Abstract
Introduction
Chronic kidney disease of uncertain etiology (CKDu), an emerging chronic kidney disease (CKD) subtype, contributes to significant morbidity and mortality in certain tropical countries. Although several indicators of CKDu have been previously suggested, sensitive and specific tests to detect early disease or predict disease progression are currently unavailable. This study focused on evaluating 8 renal urinary markers, namely neutrophil gelatinase-associated lipocalin (NGAL), Kidney Injury Molecule-1 (KIM1), cystatin C (CST3), beta 2 microglobulin (B2M), osteopontin (OPN), alpha 1 microglobulin (A1M), tissue inhibitor of metalloproteinase 1 (TIMP1), and retinol binding protein 4 (RBP4), with the hypothesis that these have distinct expression patterns in patients with CKDu.
Methods
A cross-sectional study was conducted with 5 study groups comprising subjects from CKDu, endemic CKD, nonendemic CKD, and endemic healthy and nonendemic healthy controls. The urinary levels of the 8 selected renal biomarkers were quantified using multiplex biomarker assay, and the data were subjected to systematic analysis using logistic regression algorithm aiming to extract the best marker combination that could distinctly identify the disease groups noninvasively from the healthy controls.
Results
A 3-marker signature panel comprising A1M, KIM1, and RBP4 was identified to represent the best minimum marker combination for differentiating all CKD categories, including CKDu, from healthy controls with an overall sensitivity of ≥0.867 and specificity ≥0.765. The marker combination comprising OPN, KIM1, and RBP4 showed high predictive performance for distinguishing patients with CKDu from patients with CKD with both sensitivity and specificity ≥0.93, which was superior to any existing noninvasive indicator.
Conclusion
In all, our systematic evaluation of urinary markers previously linked to CKD, in general, allowed identification of exclusive marker panel combination for early diagnosis and confirmation of CKDu.
Keywords: chronic kidney disease of uncertain etiology, early diagnosis, urinary biomarkers
CKD, also called chronic kidney failure, is characterized by structural and functional abnormalities of the kidney that often progress to end-stage renal failure. Recent epidemiological studies have suggested that CKD is more prevalent in Asian countries than Western countries.1, 2 Although the global prevalence of CKD is on the rise due to changes in lifestyle, another subgroup of CKD is emerging among some agricultural communities. Interestingly, this environmental interstitial nephropathy was initially identified as localized outbreaks of CKD without evidence of etiology in Sri Lanka, India, Nicaragua, Costa Rica, and Central American states.3, 4, 5, 6, 7 Among them, Sri Lanka reports the highest occurrence of CKDu, in the rural dry zone where extensive farming is carried out. Risk factors of this enigmatic disease could be related to environmental toxin exposure, and there is evidence that shows the association between CKDu and dehydration-prone behavior, smoking, drinking alcohol, and chewing betel.8, 9, 10
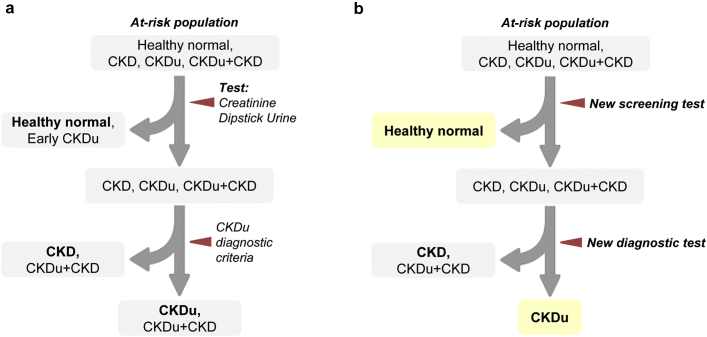
CKDu is primarily an interstitial disease typically associated with tubular atrophy, interstitial mononuclear cell infiltration, and interstitial fibrosis leading to renal impairment, often with no or minimal proteinuria in early stages.11 Although the diagnosis of CKD is straightforward and well-established, identification of CKDu is based on exclusion of known causes of CKD. Figure 1a depicts the summary of the current screening pipeline. In the absence of sensitive and specific confirmatory markers to reliably identify CKDu, renal biopsy remains as the gold standard of diagnosis. Renal biopsy is not only invasive but also applicable to a minority of patients who have sizeable kidneys at the time of diagnosis. It is evident that the current diagnostic process lacks capability of identifying creatinine normal nonproteinuric CKDu, and also diagnosing CKDu on top of preexisting renal impairment or proteinuria due to known etiologies, like diabetes and hypertension. Thus, the epidemiological data obtained through this screening procedure are likely to be erroneous and misleading. Altogether, it is evident that a more efficient, sensitive, and specific diagnostic procedure is needed for early detection and to confirm the diagnosis of CKDu.
Figure 1.
Patient screening of chronic kidney disease (CKD) of uncertain etiology (CKDu) high-risk population. (a) Current screening and diagnostic process. (b) Ideal screening and diagnostic processes by 2-step screening strategy.
The urine dipstick method is an inexpensive and rapid point-of-care diagnostic test that is widely used in CKD screening.12 Another widely used test relies on elevated albumin creatinine ratio for identifying CKD. These tests are highly sensitive in the early stages of proteinuric CKD, and are cost-effective for community screening.13 However, the applicability of these tests is limited in diagnosing early CKDu, owing to the minimally proteinuric nature of this disease.14 Another marker, serum creatinine is a routine test for assessing renal function. Of note, previous work has shown that serum creatinine levels increase only after 40% to 50% renal parenchymal damage and therefore render both serum creatinine, and the estimating equations (estimated glomerular filtration rate) on which it is based, a late marker of renal damage.15 This suggests that the serum creatinine test may not be ideal for detection of early kidney damage. In comparison with serum creatinine and albumin creatinine ratio, serum CST3 has been reported as a good functional marker for CKDu.14 Urine is considered an excellent source for biomarkers, as markers resulting from pathophysiological processes in the kidney are directly “deposited” in the urine. Notably, a study reported, that urinary biomarkers, namely KIM1 and NGAL, are useful in determining early CKDu cases.16 Even though urinary tubular markers are attractive alternatives to traditional tests, their applicability in CKDu remains uncertain. Sensitivity of an ideal screening marker should be superior to urine albumin creatinine ratio and creatinine in detection of all forms of CKD, including CKDu. Specificity of diagnostic marker must be superior to current diagnostic criteria and comparable to a renal biopsy. A confirmatory marker could be either the etiological factor itself or must be generated by specific type or site of the insult. To this end, we investigated the performance of selected renal biomarkers in urine of adult patients with CKDu, aiming to develop an ideal screening procedure, as depicted in Figure 1b. In particular, we (i) directly compared the performance of creatinine, NGAL, KIM1, CST3, B2M, OPN, A1M, TIMP1, and RBP4 in urine of CKDu, (ii) compared them with control groups including endemic and nonendemic area, and (iii) evaluated combinations of renal biomarkers for accurate identification of CKDu.
