How cells sense direction in shallow and complex chemical gradients is not well understood. Wang et al. propose a deterministic model of yeast gradient sensing in which signaling, polarity, and trafficking proteins localize to the predetermined division site before redistributing upgradient by a directed treadmill-like mechanism.
Abstract
The mating of budding yeast depends on chemotropism, a fundamental cellular process. The two yeast mating types secrete peptide pheromones that bind to GPCRs on cells of the opposite type. Cells find and contact a partner by determining the direction of the pheromone source and polarizing their growth toward it. Actin-directed secretion to the chemotropic growth site (CS) generates a mating projection. When pheromone-stimulated cells are unable to sense a gradient, they form mating projections where they would have budded in the next cell cycle, at a position called the default polarity site (DS). Numerous models have been proposed to explain yeast gradient sensing, but none address how cells reliably switch from the intrinsically determined DS to the gradient-aligned CS, despite a weak spatial signal. Here we demonstrate that, in mating cells, the initially uniform receptor and G protein first polarize to the DS, then redistribute along the plasma membrane until they reach the CS. Our data indicate that signaling, polarity, and trafficking proteins localize to the DS during assembly of what we call the gradient tracking machine (GTM). Differential activation of the receptor triggers feedback mechanisms that bias exocytosis upgradient and endocytosis downgradient, thus enabling redistribution of the GTM toward the pheromone source. The GTM stabilizes when the receptor peak centers at the CS and the endocytic machinery surrounds it. A computational model simulates GTM tracking and stabilization and correctly predicts that its assembly at a single site contributes to mating fidelity.
Introduction
Cellular responses to chemical gradients are likely important in all eukaryotic species. The best-known gradient-stimulated cellular outputs, chemotaxis (directed movement) and chemotropism (directed growth), are required for a wide range of biological phenomena. For example, chemotaxis plays a vital role in development, immunity, wound healing, inflammation, and metastasis (Iijima et al., 2002); and chemotropism is integral to axon guidance (Hong and Nishiyama, 2010; Tojima et al., 2011), angiogenesis (English et al., 2001; Basile et al., 2004; Muñoz-Chápuli et al., 2004), pollen tube guidance (Palanivelu and Preuss, 2000; Kim et al., 2004), and fungal life cycles (Snetselaar et al., 1996; Daniels et al., 2006). Although they ultimately exhibit quite different behaviors, chemotactic and chemotropic cells face similar challenges: the responding cell must determine the direction of the gradient source by sensing small differences in chemical concentration across its surface and polarize its cytoskeleton toward it.
To date, one of the best-characterized chemotropic models is the mating response of the budding yeast Saccharomyces cerevisiae (Arkowitz, 2009). In the haploid phase of its life cycle, S. cerevisiae exists as two mating types, MATa and MATα. Each type constitutively secretes a peptide pheromone that binds to G protein–coupled receptors (GPCRs) on cells of the opposite type. Upon activation by the receptor, the mating-specific Gα protein dissociates from Gβγ, and Gβ is rapidly phosphorylated on multiple sites. Free Gβγ signals to the nucleus via the Fus3 MAPK cascade, which induces cell-cycle arrest and changes in gene expression. Gβγ also serves as a positional determinant for the chemotropic growth site (CS) by linking the receptor to the machinery that nucleates actin cables via the Far1 scaffold protein. Actin-directed delivery of secretory vesicles to the CS results in the formation of mating projections, commonly called “shmoos.” In mating mixtures, cells find and contact a partner by determining the direction of the most potent pheromone source and polarizing their growth toward it (Jackson and Hartwell, 1990). When pheromone-stimulated cells are unable to sense a gradient, they shmoo at the default polarity site (DS), adjacent to the last cytokinesis site.
The yeast pheromone receptor is uniformly distributed on the surface of vegetative cells. Like most GPCRs, the activated pheromone receptor is phosphorylated on its C-terminal tail, which triggers its ubiquitination and internalization. Subsequently, the receptor reappears on the plasma membrane (PM) as a polarized crescent (Ayscough and Drubin, 1998; Moore et al., 2008; Suchkov et al., 2010). Pheromone-induced polarization of the receptor depends on its internalization and is detectable before morphogenesis; it does not require actin-cable–dependent secretion (Suchkov et al., 2010). Importantly, receptor phosphorylation appears to play a role in chemotropism apart from triggering receptor internalization (Ismael et al., 2016; Ismael and Stone, 2017). Whereas cells expressing a mutant form of the receptor that cannot be internalized (and thereby polarized) exhibit a partial defect in orienting toward mating partners, cells expressing a mutant form of the receptor that cannot be internalized or phosphorylated show no directional response in mating mixtures (Ismael et al., 2016).
As is typical of chemosensing cells, yeast exhibit a remarkable ability to interpret pheromone gradients. It has been estimated that a 1% difference in receptor occupancy across the 5-µm diameter of a yeast cell is sufficient to elicit robust orientation (Segall, 1993). Moreover, computational modeling suggests that noise at the level of the receptor is greater than the spatial signal in physiological gradients (Lakhani and Elston, 2017). And yet, in mating mixtures, yeast cells invariably select a single partner, even when surrounded by competing suitors.
A number of models have been proposed to explain yeast gradient sensing, but none address how cells switch from the intrinsically positioned DS they use for budding to the gradient-aligned CS, despite weak and complex gradients. The discovery that Gβγ links the receptor to actin-cable nucleation via its interaction with Far1 suggested a global-sensing model in which Gβγ directs exocytosis to the CS in preference to the DS (Butty et al., 1998; Nern and Arkowitz, 1998, 1999). We have reported that Gβγ inhibits pheromone-induced phosphorylation of the receptor (Ismael et al., 2016), that Gα augments this effect by recruiting the Fus3 MAPK to phosphorylate Gβ (Metodiev et al., 2002), and that the heterotrimeric G protein cointernalizes with the receptor (Suchkov et al., 2010). Based on these observations, we proposed that two interconnected positive feedback loops locally amplify pheromone signaling at the incipient CS (Ismael et al., 2016; Ismael and Stone, 2017). A computational model demonstrated that the proposed mechanisms could mimic gradient-induced polarization toward the pheromone source, assuming a 1:1:1 stoichiometry of the receptor and G protein subunits (Tian et al., 2014; Ismael et al., 2016). However, when the level of Gβγ was lowered by as little as 10% to better approximate the estimated receptor-to-Gβγ ratio of 4:1, most of the receptors were internalized and the model failed to generate polarity. Moreover, recent studies suggest that polarity is not initially established at the CS. Rather, interacting polarity proteins collectively called the “polarity complex” (PC) were found to spontaneously assemble at random cortical positions in cells responding to artificial pheromone gradients (Dyer et al., 2013; Hegemann et al., 2015; McClure et al., 2015; Hegemann and Peter, 2017). The PC is then thought to move upgradient by “biased wandering” until it encounters sufficient Gβγ to fix its position at the CS. Similarly, a patch of polarity proteins explores the cortex during mating in Schizosaccharomyces pombe, and its rate of movement is dependent on pheromone concentration (Bendezú and Martin, 2013).
Here we show that in mating mixtures, MATa cells gain their gradient-sensing ability and orient toward their partners in four phases. During global internalization, most of the receptor and G protein are removed from the PM. During assembly, signaling and trafficking proteins are polarized to the DS, starting with the recruitment of Gβγ by Far1 and ending with the concentration of exocytic and endocytic activities upgradient and downgradient, respectively. The receptor and G protein are then incrementally redistributed to the CS, where they stabilize and trigger chemotropic growth. A modified computational model simulates gradient tracking from the DS to the CS and correctly predicts that assembly of the gradient tracking machinery at a single site contributes to mating fidelity.
Results
Gradient-sensing components concentrate at the DS before redistributing to the eventual CS
We have proposed that Gβγ inhibition of receptor phosphorylation and internalization amplifies the initially slight differential of receptor activation across the surface of gradient-stimulated cells such that the G protein concentrates at the eventual CS (Ismael et al., 2016), where it stabilizes the wandering PC (Ismael and Stone, 2017). Consistent with this, we found that polarized receptor crescents were first detectable at the CS in our early time-lapse studies of mating cells (Suchkov et al., 2010). In contrast, a reporter that binds specifically to the unphosphorylated form of the receptor (Ballon et al., 2006) polarized to the DS of mating cells before relocalizing to the CS, rather than initially appearing at the CS as expected (Ismael et al., 2016). Moreover, Hegemann et al. (2015) showed that polarity of Cdc24, the activator of the Cdc42 GTPase, was first established at the DS in a mating cell.
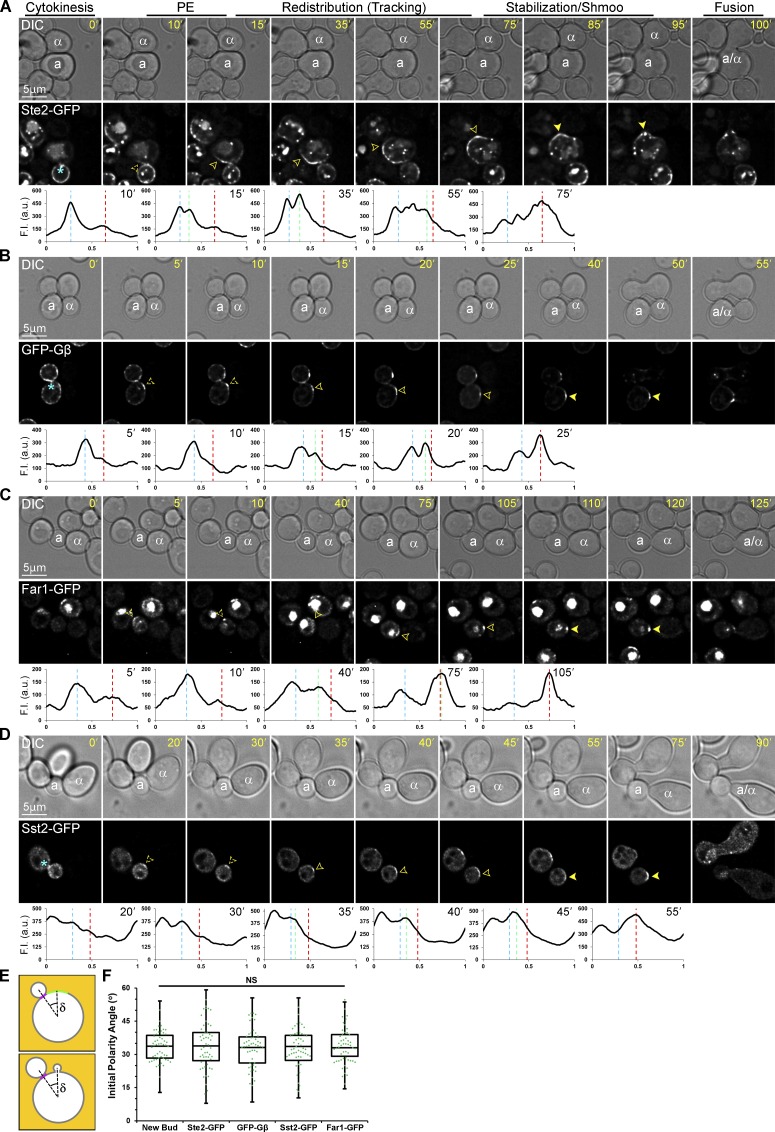
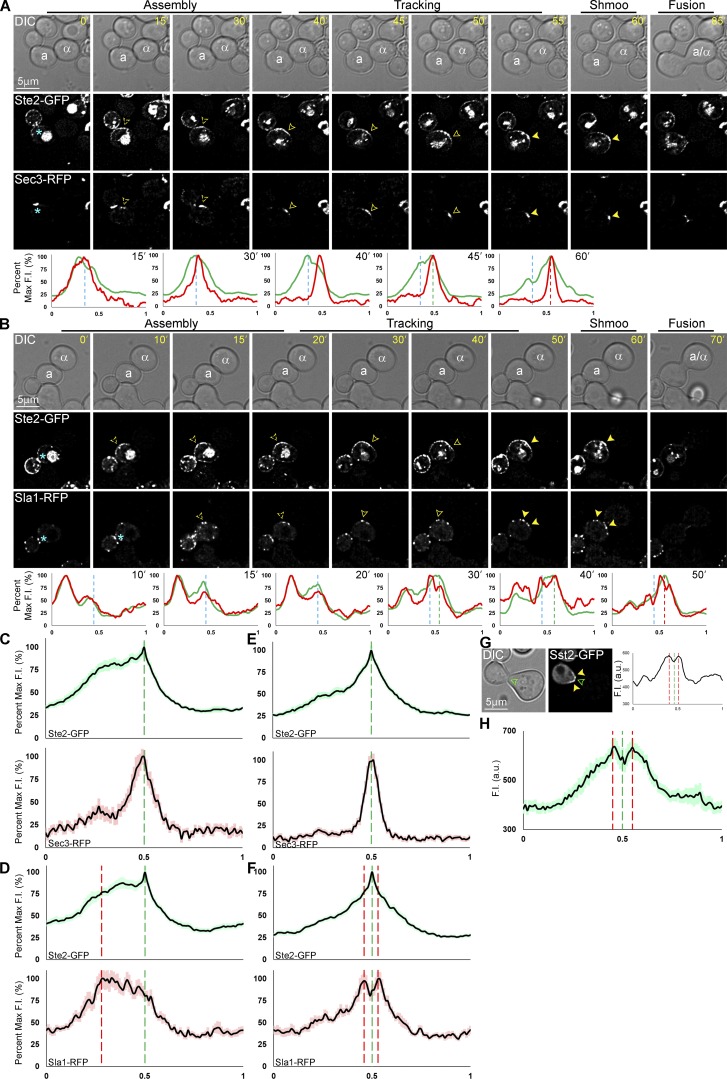
To reexamine the question of where receptor and G protein polarities are first established under physiological conditions, we took time-lapse images of MATa cells expressing receptor (Ste2-GFP) or Gβγ (GFP-Gβ) reporters in mating mixtures at 5-min intervals (Fig. 1). We followed mother and daughter cells from their separation to their fusion with mating partners. After cytokinesis, the receptor and Gβγ reporters each concentrated in a region of the PM adjacent to the division site, forming polarized crescents at the DS of both mother and daughter cells preparing to mate (Fig. 1, A and B). We infer that the initial polarity site is the DS because the receptor and Gβ crescents center at the same position relative to the cytokinesis site as the bud site in vegetative cells (Fig. 1, E and F). Remarkably, the receptor and Gβ crescents paused at the DS for 10–15 min before steadily redistributing to the CS, where they stabilized at the eventual fusion site just before shmoo emergence.
