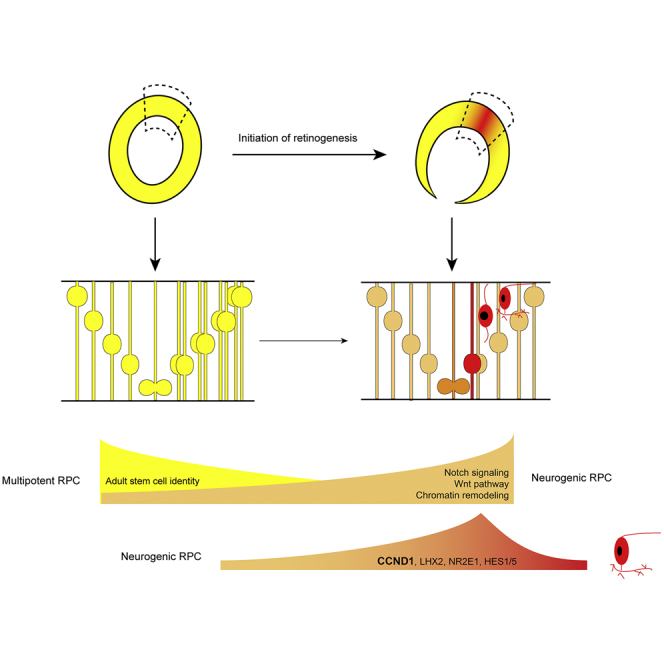
Summary
The development of the mammalian retina is a complicated process involving the generation of distinct types of neurons from retinal progenitor cells (RPCs) in a spatiotemporal-specific manner. The progression of RPCs during retinogenesis includes RPC proliferation, cell-fate commitment, and specific neuronal differentiation. In this study, by performing single-cell RNA sequencing of cells isolated from human embryonic stem cell (hESC)-derived 3D retinal organoids, we successfully deconstructed the temporal progression of RPCs during early human retinogenesis. We identified two distinctive subtypes of RPCs with unique molecular profiles, namely multipotent RPCs and neurogenic RPCs. We found that genes related to the Notch and Wnt signaling pathways, as well as chromatin remodeling, were dynamically regulated during RPC commitment. Interestingly, our analysis identified that CCND1, a G1-phase cell-cycle regulator, was coexpressed with ASCL1 in a cell-cycle-independent manner. Temporally controlled overexpression of CCND1 in retinal organoids demonstrated a role for CCND1 in promoting early retinal neurogenesis. Together, our results revealed critical pathways and novel genes in early retinogenesis of humans.
Keywords: human embryonic stem cell, retina, retinal progenitor, commitment, single-cell RNA-seq, early human retinal development
Graphical Abstract
Highlights
-
•
Fate transition occurring in RPC is concomitant with onset of retinal neurogenesis
-
•
Molecular dynamics underlying RPC commitment are dissected
-
•
CCND1 promotes retinal neurogenesis in a cell-cycle-independent manner
Retinogenesis is a complicated process involving generating distinct types of neurons from retinal progenitor cells (RPCs) in a spatiotemporal-specific manner. By taking the advantage of hESC-derived retinal organoids and single-cell RNA sequencing, Fan, Liu, Hu and colleagues show the genetic program dynamics during RPC progression. Their results revealed critical pathways and novel genes involved in early retinogenesis of humans.
Introduction
The human retina is a complicated neural tissue consisting of multiple types of neurons, which are accurately stratified in different layers during retinogenesis to ensure their proper function. All these highly specialized retinal neurons are derived from multipotent retinal progenitor cells (RPCs) (Agathocleous and Harris, 2009). Evidence at the cellular level showed that RPCs generate retinal neurons following the general rule, which is also seen in a variety of neuronal tissues. RPCs first undergo several rounds of proliferation. A subset of proliferating RPCs become neurogenic, exit the mitotic cycle, and migrate toward the correct laminar position. These committed RPCs further differentiate into mature neurons (Baye and Link, 2008).
Previous studies have focused on dissecting the molecular mechanism regulating neurogenesis of different types of retinal neurons. Despite this, our understanding of how human RPCs gain neurogenic competence before the generation of neurons is still limited. First, many prior studies use animal models such as mouse and rat retina, which may not fully recapitulate retinogenic mechanisms in humans. Second, developing retinas contain many types of heterogeneous cells, which complicates the data interpretation based on bulk genomic analysis, such as RNA sequencing (RNA-seq) data of the entire retina. To overcome these hurdles, we designed experiments by combining single-cell RNA-seq (scRNA-seq) and human embryonic stem cell (hESC)-derived three-dimensional (3D) retinal organoid, which is an in vitro model to recapitulate both morphological and molecular features of the developing human retina (Kuwahara et al., 2015, Zhong et al., 2014). Here, we report the transcriptomic analysis of 457 individual cells isolated from neuroretina of 3D retinal organoids harvested at six time points before and after the onset of retinal neurogenesis. Using systematic approaches including single-cell pseudotime analysis, single-cell trajectory reconstruction, and weighted gene coexpression network analysis (WGCNA), we discovered transcription factors (TFs), chromatin remodeling regulators, and signaling pathway that play critical roles in the commitment of multipotent RPCs to neurogenic RPCs.
Results
Combination of Single-Cell Transcriptome Analysis and Immunostaining Defined the Time Course of RPC Commitment in hESC-Derived 3D Retinal Organoids
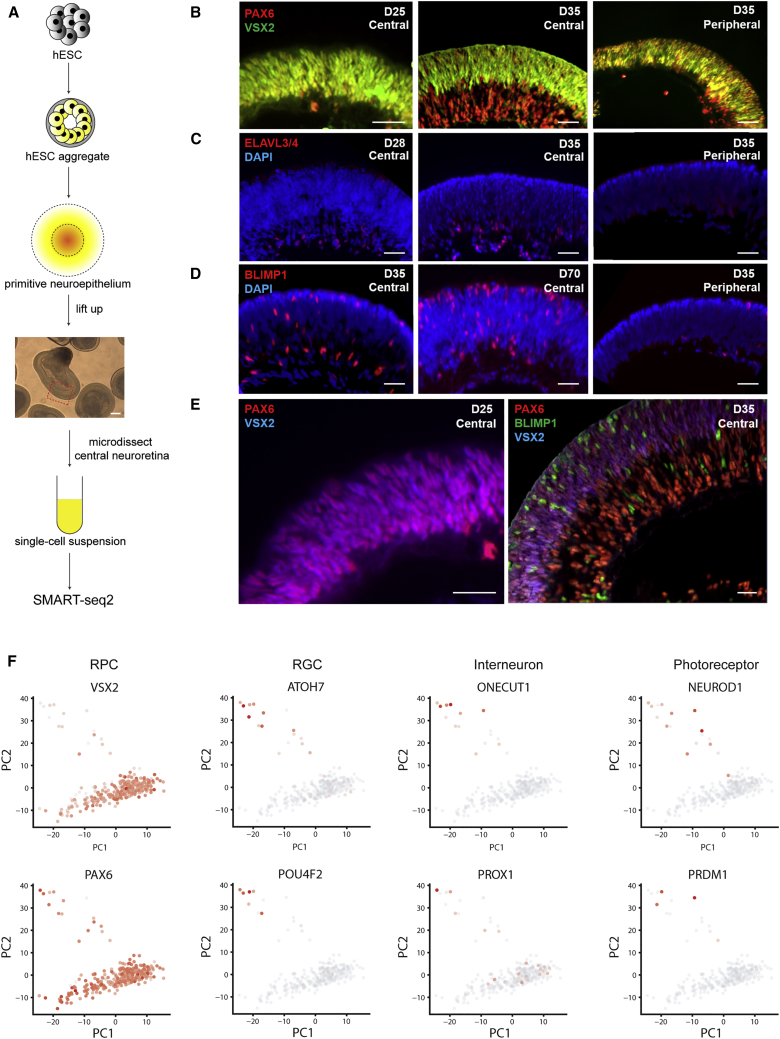
We first generated 3D retinal organoids from hESC cell line H9 using the method reported previously (Kuwahara et al., 2015, Zhong et al., 2014). In brief, hESCs were first differentiated into neuroepithelium through embryonic body formation, then treated with bone morphogenetic protein 4 (BMP4) to enhance the conversion of primitive neuroepithelium to retina. The neural retinas in the center of neuroepithelium were then mechanically dislodged and cultured in suspension to let them self-organize into 3D-retinal organoids, where production of retinal neurons will automatically occur (Figure 1A).
