Summary
Although human pluripotent stem cell-derived cardiomyocytes (hPSC-CMs) have emerged as a novel platform for heart regeneration, disease modeling, and drug screening, their immaturity significantly hinders their application. A hallmark of postnatal cardiomyocyte maturation is the metabolic substrate switch from glucose to fatty acids. We hypothesized that fatty acid supplementation would enhance hPSC-CM maturation. Fatty acid treatment induces cardiomyocyte hypertrophy and significantly increases cardiomyocyte force production. The improvement in force generation is accompanied by enhanced calcium transient peak height and kinetics, and by increased action potential upstroke velocity and membrane capacitance. Fatty acids also enhance mitochondrial respiratory reserve capacity. RNA sequencing showed that fatty acid treatment upregulates genes involved in fatty acid β-oxidation and downregulates genes in lipid synthesis. Signal pathway analyses reveal that fatty acid treatment results in phosphorylation and activation of multiple intracellular kinases. Thus, fatty acids increase human cardiomyocyte hypertrophy, force generation, calcium dynamics, action potential upstroke velocity, and oxidative capacity. This enhanced maturation should facilitate hPSC-CM usage for cell therapy, disease modeling, and drug/toxicity screens.
Keywords: cardiomyocyte maturation, metabolism, fatty acids, embryonic stem cells, induced pluripotent stem cells
Highlights
-
•
Fatty acid treatment leads to cardiomyocyte hypertrophy
-
•
Fatty acid treatment leads to enhanced contractile force generation
-
•
Fatty acid treatment leads to improved calcium transient kinetics
-
•
Fatty acid treatment leads to faster action potential maximum upstroke velocity
In this article, Yang and colleagues show that fatty acid supplements enhance the maturation of cardiomyocytes derived from human pluripotent stem cells. Fatty acid treatment induces cardiomyocyte hypertrophy and significantly increases cardiomyocyte force production. The improvement in force generation is accompanied by enhanced calcium transient kinetics and faster action potential maximum upstroke velocity.
Introduction
Promoting the maturation of the cardiomyocytes derived from human pluripotent stem cells (hPSC-CMs) has become the subject of intense research to better achieve their potential applications in heart regeneration, disease modeling, and drug screening/discovery (Yang et al., 2014a). A number of approaches have been applied to promote hPSC-CM maturation including long-term culture (Lundy et al., 2013, Sartiani et al., 2007), metabolic hormonal treatment (Birket et al., 2015, Kosmidis et al., 2015, Parikh et al., 2017, Ribeiro et al., 2015, Yang et al., 2014b), substrate stiffness (Feaster et al., 2015, Hazeltine et al., 2012), microRNAs (Kuppusamy et al., 2015, Lee et al., 2015), or tissue engineering (Nunes et al., 2013, Ronaldson-Bouchard et al., 2018, Ruan et al., 2015, Tulloch et al., 2011). These approaches have promoted some maturation in morphology, molecular, and/or functional aspects, but there is considerable room for improvement.
The role of metabolism in promoting maturation of hPSC-CMs has not been widely explored. Immediately after birth there is a major change in metabolism as the infant switches from placental nutrition to nursing. Correspondingly, while fetal cardiomyocytes mostly rely on glycolysis for ATP production, cardiomyocytes of newborns adapt to obtain their ATP predominantly through fatty acid β-oxidation (Makinde et al., 1998). Standard cell culture media principally offer glucose as a cellular energy source, and significant lipids are rarely present. For instance, in the commonly used RPMI plus B27 plus insulin (RPMI-B27-insulin) medium for hPSC-CM culture, there is <10 μM total lipids (which includes 3.5 μM oleic acid, 3.5 μM linolenic acid, and 0.2 μM lipoic acid [Brewer and Cotman, 1989]), whereas in human infants the serum fatty acid concentration is ∼300 μM (Makinde et al., 1998). In addition, polyunsaturated fatty acids (PUFAs) are important not only as energy substrates but also as ligands for some intracellular signaling pathways. Thus, the shortage of fatty acids might limit cellular functions important for maturation.
Biochemical assays demonstrate that cultured fetal and neonatal rat cardiomyocytes contain lower fatty acid contents (saturated, monounsaturated, and polyunsaturated) than native myocardium at comparable developmental stages (Karimata et al., 2013). In addition, cardiomyocytes from pythons and neonatal rats undergo hypertrophy in response to fatty acid treatments (Riquelme et al., 2011). This suggested to us that providing fatty acids to hPSC-CMs may promote a more physiological postnatal state and enhance hPSC-CM developmental maturation. In this study, we show that supplementing the culture medium with the three most abundant fatty acids in the newborn serum (palmitic, oleic, and linoleic acids) at physiological concentrations promotes hPSC-CM maturation.
Results
hPSC-CM Take Up Fatty Acids in a Carnitine-Dependent Manner
The total concentration of fatty acids in human newborn infants was estimated at ∼300 μM (Makinde et al., 1998). Among them, palmitic acid occupies ∼34% of total fatty acids, with ∼27% for oleic acid and ∼15% for linoleic acid. Therefore, in this study, to mimic the in vivo developmental fatty acid concentrations and components, we treated the hPSC-CMs with 105 μM palmitate-albumin, 81 μM oleic acid-albumin, and 45 μM linoleic acid-albumin complexes. These fatty acids were supplemented with 250 μM carnitine in RPMI-B27-insulin medium. We validated that the palmitate-albumin complex is endotoxin free and that there was no lipid toxicity by fatty acid treatment (Figures S1 and S2).
We assessed whether the hPSC-CMs are capable of taking up some of the provided fatty acids by quantifying the nonesterified fatty acids in the medium before and after 2 days of treatment in cardiomyocytes derived from IMR90 human induced IPCs (hiPSCs). We found that one million cells take up 57 ± 7 μM of the provided ∼231 μM fatty acids after two days of feeding, indicating a substantial capacity for metabolism. Similar fatty acid uptake amounts were found with cardiomyocytes derived from RUES2 human embryonic stem cells and WTC hiPSCs, suggesting that the capacity for fatty acid metabolism is a general property of hPSC-CMs. We wondered whether prolonged fatty acid feeding would enhance fatty acid uptake from the medium, but after 2 weeks of feeding, the rate of fatty acid uptake was unchanged. Since hPSC-CMs only take up about one-third of the provided fatty acids per feeding, we decided to use half (∼115.5 μM) of the original calculated fatty acid concentration in the remainder of this study. Removing carnitine reduced fatty acid uptake by nearly 50%, 31 ± 4 μM versus 57 ± 7 μM (Figure S3A), indicating this amino acid is vital to fatty acid metabolism in hPSC-CMs. Next, we measured the impact of fatty acid feeding on free fatty acid content in hPSC-CMs. At the end of the 2 weeks of fatty acid treatment, intracellular fatty acid content was increased by 46% (6,118 ± 461 nmol nonesterified fatty acid [NEFA]/g protein in control versus 8,933 ± 488 nmol NEFA/g protein in fatty acid-treated hPSC-CMs, n = 3 biological replicates, p < 0.05, Figure S3B).
