Abstract
Background
Interphase fluorescence in situ hybridization (FISH) of bone marrow cells has been confirmed to be a direct and valid method to assess the v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog (MYCN) amplification in patients with bone marrow metastatic neuroblastoma. MYCN amplification alone, however, is insufficient for pretreatment risk stratification. Chromosome band 11q23 deletion has recently been included in the risk stratification of neuroblastoma. In the present study, we aimed to evaluate the biological characteristics and prognostic impact of 11q23 deletion and MYCN amplification in patients with bone marrow metastatic neuroblastoma.
Methods
We analyzed the MYCN and 11q23 statuses of 101 patients with bone marrow metastatic neuroblastoma using interphase FISH of bone marrow cells. We specifically compared the biological characteristics and prognostic impact of both aberrations.
Results
MYCN amplification and 11q23 deletion were seen in 12 (11.9%) and 40 (39.6%) patients. The two markers were mutually exclusive. MYCN amplification occurred mainly in patients with high lactate dehydrogenase (LDH) and high neuron-specific enolase (NSE) levels (both P < 0.001), and MYCN-amplified patients had more events (tumor relapse, progression, or death) than MYCN-normal patients (P = 0.004). 11q23 deletion was associated only with age (P = 0.001). Patients with MYCN amplification had poorer outcomes than those with normal MYCN (3-year event-free survival [EFS] rate: 8.3 ± 8.0% vs. 43.8 ± 8.5%, P < 0.001; 3-year overall survival [OS] rate: 10.4 ± 9.7% vs. 63.5% ± 5.7%, P < 0.001). 11q23 deletion reflected a poor prognosis only for patients with normal MYCN (3-year EFS rate: 34.3 ± 9.5% vs. 53.4 ± 10.3%, P = 0.037; 3-year OS rate: 42.9 ± 10.4% vs. 75.9 ± 6.1%, P = 0.048). Those with both MYCN amplification and 11q23 deletion had the worst outcome (P < 0.001).
Conclusions
Chromosome band 11q23 deletion predicts poor prognosis only in bone marrow metastatic neuroblastoma patients without MYCN amplification. Combined assessment of the two markers was much superior to single-marker assessment in recognizing the patients at a high risk of disease progression.
Keywords: Neuroblastoma, MYCN amplification, 11q23 deletion, Fluorescence in situ hybridization, Bone marrow metastasis, Event-free survival, Overall survival, Neuron-specific enolase, Lactate dehydrogenase
Background
Neuroblastoma is the most common extracranial malignancy in children with remarkable heterogeneity in clinical behaviors. Neuroblastoma accounts for 10%–15% of all pediatric cancer-related deaths [1, 2]. Although some of the tumors can regress spontaneously, most patients’ diseasees progress aggressively despite multimodality treatment, which may include chemotherapy, radiotherapy, surgical resection, hematopoietic stem cell transplantation, and immunotherapy [3, 4]. In the clinic, half of neuroblastoma patients present with metastasis at diagnosis, and less than 50% survive 5 years [5, 6].
Recently, many prognostic variables have been used for risk stratification to predict the survival of patients with neuroblastoma. Among them, genetic features such as the status of the v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog (MYCN) gene [7], 11q23 allele status [8], and tumor ploidy [9, 10] are the most significant and clinically relevant factors. However, for pediatric patients with metastatic neuroblastoma who need chemotherapy prior to surgery, timely identification of genetic aberrations is not possible because of the difficulty in obtaining tumor biopsies. In addition, the results of genetic aberration identification may be inaccurate after the patients have received chemotherapy, leading to insufficient dosage and period of treatment [11, 12].
For more than two decades, a number of genetic aberrations closely associated with outcome have been identified in neuroblastoma; for the MYCN gene, these include 17q gain, loss of 3p, and 11q deletion [13]. Among them, 11q deletion is the most frequent, occurring in approximately 40% of primary neuroblastomas [14], and is associated with poor prognosis [15, 16]. However, the clinical significance of 11q deletion is unclear in neuroblastoma metastatic to bone marrow. What’s more, 11q deletion is not routinely examined when genome-wide methods are used to evaluate genetic aberrations [17]. 11q23 is the most commonly deleted 11q region, and interphase fluorescence in situ hybridization (FISH) has been previously confirmed as a reliable method for detecting MYCN status in bone marrow metastases of neuroblastoma [18, 19]. Therefore, in this study, we used FISH to simultaneously detect MYCN and 11q23 deletion in bone marrow cells in order to evaluate the biological characteristics and survival associated with these two markers.
Materials and methods
Patients and treatment protocols
Consecutive children with neuroblastoma newly diagnosed at the Hematology Oncology Center, Beijing Children’s Hospital, Capital Medical University between January 2015 and December 2016 were selected. All patients were diagnosed according to the International Neuroblastoma Staging System [20]. The criteria for patient inclusion were ≥ 20% neuroblastoma cells in the bone marrow samples and treatment according to the BCH-NB-2007 protocol (based on the Hong Kong neuroblastoma protocol) [21].