Figure 1.
Immunostaining Characterization of the Onset of Retinal Neurogenesis in hESC-Derived 3D Retinal Organoids
(A) Schematic diagram of the differentiation of hESC-derived 3D retinal organoids and the scRNA-seq library construction strategy. Scale bar, 50 μm.
(B–E) Immunostaining of representative genes in 3D retinal organoid at different time points. Scale bars, 25 μm. (B) PAX6 and VSX2 colocalized in all cells in central retina at day 25. At day 35, cells at the basal side of central retina only expressed PAX6, but not VSX2, while PAX6 and VSX2 were still colocalized in all cells of the peripheral retina at day 35. (C) At day 28, ELAVL3/4-positive cells started to present at the basal side of central retina, and increased in number over time. At the peripheral retina, the expression of ELAVL3/4 was absent from day 25 to day 35. (D) BLIMP1-positive cells first appeared in the basal side of the central retina and migrated to the apical side. However, minimal expression of BLIMP1 was observed in the peripheral retina from day 25 to day 35. (E) Staining of RPC markers and neuronal markers at day 25 and day 35. In the central retinal region, all cells were RPC, while early-born retinal neurons were generated by day 35.
(F) Expression of representative cell-type-specific genes. Red indicates high expression and gray indicates low expression. The cells in the major band-shaped cluster coexpress VSX2 and PAX6, indicating their retinal progenitor cell identity. Cells shifting away express neuron-specific genes, indicating their neuronal identity.
See also Figure S1.
We then sought to determine the time course of neurogenesis in hESC-derived 3D retinal organoids. Immunostaining showed that RPC-specific markers VSX2 and PAX6 were coexpressed throughout hESC-derived 3D optic cup before culturing on day 28 (Figure 1B). After day 28, the expression of VSX2 started to disappear at the basal side of the central retina, where the cells expressing the retinal ganglion cell (RGC) marker ELAVL3/4 began to occur at the same time, and the number of ELAVL3/4-positive cells gradually increased thereafter (Figure 1C). Photoreceptor precursors labeled by BLIMP1 first emerged at the basal side at almost the same time when RGCs were generated and gradually accumulated at the apical side, indicating that the neurogenesis was spontaneously initiated at the central part of retinal organoid at day 28 and then expanded from the center to the periphery (Figure 1D). These results were consistent with previous reports, and indicate the high reproducibility of the 3D retinal organoids culture protocol. Together, these results confirmed that neurogenesis was initiated in 3D retinal organoids at day 28.
We then tried to define the transcriptomic dynamics during RPC commitment. We isolated single cells from the central neuroretina of the 3D optic cup at six time points from day 25 to day 35, which spans RPC proliferation, RPC commitment, and onset of retinal neurogenesis (Figure 1E). We profiled transcriptome in 457 single cells, with 3 million reads per cell on average. In total 306 cells passed quality control, with 0.65 million reads (SD = 0.18 million) uniquely mapped per cell (Picelli et al., 2014). We were able to detect expression of 16,348 genes across 306 cells (genes have more than one read in at least 10% of cells) (Figures S1A and S1B). Principal component analysis (PCA) of normalized and log-transformed transcriptomic data showed that cells were clustered by cell-cycle phase, but not the culture day of the organoids (Figure S1C). Indeed, the expression of cell-cycle-related genes was highly variable, and the principal loading score showed that cell-cycle-related genes were the primary source of the heterogeneity within this cell population (Figure S1D). These results were consistent with the fact that RPCs are highly proliferative, and suggested that the transcriptomic dynamics underlying RPC commitment were largely masked by cell-cycle variation.
To reveal the transcriptomic dynamics driven by RPC commitment, we removed cell-cycle bias from transcriptome data. As expected, PCA after cell-cycle bias correction showed that cells were no longer clustered by their cell-cycle phase. Instead, a majority of cells were clustered in a band-shaped group with some cells shifting away (Figure S1E). The cells outside the band-shaped subpopulation expressed multiple neuronal lineage markers, such as ATOH7 and POU4F2 for RGC differentiation, ONECUT1 and PROX1 for interneuron differentiation, and NEUROD1 and PRDM1 for photoreceptor differentiation, indicating their neuronal identity. The cells in the band-shaped cluster were therefore annotated as RPCs due to their coexpression of RPC markers, including VSX2 and PAX6 (Figure 1F). Interestingly, we found that the RPCs collected from organoids before day 28 were separated from those obtained after day 28. Together, these results suggested a major transcriptomic transition occurred in RPCs at the onset of retinal neurogenesis.
Gene Coexpression Analysis Identified Novel Genes Promoting Neurogenic Competence in RPCs
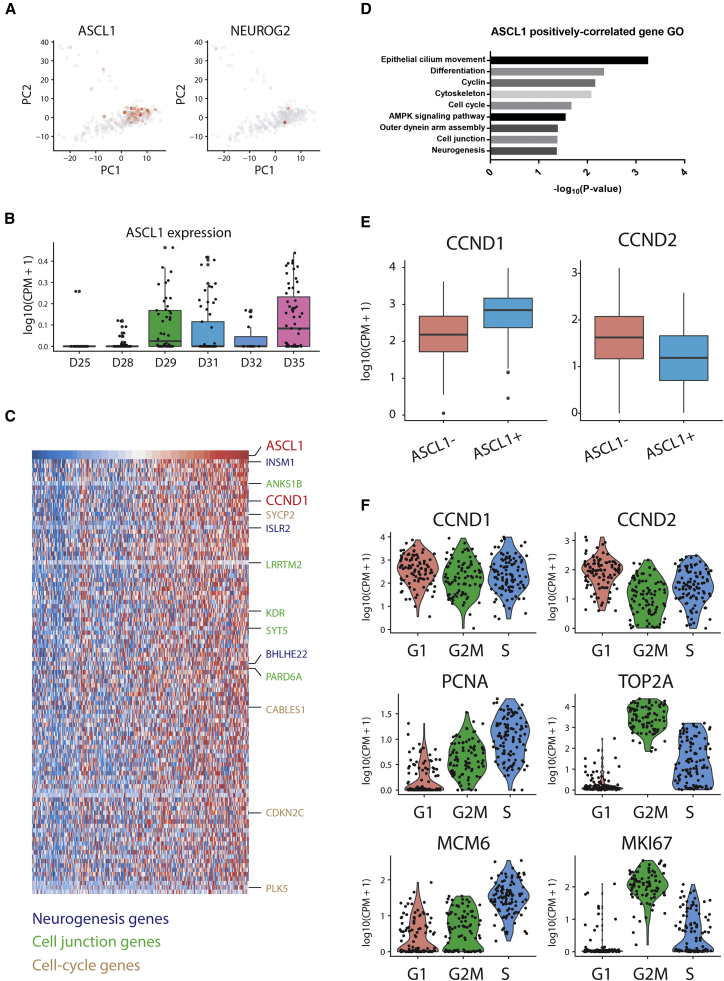
Transient expression of neuronal-fate determinant ASCL1 in RPCs can promote the neurogenic competence and direct RPC to neuronal fate (Brzezinski et al., 2011). In our data, we found that ASCL1 was exclusively expressed in the RPCs collected from organoids at day 28 and later on, which is consistent with the onset timing of RPC commitment in human retinal organoids (Figures 2A and 2B). We then sought to identify additional genes that promote neurogenic competence by performing coexpression analysis between ASCL1 and all other 16,347 expressed genes in RPCs (Figure 2C). Gene ontology analysis of the top 100 genes with the strongest correlation with ASCL1 was enriched in gene ontology terms including cell cycle, cell junction, and neurogenesis (Figures 2D and S2). We found that NGN2 was among the top genes that positively correlated with ASCL1 (r = 0.41, adjusted p < 0.05), which is consistent with its function in promoting neurogenesis during retinal development (Hufnagel et al., 2010). We also found many other genes such as INSM1, BHLHE22, and ISLR2 that are highly correlated with ASCL1 expression in RPCs (Figure 2C and Table S1). These results suggested the potential function of these ASCL1 coexpressing genes in regulating RPC commitment and retinal neurogenesis.
