3.1%; Figure 3D, Supplemental Table S1). This observed decrease was similar to the decreased proportion of HR repair events with an iwhiteΔ9 intrachromosomal donor template in the male premeiotic germline (85.4 ± 2.3%, P = 0.03, two-tailed unpaired Student’s t-test). Furthermore, males and females showed no statistical difference in the relative proportion of intrachromosomal or interhomolog HR repair events (Supplemental Table S1, P > 0.05, two-tailed Student’s t-test). Thus, both somatic and premeiotic germline tissues demonstrate a homology-dependent preference for intrachromosomal HR repair.
Discussion
Efficient repair of DSBs is critical for maintaining genomic integrity throughout a cell’s life cycle. While homologous recombination allows for accurate repair of DSBs, the factors involved in locating and discriminating between homologous donor templates remain unclear. Previous studies in Drosophila have shown that interhomolog recombination is possible (Engels et al. 1994; Rong and Golic 2003) but have not investigated whether there is a preference between allelic donor templates. Our results confirm that interhomolog recombination is possible and as frequent as intrachromosomal HR if no other sequence is available. However, given a choice between two allelic donor templates, the HR repair machinery exhibits a strong preference for intrachromosomal donor templates, which may be found elsewhere on the same chromosome (i.e., in the DR-white assay or in repetitive DNA sequence) or on the sister chromatid. Interestingly, the chromosomal preference holds even when a less diverged donor template is available on the homolog, though total recombination frequencies do decrease. We propose that, in the presence of a diverged intrachromosomal template, repair shifts to true intersister HR (e.g., repair from the identical Sce.white sequence on the sister chromatid), which cannot be distinguished phenotypically or molecularly in our assays from a no DSB or precise NHEJ event. These findings align with previous work in other eukaryotic systems that have established higher frequencies of recombination between sequences on sister chromatids (Kadyk and Hartwell 1992; Liang et al. 1998; Johnson and Jasin 2000; Engels et al. 1994).
Several models have been proposed for how the homology search is undertaken. One theory (the “null model”) suggests random sampling of DNA sequences to locate a homologous donor template (Barzel and Kupiec 2008). Given the extensive size of the eukaryotic genome and the relatively rapid kinetics of HR repair, the null model is unlikely (Barzel and Kupiec 2008). Our findings also suggest a more refined search process. Under a truly random homology search, one would not expect such a marked preference for intrachromosomal donor templates (>99% at times), especially when a less diverged donor template can be found on the homolog. The more prevailing theory is that preferences for intrachromosomal donor templates arise from the close proximity of intrachromosomal donor templates and sister chromatids (Johnson and Jasin 2000; Rong and Golic 2003; Barzel and Kupiec 2008).
The proximity theory, however, fails to explain the efficient use of interhomolog donor templates in Drosophila and ectopic sequences in yeast when no intrachromosomal donor template is available (Engels et al. 1994; Aylon and Kupiec 2003). The fact that interhomolog and ectopic recombination is even possible in these organisms implies that the HR repair machinery can effectively sample a wider share of the genome than previously suggested. One potential explanation for these differences is that homologous sequences in these organisms are paired throughout the cell cycle. Somatic pairing of homologous chromosomes in Drosophila has been long established (Metz 1916; McKee 2004). In yeast, chromosomes in interphase are arranged in specific configurations (known as Rabl or bouquet) so that allelic loci are relatively equidistant from their respective centromeric or telomeric regions (Barzel and Kupiec 2008). These global alignments of homologous sequences could explain the surprisingly efficient use of interhomolog HR in these organisms.
It is likely that proximity alone does not determine the choice of donor template. In fact, in accordance with previous work (Do et al. 2014), we consistently found a clear reduction in frequency of HR repair between slightly diverged sequences. Thus, the degree of homology between the broken and donor template remains a crucial aspect of HR repair. Interestingly, though intrachromosomal HR was preferred regardless of the identity of the intrachromosomal donor template, interhomolog HR was more pronounced when a less diverged sequence was located on the homolog. This finding suggests that the proximal donor template was rejected in favor of the allelic homologous sequence, despite being theoretically further away. A more intricate search method is therefore likely to be employed that considers both distance and homology requirements.
Recent research using single-molecule imaging has revealed a significant amount about the biochemistry of the RecA protein, a necessary component of the prokaryotic HR repair machinery. RecA, and its eukaryotic homolog Rad51 (Drosophila DmRad51/spn-A), binds extensively to the resected 3′ ssDNA overhangs to create RecA-ssDNA filaments, which can extend many kilobases (reviewed in Bell and Kowalczykowski 2016). Furthermore, the search process, taking up to 50 min in E. coli, is limited to a small volume of the cell (Bell and Kowalczykowski 2016). These studies suggest the intriguing possibility that the search machinery is able to sample multiple sequences at once along the length of the resected filament. This could explain the slightly higher proportion of interhomolog HR when the less diverged donor template is on the homolog. Rapidly-sequential sampling of both allelic sites could allow the HR repair machinery to sporadically choose the less diverged template despite the presence of a proximal donor template in cis. Indeed, when the intrachromosomal donor template is less diverged, interhomolog HR occurs <1% of the time.
