Summary
Organoid technologies have become a powerful emerging tool to model liver diseases, for drug screening, and for personalized treatments. These applications are, however, limited in their capacity to generate functional hepatocytes in a reproducible and efficient manner. Here, we generated and characterized the hepatic organoid (eHEPO) culture system using human induced pluripotent stem cell (iPSC)-derived EpCAM-positive endodermal cells as an intermediate. eHEPOs can be produced within 2 weeks and expanded long term (>16 months) without any loss of differentiation capacity to mature hepatocytes. Starting from patient-specific iPSCs, we modeled citrullinemia type 1, a urea cycle disorder caused by mutations in the argininosuccinate synthetase (ASS1) enzyme. The disease-related ammonia accumulation phenotype in eHEPOs could be reversed by the overexpression of the wild-type ASS1 gene, which also indicated that this model is amenable to genetic manipulation. Thus, eHEPOs are excellent unlimited cell sources to generate functional hepatic organoids in a fast and efficient manner.
Key words: liver, 3D organoid, citrullinemia, EpCAM, iPSC, hepatocyte, disease modelling, hepatocyte differentiation
Graphical Abstract
Highlights
-
•
iPSC-derived EpCAM+ endodermal cells generate functional hepatic organoids (eHEPOs)
-
•
eHEPOs can be stably maintained in long-term culture
-
•
An organoid-based model for a urea cycle disorder (citrullinemia) is generated
-
•
Genetic manipulation of disease-specific eHEPOs rescues disease phenotype
Erdal and colleagues generated a hepatic organoid (eHEPO) culture system using human iPSC-derived EpCAM-positive endodermal cells. eHEPOs can be produced within 2 weeks and expanded long term (>16 months) without any loss of differentiation capacity to mature hepatocytes. Starting from patient-specific iPSCs, a urea cycle disorder, citrullinemia, was modeled and disease phenotype was rescued via overexpression of wild-type ASS1 gene.
Introduction
Cellular therapies provide promising treatment options for liver pathologies ranging from cancer to genetic metabolic diseases. One of the main difficulties associated with cellular therapy approaches is the generation and expansion of mature autologous hepatocytes. While the ability to derive patient-specific induced pluripotent stem cells (iPSCs) have solved the problem of generating autologous cells, differentiation and expansion of mature patient-specific hepatocytes remain a challenge.
Hepatocyte-like cells can be derived from human iPSCs (hiPSCs) via their stepwise differentiation. Initial studies focused on developing differentiation protocols to achieve hepatocyte-like cells in standard two-dimensional (2D) conditions (Chen et al., 2012, Liu et al., 2011, Si-Tayeb et al., 2010). Such stepwise protocols utilize cocktails of growth factors/cytokines to recapitulate embryonic liver development in vitro, from definitive endoderm to hepatic progenitors and then to functional hepatocytes. However, the cell populations produced by these different protocols vary considerably in their maturation status and, in most cases, represent immature hepatocytes. This is in part due to the absence of tissue-specific architecture, mechanical and biochemical cues, and cell-cell communication under 2D conditions (Luo et al., 2018, Pampaloni et al., 2007). To overcome these limitations, an alternative approach has been utilized to induce human iPSCs into hepatic endodermal cells and then mix them in a three-dimensional (3D) co-culture system with stromal cells to generate liver bud-like aggregates (Takebe et al., 2013, Takebe et al., 2017). This liver bud-like structure is able to vascularize and perform hepatic functions after transplantation into mice. However, the long-term stability and expansion of such structures is currently unknown.
A 3D organoid culture system of adult hepatic stem/progenitor cells has been recently developed. These cultures are derived from Lgr5-positive cells sorted from injured murine liver and epithelial cell adhesion molecule (EpCAM)-positive ductal cells from intact human liver (Broutier et al., 2016, Huch and Dolle, 2016, Huch et al., 2013, Huch et al., 2015). During liver development and homeostasis, EpCAM has a dynamic expression pattern, and its expression in immature cells is gradually lost upon their maturation into hepatocytes (Schmelzer et al., 2007). EpCAM is expressed in mouse liver bud at embryonic day 9.5 (E9.5), and EpCAM+Dlk1+ hepatoblasts with high proliferation potential have the capacity to differentiate into bile duct and hepatocytes (Tanaka et al., 2009). Additionally, in zebrafish only EpCAM+ endodermal cells are able to license hepatic development by sequestering Kremen1 on the cell membrane and allowing the formation of Lrp6 signalosomes to activate Wnt2bb target genes (Lu et al., 2013). Whether human EpCAM+ endodermal cells that arise during development can form liver organoids remains unknown.
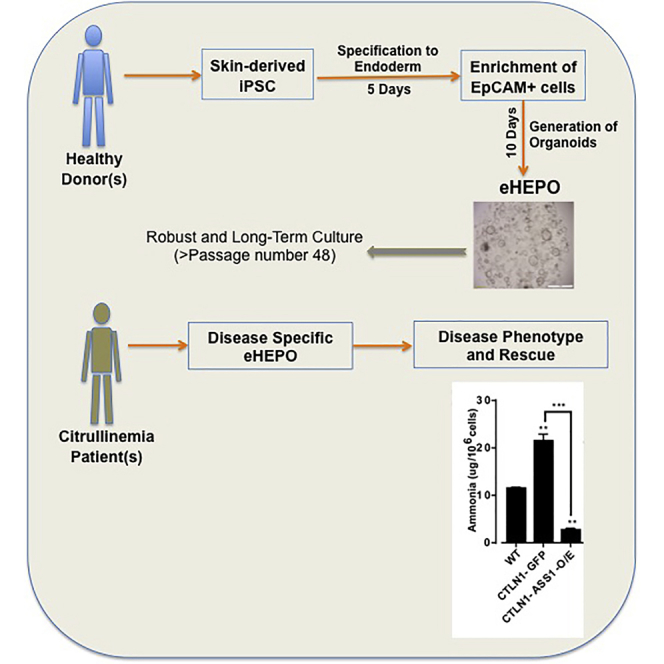
Here, we report an hiPSC-derived hepatic organoid culture system for producing functional hepatocytes using EpCAM-positive endodermal cells as an intermediate. These endoderm-derived hepatic organoids (eHEPOs) could be produced within 14 days and expanded for more than 1 year without any loss in culture efficiency. Using eHEPOs, we modeled the urea cycle disorder citrullinemia, demonstrating the power of this system for disease modeling. Finally, we performed ectopic expression of wild-type allele of the disease-causing gene, ASS1, in eHEPOs, which rescued the phenotype of the disease and showed that eHEPOs are amenable to genetic manipulation.
Results
Endodermal Progenitors Generated from hiPSCs
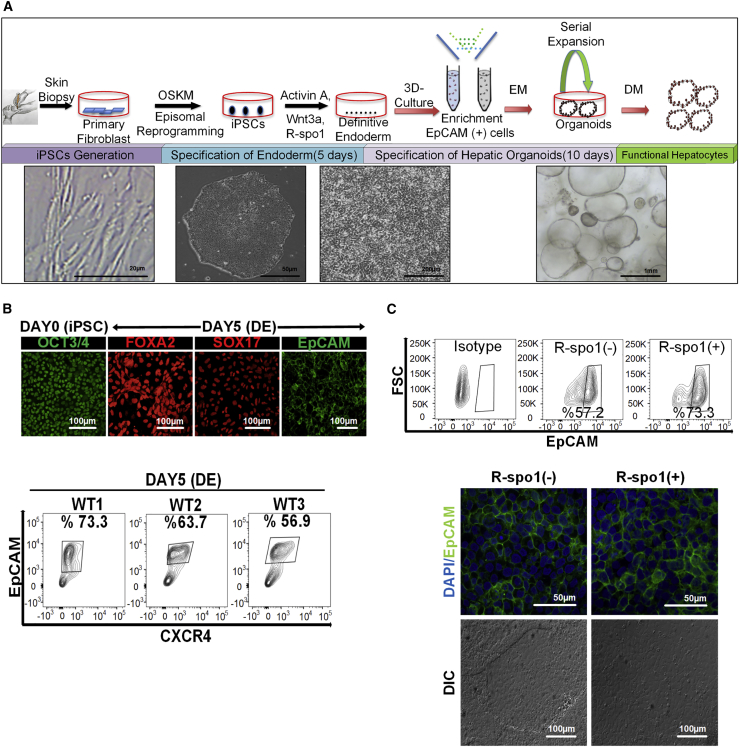
To test whether hepatic organoids can be generated from iPSCs, we first derived transgene-free iPSCs from a healthy donor via non-integrative episomal plasmid-based reprogramming of dermal fibroblasts (Fidan et al., 2015). Teratoma formation in immunocompromised mice confirmed the pluripotency of the iPSCs (Figure S1). iPSCs expanded in feeder-free culture were incubated with modified endodermal induction medium containing activin A, Wnt3a, and R-spo1 (Chen et al., 2012, Si-Tayeb et al., 2010). After 5 days, cells no longer exhibited typical embryonic stem cell morphology and adopted a polygonal shape (Figure 1A). Immunostaining revealed that almost all of the cells were positive for the endoderm markers SOX17, FOXA2, and EpCAM at day 5, indicating that the hiPSCs efficiently differentiated into definitive endoderm during the 2D monolayer endodermal induction period (Figure 1B). Additionally, the flow-cytometry analyses showed that only about 65% of populations are CXCR4+/EpCAM+ (Figure 1B) and approximately 75% of populations are FOXA2+/SOX17+ at day 5 (Figure S2). Notably, percentages of CXCR4+/EpCAM+ endodermal cells derived from three independent iPSC lines (WT1, WT2, and WT3) were not significantly different from each other after independent differentiations, which indicated the reproducibility of endodermal induction step of our protocol (Figure 1B). Notably, modification of the endoderm induction medium by adding R-spondin1, a well-known Wnt signal potentiator (Carmon et al., 2011, de Lau et al., 2011, Planas-Paz et al., 2016), caused a 15%–20% increase in the number of EpCAM+ cells differentiated from three independent iPSC lines from healthy donors (Figure 1C). Moreover, there were no morphological changes among these EpCAM+ cell populations (Figure 1C). Thus, our modified protocol allows for efficient generation of endodermal progenitor populations.
