Abstract.
Stimulated emission depletion (STED) microscopy is a powerful bioimaging technique that theoretically provides molecular spatial resolution while preserving the most important assets of fluorescence microscopy. When combined with two-photon excitation (2PE) microscopy (2PE-STED), subdiffraction resolution may be achieved for thick biological samples. The most straightforward implementation of 2PE-STED microscopy entails introduction of an STED beam operating in continuous wave (CW) into a conventional Ti:sapphire-based 2PE microscope (2PE CW-STED). In this implementation, resolution enhancement is typically achieved using time-gated detection schemes, often resulting in drastic signal-to-noise/-background ratio (SNR/SBR) reductions. Herein, we employ a pixel-by-pixel phasor approach to discard fluorescence photons lacking super-resolution information to enhance image SNR/SBR in 2PE CW-STED microscopy. We compare this separation of photons by lifetime tuning approach against other postprocessing algorithms and combine it with image deconvolution to further optimize image quality.
Keywords: multiphoton microscopy, stimulated emission depletion microscopy, super-resolution, separation of photons by lifetime tuning
1. Introduction
Stimulated emission depletion (STED) microscopy1–3 is a powerful fluorescence imaging technique for nanoscale visualization of biological processes. When coupled with two-photon excitation (2PE) microscopy,4 nanoscale spatial resolution may be achieved at depths far greater than otherwise possible using single-photon excitation approaches. The fundamental mechanism underlying STED microscopy entails spatially modulated fluorescence depletion to enable effective excitation/fluorescent volumes with size below the diffraction limit of the light. The fluorescence depletion may be achieved using continuous wave (CW) or pulsed lasers. The first implementations of 2PE-STED microscopy employed CW lasers for depletion and a mode-locked Ti:Sapphire laser for 2PE.5–7 While the use of a CW laser for STED (CW-STED) avoids synchronization challenges and mitigates costs inherent to higher peak-power pulsed lasers, the resulting less effective depletion limits the achievable spatial resolution.6,7 In fact, a higher resolution can be achieved using pulsed STED (pSTED) approaches,8–10 which in the cases of 2PE typically require synchronization of two mode-locked ultrafast lasers at different wavelengths to provide sufficient peak power for both the excitation and the depletion beams. From now on, we refer to these implementations as 2PE-pSTED microscopy.
Several solutions have been proposed to circumvent the archetypal trade-off between the spatial resolution and the cost and complexity of the STED microscope. Single wavelength 2PE-pSTED has been reported for specific orange and red dyes,11,12 by splitting the output of a single ultrafast laser to serve both as excitation and depletion (or STED) beam—with subsequent temporal stretching of the STED beam. However, the use of the same wavelength for excitation and depletion prevents independent spectral optimization of the 2PE and stimulated emission processes, greatly limiting the applicability of this technique. Recently, a 2PE-pSTED microscopy system based on electrically controllable devices allowing simpler and more compact implementation has been developed.13 Likewise, time-gated detection has been employed to reduce the complexity and enhance the resolution in CW-STED microscopy.14,15 This approach, known as gated CW-STED (gCW-STED) microscopy,16,17 allows the reduction of the STED beam intensity, otherwise required to achieve super-resolution, enabling efficient depletion despite the relative low intensity of the CW depletion beam. A major disadvantage of time-gated detection is the reduction of the signal and hence of signal-to-noise/background ratios (SNR and SBR). Even if such loss can be tolerable for single-photon excitation, where the fluorescent signal is stronger (i.e., higher photons flux and budget), it is problematic for 2PE, usually characterized by weaker fluorescence signal. Furthermore, the relatively high anti-Stokes emission background, directly induced by the depletion beam,17 can deteriorate the contrast of the STED image up to completely hiding the expected resolution enhancement. This situation is complicated by the fact that, in contrast to single-photon excitation with confocal detection, the optical sectioning is inherently obtained due to the “nonlinearity” of the excitation process; thus the pinhole can be removed, such as in the nondescanned 2PE microscopy implementations. However, removing the pinhole in the case of 2PE-STED microscopy means collecting the non-negligible out-of-focus anti-Stokes emission background. While various synchronous (i.e., lock-in) detection approaches18–21 have been proposed to “subtract” anti-Stokes emission background, such strategies amplify noise and may fail in conditions of low photon-budget such as 2PE microscopy.
Here we demonstrate how the separation of photons by lifetime tuning (SPLIT) method can enhance the resolution of 2PE CW-STED microscopy. Application of SPLIT requires a time-resolved STED image, i.e., an STED image where the nanosecond fluorescence intensity decay (i.e., the fluorescence lifetime) is recorded at each pixel.22 In CW-STED, the lifetime of the fluorophores in the periphery of the point spread function (PSF)/fluorescent volume is always shorter than the lifetime of those located in the center of the PSF. Thus, an improvement of spatial resolution can be obtained by the extraction, at each pixel, of the longer fluorescence lifetime component of the signal. To extract this component, SPLIT uses the well-known phasor approach to fluorescence lifetime imaging analysis.23 In the phasor approach, the fluorescence temporal decay of each pixel of the image is transformed from time to frequency domain and represented as a vector known as a phasor. Owing to the inherent linearity of the phasor plot, the phasor at each pixel is expressed as the combination of a phasor of longer lifetime (corresponding to the center of the PSF) and a phasor of shorter lifetime (corresponding to the periphery of the PSF). Thus, the SPLIT approach enables a robust sorting of the photons emitted from fluorophores located at the center of the PSF, based on differences in the fluorescence lifetime. This method of frequency domain sorting avoids the need for fitting procedures inherent to photon sorting in the time domain.
