Abstract
Background
Chinese hickory (Carya cathayensis) is a popular nut plant having high economic value. Grafting is applied to accelerate the transition from vegetative phase to reproductive phase. Lysine succinylation occurs frequently in the proteins associated with metabolic pathways, which may participate in the regulation of the grafting process. However, the exact regulatory mechanism underlying grafting process in Chinese hickory has not been studied at post-translational modification level.
Results
A comprehensive proteome-wide lysine succinylation profiling of Chinese hickory was explored by a newly developed method combining affinity enrichment and high-resolution LC-MS/MS. In total, 259 succinylation sites in 202 proteins were identified, representing the first comprehensive lysine succinylome in Chinese hickory. The succinylation was biased to occur in the cytosolic proteins of Chinese hickory. Moreover, four conserved succinylation motifs were identified in the succinylated peptides. Comparison of two grafting stages of Chinese hickory revealed that the differential expressed succinylated proteins were mainly involved in sugar metabolism, carbon fixation, amino acid metabolism and plant-pathogen interaction. Besides, seven heat shock proteins (HSPs) with 11 succinylation sites were also identified, all of which were observed to be up-regulated during the grafting process.
Conclusions
Succinylation of the proteins involved in amino acid biosynthesis might be required for a successful grafting. Succinylated HSPs might play a role in stress tolerance of the grafted Chinese hickory plants. Our results can be a good resource for functional validation of the succinylated proteins and a starting point for the investigation of molecular mechanisms during lysine succinylation occurring at grafting site.
Keywords: Chinese hickory, Post-translational modification, Succinylation, Grafting, Heat shock proteins
Background
Chinese hickory (Carya cathayensis Sarg.) is an economically important plant in China which produces nuts with considerable amounts of nutritious components [1]. A mature Chinese hickory nut usually contains not only an extremely high proportion of unsaturated fatty acids (over 70%), including oleic acid, palmitic and linolenic acid, but also several antitumor compounds, like phenolics, carayensin-A, carayensin-B and carayensin-C [2–4]. Limited to its long juvenile phase, the breeding efficiency and yield of Chinese hickory are largely reduced, which has limited the long-term development of Chinese hickory horticultural industry [5].
Grafting is an ancient technique that can provide an effective way to accelerate the transition from the vegetative phase to reproductive phase in Chinese hickory [6]. A successful grafting process is controlled by both intrinsic hereditary determinants and external environmental factors [7, 8]. Recent studies have revealed the response of important genes and proteins involved in the grafting process of Chinese hickory. For example, 12 cambium formation and cell growth-related genes were up-regulated during the grafting process [6]. Transcriptomic analysis of Chinese hickory uncovered a great number of auxin- and cytokinin-related differentially expressed genes, suggesting a role of hormone signaling pathways during the graft process of Chinese hickory [5]. Proteomic analysis of the graft unions in Chinese hickory showed that five flavonoid biosynthesis-related proteins, including flavanone-3-hyfroxylase, cinnamate-4-hydroxylase, dihydroflavonol-4-reductase, chalcone synthase and chalcone isomerase, were significantly up-regulated after grafting, indicating the involvement of secondary metabolism during the grafting process [9]. Recently, a grafting response gene, CcPIP1;2, was cloned and functionally characterized. Over-expression of CcPIP1;2 in Arabidopsis could increase the resistance to abiotic stresses by regulating the expression of ABA-related genes as well as cell wall expansion genes [8]. However, the molecular mechanism underlying the post-translational modifications (PTMs) of grafting-induced proteins is largely unknown.
PTM of proteins is an efficient biological mechanism controlling many biological processes, such as transcription, metabolism, and aging [10]. In both eukaryotic and prokaryotic cells, PTMs expand the functional and structural diversity of a limited number of proteins [11]. In the past few decades, many studies have focused on reversible acetylation at lysine residues [12]. In addition to acetylation, lysine residues can be modified by various short-chain acylations, such as propionylation, butyrylation, crotonylation, malonylation, succinylation and glutarylation [12–14]. Lysine succinylation has been studied in a number of organisms, including microbes (such as Mycobacterium tuberculosis and Saccharomyces cerevisiae), mammals (such as Rattus norvegicus and Mus musculus), and plants (Taxus × media and Dendrobium officinale) [15–18].
Lysine succinylation occurs frequently in the proteins associated with metabolic pathways, which may participate in the regulation of the grafting process [19, 20]. Thus, identifying succinylated proteins may provide useful information on how PTMs are involved in the grafting process of Chinese hickory. Previous studies focused on the physiological and transcriptional aspects in Chinese hickory during the grafting process [1, 2, 21–27]. However, in best of our knowledge, no report on PTMs are available till date in Chinese hickory. Here, we report the first proteomics-based quantification of lysine succinylation in Chinese hickory graft union during the grafting process. Current study provides new insight into the molecular mechanism underlying the grafting process at the PTM level.
Results
Proteome-wide analysis of lysine succinylation sites of Chinese hickory
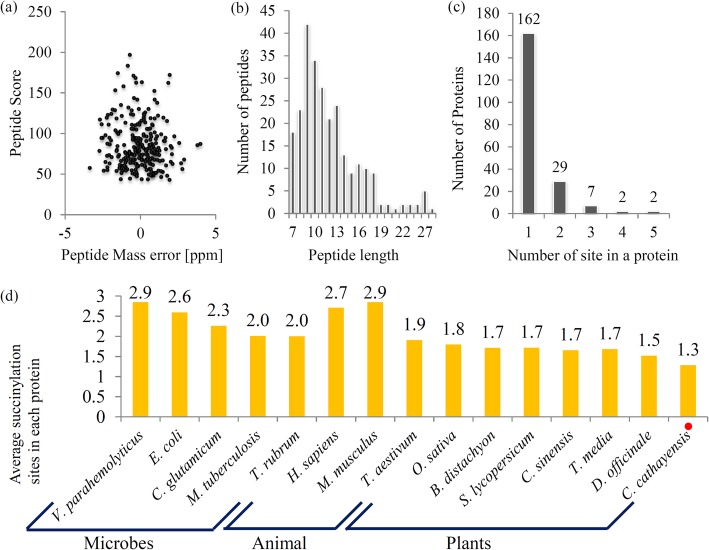
Lysine succinylation is a widespread PTM in both prokaryotic and eukaryotic cells, however, the succinylome of woody plants has not been reported previously [16]. A flow chart of the succinyl proteomics analysis has been provided (Additional file 1). Two important parameters, mass error and peptide length, were calculated. The mass errors of the most succinylated peptides were under 0.02 Da and the lengths of most succinylated peptides varied from 7 to 27 amino-acid residues (Fig. 1a and b).
