Liu et al. show that Yeats4 recruits the Dot1l–RNA Pol II complex onto the Lmo4 promoter by recognizing H3K27ac modification to initiate Lmo4 transcription in α4β7+ CLPs, leading to ILC lineage commitment.
Abstract
Innate lymphoid cells (ILCs) play critical roles in defending infections and maintaining mucosal homeostasis. All ILCs arise from common lymphoid progenitors (CLPs) in bone marrow. However, how CLPs stratify and differentiate into ILC lineages remains elusive. Here, we showed that Yeats4 is highly expressed in ILCs and their progenitors. Yeats4 conditional KO in the hematopoietic system causes decreased numbers of ILCs and impairs their effector functions. Moreover, Yeats4 regulates α4β7+ CLP differentiation toward common helper ILC progenitors (CHILPs). Mechanistically, Yeats4 recruits the Dot1l–RNA Pol II complex onto Lmo4 promoter through recognizing H3K27ac modification to initiate Lmo4 transcription in α4β7+ CLPs. Additionally, Lmo4 deficiency also impairs ILC lineage differentiation and their effector functions. Collectively, the Yeats4–Lmo4 axis is required for ILC lineage commitment.
Introduction
Innate lymphoid cells (ILCs) are a group of recently identified innate immune cells that play critical functions in defending infections and maintaining mucosal homeostasis (Diefenbach et al., 2014; Artis and Spits, 2015; Vivier et al., 2018). Similar to CD4+ helper T cell subsets, ILCs previously could be categorized into three subgroups, namely group 1 ILC (ILC1s), group 2 ILC (ILC2s), and group 3 ILC (ILC3s), according to featured cytokine profiles and fate-decision transcription factors (TFs; Eberl et al., 2015; Serafini et al., 2015). We recently defined a new regulatory subpopulation of ILCs named ILCregs (Wang et al., 2017b). ILC1s are characterized by expressing T-bet and producing type 1 cytokine IFN-γ, which is essential for clearance of intracellular microbial infections (Bernink et al., 2015). ILC2s require Gata3 for their differentiation and maintenance (Hoyler et al., 2012; Mjösberg et al., 2012). After activation by IL-25 and IL-33, ILC2s generate type 2 cytokines, such as IL-5 and IL-13, to promote resolution of helminth infections (Brestoff et al., 2015) and participate in pathogenesis of asthma (Sui et al., 2018). ILC3s are a heterogeneous lineage including lymphoid tissue inducer (LTi) cells, NKp46+ ILC3s, and CCR6−NKp46− ILC3s (Serafini et al., 2015). ILC3s are defined by RORγt expression and are a main source of type 17 cytokines, such as IL-22 and IL-17, which are extremely critical for resistance against bacterial infections (Diefenbach et al., 2017). ILCregs are a group of IL-10–producing cells that suppress activation of ILC1s and ILC3s to regulate the inflammatory response.
All ILCs, as well as natural killer (NK), B, and T cells, are derived from common lymphoid progenitors (CLPs) in bone marrow (BM; Diefenbach et al., 2014). CLPs can differentiate into α-lymphoid progenitors (αLPs; CXCR+ integrin α4β7–expressing CLPs; Yu et al., 2014), which lack the potential to generate B and T cells. A recent study showed that αLPs are quite heterogeneous and consist of early innate lymphoid progenitors, common helper ILC progenitors (CHILPs), and ILC precursors (ILCPs; Seillet et al., 2016). CHILPs can differentiate into downstream ILCPs that give rise to all ILCs but without LTi cells and ILCregs (Constantinides et al., 2014; Wang et al., 2017b). Besides cytokine signaling, ILC lineage differentiation is delicately regulated by fate-decision TFs (Vivier et al., 2018). For instance, Tox, Nfil3, and Tcf1 are required for CLP differentiation toward αLPs (Yu et al., 2014; Seehus et al., 2015; Yang et al., 2015). Id2 participates in the generation of CHILPs, and PLZF directs ILCP production (Constantinides et al., 2014; Klose et al., 2014). Id3 drives the differentiation of CHILPs toward ILCregs (Wang et al., 2017b). However, how CLPs stratify and differentiate into ILC lineages still requires further investigation.
Chromatin structures are extremely dynamic for genetic reading and transcription and can be modified by several means, including ATP-dependent chromatin remodeling, incorporation of histone variants, DNA methylation, and histone modifications (Zhou et al., 2016). The best-defined histone modifications are composed of acetylation, phosphorylation, methylation, ubiquitination, and sumoylation (Zentner and Henikoff, 2013). Yeats domain–containing proteins are conserved from yeast to human and include Yeats4 (also named as GAS41), ENL, Yeats2, and AF9 in human (Hsu et al., 2018; Wang et al., 2018). These Yeats domain–containing proteins are involved in histone acetylation recognition and gene transcription by remodeling chromatin structures (Schulze et al., 2009; Li et al., 2014). It has been reported that Yeats4 can recognize histone acetylation and facilitate lung cancer progression (Hsu et al., 2018; Klein et al., 2018). ENL also recognizes histone acetylation to promote oncogenic gene expression (Erb et al., 2017; Wan et al., 2017). Yeats2 acts as an H3K27ac reader to regulate gene transcription (Mi et al., 2017). However, how YEATS domain–containing proteins regulate ILC development and differentiation is unclear. Here, we show that Yeats4 is required for α4β7+ CLP differentiation toward ILCs. Yeats4 recognizes H3K27ac to recruit the Dot1–RNA polymerase (Pol) II complex as a histone acetylation reader and initiates TF Lmo4 expression for ILC lineage commitment.
Results
Yeats4 deficiency causes a reduction in ILCs
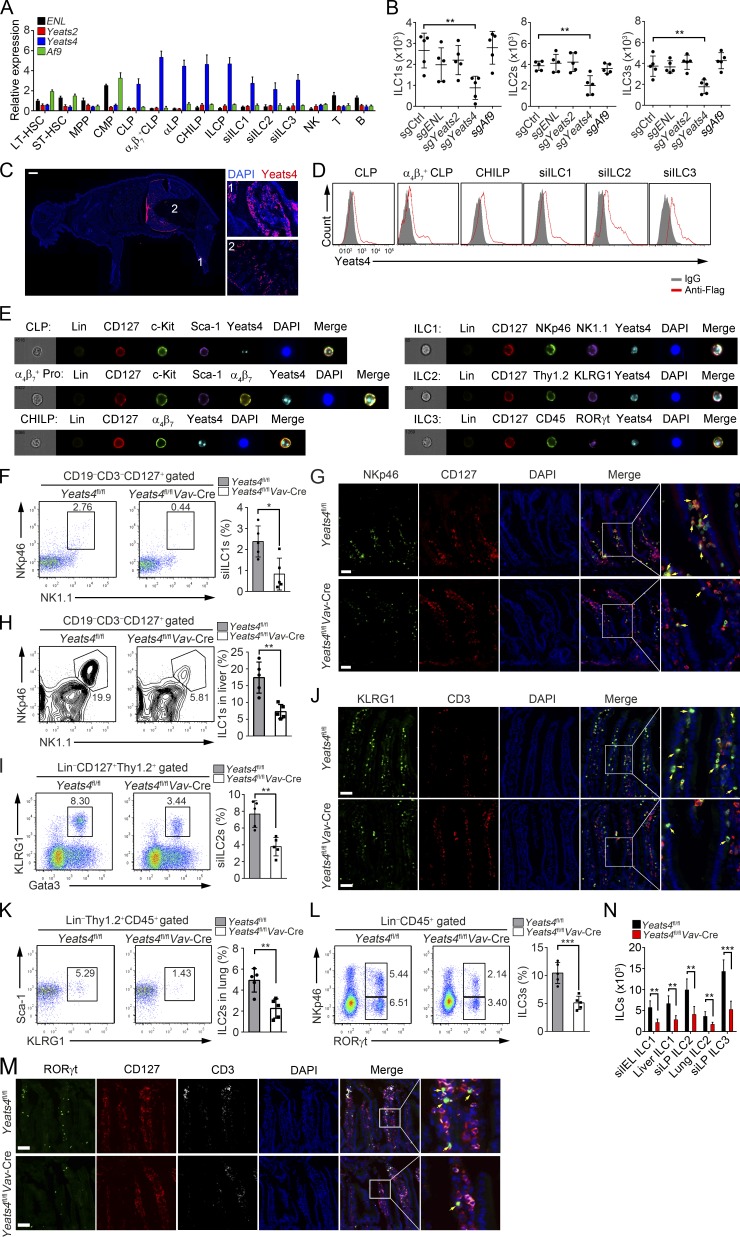
Yeats domain–containing proteins are important chromatin remodeling molecules that are implicated in the oncogenesis of acute myeloid leukemia (Erb et al., 2017; Wan et al., 2017). However, whether Yeats domain–containing proteins regulate development and functions of ILCs is still unclear. We examined expression levels of Yeats domain–containing proteins in hematopoietic cells and found that Yeats4 levels were the highest level in ILCs and their respective progenitors among all Yeats domain–containing members (Fig. 1 A and Fig. S1, A–H). We then deleted ENL, Yeats2, Yeats4, or Af9 in α4β7+ CLPs using CRISPR/Cas9 technology (Fig. S2 A), followed by an in vitro differentiation assay. We observed that Yeats4 deletion impaired ILC lineage differentiation (Fig. 1 B). By contrast, deletion of ENL, Yeats2, or Af9 did not affect ILC lineage commitment (Fig. 1 B). These results suggest that Yeats4 is involved in the modulation of the development and differentiation of ILCs. We next generated Yeats4Flag-knockin reporter mice (Fig. S2 B) and found that Yeats4 displayed high expression levels in BM and intestine (Fig. 1 C), where ILCs and their progenitors reside. High expression levels of Yeats4 in ILCs and their progenitors were further validated by FACS and imaging flow cytometry (Fig. 1, D and E).
Figure 1.
Yeats4 deficiency causes a reduction in ILC numbers. (A) Relative mRNA expression levels of ENL, Yeats2, Yeats4, and Af9 in different hematopoietic cells were examined by qPCR. All hematopoietic cells were isolated by FACS, and total RNA was extracted for qPCR. Fold changes were normalized to endogenous Actb. LT-HSCs (Lin−c-Kit+Sca-1+CD150+CD48−), ST-HSCs (Lin−c-Kit+Sca-1+CD150−CD48−), MPPs (Lin−c-Kit+Sca-1+CD150−CD48+), CMPs (Lin−c-Kit+Sca-1−CD34+CD16/32−), CLPs (Lin−CD25−CD127+c-KitintSca-1intFlt3+α4β7−), α4β7+ CLPs (Lin−CD25−CD127+c-KitintSca-1intFlt3+α4β7+), αLP (Lin−CD25−CD127+c-KitintSca-1intFlt3−α4β7+), CHILPs (Lin−CD25−CD127+Flt3−α4β7+Id2+), and ILCPs (Lin−CD127+Flt3−c-Kit+α4β7+PLZF+) were isolated from BM cells. ILC1s (CD3−CD19−CD127+NK1.1+NKp46+), ILC2s (Lin−CD127+Thy1.2+KLRG1+Gata3+), and ILC3s (Lin−CD45+RORγt+) were isolated from the small intestine. NK cells (NK1.1+) and B cells (CD19+) were isolated from the spleen. T cells (CD3+) were isolated from the thymus. (B) α4β7+ CLPs (Lin−CD25−CD127+c-KitintSca-1intFlt3+α4β7+) were isolated from 129-Gt(ROSA)26Sortm1(CAG-cas9*,-EGFP)FezhVav-Cre mice and infected with lentiCRISPRv2 containing sgRNA lentiviruses against ENL, Yeats2, Yeats4, or Af9 for gene deletion, followed by an in vitro differentiation assay. (C) 7-d-old Yeats4Flag mice were sacrificed for longitudinal sections and stained with anti-Flag antibody. A global view of section is shown in the left panel, and the indicated tissues are presented in the right panels. 1, BM; 2, intestine. Scale bar, 1,000 µm. (D) Yeats4Flag knockin mice were analyzed by FACS. (E) Imaging flow cytometry analysis for Yeats4 expression in indicated ILCs and their progenitors from Yeats4Flag knockin mice. For CHILP staining, Lin− populations contained both Flt3- and CD25-negative cells. Lineage cocktail antibodies contained anti-CD3 and anti-CD19 for ILC1 staining. α4β7+ Pro indicates α4β7+ progenitors, consisting of α4β7+ CLPs and αLPs. (F) Percentages of ILC1s (CD3−CD19−CD127+NK1.1+NKp46+) in small intestines from Yeats4fl/fl and Yeats4fl/flVav-Cre mice were analyzed by FACS. Percentages of indicated cells were calculated and shown in the right panel. n = 5 per group. (G) ILC1s (CD127+NKp46+) in small intestines from Yeats4fl/fl and Yeats4fl/flVav-Cre mice were visualized by in situ immunofluorescence staining. Arrows denote ILC1 cells. Scale bars, 50 µm. (H) Percentages of ILC1s in livers derived from Yeats4fl/fl and Yeats4fl/flVav-Cre mice were examined by FACS. Percentages of indicated cells were calculated and shown in the right panel. n = 5 per group. (I) Percentages of ILC2s (Lin−CD127+Thy1.2+KLRG1+Gata3+) in small intestines from Yeats4fl/fl and Yeats4fl/flVav-Cre mice were examined by FACS. Percentages of indicated cells were calculated and shown in the right panel. n = 5 per group. (J) ILC2s (CD3−KLRG1+) in small intestines from Yeats4fl/fl and Yeats4fl/flVav-Cre mice were visualized by in situ immunofluorescence staining. Arrows denote ILC2 cells. Scale bars, 50 µm. (K) Percentages of ILC2s (Lin−CD45+Thy1.2+KLRG1+Sca-1+) in lungs from Yeats4fl/fl and Yeats4fl/flVav-Cre mice were examined by FACS. Percentages of indicated cells were calculated and shown in the right panel. n = 5 per group. (L) Percentages of ILC3s (Lin−CD45+RORγt+) in small intestines from Yeats4fl/fl and Yeats4fl/flVav-Cre mice were examined by FACS. Percentages of indicated cells were calculated and shown in the right panel. n = 5 per group. (M) ILC3s (CD3−CD127+RORγt+) in small intestines from Yeats4fl/fl and Yeats4fl/flVav-Cre mice were visualized by in situ immunofluorescence staining. Arrows denote ILC3 cells. Scale bars, 50 µm. (N) Numbers of indicated ILCs were calculated. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by two-tailed unpaired Student’s t test. All data are representative of at least three independent experiments and are expressed as mean ± SD.
