The persistence of chemotherapy-induced senescent cancer cells is a barrier to achieving complete response. Tonnessen-Murray et al. reveal that these senescent cells acquire a phagocytic gene expression program and completely engulf and degrade whole cells in the lysosome, enhancing their survival.
Abstract
In chemotherapy-treated breast cancer, wild-type p53 preferentially induces senescence over apoptosis, resulting in a persisting cell population constituting residual disease that drives relapse and poor patient survival via the senescence-associated secretory phenotype. Understanding the properties of tumor cells that allow survival after chemotherapy treatment is paramount. Using time-lapse and confocal microscopy to observe interactions of cells in treated tumors, we show here that chemotherapy-induced senescent cells frequently engulf both neighboring senescent or nonsenescent tumor cells at a remarkable frequency. Engulfed cells are processed through the lysosome and broken down, and cells that have engulfed others obtain a survival advantage. Gene expression analysis showed a marked up-regulation of conserved macrophage-like program of engulfment in chemotherapy-induced senescent cell lines and tumors. Our data suggest compelling explanations for how senescent cells persist in dormancy, how they manage the metabolically expensive process of cytokine production that drives relapse in those tumors that respond the worst, and a function for their expanded lysosomal compartment.
Introduction
In response to chemotherapy, TP53 wild-type human breast tumors rarely undergo pathological complete response (Bertheau et al., 2002, 2007; Chen et al., 2012; Esserman et al., 2012; Nakamura et al., 2012; Wang et al., 2016; Goetz et al., 2017), and these patients have poor survival (Ungerleider et al., 2018). Further research using human patient samples and mouse models has demonstrated that p53 wild-type breast tumors respond to chemotherapy by entering cell cycle arrest and senescence with concomitant expression of cytokines and chemokines of the senescence-associated secretory phenotype (SASP; te Poele et al., 2002; Jackson et al., 2012; Tonnessen-Murray et al., 2018). The SASP can promote tumorigenic properties such as proliferation, survival, stemness, immune evasion, and metastasis (Rodier et al., 2009; Achuthan et al., 2011; Cahu et al., 2012; Canino et al., 2012; Toso et al., 2014; Rao and Jackson, 2016).
At present, it is not clear what imbues the capabilities of survival, persistence, and the metabolically expensive process of SASP production. Here, we show that chemotherapy-induced senescent breast cancer cells are highly enriched for gene expression programs related to macrophages and phagocytosis. Senescent cells engulf neighboring cells and process them to their expanded lysosome compartment, suggesting an abundant source of energy and building blocks for these cells that then drive relapse and poor survival.
Results
Cell-in-cell structures are evident in chemotherapy-induced senescent tumors and cell lines
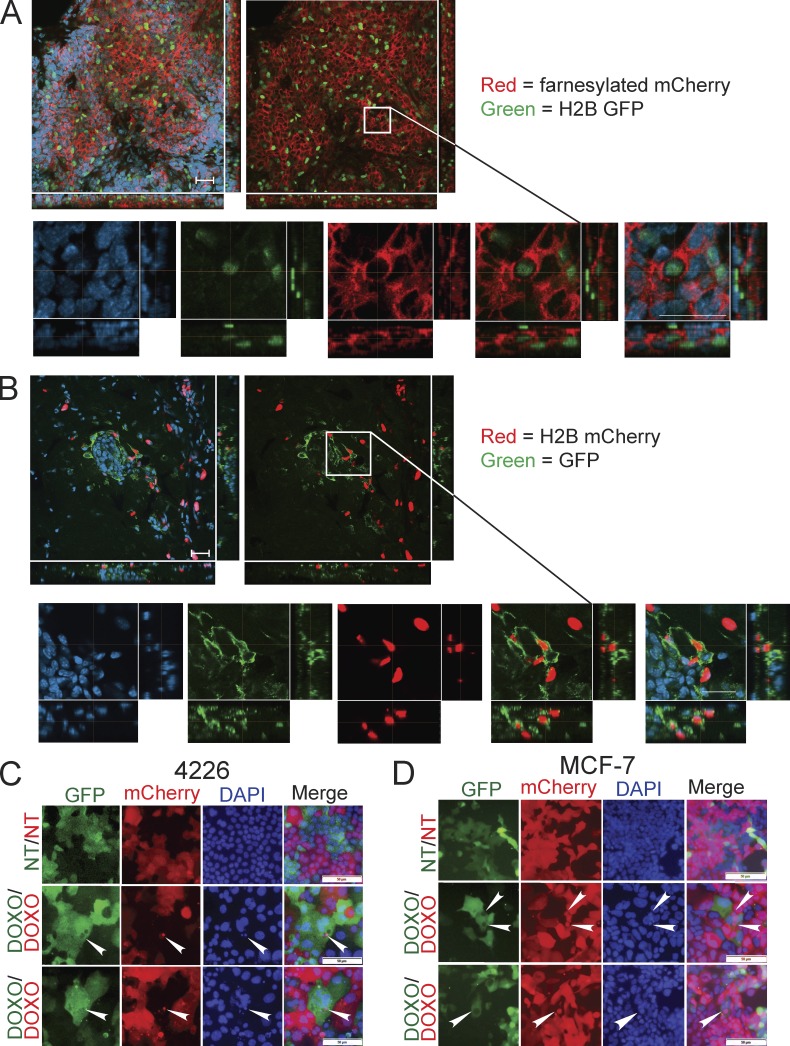
When mice bearing mouse mammary tumor virus (MMTV)–Wnt1 mammary tumors are treated with chemotherapy, senescence and SASP are induced, and tumors have regions where senescent, nonproliferating cells can be extensive and homogeneous, or adjacent to relapsing, Ki67-positive cells (Jackson et al., 2012). To examine interactions among cells in treated mammary tumors, we transplanted p53 wild-type MMTV-Wnt1 tumors that were transduced with lentiviruses expressing various GFP- and mCherry-conjugated markers. After tumors formed, mice were treated with doxorubicin to induce arrest and senescence as previously shown (Jackson et al., 2012; Tonnessen-Murray et al., 2018) and then harvested during the response. Using confocal microscopy of sections, we observed structures consistent with cells engulfed within other cells in the treated tumors. Among these was the red membrane of a cell expressing farnesylated mCherry completely surrounding the nucleus of another cell expressing histone H2B-GFP as viewed by z-stack imaging (Fig. 1 A). In another example, an H2B-mCherry cell appeared to be entirely encapsulated within a cytoplasmic GFP-expressing cell (Fig. 1 B). Cells were determined to be within the other cell when the engulfing “predator” cell completely surrounded the engulfed “prey” cell from all angles. In both examples, the DAPI-stained nuclei of both cells are visible. Examples of images considered negative (appearing engulfed within another cell on one plane of view but not another) are shown in Fig. S1.
Figure 1.
Cell-in-cell structures are observed in doxorubicin-treated mouse mammary tumors and breast cancer cell lines. (A) Cells from an MMTV-Wnt1 mammary tumor were plated and infected ex vivo in separate dishes with lentiviruses expressing either H2B-GFP or farnesylated-mCherry. Cells were orthotopically transplanted into the #4 mammary fat pad of C57BL/6j mice, and after tumors formed, cells were mixed and transplanted again in more mice. After these mosaic tumors formed with cells expressing two different markers, mice were treated with doxorubicin. Tumors were harvested during senescent response, sectioned, DAPI stained, imaged on a confocal microscope in three channels, and merged. Z-stack projections display 14 images taken 2 µm apart with a total z range of 24.12 µm. (B) Cells from a second MMTV-Wnt1 tumor were separately infected with lentiviruses expressing GFP and H2B-mCherry as in A, and mosaic transplants composed of these cells were treated, harvested, and imaged. Z-stack projections display 21 images taken 2 µm apart with a total z range of 38.82 µm. Scale bars in A and B represent 50 and 25 µm for wide-field and zoomed images, respectively. Representative images of cell-in-cell structures are depicted. (C and D) Populations of breast cancer cell lines expressing GFP or mCherry were mixed and treated with doxorubicin (DOXO) for 24 h as stated in Materials and methods. 8 d after DOXO addition, 4226 (C) and MCF-7 (D) cells were fixed and DAPI stained, imaged on a fluorescent microscope in three channels, and merged. White arrowheads denote mCherry vesicles present within GFP-positive, senescent cells. Scale bar represents 50 µm.
Intrigued by these observations, and because of difficulty in quantifying sections of 3D tumors for cell-in-cell structures, we further investigated this engulfment phenotype in mouse and human breast cancer cell lines treated with chemotherapy drugs. 8 d after doxorubicin treatment, MCF-7 and 4226 (a cell line created from a p53 wild-type MMTV-Wnt1 tumor) were ∼70 and 97% positive, respectively, for senescence-associated β-galactosidase (SA-βGAL), a widely used marker of senescence (Dimri et al., 1995; Sharpless and Sherr, 2015), and had increased expression of multiple cytokines and chemokines of the SASP (Fig. S2, A–D). Mixed populations of GFP- and mCherry-expressing cells were plated together for both 4226 and MCF-7 cell lines and then treated with or without doxorubicin for 24 h. 8 d later, remarkably consistent with our observations in vivo (Fig. 1, A and B), we found that large, senescent cells of one color contained large vesicles of the other color. GFP-positive tumor cells containing not just mCherry vesicles but also large, mCherry cells with intact, DAPI-positive nuclei were frequently observed (Fig. 1, C and D, white arrowheads). These vesicles were rarely present in nontreated (NT) proliferating cells (Fig. 1, C and D). These data demonstrate that chemotherapy-induced senescent cells have large vesicles in them that have evidently originated from other, neighboring cells in the culture or tumor.
Chemotherapy-induced senescent cells completely engulf neighboring cells at a high frequency
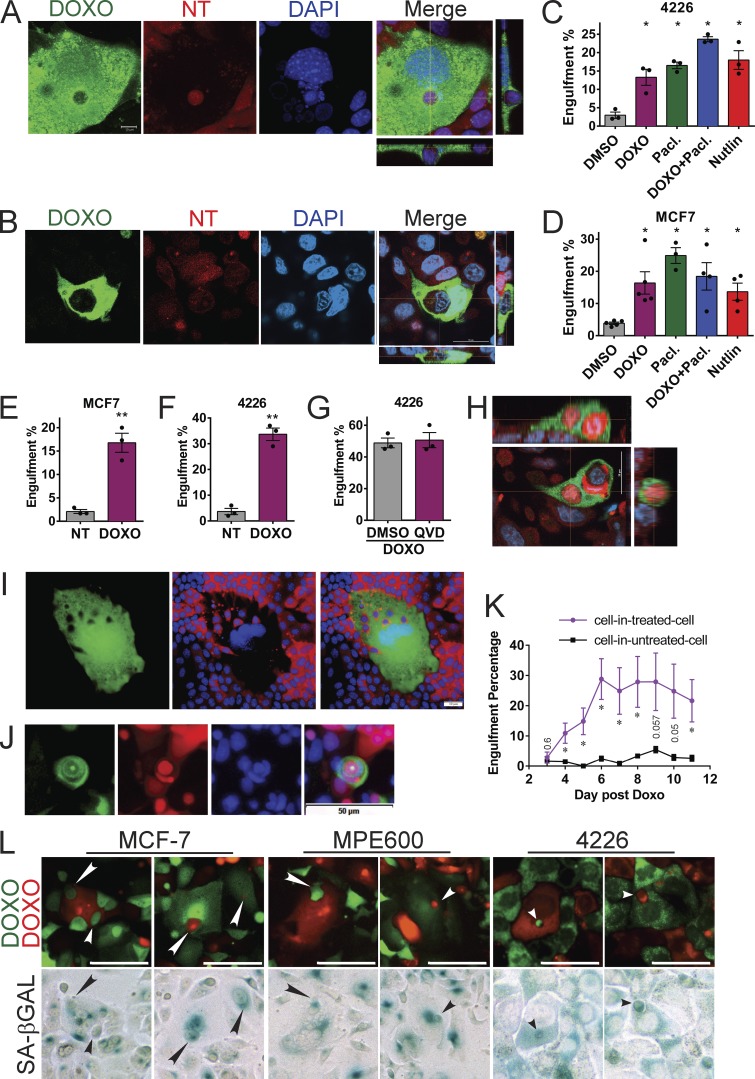
To determine more rigorously if cells that appeared to be engulfed were actually completely within the engulfing cell, we used confocal microscopy. Examining z-stack images, we found that mCherry/DAPI-positive vesicles were indeed completely within senescent, GFP-positive cells, as they were surrounded entirely by GFP-positive cytoplasm, for both 4226 (Fig. 2 A and Video 1) and MCF-7 (Fig. 2 B) cells. To better characterize the origin of the cell-in-cell structures, we captured time-lapse videos of senescent cells mixed with NT cells. We found numerous instances of 4226 (Videos 2, 3, 4, and 5, as indicated in Table S1, video guide) and MCF-7 (Videos 6 and 7) GFP-expressing senescent cells engulfing nonsenescent mCherry cells over the course of 30–92 h. Engulfed mCherry cells were broken down over time and often completely degraded in 2–3 d. In some instances, remnants persisted as smaller mCherry-positive vesicles for up to a week.