Fatty Acid Supplementation Enhances Structural Maturation
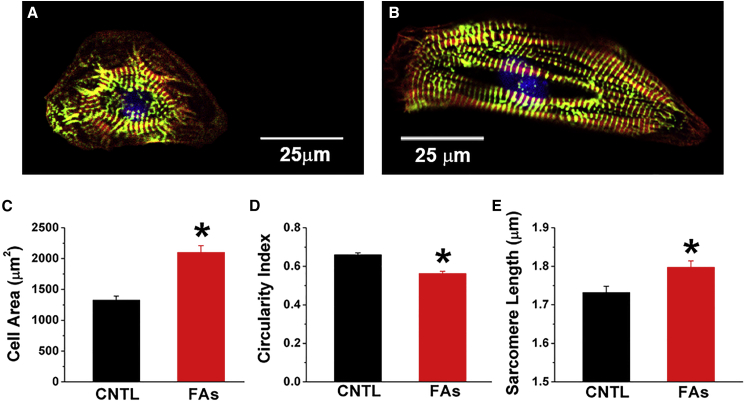
To characterize the effects of fatty acids on morphology and gene expression of hPSC-CMs, we stained cells for F-actin with phalloidin and for α-actinin (a Z-disk protein) with an antibody. Control cells were small and round-to-polygonal in shape. Fatty acid treatment for 2 weeks increased cell area by 59% (1,326 ± 63 μm2 versus 2,104 ± 105 μm2, p < 0.001) and modestly decreased circularity index (0.63 ± 0.01 versus 0.58 ± 0.01, p < 0.001). This corresponds to a more mature cardiomyocyte phenotype with bigger cells and anisotropy (Figures 1A–1D). Sarcomere length increased significantly from 1.73 ± 0.02 μm in control hPSC-CMs to 1.80 ± 0.02 μm after fatty acid treatment (p < 0.005) (Figure 1E). In addition to the cardiomyocytes derived from IMR90 hiPSCs, fatty acids also significantly increased the size of cardiomyocytes derived from RUES2 embryonic stem cells, indicating that this is a general property of hPSC-CMs (Figure S4A).
Figure 1.
Fatty Acid Treatment Leads to hPSC-CM Morphological and Molecular Changes
Representative control (A) and fatty acid-treated (B) cells were stained with α-actinin antibody (green), phalloidin (F-actin in red), and Hoechst 33342 for nuclei (blue). Scale bar, 25 μm. Compared with control hPSC-CMs, fatty acid-treated cells exhibited significant changes in cell area (C), circularity index (D), and sarcomere length (E). n > 150 from three different cardiomyocyte differentiation runs. Data are presented as mean ± SEM. ∗p < 0.05. See also Figure S4.
Fatty Acid Supplementation Enhances Calcium Transient Kinetics
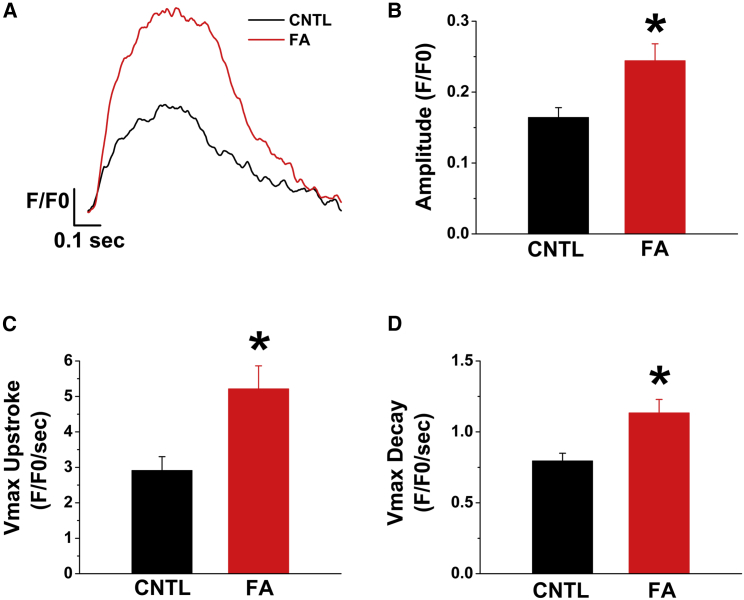
To investigate potential effects of fatty acids on calcium transient kinetics, we used the intracellular ratiometric calcium dye Fura-2 AM and compared control versus fatty acid-treated hPSC-CMs under 1-Hz electrical stimulation. Representative traces are shown in Figure 2A. Fatty acid treatment leads to significant increase in calcium peak transient amplitude (0.16 ± 0.01 versus 0.24 ± 0.02 F/F0, p < 0.005, Figure 2B). Also, the maximal upstroke and decay velocities were significantly higher in fatty acid-treated hPSC-CMs. Specifically, Vmax upstroke was faster (2.91 ± 0.39 versus 5.21 ± 0.65 F/F0/s, p < 0.005, Figure 2C) as well as the calcium transient decay velocity (0.80 ± 0.05 versus 1.13 ± 0.09 F/F0/s, p < 0.005, Figure 2D).
Figure 2.
Fatty Acid-Treated hPSC-CMs Exhibit Improved Calcium Transient Kinetics
Calcium transients were evaluated by loading the hPSC-CMs with the intracellular calcium ratiometric indicator Fura-2 AM and were stimulated at 1 Hz.
(A) Representative transients from control and fatty acid-treated hPSC-CMs. Note the higher amplitude, faster upstroke, and decay of the Ca2+ transient in the treated cells.
(B–D) Calcium transient amplitude magnitudes were significantly higher in fatty acid-treated hPSC-CM (B), as indicated by increases in maximal upstroke (C) and decay (D) velocities. n = 10–16 cells from three separate cardiomyocyte differentiation runs. Data are presented as mean ± SEM. F/F0 is the ratio of F340/F380. ∗p < 0.05 versus control hPSC-CMs.
Fatty Acids Improve Contractile Force
There are significant differences in contractile force between immature and mature cardiomyocytes (Yang et al., 2014a). We used our previously reported elastomeric micropost system (Rodriguez et al., 2014, Yang et al., 2014b) to assess the contractile force of individual control or fatty acid-treated hPSC-CMs. Figure 3A shows representative traces of the total force generated by individual cardiomyocytes. Control cells exhibited a twitch force of 10.1 ± 0.5 nN/cell. Fatty acid-treated cells showed a significantly higher twitch force of 13.4 ± 0.8 nN/cell (p < 0.0001) as shown in Figure 3B (n = 91 for control hPSC-CMs and n = 91 for fatty acid-treated hPSC-CMs). It is also worth noting that cardiomyocyte sizes were significantly increased after fatty acid treatment on the microposts (202.6 ± 4.1 μm2 for control cells versus 238.8 ± 6.6 μm2 after treatment, p < 0.0001), as shown in Figure 3C. The contractile force assay was performed in cardiomyocytes derived from RUES2 embryonic stem cells, and similar results were acquired (Figure S4B).
Figure 3.
Fatty Acid Treatment Significantly Increases hPSC-CM Contractile Force
(A–C) Representative force traces generated by control and fatty acid-treated hPSC-CMs (A). The statistical analysis results are shown in (B). Control and fatty acid-treated cardiomyocyte area on microposts is shown in (C). n = 91 for control hPSC-CMs and n = 91 for fatty acid-treated hPSC-CMs from four different cardiomyocyte differentiation runs. Data are presented as mean ± SEM. ∗p < 0.05 versus control hPSC-CMs. See also Figure S4.