Figure 1.
Generation of EpCAM+ Progenitors from hiPSCs as an Intermediate of Endoderm-Derived Hepatic Organoids
(A) Schematic representation and timeline of organoid culture establishment.
(B) Upper panel: expression of OCT3/4 (pluripotency marker), FOXA2, SOX17, and EpCAM (endoderm markers), in the different stages by immunostaining (20× magnification). Lower panel: flow-cytometric analysis of EpCAM and CXCR4 to assess the efficiency of endoderm generation at day 5. Three independent healthy iPSC lines (wild-type 1 [WT1], WT2, WT3) were tested for endoderm induction based on EpCAM and CXCR4 positivity (n = 3 for each experiment).
(C) Upper panel: representative flow-cytometric analysis with isotype control demonstrating the increase in EpCAM-expressing cell number with addition of R-spo1. Experiments were performed in triplicates for three different iPSC lines derived from healthy donors. Lower panel: effect of R-spo1 on the morphology of iPSC derived endodermal cells on 2D culture. DIC, differential interference contrast.
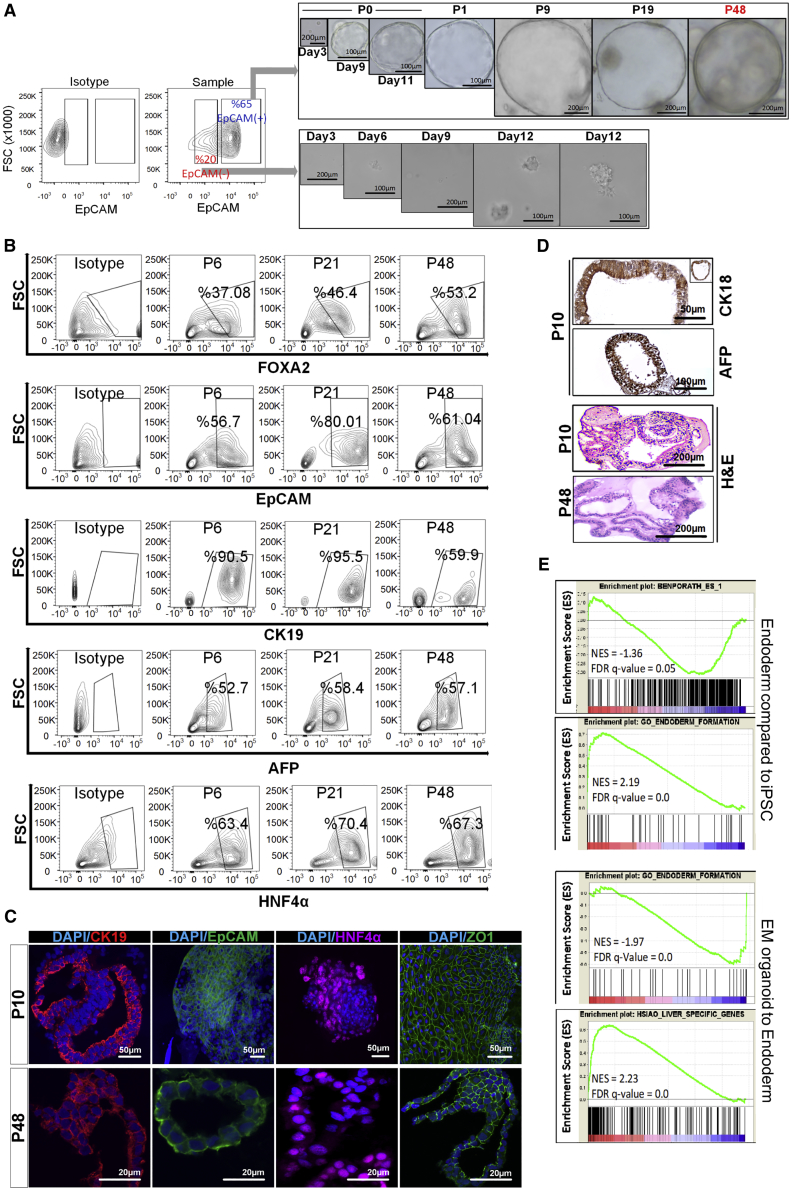
Generation of Organoids from Endodermal EpCAM+ Cells
We inoculated fluorescence-activated cell sorting (FACS)-purified EpCAM+ or EpCAM−endodermal progenitors in the liver organoid expansion medium as defined in adult liver organoid cultures. As early as 1 week, EpCAM+ endodermal cells generated 3D hollow structures similar to the adult stem cell-derived liver organoids, while EpCAM− cells lacked this ability. When culturing EpCAM+-derived organoids, structures with round-shaped organoid morphology, approximately 100 nm in diameter with distinct edges, were observed (Figure 2A). Notably, organoid cultures could be passaged every 7–10 days at 1:5 ratio and could be expanded for >9 months without any loss of phenotypic characteristics and differentiation capacity at late passages (passage >30) (Figure 2A). To determine the stability of EpCAM+ endoderm cell-derived organoids cultured in expansion medium (EM) conditions, we analyzed the phenotypic characteristics for different markers (AFP+, HNF4α+, FOXA2+, EpCAM+, and CK19+) by flow cytometry at various time points (passages 6, 21, and 48). Percentages of subpopulations expressing EpCAM and FOXA2 (endoderm markers), α-fetoprotein (AFP) (fetal liver marker), and hepatocyte nuclear factor 4α (HNF4α) (hepatic marker) did not change significantly at different passages including young and old organoids, while CK19 (hepatoblast/cholangiocyte marker) expression decreased in old organoids in EM conditions (Figure 2B). These findings supported that EM mostly maintains stable status of progenitor cells in organoid culture over the long term. Concerning structural organization, the organoids from early passage comprised CK19+ cells surrounding tightly self-organized ductal-like structures, indicating that cells with hepatoblast and/or bile duct progenitor characteristics persist in these structures. For the late-passage organoids, CK19+ cells are more likely distributed evenly throughout the organoid as well as ductal specific locations (Figure 2C). In addition, EpCAM+ cells were still present in the organoids both from early and late passages, showing the persistence of precursor cells of liver. Notably, HNF4α staining showed that organoids have already committed toward hepatic lineage both in whole-mount and frozen-section staining of early and late passages, respectively. Both early- and late-passage organoids have cubical/polyhedral epithelial cells expressing ZO-1 in a similar pattern, providing evidence for the presence of cell-cell interactions via tight junctions (Figure 2C). Of note, cell density of late-passage organoids was rather less than that of their young counterparts.
Figure 2.
Establishment of Endoderm-Derived Hepatic Organoids
(A) Organoid formation potency of sorted EpCAM+ and EpCAM− cells and bright-field images of organoid culture from various passages, where P indicates passage number.
(B) Phenotypic characterization of endoderm-derived WT organoid culture at different passage numbers (p6, p21, and p48) during expansion on EM.
