It is thought that the Nlrp6 inflammasome regulates formation of the inner mucus layer (IML) barrier that prohibits contact between the microbiota and colonic epithelial cells. Using microbiota-controlled mice and combined ex vivo/in vivo IML analytical approaches, Volk et al. delineate the relative roles of the inflammasome and microbiota in shaping IML formation and function.
Abstract
The inner mucus layer (IML) is a critical barrier that protects the colonic epithelium from luminal threats and inflammatory bowel disease. Innate immune signaling is thought to regulate IML formation via goblet cell Nlrp6 inflammasome activity that controls secretion of the mucus structural component Muc2. We report that isolated colonic goblet cells express components of several inflammasomes; however, analysis of IML properties in multiple inflammasome-deficient mice, including littermate-controlled Nlrp6−/−, detect a functional IML barrier in all strains. Analysis of mice lacking inflammasome substrate cytokines identifies a defective IML in Il18−/− mice, but this phenotype is ultimately traced to a microbiota-driven, Il18-independent effect. Analysis of phenotypic transfer between IML-deficient and IML-intact mice finds that the Bacteroidales family S24-7 (Muribaculaceae) and genus Adlercrutzia consistently positively covary with IML barrier function. Together, our results demonstrate that baseline IML formation and function is independent of inflammasome activity and highlights the role of the microbiota in determining IML barrier function.
Introduction
The intestinal mucosa are protected from luminal threats by epithelial cells overlaid with a layer of mucus that is structurally dependent on the polymeric protein Muc2. This protein is secreted by intestinal goblet cells and forms a barrier that separates the microbiota from the epithelial surface (Johansson et al., 2008). Disruption of this barrier increases susceptibility to infectious pathogens and the risk of inflammatory bowel disease (Bergstrom et al., 2010; Johansson et al., 2014; Desai et al., 2016) The structure and properties of the mucus layer vary depending on location along the gastrointestinal tract; in the distal colon, there is an inner mucus layer (IML) that is attached to the epithelial surface and is impenetrable to bacteria-sized objects. This defensive structure is necessitated by the vast number of bacteria that comprise the colonic microbiota. The distal colonic IML is converted to an unattached outer mucus layer that serves as a habitat and nutritional substrate for elements of the microbiota (Johansson et al., 2008).
Factors that can influence IML formation and function have been identified. Bacterial colonization appears to regulate formation of the IML as germ-free mice lack a functioning IML (Johansson et al., 2015; Desai et al., 2016). The configuration of the microbiota also influences IML function, as genetically identical mice from different breeding colonies show microbiota-dependent IML variation (Jakobsson et al., 2015). Several endogenous factors that promote IML function have also been identified. These include the core O-glycan glycosyltransferases that sheath the Muc2 peptide backbone with a protective layer of glycans (Fu et al., 2011; Bergstrom et al., 2017), as well as goblet cell–secreted proteins (e.g., Zg16 and Relmβ) that protect the IML from infiltration by specific bacterial taxa (Bergström et al., 2016; Propheter et al., 2017). However, despite recent advances in this area, our understanding of the mechanistic processes that regulate both the formation of the IML and its barrier capacity remains incomplete.
The proposed role of the microbiota in shaping the IML indicates that a promising area of IML regulatory research is the innate immune system, as this incorporates different signaling platforms that can detect multiple microbe-associated molecular patterns. One such platform is the inflammasome, a protein complex that links sensing of microbial molecules or cellular stress indicators by nucleotide-binding domain, leucine-rich repeat–containing proteins (NLRs) to activation of the canonical and noncanonical effector proteases caspase 1 (Casp1) and Casp11. Inflammasomes are primarily studied in macrophages, where their assembly drives maturation of proinflammatory cytokines and pyroptosis; however, recent research has highlighted the defensive role of inflammasomal activity in the intestinal epithelium (Knodler et al., 2014; Sellin et al., 2014; Birchenough et al., 2016; Zhu et al., 2017).
In a recent investigation, Wlodarska et al. (2014) examined the role of the Nlrp6 inflammasome during Citrobacter rodentium infection and found that Nlrp6−/− and Casp1/11−/− mice were more susceptible to infection. As the severity of C. rodentium infection can be linked to IML function (Bergstrom et al., 2010), it was speculated that the Nlrp6 inflammasome might regulate the formation of the IML. By analyzing fixed colonic tissue sections, they found that Nlrp6−/− mice lacked a detectable IML. This was phenocopied by Casp1/11−/− mice, thus suggesting that active epithelial Nlrp6 inflammasomes were critical regulators of Muc2 secretion, thereby controlling IML formation.
The results presented by Wlodarska et al. (2014) show clear differences between the mouse strains they examined; however, the reliance on histology to provide quantitative IML data are potentially problematic, as mucus preservation in tissue sections is highly variable. Additionally, use of nonlittermate-controlled mice means variations in the microbiota may also have led to the misinterpretation of genotype-independent IML phenotypes (Stappenbeck and Virgin, 2016). This is relevant given the role of the microbiota in shaping the IML (Jakobsson et al., 2015). We therefore sought to confirm the role of inflammasome signaling in controlling baseline IML formation using a range of different knockout mice and a combination of ex vivo and in vivo methods that can provide highly reproducible quantitative IML data. In addition, we have also applied these methods to littermate-controlled Nlrp6−/− animals.
These investigations found that mice lacking Nlrp6, Casp1, or Casp11 formed an IML that was functionally indistinguishable from WT animals. Surprisingly, we did identify a dysfunctional IML phenotype in mice lacking the inflammasome substrate cytokine Il18; however, careful examination of this phenotype revealed it to be microbiota dependent and unlikely to be driven by Il18 deficiency. Our results therefore argue against the concept that Nlrp6 or inflammasome activity is involved in IML formation or function and further highlight the absolute requirement for microbiota-controlled experiments in this field.
Results
Colonic goblet cell inflammasome expression
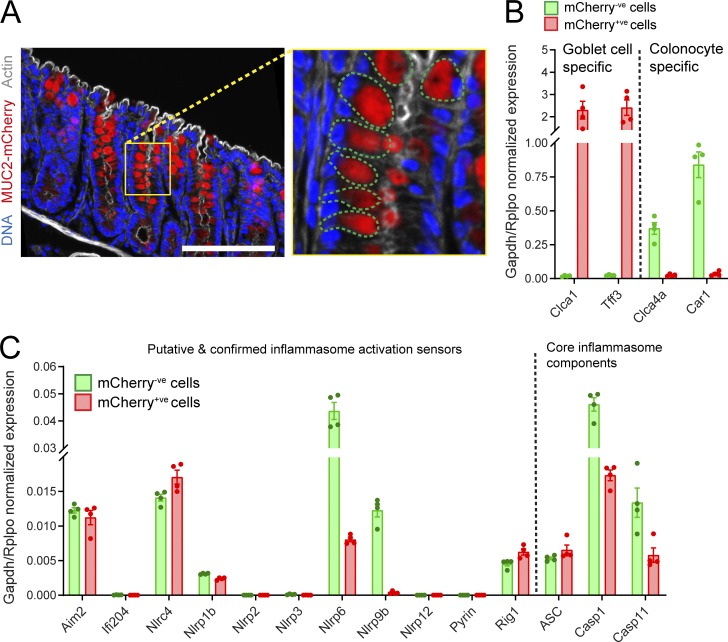
To explore the potential role of different inflammasomes in regulating mucus secretion and IML formation, we first examined the expression of inflammasome component genes in isolated distal colonic goblet cells. This was achieved by acquiring the total distal colonic epithelium from transgenic mice expressing mCherry-tagged MUC2 (RedMUC298trTg) and sorting the goblet cell (mCherry+ve) and remaining epithelial cell (mCherry−ve) populations (Fig. 1 A). RNA was extracted from each population and analyzed by quantitative RT-PCR (qRT-PCR). Sorted cell population purity was assessed by examining transcripts coding for goblet cell–specific (Clca1 and Tff3) and colonocyte-specific (Clca4a and Car1) genes, which demonstrated that the two major epithelial cell populations had been effectively separated (Fig. 1 B). Investigation of inflammasome expression in the goblet cell population detected transcripts encoding several putative or confirmed inflammasome activation sensors. Comparatively high expression of Aim2, Nlrc4, and Nlrp6 transcripts was detected. Nlrp1b, Nlrp9b, and Rig1 transcripts were also detected, albeit at lower levels (Fig. 1 C). Conversely, we detected negligible expression of Ifi204, Nlrp2, Nlrp3, Nlrp12, or Pyrin mRNA. Goblet cells also expressed transcripts for the conserved canonical inflammasome components Asc and Casp1, as well as Casp11. As inflammasome activity in other epithelial cells may influence goblet cell mucus secretion and IML formation, we also examined mRNA expression of inflammasome components in the mCherry−ve population (Fig. 1 C). With the exception of Nlrp9b, the mRNA expression pattern in this population was virtually identical to that observed in goblet cells. These results demonstrated that distal colonic goblet cells and other epithelial cells express transcripts encoding the essential components of several specific inflammasomes, all of which could therefore potentially play a role in distal colonic IML formation.
Figure 1.
Inflammasome expression in the distal colonic epithelium. (A) Confocal micrograph of cryosection from RedMUC298trTg distal colon; magnified upper crypt (yellow box) highlighting individual goblet cells (green dashed lines), DNA (blue), mCherry-MUC2 (red), and actin (gray). Scale bar, 100 μm. (B) Expression of goblet cell or colonocyte-specific genes detected by qRT-PCR of sorted epithelial cell RNA; goblet cells/mCherry+ve (red) and remaining epithelial cells/mCherry−ve (green). (C) Expression of inflammasome components detected in sorted epithelial cells; error bars represent SEM of qRT-PCR data from n = 4 animals. All data are pooled from four independent experiments (one animal per experiment).
