In the title compound, intermolecular C—H⋯O, O—H⋯O and N—H⋯O hydrogen bonds link the molecules into a three-dimensional supramolecular network.
Keywords: crystal structure, hydrogen bonding, Hirshfeld surface analysis, pyridazine
Abstract
In the title compound, C13H14N2O3, the dihydropyridazine ring (r.m.s. deviation = 0.166 Å) has a screw-boat conformation. The dihedral angle between its mean plane and the benzene ring is 0.77 (12)°. In the crystal, intermolecular O—H⋯O hydrogen bonds generate C(5) chains and N—H⋯O hydrogen bonds produce R
2
2(8) motifs. These types of interactions lead to the formation of layers parallel to (12 ). The three-dimensional network is achieved by C—H⋯O interactions, including R
2
4(8) motifs. Intermolecular interactions were additionally investigated using Hirshfeld surface analysis and two-dimensional fingerprint plots. The most significant contributions to the crystal packing are by H⋯H (43.3%), H⋯C/C⋯H (19.3%), H⋯O/H⋯O (22.6%), C⋯N/N⋯C (3.0%) and H⋯N/N⋯H (5.8%) contacts. C—H⋯π interactions and aromatic π–π stacking interactions are not observed.
). The three-dimensional network is achieved by C—H⋯O interactions, including R
2
4(8) motifs. Intermolecular interactions were additionally investigated using Hirshfeld surface analysis and two-dimensional fingerprint plots. The most significant contributions to the crystal packing are by H⋯H (43.3%), H⋯C/C⋯H (19.3%), H⋯O/H⋯O (22.6%), C⋯N/N⋯C (3.0%) and H⋯N/N⋯H (5.8%) contacts. C—H⋯π interactions and aromatic π–π stacking interactions are not observed.
Chemical context
For decades the chemistry of pyridazinones has been an interesting field. This nitrogen heterocycle became a scaffold of choice for the development of potential drug candidates (Akhtar et al., 2016 ▸; Dubey & Bhosle, 2015 ▸) because pyridazinone and its substituted derivatives are important pharmacophores possessing many different biological applications (Asif, 2014 ▸). Such compounds are used as anti-HIV (Livermore et al., 1993 ▸), antimicrobial (Sönmez et al., 2006 ▸), anticonvulsant (Partap et al., 2018 ▸), antihypertensive (Siddiqui et al., 2011 ▸), antidepressant (Boukharsa et al., 2016 ▸), analgesic (Gökçe et al., 2009 ▸), anti-inflammatory (Barberot et al., 2018 ▸), antihistaminic (Tao et al. 2012 ▸), cardiotonic (Wang et al., 2008 ▸) and herbicidal agents (Asif, 2013 ▸) or as glucan synthase inhibitors (Zhou et al., 2011 ▸).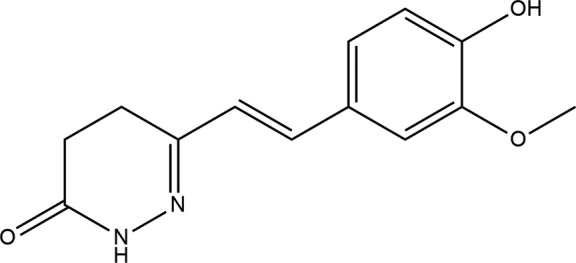
In continuation of our studies related to molecular structures and Hirshfeld surface analysis of new heterocyclic derivatives (Daoui et al., 2019a ▸,b ▸; El Kalai et al., 2019 ▸; Karrouchi et al., 2015 ▸), we report herein on the synthesis, molecular and crystal structures of (E)-6-(4-hydroxy-3-methoxystyryl)-4,5-dihydropyridazin-3(2H)-one, as well as an analysis of the Hirshfeld surfaces.
Structural commentary
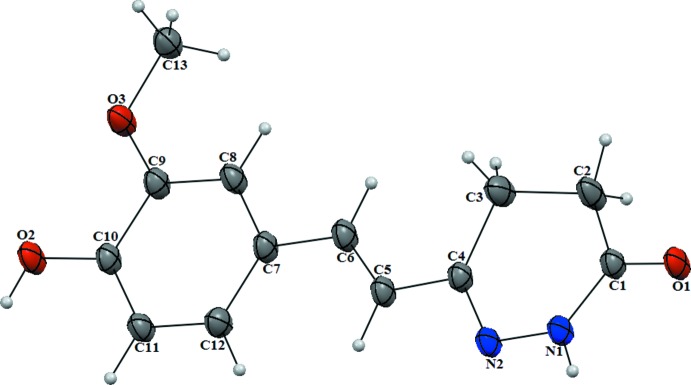
In the title molecule (Fig. 1 ▸), the configuration relative to the double bond at C5 and C6 is E. The dihydropyridazine ring has a screw-boat conformation, with an r.m.s. deviation of 0.166 Å for the ring atoms, with the maximum deviation from the ring being 0.178 (3) Å for the C3 atom; the C2 atom lies −0.177 (3) Å out of the plane in the opposite direction relative to the C3 atom. The dihedral angle between the dihydropyridazine ring mean plane and the benzene ring (C7–C12) is 0.77 (12)°, indicating an almost planar conformation of the molecule favouring delocalization over the C4—C5=C6—C7 bridge.