Fig. 1.
The basic information of LC-MS/MS data. a The peptide score of LC-MS/MS data. b Length distribution of succinylated peptides based on their length. c The number of proteins with different number of succinylated sites. d Average succinylation sites on each succinylated protein from various organisms
Altogether, 259 succinylation sites in 202 proteins were identified, out of which 177 succinylation sites in 143 proteins were quantified (Additional file 2). Most of the succinylated proteins (162 proteins) contained only one succinylation site. Twenty nine succinylated proteins contained two succinylation sites, and 11 succinylated proteins contained more than three succinylation sites (Fig. 1c). Interestingly, two proteins, including phosphoglycerate kinase (CS_87744_c0) and protein disulfide-isomerase (CS_93322_c0), contained five succinylation sites. To analyze the number of succinylated proteins and the density of succinylation sites per protein, the succinylome of Chinese hickory was compared with those published in other species. The number of succinylated proteins in Chinese hickory was recorded significantly less than those in microbes and animals, including E. coli (990 proteins), V. parahemolyticus (750 proteins), M. tuberculosis (686 proteins), H. sapiens (738 proteins) and M. musculus (750 proteins), but almost similar to those in plants, such as T. aestivum (173 proteins), S. lycopersicum (202 proteins), T. media (193 proteins), D. officinale (207 proteins) [17, 18, 28–30] (Additional file 3). Meanwhile, it is observed that the average number of succinylation sites per protein in Chinese hickory was less than that in most reported species (Fig. 1d).
Characterization of the Chinese hickory lysine succinylome
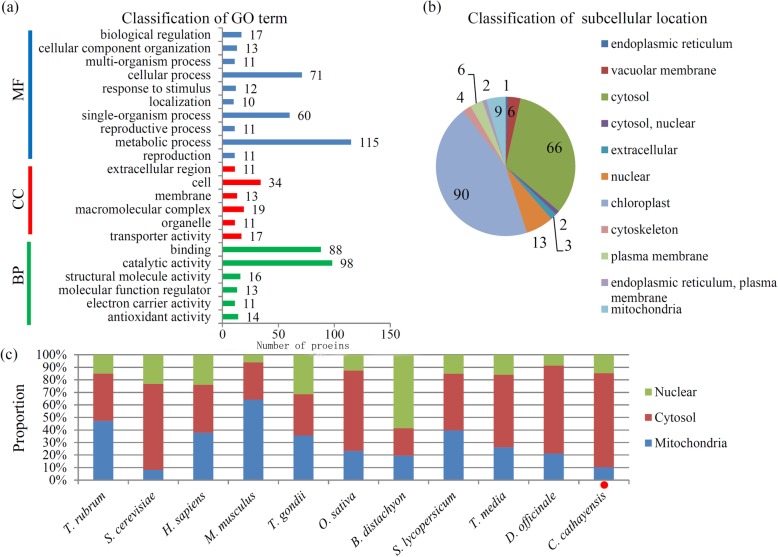
To predict the possible functions, all of identified succinylated proteins were classified into different GO categories. In ‘biological process’ category, most of the succinylated proteins were classified into the ‘binding’ (88 proteins) and ‘catalytic activity’ (98 proteins) terms; in ‘cellular component’ category, the largest group of succinylated proteins belonged to the ‘cell’ (34 proteins) and ‘macromolecular complex’ (19 proteins) terms; and in ‘molecular function’ category, the main terms were ‘metabolic process’ (115 proteins) and ‘cellular process’ (71 proteins) (Fig. 2a).
Fig. 2.
Bioinformatic analysis of lysine succinylation sites and succinylated proteins in Chinese hickory. a GO classifications for lysine succinylated proteins in Chinese hickory. b Subcellular locations of lysine succinylated proteins in Chinese hickory. c The proportions of succinylated proteins in mitochondria, cytoplasm and nucleus in different organisms
There is a close relativity between protein function and subcellular localization [30, 31]. With regard to the subcellular locations of all the identified succinylated proteins in Chinese hickory, the largest group was chloroplast-located proteins, followed by cytosol-located, nuclear-located, and mitochondria-located proteins, the number of which accounted for 44.6, 32.7, 6.4 and 4.5% for all of the identified succinylated proteins, respectively (Fig. 2b). Additionally, the relative proportions of succinylated proteins in the organelles like nuclei, cytosol and mitochondria in C. cathayensis were compared with other published organisms, including T. rubrum, S. cerevisiae, H. sapiens, M. musculus, T. gondii, O. sativa, B. distachyon, S. lycopersicum, T. media and D. officinale [16–18, 30–32]. Among these organisms, Chinese hickory possessed the highest proportion of cytosol-located succinylated proteins (75.0%), but the proportion of mitochondria-located succinylated proteins was only 10.2%, lower than that of all organisms mentioned above, excepting S. cerevisiae [16] (Fig. 2c).
Motif analysis of identified lysine-succinylated peptides
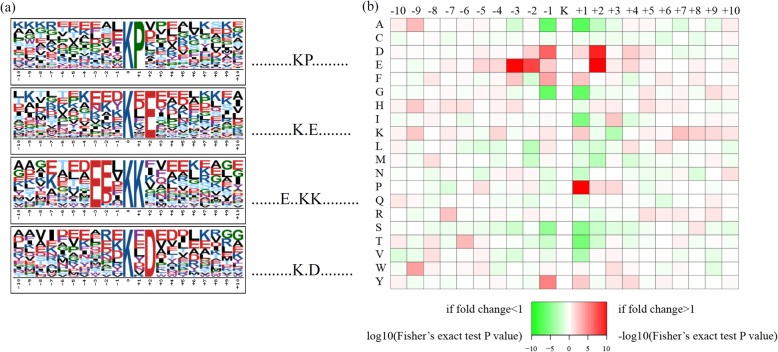
The patterns of amino acids surrounding succinylation sites are diverse among organisms but regular in a given organism [16, 18]. In the current study, the amino acid sequences of all succinylated peptides were extracted to analyze the typical motifs in the identified succinylated proteins of Chinese hickory. Several representative amino acids, such as proline (P), glutamic acid (E), lysine (K) and aspartic acid (D), were enriched downstream of the succinylated-lysine sites. In total, four preferred sequence patterns, including KsucP, Ksuc·E, E··KsucK, and Ksuc·D, were identified using software Motif-X with P at 0.000001 (Fig. 3). After comparison of the four motifs identified in Chinese hickory with those of the previously published succinylomes, it was found that these motifs were not unique but shared by both of C. cathayensis and Camellia sinensis [33].
