Abstract
Poecilia reticulata is one of the most popular ornamental fish species with a higher demand for males due to their large colorful fins. The objectives of this study were mapping of the sex-determining (SD) region on linkage group 12 of guppy, and identification of a sex specific marker. We generated eight polymorphic microsatellite markers distributed along the distal 5.4 Mbp sequence of the previously identified SD region on linkage group (LG) 12. The markers were tested for association with sex in 156 individuals of the Red Blonde and Flame strains, and 126 progeny of four full-sibs Red Blonde families. A male-specific allele was found for microsatellite gu1066 at position of 25.3 Mbp on LG12 for both strains, and gu832 at position 24.4 Mbp for the Flame strain. Thus, a region of 0.9 Mbp between these markers, harboring 27 annotated genes, was selected for analysis. Based on association of copy number variation and sex determination we mapped a duplicated region between LGs 9 and 12, of 1.26 Mbp, containing 59 genes on LG12. The common interval between the segment bounded by gu1066 and gu832, and the duplicated region of 0.43 Mbp on LG12 harbors 11 genes of which 6 have multiple copies (54%). Growth arrest and DNA damage inducible gamma-like (GADD45G-like) is a plausible positional and functional candidate gene for its role in male fertility. We characterized the genomic structure of the gene in guppy, and found two isoforms; but no sex-biased differences were evident in genomic sequence and gene expression.
Keywords: Poecilia reticulata, sex determination, sex specific marker, mapping, Genetics of Sex
There are over 34,000 fish species according to FishBase (www.fishbase.de), but only a few hundred species have been studied for environmental, epigenetic and genetic effects on SD (Devlin and Nagahama 2002; Mank et al. 2006). In many cichlid species of the genus Apistogramma, SD is affected by temperature (Ospina-Álvarez and Piferrer 2008). Epigenetic mechanisms have also been suggested to affect the establishment and maintenance of the gonad differentiation pathways of the half-smooth tongue sole (Cynoglossus semilaevis) (Chen et al. 2014; Martínez et al. 2014). Genetic mechanisms affecting SD include XX/XY sex chromosome system, with the male being heterogametic in medaka (Oryzias latipes), Nile tilapia (Oreochromis niloticus) and rainbow trout (Oncorhynchus mykiss) (Matsuda et al. 2002; Eshel et al. 2014; Yano et al. 2012), or ZZ/ZW system with female being heterogametic in Blue tilapia (Oreochromis aureus) or in the half-smooth tongue sole (Don and Avtalion 1988; Chen et al. 2008).
In some species like the half-smooth tongue sole or in the Bordallo (Squalius carolitertii) there are distinct sex chromosomes (Chen et al. 2008; Collares-Pereira et al. 1998), while in other species there are no observed sex chromosomes in the karyotype. In medaka and in the Patagonian pejerrey (Odontesthes hatcheri) the master sex determining (MSD) genes e.g., DMY and AMHY are male-specific duplications of regular DMRT1 and AMH on different chromosomes, respectively (Matsuda et al. 2002; Nanda et al. 2002; Hattori et al. 2012). However, in chicken and Nile tilapia DMRT1 and AMH duplications are positioned in a tandem manner (Smith et al. 2009; Eshel et al. 2014; Li et al. 2015). The central role of AMH duplication in SD was also demonstrated for Lingcod (Ophiodon elongates) (Rondeau et al. 2016) and Northern pike (Esox lucius) (Pan et al. 2019). SRY is a mammalian MSD gene that may have evolved from SOX3 (Waters et al. 2007). SOX3 was detected as an MSD gene in medaka-related fish (Oryzias dancena) (Takehana et al. 2014). Nevertheless, in many taxonomic groups of animals the duplication of one of the six MSD genes e.g., DMRT1, AMH, SOX3, AMHR2, GSDF and IRF9 initiates the SD pathway (Matsuda 2018; Li and Gui 2018). Thus, it was hypothesized that the molecular pathway responsible for SD is conserved among animals, and the MSD gene in the critical SD region of guppy could be deduced based on mammalian studies and human disorders (Shirak et al. 2006; Warr and Greenfield 2012; Cutting et al. 2013). This approach has been successfully used in our previous study of AMH duplication detection in the sex determining region of Nile tilapia (Shirak et al. 2006; Eshel et al. 2010, 2012, 2014).
The guppy (Poecilia reticulata) is a small viviparous poeciliid native to small clear streams in northeastern South America and adjacent islands (Endler 1995). Like other live-bearing species, guppy presents a marked sexual dimorphism in which the male body is much more pigmented and with larger fins than females, with almost continuous color pattern polymorphism (Petrescu-Mag and Bourne 2008). This species is one of the most popular ornamental fishes with a great importance in the global tropical fish trade market (Monticini 2010). However, there is a greater demand for males rather than females, due to their colorful pigmentation (Basavaraja et al. 2012). Fish are sorted by hand according to sex identification; which is time consuming, expensive and stressful; thus resulting in sickness and mortality. Our long-term objective is to produce genetically all-male populations. Guppies were characterized with an XY sex chromosome system (Winge 1922). Coloring pattern and fin-shape polymorphism are inherited in a sex linked manner (Winge 1923; Kottler and Schartl 2018). Guppy sex chromosomes are morphologically distinct, containing a pseudo autosomal and unique Y non-recombining region. Thus, the Y chromosome is significantly larger than the X chromosome in most populations of Endler’s guppy (Poecilia winger) (Nanda et al. 2014) with less pronounced difference in Trinidadian guppy (Poecilia reticulata) (Morris et al. 2018). Although the characterization of XY sex-chromosome system in guppies was determined some 100 years ago (Winge 1922) the MSD gene is still unknown. The SD regulating genomic region has been identified on linkage group (LG) 12 to its distal end between 22 to 25 Mbp (Tripathi et al. 2009; Wright et al. 2017).
Feeding of pregnant dams by estrogens 5-10 days before parturition resulted in all-female progeny (Kavumpurath and Pandian 1993). In order to distinguish between sex reversal and natural females, a sex specific marker is required. However, no genetic marker specific for the Y-chromosome and common to all guppy strains has been identified (Kottler and Schartl 2018). The objectives of this study are: 1. mapping of the sex determining region on linkage group 12 of Guppy Red Blonde and Flame, and 2. identification of a sex specific marker that will be used to identify sex reversed females as genetically males for mating with normal males in order to derive YY males.
Material and Methods
Guppy strains and families
All fish were maintained and handled in the aquaculture department at the Central and Northern Arava Research and Development center (Israel). Four full-sib families of the Red blonde line (RB) (obtained from Ginat ornamental fish farm, Moshav Ein-Yahav, Arava, Israel) were produced as follows: females were detected by sex specific features such as pigmentation, fin to body size ratio, body size and shape, gravid spot/gonopodium and behavior at about 3 months post-spawning. Each female was separated and raised in a separate tank, and later a single male was added to the tank. The fingerlings of every family were raised in separate tanks until reaching maturity, then sex was determined and the fish were fin-clipped for DNA extraction. In addition, five batches of Red Blonde and Flame strains (obtained from Ginat ornamental fish farm and Manor farm, Hatzeva, Israel) from three different ornamental fish farms in Hatzeva Israel were used for association study of microsatellites with sex (Table 1).