Figure 2.
Identification of Novel Neurogenic Regulators in hESC Retinoids
(A) PCA plot showing the expression of ASCL1 and NGN2, two proneuronal transcription factors (TFs) regulating RPC commitment and neurogenesis.
(B) Box plot showing ASCL1 expression in RPCs collected at different time points. ASCL1 started to express in RPCs at day 28 when neurogenesis in retinal organoids begins.
(C) Heatmap showing the expression of top 100 genes with the strongest positive correlation with ASCL1 in RPCs. Correlation analysis identified genes that coexpress with ASCL1 in RPCs. Genes with known function in neurogenesis, cell-cycle junction, and cell cycle are highlighted by different colors.
(D) Gene ontology analysis showed the functional enrichment of the top 100 genes with the strongest positive correlation with ASCL1 in RPCs.
(E) CCND1 showed a strong positive correlation with ASCL1 in RPCs. ASCL1-positive RPCs showed higher expression of CCND1. On the contrary, ASCL1-positive RPCs have lower expression of CCND2, the homolog of CCND1.
(F) Violin plot shows the expression of CCND1 and other representative cell-cycle-related genes in RPCs at different cell-cycle phases. CCND1 expression does not show a significant difference between different cell-cycle phases.
See also Figure S2.
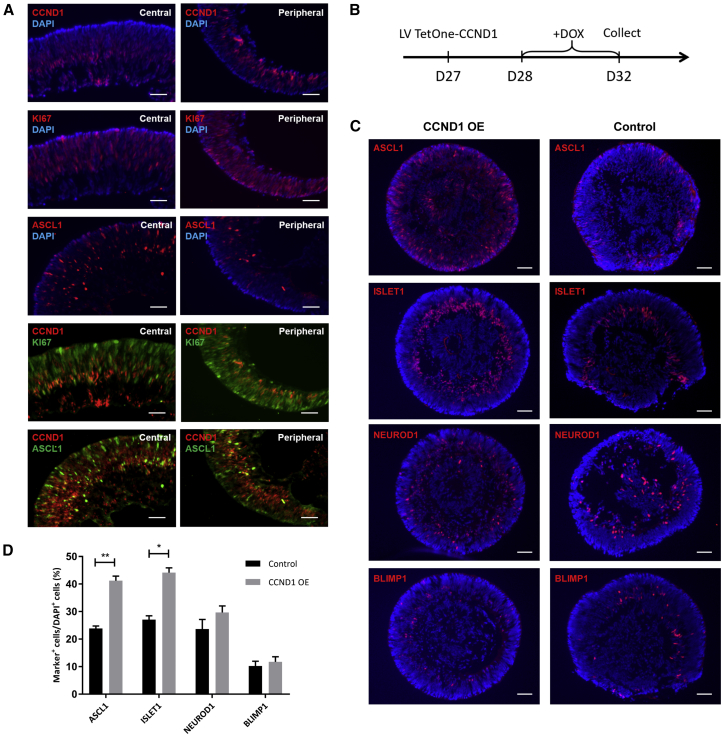
Among ASCL1 coexpressing genes, we found that Cyclin D1 (CCND1) exhibited the 10th top strongest positive correlation with ASCL1 (Figure 2C). Indeed, ASCL1-positive RPCs have higher expression level of CCND1 compared with ASCL1-negative RPCs (Figures 2E and 3A). Surprisingly, although CCND1 is conventionally known as a G1 cell-cycle regulator and exhibits cell-cycle specific expression at G1 phase, we found that in RPCs, CCND1 is expressed in a cell-cycle-independent manner. This is in stark contrast to other classical cell-cycle regulators, such as CCND2 for G1 phase, TOP2A and Ki67 for G2/M phase and PCNA and MCM6 for S phase, which were specifically expressed in their corresponding cell-cycle phases (Figure 2F). Additionally, immunostaining of day-35 retinal organoids showed that expression of CCND1 was higher in the basal side of central neuroretina, where Ki67, the pan-cell-cycle marker, was low (Figure 3A). Together, these results indicated that CCND1 could be involved in RPC commitment in a cell-cycle-independent manner.
Figure 3.
Characterization of CCND1 Function in RPC Commitment
(A) Immunostaining of CCND1, Ki67, and ASCL1 in the central and peripheral day-35 retinal organoids. Scale bars, 25 μm.
(B) Schematic presentation of CCND1 inducible overexpression experiment. Retinal organoids were transduced by lentiviral particles carrying a Tet-On CCND1 overexpression system that can overexpress ectopic CCND1 upon the addition of doxycycline (Dox). We specifically overexpress CCND1 in retinal organoids from day 28 to day 32 by adding Dox at day 28 and removing Dox after day 32.
(C) Expression patterns of ASCL1, ISLET1, NEUROD1, and BLIMP1 in CCND1-overexpressing (CCND1 OE) retinal organoids and control organoids at day 32. Scale bars, 50 μm.
(D) Quantification of Marker+ cells in CCND1 OE retinal organoids and control organoids (mean ± SD; n ≥ 5 organoids from two differentiations) at day 32. ∗p < 0.05, ∗∗p < 0.001 (unpaired t test).
See also Figure S3.
We then examined the function of CCND1 in RPC commitment, using a Tet-On inducible system, which can overexpress ectopic CCND1 upon the addition of doxycycline. Using this system, we specifically overexpressed CCND1 in retinal organoids from day 28 to day 32. This temporal-specific overexpression of CCND1 allows us to accurately assess the function of CCND1 during early retinogenesis (Figure 3B). We found that CCND1 overexpression greatly increased the number of neuronal cells labeled by ISLET1 at day 32, while the proportion of photoreceptor precursors labeled by BLIMP1 appeared to be unaltered (Figures 4A–4C). These results indicated that CCND1 overexpression induced more cells to leave the cell cycle and commit to certain neuronal fate. Notably, overexpression of CCND1 also increased ASCL1-expressing cells, indicating that CCND1 may promote the expression of ASCL1 directly or indirectly (Figure 3C). Similar results were obtained in other hESC lines (Figure S3). Together, these results demonstrated a novel function of CCND1 in promoting RPC commitment and subsequent neurogenesis in a cell-cycle-independent manner.
Figure 4.