To determine the physiological role of Yeats4, we generated Yeats4 KO mice with a frameshift mutation using CRISPR/Cas9 (Fig. S2 C). We noticed that Yeats4 full KO caused early embryonic lethality. We then established Yeats4flox/flox mice via CRISPR/Cas9 (Fig. S2 D) and generated Yeats4flox/floxVav-Cre conditional KO mice in the hematopoietic system (hereafter called Yeats4fl/flVav-Cre) via crossing Yeats4flox/flox mice with Vav-Cre mice. We found that Yeats4 was completely deleted in BM hematopoietic cells (Fig. S2 D). Of note, Yeats4fl/flVav-Cre mice showed decreased ILC1s in small intestine intraepithelial lymphocytes compared with their Yeats4flox/flox littermates (hereafter called Yeats4fl/fl; Fig. 1 F). This observation was further confirmed by in situ fluorescence staining (Fig. 1 G). In addition, liver ILC1s were also reduced in Yeats4fl/fVav-Cre mice (Fig. 1 H). Similar to ILC1s, ILC2s were also decreased in the small intestine lamina propria and lungs of Yeats4fl/flVav-Cre mice (Fig. 1, I–K). In parallel, ILC3s were also reduced in small intestine lamina propria of Yeats4fl/flVav-Cre mice (Fig. 1, L and M). ILC3s are quite heterogeneous, including CCR6+ LTi-like, T-bet+, and CCR6−T-bet− ILC3s (Gury-BenAri et al., 2016). The numbers of these three major ILC3 subpopulations were all decreased after Yeats4 deletion (Fig. S2 E). Finally, Yeats4fl/flVav-Cre mice caused reduced numbers of all ILC lineages (Fig. 1 N). Furthermore, we also measured the effects of Yeats4 on T cells, B cells, NK cells, granulocytes, and monocytes. We found that Yeats4 deficiency had no obvious effect on these cells (Fig. S2, F–I). Altogether, Yeats4 is highly expressed in ILCs and their progenitors, and Yeats4 deficiency causes reduced ILC numbers.
Yeats4 deletion impairs effector functions of ILCs
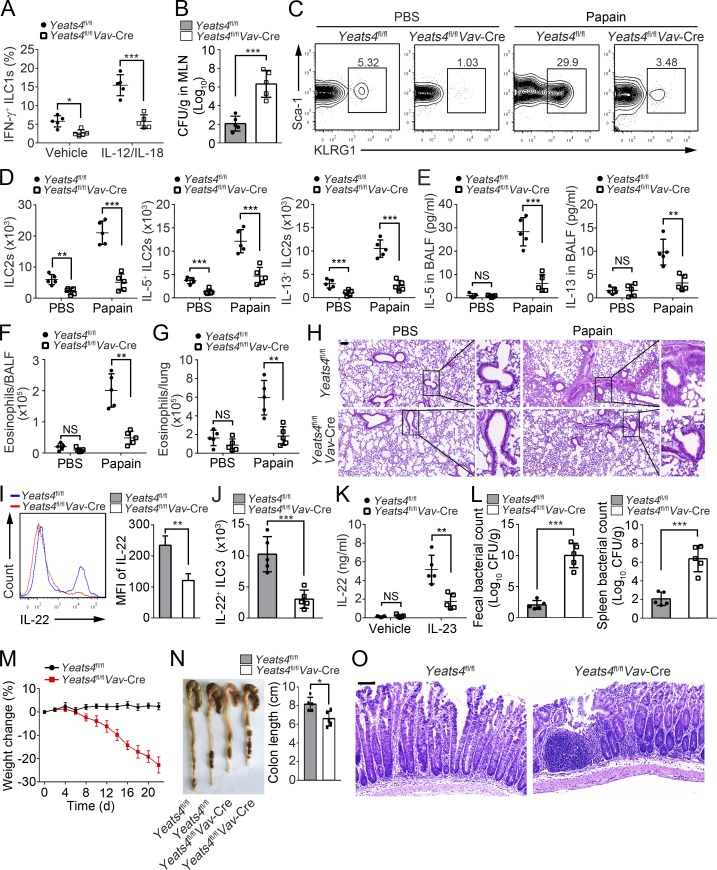
ILCs reside in mucosal surfaces that initiate immune responses, maintain mucosal integrity and facilitate lymphoid organogenesis (Vivier et al., 2018). To test whether Yeats4fl/flVav-Cre mice affect ILC functions, we treated Yeats4fl/fl and Yeats4fl/flVav-Cre mice with Salmonella typhimurium, papain, or Citrobacter rodentium and determined their resistance against infections and tissue inflammation. We found that IFN-γ+ ILC1s were dramatically decreased in the mesenteric LNs (MLNs) of Yeats4fl/flVav-Cre mice (Figs. 2 A and S2 J). Moreover, a higher number of S. typhimurium CFUs were detected in the MLNs of Yeats4fl/flVav-Cre mice (Fig. 2 B). In addition, Yeats4 deletion impaired production of IFN-γ by ILC1s (Fig. S2 K). These data indicate that Yeats4 KO impaired the antibacterial function of ILC1s.
Figure 2.
Yeats4 deletion impairs effector functions of ILCs. (A) Frequencies of IFN-γ+ ILC1s in MLNs from Yeats4fl/fl or Yeats4fl/flVav-Cre mice infected with S. typhimurium were analyzed on day 5 after injection. n = 5 per group. (B) CFUs of S. typhimurium were measured in MLNs from Yeats4fl/fl and Yeats4fl/flVav-Cre mice on day 5 after infection. n = 5 per group. (C) FACS analysis of ILC2s (Lin−CD45+Thy1.2+KLRG1+Sca-1+) in lungs from Yeats4fl/fl and Yeats4fl/flVav-Cre mice intranasally treated with 25 µg of papain or PBS on days 0, 1, and 3. n = 5 per group. (D) Total numbers of ILC2s and IL-5+ or IL-13+ ILC2s were analyzed in lungs from Yeats4fl/fl and Yeats4fl/flVav-Cre mice after challenge. n = 5 per group. (E) Concentrations of IL-5 and IL-13 in BALF were tested by ELISA. n = 5 per group. (F and G) FACS analysis of eosinophils (CD45+CD11c−Siglec-F+) in BALF (F) and lungs (G). n = 5 per group. (H) H&E staining of lung sections from Yeats4fl/fl and Yeats4fl/flVav-Cre mice treated with papain. Scale bars, 100 µm. (I and J) FACS analysis of IL-22+ ILC3s in small intestines from Yeats4fl/fl and Yeats4fl/flVav-Cre mice challenged with 5 × 109 C. rodentium. n = 5 per group. (K) Secreted IL-22 proteins by LPLs from challenged Yeats4fl/fl and Yeats4fl/flVav-Cre mice was tested by ELISA after being stimulated with IL-23 for 24 h. (L) CFUs in feces were measured. n = 5 per group. (M) Body weight changes of Yeats4fl/fl and Yeats4fl/flVav-Cre mice after C. rodentium infection. n = 8 per group. (N) Colon lengths were measured after challenged with C. rodentium. n = 5 per group. (O) Colons from challenged Yeats4fl/fl and Yeats4fl/flVav-Cre mice were examined by H&E staining. Scale bars, 50 µm. n = 5 per group. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by two-tailed unpaired Student’s t test. All data are representative of at least three independent experiments and are expressed as mean ± SD.
Upon intranasal administration of papain, total ILC2s and IL-5– or IL-13–expressing ILC2s in lungs of Yeats4fl/flVav-Cre mice were remarkably reduced (Fig. 2, C and D; and Fig. S2 L). Consistently, concentrations of IL-5 and IL-13 in bronchoalveolar lavage fluid (BALF) were also reduced in Yeats4fl/flVav-Cre mice compared with Yeats4fl/fl mice (Fig. 2 E). It has been reported that ILC2s can recruit eosinophils to promote lung inflammation upon papain treatment (Li et al., 2018). We observed that eosinophil infiltration in BALF and lungs of Yeats4fl/flVav-Cre mice was substantially reduced (Fig. 2, F and G). In addition, leukocyte infiltration was also reduced in lungs of Yeats4fl/flVav-Cre mice (Fig. 2 H). These data indicate that Yeats4 deficiency leads to impaired functions of lung ILC2s.
ILC3s play an important role in the defense against C. rodentium infection in the intestine via secretion of IL-22 (Bando et al., 2018). Thus, the C. rodentium infection model was used to test the function of ILC3s. We noticed that IL-22–producing ILC3s were markedly decreased in small intestines of Yeats4fl/flVav-Cre mice compared with those of Yeats4fl/fl littermate control mice (Fig. 2, I and J). In addition, IL-22 secreted by ILC3s in lamina propria lymphocytes (LPLs) of Yeats4fl/flVav-Cre mice was substantially reduced (Fig. 2 K). Of note, bacterial CFUs were much higher in feces and spleen of Yeats4fl/flVav-Cre mice (Fig. 2 L). Moreover, compared with WT control mice, Yeats4fl/flVav-Cre mice displayed accelerated weight loss after C. rodentium infection and shortened colon length (Fig. 2, M and N), which were accompanied by persistent intestinal damage (Fig. 2 O). These observations suggest that Yeats4fl/flVav-Cre mice are more susceptible to C. rodentium infection. Taken together, Yeats4 deficiency impairs effector functions of ILCs.