(C) Confocal images of organoids for EpCAM, CK19, and HNF4α. Nuclei were co-stained with DAPI. In the dataset for p10 organoids, EpCAM∖HNF4α and ZO-1 were stained in the whole-mount organoids. The others were stained from frozen sections.
(D) Immunohistochemical staining of CK18 and AFP in organoids. H&E indicates hematoxylin-eosin staining.
(E) GSEA plots for differentially expressed genes during iPSC to endoderm induction and endoderm to organoid differentiation. NES and FDR q values are listed for each gene set analyzed.
Further immunohistochemical analysis of organoids showed that CK18+ and AFP+ cells form pseudostratified epithelial structures as observed in epithelium development (Figure 2D). Hematoxylin-eosin staining showed that the typical structural organization including pseudostratified epithelial and ductal-like structure did not change in long-term organoids at passage 48 (Figure 2D).
To gain a global understanding of eHEPO differentiation, we performed RNA sequencing of iPSCs, endodermal progenitors, and organoids cultured in EM conditions and performed an unbiased characterization of their identity at whole-transcriptome level (see Experimental Procedures). Gene set enrichment analysis (GSEA) indicated that gene sets of gastrulation, endoderm formation, and endoderm development were highly enriched upon endoderm induction from iPSCs. Conversely, pluripotency-associated genes were downregulated at the same stage (Figure 2E). Upon EM induction, endoderm specific genes were downregulated and liver-specific gene sets were induced (Figure 2E). Taken together, these data indicate that this protocol faithfully simulates the stepwise developmental process of hepatic differentiation.
In Vitro Differentiation of Organoids into Mature Hepatocytes
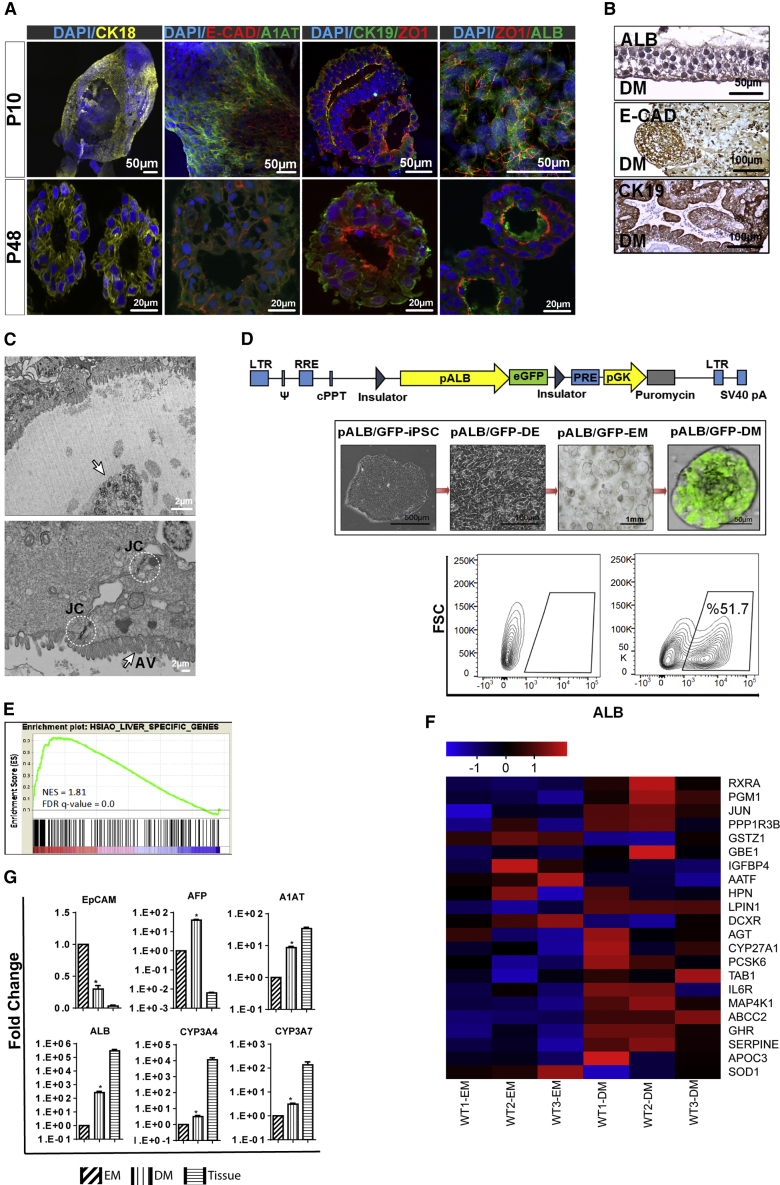
To further the maturation of the emerging hepatic cells, we cultured organoids in differentiation medium (DM) for 10–14 days (Huch et al., 2015) and analyzed the expression of liver-specific genes and structural organization by immunostaining. Here, we performed immunostaining in both early-passage (p10) and late-passage (p48) organoids at DM conditions for CK18, ZO-1, E-CAD, CK19, ALB, and A1AT. All differentiated organoids are similarly composed of ALB+ and CK18+ hepatocytes with typical polar structure and ZO-1 expression, indicating the existence of tight junctions separating apical and basolateral domains. Besides, the E-CAD staining pattern indicates the liver epithelial phenotype of organoids. ALB and A1AT staining provides evidence for hepatocyte maturation, still expressed even in late passages. Meanwhile, the presence of CK19+ cells, especially around lumen-like structures, suggests the existence of the cholangiocyte-like and/or progenitor cell population in differentiated organoids (Figure 3A). Further immunohistochemical analysis also revealed that organoids have both ALB+ and CK19+ cells, indicating mature hepatocytes and cholangiocytes in ductal-like structure, respectively. Also, E-CAD+ cells represented polygonal epithelioid structures reflecting a hepatocyte-like phenotype (Figure 3B). Ultrastructural analysis of organoids demonstrated the presence of a layer of live cells with apical and basolateral polarity and a luminal area containing the residual of apoptotic and multivesicular bodies (Figure 3C). Of note, the junctional complex between cells was defined by the characteristic of epithelial cells surrounding the lumen (Figure 3C). To further characterize the maturation of the iPSC eHEPOs, we developed an albumin-GFP reporter system. An albumin enhancer/promoter driving GFP expression was cloned into a lentiviral backbone between two flanking insulator elements and was integrated into hiPSCs, which enabled us to monitor real-time differentiation of iPSCs to mature hepatocytes in organoids. Organoids were established from reporter bearing iPSCs which, after 5 days in DM culture conditions, became GFP positive (Figure 3D). We also quantified a number of ALB+ cells within organoids from three independent differentiations starting from a single iPSC reporter line. There was no significant difference in the number of ALB+ cells (Figure 3D). For a detailed assessment of the differentiation status of organoids, we generated global expression profiles after DM induction. GSEA analysis showed that liver-specific genes were highly upregulated upon culture in DM conditions (normalized enrichment score [NES] 1.81, false discovery rate [FDR] q value = 0). This finding supports our hypothesis that liver-specific genes are further upregulated in DM conditions compared with EM conditions (Figure 3E). Most of the key enzymes and receptors involved in different aspects of liver function including glucose homeostasis (DCXR, IGFBP4, PGM1), lipid metabolism (RXRA, GHR, SOD1, APOC3, APOB, APOA1, LPIN1), and gluconeogenesis (PPP1R3B, GBE1) were induced upon DM culture (Figure 3F). qPCR validation of RNA-sequencing data confirmed the upregulation of mature hepatocyte markers such as ALB, A1AT, CYP3A7, and CYP3A4 and downregulation of the endoderm stage marker EpCAM, albeit to a lower extent when compared with freshly harvested adult human liver tissue (Figure 3G).
Figure 3.
In Vitro Differentiation of Endoderm-Derived Hepatic Organoids into Mature Hepatocytes
(A) Confocal images of organoids cultured for 10–14 days in DM conditions stained for CK18, E-cadherin, A1AT, ZO1, and ALB. Nuclei were stained with DAPI. In the dataset for p10 organoids, CK18, E-CAD/A1AT, and ZO-1/ALB were stained in the whole-mount organoids. The others were stained from frozen sections.
(B) Immunohistochemical staining of organoids for ALB, CK19, and E-cadherin.
(C) Transmission electron microscopy image of an endoderm-derived hepatic organoid (eHEPO). Arrow shows apoptotic and multivesicular bodies (upper panel). White circles and arrow indicate intercellular junctional complexes (JC) and apical villus (AV), respectively (lower panel).
(D) Lentiviral albumin promoter-GFP reporter to monitor albumin expression in organoids. Representative bright-field and fluorescence microscopy images of pALB-GFP reporter bearing iPSCs at the indicated stages of differentiation. Flow-cytometry analysis to quantify ALB+ cells within organoid.
