Super-resolution imaging combined with quantitative image analyses reveals dynamic spatial pattern formation of centriolar Plk4, a master regulator of centriole duplication. The self-organization properties of Plk4 exclusively provide the single site for centriole formation around the preexisting centriole.
Abstract
In each cell cycle, centrioles are duplicated to produce a single copy of each preexisting centriole. At the onset of centriole duplication, the master regulator Polo-like kinase 4 (Plk4) undergoes a dynamic change in its spatial pattern around the preexisting centriole, forming a single duplication site. However, the significance and mechanisms of this pattern transition remain unknown. Using super-resolution imaging, we found that centriolar Plk4 exhibits periodic discrete patterns resembling pearl necklaces, frequently with single prominent foci. Mathematical modeling and simulations incorporating the self-organization properties of Plk4 successfully generated the experimentally observed patterns. We therefore propose that the self-patterning of Plk4 is crucial for the regulation of centriole duplication. These results, defining the mechanisms of self-organized regulation, provide a fundamental principle for understanding centriole duplication.
Introduction
Centrioles are duplicated once in every cell cycle, and only a single copy is generated from each preexisting centriole in each duplication (Banterle and Gönczy, 2017; Nigg and Holland, 2018), which ensures bipolar mitotic spindle assembly and thus optimal regulation of the cell cycle. However, the mechanisms underlying the regulation of the number of centriole copies are largely unknown. In human cells, three centriolar proteins, namely, Polo-like kinase 4 (Plk4), STIL, and HsSAS6, have been identified as the core components involved in coordinating the onset of centriole duplication (Banterle and Gönczy, 2017; Nigg and Holland, 2018). In the early G1 phase, Plk4 localizes in a biased ring-like pattern surrounding the proximal periphery of the mother centriole (Ohta et al., 2014, 2018), using CEP152 as a scaffold (Cizmecioglu et al., 2010; Dzhindzhev et al., 2010; Hatch et al., 2010). Following the centriolar recruitment of STIL and HsSAS6, the ring-like pattern of Plk4 changes dynamically into a single focus containing the STIL–HsSAS6 complex through positive and negative regulation (Ohta et al., 2014, 2018; Arquint et al., 2015). Although the Plk4 focus exclusively provides the site for cartwheel assembly and subsequent procentriole formation, the significance and molecular mechanisms underlying the dynamic pattern transition of Plk4 are yet to be addressed.
A recent study revealed that Plk4 possesses intrinsic self-organization properties, such as self-assembly and the promotion of dissociation/degradation of neighboring Plk4 molecules in an autophosphorylation-dependent manner (Yamamoto and Kitagawa, 2019). This finding suggests that Plk4 itself may generate the bias in the spatial patterns around centrioles, before loading of STIL and HsSAS6. As Plk4 is the master regulator of centriole duplication (Bettencourt-Dias et al., 2005; Habedanck et al., 2005), self-organized generation of bias may play an important role in the onset of the duplication process. In this regard, quantitative imaging at higher spatial resolutions may allow the detailed analysis of the spatial patterns of centriolar Plk4. In addition, a plausible and experimentally verifiable theory to fundamentally explain the mechanisms underlying centriole duplication is yet to be developed. For example, the link between the molecular nature of Plk4 and its dynamic pattern transition in centrioles remains unclear. A number of experimental and theoretical studies have demonstrated that biological systems commonly comprise self-organization mechanisms at the cellular and tissue levels (Halatek et al., 2018; Saha et al., 2018; Sych et al., 2018; Wheeler and Hyman, 2018). Thus, it is interesting and important to reveal the implications of the self-patterning of Plk4 in the pericentriolar nanospace for centriole duplication, and consequently for cellular function. Based on previous findings related to the self-organizing properties of Plk4 (Yamamoto and Kitagawa, 2019), we here propose the first theory of centriole duplication mechanisms by integrating experimental data and mathematical modeling.
Results and discussion
The centriolar localization pattern of Plk4 is dynamic and synchronized with the cell cycle
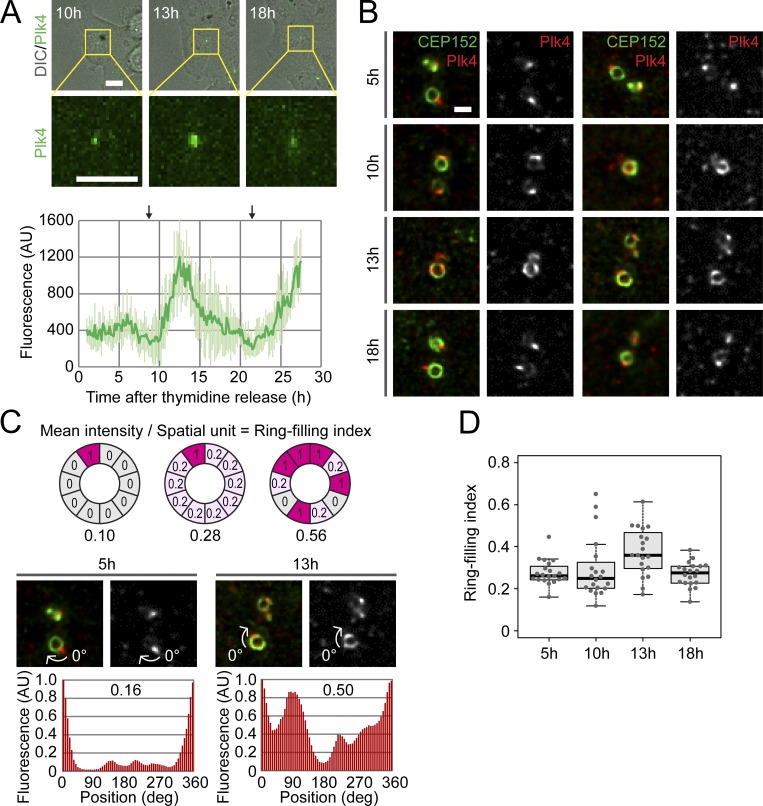
During centriole duplication, Plk4 forms a ring-like pattern around the mother centriole and, subsequently, this pattern changes into a single focus, which provides the single duplication site (Ohta et al., 2014, 2018; Arquint et al., 2015). To quantitatively confirm this observation during the actual process of the cell cycle, we monitored centriolar Plk4 dynamics in live cells and correlated them with spatial patterns in fixed cells at a higher resolution. HCT116 cells, in which endogenous Plk4 was tagged with mClover, were synchronized via thymidine arrest, then observed via spinning disc confocal microscopy after thymidine release (Fig. 1 A and Fig. S1 A). Within 1 h after mitotic exit, during the putative early G1 phase, accumulation of Plk4 around the centrioles became apparent (Fig. 1 A and Fig. S1, A and B). Centriolar Plk4–mClover signals continued to increase for 3–5 h after mitotic exit, and then began to decrease as the cells progressed toward the next mitosis (Fig. 1 A).
Figure 1.
Dynamic changes in the localization pattern of Plk4 during centriole duplication. (A) Live imaging of Plk4 endogenously tagged with mClover in HCT116 cells, using spinning-disc confocal microscopy. DIC, differential interference contrast. The graph shows the mean ± SD fluorescence of Plk4–mClover (n = 5 cells). The arrows above the graph indicate approximate time points of metaphase. Scale bars, 10 µm. (B) Confocal immunofluorescence images of endogenous Plk4, with deconvolution. Synchronized cells were fixed at the indicated time after thymidine release and stained with antibodies against Plk4 and CEP152 (centriolar marker). Two representative images are shown for each time point. Scale bar, 0.5 µm. (C and D) Quantification of the Plk4 patterns. Representative oval profiles of Plk4 and the associated ring-filling indices (C) and a plot of all ring-filling indices (D) are shown. Representative images in C are from B. Schematics in C show how ring-filling indices are calculated. AU, arbitrary units; deg, degrees.