Inflammasome deficiency does not affect IML function
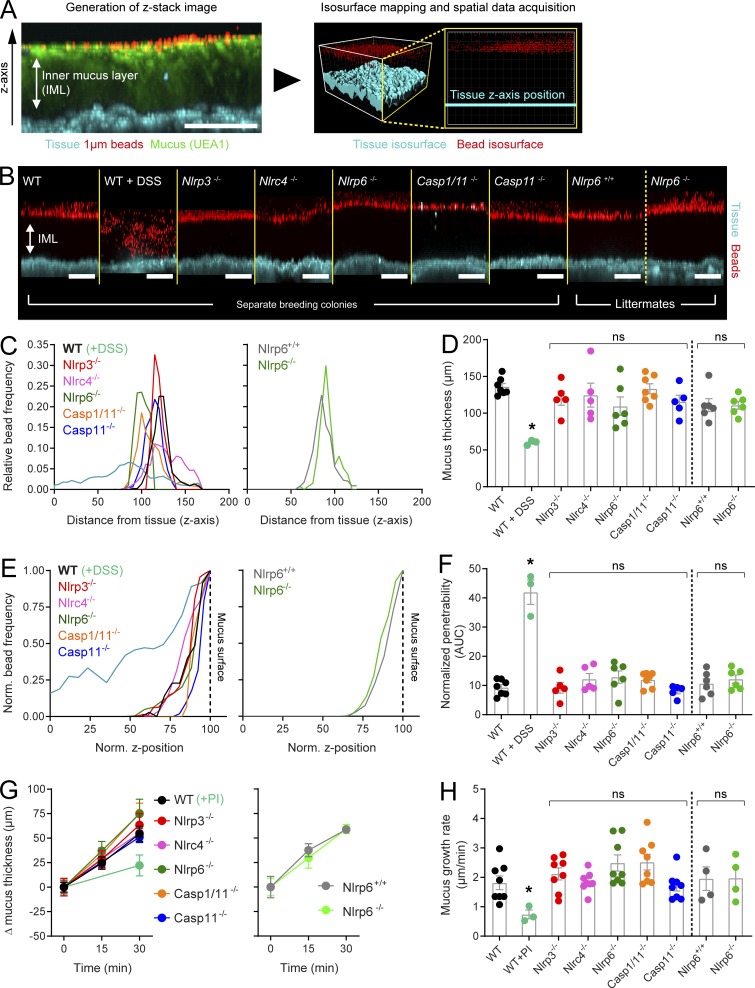
To thoroughly address the relationships among Nlrp6, inflammasome activity, and the IML, we adapted an ex vivo analysis method that allows quantitative assessment of IML function in live tissue (Gustafsson et al., 2012). The IML is normally impenetrable to bacteria-sized objects, and thus 1-µm fluorescent beads apically applied to flushed colonic tissue settle at the IML surface, as demonstrated by concurrent ex vivo staining of the IML using fluorescently labeled UEA-1 lectin (Fig. 2 A). Spatial mapping data describing bead distribution in relation to the tissue surface can consequently be used to calculate IML thickness. Penetration of beads into the mucus structure can be used to analyze IML barrier function.
Figure 2.
Ex vivo analysis of IML formation and function in inflammasome-deficient mice. (A) Ex vivo data acquisition; confocal z-stack acquired in ex vivo WT distal colon tissue showing x/z-axis cross section through the IML (left panel): tissue (blue), 1-µm beads (red), UEA-1 stained mucus (green). Isosurfaces mapped to fluorescent signal from tissue and beads (right panel) allowing spatial data collection: tissue isosurface (blue), bead isosurfaces (red). (B) Representative confocal z-stacks acquired in WT, DSS-treated, and different inflammasome knockout colonic tissue; mice from separate breeding colonies and littermates are indicated; tissue (blue), 1-µm beads (red). (C) Representative bead spatial data plots generated from z-stacks shown in B; left graph, separate breeding colony mice; right graph, littermated mice. (D) Quantification of IML thickness based on data shown in C; the dashed line separates data from separate breeding colony and littermated mice. (E) Representative normalized bead spatial data plots of separate breeding colony mice (left graph) and littermated mice (right graph); the normalized z-axis position corresponding to the mucus surface is indicated (dashed line). (F) Quantification of IML penetrability based on data shown in E; the dashed line separates data from separate breeding colony and littermated mice. (G) Mucus growth curves over 30-min ex vivo incubation in tissue from WT, protease inhibitor (PI)–treated, and different inflammasome knockout colonic tissue; left graph, separate breeding colony mice; right graph, littermated mice. (H) Mucus growth rates calculated from data shown in G. (D and F–H) Error bars are SEM of n = 3–8 animals as indicated; ns, not significant; *, P < 0.05, significance determined by Kruskal–Wallis and uncorrected Dunn’s multiple comparison. Scale bars, 100 µm. All data pooled from two independent experiments (three or four animals per experiment). AUC, area under the curve.
This method was applied to colonic tissues obtained from WT mice and strains lacking either specific inflammasome activation receptors (Nlrp3−/−, Nlrc4−/−, and Nlrp6−/−) or canonical and noncanonical inflammasome components (Casp1/11−/− and Casp11−/−). These mice were housed in the same animal facility but obtained from separate breeding colonies; however, as differences in microbiota composition can have an effect on IML properties, we also generated littermated mice from Nlrp6+/− breeding (Nlrp6+/+ and Nlrp6−/−) in order to control for microbiota variation between colonies. We specifically targeted Nlrp6 for littermate experiments due to the previous claim that Nlrp6 inflammasome activity is required for IML formation (Wlodarska et al., 2014). IML dysfunction can be induced in vivo in WT mice using the colitogenic chemical dextran sodium sulfate (DSS; Johansson et al., 2010). Therefore, IML properties were also examined in WT mice exposed to DSS in their drinking water in order to serve as a positive control for IML dysfunction in our ex vivo assay. Strikingly, under normal conditions, ex vivo experiments detected an intact IML in all strains of mice examined including the total inflammasome knockout Casp1/11−/− and both separately housed and littermate controlled Nlrp6−/− mice (Fig. 2 B). Analysis of mucus properties found that DSS treatment induced decreased mucus thickness (Fig. 2, C and D) and impaired barrier function (Fig. 2, E and F); however, all inflammasome-related knockout strains possessed an IML that was indistinguishable from the untreated WT IML. Gender-specific differences have previously been described in Nlrp6−/− mice (Lemire et al., 2017); however, we did not observe any gender-based effects on either IML thickness or barrier function (Fig. S1).
Ex vivo analysis also allows calculation of the mucus growth rate via acquisition of mucus thickness data over a period of time. Mucus growth is dependent on multiple factors but is primarily a reflection of mucus secretion and Clca1-mediated proteolytic expansion of the IML as it is converted into the outer mucus layer (Nyström et al., 2018). As both of these processes are important for continuous mucus turnover and IML maintenance, we also examined mucus growth over 30 min in the different inflammasome knockout strains. Treatment of WT tissue with a protease inhibitor significantly decreased the mucus growth rate; however, we detected no differences between untreated WT tissue and tissue from either separately housed or littermate controlled inflammasome knockout mice (Fig. 2, G and H).
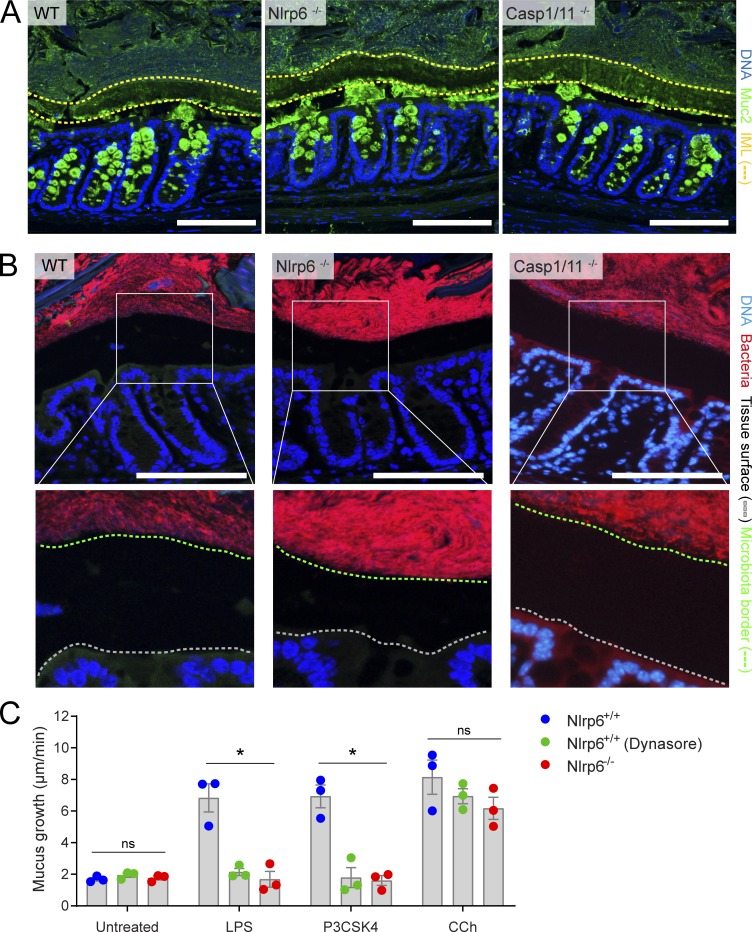
IML data obtained ex vivo were verified in vivo by immunostaining for Muc2 in tissue sections obtained from methanol-Carnoy fixed distal colons of WT, Nlrp6−/−, and Casp1/11−/− mice (Fig. 3 A). Immunostaining detected secreted Muc2 that formed a distinct IML structure in all three genotypes. Wlodarska et al. (2014) observed mucin granule–like structures in the lumen of their Nlrp6−/− mice, which were thought to occur due to dysregulated release of Muc2 from colonic goblet cells; however, we failed to detect these structures in our mice. IML barrier dysfunction can result in microbiota penetration of the mucus layer (Jakobsson et al., 2015; Desai et al., 2016), and total loss of the IML results in direct microbiota contact with the colonic epithelium, as clearly demonstrated in Muc2−/− mice (Johansson et al., 2008). Therefore, we examined the spatial distribution of the microbiota by fluorescence in situ hybridization (FISH) staining of bacterial 16S in fixed colonic tissue sections in order to confirm that our ex vivo results reflected the situation in vivo (Fig. 3 B). We observed clear separation of the luminal microbiota from the colonic epithelial surface in all three genotypes, demonstrating preservation of IML function in mice that lack any inflammasome activity.
Figure 3.
In vivo IML formation and ex vivo senGC secretory responses. (A) Micrographs of tissue sections from WT, Nlrp6−/−, and Casp1/11−/− distal colon stained to detect Muc2; DNA (blue), Muc2 (green), IML (yellow dashed lines). (B) Micrographs of the same tissues shown in A stained to detect bacterial 16S by FISH, with magnified images highlighting bacterial separation from epithelium (white boxes); DNA (blue), bacteria (red), epithelial tissue surface (gray dashed line), microbiota border (green dashed line). Scale bars, 100 µm. (C) Mucus growth rates in Nlrp6+/+ (with or without Dynasore inhibitor) and Nlrp6−/− littermate distal colon after induction of senGC-dependent (LPS and P3CSK4) or senGC-independent (carbachol [CCh]) secretory responses. Error bars represent SEM of n = 3 animals/group; *, P < 0.05, significance determined by two-way ANOVA and Dunnet’s multiple comparison. Images in A and B are representative of three independent experiments; data in C are representative of two independent experiments (three animals per experiment). ns, not significant.
Our own previous findings have indicated that the Nlrp6 inflammasome can regulate an inducible mucus secretory response driven by endocytosis of specific bacterial Tlr-ligands by sentinel goblet cells (senGCs) localized in the upper colonic crypt (Birchenough et al., 2016). However, as these findings were also partly based on comparison of nonlittermate-controlled animals, we sought to confirm that this response was conserved in littermated Nlrp6+/+ and Nlrp6−/− mice (Fig. 3 C). Ex vivo treatment with the Tlr4 ligand LPS and the Tlr2/1 ligand N-palmitoyl-5,6-dipalmitoylcysteinyl-seryl-lysine(4) (P3CSK4) induced mucus secretion in colonic tissue obtained from Nlrp6+/+ mice, but not Nlrp6−/− mice, while the senGC-independent secretory response to carbachol was maintained in both genotypes. Blockade of endocytosis using the inhibitor Dynasore prevented LPS and P3CSK4-induced secretion in Nlrp6+/+ tissues, indicating that endocytosis by senGCs remained an essential element of the inducible secretory response in these mice.