Yeats4 drives α4β7+ CLPs to ILC lineage differentiation
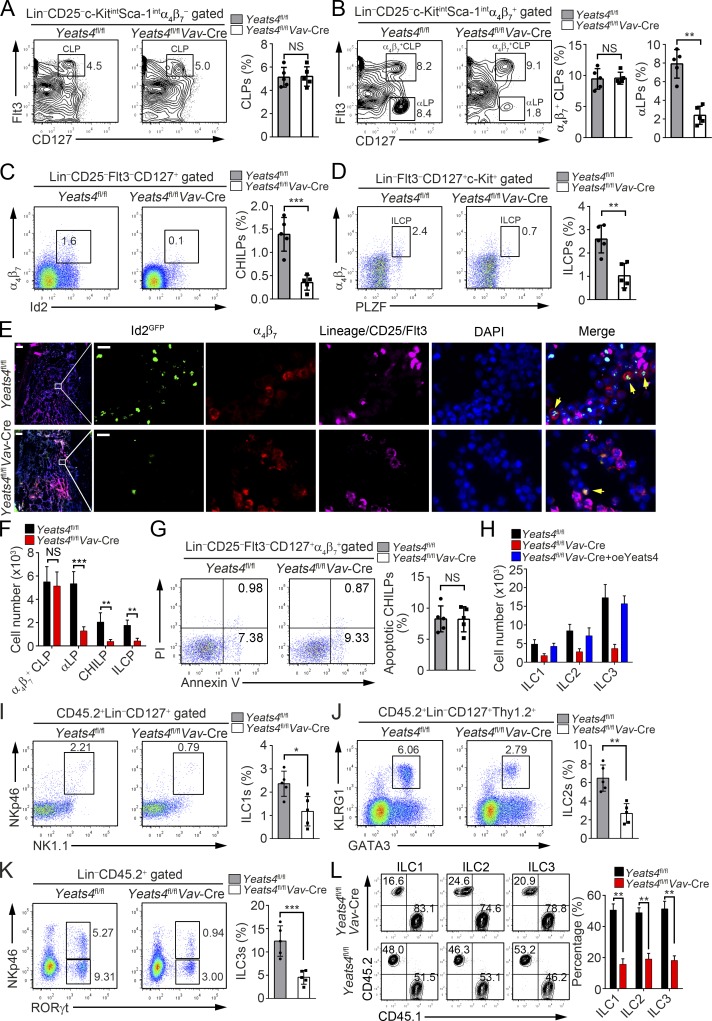
We next wanted to determine whether decreased ILC numbers and impaired effector functions caused by Yeats4 deletion were due to defective development of ILCs. We examined ILC progenitors in BM of Yeats4fl/flVav-Cre mice. First, we observed that Yeats4 deletion did not affect cell rates of long-term HSCs (LT-HSCs), short-term HSCs (ST-HSCs), and multipotent progenitors (MPPs; Fig. S3 A). Yeats4fl/flVav-Cre mice also exhibited frequencies of CLPs and α4β7+ CLPs comparable to Yeats4fl/fl littermate control mice (Fig. 3, A and B). However, the frequencies of αLPs, CHILPs, and ILCPs were significantly reduced in Yeats4fl/flVav-Cre mice (Fig. 3, B–D). Decreased numbers of CHILPs in Yeats4fl/flVav-Cre mice were further validated by in situ fluorescence staining (Fig. 3 E). As expected, total numbers of α4β7+ CLPs in BM of Yeats4fl/flVav-Cre mice were similar to those of Yeats4fl/fl littermate control mice, whereas numbers of αLPs, CHILPs and ILCPs were dramatically decreased (Fig. 3 F). Of note, CHILPs did not undergo apparent apoptosis (Fig. 3 G). Additionally, myeloid progenitors, including common myeloid progenitors (CMPs), granulocyte–macrophage progenitors, and megakaryocyte–erythrocyte progenitors, showed similar frequencies in Yeats4fl/flVav-Cre and Yeats4fl/fl mice (Fig. S3 B). These data suggest that Yeats4 deletion affects ILC lineage commitment from the α4β7+ CLPs stage.
Figure 3.
Yeats4 directs α4β7+ CLPs to ILC lineage differentiation. (A–D) Analysis of CLPs, α4β7+ CLPs, αLPs, CHILPs, and ILCPs from Yeats4fl/fl and Yeats4fl/flVav-Cre mice by FACS. Percentages of indicated cells were calculated and shown in the right panel. n = 5 per group. (E) CHILPs (Lin−CD25−Flt3−α4β7+Id2GFP) in BM from Yeats4fl/flId2+/GFP and Yeats4fl/flVav-CreId2+/GFP mice were visualized by in situ immunofluorescence staining. Arrows denote CHILP cells. Scale bars, 100 µm (left) and 10 µm (right). (F) Numbers of indicated cells from Yeats4fl/fl and Yeats4fl/flVav-Cre mice were calculated. n = 5 per group. (G) Analysis of cell apoptosis in CHILPs (Lin−CD25− Flt3−CD127+α4β7+) from Yeats4fl/fl and Yeats4fl/flVav-Cre mice. n = 5 per group. (H) 5 × 103 Yeats4-overexpressing α4β7+ CLPs isolated from Yeats4fl/flVav-Cre mice were engrafted into Rag1−/−Il2rg−/− mice. After 6 wk, ILCs in the small intestine were analyzed by FACS. (I–K) 5 × 104 CD45.2+ LSKs from Yeats4fl/fl and Yeats4fl/flVav-Cre mice with 5 × 106 CD45.1+ helper BM cells were transplanted into lethally irradiated CD45.1+ recipients. After 8 wk, percentages of ILC1s (I), ILC2s (J), and ILC3s (K) in small intestines from chimeras were tested by FACS. n = 5 for each group. (L) A 50/50 mixture of CD45.1+ WT and CD45.2+ Yeats4fl/fl or Yeats4fl/flVav-Cre BM was transplanted into lethally irradiated CD45.1+ recipients. Ratios of CD45.1+ to CD45.2+ CHILPs, ILC1s, ILC2s, or ILC3s in chimeras (n = 5) were analyzed. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by two-tailed unpaired Student’s t test. All data are representative of at least three independent experiments and are expressed as mean ± SD.
We next isolated α4β7+ CLPs from BM of Yeats4fl/flVav-Cre and Yeats4fl/fl mice and restored Yeats4 expression in Yeats4fl/flVav-Cre α4β7+ CLPs via retrovirus-mediated pMY-Yeats4-IRES-EGFP transduction. We generated Rag1−/−Il2rg−/− mice by crossing Rag1+/− mice and Il2rg+/− mice, which lacked T cells, B cells, NK cells, and ILCs and were used for adoptive transfer assay. We then transferred Yeats4fl/flVav-Cre α4β7+ CLPs and Yeats4fl/flVav-Cre α4β7+ CLPs with Yeats4 overexpression into Rag1−/−Il2rg−/− recipients. 6 wk later, we tested donor-derived ILCs in recipient mice. Expectedly, Yeats4fl/flVav-Cre α4β7+ CLPs engraftment caused impaired differentiation of ILC1–ILC3s (Fig. 3 H). However, forced expression of Yeats4 in Yeats4fl/flVav-Cre α4β7+ CLPs could restore normal numbers of ILC1-3s compared with engraftment of Yeats4flox/flox α4β7+ CLPs (Fig. 3 H). These results validate that Yeats4 drives ILC lineage commitment from the α4β7+ CLPs stage. Previous studies reported that RORγt+ ILCs are important for formation of Peyer’s patches and intestinal lymphoid follicles (Kiss et al., 2011; Seillet et al., 2014). We noticed that Yeats4 deletion also caused reduced numbers of Peyer’s patches and intestinal lymphoid follicles (Fig. S3 C).
To further determine whether Yeats4 regulated α4β7+ CLPs development in a cell-intrinsic manner, we conducted a BM transplantation assay. CD45.2+ LSK cells (Lin−Sca-1+c-Kit+) from Yeats4fl/flVav-Cre and Yeats4fl/fl mice were transplanted into lethally irradiated CD45.1+ recipients together with CD45.1+ helper BM cells. After 8 wk, ILCs were tested in small intestines of chimeras. As expected, transplantation of Yeats4fl/flVav-Cre LSK cells dramatically decreased numbers of ILC1s, ILC2s, and ILC3s (Fig. 3, I–K). In addition, we conducted competitive BM transplantation assay by injecting a 1:1 mixture of CD45.1+ WT and CD45.2+ Yeats4fl/fl or Yeats4fl/flVav-Cre BM cells into lethally irradiated recipient mice for 8 wk. Similarly, engraftment of CD45.2+ Yeats4fl/flVav-Cre BM cells produced reduced frequencies of ILC1s, ILC2s, and ILC3s (Fig. 3 L). These data suggest that Yeats4 drives α4β7+ CLPs differentiation in a cell-intrinsic manner.
Yeats4 initiates Lmo4 expression in α4β7+ CLPs
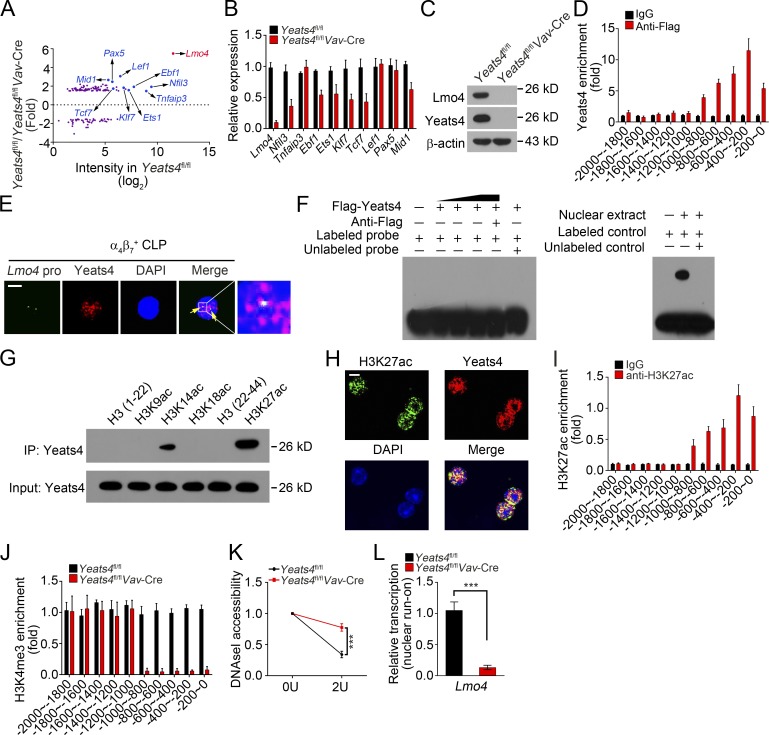
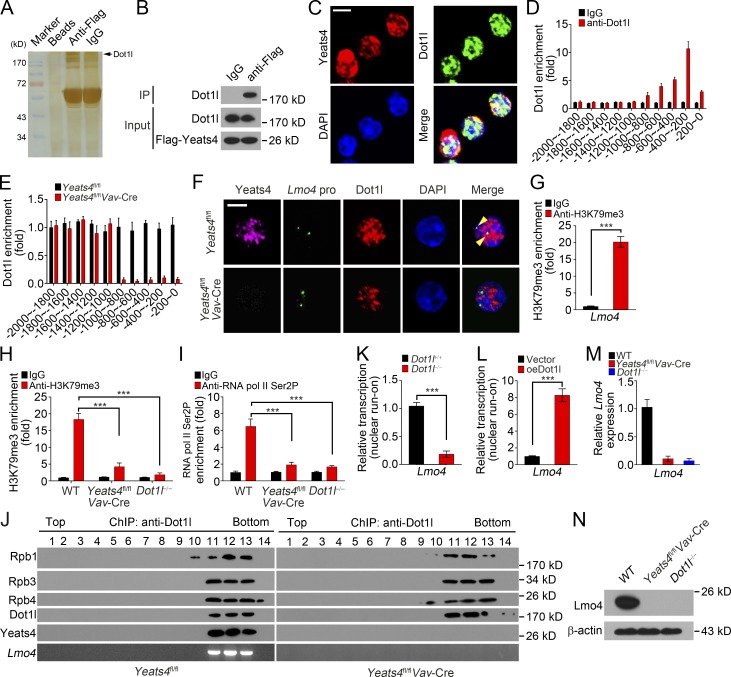
To further explore the molecular mechanism by which Yeats4 drives ILC development, we isolated Yeats4fl/fl and Yeats4fl/flVav-Cre α4β7+ progenitors (Lin−CD127+c-KitintSca-1intα4β7+) for transcriptome profile analysis. Many TFs were down-regulated in Yeats4fl/flVav-Cre α4β7+ progenitors. Among the top 10 down-regulated TFs, Lmo4 was the most down-regulated TF in Yeats4fl/flVav-Cre α4β7+ progenitors (Fig. 4 A). Moreover, Lmo4 was the most highly expressed TF in Yeats4fl/fl α4β7+ progenitors (Fig. 4 A). Down-regulation of Lmo4 was further validated in Yeats4fl/flVav-Cre α4β7+ CLPs by quantitative RT-PCR (qRT-PCR) and Western blotting (Fig. 4, B and C). We also measured gene expression levels of other TFs (including Gata3, Tox, Id2, and Zbtb16) and chemokine receptors (including Itga4, Cxcr6, Ccr6, and Cxcr5), which are important for ILC development or function. We observed that only Tox and Cxcr6 were down-regulated in Yeats4-deficient α4β7+ CLPs (Fig. S3, D and E). Using a chromatin immunoprecipitation (ChIP) assay, we found that Yeats4 is enriched in a region of the Lmo4 promoter approximately −1,000 to 0 bp from the transcription start site (Fig. 4 D). Colocalization of Yeats4 with Lmo4 promoter was confirmed by a DNA–fluorescent in situ hybridization (FISH) assay (Fig. 4 E). However, results of an electrophoretic mobility shift assay (EMSA) indicated that Yeats4 does not directly bind to the Lmo4 promoter (Fig. 4 F), suggesting that Yeats4 does not directly recognize the DNA motifs of the Lmo4 promoter.