Figure 1.
Localization of gradient-sensing proteins in mating cells. (A–D) Representative time-lapse images. MATa cells expressing reporter genes tagged with GFP in situ were mixed with an equal number of MATα cells and imaged from cytokinesis to zygote formation (fusion). The mating partners are labeled a and α in the DIC images. The fluorescent images show the localization of each reporter as the MATa cells orient toward their mating partners. The blue asterisk indicates reporter localization to the bud neck; dashed arrowheads indicate polarity establishment (PE) at the DS and mark the signal peak; closed arrowheads indicate redistribution and mark the leading peak; filled arrowheads indicate stabilization at the CS and mark the signal peak. The plots show the distribution of each reporter on the PM at the indicated time points (10-point rolling average). The x axes represent distance along the PM; the y axes indicate fluorescence intensity (F.I.). The dashed blue, green, and red lines mark the DS peak, leading peak, and CS peak, respectively. Localization of Ste2-GFP (A), GFP-Gβ (B), Far1-GFP (C), and Sst2-GFP (D). (E and F) PE position. (E) Diagram illustrating how the position of the initial polarity site was determined. δ is the angle formed by rays drawn from the middle of the cell to the center of the cytokinesis site and the center of the polarized crescent (top) or bud (bottom) when they are first detectable. (F) Box scatterplots showing δ values for each reporter. The boxes enclose the middle two quartiles with the horizontal lines indicating the means; the whiskers show the top and bottom quartiles. Mean δ ± SEM in degrees: Bud = 33.7 ± 0.9; Ste2-GFP = 33.9 ± 1.4; GFP-Gβ = 33.2 ± 1.2; Sst2-GFP = 33.6 ± 1.1; Far1-GFP = 33.0 ± 1.2. n ≥ 50 for all strains and measurements.
To determine whether polarization to the DS followed by delayed redistribution upgradient to the CS is particular to the receptor and G protein, we asked whether other proteins implicated in gradient sensing behave similarly. In addition to effecting pheromone-induced cell-cycle arrest in the nucleus, Far1 plays an essential role in chemotropism as a scaffold at the cell cortex (Butty et al., 1998; Nern and Arkowitz, 1999; Shimada et al., 2000). In pheromone-treated cells, Far1 is exported from the nucleus in complex with Cdc24 (Blondel et al., 1999; Nern and Arkowitz, 2000) and, according to the current paradigm, is recruited to the CS by direct interaction with Gβγ (Butty et al., 1998; Nern and Arkowitz, 1998, 1999). Sst2 is an RGS protein (regulator of G protein signaling). It stimulates the GTPase activity of Gα (Apanovitch et al., 1998), binds to unphosphorylated receptor (Ballon et al., 2006), and is essential for gradient sensing (Dixit et al., 2014). We found that Sst2-GFP is recruited to the PM in pheromone-treated cells (Fig. S1), presumably by direct interaction with active-unphosphorylated receptor and its substrate, Gα-GTP. MATa cells expressing GFP-tagged Far1 and Sst2 were imaged in mating mixtures as described above. Like the receptor and Gβ reporters, Far1-GFP and Sst2-GFP exhibited dynamic localization patterns: following cytokinesis, they polarized to the DS and paused before incrementally redistributing to the CS, where they stabilized just before shmoo emergence (Fig. 1, C and D).
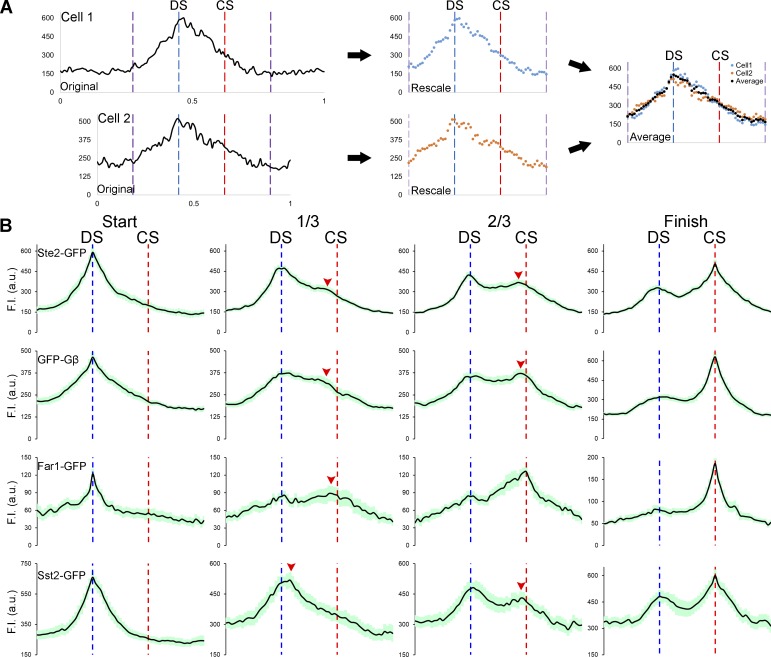
Because cells in mating mixtures vary in size and in the constellation of cells around them, we wondered whether the behavior of the particular cells we analyzed represented general characteristics of gradient sensing. A priori, the patterns of reporter redistribution from the DS to the CS could depend on local gradient conditions or on cell-specific factors. To answer this question, we followed the localization of GFP-tagged receptor, Gβ, Far1, and Sst2 in MATa cells during the formation of randomly selected zygotes, starting with cytokinesis and ending with cell fusion. Fig. 2 shows the average PM signal intensities for each reporter plotted as a function of its normalized position between the DS and CS. These plots are similar to the corresponding single-cell plots, suggesting that the kinetics of redistribution from the DS to the CS are consistent across cells for all four reporters. Moreover, the mean rates of receptor and Gβγ reporter redistribution did not differ significantly, and linear regression analyses indicate that they redistributed at a constant rate (Fig. S2).
Figure 2.
Average distribution of four gradient-sensing proteins on the PM of mating cells orienting toward a partner. MATa cells expressing Ste2 (the receptor), Gβ, Far1, and Sst2 tagged with GFP in situ were mixed with an equal number of MATα cells and imaged from cytokinesis to fusion. Identical imaging parameters were used for all reporters. (A) Illustration of the method used to normalize and average the signal distributions exhibited by multiple cells. The PM signals of cells that formed zygotes were quantified with ImageJ. 20 cells expressing each reporter were normalized by the DS-to-CS distance, and the average signal distributions were generated as described in Materials and methods. (B) The plots show the average distribution ± SEM (light green shadow) for each reporter just before initiation of redistribution from the DS (Start), after one-third and two-thirds of the time it took to complete redistribution, and having just stabilized at the CS (Finish). The dashed blue and red lines mark the DS and CS, respectively; the red arrowheads indicate the leading peaks during redistribution. F.I., fluorescence intensity.
Far1 is the first to appear at the DS, followed sequentially by Gβ, the receptor, and Sst2
The colocalization of four essential gradient-signaling proteins to the DS despite wide variation in the arrangement of potential mating partners, together with the subsequent pause at that site before redistribution to the CS, suggested that some sort of assembly process is required to enable gradient sensing. We hypothesized that the cell uses the DS to bring together a variety of signaling and trafficking proteins that make up what we call, as a working term, the gradient tracking machine (GTM). We operationally define the GTM as assembled and active when its components begin to redistribute upgradient, at which time the cell is manifestly sensing the position of potential mating partners. For convenience, we refer to the redistribution of the GTM components along the PM as tracking.
As a first step in characterizing this putative assembly process, we asked whether GTM components localize to the DS in a particular order, as indicated by when their polarity is first detectable (PE), and whether they pause for characteristic times before tracking (Fig. 3 A). MATa cells expressing Spa2-RFP and either GFP-tagged receptor, Gβ, Far1, or Sst2 were imaged at 2-min intervals from cytokinesis to the start of tracking. Spa2 is a polarity protein that moves from the mother–daughter neck to the DS upon cytokinesis (Dobbelaere and Barral, 2004; Fig. 3 B). This provides a highly reproducible marker for the M-G1 transition, the zero time in our measurement of PE. Significant differences in the mean PE of the reporters were observed in these experiments, suggesting a strict arrival sequence at the DS. The polarization of Far1-GFP was detected first, followed sequentially by GFP-Gβ, Ste2-GFP, and Sst2-GFP (Fig. 3, C–G). Notably, the mean pause of the reporters was inversely related to their PEs, with the first arrivers pausing the longest and vice versa; Far1-GFP and GFP-Gβ were an exception to this rule, as their pause times were not significantly different (Fig. 3 H). When compared according to the mean times they started tracking (PE + pause, Fig. 3 A), the reporters fell into two distinct groups: Far1-GFP and Sst2-GFP began tracking first, whereas Ste2-GFP and GFP-Gβ started tracking significantly later (Fig. 3 I). Finally, both the Ste2-GFP and GFP-Gβ signals steadily increased during the pause (Fig. 3 J). These data support the idea that the cell develops its ability to sense and respond to gradient stimulation via a regulated process in G1, which must be completed before orientation toward a mating partner can begin. Moreover, the tracking of Far1-GFP and Sst2-GFP before total receptor (Ste2-GFP) and GFP-Gβ is consistent with a relative increase in the active species of receptor and Gα (which bind Sst2) and free Gβγ (which binds Far1) upgradient.
Figure 3.
Kinetics of polarization to the DS and the initiation of tracking. (A) The time to tracking is the sum of the time it takes to detect a reporter at the DS following cytokinesis (PE) and the duration of its pause. (B) Use of Spa2-RFP as a marker for cytokinesis. MATa cells expressing in situ–tagged Spa2-RFP and GFP-Cdc3 were imaged in mating mixtures. Immediately after cytokinesis, marked by the splitting of the septin ring (filled cyan arrowhead), Spa2-RFP translocates from the bud neck to the DS (filled red arrowhead). (C–F) Representative time-lapse images. MATa cells expressing reporter genes tagged with GFP in situ were mixed with an equal number of MATα cells and imaged from cytokinesis to the initiation of tracking. The fluorescent images show the localization of each reporter as the MATa cells orient toward their mating partners. Unless indicated, the images were not deconvolved. The blue asterisk indicates reporter localization to the bud neck; dashed arrowheads indicate PE at the DS and mark the signal peak; closed arrowheads indicate redistribution (tracking) and mark the leading peak. The plots show the distribution of each reporter on the PM at the indicated time points (10-point rolling average). The dashed blue and green lines mark the DS peak and the tracking peak, respectively. Localization of Spa2-RFP and Ste2-GFP (C), GFP-Gβ (D), Far1-GFP (E), and Sst2-GFP (F; deconvolved). (G–I) Box scatterplots showing the distribution of the indicated measurements. n ≥ 50 for all strains and measurements; **, P < 0.0001; *, P < 0.002. (G) Distribution of PE values for the indicated reporters. Mean PE ± SEM in minutes: Far1-GFP = −1.9 ± 0.6; GFP-Gβ = 2.4 ± 0.4; Ste2-GFP = 6.1 ± 0.6; Sst2-GFP = 10.0 ± 0.5. (H) Distribution of Pause values for the indicated reporters. Mean Pause ± SEM in minutes: Far1-GFP = 13.5 ± 0.7; GFP-Gβ = 13.3 ± 0.8; Ste2-GFP = 9.2 ± 0.6; Sst2-GFP = 2.6 ± 0.4. (I) Distribution of times to tracking for the indicated reporters. Mean Times to tracking ± SEM in minutes: Far1-GFP = 11.6 ± 0.9; GFP-Gβ = 15.6 ± 0.8; Ste2-GFP = 15.3 ± 0.7; Sst2-GFP = 12.5 ± 0.5. (J) Signal intensity at the DS during pause. Mean intensity ± SEM, n = 25 for both reporters. F.I., fluorescence intensity.
Localization of Gβ to the DS requires Far1–Cdc24 interaction but not receptor polarization
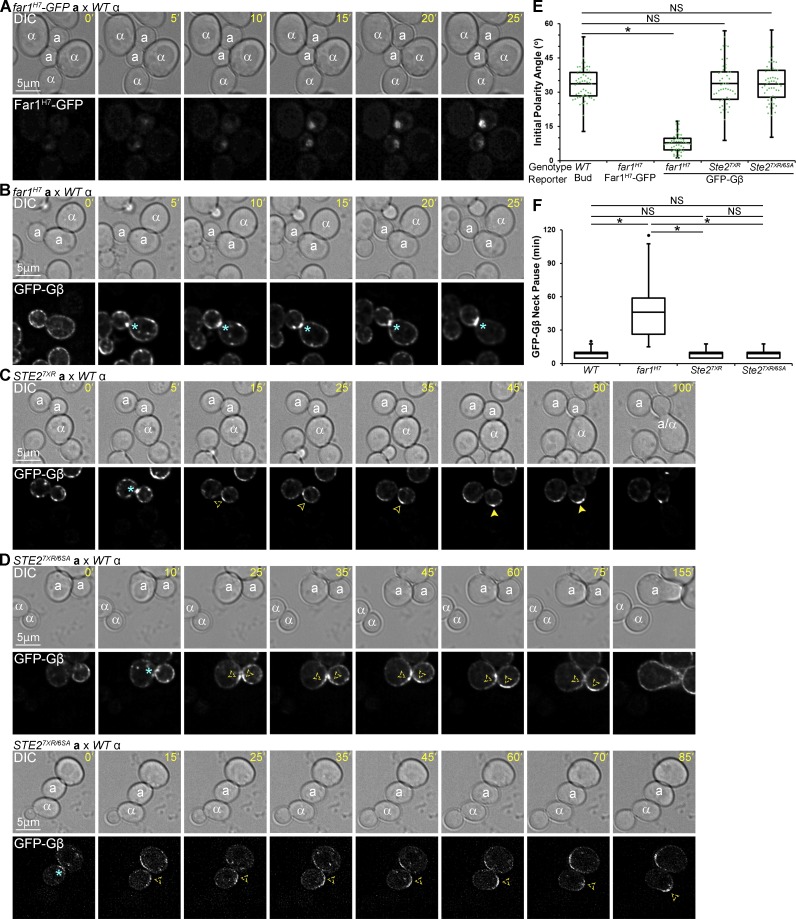
How is the gradient-sensing machinery initially recruited to the DS instead of the CS despite gradient stimulation? We hypothesized that GTM assembly begins with Far1 localization to the DS on the basis of three observations: (1) pheromone triggers the export of Far1–Cdc24 from the nucleus of cells in G1 (Blondel et al., 1999; Nern and Arkowitz, 2000); (2) Cdc24 interacts with three proteins that localize to the DS, Cdc42, the Bem1 scaffold, and the Bud1 GTPase (Park et al., 1997; Butty et al., 1998; Nern and Arkowitz, 1998, 1999, 2000; Goryachev and Pokhilko, 2008; Kang et al., 2010); and (3) Far1 appears to polarize to the DS before Gβ, the receptor, and Sst2 (Fig. 3 C). To test this hypothesis, we used Far1H7, a mutant form of Far1 that lacks the C-terminal domain required for its interaction with Cdc24 (Valtz et al., 1995). MATa cells expressing Far1H7-GFP and MATa far1-H7 mutant cells expressing GFP-Gβ were imaged at 5-min intervals in mating mixtures. Following cytokinesis, Far1H7-GFP accumulated in the nucleus, like Far1-GFP, but the mutant reporter failed to localize to the DS (Fig. 4, A and E). In the MATa far1-H7 cells, GFP-Gβ polarized to the bud neck in late M and remained there, relocalizing to the polarized growth site only after the emergence of a new bud (90% frequency) or a default shmoo (10%; Fig. 4, B, E, and F). In contrast, GFP-Gβ rapidly translocated to the DS following cytokinesis in WT cells, after transient localization to the bud neck (Figs. 1 B and 4 F). These results indicate that the premorphogenic DS-localization of both Far1 and Gβ depend on Far1–Cdc24 interaction and suggest that the DS-localization of Gβ depends on that of Far1. Conversely, localization of GFP-Gβ to the DS was as robust in cells unable to internalize and thereby polarize the receptor as in WT cells (Fig. 4, C–F). Our inference that the polarization of Gβγ to the DS is dependent on the DS-polarization of Far1 but not of the receptor is consistent with their PEs (Fig. 3 C).