We were therefore able to confirm that Nlrp6 is required for the senGC-dependent secretory response. However, using different methods and an extensive range of inflammasome knockout mice, we could not observe any evidence for differences in any IML property examined between mice lacking inflammasome activity and WT controls at steady state. These results argue against the concept that Nlrp6, or indeed any other inflammasome, has a role to play in regulating IML formation or any critical aspect of IML function under baseline conditions.
Detection of a dysfunctional IML in Il18−/− mice
Inflammasome assembly activates Casp1, resulting in proteolytic processing of numerous substrates. The best studied of these substrates are the pro forms of the inflammatory cytokines Il1β and Il18, which are cleaved by Casp1 to produce the mature, bioactive protein. As part of our investigation into the possible regulation of IML formation by inflammasome activity, we also characterized the IML of mice deficient in both of these key inflammasome substrates.
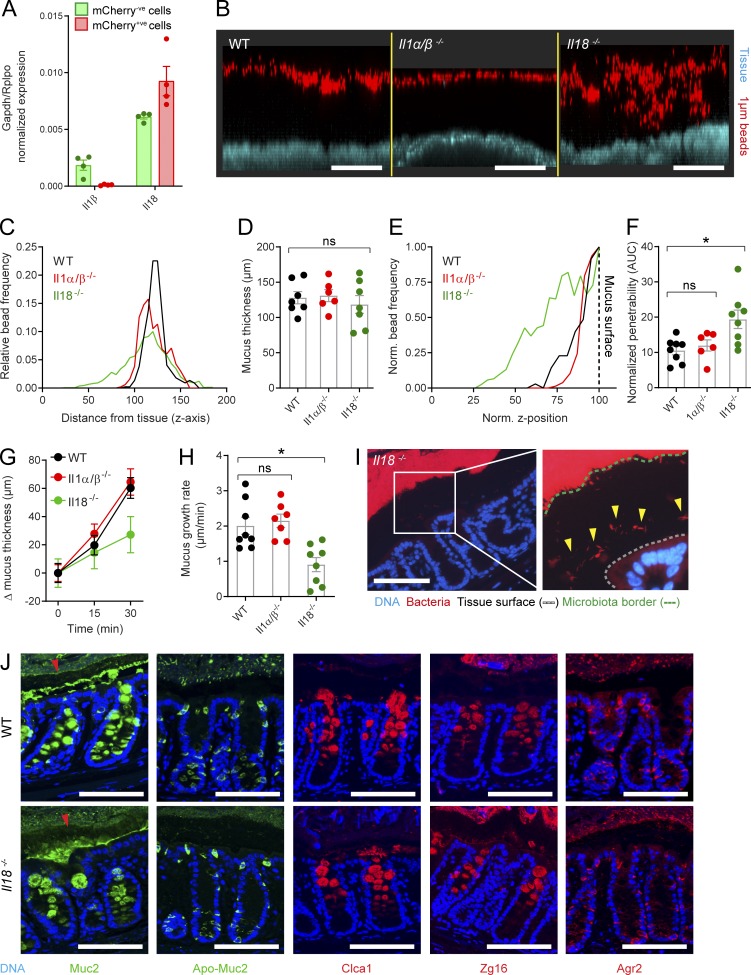
Analysis of qRT-PCR data from sorted colonic epithelial cells primarily detected transcripts coding for Il18 in both goblet cells and remaining epithelial cells (Fig. 4 A). Compared with Il18, we detected a relatively low level of Il1β expression in the mCherry−ve population, likely derived from intraepithelial lymphocytes. Nevertheless, we compared the IML of both Il1αβ−/− and Il18−/−colonic tissues to WT tissue using ex vivo analysis (Fig. 4 B). Quantification of mucus thickness identified an IML of comparable thickness to the WT IML in both cytokine knockout strains (Fig. 4, C and D); however, analysis of IML barrier function revealed a significantly more penetrable IML in Il18−/−, but not Il1α/β−/−, tissue compared with WT controls (Fig. 4, E and F). Similarly, analysis of mucus growth rates detected a significantly lower rate in the Il18−/− mice compared with both the WT and Il1αβ−/− animals (Fig. 4, G and H).
Figure 4.
Ex vivo/in vivo analysis of IML function in mice lacking inflammasome substrate cytokines. (A) Expression of inflammasome substrate cytokines detected by qRT-PCR of FACS-sorted goblet cells/mCherry+ve (red) and remaining epithelial cells/mCherry-ve (green) isolated from RedMUC298trTg distal colon (Fig. 1, A and B). (B) Representative confocal z-stacks showing x/z-axis cross sections through the IML of WT, Il1αβ−/−, and Il18−/− colonic tissue; tissue (blue), 1-µm beads (red). (C) Representative bead spatial data plots generated from z-stacks shown in B. (D) Quantification of IML thickness based on data shown in C. (E) Representative normalized bead spatial data plots; normalized z-axis position corresponding to the mucus surface is indicated (dashed line). (F) Quantification of IML penetrability based on data shown in E. (G) Mucus growth curves over 30-min ex vivo incubation in WT, Il1αβ−/−, and Il18−/− colonic tissue. (H) Mucus growth rates calculated from data shown in G. (I) Micrographs of tissue sections from Il18−/− distal colon stained to detect bacterial 16S by FISH; magnified images highlighting bacterial proximity to the epithelium (white box); DNA (blue), bacteria (red), epithelial tissue surface (gray dashed line), microbiota border (green dashed line), bacteria penetrating into IML (yellow arrows). (J) Micrographs of tissue sections from WT and Il18−/− distal colon stained to detect Muc2 and Apo-Muc2 (green) and Clca1, Zg16, and Agr2 (red); secreted Muc2 forming the IML is indicated (arrowheads). Error bars represent SEM of n = 4 (A) or n = 8 (D, F, G, and H) animals; ns, not significant; *, P < 0.05 significance determined by Kruskal–Wallis and uncorrected Dunn’s multiple comparison. Scale bars, 100 µm. Data in A are pooled from four independent experiments (one animal per experiment); data in C–H are pooled from two independent experiments (three or four animals per experiment); images in B, I, and J are representative of two independent experiments (three or four animals per experiment). AUC, area under the curve.
These results indicated that loss of Il1β had no effect on IML formation or function but that loss of Il18 had a detrimental effect on the IML that resulted in decreased barrier function and capacity to secrete and/or process mucus. We thus speculated that these defects could result in a dysfunctional mucus barrier that failed to fully separate the microbiota from the colonic epithelium. FISH staining of bacterial 16S in fixed tissue sections obtained from Il18−/− mice showed that the bulk of the microbiota remained segregated from the epithelial surface (Fig. 4 I), as observed in WT, Nlrp6−/−, and Casp1/11−/− tissues (Fig. 3 B). However, in Il18−/− tissues, a number of bacterial cells could also be observed in much closer proximity to the epithelial surface (Fig. 4 I, arrowheads). This indicated that elements of the microbiota were able to penetrate the IML in Il18−/− mice.
In vivo bacterial penetration of the IML in Il18-deficient tissue was thus likely the result of the dysfunctional IML barrier observed in ex vivo experiments. As loss of inflammasome activity had no effect on IML function, this effect was likely unrelated to inflammasomal activation of Il18; however, as Il18 can also be activated by alternate proteases to Casp1 (Omoto et al., 2006), we sought to investigate this phenotype in greater detail. To examine the potential role of Il18 in controlling IML properties by regulating colonic goblet cell function, we initially examined the synthesis and secretion of the major goblet cell–specific proteins Muc2 (and nonglycosylated Apo-Muc2), Clca1, Zg16, and Agr2 (Fig. 4 J). These proteins participate in IML barrier function or mucus processing (Johansson et al., 2008; Bergström et al., 2014, 2016; Nyström et al., 2018); however, we were unable to detect any clear differences in their expression between WT and Il18−/− tissues using this approach.
The Il18−/− IML phenotype is microbiota dependent
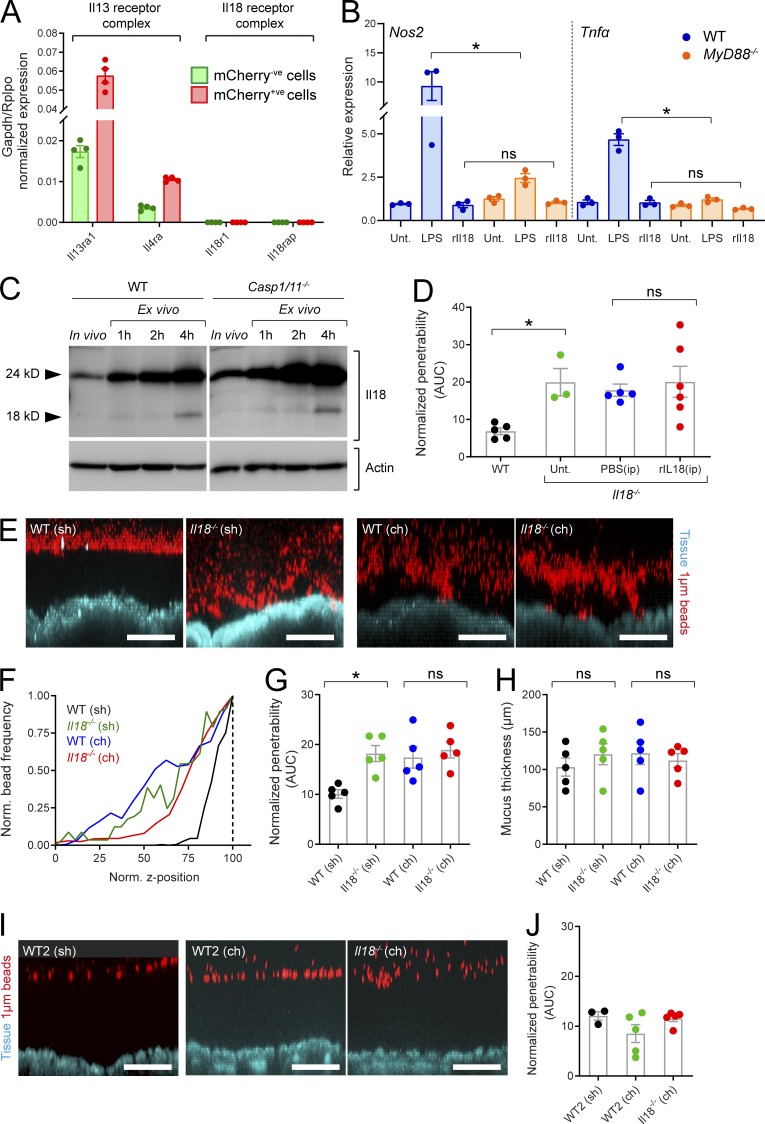
Having failed to detect differences in goblet cell phenotype between WT and Il18−/− mice, we sought to confirm the potential for Il18 signaling to affect the colonic epithelium. While we have established that colonic epithelial cells abundantly express Il18 mRNA (Fig. 4 A), reactivity to bioactive Il18 requires the presence of the Il18 heterodimeric receptor complex composed of Il18r1 and Il18rap. Another cytokine, Il13, has a well-characterized hyperplastic effect on the colonic epithelium and signals via a heterodimeric receptor complex composed of Il13ra1 and Il4ra and thus serves as a useful comparison. As expected, qRT-PCR data showed that transcripts encoding the Il13 receptor complex were highly expressed in both mCherry+ve and mCherry−ve epithelial populations; however, detection of transcripts encoding the Il18 receptor complex was negligible (Fig. 5 A). This data indicated that colonic epithelial cells lack the capacity to respond to Il18 directly. MyD88 mediates Il18r1/Il18rap signaling, and we therefore validated this concept by treating WT and MyD88−/− colonic organoids with rIl18 and quantified induction MyD88-regulated genes (Nos2 and Tnfα) by qRT-PCR (Fig. 5 B). Treatment with the Tlr4-ligand LPS induced a clear MyD88-dependent increase in Nos2 and Tnfα expression in colonic organoids; however, rIl18 had no impact on expression of either of these genes. Together, this demonstrated that Il18 is highly unlikely to directly modulate colonic epithelial cell function.