Fig. 3.
Plot shows relative abundance of amino acids flanking succinylated lysine. a Probability sequence motifs of succinylation sites consisting of 10 residues surrounding the targeted lysine residue using Motif-X. Four significantly enriched succinylation site motifs were identified. b The relative abundance was counted and schematically represented by an intensity map. The intensity map shows enrichment of amino acids in specific positions of succinylated lysine (10 amino acids upstream and downstream of the succinylation sites)
Comparison of succinylation sites on the enzymes related with the glycolysis and TCA cycle among various species
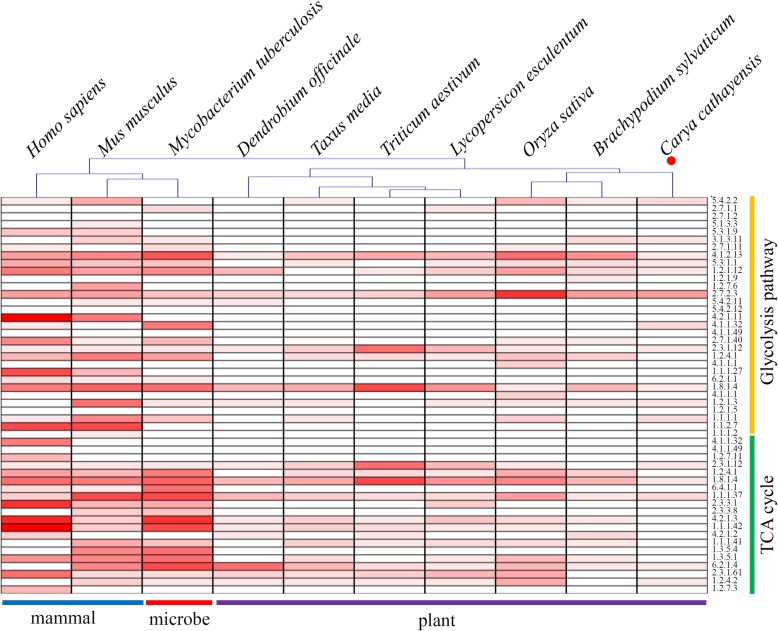
In Chinese hickory, 11 key glycolytic enzymes, including phosphoglucomutase (PGM, EC: 5.4.2.2), fructose-1,6-bisphosphatase (FBP, EC: 3.1.3.11), fructose-bisphosphate aldolase (ALDO, EC: 4.1.2.13), triosephosphate isomerase (TPI, EC: 5.3.1.1), glyceraldehyde-3-phosphate dehydrogenase (GAPDH, EC: 1.2.1.12), phosphoglycerate kinase (PGK, EC: 2.7.2.3), phosphoeno/lpyruvate carboxykinase (GTP, EC: 4.1.1.32), dihydrolipoyllysine-residue acetyltransferase (DLAT, EC: 2.3.1.12), dihydrolipoyl dehydrogenase (DLD, EC: 1.8.1.4), aldehyde dehydrogenase (ALDH, EC: 1.2.1.3) and alcohol dehydrogenase (ADH, EC: 1.1.1.1), were identified as succinylated proteins (Fig. 4 and Additional file 4). Among these enzymes, PGK contained the largest number of succinylation sites (eight sites) and four enzymes, including FBP, GAPDH, DLAT and DLD contained only one succinylation site. For the TCA cycle, five key enzymes, including dihydrolipoyllysine-residue acetyltransferase (DLAT, EC: 2.3.1.12), dihydrolipoyl dehydrogenase (DLD, EC: 1.8.1.4), malate dehydrogenase (MDH, EC: 1.1.1.37), succinate-CoA ligase (SUCLG, EC: 6.2.1.4) and oxoglutarate dehydrogenase (OGDH, EC: 1.2.4.2) were observed to be lysine-succinylated (Fig. 4 and Additional file 5).
Fig. 4.
Comparison of succinylation sites in the enzymes involved in glycolysis and TCA cycle. A heatmap showed the numbers of succinylation sites in key enzymes involved in glycolysis and TCA cycle from various organisms. Red indicates succinylation site of the corresponding enzyme. Various shades of red indicates the differences in the number of succinylation sites. Deeper red represents more succinylation sites
Differences in lysine succinylation during the grafting process of Chinese hickory
In order to investigate the differences in lysine succinylation during the grafting process, a quantitative succinylome profiling was done in the current study (Additional file 6). A total of 68 differentially expressed succinylated proteins (DESPs) were identified, including 45 up-regulated DESPs with 54 succinylation sites and 23 down-regulated DESPs with 29 succinylation sites, at 7 d after grafting as compared with the control (0 d after grafting) (Additional file 7). Among these DESPs, cytosolic glutamine synthetase β2 (CS_88158_c0) and calcyclin-binding protein (CS_48795_c0) were up-regulated over 2 folds, but only one unknown protein was down-regulated over 2 folds.
Functional enrichment-based clustering for quantitative category
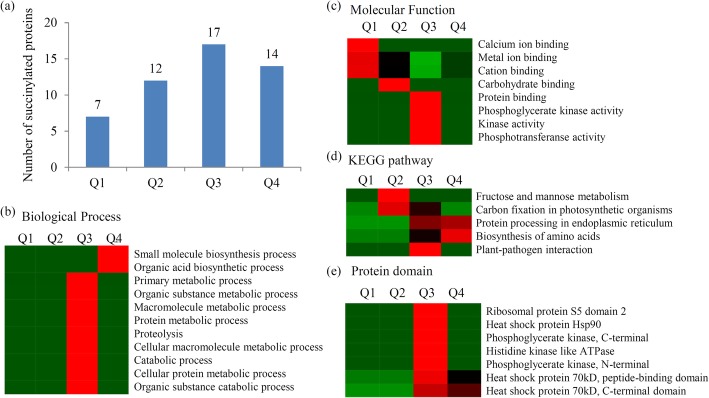
The DESPs were divided into four quantitative categories according to their D7/D0 Ratio: Q1 (0 < D7/D0 Ratio < 1/1.5), Q2 (1/1.5 < D7/D0 Ratio < 1/1.3), Q3 (1.3 < D7/D0 Ratio < 1.5) and Q4 (D7/D0 Ratio > 1.5) (Fig. 5a). The quantifiable proteins from the four categories were plotted for GO enrichment-based cluster analysis. For the ‘biological process’ category, the significantly enriched GO terms were ‘small molecule biosynthesis process’, ‘organic acid biosynthetic process’, ‘primary metabolic process’, ‘organic substance metabolic process’, ‘macromolecule metabolic process’, ‘protein metabolic process’, ‘proteolysis’, ‘cellular macromolecule metabolic process’, ‘catabolic process’, ‘cellular protein metabolic process’ and ‘organic substance catabolic process’ (Fig.5b). For the ‘molecular function’ category, the significantly enriched GO terms were ‘calcium ion binding’, ‘metal ion binding’, ‘cation binding’, ‘carbohydrate binding’, ‘protein binding’, ‘phosphoglycerate kinase activity’, ‘kinase activity’ and ‘phosphotransferanse activity’ (Fig.5c).