Figure 2.
DNA damage, microtubule stress, and ectopic p53 activation induce senescence and engulfment of entire cells. (A and B) Doxorubicin (DOXO)-treated, GFP-expressing 4226 (A) or MCF-7 (B) cells were co-plated on a glass coverslip with proliferating, mCherry-expressing cells. 7 d later, coverslips were fixed with 4% formaldehyde and stained for DAPI. Confocal images were captured, and z-stack projections are shown adjacent; representative images are shown. Scale bar represents 10 µm (A) and 20 µm (B). Z-stack projections display 76 images taken 0.2 µm apart totaling 15 µm for A, and 14 images taken 1 µm apart with a total z range of 12.62 µm for B. (C) Mixed populations of GFP- and mCherry-expressing 4226 cells were plated simultaneously at a 1:8 ratio. When near confluence, cells were treated with DMSO, 0.75 µM doxorubicin (DOXO), 10 nM paclitaxel (pacl.), or both and washed off after 24 h. Nutlin to a concentration of 15 µM was applied every other day until fixation and imaging. Average of four independent experiments is shown, error bars indicate SEM; one-way ANOVA and Newman–Keuls posttest was used to determine significance; *, significant compared with DMSO treated. (D) MCF-7 cells were treated as in C using 0.25 µM doxorubicin, 5 nM paclitaxel, and 2.5 µM nutlin. Average of five independent experiments is shown; error bars indicate SEM; one-way ANOVA and Newman–Keuls posttest were used to determine significance; *, significant compared with DMSO treated, P < 0.05. (E and F) 4226 and MCF-7 cells were left untreated or treated with 0.75 and 0.25 µM doxorubicin, respectively, for 24 h. Either NT GFP-expressing cells were co-plated with NT mCherry cells (NT, gray bar) or doxorubicin-treated GFP-expressing cells were co-plated with NT, proliferating mCherry cells (DOXO, purple bars). 7 d after co-plating, cells were fixed and DAPI stained, and random fields were imaged by microscopy. Data shown are the percentage of GFP-expressing cells examined that had one or more DAPI-positive, red vesicles inside them. Average of three independent experiments shown; error bars indicate SEM; significance determined using unpaired t test; **, P < 0.001 (E); **, P < 0.005 (F). (G) 4226 cells were treated and plated as in F. 48 h before scoring, cultures were treated with DMSO or 20 µM QV-D-Oph, a pan-caspase inhibitor. Average of three experiments are shown; not significant by unpaired t test, P = 0.46. (H) Confocal z-stack image of MCF-7 as in B. Representative image of a cell with multiple engulfed cells is shown. Z-stack projections display 12 images taken 2 µm apart with a total z range of 21.5 µm. Scale bar represents 20 µm. (I) Representative image of a senescent 4226 cell plated with NT 4226 cells as in F showing multiple engulfed cells. Scale bar represents 10 µm. (J) Senescent MCF-7 cells were treated with doxorubicin and paclitaxel, as in D. Representative image of multiple sequential engulfments shown. Scale bar represents 50 µm. (K) GFP-expressing MCF-7 cells were treated with 0.75 µM doxorubicin or not on day 0 for 24 h, and then plated on day 1 with NT mCherry-expressing cells. On day 3, the 24-well plate was placed on an IncuCyte and imaged every 2 h until day 11. In cultures of NT mCherry cells mixed with either treated GFP cells (purple line) or NT GFP cells (black line), four different random fields of view were quantified once each day for mCherry cells within GFP cells. Error bar represents SEM; significance was determined by unpaired t test on each day; *, P < 0.05; nonsignificant P values are shown in the figure. (L) Mixed cultures of GFP- and mCherry-expressing MCF-7, MPE600, and 4226 cells were plated and doxorubicin-treated as in C and imaged for fluorescence 7 d later on a Cytation, followed by fixation, SA-βGAL staining, and repeat imaging in color bright field. Shown are examples of matching sequential images of the same cell in fluorescence and color bright field. Arrowheads indicate engulfed cells visible in both images. Scale bars represent 100 µm.
We next quantitated the frequency of engulfment by cells treated with other drugs known to induce senescent-like phenotypes (Chang et al., 1999a; Huang et al., 2009). We found that within mixed cultures of GFP- and mCherry-expressing cells made senescent by doxorubicin, paclitaxel, or their combination (Fig. S2, E and F), GFP-expressing cells had engulfed at least one DAPI-positive neighboring mCherry-expressing cell at rates ranging from 14 to 23% for 4226 cells and 16 to 25% for MCF-7 cells (Fig. 2, C and D). Untreated cultures engulfed at ∼3–4% (Fig. 2, C and D), which is consistent with the background rate of senescence in proliferating cell lines (Chang et al., 1999a; Fig. S2, G and H); however, other modes of engulfment by nonsenescent cells could also be responsible and cannot be ruled out. Interestingly, cells treated with nutlin, a small-molecule inhibitor of MDM2 that activates p53 and some senescent phenotypes in the absence of DNA damage or other stress (Vassilev et al., 2004; Fig. S2, E–H) also induced robust engulfment (Fig. 2, C and D), indistinguishable from that observed after chemotherapy-induced senescence (Fig. S2 I and Video 8). The percentages calculated in Fig. 2 (C and D) are based on counts made at a single time point, and only cells that have within them an entire cell with intact nucleus were scored positive. These counts would exclude senescent cells that had previously engulfed cells but are in later stages of breakdown and lost DAPI staining. These strict criteria result in what is certainly an underestimate of the actual frequency of engulfment by senescent cells.
Apoptotic cells are often engulfed not just by “professional” phagocytes such as macrophages, but also by other, nonprofessional phagocytic cell types as well (Gray and Botelho, 2017). Our observations and previous experiments using proliferating breast cancer cell cultures suggested a very low apoptotic rate. Further, nothing in numerous videos of engulfment by senescent cells suggested that the engulfed cells were in any observable stage of apoptosis (Videos 2, 3, 4, 5, 6, 7, 9, and 10). Breast cancer cell lines treated with chemotherapy drugs or nutlin had no evidence of cleaved caspase 3 or cleaved poly(ADP-ribose) polymerase (PARP), two markers of apoptosis (Fig. S3). To quantitatively determine the contribution of apoptotic cell engulfment to overall rate of engulfment by senescent cells, GFP-expressing cells were treated with doxorubicin to induce senescent predators and then plated with NT mCherry prey cells. Senescent GFP cells readily engulfed NT proliferating mCherry cells at high rates: 34 and 17% for 4226 and MCF-7 cells, respectively (Fig. 2, E and F). If apoptotic cells were being engulfed by senescent cells, one would predict a lower rate of engulfment when apoptosis was blocked. We treated 4226 cultures with the pan-caspase inhibitor Q-VD-Oph to block apoptosis and found no change in engulfment rate, suggesting that apoptotic cells do not contribute substantially to the presence of engulfed cells (Fig. 2 G).
In addition to a large percentage of senescent cells that had engulfed another cell, we observed many examples of senescent cells engulfing multiple entire cells (Fig. 2, H and I) or engulfing cells being engulfed by other cells (Fig. 2 J). To determine how long after doxorubicin treatment cells began to engulf, we performed time course experiments that showed engulfment only after induction of the senescence program at 4 d, reaching maximum levels at day 6 (Fig. S2, A and C; and Fig. 2 K). Before 4 d, despite cells having DNA damage, p53 activation, expression of p21, DNA damage signals, and many other phenotypes, these cells did not engulf (Fig. 2 K). To determine if engulfing predator cells were indeed senescent, we treated mixed cultures of GFP/mCherry cells with doxorubicin, captured fluorescent images to identify engulfing cells, and then stained the cells with SA-βGAL and imaged in color bright field. In sequential fluorescent/SA-βGAL images of the same cell, we found that engulfed cells were visible both in the fluorescent channel and by SA-βGAL staining (Fig. 2 L). Images from three breast cancer cell lines showed that engulfing cells were highly positive for the senescence marker SA-βGAL, consistent with the high percentage of SA-βGAL positivity in these treated cultures (Fig. S2, G and H). In sum, these data show that chemotherapy-induced senescent cells engulf neighboring senescent cells and healthy/proliferating cells at a high frequency.