Figure 4.
Effects of far1H7 and STE2 mutants on GFP-Gβ localization to the DS. (A–D) Representative time-lapse images. MATa cells expressing reporter genes tagged with GFP in situ were mixed with an equal number of MATα cells and imaged from cytokinesis to the indicated time points. Localization of Far1H7-GFP (A), GFP-Gβ in MATa far1H7 cells (B), GFP-Gβ in MATa ste27XR cells (C), and GFP-Gβ in MATa ste27XR/6SA cells (D). (E) Box scatterplots showing initial polarity angle, δ (see Fig. 1 E). Mean δ ± SEM in degrees: Bud = 33.7 ± 0.9; GFP-Gβ in far1H7 cells = 7.8 ± 0.6; GFP-Gβ in ste27XR cells = 33.8 ± 1.2; GFP-Gβ in ste27XR/6SA cells = 33.5 ± 1.1. (F) Box plots showing GFP-Gβ pause times at the bud neck in the indicated backgrounds. Mean pause ± SEM in minutes: WT = 8.9 ± 0.5; far1H7 = 46.0 ± 3.5; ste27XR = 9.0 ± 0.5; ste27XR/6SA = 8.9 ± 0.4. n ≥ 50 for all strains and measurements; *, P < 0.0001.
Gβ tracking depends on receptor phosphorylation but not on receptor internalization and polarization
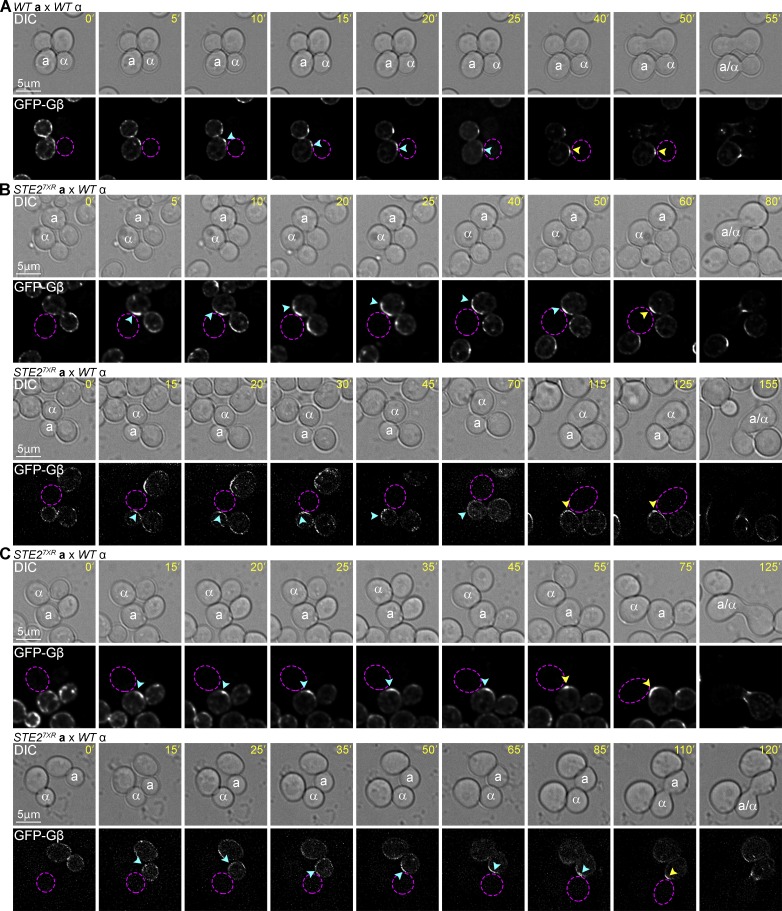
Pheromone-induced polarization of the receptor depends on receptor internalization (Suchkov et al., 2010; Ismael et al., 2016). We have previously shown that cells expressing a mutant form of the receptor that can be phosphorylated but not internalized (Ste27XR) are partially defective in gradient sensing (Ismael et al., 2016). They can establish a CS and mate, but they do not orient toward their partners as accurately as WT cells. Cells expressing a mutant form of the receptor that can be neither internalized nor phosphorylated (Ste27XR/6SA), on the other hand, show no gradient-sensing ability. They shmoo and mate only at the DS. These phenotypes correlate with the functionality of the putative GTM. In ste27XR cells, GFP-Gβ localized to the DS and tracked toward the CS (Fig. 4, C and E), although the tracking GFP-Gβ crescent either overshot or failed to reach the point of alignment with the mating partner in about two-thirds of the ste27XR cells, leading to angled zygotes (Fig. 5). In ste27XR/6SA cells, GFP-Gβ also localized to the DS but was unable to track upgradient (Fig. 4, D and E), consistent with our finding that cells unable to phosphorylate the receptor shmoo and mate only at the DS (Ismael et al., 2016). These results demonstrate that Gβ tracking from the DS to the CS requires receptor phosphorylation but not receptor polarization.
Figure 5.
ste27XR confers a defect in GFP-Gβ tracking. (A–C) Representative time-lapse images. MATa cells expressing in situ–tagged GFP-Gβ were mixed with an equal number of MATα cells and imaged from cytokinesis to fusion. The fluorescent images show the localization of GFP-Gβ as the MATa cells orient toward their mating partners (dashed purple circles). The blue arrowheads indicate the leading edge of the tracking crescent before morphogenesis. The yellow arrowheads indicate the final position of the GFP-Gβ peak and eventual site of fusion. (A) GFP-Gβ tracking in a WT MATa cell. (B and C) GFP-Gβ tracking in MATa ste27XR cells. Examples of ste27XR cells in which the tracking GFP-Gβ crescent overshot the presumptive CS before centering and intensifying at the eventual fusion site (B; 40%); failed to reach the presumptive CS, resulting in an angled zygote (C; 25%). Incidence of abnormal GFP-Gβ tracking in WT versus ste27XR cells: P < 0.0001 (χ2); n = 20.
Exocytosis concentrates toward the front and endocytosis toward the back of the tracking receptor crescent
What drives receptor redistribution from the DS to the CS in gradient-stimulated cells? Our time-lapse images and corresponding PM-signal plots (Fig. 1 A) show that as the assembly phase ends and the tracking phase begins, the polarized receptor crescent initially expands along the PM from the DS toward the CS, and subsequently diminishes along its lagging edge. A priori, this phenomenon is likely the result of directed delivery and/or biased docking of secretory vesicles to the upgradient side of the receptor crescent, along with a higher rate of receptor internalization downgradient. To test this, we first asked whether we could detect evidence of greater vesicle fusion on the GTM-tracking side of the cell. Our measurements demonstrated that mating cells grow where the receptor tracks from the DS to the CS: the mean arc increase on the tracking side of mating cells was 6.58 ± 0.59%, while the other side of the cell increased by only 0.54 ± 0.20% (P < 0.0001; n = 25). This supports the idea that vesicular docking and fusion is biased to the gradient-tracking side of the cell. In addition, we constructed MATa strains coexpressing the receptor reporter (Ste2-GFP) and either RFP-tagged Sec3, an early exocytosis marker (Finger et al., 1998), or RFP-tagged Sla1, a receptor-driven endocytosis marker (Howard et al., 2002; Goode et al., 2015). During the assembly phase at the DS in mating cells, the majority of Sec3-RFP and Sla1-RFP distributed coincident with the Ste2-GFP peak (Fig. 6, A and B). At the beginning of tracking, however, the distribution of Sec3-RFP shifted toward the front of the receptor crescent, while the bulk of Sla1-RFP remained toward the back. The Sec3 peak shifted upgradient before the receptor peak in 30% of the cells we examined and during the same 5-min interval as the receptor in the others (n = 20). The average PM signal intensities for each reporter plotted as a function of their normalized positions relative to the leading receptor peak shows that these spatial relationships were maintained during tracking (Fig. 6, C and D). These data suggest maximal exocytosis to the front and maximal receptor-driven endocytosis to the back of the tracking receptor crescent, consistent with redistribution of the receptor by a treadmill-like mechanism.
Figure 6.
Localization of Sec3, Sla1, and Sst2 relative to the receptor in mating cells. (A and B) Representative time-lapse images. MATa cells coexpressing in situ–tagged Ste2-GFP and Sec3-RFP or Sla1-RFP were mixed with an equal number of MATα cells and imaged from cytokinesis to fusion. The fluorescent images show the localization of Ste2-GFP and Sec3-RFP (A) or Sla1-RFP (B) as the MATa cell orients toward its mating partner. During tracking, closed arrowheads indicate the leading Ste2-GFP and Sec3-RFP peaks and the lagging Sla1-RFP peak. After stabilization, the filled arrowheads indicate the center Ste2-GFP and Sec3-RFP peaks and the flanking Sla1-RFP peaks. The plots show the distribution on the PM of Ste2-GFP (green) and Sec3-RFP (red) or Sla1-RFP (red), respectively, at the indicated time points. The dashed blue, green, and red lines mark the DS peak, leading peak, and CS peak, respectively. (C–F) Average PM distribution of Ste2-GFP and Sec3-RFP or Sla1-RFP in mating cells during tracking and at the prezygote stage. The PM signals of 30 cells two time points before shmoo emergence (tracking) and one time point before fusion (prezygote) were quantified with ImageJ, normalized for cell size, and averaged as described in Materials and methods. The plots show the mean signal distribution ± SEM (light shadow) of Ste2-GFP (green) and Sec3-RFP (red) or Sla1-RFP (red). The dashed green lines mark the leading receptor peak and the shmoo tip during tracking and in prezygote; the dashed red lines mark the peaks of Sla1-RFP. Average distributions of Ste2-GFP and Sec3-RFP during tracking (C), Ste2-GFP and Sla1-RFP during tracking (D), Ste2-GFP and Sec3-RFP in prezygotes (E), and Ste2-GFP and Sla1-RFP in prezygotes (F). (G and H) Sst2 surrounds the growth site after shmoo formation. Time-lapse images generated for the experiment represented in Fig. 2 were used to determine the distribution of Sst2-GFP in MATa cells at the prezygote stage. (G) Representative DIC and fluorescent images of a shmooing MATa Sst2-GFP cell and corresponding quantification of the PM signal. The filled yellow arrowheads and dashed red lines indicate the peaks of the Sst2-GFP PM signal; the closed green arrowhead and dashed green line indicate the shmoo tip. (H) Average distribution of Sst2-GFP during shmoo formation. The plot shows the average distribution of Sst2-GFP in 21 cells. The dashed green lines mark the shmoo tip; the dashed red lines mark the Sst2-GFP peaks. F.I., fluorescence intensity.
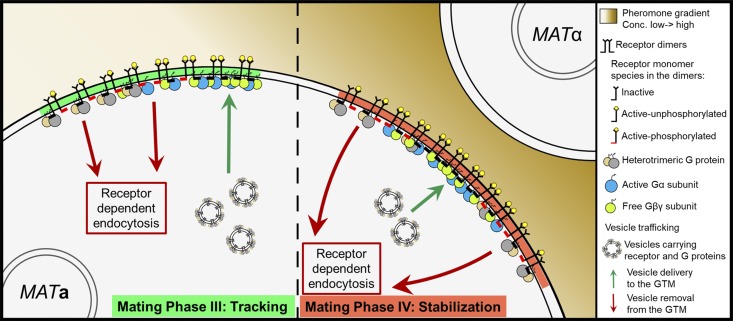
The CS is established where the sensory and secretory components stop tracking and are corralled by endocytosis
Our time-lapse imaging studies show that the tracking peaks of receptor, Gβ, Far1, Sst2, and Sec3 center and stabilize at the eventual CS before morphogenesis (Fig. 1, A–D; Fig. 2; and Fig. 6, A and E), and that the receptor-endocytosis marker Sla1 concentrates around the premorphogenic CS, where it remains until cell fusion (Fig. 6, B and F). After shmoo emergence, Sst2 also concentrates around the growth site (Fig. 6, G and H). This phenomenology strongly suggests that the stabilized GTM determines the position of the CS: while the peaks of the signaling, polarity, and secretory proteins align with each other and with the gradient source, thereby directing chemotropic growth, the negative regulators of GPCR and G protein signaling surround the shmoo tip, restricting growth to the CS by a mechanism analogous to endocytosis-based cortical corralling during budding (Jose et al., 2013).