Methods
Patient Information and Study Design
The patient group included 75 cases of definite CKDu, and 82 and 85 patients with CKD, each from endemic (ECKD) and nonendemic (NECKD) areas, respectively. A total of 79 and 85 dipstick-negative individuals with normal blood pressure, blood sugar, and creatinine levels from both the endemic (EC) and nonendemic (NEC) areas, respectively, were recruited as healthy control groups for this study. All the study subjects were enrolled after obtaining written informed consent. The ethical clearance was granted by the Ethical Review Committee of the Faculty of Medicine, University of Peradeniya (2016/EC/28). The study was carried out from June 2015 to March 2018. Subjects were defined as CKDu based on 3 criteria: (i) living in an already identified CKDu endemic area for at least 5 years, (ii) histological evidence compatible with already described features of CKDu, and (iii) known causes of CKD excluded. Patients with CKD with known etiologies (diabetes, hypertension, glomerular diseases) were recruited from both ECKD and NECKD areas. Age- and sex- matched apparently healthy individuals with no history of hypertension, diabetes, or renal diseases were recruited as controls. Nonendemic areas are those where CKDu has not been reported previously in either population screening or in surveillance programs (wet hilly middle zone of the country) (Supplementary Figure S1). In this study, we recruited NEC from “Mandaramnuwara” area in the wet middle zone farming area. CKDu, ECKD, and EC were enrolled from Girandurukotte and Wilgamuwa within the dry zone agricultural area, where a high prevalence of CKDu has been confirmed.
Clinical Sample Collection Protocol
A total of 20 ml of randomly voided, midstream urine samples were collected into a sterile container for dipstick test (for proteinuria, glucosuria), urinary biomarkers (KIM1, NGAL, B2M, A1M, CST3, OPN, and RBP4), and urine creatinine analysis. For biomarker analysis, urine samples were immediately (within half an hour) centrifuged at 3000 rpm (2016g) for 10 minutes and supernatant was separated into cryovial to store at −80 °C until analysis. Blood samples for serum creatinine were collected from peripheral veins to plain tubes and separated immediately by centrifugation at 3000 rpm for 10 minutes.
Study Sample Analysis
Serum and urine creatinine were measured in Jaffe’s method using Indiko Plus biochemical analyzer (Thermo Scientific, Vantaa, Finland). Sulfosalicylate test for urine protein was used in the initial screening of healthy controls. Urinary biomarkers were measured using Luminex MAGPIX machine (EMD Millipore Corp., Billerica, MA) using xMAP technology. Reference upper values for normal serum creatinine for male individuals was 113 μmol/L and for female individuals 96 μmol/L. Nil and trace results were taken as normal urine protein, whereas +, ++, +++, or > +++ were taken as abnormal urine protein.
MILLIPLEX Assay Analysis
Luminex xMAP technology has been previously reported for screening kidney markers.17, 18 This technology uses analyte-specific capture antibodies conjugated to xMAP beads, enabling multivariate analysis of several diseases, using minimal sample volumes. Customized biomarker bead assay kits (CERTKD-05 and HKI6/MAG-99-K-02), standards, and quality controls (QCs) were purchased from Merck Millipore (Burlington, MA). Samples were prepared with duplicates of blank (1), standards (6), QCs (2), and patients (39) on a 96-well microtiter plate, according to the manufacturer's instructions. Urine sample dilutions were 1:2 and 1:4 for CERTKD-05 and HKI6/MAG-99-K-02, respectively. Samples were analyzed on a Luminex (Austin, TX) MAGPIX analyzer. The biomarker concentrations in the samples were determined from the 5-parameter logistic fit standard curves created in nCal package in R.
Precision was evaluated with the coefficient of variation. The intra-assay percent coefficient of variation was generated from the mean of the percent coefficient of variation from 2 different concentrations of 1 experiment. Accuracy is represented by the percentage recovery for each standard concentration; acceptable recovery is within 70% to 130% the lower limit of quantification, and upper limit of quantification defines a quantitative range whereby values can be estimated within an accuracy of 80% to 120% recovery and precision below 20% intra-assay percent coefficient of variation. To establish batch to batch repeatability between experiments 1 and 2, QC materials provided by the manufacturer were measured in duplicate: QC1, a standard of known concentration within the lower concentration range, and QC2, a standard of known concentration within the higher concentration range. A concentration observed within the expected range was considered 100% recovery. Data acquisition was done using XPONENT software package.
