Su et al. show that seipin negatively regulates the production of sphingoid intermediates by binding the enzymes serine palmitoyltransferase and fatty acid elongase at discrete regions of the ER in close vicinity to lipid droplets, thereby mediating the synthesis of two major building blocks for sphingolipids.
Abstract
Seipin is known for its critical role in controlling lipid droplet (LD) assembly at the LD-forming subdomain of the endoplasmic reticulum (ER). Here, we identified a new function of seipin as a negative regulator for sphingolipid production. We show that yeast cells lacking seipin displayed altered sensitivity to sphingolipid inhibitors, accumulated sphingoid precursors and intermediates, and increased serine palmitoyltransferase (SPT) and fatty acid (FA) elongase activities. Seipin associated with SPT and FA elongase, and the interaction was reduced by inhibitors for sphingolipid synthesis in a concentration-dependent manner. We further show that the interactions of seipin with SPT and FA elongase occurred at ER–LD contacts and were likely regulated differentially. Further evidence indicated that LD biogenesis was intact when SPT activity was blocked, whereas excess sphingoid intermediates may affect LD morphology. Expression of human seipin rescued the altered sphingolipids in yeast seipin mutants, suggesting that the negative regulation of sphingolipid synthesis by seipin is likely an evolutionarily conserved process.
Introduction
Eukaryotic cells master the cellular organelle lipid droplets (LDs) to manage fat and cellular fatty acids (FAs), thereby preventing lipotoxicity. Formation of this organelle is thought to involve a coordinated synthesis of neutral lipids and phospholipids in the ER toward budding of nascent LDs from the ER outer leaflet, which subsequently grow to enlarge their size and become mature LDs (Walther et al., 2017). This process requires the ER resident protein seipin that localizes at the neck of LDs, an ER subdomain where LD biogenesis occurs. The nature of this subdomain and the molecular function of seipin are core to the field of LD biology yet only partially understood.
SEIPIN, also known as BSCL2 (Berardinelli-Seip congenital lipodystrophy type 2) is the gene linked to the most severe form of human congenital generalized lipodystrophy (Magré et al., 2001). Its deficiency in various eukaryotes results in abnormal LDs (Szymanski et al., 2007; Fei et al., 2008, 2011a; Tian et al., 2011; Liu et al., 2014). Adipose-specific seipin-knockout mice showed a severely affected metabolism of sphingolipids, phospholipids, and FAs, which suggests that seipin is crucial for lipid homeostasis (Liu et al., 2014). However, seipin deletion in yeast did not cause a drastic change in the overall lipid profile (Fei et al., 2008; Wang et al., 2014), reflecting a cell type specificity in the regulation of lipid homeostasis. Seipin is thought to directly modulate phosphatidic acid (PA) metabolism (Fei et al., 2011b; Pagac et al., 2016). Notably, abnormal PA accumulation at the exaggerated subdomain of ER has been found in yeast seipin deletion mutants, which may affect lipid bilayer tensions, causing irregular shapes of LDs emerged from the ER (Fei et al., 2011b; Grippa et al., 2015; Han et al., 2015; Wolinski et al., 2015). Seipin forms a ring-shaped oligomer, and its lumenal domain contains two adjacent four-stranded β-sheets that are folded into a hydrophobic pocket, similar to a lipid-binding domain (LBD; Sui et al., 2018; Yan et al., 2018). The protein also contains cytosolic and lumenal helices that together anchor LDs to the ER bilayer. Hence, seipin may play a dual role in facilitating lipid binding and/or sorting in addition to providing structural supports at ER–LD contacts.
Sphingolipids define a structurally distinct class of conserved membrane lipids implicated as signaling lipids involved in several human diseases (Olson et al., 2016). Its synthesis starts in the ER, where serine palmitoyltransferase (SPT) activity catalyzes the condensation of serine and CoA-activated palmitate to produce 3-ketodihydrosphingosine (3-KDS), which was subsequently converted to the basic structure of sphingolipids, termed the long-chain base (LCB), by 3-KDS reductase (Hanada, 2003; Dickson et al., 2006; Fig. 1 A). SPT contains multiple subunits and functions as the first-committed and rate-limiting step for sphingolipid synthesis (Hanada and Nishijima, 2003). In yeast, the evolutionarily conserved proteins Lcb1 and Lcb2 define the minimal SPT activity, which requires a positive regulator Tsc3 for optimal activity (Gable et al., 2000; see Fig. 3 A). On the other hand, the Orm proteins in their nonphosphorylated forms bind and inhibit SPT activity (Breslow et al., 2010). Through integrations with multiple sensors that monitor cellular sphingolipid levels, the Orm proteins act as homeostatic regulators to fine-tune SPT activity in response to various environmental cues (Han et al., 2010; Roelants et al., 2011; Liu et al., 2012; Sun et al., 2012; Gururaj et al., 2013; Shimobayashi et al., 2013).
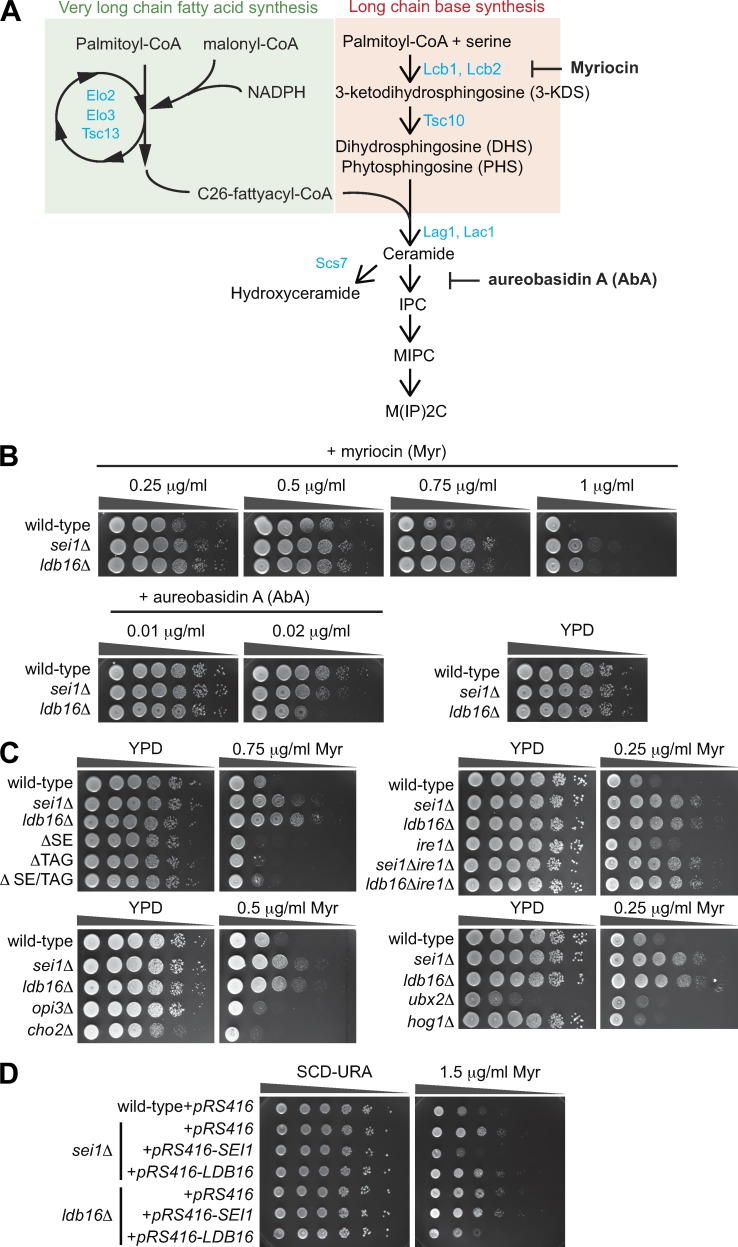
Figure 1.
Seipin deletion mutants display sphingolipid phenotypes. (A) A scheme of de novo sphingolipid biosynthetic pathway in yeast. ER-localized enzymes involved in the indicated biochemical steps are labeled in blue. Myriocin and AbA are biosynthetic inhibitors that target different steps of the pathway. (B) Wild-type, sei1Δ, and ldb16Δ cells were 10-fold serially diluted and spotted on YPD plates containing ethanol or various concentrations of myriocin (Myr) or AbA as indicated. Plates were incubated at 30°C for 2–3 d and photographed. (C) Same as B, except that different strains were analyzed: ΔSE (are1Δ are2Δ); ΔTAG (dga1Δ lro1Δ); and ΔSE/TAG (are1Δ are2Δ dga1Δ lro1Δ). (D) Wild-type cells expressing empty vector (pRS416) and sei1Δ or ldb16Δ cells expressing pRS416, pRS416-SEI1, or pRS416-LDB16, respectively, were 10-fold serially diluted and spotted on SC-URA plates containing ethanol or 1.5 µg/ml myriocin. Plates were incubated at 30°C for 2–3 d and photographed.
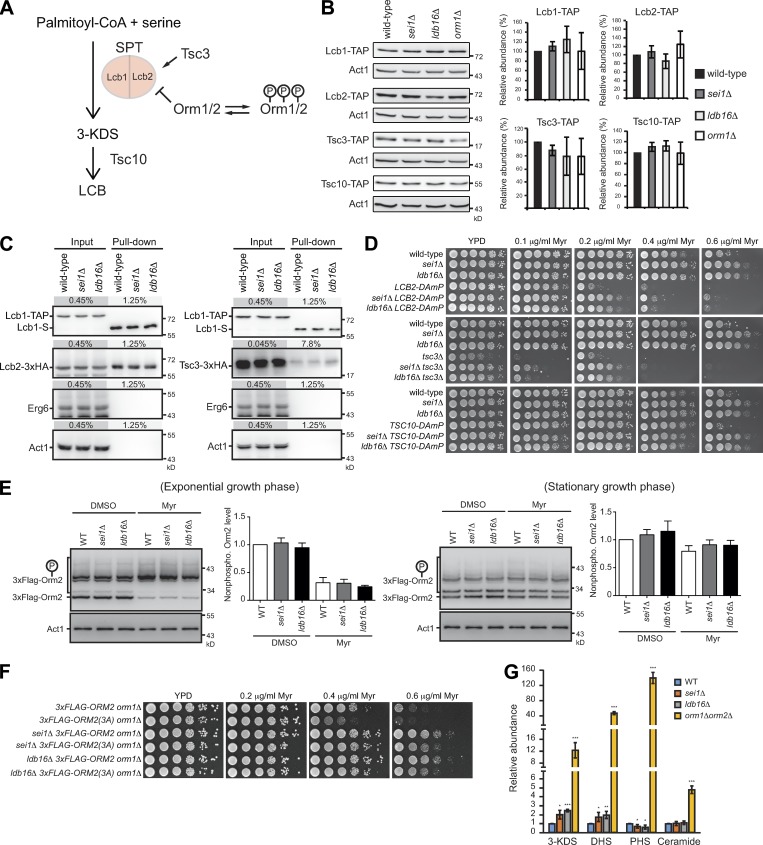
Figure 3.
The increased SPT activity in seipin mutants is regulated independently of Tsc3 and Orm. (A) A scheme of SPT regulation. (B) Wild-type, sei1Δ, ldb16Δ, and orm1Δ cells harboring Lcb1-TAP, Lcb2-TAP, Tsc3-TAP, or Tsc10-TAP at log phase were lysed, and proteins were analyzed by Western blot with peroxidase anti-peroxidase and anti-actin antibodies. After normalized to Act1 (actin), the indicated TAP-tagged protein levels were compared with that in the wild type and plotted. Data are mean ± SD from four independent experiments. (C) Wild-type, sei1Δ, and ldb16Δ cells coexpressing Lcb1-TAP and Lcb2-3xHA (left panel) or Tsc3-3xHA (right panel) were lysed, and the cleared lysates (input) were used for pull-down with IgG Sepharose, followed by TEV protease cleavage (pull-down). Input and pull-down fractions were examined by Western blot analysis with anti-S, anti-HA, anti-Erg6, and anti-actin antibodies. The percentage of load from the input and pull-down fractions are indicated. (D) Yeast strains as indicated were 10-fold serially diluted and spotted on YPD plates containing ethanol or indicated concentration of myriocin (Myr). Plates were incubated at 30°C for 2–3 d and photographed. (E) For exponential growth phase, wild-type (3xFLAG-ORM2 orm1Δ), sei1Δ (3xFLAG-ORM2 orm1Δ sei1Δ), and ldb16Δ (3xFLAG-ORM2 orm1Δ ldb16Δ) cells were grown in YPD to log phase and treated with DMSO or 0.4 µg/ml myriocin (Myr) for 1 h. For stationary growth phase, cells were grown in YPD to early log phase and treated with DMSO or 0.15 µg/ml myriocin for 16 h. Cells were lysed, and proteins were subjected to Phos-tag (for 3xFlag-Orm2) or regular (for Act1) SDS-PAGE, followed by Western blot analysis with anti-Flag or anti-actin antibodies. After normalized to Act1, the nonphosphorylated 3xFlag-Orm2 levels were compared with that in the wild type with DMSO treatment and plotted. Data are mean ± SD of three independent experiments. (F) Same as D, except that different strains were analyzed. (G) Sphingolipids were extracted from cells and their abundance was quantified. The indicated type of sphingoid intermediates in mutants were compared with that in the wild type and shown as fold change. Data are mean ± SD from four independent experiments. *, P < 0.05; ***, P < 0.001.