A computational model simulates GTM tracking and stabilization
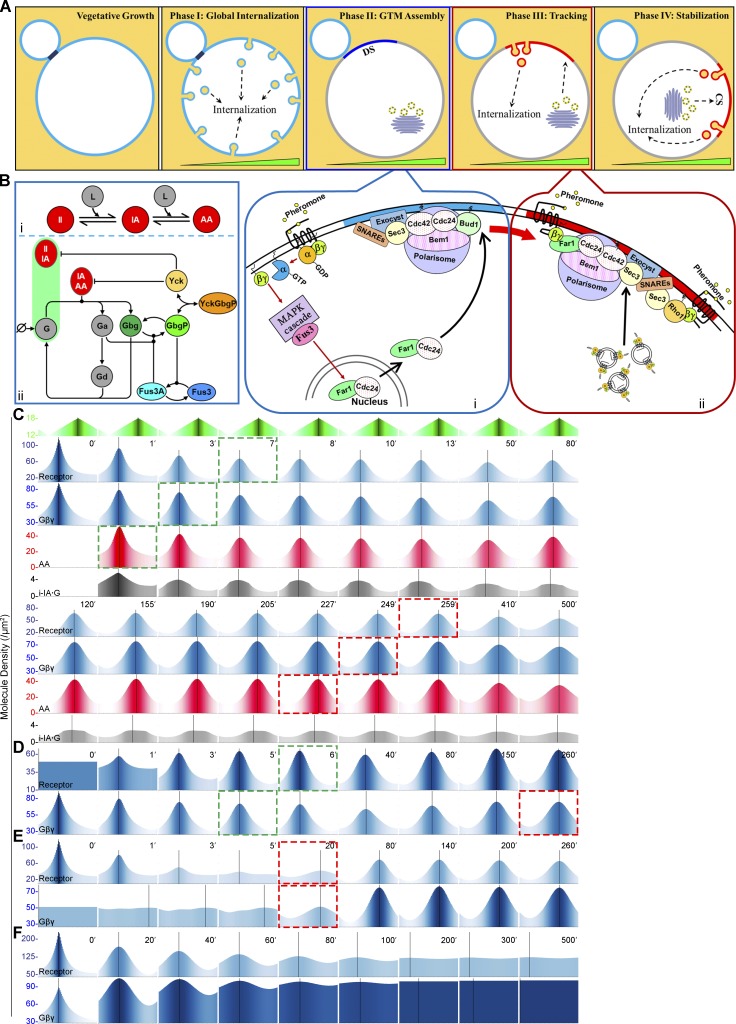
Synthesis of published results and those described here suggest a new model of yeast gradient sensing (Fig. 7 A). Following global internalization (phase I) and cytokinesis, the receptor and G protein colocalize with other components of the GTM at the DS (phase II). This assembly process begins when Far1–Cdc24 exported from the nucleus in G1 (Blondel et al., 1999; Nern and Arkowitz, 2000) is recruited to the DS via the known Cdc24–Bem1 interaction (Nern and Arkowitz, 1998; Goryachev and Pokhilko, 2008; Fig. 7 A, i and ii). Gβγ is then recruited to the DS by its interaction with Far1 (Butty et al., 1998; Nern and Arkowitz, 1999), where it inhibits receptor and G protein internalization by the positive feedback mechanisms we described in Ismael et al. (2016). As Gβγ accumulates at the DS, it contributes to the recruitment of Far1. Localization of Sec3 to the DS by direct binding to activated Cdc42 and Rho1 (Guo et al., 2001; Zhang et al., 2001; Pleskot et al., 2015) leads to assembly of the exocyst complex. Biased docking and fusion of secretory vesicles carrying the receptor, combined with local inhibition of receptor internalization, result in the emergence of the polarized receptor crescent at the DS. As the receptor and G protein polarize and colocalize at the DS, the pheromone gradient induces a corresponding gradient of activated receptors and G protein. We postulate that free Gβγ positions Sec3 via their mutual interaction with Far1–Cdc24–Cdc42 and Rho1 (Fig. 7 A, ii), while differential phosphorylation of the receptor leads to higher rates of G protein activation upgradient and receptor/G protein internalization downgradient. The GTM tracks upgradient as secretory vesicles carrying receptor and G protein are preferentially deposited near its leading edge while receptor/G protein complexes are preferentially removed toward its lagging edge (phase III). This treadmill-like mechanism moves the GTM upgradient until it reaches the maximum pheromone concentration, where coincident peaks of the signaling and secretory proteins align, and a surrounding zone of maximal receptor/G protein inactivation and internalization stabilizes the GTM at the CS (phase IV).
Figure 7.
Model and computational simulation of yeast gradient sensing. (A) Proposed four-phase model of yeast gradient sensing. In vegetative cells, the receptor and G protein are uniformly distributed on the PM. After their global internalization, the GTM is assembled at the DS. Tracking begins when a gradient of activated receptor on the cell surface induces a separation of exocytosis and endocytosis, which results in GTM redistribution upgradient (green triangle) by a treadmill-like mechanism. The GTM stops tracking at the CS and centers around the future shmoo/fusion site during the stabilization phase. The blowups show the molecular mechanisms of DS assembly (i) and biased secretion during tracking (ii). (B) Network diagram of the computational model. (i) Three dimeric forms of the receptor are explicitly modeled: II, IA, and AA. (ii) The network symbols are as follows: pheromone (L, ligand); heterotrimeric G protein (G); Gα-GTP (Ga); Gα-GDP (Gd); free Gβγ and its phosphorylated form (Gbg and GbgP); the Yck1/2 receptor kinases (Yck); and the inactive and active MAPK (Fus3 and Fus3a). The green bar represents internalization of the heterotrimeric G protein with II and IA. (C–E) Selected outputs from modeling simulations. The x-axis of each plot corresponds to the cell circumference. Vertical black lines indicate the peak values of each parameter at each time point. The green panel (top) shows the applied pheromone gradient. The dashed green and red boxes indicate the start and end of tracking, respectively. (C) Outputs from the standard model. The i-IA⋅G parameter is the internalization rate of the IA-receptor dimer bound to one heterotrimeric G protein. (D–F) In silico experiments. Outputs from modified models in which the receptor is uniformly distributed at the start of the simulation (D); Gβγ is uniformly distributed at the start of the simulation (E); and receptors are monomeric and cointernalization of the receptor and G protein cannot be induced by pheromone (F).
The reaction–diffusion computational model published in Ismael et al. (2016) (hereafter, v.1) demonstrated that two positive feedback loops, GβPγ inhibition of receptor phosphorylation and Gα-directed phosphorylation of Gβ, can locally amplify the directional signal and mimic gradient-induced polarity toward the pheromone source. However, this model did not simulate gradient tracking: polarity was initially established at the CS and was unresponsive to changes in gradient direction. Based on the findings reported here and elsewhere, we developed a second-generation computational model (hereafter, v.2) that retains the same basic network structure as v.1 (Fig. 7 B) but incorporates the following changes (see Materials and methods; Figs. S3 and S4 and Tables S3, S4, S5, S6, and S7). (1) The receptor is dimeric (Overton and Blumer, 2000; Gehret et al., 2012; Sridharan et al., 2016; Cevheroğlu et al., 2017), and gradient-stimulation generates three species: the active-active homodimer (AA), the inactive-active heterodimer (IA), and the inactive-inactive homodimer (II). This allows for pheromone-induced internalization of the G protein with the receptor (Suchkov et al., 2010) without violating the GPCR–G protein trafficking paradigm, i.e., that activation of GPCRs triggers their internalization while heterotrimeric G proteins stably interact only with inactive receptors. (2) In the initial state, the receptor and G protein are polarized to the DS in a ratio of ∼1:1, as determined by our measurements of their relative levels at the DS in mating cells (Fig. S4). (3) Secretory vesicles carrying the receptor and heterotrimeric G protein are targeted to the PM by Gβγ. This postulate is based on the well-established interactions between Gβγ and proteins that bind Sec3, Cdc42 (via Far1–Cdc24) and Rho1 (Butty et al., 1998; Nern and Arkowitz, 1998, 1999; Guo et al., 2001; Zhang et al., 2001; Bar et al., 2003; Pleskot et al., 2015), and on the observation that Gβ is essential to stabilize the position of the PC (McClure et al., 2015).
Our revised computational model mimics four key characteristics of yeast gradient sensing: the pause after the establishment of receptor and G protein polarity, tracking from the initial polarity site (the DS) to the CS, stabilization at the CS, and the ability to respond to a change in gradient direction (reorientation). Figs. 7 C and S5 and Videos 1 and 2 show selected outputs of the model, beginning with the receptor and G protein already polarized at the DS. The pause is recapitulated by the time it takes for the peak of total receptor to shift upgradient. This delay is attributable to the time required for the pheromone gradient to induce differential activation of the receptor and G protein, which ultimately results in the separation of exocytosis and endocytosis. The AA receptor dimer is the first species to shift position (1′), followed by active Gα and free Gβγ (3′), total Gβγ (4′), and finally, total receptor (7′). Notably, once the computational GTM becomes competent to sense the gradient (signified by redistribution of the total receptor and G protein), it tracks from the DS to CS and, like the actual GTM, stops when the receptor and G protein are aligned with the point source of pheromone. Like the localization of Sla1-RFP in mating cells (Fig. 6, B, D, and F), the peak rate of receptor/G protein internalization (i-IA⋅G) tracks behind the peaks of receptor and G protein but is more uniformly distributed across the GTM near the CS. Switching the direction of the gradient after the computational GTM has stabilized at the original CS causes it to resume tracking and align with the new pheromone source, capturing the ability of gradient-stimulated cells to reorient (Segall, 1993; Vallier et al., 2002; Paliwal et al., 2007; Fig. 8 and Video 3). The behaviors of the other species and reaction rates that make up v.2 are consistent with the conclusion that GTM tracking depends on inverse gradients of secretion and internalization that are ultimately determined by the spatiotemporal activity of the receptor and G protein (Fig. S5 and Videos 1 and 2).
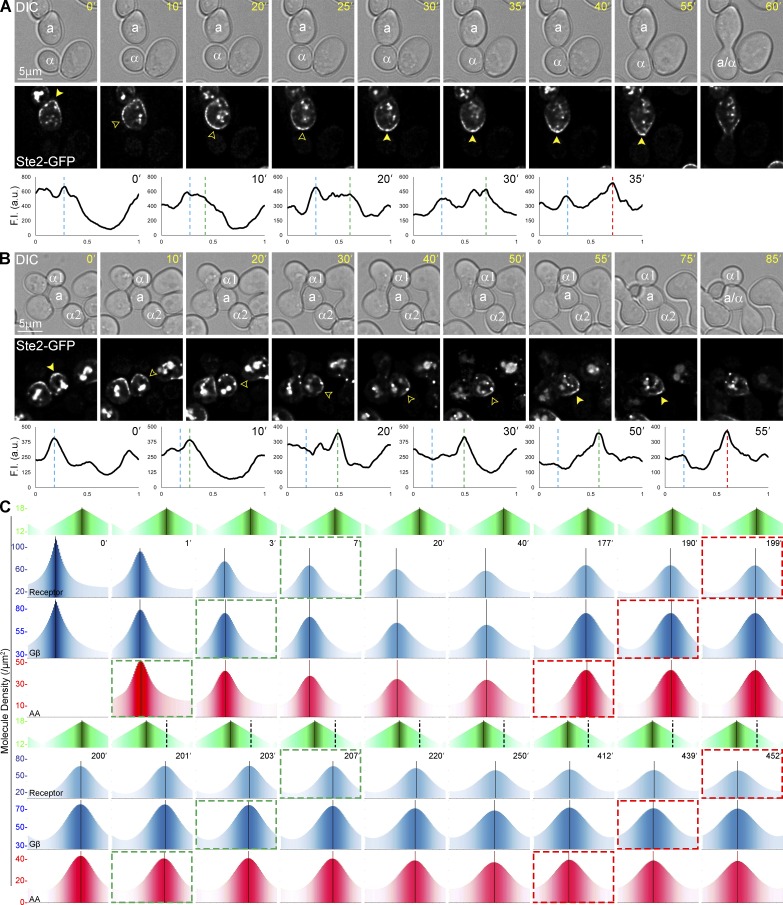
Figure 8.
Examples of reorientation and its simulation by computational model v.2. (A and B) Time-lapse images of MATa cells expressing in situ–tagged Ste2-GFP reorienting in mating mixtures. The fluorescent images show the localization of Ste2-GFP as MATa cells reorient. The filled arrowheads indicate the Ste2-GFP crescent at the initial CS (0') and after reorientation to the second CS. The closed yellow arrowheads indicate the tracking Ste2-GFP crescents. The plots show the distribution of Ste2-GFP on the PM at each time point. The dashed blue lines mark the position of the first CS; the dashed green lines mark the position of the leading peak during reorientation; the dashed red lines mark the position of the CS after reorientation. (A) Reorientation assay. MATa cells were grown to mid-log phase and treated with isotropic pheromone until they started shmooing. The treated cells were washed to remove exogenous pheromone, mixed with an equal number of MATα cells, and imaged until they formed zygotes. A representative reorienting shmoo is shown. (B) Spontaneous reorientation in vivo. An example of a MATa cell that initially oriented toward a potential partner (α1) before reorienting and mating with another (α2). (C) Outputs from the standard model challenged with a change in gradient direction. The direction of the pheromone gradient was rotated 90° at 201 min. The dashed black lines in the lower gradient panel indicate the position of the original gradient source. The dashed green boxes indicate the start of tracking and reorienting; the dashed red boxes indicate end of tracking.
Our data suggest that Gβ localizes to the DS before the receptor in mating cells, and that this depends on Far1. We hypothesize that the initial recruitment of Gβγ to the DS is integral to the assembly process because Gβγ both directs receptor delivery and inhibits its internalization. To test this in silico, we changed the initial state in our computational model from both the receptor and G protein polarized at the DS to initial states in which either the G protein is polarized to the DS while the receptor is uniformly distributed or vice versa. When the initial state is “polarized G protein/uniform receptor,” the receptor rapidly polarizes to the DS, and then the GTM tracks to the CS with kinetics identical to that of the standard simulation (Fig. 7 D). When the initial state is “polarized receptor/uniform G protein,” however, both the receptor and G protein rapidly amplify at the CS, failing to mimic the tracking behavior we observe in vivo (Fig. 7 E). These simulations support the idea that polarization of Gβγ before polarization of the receptor is essential for the assembly of the GTM. To test the importance of pheromone-induced cointernalization of the receptor and G protein in silico, we modified v.2 such that the receptors are monomeric rather than dimeric. In this version of the model, the G protein can internalize at the basal rate with the inactive monomer (I) but cannot internalize with the active monomer (A) in response to pheromone. The output from the monomeric model suggests that receptor dimers are important for gradient sensing (Fig. 7 F).