The BCH-NB-2007 protocol for newly diagnosed high-risk neuroblastoma was comprised of 7 cycles of intensive chemotherapy: CAV regimen [cyclophosphamide (70 mg/kg) on days 1–2, adriamycin (25 mg/m2) on days 1–3, and vincristine (0.033 mg/kg)] on days 1–3 for cycles 1, 2, 4, and 6; CVP regimen [cisplatin (50 mg/m2) on days 1–4 and VP16 (200 mg/m2) on days 1–3] for cycles 3, 5, and 7. Primary tumor resection was performed after 4 cycles of chemotherapy; peripheral blood stem cells (PBSCs) were then harvested for possible autologous bone marrow transplantation. Autologous PBSC transplantation (PBSCT) was performed after 7 cycles of chemotherapy. The patients received local irradiation (20–25 Gy) at the site of primary tumor as indicated [22]. At 4–5 weeks after autologous PBSCT, they received maintenance chemotherapy with cis-retinoic acid (160 mg/m2) for 14 days alternating with 14 days off for 9–12 months.
The treatment protocol for low-/intermediate-risk neuroblastoma was comprised of 2–3 cycles of CBVP regimen [carboplatin (200 mg/m2) and etoposide (150 mg/m2)] and CADO regimen [vincristine (1.5 mg/m2) and adriamycin (25 mg/m2)] used alternately. Primary tumor resection was usually performed after chemotherapy. If surgery was performed before chemotherapy, 2–3 cycles of chemotherapy for low-risk patients and 4–6 cycles for intermediate-risk patients were needed.
All patients were treated and evaluated according to the BCH-NB-2007 protocol. After the treatment, the patients were followed-up every 3 months mainly by outpatient follow-up visits (bone marrow examination and computed tomography or magnetic resonance imaging) and telephone interview. The final date for data collection was August 31, 2018. This study was approved by the Beijing Children’s Hospital Institutional Ethics Committee. Informed consent was obtained from the parents or their guardians in accordance with the Declaration of Helsinki.
Bone marrow sample processing
Bone marrow samples of all patients obtained by bone marrow puncture were collected in heparin-coated tubes at diagnosis. The sample was immediately shaken gently after collection to avoid bone marrow agglutination. Then, the cells were cultured in 8 mL RPMI-1640 (Hyclone, Beijing, China) and 2 mL fetal bovine serum medium (Invitrogen, Carlsbad, CA, USA) for 24 h. The cell density was (1 − 3) × 106/mL, and the total amount of cells was about (1 − 3) × 107. Colchicine was added at 2 h before termination of the culture. The cells were then treated with 0.075 mol/L KCl for 30 min and fixed twice in a 3:1 mixture of methanol:acetic acid. Finally, the cell suspension was stored in a fresh fixative solution at 4 °C. Bone marrow biopsies were prepared on wax blocks for morphological examination.
FISH analysis of bone marrow cells
The FISH technique was performed as previously reported [17]. The statuses of MYCN and 11q were determined with DNA probes from Vysis [N-MYC(2p24)/CEP2(2p11.1-q11.1) Dual Color Probe and LSI MLL Dual Color, Break Apart Rearrangement Probe] (cat. No. 7J72-01 and 8L57-20, Abbott Laboratories, Abbott Park, IL, USA). A Leica DM6000B microscope (Leica Microsystems GmbH, Wetzlar, Germany) was used to take fluorescence images. According to the recommendations of the European Neuroblastoma Quality Assessment group [23, 24], MYCN amplification was defined as a > fourfold increase of MYCN signals in relation to the number of chromosome 2, and 11q23 deletion was defined as only one fusion signal displayed.
Karyotype analysis of bone marrow cells
Chromosome karyotype analysis of bone marrow cells was performed on G-banded preparations. At least 20 metaphases were analyzed in detail. The karyotype results were interpreted according to the International System for Human Cytogenomic Nomenclature guidelines (2016) [25]. The clones containing at least 2 cells with the same additional chromosome or structural abnormality or at least 3 cells with the same missing chromosome were defined as abnormal clones.
Morphologic analysis and diagnostic biomarker detection
Microscopic examination of bone marrow biopsies to determine the presence of neuroblastoma cells was done by at least 2 independent laboratory experts. Serum tumor markers such as lactate dehydrogenase (LDH) and neuron specific enolase (NSE) levels were detected by radioimmunoassay and full-automatic biochemical analysis at the time of diagnosis in all patients.
Statistical analysis
All statistical analyses were performed using the SPSS software, version 16.0 (SPSS Inc., Chicago, IL, USA). Comparisons of patients with or without 11q23 deletion and MYCN amplification and the association of clinical characteristics were evaluated using the χ2 test. Event-free survival (EFS) was defined from the date of diagnosis to the date of one of the following events: progression, relapse, death, or last contact with patients in continuous complete remission (CR). Overall survival (OS) was defined from the date of diagnosis to the date of death due to any reason or the last contact with patients in continuous CR. Kaplan–Meier survival analysis and log-rank test were used to determine the differences in EFS and OS in all patients by MYCN and 11q23 statuses. Multivariate backward Cox regression analysis model was built which included only those variables reaching a P value < 0.05 in the respective univariate analysis. All tests with a P < 0.05 in two-sided distributions were considered statistically significant.