Statistical Analysis
All statistical calculations were done using the Statistical Package for the Social Sciences (SPSS) version 23 (IBM, Armonk, NY). Because the data were not normal, independent samples Mann-Whitney U test was used to find the significance between each group (Supplementary Table S1). Urine creatinine concentration was used to normalize biomarker measurements to account for the influence of urinary dilution on biomarker concentrations.19
Machine Learning
Machine learning analysis was carried out to derive candidate biomarker signature panels reported in this study. The sample group was split into a training (80%) and validation (20%) sets ensuring similar fractional distribution of test cases and controls within the sets. For the disease versus control comparisons, all the different disease categories were analyzed against NEC. For stratification of CKDu from other CKD, analyses were made using NECKD as the base control group. Based on the 8 selected protein biomarkers, logistic regression models were trained with 10-fold cross-validation. In this procedure, the training set was split into 10 random partitions, and nine-tenths of the data was used for training and one-tenth of the data was reserved for model evaluation. The whole process was iterated 10 times by randomly partitioning the training set differently during every fold and the predictive performance of the model evaluated using the subjects left-out with each iteration. In addition, the process of model training with logistic regression and cross-validation was repeated 10 times with different random partitions of the training set into 10-folds, to evaluate the stability of the model. The final model predicted with high accuracy over the 10 rounds of 10-fold cross-validation was chosen to be tested on validation set (20%). The logistic regression was performed with stepwise variable selection to iteratively remove the least contributing predictors, in this case proteins, from the final model. By doing this, we retained only those biomarkers with predictive capability for the particular comparison. In addition, to derive minimalistic signatures of protein markers, the variable importance in the final model was assessed and all possible combinations of up to 3 were systematically trained using logistic regression models with cross-validation and evaluated on the validation set. The minimalistic signature marker showing high performance and accuracy was finally chosen. Finally, the model with the highest accuracy was tested on the validation set and receiver operating characteristic (ROC) analysis was performed to determine the accuracy, sensitivity, and specificity of the final model and visualized as performance curved. The caret package in R was used for all machine learning analyses and model evaluation, and the pROC package in R was used for all ROC analyses and inference.
Results
Characterization of Renal Protein Biomarkers in Urine
Healthy controls and patients recruited in this study belonged to both endemic and nonendemic regions, and thus ideally represented all possible categories of CKD disease and healthy groups, including that represented in endemic areas. The baseline characteristics of all the study individuals are summarized in Table 1 and the stage distribution of patients in CKDu and CKD groups is shown in Table 2.
Table 1.
Baseline characteristics of all the study individuals
| Characteristic/outcome | CKDu (n = 75) | EC (n = 79) | NEC (n = 85) | ECKD (n = 82) | NECKD (n = 85) |
|---|---|---|---|---|---|
| Male, n (%) | 61 (81.3) | 55 (69.6) | 46 (54.1) | 38 (46.3) | 42 (49.4) |
| Female, n (%) | 14 (18.7) | 24 (30.4) | 39 (45.9) | 44 (53.7) | 43 (50.6) |
| Age, yr (SD) | 51 ± 10 | 38 ± 10 | 46 ± 12 | 59 ± 10 | 49 ± 13 |
| eGFR, >60, n (%) | 32 (42.7) | 79 (100) | 84 (98.8) | 15 (18.3) | 26 (30.6) |
| eGFR, <60, n (%) | 43 (57.3) | 1 (1.2) | 67 (81.7) | 59 (69.4) | |
| Comorbidity: | |||||
| DM, n (%)a | 5 (6.7) | 0 | 0 | 19 (23.2) | 37 (43.5) |
| HT, n (%)a | 26 (34.7) | 1 (1.3) | 0 | 60 (73.2) | 56 (65.9) |
CKDu, chronic kidney disease of uncertain etiology; DM, diabetes mellitus; EC, endemic control; eGFR, estimated glomerular filtration rate; ECKD, endemic chronic kidney disease; HT, hypertension; NEC, nonendemic control; NECKD, nonendemic chronic kidney disease.
These events were identified after the diagnosis of CKDu.
Table 2.
Stage distribution of patients in CKDu, ECKD, and NECKD groups
CKDu, chronic kidney disease of uncertain etiology; ECKD, endemic chronic kidney disease; NECKD, nonendemic chronic kidney disease.
These patients were limited and could not enroll more patients in stage 5 due to dialysis.
The urinary levels of 8 selected renal biomarkers, KIM1, NGAL, B2M, A1M, CST3, OPN, TIMP1, and RBP4, were quantified using Milliplex multiplexing protein assay. In addition, urine and serum creatinine levels were measured in every sample, and protein measurements were adjusted to urine creatinine level. The protein measurements after adjustment with creatinine are summarized in Table 3.
Table 3.
The median and IQR of biomarker measurements before and after adjustment with creatinine
| Biomarker |
CKDu |
EC |
NEC |
ECKD |
NECKD |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cr adjusted | Median | IQR | Median | IQR | Median | IQR | Median | IQR | Median | IQR |
| A1M, ng/g-Cr | 870.72 | 6473.07 | 78.5 | 92.52 | 53.65 | 58.39 | 617.91 | 1249.43 | 427.12 | 1272.28 |
| B2M, ng/g-Cr | 382.62 | 1518.96 | 42.64 | 51.05 | 48.12 | 69.62 | 730.18 | 1130.82 | 968.77 | 1709.75 |
| CST3, ng/g-Cr | 3.97 | 16.75 | 0.86 | 1.45 | 1.11 | 1.05 | 8.17 | 57 | 9.15 | 49.49 |
| NGAL, ng/g-Cr | 20.16 | 35.41 | 2.85 | 3.92 | 3.15 | 6.26 | 23.62 | 68.88 | 12.4 | 41.89 |
| OPN, ng/g-Cr | 67.9 | 98.61 | 38.18 | 42.82 | 50.15 | 39.66 | 25.95 | 31.81 | 39.35 | 51 |
| RBP4, ng/g-Cr | 478.01 | 550.93 | 169.21 | 261.9 | 187.72 | 200.99 | 558.34 | 440.56 | 804.69 | 864.18 |
| KIM 1, ng/g-Cr | 0.05 | 0.08 | 0.06 | 0.05 | 0.05 | 0.06 | 0.09 | 0.13 | 0.09 | 0.15 |
| TIMP 1, ng/g-Cr | 0.61 | 1.44 | 0.31 | 0.42 | 0.41 | 0.79 | 1.12 | 2.66 | 1.25 | 2.77 |
A1M, alpha 1 microglobulin; B2M, beta 2 microglobulin; CKDu, chronic kidney disease of uncertain etiology; Cr, creatinine; CST3, cystatin C; EC, endemic control; ECKD, endemic chronic kidney disease; IQR, interquartile range; KIM1, kidney injury molecule 1; NEC, nonendemic control; NECKD, nonendemic chronic kidney disease; NGAL, neutrophil gelatinase-associated lipocalin; OPN, osteopontin; RBP4, retinol binding protein 4; TIMP1, tissue inhibitor of metalloproteinase 1.