Different cell types engulf after doxorubicin-induced senescence
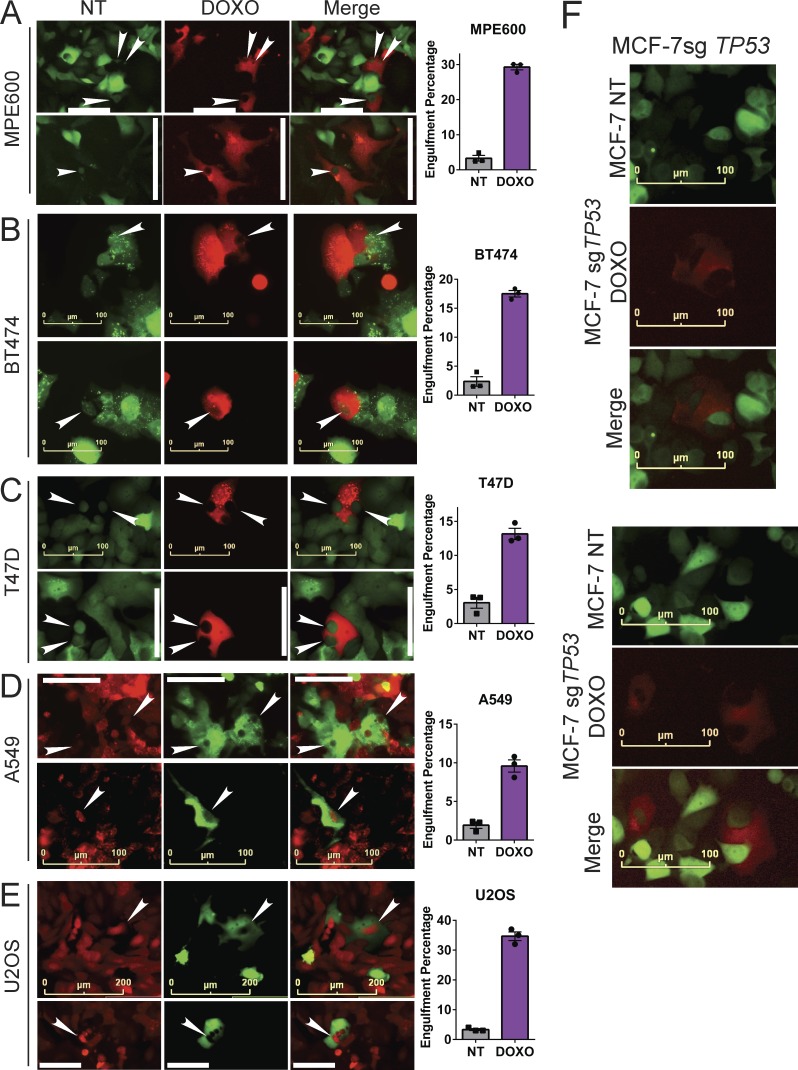
After discovering that doxorubicin-treated MCF-7 and 4226 cells engulf when senescent, we observed that another TP53 wild-type breast cancer cell line, MPE600, also engulfed at a high rate (Fig. 3 A). We next tested if TP53 mutant cells acquired the engulfment phenotype. TP53 mutant cells are more sensitive to DNA damage (Brown and Wouters, 1999; Bunz et al., 1999), but cells that survive can undergo p53-independent senescence in culture (Chang et al., 1999b). Indeed, we found that in two TP53 mutant breast cancer cell lines shown to become senescent after chemotherapy, BT474 (Rosemblit et al., 2018) and T47D (Rastogi et al., 2006), cells that survived chemotherapy could engulf neighboring NT cells (Fig. 3, B and C). Two non–breast cell lines that are TP53 wild type and undergo chemotherapy-induced senescence, A549 lung adenocarcinoma and U2OS osteosarcoma (Litwiniec et al., 2010; Anwar et al., 2018), also engulfed neighboring NT cells 7–8 d after doxorubicin treatment (Fig. 3, D and E). Lastly, MCF-7 cells with CRISPR-Cas9–edited TP53 alleles have very few cells that survive chemotherapy, but those that do also engulfed (Fig. 3 F). The TP53-edited MCF-7 cells that survived chemotherapy were SA-βGAL positive and adopted classic morphological changes consistent with senescent cells (Fig. S2 J), consistent with p53-independent senescence observed by others (Chang et al., 1999b). These findings, taken together with previous studies, suggest that p53 promotes the near-term survival of chemotherapy-treated cells that eventually develop the engulfment phenotype but is not strictly required for the phenotype.
Figure 3.
Non–breast cancer cells and TP53 mutant cells that survive doxorubicin treatment engulf neighbors. (A–E) The indicated GFP- or mCherry-expressing cells were co-plated with cells of the opposite color as in Fig. 2 E that were either NT or doxorubicin treated (DOXO). After 7 d, images of engulfed cells were captured (left) and random fields from triplicate wells were quantitated for engulfment (graphs, right). More than 100 treated cells were scored for engulfment in each well and percentages determined. Error bars represent SEM. (F) mCherry-expressing MCF-7 cells with both alleles of TP53 edited by CRISPR-Cas9 were doxorubicin treated as in A and co-plated with GFP-expressing MCF-7 cells. After 7 d, images were captured of representative remaining TP53 edited cells that had engulfed.
Chemotherapy-induced senescent cells process engulfed cells to the lysosome
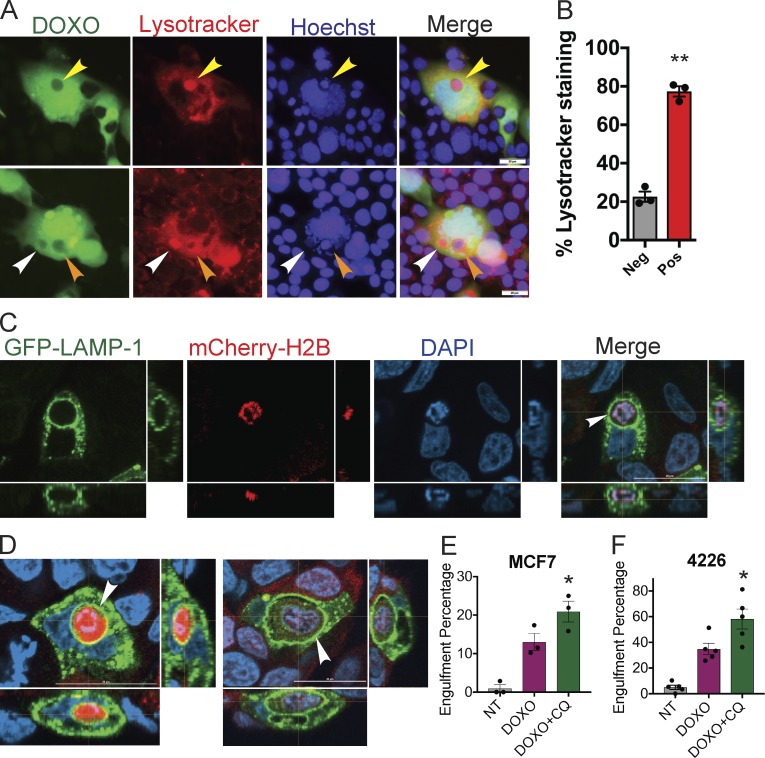
Senescent cells have a vastly expanded lysosomal compartment that is detected by the SA-βGAL assay (Kurz et al., 2000; Lee et al., 2006), which is used widely as a marker for senescence (Dimri et al., 1995; Sharpless and Sherr, 2015). We showed that engulfing cells were SA-βGAL positive (Fig. 2 L); thus, we surmised that the expanded lysosomal compartment of senescent cells might be involved in processing engulfed cells. To determine the fate of engulfed cells, we examined cultures of senescent, GFP-positive breast cancer cells mixed with NT “clear” cells that express no fluorescent protein. Consistent with increased lysosomal activity detected by SA-βGAL, LysoTracker staining was much brighter in senescent cells compared with surrounding NT proliferating cells (Fig. 4 A). Engulfed cells were in various stages of dissolution within senescent cells. Mostly intact engulfed cells stained weakly for LysoTracker and positive for Hoechst (Fig. 4 A, orange arrowhead); however, as engulfed cells became smaller, Hoechst staining fragmented and the LysoTracker signal became stronger (Fig. 4 A, yellow arrowhead), leading to DAPI-negative and strong LysoTracker staining (Fig. 4 A, white arrowhead). In 4226 cells, ∼80% of engulfed cells were positive for LysoTracker (Fig. 4 B).
Figure 4.
Engulfed cells are processed to the lysosome. (A) GFP-expressing 4226 cells were treated with 0.75 µM doxorubicin (DOXO) for 24 h and then co-plated with clear (uninfected), NT 4226 cells. After 7 d, LysoTracker and Hoechst dye was added to cocultures, followed by imaging. Arrowheads indicate clear cells within GFP-expressing senescent cells in various states of Hoechst and LysoTracker positivity. Scale bar represents 20 µm. (B) LysoTracker staining was quantitated in three separate 4226 engulfment experiments. A total of 103 senescent cells were found to have engulfed another cell and were counted. LysoTracker-positive vesicles that were both DAPI positive and completely surrounded by GFP cytoplasm from the engulfing cell were counted as positive, while vesicles that were also completely surrounded by GFP and were DAPI positive but were not LysoTracker positive were counted as negative. Significance determined by unpaired t test; **, P < 0.001. (C and D) MCF-7 cells expressing LAMP1-GFP were treated with doxorubicin and then co-plated with NT mCherry cells. 7 d later, cells were fixed and confocal microscope images were captured in three channels and merged. Z-stack projections are shown adjacent and below. Representative images shown. Scale bar represents 20 µm. Z-stack projections display 15 images taken 2 µm apart with a total z range of 28.02 µm for C, 22 images taken 1 µm apart for a total of 20.48 µm (D, left), and 33 images taken 1 µm apart totaling 31.72 µm (D, right). (E and F) Cells expressing GFP were doxorubicin treated or not for 24 h and then co-plated as in Fig. 2 (E and F). 5 d after plating, vehicle or chloroquine was added to final concentration of 20 or 10 µM for MCF-7 and 4226, respectively, and 2 d later MCF-7 (E) and 4226 (F) were fixed and DAPI stained. Engulfment percentage was determined for NT cells mixed with NT cells (gray bar), doxorubicin-treated cells mixed with NT cells (DOXO, purple bars), and doxorubicin-treated cells mixed with NT cells followed by chloroquine treatment (DOXO+CQ, green bars). Three (E) and five (F) individual experiments were performed; *, P < 0.05 determined by unpaired t test between doxorubicin treated with or without chloroquine. Errors bars represent SEM.
To confirm these findings, we expressed GFP-tagged LAMP1, a component of the lysosomal membrane, in MCF-7 cells. Costaining with LysoTracker dye on LAMP1-GFP MCF-7 cells showed nearly complete colocalization, confirming efficient marking of the lysosome (Fig. S4). After treatment, we observed that mCherry-H2B cells were enveloped within membranes containing LAMP1-GFP expressed by the treated, senescent cell (Fig. 4, C and D). Time-lapse videos show that LAMP1-GFP membranes formed around engulfed cells (Videos 9 and 10).
Blocking the acidification and activity of lysosomes with the drug chloroquine resulted in increased frequency of cells within cells in both senescent 4226 and MCF-7 cells, consistent with an impairment in ability to degrade engulfed cells (Fig. 4, E and F). Taken together, these data show that engulfed cells are processed to the lysosome, and lysosome acidification is necessary for the complete degradation of engulfed cells.
Chemotherapy-induced senescent cells develop phagocytosis proficiency but not entosis
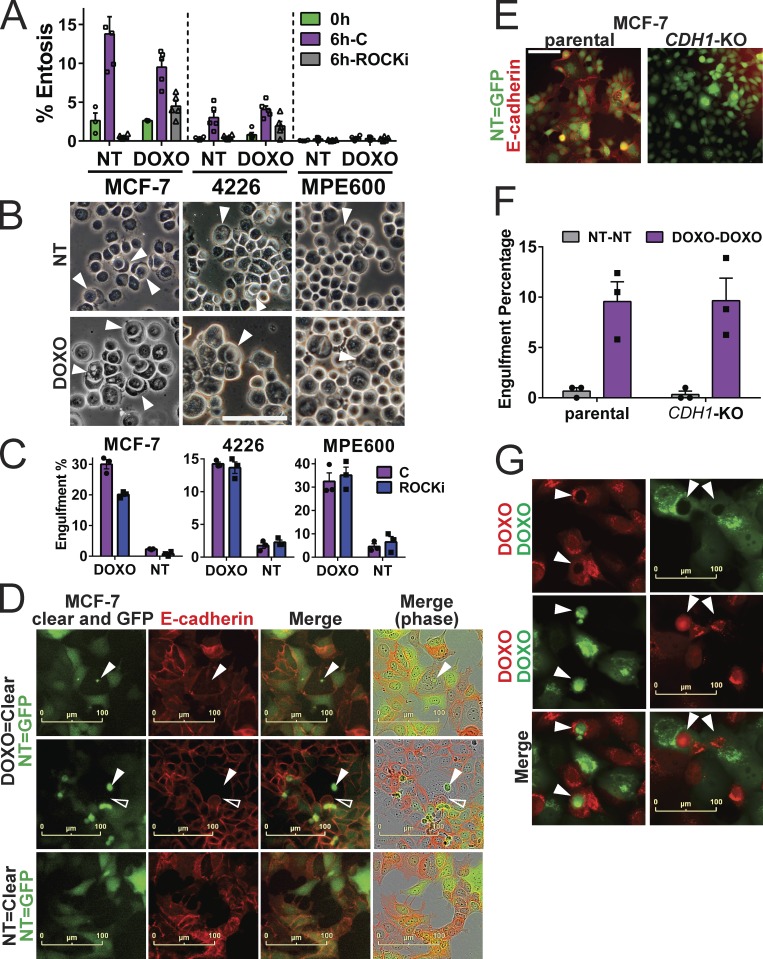
Two modes of cell engulfment are entosis and phagocytosis. We first examined entosis in chemotherapy-induced senescent cells. As shown by others (Overholtzer et al., 2007), entosis was frequent and easily identified in MCF-7 cells using standard assays (Sun and Overholtzer, 2013). Entosis rates were not increased, however, by doxorubicin treatment and induction of senescence (Fig. 5, A and B), despite engulfment in monolayer cultures being elevated 5- to 10-fold after doxorubicin treatment (Fig. 2, C–F; and Fig. 3). Further, while senescent MPE600 and 4226 cells each engulf in monolayer at rates higher than MCF-7 (Fig. 2, E and F; and Fig. 3 A), these cells underwent entosis at much lower rates and showed no increase when senescent. As others have shown (Overholtzer et al., 2007), inhibition of Rho-associated kinases (ROCK) 1 and 2 using Y-27632 effectively blocked entosis in MCF-7 (Fig. 5 A), but had little or no effect on engulfment rates by senescent cells in monolayer (Fig. 5 C). Lastly, E-cadherin (coded by the CDH1 gene) is required for and forms concentrated foci at contact points of entosis (Overholtzer et al., 2007; Sun et al., 2014a). We did not observe focal concentration of E-cadherin at contact points between senescent predator cells that were actively engulfing and prey cells compared with contacts formed by NT cells that do not engulf (Fig. 5 D). Further, CDH1 knockout MCF-7 cells (Bajrami et al., 2018) engulfed at the same rate as parental MCF-7 cells (Fig. 5, E–G).