Fig. 5.
Differences in lysine succinylation during the grafting process of Chinese hickory. a Number of DESPs during the grafting process of Chinese hickory. Enrichment analysis of DESPs in Chinese hickory during grafting process. Significantly enriched GO terms of DESPs concerning biological process (b) and molecular function (c). d Significantly enriched KEGG terms of DESPs. e Significantly enriched protein domains of DESPs. For hierarchical clustering, protein categories that were enriched in at least one of the clusters with a P-value < 0.05 were transformed by the x = −log10 (P-value) function. These x-values were z-transformed and clustered by one-way hierarchical clustering basing on the Euclidean distance. The quantifiable proteins in this study were divided into four quantitative categories according to their D7/D0 Ratio: Q1 (0 < D7/D0 Ratio < 1/1.5), Q2 (1/1.5 < D7/D0 Ratio < 1/1.3), Q3 (1.3 < D7/D0 Ratio < 1.5) and Q4 (D7/D0 Ratio > 1.5)
The DESPs were enriched in five KEGG metabolic pathways, including ‘fructose and mannose metabolism’, ‘carbon fixation in photosynthetic organisms’, ‘protein processing in endoplasmic reticulum’, ‘biosynthesis of amino acids’ and ‘plant-pathogen interaction’ (Fig.5d).
Protein domain enrichment analysis revealed that the seven protein domains, including ‘ribosomal protein S5 domain 2’, ‘heat shock protein Hsp90’, ‘phosphoglycerate kinase, C-terminal’, ‘histidine kinase like ATPase’, ‘phosphoglycerate kinase, N-terminal’, ‘heat shock protein 70kD, peptide-binding domain’ and ‘heat shock protein 70kD, C-terminal domain’, were enriched in the DESPs (Fig. 5e and Additional file 8).
Discussion
Chinese hickory is a popular nut-producing tree species with high economic value in China [9]. Grafting is one of the most frequently applied technique in asexual propagation of commercially grown Chinese hickory trees [34]. Grafting involves a number of physiological and biochemical processes and exact mechanism behind this technique yet to be explored in much detail [35]. Lysine succinylation is a widely identified PTM involved in diverse protein functions [16]. However, limited information on the changes in the succinylated proteins during the grafting process of Chinese hickory has been obtained.
In our study, the average numbers of succinylation sites in Chinese hickory and other reported organisms were counted (Fig. 1d). Data showed that the average succinylation sites in the reported organisms were more than that in Chinese hickory, indicating a low degree of succinylation in this nut plant. The role of highly diversified succinylation degrees in plants was needed to be addressed in the future.
Cytosol is the main area of cellular metabolism, whereas nucleus is the main repository of genetic information, and mitochondria is the main organelle required for the biosynthesis of succinyl-CoA and succinate [36–38]. In Chinese hickory, the succinylation was biased to occur in the cytosolic proteins, indicating a possible effect of succinylation on the activities of enzymes associated with cellular metabolism [17]. A great number of succinylated proteins were predicted to be mitochondria-localized proteins, suggesting that succinylation might play roles in maintaining and regulating metabolic function of Chinese hickory [39].
Many studies revealed that a large number of succinylated proteins are involved in various metabolic processes [28]. Several published succinyl-proteomes, including two mammals (H. sapiens and M. musculus), five microbes (V. parahemolyticus, E. coli, C. glutamicum, M. tuberculosis, T. rubrum), and six plants (D. officinale, T. media, O. sativa, S. lycopersicum, C. sinensis and B. distachyon) were extracted to evaluate the potential conservation of succinylation in the regulation of central metabolism [15, 18, 30, 40, 41]. The numbers of succinylation sites in the enzymes involved in glycolysis and TCA cycle were counted in the ten selected organisms (Fig. 4). Our data showed that the average number of succinylation sites in Chinese hickory was less than those in the ten representative organisms. Interestingly, the average number of succinylation sites in mammals and microbes was larger than those in plants, indicating a varied frequency of lysine succinylation among organisms.
In plants, such as grapevine, graft union formation affected the expression of the secondary metabolism-related genes [42]. In ‘Gold Finger’ grape berry, the contents of fructose, glucose and sucrose were affected by different rootstocks [43]. In our study, several fructose and mannose metabolism-related proteins, including fructose-bisphosphate aldolase-like protein, L-iditol 2-dehydrogenase and latex plastidic aldolase-like protein, were identified as succinylated proteins. These fructose and mannose metabolism-related proteins were observed to be significantly down-regulated during the grafting process of Chinese hickory, suggesting an important role of fructose and mannose in the grafting process (Fig. 5d). Systemic resistance in citrus was transmitted by grafting and mediated by mobile amino acids [44]. In the current study, eight amino acid biosynthesis-related proteins were identified as succinylated proteins, and most of them were significantly up-regulated during the grafting process (Fig. 5d), indicating that enhanced biosynthesis of amino acids might be required for a successful grafting of Chinese hickory.
HSPs exist ubiquitous in eukaryotes and increase in the expression of HSP genes were related with the enhanced resistance to environmental stresses in various plants [45, 46]. In total, seven HSPs with 11 succinylation sites were identified in Chinese hickory, including two HSP90s and five HSP70s (Additional file 8). Abscisic acid-induced HSP70 accumulation played a role in the heat tolerance of grafted cucumber plants [47]. Interestingly, all these HSPs were up-regulated during the grafting process of Chinese hickory, suggesting that over-accumulated HSPs might play a role in the tolerance of grafted Chinese hickory plants.
Conclusions
In the current study, a comprehensive succinyl-proteome of Chinese hickory was presented. Quantitative analysis identified a number of differentially expressed succinylated proteins during the grafting process. Our data can act as important basic resource for the functional validation of succinylated proteins and a starting point for investigations into the molecular basis of lysine succinylation in the grafting process.