Derivation of CKD Urinary Markers Distinguishing Disease and Control Population
One of the main aims of this study was to identify a possible marker or markers from existing renal disease markers that can differentiate disease and healthy populations. For this, we assessed the distribution of protein markers across different categories. We observed that some markers, such as A1M and OPN, were specifically upregulated only in the CKDu group, whereas others were elevated across all disease groups (Supplementary Figure S2). Interestingly, most markers showed differential levels among the categories, especially between the CKDu and control populations (Supplementary Table S1). This observation motivated us to explore the diagnostic potential of the renal markers for stratifying patients with CKD. For this purpose, we first used ROC analysis to evaluate the performance of individual protein markers in distinguishing disease and healthy populations. ROC analysis was performed for categorical comparison among all 3 disease groups with the control. Although our control group was represented by both endemic and nonendemic regions, only the NEC group was considered for this analysis. Currently, the identification of early CKDu is challenging and we postulate that despite our initial screening, some of the patients with early CKDu may be represented within the EC group. For this reason, and to perform robust comparisons between disease and control groups, we excluded the EC group in our analysis. From the evaluation of the markers across all comparisons (Table 4), we identified A1M as the single best candidate marker with the highest performance. The areas under the curve (AUCs) were comparably high across all 3 comparisons, with the highest sensitivity (92%) achieved for CKDu. CST3 also performed well in differentiating the disease from the control group. Other markers such as B2M showed higher predictive power in ECKD and NECKD categories, but displayed poor sensitivity for the CKDu group.
Table 4.
Diagnostic performance of candidate markers in distinguishing CKDu, ECKD, and NECKD from NEC
| Parameter | AUC (95% CI) | % (95% CI) |
P value | |
|---|---|---|---|---|
| Sensitivity | Specificity | |||
| CKDu vs. NEC | ||||
| A1M | 0.914 (0.863–0.966) | 92.0 (83.2–96.5) | 84.7 (75.4–90.9) | <0.0001 |
| B2M | 0.696 (0.604–0.787) | 53.3 (42.2–64.2) | 92.9 (85.1–97.0) | <0.0001 |
| CST3 | 0.817 (0.749–0.885) | 69.3 (58.1–78.6) | 84.7 (75.4–90.9) | <0.0001 |
| NGAL | 0.827 (0.759–0.894) | 82.7 (72.4–89.7) | 72.9 (62.6–81.2) | <0.0001 |
| OPN | 0.585 (0.486–0.684) | 44.0 (33.3–55.3) | 84.7 (75.4–90.9) | 0.093 |
| RBP4 | 0.735 (0.650–0.819) | 81.3 (70.9–88.6) | 63.5 (52.9–73.0) | <0.0001 |
| KIM1 | 0.520 (0.422–0.617) | 38.7 (28.5–50.0) | 72.9 (62.6–81.2) | 0.694 |
| TIMP1 | 0.620 (0.527–0.713) | 40.0 (29.7–51.3) | 84.7 (75.4–90.9) | 0.011 |
| ECKD vs. NEC | ||||
| A1M | 0.913 (0.867–0.959) | 87.8 (78.7–93.4) | 84.7 (75.4–90.9) | <0.0001 |
| B2M | 0.832 (0.763–0.901) | 74.4 (63.9–82.6) | 89.4 (80.8–94.5) | <0.0001 |
| CST3 | 0.843 (0.779–0.906) | 72.0 (61.3–80.5) | 90.6 (82.2–95.3) | <0.0001 |
| NGAL | 0.797 (0.728–0.865) | 75.6 (65.2–83.6) | 75.3 (65.1–83.3) | <0.0001 |
| OPN | 0.756 (0.681–0.832) | 48.8 (38.3–59.4) | 95.3 (88.0–98.5) | <0.0001 |
| RBP4 | 0.804 (0.731–0.877) | 85.4 (75.9–91.5) | 74.1 (63.8–82.3) | <0.0001 |
| KIM1 | 0.684 (0.602–0.766) | 80.5 (70.5–87.7) | 50.6 (40.2–60.9) | <0.0001 |
| TIMP1 | 0.738 (0.664–0.813) | 54.9 (44.1–65.2) | 84.7 (75.4–90.9) | <0.0001 |
| NECKD vs. NEC | ||||
| A1M | 0.891 (0.843–0.938) | 77.6 (67.6–85.2) | 91.8 (83.6–96.2) | <0.0001 |
| B2M | 0.852 (0.795–0.909) | 77.6 (67.6–85.2) | 82.4 (72.7–89.1) | <0.0001 |
| CST3 | 0.851 (0.791–0.910) | 70.6 (60.1–79.2) | 90.6 (82.2–95.3) | <0.0001 |
| NGAL | 0.767 (0.697–0.837) | 69.4 (58.9–78.2) | 75.3 (65.1–83.3) | <0.0001 |
| OPN | 0.586 (0.499–0.674) | 54.1 (43.6–64.3) | 69.4 (58.9–78.2) | 0.053 |
| RBP4 | 0.862 (0.805–0.919) | 91.8 (83.6–96.2) | 76.5 (66.3–84.2) | <0.0001 |
| KIM1 | 0.716 (0.639–0.792) | 78.8 (68.9–86.2) | 55.3 (44.7–65.4) | <0.0001 |
| TIMP1 | 0.747 (0.674–0.820) | 55.3 (44.7–65.4) | 84.7 (75.4–90.9) | <0.0001 |
A1M, alpha 1 microglobulin; AUC, area under the curve; B2M, beta 2 microglobulin; CI, confidence interval; CKDu, chronic kidney disease of uncertain etiology; CST3, cystatin C; ECKD, endemic chronic kidney disease; KIM1, kidney injury molecule 1; NEC, nonendemic control; NECKD, nonendemic chronic kidney disease; NGAL, neutrophil gelatinase-associated lipocalin; OPN, osteopontin; RBP4, retinol binding protein 4; TIMP1, tissue inhibitor of metalloproteinase 1.