Figure 1.
The molecular structure of the title compound. Displacement ellipsoids are drawn at the 50% probability level.
Supramolecular features
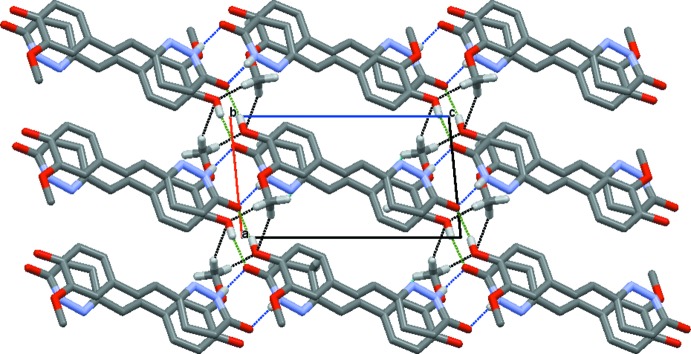
In the crystal, molecules are stacked in rows parallel to [100]. Notably, no significant C—H⋯π or π–π interactions are observed. O2—H2⋯O1i hydrogen bonds between the phenolic OH group and the carbonyl O atom of a neighbouring molecule generate C(5) chains extending parallel to [101]. Likewise, N1—H1⋯O1ii hydrogen bonds between the N—H function of the dihydropyridazine ring and the carbonyl O atom generate centrosymmetric dimers with an 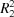 (8) motif. The two types of hydrogen bonding result in the formation of layers parallel to (12
(8) motif. The two types of hydrogen bonding result in the formation of layers parallel to (12 ). A three-dimensional supramolecular network is eventually formed through intermolecular C13—H13A⋯O2iii and C13—H13C⋯O2iv hydrogen bonds with
). A three-dimensional supramolecular network is eventually formed through intermolecular C13—H13A⋯O2iii and C13—H13C⋯O2iv hydrogen bonds with 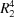 (8) motifs (Fig. 2 ▸ and Table 1 ▸).
(8) motifs (Fig. 2 ▸ and Table 1 ▸).
Figure 2.
The crystal packing of the title compound, with N—H⋯O, O—H⋯O and C—H⋯O interactions shown as blue, green and black dashed lines, respectively.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O2—H2⋯O1i | 0.82 | 1.86 | 2.671 (2) | 168 |
| N1—H1⋯O1ii | 0.86 | 2.02 | 2.875 (3) | 170 |
| C13—H13A⋯O2iii | 0.96 | 2.51 | 3.465 (3) | 172 |
| C13—H13C⋯O2iv | 0.96 | 2.57 | 3.489 (4) | 159 |
Symmetry codes: (i) 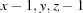 ; (ii)
; (ii) 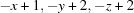 ; (iii)
; (iii) 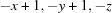 ; (iv)
; (iv) 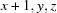 .
.
Database survey
A search of the Cambridge Structural Database (CSD, Version 5.40, update November 2018; Groom et al., 2016 ▸) revealed two structures containing a similar pyridazinone moiety as in the title structure but with different substituents, viz. 6-phenyl-4,5-dihydropyridazin-3(2H)-one (CSD refcode TADQUL; Abourichaa et al., 2003 ▸) and (R)-(−)-6-(4-aminophenyl)-5-methyl-4,5-dihydropyridazin-3(2H)-one (ADIGOK; Zhang et al., 2006 ▸). In the structure of TADQUL, the dihydropyridazine ring adopts a half-chair conformation, with atoms C1, N2, N3 and C4 in a common plane, with C5 0.222 (2) Å and C6 0.262 (2) Å on opposite sides of this plane. The plane is almost coplanar with the 4-aminophenyl ring, the dihedral angle between the two planes being 1.73 (9) Å. In the crystal, hydrogen-bonded centrosymmetric dimers are observed. The O1=C1 bond length is 1.2316 (14) Å. The N3—C4, N2—N3 and N2—C1 bond lengths are 1.3464 (15), 1.3877 (14) and 1.2830 (15) Å, respectively. In the structure of ADIGOK, the asymmetric unit consists of two molecules of the same enantiomer, and the crystal packing is stabilized by intermolecular N—H⋯O hydrogen bonds.