Very long chain FAs (VLCFAs) synthesized by FA elongase define the other major building block of sphingolipids. The LCBs, dihydrosphingosine (DHS) and phytosphingosine, are condensed with a CoA-activated VLCFA to produce the general precursors of sphingolipids termed ceramides in the ER by ceramide synthase (Guillas et al., 2001; Schorling et al., 2001; Kihara, 2012; Sassa and Kihara, 2014). Excess ceramides are toxic (Eisenberg and Büttner, 2014), and cells prevent their accumulation by efficient sorting via vesicular or nonvesicular trafficking pathways to form a set of complex sphingolipids (Funato and Riezman, 2001). Alternatively, ceramides are metabolized to other lipids, such as acylceramides stored within LDs (Voynova et al., 2012; Senkal et al., 2017). How cells monitor ceramides and integrate their sorting into various organelles remains poorly understood.
Here, we identify a novel function for seipin. Yeast cells lacking seipin showed sphingolipid-perturbed phenotypes that can be complemented by expressing human seipin. Lipidomic profiling data revealed an accumulation of sphingoid intermediates and VLCFAs, which agreed with the increased SPT and FA elongase activities observed in the mutants. We unravel an inhibitory interaction of seipin with the SPT and FA elongase enzymes, which likely coordinates to balance ceramide synthesis. We further uncover that these interactions converged at ER–LD contacts and that sphingolipid synthesis at the subdomain is not a prerequisite for LD biogenesis. Together, we propose that ER–LD contacts where seipin resides to mediate sphingolipid homeostasis are one of the sphingolipid synthesis and sorting centers in the cell.
Results
Seipin mutants show altered sphingolipid regulation
In budding yeast, Sei1 and Ldb16 form a complex at the ER–LD contacts to exert the functional form of human seipin (Wang et al., 2014). We performed Sei1-TAP purification to investigate the yeast seipin-interacting network. In addition to the previously reported Sei1-interacting factors, we identified proteins involved in several biological processes, one of which covered various aspects of lipid metabolism (Fig. S1 and Table S2). Intriguingly, sphingolipid enzymes constituted a major subgroup. Sphingolipids do not build upon a glycerol backbone and thus use separate pathways for their synthesis and metabolism. Although seipin has never been proposed to function with this specific lipid species, seipin might play a yet-unexplored role in sphingolipid metabolism.
To tackle the role of seipin in sphingolipid metabolism, we first asked whether sei1Δ and ldb16Δ altered the sensitivity to the sphingolipid biosynthesis inhibitors myriocin and aureobasidin A (AbA; Fig. 1 A). Both strains were resistant to myriocin and sensitive to AbA (Fig. 1 B). Further analyses indicated that the phenotypes were not an indirect consequence of neutral lipid perturbation or LD defects, because cells lacking either the neutral lipid triacylglycerol (TAG) or sterol ester (SE) or LDs did not show myriocin resistance (Fig. 1 C). We also did not observe myriocin resistance in opi3Δ and cho2Δ strains that harbor supersized LDs as seipin mutants (Fig. 1 C). Although ER stress and lipid imbalance may cause global impacts on lipid pathways, we did not observe myriocin resistance in cells lacking the ER stress sensor Ire1, cells severely reduced Δ9-desaturase (ubx2Δ; Surma et al., 2013), or cells previously known to confer tunicamycin sensitivity (hog1Δ; Torres-Quiroz et al., 2010; Fig. 1 C), implying a unique role of seipin in sphingolipid metabolism. The myriocin resistance of sei1Δ was complemented by Sei1, but not Ldb16, and Ldb16 complemented only ldb16Δ but not sei1Δ (Fig. 1 D). Therefore, yeast cells require intact seipin to maintain sphingolipid homeostasis.
Inhibitory functions of seipin in LCB and VLCFA productions
To explore the sphingolipid defects in seipin mutants, we first hypothesize that the myriocin resistance observed in seipin mutants is caused by sphingolipid overproduction, as myriocin specifically inhibits the first committed and rate-limiting enzyme SPT for sphingolipid synthesis. On the other hand, AbA inhibits complex sphingolipid synthesis, resulting in ceramide accumulation (Cerantola et al., 2009). As ceramide is cytotoxic, the higher sensitivity of seipin mutants to AbA is consistent with the hypothesis of sphingolipid overproduction. To test this hypothesis, we compared the level of sphingolipids produced in cells using a radioisotope-labeled serine incorporation assay (Yamaji-Hasegawa et al., 2005). Under the linear incorporation range for 3 h, we consistently observed more radioisotope counts in the seipin mutants than the wild type (Fig. 2 A), which supports sphingolipid overproduction in seipin mutants.
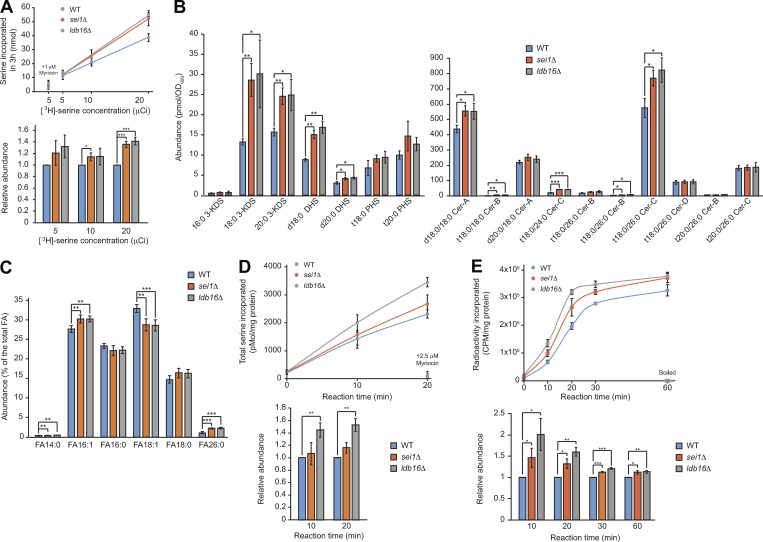
Figure 2.
Seipin deletion mutants produce more sphingoid precursors and intermediates and VLCFA, correlated with higher SPT and FA elongase activities. (A) Cells grown in SCD to early log phase were treated with indicated concentration of [3H]-serine and incubated at 30°C for 3 h. Top: Lipids were extracted from cells, and radioactivity was measured and plotted. Data are mean+/−SD from three independent experiments. As the negative control, 1 µM myriocin was added to cells at OD 0.2 before the reaction. Bottom: The radioactivity was normalized to that of the wild type and plotted. *, P < 0.05; ***, P < 0.001. (B) Sphingolipids were extracted from cells, and the abundance of indicated lipid species analyzed by LC/MS was quantified and plotted. Data are mean ± SD from four independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) FA methyl esters prepared from cells were analyzed by GC/MS and the relative levels of FA species are plotted. Data are mean ± SD from four independent experiments. **, P < 0.01; ***, P < 0.001. (D) In vitro SPT activity assay. Membrane fractions prepared from cells grown in YPD to log phase were incubated with [3H]-serine and palmitoyl-CoA at 30°C for the time indicated. Top: Lipids were extracted and the incorporation of serine was compared. Data are mean ± SD from three independent experiments. The reactions containing 2.5 µM myriocin served as negative controls. Bottom: The data were normalized to that of the wild type at the indicated reaction time and plotted. **, P < 0.01. (E) In vitro FA elongase activity assay. Membrane fractions prepared from cells grown in YPD to log phase were incubated with [2-14C]-malonyl-CoA, NADPH, and palmitoyl-CoA at 30°C for the time indicated. Top: Lipids were extracted and the radioactivity was compared. Data are mean ± SD from three independent experiments. The reactions using boiled membranes served as negative controls. Bottom: The data were normalized to that of the wild-type at the indicated reaction time and plotted. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Next, we analyzed the cellular sphingolipid profile by liquid chromatography (LC)/mass spectrometry (MS). Interestingly, we observed that sei1Δ and ldb16Δ had more sphingoid precursors and intermediates, particularly the early products along the de novo pathway, namely, 3-KDS, DHS, and ceramides, than did the wild type (Fig. 2 B). Increased levels of sphingoid intermediates were also observed in the previous study of adipose-specific seipin-knockout mice (Liu et al., 2014). However, the levels of complex sphingolipids in sei1Δ and ldb16Δ were not significantly changed (not shown). Ceramides are formed by acylation of LCB with a VLCFA. Yeast cells synthesize sphingolipids with a monounsaturated VLCFA of more than C18 in length, mainly C26 (Kihara, 2012; Sassa and Kihara, 2014). The FA26:0 content was significantly higher in seipin mutants than the wild type as analyzed by gas chromatography (GC)/MS (Fig. 2 C). Thus, we suspect that the sphingolipid phenotype observed in seipin mutants is likely associated with overproduction of the major sphingolipid building blocks, LCBs and VLCFAs.
LCBs and VLCFAs are synthesized by two key enzymes, SPT and FA elongase, respectively. We next measured their activities in vitro using membranes isolated from cells. Upon the supply of the substrates [3H]-serine and palmitoyl-CoA, we observed a higher SPT activity in ldb16Δ and a slightly higher activity in sei1Δ compared with the wild-type cells (Fig. 2 D). Importantly, the activity was abolished by 2.5 µM myriocin, showing specificity of the assay. Similarly, when we measured FA elongase activities using palmitoyl-CoA, NADPH, and [2-14C] malonyl-CoA as the substrates (Dittrich et al., 1998; Paul et al., 2006), we found a higher FA elongase activity in sei1Δ and ldb16Δ mutants than the wild type (Fig. 2 E), which explains the increased amount of VLCFAs detected in the seipin mutants. Overall, these data imply a negative role of seipin in modulating SPT and FA elongase activities.
Seipin mediates sphingolipid production independently of the known SPT regulators Tsc3 and Orm
To further determine how seipin inhibits SPT activity, we first considered that seipin might affect SPT components or its complex formation (Fig. 3 A). In seipin mutants, similar to orm1Δ, the levels of the three SPT complex subunits, Lcb1, Lcb2, and Tsc3, were unaffected (Fig. 3 B). In addition, the SPT complex remained intact in seipin mutants, judged by the same level of Lcb2-3xHA and Tsc3-3xHA being pulled down by Lcb1-TAP (Fig. 3 C), ruling out the possibility that the higher SPT activity in seipin mutants is due to enhanced Tsc3 association with SPT. Seipin deletion rescued myriocin sensitivity of the decreased abundance by mRNA perturbation (DAmP) of essential SPT enzymes (Lcb1 or Lcb2) or the 3-KDS reductase Tsc10, supporting a negative role of seipin in SPT regulation (Fig. 3 D). Additionally, the growth defect and the myriocin hypersensitivity of tsc3Δ cells were rescued by seipin deletion, suggesting that seipin negatively regulates sphingolipid production independently of Tsc3 (Fig. 3 D).
We next asked whether seipin regulates SPT through the known SPT inhibitor Orm proteins. We used a previously described strain expressing only the major yeast Orm protein 3xFlag-tagged Orm2 with Orm1 deletion (Sun et al., 2012). Because nonphosphorylated Orm proteins inhibit SPT activity (Breslow et al., 2010), we compared the steady-state 3xFlag-Orm2 phosphorylation profile using Phos-tag acrylamide gel electrophoresis assay. The levels of nonphosphorylated 3xFlag-Orm2 in seipin mutants was similar to that in the wild type in both log and stationary growth phases (Fig. 3 E). When myriocin was added to the culture to inhibit SPT activity (Sun et al., 2012; Gururaj et al., 2013), cells reduced nonphosphorylated Flag-Orm2, likely as a compensatory response to boost sphingolipid synthesis (Breslow et al., 2010; Davis et al., 2018). The effect was also observed in seipin mutants, and the 3xFlag-Orm2 phosphorylation levels were comparable between wild-type and seipin mutants (Fig. 3 E). Moreover, seipin mutations suppressed the myriocin sensitivity of the nonphosphorylable Flag-Orm2 3A mutation that dominantly inhibits SPT (Sun et al., 2012; Fig. 3 F). Thus, SPT activity is negatively regulated by seipin through a pathway parallel with Orm proteins. Of note, the extent of the accumulation of sphingoid precursors and intermediates in seipin mutants was much lower compared with that in orm1Δorm2Δ cells (Fig. 3 G). Therefore, Orm proteins act as the major regulator for sphingolipid production, whereas seipin might limit sphingolipid production in a minor and possibly restricted manner.