A biomarker signature panel, representing diverse CKD scenarios, is ideal instead of a single marker to improve the efficiency of CKD diagnosis in community screening. Toward this goal, logistic regression models were trained using the most discriminated proteins that can stratify the disease from the control group for all 3 comparisons as described in the methods section. Implementing a stepwise regression on the training set with 10-fold cross-validation, we derived a high-performing diagnostic signature containing the 5 most discriminant proteins that can best differentiate CKDu from controls. The best performing model inferred from the training set included A1M, OPN, NGAL, KIM1, and RBP4 as the desired marker panel to differentiate CKDu from control with high accuracy (training model AUC = 0.934). Finally, on evaluating this biomarker signature panel on the validation set, we observed that the model performed well with high sensitivity and specificity (AUC = 0.922; Supplementary Figure S3). On repeating the described procedure for the other disease groups NECKD and ECKD against the healthy controls, we found that the 5-protein biomarker signature performs equally well in the validation set with an AUC of 0.903 for NECKD and 0.890 for ECKD against the controls (Supplementary Figure S3). On assessing the performance of individual proteins against the marker panel, we observed that the combination panel displayed the highest area under the ROC curves for both NECKD and CKDu disease groups compared with any individual protein biomarker included within the panel. However, for ECKD subjects, the protein biomarker signature was comparable only to the predictive ability of single marker A1M in differentiating from healthy controls. To rule out the influence of possible endemic factors, we also assessed the predictive performance of the biomarker signature in distinguishing the EC group with endemic factors from the NEC group, and found that the combination panel had no predictive power in distinguishing these 2 groups, thus affirming its specificity in stratifying the disease from the control groups (Supplementary Figure S4).
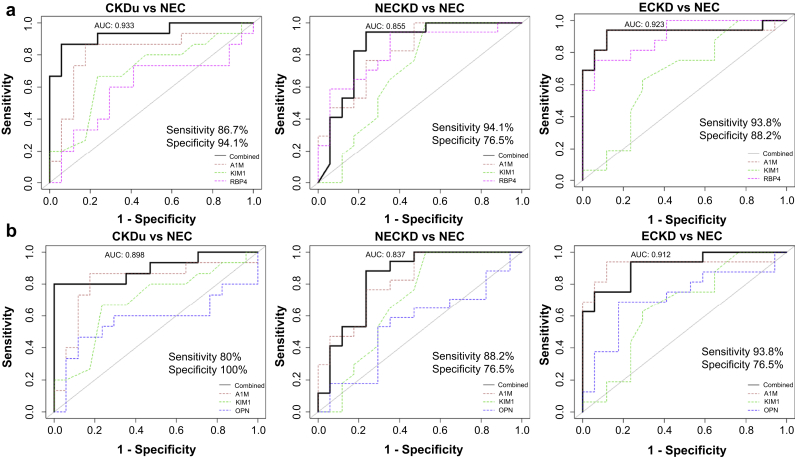
Although the 5-marker signature showed excellent capability in differentiating CKD from healthy subjects, in an actual clinical setting, screening for fewer markers with high discriminating power would be practical. Thus, we assessed the variable importance of the chosen 5 markers in the previously described panel using ranking, and enumerated combinations up to 3 proteins to derive at a model performing equally well as the 5-protein signature using logistic regression on the training set coupled with 10 iterations of 10-fold cross-validation. Through this analysis, we identified 2 different marker combinations that can discriminate the disease group from the healthy controls (Figure 2). Among them, the A1M+KIM1+RBP4 combination showed higher sensitivity than serum creatinine for all disease (CKDu, NECKD, and ECKD) against the NEC healthy control group and displayed a high AUC of 0.903 on an average across all comparisons in the validation set (Figure 2 and Supplementary Table S2). Notably, the predictive performance of this marker combination dropped on comparing CKDu against the ECKD (AUC = 0.525) or the NECKD (AUC = 0.592) group (Supplementary Figure S5A). This suggests that the marker signature has no power in differentiating CKDu from CKD of other known causes but is suited for stratifying CKD disease from healthy controls, regardless of their origin, thus demonstrating its selectivity for the disease groups. The second marker combination A1M+KIM1+OPN also showed comparable power in discriminating the disease from healthy subjects, however with lower sensitivity than A1M+KIM1+RBP4 (Figure 2). However, this marker signature displayed high predictive ability in differentiating between the disease groups (CKDu vs. NECKD and CKDu vs. ECKD) unlike the A1M+KIM1+RBP4 marker panel (Supplementary Figure S5B), thus limiting its potential in a clinical scenario. In addition, we tested the performance of 2 marker combinations among the chosen signatures (Supplementary Table S2). Nevertheless, dropping one marker from the 3-marker signature compromised the sensitivity and specificity, indicating that 3-marker combination is required for the best performance across all CKD disease groups.
Figure 2.
Best 3-marker signature to differentiate chronic kidney disease (CKD) groups from absolute healthy controls (NEC). (a) Predictive performances of A1M+KIM1+RBP4 and (b) A1M+KIM1+OPN as a combined panel and as individual protein markers in distinguishing CKD disease groups from healthy controls are shown. Area under the curve (AUC) reported based on the validation set. A1M, alpha 1 microglobulin; CKDu, chronic kidney disease of uncertain etiology; ECKD, endemic chronic kidney disease; KIM1, kidney injury molecule-1; NECKD, nonendemic chronic kidney disease; OPN, osteopontin; RBP4, retinol binding protein 4.