Hirshfeld surface analysis
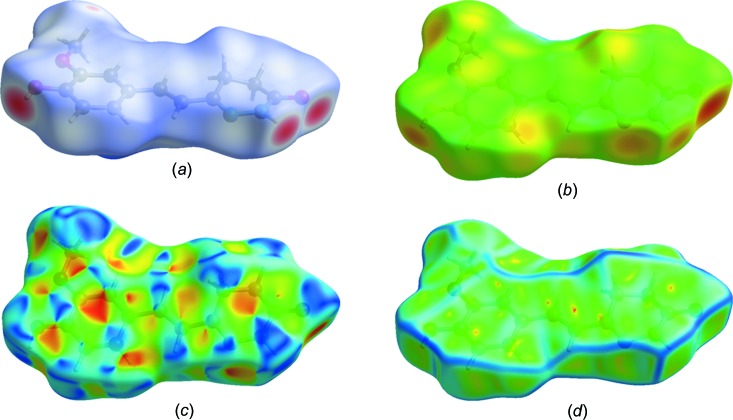
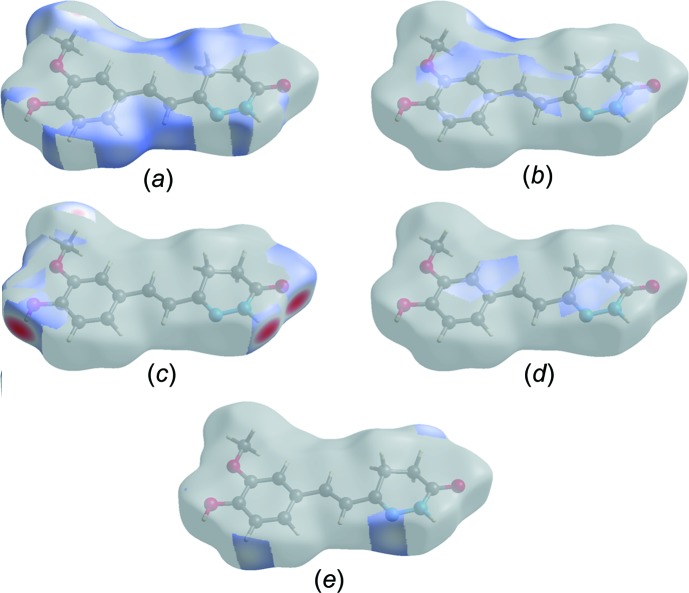
Hirshfeld surface analysis was used to quantify the intermolecular interactions of the title compound, using CrystalExplorer17.5 (Turner et al., 2017 ▸). The Hirshfeld surface analysis was planned using a standard (high) surface resolution with the three-dimensional d norm surfaces plotted over a fixed colour scale of −0.7021 (red) to 2.2382 a.u. (blue). The surfaces mapped over relevant intermolecular contacts are illustrated in Fig. 3 ▸. The Hirshfeld surface representations with the function d norm plotted onto the surface are shown for the H⋯H, H⋯C/C⋯H, H⋯O/O⋯H, C⋯N/N⋯C and H⋯N/N⋯H interactions in Figs. 4 ▸(a)–(e), respectively. The overall two-dimensional fingerprint plot and those delineated into H⋯H, H⋯C/C⋯H, H⋯O/O⋯H, C⋯N/N⋯C and H⋯N/N⋯H contacts are illustrated in Figs. 5 ▸(a)–(f), respectively. The largest interaction is that of H⋯H, contributing 43.3% to the overall crystal packing. H⋯C/C⋯H contacts add a 19.3% contribution to the Hirshfeld surface, with the tips at d e + d i ∼ 2.72 Å. H⋯O/O⋯H contacts make a 22.6% contribution to the Hirshfeld surface and are represented by a pair of sharp spikes in the region d e + d i ∼ 2.70 Å in the fingerprint plot. H⋯O/O⋯H interactions arise from intermolecular O—H⋯O hydrogen bonding and C—H⋯O contacts. The contributions of the other contacts to the Hirshfeld surface are negligible, i.e. C⋯N/N⋯C of 3.0% and H⋯N/N⋯H of 5.8%.
Figure 3.
(a) d norm mapped on the Hirshfeld surface for visualizing the intermolecular interactions, (b) d e mapped on the surface, (c) shape-index map of the title compound and (d) curvedness map of the title compound using a range from −4 to 4 Å.
Figure 4.
The Hirshfeld surface representations with the function d norm plotted onto the surface for (a) H⋯H, (b) H⋯C/ C⋯H, (c) H⋯O/O⋯H, (d) C⋯N/N⋯C and (e) H⋯N/N⋯H interactions.
Figure 5.