Analysis of Marker Genes in Multipotent and Neurogenic RPCs
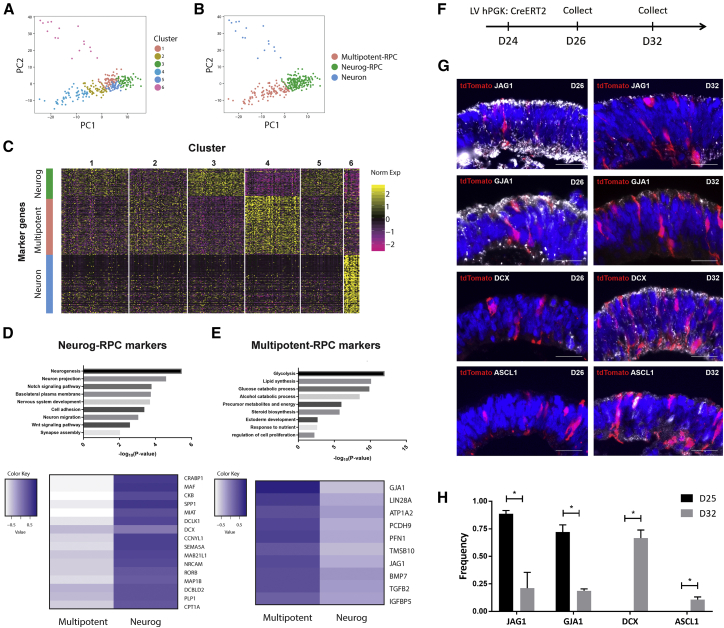
(A) Unbiased clustering initially classified cells into six clusters. Cluster 6 contains all neurons. Clusters 1 through 5 collectively contain all RPCs.
(B) PCA plot showing the biological meaningful cell subgroups. Although six clusters were initially identified, there were only three biologically meaningful subgroups, each of which express a unique set of marker genes compared with others, which is shown in (C).
(C) Heatmap showing the marker genes identified for each of the six clusters. Cluster 6 expresses a unique set of marker genes related to neuronal identity. Clusters 1, 3, and 5 express the same set of marker genes, whereas clusters 2 and 4 express the other set of markers.
(D and E) Top: functional enrichment of genes significantly highly expressed in neurogenic RPCs (D) and multipotent RPCs (E). Bottom: heatmaps showing the average expression of representative marker genes in neurogenic RPCs (D) and multipotent RPCs (E).
(F) Schematic presentation of RPC lineage tracing. AAVS1-loxp-3Stop-loxp-tdTomato H9 ESC-derived retinal organoids were transduced with hPGK:CreERT2-expressing lentivirus in the combination of low-dose 4-hydroxytamoxifen to achieve sparse labeling of multipotent RPC on day 24 and collected on days 26 and 32, respectively.
(G) Coimmunostaining of tdTomato with multipotent RPC markers (JAG1 and GJA1) and neurogenic RPC markers (DCX and ASCL1) at day 26 and day 32, respectively. Scale bars, 25 μm.
(H) Quantification of Marker+ cells among tdTomato+ cells (mean ± SD; n ≥ 5 organoids from two differentiations). ∗p < 0.001 (unpaired t test).
See also Figure S3.
Unbiased Clustering Identified Transcriptomic Transition Associated with Neurogenic Competence in RPCs during Early Retinogenesis
We have shown that RPCs before and at day 28 were separated from RPCs collected after day 28, indicating that a major transcriptomic transition occurs at day 28. We hypothesize that cell subgroups within the RPC population could correspond to different phases of RPC progression. To identify the subgroups within RPCs, we performed unbiased clustering using a shared nearest-neighbor modularity optimization-based clustering algorithm on all 309 single cells, and identified six clusters (Figure 4A and Table S2). A biologically meaningful cell subgroup should express a unique set of marker genes compared with other cells. Cluster 6 contains the cells shifted out from the RPC population, and expressed a distinctive set of marker genes compared with the other five clusters. Functional enrichment of these marker genes showed that they are highly associated with neuronal identity, suggesting that cluster-6 cells are postmitotic neurons. The other five clusters collectively resemble RPCs. However, from these five clusters, we can identify two distinct sets of marker genes, indicating that there are only two biologically meaningful subpopulations within RPCs (Figures 4B and 4C). The majority of RPCs in clusters 1, 3, and 5 were harvested from the central retina after day 28 when retinal neurogenesis was initiated (Figure S4A). Gene ontology of marker genes of these three clusters were enriched in neurogenesis, neuron projection, and nervous system development (Figure 4D), and many genes related to the neuronal development network, such as ASCL1, RTN4, GPM6A, EFNB2, GPM6B, FZD3, RUFY3, and ISLR2, were found to be the shared marker genes for these three clusters. We also found that many marker genes of these three clusters, such as DCX, DCLK1, SEMA5A, and NRCAM (Figure 4D), were also highly expressed in neuronal cells. Together, these results suggest the neurogenic competence of these RPCs. We therefore merged clusters 1, 3, and 5 into one subtype and termed them neurogenic RPCs. Interestingly, ASCL1 and NGN2 were expressed in a subset of these neurogenic RPCs, suggesting a transition from initial neurogenic RPCs to an intermediate state primed for neurogenesis.
Clusters 2 and 4 only contained RPCs collected before and on day 28, and shared another set of marker genes. These marker genes were strongly enriched in metabolic process-associated terms, such as glycolysis and lipid synthesis (Figure 4E). The high expression of metabolism-associated genes is also reported in quiescent adult neural stem cells (NSCs), and metabolism-associated genes were downregulated upon the activation of quiescent NSCs (Dulken et al., 2017). We also found that known marker genes in NSCs, such as GJA1 and ATP1A2, were highly expressed in these RPCs compared with neurogenic RPCs (Figure 4E). The adhesion molecule PCDH9 and cytoskeletal proteins PFN1 and TMSB10, molecules involved in the balance of NSC quiescence and activation, were also specifically expressed in these RPCs (Morizur et al., 2018, Saffary and Xie, 2011) (Figure 4E). Contrary to quiescent NSCs, self-renewal genes, such as LIN28A and LIN28B, were highly expressed in these RPCs. Notably, secreted signaling molecules, including JAG1, BMP7, TGFβ2, and IGFBP5, were also among the marker genes of these clusters, which were reported to be exclusively localized in the peripheral retina and ciliary margin (Trimarchi et al., 2009). Together, these results indicated the molecular resemblance of these RPCs to adult NSCs and the non-neurogenic RPCs in the peripheral retina. We therefore annotated this RPC subgroup as multipotent RPCs.
To clarify the transition between multipotent RPC and neurogenic RPCs, we used an AAVS1-loxp-3Stop-loxp-tdTomato hESC line for lineage tracing. We transduced hPGK:CreERT2-expressing lentivirus in the combination of low-dose 4-hydroxytamoxifen to achieve sparse labeling of multipotent RPCs at day 24 (Figure 4F). At day 26, the majority of labeled cells were marked by multipotent RPC markers, such as JAG1 and GJA1. By day 32, almost all tdTomato+ cells were doublecortin (DCX) positive, while JAG1 and GJA1 were greatly downregulated, indicating the molecular transition from multipotent RPCs to neurogenic RPCs. In addition, a subset of labeled cells started to express ASCL1, which primes for further differentiation (Figures 4G and 4H). Notably, the neurogenic RPC marker distribution exhibited a central-to-peripheral downgraded pattern, while multipotent markers were preferentially located at the peripheral domain, also suggesting the more advanced state of neurogenic RPCs than multipotent RPCs along the neurogenic path (Figure S4B).
WGCNA Analysis Identified Gene-Regulatory Networks in Early Retinogenesis
To systematically understand the dynamics of the genetic program in early retinogenesis, we performed the WGCNA on our scRNA-seq data. WGCNA is an unsupervised method that can identify coexpressed gene modules that consist of genes that are likely involved in similar biological progress (Langfelder and Horvath, 2008). We identified three modules that are composed of genes specifically upregulated in multipotent RPCs, neurogenic RPCs, and neurons, respectively (Figure S5A). Using the WGCNA measure of intramodular gene connectivity, we found different hub genes across the three stage-specific modules. Hub genes are the genes centrally located within a gene module. They have a tight coexpression relationship between many genes, and therefore could have important regulatory roles. GJA1 and ENO1 were identified as hub genes in multipotent RPC module (Figure S5C). Many known neurogenic TFs, including ATOH7, ONECUT2, and DLX2, were among the intramodular hub genes for the neuronal module (Figure S5D). Interestingly, for the neurogenic RPC module, FZD5 was highlighted as the hub gene in the network (Figure S5B). Previous results showed that FZD5 has different functions through different signaling pathways in different species. For example, FZD5 homolog Fz5 in Xenopus laevis mediates canonical Wnt signaling, and its knockdown causes RPC hypoproliferation. However, in zebrafish, FZD5 homolog mediates noncanonical Wnt signaling and promotes eye field formation (Agathocleous and Harris, 2009). In mice, Fzd5 does not influence RPC proliferation but instead regulates cell survival and neurogenesis (Liu et al., 2012). Together, these results suggested the possible role for FZD5 in regulating RPC commitment specifically in human retinogenesis.