(E) GSEA analysis of differentially regulated genes in DM versus EM conditions. NES and FDR q value are listed for the liver-specific gene set analyzed.
(F) Heatmap showing the expression of EM- and DM-related genes.
(G) qPCR-based mRNA expression analysis of indicated genes in EM and DM organoids as well as human liver tissue. Fold changes were calculated as DM/EM and/or tissue/EM (n = 4 for each of three separate differentiation) (∗p ≤ 0.05).
In Vitro and In Vivo Functionality of eHEPOs
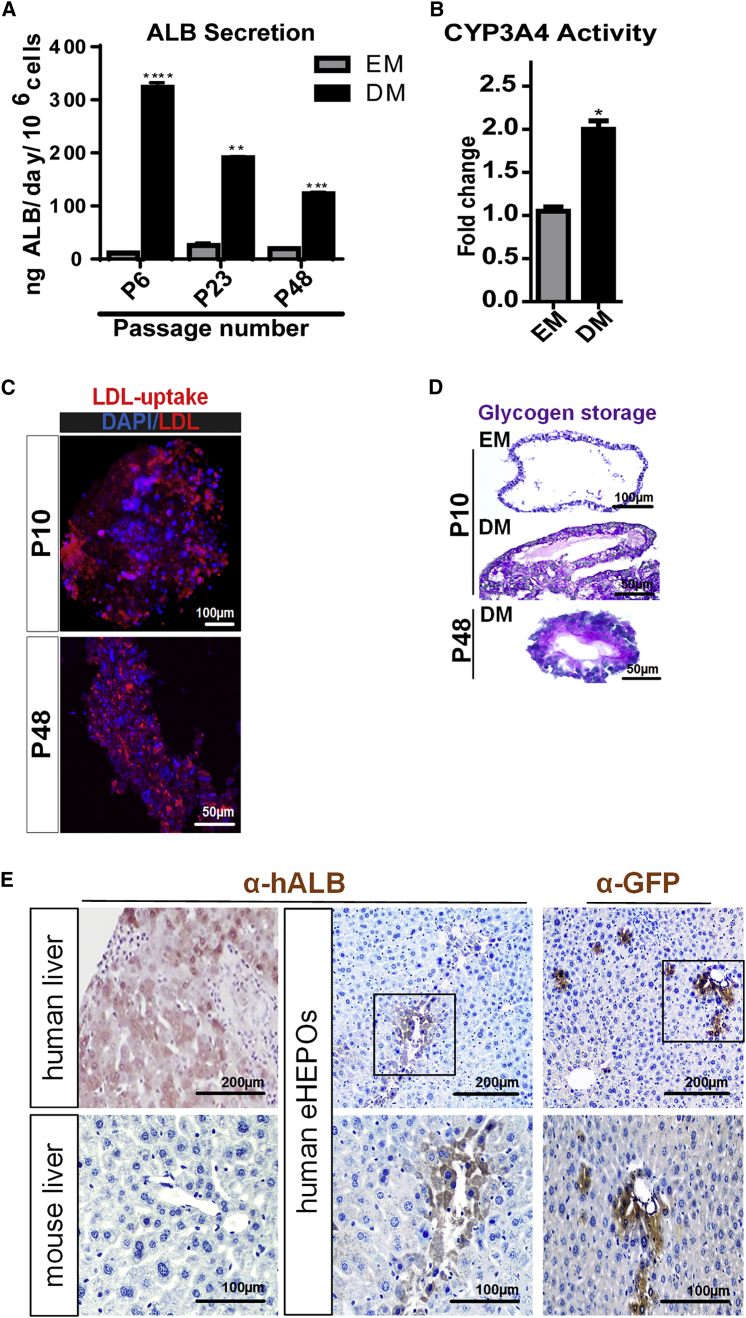
We showed that mature eHEPOs in DM from various passages secreted significant amounts of albumin to the medium, indicative of hepatocyte functionality. However, the secreted albumin level in DM conditions was gradually decreased depending on the age of organoid in the culture (Figure 4A). Upon differentiation, organoids also gained mature hepatocyte functions such as CYP3A4 activity, low-density lipoprotein (LDL) uptake, and glycogen storage (Figures 4B–4D). The late-passage eHEPOs (p48) still demonstrated liver functions such as LDL uptake and glycogen storage, similar to their early-passage counterparts (Figures 4C and 4D).
Figure 4.
In Vitro and In Vivo Functionality of Endoderm-Derived Hepatic Organoids
(A) Albumin secretion of the WT organoids from different passage numbers (p6, p23, p48) cultured in EM and DM conditions as measured by ELISA. Data are shown as mean ± SD of n = 3, given as ngALB/day/million cells.
(B) CYP3A4 activity in organoids cultured in EM and DM conditions expressed as RLU/mL/million cells.
(C) Uptake of low-density lipoprotein (LDL) detected on day 14 by immunofluorescence staining in WT DM organoids from p10 and p48.
(D) Glycogen storage function of WT DM organoids from p10 and p48 by periodic-acid-Schiff (PAS) staining (magenta). EM condition was used as undifferentiated control.
(E) Immunohistochemistry with anti-GFP and anti-human albumin antibodies in DMN-treated NSG mouse liver sections transplanted with mature eHEPOs. The presence of GFP+ and human ALB+ cells demonstrates engraftment of hepatocytes into mouse liver.
Error bars in (A) and (B) denote ±SD of three independent experiments (∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001).
To test the ability of the human mature eHEPOs to engraft as functional hepatocytes in vivo, we first produced eHEPOs from healthy donor iPSCs stably transduced with a GFP expression vector. NSG mice then were treated with dimethylnitrosamine (DMN) for 14 days to induce acute liver damage. Finally, we injected 2 million eHEPO cells intrasplenically to the mice with damaged liver, whereby 32 days after transplantation, engraftment of human cells was demonstrated by immunostaining of GFP and human-specific albumin antibodies separately (Figures 4E and S3). Human ALB+ cells were located around the interlobular veins as well as throughout the parenchyma. These results indicate that the mature, functional hepatocytes from eHEPO cultures have the ability to engraft in the mouse liver.
Disease-Specific eHEPOs
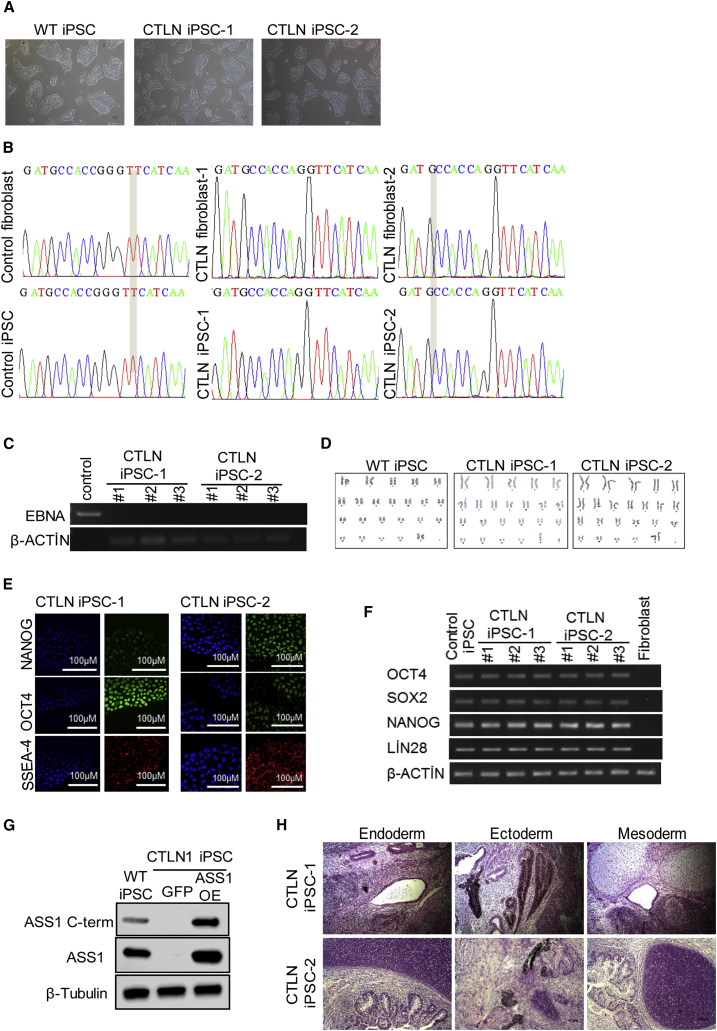
We next investigated whether the eHEPO system can be utilized for disease modeling. To this end, we generated iPSC lines from two patients who presented with neonatal hyperammonemia and were clinically diagnosed with classical citrullinemia type 1 (CTLN1). CTLN1 is an autosomal recessive urea cycle disorder caused by defects in the argininosuccinate synthetase (ASS) enzyme due to mutations in the ASS1 gene (Quinonez and Thoene, 2016). An impairment of ASS1 function can lead to a wide spectrum of phenotypes, from life-threatening neonatal hyperammonemia to a later onset with mild symptoms. Patient-specific iPSCs were grown under feeder-free conditions and displayed typical PSC morphology (Figure 5A). PCR amplification and sequencing of all coding exons of ASS1 indicated that both the patients' fibroblasts and iPSCs harbored homozygous G390R mutations in exon 15, which is one of the most common mutations in classical citrullinemia (Figures S4 and 5B) (Kose et al., 2017, Quinonez and Thoene, 2016). Established iPSC lines were devoid of episomal vector sequences as shown by genomic DNA PCR (Figure 5C). One iPSC clone from each patient was further analyzed by chromosomal G-banding and was confirmed to have a normal karyotype (Figure 5D). CTLN-iPSCs were positive for pluripotency markers OCT4, NANOG, and SSEA-4 by immunofluorescence (Figure 5E). RT-PCR analyses indicated a high expression of OCT4, SOX2, NANOG, and LIN28 mRNA in patient-derived iPSCs, but not in the original dermal fibroblasts (Figure 5F). As expected, ASS1 protein expression was detected in the healthy donor-derived iPSCs but was undetectable in patient-specific iPSCs (Figure 5G). Finally, both of the CTLN-iPSC lines formed well-differentiated teratomas containing cells derived from all three germ layers (Figure 5H). Taken together, these data confirm the pluripotency of citrullinemia patient-derived iPSCs.