Figure 4.
Yeats4 initiates Lmo4 expression in α4β7+ CLPs. (A) Differentially expressed TFs in α4β7+ progenitors (Lin−CD127+c-KitintSca-1intα4β7+) from Yeats4fl/fl and Yeats4fl/flVav-Cre mice by microarray analysis. (B) qRT-PCR analysis for mRNA levels of indicated top 10 down-regulated TFs in α4β7+ CLPs from Yeats4fl/fl and Yeats4fl/flVav-Cre mice. (C) Protein levels of Lmo4 were tested by immunoblotting in α4β7+ progenitors (Lin−CD127+c-KitintSca-1intα4β7+) from Yeats4fl/fl and Yeats4fl/flVav-Cre mice. (D) Yeats4 enrichment on Lmo4 promoter was determined. α4β7+ progenitors (Lin−CD127+c-KitintSca-1intα4β7+) were isolated from Yeats4fl/fl and Yeats4fl/flVav-Cre mice, followed by ChIP assay. (E) Yeats4 was colocalized with the Lmo4 promoter by FISH assay. Scale bar, 5 µm. (F) EMSA for detection of direct interaction between Yeats4 and the Lmo4 promoter. The Lmo4 promoter was biotin labeled. Positive control probes interacted with nuclear extract (right panel). (G) Pull-downs were performed using Flag-Yeats4 and biotinylated histone peptides with different modifications, followed by immunoblotting with anti-Flag antibody. IP, immunoprecipitation. (H) Colocalization between Yeats4 and H3K27ac in α4β7+ CLPs was assessed by immunofluorescence staining. Nuclei were stained with DAPI. Scale bar, 5 µm. (I) Analysis of H3K27ac enrichment on Lmo4 promoter using ChIP-qPCR assay. (J) Enrichment of H3K4me3 on Lmo4 promoter in αLPs was analyzed by ChIP analysis. (K) DNase I accessibility assay of Lmo4 promoter in α4β7+ progenitors (Lin−CD127+c-KitintSca-1intα4β7+) from Yeats4fl/fl and Yeats4fl/flVav-Cre mice. (L) α4β7+ progenitors (Lin−CD127+c-KitintSca-1intα4β7+) from Yeats4fl/fl and Yeats4fl/flVav-Cre mice were subjected to nuclear run-on assay, followed by RT-PCR analysis for Lmo4 transcription. ***, P < 0.001 by two-tailed unpaired Student’s t test. All data are representative of at least three independent experiments and are expressed as mean ± SD.
It has been reported that Yeats4 can recognize H3K27ac and H3K14ac as a histone acetylation reader (Hsu et al., 2018). We observed that Yeats4 mainly interacted with H3K27ac through a pull-down assay (Fig. 4 G). Moreover, Yeats4 was colocalized with H3K27ac in α4β7+ CLPs (Fig. 4 H). Notably, we found that H3K27ac modifications were enriched at the same region of the Lmo4 promoter as Yeats4 (Fig. 4 I), suggesting that Yeats4 recognizes H3K27ac to associate with the Lmo4 promoter. Yeats domain–containing proteins function as chromatin remodelers (Schulze et al., 2009; Li et al., 2014). We then wanted to determine whether Yeats4 could regulate transcriptional accessibility of the Lmo4 gene. We noticed that Yeats4 deletion abrogated active H3K4me3 enrichment on the Lmo4 promoter (Fig. 4 J). A DNase I accessibility assay showed that the Lmo4 promoter in Yeats4fl/flVav-Cre α4β7+ progenitors was more resistant to DNase I digestion (Fig. 4 K). Consequently, Yeats4 deficiency markedly suppressed Lmo4 transcription via a nuclear run-on assay (Fig. 4 L). Collectively, Yeats4 associates with the Lmo4 promoter by recognizing H3K27ac and facilitates its expression.
Yeats4 recruits the Dot1l–RNA Pol II complex onto the Lmo4 promoter to initiate its transcription
We next sought to explore how Yeats4 regulated Lmo4 transcription. We performed an immunoprecipitation assay with anti-Flag and protein A/G agarose in Yeats4Flag-knockin BM lysates. Interestingly, an ∼190-kD differential band was identified to be Dot1l (Figs. 5 A and S4 A) as a candidate interactor for Yeats4. The interaction between Yeats4 and Dot1l was verified using a coimmunoprecipitation assay (Fig. 5 B). Moreover, Yeats4 was colocalized with Dot1l in the nuclei of α4β7+ CLPs (Fig. 5 C). Of note, Dot1l was enriched on the same region of the Lmo4 promoter as Yeats4 (Fig. 5 D). Importantly, Yeats4 deletion abolished the association between Dot1l and the Lmo4 promoter (Fig. 5 E), which was further validated by a DNA-FISH assay (Fig. 5 F). These data indicate that Yeats4 interacts with Dot1l and is enriched on the Lmo4 promoter.
Figure 5.
Yeats4 recruits the Dot1l–RNA Pol II complex onto the Lmo4 promoter through recognizing H3K27Ac. (A) Pull-downs were performed in BM cells from Yeats4Flag knockin mice using anti-Flag or IgG. Eluted fractions were resolved by SDS-PAGE, followed by silver staining and mass spectrometry. (B) Flag-Yeats4 precipitated Dot1l in BM cell lysates. IP, immunoprecipitation. (C) Yeats4 and Dot1l were visualized in α4β7+ CLPs from Yeats4fl/fl mice by immunofluorescence staining. Red, Yeats4; green, Dot1l. Nuclei were counterstained by DAPI. Scale bar, 5 µm. (D) Dot1l enrichment on Lmo4 promoter was analyzed via ChIP-qPCR analysis. (E) Dot1l enrichment on Lmo4 promoter was analyzed in α4β7+ progenitors (Lin−CD127+c-KitintSca-1intα4β7+) from Yeats4fl/fl and Yeats4fl/flVav-Cre mice. (F) α4β7+ CLPs cells from Yeats4fl/fl and Yeats4fl/flVav-Cre mice were in situ hybridized with probes against the Lmo4 promoter, followed by staining with antibodies against Yeats4 and Dot1l. Arrowheads indicate Lmo4 promoters colocalized with Dot1l. Scale bar, 5 µm. (G) Enrichment of H3K79me3 on the Lmo4 promoter was tested using ChIP-qPCR analysis. (H) Enrichment of H3K79me3 on the Lmo4 promoter in WT, Yeats4-deficient, or Dot1l-deficient α4β7+ progenitor (Lin−CD127+c-KitintSca-1intα4β7+) cells was examined. (I) Enrichment of RNA Pol II Ser2P on the Lmo4 promoter in WT, Yeats4-deficient or Dot1l-deficient α4β7+ progenitor (Lin−CD127+c-KitintSca-1intα4β7+) cells was analyzed. (J) BM cells from Yeats4fl/fl and Yeats4fl/flVav-Cre mice were lysed and treated with 1% formaldehyde for cross-linking. Then, anti-Dot1l antibody was incubated with treated lysates for ChIP assays, followed by size fractionation with sucrose gradient ultracentrifugation. Eluate gradients were examined by Western blotting and PCR assays. (K) Dot1l−/− or control α4β7+ progenitors (Lin−CD127+c-KitintSca-1intα4β7+) were subjected to nuclear run-on assay, followed by RT-PCR analysis for Lmo4 transcription. (L) Dot1l-overexpressing or control α4β7+ progenitors (Lin−CD127+c-KitintSca-1intα4β7+) were subjected to nuclear run-on assay, followed by RT-PCR analysis for Lmo4 transcription. (M and N) Relative mRNA (M) and protein (N) levels of Lmo4 in Yeats4-deficient, Dot1l-deficient, or control α4β7+ progenitors (Lin−CD127+c-KitintSca-1intα4β7+) were analyzed. ***, P < 0.001 by two-tailed unpaired Student’s t test. All data are representative of at least three independent experiments and are expressed as mean ± SD.
Dot1l, a histone methyltransferase, catalyzes H3K79me3, which further recruits RNA Pol II to initiate gene transcription (Kim et al., 2012). Using a ChIP assay, we found that H3K79me3 is enriched on the Lmo4 promoter in α4β7+ progenitors (Fig. 5 G). Expectedly, Yeats4 deletion abrogated enrichment of H3K79me3 and active RNA Pol II on Lmo4 promoter in α4β7+ progenitors (Fig. 5, H and I). We also deleted Dot1l in α4β7+ progenitors using CRISPR/Cas9 technology (Fig. S3 B). Similar to Yeats4 deletion, Dot1l KO also disrupted enrichment of H3K79me3 and active RNA Pol II on the Lmo4 promoter in α4β7+ progenitors (Fig. 5, H and I). Through cross-linking and sucrose density gradient centrifugation, we found that Yeats4 coeluted with Dot1l and RNA Pol II complex components along with the Lmo4 promoter in BM lysates (Fig. 5 J). By contrast, Yeats4 deletion did not enrich the Lmo4 promoter in the Dot1l–RNA Pol II complex (Fig. 5 J). Of note, we noticed that Yeats4 precipitated the Lmo4 promoter by recognizing H3K27ac (Fig. 4, F–H). A CBP/P300 inhibitor could block H3K27ac modifications (Fig. S4 C) and consequently abolished the binding of the Lmo4 promoter with the Dot1l–RNA Pol II complex (Fig. S4 D). In addition, Dot1l deletion also dramatically suppressed Lmo4 transcription via a nuclear run-on assay (Fig. 5 K), whereas Dot1l overexpression could augment Lmo4 transcription (Fig. 5 L). Consequently, Dot1l deletion blocked Lmo4 expression (Fig. 5, M and N). Altogether, Yeats4 recruits the Dot1l–RNA Pol II complex onto the Lmo4 promoter by recognizing H3K27ac to initiate Lmo4 transcription in α4β7+ CLPs.