Given the timing of the appearance of the Plk4 ring pattern (early G1 phase, before the formation of the daughter centriole) and the single-focus pattern (following the entry of STIL and HsSAS6), the increase and subsequent decrease in the Plk4–mClover signal may reflect the formation and disappearance of the ring pattern, respectively. Indeed, when observed at higher resolution via immunofluorescence confocal microscopy, the Plk4 ring pattern most frequently appeared 13 h after thymidine release. This corresponds to the time when the peaks in the Plk4–mClover signal were most frequently detected in the live cells (Fig. 1, A and B). At the onset of the increase in the Plk4–mClover signal in the live cells (10 h), only a small proportion of the fixed cells showed the ring pattern at the higher resolution. When the Plk4–mClover signal decreased as the cells progressed toward mitosis (5 or 18 h), the single-focus pattern became more dominant.
Interestingly, through careful observation, we found that the Plk4 rings in the putative early G1 phase were frequently incomplete. Some resembled Landolt rings (Fig. 1 B, 10 h and 13 h, right panels), while others resembled partial arcs (Fig. 1 B, 10 h, left panel). Even in the single-focus pattern, the Plk4 sometimes localized weakly in rings (Fig. 1 B, 18 h). Therefore, instead of merely classifying the patterns into ring or single-focus, we defined a parameter that describes the degree of ring formation and termed it the “ring-filling index” (Fig. 1 C). Consequently, we confirmed that these indices reached their peak 13 h after thymidine release and subsequently decreased (Fig. 1 D). Combined, these findings demonstrate in a quantitative manner that Plk4 changes its spatial patterns around centrioles dynamically, in synchronization with the cell cycle.
Stimulated emission depletion (STED) super-resolution microscopy revealed discrete and periodic patterns of centriolar Plk4 localization
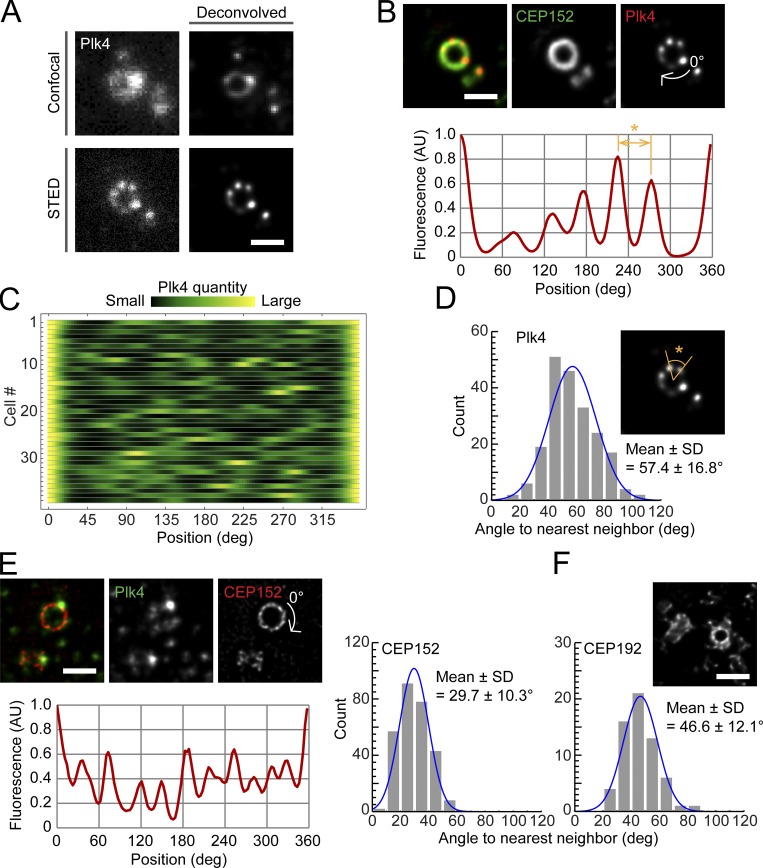
We used STED super-resolution microscopy to further investigate the incomplete ring patterns of Plk4 at higher resolution. In STED observations, to our surprise, we detected discontinuity in the ring patterns (Fig. 2 A). Instead of the expected continuous ring, we observed a chain of foci forming a discrete ring (resembling a pearl necklace; Fig. 2 A). It is unlikely that these discrete patterns merely reflect transition states between a continuous ring and a single focus, as none of our STED images acquired in this experiment captured continuous rings.
Figure 2.
Discrete and periodic Plk4 ring patterns at the super-resolution scale. (A and B) Comparison of the resolution of conventional confocal and STED images of centriolar Plk4 (from different samples) with or without deconvolution (A) and the oval profile of Plk4 (B). Scale bars, 0.5 µm. (C) An array plot of all STED profiles of Plk4 (n = 37 cells). (D) Histogram of angles between nearest-neighbor pairs of Plk4 foci in discrete ring patterns. The angle shown in the inset STED image (*) corresponds to the distance indicated in yellow (*) in the profile in B. (E) Representative STED images and quantification of CEP152 spatial patterns. Details as for B and D. n = 30 cells. Scale bar, 0.5 µm. (F) Representative STED image and quantification of CEP192 spatial patterns. n = 10 cells. Scale bar, 0.5 µm.
We then extracted oval fluorescence intensity profiles to quantitatively analyze the Plk4 ring patterns (Fig. 2, B and C). In line with previous reports (Ohta et al., 2014, 2018), in most cases we found uneven distribution patterns with single prominent peaks. In addition, as illustrated in Fig. 2 B, the profiles exhibited spatial periodicity. We detected the peaks of the profiles and measured the angle (the distance in the profile plots) from each peak to its nearest neighbor to quantify the periodicity of the discrete ring patterns. Similar methods have been used successfully to analyze protein localization patterns in the ciliary transition zone or centriole distal appendages (Shi et al., 2017; Jana et al., 2018; Yang et al., 2018). The average angle between nearest-neighbor pairs was ∼60° (Fig. 2 D). In other words, the discrete ring patterns tended to show a sixfold rotational symmetry. Autocorrelation analysis similarly demonstrated that the period of the discrete pattern was 65.0 ± 16.4° (Fig. S1 C). This was surprising, given the ninefold rotational symmetry of the centriole core architecture and the triplet microtubules. Therefore, an interpretation other than that based on the ninefold symmetry of the centriole architecture is required to explain the periodicity of the discrete ring patterns of Plk4.
The scaffold of Plk4 potentially has double its rotational symmetry and number of slots
We applied the super-resolution analysis to CEP152, the main scaffold of centriolar Plk4, to identify the factor generating the sixfold symmetry of the discrete ring patterns of Plk4. Via STED observations using an antibody to the N-terminal region of CEP152, we found that CEP152 also showed periodic patterns (Fig. 2 E). Unlike Plk4, we did not find obvious bias in the periodic patterns of CEP152, which was consistent with previous observations (Ohta et al., 2018). Interestingly, the patterns of CEP152 showed higher spatial frequencies than those of Plk4. The average angle between nearest-neighbor peaks was ∼30° (Fig. 2 E). CEP152 therefore exhibits a rotational symmetry of order 12, which is double that of Plk4. Since the periodicity of the CEP152 localization patterns was not entirely clear, we conducted a further analysis for verification (Fig. S1, D and E). Combined, these results suggest that the scaffold CEP152 provides 12 slots for Plk4, but Plk4 appears to preferentially select six of them.