Figure 5.
Derivation and Characterization of Citrullinemia-Specific hiPSCs
(A) Morphology of WT iPSC, CTLN iPSC-1, and CTLN iPSC-2.
(B) Sequence analysis of ASS1 exon15 mutations in healthy and patient-derived fibroblasts and the iPSCs.
(C) PCR-based integration analysis of the episomal reprogramming vectors.
(D) Karyotype analysis of WT iPSC, CTLN iPSC-1, and CTLN iPSC-2.
(E) Immunofluorescence images of iPSCs stained for NANOG, OCT4, and SSEA-4. Cell nuclei were counterstained with Hoechst. Scale bars, 100 μm.
(F) mRNA expression levels of pluripotency-related genes.
(G) Western blot for ASS1 using two independent antibodies in WT iPSCs, CTLN1 iPSC-overexpressing GFP, or ASS1 cDNA (OE).
(H) Teratoma formation of CTLN iPSC-1 and CTLN iPSC-2 in SCID mice. Scale bars, 100 μm.
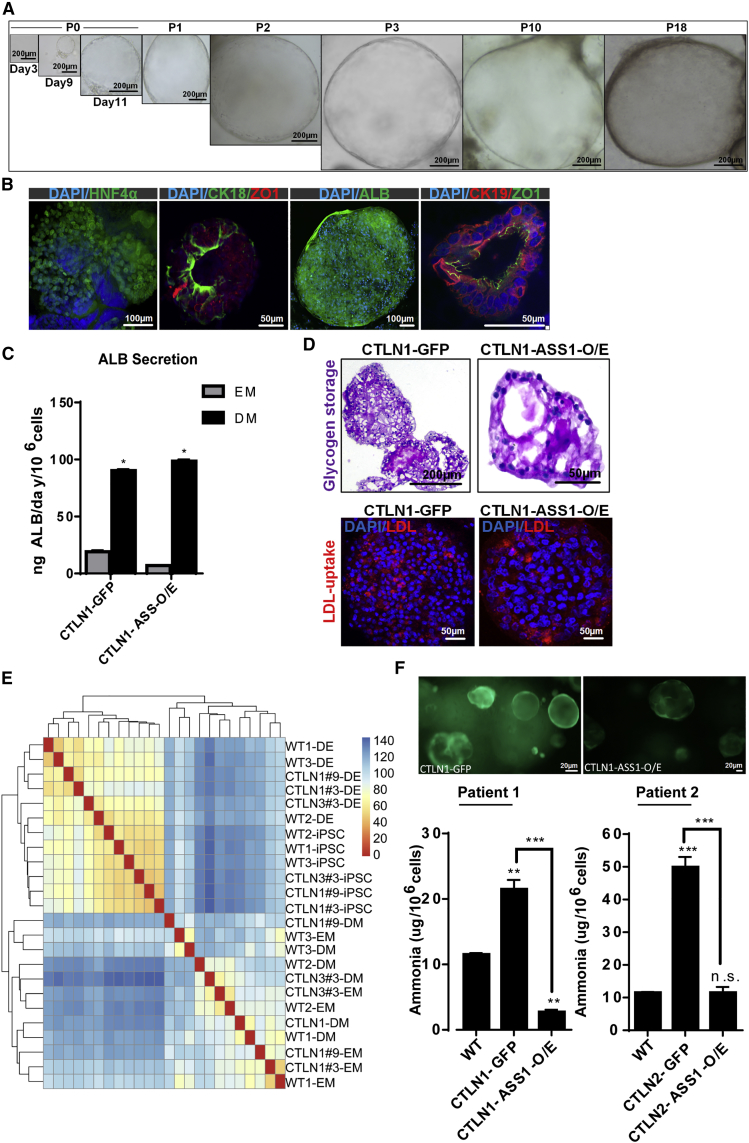
Following the protocol described above, we successfully generated CTLN organoids that could be passaged for more than 6 months in culture (Figure 6A). The comparison of CTLN and the wild-type organoids with respect to the internal structural organization revealed that both have similar structure resembling typically spherical shape(s) containing a series of lumen structures (Figure S5A). They have single-layer and/or pseudostratified epithelium as well as CK19+ cells lining mainly lumens (Figure S5B). To analyze the impact of ASS1 mutations in eHEPOs, we cloned full-length ASS1 cDNA into a GFP-containing lentiviral vector and transduced it to patient-derived iPSCs. In parallel, patient-derived iPSCs were transduced by an empty GFP vector to produce CTLN-GFP organoids as control. For phenotypic characterizations of eHEPO clones, immunofluorescence staining of HNF4α, ZO-1, ALB, CK18, and CK19 demonstrated a similar pattern/structure between healthy donor-derived organoids and CTLN organoids (Figure 6B).
Figure 6.
Endoderm-Derived Hepatic Organoids for Modeling Citrullinemia
(A) Bright-field image of citrullinemia patient-derived hepatic organoid cultures.
(B) Confocal images of CTLN organoids stained for HNF4α, CK18, ZO-1, CK19, and ALB. Nuclei were stained with DAPI. In the dataset for CTLN organoids, HNF4α and ALB were stained in the whole-mount organoids. The others were stained from frozen sections.
(C) In vitro functionality of CTLN-GFP and CTLN-ASS-O/E organoids. Albumin secretion of CTLN1-GFP and CTLN1-ASS-O/E organoids at p10 cultured in EM and DM conditions measured by ELISA. Error bars denote ±SD of three independent experiments.
(D) Glycogen storage function of CTLN1-GFP and CTLN1-ASS-O/E organoids at p10 in DM condition by PAS staining. LDL uptake of CTLN1-GFP and CTLN1-ASS-O/E organoids at p10 in DM condition.
(E) Cluster heatmap of WT and CTLN-iPSCs endoderm and organoids.
(F) Ammonia detoxification capacity of healthy donor (WT indicates organoids with wild-type ASS1 gene) and patient-derived mature eHEPOs expressing either GFP (control) or ASS1-O/E as determined by ammonia elimination assay. Data are shown as ±SD of three independent experiments in two different patient-derived eHEPOs given as μg/day/million cells (∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001).
To understand hepatic maturation efficiency of CTLN-GFP and CTLN-ASS1-O/E organoids, we tested liver functions defined for wild-type organoids (from healthy donors). For both eHEPO clones, DM-mediated maturation induced a significant increase in albumin secretion, and albumin levels were comparable with each other (Figure 6C). Furthermore, CTLN-GFP and CTLN-ASS1-O/E eHEPOs had capacity for LDL uptake and glycogen storage in DM conditions (Figure 6D).