These results highlight a broader implication in genome editing. As Cas9-mediated genome editing technology becomes more widespread, the exact mechanisms underlying endonuclease-mediated DSB repair will need to be explained to ensure efficient and predictable repair outcomes (reviewed in Doudna and Charpentier 2014). In particular, researchers hoping to reverse pathogenic mutations via CRISPR/Cas9 using homology directed repair from ectopic sequences will need to consider both the necessary homology requirements as well as the potential for chromosomal rearrangements. A recent study in Drosophila demonstrated that up to 39% of Cas9-mediated DSBs result in recombination between homologous chromosome arms (Brunner et al. 2019). Thus, the possibility of unwanted repair events in diploid organisms cannot be ignored. Our own findings indicate that, despite stringent homology requirements and the possibility of interhomolog recombination, HR occurs preferentially between sequences located on the same chromosome, potentially reducing the efficiency of ectopic genetic transformations.
Acknowledgments
This research was supported by the Georgetown University Medical Center Toulmin Pilot Project Award (J.R.L.) and National Institutes of Health grant 1R15GM129628 (J.R.L.). We would like to thank members of the LaRocque lab for thoughtful discussion and review of the manuscript.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25387/g3.9271007.
Communicating editor: K. McKim
Literature Cited
- Anand R. P., Tsaponina O., Greenwell P. W., Lee C. S., Du W. et al. , 2014. Chromosome rearrangements via template switching between diverged repeated sequences. Genes Dev. 28: 2394–2406. 10.1101/gad.250258.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y., and Kupiec M., 2003. The checkpoint protein Rad24 of Saccharomyces cerevisiae is involved in processing double-strand break ends and in recombination partner choice. Mol. Cell. Biol. 23: 6585–6596. 10.1128/MCB.23.18.6585-6596.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzel A., and Kupiec M., 2008. Finding a match: how do homologous sequences get together for recombination? Nat. Rev. Genet. 9: 27–37. 10.1038/nrg2224 [DOI] [PubMed] [Google Scholar]
- Bell J. C., and Kowalczykowski S. C., 2016. Mechanics and Single-Molecule Interrogation of DNA Recombination. Annu. Rev. Biochem. 85: 193–226. 10.1146/annurev-biochem-060614-034352 [DOI] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., and Basler K., 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104: 3312–3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman E. K., Chen T., Amendola M., and van Steensel B., 2014. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42: e168 10.1093/nar/gku936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner E., Yagi R., Debrunner M., Beck-Schneider D., Burger A. et al. , 2019. CRISPR-induced double-strand breaks trigger recombination between homologous chromosome arms. Life Sci. Alliance 2 e201800267: 1–11. 10.26508/lsa.201800267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burma S., Chen B. P., and Chen D. J., 2006. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst.) 5: 1042–1048. 10.1016/j.dnarep.2006.05.026 [DOI] [PubMed] [Google Scholar]
- Chen W., and Jinks-Robertson S., 1999. The role of the mismatch repair machinery in regulating mitotic and meiotic recombination between diverged sequences in yeast. Genetics 151: 1299–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A., Adjiri A., New L., Crouse G. F., and Jinks-Robertson S., 1996. Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 1085–1093. 10.1128/MCB.16.3.1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delabaere L., Ertl H. A., Massey D. J., Hofley C. M., Sohail F. et al. , 2017. Aging impairs double-strand break repair by homologous recombination in Drosophila germ cells. Aging Cell 16: 320–328. 10.1111/acel.12556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do A. T., and LaRocque J. R., 2015. The role of Drosophila mismatch repair in suppressing recombination between diverged sequences. Sci. Rep. 5: 17601 10.1038/srep17601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do A. T., Brooks J. T., Le Neveu M. K., and LaRocque J. R., 2014. Double-strand break repair assays determine pathway choice and structure of gene conversion events in Drosophila melanogaster. G3 (Bethesda) 4: 425–432. 10.1534/g3.113.010074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoho G., Jasin M., and Berg P., 1998. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol. Cell. Biol. 18: 4070–4078. 10.1128/MCB.18.7.4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J. A., and Charpentier E., 2014. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1258096 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- Elliott B., Richardson C., Winderbaum J., Nickoloff J. A., and Jasin M., 1998. Gene conversion tracts from double-strand break repair in mammalian cells. Mol. Cell. Biol. 18: 93–101. 10.1128/MCB.18.1.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels W. R., Preston C. R., and Johnson-Schlitz D. M., 1994. Long-range cis preference in DNA homology search over the length of a Drosophila chromosome. Science 263: 1623–1625. 10.1126/science.8128250 [DOI] [PubMed] [Google Scholar]