Methods
Plant samples and protein extraction
Chinese hickory (Carya cathayensis Sarg.) plants were planted in the plantation area at the campus of Zhejiang A&F University, Hangzhou, China. Plants were provided by the plantation (Zhejiang A&F University) where the formal identification of the plant material was undertaken by Dr. Daoliang Yan (Zhejiang A&F University). A voucher specimen of this material has not been deposited in a publicly available herbarium. All the plants were watered twice a week with a nutrient solution (1.425 mM NH4NO3, 0.323 mM NaH2PO4, 0.513 mM K2SO4, 0.998 mM CaCl2, 1.643 mM MgSO4, 0.009 mM MnCl2, 0.075 mM (NH4)6Mo7O24, 0.019 mM H3BO3, 0.155 mM CuSO4, 1 mM FeCl3, 0.070 mM citric acid and 0.152 mM ZnSO4). Plant samples were collected from the graft unions, including the cutting surfaces of the rootstocks (2 years old) and scions (1 year old), at 7 d after grafting. The samples from graft unions at the first day of grafting were used as the controls.
Plant samples were first ground with liquid N2. The cell powder was then transferred into a centrifuge tube (5 mL) and sonicated five times using a high intensity ultrasonic processor (Scientz, Ningbo, China) in pre-cooled lysis buffer containing urea (2 M), ethylenediamine tetraacetic acid (2 mM), dithiothreitol (10 mM) and Protease Inhibitor Cocktail VI (1%, MedChen Express, Monmouth Junction, USA). The remaining debris was discarded by centrifugation at 20,000 g at 4 °C for 10 min. The protein samples were precipitated with pre-cooled 15% TCA buffer for 2 h at − 20 °C. The supernatant was removed by centrifugation at 4 °C for 10 min. The remaining precipitates were washed with pre-cooled acetone for five times and then were re-dissolved in buffer containing 8 M urea and 100 mM TEAB (pH 8.0). The concentration of the final protein solution was determined by 2-D Quant kit according to manufacturer’s instructions (GE Healthcare, Little Chalfont, Buckinghamshire, UK) [48].
Trypsin digestion and TMT labeling
The protein sample solution was reduced with 10 mM DTT for 1 h at 37 °C and alkylated with 20 mM IAA for 45 min at 25 °C in darkness. For trypsin digestion, the protein solution was diluted to the final concentration of less than 2 M by adding 100 mM triethylamine borane buffer. Trypsin was added at a trypsin:protein mass ratio of 1:50 for the first round overnight-digestion, followed by a second round 4 h-digestion at a trypsin:protein mass ratio of 1:100.
Then, sample peptides were desalted using a Strata X C18 SPE column (Phenomenex, Torrance, CA, USA). The resulting peptides were vacuum dried with centrifugation. Peptides were reconstituted in 0.5 M TEA buffer and processed by 6-plex TMT kit according to the manufacturer’s instructions (Thermo scientific, Shanghai, China). Briefly, one unit of TMT reagent, together with 100 μg of sample peptides, were mixed and reconstituted in acetonitrile solution. After incubation for 2 h at 25 °C, the resulting peptide mixture was desalted and vacuum-dried again.
Affinity enrichment
To enrich Ksucc peptides, tryptic peptides dissolved in NETN buffer containing 100 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl and 0.5% NP-40 (pH 8.0), were incubated with pre-washed antibody beads (PTM Biolabs, Hangzhou, China) at 4 °C overnight with gentle shaking. The beads were washed five times with NETN buffer and twice with ddH2O. The bound peptides were eluted from the beads with 0.1% trifluoroacetic acid. After combined and vacuum-dried by centrifugation, the eluted peptides were cleaned with C18 ZipTips (Millipore, Shanghai, China) according to its instructions before LC-MS/MS analysis.
LC-MS/MS analysis
The LC-MS/MS analysis was carried out according to the procedure described previously [17]. Peptides were dissolved in 0.1% formic acid, directly loaded onto a reversed-phase pre-treated Acclaim PepMap 100 column (Thermo scientific, Shanghai, China). Peptide separation was carried out using a reversed-phase analytical Acclaim PepMap RSLC column (Thermo scientific, Shanghai, China). The resulting peptides were analyzed by Q Exactive™ plus hybrid quadrupole-Orbitrap MS (Thermo scientific, Shanghai, China).
The peptides were subjected to NSI source followed MS/MS in Q Exactive™ plus coupled online to the UPLC system (Thermo scientific, Shanghai, China). Intact peptides were detected in the Orbitrap at a high resolution of 70,000 and ion fragments were detected in the Orbitrap at a low resolution of 17,500. For MS scans, the m/z scan range was 350 to 1800, and the first mass was set as 100 m/z. The mass spectrometry proteomics data have already been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD009584 (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD009584).
Database search and protein annotation
The resulting MS/MS data was processed using MaxQuant integrated with Andromeda searching engine ver. 1.4.1.2. MS/MS were searched against Carya concatenated with reverse decoy database. Trypsin/P was used as cleavage enzyme allowing up to 4 missing cleavages, five modifications/peptide, and five charges. Mass errors were set to 10 ppm and 0.02 Da for precursor ions and fragment ions, respectively. The minimum number of peptides identified per protein was 1. Carbamido methylation on Cys was treated as fixed modification and succinylation on Lys was treated as variable modification. The false discovery rate (FDR) analysis was carried out according to the procedure described previously [17].
Annotation and enrichment analysis
For Gene Ontology (GO) annotation, all succinylated proteins were searched against the UniProt-GOA database (http://www.ebi.ac.uk/GOA/). If a succinylated protein was not annotated by the UniProt-GOA database, InterProScan database (http://www.ebi.ac.uk/interpro/) was used for further annotation. For Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation, all succinylated proteins were searched the database using the online tool KEGG Automatic Annotation Server (https://www.genome.jp/tools/kaas/). The results were mapped to different KEGG pathways using the online service tool KEGG Mapper (https://www.genome.jp/kegg/mapper.html). For subcellular localization prediction, all succinylated proteins were searched using the software ‘wolfpsort’ (http://psort.hgc.jp/). Enrichment analyses of GO, KEGG and protein domain were carried out using a two-tailed Fisher’s exact test.
For hierarchical clustering, protein categories that were enriched in at least one of the clusters with a P-value < 0.05 were transformed by the x = −log10 (P-value) function. The P-value was transformed into Z-score after log transformation:
These x-values were z-transformed and clustered by one-way hierarchical clustering (Euclidean distance, average linkage clustering) in Genesis and visualized by a heat map using the MeV software. The quantifiable proteins in this study were divided into four quantitative categories according to their D7/D0 Ratio: Q1 (0 < D7/D0 Ratio < 1/1.5), Q2 (1/1.5 < D7/D0 Ratio < 1/1.3), Q3 (1.3 < D7/D0 Ratio < 1.5) and Q4 (D7/D0 Ratio > 1.5). For the GO, KEGG and protein domain enrichment analyses, all of the proteins in each database were used as the background.