Besides the CEP152 scaffold, CEP192 serves as a scaffold of Plk4 at centrioles, and it is suggested that Plk4 switches its scaffold protein from CEP192 to CEP152 during centriole duplication processes (Kim et al., 2013; Park et al., 2014). We therefore similarly quantified the spatial pattern of centriolar CEP192 by STED analysis. Interestingly, centriolar CEP192 showed periodic patterns at intervals close to 40° (Fig. 2 F). As the diameter of centriolar CEP192 is much smaller than those of CEP152 and Plk4 (Fig. 2, B, E, and F), it is unclear whether the spatial pattern of centriolar CEP192 affects that of Plk4 through a direct interaction.
Simulation models based on the self-organization of Plk4 via a lateral inhibition (LI) effect reproduce its observed spatial patterns around centrioles
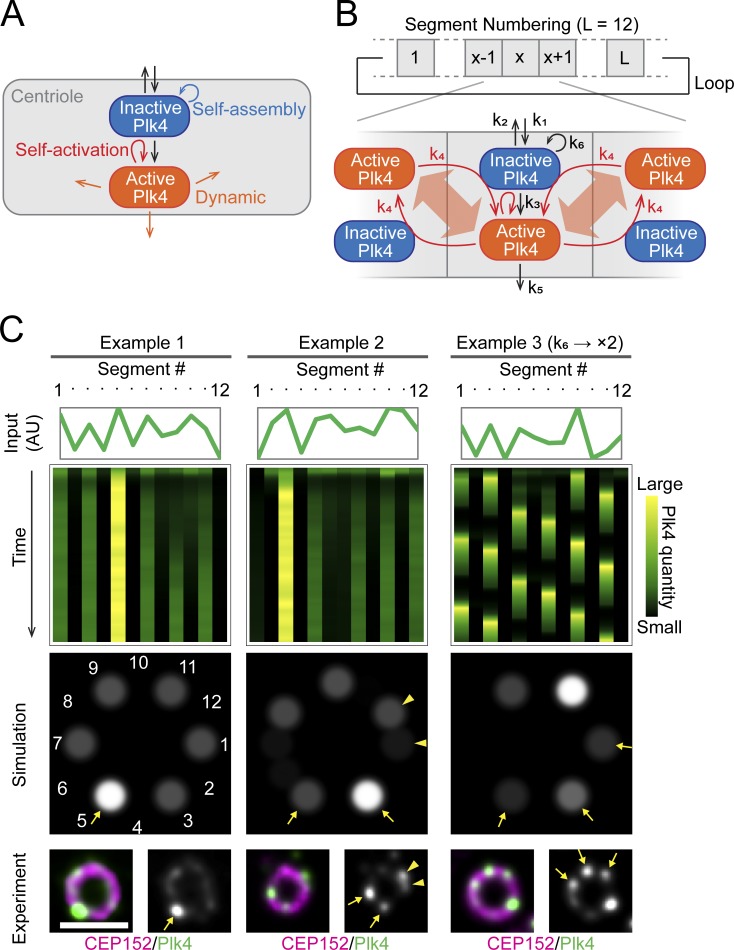
We then investigated how the discrete ring patterns of Plk4, with six uneven foci, are generated from the 12 slots of CEP152. To address this, we reasoned that the molecular nature of Plk4, which has recently been revealed (Yamamoto and Kitagawa, 2019), may provide a clue. First, Plk4 self-assembles into a macromolecular complex (or protein aggregate). Second, its properties can switch, depending on its autophosphorylation state. Of note, the phosphorylated (“active”) form of Plk4 is more dynamic in dissociating from centrioles than the nonphosphorylated (“inactive”) form. In addition, the active form of Plk4 promotes the dissociation of inactive Plk4 through self-activation (trans-autophosphorylation) feedback. These properties of Plk4 suggest that it may form the discrete ring patterns by excluding neighboring Plk4 molecules in an autophosphorylation-dependent manner, whereas existing Plk4 complexes gain their mass through self-assembly. This mutual relationship between the active and inactive forms of Plk4 is similar to that of the reaction–diffusion (RD) system (also known as the Turing model) or its analogue, the LI system, which forms periodic patterns at the cellular or tissue level (Nakamura et al., 2006; Kondo and Miura, 2010; Barad et al., 2011; Liao and Oates, 2017). Accordingly, here we propose the first verifiable theory explaining the mechanism by which the self-organization properties of Plk4 form dynamic spatial patterns and exclusively provide the single duplication site during centriole duplication. The core concept of this theory, which is based on a recently proposed speculative model (Yamamoto and Kitagawa, 2019), is shown in Fig. 3 A.
Figure 3.
Simulations of the pattern formation of Plk4 based on the LI model. (A) The core concept of the theoretical model. The self-organization properties of Plk4 result in its lateral-inhibition self-patterning behavior. (B) Schematic drawing of the LI model. (C) Examples of simulation results. Array plots showing the time course of pattern formation, with the initial quantities of Plk4 indicated in the line graphs above (Input), and the spatial pattern at the last time point (Simulation) are shown for each example. Representative STED images resembling the simulation results are shown at the bottom (Experiment). The arrows and arrowheads indicate the similarities in the patterns between the simulation and the actual observation. The basic parameter setting was used for examples 1 and 2, but k6 was doubled for example 3. Scale bar, 0.5 µm.
Our theory assumes that the active form of Plk4 is dynamic and mobile within the periphery of centrioles. These properties render the active Plk4 capable of reaching and interacting with distant Plk4 molecules along the centriole periphery. This can cause a LI effect, in which active Plk4 molecules repel nearby inactive Plk4 molecules by promoting activation and subsequent dissociation. Assuming this LI effect, we constructed a mathematical model to simulate the pattern formation of Plk4 (Fig. 3 B; see Materials and methods for details). Simulations starting with random patterns resulted in the formation of approximately six uneven foci of Plk4 that tended to occupy alternate slots to form the distinct “pearl necklace” pattern (Fig. 3 C). Notably, most of the discrete ring patterns generated in the simulations had single prominent foci, similarly to those in the STED images. In the model, the balanced influx and efflux result in the limited pool of centriolar Plk4. In addition, the model assumed that the dissociation of centriolar Plk4 is attenuated once a certain quantity of Plk4 aggregate is formed in a given segment. This assumption is based on the concept that molecules on the surface are energetically more diffusible than the ones present inside a mass. The attenuation function was defined as a sigmoid that limits the dissociation rate of Plk4, depending on its quantity. The strength of this attenuation effect significantly contributed to the generation of the bias, while the sharpness of the effect did not (Fig. S2 A). Assuming these properties of Plk4, once one of the Plk4 foci becomes prominent at random, the focus accumulates more and more Plk4 molecules and wins the race of LI.
The pattern may be more dynamic, depending on the parameters used. For example, when the k6 value was twice that used in examples 1 and 2 (Fig. 3 C), the spatiotemporal patterning of Plk4 became more dynamic and resulted in a multiple oscillation mode (example 3). Nevertheless, it still exhibited discrete patterns with a prominent peak at each time point. This variation in the spatial and temporal patterns simulated may explain the variety of the spatial patterns of Plk4 observed in our STED analysis. The LI model proposed in this study thus reproduces the observed Plk4 patterns at centrioles well.