Seipin interacts with SPT and the FA elongase subunit Tsc13
The above data support a rather unique mechanism of seipin for maintaining sphingolipid homeostasis. One possibility concerns that seipin might associate with SPT to regulate its activity. In fact, several components of the SPT complex were detected as putative interacting factors in our Sei1-TAP purification results (Fig. S1). We provide several lines of biochemical evidence to strengthen this notion. First, Sei1-TAP pulled down all three of the SPT complex subunits, including Lcb1, Lcb2, and Tsc3, with or without the addition of cross-linker during purification (Fig. 4 A). Second, Lcb1-TAP (or Lcb2-TAP; not shown) pulled down Sei1-13xMyc and Ldb16-13xMyc at the endogenous levels, but not the LD protein Erg6 or Act1, which indicates specificity of the interaction (Fig. 4 B). These data suggest a possibility that seipin, like Orm, may modulate SPT activity through inhibitory interactions.
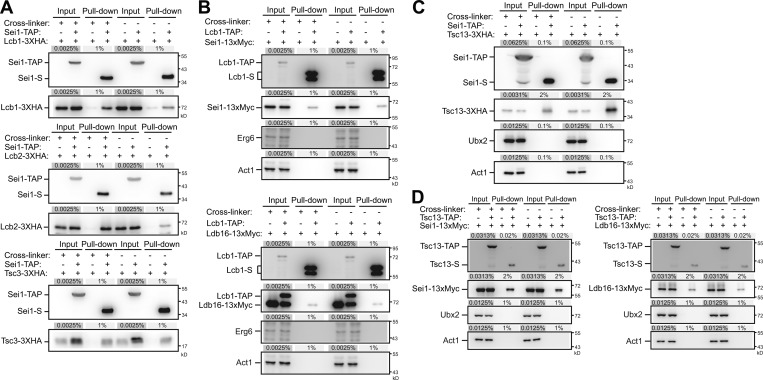
Figure 4.
Seipin interacts with SPT and FA elongase subunits. (A) Cells coexpressing Sei1-TAP(+) or Sei1(−) and Lcb1-3xHA, Lcb2-3xHA, or Tsc3-3xHA as indicated were grown in SCD medium to log phase, converted to spheroplasts, and then treated with DMSO(−) or the cross-linker dithiobis(succinimidyl propionate)(+). Spheroplasts were lysed, and the cleared lysates (input) were used for pull-down with IgG Sepharose, followed by TEV protease cleavage (pull-down). Input and pull-down fractions were examined by Western blot analysis with anti-S and anti-HA antibodies. The percentage of load from the input and pull-down fractions are indicated. (B) Same as A, except that cells expressed Lcb1-TAP(+) or Lcb1(−) and Sei1-13xMyc (top) or Ldb16-13xMyc (bottom) as indicated, and Western blot analysis was performed with anti-S, anti-Myc, anti-Erg6, and anti-actin antibodies. (C) Same as A, except that cells expressed Sei1-TAP(+) or Sei1(−) and Tsc13-3xHA as indicated, and Western blot analysis was performed with anti-S, anti-HA, anti-Ubx2, and anti-actin antibodies. (D) Same as A, except that cells coexpressed Tsc13-TAP(+) or Tsc13(−) and Sei1-13xMyc (left panel) or Ldb16-13xMyc (right panel) as indicated, and the Western blot analysis was performed with anti-S, anti-Myc, anti-Ubx2, and anti-actin antibodies.
The FA elongase enzymes Elo2, Elo3, and Tsc13 catalyze the step-wise FA elongation to form VLCFAs (Denic and Weissman, 2007; Kihara, 2012). We found that Elo3 levels appeared high in seipin mutants, similar to that in the orm1Δ strain, whereas the levels of Elo2 and Tsc13 were unaffected in any of these mutants (not shown). Several FA elongase components were detected as putative interacting factors in our Sei1-TAP purification results, with Tsc13 being the top candidate (Fig. S1). Indeed, the pull-down assay confirmed the interaction between Sei1-TAP and Tsc13-3xHA (Fig. 4 C) and the interactions between Tsc13-TAP and Sei1-13xMyc and Ldb16-13xMyc at the endogenous levels (Fig. 4 D). Given that Elo2, Elo3, and Tsc13 function together as a complex (Denic and Weissman, 2007), seipin may modulate VLCFA production by interacting with Tsc13 to inhibit FA elongase activity.
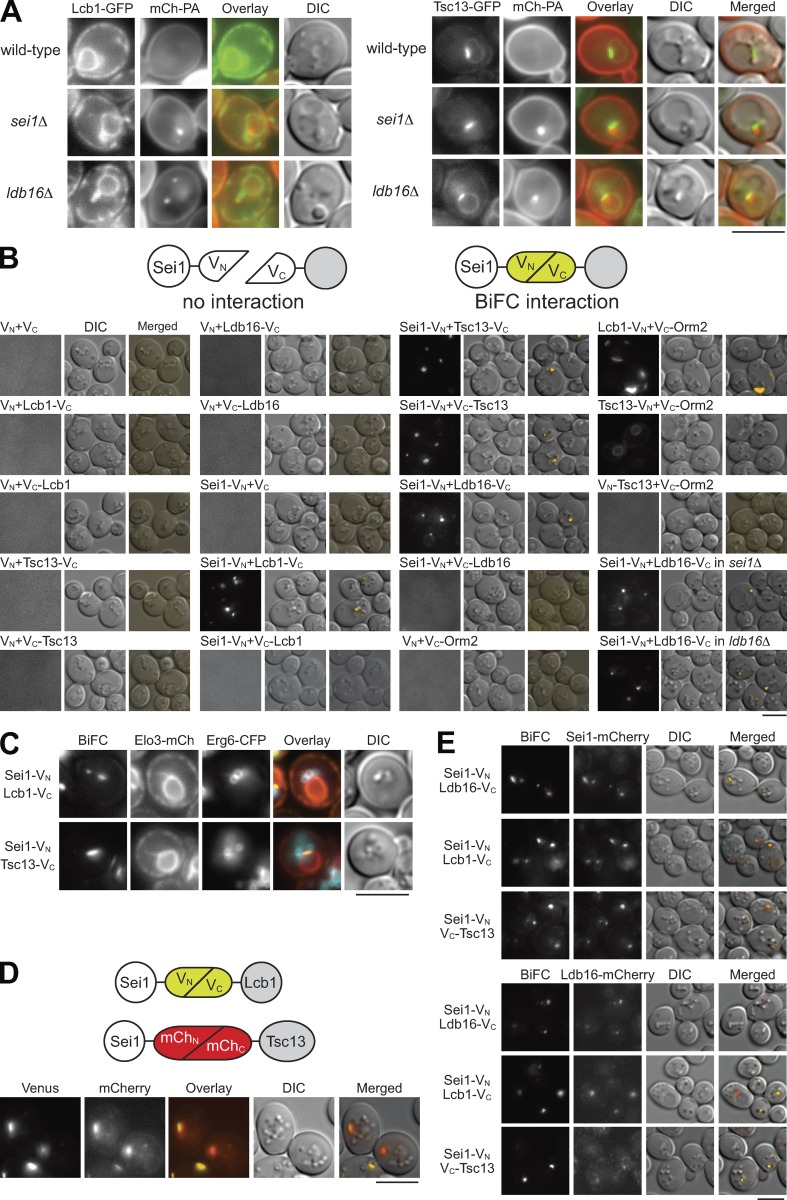
Seipin interacts with SPT and FA elongase at ER–LD contacts
The SPT component Lcb1-GFP localizes throughout the entire ER, whereas the FA elongase component Tsc13-GFP resides at nucleus–vacuole junctions (Kohlwein et al., 2001; Fig. 5 A). However, seipin is an ER protein predominantly partitioning into ER–LD junctions. Without seipin, PA accumulated at the exaggerated ER subdomain that could be detected by the PA sensor mCherry-Spo20(51–91) (Grippa et al., 2015; Han et al., 2015; Wolinski et al., 2015). In the seipin deletion strains, Lcb1-GFP and Tsc13-GFP remained at the dispersed ER and ER–vacuole junction, respectively, but the signals always overlaid at least partially with mCherry-Spo20(51–91) (Fig. 5 A).
Figure 5.
Seipin interacts with Lcb1 and Tsc13 at ER–LD contacts. (A) Wild-type, sei1Δ, and ldb16Δ cells coexpressing mCherry-Spo2051-91 (mCh-PA) and Lcb1-GFP (left panel) or Tsc13-GFP (right panel) were grown in SCD-URA medium to the diauxic shift and imaged by fluorescence microscopy. Scale bar, 5 µm. (B) The diagram depicts the BiFC assay. Wild-type cells expressing VN or Sei1-VN were transformed with various VC-containing plasmids expressing proteins as indicated. Cells were grown in SCD medium to the diauxic shift and imaged by fluorescence microscopy. Sei1-VN and Ldb16-VC coexpressed in sei1Δ and ldb16Δ cells are also shown. Scale bar, 5 µm. (C) Wild-type cells coexpressing the indicated BiFC constructs, Elo3-mCherry (ER marker), and Erg6-CFP (LD marker) were grown in SCD medium to the diauxic shift and imaged by fluorescence microscopy. Scale bar, 5 µm. (D) The diagram depicts the Venus- and mCherry-based BiFC assay. Cells coexpressing Sei1-VN, Lcb1-VC, Sei1-mChN, and Tsc13-mChC were grown in SCD medium to the diauxic shift and imaged by fluorescence microscopy. Scale bar, 5 µm. (E) Sei1-mCherry or Ldb16-mCherry strains were transformed with the indicated BiFC constructs. Cells were grown in SCD medium to the diauxic shift and imaged by fluorescence microscopy. Scale bar, 5 µm.
To learn about the exact place where seipin controls sphingolipid production, we used the bimolecular fluorescence complementation (BiFC) assay with the N-terminus of the fluorescent protein Venus (VN) fused to the C-terminus of Sei1 to test for the fluorophore complementation with the C-terminus of Venus (VC) fused to the N- or C-terminus of Sei1-interacting proteins (Sung and Huh, 2007; Fig. 5 B). Ldb16 is the most well-established Sei1-interacting protein. Indeed, the BiFC assay supported a positive interaction between Sei1-VN and Ldb16-VC (Fig. 5 B). We also demonstrated that Sei1-VN interacted with Lcb1-VC, but not with VC-Lcb1 that exposed the VC portion into the ER lumen. In addition, Sei1-VN interacted with Tsc13-VC or VC-Tsc13, whereas a collection of negative controls did not show any fluorescence (Fig. 5 B). The known Lcb1–Orm2 interaction was seen as large patches near the cell periphery, in contrast to very weak if any interaction between Orm2 and Tsc13 (Fig. 5 B).
Intriguingly, the BiFC signals for Sei1-Lcb1 or Sei1-Tsc13 were detected as one to three puncta in cells, similar to the Sei1-Ldb16–interacting pattern (Fig. 5 B). By colocalizing these structures with the ER marker Elo3-mCherry and the LD marker Erg6-CFP, we revealed that the BiFC signals for Sei1-Lcb1 or Sei1-Tsc13 were within a discrete region of the ER in close vicinity to LDs (Fig. 5 C). To address whether Sei1 interacts with Lcb1 and Tsc13 at the same site, we generated another version of BiFC constructs with divided mCherry for fluorophore complementation (Fan et al., 2008; Fig. 5 D). By coexpressing Sei1-VN, Lcb1-VC, Sei1-mChN, and Tsc13-mChC in one cell, we found the BiFC signals of Sei1-Lcb1 (Venus) and Sei1-Tsc13 (mCherry) colocalized (Fig. 5 D) Finally, the BiFC signals of Sei1-Lcb1 and Sei1-Tsc13 perfectly colocalized with the ER–LD contact site markers Sei1-mCherry and Ldb16-mCherry expressed at the endogenous levels (Fig. 5 E), in agreement with the concept that seipin interacts with sphingolipid enzymes at ER–LD contacts.