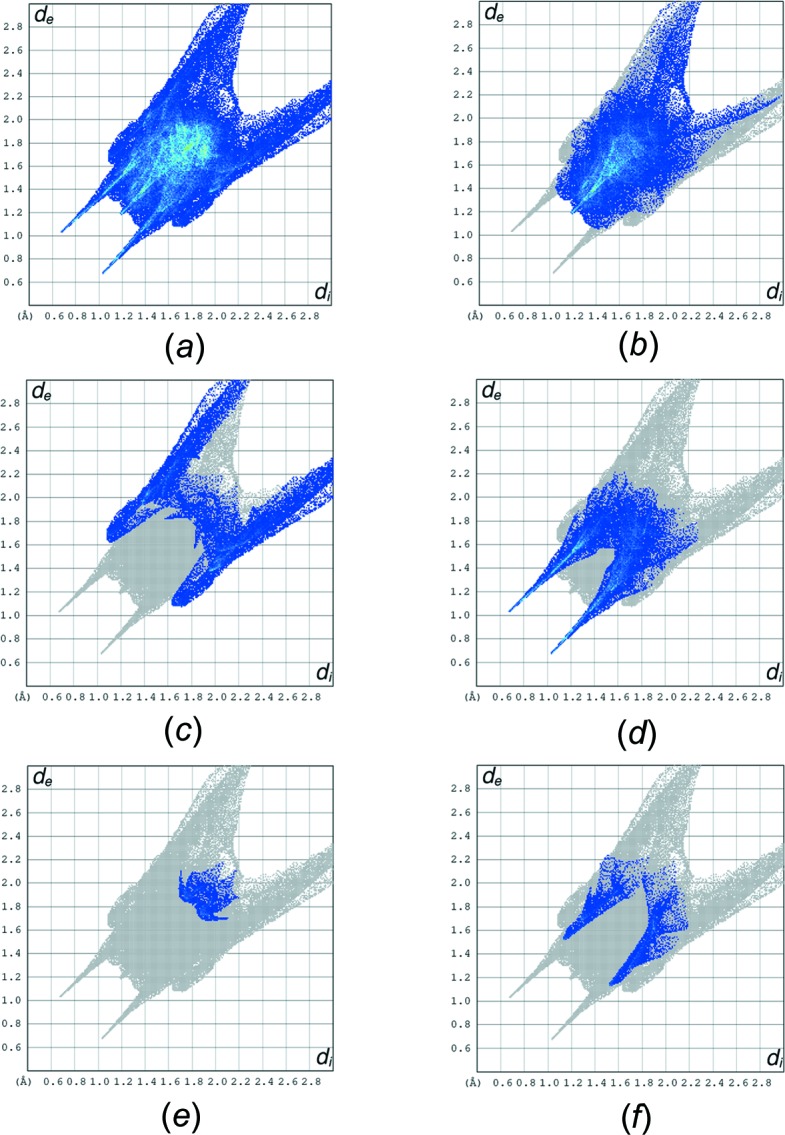
The full two-dimensional fingerprint plots for the title compound, showing (a) all interactions, and delineated into (b) H⋯H, (c) H⋯C/ C⋯H, (d) H⋯O/O⋯H, (e) C⋯N/N⋯C and (f) H⋯N/N⋯H interactions.
Synthesis and crystallization
To a solution of 6-(4-hydroxy-3-methoxyphenyl)-4-oxohex-5-enoic acid (0.25 g, 1 mmol) in 20 ml of ethanol, an equimolar amount of hydrazine hydrate was added. The mixture was maintained under reflux until thin-layer chromatography (TLC) indicated the end of the reaction. After cooling, the precipitate which formed was filtered off, washed with ethanol and recrystallized from ethanol. Slow evaporation at room temperature led to the formation of single crystals of the title compound.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. H atoms on C atoms were placed in idealized positions and refined as riding, with C—H = 0.93–0.97 Å and U iso(H) = 1.5U eq(C) for methyl H atoms and 1.2U eq(C) otherwise. The NH and OH hydrogens were located in a difference Fourier map and were constrained with N—H = 0.86 Å and U iso(H) = 1.2U eq(N), and O—H = 0.86 Å and U iso(H) = 1.5U eq(O), using a riding model.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C13H14N2O3 |
| M r | 246.26 |
| Crystal system, space group | Triclinic, P
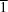
|
| Temperature (K) | 293 |
| a, b, c (Å) | 6.0828 (9), 9.4246 (13), 11.1724 (16) |
| α, β, γ (°) | 75.838 (11), 83.099 (12), 84.059 (11) |
| V (Å3) | 614.70 (16) |
| Z | 2 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.10 |
| Crystal size (mm) | 0.72 × 0.39 × 0.16 |
| Data collection | |
| Diffractometer | Stoe IPDS 2 |
| Absorption correction | Integration (X-RED32; Stoe & Cie, 2002 ▸) |
| T min, T max | 0.944, 0.989 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 6563, 2426, 1506 |
| R int | 0.054 |
| (sin θ/λ)max (Å−1) | 0.617 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.056, 0.147, 1.00 |
| No. of reflections | 2426 |
| No. of parameters | 165 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.17, −0.17 |
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989019014130/wm5521sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989019014130/wm5521Isup3.hkl
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors acknowledge the Faculty of Arts and Sciences, Ondokuz Mayıs University, Turkey, for the use of the Stoe IPDS 2 diffractometer (purchased under grant F.279 of the University Research Fund).