Results
Clinical characteristics of 101 patients with bone marrow metastatic neuroblastoma
Among the 161 patients with newly diagnosed neuroblastoma, 45 had no bone marrow metastasis, 13 gave up treatment after diagnosis, and 2 were lost to follow-up. A total of 101 patients with stage IV neuroblastoma were thereby analyzed, including 57 males and 44 females in a ratio of 1.3:1. The patients were aged 7–105 months (median, 36 months), and 85 (84.2%) were older than 18 months. The median white blood cell (WBC) count was 5.6 × 109/L (range 1.7–42.5 × 109/L) with 60 (59.4%) patients having normal WBC. Only 8 (7.9%) patients had hemoglobin > 120 g/L. The rest had different degrees of anemia, but most cases were mild. The median hemoglobin level was 94 g/L (range 58–135 g/L). The amount of platelets ranged from 37 × 109/L to 622 × 109/L (median, 275 × 109/L). Almost all patients (92.1%) were grouped as having high-risk neuroblastoma. The primary tumor sites were the abdomen for 88 patients, thorax for 12 patients, and neck for 1 patient. 11q23 deletion was seen in 40 (39.6%) patients, and MYCN amplification in 12 (11.9%). In terms of clinical serum tumor markers, only 22 (21.8%) patients had LDH levels ≥ 1500 IU/L, and 41 (40.6%) had NSE levels ≥ 370 ng/mL.
Each patient received a total of 7 cycles of chemotherapy except for those who died during therapy. During follow-up, 39 (38.6%) patients died, and 13 (12.9%) suffered from tumor relapse or progression; the other 49 (48.5%) had stable disease. Among the patients who died, 11 died of tumor progression during intensive chemotherapy, 24 died of tumor progression during maintenance treatment, and 4 died of tumor recurrence after the completion of chemotherapy. Of note, 4 of the 24 patients who died during maintenance treatment had intracranial metastasis. The main clinical characteristics of the 101 patients are shown in Table 1.
Table 1.
Baseline characteristics of the 101 patients with bone marrow metastatic neuroblastoma
| Characteristic | Total [cases (%)] |
|---|---|
| Age (months) | |
| < 18 | 16 (15.8) |
| ≥ 18 | 85 (84.2) |
| Sex | |
| Male | 57 (56.4) |
| Female | 44 (43.6) |
| WBC (× 109/L) | |
| > 10 | 17 (16.8) |
| 4–10 | 60 (59.4) |
| < 4 | 24 (23.8) |
| Hemoglobin (g/L) | |
| > 120 | 8 (7.9) |
| 90–120 | 59 (58.4) |
| 60–89 | 33 (32.7) |
| < 60 | 1 (1.0) |
| Platelet (× 109/L) | |
| > 300 | 47 (46.5) |
| 100–300 | 48 (47.5) |
| < 100 | 6 (6.0) |
| Risk stratification | |
| Low-risk | 1 (1.0) |
| Intermediate-risk | 7 (6.9) |
| High-risk | 93 (92.1) |
| Primary tumor site | |
| Abdomen | 88 (87.1) |
| Thorax | 12 (11.9) |
| Neck | 1 (1.0) |
| 11q23 deletion | |
| Yes | 40 (39.6) |
| No | 61 (60.4) |
| MYCN amplification | |
| Yes | 12 (11.9) |
| No | 89 (88.1) |
| LDH (IU/L) | |
| < 1500 | 79 (78.2) |
| ≥ 1500 | 22 (21.8) |
| NSE (ng/mL) | |
| < 370 | 60 (59.4) |
| ≥ 370 | 41 (40.6) |
| Event | |
| No event | 49 (48.5) |
| Relapse/progression | 13 (12.9) |
| Death | 39 (38.6) |
WBC white blood cell, MYCN the v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog, LDH lactate dehydrogenase, NSE neuron-specific enolase
Genetic markers of MYCN and 11q23 in bone marrow cells
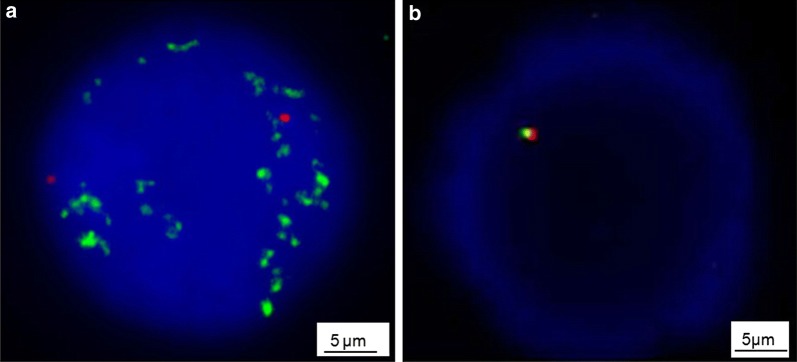
In all patients, interphase FISH detected the MYCN gene and 11q23 status of bone marrow cells. Chromosome karyotype analysis was performed in 55 patients. MYCN amplification was seen in 12 (11.9%) patients, with > 30 gene copies/cell (Fig. 1a). MYCN gain was seen in 21 (20.8%) patients, with 2–9 signal copies. Among the 101 patients, 40 (39.6%) had 11q23 deletion (Fig. 1b). Only 2 (2.0%) patients had both MYCN amplification and 11q23 deletion. MYCN amplification and 11q23 deletion were not associated with higher percentages of tumor cells in bone marrow or more severe hematopoietic suppression (both P > 0.05).