Fatty Acids Improve Action Potential Upstroke Velocity and Increase Membrane Capacitance
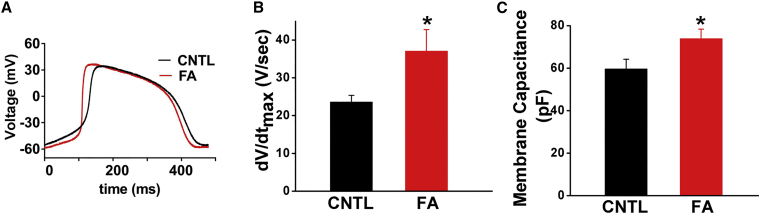
Immature cardiomyocytes display slower action potential upstroke velocity than their adult counterparts. To investigate whether fatty acid treatment improves electrophysiological maturation, we performed whole-cell patch clamping to record the cardiomyocyte spontaneous action potentials. Fatty acids increase action potential maximum upstroke velocity, dV/dtmax, by 57% (23.5 ± 1.8 V/s in control [n = 24] versus 37.0 ± 5.7 V/s after treatment [n = 25], p = 0.03, Figure 4A). Furthermore, fatty acid treatment increases membrane capacitance by 24%, indicating that cell surface area is significantly increased (59.6 ± 4.7 pF in control [n = 14] versus 73.7 ± 4.6 pF [n = 15] after fatty acid treatment, p = 0.04, Figure 4B). Fatty acid treatment does not affect spontaneous beating rate, maximal diastolic potential, action potential amplitude, action potential duration at 50% repolarization, and action potential duration at 90% repolarization (Table S2).
Figure 4.
Fatty Acid Treatment Improves Cardiomyocyte Action Potential Maximum Upstroke Velocity and Increases Cardiomyocyte Membrane Capacitance
(A) Representative action potential traces from control and fatty acid-treated cells.
(B) Statistical analysis of action potential maximum upstroke velocity.
(C) Analysis of cardiomyocyte membrane capacitance.
Data were obtained from three different cardiomyocyte differentiation runs. n = 24 for control group and n = 25 for fatty acid-treated group. ∗p < 0.05 versus control hPSC-CMs. Data are presented as mean ± SEM. See also Table S2.
Effect of Fatty Acids on Mitochondrial Respiratory Reserve Capacity
The Seahorse XF 96 extracellular flux analyzer was used to characterize mitochondrial function as previously reported (Yang et al., 2014b). In this assay, the mitochondrial respiratory reserve capacity can be determined by the difference between the normalized oxygen consumption rate (OCR) value before and after the protonophore uncoupler, FCCP (carbonyl cyanide-p-trifluoromethoxyphenylhydrazone), using glucose as the metabolic substrate. Figure 5A shows representative traces with or without fatty acid treatment, and Figure 5B shows the statistical difference between groups. Due to variations in the absolute magnitude of OCR measurements in different experimental runs, we report the ratio of fatty acid-treated/untreated OCRs for statistical analysis (n = 6). The maximal mitochondrial respiratory capacity after the FCCP treatment was enhanced by ∼38% after fatty acid treatment.
Figure 5.
The Effect of Fatty Acids on Mitochondrial Function
(A) Representative traces for control and fatty acid-treated hPSC-CMs responding to the ATP synthase inhibitor oligomycin, the respiratory uncoupler FCCP, and the respiratory chain blockers rotenone and antimycin A. Note the higher maximal OCR in the fatty acid-treated cells.
(B) Statistical analysis of the differences in respiratory reserve capacity. The OCR values were normalized to the number of cells present in each well, as described in Experimental Procedures.
Data were from six different cardiomyocyte differentiation runs. ∗p < 0.05 versus control hPSC-CMs. Data are presented as mean ± SEM.
Fatty Acids Activate Genes Involved in β-Oxidation and Suppress Genes Involved in Lipid Synthesis
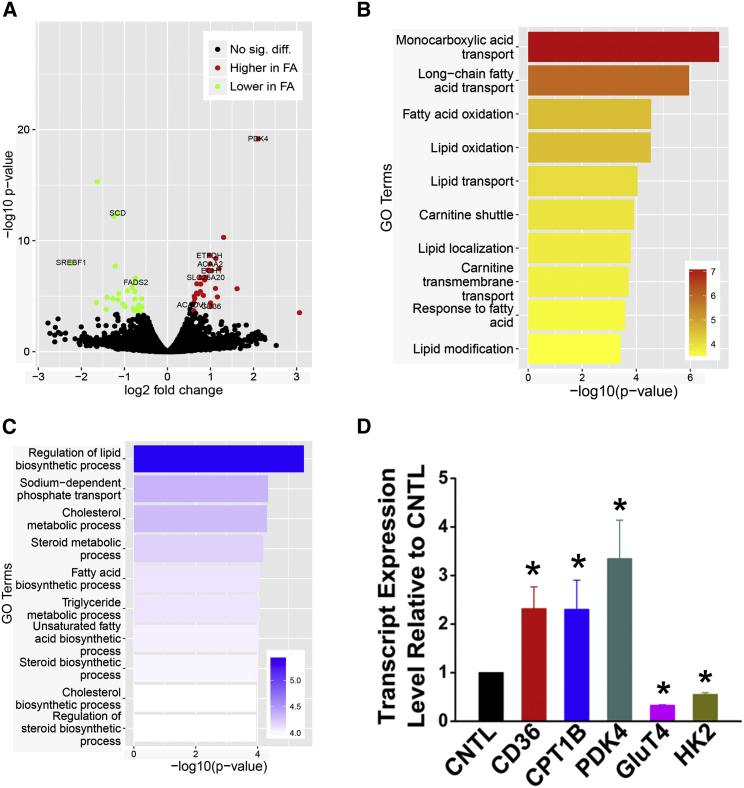
To determine the genome-wide effects of fatty acid treatment, we sequenced RNA from control and fatty acid-treated cardiomyocytes. We found 61 differentially expressed genes, as shown in the volcano plot in Figure 6A. Not surprisingly, gene ontology (GO) enrichment analysis showed that the most significant GO terms are all related to fatty acid/lipid metabolism. The upregulated genes are enriched for fatty acid and lipid oxidation (Figure 6B); the downregulated genes are enriched in lipid biosynthesis (Figure 6C). For example, stearoyl-coenzyme A (CoA) desaturase (SCD) and fatty acid desaturase 2 (FADS2) are downregulated after fatty acid treatment. Sterol regulatory element binding transcription factor 1 (SREBF1; also known as SREBP1) is a transcriptional activator of fatty acid and cholesterol synthesis. Its expression is also significantly suppressed, suggesting that cells are shutting down de novo synthesis when external fatty acid is abundant. On the other hand, the genes involved in fatty acid oxidation are significantly upregulated, such as CD36, also known as fatty acid translocase, which transfers fatty acids from extracellular space into the cytosol. Also upregulated is carnitine palmitoyltransferase 1A (CPT-1A), which transports long-chain fatty acyl-CoAs from the cytoplasm into the mitochondria and is the rate-controlling enzyme of the long-chain fatty acid β-oxidation pathway. The expression of SLC25A20, another cytosolic-mitochondrial fatty acid transporter, is also enhanced. Other genes involved in fatty acid β-oxidation (acyl-CoA dehydrogenase very long chain [ACADVL], enoyl-CoA hydratase 1 [ECH1], and acetyl-CoA acyltransferase 2 [ACAA2]) are also significantly upregulated after fatty acid feeding. The expression of pyruvate dehydrogenase kinase 4 (PDK4), which inhibits glucose and lactate metabolism, was upregulated. Electron-transfer flavoprotein dehydrogenase (ETFDH), a component of the mitochondria electron transport chain, has higher expression level in fatty acid-treated samples. Some of the aforementioned gene-expression changes were verified by performing traditional qPCR (Figure 6D). The increase in transcripts for fatty acid metabolism was accompanied by a decrease in transcripts for glucose metabolism, such as glucose transporter 4 (SLC2A4) and hexokinase 2 (HK2; Figure 6D). Because some of the cardiac differentiation runs had up to 20% noncardiomyocytes, we repeated these qPCR experiments with high-purity cardiomyocytes (over 95% positive cardiac troponin T). Similar results were observed, indicating that these gene-expression changes reflected cardiomyocytes and not the nonmyocytes (Figure S5).
Figure 6.
Genome-wide Effects of Fatty Acid Treatment Assessed by RNA Sequencing
(A) Volcano plot shows the differential expressed genes after fatty acid treatment.