Figure 5.
Analysis of factors contributing to the IML phenotype in Il18−/− mice. (A) Expression of Il13 and Il18 receptor complex components detected by qRT-PCR of FACS-sorted goblet cells/mCherry+ve (red) and remaining epithelial cells/mCherry-ve (green) isolated from RedMUC298trTg distal colon (Fig. 1, A and B). (B) Relative expression of Nos2 and Tnfα in WT and MyD88−/− colonic organoids treated with LPS or rIl18 quantified by qRT-PCR; all data relative to untreated (Unt.) control samples. (C) Western blots of SDS-PAGE resolved protein extracted from WT or Casp1/11−/− fresh (in vivo) distal colonic tissue or tissue incubated for 1, 2, or 4 h ex vivo; anti-Il18 (upper blot), anti-actin (lower blot). (D) Quantification of IML penetrability (D) in tissue from WT and Il18−/− mice with or without intraperitoneal (ip) treatment with rIl18 or PBS. (E) Representative confocal z-stacks showing x/z-axis cross sections through the IML of tissue obtained from separately housed (sh) or cohoused (ch) WT and Il18−/− mice; tissue (blue), 1-µm beads (red). (F) Representative normalized bead spatial data plots generated from z-stacks shown in E; normalized z-axis position corresponding to the mucus surface is indicated (dashed line). (G) Quantification of IML penetrability based on data shown in D. (H) Quantification of IML thickness based on bead spatial data acquired from z-stacks shown in C. (I) Representative confocal z-stacks showing x/z-axis cross sections through the IML of tissue obtained from separately housed (sh) or cohoused (ch) WT colony 2 (WT2) and Il18−/− mice; tissue (blue), 1-µm beads (red). (J) Quantification of IML penetrability based on normalized bead spatial data acquired from z-stacks shown in G. Error bars are SEM of n = 4 (A) or n = 3–5 (C, D, G, H, and J) animals or organoid cultures as indicated; ns, not significant; *, P < 0.05, significance determined by Kruskal–Wallis and uncorrected Dunn’s multiple comparison. Scale bars, 100 µm. Data in A are pooled from four independent experiments (one animal per experiment); data in B are pooled from three independent experiments (one culture per experiment); images in C representative of two independent experiments (two animals per experiment); data in D, F–H, and J are pooled from two independent experiments (two or three animals per experiment); images in E and I are representative of two independent experiments (two or three animals per experiment). AUC, area under the curve.
Bioactive Il18 generated by inflammasome-mediated proteolysis in epithelial cells may indirectly affect the epithelium via feedback mechanisms based on signaling to nonepithelial cells, which in turn produce epithelium-modulating factors. To address this possibility, we assayed the presence of bioactive Il18 in colonic tissues from WT and Casp1/11−/− mice by SDS-PAGE and Western blot of colonic proteins in order to resolve and detect the Il18 pro form (24 kD) and the bioactive (18 kD) protein (Fig. 5 C). In freshly isolated colonic tissue (in vivo sample), pro-Il18 was clearly detected; however, the bioactive form was virtually undetectable. Incubation of colonic tissue in ex vivo conditions for ≤4 h generated a rapid increase in pro-Il18 and clear detection of bioactive Il18 by the 4-h incubation time. Surprisingly, we detected no difference between WT and Casp1/11−/− tissue. This indicated that bioactive Il18 could be generated by inflammasome-independent processing in colonic tissue under ex vivo conditions but that this active form was not normally present at detectable levels in the colonic tissue of mice housed in our facility. Nonetheless, we further tested the possibility that Il18 directly regulated IML barrier function via intraperitoneal injection of bioactive recombinant Il18 (rIl18) to Il18−/− mice; however, rIl18 treatment had no effect on IML barrier properties in these animals (Fig. 5 D). Thus, the possibility of bioactive Il18 regulating epithelial cells and IML properties under normal conditions appeared unlikely.
Another potential cause of IML dysfunction in Il18−/− mice would be the presence of an IML-modulating microbiota. To assess this possibility, we cohoused Il18−/− mice with WT mice for 4 wk in order to allow microbiota normalization between the two strains and then subjected tissues to ex vivo analysis (Fig. 5 E). Analysis of ex vivo data found that cohoused WT animals acquired the dysfunctional IML barrier phenotype found in the Il18−/− mice (Fig. 5, F and G) without altering overall mucus thickness (Fig. 5 H). Consequently, this indicated that the Il18−/− IML dysfunction phenotype was likely driven by the microbiota. To assess the reproducibility of this effect, we also cohoused Il18−/− mice with a different colony of WT animals (designated WT2) that was derived from mice recently acquired from a commercial vendor (Fig. 5 I). In this case, we observed the opposite transfer effect, as the cohoused Il18−/− mice acquired the impenetrable IML phenotype from the WT2 mice (Fig. 5 J). These results demonstrated that the microbiota-driven Il18−/− IML phenotype was only conditionally dominant.
Together, this provided evidence that the dysfunctional IML phenotype in Il18−/− mice is unlikely to be Il18 regulated, as the presence of bioactive Il18 in the colon is negligible, colonic epithelial cells lack the Il18 receptor complex, and we found no evidence of alterations in Il18−/− colonic goblet cells. The transfer or loss of the Il18−/− IML phenotype upon cohousing with different WT mice instead implied that this was dependent on elements of the microbiota.
Cosegregation of microbiota components with IML phenotype
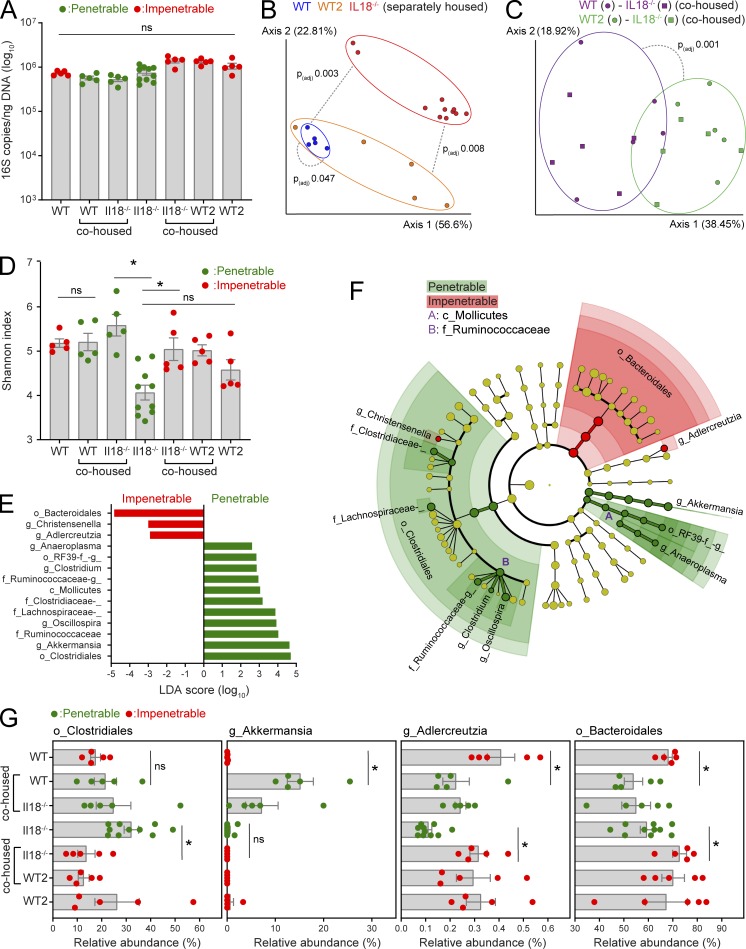
To identify specific taxonomic groups that cosegregated with the functional (impenetrable) or dysfunctional (penetrable) IML phenotypes, microbiota configuration was analyzed by 16S rRNA gene sequencing and qPCR of fecal DNA obtained from separately housed and cohoused WT, WT2, and Il18−/− mice. No significant differences in total bacterial load were detected (Fig. 6 A); however, the composition of the microbiota of all three separately housed groups were significantly divergent (Fig. 6 B). Importantly, the microbiota of cohoused WT-Il18−/− and WT2-Il18−/− mice normalized but was significantly different between cohousing experiments (Fig. S2, A and B; and Fig. 6 C). Together, this demonstrated that microbiota normalization between cohoused animals was largely effective and that separately and cohoused mice with different IML phenotypes had dissimilar microbiota configurations.
Figure 6.
Microbiota profiling in mice with divergent IML phenotypes. Total bacterial load and relative abundance of bacterial taxa were determined by analysis of 16S rRNA genes in fecal DNA extractions from separately housed and cohoused WT, WT2, and Il18−/− mice with a penetrable (red) or impenetrable (green) IML phenotype. (A) Quantification of total bacterial 16S rRNA gene copy number by qPCR. ns, not significant. (B and C) Principal component analysis of β-diversity (weighted UniFrac) from bacterial communities in separately housed (B) or cohoused (C) mice. (D) α-diversity (Shannon index) of bacterial communities. ns, not significant. (E) Linear discriminant analysis (LDA) effect size identification of bacterial taxa cosegregating with IML phenotype. (F) Cladogram highlighting taxa identified in E. (G) Relative abundance of specific taxa identified by LEfSe in different experimental groups. (A, D, and G) Error bars represent SEM of n = 5–10 mice; *, P < 0.05, significance determined by Kruskal–Wallis and uncorrected Dunn’s multiple comparison. (B and C) Dashed lines indicate statistical comparisons between groups, analysis is described in experimental procedures. All data are pooled from two independent experiments (five animals per experiment).