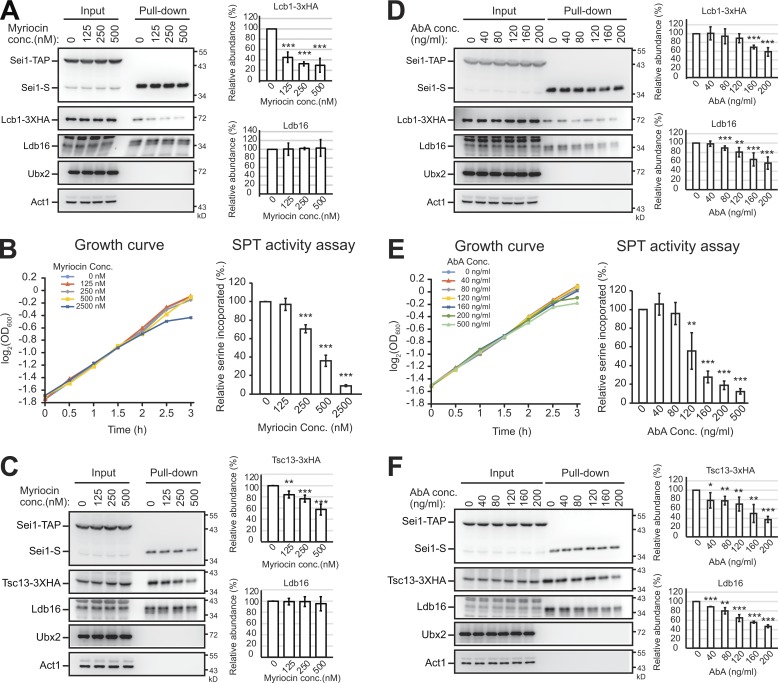
Sphingolipid inhibitors modulate the interactions of seipin with SPT and FA elongase
Sphingolipid homeostasis is regulated by complicated sensing and feedback mechanisms. To further validate that seipin interacting with SPT and FA elongase plays a negative regulatory role, we asked whether the interactions were affected by myriocin or AbA that perturb cellular sphingolipid levels. The amount of Lcb1-3xHA pulled down by Sei1-TAP gradually decreased with increasing concentrations of myriocin in the media (Fig. 6 A). Under these treatments, cell growth was not affected as judged by the growth curve (Fig. 6 B) and cell morphology (not shown). Interestingly, Sei1’s dissociation from Lcb1 occurred even at the lowest concentration of myriocin used, under which the SPT activity was unaffected (Fig. 6 B), supporting that cells were able to resolve the seipin–SPT interaction to boost sphingolipid synthesis to maintain homeostasis. Similarly, addition of myriocin in culture reduced the amount of Tsc13-3xHA, but not Ldb16, in the Sei1-TAP pull-down (Fig. 6 C). Therefore, the seipin–SPT and seipin–FA elongase interactions are subjected to regulation by sphingolipid levels.
Figure 6.
Seipin interacting with Lcb1 and Tsc13 is sensitive to sphingolipid inhibitors. (A) Cells expressing Sei1-TAP and Lcb1-3xHA were grown in SCD medium to early log phase and then treated with indicated concentrations of myriocin for 3 h. Cells were lysed by spheroplasting, and the cleared lysates (input) were used for a pull-down assay with IgG Sepharose, followed by TEV protease cleavage (pull-down). Input and pull-down fractions were examined by Western blot analysis with anti-S, anti-HA, anti-Ldb16, anti-Ubx2, and anti-actin antibodies. After normalized to Sei1-S, the levels of Lcb1-3xHA and Ldb16 in pull-down were compared with that in the control (0 nM myriocin) and plotted. Data are mean ± SD from three independent experiments. ***, P < 0.001. (B) The growth curve was measured for cells under the growth conditions used in A. For the SPT activity assay, cells were grown in SCD medium to early log phase and treated with the indicated concentrations of myriocin as in A. After 1-h myriocin treatment, 20 µCi [3H]-serine was added to the culture for another 2 h. Lipids were extracted from cells, and radioactivity was measured. Data were compared with the control (0 nM myriocin) and plotted. ***, P < 0.001. (C) Same as A, except that cells expressing Sei1-TAP and Tsc13-3xHA were used. **, P < 0.01; ***, P < 0.001. (D) Same as A, except that various concentrations of AbA instead of myriocin were used. **, P < 0.01; ***, P < 0.001. (E) Same as B, except for various concentrations of AbA instead of myriocin. **, P < 0.01; ***, P < 0.001. (F) Same as D, except that cells expressed Sei1-TAP and Tsc13-3xHA. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Similar to myriocin, we observed that AbA treatments dissociated Lcb1-3xHA and Tsc13-3xHA from Sei1-TAP (Fig. 6, D and F). We found that cell growth was not affected with AbA up to 160 ng/ml, and SPT activity was intact with up to 80 ng/ml AbA in culture (Fig. 6 E). Intriguingly, AbA treatment reduced the Sei1-TAP and Ldb16 association (Fig. 6, D and F), which was not seen with myriocin (Fig. 6, A and C). Together, these data support a feedback mechanism that controls the interactions between seipin and sphingolipid enzymes, consistent with the notion that seipin is a negative regulator for sphingolipid production.
Sphingolipid synthesis is not a prerequisite for LD biogenesis, although sphingoid intermediates may affect LD morphology
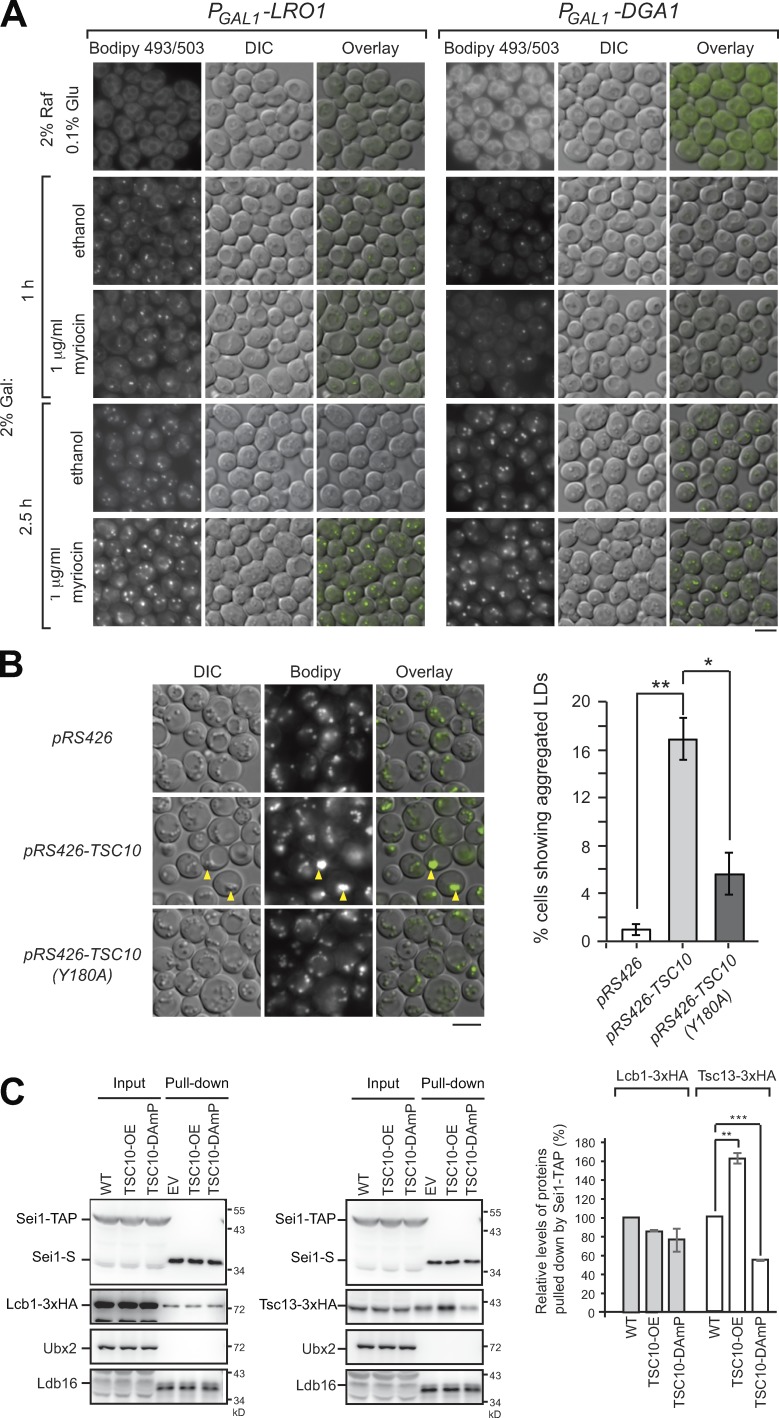
The interaction of seipin with SPT and FA elongase at ER–LD contacts raises the question of whether sphingolipid synthesis may affect LD biogenesis. By staining cells with Bodipy 493/503, we found little if any difference in LD morphology in various strains with deletion or DAmP mutation of sphingolipid enzymes (Fig. S2). To directly determine whether sphingolipid synthesis is required for LD biogenesis, we generated yeast strains harboring only one neutral lipid synthesis enzyme, Lro1 or Dga1, expressed under the control of the inducible GAL1 promoter (Jacquier et al., 2011). When these strains were grown in medium containing 2% raffinose and 0.1% glucose, LDs were absent in the cells (Fig. 7 A). Upon 2% galactose addition for 1 h, nascent LDs rapidly formed, and the size increased over time. The galactose-induced LDs formed as efficiently with myriocin treatment as with the control (Fig. 7 A), indicating that inhibiting sphingolipid synthesis did not affect LD biogenesis and assembly.
Figure 7.
Blocking sphingolipid synthesis does not affect LD biogenesis, but LD morphology is affected by sphingoid intermediates. (A) Yeast strains expressing the sole neutral lipid enzyme driven by the GAL1 promoter, PGAL1-LRO1 or PGAL1-DGA1, were grown in SCD medium containing 2% raffinose (Raf) and 0.1% glucose (Glu) and then shifted to SCD medium containing 2% galactose (Gal) with 1 µg/ml myriocin or an equal volume of ethanol for the time indicated. Cells were stained with Bodipy 493/503 followed by fluorescence microscopy. Scale bar, 5 µm. (B) Wild-type cells harboring pRS426, pRS426-TSC10, and pRS426-TSC10 (Y180A) plasmids were grown in SCD-URA medium to the stationary phase. Left: Cells were stained with Bodipy 493/503 and imaged by fluorescence microscopy. Arrowheads denote aggregated LDs. Scale bar, 5 µm. Right: The percentage of cells (n > 233) showing LD aggregation was quantified. Data are mean ± SD from three independent experiments. *, P < 0.05; **, P < 0.01. (C) Yeast strains expressing Sei1-TAP and Lcb1-3xHA (left) or Tsc13-3xHA (right) were transformed with pRS426 (WT) and pRS426-TSC10 (TSC10-OE). The TSC10-DAmP strains expressing Sei1-TAP and Lcb1-3xHA or Tsc13-3xHA were transformed with pRS426 (TSC10-DAmP). Cells grown in SCD-URA medium to log phase were lysed by spheroplasting and the cleared lysates (input) underwent pull-down with IgG Sepharose followed by TEV protease cleavage (pull-down) and then examined by Western blot analysis with anti-S, anti-HA, anti-Ubx2, and anti-Ldb16 antibodies. After being normalized to Sei1-S in the pull-down fractions, the level of Lcb1-3xHA or Tsc13-3xHA in the pull-down fractions of TSC10-OE and TSC10-DAmP was compared with that in wild type and plotted. Data are mean ± SD from three independent experiments. **, P < 0.01; ***, P < 0.001.
To investigate whether sphingoid intermediates affect LD assembly, we overexpressed several ER-localized sphingolipid enzymes in wild-type cells (Fig. S2). We found ∼15% of cells overexpressing Tsc10 showed aggregated LDs (Fig. 7 B). The phenotype is dependent on the 3-KDS reductase activity of Tsc10, because overexpression of a Tsc10 active-site mutant, Y180A, did not generate aggregated LDs (Fig. 7 B). Thus, the sphingoid intermediates downstream of the Tsc10 activity, such as LCB or ceramides produced in the ER, might intervene in LD assembly, causing LD aggregation.
Among all sphingolipid biosynthetic enzymes, Tsc10 localizes specifically to LDs with no functional implication (Currie et al., 2014). Tsc10 overexpression increases LCB and ceramide levels (Kim et al., 2010), whereas TSC10-DAmP mutant accumulated 3-KDS (Ren et al., 2018). Manipulating Tsc10 level by these approaches had a profound and opposite impact on the seipin-Tsc13 interaction, without an apparent effect on the seipin-Lcb1 interaction (Fig. 7 C). Thus, the sphingolipid control systems involving seipin are likely regulated by different modes.