The SPLIT approach has already been applied to enhance resolution in single-photon CW-STED microscopy24,25 and fluctuation correlation spectroscopy26 and, more recently, to pSTED microscopy.27 Herein, we explore via the SPLIT method the spatially controlled gradients in fluorescence lifetime generated by the doughnut-shaped STED beam in a 2PE CW-STED microscope. In particular, to enhance SNR and SBR, the SPLIT approach implements an intelligent algorithm to simultaneously remove the STED background signal as well as the early photons originating from molecules at the periphery of the effective fluorescent volume, thus not effectively depleted/quenched. Early photons stemming from the center of the excitation volume (useful because carrying super-resolved information content), which would partially be discarded using time-gated detection, are retained. We exemplify this method through imaging subcellular structures and comparing the algorithm to other time-resolved STED microscopy approaches (i.e., based on photon-arrival times) for 2PE-STED optimization. Image quality is further enhanced through subsequent deconvolution28 on the SPLIT image to maximally exploit temporal and spatial information.
2. Methods
2.1. Two-Photon Excitation Continuous Wave-Stimulated Emission Depletion Microscope
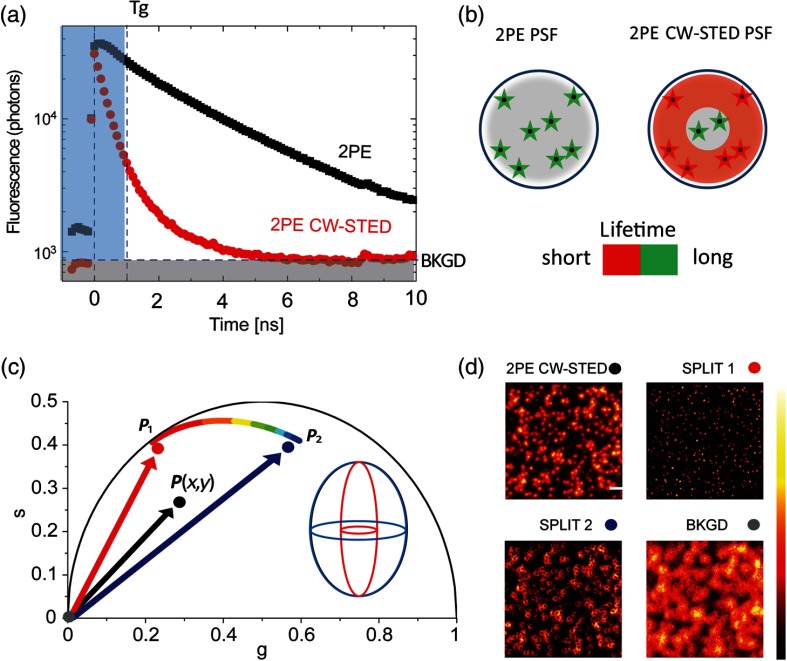
The experimental microscopy setup was described in a previous work.29,30 Briefly, a custom single-photon excitation STED microscope was adapted for 2PE. The doughnut-shaped intensity focal distribution of the STED beam was coaligned with the classical Gaussian-shaped focus of the excitation (single- or two-photon) beam of a scanning microscope. Axial compensation using beams collimators was needed due to longer wavelength separations between the 2PE and STED beams. The role of the STED beam is to de-excite, via stimulated emission, all the fluorophores that have been previously excited by the excitation beam, except those located near the doughnut’s center—or “zero”-intensity point. This condition permits the observation of a subdiffracted volume within the sample. Scanning this volume across the sample generates images with spatial resolution beyond the Abbe’s diffraction limit. The resolution of an STED microscope is function of the intensity of the STED beam: the higher the peak intensity, the higher the fluorescent confinement and the subsequent spatial resolution. In CW-STED microscopy, the resolution enhancement may also be achieved through the analysis of fluorescence dynamics (or fluorescence lifetime), without increasing STED beam intensity. Figure 1(a) illustrates temporal decay on fluorescent beads of 2PE and 2PE CW-STED fluorescence signals acquired with a time-correlated single-photon counting (TCSPC) card. In particular, the figure shows the histogram of the photon-arrival times for all the photons collected during the scanning. While, the 2PE signal is well described by a single-exponential decay, the 2PE CW-STED counterpart is more convoluted. This behavior depends on the fluorescence lifetime distributions of the fluorophore within the PSF. Indeed, the fluorescence lifetime is uniform along the PSF in 2PE microscopy while a gradient occurs with 2PE CW-STED microscopy, with the shortest fluorescence lifetime occurring in the periphery of the PSF corresponding with the maximum STED intensity in the doughnut profile [Fig. 1(b)]. A longer lifetime occurs in the center of PSF, corresponding to the zero intensity of the STED beam. The temporal dynamics in the center of the PSF of the 2PE CW-STED microscope are similar to those obtained with conventional 2PE microscopy. Such temporal information may be exploited to enhance resolution in gated STED microscopy.16 By discarding (filtering) early arriving photons, which mostly arise from the periphery where the fluorescent lifetime is short, it is possible to specifically select the fluorescent signal emanating from the center of the PSF (where the fluorescent lifetime is longer), effectively narrowing the PSF.