Figure 5.
Engulfment by doxorubicin (DOXO)-induced senescent cells is distinct from e-cadherin–dependent entosis. (A and B) MCF-7, 4226, and MPE600 cells, proliferating and 8 d after doxorubicin treatment, were placed in suspension in low-adhesion plates in the presence or absence of 10 µM ROCK inhibitor Y-27632 (ROCKi) for 6 h, followed by cytospin, fixation, and imaging, along with cells cytospun immediately after suspension (0 h). (A) Number of entosis cells and total number of cells from high-power phase fields were quantified and percentages were graphed. Individual data points are shown; error bars represent SEM. Data represent at least two separate experiments. (B) Representative examples of NT cells and cells 8 d after doxorubicin (DOXO) treatment undergoing entosis, denoted by white arrowheads. Scale bar represents 50 µm. (C) Indicated cell lines were doxorubicin treated and plated for engulfment assays in triplicate as in Figs. 2 E and 3 A. In an additional group, 10 µM ROCK inhibitor Y-27632 was added every other day. Cultures were followed up to 8 d after doxorubicin on an IncuCyte, and cell-in-cell structures were quantified and graphed on day 8. Graphs show mean percentages from triplicate wells (all three data points are shown) of GFP-expressing cells (NT or DOXO) with an NT mCherry-expressing cell inside them. Error bars represent SEM. Data shown are representative of at least two independent experiments. (D) Untreated GFP-expressing MCF-7 cells (NT) were plated as in Fig. 2 E with NT or doxorubicin-treated (DOXO) clear MCF-7 parental cells. Cultures were followed for 5 d on IncuCyte, and cells were fixed, stained for e-cadherin (CDH1), and imaged on IncuCyte again. Shown are representative images of senescent clear cells that had previously engulfed another cell (indicated by arrowheads, verified in time lapse to be a live cell engulfment) making contact with NT cells (top two panels; open arrowhead indicates an NT cell intruding into the senescent cell). Lower panel shows e-cadherin staining of contacts made between NT clear and NT GFP-expressing cells. (E) MCF-7 cells with knockout of the gene for e-cadherin (CDH1-KO) and the parental line from which they were derived were stained for E-cadherin. Scale bar represents 50 µm. (F) GFP- and mCherry-expressing MCF-7 parental and CDH1-KO cell lines were plated and doxorubicin treated in triplicate as in Fig. 2 C. Cultures were imaged on IncuCyte, and 6 d after doxorubicin, cell-in-cell structures were quantified and graphed to show mean percentages of GFP-expressing cells (NT-NT or DOXO-DOXO) with an mCherry-expressing cell inside them (all three data points are shown). Error bars represent SEM. Data shown are representative of two independent experiments. (G) Representative images of engulfments occurring in cultures from F. Arrowheads indicate engulfed treated cells within other treated cells from both MCF-7 parental and CDH1-KO cells. Scale bar represents 100 µm. Data shown are representative of at least two independent experiments.
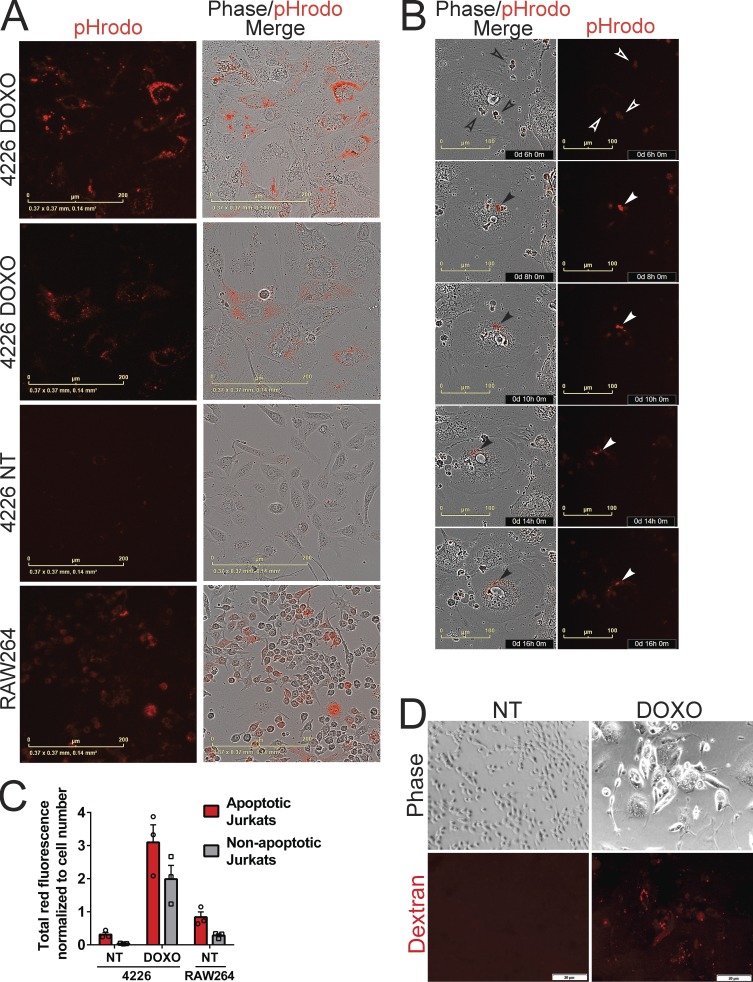
To determine if senescent cells gained proficiency in phagocytosis, we performed an established assay using apoptotic Jurkat cells labeled with pHrodo dye that become fluorescent in the acidic pH of the lysosome following phagocytosis (Aziz et al., 2013). We found that doxorubicin-induced senescent 4226 cells, but not NT cells, engulfed apoptotic Jurkat cells (and, to a lesser extent, nonapoptotic Jurkat cells). Senescent cell engulfment and lysosomal transport of Jurkat cells occurred at levels that exceeded even that of RAW264 macrophages at the 48-h time point (Fig. 6, A–C; and Fig. S5), perhaps at least partially due to the large surface area of senescent cells (Fig. 6 A). To test if senescent cells could engulf substrates other than senescent cells, proliferating cells, and Jurkat cells, we applied fluorescent dextran to cultures. We found a qualitative increase in dextran uptake by senescent 4226 cells (Fig. 6 D). In sum, our data show that entosis is unlikely to be the cause of the increased frequency of cell-in-cell structures in chemotherapy-induced senescent cells, and senescent cells gain proficiency in well-established assays measuring phagocytosis.
Figure 6.
Doxorubicin (DOXO)-induced senescent cells are proficient in phagocytosis. (A–C) pHrodo-labeled Jurkat cells that were NT (nonapoptotic) or treated with doxorubicin for 24 h (apoptotic) were placed onto cultures of RAW264 macrophages and proliferating and senescent 4226 cells. After 2 d, Jurkats were washed off and plates were imaged and quantitated on IncuCyte for the red fluorescence emitted after the phagocytosis of Jurkat cells and their transport to lysosome. (A) Representative images of doxorubicin-treated, senescent 4226 cells (DOXO), NT 4226 cells, and RAW264 macrophage cell line. Scale bar represents 200 µm. (B) Representative phase and red fluorescence images were captured over a time course of a senescent 4226 cell engulfing pHrodo-labeled cells and processing them to the lysosome. Scale bar represents 100 µm; time of capture after addition of pHrodo Jurkats is shown. Open arrowheads indicate pHrodo Jurkats before engulfment, and closed arrowheads indicate pHrodo fluorescence activation at low pH, such as occurs in the lysosome. (C) Total red fluorescence in each well was quantified and normalized to the cell number in each well. The mean and individual data points from triplicate wells were plotted; error bar represents SEM. (D) Proliferating and senescent 4226 cells were serum starved for 1 h, 60 µg/ml of 70-kD Texas Red Dextran was added, 30 min later cells were washed and fixed with 4% formaldehyde, and images were captured. Representative images shown. Scale bar represents 20 µm. Data shown are representative of at least two independent experiments.