Previous research has associated decreased microbiota diversity with dysbiosis that results in IML dysfunction (Schroeder et al., 2018). Accordingly, we assessed microbiota α-diversity in our different mouse groups (Fig. 6 D). We detected significantly lower diversity in separately housed Il18−/− compared with WT mice, and this was normalized to WT levels by cohousing. However, diversity increased in Il18−/− mice after cohousing with both WT and WT2 mice, demonstrating that it did not covary with IML barrier function. To detect specific bacteria that covaried with IML phenotype, we subsequently employed linear discriminant analysis effect size (LEfSe), which identified 14 significant operational taxonomic units (OTUs) associated with either the impenetrable (3 OTUs) or penetrable (11 OTUs) IML phenotypes (Fig. 6, E and F). Strikingly, these included the orders Bacteroidales and Clostridiales, which were associated with the impenetrable and penetrable IML phenotypes respectively and together accounted for ∼85% of bacteria detected in all samples, indicating that broad shifts in microbiota configuration correlated with IML barrier function.
We next sought to determine if the OTUs identified by LEfSe consistently cosegregated with IML phenotype in different experimental settings. This was achieved by examining shifts in abundance in mice where the IML phenotype switched between separately housed and cohoused mice from the same genetic background (i.e., WT mice before and after cohousing with Il18−/− mice and Il18−/− mice before and after cohousing with WT2 mice; Figs. 6 G and S2 C). This analysis found that differences in all of the OTUs associated with the penetrable IML phenotype; e.g., the Clostridiales and the genus Akkermansia were primarily driven by changes in one, but not both, experimental groups. However, two of the OTUs associated with the impenetrable IML phenotype, the order Bacteroidales (>99% composed of the family S24-7/Muribaculaceae in our data) and the genus Adlercreutzia, were consistent in their behavior across experimental groups in that they were both more abundant in mice with a functional IML but decreased when IML barrier function was compromised.
Finally, to contextualize our microbiota data, we compared it to previous findings comprising a limited number of earlier investigations have simultaneously measured IML properties and microbiota composition. We examined this pool of published datasets and identified bacterial taxons that were positively or negatively associated with IML thickness or barrier function (Fig. S3). Most bacterial taxon-IML associations were only represented by single observations or were found to be opposite in in different datasets; however, several taxons appeared to behave relatively consistently across different studies. Notably, both Adlercretuzia and the Muribaculaceae have been positively correlated to IML barrier function by several previous investigations, thus indicating that the abundance of these bacteria is associated with IML barrier function.
The fact that the microbiota both cosegregates with and alters the IML phenotype independently of the presence of Il18 indicates that the Il18−/− phenotype is likely to be a genotype-independent false positive with respect to the detection of a dysfunctional IML barrier. However, detailed analysis of microbiota and IML cosegregation in these mice has allowed us to identify microbiota components that might be linked to modulation of IML barrier properties.
Discussion
In the current investigation, we have applied combined ex vivo/in vivo IML analysis methods to multiple inflammasome knockout mice, including littermate-controlled Nlrp6−/−, to verify the claim that Nlrp6 and inflammasome activity is required for IML formation and barrier function. Using this approach, we found no effect of Nlrp6 or inflammasome deficiency on IML thickness, barrier function, or mucus processing. These results conclusively argue against the concept that Nlrp6 regulates IML formation or function. Data acquired from other inflammasome knockout strains were not littermate controlled, raising the possibility that microbiota-dependent effects might influence our approach. Nonetheless, our data demonstrates that the IML is at minimum both present and functional in all inflammasome knockout strains examined. Consequently, while we cannot rule out minor effects, our data support the conclusion that the inflammasome has no major role to play in regulating the IML properties we have investigated.
An IML deficiency in Nlrp6−/−, Asc−/−, and Casp1/11−/− mice previously indicated that epithelial Nlrp6 inflammasome activity regulated goblet cell Muc2 secretion and IML formation (Wlodarska et al., 2014). This investigation was the first to provide compelling evidence of a link between innate immune signaling and the IML. However, a reliance on histology to quantitatively assess IML formation and comparison of nonlittermate WT and null mutant mouse colonies raised the possibility that these observations were driven by genotype-independent factors. Although histological staining of methanol-Carnoy–fixed tissue sections is the most commonly used method of IML assessment, it is highly susceptible to artifacts caused by IML shrinkage during fixation or damage during sectioning. Thus, while histology can provide indicative information regarding the IML, it is not always suitable for quantitative analysis and should be supported by other experimental approaches where possible. The IML is in direct contact with the colonic microbiota, and thus, its properties can be strongly influenced by microbiota configuration (Jakobsson et al., 2015). Previous research investigating the same Nlrp6−/− mouse colony used by Wlodarska et al. (2014) identified a dysbiotic microbiota that could colonize deep within the colonic crypts, indicating the presence of microbes with the capacity to disrupt or circumvent the IML (Elinav et al., 2011). While the causal role of Nlrp6 in driving microbiota alterations is disputed (Lemire et al., 2017; Mamantopoulos et al., 2017), it remains possible that the IML dysfunction in these mice is microbiota induced as opposed to directly caused by Nlrp6 deficiency. However, without closer investigation of the specific mice in question, we can only speculate as to the extent that these factors, or others, may be responsible for the discrepancy between our findings and those previously reported.
Screening inflammasome-related knockout mouse strains identified a dysfunctional IML in mice lacking the inflammasome substrate Il18. This proinflammatory cytokine is an important regulator of intestinal homeostasis (reviewed by Maloy and Powrie, 2011), and a link to regulation of IML barrier function would therefore be of significance. We thus sought to determine the capacity of Il18 to regulate IML function but found no evidence of epithelial Il18 receptor complex expression or the presence of bioactive Il18 in freshly isolated colonic tissue from mice housed in our facility. Similar experiments in both mice (Wirtz et al., 2002; Nishida et al., 2009; Muñoz et al., 2015; De Arcangelis et al., 2017) and humans (Monteleone et al., 1999; Pizarro et al., 1999) have also indicated that bioactive Il18 is not present in the healthy intestine. This observation was unexpected, given that the cytokine appears to control colonic microbiota composition via homeostatic regulation of antimicrobial peptide expression (Levy et al., 2015); however, it is possible that the activation state of Il18 under otherwise healthy conditions varies between WT mice housed at different locations. In addition, our results clearly show that ex vivo manipulation of intestinal tissue results in activation of Il18, a phenomenon that has been remarked upon by other investigators (Muñoz et al., 2015). This observation is consistent with Il18’s role as an alarmin-type cytokine that is activated in response to tissue damage. However, this activation sensitivity must be taken into account when designing experiments that assess the activation state of Il18 in vivo.
These observations argued against regulation of the IML by either direct Il18 signaling to the epithelium or indirect effects mediated by Il18-sensitive nonepithelial cells. The transfer or suppression of the dysfunctional IML phenotype observed in Il18−/− mice upon cohousing with WT mice clearly implicated a microbiota-dependent effect on the IML that was not dependent on the presence or absence of Il18. This was supported by microbiota analysis, which identified a clear correlation of a limited number of bacterial taxa with IML phenotype. Based on this evidence, it is likely that our Il18−/− mice have a microbiota configuration that negatively impacts on IML barrier function. While this finding is somewhat less significant than the invalidated concept that Il18 might regulate IML function, it does provide clear illustration of the capacity of the microbiota to affect the IML in a genotype-independent manner. Thus, it also serves as an excellent example of how even combined ex vivo/in vivo analysis methods may generate false–positive results in the absence of appropriate controls.
Previous investigations that have simultaneously examined IML barrier function and microbiota composition have found a number of significant correlations, primarily consisting of taxa that increase in abundance when the IML is dysfunctional (Chassaing et al., 2015; Jakobsson et al., 2015). However, these have largely employed comparison of two groups in a confounding experimental setting (e.g., different diets), making it difficult to identify bacteria that might have a causal or consistent relationship with IML barrier function. Conversely, our study exploited three groups of mice with distinctive initial microbiota configurations and transmissible IML phenotypes that permitted identification of two bacterial taxa that consistently and positively correlated with IML function. Of these, the Bacteroidales, almost exclusively composed of the Muribaculaceae/S24-7 family, demonstrated the largest absolute change in abundance. However, the present lack of taxonomic resolution in this clade means we are currently unable to determine if these changes relate to specific subfamily taxonomic groups. The alterations in Adlercreutzia abundance were less dramatic, as they comprise <0.5% of the total microbiota in most samples; however, as this genus is a known producer of at least one bioactive metabolite (equol), it has the potential to modulate host intestinal functions. Importantly, both Muribaculaceae/S24-7 and Adlercrutzia have been correlated with improved IML barrier function in previous studies (Wlodarska et al., 2011; Chassaing et al., 2015; Schroeder et al., 2018), which reinforces their potential for future investigations seeking to identify bacteria that have a positive impact on the IML barrier. One drawback of the current study is that we have only analyzed the fecal microbiota. While this approach would have incorporated bacteria that are directly associated with the mucus layer, specific analysis of this community may have revealed microbiota alterations with a greater potential to modulate IML properties.
It is evident that specific inflammasomes are highly expressed by the major intestinal epithelial cell lineages. Current research largely supports a role for these signaling platforms as cytosolic sensors that detect microbial molecules during viral or bacterial intracellular infections (Knodler et al., 2014; Sellin et al., 2014; Zhu et al., 2017). Inflammasome activation during epithelial infection drives maturation of proinflammatory cytokines and pyroptotic expulsion of the infected cell, thereby maintaining epithelial integrity and alerting the immune system to the presence of microbial intruders. The suggestion that the Nlrp6 inflammasome regulated IML formation independently of cytokine maturation marked a significant departure from this pattern and suggested that active inflammasomes could have supplementary functions to the generation of proinflammatory signals. Indeed, our own research has found that the Nlrp6 inflammasome is an essential component of a signaling pathway that regulates compound Muc2 exocytosis in colonic senGCs after exposure to an abnormally high concentration of bacterial molecules (Birchenough et al., 2016; McGuckin and Hasnain, 2017). We have hypothesized that this mechanism acts as a defensive secretory response that is dormant under healthy conditions and independent of the Muc2 secretion that results in baseline IML formation. Consequently, while our previous work absolutely supports the fundamental concept posited by Wlodarska et al. (2014) that epithelial Nlrp6 inflammasome activity does have a novel role to play in regulating Muc2 secretion, this function is likely only active under certain circumstances. The results we have acquired during the present study do not support the hypothesis that inflammasome activity regulates normal IML formation.
In conclusion, this study illustrates how accurate analysis of IML properties requires the use of a combination of methods and comparison of tissues from microbiota-controlled animals. While our investigation does not support a role for the inflammasome in regulating IML formation, this does not mean that other innate immune signaling pathways have no role to play in this process. We therefore hope that this investigation will inform future research in this field and motivate investigation of alternate IML regulatory mechanisms.