(B and C) Gene ontology (GO) enrichment analysis illustrating the upregulated and downregulated pathways after fatty acid treatment.
(D) qPCR verified some of the candidate genes in RNA sequencing that involved fatty acid transportation, and also the genes involved in glucose metabolism. ∗p < 0.05 versus control hPSC-CMs.
See also Table S1.
Fatty Acids Activate Multiple Intracellular Signaling Pathways
There are reports that free fatty acids (FFAs) acutely stimulate protein phosphorylation in immortalized cell lines, suggesting a diverse role in signal transduction (Sanchez-Reyes et al., 2014, Watt et al., 2006). Indeed, some G-protein-coupled receptors have been identified as FFA receptors (Watson et al., 2012). To investigate possible signal pathways involved in fatty acid treatment of hPSC-CMs, we assessed multiple phosphokinases. AMP-activated protein kinase (AMPK) is activated by phosphorylation of Thr172 within the α subunit by LKB1. AMPK is described as a cellular “energy sensor” because its activity is increased when AMP levels are elevated, resulting in increased catabolism and ATP regeneration. Fifteen minutes and 30 min after fatty acid treatment, we found that AMPK Thr172 phosphorylation is upregulated markedly, which would be expected to lead to a feedforward activation of fatty acid oxidation pathway (Figure 7). Acetyl-CoA carboxylase (ACC) was phosphorylated by AMPK and then facilitated fatty acid transport into mitochondria, thus inhibiting fatty acid biosynthesis. After fatty acid treatment, the phospho-ACC level also was upregulated. In contrast, the level of phospho-Akt (Ser473; part of the insulin response pathway) was decreased by fatty acid treatment. We also found that phospho-ERK and phospho-p38 MAPK levels were upregulated by acute FFA treatment (Figure 7).
Figure 7.

Fatty Acid Treatment Activates Multiple Intracellular Signal Pathways
Western blots for the phosphoprotein level in control, 15-min, and 30-min fatty acid-treated hPSC-CMs.
Discussion
Human PSC-CMs provide an attractive cell source for heart regeneration, disease modeling, drug screening, and toxicity testing. A significant limitation of these hPSC-CMs is that they exhibit immature properties (Yang et al., 2014a), which limits their utility in disease modeling and therapeutic applications. Although some progress has been made to obtain more mature hPSC-CMs, there is much room for improvement. Because of the marked postnatal changes in cardiac metabolism (Makinde et al., 1998), we hypothesized that altering metabolic substrate to fatty acids would drive maturation.
Riquelme et al. (2011) showed that fatty acids stimulate the postprandial growth of the python heart, and that adding three of the constituent fatty acids (myristate C14:0, palmitate C16:0, and palmitoleate C16:1) was sufficient to induce hypertrophy in cultured neonatal rat cardiomyocytes. In pilot studies, we verified that these fatty acids caused the expected changes in gene expression in neonatal rat cardiomyocytes, including increased MYH6, and decreased MYH7 and NPPA (data not shown). However, treating the hPSC-CMs with this fatty acid cocktail does not lead to the same gene-expression changes. We then hypothesized that human cardiomyocytes would be fatty acid responsive, but they require a different cocktail than either pythons or rats. We therefore designed a cocktail based on breastfed human infant serum (Hardell and Walldius, 1980) and breast milk (Gibson and Kneebone, 1981), and found that palmitate, oleate, and linoleate could induce hPSC-CM hypertrophy. The specificity of the fatty acids suggests a role beyond metabolic flux in mediating hypertrophic growth, supported by our observations that these molecules activate multiple signaling pathways (Figure 7).
Although fatty acid treatment clearly promoted hypertrophic growth and increased myofibril content (Figure 1), we were surprised that this was not accompanied by an increase in mRNA transcripts for myofibril genes. It is possible that post-transcriptional pathways such as translation efficiency or protein stability account for this. Conversely, since our RNA-sequencing experiments were normalized to total transcript abundance (FPKM [fragments per kilobase per million reads mapped]), it is possible that a “more of everything per cell” state would not be detected by this approach.
In contrast, RNA sequencing showed that fatty acid treatment upregulates genes involved in fatty acid β-oxidation and downregulates genes in lipid synthesis and glucose metabolism—specifically, the mRNA level of CD36, CPT-1B, and PDK4, which likely enhance the capacity to oxidize fatty acids. Concomitantly, the mRNAs encoding proteins involved in glucose handling (SLC2A4 and HK2) are downregulated, in agreement with the Randle cycle (Hue and Taegtmeyer, 2009), in which fatty acid oxidation inhibits glucose utilization. As far as the mechanism is concerned, evidence is accumulating for the existence of fatty acid-mediated transcriptional regulation of various genes involved in lipid metabolism. In a number of cases the responsive cis-regulatory element has been identified as a peroxisome proliferator responsive element, which binds peroxisome proliferator activated receptors (PPARs), members of the nuclear hormone receptor family. Studies have provided strong evidence that long-chain fatty acids act as natural ligands for PPARs (Amri et al., 1995, Kliewer et al., 1997). Indeed, the aforementioned FAT/CD36 and CPT-1B are PPAR responsive.
In addition to long-term changes in gene expression, fatty acids have short-term physiological effects on the heart. In Langendorff-perfused rat hearts, low doses of fatty acids (sodium palmitate at 0.12 mM) increased ventricular developed pressure and lowered heart rate and glucose uptake, while high doses of fatty acids (sodium palmitate from 0.3 to 1.5 mM) decreased pressure development (Guarner et al., 2002). Taken together with our single-cell contractility data, these findings indicate that fatty acids can augment cardiac contractile function. Our data add to these observations by demonstrating that enhanced calcium dynamics likely underlie at least part of this boost in contractility (Figure 4).
We observed a significant set of protein phosphorylation events after fatty acid feeding. Fatty acids acutely activate the AMPK pathway, which successively leads to ACC phosphorylation and the promotion of fatty acid oxidation. Interestingly, fatty acids also lead to ERK and p38-MAPK phosphorylation, suggesting that fatty acids not only serve as cellular energy sources but also ligands to activate their own downstream signal pathways. A previous report (Miller et al., 2005) indicated that treating neonatal rat cardiomyocytes with lipotoxic levels of palmitate (0.5 mM) for 18 h increased phosphorylation of ERK and p38. This phosphorylation was implicated in the cytotoxicity of palmitate, since cotreatment of the cells with 0.1 mM oleate prevented palmitate-induced ERK and p38 activation. Since no lipid cytotoxicity was observed in this study, we hypothesize that ERK and p38 transient activation by fatty acid treatment leads to signal cascades that promote hPSC-CM growth and hypertrophy (Maillet et al., 2013).
The effect of fatty acid feeding appears to be robust, in that we observed similar maturation effects in cardiomyocytes derived from three different hPSC lines. IMR90 cardiomyocytes were the major line used for all of the endpoints. Cardiomyocytes derived from an embryonic stem cell line, RUES2, were used to test the major findings of this study including the amount of fatty acids taken up by the cells, cellular hypertrophy, and contractile force after fatty acid exposure. We also investigated fatty acid uptake by cardiomyocytes derived from another iPSC line, WTC-11. After 2 days of feeding, one million WTC-11-derived cardiomyocytes take up 50 ± 7 μM fatty acid. Thus, fatty acids regulate cardiac maturation in both human iPSC- and embryonic stem cell-derived cardiomyocytes.