Derivation of CKD Urinary Markers Distinguishing CKDu and CKD Population
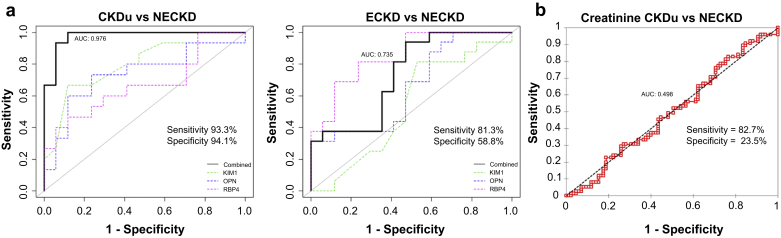
Our second aim was to identify possible marker or a marker panel from existing renal markers that can differentiate CKDu and CKD of other known causes. ECKD and NECKD together would also include the actual endemic population, and thus provide the ideal CKD background for assessing the predictive ability for CKDu. However, ECKD subjects may manifest dual pathologies as CKD+CKDu, and we posit that including such subjects for the analysis would limit identification of true markers capable of confirming CKDu. Despite compromising on the endemic factors by excluding ECKD, at this stage we were more determined to derive the marker panel with the best diagnostic performance in ascertaining CKDu from true CKD. We first evaluated if single protein biomarkers have distinguishing potential to identify CKDu from NECKD. However, the predictive ability was not promising with the observed AUCs in the range 0.546 to 0.721 (Table 5), precluding the ability of these single markers to reliably identify CKDu. Thus, in a procedure similar to the one described earlier, we trained logistic regression models with 10-fold cross-validation coupled with stepwise variable selection on the training set, which represented 80% of the subjects from CKDu and NECKD groups. A panel containing 6 biomarkers with the exclusion of B2M and TIMP1 was inferred from the training set with high accuracy to differentiate CKDu from NECKD (Supplementary Figure S6). To derive a minimalistic marker signature, we assessed combinations of up to 3 markers that performed equally well without much compromise on the sensitivity, specificity, and accuracy. We also required that the marker signature display high predictive performance only against the NECKD disease group but not against the control group. With such a constraint, we identified a minimalistic panel consisting of OPN+KIM1+RBP4 with excellent predicting ability (AUC = 0.976) on the validation set complemented with high accuracy (93.7%), sensitivity (93.3%), and specificity (94.1%) for distinguishing CKDu from NECKD subjects (Figure 3a). The predictive performance of this model suffered a setback (AUC = 0.69) when stratifying patients with CKDu from healthy controls (NEC) with the specificity particularly dropping to 58.8% (Supplementary Figure S7 and Supplementary Table S3), thus affirming its selectivity for differentiating between the disease groups. Although both ECKD and CKDu have endemic factor influence, the 3-marker signature is highly specific for CKDu while its performance dropped (AUC = 0.735) on comparing ECKD and NECKD groups with a huge decrease in specificity by almost 35% (Figure 3). This demonstrates that the marker panel is selective for CKDu and is not predictive based on endemic factors only. It is worthy to note that although serum creatinine still proves useful for differentiating CKD from healthy controls (Supplementary Figure S8), its predictive ability in distinguishing CKDu from other CKD (NECKD) is poor (AUC = 0.498) (Figure 3b). This accentuates the need for developing a high-performing new marker signature that can accurately identify subjects with CKDu for disease stratification and proper patient management, thus emphasizing on the usefulness of our proposed 3-marker signature.
Table 5.
Diagnostic performance of candidate markers in distinguishing CKDu, and ECKD from NECKD
| Parameter | AUC (95% CI) | % (95% CI) |
P value | |
|---|---|---|---|---|
| Sensitivity | Specificity | |||
| CKDu vs. NECKD | ||||
| A1M | 0.621 (0.527–0.716) | 36.0 (26.1–47.3) | 97.6 (91.2–99.8) | 0.012 |
| B2M | 0.624 (0.53–0.717) | 45.3 (34.6–56.6) | 77.6 (67.6–85.2) | 0.009 |
| CST3 | 0.585 (0.49–0.679) | 80.0 (69.4–87.5) | 36.5 (27.0–47.1) | 0.078 |
| NGAL | 0.546 (0.452–0.64) | 93.3 (84.9–97.4) | 22.4 (14.8–32.4) | 0.333 |
| OPN | 0.627 (0.533–0.721) | 48.0 (37.1–59.1) | 78.8 (68.9–86.2) | 0.008 |
| RBP4 | 0.688 (0.602–0.775) | 58.7 (47.4–69.1) | 70.6 (60.1–79.2) | <0.0001 |
| KIM1 | 0.721 (0.637–0.805) | 44.0 (33.3–55.3) | 91.8 (83.6–96.2) | <0.0001 |
| TIMP1 | 0.633 (0.540–0.725) | 42.7 (32.1–54.0) | 80.0 (70.1–87.2) | 0.005 |
| ECKD vs. NECKD | ||||
| A1M | 0.527 (0.437–0.616) | 92.7 (84.6–96.8) | 17.6 (10.9–27.3) | 0.562 |
| B2M | 0.561 (0.472–0.650) | 75.6 (65.2–83.6) | 44.7 (34.6–55.3) | 0.176 |
| CST3 | 0.507 (0.417–0.597) | 37.8 (28.1–48.6) | 71.8 (61.3–80.2) | 0.884 |
| NGAL | 0.557 (0.467–0.646) | 58.5 (47.7–68.6) | 58.8 (48.2–68.7) | 0.216 |
| OPN | 0.677 (0.595–0.759) | 41.5 (31.4–52.3) | 89.4 (80.8–94.5) | <0.0001 |
| RBP4 | 0.672 (0.590–0.754) | 89.0 (80.2–94.3) | 42.4 (32.4–53.0) | <0.0001 |
| KIM1 | 0.538 (0.448–0.628) | 62.2 (51.4–71.9) | 47.1 (36.8–57.6) | 0.407 |
| TIMP1 | 0.511 (0.422–0.601) | 30.5 (21.6–41.2) | 78.8 (68.9–86.2) | 0.804 |
A1M, alpha 1 microglobulin; AUC, area under the curve; B2M, beta 2 microglobulin; CI, confidence interval; CKDu, chronic kidney disease of uncertain etiology; CST3, cystatin C; ECKD, endemic chronic kidney disease; KIM1, kidney injury molecule 1; NEC, nonendemic control; NECKD, nonendemic chronic kidney disease; NGAL, neutrophil gelatinase-associated lipocalin; OPN, osteopontin; RBP4, retinol binding protein 4; TIMP1, tissue inhibitor of metalloproteinase 1.
Figure 3.
Best 3-marker interdisease signature to distinguish chronic kidney disease (CKD) of uncertain etiology (CKDu) from CKD. (a) Receiver operating characteristic analysis of 3-marker combination panel OPN+KIM1+RBP4 in distinguishing CKDu from nonendemic chronic kidney disease (NECKD) and endemic chronic kidney disease (ECKD) are shown along with the performance of individual protein markers. Area under the curve (AUC) reported based on the validation set. (b) Performance of serum creatinine in distinguishing CKDu from NECKD is shown. KIM1, kidney injury molecule-1; OPN, osteopontin; RBP4, retinol binding protein 4.