Pseudotime Analysis Identified Genes Dynamically Changed during the Progression of RPC Competence
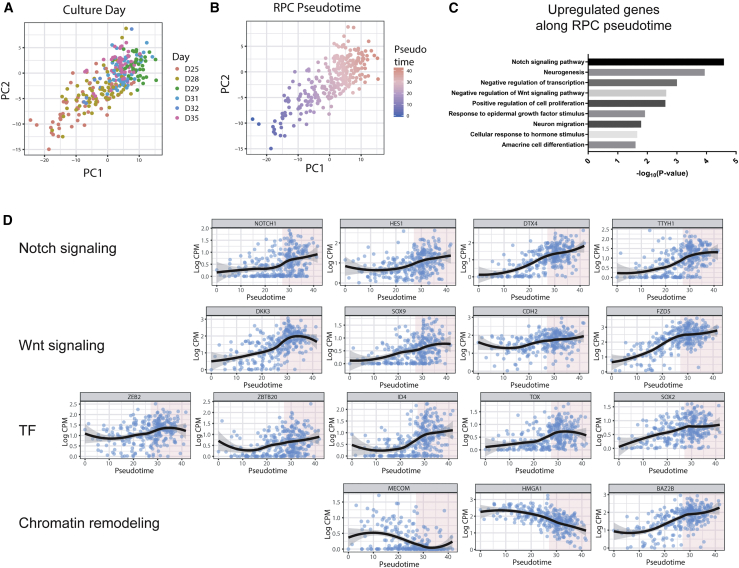
To further delineate the mechanism underlying the RPC progression, we reconstructed the development pseudotime for each RPC by applying a principle curve analysis to RPCs and identifying genes whose expression exhibit progressive change along the pseudotime using Spearman’s correlation (Figures 5A, 5B, and S6). Gene set enrichment of top 100 correlated genes showed association to Notch signaling and canonical Wnt pathway (Figure 5C). Notch components (NOTCH1, HES1) and positive regulators (DTX4, TTYH1) increased gradually during RPC progression (Figure 5D). Notch signaling is a highly conservative pathway functioning in lateral inhibition of neurogenesis and progenitor maintenance (Zhang et al., 2018). Thus, the enhancement of Notch signaling at the initiation of retinal neurogenesis indicated its noncanonical role in promoting the neurogenic competence of RPCs, which is consistent with the observation of a dramatic loss of NGN2+ cells in the central retina of Notch signaling DNA-binding protein Rbpj mutant (Maurer et al., 2014). In contrast, Wnt/β-catenin pathway was negatively regulated by the upregulation of Wnt signaling inhibitor of DKK3 and negative targets (ERG1, SOX9, and CDH2) (Figure 5D). Consistently in previous studies, β-catenin expression decreased along the peripheral to central gradient, and the inhibition of canonical Wnt signaling by Dkk3 was also found to promote the retinogenic process (Bharti et al., 2012).
Figure 5.
Pseudotime Analysis of RPC Progression Using Principal Curve Method
(A and B) To focus on the expression dynamics in RPCs, we inferred developmental pseudotime (B) from culture days (A) using RPCs by the principle curve (PC) method.
(C) Enriched gene ontology terms and p values of positively correlated genes with pseudotime.
(D) Expression change of genes that were dynamically regulated in RPCs during developmental pseudotime. These genes were related to Notch signaling, Wnt signaling, and chromatin remodeling, and some of them were TFs.
See also Figure S6.
We also found many TFs that were correlated with the RPC progression pseudotime. For example, TFs belonging to transcriptional repressors, such as ZEB2, ZBTB20, and ID4 (Figure 5D), exhibited upregulated expression. Zeb2 was reported to promote the timely differentiation of retinal interneurons, possibly through the repression of the TGFβ/Smad pathway, while Zbtb20 and Id4 determined the balance between neurogenic and gliogenic output by preventing premature neurogenesis (Bedford et al., 2005, Menuchin-Lasowski et al., 2016, Nagao et al., 2016). Another family of TFs, which contains the high-mobility group (HMG) box that maintains proliferative progenitors in the neurogenic region and controls the temporal regulation of neurogenesis, such as TOX, SOX2, and SOX9, was also upregulated in the RPC lineage (Artegiani et al., 2015, Poche et al., 2008). Interestingly, several chromatin structure regulators were also dynamically changed during RPC progression (Figure 5D). For example, HMGA1 is required for the maintenance of an open chromatin state and was downregulated along RPC progression (Kishi et al., 2012). BAZ2B, which is a component of the chromatin remodeling complex and has been demonstrated to play a role in transcriptional activation via histone acetylation, was significantly upregulated (Oppikofer et al., 2017). MECOM is widely involved in the chromatin remodeling that leads to transcriptional repression and is downregulated during RPC progression (Cattaneo and Nucifora, 2008). These results suggested that chromatin remodeling is actively involved in early retinogenesis.
Single-Cell Trajectory Analysis Identified Genes Transiently Activated along the Trajectory of RPC Commitment to Early-Born Retinal Subtypes
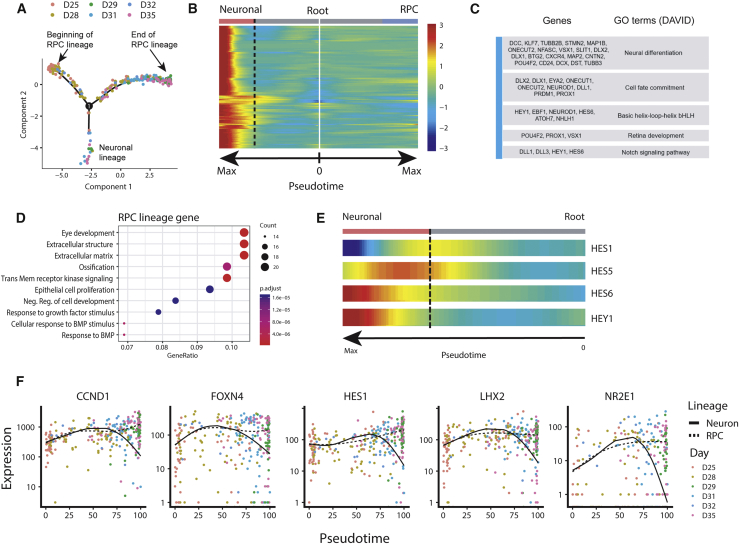
So far, the analysis of RPC progression has been done by comparing transcriptomes between discrete cell populations. To reveal gene-expression dynamics over the continuous development process, we reconstructed a cell-developmental trajectory using Monocle2 and analyzed the gene-expression change along pseudotime (Qiu et al., 2017). Monocle2 identified a bifurcating trajectory with three branches. One branch specifically expressed neuronal-fate TFs, such as ATOH7, PRDM1, ONECUT1, and PROX1, indicating their neuronal identity (Figures 6A and S7A). The other two branches showing high expression of VSX2 and PAX6, but different expression of ASCL1, were therefore annotated as multipotent and neurogenic RPCs. These two RPC branches collectively represented the whole RPC lineage. We found that GJA1 and JAG1, two multipotent RPC markers, were downregulated with pseudotime progress in RPC lineage; the markers of neurogenic RPCs such as ASCL1, NGN2, and DCX were gradually upregulated (Figures S7B and S7C).