During the revision process of this article, three other reports (Correia et al., 2017, Hu et al., 2018, Mills et al., 2017) were published in which fatty acids were found to promote hPSC-CM maturation. Mills et al. (2017) used engineered heart tissues in a 96-well device for medium-throughput screening of factors (including extracellular matrix, metabolic substrate, and growth factor conditions) that promote functional maturation. The screen revealed that a switch to fatty acid metabolism is a central driver for cardiac maturation, including increasing hypertrophy, contractility, and reducing cell-cycle activity. Correia et al. (2017) reported that shifting medium from glucose-containing to glucose-free, with the addition of galactose and fatty acid (oleate + palmitate), promotes hPSC-CM maturation, resulting in cardiomyocytes with higher oxidative metabolism, transcriptional signatures closer to those of adult tissue, improved calcium handling, and enhanced contractility. These authors did not indicate that their fatty acids were loaded onto albumin, and this may explain why lipotoxicity was observed in their experiments. Most recently, Hu et al. (2018) cultured cells in medium containing 100 μM oleate and 50 μM palmitate with or without glucose and found no functional benefits when culturing cells in the medium containing fatty acids in the presence of high glucose for 1 week, while in glucose-free medium containing fatty acids, hPSC-CM exhibited greater force, and enhanced calcium dynamics and greater mitochondrial respiration rates. In this study, the fatty acids were not bound with albumin. Our current study reinforces these findings and is unique in that we used a developmentally related, albumin-bound, fatty acids cocktail at a physiological stoichiometry, whereby we identified that increased calcium dynamics underlie increases in contractility, demonstrated the need for carnitine to promote maximal cellular fatty acid uptake, and showed increased fatty acid content in hPSC-CMs. In addition, we investigated the signal pathways that are involved after the cells exposed to fatty acids (including AMPK, ERK, and p38 MAPK). Taken together, these studies provide strong evidence that the metabolic switch from glucose to fatty acids is a driver of hPSC-CM maturation.
Although treating hPSC-CMs with physiologically and developmentally relevant concentrations of long-chain fatty acids leads to a more mature morphological and functional hPSC-CM phenotype, these cells are still immature compared with adult cardiomyocytes (Yang et al., 2014a). Since developing cardiac cells in vivo are exposed to the combined effects of diverse cues including mechanical signals, extracellular matrix, soluble factors, substrate stiffness, and electrical fields, it seems likely that a combinatorial approach will lead to better hPSC-CM maturation.
Experimental Procedures
Making Palmitate-Albumin Complexes
Palmitate-BSA stock was prepared at a final concentration of 4 mM palmitate with 12% fatty acid-free BSA (molecular ratio 2.2:1) (both purchased from Sigma-Aldrich) in glucose/pyruvate-free Krebs-Henseleit buffer (KHB) with modifications as described previously (Bakrania et al., 2016). In brief, palmitate was dissolved in 38% ethanol in 0.5 mM Na2CO3 at 60°C under constant nitrogen gas. Upon dissolving, ethanol was boiled off by increasing the temperature to 80°C–90°C. BSA was dissolved in KHB at 37°C. BSA solution and palmitate solution were mixed together under constant stirring at 37°C. Successful complexing of palmitate to BSA was monitored when no clouding of the solution occurred. The BSA-palmitate solution was stirred for an additional 5 min before being dialyzed overnight and sterile filtered using 0.45-μm pore size membrane filter. Aliquots were stored at −20°C. Oleic acid-albumin and linoleic acid-albumin complexes were purchased from Sigma.
Cell Culture
Undifferentiated human IMR90-induced pluripotent stem cells, originally derived from lung fibroblasts (Yu et al., 2007) (James A. Thomson, University of Wisconsin-Madison), were expanded using mouse embryonic fibroblast-conditioned medium supplemented with 5 ng/mL basic fibroblast growth factor. Cardiomyocytes were obtained using a protocol based on our previously reported directed differentiation method that involves the serial application of activin A and bone morphogenetic protein 4 (BMP4) under serum-free, monolayer culture conditions (Hofsteen et al., 2016). The cultures were also supplemented with the Wnt agonist CHIR 99021 in the early stages of differentiation followed by the Wnt antagonist Xav 939. After 20 days of in vitro differentiation, the cells were dispersed using 0.025% trypsin-EDTA and replated. Cultures were fed every other day thereafter with serum-free RPMI-B27 plus L-glutamine. Only cell preparations containing >80% cardiac troponin T-positive cardiomyocytes were used for the current study. After 20 days of differentiation, the cells were treated with half dosage (52.5 μM palmitate-BSA, 40.5 μM oleate-BSA, and 22.5 μM linoleate-BSA) of fatty acid-albumin complexes in RPMI-B27-insulin medium for 2 weeks. Note that glucose was maintained at its normal media levels (11 mM). L-Carnitine was added to the medium at 120 μM, because this dipeptide is essential for fatty acid transport into mitochondria. Control hPSC-CMs were fed with the same basal medium (RPMI-B27-insulin) supplemented with 50 μM fatty acid-free albumin and 120 μM carnitine. Media were changed every other day. This study was mainly performed with cardiomyocytes derived from the IMR90 hiPSC line. However, cardiomyocytes from another hiPS cell line, WTC-11 (Miyaoka et al., 2014) (Wild Type C, Gladstone Institute of Cardiovascular Disease, University of California San Francisco), and an embryonic stem cell line, RUES2 (Rockefeller University, New York, NY), were also used to confirm that major findings were generally applicable. RUES2 and WTC-11 undifferentiated cells were cultured with mTeSR medium and differentiation protocols similar to those for IMR90 were used to obtain cardiomyocytes from these two cell lines. Only cell preparations containing >80% cardiac troponin T-positive cardiomyocytes (by flow cytometry) were used for the current investigation.
Fatty Acid Concentration Measurements
Fatty acid concentration in the medium before and 48 h after treating the cells was determined using the HR Series NEFA-HR (Kanno et al., 2004) kit from Wako Life Sciences according to the manufacturer’s instructions. In brief, in the presence of ATP and CoA, the nonesterified fatty acids were treated with acyl-CoA synthase, which leads to the production of acyl-CoA. Subsequently, the acyl-CoA oxidase is added to oxidize acyl-CoA to produce hydrogen peroxide. In the presence of peroxidase, hydrogen peroxide allows for the oxidative condensation of 3-methyl-N-ethyl-N-(β-hydroxyethyl)-aniline with 4-aminoantipyrine to form a purple-colored end product with an absorption maximum at 550 nm. Therefore, the amount of fatty acids in the medium can be determined from the optical density measured at 550 nm.
For measurement of intracellular fatty acid content, cardiomyocytes were grown in 10 cm2 plates, treated with fatty acids for 2 weeks, collected in PBS, and homogenized at 4°C with Bullet Blender at maximum speed. A small aliquot was taken for protein concentration measurement. Lipids were extracted with 2:1 chloroform/methanol. The organic fraction was evaporated under nitrogen and reconstituted in isopropanol with 0.5% Triton X-100. Non-esterified fatty acids were measured with the WAKO NEFA-HR (Kanno et al., 2004) assay according to manufacturer’s instructions. The fatty acid content was normalized to protein concentration.
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde for 15 min followed by a PBS wash. The fixed cells were blocked with 1.5% normal goat serum for 1 h at room temperature and incubated overnight at 4°C with primary antibody (mouse anti-α-actinin from Sigma, #A7811). The cells were then washed with PBS and incubated with a goat-anti-mouse secondary antibody for 1 h at room temperature. Samples subjected to F-actin staining were incubated with TRITC-labeled phalloidin (Thermo Fisher Scientific, R415) for 20 min at room temperature. Nuclei were stained with Hoechst 33342.
Imaging and Morphological Analysis
Fluorescent images were acquired using a Zeiss AxioCam mounted on a Zeiss AxioObserver microscope, and confocal images were processed and quantified using NIS Elements. Each cell was analyzed for cell size and circularity index. To calculate sarcomere length, we selected myofibrils with at least ten continuous well-recognized α-actinin-positive bands and divided the length value by the number of sarcomeres.