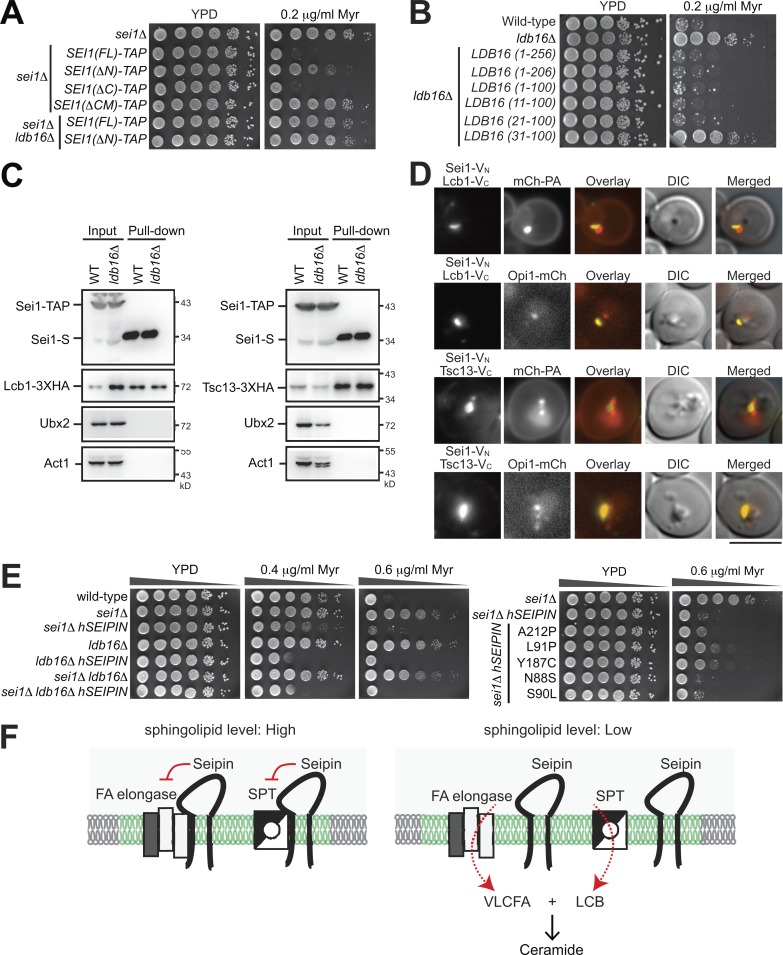
The sphingolipid regulatory function of seipin is conserved throughout evolution
Sei1 has two transmembrane domains and adopts a conformation in the ER by exposing both the N- and C-terminus toward the cytoplasm (Cartwright and Goodman, 2012). The strain harboring Sei1(ΔN)-TAP conferred the myriocin-resistant phenotype (Fig. 8 A), which indicates that the N-terminus 14 amino acids of Sei1, shown to bind the LD surface (Sui et al., 2018), play a crucial role in sphingolipid regulation. Sei1(ΔN)-TAP pulled down less Lcb1 and Tsc13 compared with the wild type (Fig. S3). In addition, the N-terminus of Sei1 was needed for stabilizing Ldb16. By contrast, the C-terminus of Sei1 was dispensable for sphingolipid regulation (Fig. 8 A).
Figure 8.
Human seipin expression maintains sphingolipid homeostasis in yeast seipin mutants. (A and B) Yeast strains were 10-fold serially diluted and spotted on YPD plates containing ethanol or myriocin (Myr) in ethanol. Plates were incubated at 30°C for 2–3 d and photographed. (C) Wild-type and ldb16Δ cells coexpressing Sei1-TAP and Lcb1-3xHA (left panel) or Tsc13-13xHA (right panel) were grown in SCD medium to log phase and lysed by spheroplasting; the cleared lysates (input) underwent pull-down with IgG Sepharose, followed by TEV protease cleavage (pull-down). Western blot analysis was performed with anti-S, anti-HA, anti-Ubx2, and anti-actin antibodies. (D) ldb16Δ cells coexpressing the indicated BiFC constructs and mCherry-Spo2051-91 (mCh-PA) or Opi1-mCherry were grown in SCD medium to the diauxic shift and imaged by fluorescence microscopy. Scale bar, 5 µm. (E) Wild-type, sei1Δ, and ldb16Δ cells, alone or expressing various forms of human Seipin (hSEIPIN), were subjected to dilation plate analysis as in A. (F) A working model of how seipin mediates sphingolipid synthesis. With sufficient sphingolipids, seipin binds both SPT and FA elongase at ER–LD contacts (green zone) to block LCB and VLCFA production. With low sphingolipid levels, seipin dissociates from SPT and FA elongase, thus allowing the enzymes to synthesize LCB and VLCFA, which combine to form ceramide, at ER–LD contacts.
Ldb16 is essential for sphingolipid homeostasis (Fig. 1 B). We mapped down to only a small portion of Ldb16 (residues 21–100) sufficient for regulating sphingolipid homeostasis (Fig. 8 B), which basically covers the two transmembrane helices with a short ER luminal loop, previously known to be critical for interacting with Sei1 and for controlling the LD size (Wang et al., 2014). Intriguingly, Sei1 interacted with Lcb1 and Tsc13 in the absence of Ldb16 at the same level as in the wild type (Fig. 8 C), which indicates that the yeast seipin complex interacts with SPT and FA elongase via Sei1. Consistent with this notion, the BiFC interactions of Sei1-Lcb1 and Sei1-Tsc13 remained in ldb16Δ, and the signal partially overlapped with the PA puncta marker mCherry-PA and the LD biogenesis marker Opi1-mCherry (Fig. 8 D).
Seipin is conserved throughout evolution. Human seipin expressed in yeast also complemented the altered sphingolipid phenotypes of sei1Δ and ldb16Δ strains (Fig. 8 E), supporting that the sphingolipid regulatory role of seipin is evolutionarily conserved. Additionally, some of the human seipin lipodystrophy (A212P, L91P, and Y187P), but not seipinopathy (N88S and S90S), mutations showed partial sphingolipid resistance in yeast, which implies a potential contribution of sphingolipid regulation to the etiology of seipin lipodystrophy mutations.
We propose a model to depict the seipin-mediated sphingolipid regulatory mechanism (Fig. 8 F). With sufficient sphingolipid supply, seipin binds both SPT and FA elongase to inhibit their activities, thus blocking LCB and VLCFA synthesis at ER–LD contacts. Dissociation of seipin from SPT and FA elongase occurs when a low sphingolipid level is sensed and signaled through unknown factors to ER–LD contacts, thus enabling LCB and VLCFA synthesis there for coordinated condensation to form ceramide. Based on our findings that Sei1, the sequence homologue of human seipin in yeast, binds with SPT and FA elongase, we hypothesize that human seipin mediates sphingolipid homeostasis through a similar mechanism.
Discussion
In this study, we document a new sphingolipid regulatory function for the key LD biogenesis protein seipin at ER–LD contacts. We found that seipin associates with two key ER enzymes, SPT and FA elongase, and negatively regulates their activity for sphingolipid production (Figs. 4, 5, and 6). Unlike the well-established SPT negative-regulator Orm proteins that control the major pool of sphingolipid synthesis (Fig. 3 G), seipin restricts LCB and ceramide synthesized at discrete ER subdomains. LCB and VLCFA are two major building blocks of ceramide. However, how cells coordinate VLCFA with LCB for ceramide synthesis is not known. In addition, the regulation of VLCFA production is poorly understood, even though VLCFA may contribute to the essential function of sphingolipids (Nagiec et al., 1993). The identification that seipin interacting with SPT and FA elongase is subjected to regulation by sphingolipids reveals a mechanism of how LCB and VLCFA synthesis could be regulated simultaneously.
Our findings that the inhibitory interactions of seipin with sphingolipid enzymes are organized at the ER–LD contacts are of particular interest, since it implies ER–LD contacts being one of the places where the sphingoid intermediates are likely synthesized and sorted (Fig. 5). Indeed, sphingoid intermediates have been identified in isolated LDs (Schorling et al., 2001). In addition, TAG-synthesizing enzymes, including yeast Dga1 and mammalian DGAT2, catalyze the acylation of ceramides for acylceramides stored within LDs (Voynova et al., 2012; Senkal et al., 2017). These observations, together with our finding, provide the evidence of a link between the synthesis of sphingoid intermediates and LDs. As the core of ER–LD contacts, seipin may function as a lipid synthesis and sorting mediators. In seipin mutants, locally produced sphingoid intermediates may be trapped around LDs, perhaps at or adjacent to the previously described PA puncta (Grippa et al., 2015; Han et al., 2015; Wolinski et al., 2015). The sphingoid intermediates may readily create ER subdomains, blocking sorting in or out of factors for lipid synthesis and/or LD assembly, resulting in LD assembly defects. Our findings that LD aggregated upon overexpression of the catalytically active Tsc10 supports that notion (Fig. 7 B).
Seipin functions in sphingolipid homeostasis. The seipin–SPT and seipin–FA elongase interactions are likely mediated by some other proteins at ER–LD contacts. The most intriguing findings in our analyses are the regulation of these interactions by sphingolipid levels (Fig. 6). Unlike myriocin treatment, which caused a dissociation of Lcb1 and Tsc13 from the intact seipin complex, AbA treatment resulted in seipin complex breakdown, even at the low dosage used, which did not affect SPT activity (Fig. 6, D and F). These results raise an interesting possibility that sphingolipid homeostatic signaling may regulate seipin complex assembly. Alternatively, AbA treatment, which is known to increase sphingoid intermediate levels in the ER, may have a pleiotropic effect on the assembly of seipin complex. Further studies with the use of an inducible system for sphingolipid enzymes or a direct supply of sphingolipid intermediates could provide more insights.
Another interesting, yet unanswered, question concerns the nature of sphingolipids, the synthesis of which is regulated by seipin. In addition to structural and signaling roles, sphingolipids in yeast have been shown to facilitate asymmetric membrane protein transfer to the bud (Singh et al., 2017). The sphingoid intermediate phytosphingosine is known to activate the ER stress surveillance pathway that ensures inheritance of a functional ER in daughter cells (Piña et al., 2018). It would be interesting to determine whether any of these processes involve seipin. In addition, sphingolipid synthesized by seipin may be channeled directly to acylceramide synthesis. In yeast, Orm phosphorylation is mediated by target of rapamycin complex pathways, and the pools of sphingolipid regulated by Orm are involved in a wide range of cellular events (Han et al., 2010; Liu et al., 2012; Gururaj et al., 2013; Shimobayashi et al., 2013; Chauhan et al., 2017). The contribution of the sphingolipid pool regulated by seipin, its physiological impact, and how it is monitored by the system remains to be determined.
In the context of pathologies of metabolic diseases, perturbed sphingolipid metabolism has been shown to concurrently affect LDs (Bikman and Summers, 2011). In contrast, various types of sphingolipids, in parallel with TAGs, were accumulated in various tissues of obese animals (Chaurasia and Summers, 2015; Iqbal et al., 2017). However, a discrepancy remains regarding the role of sphingolipid synthesis in LD formation (Loizides-Mangold et al., 2014; Tong et al., 2014; Xia et al., 2015; Alexaki et al., 2017). We took advantage of the LD induction system in yeast and found that LD biogenesis occurred when myriocin was added (Fig. 7 A), suggesting that sphingolipid synthesis does not directly contribute to LD biogenesis. However, we cannot exclude the possibility that a tissue-specific lipid regulatory loop may exist between sphingolipids and glycerolipids.
The exact molecular function of seipin and the nature and physiological importance of ER–LD contacts remain key questions in LD biology. Based on the seipin-interacting network, we were able to assign a new function of seipin in sphingolipid metabolism. Our work provides a foundation toward a better understanding of this molecule for its contribution to lipid metabolism and how its connection to sphingolipid synthesis may have any relevance to human diseases.
Materials and methods
Yeast strains and growth conditions
Table S1 summarizes the strains, plasmids, and primers used in this study. Yeast strains were generated by a plasmid- or PCR-based transformation approach (Longtine et al., 1998), and transformants on selection plates were further confirmed by colony PCR or Western blot analysis. We grew yeast cells in YPD (1% yeast extract, 2% peptone, and 2% glucose) or SCD (0.67% yeast nitrogen base, amino acids, and 2% glucose) at 30°C. For LD-induction experiments, cells were grown first in SC complete medium (0.67% yeast nitrogen base without amino acids and complete amino acids) containing 2% raffinose and 0.1% glucose and then shifted to SC complete medium containing 2% galactose. For the dilution plate assay, yeast cells grown in YPD or SCD were diluted with sterile water and spotted on YPD or SCD plates with or without various concentrations of inhibitors, and plates were incubated at 30°C for 2–3 d.
Reagents
Anti-Erg6, Ubx2, and Ldb16 were homemade antibodies that have been described previously (Wang and Lee, 2012; Wang et al., 2014). The anti-S tag antibody (ab24838, mouse monoclonal) was from Abcam, anti-Act1 (actin) antibody (MAB1501, mouse monoclonal) was from Millipore, peroxidase anti-peroxidase (323-005-024, rabbit polyclonal) was from Jackson ImmunoResearch Laboratories, and anti-Flag antibody (F3165, mouse monoclonal) was from Sigma-Aldrich. Myriocin was from Sigma-Aldrich, and AbA was from Takara. IgG Sepharose 6 Fast Flow was from GE, Sepharose CL-6B was from Sigma-Aldrich, AcTEV protease was from Invitrogen, the Pierce silver staining kit was from Thermo Fisher Scientific, NADPH was from Sigma-Aldrich, palmitoyl-CoA was from Avanti Polar Lipids, and [2-14C] malonyl-CoA and [3H]-serine were from PerkinElmer. Complete EDTA-free protease inhibitor cocktail was from Roche. Phos-tag was from Wako Pure Chemical Industries. Bodipy 493/503 was from Invitrogen. All other common chemical reagents were from Sigma-Aldrich. Restriction and modifying enzymes for cloning and PCR were from Thermo Fisher Scientific.