supplementary crystallographic information
Crystal data
| C13H14N2O3 | Z = 2 |
| Mr = 246.26 | F(000) = 260 |
| Triclinic, P1 | Dx = 1.330 Mg m−3 |
| a = 6.0828 (9) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 9.4246 (13) Å | Cell parameters from 13077 reflections |
| c = 11.1724 (16) Å | θ = 2.2–30.7° |
| α = 75.838 (11)° | µ = 0.10 mm−1 |
| β = 83.099 (12)° | T = 293 K |
| γ = 84.059 (11)° | Prism, yellow |
| V = 614.70 (16) Å3 | 0.72 × 0.39 × 0.16 mm |
Data collection
| Stoe IPDS 2 diffractometer | 1506 reflections with I > 2σ(I) |
| Detector resolution: 6.67 pixels mm-1 | Rint = 0.054 |
| rotation method scans | θmax = 26.0°, θmin = 2.2° |
| Absorption correction: integration (X-RED32; Stoe & Cie, 2002) | h = −7→7 |
| Tmin = 0.944, Tmax = 0.989 | k = −11→11 |
| 6563 measured reflections | l = −13→13 |
| 2426 independent reflections |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.056 | H-atom parameters constrained |
| wR(F2) = 0.147 | w = 1/[σ2(Fo2) + (0.0747P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.00 | (Δ/σ)max < 0.001 |
| 2426 reflections | Δρmax = 0.17 e Å−3 |
| 165 parameters | Δρmin = −0.17 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.7518 (3) | 0.88682 (19) | 0.99156 (15) | 0.0601 (5) | |
| O3 | 0.4960 (3) | 0.58008 (19) | 0.11563 (16) | 0.0614 (5) | |
| O2 | 0.1290 (3) | 0.7317 (2) | 0.05778 (16) | 0.0636 (5) | |
| H2 | 0.016474 | 0.786724 | 0.044599 | 0.095* | |
| N1 | 0.4947 (3) | 0.9323 (2) | 0.85757 (17) | 0.0525 (5) | |
| H1 | 0.429651 | 0.995567 | 0.896673 | 0.063* | |
| N2 | 0.3918 (3) | 0.9139 (2) | 0.75870 (17) | 0.0519 (5) | |
| C9 | 0.3982 (4) | 0.6576 (2) | 0.1986 (2) | 0.0497 (6) | |
| C5 | 0.3927 (4) | 0.8291 (3) | 0.5807 (2) | 0.0506 (6) | |
| H5 | 0.252921 | 0.878478 | 0.572201 | 0.061* | |
| C1 | 0.6829 (4) | 0.8631 (3) | 0.8987 (2) | 0.0492 (6) | |
| C4 | 0.5043 (4) | 0.8412 (2) | 0.6851 (2) | 0.0472 (6) | |
| C8 | 0.4803 (4) | 0.6645 (3) | 0.3066 (2) | 0.0521 (6) | |
| H8 | 0.616062 | 0.614251 | 0.326018 | 0.062* | |
| C11 | 0.0780 (4) | 0.8118 (3) | 0.2494 (2) | 0.0538 (6) | |
| H11 | −0.059111 | 0.860442 | 0.230853 | 0.065* | |
| C7 | 0.3664 (4) | 0.7445 (2) | 0.3875 (2) | 0.0496 (6) | |
| C10 | 0.1954 (4) | 0.7364 (2) | 0.1681 (2) | 0.0487 (6) | |
| C6 | 0.4707 (4) | 0.7544 (3) | 0.4962 (2) | 0.0549 (6) | |
| H6 | 0.607897 | 0.702171 | 0.507391 | 0.066* | |
| C12 | 0.1607 (4) | 0.8164 (3) | 0.3580 (2) | 0.0552 (6) | |
| H12 | 0.078822 | 0.867751 | 0.411814 | 0.066* | |
| C13 | 0.7007 (4) | 0.4965 (3) | 0.1419 (3) | 0.0614 (7) | |
| H13A | 0.745228 | 0.441469 | 0.079994 | 0.092* | |