RNA sequencing of patient-derived cultures and k-means clustering of Pearson correlation of whole transcriptomes showed clear separation of organoids from less differentiated cell types (Figure 6E). While iPSCs and endodermal cells were similar in molecular identity, they still clustered separately, indicating proper discrimination of cell types (Figure 6E). Most importantly, patient-derived cells were almost indistinguishable from their healthy counterparts. DM cultures of healthy and CTLN patient-derived organoids were almost identical (two genes with FDR < 0.01), indicating that CTLN disease-specific mutation does not affect the differentiation capacity of patient-derived cells.
In patients, ASS1 mutation causes accumulation of ammonia and decreases ureagenesis. We next examined these phenotypes in patient-derived eHEPOs and asked whether the re-expression of wild-type ASS1 can rescue disease-related phenotypes in our hepatic organoid model. The healthy donor-derived organoids had significantly less ammonia compared with CTLN patient organoids while re-expression of wild-type ASS1 in CTLN organoids rescued this defect (Figure 6F). In parallel, we also measured urea levels in media and observed that CTLN1-GFP organoids had lower levels of ureagenesis compared with healthy organoids and, importantly, ASS1 overexpression partially rescued this phenotype (Figure S6). Taken together, these data indicate that hepatic organoids can faithfully recapitulate the urea cycle-related disease phenotype, and restoration of gene function can be carried out in the eHEPO model.
Discussion
Hepatocyte-like cells differentiated from iPSCs have been used to model several liver-related diseases, including α1-antitrypsin deficiency, citrin deficiency, and CTLN1, highlighting the pathophysiology to pursue new medical strategies (Kim et al., 2016, Yoshitoshi-Uebayashi et al., 2017, Yusa et al., 2011). Although these models represent the phenotype of disease in a dish, the major limitation is the lack of protocols that produce fully functional mature hepatocytes (Baxter et al., 2015, Gieseck et al., 2014, Godoy et al., 2015). Additionally, to our knowledge long-term expansion/maintenance of these cells without losing function is very limited, and freeze/thawing of these cells in 2D culture is another major challenge in the field.
The 3D cell-culture models have recently become important for hepatic functional studies because they often induce levels of cell differentiation and tissue organization with proper cell-cell contact not observed in conventional 2D culture systems (Duval et al., 2017, Pampaloni et al., 2007). Studies have shown that the gene expression profiles (Godoy et al., 2015) as well as the responses to drug toxicity (Takayama et al., 2013) in the multicellular spheroid 3D models resemble more closely the in vivo situation.
The recent availability of stem cell-derived organoid systems to provide 3D self-organized tissue models provides a compelling new class of biological model to serve as both tissue and organ proxies (Lancaster and Knoblich, 2014). Organoids recapitulate a large number of biological parameters including the spatial organization of heterogeneous tissue-specific cells, cell-cell interactions, cell-matrix interactions, and certain physiological functions generated by tissue-specific cells within the organoid. Researchers are toiling to refine organoid culture systems to make them more complex, more mature, and more reproducible.
Here, we developed a novel hiPSC-derived 3D organoid culture system for producing functional hepatocytes using EpCAM+ endodermal cells as an intermediate. EpCAM has been identified as a surface marker on human hepatic stem/progenitor cells (Dollé et al., 2014, Schmelzer et al., 2006, Yoon et al., 2011), and freshly isolated EpCAM+ cells from either fetal or postnatal livers have been shown to be expandable in culture and have the ability to form human liver tissue after transplantation into livers of NOD/SCID mice (Schmelzer et al., 2007). Moreover, endoderm-specific EpCAM is a key regulator of licensing hepatic commitment in zebrafish embryos (Lu et al., 2013). EpCAM+ liver stem cells isolated from adult liver can be expanded in long-term organoid culture in a genome-stable manner. Based on these findings, we hypothesized that freshly isolated EpCAM+ progenitor cells from iPSC-derived endoderm may be used as cell sources to generate hepatic organoids in a more simple and reproducible manner, yielding more mature cell types. To test this, we sorted EpCAM+ cells after endoderm induction from iPSCs and used them to make 3D organoids. At the initial step of our protocol, we added R-spo1 as a secreted Wnt pathway agonist to the endoderm induction culture consisting of activin A and Wnt3a to obtain increased numbers of EpCAM+ cells. We showed that only freshly isolated EpCAM+ endodermal cells can readily make 3D organoids within 8–10 days in EM culture conditions while their EpCAM− counterparts cannot do so. Of note, these eHEPOs have been expanded over a longer term in culture (up to passage 48) and still produced albumin after differentiation by Huch's DM. Notably, we showed that organoids that originated from endoderm could be produced in Huch's medium, which was originally defined only for organoids derived from adult stem cells. We also transplanted eHEPOs into NSG mice and showed human-specific ALB+ hepatocytes embedded in mouse liver. Finally, we used this technology to model liver metabolic disease—citrullinemia—and showed the pathophysiological phenotype of disease as evidenced by a decrease in ammonia detoxification capacity. The rescue of disease phenotype via overexpression of the wild type of the ASS1 gene also demonstrates the potential of eHEPOs in the treatment of disease via genetic manipulation.
Recently, Guan et al. developed a model system whereby iPSC lines differentiate into 3D human hepatic organoids, and introduced certain known mutations to model genetic diseases such as Alagille syndrome and tetralogy of Fallot (Guan et al., 2017). In this model, iPSCs were first differentiated to hepatoblasts by a modified Hannan 2D hepatocyte differentiation protocol for 9 days. Cells were then cultured in suspension for an additional 12 days until large and complex structures with limited oxygen and nutrient availability in their centers formed. Since these hepatic organoid (HO1) structures were not suitable for further differentiation, single-cell dissociation was obligatory in this protocol. The dissociated cells could be grown further and differentiated toward hepatic organoids (HO2) for an additional 30 days while maintaining their ability to form organoids with luminal structures over a 4-month period. In our protocol, eHEPOs expressed CK18 (hepatocyte-like cell marker) and CK19 (bile duct-like cell) as well as bipotent markers such as AFP and HNF4α at an early time point starting at day 10. However, in Guan's protocol, only parenchymal type HO1, the minor type, expressed mature hepatocyte markers such as A1AT and albumin after day 50. eHEPOs offer the possibility to obtain more clonal parenchymal types from hepatic organoids in an efficient way. In addition, the long duration of organoid formation from earlier 3D models may render them inconvenient in comparison with our robust eHEPO model to procure functional hepatocytes from hiPSCs. Furthermore, our extensive transcriptomic analyses further validate the fidelity of differentiation from eHEPOs at various steps. We anticipate that generation of eHEPOs from a diverse set of genetic metabolic and chronic diseases will significantly accelerate the development of new therapeutic strategies against such diseases.
Experimental Procedures
Derivation of Human Fibroblasts and iPSC Generation
After informed consent was obtained from citrullinemia patients and healthy donors, skin-punch biopsies were taken according to a protocol approved by the relevant Institutional Review Boards of Dokuz Eylul University and Koç University. Human primary fibroblast cultures and iPSCs were generated and characterized as described in Supplemental Information.
Mutation Analysis of iPSCs
The CTLN1 patients whose fibroblasts were reprogrammed carry homozygous ASS-p∼G390R (c.1168G>A) mutations in the ASS1 gene. Genomic DNAs were amplified by PCR using the following primers: Exon15-Fwd, CAG TCC TCC CTT CAA GCA GA; Exon15-Rev, GCA GTC AAG GTC GCA TCA AA. Sanger sequencing was performed, which confirmed the presence of homozygous single-nucleotide mutation that leads to ASS-p∼G390R (c.1168G>A).
In Vitro Differentiation of Human iPSCs into Definitive Endoderm
When iPSCs had attained a confluence of 70%, the mouse embryonic fibroblast-conditioned medium was replaced with Roswell Park Memorial Institute/B27 with 100 ng/mL activin A (PeproTech), 50 ng/mL Wnt3a (R&D Systems), and 5 ng/mL R-spo1 (R&D Systems) for 5 days.
Organoid Culture of EpCAM+ Hepatic Progenitors
After 4 days of 2D monolayer induction of definitive endoderm, cells were harvested and trypsinized by TrypLE (Gibco) for 3–4 min at 37°C. The dissociated cells were washed with PBS and stained with anti-human CD326 (EpCAM) (Milteny) antibody and sorted on a BD FACSARIA III sorter. EpCAM+ cells were mixed with Matrigel (BD Biosciences) and seeded onto 24-well plates at ratios of 3,000, 5,000, or 10,000 cells per well. Non-attachment plates were used in all procedures (Sarsted). After Matrigel had solidified, culture medium was added. Culture medium termed EM comprised AdDMEM/F12 (Invitrogen) supplemented with 1% B27 (Gibco), 1.25 mM N-acetylcysteine (Sigma), 10 nM gastrin (Sigma), and the growth factors: 50 ng/mL epidermal growth factor (EGF) (Peprotech), 10% R-spo1 conditioned medium (home-made), 100 ng/mL fibroblast growth factor 10 (FGF10) (Peprotech), 25 ng/mL hepatocyte growth factor (HGF) (Peprotech), 10 mM nicotinamide (Sigma), 5 μM A83.01 (Tocris), and 10 μM forskolin (Tocris). Only for the first 3 days, the medium was supplemented with 5% Noggin and 30% Wnt conditioned medium (both prepared as described by Barker et al., 2010), and 10 μM Y27632 (Sigma-Aldrich). After 10–11 days, organoids were removed from the Matrigel and mechanically dissociated into small fragments before transferring to fresh Matrigel-containing dishes. In each passage, split ratio was 1:4 to 1:8, which was performed in once every 7–10 days for 9–12 months.