Fig. 1.
Principle of 2PE CW-STED SPLIT microscopy. (a) Temporal information demonstrating the fluorescence lifetime of a fluorescent sample following excitation by means of 2PE or 2PE CW-STED. Early photons (blue) and background (BKGD) (gray) are usually discarded. (b) Schematic representation of the spatial distribution of fluorescence lifetimes in the 2PE PSF (left) and in the 2PE CW-STED PSF (right). (c) Principle of the SPLIT illustrated using a phasor plot. Photons are separated based on their dynamics, represented as vectors in the phasor plot. The experimental phasor , describing the temporal information of the signal at each pixel, is used to obtain the final image. The first component (red) corresponds to the center of the PSF and contains super-resolution information, whereas the second component (blue) corresponds to the PSF periphery. The background signal is located at the origin. (d) Representation of the SPLIT approach. The images of SPLIT, respectively, represent raw 2PE CW-STED image (super-resolution information + early photons + background signal), SPLIT 1 (super-resolution information), SPLIT 2 (information discarded), and background. Color dots correspond to the definitions mentioned in panel (c).
Through a similar reasoning, temporal dynamics may also be exploited to remove anti-Stokes emission background. Indeed, in a CW-STED microscope, the anti-Stokes emission background is uncorrelated with respect to the fluorescence signal generated by the excitation beam. Such unwanted signal may arise at the periphery of the PSF where the STED beam has a non-negligible probability of exciting the fluorophores, effectively deteriorating the final image resolution. In CW-STED, the anti-Stokes emission adds to the fluorescence temporal decay as uncorrelated background uniformly distributed across the pulse-to-pulse excitation interval. This background may be readily removed, provided the photon arrival times are known.20
2.2. Time-Resolved Two-Photon Excitation Continuous Wave-Stimulated Emission Depletion Microscope with Separation of Photons by Lifetime Tuning
The elegant realization of time-gated detection and background subtraction to enhance the contrast is limited by the reduction of fluorescence signal. For this reason, a more robust implementation is needed. However, decoding spatial information in STED microscopy through arrival-time-based photon sorting is challenging in the time domain. Such decoding may be more readily achieved in the frequency domain via fast Fourier transformation (FFT). Herein, the challenge of increasing the spatial resolution of an optical microscope is effectively addressed within the realm of molecular spectroscopy.
In SPLIT−STED, a significant enhancement of spatial resolution is achieved by the frequency-domain analysis of the temporal decay at each pixel. The SPLIT method entails the separation of time-resolved fluorescence components arranging photons according to the spatial position from which they originated. Given that a different position within the (excitation) PSF corresponds to different phasor, herein we use the linear properties of the phasors to (1) extract components having longer lifetimes, as they are likely to be originated from the center of the volume under investigation, and (2) to remove uncorrelated background signal. In this way, photons emitted from the center of the PSF are separated from the rest without losses. In contrast, time-gated detection and background subtraction always result in some discard of fluorescent photons from the center of the PSF, reducing the SNR of the final super-resolved image.
We have already demonstrated the general theory of SPLIT for resolution enhancement in conventional (i.e., single-photon excitation) STED microscopy.24 This approach can be readily extended to 2PE CW-STED microscopy, as it depends solely on fluorescence temporal decay and not on the excitation mechanism. Here, the principal equations governing SPLIT implementation in CW-STED microscopy are summarized. The time-dependent fluorescence signal at each pixel is obtained by integrating the contribution of all fluorophores located in the effective PSF (E-PSF) centered at pixel position .24 The expression for the fluorescence signal reads as
| (1) |
where describes the concentration of the fluorophores in a concentric region of radius around the pixel position, is the decay rate, is the waist of the E-PSF, and is a constant that depends on the quantum yield of the fluorophore, the maximum of the excitation intensity, and the detection efficiency. The term represents the contribution of uncorrelated background (i.e., anti-Stokes emission) to the fluorescence signal. Owing to the doughnut-shaped (approximated by a parabolic function) STED beam, the decay rate distribution is given as
| (2) |
where is the unperturbed excited-state lifetime and is the ratio between (the value of STED intensity at radial position ) and the saturation value for which the probability of decay due to stimulated and spontaneous emissions are equal. Notably is a constant that quantifies the relative variation of decay rate values within the E-PSF of the 2PE CW-STED microscope, at a given STED power. For instance, if , the lifetime at radial position is , whereas at the center () is .
To find an approximated, explicit expression for the temporal decay of fluorescence emitted from the center of the detection volume, we split the integral into parts (SPLIT 1 and SPLIT 2).
| (3) |
where and represent the two temporal decay patterns associated with the center and with the periphery of the PSF, respectively, and and are the intensity fractions corresponding to the center and to the periphery of the PSF, respectively. The temporal decay patterns and can be obtained from knowledge of the decay rate distribution [Eq. (2)], i.e., from knowledge of the unperturbed excited-state lifetime and the constant .24 The parameter depends on the specific fluorophore and can be measured from the sample. The parameter is proportional to the STED beam power, and its value depends on the optical configuration and on the properties of the sample. We have shown that, within the same theoretical framework, we can estimate the value of from the very same image by considering the average time-resolved decay of all the pixels of an image,24
| (4) |
where and and are constants. This equation is an approximation of the average STED decay and the nonexponential behavior is due to the continuous distribution of decay rates induced by the STED beam.