Fig. 1.
Typical fluorescence in situ hybridization (FISH) images of bone marrow cells from patients with bone marrow metastatic neuroblastoma. a The status of v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog (MYCN) amplification was detected using a dual-color probe. Green signals represent the specific probe for MYCN, and red signals stand for centromeric chromosome 2 probes. MYCN signals show more than 10 copies within the nuclei. b The status of 11q23 deletion was also detected using a dual-color probe. Normal cells show a two orange/green fusion signal pattern. In a cell with 11q23 deletion, the pattern is one orange/green fusion signal
Relationship of MYCN and 11q23 statuses with clinicobiological characteristics of patients with bone marrow metastatic neuroblastoma
We observed no association of MYCN amplification with age, sex, or primary tumor site (Table 2). However, the primary tumor site of all patients with MYCN amplification was in the abdomen. We found that MYCN amplification mainly occurred in those with high LDH and NSE levels (both P < 0.001), and more events were observed in patients with MYCN amplification than in those with normal MYCN (P = 0.004).
Table 2.
Clinical characteristics of bone marrow metastatic neuroblastoma patients according to MYCN and 11q23 statuses
| Variable | MYCN status [cases (%)] | P value* | 11q23 status [cases (%)] | P value* | ||
|---|---|---|---|---|---|---|
| Amplified | Normal | Deleted | Normal | |||
| Total | 12 | 89 | 40 | 61 | ||
| Sex | ||||||
| Male | 7 (58.3) | 50 (56.2) | 0.536 | 24 (60.0) | 33 (54.1) | 0.682 |
| Female | 5 (41.7) | 39 (43.8) | 16 (40.0) | 28 (45.9) | ||
| Age (months) | ||||||
| < 18 | 4 (33.3) | 12 (13.5) | 0.095 | 2 (5.0) | 14 (23.0) | 0.024 |
| ≥ 18 | 8 (66.7) | 77 (86.5) | 38 (95.0) | 47 (77.0) | ||
| Primary tumor site | ||||||
| Abdomen | 12 (100.0) | 76 (85.4) | 0.436 | 33 (82.5) | 55 (90.2) | 0.339 |
| Thorax | 0 (0) | 12 (13.5) | 6 (15.0) | 6 (9.8) | ||
| Neck | 0 (0) | 1 (1.1) | 1 (2.5) | 0 (0) | ||
| LDH (IU/L) | ||||||
| < 1500 | 1 (8.3) | 78 (87.6) | <0.001 | 32 (80.0) | 47 (77.0) | 0.808 |
| ≥ 1500 | 11 (91.7) | 11 (12.4) | 8 (20.0) | 14 (23.0) | ||
| NSE (ng/mL) | ||||||
| < 370 | 1 (8.3) | 59 (66.3) | < 0.001 | 21 (52.5) | 39 (63.9) | 0.302 |
| ≥ 370 | 11 (91.7) | 30 (33.7) | 19 (47.5) | 22 (36.1) | ||
| Event | ||||||
| No | 1 (8.3) | 48 (53.9) | 0.004 | 16 (40.0) | 33 (54.1) | 0.118 |
| Yes | 11 (91.7) | 41 (46.1) | 24 (60.0) | 28 (45.9) | ||
MYCN the v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog, LDH lactate dehydrogenase, NSE neuron-specific enolase
* The χ2 test was used for statistical analysis
Unlike the MYCN gene, 11q23 deletion was associated with age (P = 0.024). We found 38 (95.0%) cases of 11q23 deletion occurred in children older than 18 months. The median age at diagnosis was 41 months for patients with 11q23 deletion, but 24.5 months for those with MYCN amplification. Although 24 (60.0%) patients with 11q23 deletion had events occurred, no significant difference was observed between patients with 11q23 deletion and normal 11q23 (P = 0.118).