Lmo4 deficiency impairs ILC commitment and their effector functions
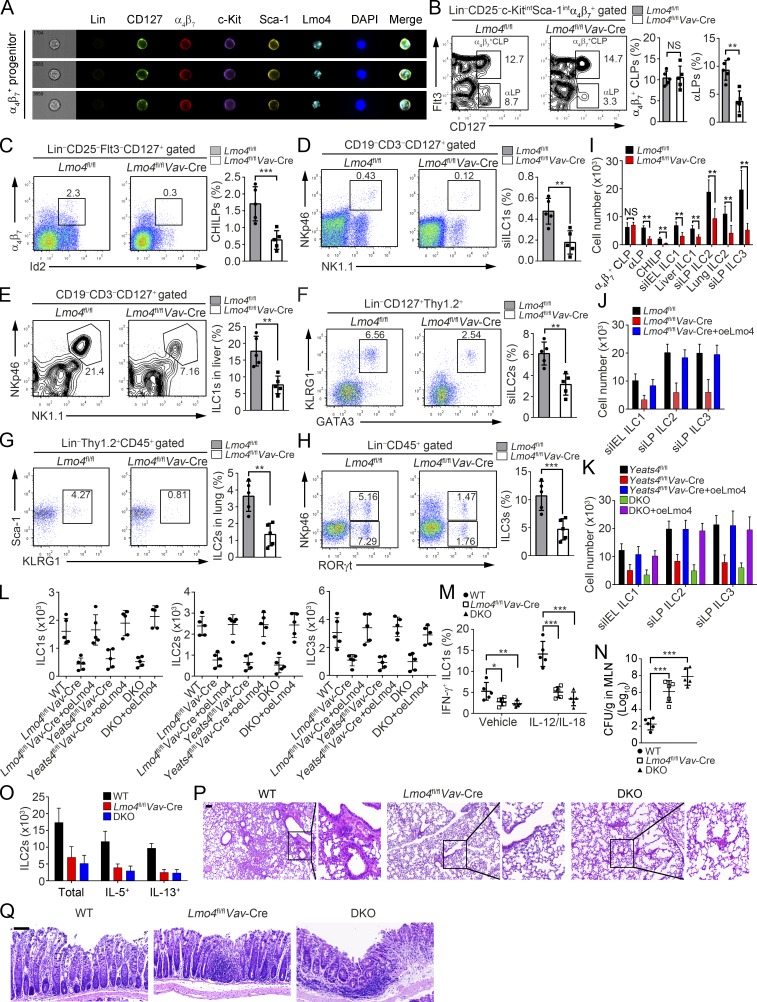
We found that Lmo4 was highly expressed in ILCs and their progenitor α4β7+ CLPs, αLPs, and CHILPs through qRT-PCR and imaging flow cytometry (Figs. 6 A and S5 A). We then crossed Lmo4flox/flox mice with Vav-Cre mice to generate Lmo4flox/floxVav-Cre mice (hereafter referred to as Lmo4fl/flVav-Cre). Lmo4 was completely deleted in BM hematopoietic cells of Lmo4fl/flVav-Cre mice (Fig. S5 B). We observed that Lmo4fl/flVav-Cre mice displayed similar frequencies of α4β7+ CLPs compared with Lmo4flox/flox littermate control mice (hereafter called Lmo4fl/fl; Fig. 6 B), but Lmo4fl/flVav-Cre mice had decreased frequencies of αLPs, CHILPs, and ILC1s in the small intestine and liver, ILC2s in the small intestine and lung, and ILC3s in the small intestine (Fig. 6, B–H). Consistently, Lmo4fl/flVav-Cre mice showed reduced total numbers of these cells (Fig. 6 I). In addition, Lmo4 deficiency decreased the number of Peyer’s patches and intestinal lymphoid follicles (Fig. S5 C). However, NK cells in the spleen and blood were not affected after Lmo4 deletion (Fig. S5 D). Then we injected α4β7+ CLPs isolated from Lmo4fl/fl and Lmo4fl/flVav-Cre mice into Rag1−/−Il2rg−/− recipients. After 6 wk, engraftment of Lmo4fl/flVav-Cre α4β7+ CLPs produced fewer numbers of ILCs (Fig. 6 J). However, transplantation of Lmo4fl/flVav-Cre α4β7+ CLPs with Lmo4 overexpression could restore normal numbers of ILCs (Figs. 6 J and S5 E). We next generated Yeats4flox/floxLmo4flox/floxVav-Cre mice (hereafter called DKO; Fig. S5 F). We performed an in vivo differentiation assay by engrafting Yeats4fl/fl, Yeats4fl/flVav-Cre, or DKO α4β7+ CLPs into Rag1−/−Il2rg−/−recipients. We observed that engraftment of DKO α4β7+ CLPs produced much fewer ILCs than that of Yeats4fl/fl ones (Fig. 6 K). However, restoration of Lmo4 in Yeats4fl/flVav-Cre or DKO α4β7+ CLPs could rescue normal numbers of ILCs (Fig. 6 K). In addition, we also conducted in vitro differentiation assay by culturing α4β7+ CLPs with OP9 feeder cells. In parallel, the differentiation ability of Yeats4fl/flVav-Cre and Lmo4fl/flVav-Cre α4β7+ CLPs was remarkably suppressed, whereas the differentiation ability of DKO α4β7+ CLPs was almost impaired (Fig. 6 L). Restoration of Lmo4 in Yeats4fl/flVav-Cre, Lmo4fl/flVav-Cre, or DKO α4β7+ CLPs could rescue their differentiation capacity (Fig. 6 L). These data indicate that Yeats4 and Lmo4 are involved in the regulation of α4β7+ CLP differentiation.
Figure 6.
Lmo4 deficiency impairs ILC commitment and their effector functions. (A) Imaging flow cytometry analysis for Lmo4 expression in α4β7+ progenitors. (B–H) Percentages of α4β7+ CLPs (B), αLPs (B), CHILPs (C), siILC1s (D), liver ILC1s (E), siILC2s (F), lung ILC2s (G), and siILC3s (H) in Lmo4fl/fl and Lmo4fl/flVav-Cre mice were tested by FACS. n = 5 per group. (I) Numbers of indicated cells in Lmo4fl/fl and Lmo4fl/flVav-Cre mice were calculated. (J) Lmo4-overexpressing α4β7+ CLPs isolated from Lmo4fl/flVav-Cre mice were engrafted into Rag1−/−Il2rg−/− mice. After 6 wk, ILCs were analyzed by FACS. (K) Lmo4-overexpressing α4β7+ CLPs isolated from Yeats4fl/flVav-Cre or DKO mice were injected into Rag1−/−Il2rg−/− mice and assayed in J. (L) α4β7+ CLPs from indicated mice were isolated and used for in vitro differentiation assay with OP9 feeder cells. After 12-d culture, ILC1s, ILC2s, and ILC3s were analyzed. (M) Frequencies of IFN-γ+ ILC1s in MLNs from WT, Lmo4fl/flVav-Cre, or DKO mice infected with S. typhimurium were analyzed on day 5 after injection. n = 5 per group. (N) CFUs of S. typhimurium were measured in MLNs from WT, Lmo4fl/flVav-Cre, or DKO mice on day 5 after infection. n = 5 per group. (O) Total numbers of ILC2s and IL-5+ or IL-13+ ILC2s were analyzed in lungs from WT, Lmo4fl/flVav-Cre, or DKO mice after papain challenge. n = 5 per group. (P) H&E staining of lung sections from WT, Lmo4fl/flVav-Cre, or DKO mice treated with papain. Scale bars, 100 µm. (Q) Colons from challenged WT, Lmo4fl/flVav-Cre, or DKO mice were analyzed by H&E staining. Scale bars, 50 µm. n = 5 per group. **, P < 0.01; ***, P < 0.001 by two-tailed unpaired Student’s t test. All data are representative of at least three independent experiments and are expressed as mean ± SD.
We next challenged WT, Lmo4fl/flVav-Cre, or DKO mice with S. typhimurium, papain, or C. rodentium and tested their resistance to infection and tissue inflammation. Lmo4fl/flVav-Cre or DKO mice showed a reduced number of IFN-γ+ ILC1s and a higher number of S. typhimurium CFUs in MLNs (Fig. 6, M and N). Of note, Lmo4fl/flVav-Cre or DKO mice displayed a decreased number of ILC2s in the lung (Fig. 6 O) and a reduced number of IL-5– and IL-13–secreting ILC2s in the lung (Fig. 6 O). Consequently, leukocyte infiltration was also attenuated in lungs of Lmo4fl/flVav-Cre and DKO mice (Fig. 6 P). Consistently, Lmo4fl/flVav-Cre and DKO mice displayed a higher number of bacterial CFUs in feces (Fig. S5 G), weight loss (Fig. S5 H), and shrinking colon length (Fig. S5 I), which were accompanied by more severe intestinal damage (Fig. 6 Q). Taken together, Lmo4 is required for ILC lineage commitment.
Discussion
ILCs exert critical roles in innate immune responses by communicating with hematopoietic or nonhematopoietic cells (Artis and Spits, 2015). ILC lineage specification is a multistep process that is finely regulated (Vivier et al., 2018). In this study, we showed that Yeats4 is highly expressed in ILCs and their progenitors, including αLPs, CHILPs, and ILCPs. Yeats4 conditional KO in the hematopoietic system causes decreased numbers of ILCs and impaired effector functions. Moreover, we demonstrated that Yeats4 regulates α4β7+ CLPs differentiation toward CHILPs. Mechanistically, Yeats4 recruits the Dot1l–RNA Pol II complex onto the Lmo4 promoter by recognizing H3K27ac modification to initiate Lmo4 transcription in α4β7+ CLPs. Importantly, Lmo4 deficiency also impairs ILC lineage commitment and their effector functions. Collectively, Yeats4 drives α4β7+ CLPs to ILC lineage commitment via activation of Lmo4 transcription.
All ILCs are derived from CLPs residing in the BM. Besides cytokine signaling pathways, fate-decision TFs play critical roles in the regulation of ILC lineage differentiation. For instance, CLPs differentiate into αLPs under several fate-decision TFs, such as Nfil3 and Tox (Yu et al., 2014; Seehus et al., 2015). A subpopulation of CHILPs was identified by Id2 reporter mice, suggesting Id2 directs CHILP differentiation (Klose et al., 2014). CHILPs generate multiple ILC lineages, including LTi cells, and include a subpopulation of PLZFhi cells (Vivier et al., 2018). PLZF is linked to the function of NK T cells (Mao et al., 2016), and directs a subset of ILC lineage-specific progenitor cells termed ILCPs. ILCPs express the integrin α4β7 and can generate all ILCs except LTi cells and conventional NK cells (Constantinides et al., 2014). However, whether other factors besides Id2 regulate CHILP differentiation remains elusive. Here, we showed that deletion of Yeats4 or Lmo4 impairs ILC lineage commitment and their effector functions. We conclude that Yeats4 or Lmo4 is required for the CHILP differentiation from α4β7+ CLPs.
Both genetic and epigenetic modulations are involved in the regulation of ILC lineage differentiation (Antignano et al., 2016). Chromatin remodeling is a prerequisite for eukaryotic gene transcription, which depends on ATP-dependent remodeling complexes (also called remodelers). These remodelers modulate chromatin structures through multiple means, including histone modification, DNA methylation, and incorporation of histone variants. We previously showed that the SRCAP remodeling complex regulates lymphoid lineage commitment by Pcid2 (Ye et al., 2017). We also demonstrated that the nucleosome remodeling factor remodeling complex participates in the regulation of in ILC3 maintenance (Liu et al., 2017a). Yeats4 was identified as a subunit of the SRCAP complex and the NuA4 complex and regulates gene expression as a remodeler (Hsu et al., 2018). Of note, Yeats4 is a YEATS domain–containing protein with high conservation among species. The YEATS domain proteins were recently identified as a group of histone acetyl-lysine readers (Li et al., 2014). As a member of histone acetylation reader, Yeats4 is implicated in the oncogenesis of lung cancer (Hsu et al., 2018; Klein et al., 2018). However, how Yeats4 regulates ILC development still unknown. Herein, we showed that Yeats4 can recognize H3K27ac as a histone acetylation reader and recruits the Dot1l–RNA Pol II complex onto the Lmo4 promoter to initiate Lmo4 transcription in α4β7+ CLPs, leading to differentiation toward CHILPs.
H3K27ac was identified as an active mark for gene transcription (Hsu et al., 2018). Dot1l, a methyltransferase, can catalyze H3K79 methylation (Min et al., 2003). The Dot1l-mediated H3K79 methylation links to active transcription by directly interacting with the RNA Pol II complex (Kim et al., 2012). Previous reports showed that Dot1l is implicated in the tumorigenesis of acute myeloid leukemia (Riedel et al., 2016; Okuda et al., 2017). In addition, suppression of DOT1L can alleviate graft-versus-host disease via inhibiting T cell activation (Kagoya et al., 2018). These studies suggest that Dot1l may regulate the development of hematopoietic cells. In this study, we showed that Dot1l interacts with Yeats4 in α4β7+ CLPs and links to the RNA Pol II complex on the Lmo4 promoter, leading to H3K79 methylation of the Lmo4 promoter region. To our knowledge, we are the first to report that Yeats4 recruits the Dot1l–RNA Pol II complex onto the Lmo4 promoter by recognizing H3K27ac to initiate Lmo4 transcription in α4β7+ CLPs, which drives downstream CHILP differentiation.