Table 1. Distribution of genotypes of males and females by gu1066 genotypes in the Red Blonde (RB) and Flame (FL) lines from 3 farms, and in 4 full-sibs families of Red Blond from the Arava research center.
| Farma | Lineb | Batch/ Familyc | Males | Females | Nominal probabilityd | Experiment-wise probabilitye | ||
|---|---|---|---|---|---|---|---|---|
| XY | XX | XY | XX | |||||
| Ginat | FL | 23 | 0 | 0 | 23 | 5.7E-10 | 5.1E-09 | |
| Ginat | RB | 1 | 23 | 0 | 0 | 23 | 5.7E-10 | 5.1E-09 |
| Ginat | RB | 2 | 6 | 4 | 0 | 10 | 1.5E-02 | NS |
| Manor | RB | 8 | 2 | 0 | 10 | 3.5E-03 | 3.1E-02 | |
| Ran | RB | 6 | 4 | 0 | 10 | 1.5E-02 | NS | |
| Arava | RB | A | 9 | 4 | 0 | 3 | 1.8E-01 | NS |
| Arava | RB | B | 20 | 3 | 0 | 15 | 4.5E-06 | 4.0E-05 |
| Arava | RB | C | 16 | 2 | 0 | 21 | 5.5E-07 | 4.9E-06 |
| Arava | RB | D | 11 | 2 | 0 | 17 | 3.6E-05 | 3.2E-04 |
| Total | 122 | 21 | 0 | 132 | ||||
Genotypes of gu1066 were designated “XY” and “XX”, where “Y” is male specific allele of 212 bp (206 or 212 bp in Manor), and “X” - all other alleles.
FL - Flame; RB - Red Blonde.
Full-sibs families A to D.
Significance values of Chi-squared test for association between gu1066 segregating microsatellite and sex, based on detection of male-specific allele of gu1066.
With application of the Bonferroni correction for multiple comparisons, NS - Not significant.
DNA extraction and amplification
DNA was extracted from fins or whole body samples using the MasterPure DNA Purification Kit (Epicentre Biotechnologies, Madison, WI, USA) following the manufacturer’s recommended protocol. DNA concentration was quantified using a NanoDrop 1000 Spectrophotometer, and DNA samples diluted to 20 ng/µl were distributed into 96-well PCR plates. Microsatellites were amplified by a two-step PCR following Dor et al. (2014), except that the annealing temperature was 59° and 61° for the first and second PCR steps, respectively. After detection of an association between marker gu1066 and SD, a specific fluorescently-labeled forward primer was developed for gu1066 and used in one step PCR amplification with annealing temperature of 61°.
Marker development
A previous study indicated the localization of a sex determining region of P. reticulata in the terminal region of LG12 (Tripathi et al. 2009). Therefore, the last 5.4 Mbp sequence of female LG12 was downloaded from NCBI (assembly GCF_000633615.1). The sequence was masked for repeats with CENSOR (Kohany et al. 2006) and Tilapia Repeat Masker (Shirak et al. 2010) and scanned for the following microsatellite repeat motifs: (CA)9NNN/(AC)9NNN, (AT)10NNN/(TA)10NNN, (TG)10NNN/(GT)10NNN. Evenly spread motifs along the sequence were selected as potential markers and primers were designed using Primer3 (Untergasser et al. 2012). Marker names were designated “gu” followed by a three or four-digit number, e.g., gu001. The primer sequences are presented in Supplemental Table S1.
Genotyping of microsatellite markers
The fluorescently labeled PCR products were detected by an ABI3130 Genetic Analyzer and automatically genotyped by GeneMapper software v.4.0 using GeneScan-500 LIZ size standard (Applied Biosystems, Foster City, CA, USA).
Statistical analysis
Chi squared test was used to test for divergence of sex distribution from equality between SD genotypes. Fisher’s exact test was computed using PROC FREQ in SAS (version 9.2; SAS Institute Inc., Cary, NC) for association study of microsatellites and sex. The nominal probability of significance was adjusted to account for multiple comparisons using the Bonferroni correction by dividing the nominal probability by the number of comparisons (Hochberg 1988).
Genomic analysis
Using Ensembl with Biomart option (www.ensembl.org) and NCBI, the female guppy genome was searched for all annotated genes in the genomic region on LG12 between markers gu832 and gu1066. Genes without annotation in Guppy were Blastp searched against “Teleostei”. After detection of similarity between a region on LG9 and the SD region on LG12 duplicate genes on both LGs were identified.
Based on medaka as a reference genome, we searched for duplicated genes on LG9 that belong to the group of six MSD genes or their interacting genes. Boundaries of the region were marked if no additional duplicate genes were identified in the marginal 1 Mbp sequences. The gene list was plotted using MapChart software (Voorrips 2002).
In view of the partial GADD45G-like annotation for P. reticulata, the P. formosa sequence of scaffold NW_006800603.1 (from 35,000 to 41,000 bp) was downloaded and masked using GIRI (Kohany et al. 2006). This sequence contains two copies of GADD45G-like gene (LOC103130705 and LOC103130706). DNA-seq data of guppy Aripo female (ERX2156226) and male (ERX2156238) embryos (PRJEB22221 project database; Bergero et al. 2018) was downloaded and aligned to the P. formosa sequence using Gap5 software (Bonfield and Whitwham 2010). The annotated nucleotide sequence of this directed assembly of GADD45G-like, including the two orthologous genes present in P. reticulata locus is provided in Supplemental Figure S1.
RNA seq analysis
The genomic guppy GADD45G-like sequence (LOC103474023) was BLAST searched using a word size of 48 bp against RNA-seq data (12 expression libraries) of female (SRX388849) and male (SRX388850) guppy embryos and organs (PRJNA230881 project database; Sharma et al. 2014). The resulting reads were aligned to reference transcripts of P. formosa genes (LOC103130705 and LOC103130706) using Gap5. The number of hits were compared between spliced isoforms, normalized and compared between males and females.
Sex reversal treatment
Each female was introduced with a single male into a separate container. After the first spawn, each female was fed continuously with 30% protein pellet containing β-estradiol, at concentration of 200, 400 or 600 mg/kg, in order to induce female phenotype of fingerlings of subsequent spawns. All fries of second spawns were collected and reared separately until maturity while given dry feed containing β-estradiol continuously. Ultimately, individual fingerlings of 11 “sex reversed” families were identified for sex and fin clipped for DNA extraction.
Histopathology
Histological analyses of 98 treated and control fish were examined of which 47 were sex reversed females. Fish were anesthetized, a cut in the abdomen was performed and the tail fin was cut off. The body was then immediately fixed in neutral buffered formalin and transferred to 70% ethanol after 48 h. Processing was performed after decalcification (44% formic acid and 12.5% sodium citrate for 12 h) using a microwave histo-processor (RHS-1, Milstone, Italy), after which the samples were embedded in paraffin blocks and sectioned at 4 μm. Sections were stained with hematoxylin and eosin (H&E). Samples were analyzed under a light microscope with 400× magnification for identification of the gonad and its condition thus determining the sex of the fish and the stage of sexual development.