Western Blotting
Total protein was acquired from control cardiomyocytes or cardiomyocytes after 15 and 30 min of fatty acid treatment and subjected to SDS-PAGE. The lanes were loaded with equal amount of protein and were checked by Ponceau S staining. After blocking with milk, the membranes were incubated with anti-phospho-AMPK (2535S), anti-phospho-ACC (3661S), anti-phospho-Akt (4051S), anti-phospho-ERK (9106S), anti-phospho-p38 MAPK antibodies (9211S) (Cell Signaling Technologies), or anti-GAPDH mouse monoclonal antibody (Abcam, ab8245) overnight while shaking at 4°C. After incubation with anti-mouse (for phospho-Akt, phospho-ERK, and GAPDH) or anti-rabbit (for phospho-AMPK, phospho-p38-MAPK, and phospho-ACC) horseradish peroxidase (HRP)-coupled secondary antibody (Santa Cruz Biotechnology, sc-2031 for goat anti-mouse immunoglobulin G [IgG]-HRP and sc-2004 for goat anti-rabbit IgG-HRP), bands were visualized with SuperSignal West Femto Trial Kit (Thermo Scientific).
Contractile Force Measurement
Arrays of silicone microposts were fabricated by casting polydimethylsiloxane (PDMS) from a silicon wafer with patterned SU8 features as previously described (Tan et al., 2003). The microposts used in this study were 6.45 μm in height and 2.3 μm in diameter, and the center-to-center spacing between adjacent microposts was 6 μm. The stiffness of each micropost, which was based on the dimensions of the microposts and the material properties of PDMS, was 38.4 nN/μm. To enable cell attachment, we stamped the tips of these microposts with 50 μg/mL of mouse laminin (Life Technologies) via microcontact printing, while the remaining surfaces of the micropost array were fluorescently stained with BSA conjugates with Alexa Fluor 594 and blocked with 0.2% Pluronic F-127 (in PBS) (Sniadecki and Chen, 2007). Twenty days following differentiation, hiPSC-derived cardiomyocytes were seeded onto the arrays at a density of 250,000/cm2. One week after fatty acid treatment, individual cardiomyocyte twitch forces were recorded under phase light using high-speed video microscopy as previously described (Rodriguez et al., 2014). Only the contractile forces of single cardiomyocytes (no junctions with adjacent cells) with obvious beating activity were assessed. The experiments were performed in a live cell chamber at 37°C with 15 mM HEPES-containing medium. Post deflections were optically measured at 100–150 frames/s using phase-contrast microscopy on a Nikon Ti-E upright microscope with a 60× water immersion objective. A custom-written MATLAB code was used to compare each time frame of the video with a reference fluorescent image of the base plane of the posts. Twitch forces were subsequently calculated by multiplying the deflection of the posts by the bending stiffness of the microposts:
| (Equation 1) |
where F is the force at a single micropost, k is the post's bending stiffness (38.4 nN/μm), and δ is the horizontal distance between the centroid of the post's tip and the centroid of the post's base. The total twitch force was then determined by adding together the forces measured at each post beneath the individual cardiomyocytes.
Calcium Imaging
Intracellular calcium content was measured using the ratiometric indicator dye Fura-2 AM as described previously (Korte et al., 2011). For this assay, cardiomyocytes were replated onto fibronectin-coated glass slides, after which the cardiomyocytes were subsequently treated for 2 weeks. On the experimental day, cells were incubated in 0.2 μM Fura-2 AM dye for 20 min at 37°C and washed with PBS. Stimulated calcium transients (1 Hz) were then recorded with the Ionoptix Stepper Switch system coupled to a Nikon inverted fluorescence microscope. The fluorescence signal was acquired using a 40× Olympus objective and passed through a 510-nm filter, and the signal was quantified using a photomultiplier tube. The experiments were done at 37°C with Tyrode's solution containing 0.6 mM calcium chloride to more closely match the calcium concentration in RPMI medium.
Mitochondria Functional Assay
The Seahorse XF96 extracellular flux analyzer was used to assess mitochondria function. The plates were pretreated with 0.1% gelatin. At around 20 days after differentiation, the cardiomyocytes were seeded onto the plates with a density of 30,000 per XF96 well (2,500/mm2). The cells were treated with fatty acids for 2 weeks in the Seahorse plates before the assay. Culture medium was exchanged for base medium (unbuffered DMEM, Sigma D5030, supplemented with 2 mM glutamine) without fatty acids 1 h before the assay and for the duration of the measurement. Substrates and selective inhibitors were injected during the measurements to achieve final concentrations of glucose at 25 mM, oligomycin at 2.5 μM, FCCP at 1 μM, rotenone at 2.5 μM, and antimycin A at 2.5 μM. The OCR values were further normalized to the number of cells present in each well, quantified by the Hoechst staining (Hoechst 33342; Sigma-Aldrich) as measured using fluorescence at 355 nm excitation and 460 nm emission. The mitochondrial respiratory reserve capacity was calculated by the difference between FCCP and baseline OCR values. Due to variations in the absolute magnitude of OCR measurements in different experiments, the relative fatty acid-treated/untreated control levels were used to compare and summarize independent biological replicates (n = 6).
mRNA-Sequencing Analysis
RNA was extracted using an RNeasy mini kit (Qiagen). The RNA library was prepared using TruSeq library preparation kits (Illumina) following the manufacturer's protocols. The samples were sequenced on 20–25 million coverage. Raw sequences were first trimmed for any adapter and mapped to hg19 reference genome. RNA-sequencing results were aligned to Ensembl GRCh37 using TopHat (Trapnell et al., 2009) (version 2.0.13). Gene-level read counts were quantified using htseq-count (Anders et al., 2015) using Ensembl GRCh 37 gene annotations. Differential gene-expression analysis was performed using DESeq (Anders and Huber, 2010) and topGO R package (Alexa et al., 2006), with Fisher's exact test used for GO enrichment analysis.
qRT-PCR
Total RNA was isolated using the Qiagen RNeasy kit, and mRNA was reverse transcribed using the Superscript III first-strand cDNA synthesis kit (Invitrogen). All primers were purchased from Real Time Primers, and qRT-PCR was performed using SYBR Green chemistry and an ABI 7900HT instrument. Samples were normalized using HPRT (hypoxanthine-guanine phosphoribosyltransferase) as a housekeeping gene. The sequences of the primers are listed in Table S1.
Whole-Cell Current-Clamp Recording
At 20 days following cardiac differentiation, hPSC-CMs were dispersed using trypsin-EDTA and replated at low density onto fibronectin-coated glass coverslips. After 2 weeks of fatty acid treatment, the spontaneously generated action potentials were recorded using an Axopatch 200B amplifier (Axon Instruments, Foster City, CA, USA), operated in current-clamp mode. Temperature was maintained at 36°C ± 1°C by a Warner Instruments TC-324B Single Channel Automatic Heater Controller. The bath solution for recordings contained 136 mM NaCl, 4 mM KCl, 1 mM CaCl2, 2 mM MgCl2, 10 mM HEPES, and 10 mM glucose (pH 7.40 with NaOH); the pipette solution contained 135 mM KCl, 1 mM MgCl2, 10 mM EGTA, 10 mM HEPES, and 5 mM glucose (pH 7.20 with KOH). Patch electrodes were pulled from borosilicate glass capillaries using a Puller (PC-10, Narishige, Tokyo, Japan). Pipette resistance ranged from 2 to 4 MΩ when filled with the internal solution. The experiments were controlled by using Clampex 10.3 software (Axon Instruments). Data analysis was done by using Clampfit 10.3 (Axon Instruments). Parameters for data analysis included APD50 (action potential duration at 50% repolarization), APD90 (action potential duration at 90% repolarization), mean diastolic potential (MDP), APA (action potential amplitude), and maximal upstroke velocity. Cell membrane capacitance was measured by applying a ±10 mV pulse starting from a holding potential of −70 mV.