Impact of MYCN and 11q23 on prognosis of bone marrow metastatic neuroblastoma
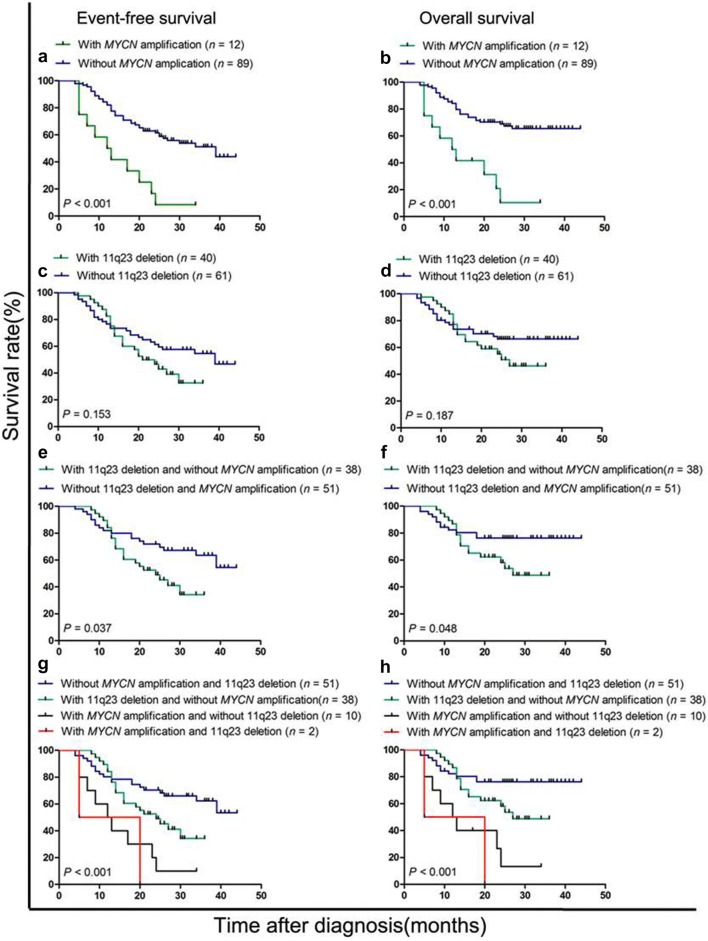
The 3-year EFS and OS rates of the 101 patients were 45.9 ± 5.5% and 59.1 ± 5.2%, with a median follow-up time of 23 months (range, 4–44 months). The estimated 3-year EFS and OS rates of patients with MYCN amplification were obviously lower than those of patients with normal MYCN (8.3 ± 8.0% vs. 43.8 ± 8.5% and 10.4 ± 9.7% vs. 63.5% ± 5.7%, both P < 0.001) (Fig. 2a, b). The difference in survival was not significant between patients with and without 11q23 deletion (Fig. 2c, d). The median EFS of patients with and without MYCN amplification was 12.5 and 25 months, respectively. The median EFS of patients with and without 11q23 deletion was 20.5 and 26 months, respectively.
Fig. 2.
The prognostic significance of MYCN amplification and 11q23 deletion in patients with bone marrow metastatic neuroblastoma. a, b Event-free survival (EFS) and overall survival (OS) of the 101 patients stratified by MYCN gene. c, d EFS and OS of the 101 patients stratified by 11q23 status. e, f EFS and OS of the 89 patients without MYCN amplification stratified by 11q23 status. g, h EFS and OS of the 101 patients stratified by combined assessment of MYCN and 11q23 statuses
To better understand the effect of 11q23 deletion on prognosis of bone marrow metastatic neuroblastoma, we performed further survival analysis of the 89 patients without MYCN amplification. We surprisingly found that 11q23 deletion was a powerful prognostic factor if the influence of the MYCN gene was excluded. The estimated 3-year EFS and OS rates of the 38 patients with 11q23 deletion were significantly lower than those of the 51 patients without 11q23 deletion (34.3 ± 9.5% vs. 53.4 ± 10.3%, P = 0.037; 42.9 ± 10.4% vs. 75.9 ± 6.1%, P = 0.048) (Fig. 2e, f).
We then investigated the prognostic value of the combination of MYCN and 11q23. The 101 patients were stratified into four groups according to the statuses of MYCN and 11q23. The four groups differed significantly in survival (both P < 0.001) (Fig. 2g, h). 51 patients without MYCN amplification and 11q23 deletion had the best prognosis (3-year EFS and OS rates were 53.4 ± 10.3% and 73.0 ± 6.2%). The 2 patients with both MYCN amplification and 11q23 deletion had the worst outcome (3-year EFS and OS rates were both 0). Multivariate analysis showed that only MYCN amplification was an independent prognostic factor of EFS for all patients (Table 3). 11q23 deletion was a prognostic factor in univariate analysis only for patients without MYCN amplification, but it was not an independent prognostic factor in the multivariate analysis.
Table 3.
Univariate and multivariate analysis of event-free survival (EFS) of children with bone marrow metastatic neuroblastoma
| Variable | Total patients (n = 101) | Patients without MYCN amplification (n = 89) | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariate* | Multivariate# | Univariate* | Multivariate# | |||||
| P | HR | 95% CI | P | P | HR | 95% CI | P | |
| MYCN status (amplification vs. normal) | < 0.001 | 2.399 | 1.113–5.169 | 0.032 | – | – | – | – |
| LDH (≥ 1500 vs. < 1500 IU/L) | < 0.001 | 1.543 | 0.622–3.83 | 0.351 | 0.040 | 1.488 | 0.564–3.926 | 0.422 |
| NSE (≥ 370 vs. < 370 ng/mL) | 0.001 | 1.829 | 0.972–3.44 | 0.068 | 0.025 | 2.016 | 1.074–3.784 | 0.033 |
| 11q23 status (deletion vs. normal) | 0.153 | – | – | – | 0.037 | 1.674 | 0.869–3.225 | 0.124 |
MYCN the v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog; LDH, lactate dehydrogenase, NSE neuron-specific enolase, HR hazard ratio, CI confidence interval
* Univariate analysis for EFS was performed using the Kaplan–Meier log-rank test. All factors with P < 0.05 in the univariate analysis are shown
#All factors with P < 0.05 in the univariate analysis were selected in the backward Cox regression model for multivariate analysis of EFS
Discussion
In recent years, the pretreatment risk stratification for neuroblastoma patients has become increasingly necessary [26, 27]. Currently, the identified biological factors closely related to prognosis include age, histology, ploidy, MYCN gene and specific segmental chromosomal aberrations [28, 29]. In the present study, we evaluated the prognostic value of 11q23 deletion with or without MYCN amplification in bone marrow metastatic neuroblastoma. We found that 11q23 deletion was a prognostic marker only for patients without MYCN amplification and that 11q23 deletion was closely associated with age. Almost all cases of 11q23 deletion occurred in children older than 18 months. Multivariate analyses for EFS indicated that MYCN amplification was an independent prognostic factor, but 11q23 deletion was not. Combined assessment of MYCN and 11q23 deletion was much better than single-marker assessment in identifying the patients at a high risk of disease progression in bone marrow metastatic neuroblastoma.