Materials and methods
Animals
All WT, knockout, and transgenic mice were 6–14 wk old on the C57BL/6 background and bred in-house. The origin of each mouse strain is listed in Table S1. For littermate-controlled experiments, animals were generated from heterozygous breeding pairs and genotyped to determine genetic status. Littermated WT and knockout mice were cohoused until sacrifice. For cohousing experiments, female mice of different genotypes were cohoused for 4 wk before analysis. All experimental comparisons were between gender- and age-matched animals. Animals were anesthetized using isoflurane and killed by cervical dislocation before collection of samples. For DSS treatments, DSS (TdB Consultancy) was dissolved in sterile drinking water to 3% (wt/vol). Samples were collected from DSS-treated mice 84 h after initial exposure. For some experiments, mice received daily intraperitoneal injections of 200 µl recombinant mouse Il18 (5 µg/ml; R&D Systems) dissolved in sterile PBS for 5 d before sample collection. All animal experimental procedures were approved by the Swedish Laboratory Animal Ethical Committee in Gothenburg.
Colonic epithelial cell isolation and sorting
Freshly collected distal colonic tissue from WT and RedMUC298trTg mice was flushed with ice-cold HBSSwo (HBSS without Mg2+/Ca2+ supplemented with 10 mM Hepes, pH 7.2) to remove luminal content. The colonic tissue was inverted and inflated by injection of HBSSwo. Epithelial isolation was performed twice by incubation of tissue for 30 min in 20 ml predigestion buffer (HBSSwo, 5 mM EDTA, 5% vol/vol FCS, and 1 mM dithiothreitol) in a shaking incubator set to 37°C and 140 revolutions/min. The epithelium was detached by vortexing the tube for 30 s, and the remaining tissue was discarded. The isolated epithelial cells were centrifuged at 400 RCF (relative centrifugal force; g) for 10 min at 4°C, resuspended in 2 ml digestion buffer (HBSS with Mg2+/Ca2+ supplemented with 2 mg/ml collagenase type I and 40 U/ml DNase I), and incubated at 37°C for 30 min, with mixing every 10 min. Cells were washed and resuspended in 1 ml ice-cold PBS. Cells were stained with Fixable Viability Dye eFluor 780 (1:1,000; eBioscience) for 30 min, on ice, and washed in 25 ml ice-cold HBSSwo before resuspension in 1 ml FACS buffer (HBSSwo, 2% vol/vol FCS, and 5 mM EDTA) before FACS. Goblet cells and remaining epithelial cells were sorted using a FACS Jazz (Becton Dickinson) according to the presence or absence of mCherry signal. Dead cells were excluded from the sorting using the Fixable Viability Dye eFluor 780. In the living population, discrimination of doublets or aggregated cells was consecutively made for side-scatter (SSC-Width [W]/SSC-Height [H] plot) and for forward scatter (FSC-W/FSC-H plot). The purity of the mCherry+ve and mCherry−ve cell populations was >96%. Cells were sorted into cold FACS buffer, and RNA was isolated immediately afterwards. Gating and sorting strategies are shown in Fig. S4.
RNA extraction and qRT-PCR
Sorted cells were pelleted at 400 RCF for 5 min and immediately resuspended in 350 µl RLT buffer (Qiagen) supplemented with β-merchaptoethanol (Gibco). Samples were homogenized using QIAshredder columns (Qiagen) according to manufacturer’s instructions. Goblet cell lysates were loaded onto RNeasy MinElute column (Qiagen) and remaining epithelial cell lysates onto RNeasy Mini column (Qiagen). The columns were washed according to manufacturer’s instructions, and RNA was eluted with 18 µl (goblet cells) or 35 µl (remaining epithelial cells) RNase-free H2O (Qiagen). The quality of isolated RNA was determined using an Experion Automated Electrophoresis platform (Bio-Rad), and samples were kept at −80°C until further analysis. Expression of specific genes was analyzed by qRT-PCR of cDNA prepared from 600 ng RNA extractions using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher). PCRs (20 µl) were prepared using SsoFast EvaGreen Supermix (Bio-Rad), 450 nM forward and reverse primers, and 10 ng cDNA. PCR cycling conditions were 95°C for 2 min and 40× cycles of 95°C for 5 s and 58°C for 30 s. Reactions were monitored using a CFX96 platform (Bio-Rad) and analyzed using CFX Manager software (v. 3.1; Bio-Rad). Gene expression was quantified using the ΔΔCq method with data normalized to the reference genes Gapdh and Rplpo. Unless otherwise indicated, all primers were prevalidated and supplied by the PrimePCR (Bio-Rad) or KiCqStart (Sigma-Aldrich) services. Specific primer information is provided in Table S2.
Preparation of tissue for ex vivo analysis
Tissue was prepared for ex vivo analysis as previously described (Gustafsson et al., 2012). Freshly dissected tissue was flushed with ice-cold oxygenated Krebs buffer (116 mM NaCl, 1.3 mM CaCl2, 3.6 mM KCl, 1.4 mM KH2PO4, 23 mM NaHCO3, and 1.2 mM MgSO4, pH 7.4) to remove luminal content and the loose mucus layer. The mesenteric tissue was removed, the colon was opened longitudinally, and the muscle layer was removed by microdissection. The tissue was then mounted mucosa-side up in a custom-made horizontal perfusion chamber with oxygenated Krebs-glucose buffer (10 mM) in the basolateral chamber and oxygenated Krebs-mannitol (10 mM) in the apical chamber.
Quantification of IML thickness and penetrability
Distal colonic IML barrier formation and function was assessed by quantifying IML thickness and penetrability to fluorescent microbeads. The tissue was stained by adding Calcein violet AM (1 µg/ml; Thermo Fisher) to the basolateral Krebs-glucose buffer, and the apical mucosal surface was overlaid with 1-µm-diameter Fluosphere crimson microbeads (1:10; Thermo Fisher) diluted in 10 µl Krebs-mannitol buffer. Microbeads were allowed to sediment onto the mucus surface for 5 min, and then the mucosal surface was gently washed with 0.5 ml Krebs-mannitol to remove excess microbeads. For some experiments, the mucus was stained by adding rhodamine-conjugated UEA-1 lectin (50 µg/ml; Vector Labs) to the bead solution. The apical perfusion chamber compartment was then filled with 2 ml Krebs-mannitol, and the perfusion chamber was transferred to an LSM700 confocal imaging system (Carl Zeiss). Tissue and microbeads were visualized by acquiring confocal z-stacks using a ×20 water-immersion objective lens, 405/639-nm lasers, and Zen acquisition software (Carl Zeiss). Zen files were imported into Imaris software (Bitplane), and isosurfaces were mapped to Calcein violet (tissue) and Fluosphere (microbeads) fluorescent signals. Data describing the z-axis position of the tissue and individual microbeads were extracted, and the mucus layer thickness was quantified by calculating the average tissue-microbead z-axis distance. Mucus penetrability was quantified by analysis of microbead distribution within the mucus layer. Initially, a frequency distribution curve of tissue-microbead z-axis distance data was generated for each z-stack using Prism 6 software (GraphPad). To allow comparison of distribution curves acquired from different z-stacks, curves were first normalized to maximum frequency values in order to correct for differences in the absolute number of microbeads detected in each z-stack. Subsequently, curves were normalized to the position of the mucus surface (the z-axis position with the maximum microbead frequency) in order to correct for variable mucus layer thickness, and cropped to exclude data from microbeads above the mucus surface. Area under the curve data were generated for each normalized distribution curve and expressed as normalized penetrability in order to allow quantitative comparison of microbead penetration into the mucus layers of different samples.
Ex vivo analysis of IML growth rate
Distal colonic IML secretion and processing was assessed by quantifying the ex vivo mucus growth rate as previously described (Gustafsson et al., 2012). Tissue was chamber mounted, basolateral perfusion was set to 5 ml/h, and chambers were heated to 37°C. The mucus surface was visualized by apical application of 10-µm-diameter black microbeads (1:20; Polybead) diluted in Krebs-mannitol. Microbeads were allowed to sediment for 5 min and subsequently the apical chamber was filled with Krebs-mannitol. For some experiments the metalloprotease inhibitor EGTA (10 mM; Sigma-Aldrich) or the endocytosis inhibitor Dynasore (100 µM; Sigma-Aldrich) were added to Krebs-mannitol. For experiments examining senGC-dependent/independent mucus secretory responses, tissue was apically treated with P3CSK4 (50 µg/ml; Invivogen) or ultrapure LPS from Escherichia coli 0111:B4 (200 µg/ml; Invivogen) or basolaterally treated with carbachol (1 mM; Sigma-Aldrich). The mucus and tissue were observed under a stereomicroscope (Leica MZ125) and the mucus thickness measured using a 5-µm-diameter micropipette attached to a micrometer (Mitotoyo). Mucus thickness measurements were acquired at over a 30–60-min period and mucus growth rates expressed as micrometers per minute.
Immunohistochemical staining
Distal colon tissue containing a fecal pellet was fixed by submersion in methanol-Carnoy solution for at least 24 h. Fixed tissue was paraffin embedded and cut into 5-µm sections. Tissue sections were deparaffinized by sequential washing in Xylene substitute (20 min at 60°C; Sigma-Aldrich) and 100% (5 min), 95% (5 min), 70% (5 min), and 30% (5 min) ethanol. Antigen retrieval was performed by immersion in 10 mM citrate buffer (95°C, 30 min). Sections were washed in PBS, permeabilized for 5 min using 0.1% vol/vol Triton X-100 (Sigma-Aldrich), and blocked using 5% vol/vol FCS. Sections were incubated overnight at 4°C with primary antibody solutions to stain Muc2 (Muc2C3, 1:500; Johansson et al., 2008), Apo-Muc2 (PH497, 1:1,000; Hansson et al., 1994), Clca1 (ab46512, 1:2,000; Abcam), Zg16 (anti-ZG16, 1:600; Rodríguez-Piñeiro et al., 2013), and Agr2 (H00010551-M01, 1:2,000; Abnova). Sections were washed in PBS, and primary antibodies were detected by incubation with goat anti-rabbit Alexa Fluor 488– or Alexa Fluor 555–conjugated secondary antibodies (1:2,000; Thermo Fisher) for 2 h at room temperature. Finally, slides were rinsed in double-distilled water and counterstained with Hoechst dye (5 µg/ml; Sigma-Aldrich) for 5 min. Stained slides were subsequently imaged using an LSM700 confocal microscope (Zeiss).