Singularity contributes to mating fidelity
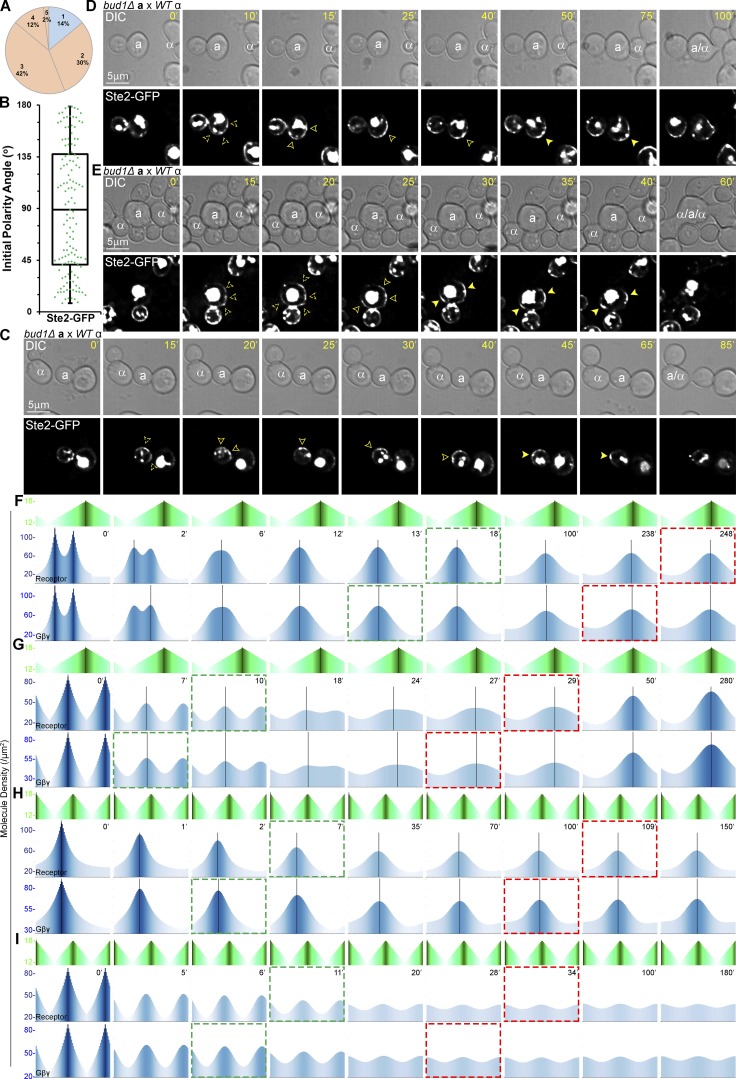
In both mother and daughter cells exposed to natural pheromone gradients, the GTM components we followed invariably polarized to the DS before relocalizing to the CS. To determine whether polarization to the DS is required to enable gradient sensing, we assayed the localization of Ste2-GFP in mating MATa cells lacking Bud1, a cortical marker essential for the establishment of the DS during vegetative growth. Bud sites arise spontaneously by symmetry breaking and are randomly positioned in bud1Δ cells (Kozubowski et al., 2008). Howell et al. (2009) showed that two or more polarity sites compete early in this process before one predominates and becomes the singular bud site. Similarly, we found that in mating mixtures, MATa bud1Δ cells often formed multiple, randomly positioned Ste2-GFP polarity patches that ultimately merged into one (Fig. 9, A–E). The behavior of the Ste2-GFP patches depended on their initial positions relative to the eventual mating partner. If on the same side of the MATa bud1Δ cell, the patches merged and then tracked to the CS (Fig. 9, C and E); if on opposite sides, they tracked independently to the CS (Fig. 9, D and E). Our computational model faithfully reproduced both of these behaviors (Fig. 9, F and G; Videos 4 and 5). The failure of the receptor to polarize directly to the CS, even in cells lacking intrinsic polarity, supports our conclusion that GTM assembly is required to enable gradient sensing and, further, demonstrates that this process can be initiated at random positions by symmetry breaking.
Figure 9.
Effect of disrupting default polarity on gradient sensing and mating. MATa bud1Δ cells expressing in situ–tagged Ste2-GFP were mixed with an equal number of MATα cells and imaged from cytokinesis to fusion. (A) Pie graph showing the proportion of cells with the indicated number of Ste2-GFP polarity patches at the first time point. n = 50 in two trials. (B) Box scatterplot showing the initial angles of Ste2-GFP polarity patches relative to the cytokinesis site (δ values in Fig. 1 E). n = 130 from the 50 cells quantified in A. Mean angle ± SEM = 89.0° ± 4.8°. (C–E) Representative images of mating MATa bud1Δ cells. The fluorescent images show the localization of Ste2-GFP as the MATa cells orient toward their mating partners. (C) A cell with two initial polarity patches. (D) A cell with three initial polarity patches. (E) A cell with four initial polarity patches that mated with two MATα cells. (F–H) Modeling simulations of bud1∆ cells. Outputs from a modified model in which there are two initial polarity sites on the same side of the cell relative to the gradient source (F); two initial polarity sites on opposite sides of the cell relative to the gradient source (G). (H and I) Modeling simulations of partner choice in WT and bud1∆ cells. (H) Outputs from the standard model challenged by two equal gradients with the DS closer to one source. (I) Outputs from a modified model in which a cell with two initial polarity sites is challenged by two equal gradients. Each polarity site is positioned closer to one of the two gradient sources, duplicating the spatial relationships used in the WT simulation shown in H.
In rare cases (∼1%), when Ste2-GFP happened to polarize in patches proximal to two potential mating partners, we observed double matings: a single MATa bud1Δ cell fused with two MATα cells (Fig. 9 E). We have never seen this in WT mating mixtures. This suggests that, although gradient sensing does not depend on the DS in particular, assembly of the GTM at a unique site contributes to mating fidelity. To further evaluate this possibility, we asked whether the number of initial polarity sites affects the ability of our v.2 computational cell to choose a single partner when challenged with competing gradient sources. Whereas our standard model (initial condition: one polarity site) decisively tracked and rapidly polarized toward the nearer of two equal pheromone sources (Fig. 9 H), the “bud1∆” version of the model (initial condition: two polarity sites) polarized weakly to both sources (Fig. 9 I and Videos 6 and 7), consistent with our experimental results.
Discussion
The discovery that Gβγ directly interacts with Far1 suggested a global mechanism for yeast gradient sensing: Gβγ serves as an internal cortical landmark that recruits Far1–Cdc24 to direct exocytosis where the external pheromone concentration is highest (Butty et al., 1998; Nern and Arkowitz, 1998, 1999). Although appealing in its simplicity, this model failed to explain how the vanishingly small initial differential of free Gβγ, reflecting the estimated 1% pheromone gradient, could accurately position the CS, especially given the strong intrinsic polarity of the DS. Recently, we described two interconnected positive feedback loops expected to amplify the spatial signal. A computational model (v.1) showed that these feedback loops could underlie a global gradient-sensing mechanism but required a fourfold higher level of Gβγ than has been reported. Moreover, two groups suggested that yeast cells use a local, rather than a global, gradient-sensing mechanism.
Lew and colleagues (Dyer et al., 2013; McClure et al., 2015) and Peter and colleagues (Hegemann et al., 2015; Hegemann and Peter, 2017) have proposed stochastic local sampling models with the following common elements. (1) Polarity is not initially established at the CS in gradient-stimulated cells. Rather, the Cdc42-Cdc24-Bem1 positive feedback loop catalyzes assembly of the PC at sites randomly positioned with respect to the gradient. Consistent with our results, the Peter group showed one example of a mating MATa cell in which Cdc24 initially polarized at the DS (Hegemann et al., 2015). (2) From PE to the onset of morphogenesis, the PC is mobile, driven stochastically along the PM by vesicle delivery (Dyer et al., 2013). (3) PC mobility is inversely related to receptor and G protein activity. The PC can move in any direction; however, its mobility decreases as it moves toward the pheromone source (Dyer et al., 2013; Hegemann et al., 2015; McClure et al., 2015; Hegemann and Peter, 2017), biasing its displacement upgradient.
The stochastic local sampling models (hereafter, biased wandering) were largely based on how PC reporters behaved in cells exposed to artificial pheromone gradients and in isotropic dose–response experiments. Although our assays of the receptor, Gβ, Far1, Sst2, Sec3, and Sla1 localization in mating cells are consistent with the core principle of these models, i.e., that polarity is established at random positions with respect to the gradient and then incrementally moved to the position of highest pheromone concentration, three of our findings conflict with their predictions. First, we observed a consistent and easily measured pause between PE and tracking. There is no reason to expect such a pause if mobility is the direct result of vesicle fusion. Second, the GTM reporters we followed in cells that formed zygotes invariably tracked directly from the DS to the CS by the shortest route possible. If the initial direction of PC movement is determined by off-center vesicle delivery (Dyer et al., 2013), circuitous paths to the CS would be common. Third, we found that the receptor and Gβ tracked from the DS to the CS at a constant rate. This contradicts the prediction of the biased wandering models that the mobility of the PC gradually decreases as it moves upgradient.
Based on our previous work (Metodiev et al., 2002; Suchkov et al., 2010; Ismael et al., 2016) and the evidence presented here, we propose a model that resolves the contradictions discussed above by replacing biased wandering with receptor-directed tracking. The central concept of the model is that a rapidly changing pattern of dimeric receptor species segregates anterograde and retrograde vesicle trafficking, effecting redistribution of the GTM toward the pheromone source, and stabilization at the CS (Fig. 10). An essential aspect of our model is that Gβγ inhibits receptor phosphorylation, thus steepening the gradient of signaling-competent receptors relative to that generated by the pheromone gradient alone. The uncoupling of GPCRs from their G proteins by phosphorylation of their C-terminal tails is paradigmatic (Tobin, 2008), although this has not yet been demonstrated in yeast. A “bare bones” computational model, which includes differential protection of the receptor by the G protein, simulates the postulated treadmill-like mechanism and demonstrates the feasibility of deterministic gradient sensing.
Figure 10.
Deterministic model of local gradient sensing. Feedback interactions between the receptor, its kinases, and its G protein steepen the gradient of signaling-competent (active-unphosphorylated) and signaling-incompetent (active-phosphorylated and inactive) receptors on the cell surface. (A) During tracking, signaling-competent receptors concentrate vesicle delivery upgradient, while signaling-incompetent receptors concentrate receptor-driven endocytosis downgradient, thereby directing the GTM toward the pheromone source. (B) During stabilization, signaling-competent receptors concentrate vesicle delivery at the center of the incipient CS, while signaling-incompetent receptors concentrate endocytosis around it, thereby directing chemotropic growth toward the mating partner. Because the GTM is positioned by the dynamic pattern of receptor species, it rapidly reflects changes in gradient profile and direction.
Differential receptor phosphorylation directs the PC upgradient and promotes stabilization at the CS
Global internalization and assembly
Global internalization of the receptor and G protein is a rapid, cell cycle–independent response to pheromone stimulation. In contrast, cells acquire the ability to decode pheromone gradients in G1 only. What purpose does global internalization of the signaling machinery serve, and why is the development of gradient-sensing competence restricted to G1?
Our observation that signaling components polarize to the DS in a strict order, after which they pause for reproducible intervals before tracking, suggested that the regulated assembly of one or more complexes is required to enable gradient sensing. Of the proteins we followed in this study, Far1 was the first to accumulate at the DS, consistent with the establishment of polarity by the Cdc42-Cdc24-Bem1 module. The subsequent polarization of Gβγ, which is dependent on the DS localization of Far1, promotes retention of newly delivered receptor by inhibiting its phosphorylation (Ismael et al., 2016). We suggest that global internalization and relocalization of the receptor and G protein to the DS facilitates gradient sensing by optimizing the relative amounts of receptor and Gβγ for tracking, as their stoichiometry changes from 4:1 in vegetative cells to ∼1:1 in the GTM. Although our computational model v.2 does not explicitly consider Far1 and other components of the PC, simulation of tracking nevertheless required initial polarization of the G protein to the DS. Given the essential role that Far1 plays in this process, it is not surprising that cells become gradient sensitive in G1, when Far1–Cdc24 is exported from the nucleus.
It is notable that Far1-GFP, a marker for the PC, pauses for ∼13 min after polarizing to the DS before beginning to redistribute upgradient. Directed vesicle delivery clearly occurs during this time, as the Ste2-GFP signal steadily increases between PE and the initiation of tracking. If PC mobility results from vesicle delivery, as proposed by the biased wandering models, we would expect the polarization of Far1 and its movement along the PM to be coincident. Why the delay? The behavior of the exocytosis marker, Sec3, may provide the answer. Early in assembly, Sec3 polarizes in a sharp peak that aligns with the peak of receptor. Just before tracking begins, the Sec3 peak shifts upgradient, and this shift precedes (or is concomitant with) that of the receptor.
What causes Sec3 to shift its position upgradient? We hypothesize that the end of assembly is marked by a maturing of the receptor/G protein distribution such that signaling-competent (active-unphosphorylated) receptors and active G proteins (Gα-GTP and free Gβγ) predominate on the upgradient side of the GTM, while signaling-incompetent (active-phosphorylated and inactive) receptors and inactive G proteins predominate downgradient (Fig. 10 A). The PC is then recruited toward the peak of signaling-competent receptor by the Gβγ–Far1 interaction, Sec3 shifts upgradient, and tracking begins. The kinetics of Sst2-GFP localization during assembly support this hypothesis. Although Sst2-GFP polarizes to the DS significantly later than the receptor, it begins tracking significantly earlier. This suggests that those species known to interact with Sst2 on the PM, the active-unphosphorylated form of the receptor and active Gα, accumulate shortly before tracking begins, and that the peaks of active receptor and G protein move upgradient ahead of the total receptor. Our computational model v.2, which allows us to simulate the dynamics of all receptor and G protein species, as well as their delivery to and removal from the PM, is corroborative: the peak of the AA dimeric form of the receptor moves upgradient first, followed by the peaks of the activated G protein subunits, and finally, the peak of total receptor.