Figure 6.
Single-Cell Developmental Trajectory Analysis Using Monocle2
(A) Using both RPCs and neuronal data to construct lineage bifurcation in hESC retinoids. Two lineages, representing RPC lineage and neuronal lineage, were identified.
(B) Heatmap showing the expression change of neuron-specific genes (genes classified into the first cluster) in both neuronal lineage and RPC lineage.
(C) Representative genes in the neuron-specific gene set and their functions.
(D) Functional enrichment of genes upregulated over pseudotime in RPC lineage.
(E) Expression of HES1, HES5, HES6, and HEY1 in a neuronal lineage over pseudotime.
(F) Expression of genes that were transiently upregulated in RPC lineage. Dashed lines and solid lines show the smoothed expression over pseudotime in RPC lineage and neuronal lineage, respectively.
See also Figures S7 and S8.
We then identified genes whose expression was dynamically changed over pseudotime in any branches. Unbiased hierarchical clustering classified 384 dynamically changed genes into four major clusters, each of which contains a set of genes showing similar expression pattern along the trajectory (Figure S8A). The first cluster consists of all genes that were specifically upregulated in neuronal branches and showed no significant change in RPC lineage (Figure 6B). Gene ontology analysis showed that these genes were enriched in biological processes such as “neuron differentiation” and “cell fate commitment,” suggesting their function in committing RPCs to retinal neurons (Figure 6C). Genes in cluster II maintained consistent expression in RPC and showed a transiently upregulated pattern at the branching point, enriched for the gene ontology terms, such as “nucleosome assembly,” “DNA replication,” and “chromosome segregation” (Figure S8B). A subset of linker histone H1 genes were also accumulated in this cluster, indicating the involvement of chromatin compaction prior to neuronal differentiation. Genes in the third and fourth clusters were upregulated in neurogenic RPCs compared with multipotent RPCs and were repressed in the neuronal lineage after the cell-fate branching point. Many genes within these two clusters were related to cell proliferation, metabolic process, and embryonic morphogenesis, suggesting the function of these genes in maintaining retinal progenitor cell identity (Figures 6D and S8B). These results suggested that the single-cell trajectory recapitulated the developmental trajectory during human early retinogenesis.
Genes transiently upregulated at the bifurcation point where neuronal lineage separated from RPC lineage could have a critical function in promoting the commitment of RPCs. Using Monocle2, we found that many TFs, including FOXN4, LHX2, NR2E1, and HES1, were transiently upregulated directly before the lineage bifurcation point and then repressed in neuronal lineage (Figure 6F). Previous studies showed that Foxn4 directs RPCs to amacrine or horizontal fate (Li et al., 2004). LHX2 and NR2E1 have been conventionally reported to maintain the proliferative competence and developmental pluripotency in the developing retina, but their function in RPC fate commitment during early retinogenesis is not known (Gordon et al., 2013, Miyawaki et al., 2004). Previous research showed that Hes1 and Hes5 are expressed in RPCs and can block neurogenic programming in RPCs by suppressing proneuronal basic helix-loop-helix gene expression, while Hes6 was silent in RPCs and upregulated in terminally differentiated neurons (Zhang et al., 2018). Consistently in our data, we found that HES1 and HES5 were transiently activated in RPCs and then repressed in terminally differentiated neurons, and HES6 were persistently upregulated in terminally differentiated neurons after the lineage bifurcation point (Figure 6E). Notably, our data showed that HES1 was activated much earlier than HES5, which is confirmed by a recent study showing that Hes1 was initially coexpressed with both Ccnd1 and Hes5 in the central developing retina, but only the expression of Hes5 and not Hes1 persists in the majority of differentiating RGCs (Riesenberg et al., 2018). Together, our data therefore indicated a sequential activation of HES1 and then HES5 during RPC commitment. It would be particularly interesting to elucidate the unique function of HES1 and HES5 separately in RPC commitment during early retinogenesis.
Discussion
The study of early retinal development in humans has been greatly hampered by the difficulty in obtaining early human retina and the heterogeneity of retinal tissue. Although the studies from model organisms have provided us with some hints about the retinal development in mammals, these results must be interpreted with caution because of the potential species difference in retinal development. To overcome these challenges, our study took advantage of hESC-derived 3D retinal culture, which allows us to study the very early retinal development with ultrahigh temporal and spatial resolution. With the power of scRNA-seq, we were able to directly measure the transcriptome dynamics within different cell types.
In this study we profiled transcriptional heterogeneity of RPCs during the onset of retinal neurogenesis, and identified a previous unknown transcriptomic transition in RPCs during the onset of retinal neurogenesis. Single-cell analysis has revealed the involvement of the Wnt pathway in the regulation of this transition. The function of the canonical Wnt pathway was revealed long ago, which biases the RPC fate to retinal pigment epithelium and ciliary margin but not the neuroretina (Fujimura, 2016). Consistent with previous findings, in our data the canonical Wnt signaling was negatively regulated via the upregulation of DKK3 along RPC progression. Interestingly, the Wnt receptor FZD5 was revealed as the master hub gene in the gene-regulatory network of neurogenic RPCs, and undergoes a progressive upregulation during RPC progression in our dataset using a human model. However, the functional studies of FZD5 revealed significant variance among different species. Therefore, our data of hESC-derived retinal organoids support the role for FZD5 in regulating human RPC commitment, which is not seen in model organisms such as mice and zebrafish.
Through gene coexpression analysis, novel genes correlated with the expression of ASCL1 were identified, which have the potential to promote the neurogenic competence of RPCs. Intriguingly, CCND1, a progenitor-associated gene that positively regulates cell-cycle progression, was found to be upregulated in ASCL1+ RPCs, and its overexpression significantly activate ASCL1 expression in RPCs and promote downstream neurogenesis in a cell-cycle-independent way. Surprisingly, the phenotype of CCND1 overexpression closely mirrored that in Ccnd1 knockout mice, which exhibited the precocious cell-cycle exit of RPCs and the overproduction of RGCs (Das et al., 2009). These results indicated that CCND1 was not only necessary for the progenitor maintenance but also essential for promoting neurogenesis, which is consistent with the finding in developing mouse spinal cord (Lukaszewicz and Anderson, 2011). The dual function was also implicated in other canonical progenitor genes, such as LHX2, NR2E1, and HES5, which were shown as transiently upregulated genes at the fate bifurcation point. Their neurogenic function has been revealed in many other neural systems by overexpression (Elmi et al., 2010, Subramanian et al., 2011). It would be of particular interest to also investigate their potential function in RPC commitment.
Regeneration of retinal neurons from Müller cells with adult stem cell characteristics is a promising therapy for retinal damage. However, recent strategies to enhance reprogramming of Müller cells into neurons were largely based on the mechanism of zebrafish Müller cell reprogramming after injury, which may not be suitable for the mammalian system. We found that the multipotent RPCs characterized in this study closely resembled adult NSCs but exhibited strong proliferative competence. We suggest that the comparison of the molecular properties between multipotent RPCs and Müller cells in a human model could provide hints about how to reactivate human retinal stem cells to enhance in situ regeneration of retinal neurons in the future.