In the present study, MYCN amplification was present in 11.9% of the patients, which was lower than those in our previous study [17] and other reports [21, 30]. 11q23 deletion was found in 39.6% of the patients, a rate close to those reported by other groups [11, 31]. Similar to another group [31], we here found that 11q23 deletion occurred commonly in patients with normal MYCN. In fact, the two markers were mutually exclusive; only 2 patients presented with both. We thought that MYCN amplification and 11q23 deletion were distinct genetic aberrations with different clinicobiological characteristics. MYCN amplification was associated with LDH and NSE levels, whereas 11q23 deletion was associated with age. There was a marked difference in age at diagnosis between the two groups, with a median age of 24.5 months in the MYCN amplification group and 41 months in the 11q23 deletion group.
We further confirmed that 11q23 deletion constituted a distinct group of patients with unfavorable neuroblastoma. In the present study, MYCN amplification was confirmed as an independent prognostic factor. We found no obvious differences in EFS and OS between the 11q23 deletion and the 11q23 normal groups. However, we found that 11q23 deletion was a powerful marker for poor prognosis in patients without MYCN amplification. Among the 89 patients without MYCN amplification, the 3-year EFS and OS rates were significantly lower in patients with 11q23 deletion than in those without the deletion (both P < 0.05).
It was reported that 11q23 deletion may be involved in the dysregulation of important genes, including tumor suppressor genes and oncogenes [32]. One candidate gene is forkhead box R1 (FOXR1), which is located in 11q23.3. 11q23.3 deletion results in either mixed-lineage leukemia (MLL) or platelet activating factor acetylhydrolase 1b catalytic subunit 2 (PAFAH1B2) fusing to the proximal side of FOXR1. The new fusion transcripts stimulate the overexpression of FOXR1. Functional analyses have shown that FOXR1 overexpression could promote cell proliferation and may be a tumor-driving event [33]. Another important tumor suppressor gene is h2a histone family member X (H2AFX), which resides in the 11q23.2-q23.3 region. H2AFX participates in DNA double-strand breaks (DSBs) through phosphorylation. The loss of H2AFX promotes tumorigenesis due to defective DSB repair, increased radiation sensitivity, and genomic instability [10]. It was reported that more chromosomal breaks were seen in neuroblastoma patients with 11q23 deletion, which may be explained by genomic instability related to the loss of H2AFX gene [34]. This hypothesis also implied that 11q23 deletion occurred early in tumorigenesis. However, in the present study, the median age at diagnosis was 41 months for patients with 11q23 deletion and 24.5 months for those with MYCN amplification. The definitive role of 11q23 deletion in tumorigenesis of neuroblastoma still needs to be elucidated.
The present study showed that a large number of patients with MYCN amplification or 11q23 deletion had unfavorable prognosis. The survival rate was higher in patients with 11q23 deletion than in those with MYCN amplification. However, OS has been reported to be similar for patients with the two aberrations after 8 years of follow-up [10]. We further evaluated the prognostic value of combined assessment of MYCN and 11q23 statuses and found that the combined assessment was much superior to single-marker assessment in recognizing the patients at a high risk of disease progression.
The present study investigated the outcomes of bone marrow metastatic neuroblastoma patients with 11q23 deletion and of the rare patients with both MYCN amplification and 11q23 deletion. However, there were some limitations of this study. First, it is unclear how the results would be used to affect treatment. Second, the cases of both MYCN amplification and 11q23 deletion were limited. In our future study, we will focus on how further subdivision of these patients would alter treatment.
Conclusions
In the present study, we found that for patients with bone marrow metastatic neuroblastoma, 11q23 deletion predicted poor prognosis only in those without MYCN amplification. Combined assessment of the two markers was superior to single-marker assessment in identifying patients at a high risk of disease progression.
Acknowledgements
Not applicable.
Abbreviations
- MYCN
V-Myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog
- FISH
fluorescence in situ hybridization
- HR
high-risk
- PBSC
peripheral blood stem cells
- PBSCT
peripheral blood stem cell transplantation
- LDH
lactate dehydrogenase
- NSE
neuron specific enolase
- EFS
event-free survival
- CR
complete remission
- OS
overall survival
- WBC
white blood cell
Authors’ contributions
YZX and MXL designed the study. YZX performed the experiments, collected the data, and wrote the manuscript. XTY, GC and LSG helped to collect the samples. ZW, ZQ, WXS and JM recruited the patients. GC and LSG participated in the statistical analysis. MXL reviewed the final manuscript and take primary responsibility for the article. All authors read and approved the final manuscript.