| H13B | 0.682204 | 0.430200 | 0.222071 | 0.092* | |
| H13C | 0.812721 | 0.561367 | 0.141331 | 0.092* | |
| C2 | 0.7972 (5) | 0.7565 (3) | 0.8299 (3) | 0.0694 (8) | |
| H2A | 0.761992 | 0.658432 | 0.875007 | 0.083* | |
| H2B | 0.956271 | 0.761159 | 0.827335 | 0.083* | |
| C3 | 0.7375 (4) | 0.7807 (3) | 0.7002 (2) | 0.0673 (8) | |
| H3A | 0.836608 | 0.847839 | 0.645178 | 0.081* | |
| H3B | 0.760060 | 0.688125 | 0.675416 | 0.081* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0632 (11) | 0.0804 (12) | 0.0479 (10) | 0.0136 (8) | −0.0292 (8) | −0.0331 (8) |
| O3 | 0.0642 (11) | 0.0739 (11) | 0.0579 (10) | 0.0218 (8) | −0.0273 (9) | −0.0392 (9) |
| O2 | 0.0633 (12) | 0.0859 (13) | 0.0537 (10) | 0.0146 (9) | −0.0326 (9) | −0.0349 (9) |
| N1 | 0.0536 (12) | 0.0686 (13) | 0.0448 (11) | 0.0094 (9) | −0.0204 (9) | −0.0299 (10) |
| N2 | 0.0510 (12) | 0.0670 (13) | 0.0450 (11) | 0.0068 (9) | −0.0213 (9) | −0.0237 (10) |
| C9 | 0.0569 (14) | 0.0515 (13) | 0.0479 (13) | 0.0029 (11) | −0.0190 (11) | −0.0217 (11) |
| C5 | 0.0550 (14) | 0.0602 (14) | 0.0422 (12) | 0.0029 (11) | −0.0196 (11) | −0.0185 (11) |
| C1 | 0.0539 (14) | 0.0552 (14) | 0.0427 (12) | 0.0056 (11) | −0.0182 (11) | −0.0172 (11) |
| C4 | 0.0524 (14) | 0.0522 (13) | 0.0416 (12) | 0.0007 (10) | −0.0152 (11) | −0.0168 (10) |
| C8 | 0.0533 (14) | 0.0584 (14) | 0.0511 (14) | 0.0074 (11) | −0.0248 (12) | −0.0207 (11) |
| C11 | 0.0495 (13) | 0.0675 (15) | 0.0506 (14) | 0.0090 (11) | −0.0211 (11) | −0.0237 (12) |
| C7 | 0.0579 (14) | 0.0565 (14) | 0.0412 (12) | 0.0011 (11) | −0.0181 (11) | −0.0204 (11) |
| C10 | 0.0540 (14) | 0.0560 (14) | 0.0432 (13) | −0.0004 (11) | −0.0192 (11) | −0.0202 (11) |
| C6 | 0.0601 (15) | 0.0640 (15) | 0.0467 (13) | 0.0033 (12) | −0.0229 (12) | −0.0197 (12) |
| C12 | 0.0558 (15) | 0.0681 (15) | 0.0490 (14) | 0.0056 (12) | −0.0149 (12) | −0.0274 (12) |
| C13 | 0.0656 (16) | 0.0634 (15) | 0.0600 (16) | 0.0144 (12) | −0.0203 (13) | −0.0248 (13) |
| C2 | 0.0734 (18) | 0.0867 (19) | 0.0599 (16) | 0.0316 (14) | −0.0365 (14) | −0.0406 (14) |
| C3 | 0.0576 (16) | 0.098 (2) | 0.0593 (16) | 0.0136 (14) | −0.0231 (13) | −0.0423 (15) |
Geometric parameters (Å, º)
| O1—C1 | 1.241 (3) | C8—H8 | 0.9300 |
| O3—C9 | 1.362 (3) | C11—C10 | 1.377 (3) |
| O3—C13 | 1.424 (3) | C11—C12 | 1.379 (3) |
| O2—C10 | 1.355 (2) | C11—H11 | 0.9300 |
| O2—H2 | 0.8200 | C7—C12 | 1.396 (3) |
| N1—C1 | 1.331 (3) | C7—C6 | 1.462 (3) |
| N1—N2 | 1.387 (2) | C6—H6 | 0.9300 |
| N1—H1 | 0.8600 | C12—H12 | 0.9300 |
| N2—C4 | 1.288 (3) | C13—H13A | 0.9600 |
| C9—C8 | 1.378 (3) | C13—H13B | 0.9600 |
| C9—C10 | 1.406 (3) | C13—H13C | 0.9600 |
| C5—C6 | 1.327 (3) | C2—C3 | 1.493 (3) |