Data availability
Supplemental Table S1 contains the sequences of primers for 8 polymorphic markers on LG12. Supplemental Figure S1 contains the assembled genomic sequences of GADD45G-like gene locus of P. reticulata. Supplemental material available at FigShare: https://doi.org/10.25387/g3.9841160.
Results
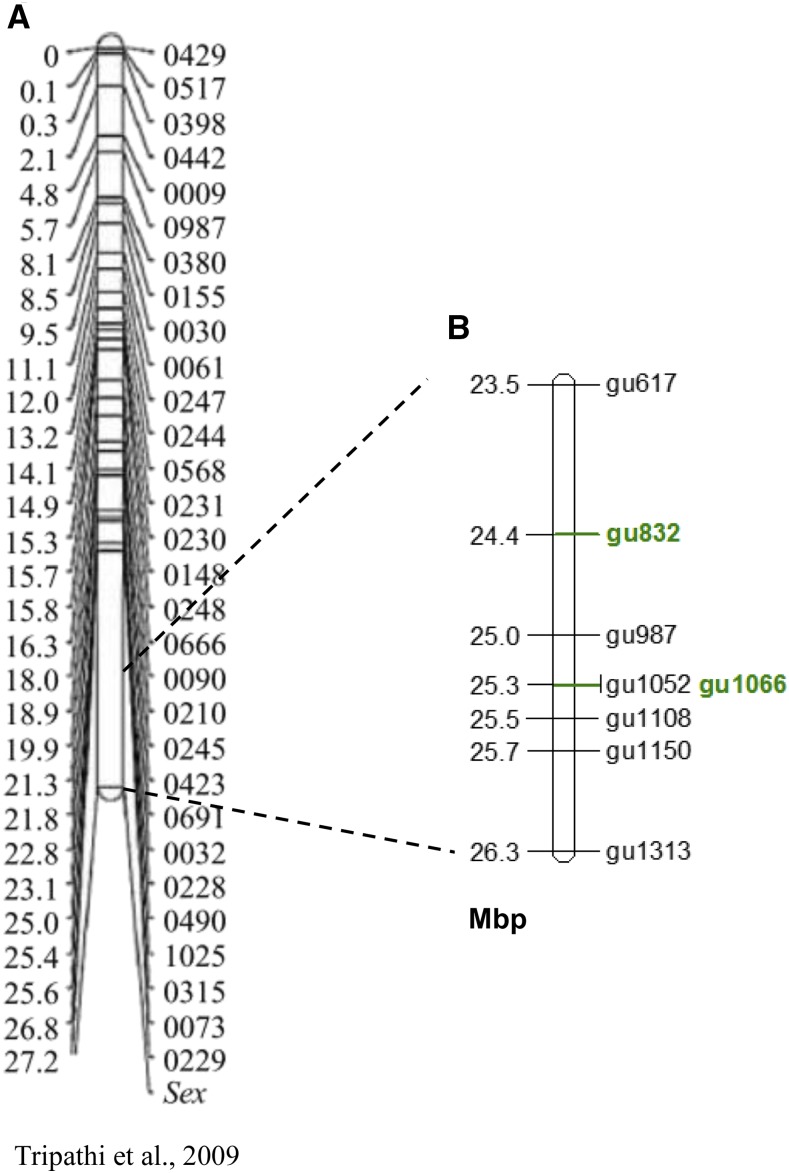
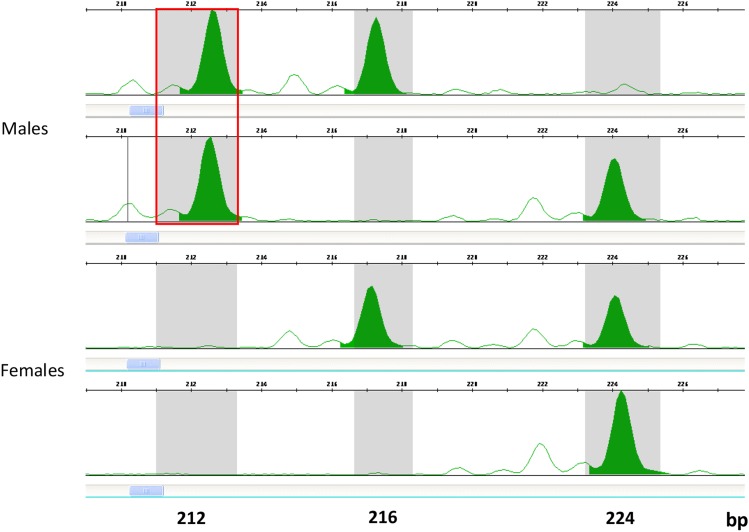
Of 15 microsatellite markers designed in several locations along the terminal 5.4 Mbp genomic region on LG12, 7 were polymorphic in populations of Red Blonde and Flame with 2 to 6 alleles per marker. Five markers were polymorphic in progeny of 4 families with 2 to 4 alleles per marker. The locations of 8 polymorphic markers on LG12 in Mbp are displayed on Figure 1B, along with the previous map of the sex determining region on LG12 (Figure 1A). Association study for sex based on 46 individuals of Red Blonde and Flame strains showed complete concordance for only the gu1066 microsatellite (P < 5.7E-10, Table 1). Smaller batches of these populations and full-sib progeny showed lower significance in the association study. Nevertheless, a male-specific allele for gu1066 was evident in both strains (Table 1, Figure 2). The length of the male-specific chromosomal segment was 212 bp in all strains and families; as compared to either 206 or 212 bp for the Red Blonde batch in Manor Farm. In 21 out of 143 males (14.7%) the male-specific allele was not identified. Nevertheless, this allele was not found in any of the 132 tested females (Table 1). Thus, presence of the specific allele indicates male determination.
Figure 1.
Mapping of sex determining region on LG12. A. Mapping of the sex region on LG12 in guppy as presented in Tripathi et al. (2009); B. Location of polymorphic microsatellites on LG12 terminal region, with the significant microsatellites associated with sex marked in green, Units are in Mbp.
Figure 2.
A typical genotyping results of two guppy males and two guppy females for microsatellite gu1066. Allele size is marked on the X-axis (in bp). The fluorescent intensity is marked on the Y-axis. Red square marks the male-specific allele representing “Y” (212 bp), whereas all other alleles represent “X” (216 or 224 bp).