Chemotherapy-induced senescent cells are highly enriched for macrophage- and phagocytosis-related genes
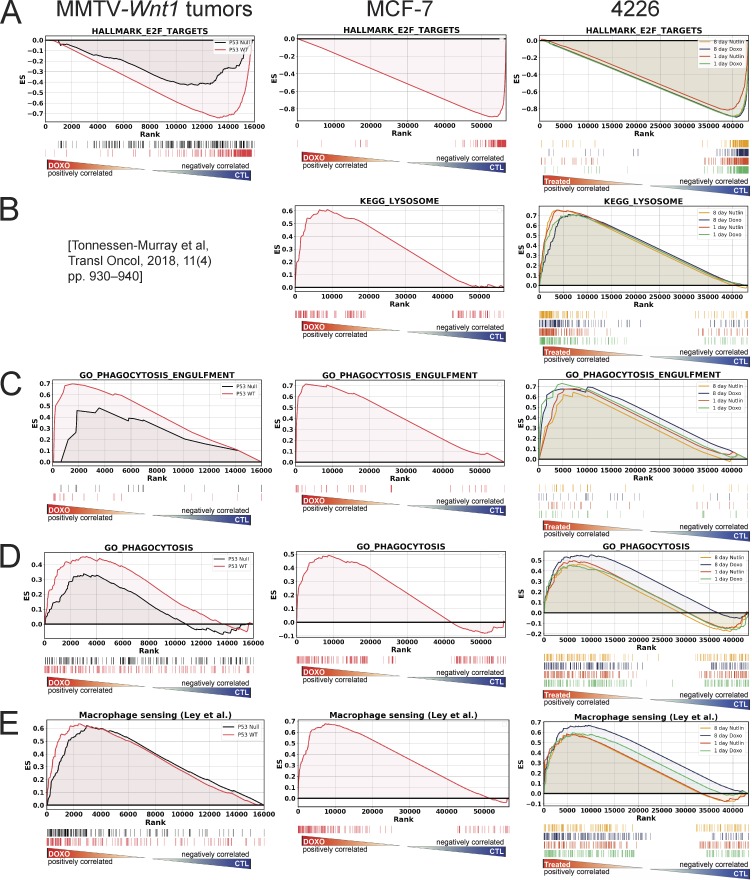
To understand the molecular mechanisms altered in senescent cells, RNA sequencing (RNA-seq) and gene set enrichment analysis (GSEA) were performed on MMTV-Wnt1 tumors (Tonnessen-Murray et al., 2018) and two cell lines, MCF-7 and 4226. Strikingly, a potential program of engulfment was revealed. We previously showed that MMTV-Wnt1 tumors that are p53 mutant fail to arrest after chemotherapy and undergo cell death and tumor regression, while p53 wild-type tumors undergo senescence and relapse quickly (Jackson et al., 2012). We found that the NT p53 wild-type MMTV-Wnt1 tumors, MCF-7 cells, and 4226 cells were highly enriched for expression of cell cycle genes (Fig. 7 A), consistent with the arrest induced in treated tumors and cells (Jackson et al., 2012; Tonnessen-Murray et al., 2018). Lysosome signature was previously shown to be enriched in treated tumors (Tonnessen-Murray et al., 2018) and was found to be highly enriched in both senescent MCF-7 and 4226 (Fig. 7 B). Remarkably, treated p53 wild-type tumors, MCF-7, and 4226 had strong enrichment for gene ontology categories GO_PHAGOCYTOSIS and GO_PHAGOCYTOSIS_ENGULFMENT signatures (Fig. 7, C and D), as well as a signature of genes related to how macrophages sense their environment (Ley et al., 2016; Fig. 7 E). p53 mutant tumors had a more modest enrichment (Fig. 7, C–E), consistent with other senescence-related gene signatures in this genotype (Tonnessen-Murray et al., 2018) and reflective of cell death occurring in p53 mutant cells that might have incurred enough cell damage to activate the engulfment pathway (Tonnessen-Murray et al., 2018). Cell engulfment, SA-βGAL–positive staining, and morphological changes become evident ∼3–5 d after doxorubicin treatment (Figs. 2 K and S2, A and C); thus we performed RNA-seq on treated 4226 cells before and after senescent/engulfment phenotypes appeared (24 h and 8 d after doxorubicin treatment). Interestingly, the lysosome, cell cycle, and phagocytosis signatures were strongly enriched even at the 24-h time point, thus anticipating phenotypes that are not present for approximately three or four more days. This delay is reminiscent of the amount of time required for phenotypic differentiation of neurons driven by acute gene expression changes induced by retinoic acid (Encinas et al., 2000). Cell cycle, lysosome, and phagocytic signatures were also evident after nutlin treatment (Fig. 7 C), consistent with the engulfment phenotype induced (Fig. 2, C and D; Fig. S2 I; and Video 8).
Figure 7.
Gene expression analysis of doxorubicin-treated tumors and cell lines reveals enrichment of phagocytosis signatures. (A–E) GSEA was performed with RNA-seq data from MMTV-Wnt1 mammary tumors (p53 wild type, red line; p53-null, black line) for NT (Ctl) versus doxorubicin-treated cells harvested 24 h after the final treatment (left panels); from control MCF-7 cells and MCF-7 cells 8 d after doxorubicin (0.5 µM) treatment (middle panels); and from NT 4226 cells versus cells 24 h (green line) and 8 d (blue line) after doxorubicin (0.75 µM) or treated for 24 h (red line) and 8 d (yellow line) with nutlin (15 µM every other day), as indicated. RNA-seq was performed on MCF-7 and 4226 using samples from three independent experiments each. RNA was extracted from six doxorubicin-treated and six NT MMTV-Wnt1 tumor samples and sent for sequencing. GSEA was performed for “HALLMARK_E2F_TARGETS” (A); KEGG_LYSOSOME (B); GO_PHAGOCYTOSIS_ENGULFMENT (C); GO_PHAGOCYTOSIS (D); and genes involved in macrophage sensing their environment as listed in Ley et al. (2016) (E). Enrichment scores (ES) were plotted. Statistical analysis using GSEA software is provided as false discovery rate q values (significance at false discovery rate q < 0.25) in Table S2. GO, gene ontology.
Chemotherapy-induced senescent cells that have engulfed have a survival advantage
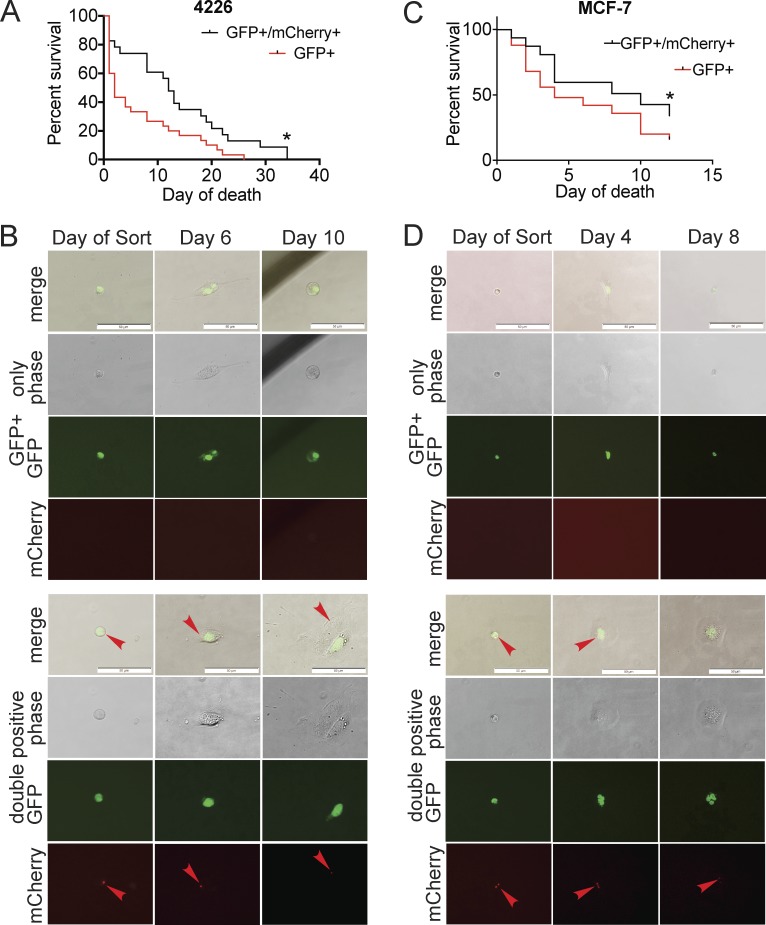
Survival and persistence are characteristics of senescent cells that likely contribute to their promotion of relapse (Tonnessen-Murray et al., 2016). To determine whether senescent cells that have engulfed other cells have superior persistence, MCF-7 and 4226 cells expressing GFP-tagged H2B were treated with doxorubicin to induce senescence and then plated with NT cells expressing mCherry-H2B. 7 d later, live cells were single-cell sorted into wells of a 96-well plate and confirmed microscopically to have only one cell per well. Each cell was scored microscopically as either GFP and mCherry positive (engulfed) or GFP only (has not engulfed). Cells were followed over time and marked as dead when they became rounded and/or fragmented for >48 h (Fig. 8, B and D). Both 4226 and MCF-7 cells that were double positive, and therefore had engulfed another cell, lived longer in culture than those that had not (Fig. 8). These data suggest that cell engulfment is beneficial to senescent cancer cell viability. While only two cell lines were tested, the survival phenotype after engulfment (Fig. 8, A and B) is consistent with the ability of breast cancer cells that undergo senescence in response to chemotherapy to avoid death, resulting in tumor persistence and poor patient survival.
Figure 8.
Senescent cells that have engulfed a cell have a survival advantage. (A–D) H2B-GFP–expressing senescent 4226 (A and B) or MCF-7 (C and D) cells were co-plated with NT H2B-mCherry–expressing 4226 and MCF-7 cells, respectively. 7 d after coplating, cells were trypsinized and sorted one cell per well of a 96-well plate. Each cell was confirmed microscopically and marked as double positive (engulfed, black line) or GFP positive only (not engulfed, red line) and followed over time. (A) Four individual experiments were sorted; a total of n = 23 double-positive and n = 30 GFP-only 4226 cells were followed. Significance was determined by log-rank Mantel–Cox test, *, P < 0.05. (C) One experiment was sorted; a total of n = 47 double-positive and n = 50 GFP-only cells were followed. Experiment terminated at 12 d. Significance was determined by log-rank Mantel–Cox test, *, P < 0.05. Examples of GFP+-only cells that died are shown in upper panels of B and D; examples of surviving GFP-expressing cells with an mCherry vesicle within them (red arrowhead) are shown in lower panels of B and D. Scale bars represent 50 µm.
Discussion
Breast cancers are frequently wild type for TP53 (Shahbandi and Jackson, 2019), and compelling evidence from our laboratory and others shows that chemotherapy-treated breast cancers that are TP53 wild type are more likely to have residual disease (Bertheau et al., 2002, 2007; Chen et al., 2012; Esserman et al., 2012; Nakamura et al., 2012; Wang et al., 2016; Goetz et al., 2017), with senescent cells present (te Poele et al., 2002; Jackson et al., 2012; Tonnessen-Murray et al., 2018), resulting in markedly worse survival (Ungerleider et al., 2018). The present study reveals a previously unknown ability of chemotherapy-induced senescent cancer cells to engulf and degrade nearby cells, resulting in enhanced survival and providing compelling explanations for well-known but poorly understood senescent cell phenotypes such as SASP and SA-βGAL staining.
Positive staining for SA-βGAL activity, caused by an expanded lysosomal compartment (Kurz et al., 2000; Lee et al., 2006), is a well-described feature of many senescent cell types, both in vivo and in vitro, in normal and cancer cells (Jackson et al., 2012; Sharpless and Sherr, 2015), including chemotherapy-treated breast cancers (te Poele et al., 2002; Jackson et al., 2012; Tonnessen-Murray et al., 2018). The function for this near-ubiquitous feature of senescent cells was previously unclear, but our observation of entire engulfed cells being processed to the lysosome suggests an obvious purpose for lysosome expansion. Indeed, we observed entire engulfed cells encapsulated in lysosomal membrane (Fig. 4, C and D; and Videos 9 and 10).
Arrested senescent cells are highly metabolically active (Quijano et al., 2012). Furthermore, these cells are metabolically very different from nonsenescent cells (Zwerschke et al., 2003; Erol, 2010; Kim et al., 2010, 2011; Quijano et al., 2012; Gey and Seeger, 2013), producing and secreting cytokines and chemokines that contribute to relapse (Jackson et al., 2012; Velarde et al., 2013; Rao and Jackson, 2016). Although the mechanisms employed by senescent cells that allow them to persist, survive, and accommodate the energy-intensive processes involved in translation, processing, and secretion of cytokines are not completely understood, the engulfment and breakdown of other cells is likely involved. We propose this process as a potential source of energy and building blocks for the secretory phenotype and enhanced survival, and thus as a source for therapeutic opportunities. This is supported by our finding that senescent cells that have not engulfed another cell die earlier in culture.