| C5—C4 | 1.451 (3) | C2—H2A | 0.9700 |
| C5—H5 | 0.9300 | C2—H2B | 0.9700 |
| C1—C2 | 1.480 (3) | C3—H3A | 0.9700 |
| C4—C3 | 1.486 (3) | C3—H3B | 0.9700 |
| C8—C7 | 1.394 (3) | ||
| C9—O3—C13 | 117.98 (17) | O2—C10—C11 | 124.3 (2) |
| C10—O2—H2 | 109.5 | O2—C10—C9 | 116.1 (2) |
| C1—N1—N2 | 127.3 (2) | C11—C10—C9 | 119.54 (19) |
| C1—N1—H1 | 116.4 | C5—C6—C7 | 127.7 (2) |
| N2—N1—H1 | 116.4 | C5—C6—H6 | 116.1 |
| C4—N2—N1 | 117.42 (18) | C7—C6—H6 | 116.1 |
| O3—C9—C8 | 126.0 (2) | C11—C12—C7 | 120.5 (2) |
| O3—C9—C10 | 115.23 (18) | C11—C12—H12 | 119.8 |
| C8—C9—C10 | 118.8 (2) | C7—C12—H12 | 119.8 |
| C6—C5—C4 | 126.4 (2) | O3—C13—H13A | 109.5 |
| C6—C5—H5 | 116.8 | O3—C13—H13B | 109.5 |
| C4—C5—H5 | 116.8 | H13A—C13—H13B | 109.5 |
| O1—C1—N1 | 120.4 (2) | O3—C13—H13C | 109.5 |
| O1—C1—C2 | 123.3 (2) | H13A—C13—H13C | 109.5 |
| N1—C1—C2 | 116.36 (19) | H13B—C13—H13C | 109.5 |
| N2—C4—C5 | 115.5 (2) | C1—C2—C3 | 114.4 (2) |
| N2—C4—C3 | 122.97 (19) | C1—C2—H2A | 108.7 |
| C5—C4—C3 | 121.5 (2) | C3—C2—H2A | 108.7 |
| C9—C8—C7 | 122.0 (2) | C1—C2—H2B | 108.7 |
| C9—C8—H8 | 119.0 | C3—C2—H2B | 108.7 |
| C7—C8—H8 | 119.0 | H2A—C2—H2B | 107.6 |
| C10—C11—C12 | 121.0 (2) | C4—C3—C2 | 113.3 (2) |
| C10—C11—H11 | 119.5 | C4—C3—H3A | 108.9 |
| C12—C11—H11 | 119.5 | C2—C3—H3A | 108.9 |
| C8—C7—C12 | 118.02 (19) | C4—C3—H3B | 108.9 |
| C8—C7—C6 | 118.9 (2) | C2—C3—H3B | 108.9 |
| C12—C7—C6 | 123.1 (2) | H3A—C3—H3B | 107.7 |
| C1—N1—N2—C4 | −12.5 (4) | O3—C9—C10—O2 | −3.3 (3) |
| C13—O3—C9—C8 | 0.8 (3) | C8—C9—C10—O2 | 176.6 (2) |
| C13—O3—C9—C10 | −179.3 (2) | O3—C9—C10—C11 | 176.6 (2) |
| N2—N1—C1—O1 | −177.3 (2) | C8—C9—C10—C11 | −3.5 (4) |
| N2—N1—C1—C2 | 1.2 (4) | C4—C5—C6—C7 | −177.5 (2) |
| N1—N2—C4—C5 | −177.97 (19) | C8—C7—C6—C5 | 177.4 (3) |
| N1—N2—C4—C3 | −1.5 (3) | C12—C7—C6—C5 | 0.0 (4) |
| C6—C5—C4—N2 | −176.7 (3) | C10—C11—C12—C7 | 0.2 (4) |
| C6—C5—C4—C3 | 6.8 (4) | C8—C7—C12—C11 | −2.2 (4) |
| O3—C9—C8—C7 | −178.7 (2) | C6—C7—C12—C11 | 175.2 (2) |
| C10—C9—C8—C7 | 1.4 (4) | O1—C1—C2—C3 | −159.6 (3) |
| C9—C8—C7—C12 | 1.4 (4) | N1—C1—C2—C3 | 21.9 (4) |
| C9—C8—C7—C6 | −176.1 (2) | N2—C4—C3—C2 | 23.8 (4) |
| C12—C11—C10—O2 | −177.4 (2) | C5—C4—C3—C2 | −160.0 (2) |
| C12—C11—C10—C9 | 2.7 (4) | C1—C2—C3—C4 | −32.7 (4) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H2···O1i | 0.82 | 1.86 | 2.671 (2) | 168 |
| N1—H1···O1ii | 0.86 | 2.02 | 2.875 (3) | 170 |
| C13—H13A···O2iii | 0.96 | 2.51 | 3.465 (3) | 172 |
| C13—H13C···O2iv | 0.96 | 2.57 | 3.489 (4) | 159 |
Symmetry codes: (i) x−1, y, z−1; (ii) −x+1, −y+2, −z+2; (iii) −x+1, −y+1, −z; (iv) x+1, y, z.