- Ertl H. A., Russo D. P., Srivastava N., Brooks J. T., Dao T. N. et al. , 2017. The role of Blm helicase in homologous recombination, gene conversion tract length, and recombination between diverged sequences in Drosophilamelanogaster. Genetics 207: 923–933. 10.1534/genetics.117.300285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor G. B., Preston C. R., Johnson-Schlitz D. M., Nassif N. A., Phillis R. W. et al. , 1993. Type I repressors of P element mobility. Genetics 135: 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakem R., 2008. DNA-damage repair; the good, the bad, and the ugly. EMBO J. 27: 589–605. 10.1038/emboj.2008.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W. D., Ehmsen K. T., and Liu J., 2010. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44: 113–139. 10.1146/annurev-genet-051710-150955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S. P., and Bartek J., 2009. The DNA-damage response in human biology and disease. Nature 461: 1071–1078. 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A., Breuer G. A., Brinkman E. K., van der Meulen A. I., Borden S. V. et al. , 2016. A single double-strand break system reveals repair dynamics and mechanisms in heterochromatin and euchromatin. Genes Dev. 30: 1645–1657. 10.1101/gad.283028.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A., and Medema R. H., 2013. Genetic instability: tipping the balance. Oncogene 32: 4459–4470. 10.1038/onc.2012.576 [DOI] [PubMed] [Google Scholar]
- Jasin M., and Rothstein R., 2013. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 5: a012740 10.1101/cshperspect.a012740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. D., and Jasin M., 2000. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 19: 3398–3407. 10.1093/emboj/19.13.3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk L. C., and Hartwell L. H., 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132: 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna K. K., and Jackson S. P., 2001. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27: 247–254. 10.1038/85798 [DOI] [PubMed] [Google Scholar]
- LaRocque J. R., and Jasin M., 2010. Mechanisms of recombination between diverged sequences in wild-type and BLM-deficient mouse and human cells. Mol. Cell. Biol. 30: 1887–1897. 10.1128/MCB.01553-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. S., Wang R. W., Chang H. H., Capurso D., Segal M. R. et al. , 2016. Chromosome position determines the success of double-strand break repair. Proc. Natl. Acad. Sci. USA 113: E146–E154. 10.1073/pnas.1523660113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F., Han M., Romanienko P. J., and Jasin M., 1998. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl. Acad. Sci. USA 95: 5172–5177. 10.1073/pnas.95.9.5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M., and Haber J. E., 1989. Position effects in ectopic and allelic mitotic recombination in Saccharomyces cerevisiae. Genetics 123: 261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A., Ivanov E. L., and Haber J. E., 1996. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. USA 93: 7131–7136. 10.1073/pnas.93.14.7131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazón G., Mimitou E. P., and Symington L. S., 2010. SnapShot: homologous recombination in dna double-strand break repair. Cell 142: 648.e1–648.e2. 10.1016/j.cell.2010.08.006 [DOI] [PubMed] [Google Scholar]
- McKee B. D., 2004. Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim. Biophys. Acta 1677: 165–180. 10.1016/j.bbaexp.2003.11.017 [DOI] [PubMed] [Google Scholar]
- Metz C. W., 1916. Chromosome studies on the Diptera. II. The paired association of chromosomes in the Diptera, and its significance. J. Exp. Zool. 21: 213–279. 10.1002/jez.1400210204 [DOI] [Google Scholar]
- Mimitou E. P., and Symington L. S., 2011. DNA end resection–Unraveling the tail. DNA Repair (Amst.) 10: 344–348. 10.1016/j.dnarep.2010.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif N., and Engels W. R., 1993. DNA homology requirements for mitotic gap repair in Drosophila. Proc. Natl. Acad. Sci. USA 90: 1262–1266. 10.1073/pnas.90.4.1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S., Gorgoulis V. G., and Halazonetis T. D., 2010. Genomic instability - an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 11: 220–228. 10.1038/nrm2858 [DOI] [PubMed] [Google Scholar]
- Port F., Chen H. M., Lee T., and Bullock S. L., 2014. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA 111: E2967–E2976. 10.1073/pnas.1405500111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C., Moynahan M. E., and Jasin M., 1998. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 12: 3831–3842. 10.1101/gad.12.24.3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y. S., and Golic K. G., 2003. The homologous chromosome is an effective template for the repair of mitotic DNA double-strand breaks in Drosophila. Genetics 165: 1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J., Sung P., and Klein H., 2008. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77: 229–257. 10.1146/annurev.biochem.77.061306.125255 [DOI] [PubMed] [Google Scholar]
- Shrivastav M., De Haro L. P., and Nickoloff J. A., 2008. Regulation of DNA double-strand break repair pathway choice. Cell Res. 18: 134–147. 10.1038/cr.2007.111 [DOI] [PubMed] [Google Scholar]
- So A., Le Guen T., Lopez B. S., and Guirouilh-Barbat J., 2017. Genomic rearrangements induced by unscheduled DNA double-strand breaks in somatic mammalian cells. FEBS J. 284: 2324–2344. 10.1111/febs.14053 [DOI] [PubMed] [Google Scholar]
- Sullivan W., Ashburner M., and Hawley R. S., 2000. Drosophila Protocols, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. Supplemental Table S1, the R script algorithm (Supplemental File S1), and sequence files (Supplemental Files S2-S26) used for TIDE analyses are available on FigShare. https://doi.org/10.25387/g3.9271007.