Given the somewhat unclear periodicity of the CEP152 spatial patterns, we tested the robustness of the model with respect to the periodicity of the scaffolds. All simulations using different numbers of slots (9, 10, 11, or 13) resulted in the formation of pearl necklace patterns (Fig. S2 B), as in the simulations using 12 slots (Fig. 3, B and C). The LI effect of Plk4 is thus not strictly dependent on the number of slots, and this model allows flexibility in scaffold conformation. Furthermore, when we more explicitly considered the lateral diffusion of Plk4 within the periphery of the centriole (Fig. S2 C; see Materials and methods for details), simulations also resulted in the formation of discrete ring patterns, regardless of the number of segments (Fig. S2 D). Together, these results further support our theory that Plk4 can self-organize into the patterns around centrioles by its intrinsic properties (Fig. 3 A). Hereafter, we adopt and further verify the LI model (Fig. 3 B) as a tentative model.
The LI model reproduces centriole duplication under physiological and perturbed conditions
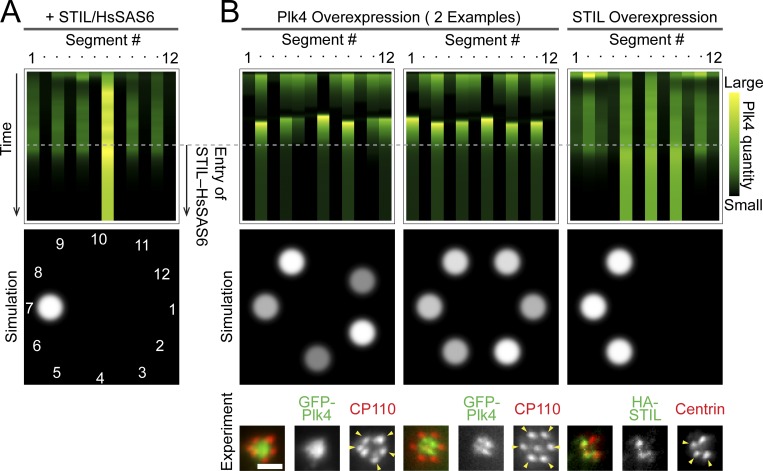
Using the LI model, we then simulated the pattern transition of Plk4 after the centriolar entry of STIL and HsSAS6 at the onset of procentriole formation (Ohta et al., 2014, 2018). Since the mechanism through which STIL and HsSAS6 cooperatively regulate the pattern transition to restrict the duplication site remains unclear, in the model, we only included the centriolar entry of STIL, using it to represent the STIL–HsSAS6 complex. The time-evolved simulations were set as follows: (1) initially, the random seeds of Plk4 evolved to form discrete ring patterns via the LI effect, as shown in Fig. 3 C; (2) next, the expression level of STIL began to increase, as observed in the late G1 to S phases in human cells (Izraeli et al., 1997; Tang et al., 2011; Arquint et al., 2012; Arquint and Nigg, 2014); (3) finally, STIL entered the Plk4 slots in which the quantity of active (phosphorylated) Plk4 was above a given threshold as the centriolar loading of STIL has been reported to depend on Plk4-mediated phosphorylation (Dzhindzhev et al., 2014, 2017; Ohta et al., 2014; Moyer et al., 2015; McLamarrah et al., 2018). The other condition we considered in the simulations was that, once the cytosolic level of STIL had begun to increase, the cytosolic level of Plk4 was assumed to decrease, as increased quantities of STIL in the cytoplasm have been suggested to promote the degradation of Plk4 (Arquint et al., 2015; Moyer et al., 2015; Ohta et al., 2018).
Consequently, simulations using the LI model reproduced the Plk4 pattern transition from the discrete ring into the single focus upon centriolar loading of the STIL–HsSAS6 complex (Fig. 4 A). The most prominent focus of Plk4 in the rings tended to remain after STIL–HsSAS6 loading. In addition, simulations including negative-feedback regulation mediated by the Plk4–STIL interaction (Ohta et al., 2018) yielded similar results but with slightly faster transition from the discrete ring into the single focus after centriolar loading of the STIL–HsSAS6 complex (Fig. S2 E).
Figure 4.
Simulations using centriolar entry of STIL–HsSAS6 and exogenous perturbation. (A) Representative simulation results using the entry of the STIL–HsSAS6 complex. (B) Representative simulation results using overexpression of Plk4 or STIL. Representative experimental data showing similar patterns are shown at the bottom. The arrowheads in the images indicate the duplicated centrioles. Scale bar, 1 µm.
Next, we simulated a perturbation of centriole duplication through overexpression of the components and compared the simulated results with experimental results to further verify this model. Overexpression of Plk4, STIL, or HsSAS6 is known to induce overduplication of centrioles (Habedanck et al., 2005; Kleylein-Sohn et al., 2007; Strnad et al., 2007; Tang et al., 2011; Arquint et al., 2012; Vulprecht et al., 2012). In simulations using overexpression of Plk4 (the cytosolic level of Plk4 was set five times higher), multiple Plk4 foci tended to remain even after STIL–HsSAS6 loading. This was consistent with the experimental results (Fig. 4 B). This is due to increased influx of Plk4 into the centriole, resulting in multiple foci retaining Plk4 quantities above the threshold of STIL binding. Despite this increased influx, the LI effect remained. Interestingly, under overexpression of Plk4, the simulated Plk4 patterns were relatively unstable throughout the time course (Fig. 4 B), until they were stabilized via the entry of STIL. Overexpression of STIL was then simulated by setting a lower threshold (one fifth) of STIL binding to Plk4 foci. Under this condition, several Plk4 foci tended to remain after STIL–HsSAS6 loading, which was also consistent with our experimental observations (Fig. 4 B). Interestingly, the numbers of stabilized Plk4 foci in the overexpression simulations did not exceed six, potentially explaining the limitation in the maximum number of overduplicated daughter centrioles. Indeed, most previous studies have convincingly demonstrated that six is the maximum number of daughter centrioles that can be formed on a single mother centriole (Habedanck et al., 2005; Kleylein-Sohn et al., 2007; Strnad et al., 2007; Tang et al., 2011; Arquint et al., 2012; Vulprecht et al., 2012), although it is also possible that this may be merely because of space limitations. Importantly, we provide the first theoretical explanation and prediction of the mechanisms involved in centriole overduplication.
Together, we conclude that the model successfully reproduced our observations regardless of experimental conditions. This further supports our self-patterning theory (Fig. 3 A).
Inhibition of Plk4 activity resulted in unexpected patterns, implying flexibility in the pattern of the Plk4 scaffold
Assuming that Plk4 is converted into its active form via autophosphorylation, inhibition of its kinase activity may decelerate the activation of Plk4 and diminishes the LI effect (Fig. 3 A). Using lower activation rates, our model predicted that Plk4 would occupy all of the available slots and exhibit 12 foci symmetrically arranged around a centriole (data not shown). In contrast, upon treatment with the Plk4 inhibitor centrinone (Wong et al., 2015), Plk4 exhibited ninefold rotationally symmetrical spatial patterns in our STED observations (Fig. S3 A). Interestingly, in the presence of centrinone, the middle part of CEP152 exhibited patterns closer to ninefold symmetry (40° intervals), whereas the N-terminal part continued to hold ∼12 slots (30° intervals) at the centriole (Fig. S3 B). Collectively, these findings suggest that the inhibition of kinase activity changes the mode of Plk4–CEP152 interaction. Considering the somewhat unclear periodicity of CEP152, further investigation using electron microscopy or perhaps expansion microscopy (Chen et al., 2015; Chang et al., 2017), besides STED microscopy, may provide more information about the relationship between the activity of Plk4 and its scaffold.
