Summary
Cellular responses to transforming growth factor β (TGF-β) depend on cell context. Here, we explored how TGF-β/nodal signaling crosstalks with the epigenome to promote mesendodermal differentiation. We find that expression of a group of mesendodermal genes depends on both TRIM33 and nodal signaling in embryoid bodies (EBs) but not in embryonic stem cells (ESCs). Only in EBs, TRIM33 binds these genes in the presence of expanded H3K18ac marks. Furthermore, the H3K18ac landscape at mesendodermal genes promotes TRIM33 recruitment. We reveal that HDAC1 binds to active gene promoters and interferes with TRIM33 recruitment to mesendodermal gene promoters. However, the TRIM33-interacting protein p300 deposits H3K18ac and further enhances TRIM33 recruitment. ATAC-seq data demonstrate that TRIM33 primes mesendodermal genes for activation by maintaining chromatin accessibility at their regulatory regions. Altogether, our study suggests that HDAC1 and p300 are key factors linking the epigenome through TRIM33 to the cell context-dependent nodal response during mesendodermal differentiation.
Keywords: nodal signaling, histone acetylation, chromatin accessibility, TRIM33, mesendodermal differentiation, embryonic stem cells
Highlights
-
•
The H3K18ac landscape changes at gene promoters during mesendodermal differentiation
-
•
Histone acetylation facilitates TRIM33 recruitment to the chromatin
-
•
HDAC1 and p300 are the key factors for nodal signaling crosstalk with epigenome
-
•
Chromatin accessibility at mesendodermal genes depends on TRIM33
Xi and colleagues explored how nodal signaling crosstalks with epigenome to specifically promote mesendodermal differentiation. They showed that TRIM33 specifically regulates the expression of a group of mesendodermal genes upon nodal signaling by maintaining the H3K18ac mark, and that HDAC1 and p300 are the key factors linking epigenome to the cell context-dependent nature of nodal responses during mesendodermal differentiation.
Introduction
The transforming growth factor β (TGF-β) signaling pathway plays important roles in embryogenesis, tissue homeostasis, immune responses, and wound healing (Chen and Ten Dijke, 2016, David and Massague, 2018, Flavell et al., 2010, Massague, 2008, Sanjabi et al., 2017, Shen, 2007). Defective TGF-β signaling often causes developmental diseases and cancer (David and Massague, 2018, van der Kraan, 2017). Moreover, depletion of key components of the TGF-β pathway leads to embryonic lethality in knockout mouse models, demonstrating an essential role of the pathway in development (Arnold and Robertson, 2009, Derynck and Akhurst, 2007). For example, Nodal, a secreted protein from the TGF-β superfamily, promotes mesendodermal gene expression during gastrulation and embryonic stem cell (ESC) differentiation in vitro (Arnold and Robertson, 2009, Hill, 2017, Robertson, 2014, Wang et al., 2017b, Watabe and Miyazono, 2009, Wei and Wang, 2017). TGF-β signaling is executed by activating receptor-activated Smad (R-Smad) transcription factors, which in turn modulate the expression of specific target genes (Gaarenstroom and Hill, 2014, Massague, 2012). R-Smad, as the central transcription factor of TGF-β signaling, not only recruits cell-type-specific master transcription factors and coactivators but also interacts with epigenetic regulators to shape the transcriptome (Massague et al., 2005, Mullen et al., 2011). Many studies have shown that epigenetic regulation is an integral part of the TGF-β pathway that orchestrates transcription factor R-Smads and the epigenome to regulate development (Bai and Xi, 2018, Mullen and Wrana, 2017, Oshimori and Fuchs, 2012).
A specific cell type is associated with a unique epigenetic status during embryogenesis according to the epigenetic landscape model of development proposed by Waddington (1957). The epigenetic landscape or epigenome of a cell type generally refers to DNA methylation, histone modifications, and nucleosome positions on its chromatin. The epigenome is established and maintained by chromatin modifiers and remodelers, which in general are recruited to the chromatin by specific transcriptional factors in response to cell signaling (Badeaux and Shi, 2013). The resulting epigenomic alterations cause a dramatic change in chromatin accessibility, which in turn determines the expression of a specific set of genes. How TGF-β signaling crosstalks with the epigenome to regulate gene expression during development has not been well elucidated.
TRIM33 is a nodal signaling-specific chromatin reader and has been found to bind histone marks H3K9me3 and H3K18ac through its PHD-Bromo cassette to regulate the expression of mesendodermal master regulator genes (Xi et al., 2011). Therefore, we posit that TRIM33 interacts specifically with the epigenetic landscape at mesendodermal genes in response to nodal and promotes mesendodermal differentiation. However, TRIM33 is ubiquitously expressed in all tissues, and has been shown to be involved in diverse processes such as transcriptional activation, repression, elongation, and post-translational modification of proteins (Agricola et al., 2011, Bai et al., 2010, Dupont et al., 2005, Tanaka et al., 2018, Wang et al., 2015). The factors that determine this cell-context-dependent nodal response are still elusive.