Tracking
Our data indicate that the GTM architecture established during assembly is maintained during tracking. The peaks of Sec3 and Sla1 lead and lag the peak of total receptor (Ste2-GFP), respectively, reflecting the predominance of signaling-competent receptors and active G proteins upgradient and signaling-incompetent receptors and inactive G proteins downgradient. The separation of exocytosis and endocytosis leads to the incremental redistribution of the GTM as secretory vesicles carrying receptor and G protein are preferentially deposited near its leading edge, while receptor/G protein complexes are preferentially removed along its lagging edge (Fig. 10 A). This is a deterministic and iterative process whereby the peak of signaling-competent receptors continually tracks ahead of the peak of total receptor, recruiting the PC upgradient via the activated G protein. Output from our computational model v.2 demonstrates that redistribution of the receptor and G protein upgradient can emerge from a relatively simple system in which free Gβγ antagonizes receptor internalization and positions the center of vesicle delivery. Tracking simulations recapitulate the expected order in which the peaks of the various species redistribute: the AA dimeric receptor leads, the activated G protein subunits follow, and the total receptor is last.
That the pattern of specific receptor dimers ultimately guides the PC toward the pheromone source is supported by our analysis of receptor mutants. We have found that internalization and polarization of the receptor contribute to the accuracy of cell orientation (Ismael et al., 2016) and GFP-Gβ tracking but are not essential for gradient sensing. In contrast, receptor phosphorylation is necessary for gradient sensing: cells unable to phosphorylate the receptor cannot orient toward mating partners (Ismael et al., 2016), and in such cells, GFP-Gβ polarizes to the DS but does not track. These results indicate that receptor phosphorylation plays an essential role in gradient sensing apart from triggering receptor internalization, consistent with the idea that differential phosphorylation of the receptor steepens the signaling gradient.
Stabilization
In mating cells, the tracking receptor and Gβ crescents move at a constant rate from the DS to the CS, then appear to stop abruptly upon centering around the future shmoo site. Notably, the transition from tracking to stabilization correlates with a change in the architecture of the GTM. During tracking, Sec3 and Sla1 concentrate toward the front and back of the GTM, respectively, whereas in the stabilization phase, the Sec3 and receptor peaks align at the center of the incipient shmoo site, while Sla1 assumes a corralling distribution. This spatial rearrangement of GTM trafficking functions can be explained as follows. The tracking receptor is asymmetrically distributed and subjected to a directional gradient; during stabilization, the receptor is distributed symmetrically around the CS and subjected to a radial gradient. The patterns of dimeric receptor species are consequently different in the two phases: signaling-competent receptors track ahead of signaling-incompetent receptors (Fig. 10 A), whereas signaling-competent receptors are surrounded by signaling-incompetent receptors during stabilization (Fig. 10 B). In essence, the receptor senses decreasing pheromone concentrations beyond the CS in all directions. Feedback between the receptor and G protein then patterns the downstream trafficking functions to effect focused growth. Receptor-directed organization of the GTM not only serves to accurately position and stabilize the PC but also allows for rapid rearrangement in response to dynamic gradients. Although we have not yet visualized the putative dimeric receptor subspecies, output from our computational model v.2 is consistent with the proposed mechanisms. The computational model also demonstrates that the GTM can readily reorient in response to a change in gradient direction, as in real cells.
Default polarity and gradient tracking in other organisms
Our studies of yeast gradient sensing have yielded two insights that may be relevant to other systems. First, we concluded that Gβγ inhibition of the yeast receptor kinases synergizes with the pheromone gradient to steepen the asymmetric distribution of signaling competent (unphosphorylated) and incompetent (phosphorylated) receptors (Ismael et al., 2016). Given the many well-established examples of Gβγ/receptor–kinase interactions in higher eukaryotes, it is plausible that differential GPCR phosphorylation plays a role in the polarization of other gradient-sensing cell types (Ribas et al., 2007). Second, we found that even yeast cells lacking a DS cannot orient directly toward the gradient source under physiological conditions. Assembly of the GTM, whether at an intrinsic site or by symmetry breaking, appears to be a mandatory process that primes the cell’s response to the gradient. Similarly, the polarization of axonal and dendritic zones is an obligatory step that prepares differentiating neurons to respond to morphogens (Yogev and Shen, 2017). Like yeast, metazoan cells commonly inherit cell division–associated polarity (Sohrmann and Peter, 2003; Wedlich-Soldner and Li, 2003; Kimmel and Firtel, 2004; Krishnan and Iglesias, 2007; Millius et al., 2009; Yogev and Shen, 2017). It remains to be seen whether the use of intrinsic polarity to prime shaping by the environment is a widespread strategy employed by gradient-sensing cells. As the yeast GTM comprises highly conserved proteins that mediate processes ubiquitous in eukaryota, analogous mechanisms are likely to be discovered in other species.
Materials and methods
Molecular and microbiological techniques
Standard methods were used for microbial culture and molecular manipulation, which were performed as described previously (Sherman et al., 1986; Ausubel et al., 1994; Guthrie and Fink, 2002).
Yeast strain construction
The yeast strains used in this study are listed in Table S1. Unless noted otherwise, they were derived by transformation of strain 15Dau (MATa ade1 his2 leu2-3,-112 trp1 ura3Δ), which is congenic with strain BF264-15D (Reed et al., 1985). Strain XWY065 was generated by integrating HpaI-cut LHP1921 into strain DSY129. Strain XWY096 was generated by integrating HpaI-cut MCB003 into DSY129. Strains XWY086 and XWY097 were generated by integrating BamHI-cut XWB087 and XWB097, respectively, into strain XWY065. The DNA fragment for FAR1-GFP in situ tagging was PCR amplified from pFA6A-GFP(S65T)-kanMX6 with the oligomers 5′-GCAGCAAAGAATTCATCAGACCCTGGAAGTTCCCAACCTCAGTAAAGGAGAAGAACTTTTC-3′ and 5′-GAAAAGCAAAAGCCTCGAAATACGGGCCTCGATTCCCGAACTAGACAGCAGTATAGCGACCAG-3′, in which the underlined sequences direct recombination at the 3′-end of the FAR1 gene. Strain BBY009 was generated by GFP tagging FAR1 in situ in strain DSY129 using the PCR fragment described above. The DNA fragment for tagging SST2 with GFP in situ was PCR amplified from pFA6A-GFP with the oligomers 5′-CAAAGATGCTAGCGCTTTAATAGAAATCCAAGAAAAGTGCAGTAAAGGAGAAGAACTTTTC-3′ and 5′-GTGCAATTGTACCTGAAGATGAGTAAGACTCTCAATGAAAGACAGCAGTATAGCGACCAG-3′, in which the underlined sequences direct recombination at the 3′-end of the SST2 gene. Strain BSY008 was generated by GFP tagging SST2 in situ in strain DSY129 using the PCR fragment described above. The DNA fragment for tagging SPA2 with RFP in situ was PCR amplified from XWB087 with the oligomers 5′-GTTTGAAGGACGAAATAAATTATTTGAATTCGAAGTTGAAGATGGTTTCAAAAGGTGAAGAAG-3′ and 5′-CTTTGTCTTCCTTTTCTTTCTCCTCTAGATACTACTAACTTTCAATTCAATTCATCATTTTTTTTTTATTC-3′, in which the underlined sequences direct recombination at the 3′-end of the SPA2 gene. Strains XWY105, XWY108, XWY137, BBY015, and BSY031 were generated by RFP tagging SPA2 in situ in strains XWY096, XWY065, DSY129, BBY009, and BSY008, respectively, using the PCR fragment described above. Strains XWY109 and XWY114 were generated by integrating Cla1-cut DLB3784 and DLB3850, respectively, into strain XWY096. Strain XWY117 was generated by integrating BamHI-cut XWB043 into strain XWY096. Strain XWY147 was generated by integrating BamHI-cut XWB125 into strain XWY137. The DNA fragment for far1H7-GFP was PCR amplified from pFA6a-GFP(S65T)-kanMX6 with the oligomers 5′-TTCGCAACGCCGCATGACTCCATTGAACGCTAGCATTAAGAGTAAAGGAGAAGAACTTTTC-3′ and 5′-GAAAAGCAAAAGCCTCGAAATACGGGCCTCGATTCCCGAACTAGACAGCAGTATAGCGACCAG-3′, in which the underlined sequences direct recombination such that GFP is inserted after FAR1 codon 756. Strain BBY019 was generated by integrating the DNA fragment above into DSY129. The bud1Δ fragment was PCR amplified from YIplac211 with the oligomers 5′-ATGAGAGACTATAAATTAGTAGTATTGGGTGCTGGTGGTGCCAGCTTTTCAATTCAATTC-3′ and 5′-ACTATAGAATAGTGCAAGTGGAAGCGTTTTTCTTCTTTTTTTAGTTTTGCTGGCCGCATC-3′, in which the underlined sequences direct recombination at the 5′- and 3′-end of the BUD1 gene, respectively. Strain XWY121 was generated by replacing the BUD1 gene with the PCR fragment described above in strain XWY065. All genomic modifications were confirmed by sequencing (University of Illinois at Chicago Research Resource Center Sequencing Core).
Plasmid construction
The plasmids used in this study are listed in Table S2. YIplac211-FAR11,549–2,268 was constructed by PCR amplifying FAR1 bases 1,549–2,268 from DSY129 genomic DNA with the oligomers 5′-ATGCAAGCTTTGAGGACGTACTACTTTGTAGTTTG-3′ and 5′-ATGCGGTACCTTAGCTAGCGTTCAATGGAGTCATG-3′. The HindIII- and KpnI-digested PCR product was inserted into YIplac211. YIplac211-SLA12,491–3,732-RFP was constructed by PCR amplifying SLA1 bases 2,491–3,732 from DSY129 genomic DNA with the oligomers 5′-ATCGAAGCTTACTGGCGGCACCACTGTTCC-3′ and 5′-ATCGGGTACCGAATCCAAACGGATTTGATGCAGTAGCATTG-3′, and RFP from pRS406/RFP-BUD1 (DSB405) with the oligomers 5′-ATCGGGTACCATGGTTTCAAAAGGTGAAGAAGATAATATG-3′ and 5′-ATCAGAATTCTTATTTATATAATTCATCCATACCACCAGTTG-3′. The HindIII- and KpnI-digested SLA1 PCR product and the KpnI- and EcoRI-digested RFP PCR product were inserted into YIplac211. YIplac211-SEC33,320–4,008-RFP was constructed by PCR amplifying SEC3 bases 3,320–4,008 from DSY129 genomic DNA with the oligomers 5′-ATCGAAGCTTAAGATACCGATGAAGGCTACGAG-3′ and 5′-ATCGGGTACCGGCATTCTTGTATTCCTCGAACGC-3′, and RFP as described above. The HindIII- and KpnI-digested SEC3 PCR product and the KpnI- and EcoRI-digested RFP PCR product were inserted into YIplac211. YIplac128-Pcdc3-GFP-CDC31–194,196–400 was constructed by PCR amplifying the CDC3 promoter with the oligos 5′-ATCAGAATTCATCATCCAGAATCTGCAAACGC-3′ and 5′-ATCAGGTACCGGCAACGTATTATAGGCCGATG-3′, GFP with the oligos 5′-ATCAGGTACCATGAGTAAAGGAGAAGAACTTTTCAC-3′ and 5′-ATCAGTCGACTTTGTATAGTTC-ATCCATGCCATG-3′, CDC3 bases 1–194 with the oligos 5′-ATCAGTCGACATGAGTTTAA-AGGAGGAACAAGTGTCC-3′ and 5′-ATCGGGATCCACCTTTACATCGGATTCAGC-3′, and CDC3 bases 196–400 with the oligos 5′-ATCGGGATCCCGGCCTCGGTATGGGCATC-3′ and 5′-CAGTAAGCTTAAATACCATCAGGGCCGACAC-3′. The EcoRI- and KpnI-digested Pcdc3 PCR product, the KpnI- and SalI-digested GFP PCR product, the SalI- and BamHI-digested CDC31–194 PRC product, and the BamHI- and HindIII-digested CDC3196–400 PCR product were inserted into YIplac128. YIplac128-PSTE4-GFP-STE41–547 was constructed by PCR amplifying the STE4 promoter and GFP fused in-frame to the N-terminal half of STE4 from PRS316-PSTE4-GFP-STE41–1,272 (DSB156) with the oligomers 5′-GGGGAGCTCAAATCATACATATTTATCGCACCATTGTCACTTCCTG-3′ and 5′-GTGGAATTCAATGTTTCAGGAAGAGATACTGCGTAAAAAAAAGAC-3′. The SalI- and EcoRI-digested PCR product was inserted into YIplac211. All constructions were confirmed by sequencing.
Time-lapse imaging of mating mixtures
WT MATα cells and experimental MATa cells were grown to mid-log phase in synthetic 2% dextrose medium, mixed 1:1, and spread at a density of 14,000 cells/mm2 on agarose pads made from synthetic dextrose medium. Mating mixtures were maintained at 30°C using a DeltaVision environment control chamber. Differential interference contrast (DIC) and fluorescent images were acquired from 12 fields at 2- or 5-min intervals using a DeltaVision Elite Deconvolution Microscope (GE Healthcare Biosciences) with a 60× oil-immersion objective and a Front Illuminated sCMOS camera. To minimize phototoxicity, 5 z-sections 0.5 µm apart were acquired around the center slice of each cell at each time point. Identical light-emitting diode intensities and exposure times were used to image cells expressing the GFP-tagged reporters (10% maximum intensity at 461–489 nm for 150 ms) and the RFP-tagged reporters (10% maximum intensity at 529–556 nm for 300 ms). Representative fluorescent images were deconvolved using Huygens Essential software (Scientific Volume Imaging) in standard mode except as noted.