Funding
This work was supported by Capital’s Funds for Health Improvement and Research (2018-2-2095).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was approved by the Beijing Children’s Hospital Institutional Ethics Committee. Informed consents were obtained from the parents or their guardians in accordance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Zhi-Xia Yue, Email: yuezhixia168@126.com.
Tian-Yu Xing, Email: student_xingty@yeah.net.
Chao Gao, Email: gaochaoconnie@live.cn.
Shu-Guang Liu, Email: shuguangliubch@126.com.
Wen Zhao, Email: zhaowen7534@sina.com.
Qian Zhao, Email: bj_zhaoqian@sohu.com.
Xi-Si Wang, Email: xday_115@163.com.
Mei Jin, Email: meijin1975@sina.com.
Xiao-Li Ma, Phone: 86-10-59617600, Email: mxl1123@vip.sina.com.
References
- 1.Irwin MS, Park JR. Neuroblastoma: paradigm for precision medicine. Pediatr Clin North Am. 2015;62(1):225–256. doi: 10.1016/j.pcl.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 2.Davidoff AM. Neuroblastoma. Semin Pediatr Surg. 2012;21(1):2–14. doi: 10.1053/j.sempedsurg.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brodeur GM, Bagatell R. Mechanisms of neuroblastoma regression. Nat Rev Clin Oncol. 2014;11(12):704–713. doi: 10.1038/nrclinonc.2014.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hald ØH, Olsen L, Gallo-Oller G, Elfman LHM, Løkke C, Kogner P, et al. Inhibitors of ribosome biogenesis repress the growth of MYCN-amplified neuroblastoma. Oncogene. 2019;38(15):2800–2813. doi: 10.1038/s41388-018-0611-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berthold F, Spix C, Kaatsch P, Lampert F. Incidence, survival, and treatment of localized and metastatic neuroblastoma in Germany 1979–2015. Paediatr Drugs. 2017;19(6):577–593. doi: 10.1007/s40272-017-0251-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stigliani S, Coco S, Moretti S, Oberthuer A, Fischer M, Theissen J, et al. High genomic instability predicts survival in metastatic high-risk neuroblastoma. Neoplasia. 2012;14(9):823–832. doi: 10.1593/neo.121114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoda H, Inoue T, Shinozaki Y, Lin J, Watanabe T, Koshikawa N, et al. Direct targeting of MYCN gene amplification by site-specific DNA alkylation in neuroblastoma. Cancer Res. 2019;79(4):830–840. doi: 10.1158/0008-5472.CAN-18-1198. [DOI] [PubMed] [Google Scholar]
- 8.Fusco P, Esposito MR, Tonini GP. Chromosome instability in neuroblastoma. Oncol Lett. 2018;16(6):6887–6894. doi: 10.3892/ol.2018.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domingo-Fernandez R, Watters K, Piskareva O, Stallings RL, Bray I. The role of genetic and epigenetic alterations in neuroblastoma disease pathogenesis. Pediatr Surg Int. 2013;29(2):101–119. doi: 10.1007/s00383-012-3239-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carén H, Kryh H, Nethander M, Sjöberg RM, Träger C, Nilsson S, et al. High-risk neuroblastoma tumors with 11q-deletion display a poor prognostic, chromosome instability phenotype with later onset. Proc Natl Acad Sci USA. 2010;107(9):4323–4328. doi: 10.1073/pnas.0910684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yagyu S, Iehara T, Gotoh T, Miyachi M, Katsumi Y, Kikuchi K, et al. Preoperative analysis of 11q loss using circulating tumor-released DNA in serum:a novel diagnostic tool for therapy stratification of neuroblastoma. Cancer Lett. 2011;309(2):185–189. doi: 10.1016/j.canlet.2011.05.032. [DOI] [PubMed] [Google Scholar]
- 12.Tolbert VP, Matthay KK. Neuroblastoma:clinical and biological approach to risk stratification and treatment. Cell Tissue Res. 2018;372(2):195–209. doi: 10.1007/s00441-018-2821-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mlakar V, Jurkovic Mlakar S, Lopez G, Maris JM, Ansari M, Gumy-Pause F. 11q deletion in neuroblastoma:a review of biological and clinical implications. Mol Cancer. 2017;16(1):1–12. doi: 10.1186/s12943-017-0686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schleiermacher G, Janoueix-Lerosey I, Delattre O. Recent insights into the biology of neuroblastoma. Int J Cancer. 2014;135(10):2249–2261. doi: 10.1002/ijc.29077. [DOI] [PubMed] [Google Scholar]
- 15.Maris JM, Guo C, White PS, Hogarty MD, Thompson PM, Stram DO, et al. Allelic deletion at chromosome bands 11q14-23 is common in neuroblastoma. Med Pediatr Oncol. 2001;36(1):24–27. doi: 10.1002/1096-911X(20010101)36:1<24::AID-MPO1007>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 16.Spitz R, Hero B, Simon T, Berthold F. Loss in chromosome 11q identifies tumors with increased risk for metastatic relapses in localized and 4S neuroblastoma. Clin Cancer Res. 2006;12(11):3368–3373. doi: 10.1158/1078-0432.CCR-05-2495. [DOI] [PubMed] [Google Scholar]