Lmo4, a member of the LIM-only (LMO) subfamily of LIM domain–containing TFs, was first identified to participate in embryogenesis (Kenny et al., 1998). Some studies showed that Lmo4 is required for neural tube closure (Lee et al., 2005) and assembly of the thalamocortical somatosensory circuit (Wang et al., 2017a). It has been reported that Lmo4 is expressed in the thymus and is implicated in the oncogenesis of T cell acute leukemia (Grutz et al., 1998). In addition, Lmo4 can inhibit differentiation of mammary epithelial cells, and its high expression promotes breast cancer by repressing tumor suppressor BRCA1-mediated transcriptional activation (Visvader et al., 2001; Sum et al., 2002). Lmo4 is also involved in the tumorigenesis of pancreatic cancer (Yu et al., 2008) and non–small cell lung cancer (Wang et al., 2016). Several studies reported that Lmo4 is highly expressed in ILCs and their progenitors (Robinette et al., 2015; Gury-BenAri et al., 2016; Seillet et al., 2016; Harly et al., 2018). Of note, Lmo4 can interact with LIM domain binding 1 to form a dimer that regulates gene transcription (Deane et al., 2004). However, whether Lmo4 regulates ILC development is unclear. Here, we showed that Lmo4 is highly expressed in ILCs and their progenitors. Lmo4 deficiency impairs ILC lineage commitment and their effector functions. We conclude that Lmo4 is required for ILC lineage differentiation. In sum, Yeats4 and Lmo4 are highly expressed in ILCs and their progenitors. Yeats4 recruits the Dot1l–RNA Pol II complex onto the Lmo4 promoter to initiate Lmo4 transcription, which drives differentiation from α4β7+ CLPs to CHILPs, leading to ILC lineage commitment.
Materials and methods
Antibodies and reagents
Anti-H3K4me3 (catalog number 9751), anti-H3K27ac (catalog number 8173), anti-H3K79me3 (catalog number 4260), anti-H3K27me3 (catalog number 9733), anti-RNA Pol II Ser2P (catalog number 13499), anti-Rpb1 (catalog number 2629), and anti–β-actin (catalog number 3700) were from Cell Signaling Technology. Anti-Rpb3 (catalog number ab186867), anti-AF9 (catalog number ab154492), and anti-Rpb4 (catalog number ab186868) were from Abcam. Anti-ENL (catalog number sc-393196) was from Santa Cruz Biotechnology. Anti-Dot1l (catalog number NB100-40845) was from Novus. Anti-Lmo4 (catalog number PA5-24248) and anti-Yeats2 (catalog number PA5-36939) were from Invitrogen. Anti-Yeats4 (catalog number LS-C155530) was from LifeSpan BioScience. The antibody against Flag-tag and CBP/P300 inhibitor (C646; catalog number SML0002) were purchased from Sigma-Aldrich. Anti-CD127 (A7R34), c-Kit (2B8), anti-CD3 (17A2), anti-CD4 (GK1.5), anti-CD19 (1D3), anti-NK1.1 (PK136), anti-CD150 (mShad150), anti-CD34 (RAM34), anti-CD11c (N418), anti-CD45 (30-F11), anti-CD90 (HIS51), anti–IL-22 (IL22JOP), anti-CD45.2 (104), Sca-1 (D7), CD25 (PC61.5), anti-CD45.1 (A20), anti-Flt3 (A2F10), anti-α4β7 (DATK32), anti-RORγt (AFKJS-9), anti-NKp46 (29A1.4), anti-Gata3 (TWAJ), anti-KLRG1 (2F1), anti-PLZF (Mags.21F7), Lineage cocktail (88–7772-72), anti-CD48 (HM48-1), anti-CD16/32 (93), anti-CD8 (53–6.7), anti–IFN-γ (XMG1.2), anti–IL-5 (TRFK5), anti–IL-13 (eBio13A), and anti–Siglec-F (1RNM44N) were purchased from eBioscience. Paraformaldehyde (PFA) and DAPI were from Sigma-Aldrich. IL-5, IL-13, and IL-22 ELISA kits were purchased from eBioscience.
Generation of Yeats4Flag knockin and Yeats4flox/flox mice by CRISPR/Cas9 technology
For generation of Yeats4Flag knockin mice, the CRISPR-mediated single-stranded oligodecxynucleotide donor was synthesized and in-frame knocked into the endogenous Yeats4 locus immediately before the stop codon. Injected zygotes were transferred into the uterus of pseudopregnant ICR females. Knockin Flag was identified by PCR screening and DNA sequencing. Single-guide RNA (sgRNA) sequences are listed in Table S1. For generation of Yeats4flox/flox mice, two loxP elements were flanked on exon 2 of the Yeats4 gene using CRISPR/Cas9 approaches as described previously (Zhu et al., 2014). Approximately 250 zygotes from C57BL/6 mice were injected with sgRNAs and subsequently transferred to the uterus of pseudopregnant ICR females from which viable founder mice were obtained. loxP knockin was identified by PCR screening and DNA sequencing. Vav-Cre, PLZFGFPcre, B6.129S4-Il2rgtm1Wjl/J;Rorc(γt)+/GFP, and Id2+/GFP mice were purchased from The Jackson Laboratory. Lmo4flox/flox mice were kindly provided by Drs. Sam Pfaff (Salk Institute for Biological Studies, La Jolla, CA), Soo-Kyung Lee (Oregon Health and Science University, Portland, OR), and Shen-Ju Chou (Institute of Cellular and Organismic Biology, Academia Sinica, Taipei, Taiwan). Yeats4flox/floxVav-Cre mice were obtained by crossing Yeats4flox/flox mice with Vav-Cre mice. Lmo4flox/floxVav-Cre mice were obtained by crossing Lmo4flox/flox mice with Vav-Cre mice. Rag1−/−Il2rg−/− mice were generated by crossing Rag1+/− mice (from the Model Animal Research Center, Nanjing University, Nanjing, China) with Il2rg+/− mice. All the mouse strains were C57BL/6 background and maintained under specific pathogen–free conditions with approval by the Institutional Committee of Institute of Biophysics, Chinese Academy of Sciences. The study is compliant with all relevant ethical regulations regarding animal research.
Gene KO by CRISPR/Cas9-mediated genome editing in vitro
In vitro deletion was performed using CRISPR/Cas9-mediated genome editing as previously described (Liu et al., 2017b). In brief, α4β7+ CLPs (Lin−CD25−CD127+c-KitintSca-1intFlt3+α4β7+) isolated by FACS from 129-Gt(ROSA)26Sortm1(CAG-cas9*,-EGFP)FezhVav-Cre mice were infected with LentiCRISPRv2 virus containing sgRNAs targeting ENL, Yeats2, Yeats4, AF9, Dot1l, or control.
Intestinal lymphocyte separation
The protocol for isolating lymphocytes from small intestines has been described previously (Liu et al., 2017a). Briefly, mice were sacrificed, and small intestines were isolated. After removal of Peyer’s patches, small intestines were cut open longitudinally and washed five times using PBS. Then, intestines were cut into pieces and washed five times using solution I buffer (10 mM HEPES and 5 mM EDTA in Hanks’ balanced salt solution). Afterwards, intestine fragments were digested into LPLs with solution II buffer (DNase I, 5% FBS, and 0.5 mg/ml collagenase II and collagenase III). LPL cells were then sifted through 70-µm strainers and used for experiments.
Flow cytometry
BM cells were flushed out from femurs in PBS (containing 5% FBS) buffer and sifted through 70-µm cell strainers. LPL cells were isolated as described above. For flow cytometric analysis, LT-HSCs (Lin−Sca-1+c-Kit+CD150+CD48−), ST-HSCs (Lin−Sca-1+c-Kit+CD150−CD48−), MPPs (Lin−Sca-1+c-Kit+CD150−CD48+), CMPs (Lin−Sca-1−c-Kit+CD34+CD16/32−), CLPs (Lin−CD127+c-KitintSca-1intFlt3+α4β7−), α4β7+ CLPs (Lin−CD127+c-KitintSca-1intFlt3+α4β7+), αLPs (Lin−CD25−CD127+c-KitintSca-1intFlt3−α4β7+), CHILPs (Lin−CD25−CD127+Flt3−α4β7+Id2+), ILCPs (Lin−CD127+Flt3−c-Kit+α4β7+PLZF+), siILC1s (CD3−CD19−CD127+NK1.1+NKp46+), liver ILC1s (CD3−CD19−CD127+NK1.1+NKp46+), siILC2s (Lin−CD127+CD90+KLRG1+Gata3+), lung ILC2s (Lin−CD45+CD90+KLRG1+Sca-1+), siILC3s (Lin−RORγt+CD45+), and NK1.1+ NK, CD19+ B, and CD3+ T cell populations were analyzed or sorted with a FACSAria III instrument (BD Biosciences). PLZFGFPcre mice were used for ILCP (Lin−CD127+α4β7+PLZFGFP) isolation, and Id2+/GFP mice were used for CHILP isolation by FACS. For imaging flow cytometry, cells were further stained with DAPI for nuclei visualization. Then cells were analyzed by imaging flow cytometer (Amnis ImageStream MakII; Merck), and data were analyzed using IDEAS software (Merck).
Immunofluorescence staining
α4β7+ CLPs (Lin−CD127+c-KitintSca-1intFlt3+α4β7+) isolated by FACS or intestinal frozen sections were fixed with 4% PFA (Sigma-Aldrich) for 20 min at room temperature, perforated with PBS containing 1% Triton X-100 for 20 min, blocked with 5% donkey and 5% rat serum for 1 h at room temperature, incubated with appropriate primary antibodies at 4°C overnight, and then incubated with fluorescence-conjugated secondary antibodies. DAPI was used for nucleus staining. Cells were visualized with an Olympus FV1200 laser scanning confocal microscopy. Intestinal frozen sections were visualized with an AxioImagerM2 upright microscope (Zeiss).
DNA FISH
α4β7+ CLPs (Lin−CD127+c-KitintSca-1intFlt3+α4β7+) were isolated by FACS and fixed with 4% PFA containing 10% acetic acid for 20 min at room temperature. Then cells were treated with 70% ethanol at −20°C and incubated in buffer containing 100 mM Tris-HCl (pH 7.5) and 150 mM NaCl, followed by cytoplasm digestion in 0.01% pepsin/0.01 N HCl for 3 min at 37°C. Cells were further fixed with 3.7% PFA and replaced with ethanol to a final concentration of 100%. Afterwards, cells were air dried and washed using 2× saline-sodium citrate and blocked using buffer containing 100 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.05% Tween 20, and 3% BSA for 20 min. Then, cells were denatured in 70% formamide/2× saline-sodium citrate and incubated with fluorescence-labeled DNA probes overnight. After staining with DAPI for nucleus, cells were visualized by Olympus FV1200 laser scanning confocal microscopy.
qRT-PCR
Total RNA was isolated from cell populations using an RNA Miniprep Kit (Tiangen) according to the manufacturer’s protocol as previously described (Liu et al., 2017b). Then, cDNA was synthesized with M-MLV reverse transcription (Promega). mRNA transcripts were analyzed with the ABI 7300 quantitative PCR (qPCR) system using the specific primer pairs listed in Table S2. Relative expression levels were calculated and normalized to endogenous Actb.
ChIP assay
α4β7+ progenitors (Lin−CD127+c-KitintSca-1intα4β7+; 1 × 105) were isolated by FACS and cross-linked with 1% formaldehyde at 37°C for 10 min. Then, cells were washed twice with PBS, lysed with SDS lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris) and sonicated to make 200–500-bp DNA fragments. Lysates were precleared with protein A agarose/salmon sperm DNA (50% slurry) and then incubated with 4 µg antibody overnight at 4°C. Then, protein A agarose/salmon sperm DNA (50% slurry) beads were added and incubated for 4 h. After washing, DNA was eluted from beads and purified. DNA fragments were analyzed using primer pairs listed in Table S2. For ChIP immunoblotting assay, BM cells were cross-linked with 1% formaldehyde and lysed with SDS lysis buffer and sonicated to 200–500 bp. Lysates were precleared with protein A agarose/salmon sperm DNA (50% slurry). Then, 4 µg antibody was incubated with treated lysates for ChIP assays, followed by size fractionation with sucrose gradient ultracentrifugation. 500 µl eluate was put onto 30 ml 5–30% (vol/vol) sucrose gradient followed by ultracentrifugation at 55,000 g with an S Beckman SW28 rotor. Eluent gradients were concentrated and reversely cross-linked. Proteins were examined by Western blotting. DNA was extracted and examined by PCR assays.
DNase I accessibility assay
DNase I digestion assay was described previously (Ye et al., 2017). In brief, nuclei were purified from α4β7+ progenitors (Lin−CD127+c-KitintSca-1intα4β7+) according to the manufacturer’s protocol with a nuclei isolating kit (Sigma-Aldrich). Then, nuclei were resuspended with DNase I digestion buffer and treated with indicated units of DNase I (Sigma-Aldrich) at 37°C for 5 min 2 × DNase I stop buffer (20 mM Tris, pH 8.0, 4 mM EDTA, and 2 mM EGTA) was added to stop reactions. DNA was extracted and examined by qPCR.