Hepatocyte Differentiation and In Vitro Functional Studies
To induce differentiation, we cultured organoids for 3–5 days with EM in fresh Matrigel as described above, supplemented with bone morphogenetic protein 7 (BMP7) (25 ng/mL). Medium was then replaced with DM, which comprised AdDMEM/F12 medium supplemented with 1% B27 and containing EGF (50 ng/mL), gastrin (10 nM, Sigma), HGF (25 ng/mL), FGF19 (100 ng/mL), A8301 (500 nM), DAPT (10 μM), BMP7 (25 ng/mL), and dexamethasone (30 μM). This medium was refreshed every 3 days for 10 days.
Transplantation
Immunodeficient NSG mice (NOD.Cg-Prkdcscid Il2rgtm1WjI/SzJ) were purchased from Jackson Laboratories (Bar Harbor, ME, USA). Animals were housed and maintained under specific pathogen-free conditions in accordance with institutional guidelines under approved protocols in IBG-Vivarium Mouse Facility.
Six- to 8-week-old NSG mice received DMN (7.5 mg/kg, Sigma; 1.0% dissolved in saline, intraperitoneally) for 3 consecutive days per week for 2 weeks. After a 2-week treatment, 1 day after the final DMN injection the mice were anesthetized (ketamine 100 mg/kg, xylazine 8 mg/kg), and 2 × 106 iPSC-derived organoid cells transduced by a GFP containing vector in 30 μL of cell suspension were transplanted intrasplenically to DMN-treated mice. Mice were sacrificed, and liver samples were obtained 10 days and 32 days after transplantation.
Messenger RNA Sequencing
Messenger RNA sequencing was performed using the CEL-Seq2 method (Basak et al., 2017, Hashimshony et al., 2016). In brief, total RNA was extracted with an RNeasy mini kit (Qiagen). Ten nanograms of total RNA per sample was precipitated with 2 μg of GlycoBlue (Ambion) overnight at −80°C. The pellet was dissolved in 4-UMI barcode containing primers and incubated at 70°C for 2 min. Deoxynucleotide triphosphates (Promega) were added and the mRNA was reverse transcribed using the SuperScript II reverse transcription kit (Thermo Fisher Scientific). The second strand was synthesized using the DNA ligase I (Thermo Fisher Scientific). Samples were pooled for in vitro transcription through the T7 binding sequence (Ambion Megascript kit). Amplified RNA was converted to cDNA using the SuperScript II reverse transcription kit (Thermo Fisher Scientific). DNA libraries were generated using Phusion Polymerase (NEB) and the Illumina TruSeq small adapter primers (Hashimshony et al., 2016). Ampure XP beads (Beckman Coulter) were used for nucleic acid clean-up of double stranded DNA, amplified RNA, and the final DNA library. Libraries were sequenced on an Illumina HiSeq4000 using 150-bp paired-end sequencing at GenomeScan. All the samples were sequenced in a single run to an average depth of ∼10 million reads per sample. Reads were mapped to the latest human coding transcriptome and normalized as described elsewhere (Basak et al., 2017). The data were processed using the DESeq2 package (Love et al., 2014). For heatmaps, the values of the variance stabilizing transformation of the data are displayed (see DESeq2 manual). For the cluster heatmap, k-means clustering of the Pearson correlation of the whole transcriptome of the samples is displayed.
For additional and detailed experimental information about in vitro functional assays (periodic acid-Schiff staining, LDL uptake, measurement of CYP3A4 activity, RNA isolation and real-time PCR, ammonia elimination, determination of albumin secretion, immunostaining, transmission electron microscopy, and flow-cytometry analysis), please refer to Supplemental Information.
Data Analysis
Statistical analysis was carried out using GraphPad Prism (GraphPad Software, CA, USA). Differences between groups were considered significant at ∗p < 0.05, ∗∗p < 0.001, and ∗∗∗p < 0.0001.
Author Contributions
E. Erdal, T.T.O., and S.A. conceived the presented idea, and designed and analyzed experiments. G.G.S. performed the experiments for the production and characterization of iPSCs. B.S. and B.O. assisted in iPSC experiments. S.A. designed and performed all experiments related to 3D liver organoid culture and disease modeling including FACS, immunostaining, confocal imaging, and in vitro assays. A.B. and N.E. performed organoid transplantation in the DMN induced animal model and all in vivo assays. S.A. and K.K. performed in vitro functional assays and analyzed the data. E. Enustun prepared the ALB promoter GFP reporter plasmid. S.A., K.K., and N.E. also performed all revision experiments. E.O. prepared the ASS1 lentiviral expression vector. S.A., T.T.O., and E. Erdal together with O.B. designed RNA-sequencing experiments. O.B. supervised next-generation sequencing, S.A. prepared RNAs. O.B. and K.S. performed bioinformatics analyses. N.A. provided human donor biopsies and patient material. S.A. cultured biopsy material and performed primary fibroblast culture. S.A. and K.K. performed patient phenotype rescue assays. G.G.S., S.A., and T.T.O. prepared all figures. E. Erdal, T.T.O., S.A., G.G.S., and O.B wrote the paper. All authors commented on the manuscript.
Acknowledgments
We would like to thank to Prof. Dr. Hans Clevers (Hubrecht Institute/KNAW, Utrecht, the Netherlands) for hosting S.A. in the Hubrecht Institute (Utrecht, the Netherlands) and for the kind gift of L-Wnt3a and Noggin cell lines. We are also immensely grateful to the members of Clevers Lab for sharing their experience about organoid culture. Finally, we would gratefully like to thank Melek Ucuncu and Muge Ozkan for their help in interpreting the imaging and flow-cytometry data, respectively. This research was supported by TUBİTAK (The Scientific and Technological Research Council of Turkey) via projects SBAG-115S465 and SBAG-213S182 and by an EMBO installation grant no. 2543 (T.T.O.). K.S. was supported by a TUBITAK BIDEB Scholarship. The authors gratefully acknowledge use of the services and facilities of the Koç University Research Center for Translational Medicine (KUTTAM) and of the Izmir Biomedicine and Genome Center, both funded by the Republic of Turkey Ministry of Development. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Ministry of Development.
Published: September 12, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.08.007.
Contributor Information
Tamer Tevfik Önder, Email: tonder@ku.edu.tr.
Esra Erdal, Email: esra.erdal@ibg.edu.tr.
Accession Numbers
The datasets generated in the current study are available in the GEO repository, accession number GEO: GSE120145.