Discussion
In this study, we took advantage of the renal biomarkers previously reported for CKD and assessed their predictive performance in the context of CKDu. Through a machine leaning approach, we identified potential marker combinations that can be used to distinguish the CKD population from healthy controls and also to stratify patients with CKDu from all subjects diagnosed with CKD.
The primary aim of this study was to distinctly identify the CKDu group using a highly predictive biomarker signature. Although it may be straightforward to establish a unique biomarker panel that can exclusively identify CKDu from rest of the categories (irrespective of disease or control status), this is not rational or ethical in population screening. Hence, we propose a 2-step screening strategy wherein the disease groups are reliably identified from the healthy controls in the first step, and among the patients diagnosed with CKD, those who belong to CKDu are identified in the second step (Figure 1b). Because the diagnostic signatures we propose in this study are based on urine biomarkers, the screening process is noninvasive and economical to be implemented in at-risk populations.
Currently, the identification of CKDu relies on biomarkers used to diagnose CKD in general, which includes serum creatinine and urinary protein. Recently, several surrogate markers have been reported to perform better in diagnosing CKD and CKDu. For example, CST3 is reported to better predict the clinical outcomes of CKD than creatinine.20 More recently, CST3 was also reported to show a higher efficacy in detecting CKDu in endemic regions.14 On screening a set of 8 selected biomarkers, we found that not all markers perform uniformly well across all groups. A1M marker stood out as the single strong candidate marker that was highly specific in identifying CKD from healthy controls across all disease comparisons. CKDu characteristically affects the tubulointerstitium and notably, A1M is an indicator of renal tubular function. In comparison with using a single marker, our analysis suggested higher predictive performance of combination biomarker signature consisting of A1M+KIM1+RBP4 in accurately identifying the disease groups, particularly CKDu and NECKD. Serum RBP4 level has been associated with renal tubular dysfunction and CKD, and KIM1 is a marker for renal proximal tubular damage and predictor of kidney function decline.21, 22 Although for ECKD, the predictive ability of the marker signature was comparable only to that of A1M alone, we propose that the marker signature may still prove to be a better option for making diagnostic decisions than reliance on a single marker.
Although current disease management holds promise for identifying patients affected with CKD from healthy individuals, distinctly diagnosing CKD disease groups still remains a major challenge. Although serum creatinine shows excellent performance in identifying the disease groups irrespective of their etiology, it offers no predictive ability for interdisease comparisons. Also, of the 8 biomarkers screened in this study, none of them showed selective advantage in identifying CKDu from other causes of CKD. Strikingly, a 3-marker signature constituting OPN+KIM1+RBP4 accurately predicted CKDu with high performance from a CKD background. Of note, this marker panel was found to be uniquely predictive for CKDu and not for ECKD. CKDu and ECKD both occur in the endemic region, and because of the unidentified etiology, patients with CKDu are often misdiagnosed as diabetic or with hypertensive kidney disease if they have concomitant diseases. The novel biomarker signature thus holds great potential in improving clinical decisions, leading to better patient management and clinical care.
Here we report exclusive marker combinations that are useful in both disease identification and stratification. Even though most of these markers have been linked previously to CKD in general, by systematically evaluating them, we show their improved diagnostic potential in efficiently identifying those patients with CKDu. This work was done on an exploratory basis using smaller sample groups to uncover marker signatures to distinctly identify CKD of any cause from healthy individuals and also differentiate CKDu from CKD of known causes. Although the reported biomarker signatures hold promise, the performance of these urinary markers will need to be tested on a larger independent validation cohort to prove their clinical utility and applicability in CKD diagnosis. In deriving the optimal biomarker signatures, only the healthy and the disease groups representing the nonendemic regions (NEC and NECKD) were considered for comparisons. Although inclusion of EC along with NEC would be ideal for deriving a disease-specific signature against healthy controls, the possibility of at-risk patients with early CKD among the EC controls cannot be ruled out. Also, ECKD patients invariably display dual pathologies of CKD and CKDu, which is not well defined in most cases. Thus, to limit confounding factors from such a heterogeneous population toward biomarker discovery, our study at this stage focused only on homogeneous patient groups. The proposed biomarker signatures should be tested on well-defined patient cohorts, particularly from endemic regions, to prove its clinical usefulness. Also, apart from the existing biomarkers proposed in this study, the complexity of CKDu as a disease with unknown etiology and severe manifestations, calls for the need to explore new biomarkers to accurately define various groups and various stages of the disease.
Disclosure
All the authors declared no competing interests.
Acknowledgments
The authors are thankful to Dr. Lishantha Gunarathna and the staff of the Renal Center at Girandurukotte, Sri Lanka, and the staff of the Renal Unit, Teaching Hospital, Kandy, Sri Lanka. We appreciate the contribution of Dr. Hannah L.F. Swa, Institute of Molecular Biology, A ∗STAR, Singapore in data analysis. We also thank the President’s Task Force for their contribution in facilitating the research. This work was funded by the Ministry of Health and National Research Council (Grant No. TO 14-05), Sri Lanka. Asfa Alli-Shaik and Jayantha Gunaratne are funded by the Agency for Science, Technology, and Research (A*STAR), Singapore.
Author Contributions
NN, ZB, RKDH, and BNTWF conceived the study. BNTWF, ZB, RKDH, TDJA, AW, SR, and TWH contributed to sample collection and sample processing. BNTWF and RKDH performed all the experiments and collected the data. BNTWF, HTKA, and RKDH contributed to data analysis. AAS and JG designed the statistical data analysis pipeline. AAS performed the machine learning and statistical analysis. AAS, JG, and NN interpreted the data. BNTWF, AAS, JG, and NN wrote the manuscript. JG and NN critically reviewed the manuscript. NN supervised the overall study.
Footnotes
Table S1. Differential expression of biomarkers between CKDu and other groups.
Table S2. Performance of the marker combination to differentiate CKDu/CKD from NEC.
Table S3. Performance of the marker combination to differentiate CKDu from CKD.
Figure S1. Endemic regions of CKDu in Sri Lanka.
Figure S2. Biomarker expression among different categories.
Figure S3. Performance of 5-marker signature panel across disease groups.