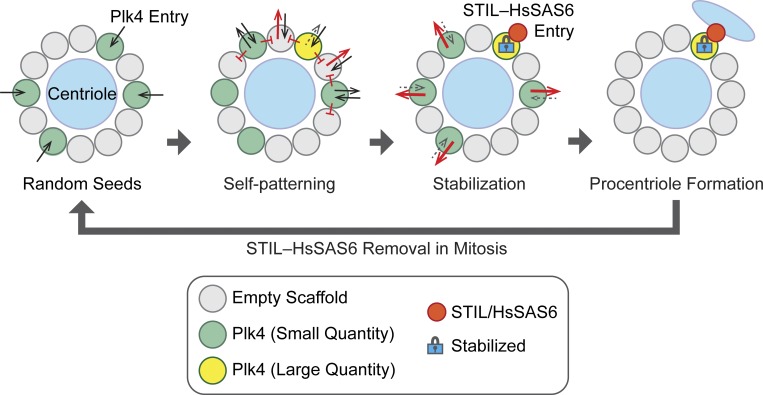
The self-patterning theory of centriole duplication
As schematically shown in Fig. 5, our theory explains the involvement of molecular dynamics in the determination of centriole duplication sites. In this model, Plk4 first forms randomly distributed seeds, and subsequent competition via the LI effect results in the appearance of discrete ring patterns. The model also includes the self-assembly property of Plk4 (Yamamoto and Kitagawa, 2019), by which condensed Plk4 attenuates its dissociation rate. Therefore, the slot that recruits the greatest quantity of Plk4 at the onset is favored, in that it prevents neighboring slots from recruiting Plk4 molecules and tends to survive as the largest Plk4 focus via first-come-first-served and the-rich-grow-richer processes (“Self-patterning” in Fig. 5). Through stochastic means, STIL and HsSAS6 preferentially bind to the largest Plk4 focus in the discrete ring, leading to stabilization. This assumption is reasonable, given that stochastic interactions between Plk4 and STIL–HsSAS6 are probably biased such that the more Plk4 there is in a focus, the more frequently the molecules interact with each other. Increasing expression levels of STIL in turn promote the degradation of cytosolic Plk4, resulting in decreased entry of Plk4 into the centriole. Given that the population of phosphorylated Plk4 in centrioles increases after centriolar loading of the STIL–HsSAS6 complex (Ohta et al., 2018; Yamamoto and Kitagawa, 2019), it is also conceivable that the negative-feedback regulation based on the Plk4–STIL interaction promotes the dissociation/degradation of Plk4 around the mother centriole wall, except at the duplication site (Ohta et al., 2018). Consequently, the largest Plk4 focus remains as the only site of centriole duplication.
Figure 5.
Model of centriole duplication. The self-patterning theory explains how the self-organization properties of Plk4 result in the formation of the biased discrete ring patterns to provide the single centriole duplication sites.
According to our self-patterning theory, Plk4 can independently form periodic discrete patterns, i.e., potential duplication sites. Most importantly, because of its intrinsic properties, Plk4 alone can generate bias from random seeds in the periphery of preexisting centrioles, suggesting that this is the first spontaneous symmetry breaking in centriole duplication. However, the self-patterning of Plk4 exhibits fluctuation and plasticity until it is stabilized by STIL and HsSAS6. Indeed, recent theoretical work has suggested the importance of a STIL-mediated mechanism in the symmetry breaking of Plk4 (Leda et al., 2018). Their model has similarities with ours in autoamplification and competition mechanisms, while the scope is different. The core concept of our models is that the intrinsic properties of Plk4 can drive the symmetry breaking, which is essential as the biased pattern of Plk4 can be observed before centriolar loading of STIL, and even in STIL-depleted cells (Ohta et al., 2014; Yamamoto and Kitagawa, 2019). However, apart from the initial symmetry-breaking event, both models may come to the same conclusion, that the STIL-mediated mechanism plays a critical role in completing the site selection for centriole duplication. Such a two-step process—the self-patterning of Plk4 and the subsequent stabilization by STIL and/or HsSAS6—may serve as backup and buffer mechanisms to ensure the precise regulation of centriole duplication. In biological systems, it is commonly observed that initial symmetry-breaking steps induce weak bias, and subsequent feedback mechanisms robustly fix the asymmetry (Goryachev and Leda, 2017; Chen et al., 2018; Kim et al., 2018). Instead of forming the definitive duplication site from the beginning, the weakly biased ring patterns of Plk4 may provide backup sites, while the entry of STIL–HsSAS6 ensures that only one site is determined for centriole duplication. In this regard, it is interesting that the spatiotemporal patterning of Plk4 may be more dynamic, depending on the parameters used in simulations (Fig. 3 C, Example 3). Thus, our theory predicts the possibility that potential centriole duplication sites switch dynamically within the periphery of centrioles, although such a system may be too unstable. The development of future techniques enabling live-cell super-resolution imaging may provide the answer. Alternatively, addition of other concepts, such as liquid–liquid phase separation (Woodruff et al., 2017), may significantly improve our models. While our theory sheds light on numerous experimental observations, further research based on both experimental data and further improvement of the present models may assist in reaching definitive conclusions.
Materials and methods
Key resources
Antibodies
Antibodies used were Plk4 (1:250; MABC544; Merck), CEP152-N (1:1,000; A302-479A; Bethyl), CEP152-Mid (1:1,000; A302-480A; Bethyl), GFP (1:500; A11120; Invitrogen), HA (1:500; ab9110; Abcam), CP110 (1:1,000; 12780–1-AP; Proteintech), and centrin (1:1,000; 04–1624; Merck).
Chemicals
Chemicals used were thymidine (T1895; Sigma-Aldrich) and centrinone (HY-18682; MedChem Express).
Experimental model and subject details
Cell lines
HCT116 cells were cultured in McCoy’s 5A medium (GE Healthcare) supplied with 10% FBS, 1% glutamine, and 1% penicillin/streptomycin. U2OS cells were cultured in DMEM supplied with 10% FBS and 1% penicillin/streptomycin. For synchronization of the cell cycle, 2 mM thymidine was added to the medium, as indicated in Fig. S1 A. For Plk4 inhibition, cells were incubated for 4 h in the presence of 200 nM centrinone before fixation.
Cells were transfected with DNA or siRNA using Lipofectamine 2000 or Lipofectamine RNAiMAX, according to the manufacturer’s instructions.
The HCT116 Plk4–mClover cell line was produced via CRISPR-Cas9 genome editing. The mClover sequence was inserted into the 3′ region of the Plk4 gene, and cell clones were selected using hygromycin. Cloned cells were genotyped using PCR, and the proper localization of expressed Plk4–mClover was verified via immunofluorescence. We were only able to obtain a monoallelic cell line, which we therefore used in this study.
Method details
Immunofluorescence
Cells cultured on coverslips were fixed using cold methanol at −20°C for 5 min. The fixed cells were washed three times with PBS and incubated in blocking buffer (1% BSA and 0.05% Triton X-100 in PBS) for 20 min at RT. The cells were then incubated with primary antibodies in blocking buffer at 4°C overnight, washed three times with PBS, and incubated with secondary antibodies in blocking buffer for 1 h at RT. The cells were stained with Hoechst 33258 (DOJINDO) in PBS for 5 min at RT, washed three times with PBS, and subsequently mounted with ProLong Gold (P36930; Thermo Fisher Scientific).