In practice, applications of SPLIT require measurement of only two parameters ( and ). The fluorescence lifetime for a given fluorophore is often known or may be readily measured from 2PE images of the sample by switching off the STED beam, and the constant can be retrieved from a fit of the average temporal decay of the STED image.24 Finally, we make use of the phasor analysis of fluorescence lifetime to identify temporal dynamics associated with photons emanating from the center and periphery of the PSF, as two vectors in the phasor plot. The parameters and are sufficient to calculate the positions of the phasors and , which correspond to the temporal decays and associated with the center and periphery of the PSF, respectively [Fig. 1(c)]. Then the dynamics at each pixel, represented as a phasor , is used to calculate the fractions of photons associated with the center of the PSF, with its periphery, and with the background. Owing to the linear properties of the phasors, photons emitted from the center of the PSF (i.e., ) may be efficiently sorted from those lacking super-resolution information. In comparison to gated detection alone, this approach enables automatic removal of noncorrelated background, together with improved spatial resolution without attenuation of the super-resolution signal [Fig. 1(d)].
A well-known approach to increase the SNR of a microscope image is to process it with a deconvolution algorithm.31 But, in contrast to deconvolution approaches that use spatial information to improve resolution, SPLIT does not require prior knowledge of the effective PSF of the CW-STED system. However, further image enhancement may be achieved when both spatial information and temporal information are combined (i.e., deconvolution and SPLIT). The fact that SPLIT preserves linearity and space invariantly on the resulting imaging system makes it possible to successively apply image deconvolution. To this end, we deconvolve the SPLIT images using the well-known iterative Richardson–Lucy algorithm:
| (5) |
where denotes the restored images at the ’th iteration; denotes the image; is the notation for the discretization of the convolution operator associated to the PSF . In this scenario, good results were obtained with a Gaussian PSF with full width at half maximum extracted directly from the image using a method based on the Fourier ring correlation (FRC), which was previously described in Refs. 32 and 33. Finally, the software used for all the image processing was developed in MATLAB (MathWorks) using the built-in implementation of the FFT to speed up the processing time.
3. Results and Discussion
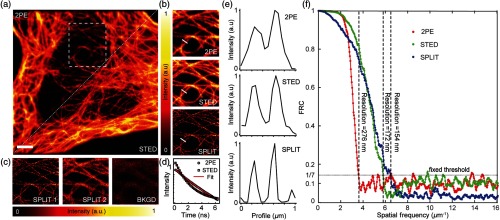
The 2PE and 2PE CW-STED images were obtained from fluorescent beads [Fig. 1(d)] and biological samples. Figure 2 demonstrates 2PE and 2PE CW-STED images with and without enhanced resolution using temporal information (SPLIT). Background signal induced by the STED beam is noted to negatively impact the quality of 2PE CW-STED images. Background-free super-resolution images of subcellular structures were subsequently obtained using the SPLIT method. Intensity profiles along close-packed structures in the sample clearly show enhancement in the resolution [Fig. 2(e)]. Herein, super-resolution enhancement is achieved using only the temporal information at each pixel, with concurrent isolation of uncorrelated background signal.
Fig. 2.
Comparison between 2PE and 2PE CW-STED imaging of microtubules in a fixed HeLa cell. (a) Basal membrane imaged with 2PE (left-top corner) and 2PE CW-STED (right-bottom corner) microscopy. (b) Magnified views of the boxed area in (a) with respective intensity profiles corresponding to the white bars. (c) Images corresponding to the PSF center (SPLIT 1), PSF periphery (SPLIT 2), and uncorrelated background (BKGD) obtained by application of the SPLIT algorithm. (d) Average fluorescence intensity decay for the 2PE and 2PE-STED data. The solid lines are fit of the data to Eq. (4) (2PE, , ; 2PE-STED, , ). (e) Intensity profiles along the dashed line in (e). (f) FRC of 2PE, 2PE CW-STED, and 2PE CW-STED SPLIT images in (a). This method is based on the acquisition of two independent images. These were obtained by sorting photons according to their arrival times. , , , . Laser powers were measured in the back focal plane of the objective lens. Scale bars: .
The advantages of the SPLIT method include resolution enhancement without the need for higher STED beam intensity and reduced system costs and complexity, compared to pSTED setups. Nonetheless, the TCSPC hardware to track photon arrival times is required to perform the SPLIT method; it is also necessary to collect enough photons to allow the subsequent splitting of the obtained STED images. In theory, the spatial resolution can be increased indefinitely by increasing the number of resolved components up to a maximum, predictable number, only limited by the amount of noise. In practice, usually the collected signal is sufficient to separate the STED image only in two components (SPLIT 1 and SPLIT 2). Also, pixel size and bin time were carefully selected in our experiment for the same reason. One common limitation of postprocessing techniques is the formation of artifacts. In our case, the absence of artifacts was verified with fluorescent beads and qualitative side-by-side comparison between the 2PE and SPLIT 2PE CW-STED images [Fig. 2(b)]. To estimate the performance of the SPLIT 2P-CW-STED system, we calculated image resolution using FRC. The FRC approach estimates image resolution by measuring the degree of similarity of two independent reconstructions of the same object.32,34 In our case, the two independent reconstructions were obtained from a given time-resolved STED image, by considering either even or odd temporal bins. This resulted in two independent, time-resolved STED images of the same object.