Image analysis
With the exception of Fig. 3, mating MATa cells were randomly chosen for analysis by scanning the time-lapse images for cells that completed a division cycle and formed a zygote. For the analysis of assembly shown in Fig. 3, MATa cells were randomly chosen by scanning the time-lapse images for cells that completed a division cycle and initiated tracking. A maximum of three cells were selected per field. PM fluorescent signal intensities were quantified by tracing the circumference of the center-slice DIC images using the segmented line tool of ImageJ (National Institutes of Health); pixel values were determined from the corresponding raw fluorescent images after subtracting the background. The ImageJ plots were used to determine initial localization of the reporters to the DS (PE) and the initiation of their redistribution (tracking) to the CS: PE was defined as the de novo appearance of a peak at the DS or as the shift of an existing peak from the bud neck to the DS, and tracking was defined as the appearance of a distinct peak or shoulder upgradient from the DS. Because the ImageJ plots are based on the raw data, they do not always mirror the corresponding display images. Initial PE positions were measured using the ImageJ angle tool as shown in Fig. 1 E. The normalized and averaged plots of reporter signals on the PM during tracking and stabilization (Fig. 2 B) were generated as follows. For each mating cell, the distance tracked (DS → CS) in pixels was consolidated to 20 evenly spaced points, with the signal value for each point determined by the original curve. Equal-sized PM intervals on either side of the tracking region were treated in the same way, producing a 60-point plot that displays the full DS and CS peaks normalized for tracking distance. The average plots show the mean value ± SEM at each of the 60 points for 20 cells normalized in this way (see Fig. 2 A). For the two-reporter plots (Fig. 6, C–F and H), cell circumferences were normalized to 100 points. After normalization, the mean distribution of the receptor was generated by aligning the leading Ste2-GFP peaks with each other during tracking and with the center of the fusion site at the prezygote stage. To measure the Ste2-GFP and GFP-Gβ signal increase during the pause, PM fluorescent signal intensities for the first five 2-min time points from PE were quantified by tracing the center-slice DS region using the segmented line tool of ImageJ; pixel values were determined from the raw fluorescent images after subtracting the background. The mean signals from 25 cells were averaged at each time point and plotted against time (Fig. 3 F). The pause time for GFP-Gβ at the bud neck (Fig. 4 F) was scored visually; it reflects the number of 5-min time points that the reporter was detectable at the mother–daughter neck.
Computational modeling
Reaction network
To model interactions among molecular species of the yeast GTM, we created a biochemical reaction network that includes only the most basic and best-characterized components. The network comprises receptor–pheromone interaction, the G protein cycle, and Yck-dependent internalization of the receptor and G protein (Fig. 7 B and Table S3). To allow for pheromone-induced internalization of the G protein with the receptor (Suchkov et al., 2010) without violating the GPCR–G protein trafficking paradigm, pheromone receptors were modeled as dimers. The existence of both Ste2 homo- and hetero-oligomers has been well established (Overton and Blumer, 2000; Sridharan et al., 2016; Cevheroğlu et al., 2017). Pheromone (L, ligand)–receptor association and dissociation generate a dynamic population of inactive, partially active, and fully active dimers (II, IA, and AA, respectively). Any receptor dimer with an inactive monomer (II or IA) can associate with a heterotrimeric G protein (G) and cause its cointernalization. Any receptor dimer with an active monomer (IA or AA) can activate both predocked and uncoupled heterotrimeric G proteins; the latter are activated by collision (Tolkovsky and Levitzki, 1978; Bünemann et al., 2001; Brinkerhoff et al., 2008; Vilardaga et al., 2009; van Hemert et al., 2010; Xu et al., 2010). Activation of the heterotrimeric G protein results in the dissociation of Gα-GTP (Ga) from Gβγ (Gbg). Upon hydrolysis of its bound nucleotide, Gα-GTP returns to its inactive conformation, Gα-GDP (Gd), and reassociates with Gβγ to reform the heterotrimer. Pheromone induces the phosphorylation of Gβ by active Fus3 (Fus3A), and the generation of phosphorylated Gβγ (GbgP) is increased by the recruitment of active Fus3 to the cell cortex by active Gα. Interaction of Yck1/2 (Yck) with partially and fully active receptor dimers (IA and AA) causes immediate loss of these receptor species from the PM (internalization). The basal rate of inactive-receptor (II) internalization results from Yck interaction with receptor monomers that spontaneously activate (Hicke et al., 1998). Phosphorylated Gβγ binds directly to Yck (Yck-GbgP) and thereby inhibits the Yck-dependent internalization of receptor dimers and heterotrimeric G proteins. For the monomeric-receptor version of the model (Fig. 7 F), the initial receptor abundance was set to equal the number of total monomers in the standard dimeric-receptor model, and the heterotrimeric G protein was allowed to internalize at the basal rate. All other initial conditions were the same (Table S4).
Spatial model
To study the spatiotemporal dynamics of the GTM, we modeled the yeast PM as a 2D ring with radius r, discretized uniformly into n wedges (Fig. S3). The center of each wedge is used to represent its position. The surface distance d between neighboring wedges is given by Eq. 1 in Table S5. Because the pheromone gradient is aligned along the x axis, and the pheromone concentration is assumed to change linearly with x, only the x coordinate of the ith wedge is needed to determine the local pheromone concentration [L]i (Eq. 2 in Table S5). All proteins are assumed to diffuse laterally, as only the PM is modeled in this analysis. If the effect of surface curvature is ignored, the diffusion of each molecular species can be obtained from Eq. 3 (Table S5).
Reaction–diffusion system
The spatiotemporal dynamics of this reaction–diffusion system were simulated using the partial differential equations in Table S6. For simplicity, i subscripts were included only when necessary. Because the total amounts of receptor and Gβ did not change significantly over the course of our time-lapse mating experiments, we modeled the total synthetic rates of receptor dimers and heterotrimeric G proteins (krs_tot and kgs_tot, respectively) as functions of their total amounts (Eqs. 20–27 in Table S7). When the total amounts of receptor and G protein are low, the synthetic rate of the whole cell (krs_tot and kgs_tot) is increased up to two times the basal rate (krs_bas and kgs_bas); when the total amounts are high, synthesis rates are decreased. When the total levels of receptor and G protein reach two times their basal levels, their synthesis stops (i.e., krs_tot = 0 and kgs_tot = 0). In addition, we found that secretion to the PM is not uniform. Based on the well-established interactions between Gβγ and proteins that bind Sec3, Cdc42 (via Far1–Cdc24), and Rho1 (Butty et al., 1998; Nern and Arkowitz, 1998, 1999; Guo et al., 2001; Zhang et al., 2001; Bar et al., 2003; Pleskot et al., 2015), and on the observation that Gβ is essential to stabilize the position of the PC (McClure et al., 2015), we modeled the local amount of Gβγ as a positive determinant of the local delivery rate, krs(i) and kgs(i) (Eqs. 28 and 29 in Table S7). In the initial state of the standard model, inactive receptor dimers and heterotrimeric G proteins are polarized to the DS. The total amount of inactive receptor dimers on the membrane is 5,000 (Yi et al., 2003). Based on the relative PM signal intensity of Ste2-GFP and GFP-Gβ at the DS of mating cells, the ratio between the total amounts of inactive receptor dimers and heterotrimeric G proteins is ∼1. The distributions of the receptor dimers and heterotrimeric G proteins were fitted to the experimentally measured distributions (Fig. S4).
Variables and parameters
A description of variables and values of parameters can be found in Table S8. The model simulates tracking and polarization across a range of values for key unpublished parameters (Table S9).
Online supplemental material
Fig. S1 shows the translocation of Sst2-GFP from the cytoplasm to the PM in response to isotropic pheromone treatment. Fig. S2 shows the tracking rates of Ste2-GFP and GFP-Gβ in mating cells. Fig. S3 shows the spatial parameters used in the computational model. Fig. S4 shows the initial distributions of inactive receptor dimers and heterotrimeric G proteins used in the standard computational model compared with the experimentally determined distributions of the receptor and Gβ. Fig. S5 shows outputs of the key parameters that underlie tracking in the standard model. Table S1 lists the yeast strains used in this study. Table S2 lists the plasmids used in this study. Table S3 lists the reaction formulae used in the standard computational model. Table S4 lists the reaction formulae used in the monomeric-receptor variation of the computational model. Table S5 lists the equations corresponding to Fig. S3. Table S6 lists the reaction–diffusion equations used in the computational model. Table S7 lists the equations that describe the local delivery rates of receptor dimers and heterotrimeric G proteins in the computational model. Table S8 lists the variables and parameters used in the computational model. Table S9 shows the sensitivity of model performance to the most critical unpublished parameters. Video 1 is an animation of receptor, Gβγ, AA, and i-IA⋅G outputs from the standard computational model. Video 2 is an animation of receptor, active Gα, and inactive Gα outputs from the standard computational model. Video 3 is an animation of representative outputs from the reorientation simulation. Video 4 is an animation of representative outputs from the modified computational model with two initial polarity sites on the same side of the cell relative to the gradient source. Video 5 is an animation of representative outputs from the modified computational model with two initial polarity sites on opposite sides of the cell relative to the gradient source. Video 6 is an animation of representative outputs from the standard computational model challenged by two equal gradients. Video 7 is an animation of representative outputs from a modified computational model with two initial polarity sites challenged by two equal gradient sources.
Supplementary Material
Acknowledgments
We thank Rob Arkowitz, Holly Stratton, Madhu Sukumar, Ankur Saxena, and members of the Stone laboratory for helpful discussions and critical reading of the manuscript.
This work was supported by National Science Foundation grants 1415589 and 1818067 (D.E. Stone) and National Institutes of Health grants R35GM127084, R01CA204962, and R21AI126308 (J. Liang).
The authors declare no competing financial interests.
Author contributions: X. Wang and D.E. Stone conceived the project, designed the experiments, analyzed the data, and wrote the manuscript. B.T. Banh and B.-M. Statler constructed the strains and performed the Far1-GFP and Sst2-GFP experiments, respectively. X. Wang performed all of the other experiments, developed the data analysis methods, and made all of the figures. Under the supervision of J. Liang and D.E. Stone, W. Tian created the computational model, generated the modeling output, and performed the in silico experiments.
References
- Apanovitch D.M., Slep K.C., Sigler P.B., and Dohlman H.G.. 1998. Sst2 is a GTPase-activating protein for Gpa1: purification and characterization of a cognate RGS-Galpha protein pair in yeast. Biochemistry. 37:4815–4822. 10.1021/bi9729965 [DOI] [PubMed] [Google Scholar]
- Arkowitz R.A. 2009. Chemical gradients and chemotropism in yeast. Cold Spring Harb. Perspect. Biol. 1:a001958 10.1101/cshperspect.a001958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., and Struhl K.. 1994. Current Protocols in Molecular Biology. John Wiley and Sons, Hoboken, NJ. [Google Scholar]
- Ayscough K.R., and Drubin D.G.. 1998. A role for the yeast actin cytoskeleton in pheromone receptor clustering and signalling. Curr. Biol. 8:927–930. 10.1016/S0960-9822(07)00374-0 [DOI] [PubMed] [Google Scholar]
- Ballon D.R., Flanary P.L., Gladue D.P., Konopka J.B., Dohlman H.G., and Thorner J.. 2006. DEP-domain-mediated regulation of GPCR signaling responses. Cell. 126:1079–1093. 10.1016/j.cell.2006.07.030 [DOI] [PubMed] [Google Scholar]
- Bar E.E., Ellicott A.T., and Stone D.E.. 2003. Gbetagamma recruits Rho1 to the site of polarized growth during mating in budding yeast. J. Biol. Chem. 278:21798–21804. 10.1074/jbc.M212636200 [DOI] [PubMed] [Google Scholar]
- Basile J.R., Barac A., Zhu T., Guan K.L., and Gutkind J.S.. 2004. Class IV semaphorins promote angiogenesis by stimulating Rho-initiated pathways through plexin-B. Cancer Res. 64:5212–5224. 10.1158/0008-5472.CAN-04-0126 [DOI] [PubMed] [Google Scholar]
- Bendezú F.O., and Martin S.G.. 2013. Cdc42 explores the cell periphery for mate selection in fission yeast. Curr. Biol. 23:42–47. 10.1016/j.cub.2012.10.042 [DOI] [PubMed] [Google Scholar]
- Blondel M., Alepuz P.M., Huang L.S., Shaham S., Ammerer G., and Peter M.. 1999. Nuclear export of Far1p in response to pheromones requires the export receptor Msn5p/Ste21p. Genes Dev. 13:2284–2300. 10.1101/gad.13.17.2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkerhoff C.J., Traynor J.R., and Linderman J.J.. 2008. Collision coupling, crosstalk, and compartmentalization in G-protein coupled receptor systems: can a single model explain disparate results? J. Theor. Biol. 255:278–286. 10.1016/j.jtbi.2008.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bünemann M., Bücheler M.M., Philipp M., Lohse M.J., and Hein L.. 2001. Activation and deactivation kinetics of alpha 2A- and alpha 2C-adrenergic receptor-activated G protein-activated inwardly rectifying K+ channel currents. J. Biol. Chem. 276:47512–47517. 10.1074/jbc.M108652200 [DOI] [PubMed] [Google Scholar]