Microscopy
For general observations, an upright epifluorescence microscope (Axio Imager 2; Zeiss) with a 100× oil-immersion objective (NA 1.4) and an AxioCam HRm camera, or an inverted confocal microscope (Leica TCS SP8) equipped with a 63× oil-immersion objective (NA 1.4), was used. Z-stacked confocal images were obtained at 0.13-µm intervals.
For live-cell imaging, a spinning disc–based confocal microscope (CV1000; Yokogawa) equipped with a 60× oil-immersion objective (NA 1.35), a back-illuminated electron-multiplying charge-coupled device camera, and a stage incubator supplied with 5% CO2 was used. Typically, 10–30 fields of view were recorded every 10 min for up to 30 h in a single experiment, and each field contained 25 z-slices with 1.3-µm intervals, subsequently max-projected using ImageJ software.
For STED observations, a STED system based on an inverted confocal microscope (TCS SP8 STED; Leica) equipped with 592/660 STED laser lines and a 100× oil-immersion objective (NA 1.4) was used. The interval of optical sectioning was 0.18 µm. In STED observations, U2OS cells were used instead of HCT116 cells, because they provided better immunofluorescence images and there were no obvious differences in the analyzed data (data not shown).
The Huygens Essential or Professional image processing software was used for image deconvolution, and ImageJ software was used for image processing and analyses.
Mathematical modeling
Our mathematical models were based on the molecular nature of Plk4 (Fig. 3 A), which has recently been revealed (Yamamoto and Kitagawa, 2019). Given the periodicity of CEP152 scaffolds (Fig. 2 E), we typically placed 12 segments as Plk4 slots around the centriole. The last segment (x = 12) was connected to the first (x = 1) to form a closed-loop centriolar ring (Fig. 3 B). As described above, Plk4 can take two distinct forms: active and inactive. The inactive form influxes from the cytosol (with the kinetic constant k1) to the centriolar segments, and both forms dissociate from the centriole (with the distinct kinetic constants k2 and k5, respectively). The active form was set to be more dynamic in its dissociation (k5 > k2) and mobile in its interaction with other Plk4 molecules in the neighboring segments (bold orange arrows in Fig. 3 B). The inactive form self-assembles (k6) to cause a positive-feedback effect and turns into the active form through autophosphorylation (k3). The self-assembly rate is proportional to the concentration of Plk4 in each segment and in the cytosol. Therefore, all Plk4 influx into each segment was included in the self-assembly term. The active form of Plk4 promotes the activation of adjacent inactive Plk4 in its own segment and in neighboring segments (k4). This promotes the dissociation of activated Plk4, generating the LI effect. Importantly, when the total amount of Plk4 in a segment reaches a given threshold, the molecules are assumed to form a stable complex (aggregate; Yamamoto and Kitagawa, 2019), which reduces the dissociation rates of both forms of Plk4. A random fluctuation (±10%) was added to the self-assembly term, so the results differ in each simulation. The initial seeds (input) were provided as random real numbers between 0 and 1. It should be noted that the model does not distinguish between dissociation and degradation, so the observed dissociation may be partly degradation. The units for the kinetic constants and time are arbitrary.
To simulate centriole duplication, we constructed two mathematical models as follows. For convenience, here the models are termed the LI and the RD models. Both models are based on the intrinsic properties of Plk4 that allow it to self-assemble and promote the dissociation/degradation of neighboring molecules following its activation. The units, including that used for time, are arbitrary (relative). Although no actual data are available at present, the parameter sets used in this study may be realistic when standard units are supposed; for example, micrometers for the length unit and s−1 for the kinetic constants given the data reported for other systems, e.g., phosphorylation by the serine-threonine protein kinase S6K1 (Keshwani and Harris, 2008). As schematically shown in Fig. 3 B and Fig. S2 C, there are only slight differences between the models. Specifically, the RD model more explicitly considers the lateral diffusion of Plk4 molecules within the periphery of the centriole. The periphery of the centriole (the scaffold of Plk4) is divided into segments (x), such that the last segment (x = L) is connected to the first (x = 1) to form a closed loop. L was typically assigned a value of 12. Plk4 can exist in the active (autophosphorylated) or the inactive form. In the LI model, the rate of change in the concentration of Plk4 at segment x and time t (Ix(t) for the inactive form and Ax(t) for the active form) is expressed using the following partial differential equations:
and
where N is a noise parameter, Icyto(t) is the cytosolic concentration of inactive Plk4, limits the centriolar influx of Plk4, and and express the sigmoidal deceleration of Plk4 dissociation (termed here “the dissociation-attenuation function”) caused by the decreased surface-to-volume ratio upon self-assembly. Representative plots of the dissociation-attenuation function and the dependency of the model on the function are shown in Fig. S2 A. The parameter ks(x) decreases as STIL enters the segment and stabilizes Plk4. Notably, in this model, the centriolar loading of STIL was biased according to the Plk4 bias within the discrete ring pattern. In other words, Plk4–STIL interactions at centrioles were assumed to occur stochastically, as in general protein–protein interactions. Specifically, in the presence of cytosolic STIL, when the quantity of active Plk4 in segment x, Ax(t) exceeds a given threshold (typically 0.5), STIL enters the segment and ks(x) decreases by 25%. Icyto(t) is kept constant in the absence of STIL and starts exponential decrease (time constant = 50) once the centriolar loading of STIL starts. The prominent Plk4 focus contained the active form of Plk4 most abundantly among the segments as previously observed for phosphorylated Plk4 at centrioles (Yamamoto and Kitagawa, 2019), while simulation data only for total quantity of Plk4 (inactive + active) are shown for simplicity (e.g., Fig. 3 C). The initial values of the inactive form, Ix(0), are provided as a list of random real numbers between 0 and 1, whereas those of the active form Ax(t) are set to 0. The basic parameter settings are listed in Table S1.
Next, to test the validity of the core concept that the self-organization properties of Plk4 alone can cause the discrete ring patterns to form, we tested how altering the mobility of Plk4 would affect the model. For this alternative mathematical model, the RD model, we used the core concept shown in Fig. 3 A, but more explicitly considered the lateral diffusion of Plk4 within the periphery of the centriole (Fig. S2 C). This is possible since the pericentriolar space is filled with a number of proteins and therefore serves as a diffusion trap to tether Plk4 around the centrioles. Similarly to the LI model (Fig. 3 B), the active form of Plk4 was assumed to be more dynamic (i.e., larger diffusion coefficient) than the inactive form (Fig. S2 C and Table S2). The active form repels the inactive form through self-activation feedback, although only within the same segment (Fig. S2 C). Given that the RD model explicitly includes diffusion formulae, the concentration changes are expressed as:
and
where DI and DA are the diffusion coefficients for the inactive and active forms of Plk4, respectively, and dx is the distance between each segment. The basic parameter settings are listed in Table S2.
The simultaneous differential equations were numerically solved using our original Mathematica program. Specifically, the total time course was divided into 50 time windows, and the equations were sequentially solved in each time window. The noise parameter N (a random real number between 0.9 and 1.1) and ks(x) were updated at the onset of each time window. The initial value of ks(x) was 1 in all segments. Ax(t) was monitored at the onset of every time window and if Ax(t) > 0.5, then ks(x) was updated such that ks(x) = 0.75 ⋅ ks(x). The numerical solutions of each time window were inherited as the initial values to the next time window. The program also generated the time-evolved simulation graphs and the simulated spatial distributions shown in Figs. 3, 4, and S2. For simplicity, the representative results illustrated show the total quantities of Plk4 (i.e., both inactive and active), even though each focus typically included a certain proportion of active Plk4.