In this report, we first identified a group of mesendodermal genes whose expression depends on TRIM33-directed nodal signaling. We showed that in embryoid bodies (EBs), which differentiate from ESCs, but not in the ESCs themselves, TRIM33 binds to promoters of target genes and attract to these positions nodal-activated Smad2/3 to form the TRIM33-Smad2/3 complex for transcription activation. We then demonstrated that histone acetylation at histone H3 lysine 18 facilitates TRIM33 recruitment to its target genes. We identified histone deacetylase 1 (HDAC1) and p300 as TRIM33-interacting proteins by affinity purification, and present evidence that they play opposite roles in TRIM33 recruitment by regulating H3K18ac levels. Thus, TRIM33's role in promoting mesendodermal differentiation is facilitated by p300 and suppressed by HDAC1. Finally, we showed that TRIM33-dependent changes in chromatin accessibility are critical for mesendodermal differentiation of mouse ESCs (mESCs). These results strongly suggest that histone modifiers HDAC1 and p300 are the key factors linking TRIM33 to the cell-context-dependent nature of nodal responses during mesendodermal differentiation.
Results
TRIM33 and Smad2/3 Colocalize at Chromatin of Mesendodermal Genes in EBs but Not in ESCs
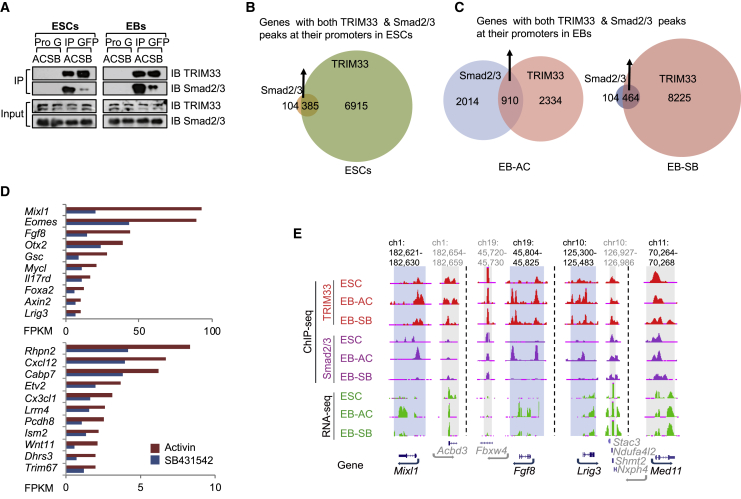
Nodal (TGF-β) signaling is one of the most important signaling pathways that regulates mesendodermal differentiation. Activation of mesendodermal genes generally requires the recruitment of Smad2/3 to their promoters (Estaras et al., 2015, Wang et al., 2017a, Wang et al., 2017b). We found that TRIM33 forms a non-canonical complex with Smad2/3 upon nodal signaling in ESCs and during ESC differentiation to EBs (Figure 1A). To study this further, we used activin A (hereafter, activin) for TGF-β receptor activation, as this ligand is more readily available from mammalian sources than nodal, and a small-molecule inhibitor SB431542 to inhibit TGF-β/activin/nodal pathway (Inman et al., 2002). We then mapped TRIM33 chromatin binding by performing TRIM33 chromatin immunoprecipitation sequencing (ChIP-seq) in ESCs, in activin-treated EBs (hereafter, EB-AC), and in SB431542-treated EBs (hereafter, EB-SB) at day 2.5 (Figures S1A and S1B), and compared them with Smad2/3 ChIP-seq (Wang et al., 2017b). Genome-wide analysis of TRIM33 and Smad2/3 binding revealed largely different cobinding profiles at gene promoter regions (transcription start site [TSS] ±2.5 kb) between EBs and ESCs, with fewer overlapping binding sites in ESCs (385 genes) compared with EB-AC (910 genes) (Figures 1B and 1C; Tables S1 and S2). Almost half of the Smad2/3 and TRIM33 cobinding sites at promoters in EB-AC lost Smad2/3 binding in EB-SB (Figures 1C and S1C), thus their co-occupancy at these sites depended on activin treatment, i.e., activation of TGF-β signaling (Figure 1C).
Figure 1.