References
- Abourichaa, S., Benchat, N., Anaflous, A., Melhaoui, A., Ben-Hadda, T., Oussaid, B., El Bali, B. & Bolte, M. (2003). Acta Cryst. E59, o802–o803.
- Akhtar, W., Shaquiquzzaman, M., Akhter, M., Verma, G., Khan, M. F. & Alam, M. M. (2016). Eur. J. Med. Chem. 123, 256–281. [DOI] [PubMed]
- Asif, M. (2013). Mini-Rev. Org. Chem. 10, 113–122.
- Asif, M. (2014). Rev. Med. Chem. 14, 1093–1103. [DOI] [PubMed]
- Barberot, C., Moniot, A., Allart-Simon, I., Malleret, L., Yegorova, T., Laronze-Cochard, M., Bentaher, A., Médebielle, M., Bouillon, J. P., Hénon, E., SAPI, J., Velard, F. & Gérard, S. (2018). Eur. J. Med. Chem. 146, 139–146. [DOI] [PubMed]
- Boukharsa, Y., Meddah, B., Tiendrebeogo, R. Y., Ibrahimi, A., Taoufik, J., Cherrah, Y., Benomar, A., Faouzi, M. E. A. & Ansar, M. (2016). Med. Chem. Res. 25, 494–500.
- Daoui, S., Cinar, E. B., El Kalai, F., Saddik, R., Karrouchi, K., Benchat, N., Baydere, C. & Dege, N. (2019b). Acta Cryst. E75, 1352–1356. [DOI] [PMC free article] [PubMed]
- Daoui, S., Faizi, M. S. H., Kalai, F. E., Saddik, R., Dege, N., Karrouchi, K. & Benchat, N. (2019a). Acta Cryst. E75, 1030–1034. [DOI] [PMC free article] [PubMed]
- Dubey, S. & Bhosle, P. A. (2015). Med. Chem. Res. 24, 3579–3598.
- El Kalai, F., Baydere, C., Daoui, S., Saddik, R., Dege, N., Karrouchi, K. & Benchat, N. (2019). Acta Cryst. E75, 892–895. [DOI] [PMC free article] [PubMed]
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Gökçe, M., Utku, S. & Küpeli, E. (2009). Eur. J. Med. Chem. 44, 3760–3764. [DOI] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Karrouchi, K., Ansar, M., Radi, S., Saadi, M. & El Ammari, L. (2015). Acta Cryst. E71, o890–o891. [DOI] [PMC free article] [PubMed]
- Livermore, D., Bethell, R. C., Cammack, N., Hancock, A. P., Hann, M. M., Green, D., Lamont, R. B., Noble, S. A., Orr, D. C. & Payne, J. J. (1993). J. Med. Chem. 36, 3784–3794. [DOI] [PubMed]
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Partap, S., Akhtar, M. J., Yar, M. S., Hassan, M. Z. & Siddiqui, A. A. (2018). Bioorg. Chem. 77, 74–83. [DOI] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Siddiqui, A. A., Mishra, R., Shaharyar, M., Husain, A., Rashid, M. & Pal, P. (2011). Bioorg. Med. Chem. Lett. 21, 1023–1026. [DOI] [PubMed]
- Sönmez, M., Berber, I. & Akbaş, E. (2006). Eur. J. Med. Chem. 41, 101–105. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Stoe & Cie (2002). X-AREA and X-RED32. Stoe & Cie GmbH, Darmstadt, Germany.
- Tao, M., Aimone, L. D., Gruner, J. A., Mathiasen, J. R., Huang, Z., Lyons, J., Raddatz, R. & Hudkins, R. L. (2012). Bioorg. Med. Chem. Lett. 22, 1073–1077. [DOI] [PubMed]
- Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Spackman, P. R., Jayatilaka, D. & Spackman, M. A. (2017). CrystalExplorer17. University of Western Australia. http://hirshfeldsurface.net.
- Wang, T., Dong, Y., Wang, L.-C., Xiang, B.-R., Chen, Z. & Qu, L.-B. (2008). Arzneimittelforschung, 58, 569–573. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Zhang, C.-T., Wu, J.-H., Zhou, L.-N., Wang, Y.-L. & Wang, J.-K. (2006). Acta Cryst. E62, o2999–o3000.
- Zhou, G., Ting, P. C., Aslanian, R., Cao, J., Kim, D. W., Kuang, R., Lee, J. F., Schwerdt, J., Wu, H., Jason Herr, R., Zych, A. J., Yang, J., Lam, S., Wainhaus, S., Black, T. A., McNicholas, P. M., Xu, Y. & Walker, S. S. (2011). Bioorg. Med. Chem. Lett. 21, 2890–2893. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989019014130/wm5521sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989019014130/wm5521Isup3.hkl
Additional supporting information: crystallographic information; 3D view; checkCIF report