There are many observed types of cell engulfment, including phagocytosis and efferocytosis of apoptotic or dying cells by macrophages or other nonprofessional phagocytes (Gray and Botelho, 2017), emperipolesis, and entosis (Overholtzer et al., 2007; Krishna and Overholtzer, 2016; Durgan et al., 2017; Hamann et al., 2017). Our data show that senescent cells acquire a phagocytic phenotype ultimately targeting nonapoptotic cancer cells, superficially similar to entosis. Entosis was described as cannibalism by cancer cells in culture after loss of substrate adherence, sometimes resulting in death of the engulfing cell and sometimes lysosomal degradation of the engulfed cell (Overholtzer et al., 2007; Krishna and Overholtzer, 2016). Rates of entosis can be increased by glucose starvation (Hamann et al., 2017) or 12–48 h after radiation/chemotherapy treatment (Martins et al., 2018). Breast cancer cells in 3D culture were shown to engulf bone marrow–derived mesenchymal stem/stromal cells. Following the engulfment, dormancy and senescent phenotypes were then induced in the engulfing breast cancer cell (Bartosh et al., 2016).
The phenotype we show here shares some attributes with entosis and may be related, but it differs in important ways. Breast cancer cells showed no increase in entosis rates after induction of senescence following chemotherapy treatment, even though these cells show markedly elevated rates of engulfment in monolayer. Further, breast cancer cells that did not undergo entosis could still engulf when senescent, and engulfment was independent of E-cadherin (Fig. 5), unlike entosis (Overholtzer et al., 2007; Sun et al., 2014a). Our phenotype was induced over time by chemotherapy treatment and senescence, not acutely after loss of substrate adherence (Overholtzer et al., 2007) or mitotic rounding (Durgan et al., 2017). Rather than increasing engulfment relatively quickly due to physical changes caused by loss of substrate adherence that result in cells that happen to be highly deformable in suspension ingesting cells that are less deformable (Sun et al., 2014b), our phenotype required days to develop and was driven by a macrophage/phagocytosis gene signature in vitro and in vivo. Evidence we present here suggests that entosis and senescent cell engulfment, while similar in some ways, are fundamentally different.
Engulfment by senescent cells increased their survival and could provide building blocks that might enable SASP to drive relapse. These factors suggest immediate clinical relevance, as p53-mediated induction of senescence contributes to poor response (Bertheau et al., 2002; Jackson et al., 2012) and poor survival (Ungerleider et al., 2018) of breast cancer patients.
Materials and methods
Mice
All experiments were approved by the Tulane Institutional Animal Care and Use Committee, Protocol ID# 4381, and conformed to the guidelines of the United States Animal Welfare Act and the National Institutes of Health. For tumor transplants performed as previously described (Tonnessen-Murray et al., 2018), ∼4 × 106 cells from cryopreserved MMTV-Wnt1 tumors were resuspended in Matrigel (Corning) and injected into the #4 mammary fat pad. Tumors formed after 2–3 wk, and when they became ∼1.25 cm, they were excised, minced, trypsinized for 10 min, and filtered through a 100-µm filter. Multiple concentrations of cells ranging from 2 × 105 to 2 × 106 were plated in a well of a 6-well plate (many of these cells just extracted from the tumor were nonviable). After transduction and FACS to purify a fluorescent population (Tonnessen-Murray et al., 2018), cells were injected into the mammary fat pad of C57BL/6j mice for passaging or doxorubicin treatment as previously described (Jackson et al., 2012; Tonnessen-Murray et al., 2018). For Fig. 1, a total of six MMTV-Wnt1 doxorubicin-treated, mixed color tumors were harvested. 10 random fields of view were imaged for each sample.
Lentiviral infections
Fluorescent protein inserts (cytoplasmic GFP and mCherry, in the backbone CD502A-1 [pCDH-EF1α-MCS cDNA Single Promoter Cloning and Expression Lentivector] from Systems Biosciences; LAMP-GFP [34831]; mCherry-Farnesyl for membranes [55045]; H2B-GFP [21210]; and H2B-mCherry [21217] from Addgene) were cloned into plasmids used in our laboratory previously (Tonnessen-Murray et al., 2018). Lentivirus was produced using 293T cells and a standard calcium phosphate transfection protocol, or with Lipofectamine 2000 according to the manufacturer’s instructions (Thermo Fisher Scientific). Viral supernatants were collected 48 to 72 h after transfection and filtered (0.45 µm), and then cells (ex vivo tumors or cell lines) were infected in the presence of polybrene (8 µg/ml). Positive cells were FACS sorted using a BD FACSAriaII as previously published (Tonnessen-Murray et al., 2018).
Cell culture
MCF-7 cells were obtained from ATCC and cultured in complete Eagle’s minimum essential medium (RRID: CVCL_0031; ATCC, HTB-22). MPE600 cells (RRID: CVCL_9875) were a gift from Joe Gray (Oregon Health Sciences University, Portland, OR) and were cultured in complete DMEM. 4226 cells were created by excision of a spontaneous MMTV-Wnt1 tumor, mincing, trypsinizing, and plating, followed by passaging in complete DMEM and eventual clonal outgrowth of the cell line. A549 (RRID: CVCL_0023) was a gift from Joseph Lasky (Tulane School of Medicine, New Orleans, LA); U2OS (RRID: CVCL_0042) was a gift from Sean Lee (Tulane School of Medicine); RAW264 cells (RRID: CVCL_4478) were a gift from Heather Machado (Tulane School of Medicine); and each was cultured in complete DMEM. BT474 (RRID: CVCL_0179) and T47D (RRID: CVCL_0553) were obtained from ATCC (HTB-20 and HTB-133, respectively) and were cultured in complete RPMI 1640. Creation of MCF-7 CRISPR-Cas9 TP53 edited cells has been described (Ungerleider et al., 2018), and data shown here are from a single cell clone with biallelic editing and verified by sequencing to be TP53 c.[884_892delATCCACTAC; 885_892delTCCACTAC]. MCF-7 CDH1 KO cells have been described (Bajrami et al., 2018) and were compared with the parental MCF-7 from which they were derived. Cell lines were routinely confirmed to be mycoplasma free by DAPI staining. Cell lines were treated with doxorubicin for 24 h (0.25 µM for MCF-7, BT474, T47D, A549, and U2OS; 0.75 µM for 4226; 0.50 µM for MPE600), followed by PBS washes and medium replenishment. For engulfment experiments, dishes were washed four times and trypsinized, and 75,000 cells were added to each well of a 6-well dish that already had respective NT cells plated the day before. Medium was changed every other day. 7 d after co-plating, dishes were imaged. In some experiments, chloroquine (Sigma-Aldrich; C6628-25G), 10 µM for 4226 and 20 µM for MCF-7, was added 48 h before collection. 70 kD Dextran (Thermo Fisher Scientific; D1864) was used at 60 µg/ml and LysoTracker Red DND-99 (Thermo Fisher Scientific; L7528) was added to 75 nM 1 h before collection. Paclitaxel (Apex Bio) was used at 5 and 10 nM for MCF-7 and 4226 cells, respectively. Nutlin-3a (Sigma-Aldrich; 444152) was refreshed every other day for the length of experiment, at concentrations of 2.5 and 15 µM for MCF-7 and 4226, respectively.
Cell engulfment
On the day indicated in Fig. 2 (C–G), cells were fixed with 4% formaldehyde, permeabilized with Triton X-100, DAPI stained, and imaged by epifluorescence or confocal microscopy. Cells were judged to have engulfed another cell if the engulfed vesicle was positive for DAPI staining and completely surrounded by the cytoplasm/membrane of the engulfing cell. For quantitative experiments, at least three individual experiments were performed. Random fields of view were imaged, and ≥50 senescent cells were counted as engulfed or not engulfed for each condition per experiment. For Figs. 2 K, 3, and 5, GFP- and mCherry-expressing cells were treated and combined as detailed in the figure legends, and images were captured on IncuCyte. Engulfment was determined by presence of an mCherry structure inside a vesicle that was observable in the GFP channel of a GFP cell. In Fig. 2 K, cells were plated and imaged over a time course on the IncuCyte, and engulfment percentage was determined as detailed in the figure legend. Similar results were found in repeated experiments using FACS sorting of double-positive (engulfed) versus single-positive (not engulfed) senescent cells at days 4, 6, and 8 after doxorubicin addition. At least 45,000 events were counted per time point per condition. Double-positive events were collected and confirmed to be engulfed (rather than stuck together) microscopically (≥50 counted per sample). ROCK inhibitor Y-27632 (10 µM; Apex Bio) was added to cells the day after plating and was changed every other day.
Entosis
Entosis was assayed as previously described (Sun and Overholtzer, 2013). Briefly, cells were trypsinized, washed, and resuspended in complete medium at 4 × 105 cells per milliliter. For entosis after 6 h, 0.5 ml was plated in a well of a Costar 24-well Ultra-Low attachment plate (Corning; 3473). After 6-h incubation, cells were centrifuged and thoroughly resuspended in 400 µl PBS, and then 200 µl was Cytospun onto a glass slide for 5 min at 500 rpm. Cells on slides were fixed in 2% formaldehyde (diluted from 16% methanol-free formaldehyde; Thermo Fisher Scientific; 28908), and then mounted with a coverslip in PBS. For the 0-h time point of entosis, cells were processed for cytospin as above immediately after trypsinization. High-power 20× fields (comprising ∼300 to 1,000 total cells each) were captured on a phase-contrast Olympus microscope (below). Cells undergoing entosis (a positive was scored as any cell with one half or more of another cell body internalized inside of it) and total cells were scored as previously described (Sun and Overholtzer, 2013) and are shown as individual data points. ROCK inhibitor Y-27632 (10 µM; Apex Bio) was added to cells at the time of plating in low-adhesion plates.
Phagocytosis assay
Phagocytosis of pHrodo-labeled Jurkat cells was performed essentially according to the manufacturer’s instructions (Essen BioScience). Briefly, 4226 cells were made senescent as in Fig. 2 C. RAW264 cells were plated at 50,000 cells/48 wells. Jurkat cells were plated in a 6-well plate at 106 cells/ml and then treated or not with 1 µM doxorubicin for 24 h, and apoptosis was confirmed by Annexin V staining. Apoptotic or nonapoptotic Jurkats were then stained with pHrodo dye according to the manufacturer’s instructions (Essen BioScience), and then 200,000 cells were plated on top of RAW264 or proliferating 4226 cells plated the day before or senescent 4226 cells. 2 d later, Jurkats were washed off, fresh medium was replenished, and images were captured at 20× on IncuCyte for red fluorescence and phase. From IncuCtye images, cell number was determined, and then total red object integrated intensity (red-calibrated unit × μm2/image) was calculated for each well after masking using a red-calibrated unit threshold of 0.2 and an area threshold of 1,000. The total red object intensity was then divided by cell number to normalize the intensity in a well for the number of cells in that well. Data are representative of at least two independent experiments.
Microscopes
Cells were imaged on an Olympus inverted immunofluorescence microscope (IX-71) with DP-70 camera using cellSens 1.12 Standard Imaging Software at room temperature using a 20× or 40× objective. Confocal imaging was performed on tissue sections or cells mounted in Prolong Diamond Antifade Mountant (P36965; Thermo Fisher Scientific) on glass coverslips, using an Olympus FV1000 with a Plan Apochromat N 60× oil objective. Confocal microscopy was also performed using a Nikon TiE-2 Inverted Research Microscope and Nikon A1rSi Laser Point Scanning Confocal, with a Plan Apochromat λ 60× oil objective for cell lines and Plan Fluor 40× oil for tissue sections, and NIS-Elements software. Videos were captured on a Carl Zeiss Cell Observer inverted microscope with an LD Plan-Neofluar 20× objective using Axiovision software and an Olympus Viva-view LCV110U with a Hamamatsu C10600 Orca-R2–controlled cooled charge-coupled device camera, using 10× and 20× objectives, at 37°C and 5% CO2 in full growth medium. Cell images, fluorescence data for pHrodo, and time course images were captured on an IncuCyte S3 system using 2018A software version, at 37°C and 5% CO2 in full growth medium at 10× or 20× magnification. Cell images (37°C and 5% CO2), DAPI counts, and color bright-field images for SA-βGAL stains (at 25°C and atmospheric CO2 in PBS at 20× magnification) were captured on a Cytation5 using Gen5 software version 3.05. Videos were cropped using Axiovision or ImageJ (National Institutes of Health). Image brightness and background levels were adjusted uniformly for whole images using Photoshop for visual clarity in publication where necessary.