- Butty A.C., Pryciak P.M., Huang L.S., Herskowitz I., and Peter M.. 1998. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science. 282:1511–1516. 10.1126/science.282.5393.1511 [DOI] [PubMed] [Google Scholar]
- Cevheroğlu O., Kumaş G., Hauser M., Becker J.M., and Son C.D.. 2017. The yeast Ste2p G protein-coupled receptor dimerizes on the cell plasma membrane. Biochim Biophys Acta Biomembr. 1859:698–711. 10.1016/j.bbamem.2017.01.008 [DOI] [PubMed] [Google Scholar]
- Daniels K.J., Srikantha T., Lockhart S.R., Pujol C., and Soll D.R.. 2006. Opaque cells signal white cells to form biofilms in Candida albicans. EMBO J. 25:2240–2252. 10.1038/sj.emboj.7601099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit G., Kelley J.B., Houser J.R., Elston T.C., and Dohlman H.G.. 2014. Cellular noise suppression by the regulator of G protein signaling Sst2. Mol. Cell. 55:85–96. 10.1016/j.molcel.2014.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J., and Barral Y.. 2004. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 305:393–396. 10.1126/science.1099892 [DOI] [PubMed] [Google Scholar]
- Dyer J.M., Savage N.S., Jin M., Zyla T.R., Elston T.C., and Lew D.J.. 2013. Tracking shallow chemical gradients by actin-driven wandering of the polarization site. Curr. Biol. 23:32–41. 10.1016/j.cub.2012.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English D., Garcia J.G., and Brindley D.N.. 2001. Platelet-released phospholipids link haemostasis and angiogenesis. Cardiovasc. Res. 49:588–599. 10.1016/S0008-6363(00)00230-3 [DOI] [PubMed] [Google Scholar]
- Finger F.P., Hughes T.E., and Novick P.. 1998. Sec3p is a spatial landmark for polarized secretion in budding yeast. Cell. 92:559–571. 10.1016/S0092-8674(00)80948-4 [DOI] [PubMed] [Google Scholar]
- Gehret A.U., Connelly S.M., and Dumont M.E.. 2012. Functional and physical interactions among Saccharomyces cerevisiae α-factor receptors. Eukaryot. Cell. 11:1276–1288. 10.1128/EC.00172-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode B.L., Eskin J.A., and Wendland B.. 2015. Actin and endocytosis in budding yeast. Genetics. 199:315–358. 10.1534/genetics.112.145540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goryachev A.B., and Pokhilko A.V.. 2008. Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Lett. 582:1437–1443. 10.1016/j.febslet.2008.03.029 [DOI] [PubMed] [Google Scholar]
- Guo W., Tamanoi F., and Novick P.. 2001. Spatial regulation of the exocyst complex by Rho1 GTPase. Nat. Cell Biol. 3:353–360. 10.1038/35070029 [DOI] [PubMed] [Google Scholar]
- Guthrie C., and Fink G.R.. 2002. Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego, CA; 325–456. [Google Scholar]
- Hegemann B., and Peter M.. 2017. Local sampling paints a global picture: Local concentration measurements sense direction in complex chemical gradients. BioEssays. 39:1600134 10.1002/bies.201600134 [DOI] [PubMed] [Google Scholar]
- Hegemann B., Unger M., Lee S.S., Stoffel-Studer I., van den Heuvel J., Pelet S., Koeppl H., and Peter M.. 2015. A Cellular System for Spatial Signal Decoding in Chemical Gradients. Dev. Cell. 35:458–470. 10.1016/j.devcel.2015.10.013 [DOI] [PubMed] [Google Scholar]
- Hicke L., Zanolari B., and Riezman H.. 1998. Cytoplasmic tail phosphorylation of the alpha-factor receptor is required for its ubiquitination and internalization. J. Cell Biol. 141:349–358. 10.1083/jcb.141.2.349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K., and Nishiyama M.. 2010. From guidance signals to movement: signaling molecules governing growth cone turning. Neuroscientist. 16:65–78. 10.1177/1073858409340702 [DOI] [PubMed] [Google Scholar]
- Howard J.P., Hutton J.L., Olson J.M., and Payne G.S.. 2002. Sla1p serves as the targeting signal recognition factor for NPFX(1,2)D-mediated endocytosis. J. Cell Biol. 157:315–326. 10.1083/jcb.200110027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell A.S., Savage N.S., Johnson S.A., Bose I., Wagner A.W., Zyla T.R., Nijhout H.F., Reed M.C., Goryachev A.B., and Lew D.J.. 2009. Singularity in polarization: rewiring yeast cells to make two buds. Cell. 139:731–743. 10.1016/j.cell.2009.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima M., Huang Y.E., and Devreotes P.. 2002. Temporal and spatial regulation of chemotaxis. Dev. Cell. 3:469–478. 10.1016/S1534-5807(02)00292-7 [DOI] [PubMed] [Google Scholar]
- Ismael A., and Stone D.E.. 2017. Yeast chemotropism: A paradigm shift in chemical gradient sensing. Cell. Logist. 7:e1314237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismael A., Tian W., Waszczak N., Wang X., Cao Y., Suchkov D., Bar E., Metodiev M.V., Liang J., Arkowitz R.A., and Stone D.E.. 2016. Gβ promotes pheromone receptor polarization and yeast chemotropism by inhibiting receptor phosphorylation. Sci. Signal. 9:ra38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C.L., and Hartwell L.H.. 1990. Courtship in S. cerevisiae: both cell types choose mating partners by responding to the strongest pheromone signal. Cell. 63:1039–1051. 10.1016/0092-8674(90)90507-B [DOI] [PubMed] [Google Scholar]
- Jose M., Tollis S., Nair D., Sibarita J.B., and McCusker D.. 2013. Robust polarity establishment occurs via an endocytosis-based cortical corralling mechanism. J. Cell Biol. 200:407–418. 10.1083/jcb.201206081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P.J., Béven L., Hariharan S., and Park H.O.. 2010. The Rsr1/Bud1 GTPase interacts with itself and the Cdc42 GTPase during bud-site selection and polarity establishment in budding yeast. Mol. Biol. Cell. 21:3007–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Dong J., and Lord E.M.. 2004. Pollen tube guidance: the role of adhesion and chemotropic molecules. Curr. Top. Dev. Biol. 61:61–79. 10.1016/S0070-2153(04)61003-9 [DOI] [PubMed] [Google Scholar]
- Kimmel A.R., and Firtel R.A.. 2004. Breaking symmetries: regulation of Dictyostelium development through chemoattractant and morphogen signal-response. Curr. Opin. Genet. Dev. 14:540–549. 10.1016/j.gde.2004.08.001 [DOI] [PubMed] [Google Scholar]
- Kozubowski L., Saito K., Johnson J.M., Howell A.S., Zyla T.R., and Lew D.J.. 2008. Symmetry-breaking polarization driven by a Cdc42p GEF-PAK complex. Curr. Biol. 18:1719–1726. 10.1016/j.cub.2008.09.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan J., and Iglesias P.A.. 2007. Receptor-mediated and intrinsic polarization and their interaction in chemotaxing cells. Biophys. J. 92:816–830. 10.1529/biophysj.106.087353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhani V., and Elston T.C.. 2017. Testing the limits of gradient sensing. PLOS Comput. Biol. 13:e1005386 10.1371/journal.pcbi.1005386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure A.W., Minakova M., Dyer J.M., Zyla T.R., Elston T.C., and Lew D.J.. 2015. Role of Polarized G Protein Signaling in Tracking Pheromone Gradients. Dev. Cell. 35:471–482. 10.1016/j.devcel.2015.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metodiev M.V., Matheos D., Rose M.D., and Stone D.E.. 2002. Regulation of MAPK function by direct interaction with the mating-specific Galpha in yeast. Science. 296:1483–1486. 10.1126/science.1070540 [DOI] [PubMed] [Google Scholar]
- Millius A., Dandekar S.N., Houk A.R., and Weiner O.D.. 2009. Neutrophils establish rapid and robust WAVE complex polarity in an actin-dependent fashion. Curr. Biol. 19:253–259. 10.1016/j.cub.2008.12.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T.I., Chou C.S., Nie Q., Jeon N.L., and Yi T.M.. 2008. Robust spatial sensing of mating pheromone gradients by yeast cells. PLoS One. 3:e3865 10.1371/journal.pone.0003865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Chápuli R., Quesada A.R., and Angel Medina M.. 2004. Angiogenesis and signal transduction in endothelial cells. Cell. Mol. Life Sci. 61:2224–2243. 10.1007/s00018-004-4070-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A., and Arkowitz R.A.. 1998. A GTP-exchange factor required for cell orientation. Nature. 391:195–198. [DOI] [PubMed] [Google Scholar]
- Nern A., and Arkowitz R.A.. 1999. A Cdc24p-Far1p-Gbetagamma protein complex required for yeast orientation during mating. J. Cell Biol. 144:1187–1202. 10.1083/jcb.144.6.1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A., and Arkowitz R.A.. 2000. Nucleocytoplasmic shuttling of the Cdc42p exchange factor Cdc24p. J. Cell Biol. 148:1115–1122. 10.1083/jcb.148.6.1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overton M.C., and Blumer K.J.. 2000. G-protein-coupled receptors function as oligomers in vivo. Curr. Biol. 10:341–344. [DOI] [PubMed] [Google Scholar]
- Palanivelu R., and Preuss D.. 2000. Pollen tube targeting and axon guidance: parallels in tip growth mechanisms. Trends Cell Biol. 10:517–524. [DOI] [PubMed] [Google Scholar]
- Paliwal S., Iglesias P.A., Campbell K., Hilioti Z., Groisman A., and Levchenko A.. 2007. MAPK-mediated bimodal gene expression and adaptive gradient sensing in yeast. Nature. 446:46–51. [DOI] [PubMed] [Google Scholar]
- Park H.O., Bi E., Pringle J.R., and Herskowitz I.. 1997. Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc. Natl. Acad. Sci. USA. 94:4463–4468. 10.1073/pnas.94.9.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleskot R., Cwiklik L., Jungwirth P., Žárský V., and Potocký M.. 2015. Membrane targeting of the yeast exocyst complex. Biochim. Biophys. Acta. 1848:1481–1489. 10.1016/j.bbamem.2015.03.026 [DOI] [PubMed] [Google Scholar]
- Reed S.I., Hadwiger J.A., and Lörincz A.T.. 1985. Protein kinase activity associated with the product of the yeast cell division cycle gene CDC28. Proc. Natl. Acad. Sci. USA. 82:4055–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas C., Penela P., Murga C., Salcedo A., García-Hoz C., Jurado-Pueyo M., Aymerich I., and Mayor F. Jr. 2007. The G protein-coupled receptor kinase (GRK) interactome: role of GRKs in GPCR regulation and signaling. Biochim. Biophys. Acta. 1768:913–922. 10.1016/j.bbamem.2006.09.019 [DOI] [PubMed] [Google Scholar]
- Segall J.E. 1993. Polarization of yeast cells in spatial gradients of alpha mating factor. Proc. Natl. Acad. Sci. USA. 90:8332–8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink G.R., and Hicks J.B., editors. 1986. Laboratory Course Manual For Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Shimada Y., Gulli M.P., and Peter M.. 2000. Nuclear sequestration of the exchange factor Cdc24 by Far1 regulates cell polarity during yeast mating. Nat. Cell Biol. 2:117–124. 10.1038/35000073 [DOI] [PubMed] [Google Scholar]
- Snetselaar K.M., Bolker M., and Kahmann R.. 1996. Ustilago maydis Mating Hyphae Orient Their Growth toward Pheromone Sources. Fungal Genet. Biol. 20:299–312. 10.1006/fgbi.1996.0044 [DOI] [PubMed] [Google Scholar]
- Sohrmann M., and Peter M.. 2003. Polarizing without a c(l)ue. Trends Cell Biol. 13:526–533. 10.1016/j.tcb.2003.08.006 [DOI] [PubMed] [Google Scholar]
- Sridharan R., Connelly S.M., Naider F., and Dumont M.E.. 2016. Variable Dependence of Signaling Output on Agonist Occupancy of Ste2p, a G Protein-coupled Receptor in Yeast. J. Biol. Chem. 291:24261–24279. 10.1074/jbc.M116.733006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchkov D.V., DeFlorio R., Draper E., Ismael A., Sukumar M., Arkowitz R., and Stone D.E.. 2010. Polarization of the yeast pheromone receptor requires its internalization but not actin-dependent secretion. Mol. Biol. Cell. 21:1737–1752. 10.1091/mbc.e09-08-0706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W., Cao Y., Ismael A., Stone D., and Liang J.. 2014. Roles of regulated internalization in the polarization of cell surface receptors. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2014:1166–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin A.B. 2008. G-protein-coupled receptor phosphorylation: where, when and by whom. Br. J. Pharmacol. 153(Suppl 1):S167–S176. 10.1038/sj.bjp.0707662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojima T., Hines J.H., Henley J.R., and Kamiguchi H.. 2011. Second messengers and membrane trafficking direct and organize growth cone steering. Nat. Rev. Neurosci. 12:191–203. 10.1038/nrn2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolkovsky A.M., and Levitzki A.. 1978. Mode of coupling between the beta-adrenergic receptor and adenylate cyclase in turkey erythrocytes. Biochemistry. 17:3795–3810. 10.1021/bi00611a020 [DOI] [PubMed] [Google Scholar]
- Vallier L.G., Segall J.E., and Snyder M.. 2002. The alpha-factor receptor C-terminus is important for mating projection formation and orientation in Saccharomyces cerevisiae. Cell Motil. Cytoskeleton. 53:251–266. 10.1002/cm.10073 [DOI] [PubMed] [Google Scholar]
- Valtz N., Peter M., and Herskowitz I.. 1995. FAR1 is required for oriented polarization of yeast cells in response to mating pheromones. J. Cell Biol. 131:863–873. 10.1083/jcb.131.4.863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hemert F., Lazova M.D., Snaar-Jagaska B.E., and Schmidt T.. 2010. Mobility of G proteins is heterogeneous and polarized during chemotaxis. J. Cell Sci. 123:2922–2930. 10.1242/jcs.063990 [DOI] [PubMed] [Google Scholar]
- Vilardaga J.P., Bünemann M., Feinstein T.N., Lambert N., Nikolaev V.O., Engelhardt S., Lohse M.J., and Hoffmann C.. 2009. GPCR and G proteins: drug efficacy and activation in live cells. Mol. Endocrinol. 23:590–599. 10.1210/me.2008-0204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Soldner R., and Li R.. 2003. Spontaneous cell polarization: undermining determinism. Nat. Cell Biol. 5:267–270. 10.1038/ncb0403-267 [DOI] [PubMed] [Google Scholar]
- Xu X., Meckel T., Brzostowski J.A., Yan J., Meier-Schellersheim M., and Jin T.. 2010. Coupling mechanism of a GPCR and a heterotrimeric G protein during chemoattractant gradient sensing in Dictyostelium. Sci. Signal. 3:ra71 10.1126/scisignal.2000980 [DOI] [PubMed] [Google Scholar]
- Yi T.M., Kitano H., and Simon M.I.. 2003. A quantitative characterization of the yeast heterotrimeric G protein cycle. Proc. Natl. Acad. Sci. USA. 100:10764–10769. 10.1073/pnas.1834247100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev S., and Shen K.. 2017. Establishing Neuronal Polarity with Environmental and Intrinsic Mechanisms. Neuron. 96:638–650. 10.1016/j.neuron.2017.10.021 [DOI] [PubMed] [Google Scholar]
- Zhang X., Bi E., Novick P., Du L., Kozminski K.G., Lipschutz J.H., and Guo W.. 2001. Cdc42 interacts with the exocyst and regulates polarized secretion. J. Biol. Chem. 276:46745–46750. 10.1074/jbc.M107464200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.