Figure 2(f) illustrates the respective FRC curves of images shows in Fig. 2(a). When STED is applied, higher spatial frequencies than those present in the 2PE image occur. In this example, the cutoff frequency and effective resolution are 276, 172, and 154 nm, respectively, for 2PE, 2PE CW-STED, and 2PE CW-STED SPLIT microscopes. Higher frequencies for 2PE CW-STED images are encoded in the temporal channel and are recovered using the SPLIT approach. It is worth noting that, each of the two time-resolved STED images used for FRC contains only half the photon counts compared to the original image. Thus, as the FRC method is applied on images containing half of the photons of the original image, it may underestimate the effective cutoff frequency (i.e., resolution). Further, the SPLIT method is also very sensitive to the SNR of the input image, and herein it is applied on the (half-photon) images separately.
It is well known that various postprocessing techniques may be employed to enhance effective resolution in STED microscopy.3,30 Such approaches improve the resolution but often require considerable postprocessing time. One must balance the degree of effective image resolution improvement against the cost, speed, and complexity of various image enhancement approaches. Herein, we provide a survey of existing postprocessing techniques that exploit photon arrival time to enhance STED microscopy images. Figure 3 demonstrates a panel with several postprocessing images of sparse cytoskeletal structures, highlighting the utility of temporal dynamics in STED microscopy. It is interesting to compare this SPLIT approach with time-gated detection and filtering approaches, where the image is obtained by removing the early arriving photons and subtracting the background (properly weighted).20 Even if the early photons and background signal may have useful informative content, the usage of these latter approaches requires them to be completely discarded to obtain the final super-resolved image. This can result in unsuitable final images due to a very poor SNR. In this scenario, the SPLIT method leads to superior results compared to what obtained with gCW-STED microscopy and filtered gCW-STED microscopy.20 Similar results were obtained using SPLIT and multi-image deconvolution (MID),29,35 due to the ability of these methods to recover the high frequencies contained solely in the late-photons image, without completely discarding the information contained in early ones. While gCW-STED microscopy with or without filtering may be implemented using a purely hardware approach,21,36 MID and SPLIT approaches rely exclusively on postprocessing. Table 1 summarizes the advantages and limitations of the approaches discussed herein.
Fig. 3.
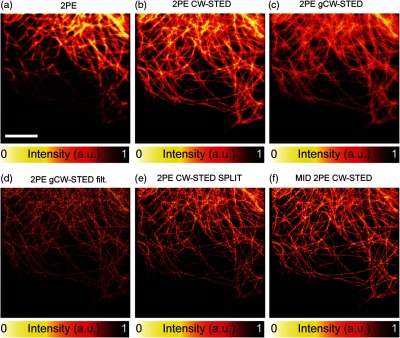
(a)–(f) Examples of different postprocessing techniques on microtubules in a fixed HeLa cell labeled with Alexa 488. The presence of STED-induced background (uncorrelated) negatively impacts the 2PE CW-STED image; the improvement is effective when this background is efficiently removed. The SPLIT technique has the best performance, similar to that obtained with MID. and . Scale bars: .
Table 1.
Comparison of different postprocessing techniques in the context of CW-STED microscopy.
| CW-STED | gCW-STED | gCW-STED filt. | MID CW-STED | SPLIT CW-STED | |
|---|---|---|---|---|---|
| Principle | PSF shrinking by stimulated emission depletion | PSF shrinking by stimulated emission depletion and time-gating detection | PSF shrinking by stimulated emission depletion and time-gating detection; filtering for background removal | PSF shrinking by stimulated emission depletion and MID (background is taken into account and removed) | PSF shrinking by stimulated emission depletion and SPLIT (background is estimated and removed) |
| Lateral resolution | 20 to 50 nm3 | 17 | 20 | 35 | 24 |
| Speed | High | High | High | Middle | Middle |
| Power required for the sample (mW) | |||||
| Advantages | Simplicity and cost | Enhanced on–off contrast using moderate light intensities | Enhanced on–off contrast using moderate light intensities | Enhanced SNR | Enhanced SNR |
| Drawbacks | High photobleaching | Low SNR may negate benefit | Low SNR may negate benefit | Complex data processing with possible artifacts | Complex data processing with possible artifacts |
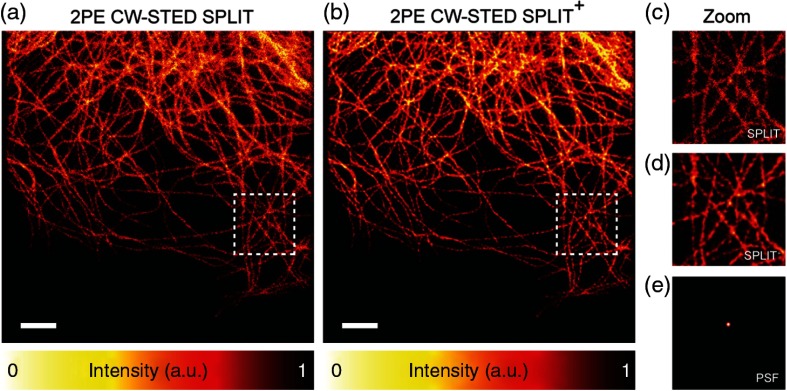
As said before, a further enhancement of SPLIT images is possible by exploiting spatial information through deconvolution algorithms. Deconvolution is a well-established technique to reduce blurring and improve SNR and hence effective resolution of STED microscopy. In contrast to subtractive methods, the solution is constrained to non-negative values and the noise, if properly modeled and under the right conditions, is not amplified. Figure 4 demonstrates the value added by deconvolution to SPLIT-STED images, exemplifying the utility of exploiting simultaneously temporal and spatial information in postprocessing optimization schemes. The application of deconvolution immediately after the SPLIT method is possible as this last method preserves the properties of linearity and space invariance of the system, and thus a specific PSF can be assigned the resulting image.24 Future work to improve SNR on CW-STED microscopy should directly employ the SPLIT method to generate a temporal PSF or using SPLIT 1 and SPLIT 2 images into a more efficient MID method described in Ref. 35.