FISH
FISH staining for bacterial 16S was performed using the tissue sections described above. Sections were deparaffinized by sequential washing in Xylene substitute (20 min at 60°C; Sigma-Aldrich), 100% ethanol (5 min), and 95% ethanol (5 min). Slides were air dried and flooded with hybridization buffer (40% vol/vol formamide, 0.1% wt/vol SDS, 0.9 M NaCl, and 20 mM Tris, pH 7.4) supplemented with Alexa Fluor 555–labeled universal bacterial FISH probe EUB338 (1 mM). Slides were incubated at 37°C overnight in humidity chambers containing 40% vol/vol formamide solution, subsequently submerged in wash buffer (0.9 M NaCl and 25 mM Tris, pH 7.4), and incubated at 50°C for 20 min. Finally, slides were rinsed in double-distilled water and counterstained with Hoechst dye (5 µg/ml; Sigma-Aldrich) for 5 min. Stained slides were imaged using an LSM700 confocal microscope (Zeiss).
SDS-PAGE and Western blot
Distal colonic tissue lysates were prepared from freshly dissected tissue or tissue incubated in Krebs buffer ex vivo for 1, 2, or 4 h at 37°C. Approximately 3 cm of flushed tissue was lysed using an Ultra-Turrax rotor-stator homogenizer (IKA Werke) in 0.4 ml ice-cold lysis buffer (PBS supplemented with 1% vol/vol Triton X-100, 1 mM EDTA, and 2× Complete protease inhibitor cocktail). Lysates were clarified by centrifugation at 2,000 RCF for 10 min at 4°C and supernatants collected and stored at −20°C until analysis. Lysate protein concentration was determined using a BCA protein assay kit (Pierce). For SDS-PAGE analysis, 80 µg protein for each sample was incubated at 95°C in Laemmli Sample Buffer with 1.25% vol/vol 2-mercaptoethanol for 5 min and then resolved on 15% polyacrylamide gels in a Mini-PROTEAN Tetra Cell system (Bio-Rad). Samples were resolved concurrently with MagicMark XP Western Protein Standards (Thermo Fisher). Resolved proteins were transferred to 0.2-µm-pore-diameter polyvinylidene difluoride membranes using a Transblot Turbo system and RTA transfer kit (Bio-Rad). Membranes were blocked in 5% wt/vol milk in PBS-T for 20 min. Membranes were incubated overnight at 4°C with rabbit anti-Il18 (ab71495, 1:500; Abcam) or mouse anti-actin (MAB1501, 1:20,000; Merck). Membranes were washed three times in PBS-T and incubated with goat-anti rabbit IgG-HRP (4030–05, 1:10,000; Southern Biotech) or goat-anti mouse IgG-HRP (1030–05, 1:20,000; Southern Biotech) for 2 h. Membranes were washed a further three times in PBS-T and then developed for enhanced chemiluminescent imaging using Immobilon Western reagent. Enhanced chemiluminescent signals were detected using a LAS-4000 imaging system (Fujifilm).
Microbiota profiling by 16S rRNA gene qPCR and sequencing
Fecal pellets were collected from mouse distal colon using sterilized equipment and flash frozen in liquid nitrogen. Total DNA was extracted from frozen pellets using a QIAamp Powerfecal DNA Kit (Qiagen) and quantified using a Nanodrop spectrophotometer (Thermo Fisher). DNA extractions were analyzed by qPCR using SsoFast EvaGreen Supermix (Bio-Rad) with 0.3 µM universal primers 926f (5′-AAACTCAAAKGAATTGACGG-3′) and 1062r (5′-CTCACRRCACGAGCTGAC-3′) with 45 ng template DNA. Reactions were performed and monitored using a CFX96 platform (Bio-Rad). Absolute bacterial 16S copy number was quantified using standard curves generated from qPCR of whole 16S gene amplicons purified from E. coli.
Microbiota community structure was profiled by sequencing of the V4 region of the 16S rRNA gene. Sequencing was performed on an Illumina MiSeq (Illumina RTA v1.17.28; MCS v2.5) using 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) primers designed for dual indexing and the V2 kit (2 × 250–bp paired-end reads). Samples were amplified in duplicates in reaction volumes of 25 µl containing 1× Five Prime Hot Master Mix (Quantabio), 200 nM of each primer, 0.4 mg/ml BSA, 5% DMSO, and 20 ng of genomic DNA. PCR was performed under the following conditions: initial denaturation for 3 min at 94°C, followed by 25 cycles of denaturation for 45 s at 94°C, annealing for 60 s at 52°C and elongation for 90 s at 72°C, and a final elongation step for 10 min at 72°C. Replicates were combined, purified with the NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel), and quantified using the Quant-iT PicoGreen dsDNA kit (Thermo Fisher). Equal amounts of purified PCR products were pooled, and the pooled PCR products were purified again using Ampure magnetic purification beads (Agencourt) to remove short amplification products.
Illumina paired-end reads were merged using PEAR (paired-end read merger) and quality filtered to remove reads that had at least one base with a q-score <20 and that were <220 nt or >350 nt. Sequences were clustered into OTUs at a 97% identity threshold using an open-reference OTU picking approach in QIIME 1 (version 1.9.1) with UCLUST against the Greengenes reference database (13_8 release). All sequences that failed to cluster when tested against the Greengenes database were used as input for picking OTUs de novo. Representative sequences for the OTUs were Greengenes reference sequences or cluster seeds and were taxonomically assigned using the Greengenes taxonomy and the Ribosomal Database Project Classifier. Representative OTUs were aligned using PyNAST and used to build a phylogenetic tree with FastTree. Chimeric sequences were identified with ChimeraSlayer and excluded from all downstream analyses. Similarly, sequences that could not be aligned with PyNAST, singletons, and very low-abundant sequence rRNA genes (relative abundance <0.005%) were also excluded. To correct for differences in sequencing depth, the same amount of sequences was randomly subsampled for each group of samples (rarefaction; maximum depth, 27.970).
The software package QIIME 2 (version 2018.8) was used to compute α- and β-diversity of the samples, and statistical differences between groups were calculated using PERMANOVA and 999 permutations. Genus-level OTU data were correlated to IML phenotype using the LEfSe algorithm (Segata et al., 2011).
Identification of microbiota-IML associations in published datasets
Source articles were identified by searching Scopus (Elsevier) for primary research that simultaneously quantified colonic IML thickness or barrier function and the abundance of bacterial taxons in the microbiota. Each article was assessed to identify positive, mixed, or negative associations between bacterial taxon abundance and IML properties. Associations were subclassed as correlative or causative based on the type of experiments used in each article. Only associations identified in WT animals were analyzed. The phylogenetic relationships between identified taxons was visualized using the Interactive Tree of Life tool (Letunic and Bork, 2019). All source data references and identified bacterial taxon–IML associations are listed in Table S3.
Culture and treatment of colonic organoids
Primary colonic organoid cultures were established from isolated crypts of Myd88−/− and WT C57BL/6 mice and maintained as previously described (Miyoshi and Stappenbeck, 2013). Experiments were performed on spheroid cultures embedded in Matrigel (Corning) cultured for 24 h in 50% L-WRN (L-cell derived Wnt3a, R-spondin 3, and Noggin) conditioned media, followed by 72 h in differentiation media composed of DMEM/F12 advanced media complemented with 2 mM GlutaMAX, 50 ng/ml EGF (Gibco), 1 µg/ml R-spondin (Peprotech), 100 ng/ml Noggin (Sigma-Aldrich), 0.5 µM A 83–01 (Tocris), and 10 µM Y-27632 (Tocris). Colonoid cultures were treated for 4 h with 50 ng/ml mouse rIl18 (9139-IL; R&D Systems) or 1 µg/ml lipopolysaccharide (L2880; Sigma-Aldrich). Cells were directly lysed on the plate in RLT buffer (Qiagen) and directly processed for mRNA isolation and subsequent qRT-PCR analysis as described above.
Data availability
Microbiota 16S rRNA gene sequencing results have been deposited in the European Nucleotide Archive sequence read archive with accession no. PRJEB30844 (http://www.ebi.ac.uk/ena/data/view).
Online supplemental information
Fig. S1 shows that there is no difference in IML phenotypes between male and female Nlrp6+/+ and Nlrp6−/− littermates. Fig. S2 shows additional microbiota community analysis in cohoused WT and Il18−/− mice. Fig. S3 shows causative and correlative relationships between bacterial taxon abundance and IML phenotype. Fig. S4 shows the sorting strategy for FACS isolation of colonic goblet cells. Tables S1 and S2 respectively list all mouse strains and PCR primers used in this study. Table S3 lists the source references for all bacterial–IML relationships shown Fig. S3.
Supplementary Material
Acknowledgments
We thank the Gothenburg Centre for Cellular Imaging for technical help and Valentina Tremaroli and Fredrik Bäckhed for assistance with microbiota profiling.
This work was supported by the Swedish Research Council, Swedish Cancer Foundation, Knut and Alice Wallenberg Foundation, Lundberg Foundation, Sahlgren’s University Hospital (Avtal om Läkarutbildning och Forskning), Torsten Söderbergs Stiftelse, National Institute of Allergy and Infectious Diseases (grant U01AI095473), and Swedish Foundation for Strategic Research. B.O. Schroeder is supported by the Human Frontier Science Program (long-term fellowship LT000109/2014). S. van der Post is supported by the Swedish Research Council (2015-00656).
The authors declare no competing financial interests.
Author contributions: Conceptualization, J.K. Volk and G.M.H. Birchenough; Methodology, G.M.H. Birchenough, B.M. Abad, E.E.L. Nyström, F. Svensson, and S. van der Post; Investigation: J.K. Volk, B.M. Abad, S. van der Post, E.E.L. Nyström, B.O. Schroeder, Å. Johansson, S. Jäverfelt, and G.M.H. Birchenough; Original draft, G.M.H. Birchenough; Draft editing, J.K. Volk, B.M. Abad, E.E.L. Nyström, G.C. Hansson, and M.E.V. Johansson; Supervision, G.M.H. Birchenough; Funding: G.C. Hansson, G.M.H. Birchenough, and M.E.V. Johansson.