Gene expression analysis
Gene expression analysis of MMTV-Wnt1 tumors has been described previously (Tonnessen-Murray et al., 2018). Briefly, spontaneous tumors from mice that were treated or not with doxorubicin were harvested (Jackson et al., 2012), RNA was prepared with Trizol (Thermo Fisher Scientific) reagent, and RNA-seq was performed by the MD Anderson Cancer Center Sequencing and Microarray Facility (Houston, TX) as described previously (Tonnessen-Murray et al., 2018). Cell lines were plated, treated, and then harvested at time points as indicated in the figures, and RNA was prepared using Trizol reagent, as previously described (Jackson and Pereira-Smith, 2006). RNA-seq was performed by BGI (Shenzhen, China) using BGISEQ-500, single-end 50-bp, 20 million reads per sample (4226) and Hiseq 4000, paired-end 100-bp, one lane 100PE sequencing (MCF-7). Gene expression analysis using GSEA software (http://software.broadinstitute.org/gsea/index.jsp) was performed using the gene lists indicated in the figures, as previously described (Tonnessen-Murray et al., 2018). Real-time PCR was performed on a QuantStudio6 (Applied Biosystems) using Sybr Green (Bio-Rad) and analysis as per Pfaffl (2001), as previously described (Jackson et al., 2012).
Immunoblotting
Cell lines were treated and harvested as indicated in the figures. Lysates were prepared using TNESV buffer (1% NP-40, 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 2 mM EDTA, and 10 mM NaVO4; Jackson et al., 1998), and 50 µg of total protein was separated by 10% SDS-PAGE and then transferred to nitrocellulose membranes using Bio-Rad reagents and apparatus. Immunoblotting was performed as previously described (Jackson and Yee, 1999) for cleaved caspase 3 (rabbit polyclonal, 9661S; Cell Signaling), PARP (rabbit polyclonal, 9542S; Cell Signaling), and actin (mouse monoclonal, MA5-15739; Invitrogen). All antibodies were used at 1:1,000 dilution.
Videos
Video 1 was reconstructed from images taken on an Olympus FV100 confocal microscope using a Plan Apochromat N 60× oil objective. Videos 2, 6, 7, 9, and 10 were imaged on an Olympus Viva-view LCV110U using a 10× objective for 2, 6, 9, and 10 and a 20× objective for 7. Videos 3, 4, and 5 were imaged on a Carl Zeiss cell observer inverted microscope with an LD Plan-Neofluar 20× objective. Video 8 was reconstructed using NIS-Elements software from images taken on a Nikon A1rSi laser point scanning confocal microscope with a Plan Apochromat λ 60× oil objective. All videos began 6–7 d after doxorubicin treatment, unless otherwise noted. Videos were created of senescent 4226 cells engulfing NT cells in seven independent experiments, totaling 199 separate videos. Representative videos of complete cell engulfment captured are shown. Videos were created of senescent MCF-7 cells engulfing NT cells in three separate experiments. One experiment was captured on an IncuCyte and two other experiments were captured on other microscopes mentioned above, totaling 35 videos.
Online supplemental material
Fig. S1 shows confocal images of MMTV-Wnt1 tumor sections depicting examples of structures that would not be counted as cells within cells. Fig. S2 shows images and quantifications of SA-βGAL staining of 4226 and MCF-7 cells, real-time PCR of SASP genes, and images of nutlin-induced senescent engulfment. Fig. S3 contains images of Western immunoblots of PARP and cleaved caspase 3 in chemotherapy-treated, senescent cultures. Fig. S4 shows fluorescent images of cell engulfment by senescent LAMP1-GFP MCF-7 cells also stained with LysoTracker. Fig. S5 shows two additional examples of individual, senescent 4226 cells engulfing apoptotic Jurkat cells and processing them to the lysosome where pHrodo fluorescence can be imaged. Video 1 shows a 3D reconstruction of an engulfed 4226 cell; Videos 2, 3, 4, and 5 show engulfment by doxorubicin-treated 4226 cells; Videos 6 and 7 show engulfment by doxorubicin-treated MCF-7 cells; Video 8 depicts 3D reconstruction of nutlin-induced senescence engulfment in 4226 cells; and Videos 9 and 10 show doxorubicin-induced senescence engulfment by LAMP1-GFP MCF-7 cells. Table S1 is a guide to the supplemental videos. Table S2 contains the statistical analyses corresponding to gene expression data in Fig. 7.
Supplementary Material
Acknowledgments
The authors acknowledge flow cytometry support from Dorota Wyczechowska, PhD; confocal microscopy support from Luis Marrero, PhD; and technical support from Julie Nguyen.
This study was supported by the U.S. Department of Defense Breast Cancer Research Program (grant number W81XWH-14-1-0216 to J.G. Jackson). The Cellular Immunology and Immune Metabolism Core at the Louisiana Cancer Research Consortium is supported by the National Institutes of Health National Institute of General Medical Sciences (grant number 1P30GM114732-01). The Louisiana Clinical and Translational Science Center is supported in part by the National Institute of General Medical Sciences (grant U54 GM104940). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare no competing financial interests.
Author contributions: C.A. Tonnessen-Murray conceived the study; created fluorescent tagged cell lines; performed engulfment, time course, survival, SA-βGAL, and lysosome experiments; performed microscopy, quantification, and statistical analyses; performed mRNA isolation and analyses; prepared figures; and wrote the manuscript draft. W.D. Frey performed time course experiments, gene expression analyses, entosis experiments, and engulfment experiments with drugs and edited the manuscript. S.G. Rao performed mouse studies, immunoblotting, and engulfment experiments. A. Shahbandi performed lysosome analysis, engulfment experiments, phagocytosis assays, and E-cadherin staining. N.A. Ungerleider performed gene expression analyses and prepared figures. J.O. Olayiwola created fluorescent tag–expressing cell lines. L.B. Murray performed engulfment experiments. B.T. Vinson, D.B. Chrisey, and C.A. Tonnessen-Murray generated time-lapse videos. C. Lord generated the CDH1 knockout cell line. J.G. Jackson supervised the study; prepared figures; performed engulfment, entosis, and phagocytosis experiments and analyses; and wrote the manuscript.
References
- Achuthan S., Santhoshkumar T.R., Prabhakar J., Nair S.A., and Pillai M.R.. 2011. Drug-induced senescence generates chemoresistant stemlike cells with low reactive oxygen species. J. Biol. Chem. 286:37813–37829. 10.1074/jbc.M110.200675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar T., Sen B., Aggarwal S., Nath R., Pathak N., Katoch A., Aiyaz M., Trehanpati N., Khosla S., and Ramakrishna G.. 2018. Differentially regulated gene expression in quiescence versus senescence and identification of ARID5A as a quiescence associated marker. J. Cell. Physiol. 233:3695–3712. [DOI] [PubMed] [Google Scholar]
- Aziz M., Yang W.L., and Wang P.. 2013. Measurement of phagocytic engulfment of apoptotic cells by macrophages using pHrodo succinimidyl ester. Curr. Protoc. Immunol. Chapter 14:Unit 14.31. [DOI] [PubMed] [Google Scholar]
- Bajrami I., Marlow R., van de Ven M., Brough R., Pemberton H.N., Frankum J., Song F., Rafiq R., Konde A., Krastev D.B., et al. 2018. E-Cadherin/ROS1 Inhibitor Synthetic Lethality in Breast Cancer. Cancer Discov. 8:498–515. 10.1158/2159-8290.CD-17-0603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosh T.J., Ullah M., Zeitouni S., Beaver J., and Prockop D.J.. 2016. Cancer cells enter dormancy after cannibalizing mesenchymal stem/stromal cells (MSCs). Proc. Natl. Acad. Sci. USA. 113:E6447–E6456. 10.1073/pnas.1612290113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertheau P., Plassa F., Espié M., Turpin E., de Roquancourt A., Marty M., Lerebours F., Beuzard Y., Janin A., and de Thé H.. 2002. Effect of mutated TP53 on response of advanced breast cancers to high-dose chemotherapy. Lancet. 360:852–854. 10.1016/S0140-6736(02)09969-5 [DOI] [PubMed] [Google Scholar]
- Bertheau P., Turpin E., Rickman D.S., Espié M., de Reyniès A., Feugeas J.P., Plassa L.F., Soliman H., Varna M., de Roquancourt A., et al. 2007. Exquisite sensitivity of TP53 mutant and basal breast cancers to a dose-dense epirubicin-cyclophosphamide regimen. PLoS Med. 4:e90 10.1371/journal.pmed.0040090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.M., and Wouters B.G.. 1999. Apoptosis, p53, and tumor cell sensitivity to anticancer agents. Cancer Res. 59:1391–1399. [PubMed] [Google Scholar]
- Bunz F., Hwang P.M., Torrance C., Waldman T., Zhang Y., Dillehay L., Williams J., Lengauer C., Kinzler K.W., and Vogelstein B.. 1999. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J. Clin. Invest. 104:263–269. 10.1172/JCI6863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahu J., Bustany S., and Sola B.. 2012. Senescence-associated secretory phenotype favors the emergence of cancer stem-like cells. Cell Death Dis. 3:e446 10.1038/cddis.2012.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canino C., Mori F., Cambria A., Diamantini A., Germoni S., Alessandrini G., Borsellino G., Galati R., Battistini L., Blandino R., et al. 2012. SASP mediates chemoresistance and tumor-initiating-activity of mesothelioma cells. Oncogene. 31:3148–3163. 10.1038/onc.2011.485 [DOI] [PubMed] [Google Scholar]
- Chang B.D., Broude E.V., Dokmanovic M., Zhu H., Ruth A., Xuan Y., Kandel E.S., Lausch E., Christov K., and Roninson I.B.. 1999a A senescence-like phenotype distinguishes tumor cells that undergo terminal proliferation arrest after exposure to anticancer agents. Cancer Res. 59:3761–3767. [PubMed] [Google Scholar]
- Chang B.D., Xuan Y., Broude E.V., Zhu H., Schott B., Fang J., and Roninson I.B.. 1999b Role of p53 and p21waf1/cip1 in senescence-like terminal proliferation arrest induced in human tumor cells by chemotherapeutic drugs. Oncogene. 18:4808–4818. 10.1038/sj.onc.1203078 [DOI] [PubMed] [Google Scholar]
- Chen M.B., Zhu Y.Q., Xu J.Y., Wang L.Q., Liu C.Y., Ji Z.Y., and Lu P.H.. 2012. Value of TP53 status for predicting response to neoadjuvant chemotherapy in breast cancer: a meta-analysis. PLoS One. 7:e39655 10.1371/journal.pone.0039655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri G.P.X., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O., et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA. 92:9363–9367. 10.1073/pnas.92.20.9363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durgan J., Tseng Y.Y., Hamann J.C., Domart M.C., Collinson L., Hall A., Overholtzer M., and Florey O.. 2017. Mitosis can drive cell cannibalism through entosis. eLife. 6:e27134 10.7554/eLife.27134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encinas M., Iglesias M., Liu Y., Wang H., Muhaisen A., Ceña V., Gallego C., and Comella J.X.. 2000. Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J. Neurochem. 75:991–1003. 10.1046/j.1471-4159.2000.0750991.x [DOI] [PubMed] [Google Scholar]