Fig. 4.
(a), (b) 2PE CW-STED SPLIT imaging of -tubulin cells labeled with Alexa 488. The quality of the images is further enhanced applying deconvolution (Richardson–Lucy) on the SPLIT image. (c)–(e) The magnified views of the marked area. (e) The PSF imaging used on the deconvolution. Scale bars: .
In this work, a high numerical aperture objective lens designed for STED microscopy was used to achieve super-resolution imaging. Enhancement of resolution was observed on the apical membrane of the cells above the basal membrane. However, imaging with a lower numerical aperture objective lens suitable for water immersion is preferable in case of applications were imaging deeper on thick samples like tissue is needed. A good balance between high resolution and sample penetration may be achieved with a glycerol objective with correction collar.37 The correction collar is used to compensate differences in refractive indices between the coverslip/immersion liquid/samples as the relative contributions of these changes when the imaging -depth is changed. When working in aqueous medium, the use of water immersion objectives in combination with a spatial light modulator (SLM) allows STED imaging at great depths. Further, the use of SLM can correct aberration and alleviate degradation of the PSF deeper into the sample.38
The SPLIT technique provides a strategy to obtain optimized 2PE CW-STED images, but this might also result in useful postprocessing optimization of images obtained using 2PE pSTED approaches.24 Further studies will focus in the application of the SPLIT approach with the use of new emerging high photon collection efficiency () time-resolved card without the dead time typical of conventional TCSPC approach.24 Further, relatively inexpensive externally synchronized subnanosecond fiber lasers with low peak power may be employed in 2PE pSTED microscopy; their use has been shown to reduce photobleaching in pSTED.39,40
Even though STED microscopes have been disseminated by laboratories throughout the world, the progress in 2PE-STED microscopy is still limited. Further, no commercial implementation of 2PE STED microscopy is currently available on the market, in part due to high cost and conflicting demands of optical coatings optimized for the visible range for the STED beam and those optimized for the infrared for the 2PE beam. Our aim in this work is to contribute to the development of more efficient multiphoton nanoscopes. We eventually foresee a commercially available 2PE-STED microscope.
4. Conclusions
The spatial resolution of a 2PE CW-STED microscope can be increased by unmixing the detected fluorescence signal into components of different fluorescence lifetime. The SPLIT technique exploits the spatiotemporal information contained in a time-resolved STED image to enhance the performance of 2PE CW-STED microscopes. Herein, the quality of 2PE CW-STED images was improved due to the phasor analysis of fluorescence lifetime variations induced by the donut-shaped STED beam within the detection volume. Super-resolution information was efficiently separated from the signal originating from the periphery of the depletion beam and temporally noncorrelated background signal. Image quality was further enhanced with the use of image deconvolution techniques. The utility of pairing 2PE CW-STED microscopy with SPLIT by imaging of subcellular structures has been demonstrated herein. The improved efficiency of SPLIT enables the use of a lower power depletion beam, compared to conventional CW-STED approaches, potentially decreasing the photobleaching rate. This approach may encourage a broader implementation of 2PE STED microscopy in biology for deep super-resolution imaging of thick samples and could be achieved by modification of existing commercial 2PE microscopes. To achieve even greater imaging depths, hardware modifications, including adaptive optics and objective lenses with long working distances will be required.
Acknowledgments
This study was supported by a generous donation from the Berthiaume Family (I.C.H and N.J.).
Biographies
Iván Coto Hernández studied nuclear physics at the Higher Institute of Technologies and Applied Sciences, Cuba. Later, he enrolled the PhD program in the Italian Institute of Technology (Italy) and a postdoc in the Université Paris Sud, France. Currently, he works as an instructor at Massachusetts Eye and Ear and Harvard Medical School. His main interests include super resolution and nonlinear microscopy.