Figure S4. Performance of 5-marker signature in distinguishing healthy controls.
Figure S5. Performance of 3-marker signatures in distinguishing CKD disease groups.
Figure S6. Identification of marker signature for distinguishing CKDu from NECKD.
Figure S7. Performance of interdisease 3-marker signature against healthy controls.
Figure S8. Serum creatinine for distinguishing disease group from control.
Supplementary Material
References
- 1.Perkovic V., Cass A., Patel A.A. High prevalence of chronic kidney disease in Thailand. Kidney Int. 2008;73:473–479. doi: 10.1038/sj.ki.5002701. [DOI] [PubMed] [Google Scholar]
- 2.Imai E., Horio M., Watanabe T. Prevalence of chronic kidney disease in the Japanese general population. Clin Exp Nephrol. 2009;13:621–630. doi: 10.1007/s10157-009-0199-x. [DOI] [PubMed] [Google Scholar]
- 3.Rao D.S., Shih M.S., Mohini R. Effect of serum parathyroid hormone and bone marrow fibrosis on the response to erythropoietin in anaemia. N Engl J Med. 1993;328:171–175. doi: 10.1056/NEJM199301213280304. [DOI] [PubMed] [Google Scholar]
- 4.Torres C., Aragon A., Gonzalez M. Decreased kidney function of unknown cause in Nicaragua: a community-based survey. Am J Kidney Dis. 2010;55:485–496. doi: 10.1053/j.ajkd.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 5.Cerdas M. Chronic kidney disease in Costa Rica. Kidney Int. 2005;68:31–33. doi: 10.1111/j.1523-1755.2005.09705.x. [DOI] [PubMed] [Google Scholar]
- 6.Pan American Health Organization 2014. Resolution CD52.R1. Chronic kidney disease in agricultural communities in Central America. Washington, DC. 2013. http://www.paho.org/hq/index.php?option=com_content&view=article&id=8833&Itemid=40033&lang=en Available at:
- 7.Aturaliya TNC, Abeysekara DTDJ, Amerasinghe PH, et al. Towards understanding of CKD of North Central Province. In: Proceedings of Annual Scientific Sessions of Sri Lanka Medical Association. Colombo, Sri Lanka: Sri Lanka Medical Association; 2006.
- 8.Athuraliya N.T.C., Abeysekara T.D.J., Amarasinghe P.H. Uncertain etiologies of proteinuric-chronic kidney disease in rural SriLanka. Kidney Int. 2011;80:1212–1221. doi: 10.1038/ki.2011.258. [DOI] [PubMed] [Google Scholar]
- 9.Nanayakkara S., Senevirathna S., Abeysekara T. An integrative study of the genetic, social and environmental determinants of chronic kidney disease characterized by tubulointerstitial damages in the north central region of SriLanka. J Occup Health. 2014;56:28–38. doi: 10.1539/joh.13-0172-oa. [DOI] [PubMed] [Google Scholar]
- 10.Wanigasuriya K., Peiris-John R.J., Wickremasinghe R. Chronic kidney disease of unknown etiology in Srilanka: is cadmium a likely cause? BMC Nephrol. 2011;12:32. doi: 10.1186/1471-2369-12-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wijetunge S., Ratnatunga N.V., Abeysekera T.D. Endemic chronic kidney disease of unknown etiology in Sri Lanka: correlation of pathology with clinical stages. Indian J Nephrol. 2015;25:274–280. doi: 10.4103/0971-4065.145095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kashif W., Siddiqi N., Dincer A.P. Proteinuria: how to evaluate an important finding. Cleve Clin J Med. 2003;70:535–537. doi: 10.3949/ccjm.70.6.535. [DOI] [PubMed] [Google Scholar]
- 13.Levey A.S., Coresh J. Chronic kidney disease. Lancet. 2012;378:165–180. doi: 10.1016/S0140-6736(11)60178-5. [DOI] [PubMed] [Google Scholar]
- 14.Ratnayake S., Badurdeen Z., Nanayakkara N. Screening for CKDu in SriLanka: usability of surrogate biomarkers over dipstick proteinuria. BMC Nephrol. 2017;18:199. doi: 10.1186/s12882-017-0610-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steubl D., Block M., Herbst V. Plasma uromodulin correlates with kidney function and identifies early stages in chronic kidney disease patients. Medicine. 2016;95 doi: 10.1097/MD.0000000000003011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Silva P.M.C.S., Mohammed Abdul K.S., Eakanayake E.M.D.V. Urinary biomarkers KIM-1 and NGAL for detection of chronic kidney disease of uncertain etiology (CKDu) among agricultural communities in Sri Lanka. PLoS Negl Trop Dis. 2016;10 doi: 10.1371/journal.pntd.0004979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaidya V.S., Ferguson M.A., Bonventre J.V. Biomarkers of acute kidney injury. Annu Rev Pharmacol Toxicol. 2008;48:463–493. doi: 10.1146/annurev.pharmtox.48.113006.094615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs T.C., Frick K., Emde B. Evaluation of novel acute urinary rat kidney toxicity biomarker for subacute toxicity studies in preclinical trials. Toxicol Pathol. 2012;40:1031–1048. doi: 10.1177/0192623312444618. [DOI] [PubMed] [Google Scholar]
- 19.Waiker S.S., Sabbisetti V.S., Bonventre J.V. Normalization of urinary biomarkers to creatinine during changes in glomerular filtration rate. Kidney Int. 2010;78:486–494. doi: 10.1038/ki.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu X., Liu C., Ye Y. The diagnostic value of serum creatinine and cystatin c in evaluating glomerular filtration rate in patients with chronic kidney disease: a systematic literature review and meta-analysis. Oncotarget. 2017;8:72985–72999. doi: 10.18632/oncotarget.20271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xun C., Zhao Y., Wang W. Circulating RBP4 increase and its diagnosis of chronic kidney disease. Ann Clin Lab Sci. 2018;48:205–207. [PubMed] [Google Scholar]
- 22.Han W.K., Bailly V., Abichandani R. Kidney Injury Molecule-1 (KIM-1): a novel biomarker for human renal proximal tubule injury. Kidney Int. 2002;62:237–244. doi: 10.1046/j.1523-1755.2002.00433.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.