Fluorescence microscopy
To image LDs, cells were stained by 0.1 µg/ml Bodipy 493/503 followed by two washes in 50 mM Tris, pH 7.5, and subjected directly to fluorescence microscopy. All fluorescence images of cells were captured by using an Olympus IX81 inverted microscope equipped with a 100× Plan-Apochromat objective lens (NA = 1.4) and an ORCA Flash 4.0 cMos v2 camera (Hamamatsu) with MetaMorph software (Molecular Devices). The fluorescence filters used in this study were the GFP filter (excitation filter BP460-480, emission filter BP495-540), mCherry filter (excitation filter BP535-555, emission filter BA570-625), YFP filter (excitation filter BP490-500, emission filter BP515-560), and CFP filter (excitation filter BP425-445, emission filter BA460-510). Images were processed by using MetaMorph and/or Photoshop CS4 (Adobe).
Protein analyses
Protein analysis of total cell lysates
To analyze proteins by immunoblotting, cultured yeast cells were treated with 10% TCA on ice for 30 min, washed twice with cold acetone, and lysed by glass beads in MURB buffer (50 mM sodium phosphate, 25 mM MES, pH 7.0, 1% SDS, 3 M urea, and 5% β-mercaptoethanol). Samples were heated to 55°C for 10 min and subjected to SDS-PAGE gel electrophoresis followed by Western blot analysis with specific antibodies. The UVP ChemiDoc-It imager and software were used for data acquisition and quantification. Data from several repeats were exported to Microsoft Excel for further statistical analysis and chart output. We followed the manufacturer’s suggestion for silver staining analysis.
TAP purification
Yeast cells were grown in SCD to OD600 = 0.8. Cells were first treated with 0.1 M Tris-SO4, pH 9.4, and 10 mM DTT for 10 min at 30°C and then converted to spheroplasts by lyticase treatment in 50 mM Hepes, pH 7.4, and 1.2 M sorbitol in 0.2X YPD for 30 min at 30°C. The spheroplasts were recovered and resuspended in X-link buffer (20 mM Hepes, pH 7.4, 700 mM sorbitol, and 100 mM KOAc) with the addition of 10 mM dithiobis(succinimidyl propionate) in DMSO to a final concentration of 0.25 mM or DMSO alone. After 30-min incubation at room temperature, 1 M Tris, pH 7.4, was added to quench the reaction. The spheroplasts recovered by 3,000 rpm spin for 3 min were resuspended in cold lysis buffer (20 mM Hepes, pH 7.4, 150 mM NaCl, and 1% Triton X-100) containing protease inhibitor cocktail. After Dounce homogenization on ice, lysates were centrifuged at 13,000 rpm for 10 min, the corresponding supernatant was transferred to a CL6B Sepharose column, and the flow through was collected for binding with IgG Sepharose at 4°C overnight with gentle rotation. The beads were recovered and washed four times with cold lysis buffer and then once with TEV buffer (50 mM Tris, pH 8.0, 0.5 mM EDTA, and 1 mM DTT). The beads were treated with AcTEV protease in TEV buffer at 10°C for 3 h, followed by a centrifugation step at 1,000 rpm for 30 s to collect the supernatant for LC/MS/MS. Alternatively, samples were adjusted to a final concentration of 1× MURB buffer , heated at 55°C for 10 min, and subjected to SDS-PAGE followed by immunoblot analysis as described above.
LC/MS/MS
For proteomic analysis, we followed a previously described method for sample treatment with trypsin followed by desalting on a C18 solid-phase extraction cartridge (Wiśniewski et al., 2009). The dried sample was dissolved in 10 µl of 0.1% formic acid for LC/MS/MS analysis with the use of the Thermo Fisher Scientific LC-nESI-Q Exactive mass spectrometer coupled with online nanoUHPLC (Dionex UltiMate 3000 Binary RSLCnano). An Acclaim PepMap 100 C18 trap column (75 µm × 2.0 cm, 3 µm, 100 Å; Thermo Fisher Scientific) and an Acclaim PepMap RSLC C18 nano LC column (75 µm × 25 cm, 2 µm, 100 Å) were used to deliver solvent and separate tryptic peptides with a linear gradient from 5% to 35% acetonitrile in 0.1% (vol/vol) formic acid for 90 min at a flow rate of 300 nl/min. The acquisition cycle of the MS data were performed with the data-dependent mode with a full-survey MS scan followed by 10 MS/MS scans of the top 10 precursor ions from the MS scan. The MS scan was performed with a resolving power of 70,000 over the mass-to-charge (m/z) range of 350 to 1,600 and dynamic exclusion enabled. The data-dependent MS/MS acquisitions were performed with a 2-m/z isolation window, 27% normalized collision energy, and 17,500 resolving power. Finally, the MS data were analyzed using Proteome Discoverer 2.1 SP1 (Thermo Fisher Scientific).
Analysis of the putative Sei1-TAP–interacting protein
To rank the confidence level of putative interacting proteins identified by the Sei1-TAP pull-down assay, we used a method previously described by the Olzmann group (Bersuker et al., 2018). In brief, the normalized confidence score (CS(N)) was calculated by multiplying Eq. 1 by Eq. 2 (the equations can be found in Fig. S1 C of Bersuker et al., 2018). Eq. 1 represented the number of times the protein pulled down by Sei1-TAP identified in the experimental replicates. Eq. 2 represented the sum of the spectral abundance factor (SAF) score of the protein pulled down by Sei1-TAP minus the SAF score of the protein pulled down by Sei1 alone. The SAF is calculated by dividing the total spectral counts by the number of amino acids in the protein, multiplied by 10. The normalized SAF score was calculated by dividing the SAF by the average SAF of each set of sample if SAF > 0. Finally, the CS(N) is calculated by dividing the confidence score by the confidence score of the known Sei1 interactor Ldo45. A heat map was generated by using Multiple Experiment Viewer (4.9.0). The 712 proteins with CS(N) ≥ 1 were subjected to Gene Ontology (GO) analysis with ClueGO (Bindea et al., 2009; http://apps.cytoscape.org/apps/cluego) and were assigned to 176 fused GO/pathway terms and 24 fused GO/pathway term groups (P ≤ 0.05).
Phos-tag gel electrophoresis and immunoblotting
To assay the phosphorylation status of 3xFlag-Orm2 in exponentially growing yeast strains, 5 ml YPD medium was inoculated in duplicate with overnight precultures to OD600 = 0.1 and grown to OD600 = 0.6. One of the duplicate cultures was treated with 400 ng/ml myriocin, and both cultures were cultured for an additional 60 min. To investigate the phosphorylation status of 3xFlag-Orm2 in stationary phase, 5 ml YPD medium was inoculated in duplicate with OD600 = 0.1 for each strain; one of the duplicate cultures was treated with 150 ng/ml myriocin, and both cultures were cultured for an additional 16 h. Ice-cold TCA was added to 1 OD600 of each culture to a final concentration of 10% (vol/vol) and incubated on ice for 60 min. After centrifugation at 4°C for 10 min, the pellet was washed in cold acetone and air-dried for 5–10 min. The pellets were dissolved by vortex with glass beads in 400 µl 1× MURB buffer for 10 min at room temperature, followed by heating at 65°C for 20 min.
Phospho-dependent mobility shifts of 3xFlag-Orm2 were detected by loading equal protein amounts (0.025 OD600 equivalent of cells) onto 10% Bis-Tris SDS-PAGE gels containing 100 µM Phos-tag (Wako Pure Chemical Industries) and 200 µM ZnCl2. Before transfer onto polyvinylidene difluoride membranes, Phos-tag gels were washed twice for 15 min in transfer buffer (25 mM Tris, pH 8.3, 192 mM glycine, and 20% [vol/vol] MeOH) supplemented with 2 mM EDTA and then once for 15 min in transfer buffer without EDTA. Membranes were then probed with mouse monoclonal anti-Flag or mouse Act1 antibody. Relative nonphosphorylated Flag-Orm2 protein levels in each condition were quantified by using the UVP ChemiDoc-It imager and software.
Lipid extraction and analyses
Sphingolipid analysis
We used a previously described method to extract sphingolipids from yeast cells (Guan and Wenk, 2006). In brief, yeast cells (40 OD600) were killed by using 20 mM NaN3. Cells resuspended in 2 ml ethanol/ddH2O/diethyl ether/pyridine/ammonium hydroxide (15:15:5:1:0.018, vol/vol) with the addition of 20 µl of 100 µg/ml ceramide (17:0/18:1) as the internal controls were lysed by vortexing with glass beads for 10 min. After heating up at 60°C for 1 h, samples were subjected to a quick spin to collect the supernatant that was then concentrated by using the Thermo Fisher Scientific SpeedVac concentrator SPD111v for 1 h. The remaining ∼1 ml solution was supplemented with 400 µl of 0.2 N CHCl3/methanol/ddH2O (16:16:5, vol/vol) and then 400 µl of 0.2 N NaOH and incubated at 30°C for 45 min. We then added 400 µl of 0.5 M EDTA, 80 µl of 1 N acetic acid, and 500 µl CHCl3 to extract sphingolipids, and the organic phase was collected and dried by the Thermo Fisher Scientific SpeedVac concentrator. The extracted sphingolipids were dissolved in 150 µl CHCl3/methanol (2:1) and subjected to LC/MS analysis. Ceramide (d18:1/17:0) was used for ceramide quantification, and DHS (d18:0) was used for 3-KDS and LCB quantification. Lipid concentrations were calculated relative to the relevant standards.
We used a linear ion-trap Orbitrap mass spectrometer (Orbitrap Elite; Thermo Fisher Scientific) coupled online with an Ultra Performance Liquid Chromatography system (ACQUITY UPLC; Waters) for UPLC/MS. In brief, samples of 4 µl were separated by a High Strength Silica T3 column (1.8 µm, 2.1 mm × 100 mm, ACQUITY UPLC; Waters) at a flow rate of 0.5 ml/min using a cycle of 40–99.9% solvent B over 0–10 min (solvent A containing acetonitrile/water [40:60] with 10 mM ammonium acetate, pH 5.0; solvent B containing isopropanol/acetonitrile [90:10] with 10 mM ammonium acetate, pH 5.0), holding at 99.9% B for 2 min, and reequilibrating at 40% B. The MS was operated with positive or negative ion modes, and we set the full Fourier-transfer MS scan at m/z 100–1,200, resolution 60,000. The sphingolipid species were quantified on the basis of Guan and Wenk (2006) and Ren et al. (2018) with Xcalibur software (Thermo Fisher Scientific), and Microsoft Excel was used for statistical analysis.
FA analyses by GC/MS
We used Folch’s method of lipid extraction (Folch et al., 1957). In brief, cells were lysed using glass beads in methanol with the addition of FA17:0 as an internal control, and then chloroform was added. After centrifugation to remove the debris, the supernatant was supplemented with NaCl to separate the lower organic phase by centrifugation. The organic phase was dried with the Thermo Fisher Scientific SpeedVac concentrator SPD111v, followed by derivatization with BF3 to prepare the FA methyl esters as described previously (Morrison and Smith, 1964). In brief, lipids dissolved in 230 µl 14% BF3 in methanol were heated to 95°C for 10 min. After the samples were cooled to room temperature, 198 µl benzene was added and then heated to 95°C for 30 min. Once returned to room temperature, samples were treated with 230 µl ddH2O and 690 µl petroleum ether for phase separation. The organic phase was recovered and concentrated with the Thermo Fisher Scientific SpeedVac concentrator SPD111v. Samples were finally dissolved in CHCl3/methanol (2:1 vol/vol) and loaded for GC/MS with the Agilent Technologies 7890A GC system equipped with 5975C inert Mass Selective Dectector. FA methyl esters were separated on the DB-5MS column (30 m × 0.25 mm × 0.25 µm; Agilent Technologies) at a flow rate of 1 ml/min. The GC program was previously described (Yu et al., 2017). The data were acquired at the scan mode (50–650 m/z). Target search and data quantification involved using the Agilent Technologies chemstation data analysis software with the National Institute of Standards and Technology MS search library.
Enzyme activity assay
SPT activity assay (in vivo)
The SPT activity assay was modified from a previous report (Yamaji-Hasegawa et al., 2005). In brief, 5, 10, or 20 µCi [3H]-serine was added to 4 ml cells grown in SCD medium to early log phase (OD600 = 0.4), and the culture continued to grow at 30°C for 3 h. For the myriocin treatment control, myriocin was added to a final concentration of 1 µM when cells were grown in SCD medium to OD600 = 0.2. Cells were killed by 20 mM NaN3 and then subjected to sphingolipid extraction as described above. The organic phase after the extraction was dried by nitrogen and resuspended in 20 µl CHCl3/methanol (2:1 vol/vol). An amount of 4 µl of each sample was then diluted in 3 ml Ultima Gold (PerkinElmer), and the radioactivity was measured using the PerkinElmer liquid scintillation analyzer Tri-Carb 2910 TR.