Luca Lanzanò obtained his PhD in physics at the University of Catania. As a postdoc, he worked on the development of fluorescence-based methods to measure protein dynamics and interactions in live cells (Laboratory for Fluorescence Dynamics, Irvine). Since 2013, he has been a researcher at the Istituto Italiano di Tecnologia. His main interests include the development and application of super-resolution methods and the investigation of molecular mechanisms of cancer formation.
Giuseppe Vicidomini studied computer science at the University of Genoa, where, in 2008, he received his PhD. From 2008 to 2011, he worked as a postdoctoral fellow at the Department of NanoBiophotonics, MPI for Biophysical Chemistry. Since May 2011, he has worked at the Istituto Italiano di Tecnologia, where, in May 2016, he became principal investigator of the Molecular Microscopy and Spectroscopy research line. His research interests are primarily concentrated on theoretical design, development, and validation of novel optical and analytical tools that allow the modern biologists to peer inside living biological systems with unprecedented temporal/spatial abilities and massive information content.
Biographies of the other authors are not available.
Disclosures
No conflicts of interest are declared by the authors.
Contributor Information
Iván Coto Hernández, Email: Ivan_CotoHernandez@MEEI.HARVARD.EDU.
Marco Castello, Email: marco.castello@iit.it.
Giorgio Tortarolo, Email: giorgio.tortarolo@iit.it.
Nate Jowett, Email: Nate_Jowett@MEEI.HARVARD.EDU.
Alberto Diaspro, Email: alberto.diaspro@iit.it.
Luca Lanzanò, Email: luca.lanzano@iit.it.
Giuseppe Vicidomini, Email: Giuseppe. Vicidomini@iit.it.
References
- 1.Hell S. W., Wichmann J., “Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy,” Opt. Lett. 19, 780–782 (1994). 10.1364/OL.19.000780 [DOI] [PubMed] [Google Scholar]
- 2.Bianchini P., et al. , “STED nanoscopy: a glimpse into the future,” Cell Tissue Res. 360, 143–150 (2015). 10.1007/s00441-015-2146-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vicidomini G., Bianchini P., Diaspro A., “STED super-resolved microscopy,” Nat. Methods 15, 173–182 (2018). 10.1038/nmeth.4593 [DOI] [PubMed] [Google Scholar]
- 4.Denk W., Strickler J. H., Webb W. W., “Two-photon laser scanning fluorescence microscopy,” Science 248, 73–76 (1990). 10.1126/science.2321027 [DOI] [PubMed] [Google Scholar]
- 5.Moneron G., Hell S. W., “Two-photon excitation STED microscopy,” Opt. Express 17, 14567–14573 (2009). 10.1364/OE.17.014567 [DOI] [PubMed] [Google Scholar]
- 6.Ding J. B., Takasaki K. T., Sabatini B. L., “Supraresolution imaging in brain slices using stimulated-emission depletion two-photon laser scanning microscopy,” Neuron 63, 429–437 (2009). 10.1016/j.neuron.2009.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q., et al. , “2PE-STED microscopy with a single Ti:sapphire laser for reduced illumination,” PLoS One 9, e88464 (2014). 10.1371/journal.pone.0088464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Q., Wu S. S. H., Chou K. C., “Subdiffraction-limit two-photon fluorescence microscopy for GFP-tagged cell imaging,” Biophys. J. 97, 3224–3228 (2009). 10.1016/j.bpj.2009.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bethge P., et al. , “Two-photon excitation STED microscopy in two colors in acute brain slices,” Biophys. J. 104, 778–785 (2013). 10.1016/j.bpj.2012.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takasaki K. T., Ding J. B., Sabatini B. L., “Live-cell superresolution imaging by pulsed STED two-photon excitation microscopy,” Biophys. J. 104, 770–777 (2013). 10.1016/j.bpj.2012.12.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scheul T., et al. , “Two-photon excitation and stimulated emission depletion by a single wavelength,” Opt. Express 19, 18036–18048 (2011). 10.1364/OE.19.018036 [DOI] [PubMed] [Google Scholar]
- 12.Bianchini P., et al. , “Single-wavelength two-photon excitation-stimulated emission depletion (SW2PE-STED) superresolution imaging,” Proc. Natl. Acad. Sci. U.S.A. 109, 6390–6393 (2012). 10.1073/pnas.1119129109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otomo K., et al. , “Advanced easySTED microscopy based on two-photon excitation by electrical modulations of light pulse wavefronts,” Biomed. Opt. Express 9, 2671–2680 (2018). 10.1364/BOE.9.002671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moffitt J. R., Osseforth C., Michaelis J., “Time-gating improves the spatial resolution of STED microscopy,” Opt. Express 19, 4242–4254 (2011). 10.1364/OE.19.004242 [DOI] [PubMed] [Google Scholar]
- 15.Vicidomini G., et al. , “STED nanoscopy with time-gated detection: theoretical and experimental aspects,” PLoS One 8, e54421 (2013). 10.1371/journal.pone.0054421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vicidomini G., et al. , “Sharper low-power STED nanoscopy by time gating,” Nat. Methods 8, 571–573 (2011). 10.1038/nmeth.1624 [DOI] [PubMed] [Google Scholar]