- Erol A. 2010. Systemic DNA damage response and metabolic syndrome as a premalignant state. Curr. Mol. Med. 10:321–334. 10.2174/156652410791065282 [DOI] [PubMed] [Google Scholar]
- Esserman L.J., Berry D.A., Cheang M.C., Yau C., Perou C.M., Carey L., DeMichele A., Gray J.W., Conway-Dorsey K., Lenburg M.E., et al. I-SPY 1 TRIAL Investigators . 2012. Chemotherapy response and recurrence-free survival in neoadjuvant breast cancer depends on biomarker profiles: results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657). Breast Cancer Res. Treat. 132:1049–1062. 10.1007/s10549-011-1895-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gey C., and Seeger K.. 2013. Metabolic changes during cellular senescence investigated by proton NMR-spectroscopy. Mech. Ageing Dev. 134:130–138. 10.1016/j.mad.2013.02.002 [DOI] [PubMed] [Google Scholar]
- Goetz M.P., Kalari K.R., Suman V.J., Moyer A.M., Yu J., Visscher D.W., Dockter T.J., Vedell P.T., Sinnwell J.P., Tang X., et al. 2017. Tumor Sequencing and Patient-Derived Xenografts in the Neoadjuvant Treatment of Breast Cancer. J. Natl. Cancer Inst. 109:djw306 10.1093/jnci/djw306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M., and Botelho R.J.. 2017. Phagocytosis: Hungry, Hungry Cells. Methods Mol. Biol. 1519:1–16. 10.1007/978-1-4939-6581-6_1 [DOI] [PubMed] [Google Scholar]
- Hamann J.C., Surcel A., Chen R., Teragawa C., Albeck J.G., Robinson D.N., and Overholtzer M.. 2017. Entosis Is Induced by Glucose Starvation. Cell Reports. 20:201–210. 10.1016/j.celrep.2017.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Deo D., Xia M., and Vassilev L.T.. 2009. Pharmacologic p53 activation blocks cell cycle progression but fails to induce senescence in epithelial cancer cells. Mol. Cancer Res. 7:1497–1509. 10.1158/1541-7786.MCR-09-0144 [DOI] [PubMed] [Google Scholar]
- Jackson J.G., and Pereira-Smith O.M.. 2006. Primary and compensatory roles for RB family members at cell cycle gene promoters that are deacetylated and downregulated in doxorubicin-induced senescence of breast cancer cells. Mol. Cell. Biol. 26:2501–2510. 10.1128/MCB.26.7.2501-2510.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J.G., and Yee D.. 1999. IRS-1 expression and activation are not sufficient to activate downstream pathways and enable IGF-I growth response in estrogen receptor negative breast cancer cells. Growth Horm. IGF Res. 9:280–289. 10.1054/ghir.1999.0113 [DOI] [PubMed] [Google Scholar]
- Jackson J.G., White M.F., and Yee D.. 1998. Insulin receptor substrate-1 is the predominant signaling molecule activated by insulin-like growth factor-I, insulin, and interleukin-4 in estrogen receptor-positive human breast cancer cells. J. Biol. Chem. 273:9994–10003. 10.1074/jbc.273.16.9994 [DOI] [PubMed] [Google Scholar]
- Jackson J.G., Pant V., Li Q., Chang L.L., Quintás-Cardama A., Garza D., Tavana O., Yang P., Manshouri T., Li Y., et al. 2012. p53-mediated senescence impairs the apoptotic response to chemotherapy and clinical outcome in breast cancer. Cancer Cell. 21:793–806. 10.1016/j.ccr.2012.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.C., Han N.K., Byun H.O., Kim S.S., Ahn E.K., Chu I.S., Leem S.H., Lee C.K., and Lee J.S.. 2010. Time-dependently expressed markers and the characterization for premature senescence induced by ionizing radiation in MCF7. Oncol. Rep. 24:395–403. [DOI] [PubMed] [Google Scholar]
- Kim J.S., Kim E.J., Kim H.J., Yang J.Y., Hwang G.S., and Kim C.W.. 2011. Proteomic and metabolomic analysis of H2O2-induced premature senescent human mesenchymal stem cells. Exp. Gerontol. 46:500–510. 10.1016/j.exger.2011.02.012 [DOI] [PubMed] [Google Scholar]
- Krishna S., and Overholtzer M.. 2016. Mechanisms and consequences of entosis. Cell. Mol. Life Sci. 73:2379–2386. 10.1007/s00018-016-2207-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz D.J., Decary S., Hong Y., and Erusalimsky J.D.. 2000. Senescence-associated (beta)-galactosidase reflects an increase in lysosomal mass during replicative ageing of human endothelial cells. J. Cell Sci. 113:3613–3622. [DOI] [PubMed] [Google Scholar]
- Lee B.Y., Han J.A., Im J.S., Morrone A., Johung K., Goodwin E.C., Kleijer W.J., DiMaio D., and Hwang E.S.. 2006. Senescence-associated beta-galactosidase is lysosomal beta-galactosidase. Aging Cell. 5:187–195. 10.1111/j.1474-9726.2006.00199.x [DOI] [PubMed] [Google Scholar]
- Ley K., Pramod A.B., Croft M., Ravichandran K.S., and Ting J.P.. 2016. How Mouse Macrophages Sense What Is Going On. Front. Immunol. 7:204 10.3389/fimmu.2016.00204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwiniec A., Grzanka A., Helmin-Basa A., Gackowska L., and Grzanka D.. 2010. Features of senescence and cell death induced by doxorubicin in A549 cells: organization and level of selected cytoskeletal proteins. J. Cancer Res. Clin. Oncol. 136:717–736. 10.1007/s00432-009-0711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins I., Raza S.Q., Voisin L., Dakhli H., Allouch A., Law F., Sabino D., De Jong D., Thoreau M., Mintet E., et al. 2018. Anticancer chemotherapy and radiotherapy trigger both non-cell-autonomous and cell-autonomous death. Cell Death Dis. 9:716 10.1038/s41419-018-0747-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Oshima K., Naoi Y., Nakayama T., Kim S.J., Shimazu K., Shimomura A., Maruyama N., Tamaki Y., and Noguchi S.. 2012. 14-3-3σ expression is associated with poor pathological complete response to neoadjuvant chemotherapy in human breast cancers. Breast Cancer Res. Treat. 134:229–236. 10.1007/s10549-012-1976-x [DOI] [PubMed] [Google Scholar]
- Overholtzer M., Mailleux A.A., Mouneimne G., Normand G., Schnitt S.J., King R.W., Cibas E.S., and Brugge J.S.. 2007. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell. 131:966–979. 10.1016/j.cell.2007.10.040 [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quijano C., Cao L., Fergusson M.M., Romero H., Liu J., Gutkind S., Rovira I.I., Mohney R.P., Karoly E.D., and Finkel T.. 2012. Oncogene-induced senescence results in marked metabolic and bioenergetic alterations. Cell Cycle. 11:1383–1392. 10.4161/cc.19800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S.G., and Jackson J.G.. 2016. SASP: Tumor Suppressor or Promoter? Yes! Trends Cancer. 2:676–687. 10.1016/j.trecan.2016.10.001 [DOI] [PubMed] [Google Scholar]
- Rastogi S., Joshi B., Dasgupta P., Morris M., Wright K., and Chellappan S.. 2006. Prohibitin facilitates cellular senescence by recruiting specific corepressors to inhibit E2F target genes. Mol. Cell. Biol. 26:4161–4171. 10.1128/MCB.02142-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier F., Coppé J.P., Patil C.K., Hoeijmakers W.A., Muñoz D.P., Raza S.R., Freund A., Campeau E., Davalos A.R., and Campisi J.. 2009. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 11:973–979. 10.1038/ncb1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosemblit C., Datta J., Lowenfeld L., Xu S., Basu A., Kodumudi K., Wiener D., and Czerniecki B.J.. 2018. Oncodriver inhibition and CD4+ Th1 cytokines cooperate through Stat1 activation to induce tumor senescence and apoptosis in HER2+ and triple negative breast cancer: implications for combining immune and targeted therapies. Oncotarget. 9:23058–23077. 10.18632/oncotarget.25208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbandi A., and Jackson J.G.. 2019. Analysis across multiple tumor types provides no evidence that mutant p53 exerts dominant negative activity. npj Precision Oncol. 3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless N.E., and Sherr C.J.. 2015. Forging a signature of in vivo senescence. Nat. Rev. Cancer. 15:397–408. 10.1038/nrc3960 [DOI] [PubMed] [Google Scholar]
- Sun Q., and Overholtzer M.. 2013. Methods for the study of entosis. Methods Mol. Biol. 1004:59–66. 10.1007/978-1-62703-383-1_5 [DOI] [PubMed] [Google Scholar]
- Sun Q., Cibas E.S., Huang H., Hodgson L., and Overholtzer M.. 2014a Induction of entosis by epithelial cadherin expression. Cell Res. 24:1288–1298. 10.1038/cr.2014.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Luo T., Ren Y., Florey O., Shirasawa S., Sasazuki T., Robinson D.N., and Overholtzer M.. 2014b Competition between human cells by entosis. Cell Res. 24:1299–1310. 10.1038/cr.2014.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Poele R.H., Okorokov A.L., Jardine L., Cummings J., and Joel S.P.. 2002. DNA damage is able to induce senescence in tumor cells in vitro and in vivo. Cancer Res. 62:1876–1883. [PubMed] [Google Scholar]
- Tonnessen-Murray C., Lozano G., and Jackson J.G.. 2016. The p53 Protein: From Cell Regulation to Cancer. In Lozano G., and Levine A.J., editors. The p53 Protein: From Cell Regulation to Cancer. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 173–186. [Google Scholar]
- Tonnessen-Murray C., Ungerleider N.A., Rao S.G., Wasylishen A.R., Frey W.D., and Jackson J.G.. 2018. p53 Mediates Vast Gene Expression Changes That Contribute to Poor Chemotherapeutic Response in a Mouse Model of Breast Cancer. Transl. Oncol. 11:930–940. 10.1016/j.tranon.2018.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toso A., Revandkar A., Di Mitri D., Guccini I., Proietti M., Sarti M., Pinton S., Zhang J., Kalathur M., Civenni G., et al. 2014. Enhancing chemotherapy efficacy in Pten-deficient prostate tumors by activating the senescence-associated antitumor immunity. Cell Reports. 9:75–89. 10.1016/j.celrep.2014.08.044 [DOI] [PubMed] [Google Scholar]
- Ungerleider N.A., Rao S.G., Shahbandi A., Yee D., Niu T., Frey W.D., and Jackson J.G.. 2018. Breast cancer survival predicted by TP53 mutation status differs markedly depending on treatment. Breast Cancer Res. 20:115 10.1186/s13058-018-1044-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev L.T., Vu B.T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., et al. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 303:844–848. 10.1126/science.1092472 [DOI] [PubMed] [Google Scholar]
- Velarde M.C., Demaria M., and Campisi J.. 2013. Senescent cells and their secretory phenotype as targets for cancer therapy. Interdiscip. Top. Gerontol. 38:17–27. 10.1159/000343572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xu Y., Chen J., Ouyang T., Li J., Wang T., Fan Z., Fan T., Lin B., and Xie Y.. 2016. TP53 mutations are associated with higher rates of pathologic complete response to anthracycline/cyclophosphamide-based neoadjuvant chemotherapy in operable primary breast cancer. Int. J. Cancer. 138:489–496. 10.1002/ijc.29715 [DOI] [PubMed] [Google Scholar]
- Zwerschke W., Mazurek S., Stöckl P., Hütter E., Eigenbrodt E., and Jansen-Dürr P.. 2003. Metabolic analysis of senescent human fibroblasts reveals a role for AMP in cellular senescence. Biochem. J. 376:403–411. 10.1042/bj20030816 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.