SPT activity assay (in vitro)
The in vitro SPT activity assay was modified from previous reports (Davis et al., 2019; Han et al., 2019). In brief, yeast cells were harvested at the log phase and lysed in TEGM buffer (50 mM Tris, pH 7.5, 1 mM EGTA, and 1 mM β-mercaptoethanol) containing protease inhibitors at 1 ml/50 OD cells by vortexing with glass beads for 8 cycles of 30 s and 30 s on ice in between. After a centrifugation at 8,000 g for 10 min, the corresponding supernatant was subjected immediately to high-speed centrifugation at 100,000 g for 30 min. The P100 pellet was resuspended in TEGM buffer, and the protein concentration was determined by Bradford assay and adjusted to 8 mg/ml with the same buffer. Each reaction contained 200 µg of P100 fraction in 50 mM Hepes, pH 8.0, 2.5 mM L-serine, 50 µM pyridoxal-5′-phosphate, 20 µCi [3H]-serine, and 100 µM palmitoyl-CoA in a volume of 300 µl at 30°C. To terminate the reaction, NH4OH was added to a final concentration of 0.2 M, followed by the addition of 1.5 ml CHCl3/methanol (1:2 vol/vol). After 1 ml of CHCl3 and 2 ml of 0.5 M NH4OH addition, the samples were vortexed vigorously and centrifuged briefly. The upper aqueous phase was removed, and the lower phase was washed twice with 2 ml of 30 mM KCl. 1 ml of the sample was removed and dried under nitrogen gas, and then 3 ml Ultima Gold was added. Radioactivity measurement was performed with the PerkinElmer liquid scintillation analyzer Tri-Carb 2910 TR.
FA elongase activity assay
The FA elongase activity assay was modified from previous reports (Dittrich et al., 1998; Paul et al., 2006). In brief, yeast cells were converted to spheroplasts and lysed in HKMS buffer (20 mM Hepes, pH 7.4, 150 mM KCl, 1 mM MgCl2, and 250 mM sorbitol). The lysates were first spun at 300 g for 5 min to remove unlysed cells, followed by a 13,000-g spin at 4°C to harvest the P13 pellet. Each reaction contained 100 µg P13 fraction in 10 mM Tris/maleic acid, pH 6.7, 0.65 M mannitol, 0.36 mM EGTA, 0.9 mM MgCl2, 1.8 mM DTT, 270 µM Triton X-100, 0.9 mM NADPH, 54 µM palmitoyl-CoA, and 0.045 µCi [2-14C] malonyl-CoA. The reactions were initiated by shifting tubes from ice to 30°C. At the indicated time, 200 µl of 5 M KOH was added, and the mixture was incubated at 70°C for 1 h and then mixed with 200 µl 5 M H2SO4 in 10% malonic acid. The FA was extracted with CHCl3, washed twice with water, and then transferred to 3 ml Ultima Gold for radioactivity measurement with the PerkinElmer liquid scintillation analyzer Tri-Carb 2910 TR.
Statistical analysis
All experiments involved at least three repeats, and the data are presented as mean ± SD, with details described in the figure legends. The sample size is indicated in the figures. Statistical analysis involved two-tailed Student t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Data distribution was assumed to be normal, but this was not formally tested.
Online supplemental material
Fig. S1 shows the identification of putative seipin interacting proteins by a large-scale Sei1-TAP purification assay. Fig. S2 shows LD morphology was not much affected in various strains lacking or reducing the expression of sphingolipid enzymes, whereas overexpressing Tsc10 caused LD aggregation in cells. Fig. S3 shows that Sei1-TAP lacking the N- or C-terminus affected protein abundance and the interaction with Lcb1 and Tsc13. Table S1 lists the strains, plasmids, and primers used in this study. Table S2 lists the complete set of data for the Sei1-TAP purification screen.
Supplementary Material
Acknowledgments
The authors thank Dr. David Drubin (University of California, Berkeley, Berkeley, CA) for sharing yeast strains, Dr. Rey-Huei Chen (Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan) for help with experiments and improving the manuscript, Ms. Yu-Ching Wu at the Small Molecule Metabolomics Core Laboratory (Institute of Plant and Microbial Biology, Academia Sinica, Taipei, Taiwan) for advice on lipidomic analysis, members of the Proteomic core (Institute of Plant and Microbial Biology, Academia Sinica, Taipei, Taiwan) for proteomic analyses and discussion, and former members Mr. Tze-Yu Tsao and Ms. Jean-Yun Chang for technical assistance.
This work was supported by the Ministry of Science and Technology (grants 103-2633-B-001-003, 104-2633-B-001-001, and 108-2311-B-001-023) and Academia Sinica intramural funds and a career development award (to C.-W. Wang).
The authors declare no competing financial interests.
Author contributions: C.-W. Wang conceived and designed the experiments. W.-C. Su analyzed lipids and contributed Figs. 2 and 3 G; Fig. 6, A and D; and Fig. S1, D–F; Y.-H. Lin performed the pull-down experiments and contributed Fig. 3, B and C; Fig. 4; Fig. 6, B, C, E, and F; and Figs. 7 C, 8 C, and S3 along with C.-W. Wang. M. Pagac contributed Fig. 3, E and F, and performed cloning for some BiFC constructs. C.-W. Wang contributed data shown in Figs. 1 and 5; Fig. 7, A and B; Fig. 8, A, B, and D–F; Fig. S1, A–C; and Fig. S2. C.-W. Wang interpreted the data and wrote the paper.
References
- Alexaki A., Clarke B.A., Gavrilova O., Ma Y., Zhu H., Ma X., Xu L., Tuymetova G., Larman B.C., Allende M.L., et al. 2017. De Novo Sphingolipid Biosynthesis Is Required for Adipocyte Survival and Metabolic Homeostasis. J. Biol. Chem. 292:3929–3939. 10.1074/jbc.M116.756460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersuker K., Peterson C.W.H., To M., Sahl S.J., Savikhin V., Grossman E.A., Nomura D.K., and Olzmann J.A.. 2018. A Proximity Labeling Strategy Provides Insights into the Composition and Dynamics of Lipid Droplet Proteomes. Dev. Cell. 44:97–112. 10.1016/j.devcel.2017.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikman B.T., and Summers S.A.. 2011. Sphingolipids and hepatic steatosis. Adv. Exp. Med. Biol. 721:87–97. 10.1007/978-1-4614-0650-1_6 [DOI] [PubMed] [Google Scholar]
- Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pagès F., Trajanoski Z., and Galon J.. 2009. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 25:1091–1093. 10.1093/bioinformatics/btp101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow D.K., Collins S.R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A., Ejsing C.S., and Weissman J.S.. 2010. Orm family proteins mediate sphingolipid homeostasis. Nature. 463:1048–1053. 10.1038/nature08787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright B.R., and Goodman J.M.. 2012. Seipin: from human disease to molecular mechanism. J. Lipid Res. 53:1042–1055. 10.1194/jlr.R023754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerantola V., Guillas I., Roubaty C., Vionnet C., Uldry D., Knudsen J., and Conzelmann A.. 2009. Aureobasidin A arrests growth of yeast cells through both ceramide intoxication and deprivation of essential inositolphosphorylceramides. Mol. Microbiol. 71:1523–1537. 10.1111/j.1365-2958.2009.06628.x [DOI] [PubMed] [Google Scholar]
- Chauhan N., Han G., Somashekarappa N., Gable K., Dunn T., and Kohlwein S.D.. 2017. Regulation of sphingolipid biosynthesis by the morphogenesis checkpoint kinase Swe1. J. Biol. Chem. 292:9431 10.1074/jbc.A115.693200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaurasia B., and Summers S.A.. 2015. Ceramides - Lipotoxic Inducers of Metabolic Disorders. Trends Endocrinol. Metab. 26:538–550. 10.1016/j.tem.2015.07.006 [DOI] [PubMed] [Google Scholar]
- Currie E., Guo X., Christiano R., Chitraju C., Kory N., Harrison K., Haas J., Walther T.C., and Farese R.V. Jr. 2014. High confidence proteomic analysis of yeast LDs identifies additional droplet proteins and reveals connections to dolichol synthesis and sterol acetylation. J. Lipid Res. 55:1465–1477. 10.1194/jlr.M050229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D., Kannan M., and Wattenberg B.. 2018. Orm/ORMDL proteins: Gate guardians and master regulators. Adv. Biol. Regul. 70:3–18. 10.1016/j.jbior.2018.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D.L., Gable K., Suemitsu J., Dunn T.M., and Wattenberg B.W.. 2019. The ORMDL/Orm-serine palmitoyltransferase (SPT) complex is directly regulated by ceramide: Reconstitution of SPT regulation in isolated membranes. J. Biol. Chem. 294:5146–5156. 10.1074/jbc.RA118.007291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V., and Weissman J.S.. 2007. A molecular caliper mechanism for determining very long-chain fatty acid length. Cell. 130:663–677. 10.1016/j.cell.2007.06.031 [DOI] [PubMed] [Google Scholar]
- Dickson R.C., Sumanasekera C., and Lester R.L.. 2006. Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Prog. Lipid Res. 45:447–465. 10.1016/j.plipres.2006.03.004 [DOI] [PubMed] [Google Scholar]
- Dittrich F., Zajonc D., Hühne K., Hoja U., Ekici A., Greiner E., Klein H., Hofmann J., Bessoule J.J., Sperling P., and Schweizer E.. 1998. Fatty acid elongation in yeast--biochemical characteristics of the enzyme system and isolation of elongation-defective mutants. Eur. J. Biochem. 252:477–485. 10.1046/j.1432-1327.1998.2520477.x [DOI] [PubMed] [Google Scholar]
- Eisenberg T., and Büttner S.. 2014. Lipids and cell death in yeast. FEMS Yeast Res. 14:179–197. 10.1111/1567-1364.12105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J.Y., Cui Z.Q., Wei H.P., Zhang Z.P., Zhou Y.F., Wang Y.P., and Zhang X.E.. 2008. Split mCherry as a new red bimolecular fluorescence complementation system for visualizing protein-protein interactions in living cells. Biochem. Biophys. Res. Commun. 367:47–53. 10.1016/j.bbrc.2007.12.101 [DOI] [PubMed] [Google Scholar]
- Fei W., Shui G., Gaeta B., Du X., Kuerschner L., Li P., Brown A.J., Wenk M.R., Parton R.G., and Yang H.. 2008. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 180:473–482. 10.1083/jcb.200711136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W., Li H., Shui G., Kapterian T.S., Bielby C., Du X., Brown A.J., Li P., Wenk M.R., Liu P., and Yang H.. 2011a Molecular characterization of seipin and its mutants: implications for seipin in triacylglycerol synthesis. J. Lipid Res. 52:2136–2147. 10.1194/jlr.M017566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W., Shui G., Zhang Y., Krahmer N., Ferguson C., Kapterian T.S., Lin R.C., Dawes I.W., Brown A.J., Li P., et al. 2011b A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 7:e1002201 10.1371/journal.pgen.1002201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J., Lees M., and Sloane Stanley G.H.. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226:497–509. [PubMed] [Google Scholar]
- Funato K., and Riezman H.. 2001. Vesicular and nonvesicular transport of ceramide from ER to the Golgi apparatus in yeast. J. Cell Biol. 155:949–959. 10.1083/jcb.200105033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gable K., Slife H., Bacikova D., Monaghan E., and Dunn T.M.. 2000. Tsc3p is an 80-amino acid protein associated with serine palmitoyltransferase and required for optimal enzyme activity. J. Biol. Chem. 275:7597–7603. 10.1074/jbc.275.11.7597 [DOI] [PubMed] [Google Scholar]
- Grippa A., Buxó L., Mora G., Funaya C., Idrissi F.Z., Mancuso F., Gomez R., Muntanyà J., Sabidó E., and Carvalho P.. 2015. The seipin complex Fld1/Ldb16 stabilizes ER-lipid droplet contact sites. J. Cell Biol. 211:829–844. 10.1083/jcb.201502070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X.L., and Wenk M.R.. 2006. Mass spectrometry-based profiling of phospholipids and sphingolipids in extracts from Saccharomyces cerevisiae. Yeast. 23:465–477. 10.1002/yea.1362 [DOI] [PubMed] [Google Scholar]