- 17.Vicidomini G., et al. , “Gated CW-STED microscopy: a versatile tool for biological nanometer scale investigation,” Methods 66, 124–130 (2014). 10.1016/j.ymeth.2013.06.029 [DOI] [PubMed] [Google Scholar]
- 18.Vicidomini G., et al. , “STED with wavelengths closer to the emission maximum,” Opt. Express 20, 5225–5236 (2012). 10.1364/OE.20.005225 [DOI] [PubMed] [Google Scholar]
- 19.Ronzitti E., Harke B., Diaspro A., “Frequency dependent detection in a STED microscope using modulated excitation light,” Opt. Express 21, 210–219 (2013). 10.1364/OE.21.000210 [DOI] [PubMed] [Google Scholar]
- 20.Coto Hernández I., et al. , “A new filtering technique for removing anti-Stokes emission background in gated CW-STED microscopy,” J. Biophotonics 7, 376–380 (2014). 10.1002/jbio.v7.6 [DOI] [PubMed] [Google Scholar]
- 21.Castello M., et al. , “Removal of anti-Stokes emission background in STED microscopy by FPGA-based synchronous detection,” Rev. Sci. Instrum. 88, 053701 (2017). 10.1063/1.4983082 [DOI] [PubMed] [Google Scholar]
- 22.Lanzanò L., et al. , “Role of the pico-nano-second temporal dimension in STED microscopy,” in Perspectives on Fluorescence, Jameson D., Ed., Springer Series on Fluorescence (Methods and Applications), Vol. 17, pp. 311–329, Springer, Cham: (2016). [Google Scholar]
- 23.Digman M. A., et al. , “The phasor approach to fluorescence lifetime imaging analysis,” Biophys. J. 94, L14–L16 (2008). 10.1529/biophysj.107.120154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanzanò L., et al. , “Encoding and decoding spatio-temporal information for super-resolution microscopy,” Nat. Commun. 6, 6701 (2015). 10.1038/ncomms7701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarmento M. J., et al. , “Exploiting the tunability of stimulated emission depletion microscopy for super-resolution imaging of nuclear structures,” Nat. Commun. 9, 3415 (2018). 10.1038/s41467-018-05963-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanzanò L., et al. , “Measurement of nanoscale three-dimensional diffusion in the interior of living cells by STED-FCS,” Nat. Commun. 8, 65 (2017). 10.1038/s41467-017-00117-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tortarolo G., et al. , “Photon-separation to enhance the spatial resolution of pulsed STED microscopy,” Nanoscale 11, 1754–1761 (2019). 10.1039/C8NR07485B [DOI] [PubMed] [Google Scholar]
- 28.Bertero M., et al. , “Image deblurring with Poisson data: from cells to galaxies,” Inverse Probl. 25, 123006 (2009). 10.1088/0266-5611/25/12/123006 [DOI] [Google Scholar]
- 29.Coto Hernández I., et al. , “Two-photon excitation STED microscopy with time-gated detection,” Sci. Rep. 6, 19419 (2016). 10.1038/srep19419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coto Hernández I., et al. , “Improving multiphoton STED nanoscopy with separation of photons by lifetime tuning (SPLIT),” Proc. SPIE 10498, 104982U (2018). 10.1117/12.2286912 [DOI] [Google Scholar]
- 31.Zanella R., et al. , “Towards real-time image deconvolution: application to confocal and STED microscopy,” Sci. Rep. 3, 2523 (2013). 10.1038/srep02523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tortarolo G., et al. , “Evaluating image resolution in stimulated emission depletion microscopy,” Optica 5, 32–35 (2018). 10.1364/OPTICA.5.000032 [DOI] [Google Scholar]
- 33.Koho S. V., et al. , “Fourier ring correlation simplifies image restoration in fluorescence microscopy,” Nat. Commun. 10, 3103 (2019). 10.1038/s41467-019-11024-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banterle N., et al. , “Fourier ring correlation as a resolution criterion for super-resolution microscopy,” J. Struct. Biol. 183, 363–367 (2013). 10.1016/j.jsb.2013.05.004 [DOI] [PubMed] [Google Scholar]
- 35.Castello M., Diaspro A., Vicidomini G., “Multi-images deconvolution improves signal-to-noise ratio on gated stimulated emission depletion microscopy,” Appl. Phys. Lett. 105, 234106 (2014). 10.1063/1.4904092 [DOI] [Google Scholar]
- 36.Coto Hernández I., et al. , “Gated STED microscopy with time-gated single-photon avalanche diode,” Biomed. Opt. Express 6, 2258 (2015). 10.1364/BOE.6.002258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urban N. T., et al. , “STED nanoscopy of actin dynamics in synapses deep inside living brain slices,” Biophys. J. 101, 1277–1284 (2011). 10.1016/j.bpj.2011.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heine J., et al. , “Three dimensional live-cell STED microscopy at increased depth using a water immersion objective,” Rev. Sci. Instrum. 89, 053701 (2018). 10.1063/1.5020249 [DOI] [PubMed] [Google Scholar]
- 39.Oracz J., et al. , “Photobleaching in STED nanoscopy and its dependence on the photon flux applied for reversible silencing of the fluorophore,” Sci. Rep. 7, 11354 (2017). 10.1038/s41598-017-09902-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castello M., et al. , “Gated-STED microscopy with subnanosecond pulsed fiber laser for reducing photobleaching,” Microsc. Res. Tech. 79, 785–791 (2016). 10.1002/jemt.v79.9 [DOI] [PubMed] [Google Scholar]