Motif clustering analysis
Software Motif-X was used to analyze the amino acid model of sequences in specific positions of modity-21-mers (10 amino acids up- and down-stream of the succinylation sites). All the protein sequences in the database were used as background parameter. Firstly, enrichment analyses of all lysine succinylation sites were carried out using their significance P values. Categories were filtered out according to the criteria: P value < 0.05. Then, the P value matrices were calculated according to method reported in our previous study [17]. Heatmaps were applied to visualize the results using the ‘ggplots’ R-package (http://ggplot2.org/).
Supplementary information
Additional file 1: Figure S1. A flow chart of the succinyl proteomics analysis.
Additional file 2: Table S1. The information of succinylated sites in each succinylated protein.
Additional file 3: Figure S2. The number of succinylated proteins in different species.
Additional file 4: Table S2. The number of succinylation sites on the enzymes engaged in the glycolysis of Chinese hickory.
Additional file 5: Table S3. The number of succinylation sites on the enzymes engaged in the TCA cycle of Chinese hickory.
Additional file 6: Table S4. A quantitative succinylome profiling of Chinese hickory during the grafting process.
Additional file 7: Figure S3. The numbers of differentially expressed succinylated proteins in Chinese hickory during the grafting process.
Additional file 8: Table S5. The differentially expressed HSPs.
Acknowledgements
We are also grateful to LC Sciences company (Hangzhou, China) and LetPub company (Hangzhou, China) for technical support.
Abbreviations
- ADH
Alcohol dehydrogenase
- ALDH
Aldehyde dehydrogenase
- ALDO
Fructose-bisphosphate aldolase
- D
Aspartic acid
- DESP
Differentially expressed succinylated protein
- DLAT
Dihydrolipoyllysine-residue acetyltransferase
- DLD
Dihydrolipoyl dehydrogenase
- E
Glutamic acid
- FBP
Fructose-1,6-bisphosphatase
- FDR
False discovery rate
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- GO
Gene ontology
- GTP
Phosphoeno/lpyruvate carboxykinase
- HSP
Heat shock proteins
- K
Lysine
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LC-MS/MS
High performance liquid chromatography-mass spectrometry
- MDH
Malate dehydrogenase
- OGDH
Oxoglutarate dehydrogenase
- P
Proline
- PGK
Phosphoglycerate kinase
- PGM
Phosphoglucomutase
- PTM
Post-translational modification
- SUCLG
Succinate-CoA ligase
- TMT
Tandem Mass Tag
- TPI
Triosephosphate isomerase
Authors’ contributions
JC, YH and BZ conceived and designed the study. JC, DY and YH collected and taken care of the plant samples. JC, YY, HY, CS, DX and JW performed the experiments. DX and DY analyzed the data. CS wrote the manuscript. All authors have read and approved the manuscript, and ensure that this is the case.
Funding
This study was supported by National Key Research and Development Program of China (2018YFD1000600, 2018YFD1000604); Key Project of Zhejiang Provincial Natural Science Foundation (LZ18C160001); Zhejiang Provincial Natural Science Foundation of China (LY19C160001); Open Foundation of State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University (KF201708); Independent Research Project of State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University (ZY20180208, ZY20180308); Key Research and Development Program of Zhejiang Province (2018C02004); National Natural Science Foundation of China (31470683, 31270716, and 31070604); National High Technology Research and Development Program of China (863 Program) (2013AA102605); The First-class General Financial Grant from the China Postdoctoral Science Foundation (2017 M610377); Fruit Innovation Team Project of Zhejiang Province (2016C02052–12); Key Agricultural New Varieties Breeding Projects founded by Zhejiang Province Science and Technology Department (2016C02052–13); Zhejiang Provincial Natural Science Foundation for Distinguished Young Scholar (LR13C160001); Open Foundation of First-class Discipline of Forestry, Zhejiang Province (201703). There is no role of the funding body in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files. The mass spectrometry proteomics data have already been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD009584 (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD009584). The data will be publicly accessible after 1th November, 2019.
Ethics approval and consent to participate
This project uses plant materials and does not utilize transgenic technology. The authority responsible for the Carya cathayensis resources is the Mount Tianmu National Nature Reserve, who provided permission to collect the samples for our scientific research.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Huwei Yuan, Juanjuan Chen and Ying Yang contributed equally to this work.
Contributor Information
Huwei Yuan, Email: hwyuan@zafu.edu.cn.
Juanjuan Chen, Email: 15738825451@163.com.
Ying Yang, Email: sky_yangying@163.com.
Chenjia Shen, Email: shencj@hznu.edu.cn.
Dongbin Xu, Email: xdongbin@gmail.com.
Junfeng Wang, Email: 343781413@qq.com.
Daoliang Yan, Email: liangsie2000@163.com.
Yi He, Email: heyi@zju.edu.cn.
Bingsong Zheng, Phone: 0086 (0)571 63730936, Email: bszheng@zafu.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12870-019-2072-8.