Experimental Procedures
hESC Culture and Retinal Organoid Differentiation
Human ESC lines H9 and H1 were kindly provided by Stem Cell Bank, Chinese Academy of Sciences. hESCs were maintained in Essential-8 (E8) medium (Invitrogen) on Vitronectin (VTN-N)-coated plates and passaged twice per week. Medium was changed every day. To differentiate hESCs into 3D retinal organoid, we enzymatically lifted hESC colonies with dispase (2 mg/mL) into small cell clusters of about 100–150 μm in diameter. The medium was switched from E8 to neural induction medium (DMEM/F12 [1:1], 1% N2 supplement, MEM nonessential amino acids, penicillin-streptomycin, and 2 μg/mL heparin sulfate) gradually over the course of 4 days. On day 7, cell aggregates were attached to the plate with 10% fetal bovine serum (FBS). On day 16, neural rosettes were manually dissected from the plate and put in retinal differentiation medium (RDM: DMEM/F12 [3:1], 2% B27 supplement, MEM nonessential amino acids, and penicillin-streptomycin) to allow free-floating organoids to form. From day 30, RDM was supplemented with 10% FBS, 100 μM taurine, 2 mM GlutaMAX, and 0.5 μM retinoic acid for long-term differentiation. Recombinant human BMP4 (Stemgent) was added to the culture on day 6 to the final concentration of 50 ng/mL, which was diluted by a half medium change every third day to enhance the efficiency of retinal induction (Kuwahara et al., 2015).
Immunocytochemistry
Retinal organoids were fixed in 4% paraformaldehyde for 30 min, cryoprotected overnight at 4°C in 30% sucrose, and cryosectioned. Frozen sections were incubated in blocking buffer (10% FBS, 2% donkey serum, and 0.2% Triton X-100 in PBS) for 1 h, followed by incubation with primary antibodies overnight at 4°C. Next day, sections were incubated with secondary antibodies at 1:1,000 (Alexa Fluor, Thermo Fisher) for 45 min. After washing and mounting, sections were imaged on a fluorescence microscope (Zeiss). Antibodies against the following proteins were used at the indicated dilutions: ASCL1 (rabbit, 1:100, Abcam ab211327), BLIMP1 (rat, 1:100, Santa Cruz Biotechnology sc-47732), CCND1 (mouse, 1:100, Santa Cruz sc-8396), DCX (mouse, 1:100, Santa Cruz sc-271390), ELAVL3/4 (mouse, 1:200, Molecular Probes MP21271), GJA1 (mouse, 1:100, Santa Cruz sc-271837), HES1 (rabbit, 1:200, Abcam ab71559), ISLET1 (rabbit, 1:100, Abcam ab20670), JAG1 (mouse, 1:100, Santa Cruz sc-390177), Ki67 (mouse, 1:100, Abcam ab8191), NEUROD1 (mouse, 1:100, Santa Cruz sc-46684), NOTCH1 (rabbit, 1:100, Abcam ab27526), PAX6 (rabbit, 1:500, Covance PRB-278P), tdTomato (goat, 1:200, Biorbyt orb182397), VSX2 (sheep, 1:500, Millipore AB9016).
Lentivirus Preparation and Organoid Infection
A lentivirus carrying both Tet-On 3G transactivator and TRE3GS promoter-mediated CCND1 was purchased from HanBio to perform inducible CCND1 overexpression. To perform lentiviral transduction on retinal organoids, we added 1 million lentiviral transducing units per retinal organoid to RDM at day 27 in the presence of 8 μg/mL polybrene (Sigma). After incubation for 12 h, lentivirus particles were removed by changing the medium. To induce CCND1 overexpression at day 28, we added 200 ng/mL doxycycline to the medium and maintained it for 4 days.
Lineage Tracing
AAVS1-loxp-3Stop-loxp-tdTomato H9 ESC (Biocytogen, BCG-H9-009) was used for lineage tracing of multipotent RPC. On day 24, retinal organoids were transduced with lentivirus expressing CreERT2. One day post infection, 0.5 μM 4-hydroxytamoxifen (Sigma) was added to the medium for 24 h. We collected organoids following tamoxifen treatment after 24 h and 7 days, respectively.
scRNA-Seq Library Preparation and Analysis
The central neuroretina of retinal organoids were manually dissected and dissociated in Accutase (StemPro) for 30 min to create a single-cell suspension. Single cells were collected from two independent differentiation experiments. From the first batch, organoids were collected at day 25, day 28, day 31, and day 35; from the second batch, organoids were collected at day 26, day 29, day 32, and day 35. From each time point, 2–3 organoids were collected. This balanced sample-collection strategy can eliminate potential bias in data analysis that result from organoid differentiation batch effect. Cells were collected by centrifugation at 300 × g for 5 min and resuspended in 500 μL of RDM. We used a mouth pipette to immediately transfer single cells into prepared lysis buffer. The scRNA-seq library was constructed following a modified Smart-seq2 protocol (Picelli et al., 2014). In detail, samples were incubated at 72°C for 3 min. The first-strand cDNA was reverse-synthesized using oligo(dT) and template-switching oligonucleotides. The synthesized first-strand cDNAs were amplified by 18 cycles. After purification, 0.1 ng of cDNA was used for Nextera tagmentation (Illumina) and library construction. Sequencing was performed on Illumina HiSeq 4000.
scRNA-Seq Data Processing and Analysis
Single-cell libraries were pooled and sequenced on HiSeq 4000 aiming for 3 million reads per library on average, in pair-end 150-bp mode. Raw sequencing reads were demultiplexed, and trimmed using TrimGalore! Software. Trimmed data were aligned to Human reference Hg38 using the STAR 2.6.0 aligner with default parameters. Reads aligned to each gene were counted using featureCounts according to Gencode Hg38 gene annotation. Gene-expression data normalization, dimensional reduction, and data visualization were performed using customized R script and R package Seurat following the developer's instructions. Cell-cycle bias correction was performed using CellCycleScoring and ScaleData functions implemented in Seurat. In brief, each cell was assigned an “S Phase Score” and a “G2M Phase Score” using the strategy described by Tirosh et al. (2016), which is defined as the sum of bin-normalized expression of S-phase marker genes or G2M-phase marker genes, respectively. These two scores can be regarded as the quantitative representation of the cell-cycle phase of each single cell. The heterogeneity resulting from the cell cycle then can be removed from the data by regressing out the “S Phase Score” and a “G2M Phase Score” from the log-transformed RPM (gene read count per million mapped reads) matrix.
Author Contributions
X.M., Q.A., Y.H., and G.F. conceived the idea, designed the study, and interpreted the results. X.M. and H.X. performed the experiments and generated the data. X.Z. helped with the culture of 3D retinal organoid. Q.A. performed bioinformatics analysis. Q.A. and X.M. wrote the manuscript. X.Y., S.Y., J.W., Q.L., and G.F. revised the manuscript.
Acknowledgments
This study was supported by the National Key R&D Program of China (2017YFA0104101, 2017YFC1001300), National Natural Science Foundation of China (81870694, 81770973, 81970821), Jiangsu Provincial Natural Science Foundation of China (BK20151586), Construction Program of Jiangsu Provincial Clinical Research Center Support System (BL2014084), Six Talent Peaks Project in Jiangsu Province (WSW-027) and the “333 Project” Research Projects of the Fifth Phase of Jiangsu Province (BRA2017548), NIH grants (R01DE025474, R01EY026319), CIRM Stem Cell Genomics Centers of Excellence Award CRP2, and an unrestricted grant from the Research to Prevent Foundation to the Department of Ophthalmology at University of California Los Angeles.
Published: September 19, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.08.012.
Contributor Information
Youjin Hu, Email: huyoujin@gzzoc.com.
Qinghuai Liu, Email: liuqh@njmu.edu.cn.
Guoping Fan, Email: gfan@mednet.ucla.edu.
Accession Numbers
All scRNA-seq data reported in this paper are deposited in the Gene Expression Omnibus with accession number GEO: GSE122783.