- Guillas I., Kirchman P.A., Chuard R., Pfefferli M., Jiang J.C., Jazwinski S.M., and Conzelmann A.. 2001. C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. EMBO J. 20:2655–2665. 10.1093/emboj/20.11.2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gururaj C., Federman R.S., and Chang A.. 2013. Orm proteins integrate multiple signals to maintain sphingolipid homeostasis. J. Biol. Chem. 288:20453–20463. 10.1074/jbc.M113.472860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G., Gupta S.D., Gable K., Bacikova D., Sengupta N., Somashekarappa N., Proia R.L., Harmon J.M., and Dunn T.M.. 2019. The ORMs interact with transmembrane domain 1 of Lcb1 and regulate serine palmitoyltransferase oligomerization, activity and localization. Biochim Biophys Acta Mol Cell Biol Lipids. 1864:245–259. 10.1016/j.bbalip.2018.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Lone M.A., Schneiter R., and Chang A.. 2010. Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc. Natl. Acad. Sci. USA. 107:5851–5856. 10.1073/pnas.0911617107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Binns D.D., Chang Y.F., and Goodman J.M.. 2015. Dissecting seipin function: the localized accumulation of phosphatidic acid at ER/LD junctions in the absence of seipin is suppressed by Sei1p(ΔNterm) only in combination with Ldb16p. BMC Cell Biol. 16:29 10.1186/s12860-015-0075-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K. 2003. Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim. Biophys. Acta. 1632:16–30. 10.1016/S1388-1981(03)00059-3 [DOI] [PubMed] [Google Scholar]
- Hanada K., and Nishijima M.. 2003. Purification of mammalian serine palmitoyltransferase, a hetero-subunit enzyme for sphingolipid biosynthesis, by affinity-peptide chromatography. Methods Mol. Biol. 228:163–174. [DOI] [PubMed] [Google Scholar]
- Iqbal J., Walsh M.T., Hammad S.M., and Hussain M.M.. 2017. Sphingolipids and Lipoproteins in Health and Metabolic Disorders. Trends Endocrinol. Metab. 28:506–518. 10.1016/j.tem.2017.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier N., Choudhary V., Mari M., Toulmay A., Reggiori F., and Schneiter R.. 2011. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Sci. 124:2424–2437. 10.1242/jcs.076836 [DOI] [PubMed] [Google Scholar]
- Kihara A. 2012. Very long-chain fatty acids: elongation, physiology and related disorders. J. Biochem. 152:387–395. 10.1093/jb/mvs105 [DOI] [PubMed] [Google Scholar]
- Kim S.K., Noh Y.H., Koo J.R., and Yun H.S.. 2010. Effect of expression of genes in the sphingolipid synthesis pathway on the biosynthesis of ceramide in Saccharomyces cerevisiae. J. Microbiol. Biotechnol. 20:356–362. [PubMed] [Google Scholar]
- Kohlwein S.D., Eder S., Oh C.S., Martin C.E., Gable K., Bacikova D., and Dunn T.. 2001. Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:109–125. 10.1128/MCB.21.1.109-125.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Jiang Q., Wang X., Zhang Y., Lin R.C., Lam S.M., Shui G., Zhou L., Li P., Wang Y., et al. 2014. Adipose-specific knockout of SEIPIN/BSCL2 results in progressive lipodystrophy. Diabetes. 63:2320–2331. 10.2337/db13-0729 [DOI] [PubMed] [Google Scholar]
- Liu M., Huang C., Polu S.R., Schneiter R., and Chang A.. 2012. Regulation of sphingolipid synthesis through Orm1 and Orm2 in yeast. J. Cell Sci. 125:2428–2435. 10.1242/jcs.100578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loizides-Mangold U., Clément S., Alfonso-Garcia A., Branche E., Conzelmann S., Parisot C., Potma E.O., Riezman H., and Negro F.. 2014. HCV 3a core protein increases lipid droplet cholesteryl ester content via a mechanism dependent on sphingolipid biosynthesis. PLoS One. 9:e115309 10.1371/journal.pone.0115309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Magré J., Delépine M., Khallouf E., Gedde-Dahl T. Jr., Van Maldergem L., Sobel E., Papp J., Meier M., Mégarbané A., Bachy A., et al. BSCL Working Group . 2001. Identification of the gene altered in Berardinelli-Seip congenital lipodystrophy on chromosome 11q13. Nat. Genet. 28:365–370. 10.1038/ng585 [DOI] [PubMed] [Google Scholar]
- Morrison W.R., and Smith L.M.. 1964. Preparation of Fatty Acid Methyl Esters and Dimethylacetals from Lipids with Boron Fluoride–Methanol. J. Lipid Res. 5:600–608. [PubMed] [Google Scholar]
- Nagiec M.M., Wells G.B., Lester R.L., and Dickson R.C.. 1993. A suppressor gene that enables Saccharomyces cerevisiae to grow without making sphingolipids encodes a protein that resembles an Escherichia coli fatty acyltransferase. J. Biol. Chem. 268:22156–22163. [PubMed] [Google Scholar]
- Olson D.K., Fröhlich F., Farese R.V. Jr., and Walther T.C.. 2016. Taming the sphinx: Mechanisms of cellular sphingolipid homeostasis. Biochim. Biophys. Acta. 1861(8, 8 Pt B):784–792. 10.1016/j.bbalip.2015.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagac M., Cooper D.E., Qi Y., Lukmantara I.E., Mak H.Y., Wu Z., Tian Y., Liu Z., Lei M., Du X., et al. 2016. SEIPIN Regulates Lipid Droplet Expansion and Adipocyte Development by Modulating the Activity of Glycerol-3-phosphate Acyltransferase. Cell Reports. 17:1546–1559. 10.1016/j.celrep.2016.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S., Gable K., Beaudoin F., Cahoon E., Jaworski J., Napier J.A., and Dunn T.M.. 2006. Members of the Arabidopsis FAE1-like 3-ketoacyl-CoA synthase gene family substitute for the Elop proteins of Saccharomyces cerevisiae. J. Biol. Chem. 281:9018–9029. 10.1074/jbc.M507723200 [DOI] [PubMed] [Google Scholar]
- Piña F., Yagisawa F., Obara K., Gregerson J.D., Kihara A., and Niwa M.. 2018. Sphingolipids activate the endoplasmic reticulum stress surveillance pathway. J. Cell Biol. 217:495–505. 10.1083/jcb.201708068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Snider J., Airola M.V., Zhong A., Rana N.A., Obeid L.M., and Hannun Y.A.. 2018. Quantification of 3-ketodihydrosphingosine using HPLC-ESI-MS/MS to study SPT activity in yeast Saccharomyces cerevisiae. J. Lipid Res. 59:162–170. 10.1194/jlr.D078535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants F.M., Breslow D.K., Muir A., Weissman J.S., and Thorner J.. 2011. Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 108:19222–19227. 10.1073/pnas.1116948108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassa T., and Kihara A.. 2014. Metabolism of very long-chain Fatty acids: genes and pathophysiology. Biomol. Ther. (Seoul). 22:83–92. 10.4062/biomolther.2014.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorling S., Vallée B., Barz W.P., Riezman H., and Oesterhelt D.. 2001. Lag1p and Lac1p are essential for the Acyl-CoA-dependent ceramide synthase reaction in Saccharomyces cerevisae. Mol. Biol. Cell. 12:3417–3427. 10.1091/mbc.12.11.3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkal C.E., Salama M.F., Snider A.J., Allopenna J.J., Rana N.A., Koller A., Hannun Y.A., and Obeid L.M.. 2017. Ceramide Is Metabolized to Acylceramide and Stored in Lipid Droplets. Cell Metab. 25:686–697. 10.1016/j.cmet.2017.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimobayashi M., Oppliger W., Moes S., Jenö P., and Hall M.N.. 2013. TORC1-regulated protein kinase Npr1 phosphorylates Orm to stimulate complex sphingolipid synthesis. Mol. Biol. Cell. 24:870–881. 10.1091/mbc.e12-10-0753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Ramachandran S.K., Zhu J., Kim B.C., Biswas D., Ha T., Iglesias P.A., and Li R.. 2017. Sphingolipids facilitate age asymmetry of membrane proteins in dividing yeast cells. Mol. Biol. Cell. 28:2712–2722. 10.1091/mbc.e17-05-0335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui X., Arlt H., Brock K.P., Lai Z.W., DiMaio F., Marks D.S., Liao M., Farese R.V. Jr., and Walther T.C.. 2018. Cryo-electron microscopy structure of the lipid droplet-formation protein seipin. J. Cell Biol. 217:4080–4091. 10.1083/jcb.201809067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Miao Y., Yamane Y., Zhang C., Shokat K.M., Takematsu H., Kozutsumi Y., and Drubin D.G.. 2012. Orm protein phosphoregulation mediates transient sphingolipid biosynthesis response to heat stress via the Pkh-Ypk and Cdc55-PP2A pathways. Mol. Biol. Cell. 23:2388–2398. 10.1091/mbc.e12-03-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung M.K., and Huh W.K.. 2007. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast. 24:767–775. 10.1002/yea.1504 [DOI] [PubMed] [Google Scholar]
- Surma M.A., Klose C., Peng D., Shales M., Mrejen C., Stefanko A., Braberg H., Gordon D.E., Vorkel D., Ejsing C.S., et al. 2013. A lipid E-MAP identifies Ubx2 as a critical regulator of lipid saturation and lipid bilayer stress. Mol. Cell. 51:519–530. 10.1016/j.molcel.2013.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski K.M., Binns D., Bartz R., Grishin N.V., Li W.P., Agarwal A.K., Garg A., Anderson R.G., and Goodman J.M.. 2007. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. USA. 104:20890–20895. 10.1073/pnas.0704154104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Bi J., Shui G., Liu Z., Xiang Y., Liu Y., Wenk M.R., Yang H., and Huang X.. 2011. Tissue-autonomous function of Drosophila seipin in preventing ectopic lipid droplet formation. PLoS Genet. 7:e1001364 10.1371/journal.pgen.1001364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong M., Longato L., Ramirez T., Zabala V., Wands J.R., and de la Monte S.M.. 2014. Therapeutic reversal of chronic alcohol-related steatohepatitis with the ceramide inhibitor myriocin. Int. J. Exp. Pathol. 95:49–63. 10.1111/iep.12052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Quiroz F., García-Marqués S., Coria R., Randez-Gil F., and Prieto J.A.. 2010. The activity of yeast Hog1 MAPK is required during endoplasmic reticulum stress induced by tunicamycin exposure. J. Biol. Chem. 285:20088–20096. 10.1074/jbc.M109.063578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voynova N.S., Vionnet C., Ejsing C.S., and Conzelmann A.. 2012. A novel pathway of ceramide metabolism in Saccharomyces cerevisiae. Biochem. J. 447:103–114. 10.1042/BJ20120712 [DOI] [PubMed] [Google Scholar]
- Walther T.C., Chung J., and Farese R.V. Jr. 2017. Lipid Droplet Biogenesis. Annu. Rev. Cell Dev. Biol. 33:491–510. 10.1146/annurev-cellbio-100616-060608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.W., and Lee S.C.. 2012. The ubiquitin-like (UBX)-domain-containing protein Ubx2/Ubxd8 regulates lipid droplet homeostasis. J. Cell Sci. 125:2930–2939. 10.1242/jcs.100230 [DOI] [PubMed] [Google Scholar]
- Wang C.W., Miao Y.H., and Chang Y.S.. 2014. Control of lipid droplet size in budding yeast requires the collaboration between Fld1 and Ldb16. J. Cell Sci. 127:1214–1228. 10.1242/jcs.137737 [DOI] [PubMed] [Google Scholar]
- Wiśniewski J.R., Zougman A., Nagaraj N., and Mann M.. 2009. Universal sample preparation method for proteome analysis. Nat. Methods. 6:359–362. 10.1038/nmeth.1322 [DOI] [PubMed] [Google Scholar]
- Wolinski H., Hofbauer H.F., Hellauer K., Cristobal-Sarramian A., Kolb D., Radulovic M., Knittelfelder O.L., Rechberger G.N., and Kohlwein S.D.. 2015. Seipin is involved in the regulation of phosphatidic acid metabolism at a subdomain of the nuclear envelope in yeast. Biochim. Biophys. Acta. 1851:1450–1464. 10.1016/j.bbalip.2015.08.003 [DOI] [PubMed] [Google Scholar]
- Xia J.Y., Holland W.L., Kusminski C.M., Sun K., Sharma A.X., Pearson M.J., Sifuentes A.J., McDonald J.G., Gordillo R., and Scherer P.E.. 2015. Targeted Induction of Ceramide Degradation Leads to Improved Systemic Metabolism and Reduced Hepatic Steatosis. Cell Metab. 22:266–278. 10.1016/j.cmet.2015.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji-Hasegawa A., Takahashi A., Tetsuka Y., Senoh Y., and Kobayashi T.. 2005. Fungal metabolite sulfamisterin suppresses sphingolipid synthesis through inhibition of serine palmitoyltransferase. Biochemistry. 44:268–277. 10.1021/bi048605l [DOI] [PubMed] [Google Scholar]
- Yan R., Qian H., Lukmantara I., Gao M., Du X., Yan N., and Yang H.. 2018. Human SEIPIN Binds Anionic Phospholipids. Dev. Cell. 47:248–256. [DOI] [PubMed] [Google Scholar]
- Yu T., Zhou Y.J., Wenning L., Liu Q., Krivoruchko A., Siewers V., Nielsen J., and David F.. 2017. Metabolic engineering of Saccharomyces cerevisiae for production of very long chain fatty acid-derived chemicals. Nat. Commun. 8:15587 10.1038/ncomms15587 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.