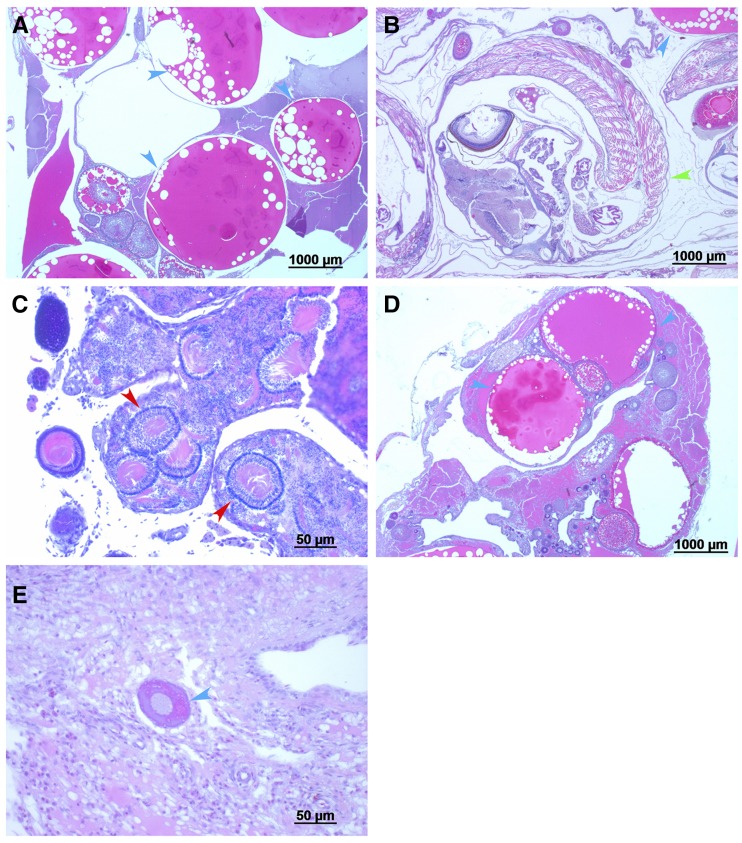
The distribution of males and females in sex reversed families (all-females) based on identification of the “Y” male-specific allele for microsatellite gu1066, is presented in Table 2. Non-significant deviation from a 1:1 ratio between females and males was evident in progeny of all families except one (Family #8). The twofold frequency of females as compared to males in this family with a nominal probability of significance of 0.03 is also non-significant when accounting for multiple comparisons (P = 0.33). Furthermore, based on the results in Table 1, the male-specific allele was identified in only 85% of males, but not in females. Thus, the observed overall distribution of 97 males and 129 females based on male-specific allele identification may be adjusted accordingly, resulting in similar proportions of males and females. Histological examination was performed on the ovaries of 47 sex reversed females and 25 genetic females. Typical histological analyses are presented in Figure 3 confirming the status of gonads. Of the sex reversed females, 19% exhibited definite matured ovaries, as compared to 64% in controls (Chisq; P < 8.9E-23).
Table 2. Distribution of genotypes in sex reversed families (all-females) based on detection of the “Y” male-specific allele of microsatellite gu1066.
| Familya | β-estradiol treatmentb | XY | XX | N/Ac |
|---|---|---|---|---|
| 1 | 400 | 6 | 8 | 2 |
| 2 | 400 | 18 | 18 | 5 |
| 3 | 200 | 12 | 18 | |
| 4 | 400 | 12 | 13 | |
| 5 | 600 | 10 | 13 | |
| 6 | 200 | 6 | 8 | |
| 7 | 400 | 7 | 13 | |
| 8 | 600 | 5 | 10 | |
| 9 | 200 | 8 | 7 | 9 |
| 10 | 400 | 6 | 10 | 2 |
| 11 | 600 | 7 | 11 | |
| Total | 97 | 129 |
Deviation from equal distribution of males and females was not significant for all families with application of Bonferroni correction for multiple comparisons (P > 0.05).
β-estradiol concentration in food (mg/kg).
N/A – Not analyzed due to unsuccessful DNA extraction.
Figure 3.
Histological examination of gonads of guppy at 10-14 months old. A. XX, female, normal ovarian tissue; B. XX, female with embryo; C. XY, male, normal testicular tissue; D. ΔXY, female, normal ovarian tissue; E. ΔXY, high proportion of undifferentiated tissue. XX and XY were determined based on genotype for gu1066 marker; Sex was defined by phenotype; Δ – β-estradiol treatment; Blue arrows – follicles in different developmental stage; Red arrows – seminiferous tubules; Green arrow - embryo. Size of ladder (in micrometer) is underlined in the lower right of each picture.
Marker gu1066 at 25.3 Mbp was associated with sex in both strains (P < 5.7E-10, Table 1). Marker gu1066 is located within the Stomatin-like 2 gene (STOML2). In Significance values for association between gu1066 and adjacent segregating microsatellites are presented in Table 3. The upstream adjacent marker e.g., gu1150, at 25.6 Mbp, was not associated with sex in both strains. However, the downstream adjacent marker gu832 at 24.4 Mbp, had the second most significant association with sex; based on the Flame strain (P < 3.9E-07). Thus, the genomic interval of 0.9 Mbp between gu832 and gu1066 markers, harboring 27 annotated genes, was selected for detailed analysis (Figure 4).
Table 3. Probability values of Fisher’s exact test for association between gu1066 and adjacent segregating microsatellites in the critical region on LG12 for sex determination.
| Markers | |||||
|---|---|---|---|---|---|
| Linea | gu617 n(23.5) | gu832 (24.4) | gu1052 (25.25) | gu1108 (25.48) | gu1150 (25.6) |
| FLb | 0.035 | 3.9E-07 | 0.046 | NSc | NS |
| RBd | NS | NS | NS | NS | NS |
Genotypes for gu1066 had complete concordance with sex. Mapping position on LG12 for each microsatellite is shown in bracket (in Mbp).
FL – Flame (n = 48).
Not significant.
RB – Red Blonde (n = 48).
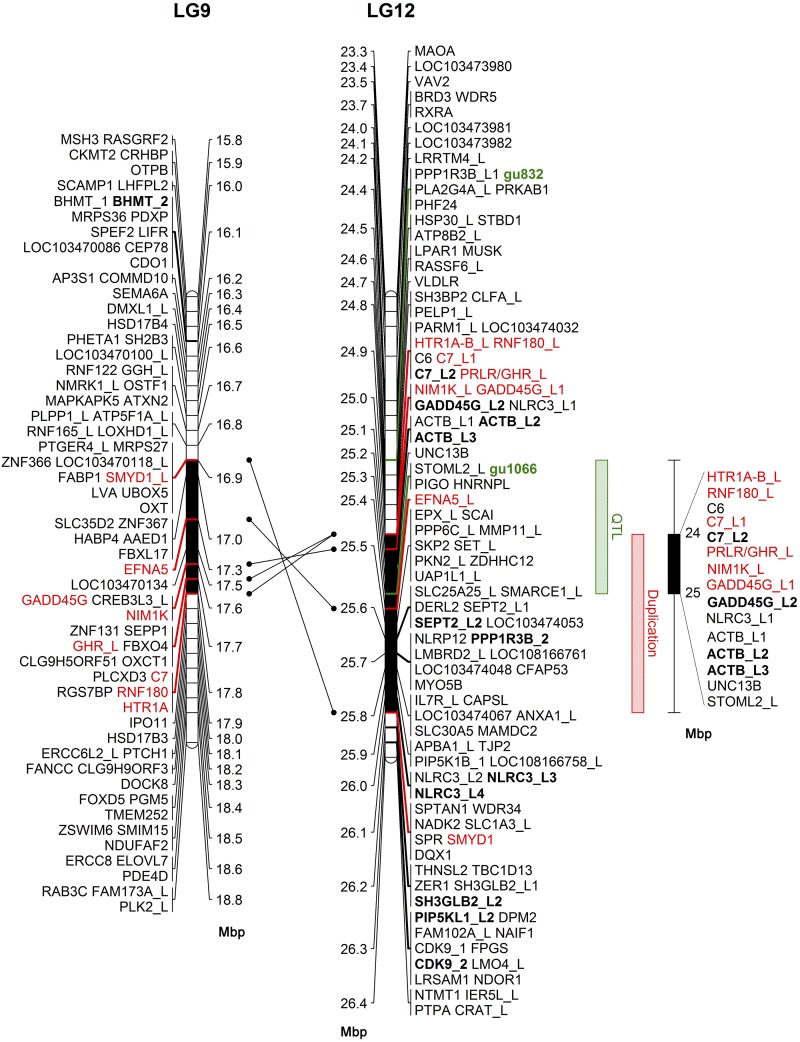
Figure 4.