References
- 1.Li J, Zeng Y, Shen D, Xia G, Huang Y, Huang Y, et al. Development of SSR markers in hickory (Carya cathayensis Sarg.) and their transferability to other species of Carya. J Appl Stat. 2014;15(5):357–379(323). doi: 10.2174/138920291505141106103734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang YJ, Zhou Q, Huang JQ, Zeng YR, Wang ZJ, Zhang QX, et al. Transcriptional profiling by DDRT-PCR analysis reveals gene expression during seed development in Carya cathayensis Sarg. Plant Physiol Biochem. 2015;91:28–35. doi: 10.1016/j.plaphy.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Wu W, Bi XL, Cao JQ, Zhang KQ, Zhao YQ. New antitumor compounds from Carya cathayensis. Bioorg Med Chem Lett. 2012;22(5):1895–1898. doi: 10.1016/j.bmcl.2012.01.062. [DOI] [PubMed] [Google Scholar]
- 4.Wang Zhengjia, Huang Ruiming, Sun Zhichao, Zhang Tong, Huang Jianqin. Identification and profiling of conserved and novel microRNAs involved in oil and oleic acid production during embryogenesis in Carya cathayensis Sarg. Functional & Integrative Genomics. 2017;17(2-3):365–373. doi: 10.1007/s10142-016-0542-8. [DOI] [PubMed] [Google Scholar]
- 5.Qiu L, Jiang B, Fang J, Shen Y, Fang Z, Rm SK, et al. Analysis of transcriptome in hickory (Carya cathayensis), and uncover the dynamics in the hormonal signaling pathway during graft process. BMC Genomics. 2016;17(1):935. doi: 10.1186/s12864-016-3182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng BS, Chu HL, Jin SH, Huang YJ, Wang ZJ, Chen M, et al. cDNA-AFLP analysis of gene expression in hickory (Carya cathayensis) during graft process. Tree Physiol. 2010;30(2):297–303. doi: 10.1093/treephys/tpp102. [DOI] [PubMed] [Google Scholar]
- 7.Saravana Kumar RM, Gao LX, Yuan HW, Xu DB, Liang Z, Tao SC, et al. Auxin enhances grafting success in Carya cathayensis (Chinese hickory) Planta. 2018;247(3):761–772. doi: 10.1007/s00425-017-2824-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar RMS, Ji G, Guo H, Zhao L, Zheng B. Over-expression of a grafting-responsive gene from hickory increases abiotic stress tolerance in Arabidopsis. Plant Cell Rep. 2018;37(3):541–552. doi: 10.1007/s00299-018-2250-4. [DOI] [PubMed] [Google Scholar]
- 9.Xu D, Yuan H, Tong Y, Zhao L, Qiu L, Guo W, et al. Comparative proteomic analysis of the graft unions in hickory (Carya cathayensis) provides insights into response mechanisms to grafting process. Front Plant Sci. 2017;8:676. doi: 10.3389/fpls.2017.00676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Witze ES, Old WM, Resing KA, Ahn NG. Mapping protein post-translational modifications with mass spectrometry. Nat Methods. 2007;4(10):798–806. doi: 10.1038/nmeth1100. [DOI] [PubMed] [Google Scholar]
- 11.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 12.Yang XJ, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26(37):5310–5318. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- 13.Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014;19(4):605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nadal S, Raj R, Mohammed S, Davis BG. Synthetic post-translational modification of histones. Curr Opin Chem Biol. 2018;45:35–47. doi: 10.1016/j.cbpa.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Yang M, Wang Y, Chen Y, Cheng Z, Gu J, Deng J, et al. Succinylome analysis reveals the involvement of lysine succinylation in metabolism in pathogenic Mycobacterium tuberculosis. Mol Cell Proteomics. 2015;14(4):796. doi: 10.1074/mcp.M114.045922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinert BT, Scholz C, Wagner SA, Iesmantavicius V, Su D, Daniel JA, et al. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013;4(4):842–851. doi: 10.1016/j.celrep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 17.Shen C, Xue J, Sun T, Guo H, Zhang L, Meng Y, et al. Succinyl-proteome profiling of a high taxol containing hybrid Taxus species (Taxus × media) revealed involvement of succinylation in multiple metabolic pathways. Sci Rep. 2016;6:21764. doi: 10.1038/srep21764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng S, Jiao K, Guo H, Jiang M, Hao J, Wang H, et al. Succinyl-proteome profiling of Dendrobium officinale, an important traditional Chinese orchid herb, revealed involvement of succinylation in the glycolysis pathway. BMC Genomics. 2017;18(1):598. doi: 10.1186/s12864-017-3978-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He D, Wang Q, Li M, Damaris RN, Yi X, Cheng Z, et al. Global proteome analyses of lysine acetylation and succinylation reveal the widespread involvement of both modification in metabolism in the embryo of germinating rice seed. J Proteome Res. 2016;15(3):879–890. doi: 10.1021/acs.jproteome.5b00805. [DOI] [PubMed] [Google Scholar]
- 20.Santos ICD, Almeida AF, Pirovani CP, Costa MGC, Silva M, Bellete BS, et al. Differential accumulation of flavonoids and phytohormones resulting from the canopy/rootstock interaction of citrus plants subjected to dehydration/rehydration. Plant Physiol Biochem. 2017;119:147–158. doi: 10.1016/j.plaphy.2017.08.019. [DOI] [PubMed] [Google Scholar]
- 21.Zheng BS, Liu L, Huang JQ, Cheng XJ, Zhu YQ, Hong quan XU. Analysis on physiological and biochemical traits of survival of Carya cathayensis grafted seedling. J Fujian College Forestry. 2002;22(4):320–4.
- 22.Wang Z, Huang J, Sun Z, Zheng B. Identification of microRNAs differentially expressed involved in male flower development. Funct Integr Genomics. 2015;15(2):225–232. doi: 10.1007/s10142-014-0409-9. [DOI] [PubMed] [Google Scholar]
- 23.Shen C, Xu Y, Huang J, Wang Z, Qiu J, Huang Y. Molecular characterization and expression analysis of the critical floral genes in hickory (Carya cathayensis Sarg.) Plant Physiol Biochem. 2014;83:142–150. doi: 10.1016/j.plaphy.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 24.Huang YJ, Liu LL, Huang JQ, Wang ZJ, Chen FF, Zhang QX, et al. Use of transcriptome sequencing to understand the pistillate flowering in hickory (Carya cathayensis Sarg.) BMC Genomics. 2013;14:691. doi: 10.1186/1471-2164-14-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji GC, Zheng BS, Li XQ, Zhu XT, Jin SH. Cloning and expression analysis of Rubisco activase genes in Carya cathayensis. Biotechnology & Biotechnological Equipment. 2016;30(5):1–8. doi: 10.1080/13102818.2016.1208060. [DOI] [Google Scholar]
- 26.Wang ZJ, Huang JQ, Huang YJ, Li Z, Zheng BS. Discovery and profiling of novel and conserved microRNAs during flower development in Carya cathayensis via deep sequencing. Planta. 2012;236(2):613–621. doi: 10.1007/s00425-012-1634-x. [DOI] [PubMed] [Google Scholar]