BM transplantation
5 × 104 CD45.2+ LSK cells (Lin−Sca-1+c-Kit+) from Yeats4+/+ and Yeats4−/− mice with 5 × 106 CD45.1+ helper cells were transplanted into lethally irradiated CD45.1+ recipients. 8 wk after transplantation, percentages of ILCs derived from donor cells were analyzed by FACS. For competitive transplantation, 1 × 106 CD45.2+ BM cells from Yeats4+/+ or Yeats4−/− mice and 1 × 106 WT CD45.1+ BM cells were injected into lethally irradiated CD45.1+ recipient mice. 8 wk after transplantation, ratios of CD45.2+ ILCs to CD45.1+ ILCs were analyzed.
In vitro ILC differentiation assay
α4β7+ CLPs were isolated from Yeats4+/+ or Yeats4−/− mice. For Yeats4 or Lmo4 overexpression, a DNA fragment encoding Yeats4 or Lmo4 was constructed into a pMY-IRES-EGFP vector and flowed by transfection into Platinum-E cells to generate recombinant retrovirus. Then α4β7+ CLPs were infected with retrovirus for 1 d, followed by additional assays. In vitro ILC differentiation assay was described previously (Constantinides et al., 2014). In brief, α4β7+ CLPs were cultured on mitomycin C–treated OP9 feeder cells supplemented with IL-7 (25 ng/ml; catalog number 217–17) and recombinant stem cell factor (25 ng/ml; catalog number 250–03) for 12 d. Then cells were collected for flow cytometry analysis.
In vivo ILC differentiation assay
5 × 103 α4β7+ CLPs were isolated from Yeats4fl/fl, Yeats4fl/flVav-Cre, Lmo4fl/fl, or Lmo4fl/flVav-Cre mice and adoptively transferred into Rag1−/−Il2rg−/− mice. 6 wk later, ILCs were analyzed using FACS.
C. rodentium and S. typhimurium infection
For C. rodentium infection, WT, Yeats4fl/flVav-Cre, and Lmo4fl/flVav-Cre mice were made fast for 8 h before being infected with 5 × 109 C. rodentium orally as described previously (Ebihara et al., 2015). C. rodentium was a gift from Dr. Baoxue Ge (Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China). Mice were sacrificed to examine colon pathology and bacterial loads on day 6 after infection. Feces and colons were collected from infected mice on day 6 after infection. Feces were weighed and homogenized, and homogenates were plated on MacConkey agar plates for analysis of bacterial counts. LPLs were isolated from small intestines of infected mice, followed by analysis of ILC3s and IL-22 production. S. typhimurium (from the Institute of Microbiology, Chinese Academy of Sciences, Beijing, China) infection was performed as previously reported (Klose et al., 2013).
Papain administration
Mice were anesthetized with isoflurane and administrated with papain (25 µg in 50 µl PBS; Sigma-Aldrich) via intranasal instillation on days 0, 1, and 3. Then, BALF and lungs were collected and analyzed on day 4.
Histology analysis
Mouse colons and lungs were removed, fixed in 4% PFA for 48 h, washed with PBS, and stored in 75% ethanol before being embedded in paraffin. Then, colons and lungs were sectioned and stained with H&E according to standard laboratory procedures.
Immunoprecipitation assay
BM cells from Yeats4Flag knockin mice were lysed, and supernatants were incubated with anti-Flag or IgG control. Then precipitated components were separated with SDS-PAGE and silver staining. Differential bands enriched by anti-Flag were analyzed by LTQ Orbitrap XL mass spectrometry or immunoblotting.
Nuclear run-on assay
Sorted cells were harvested in buffer containing 150 mM KCl, 10 mM Tris-HCl, 4 mM MgOAc with pH 7.4, followed by centrifugation to collect cell pellets. Pellets were lysed in buffer containing 150 mM KCl, 10 mM Tris-HCl, 4 mM MgOAc, and 0.5% NP-40, followed by sucrose density gradient centrifugation to prepare crude nuclei. Crude nuclei were incubated with 10 mM ATP, CTP, GTP, BrUTP, and RNase inhibitor at 28°C for 5 min. RNAs were extracted using TRIzol reagent with manufacturer’s guidelines, followed by DNA digestion with DNase I. RNA transcripts were immunoprecipitated with antibody against BrdU, followed by reverse transcription and RT-PCR analysis. 5 × 104 α4β7+ progenitors (Lin−CD127+c-KitintSca-1intα4β7+) were used for each single sample.
Microarray assay
RNAs from Yeats4fl/fl and Yeats4fl/flVav-Cre α4β7+ progenitors (Lin−CD127+c-KitintSca-1intα4β7+) were isolated using Trizol reagent (Invitrogen) and prepared for Affymetrix mRNA microarray assay by Beijing Cnkingbio Biotechnology. Microarray data have been deposited under GEO accession no. GSE134314.
EMSA
Biotin-labeled DNA fragment (approximately −400 to −200 bp from the transcription start site) of the Lmo4 promoter were synthesized by Invitrogen. Probes and Flag-Yeats4 proteins were incubated in binding buffer, and a mobility shift assay was performed using the Light Shift Chemiluminescent RNA EMSA Kit (Thermo Scientific) according to the manufacturer’s protocol.
ELISA
ILCs from treated mice were isolated and cultured for 24 h with indicated cytokines. Then supernatants were collected and cytokines were detected using ELISA kit (eBioscience) according to the manufacturer’s instructions.
Statistical analysis
For statistical evaluation, an unpaired Student’s t test for two-group comparison or one-way ANOVA with Tukey’s test for multiple comparisons was applied for calculating statistical probabilities in this study. For all panels, at least three independent experiments were performed with similar results, and representative experiments are shown. Data were analyzed by using Microsoft Excel or SPSS 22. All flow cytometry data were analyzed with FlowJo 10 (Treestar). Adobe Photoshop CC 14.0 and ImageJ 1.48 were used for figure presentation. Two-tailed unpaired Student’s t test was performed using Excel 2010. P values ≤ 0.05 were considered significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, P > 0.05).
Online supplemental material
Fig. S1 shows the gating strategies used for different hematopoietic cell populations. Fig. S2 shows the construction strategy used for Yeats4-deficient mice. Fig. S3 shows that Yeats4 deletion does not affect the frequencies of HSCs and CMPs. Fig. S4 shows that Yeats4 interacts with Dot1l. Fig. S5 shows that Lmo4 deficiency impairs effector functions of ILC3s. Table S1 lists the sgRNA sequences used in this study for gene editing by CRISPR/Cas9. Table S2 lists the primer oligonucleotides used in this study for qPCR and ChIP-qPCR assays.
Supplementary Material
Acknowledgments
We thank Dongdong Fan, Yan Teng, and Junying Jia for technical support. We also thank Jing Li (Cnkingbio Company, Beijing, China) for technical support.
This work was supported by Strategic Priority Research Programs of the Chinese Academy of Sciences (XDA19050301, XDA12020219, and XDB19030203), the National Natural Science Foundation of China (91640203, 31530093, 31728006, 31771638, 81772646, 81572433, 31870883, 31871494, and 31670886), and the Beijing Natural Science Foundation (7181006).
The authors declare no competing financial interests.
Author contributions: B. Liu and L. Yang performed experiments; B. Liu analyzed data and wrote the paper; X. Zhu and S. Meng constructed genetic mouse strains; P. Zhu, H. Li, J. Wu, T. Lu, and L. He performed some experiments; L. Zhou analyzed data; B. Ye performed experiments and analyzed data; Y. Tian initiated the study and analyzed data; and Z. Fan initiated the study and organized, designed, and wrote the paper.
References
- Antignano F., Braam M., Hughes M.R., Chenery A.L., Burrows K., Gold M.J., Oudhoff M.J., Rattray D., Halim T.Y., Cait A., et al. 2016. G9a regulates group 2 innate lymphoid cell development by repressing the group 3 innate lymphoid cell program. J. Exp. Med. 213:1153–1162. 10.1084/jem.20151646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artis D., and Spits H.. 2015. The biology of innate lymphoid cells. Nature. 517:293–301. 10.1038/nature14189 [DOI] [PubMed] [Google Scholar]
- Bando J.K., Gilfillan S., Song C., McDonald K.G., Huang S.C., Newberry R.D., Kobayashi Y., Allan D.S.J., Carlyle J.R., Cella M., and Colonna M.. 2018. The Tumor Necrosis Factor Superfamily Member RANKL Suppresses Effector Cytokine Production in Group 3 Innate Lymphoid Cells. Immunity. 48:1208–1219.e4. 10.1016/j.immuni.2018.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernink J.H., Krabbendam L., Germar K., de Jong E., Gronke K., Kofoed-Nielsen M., Munneke J.M., Hazenberg M.D., Villaudy J., Buskens C.J., et al. 2015. Interleukin-12 and -23 Control Plasticity of CD127(+) Group 1 and Group 3 Innate Lymphoid Cells in the Intestinal Lamina Propria. Immunity. 43:146–160. 10.1016/j.immuni.2015.06.019 [DOI] [PubMed] [Google Scholar]
- Brestoff J.R., Kim B.S., Saenz S.A., Stine R.R., Monticelli L.A., Sonnenberg G.F., Thome J.J., Farber D.L., Lutfy K., Seale P., and Artis D.. 2015. Group 2 innate lymphoid cells promote beiging of white adipose tissue and limit obesity. Nature. 519:242–246. 10.1038/nature14115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides M.G., McDonald B.D., Verhoef P.A., and Bendelac A.. 2014. A committed precursor to innate lymphoid cells. Nature. 508:397–401. 10.1038/nature13047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane J.E., Ryan D.P., Sunde M., Maher M.J., Guss J.M., Visvader J.E., and Matthews J.M.. 2004. Tandem LIM domains provide synergistic binding in the LMO4:Ldb1 complex. EMBO J. 23:3589–3598. 10.1038/sj.emboj.7600376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbach A., Colonna M., and Koyasu S.. 2014. Development, differentiation, and diversity of innate lymphoid cells. Immunity. 41:354–365. 10.1016/j.immuni.2014.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbach A., Colonna M., and Romagnani C.. 2017. The ILC World Revisited. Immunity. 46:327–332. 10.1016/j.immuni.2017.03.008 [DOI] [PubMed] [Google Scholar]
- Eberl G., Colonna M., Di Santo J.P., and McKenzie A.N.. 2015. Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science. 348:aaa6566 10.1126/science.aaa6566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara T., Song C., Ryu S.H., Plougastel-Douglas B., Yang L., Levanon D., Groner Y., Bern M.D., Stappenbeck T.S., Colonna M., et al. 2015. Runx3 specifies lineage commitment of innate lymphoid cells. Nat. Immunol. 16:1124–1133. 10.1038/ni.3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb M.A., Scott T.G., Li B.E., Xie H., Paulk J., Seo H.S., Souza A., Roberts J.M., Dastjerdi S., Buckley D.L., et al. 2017. Transcription control by the ENL YEATS domain in acute leukaemia. Nature. 543:270–274. 10.1038/nature21688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grutz G., Forster A., and Rabbitts T.H.. 1998. Identification of the LMO4 gene encoding an interaction partner of the LIM-binding protein LDB1/NLI1: a candidate for displacement by LMO proteins in T cell acute leukaemia. Oncogene. 17:2799–2803. 10.1038/sj.onc.1202502 [DOI] [PubMed] [Google Scholar]
- Gury-BenAri M., Thaiss C.A., Serafini N., Winter D.R., Giladi A., Lara-Astiaso D., Levy M., Salame T.M., Weiner A., David E., et al. 2016. The Spectrum and Regulatory Landscape of Intestinal Innate Lymphoid Cells Are Shaped by the Microbiome. Cell. 166:1231–1246.e13. 10.1016/j.cell.2016.07.043 [DOI] [PubMed] [Google Scholar]