Comparative analysis of annotated genes between the critical SD region on LG12 and the respective duplication region on LG9 in guppy. Annotated genes are positioned in linkage groups 9 and 12. The region on LG12 between markers gu832 and gu1066 (in green) is represented by a green box on the right. Duplicated genes across LGs 9 and 12 are in red with connecting arrows between their respective positions. The common duplication region on both LGs is in black and represented by red box shape on the right. Annotated genes with multiple copies are marked with bold and underscore followed by the copy number. Gene name annotations containing “like” are marked with underscore followed by “L” and the copy number. Intersected region of the QTL and the duplication regions are represented by black box shape on the right with gene name annotations with copy numbers separated by “/”. Units are in Mbp.
In the search for enriched region with genes with multiple copies that may be associated with SD, we identified a region of duplication between LGs 9 and 12, of 1.26 Mbp, containing 59 genes on LG12 (Figure 4). In this region 17 genes had multiple copies (29%), of which 8 genes had copies across LGs 9 and 12 (in red), and the remaining 9 genes within LG12 (in bold). The common interval of the segment bordered by gu1066 and gu832, and the duplication region ranged from 24.88 to 25.31 (0.43 Mbp). This interval included 11 genes of which 6 (54%) i.e., HTR1A, RNF180, C7, GHR, NIM1K and GADD45G like have multiple copies (black box shape, Figure 4).
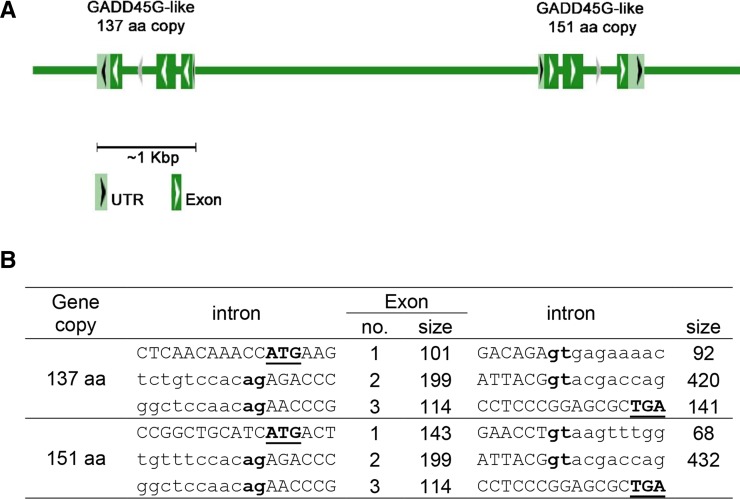
The Growth arrest and DNA damage Inducible Gamma-like gene (LOC103474023, GADD45G like) is the most attractive candidate gene for SD in this region, as it has a specific role in male fertility and testis development (Johnen et al. 2013). This gene was prioritized for analysis. Assembly of 1428 reads located by BLAST-searching project PRJNA230881, indicated two GADD45G-like spliced isoforms of 151 and 137 amino acids that have not been characterized in P. reticulata but present in P. formosa (XP_007542235 and XP_007542234, respectively). Genomic sequences of both copies of the GADD45G-like gene in guppy, were assembled in a head-to-head orientation with a ∼4 Kbp intergenic region between them (Figure 5, Supplemental Figure S1). The two assembled copies contain 3 exons of which only the first one differed in sequence and size between them. The GADD45G-like genomic sequences of females and males were explored with no significant differences between them. Mean expression was 15 fold higher in the 151 vs. 137 AA isoforms based on 12 different expression libraries of guppy (Sharma et al. 2014). However, our analysis for GADD45G-like gene expression based on the latter data showed no definite sex-biased difference.
Figure 5.
The structure of the duplicated GADD45G-Like gene in Guppy. The schematic (A) and genomic (B) structure of GADD45G-Like gene. Exon and intron sizes are given in base pairs. Intron and exon sequences are written in lowercase and uppercase letters, respectively. The first and last two bases of introns (gt and ag for donor and acceptor splice sites, respectively) are in bold type. The initiation and stop codons, and the putative poly adenylation signal (ATG, TGA) are in bold and underlined type.
Discussion
Eight of the 15 markers that were developed based on the published female guppy genome were found polymorphic with two or more alleles in Red Blonde and Flame strains and in full-sib families of Red Blonde. The association study for sex showed complete concordance for only the gu1066 microsatellite in both strains. Nevertheless, in additional small batches of samples the male-specific allele was not detected in all males, although not detected in any female. Failure to determine all males based on gu1066 genotype may be due to the occurrence of null alleles (Paetkau and Strobeck 1994), although design of alternative primers along the target sequence did not alter the genotypes (data not shown). Interestingly, additional markers that were mapped closely to gu1066, such as gu1108 and gu1150, based on the published female guppy genome, were not significantly associated to SD on LG12, possibly because of the difficulty to determine their position in duplicated regions. This ambiguity in precise locations of markers in relation to the sex region was also observed by Tripathi et al. (2009).
The second most significant associated marker in Flame, but not in the Red Blonde line, was gu832 located 0.9 Mbp from gu1066, thus validating the finding of a distal region on LG12 affecting sex determination (Wright et al. 2017), and some differences between Y chromosomes of different lines (Tripathi et al. 2009). A follow up analysis of full-sib progeny of the Red Blonde family D that segregated for both markers showed that two males out of 21 that lacked the 212 bp male-specific allele of gu1066, also lacked the 352 bp male-specific allele of gu832; thus indicating a possible occurrence of recombination outside the SD region harboring both markers and the sex determining locus. Nevertheless, these two males were progeny of a single family out of the four families tested (Family D), indicating a possible environmental effect of elevated temperature on sex reversal of these individuals (Karayucel et al. 2006). Moreover, none of the 132 tested females, including the 17 females in family D, displayed the male-specific allele as the putative reciprocal recombinant, in agreement with possible environmental effect or recombination suppression (Traut and Winking 2001; Tripathi et al. 2009; Charlesworth 2018). It may be possible that this type of recombinant is lethal, and thus was not found in adult females. Alternatively, the rare recombination may represent gene conversion that occurs in regions that do not undergo crossing over (Bergero et al. 2018; Bergero et al. 2019; Korunes and Noor 2017). Although sex is a categorical trait, a quantitative trait loci (QTL) analysis of the sex determining region may be more appropriate, accounting for aberrant crossing-over, segregation of genes in different regions and environmental effects (Eshel et al. 2010; Dor et al. 2016).