Statistics
Data are expressed as mean ± SEM. Statistical analysis was performed using Student's t test. A p value of less than 0.05 was considered significantly different.
Author Contributions
X.Y. and C.E.M. conceived experiments; X.Y., M.L.R., A.L., L.S., K.A.F., J.R., L.Z., and S.C.K. performed experiments and analyzed data; Y.W. analyzed data; N.J.S., R.T., H.R., H.X., and C.E.M. supervised experimental work; C.E.M. obtained research funding; X.Y. wrote the manuscript; M.L.R., A.L., Y.W., J.R., S.C.Z., L.P., H.R., N.J.S., R.T., H.R.B., H.X., and C.E.M. edited the manuscript.
Conflicts of Interests
C.E.M. is a scientific founder and equity holder in Cytocardia. N.J.S. is co-founder and equity holder in Stasys Medical Corporation.
Acknowledgments
We are thankful for the assistance from Dr. Stephen R. Coats (Department of Periodontics, University of Washington) for the endotoxin assay. This work was supported by the National Institutes of Health grants R01HL146868, R01HL128362, R01HL128368, U54DK107979, P01HL094374 (all to C.E.M.), P01GM081619 (to C.E.M. and H.R.-B.), and a grant from the Fondation Leducq Transatlantic Network of Excellence (to C.E.M.). X.Y. was supported by the American Heart Association post-doctoral scholarship 12POST11940060 and 14POST20310023. M.L.R. was supported by an NSF Graduate Research Fellowship 2011126228. J.R. was supported by a post-doctoral fellowship from German Research Foundation (DFG) RI 2764/1-1. N.J.S. has an NSF CAREER award CMMI-0846780. R.T. is supported by the National Institutes of Health grant R01HL129510.
Published: September 26, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.08.013.
Accession Numbers
Raw RNA-sequencing data have been deposited in the Gene Expression Omnibus (accession number GEO: GSE124057).
Supplemental Information
References
- Alexa A., Rahnenfuhrer J., Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- Amri E.Z., Bonino F., Ailhaud G., Abumrad N.A., Grimaldi P.A. Cloning of a protein that mediates transcriptional effects of fatty acids in preadipocytes. Homology to peroxisome proliferator-activated receptors. J. Biol. Chem. 1995;270:2367–2371. doi: 10.1074/jbc.270.5.2367. [DOI] [PubMed] [Google Scholar]
- Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakrania B., Granger J.P., Harmancey R. Methods for the determination of rates of glucose and fatty acid oxidation in the isolated working rat heart. J. Vis. Exp. 2016;115 doi: 10.3791/54497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birket M.J., Ribeiro M.C., Kosmidis G., Ward D., Leitoguinho A.R., van de Pol V., Dambrot C., Devalla H.D., Davis R.P., Mastroberardino P.G. Contractile defect caused by mutation in MYBPC3 revealed under conditions optimized for human PSC-cardiomyocyte function. Cell Rep. 2015;13:733–745. doi: 10.1016/j.celrep.2015.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer G.J., Cotman C.W. Survival and growth of hippocampal neurons in defined medium at low density: advantages of a sandwich culture technique or low oxygen. Brain Res. 1989;494:65–74. doi: 10.1016/0006-8993(89)90144-3. [DOI] [PubMed] [Google Scholar]
- Correia C., Koshkin A., Duarte P., Hu D., Teixeira A., Domian I., Serra M., Alves P.M. Distinct carbon sources affect structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Sci. Rep. 2017;7:8590. doi: 10.1038/s41598-017-08713-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feaster T.K., Cadar A.G., Wang L., Williams C.H., Chun Y.W., Hempel J.E., Bloodworth N., Merryman W.D., Lim C.C., Wu J.C. Matrigel mattress: a method for the generation of single contracting human-induced pluripotent stem cell-derived cardiomyocytes. Circ. Res. 2015;117:995–1000. doi: 10.1161/CIRCRESAHA.115.307580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson R.A., Kneebone G.M. Fatty acid composition of human colostrum and mature breast milk. Am. J. Clin. Nutr. 1981;34:252–257. doi: 10.1093/ajcn/34.2.252. [DOI] [PubMed] [Google Scholar]
- Guarner V., Contreras K., Carbo R. Effect of glucose and fatty acid availability on neonatal and adult heart contractility. Biol. Neonate. 2002;82:39–45. doi: 10.1159/000064151. [DOI] [PubMed] [Google Scholar]
- Hardell L.I., Walldius G. Fatty acid composition of human serum lipids at birth. Ups. J. Med. Sci. 1980;85:45–58. doi: 10.3109/03009738009179171. [DOI] [PubMed] [Google Scholar]
- Hazeltine L.B., Simmons C.S., Salick M.R., Lian X., Badur M.G., Han W., Delgado S.M., Wakatsuki T., Crone W.C., Pruitt B.L. Effects of substrate mechanics on contractility of cardiomyocytes generated from human pluripotent stem cells. Int. J. Cell Biol. 2012;2012:508294. doi: 10.1155/2012/508294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofsteen P., Robitaille A.M., Chapman D.P., Moon R.T., Murry C.E. Quantitative proteomics identify DAB2 as a cardiac developmental regulator that inhibits WNT/beta-catenin signaling. Proc. Natl. Acad. Sci. U S A. 2016;113:1002–1007. doi: 10.1073/pnas.1523930113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D., Linders A., Yamak A., Correia C., Kijlstra J.D., Garakani A., Xiao L., Milan D.J., van der Meer P., Serra M. Metabolic maturation of human pluripotent stem cell-derived cardiomyocytes by inhibition of HIF1alpha and LDHA. Circ. Res. 2018;123:1066–1079. doi: 10.1161/CIRCRESAHA.118.313249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hue L., Taegtmeyer H. The Randle cycle revisited: a new head for an old hat. Am. J. Physiol. Endocrinol. Metab. 2009;297:E578–E591. doi: 10.1152/ajpendo.00093.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno S., Kim P.K., Sallam K., Lei J., Billiar T.R., Shears L.L., 2nd Nitric oxide facilitates cardiomyogenesis in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U S A. 2004;101:12277–12281. doi: 10.1073/pnas.0401557101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimata T., Sato D., Seya D., Sato D., Wakatsuki T., Feng Z., Nishina A., Kusunoki M., Nakamura T. Fatty acid composition in fetal, neonatal, and cultured cardiomyocytes in rats. In Vitro Cell. Dev. Biol. Anim. 2013;49:798–804. doi: 10.1007/s11626-013-9668-3. [DOI] [PubMed] [Google Scholar]
- Kliewer S.A., Sundseth S.S., Jones S.A., Brown P.J., Wisely G.B., Koble C.S., Devchand P., Wahli W., Willson T.M., Lenhard J.M. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma. Proc. Natl. Acad. Sci. U S A. 1997;94:4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte F.S., Dai J., Buckley K., Feest E.R., Adamek N., Geeves M.A., Murry C.E., Regnier M. Upregulation of cardiomyocyte ribonucleotide reductase increases intracellular 2 deoxy-ATP, contractility, and relaxation. J. Mol. Cell. Cardiol. 2011;51:894–901. doi: 10.1016/j.yjmcc.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmidis G., Bellin M., Ribeiro M.C., van Meer B., Ward-van Oostwaard D., Passier R., Tertoolen L.G., Mummery C.L., Casini S. Altered calcium handling and increased contraction force in human embryonic stem cell derived cardiomyocytes following short term dexamethasone exposure. Biochem. Biophys. Res. Commun. 2015;467:998–1005. doi: 10.1016/j.bbrc.2015.10.026. [DOI] [PubMed] [Google Scholar]