Supplemental Information
References
- Agathocleous M., Harris W.A. From progenitors to differentiated cells in the vertebrate retina. Annu. Rev. Cell Dev. Biol. 2009;25:45–69. doi: 10.1146/annurev.cellbio.042308.113259. [DOI] [PubMed] [Google Scholar]
- Artegiani B., de Jesus D.A., Bragado A.S., Brandl E., Massalini S., Dahl A., Calegari F. Tox: a multifunctional transcription factor and novel regulator of mammalian corticogenesis. EMBO J. 2015;34:896–910. doi: 10.15252/embj.201490061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baye L.M., Link B.A. Nuclear migration during retinal development. Brain Res. 2008;1192:29–36. doi: 10.1016/j.brainres.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford L., Walker R., Kondo T., van Cruchten I., King E.R., Sablitzky F. Id4 is required for the correct timing of neural differentiation. Dev. Biol. 2005;280:386–395. doi: 10.1016/j.ydbio.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Bharti K., Gasper M., Ou J., Brucato M., Clore-Gronenborn K., Pickel J., Arnheiter H. A regulatory loop involving PAX6, MITF, and WNT signaling controls retinal pigment epithelium development. PLoS Genet. 2012;8:e1002757. doi: 10.1371/journal.pgen.1002757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski J.T., Kim E.J., Johnson J.E., Reh T.A. Ascl1 expression defines a subpopulation of lineage-restricted progenitors in the mammalian retina. Development. 2011;138:3519–3531. doi: 10.1242/dev.064006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo F., Nucifora G. EVI1 recruits the histone methyltransferase SUV39H1 for transcription repression. J. Cell. Biochem. 2008;105:344–352. doi: 10.1002/jcb.21869. [DOI] [PubMed] [Google Scholar]
- Das G., Choi Y., Sicinski P., Levine E.M. Cyclin D1 fine-tunes the neurogenic output of embryonic retinal progenitor cells. Neural Dev. 2009;4:15. doi: 10.1186/1749-8104-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulken B.W., Leeman D.S., Boutet S.C., Hebestreit K., Brunet A. Single-cell transcriptomic analysis defines heterogeneity and transcriptional dynamics in the adult neural stem cell lineage. Cell Rep. 2017;18:777–790. doi: 10.1016/j.celrep.2016.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmi M., Matsumoto Y., Zeng Z.J., Lakshminarasimhan P., Yang W., Uemura A., Nishikawa S., Moshiri A., Tajima N., Agren H. TLX activates MASH1 for induction of neuronal lineage commitment of adult hippocampal neuroprogenitors. Mol. Cell. Neurosci. 2010;45:121–131. doi: 10.1016/j.mcn.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Fujimura N. WNT/beta-Catenin signaling in vertebrate eye development. Front. Cell Dev. Biol. 2016;4:138. doi: 10.3389/fcell.2016.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon P.J., Yun S., Clark A.M., Monuki E.S., Murtaugh L.C., Levine E.M. Lhx2 balances progenitor maintenance with neurogenic output and promotes competence state progression in the developing retina. J. Neurosci. 2013;33:12197–12207. doi: 10.1523/JNEUROSCI.1494-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufnagel R.B., Le T.T., Riesenberg A.L., Brown N.L. Neurog2 controls the leading edge of neurogenesis in the mammalian retina. Dev. Biol. 2010;340:490–503. doi: 10.1016/j.ydbio.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi Y., Fujii Y., Hirabayashi Y., Gotoh Y. HMGA regulates the global chromatin state and neurogenic potential in neocortical precursor cells. Nat. Neurosci. 2012;15:1127–1133. doi: 10.1038/nn.3165. [DOI] [PubMed] [Google Scholar]
- Kuwahara A., Ozone C., Nakano T., Saito K., Eiraku M., Sasai Y. Generation of a ciliary margin-like stem cell niche from self-organizing human retinal tissue. Nat. Commun. 2015;6:6286. doi: 10.1038/ncomms7286. [DOI] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Mo Z., Yang X., Price S.M., Shen M.M., Xiang M. Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron. 2004;43:795–807. doi: 10.1016/j.neuron.2004.08.041. [DOI] [PubMed] [Google Scholar]
- Liu C., Bakeri H., Li T., Swaroop A. Regulation of retinal progenitor expansion by Frizzled receptors: implications for microphthalmia and retinal coloboma. Hum. Mol. Genet. 2012;21:1848–1860. doi: 10.1093/hmg/ddr616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewicz A.I., Anderson D.J. Cyclin D1 promotes neurogenesis in the developing spinal cord in a cell cycle-independent manner. Proc. Natl. Acad. Sci. U S A. 2011;108:11632–11637. doi: 10.1073/pnas.1106230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer K.A., Riesenberg A.N., Brown N.L. Notch signaling differentially regulates Atoh7 and Neurog2 in the distal mouse retina. Development. 2014;141:3243–3254. doi: 10.1242/dev.106245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menuchin-Lasowski Y., Oren-Giladi P., Xie Q., Ezra-Elia R., Ofri R., Peled-Hajaj S., Farhy C., Higashi Y., Van de Putte T., Kondoh H. Sip1 regulates the generation of the inner nuclear layer retinal cell lineages in mammals. Development. 2016;143:2829–2841. doi: 10.1242/dev.136101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki T., Uemura A., Dezawa M., Yu R.T., Ide C., Nishikawa S., Honda Y., Tanabe Y., Tanabe T. Tlx, an orphan nuclear receptor, regulates cell numbers and astrocyte development in the developing retina. J. Neurosci. 2004;24:8124–8134. doi: 10.1523/JNEUROSCI.2235-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizur L., Chicheportiche A., Gauthier L.R., Daynac M., Boussin F.D., Mouthon M.A. Distinct molecular signatures of quiescent and activated adult neural stem cells reveal specific interactions with their microenvironment. Stem Cell Reports. 2018;11:565–577. doi: 10.1016/j.stemcr.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao M., Ogata T., Sawada Y., Gotoh Y. Zbtb20 promotes astrocytogenesis during neocortical development. Nat. Commun. 2016;7:11102. doi: 10.1038/ncomms11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppikofer M., Bai T., Gan Y., Haley B., Liu P., Sandoval W., Ciferri C., Cochran A.G. Expansion of the ISWI chromatin remodeler family with new active complexes. EMBO Rep. 2017;18:1697–1706. doi: 10.15252/embr.201744011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S., Faridani O.R., Bjorklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Poche R.A., Furuta Y., Chaboissier M.C., Schedl A., Behringer R.R. Sox9 is expressed in mouse multipotent retinal progenitor cells and functions in Muller glial cell development. J. Comp. Neurol. 2008;510:237–250. doi: 10.1002/cne.21746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesenberg A.N., Conley K.W., Le T.T., Brown N.L. Separate and coincident expression of Hes1 and Hes5 in the developing mouse eye. Dev. Dyn. 2018;247:212–221. doi: 10.1002/dvdy.24542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffary R., Xie Z. FMRP regulates the transition from radial glial cells to intermediate progenitor cells during neocortical development. J. Neurosci. 2011;31:1427–1439. doi: 10.1523/JNEUROSCI.4854-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L., Sarkar A., Shetty A.S., Muralidharan B., Padmanabhan H., Piper M., Monuki E.S., Bach I., Gronostajski R.M., Richards L.J. Transcription factor Lhx2 is necessary and sufficient to suppress astrogliogenesis and promote neurogenesis in the developing hippocampus. Proc. Natl. Acad. Sci. U S A. 2011;108:E265–E274. doi: 10.1073/pnas.1101109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I., Izar B., Prakadan S.M., Wadsworth M.N., Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi J.M., Cho S.H., Cepko C.L. Identification of genes expressed preferentially in the developing peripheral margin of the optic cup. Dev. Dyn. 2009;238:2327–2329. doi: 10.1002/dvdy.21973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Engler A., Taylor V. Notch: an interactive player in neurogenesis and disease. Cell Tissue Res. 2018;371:73–89. doi: 10.1007/s00441-017-2641-9. [DOI] [PubMed] [Google Scholar]
- Zhong X., Gutierrez C., Xue T., Hampton C., Vergara M.N., Cao L.H., Peters A., Park T.S., Zambidis E.T., Meyer J.S. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun. 2014;5:4047. doi: 10.1038/ncomms5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.