The chromosomal region of 0.9 Mbp between gu1066 and gu832 includes 27 annotated genes. We searched this region for enrichment of genes with multiple copies, because of the role of gene duplication in the turnover of sex chromosomes in teleosts (Eshel et al. 2014; Matsuda et al. 2002; Nanda et al. 2002; Hattori et al. 2012; Rondeau et al. 2016; Pan et al. 2019). We identified a region containing 59 genes on LG12 that was enriched with 17 genes (29%) with multiple copies, of which 8 had copies across both LGs 9 and 12. The length of the intersected interval between the two regions is 0.43 Mbp, containing 11 genes of which six have multiple copies. GADD45G-like is the most attractive candidate gene for SD in the intersected QTL/duplication region, as it has a specific role in male fertility and testis development (Johnen et al. 2013), and functions in male sex determination by promoting p38 signaling and SRY expression (Gierl et al. 2012). Furthermore, GADD45G−/− XY mice were born as completely sex-reversed XY-females due to their gonads failure to achieve the SRY expression threshold necessary for testis differentiation, resulting in ovary and Müllerian duct development (Johnen et al. 2013). We characterized the genomic structure of the gene in guppy, and found two isoforms that share identical sequence for exons 2 and 3, but not for exon 1 (Figure 5, Supplemental Figure S1). Mean expression was 15 fold higher in the 151 vs. 137 AA isoforms based on 12 different expression libraries of guppy (Sharma et al. 2014). However, we did not find any significant difference between sex groups in either genomic sequence or gene expression.
Forty putative Y genes, including two plausible candidate genes that may be involved in sex determination, were also suggested in guppies (Morris et al. 2018). To detect the progenitor of the guppy MSD gene on LG12, we analyzed the presumptive ancient region on LG9, and did not detect strong candidate genes for MSD, except for GADD45G. As the genome reference of a female guppy was used for assembly of genes, the MSD gene might presumably be missing in the SD region. However, high similarity was found between the genes on LGs 9 and 12. Moreover, the duplicated male-specific copy of AMH in tilapia is located in tandem along with the regular gene in female (Eshel et al. 2014; Li et al. 2015). Thus, it is implicated that the female guppy genome may contain the MSD gene and/or its regulatory elements.
Comparison of female and male synaptonemal complex showed that XY synapsis occurred rarely and preferentially in the terminal positions reflecting suppression of recombination over the entire Y chromosome, except a 1-2 Mbp of the small pseudo-autosomal region (Lisachov et al. 2015). A lack of crossing over in Y chromosomes over 25 Mbp region was supported by marker analysis for most guppy populations and families (Bergero et al. 2018; Bergero et al. 2019). However, Wright et al. (2017) predicted different patterns of recombination between X and Y over a 15-22 Mbp region of high recombination. In a preliminary study we analyzed sequence data of guppy family along three generations (GenBank project PRJEB7924), and found no crossing over in a 25 Mbp region of Y chromosome (data not shown). However, in the present study we detected significant level of recombination at an interval of 21-26.4 Mbp. Analysis of recombination in tilapia sex determining regions demonstrated that rate of recombination is highly affected by mating between closely related species (McConnell et al. 2000; Shirak et al. 2006; Cnaani et al. 2008). We hypothesize that several commercial, laboratory and natural stocks are hybrids of P. reticulata and related species. The mtDNA analysis of P. wingei natural population revealed its origin as hybrid with P. reticulata (Schories et al. 2009).
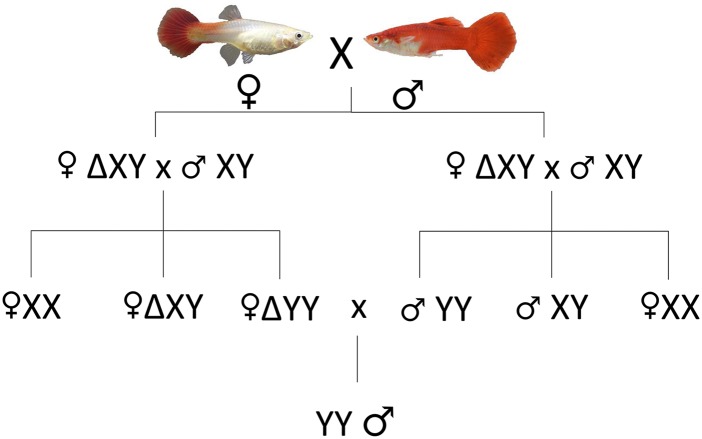
The incentive to mate sexually reversed females that are genetically males (XY) with normal males (XY) in order to derive YY males is dependent on the ability to genetically identify males using a DNA-based method. The entire scheme of mating is displayed in Figure 6. Subsequently, the YY males would be mated with normal females in order to produce XY fingerlings that will constitute the all-male commercial population (F3, Figure 6). Using the gu1066 marker we identified unequivocally 97 sex reversed females as genetically males, and used them for mating with normal males (F2, Figure 6). However, only 19% exhibited definite histologically matured ovaries, and only a single sex reversed female spawned four live fingerlings of which three were determined as XY and one as XX. Thus, further experiments are needed to assess the viability of YY genotypes of the Red Blonde and Flame strains, and the potential of this strategy to produce all-males population.
Figure 6.
Mating scheme for production of all-males population based on detection of the “Y” male-specific allele of microsatellite gu1066. Δ – Sex reversed using β-estradiol, validated by SD test.
Acknowledgments
This work was supported by the ICA Foundation and the Ministry of Agriculture and Rural Development, Israel. Grant: 93-04-0002
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25387/g3.9841160.