References
- Bergstrom K.S., Kissoon-Singh V., Gibson D.L., Ma C., Montero M., Sham H.P., Ryz N., Huang T., Velcich A., Finlay B.B., et al. 2010. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 6:e1000902 10.1371/journal.ppat.1000902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom K., Fu J., Johansson M.E., Liu X., Gao N., Wu Q., Song J., McDaniel J.M., McGee S., Chen W., et al. 2017. Core 1- and 3-derived O-glycans collectively maintain the colonic mucus barrier and protect against spontaneous colitis in mice. Mucosal Immunol. 10:91–103. 10.1038/mi.2016.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergström J.H., Berg K.A., Rodríguez-Piñeiro A.M., Stecher B., Johansson M.E., and Hansson G.C.. 2014. AGR2, an endoplasmic reticulum protein, is secreted into the gastrointestinal mucus. PLoS One. 9:e104186 10.1371/journal.pone.0104186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergström J.H., Birchenough G.M., Katona G., Schroeder B.O., Schütte A., Ermund A., Johansson M.E., and Hansson G.C.. 2016. Gram-positive bacteria are held at a distance in the colon mucus by the lectin-like protein ZG16. Proc. Natl. Acad. Sci. USA. 113:13833–13838. 10.1073/pnas.1611400113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchenough G.M., Nyström E.E., Johansson M.E., and Hansson G.C.. 2016. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science. 352:1535–1542. 10.1126/science.aaf7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing B., Koren O., Goodrich J.K., Poole A.C., Srinivasan S., Ley R.E., and Gewirtz A.T.. 2015. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. 519:92–96. 10.1038/nature14232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Arcangelis A., Hamade H., Alpy F., Normand S., Bruyère E., Lefebvre O., Méchine-Neuville A., Siebert S., Pfister V., Lepage P., et al. 2017. Hemidesmosome integrity protects the colon against colitis and colorectal cancer. Gut. 66:1748–1760. 10.1136/gutjnl-2015-310847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M.S., Seekatz A.M., Koropatkin N.M., Kamada N., Hickey C.A., Wolter M., Pudlo N.A., Kitamoto S., Terrapon N., Muller A., et al. 2016. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell. 167:1339–1353.e21. 10.1016/j.cell.2016.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E., Strowig T., Kau A.L., Henao-Mejia J., Thaiss C.A., Booth C.J., Peaper D.R., Bertin J., Eisenbarth S.C., Gordon J.I., and Flavell R.A.. 2011. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 145:745–757. 10.1016/j.cell.2011.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Wei B., Wen T., Johansson M.E., Liu X., Bradford E., Thomsson K.A., McGee S., Mansour L., Tong M., et al. 2011. Loss of intestinal core 1-derived O-glycans causes spontaneous colitis in mice. J. Clin. Invest. 121:1657–1666. 10.1172/JCI45538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson J.K., Ermund A., Johansson M.E., Schütte A., Hansson G.C., and Sjövall H.. 2012. An ex vivo method for studying mucus formation, properties, and thickness in human colonic biopsies and mouse small and large intestinal explants. Am. J. Physiol. Gastrointest. Liver Physiol. 302:G430–G438. 10.1152/ajpgi.00405.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson G.C., Baeckström D., Carlstedt I., and Klinga-Levan K.. 1994. Molecular cloning of a cDNA coding for a region of an apoprotein from the ‘insoluble’ mucin complex of rat small intestine. Biochem. Biophys. Res. Commun. 198:181–190. 10.1006/bbrc.1994.1026 [DOI] [PubMed] [Google Scholar]
- Jakobsson H.E., Rodríguez-Piñeiro A.M., Schütte A., Ermund A., Boysen P., Bemark M., Sommer F., Bäckhed F., Hansson G.C., and Johansson M.E.. 2015. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 16:164–177. 10.15252/embr.201439263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M.E., Phillipson M., Petersson J., Velcich A., Holm L., and Hansson G.C.. 2008. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. USA. 105:15064–15069. 10.1073/pnas.0803124105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M.E., Gustafsson J.K., Sjöberg K.E., Petersson J., Holm L., Sjövall H., and Hansson G.C.. 2010. Bacteria penetrate the inner mucus layer before inflammation in the dextran sulfate colitis model. PLoS One. 5:e12238 10.1371/journal.pone.0012238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M.E., Gustafsson J.K., Holmén-Larsson J., Jabbar K.S., Xia L., Xu H., Ghishan F.K., Carvalho F.A., Gewirtz A.T., Sjövall H., and Hansson G.C.. 2014. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 63:281–291. 10.1136/gutjnl-2012-303207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M.E., Jakobsson H.E., Holmén-Larsson J., Schütte A., Ermund A., Rodríguez-Piñeiro A.M., Arike L., Wising C., Svensson F., Bäckhed F., and Hansson G.C.. 2015. Normalization of Host Intestinal Mucus Layers Requires Long-Term Microbial Colonization. Cell Host Microbe. 18:582–592. 10.1016/j.chom.2015.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler L.A., Crowley S.M., Sham H.P., Yang H., Wrande M., Ma C., Ernst R.K., Steele-Mortimer O., Celli J., and Vallance B.A.. 2014. Noncanonical inflammasome activation of caspase-4/caspase-11 mediates epithelial defenses against enteric bacterial pathogens. Cell Host Microbe. 16:249–256. 10.1016/j.chom.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemire P., Robertson S.J., Maughan H., Tattoli I., Streutker C.J., Platnich J.M., Muruve D.A., Philpott D.J., and Girardin S.E.. 2017. The NLR Protein NLRP6 Does Not Impact Gut Microbiota Composition. Cell Reports. 21:3653–3661. 10.1016/j.celrep.2017.12.026 [DOI] [PubMed] [Google Scholar]
- Letunic I., and Bork P.. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47(W1):W256–W259. 10.1093/nar/gkz239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M., Thaiss C.A., Zeevi D., Dohnalová L., Zilberman-Schapira G., Mahdi J.A., David E., Savidor A., Korem T., Herzig Y., et al. 2015. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell. 163:1428–1443. 10.1016/j.cell.2015.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy K.J., and Powrie F.. 2011. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 474:298–306. 10.1038/nature10208 [DOI] [PubMed] [Google Scholar]
- Mamantopoulos M., Ronchi F., Van Hauwermeiren F., Vieira-Silva S., Yilmaz B., Martens L., Saeys Y., Drexler S.K., Yazdi A.S., Raes J., et al. 2017. Nlrp6- and ASC-Dependent Inflammasomes Do Not Shape the Commensal Gut Microbiota Composition. Immunity. 47:339–348.e4. 10.1016/j.immuni.2017.07.011 [DOI] [PubMed] [Google Scholar]
- McGuckin M.A., and Hasnain S.Z.. 2017. Goblet cells as mucosal sentinels for immunity. Mucosal Immunol. 10:1118–1121. 10.1038/mi.2016.132 [DOI] [PubMed] [Google Scholar]
- Miyoshi H., and Stappenbeck T.S.. 2013. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc. 8:2471–2482. 10.1038/nprot.2013.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteleone G., Trapasso F., Parrello T., Biancone L., Stella A., Iuliano R., Luzza F., Fusco A., and Pallone F.. 1999. Bioactive IL-18 expression is up-regulated in Crohn’s disease. J. Immunol. 163:143–147. [PubMed] [Google Scholar]
- Muñoz M., Eidenschenk C., Ota N., Wong K., Lohmann U., Kühl A.A., Wang X., Manzanillo P., Li Y., Rutz S., et al. 2015. Interleukin-22 induces interleukin-18 expression from epithelial cells during intestinal infection. Immunity. 42:321–331. 10.1016/j.immuni.2015.01.011 [DOI] [PubMed] [Google Scholar]
- Nishida K., Kamizato M., Kawai T., Masuda K., Takeo K., Teshima-Kondo S., Tanahashi T., and Rokutan K.. 2009. Interleukin-18 is a crucial determinant of vulnerability of the mouse rectum to psychosocial stress. FASEB J. 23:1797–1805. 10.1096/fj.08-125005 [DOI] [PubMed] [Google Scholar]
- Nyström E.E.L., Birchenough G.M.H., van der Post S., Arike L., Gruber A.D., Hansson G.C., and Johansson M.E.V.. 2018. Calcium-activated Chloride Channel Regulator 1 (CLCA1) Controls Mucus Expansion in Colon by Proteolytic Activity. EBioMedicine. 33:134–143. 10.1016/j.ebiom.2018.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto Y., Tokime K., Yamanaka K., Habe K., Morioka T., Kurokawa I., Tsutsui H., Yamanishi K., Nakanishi K., and Mizutani H.. 2006. Human mast cell chymase cleaves pro-IL-18 and generates a novel and biologically active IL-18 fragment. J. Immunol. 177:8315–8319. 10.4049/jimmunol.177.12.8315 [DOI] [PubMed] [Google Scholar]
- Pizarro T.T., Michie M.H., Bentz M., Woraratanadharm J., Smith M.F. Jr., Foley E., Moskaluk C.A., Bickston S.J., and Cominelli F.. 1999. IL-18, a novel immunoregulatory cytokine, is up-regulated in Crohn’s disease: expression and localization in intestinal mucosal cells. J. Immunol. 162:6829–6835. [PubMed] [Google Scholar]
- Propheter D.C., Chara A.L., Harris T.A., Ruhn K.A., and Hooper L.V.. 2017. Resistin-like molecule β is a bactericidal protein that promotes spatial segregation of the microbiota and the colonic epithelium. Proc. Natl. Acad. Sci. USA. 114:11027–11033. 10.1073/pnas.1711395114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Piñeiro A.M., Bergström J.H., Ermund A., Gustafsson J.K., Schütte A., Johansson M.E., and Hansson G.C.. 2013. Studies of mucus in mouse stomach, small intestine, and colon. II. Gastrointestinal mucus proteome reveals Muc2 and Muc5ac accompanied by a set of core proteins. Am. J. Physiol. Gastrointest. Liver Physiol. 305:G348–G356. 10.1152/ajpgi.00047.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder B.O., Birchenough G.M.H., Ståhlman M., Arike L., Johansson M.E.V., Hansson G.C., and Bäckhed F.. 2018. Bifidobacteria or Fiber Protects against Diet-Induced Microbiota-Mediated Colonic Mucus Deterioration. Cell Host Microbe. 23:27–40.e7. 10.1016/j.chom.2017.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., and Huttenhower C.. 2011. Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellin M.E., Müller A.A., Felmy B., Dolowschiak T., Diard M., Tardivel A., Maslowski K.M., and Hardt W.D.. 2014. Epithelium-intrinsic NAIP/NLRC4 inflammasome drives infected enterocyte expulsion to restrict Salmonella replication in the intestinal mucosa. Cell Host Microbe. 16:237–248. 10.1016/j.chom.2014.07.001 [DOI] [PubMed] [Google Scholar]
- Stappenbeck T.S., and Virgin H.W.. 2016. Accounting for reciprocal host-microbiome interactions in experimental science. Nature. 534:191–199. 10.1038/nature18285 [DOI] [PubMed] [Google Scholar]
- Wirtz S., Becker C., Blumberg R., Galle P.R., and Neurath M.F.. 2002. Treatment of T cell-dependent experimental colitis in SCID mice by local administration of an adenovirus expressing IL-18 antisense mRNA. J. Immunol. 168:411–420. 10.4049/jimmunol.168.1.411 [DOI] [PubMed] [Google Scholar]
- Wlodarska M., Willing B., Keeney K.M., Menendez A., Bergstrom K.S., Gill N., Russell S.L., Vallance B.A., and Finlay B.B.. 2011. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect. Immun. 79:1536–1545. 10.1128/IAI.01104-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M., Thaiss C.A., Nowarski R., Henao-Mejia J., Zhang J.P., Brown E.M., Frankel G., Levy M., Katz M.N., Philbrick W.M., et al. 2014. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 156:1045–1059. 10.1016/j.cell.2014.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Ding S., Wang P., Wei Z., Pan W., Palm N.W., Yang Y., Yu H., Li H.B., Wang G., et al. 2017. Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature. 546:667–670. 10.1038/nature22967 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microbiota 16S rRNA gene sequencing results have been deposited in the European Nucleotide Archive sequence read archive with accession no. PRJEB30844 (http://www.ebi.ac.uk/ena/data/view).