The RD model is basically similar to the LI model (Fig. 3 B) and, indeed, consistently generated the discrete ring patterns (Fig. S2 D). The LI model (Fig. 3 B) assumes that Plk4 molecules can diffuse only into the neighboring segments, which can be therefore regarded as a limited and simplified RD model. Despite the similarities and consistency in these two models, however, the LI model seems more feasible by reducing parameters as little is known about the actual properties of diffusion of Plk4 in the pericentriolar space.
Quantification and statistical analysis
All quantification and statistical analyses were performed using ImageJ, Mathematica, R, and Excel software. For live imaging, the fluorescence intensity of regions of interest of the same size was measured using ImageJ on maximum projection images, and the fluorescence intensity of a no-cell region was used for background subtraction (Fig. 1 A). The oval profiles of Plk4 and CEP152 were measured using the Oval Profile Plot plugin with the “Along Oval” option, as shown in Fig. 2, B and E. The number of sampling points was set to 64 for conventional confocal images or 128 for STED images, depending on image resolution. For the calculation of the ring-filling indices (Fig. 1 C), the profiles were exported to and processed in Excel. Using CEP152 as a marker, we determined the ring-filling indices based on normalized oval fluorescence intensity profiles of Plk4 (Fig. 1 C). The index for a complete ring was 1, and if 50% of the circumference around the centriole was filled with Plk4, the ring-filling index was 0.5. The box-and-whisker plots in Fig. 1 D were generated using R. The profiles were exported to Mathematica to generate the 2D array plot shown in Fig. 2 C. Using our Mathematica programs, the peaks in the profiles were detected by calculating the local maxima, and the angles (distances) between nearest-neighbor pairs of foci were obtained, as shown in Fig. 2, D and E. In addition, we developed a Mathematica program to obtain autocorrelation functions (Fig. S2 A). The original data were zero-padded for the calculations.
Online supplemental material
Fig. S1 shows the time course of cell synchronization and analyses for periodicity of spatial patterns of centriolar Plk4 and CEP152. Fig. S2 shows simulations with alternative parameter sets or assumptions. Fig. S3 shows the effect of the inhibition of kinase activity on the pattern formation of Plk4. Table S1 shows basic parameter settings for the LI model. Table S2 shows basic parameter settings for the RD model.
Supplementary Material
Acknowledgments
We gratefully acknowledge T. Fujiwara and Institute for Integrated Cell-Material Sciences, Kyoto University, for technical support in STED microscopy; A. Kimura and D. Miyashiro for advice on mathematical modeling; M. Ohta, K. Watanabe, S. Yoshiba, and Y. Tsuchiya for technical support in the generation of the Plk4–mClover cell line and STED imaging of Plk4; and the members of the Kitagawa laboratory for technical support, discussion, and critical review of the manuscript.
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (C [18K06233] and S [19H05651]), by the Uehara Memorial Foundation, and by the Japan Prize Foundation.
The authors declare no competing financial interests.
Author contributions: D. Kitagawa conceived the study. D. Takao and D. Kitagawa designed the study and experiments. D. Takao performed all the experiments and simulations. D. Takao analyzed the data. D. Takao and D. Kitagawa interpreted the data. D. Takao and D. Kitagawa wrote the manuscript. S. Yamamoto participated in the incubation of the core concept of the theory and reviewed the manuscript.
References
- Arquint C., and Nigg E.A.. 2014. STIL microcephaly mutations interfere with APC/C-mediated degradation and cause centriole amplification. Curr. Biol. 24:351–360. 10.1016/j.cub.2013.12.016 [DOI] [PubMed] [Google Scholar]
- Arquint C., Sonnen K.F., Stierhof Y.-D., and Nigg E.A.. 2012. Cell-cycle-regulated expression of STIL controls centriole number in human cells. J. Cell Sci. 125:1342–1352. 10.1242/jcs.099887 [DOI] [PubMed] [Google Scholar]
- Arquint C., Gabryjonczyk A.M., Imseng S., Böhm R., Sauer E., Hiller S., Nigg E.A., and Maier T.. 2015. STIL binding to Polo-box 3 of PLK4 regulates centriole duplication. eLife. 10.7554/eLife.07888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banterle N., and Gönczy P.. 2017. Centriole Biogenesis: From Identifying the Characters to Understanding the Plot. Annu. Rev. Cell Dev. Biol. 33:23–49. 10.1146/annurev-cellbio-100616-060454 [DOI] [PubMed] [Google Scholar]
- Barad O., Hornstein E., and Barkai N.. 2011. Robust selection of sensory organ precursors by the Notch-Delta pathway. Curr. Opin. Cell Biol. 23:663–667. 10.1016/j.ceb.2011.09.005 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M.K., Carmo N., Balloux F., Callaini G., and Glover D.M.. 2005. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15:2199–2207. 10.1016/j.cub.2005.11.042 [DOI] [PubMed] [Google Scholar]
- Chang J.-B., Chen F., Yoon Y.-G., Jung E.E., Babcock H., Kang J.S., Asano S., Suk H.-J., Pak N., Tillberg P.W., et al. . 2017. Iterative expansion microscopy. Nat. Methods. 14:593–599. 10.1038/nmeth.4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Tillberg P.W., and Boyden E.S.. 2015. Optical imaging. Expansion microscopy. Science. 347:543–548. 10.1126/science.1260088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Shi J., Tao Y., and Zernicka-Goetz M.. 2018. Tracing the origin of heterogeneity and symmetry breaking in the early mammalian embryo. Nat. Commun. 9:1819 10.1038/s41467-018-04155-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cizmecioglu O., Arnold M., Bahtz R., Settele F., Ehret L., Haselmann-Weiss U., Antony C., and Hoffmann I.. 2010. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J. Cell Biol. 191:731–739. 10.1083/jcb.201007107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev N.S., Yu Q.D., Weiskopf K., Tzolovsky G., Cunha-Ferreira I., Riparbelli M., Rodrigues-Martins A., Bettencourt-Dias M., Callaini G., and Glover D.M.. 2010. Asterless is a scaffold for the onset of centriole assembly. Nature. 467:714–718. 10.1038/nature09445 [DOI] [PubMed] [Google Scholar]
- Dzhindzhev N.S., Tzolovsky G., Lipinszki Z., Schneider S., Lattao R., Fu J., Debski J., Dadlez M., and Glover D.M.. 2014. Plk4 phosphorylates Ana2 to trigger Sas6 recruitment and procentriole formation. Curr. Biol. 24:2526–2532. 10.1016/j.cub.2014.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev N.S., Tzolovsky G., Lipinszki Z., Abdelaziz M., Debski J., Dadlez M., and Glover D.M.. 2017. Two-step phosphorylation of Ana2 by Plk4 is required for the sequential loading of Ana2 and Sas6 to initiate procentriole formation. Open Biol. 7:170247 10.1098/rsob.170247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goryachev A.B., and Leda M.. 2017. Many roads to symmetry breaking: molecular mechanisms and theoretical models of yeast cell polarity. Mol. Biol. Cell. 28:370–380. 10.1091/mbc.e16-10-0739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R., Stierhof Y.-D., Wilkinson C.J., and Nigg E.A.. 2005. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7:1140–1146. 10.1038/ncb1320 [DOI] [PubMed] [Google Scholar]