Communicating editor: M. Arbeitman
Literature Cited
- Basavaraja N., Chandrashekhara B. H., and Ahamad R. M., 2012. Production of an all-male population of guppy, Poecilia reticulata (Schneider). Curr. Sci. 103: 1151–1152. [Google Scholar]
- Bergero R., Gardner J., Bader B., Yong L., and Charlesworth D., 2018. Sexually dimorphic recombination can facilitate the establishment of sexually antagonistic polymorphisms in guppies. bioRxiv 10.1101/365114 [DOI]
- Bergero R., Gardner J., Bader B., Yong L., and Charlesworth D., 2019. Exaggerated heterochiasmy in a fish with sex-linked male coloration polymorphisms. Proc. Natl. Acad. Sci. USA 116: 6924–6931. 10.1073/pnas.1818486116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfield J. K., and Whitwham A., 2010. Gap5—editing the billion fragment sequence assembly. Bioinformatics 26: 1699–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., 2018. The guppy sex chromosome system and the sexually antagonistic polymorphism hypothesis for Y chromosome recombination suppression. Genes (Basel) 9: 264 10.3390/genes9050264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.-L., Deng S.-P., Ma H.-Y., Tian Y.-S., Xu J.-Y. et al. , 2008. Molecular marker-assisted sex control in half-smooth tongue sole (Cynoglossus semilaevis). Aquaculture 283: 7–12. 10.1016/j.aquaculture.2008.07.015 [DOI] [Google Scholar]
- Chen S., Zhang G., Shao C., Huang Q., Liu G. et al. , 2014. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 46: 253–260. 10.1038/ng.2890 [DOI] [PubMed] [Google Scholar]
- Cnaani A., Lee B.-Y., Zilberman N., Ozouf-Costaz C., Hulata G. et al. , 2008. Genetics of Sex Determination in Tilapiine Species. Sex Dev. 2: 43–54. 10.1159/000117718 [DOI] [PubMed] [Google Scholar]
- Collares-Pereira M. J., Próspero M. I., Biléu R. I., and Rodrigues E. M., 1998. Leuciscus (Pisces, Cyprinidae) karyotypes: Transect of Portuguese populations. Genet. Mol. Biol. 21: 63–69. 10.1590/S1415-47571998000100011 [DOI] [Google Scholar]
- Cutting A., Chue J., and Smith C. A., 2013. Just how conserved is vertebrate sex determination? Dev. Dyn. 242: 380–387. 10.1002/dvdy.23944 [DOI] [PubMed] [Google Scholar]
- Devlin R. H., and Nagahama Y., 2002. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture 208: 191–364. 10.1016/S0044-8486(02)00057-1 [DOI] [Google Scholar]
- Don J., and Avtalion R. R., 1988. Comparative study on the induction of triploidy in tilapias, using cold- and heat-shock techniques. J. Fish Biol. 32: 665–672. 10.1111/j.1095-8649.1988.tb05406.x [DOI] [Google Scholar]
- Dor L., Shirak A., Rosenfeld H., Ashkenazi I. M., Band M. R. et al. , 2016. Identification of the sex-determining region in flathead grey mullet (Mugil cephalus). Anim. Genet. 47: 698–707. 10.1111/age.12486 [DOI] [PubMed] [Google Scholar]
- Dor L., Shirak A., Gorshkov S., Ron M., and Hulata G., 2014. Development of genetic markers for the white grouper (Epinephelus aeneus). Aquaculture 420–421: S104–S110. 10.1016/j.aquaculture.2013.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler J. A., 1995. Multiple-trait coevolution and environmental gradients in guppies. Trends Ecol. Evol. 10: 22–29. 10.1016/S0169-5347(00)88956-9 [DOI] [PubMed] [Google Scholar]
- Eshel O., Shirak A., Weller J. I., Slossman T., Hulata G. et al. , 2010. Fine-mapping of a locus on linkage group 23 for sex determination in Nile tilapia (Oreochromis niloticus). Anim. Genet. 42: 222–224. 10.1111/j.1365-2052.2010.02128.x [DOI] [PubMed] [Google Scholar]
- Eshel O., Shirak A., Weller J. I., Hulata G., and Ron M., 2012. Linkage and Physical Mapping of Sex Region on LG23 of Nile Tilapia (Oreochromis niloticus). G3 (Bethesda) 2: 35–42. 10.1534/g3.111.001545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel O., Shirak A., Dor L., Band M., Zak T. et al. , 2014. Identification of male-specific amh duplication, sexually differentially expressed genes and microRNAs at early embryonic development of Nile tilapia (Oreochromis niloticus). BMC Genomics 15: 774 10.1186/1471-2164-15-774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierl M. S., Gruhn W. H., von Seggern A., Maltry N., and Niehrs C., 2012. GADD45G Functions in male sex determination by promoting p38 signaling and sry expression. Dev. Cell 23: 1032–1042. 10.1016/j.devcel.2012.09.014 [DOI] [PubMed] [Google Scholar]
- Hattori R. S., Murai Y., Oura M., Masuda S., Majhi S. K. et al. , 2012. A Y-linked anti-Müllerian hormone duplication takes over a critical role in sex determination. Proc. Natl. Acad. Sci. USA 109: 2955–2959. 10.1073/pnas.1018392109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y., 1988. A sharper Bonferroni procedure for multiple tests of significance. Biometrika 75: 800–802. 10.1093/biomet/75.4.800 [DOI] [Google Scholar]
- Johnen H., González-Silva L., Carramolino L., Flores J. M., Torres M. et al. , 2013. Gadd45g Is essential for primary sex determination, male fertility and testis development. PLoS One 8: e58751 10.1371/journal.pone.0058751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavumpurath S., and Pandian T. J., 1993. Masculinization of Poecilia reticulata by dietary administration of synthetic or natural androgen to gravid females. Aquaculture 116: 83–89. [Google Scholar]
- Karayucel, I., O. Ak, and S. Karayucel, 2006 Effect of temperature on sex ratio in guppy Poecilia reticulata (Peters 1860). Aquacult. Res. 37: 139–150. [Google Scholar]
- Kohany O., Gentles A. J., Hankus L., and Jurka J., 2006. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics 7: 474 10.1186/1471-2105-7-474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korunes K. L., and Noor M. A. F., 2017. Gene conversion and linkage: effects on genome evolution and speciation. Mol. Ecol. 26: 351–364. 10.1111/mec.13736 [DOI] [PubMed] [Google Scholar]
- Kottler V., and Schartl M., 2018. The colorful sex chromosomes of teleost fish. Genes (Basel) 9: 233 10.3390/genes9050233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Sun Y., Zhao J., Shi H., Zeng S. et al. , 2015. A Tandem Duplicate of Anti-Müllerian Hormone with a Missense SNP on the Y Chromosome Is Essential for Male Sex Determination in Nile Tilapia, Oreochromis niloticus. PLoS Genet. 11: e1005678 10.1371/journal.pgen.1005678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-Y., and Gui J.-F., 2018. An epigenetic regulatory switch controlling temperature-dependent sex determination in vertebrates. Sci. China Life Sci. 61: 996–998. 10.1007/s11427-018-9314-3 [DOI] [PubMed] [Google Scholar]
- Lisachov A. P., Zadesenets K. S., Rubtsov N. B., and Borodin P. M., 2015. Sex Chromosome Synapsis and Recombination in Male Guppies. Zebrafish 12: 174–180. 10.1089/zeb.2014.1000 [DOI] [PubMed] [Google Scholar]
- McConnell S. K., Beynon C., Leamon J., and Skibinski D. O., 2000. Microsatellite marker based genetic linkage maps of Oreochromis aureus and O. niloticus (Cichlidae): extensive linkage group segment homologies revealed. Anim. Genet. 31: 214–218. 10.1046/j.1365-2052.2000.00631.x [DOI] [PubMed] [Google Scholar]