- 17.Yue ZX, Huang C, Gao C, Xing TY, Liu SG, Li XJ, et al. MYCN amplification predicts poor prognosis based on interphase fluorescence insitu hybridization analysis of bone marrow cells in bone marrow metastases of neuroblastoma. Cancer Cell Int. 2017;17(43):1–7. doi: 10.1186/s12935-017-0412-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morandi F, Corrias MV, Pistoia V. Evaluation of bone marrow as a metastatic site of human neuroblastoma. Ann N Y Acad Sci. 2015;1335(1):23–31. doi: 10.1111/nyas.12554. [DOI] [PubMed] [Google Scholar]
- 19.Coco S, Theissen J, Scaruffi P, Stigliani S, Moretti S, Oberthuer A, et al. Age dependent accumulation of genomic aberrations and deregulation of cell cycle and telomerase genes in metastatic neuroblastoma. Int J Cancer. 2012;131(7):1591–1600. doi: 10.1002/ijc.27432. [DOI] [PubMed] [Google Scholar]
- 20.Cohn SL, Pearson AD, London WB, Monclair T, Ambros PF, Brodeur GM, et al. The International Neuroblastoma Risk Group (INRG) classification system: an INRG task force report. J Clin Oncol. 2009;27(2):289–297. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duan C, Wang H, Chen Y, Chu P, Xing TY, Gao C, et al. Whole exome sequencing reveals novel somatic alterations in neuroblastoma patients with chemotherapy. Cancer Cell Int. 2018;18(21):1–6. doi: 10.1186/s12935-018-0521-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jo JH, Ahn SD, Koh M, Kim JH, Lee SW, Song SY, et al. Patterns of recurrence after radiation therapy for high-risk neuroblastoma. Radiat Oncol J. 2019;37(3):224–231. doi: 10.3857/roj.2019.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theissen J, Boensch M, Spitz R, Betts D, Stegmaier S, Christiansen H, et al. Heterogeneity of the MYCN Oncogene in Neuroblastoma. Clin Cancer Res. 2009;15(6):2085–2090. doi: 10.1158/1078-0432.CCR-08-1648. [DOI] [PubMed] [Google Scholar]
- 24.Villamón E, Berbegall AP, Piqueras M, Tadeo I, Castel V, Djos A, et al. Genetic instability and intratumoral heterogeneity in neuroblastoma with MYCN amplification plus 11q deletion. PLoS ONE. 2013;8(1):1–10. doi: 10.1371/journal.pone.0053740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevens-Kroef M, Simons A, Rack K, Hastings RJ. Cytogenetic nomenclature and reporting. Methods Mol Biol. 2017;1541(1):303–309. doi: 10.1007/978-1-4939-6703-2_24. [DOI] [PubMed] [Google Scholar]
- 26.Wang XS, Wang LJ, Su Y, Yue ZX, Xing TY, Zhao W, et al. Plasma cell-free DNA quantification is highly correlated to tumor burden in children with neuroblastoma. Cancer Med. 2018;7(7):3022–3030. doi: 10.1002/cam4.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacFarland S, Bagatell R. Advances in neuroblastoma therapy. Curr Opin Pediatr. 2019;31(1):14–20. doi: 10.1097/MOP.0000000000000711. [DOI] [PubMed] [Google Scholar]
- 28.Molenaar JJ, Koster J, Zwijnenburg DA, vanSluis P, Valentijn LJ, vanderPloeg I, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483(7391):589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 29.Janoueix-Lerosey I, Schleiermacher G, Michels E, Mosseri V, Ribeiro A, Lequin D, et al. Overall genomic pattern is a predictor of outcome in neuroblastoma. J Clin Oncol. 2009;27(7):1026–1033. doi: 10.1200/JCO.2008.16.0630. [DOI] [PubMed] [Google Scholar]
- 30.Selmi A, deSaint-Jean M, Jallas AC, Garin E, Hogarty MD, Bénard J, et al. TWIST1 is a direct transcriptional target of MYCN and MYC in neuroblastoma. Cancer Lett. 2015;357(1):412–418. doi: 10.1016/j.canlet.2014.11.056. [DOI] [PubMed] [Google Scholar]
- 31.Guo C, White PS, Hogarty MD, Brodeur GM, Gerbing R, Stram DO, et al. Deletion of 11q23 is a frequent event in the evolution of MYCN single-copy high-risk neuroblastomas. Med Pediatr Oncol. 2000;35(6):544–546. doi: 10.1002/1096-911X(20001201)35:6<544::AID-MPO10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 32.Mlakar V, Jurkovic Mlakar S, Lopez G, Maris JM, Ansari M, Gumy-Pause F. 11q deletion in neuroblastoma: a review of biological and clinical implications. Mol Cancer. 2017;16(1):114–126. doi: 10.1186/s12943-017-0686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santo EE, Ebus ME, Koster J, Schulte JH, Lakeman A, van Sluis P, et al. Oncogenic activation of FOXR1 by 11q23 intrachromosomal deletion fusions in neuroblastoma. Oncogene. 2012;31(12):1571–1581. doi: 10.1038/onc.2011.344. [DOI] [PubMed] [Google Scholar]
- 34.Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, et al. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114(3):359–370. doi: 10.1016/S0092-8674(03)00566-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.