- Halatek J., Brauns F., and Frey E.. 2018. Self-organization principles of intracellular pattern formation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373:20170107 10.1098/rstb.2017.0107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch E.M., Kulukian A., Holland A.J., Cleveland D.W., and Stearns T.. 2010. Cep152 interacts with Plk4 and is required for centriole duplication. J. Cell Biol. 191:721–729. 10.1083/jcb.201006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izraeli S., Colaizzo-Anas T., Bertness V.L., Mani K., Aplan P.D., and Kirsch I.R.. 1997. Expression of the SIL gene is correlated with growth induction and cellular proliferation. Cell Growth Differ. 8:1171–1179. [PubMed] [Google Scholar]
- Jana S.C., Mendonça S., Machado P., Werner S., Rocha J., Pereira A., Maiato H., and Bettencourt-Dias M.. 2018. Differential regulation of transition zone and centriole proteins contributes to ciliary base diversity. Nat. Cell Biol. 20:928–941. 10.1038/s41556-018-0132-1 [DOI] [PubMed] [Google Scholar]
- Keshwani M.M., and Harris T.K.. 2008. Kinetic mechanism of fully activated S6K1 protein kinase. J. Biol. Chem. 283:11972–11980. 10.1074/jbc.M800114200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.J.Y., Korotkevich E., and Hiiragi T.. 2018. Coordination of Cell Polarity, Mechanics and Fate in Tissue Self-organization. Trends Cell Biol. 28:541–550. 10.1016/j.tcb.2018.02.008 [DOI] [PubMed] [Google Scholar]
- Kim T.-S., Park J.-E., Shukla A., Choi S., Murugan R.N., Lee J.H., Ahn M., Rhee K., Bang J.K., Kim B.Y., et al. . 2013. Hierarchical recruitment of Plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, Cep192 and Cep152. Proc. Natl. Acad. Sci. USA. 110:E4849–E4857. 10.1073/pnas.1319656110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y.-D., and Nigg E.A.. 2007. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 13:190–202. 10.1016/j.devcel.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Kondo S., and Miura T.. 2010. Reaction-Diffusion Model as a Framework for Understanding Biological Pattern Formation. Science. 329:1616–1620. 10.1126/science.1179047 [DOI] [PubMed] [Google Scholar]
- Leda M., Holland A.J., and Goryachev A.B.. 2018. Autoamplification and Competition Drive Symmetry Breaking: Initiation of Centriole Duplication by the PLK4-STIL Network. iScience. 8:222–235. 10.1016/j.isci.2018.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B.-K., and Oates A.C.. 2017. Delta-Notch signalling in segmentation. Arthropod Struct. Dev. 46:429–447. 10.1016/j.asd.2016.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLamarrah T.A., Buster D.W., Galletta B.J., Boese C.J., Ryniawec J.M., Hollingsworth N.A., Byrnes A.E., Brownlee C.W., Slep K.C., Rusan N.M., and Rogers G.C.. 2018. An ordered pattern of Ana2 phosphorylation by Plk4 is required for centriole assembly. J. Cell Biol. 217:1217–1231. 10.1083/jcb.201605106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer T.C., Clutario K.M., Lambrus B.G., Daggubati V., and Holland A.J.. 2015. Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. J. Cell Biol. 209:863–878. 10.1083/jcb.201502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Mine N., Nakaguchi E., Mochizuki A., Yamamoto M., Yashiro K., Meno C., and Hamada H.. 2006. Generation of robust left-right asymmetry in the mouse embryo requires a self-enhancement and lateral-inhibition system. Dev. Cell. 11:495–504. 10.1016/j.devcel.2006.08.002 [DOI] [PubMed] [Google Scholar]
- Nigg E.A., and Holland A.J.. 2018. Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol. 19:297–312. 10.1038/nrm.2017.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Ashikawa T., Nozaki Y., Kozuka-Hata H., Goto H., Inagaki M., Oyama M., and Kitagawa D.. 2014. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat. Commun. 5:5267 10.1038/ncomms6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Watanabe K., Ashikawa T., Nozaki Y., Yoshiba S., Kimura A., and Kitagawa D.. 2018. Bimodal Binding of STIL to Plk4 Controls Proper Centriole Copy Number. Cell Reports. 23:3160–3169.e4. 10.1016/j.celrep.2018.05.030 [DOI] [PubMed] [Google Scholar]
- Park S.Y., Park J.E., Kim T.S., Kim J.H., Kwak M.J., Ku B., Tian L., Murugan R.N., Ahn M., Komiya S., et al. . 2014. Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis. Nat. Struct. Mol. Biol. 21:696–703. 10.1038/nsmb.2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Nagy T.L., and Weiner O.D.. 2018. Joining forces: crosstalk between biochemical signalling and physical forces orchestrates cellular polarity and dynamics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373:20170145 10.1098/rstb.2017.0145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Garcia G. III, Van De Weghe J.C., McGorty R., Pazour G.J., Doherty D., Huang B., and Reiter J.F.. 2017. Super-resolution microscopy reveals that disruption of ciliary transition-zone architecture causes Joubert syndrome. Nat. Cell Biol. 19:1178–1188. 10.1038/ncb3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P., Leidel S., Vinogradova T., Euteneuer U., Khodjakov A., and Gönczy P.. 2007. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev. Cell. 13:203–213. 10.1016/j.devcel.2007.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sych T., Mély Y., and Römer W.. 2018. Lipid self-assembly and lectin-induced reorganization of the plasma membrane. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373:20170117 10.1098/rstb.2017.0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.-J.C., Lin S.-Y., Hsu W.-B., Lin Y.-N., Wu C.-T., Lin Y.-C., Chang C.-W., Wu K.-S., and Tang T.K.. 2011. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J. 30:4790–4804. 10.1038/emboj.2011.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulprecht J., David A., Tibelius A., Castiel A., Konotop G., Liu F., Bestvater F., Raab M.S., Zentgraf H., Izraeli S., and Krämer A.. 2012. STIL is required for centriole duplication in human cells. J. Cell Sci. 125:1353–1362. 10.1242/jcs.104109 [DOI] [PubMed] [Google Scholar]
- Wheeler R.J., and Hyman A.A.. 2018. Controlling compartmentalization by non-membrane-bound organelles. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373:20170193 10.1098/rstb.2017.0193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Y.L., Anzola J.V., Davis R.L., Yoon M., Motamedi A., Kroll A., Seo C.P., Hsia J.E., Kim S.K., Mitchell J.W., et al. . 2015. Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science. 348:1155–1160. 10.1126/science.aaa5111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff J.B., Ferreira Gomes B., Widlund P.O., Mahamid J., Honigmann A., and Hyman A.A.. 2017. The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell. 169:1066–1077.e10. 10.1016/j.cell.2017.05.028 [DOI] [PubMed] [Google Scholar]
- Yamamoto S., and Kitagawa D.. 2019. Self-organization of Plk4 regulates symmetry breaking in centriole duplication. Nat. Commun. 10:1810 10.1038/s41467-019-09847-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T.T., Chong W.M., Wang W.-J., Mazo G., Tanos B., Chen Z., Tran T.M.N., Chen Y.-D., Weng R.R., Huang C.-E., et al. . 2018. Super-resolution architecture of mammalian centriole distal appendages reveals distinct blade and matrix functional components. Nat. Commun. 9:2023 10.1038/s41467-018-04469-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.