Supplemental Information
References
- Barker N., Huch M., Kujala P., van de Wetering M., Snippert H.J., van Es J.H., Sato T., Stange D.E., Begthel H., van den Born M. Lgr5 +ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Basak O., Beumer J., Wiebrands K., Seno H., van Oudenaarden A., Clevers H. Induced quiescence of Lgr5+ stem cells in intestinal organoids enables differentiation of hormone-producing enteroendocrine cells. Cell Stem Cell. 2017;20:177–190.e4. doi: 10.1016/j.stem.2016.11.001. [DOI] [PubMed] [Google Scholar]
- Baxter M., Withey S., Harrison S., Segeritz C.-P., Zhang F., Atkinson-Dell R., Rowe C., Gerrard D.T., Sison-Young R., Jenkins R. Phenotypic and functional analyses show stem cell-derived hepatocyte-like cells better mimic fetal rather than adult hepatocytes. J. Hepatol. 2015;62:581–589. doi: 10.1016/j.jhep.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broutier L., Andersson-Rolf A., Hindley C.J., Boj S.F., Clevers H., Koo B.K., Huch M. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat. Protoc. 2016;11:1724–1743. doi: 10.1038/nprot.2016.097. [DOI] [PubMed] [Google Scholar]
- Carmon K.S., Gong X., Lin Q., Thomas A., Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/β-catenin signaling. Proc. Natl. Acad. Sci. U S A. 2011;108:11452–11457. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.F., Tseng C.Y., Wang H.W., Kuo H.C., Yang V.W., Lee O.K. Rapid generation of mature hepatocyte-like cells from human induced pluripotent stem cells by an efficient three-step protocol. Hepatology. 2012;55:1193–1203. doi: 10.1002/hep.24790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lau W., Barker N., Low T.Y., Koo B.-K., Li V.S., Teunissen H., Kujala P., Haegebarth A., Peters P.J., van de Wetering M. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- Dollé L., Theise N.D., Schmelzer E., Boulter L., Gires O., van Grunsven L.A. EpCAM and the biology of hepatic stem/progenitor cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2014;308:G233–G250. doi: 10.1152/ajpgi.00069.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval K., Grover H., Han L.-H., Mou Y., Pegoraro A.F., Fredberg J., Chen Z. Modeling physiological events in 2D vs. 3D cell culture. Physiology. 2017;32:266–277. doi: 10.1152/physiol.00036.2016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Fidan K., Kavaklıoğlu G., Ebrahimi A., Özlü C., Ay N.Z., Ruacan A., Gül A., Önder T.T. Generation of integration-free induced pluripotent stem cells from a patient with Familial Mediterranean Fever (FMF) Stem Cell Res. 2015;15:694–696. doi: 10.1016/j.scr.2015.10.018. [DOI] [PubMed] [Google Scholar]
- Gieseck R.L., III, Hannan N.R., Bort R., Hanley N.A., Drake R.A., Cameron G.W., Wynn T.A., Vallier L. Maturation of induced pluripotent stem cell derived hepatocytes by 3D-culture. PLoS One. 2014;9:e86372. doi: 10.1371/journal.pone.0086372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy P., Schmidt-Heck W., Natarajan K., Lucendo-Villarin B., Szkolnicka D., Asplund A., Björquist P., Widera A., Stöber R., Campos G. Gene networks and transcription factor motifs defining the differentiation of stem cells into hepatocyte-like cells. J. Hepatol. 2015;63:934–942. doi: 10.1016/j.jhep.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Xu D., Garfin P.M., Ehmer U., Hurwitz M., Enns G., Michie S., Wu M., Zheng M., Nishimura T. Human hepatic organoids for the analysis of human genetic diseases. JCI Insight. 2017;7:e94954. doi: 10.1172/jci.insight.94954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimshony T., Senderovich N., Avital G., Klochendler A., de Leeuw Y., Anavy L., Gennert D., Li S., Livak K.J., Rozenblatt-Rosen O. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 2016;17:77. doi: 10.1186/s13059-016-0938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M., Dolle L. The plastic cellular states of liver cells: are EpCAM and Lgr5 fit for purpose? Hepatology. 2016;64:652–662. doi: 10.1002/hep.28469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M., Dorrell C., Boj S.F., van Es J.H., Li V.S., van de Wetering M., Sato T., Hamer K., Sasaki N., Finegold M.J. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M., Gehart H., van Boxtel R., Hamer K., Blokzijl F., Verstegen M.M., Ellis E., van Wenum M., Fuchs S.A., de Ligt J. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160:299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Choi J.-Y., Lee S.-H., Lee B.-H., Yoo H.-W., Han Y.-M. Malfunction in mitochondrial β-oxidation contributes to lipid accumulation in hepatocyte-like cells derived from citrin deficiency-induced pluripotent stem cells. Stem Cells Dev. 2016;25:636–647. doi: 10.1089/scd.2015.0342. [DOI] [PubMed] [Google Scholar]
- Kose E., Unal O., Bulbul S., Gunduz M., Häberle J., Arslan N. Identification of three novel mutations in fourteen patients with citrullinemia type 1. Clin. Biochem. 2017;50:686–689. doi: 10.1016/j.clinbiochem.2017.01.011. [DOI] [PubMed] [Google Scholar]
- Lancaster M.A., Knoblich J.A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- Liu H., Kim Y., Sharkis S., Marchionni L., Jang Y.-Y. In vivo liver regeneration potential of human induced pluripotent stem cells from diverse origins. Sci. Transl. Med. 2011;3:82ra39. doi: 10.1126/scitranslmed.3002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Ma J., Yang Y., Shi W., Luo L. EpCAM is an endoderm-specific Wnt derepressor that licenses hepatic development. Dev. Cell. 2013;24:543–553. doi: 10.1016/j.devcel.2013.01.021. [DOI] [PubMed] [Google Scholar]
- Luo Y., Lou C., Zhang S., Zhu Z., Xing Q., Wang P., Liu T., Liu H., Li C., Shi W. Three-dimensional hydrogel culture conditions promote the differentiation of human induced pluripotent stem cells into hepatocytes. Cytotherapy. 2018;20:95–107. doi: 10.1016/j.jcyt.2017.08.008. [DOI] [PubMed] [Google Scholar]
- Pampaloni F., Reynaud E.G., Stelzer E.H. The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 2007;8:839. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- Planas-Paz L., Orsini V., Boulter L., Calabrese D., Pikiolek M., Nigsch F., Xie Y., Roma G., Donovan A., Marti P. The RSPO-LGR4/5-ZNRF3/RNF43 module controls liver zonation and size. Nat. Cell Biol. 2016;18:467. doi: 10.1038/ncb3337. [DOI] [PubMed] [Google Scholar]
- Quinonez S.C., Thoene J.G. Citrullinemia type I. In: Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Stephens K., Amemiya A., editors. GeneReviews® [Internet] University of Washington; 2016. pp. 1993–2018. [Google Scholar]
- Schmelzer E., Wauthier E., Reid L.M. The phenotypes of pluripotent human hepatic progenitors. Stem Cells. 2006;24:1852–1858. doi: 10.1634/stemcells.2006-0036. [DOI] [PubMed] [Google Scholar]
- Schmelzer E., Zhang L., Bruce A., Wauthier E., Ludlow J., Yao H.L., Moss N., Melhem A., McClelland R., Turner W. Human hepatic stem cells from fetal and postnatal donors. J. Exp. Med. 2007;204:1973–1987. doi: 10.1084/jem.20061603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K., Noto F.K., Nagaoka M., Li J., Battle M.A., Duris C., North P.E., Dalton S., Duncan S.A. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K., Kawabata K., Nagamoto Y., Kishimoto K., Tashiro K., Sakurai F., Tachibana M., Kanda K., Hayakawa T., Furue M.K. 3D spheroid culture of hESC/hiPSC-derived hepatocyte-like cells for drug toxicity testing. Biomaterials. 2013;34:1781–1789. doi: 10.1016/j.biomaterials.2012.11.029. [DOI] [PubMed] [Google Scholar]
- Takebe T., Sekine K., Enomura M., Koike H., Kimura M., Ogaeri T., Zhang R.-R., Ueno Y., Zheng Y.-W., Koike N. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- Takebe T., Sekine K., Kimura M., Yoshizawa E., Ayano S., Koido M., Funayama S., Nakanishi N., Hisai T., Kobayashi T. Massive and reproducible production of liver buds entirely from human pluripotent stem cells. Cell Rep. 2017;21:2661–2670. doi: 10.1016/j.celrep.2017.11.005. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Okabe M., Suzuki K., Kamiya Y., Tsukahara Y., Saito S., Miyajima A. Mouse hepatoblasts at distinct developmental stages are characterized by expression of EpCAM and DLK1: drastic change of EpCAM expression during liver development. Mech. Dev. 2009;126:665–676. doi: 10.1016/j.mod.2009.06.939. [DOI] [PubMed] [Google Scholar]
- Yoon S.M., Gerasimidou D., Kuwahara R., Hytiroglou P., Yoo J.E., Park Y.N., Theise N.D. Epithelial cell adhesion molecule (EpCAM) marks hepatocytes newly derived from stem/progenitor cells in humans. Hepatology. 2011;53:964–973. doi: 10.1002/hep.24122. [DOI] [PubMed] [Google Scholar]
- Yoshitoshi-Uebayashi E.Y., Toyoda T., Yasuda K., Kotaka M., Nomoto K., Okita K., Yasuchika K., Okamoto S., Takubo N., Nishikubo T. Modelling urea-cycle disorder citrullinemia type 1 with disease-specific iPSCs. Biochem. Biophys. Res. Commun. 2017;486:613–619. doi: 10.1016/j.bbrc.2017.03.037. [DOI] [PubMed] [Google Scholar]
- Yusa K., Rashid S.T., Strick-Marchand H., Varela I., Liu P.-Q., Paschon D.E., Miranda E., Ordóñez A., Hannan N.R., Rouhani F.J. Targeted gene correction of α 1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478:391. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.