- 27.Jin SH, Huang JQ, Li XQ, Zheng BS, Wu JS, Wang ZJ, et al. Effects of potassium supply on limitations of photosynthesis by mesophyll diffusion conductance in Carya cathayensis. Tree Physiol. 2011;31(10):1142–1151. doi: 10.1093/treephys/tpr095. [DOI] [PubMed] [Google Scholar]
- 28.Pan J, Chen R, Li C, Li W, Ye Z. Global analysis of protein lysine succinylation profiles and their overlap with lysine acetylation in the marine bacterium Vibrio parahemolyticus. J Proteome Res. 2015;14(10):4309–4318. doi: 10.1021/acs.jproteome.5b00485. [DOI] [PubMed] [Google Scholar]
- 29.Xie L, Liu W, Li Q, Chen S, Xu M, Huang Q, et al. First succinyl-proteome profiling of extensively drug-resistant Mycobacterium tuberculosis revealed involvement of succinylation in cellular physiology. J Proteome Res. 2015;14(1):107–119. doi: 10.1021/pr500859a. [DOI] [PubMed] [Google Scholar]
- 30.Jin W, Wu F. Proteome-wide identification of lysine succinylation in the proteins of tomato (Solanum lycopersicum) PLoS One. 2016;11(2):e0147586. doi: 10.1371/journal.pone.0147586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, Hu X, Wan Y, Xie G, Li X, Chen D, et al. Systematic identification of the lysine succinylation in the protozoan parasite Toxoplasma gondii. J Proteome Res. 2014;13(12):6087–6095. doi: 10.1021/pr500992r. [DOI] [PubMed] [Google Scholar]
- 32.Xu X, Liu T, Yang J, Chen L, Liu B, Wei C, et al. The first succinylome profile of Trichophyton rubrum reveals lysine succinylation on proteins involved in various key cellular processes. BMC Genomics. 2017;18(1):577. doi: 10.1186/s12864-017-3977-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu YX, Shen CJ, Ma JQ, Chen W, Mao J, Zhou YY et al: Quantitative succinyl-proteome profiling of Camellia sinensis cv. 'Anji Baicha' during periodic albinism. Scientific reports 2017;7(1):1873. [DOI] [PMC free article] [PubMed]
- 34.Sima X, Jiang B, Fang J, He Y, Fang Z, Saravana Kumar KM, et al. Identification by deep sequencing and profiling of conserved and novel hickory microRNAs involved in the graft process. Plant Biotechnology Reports. 2015;9(3):115–124. doi: 10.1007/s11816-015-0349-4. [DOI] [Google Scholar]
- 35.Pina A, Errea P. A review of new advances in mechanism of graft compatibility–incompatibility. Sci Hortic. 2005;106(1):1–11. doi: 10.1016/j.scienta.2005.04.003. [DOI] [Google Scholar]
- 36.Phillips D, Aponte AM, French SA, Chess DJ, Balaban RS. Succinyl-CoA synthetase is a phosphate target for the activation of mitochondrial metabolism. Biochemistry. 2009;48(30):7140–7149. doi: 10.1021/bi900725c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell. 2013;50(6):919–930. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bachs Oriol, Agell Neus. Calcium and Calmodulin Function in the Cell Nucleus. Berlin, Heidelberg: Springer Berlin Heidelberg; 1995. The Cell Nucleus: Structure and Main Functions; pp. 11–19. [Google Scholar]
- 39.Stram AR, Payne RM. Post-translational modifications in mitochondria: protein signaling in the powerhouse. Cell Mol Life Sci. 2016;73(21):4063–4073. doi: 10.1007/s00018-016-2280-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhen S, Deng X, Wang J, Zhu G, Cao H, Yuan L, et al. First comprehensive proteome analyses of lysine acetylation and succinylation in seedling leaves of Brachypodium distachyon L. Sci Rep. 2016;6:31576. doi: 10.1038/srep31576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Wang G, Song L, Mu P, Wang S, Liang W, et al. Global analysis of protein lysine succinylation profiles in common wheat. BMC Genomics. 2017;18(1):309. doi: 10.1186/s12864-017-3698-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cookson SJ, Clemente Moreno MJ, Hevin C, Nyamba Mendome LZ, Delrot S, Trossat-Magnin C, et al. Graft union formation in grapevine induces transcriptional changes related to cell wall modification, wounding, hormone signalling, and secondary metabolism. J Exp Bot. 2013;64(10):2997–3008. doi: 10.1093/jxb/ert144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin Z, Sun H, Sun T, Wang Q, Yao Y. Modifications of ‘gold finger’ grape berry quality as affected by the different rootstocks. J Agric Food Chem. 2016;64(21):4189–4197. doi: 10.1021/acs.jafc.6b00361. [DOI] [PubMed] [Google Scholar]
- 44.Agut B, Gamir J, Jaques JA, Flors V. Systemic resistance in citrus to Tetranychus urticae induced by conspecifics is transmitted by grafting and mediated by mobile amino acids. J Exp Bot. 2016;67(19):5711–5723. doi: 10.1093/jxb/erw335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun Y, Guo H, Yuan E, Ge F. Elevated CO2 increases R gene-dependent resistance of Medicago truncatula against the pea aphid by up-regulating a heat shock gene. New Phytol. 2018;217(4):1696–1711. doi: 10.1111/nph.14892. [DOI] [PubMed] [Google Scholar]
- 46.Wang X, Yan B, Shi M, Zhou W, Zekria D, Wang H, et al. Overexpression of a Brassica campestris HSP70 in tobacco confers enhanced tolerance to heat stress. Protoplasma. 2016;253(3):637–645. doi: 10.1007/s00709-015-0867-5. [DOI] [PubMed] [Google Scholar]
- 47.Li H, Liu SS, Yi CY, Wang F, Zhou J, Xia XJ, et al. Hydrogen peroxide mediates abscisic acid-induced HSP70 accumulation and heat tolerance in grafted cucumber plants. Plant Cell Environ. 2014;37(12):2768–2780. doi: 10.1111/pce.12360. [DOI] [PubMed] [Google Scholar]
- 48.Hao J, Guo H, Shi X, Wang Y, Wan Q, Song YB, et al. Comparative proteomic analyses of two Taxus species (Taxus × media and Taxus mairei) reveals variations in the metabolisms associated with paclitaxel and other metabolites. Plant Cell Physiol. 2017;58(11):1878–1890. doi: 10.1093/pcp/pcx128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. A flow chart of the succinyl proteomics analysis.
Additional file 2: Table S1. The information of succinylated sites in each succinylated protein.
Additional file 3: Figure S2. The number of succinylated proteins in different species.
Additional file 4: Table S2. The number of succinylation sites on the enzymes engaged in the glycolysis of Chinese hickory.
Additional file 5: Table S3. The number of succinylation sites on the enzymes engaged in the TCA cycle of Chinese hickory.
Additional file 6: Table S4. A quantitative succinylome profiling of Chinese hickory during the grafting process.
Additional file 7: Figure S3. The numbers of differentially expressed succinylated proteins in Chinese hickory during the grafting process.
Additional file 8: Table S5. The differentially expressed HSPs.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files. The mass spectrometry proteomics data have already been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD009584 (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD009584). The data will be publicly accessible after 1th November, 2019.