- Mank J. E., Promislow D. E. L., and Avise J. C., 2006. Evolution of alternative sex-determining mechanisms in teleost fishes. Biol. J. Linn. Soc. Lond. 87: 83–93. 10.1111/j.1095-8312.2006.00558.x [DOI] [Google Scholar]
- Martínez P., Viñas A. M., Sánchez L., Díaz N., Ribas L. et al. , 2014. Genetic architecture of sex determination in fish: applications to sex ratio control in aquaculture. Front. Genet. 5: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M., 2018. Genetic control of sex determination and differentiation in fish, Reproductive and Developmental Strategies, pp. 289–306 in Springer, Tokyo. [Google Scholar]
- Matsuda M., Nagahama Y., Shinomiya A., Sato T., Matsuda C. et al. , 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417: 559–563. 10.1038/nature751 [DOI] [PubMed] [Google Scholar]
- Monticini P., 2010. The ornamental fish trade production and commerce of ornamental fish: technical-managerial and legislative aspects. J. Fish Biol. 37: 53–59. [Google Scholar]
- Morris J., Darolti I., Bloch N. I., Wright A. E., and Mank J. E., 2018. Shared and species-specific patterns of nascent y chromosome evolution in two guppy species. Genes (Basel) 9: 238 10.3390/genes9050238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda I., Kondo M., Hornung U., Asakawa S., Winkler C. et al. , 2002. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl. Acad. Sci. USA 99: 11778–11783. 10.1073/pnas.182314699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda I., Schories S., Tripathi N., Dreyer C., Haaf T. et al. , 2014. Sex chromosome polymorphism in guppies. Chromosoma 123: 373–383. 10.1007/s00412-014-0455-z [DOI] [PubMed] [Google Scholar]
- Ospina-Álvarez N., and Piferrer F., 2008. Temperature-dependent sex determination in fish revisited: prevalence, a single sex ratio response pattern, and possible effects of climate change. PLoS One 3: e2837 10.1371/journal.pone.0002837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paetkau D., and Strobeck C., 1994. Microsatellite analysis of genetic variation in black bear populations. Mol. Ecol. 3: 489–495. 10.1111/j.1365-294X.1994.tb00127.x [DOI] [PubMed] [Google Scholar]
- Pan Q., Feron R., Yano A., Guyomard R., Jouanno E. et al. , 2019. Identification of the master sex determining gene in Northern pike (Esox lucius) reveals restricted sex chromosome differentiation. PLoS Genet. 15: e1008013 10.1371/journal.pgen.1008013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrescu-mag I. V., and Bourne G. R., 2008. Crossing-over between Y chromosomes : another possible source of phenotypic variability in the. Int. J. Bioflux Soc. 54: 1–10. [Google Scholar]
- Rondeau E. B., Laurie C. V., Johnson S. C., and Koop B. F., 2016. A PCR assay detects a male-specific duplicated copy of Anti-Müllerian hormone (amh) in the lingcod (Ophiodon elongatus). BMC Res. Notes 9: 230 10.1186/s13104-016-2030-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schories S., Meyer M. K., and Schartl M., 2009. Description of Poecilia (Acanthophacelus) obscura n. sp., (Teleostei: Poeciliidae), a new guppy species from western Trinidad, with remarks on P. wingei and the status of the Endlers guppy. Zootaxa 2266: 35–50. 10.11646/zootaxa.2266.1.2 [DOI] [Google Scholar]
- Sharma E., Künstner A., Fraser B. A., Zipprich G., Kottler V. A. et al. , 2014. Transcriptome assemblies for studying sex-biased gene expression in the guppy, Poecilia reticulata. BMC Genomics 15: 400 10.1186/1471-2164-15-400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirak A., Grabherr M., Di Palma F., Lindblad-Toh K., Hulata G. et al. , 2010. Identification of repetitive elements in the genome of Oreochromis niloticus: tilapia repeat masker. Mar. Biotechnol. (NY) 12: 121–125. 10.1007/s10126-009-9236-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirak A., Seroussi E., Cnaani A., Howe A. E., Domokhovsky R. et al. , 2006. Amh and Dmrta2 genes map to tilapia (Oreochromis spp.) linkage group 23 within quantitative trait locus regions for sex determination. Genetics 174: 1573–1581. 10.1534/genetics.106.059030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A., Roeszler K. N., Ohnesorg T., Cummins D. M., Farlie P. G. et al. , 2009. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461: 267–271. 10.1038/nature08298 [DOI] [PubMed] [Google Scholar]
- Takehana Y., Matsuda M., Myosho T., Suster M. L., Kawakami K. et al. , 2014. Co-option of Sox3 as the male-determining factor on the y chromosome in the fish Oryzias dancena. Nat. Commun. 5: 4157 10.1038/ncomms5157 [DOI] [PubMed] [Google Scholar]
- Traut W., and Winking H., 2001. Meiotic chromosomes and stages of sex chromosome evolution in fish: Zebrafish, platyfish and guppy. Chromosome Res. 9: 659–672. 10.1023/A:1012956324417 [DOI] [PubMed] [Google Scholar]
- Tripathi N., Hoffmann M., Willing E.-M., Lanz C., Weigel D. et al. , 2009. Genetic linkage map of the guppy, Poecilia reticulata, and quantitative trait loci analysis of male size and colour variation. Proc. Biol. Sci. 276: 2195–2208. 10.1098/rspb.2008.1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B. C. et al. , 2012. Primer3–new capabilities and interfaces. Nucleic Acids Res. 40: e115 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips R. E., 2002. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 93: 77–78. 10.1093/jhered/93.1.77 [DOI] [PubMed] [Google Scholar]
- Warr N., and Greenfield A., 2012. The molecular and cellular basis of gonadal sex reversal in mice and humans. Wiley Interdiscip. Rev. Dev. Biol. 1: 559–577. 10.1002/wdev.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters P. D., Wallis M. C., and Graves J. A. M., 2007. Mammalian sex—Origin and evolution of the Y chromosome and SRY. Semin. Cell Dev. Biol. 18: 389–400. 10.1016/j.semcdb.2007.02.007 [DOI] [PubMed] [Google Scholar]
- Winge O., 1923. Crossing over between the X- and the Y-chromosome in Lebistes. Genetics 13: 201–217. 10.1007/BF02983055 [DOI] [Google Scholar]
- Winge O., 1922. A peculiar mode of inheritance and its cytological explanation. J. Genet. 12: 137–144. 10.1007/BF02983077 [DOI] [Google Scholar]
- Wright A. E., Darolti I., Bloch N. I., Oostra V., Sandkam B. et al. , 2017. Convergent recombination suppression suggests role of sexual selection in guppy sex chromosome formation. Nat. Commun. 8: 14251 10.1038/ncomms14251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano A., Guyomard R., Nicol B., Jouanno E., Quillet E. et al. , 2012. An immune-related gene evolved into the master sex-determining gene in Rainbow trout, Oncorhynchus mykiss. Curr. Biol. 22: 1423–1428. 10.1016/j.cub.2012.05.045 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supplemental Table S1 contains the sequences of primers for 8 polymorphic markers on LG12. Supplemental Figure S1 contains the assembled genomic sequences of GADD45G-like gene locus of P. reticulata. Supplemental material available at FigShare: https://doi.org/10.25387/g3.9841160.