- Harly C., Cam M., Kaye J., and Bhandoola A.. 2018. Development and differentiation of early innate lymphoid progenitors. J. Exp. Med. 215:249–262. 10.1084/jem.20170832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyler T., Klose C.S., Souabni A., Turqueti-Neves A., Pfeifer D., Rawlins E.L., Voehringer D., Busslinger M., and Diefenbach A.. 2012. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 37:634–648. 10.1016/j.immuni.2012.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C.C., Shi J., Yuan C., Zhao D., Jiang S., Lyu J., Wang X., Li H., Wen H., Li W., and Shi X.. 2018. Recognition of histone acetylation by the GAS41 YEATS domain promotes H2A.Z deposition in non-small cell lung cancer. Genes Dev. 32:58–69. 10.1101/gad.303784.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagoya Y., Nakatsugawa M., Saso K., Guo T., Anczurowski M., Wang C.H., Butler M.O., Arrowsmith C.H., and Hirano N.. 2018. DOT1L inhibition attenuates graft-versus-host disease by allogeneic T cells in adoptive immunotherapy models. Nat. Commun. 9:1915 10.1038/s41467-018-04262-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny D.A., Jurata L.W., Saga Y., and Gill G.N.. 1998. Identification and characterization of LMO4, an LMO gene with a novel pattern of expression during embryogenesis. Proc. Natl. Acad. Sci. USA. 95:11257–11262. 10.1073/pnas.95.19.11257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.K., Jung I., Lee H., Kang K., Kim M., Jeong K., Kwon C.S., Han Y.M., Kim Y.S., Kim D., and Lee D.. 2012. Human histone H3K79 methyltransferase DOT1L protein [corrected] binds actively transcribing RNA polymerase II to regulate gene expression. J. Biol. Chem. 287:39698–39709. 10.1074/jbc.M112.384057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss E.A., Vonarbourg C., Kopfmann S., Hobeika E., Finke D., Esser C., and Diefenbach A.. 2011. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 334:1561–1565. 10.1126/science.1214914 [DOI] [PubMed] [Google Scholar]
- Klein B.J., Ahmad S., Vann K.R., Andrews F.H., Mayo Z.A., Bourriquen G., Bridgers J.B., Zhang J., Strahl B.D., Côté J., and Kutateladze T.G.. 2018. Yaf9 subunit of the NuA4 and SWR1 complexes targets histone H3K27ac through its YEATS domain. Nucleic Acids Res. 46:421–430. 10.1093/nar/gkx1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose C.S., Kiss E.A., Schwierzeck V., Ebert K., Hoyler T., d’Hargues Y., Göppert N., Croxford A.L., Waisman A., Tanriver Y., and Diefenbach A.. 2013. A T-bet gradient controls the fate and function of CCR6-RORγt+ innate lymphoid cells. Nature. 494:261–265. 10.1038/nature11813 [DOI] [PubMed] [Google Scholar]
- Klose C.S.N., Flach M., Möhle L., Rogell L., Hoyler T., Ebert K., Fabiunke C., Pfeifer D., Sexl V., Fonseca-Pereira D., et al. 2014. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 157:340–356. 10.1016/j.cell.2014.03.030 [DOI] [PubMed] [Google Scholar]
- Lee S.K., Jurata L.W., Nowak R., Lettieri K., Kenny D.A., Pfaff S.L., and Gill G.N.. 2005. The LIM domain-only protein LMO4 is required for neural tube closure. Mol. Cell. Neurosci. 28:205–214. 10.1016/j.mcn.2004.04.010 [DOI] [PubMed] [Google Scholar]
- Li Q., Li D., Zhang X., Wan Q., Zhang W., Zheng M., Zou L., Elly C., Lee J.H., and Liu Y.C.. 2018. E3 Ligase VHL Promotes Group 2 Innate Lymphoid Cell Maturation and Function via Glycolysis Inhibition and Induction of Interleukin-33 Receptor. Immunity. 48:258–270.e5. 10.1016/j.immuni.2017.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wen H., Xi Y., Tanaka K., Wang H., Peng D., Ren Y., Jin Q., Dent S.Y., Li W., et al. 2014. AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell. 159:558–571. 10.1016/j.cell.2014.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Ye B., Yang L., Zhu X., Huang G., Zhu P., Du Y., Wu J., Qin X., Chen R., et al. 2017a Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat. Immunol. 18:499–508. 10.1038/ni.3712 [DOI] [PubMed] [Google Scholar]
- Liu B., Ye B., Zhu X., Huang G., Yang L., Zhu P., Du Y., Wu J., Meng S., Tian Y., and Fan Z.. 2017b IL-7Rα glutamylation and activation of transcription factor Sall3 promote group 3 ILC development. Nat. Commun. 8:231 10.1038/s41467-017-00235-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao A.P., Constantinides M.G., Mathew R., Zuo Z., Chen X., Weirauch M.T., and Bendelac A.. 2016. Multiple layers of transcriptional regulation by PLZF in NKT-cell development. Proc. Natl. Acad. Sci. USA. 113:7602–7607. 10.1073/pnas.1601504113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi W., Guan H., Lyu J., Zhao D., Xi Y., Jiang S., Andrews F.H., Wang X., Gagea M., Wen H., et al. 2017. YEATS2 links histone acetylation to tumorigenesis of non-small cell lung cancer. Nat. Commun. 8:1088 10.1038/s41467-017-01173-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J., Feng Q., Li Z., Zhang Y., and Xu R.M.. 2003. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell. 112:711–723. 10.1016/S0092-8674(03)00114-4 [DOI] [PubMed] [Google Scholar]
- Mjösberg J., Bernink J., Golebski K., Karrich J.J., Peters C.P., Blom B., te Velde A.A., Fokkens W.J., van Drunen C.M., and Spits H.. 2012. The transcription factor GATA3 is essential for the function of human type 2 innate lymphoid cells. Immunity. 37:649–659. 10.1016/j.immuni.2012.08.015 [DOI] [PubMed] [Google Scholar]
- Okuda H., Stanojevic B., Kanai A., Kawamura T., Takahashi S., Matsui H., Takaori-Kondo A., and Yokoyama A.. 2017. Cooperative gene activation by AF4 and DOT1L drives MLL-rearranged leukemia. J. Clin. Invest. 127:1918–1931. 10.1172/JCI91406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel S.S., Haladyna J.N., Bezzant M., Stevens B., Pollyea D.A., Sinha A.U., Armstrong S.A., Wei Q., Pollock R.M., Daigle S.R., et al. 2016. MLL1 and DOT1L cooperate with meningioma-1 to induce acute myeloid leukemia. J. Clin. Invest. 126:1438–1450. 10.1172/JCI80825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinette M.L., Fuchs A., Cortez V.S., Lee J.S., Wang Y., Durum S.K., Gilfillan S., and Colonna M.. Immunological Genome Consortium . 2015. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 16:306–317. 10.1038/ni.3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze J.M., Wang A.Y., and Kobor M.S.. 2009. YEATS domain proteins: a diverse family with many links to chromatin modification and transcription. Biochem. Cell Biol. 87:65–75. 10.1139/O08-111 [DOI] [PubMed] [Google Scholar]
- Seehus C.R., Aliahmad P., de la Torre B., Iliev I.D., Spurka L., Funari V.A., and Kaye J.. 2015. The development of innate lymphoid cells requires TOX-dependent generation of a common innate lymphoid cell progenitor. Nat. Immunol. 16:599–608. 10.1038/ni.3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seillet C., Rankin L.C., Groom J.R., Mielke L.A., Tellier J., Chopin M., Huntington N.D., Belz G.T., and Carotta S.. 2014. Nfil3 is required for the development of all innate lymphoid cell subsets. J. Exp. Med. 211:1733–1740. 10.1084/jem.20140145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seillet C., Mielke L.A., Amann-Zalcenstein D.B., Su S., Gao J., Almeida F.F., Shi W., Ritchie M.E., Naik S.H., Huntington N.D., et al. 2016. Deciphering the Innate Lymphoid Cell Transcriptional Program. Cell Reports. 17:436–447. 10.1016/j.celrep.2016.09.025 [DOI] [PubMed] [Google Scholar]
- Serafini N., Vosshenrich C.A., and Di Santo J.P.. 2015. Transcriptional regulation of innate lymphoid cell fate. Nat. Rev. Immunol. 15:415–428. 10.1038/nri3855 [DOI] [PubMed] [Google Scholar]
- Sui P., Wiesner D.L., Xu J., Zhang Y., Lee J., Van Dyken S., Lashua A., Yu C., Klein B.S., Locksley R.M., et al. 2018. Pulmonary neuroendocrine cells amplify allergic asthma responses. Science. 360:eaan8546 10.1126/science.aan8546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sum E.Y., Peng B., Yu X., Chen J., Byrne J., Lindeman G.J., and Visvader J.E.. 2002. The LIM domain protein LMO4 interacts with the cofactor CtIP and the tumor suppressor BRCA1 and inhibits BRCA1 activity. J. Biol. Chem. 277:7849–7856. 10.1074/jbc.M110603200 [DOI] [PubMed] [Google Scholar]
- Visvader J.E., Venter D., Hahm K., Santamaria M., Sum E.Y.M., O’Reilly L., White D., Williams R., Armes J., and Lindeman G.J.. 2001. The LIM domain gene LMO4 inhibits differentiation of mammary epithelial cells in vitro and is overexpressed in breast cancer. Proc. Natl. Acad. Sci. USA. 98:14452–14457. 10.1073/pnas.251547698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier E., Artis D., Colonna M., Diefenbach A., Di Santo J.P., Eberl G., Koyasu S., Locksley R.M., McKenzie A.N.J., Mebius R.E., et al. 2018. Innate Lymphoid Cells: 10 Years On. Cell. 174:1054–1066. 10.1016/j.cell.2018.07.017 [DOI] [PubMed] [Google Scholar]
- Wan L., Wen H., Li Y., Lyu J., Xi Y., Hoshii T., Joseph J.K., Wang X., Loh Y.E., Erb M.A., et al. 2017. ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature. 543:265–269. 10.1038/nature21687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.F., Hsing H.W., Zhuang Z.H., Wen M.H., Chang W.J., Briz C.G., Nieto M., Shyu B.C., and Chou S.J.. 2017a Lhx2 Expression in Postmitotic Cortical Neurons Initiates Assembly of the Thalamocortical Somatosensory Circuit. Cell Reports. 18:849–856. 10.1016/j.celrep.2017.01.001 [DOI] [PubMed] [Google Scholar]
- Wang S., Xia P., Chen Y., Qu Y., Xiong Z., Ye B., Du Y., Tian Y., Yin Z., Xu Z., and Fan Z.. 2017b Regulatory Innate Lymphoid Cells Control Innate Intestinal Inflammation. Cell. 171:201–216.e18. 10.1016/j.cell.2017.07.027 [DOI] [PubMed] [Google Scholar]
- Wang W., Wu S., Guo M., and He J.. 2016. LMO4 is a prognostic marker involved in cell migration and invasion in non-small-cell lung cancer. J. Thorac. Dis. 8:3682–3690. 10.21037/jtd.2016.12.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Jin J., Chung M.W.H., Feng L., Sun H., and Hao Q.. 2018. Identification of the YEATS domain of GAS41 as a pH-dependent reader of histone succinylation. Proc. Natl. Acad. Sci. USA. 115:2365–2370. 10.1073/pnas.1717664115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Li F., Harly C., Xing S., Ye L., Xia X., Wang H., Wang X., Yu S., Zhou X., et al. 2015. TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nat. Immunol. 16:1044–1050. 10.1038/ni.3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B., Liu B., Yang L., Huang G., Hao L., Xia P., Wang S., Du Y., Qin X., Zhu P., et al. 2017. Suppression of SRCAP chromatin remodelling complex and restriction of lymphoid lineage commitment by Pcid2. Nat. Commun. 8:1518 10.1038/s41467-017-01788-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Ohuchida K., Nakata K., Mizumoto K., Cui L., Fujita H., Yamaguchi H., Egami T., Kitada H., and Tanaka M.. 2008. LIM only 4 is overexpressed in late stage pancreas cancer. Mol. Cancer. 7:93 10.1186/1476-4598-7-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Wang Y., Deng M., Li Y., Ruhn K.A., Zhang C.C., and Hooper L.V.. 2014. The basic leucine zipper transcription factor NFIL3 directs the development of a common innate lymphoid cell precursor. eLife. 3:e04406 10.7554/eLife.04406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner G.E., and Henikoff S.. 2013. Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Biol. 20:259–266. 10.1038/nsmb.2470 [DOI] [PubMed] [Google Scholar]
- Zhou C.Y., Johnson S.L., Gamarra N.I., and Narlikar G.J.. 2016. Mechanisms of ATP-Dependent Chromatin Remodeling Motors. Annu. Rev. Biophys. 45:153–181. 10.1146/annurev-biophys-051013-022819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Xu Y., Yu S., Lu L., Ding M., Cheng J., Song G., Gao X., Yao L., Fan D., et al. 2014. An efficient genotyping method for genome-modified animals and human cells generated with CRISPR/Cas9 system. Sci. Rep. 4:6420 10.1038/srep06420 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.