- Kuppusamy K.T., Jones D.C., Sperber H., Madan A., Fischer K.A., Rodriguez M.L., Pabon L., Zhu W.Z., Tulloch N.L., Yang X. Let-7 family of microRNA is required for maturation and adult-like metabolism in stem cell-derived cardiomyocytes. Proc. Natl. Acad. Sci. U S A. 2015;112:E2785–E2794. doi: 10.1073/pnas.1424042112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.S., Chen J.H., Lundy D.J., Liu C.H., Hwang S.M., Pabon L., Shieh R.C., Chen C.C., Wu S.N., Yan Y.T. Defined microRNAs induce aspects of maturation in mouse and human embryonic-stem-cell-derived cardiomyocytes. Cell Rep. 2015;12:1960–1967. doi: 10.1016/j.celrep.2015.08.042. [DOI] [PubMed] [Google Scholar]
- Lundy S.D., Zhu W.Z., Regnier M., Laflamme M.A. Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cell Dev. 2013;22:1991–2002. doi: 10.1089/scd.2012.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet M., van Berlo J.H., Molkentin J.D. Molecular basis of physiological heart growth: fundamental concepts and new players. Nat. Rev. Mol. Cell Biol. 2013;14:38–48. doi: 10.1038/nrm3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinde A.O., Kantor P.F., Lopaschuk G.D. Maturation of fatty acid and carbohydrate metabolism in the newborn heart. Mol. Cell. Biochem. 1998;188:49–56. [PubMed] [Google Scholar]
- Miller T.A., LeBrasseur N.K., Cote G.M., Trucillo M.P., Pimentel D.R., Ido Y., Ruderman N.B., Sawyer D.B. Oleate prevents palmitate-induced cytotoxic stress in cardiac myocytes. Biochem. Biophys. Res. Commun. 2005;336:309–315. doi: 10.1016/j.bbrc.2005.08.088. [DOI] [PubMed] [Google Scholar]
- Mills R.J., Titmarsh D.M., Koenig X., Parker B.L., Ryall J.G., Quaife-Ryan G.A., Voges H.K., Hodson M.P., Ferguson C., Drowley L. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl. Acad. Sci. U S A. 2017;114:E8372–E8381. doi: 10.1073/pnas.1707316114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaoka Y., Chan A.H., Judge L.M., Yoo J., Huang M., Nguyen T.D., Lizarraga P.P., So P.L., Conklin B.R. Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nat. Methods. 2014;11:291–293. doi: 10.1038/nmeth.2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes S.S., Miklas J.W., Liu J., Aschar-Sobbi R., Xiao Y., Zhang B., Jiang J., Masse S., Gagliardi M., Hsieh A. Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat. Methods. 2013;10:781–787. doi: 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh S.S., Blackwell D.J., Gomez-Hurtado N., Frisk M., Wang L., Kim K., Dahl C.P., Fiane A., Tonnessen T., Kryshtal D.O. Thyroid and glucocorticoid hormones promote functional T-tubule development in human-induced pluripotent stem cell-derived cardiomyocytes. Circ. Res. 2017;121:1323–1330. doi: 10.1161/CIRCRESAHA.117.311920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro M.C., Tertoolen L.G., Guadix J.A., Bellin M., Kosmidis G., D'Aniello C., Monshouwer-Kloots J., Goumans M.J., Wang Y.L., Feinberg A.W. Functional maturation of human pluripotent stem cell derived cardiomyocytes in vitro—correlation between contraction force and electrophysiology. Biomaterials. 2015;51:138–150. doi: 10.1016/j.biomaterials.2015.01.067. [DOI] [PubMed] [Google Scholar]
- Riquelme C.A., Magida J.A., Harrison B.C., Wall C.E., Marr T.G., Secor S.M., Leinwand L.A. Fatty acids identified in the Burmese python promote beneficial cardiac growth. Science. 2011;334:528–531. doi: 10.1126/science.1210558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.L., Graham B.T., Pabon L.M., Han S.J., Murry C.E., Sniadecki N.J. Measuring the contractile forces of human induced pluripotent stem cell-derived cardiomyocytes with arrays of microposts. J. Biomech. Eng. 2014;136:051005. doi: 10.1115/1.4027145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronaldson-Bouchard K., Ma S.P., Yeager K., Chen T., Song L., Sirabella D., Morikawa K., Teles D., Yazawa M., Vunjak-Novakovic G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature. 2018;556:239–243. doi: 10.1038/s41586-018-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J.L., Tulloch N.L., Saiget M., Paige S.L., Razumova M.V., Regnier M., Tung K.C., Keller G., Pabon L., Reinecke H. Mechanical stress promotes maturation of human myocardium from pluripotent stem cell-derived progenitors. Stem Cells. 2015;33:2148–2157. doi: 10.1002/stem.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Reyes O.B., Romero-Avila M.T., Castillo-Badillo J.A., Takei Y., Hirasawa A., Tsujimoto G., Villalobos-Molina R., Garcia-Sainz J.A. Free fatty acids and protein kinase C activation induce GPR120 (free fatty acid receptor 4) phosphorylation. Eur. J. Pharmacol. 2014;723:368–374. doi: 10.1016/j.ejphar.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Sartiani L., Bettiol E., Stillitano F., Mugelli A., Cerbai E., Jaconi M.E. Developmental changes in cardiomyocytes differentiated from human embryonic stem cells: a molecular and electrophysiological approach. Stem Cells. 2007;25:1136–1144. doi: 10.1634/stemcells.2006-0466. [DOI] [PubMed] [Google Scholar]
- Sniadecki N.J., Chen C.S. Microfabricated silicone elastomeric post arrays for measuring traction forces of adherent cells. In: Wang Y., Discher D.E., editors. Methods in Cell Biology: Cell Mechanics. Elsevier Inc.; 2007. pp. 313–328. [DOI] [PubMed] [Google Scholar]
- Tan J.L., Tien J., Pirone D.M., Gray D.S., Bhadriraju K., Chen C.S. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc. Natl. Acad. Sci. U S A. 2003;100:1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulloch N.L., Muskheli V., Razumova M.V., Korte F.S., Regnier M., Hauch K.D., Pabon L., Reinecke H., Murry C.E. Growth of engineered human myocardium with mechanical loading and vascular coculture. Circ. Res. 2011;109:47–59. doi: 10.1161/CIRCRESAHA.110.237206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson S.J., Brown A.J., Holliday N.D. Differential signaling by splice variants of the human free fatty acid receptor GPR120. Mol. Pharmacol. 2012;81:631–642. doi: 10.1124/mol.111.077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt M.J., Steinberg G.R., Chen Z.P., Kemp B.E., Febbraio M.A. Fatty acids stimulate AMP-activated protein kinase and enhance fatty acid oxidation in L6 myotubes. J. Physiol. 2006;574:139–147. doi: 10.1113/jphysiol.2006.107318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Pabon L., Murry C.E. Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ. Res. 2014;114:511–523. doi: 10.1161/CIRCRESAHA.114.300558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Rodriguez M., Pabon L., Fischer K.A., Reinecke H., Regnier M., Sniadecki N.J., Ruohola-Baker H., Murry C.E. Tri-iodo-l-thyronine promotes the maturation of human cardiomyocytes-derived from induced pluripotent stem cells. J. Mol. Cell. Cardiol. 2014;72:296–304. doi: 10.1016/j.yjmcc.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.