TRIM33 and Smad2/3 Colocalize at Chromatin of Mesendodermal Genes in EBs, but Not in ESCs
(A) TRIM33 and Smad2/3 form complex upon nodal signaling both in ESCs and EBs. GFP-TRIM33 overexpressing ESCs or day-2.5 EBs were treated with activin A (hereafter, EB-AC) or SB431542 (hereafter, EB-SB) for 2 h. Cell lysates were immunoprecipitated with anti-GFP-Trap affinity beads, and immune complexes were analyzed by immunoblotting using antibodies against the indicated proteins.
(B and C) Venn diagrams show the overlap of genes with both TRIM33 and Smad2/3 peaks at their promoters in ESCs (B), EB-AC, and EB-SB (C).
(D) Wild-type (WT) and Trim33 null (KO) EBs at day 2.5 were treated with activin A (AC) or SB431542 (SB) for 2 h, then the total RNA was analyzed using RNA-seq. We identified a group of genes whose expression depends on TRIM33-directed nodal signaling, namely genes depending on nodal signaling and TRIM33 (GDNT) (fold change >1.5 for AC versus SB in WT EBs, or >1.5 for WT versus KO in EB-AC, false discovery rate <0.05, fragments per kilobase per million reads [FPKM] >1 in control). Red bar depicts the FPKM of indicated genes in EB-AC. Blue bar depicts the FPKM of indicated genes in EB-SB.
(E) The UCSC browser view shows ChIP-seq signals of Smad2/3 and TRIM33 at the indicated loci and conditions. Different loci are separated by dashed lines. Mixl1, Fgf8, and Lrig3 are nodal target genes, whereas Med11 is not regulated by nodal signaling and is used here as a control. Target regions are shaded blue, control regions are shaded gray, and genes in gray are in the control region. Genome location in kilobases.
See also Figure S1.
Our previous data showed that TRIM33 only regulates a limited number of (only one gene, Lefty1) TGF-β target genes in ESCs (Xi et al., 2011), which suggests that TRIM33 does not play a major role in TGF-β signaling in ESCs. By analyzing RNA sequencing (RNA-seq) of wild-type and Trim33 null EBs, we identified a group of 21 genes that were highly expressed in EBs and whose expression was dependent on both nodal signaling and the presence of TRIM33, which we named the GDNT gene set (Genes Depended on Nodal signaling and TRIM33) (Figures 1D and S1D). Key mesendodermal genes are noted in our GDNT list (Estaras et al., 2015). Both TRIM33 and Smad2/3 were recruited to the GDNT genes, such as Mixl1, Fgf8, and Lrig3, in activin-treated EBs, but not in ESCs (Figures 1E and S1E). Moreover, inhibition of nodal signaling by SB431542 for 2 h had a relatively limited effect on TRIM33 binding to these genes in EBs, whereas it did impair Smad2/3 binding and expression of three nodal target genes, Mixl1, Fgf8, and Lrig3 (Figures 1E and S1E).
TRIM33 Colocalizes with Histone H3K18ac at the Promoters of Mesendodermal Genes
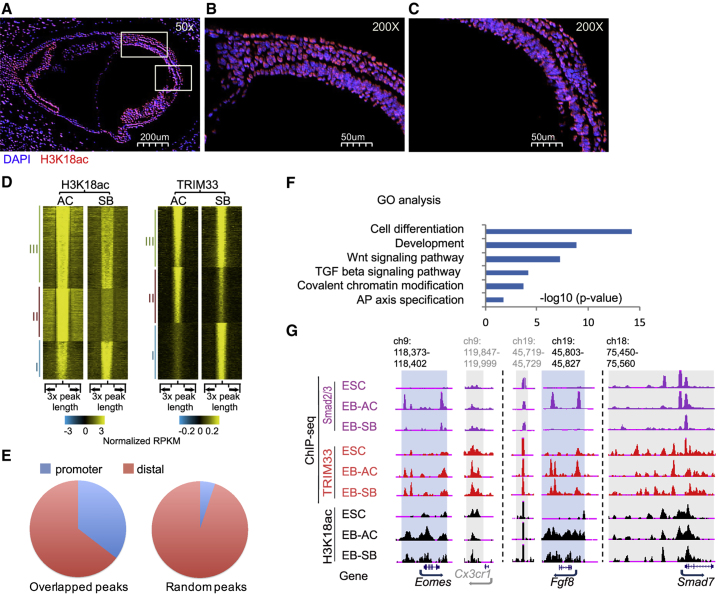
TRIM33 has been shown biochemically to recognize dual histone marks, H3K9me3-H3K18ac, and this interaction is required for activation of nodal target genes (Xi et al., 2011). Like TRIM33, H3K18ac immunostaining was present throughout all regions of embryonic day 7.5 (E7.5) embryos, especially node and primitive streak regions that undergo mesendodermal differentiation in response to nodal (Figures 2A–2C). We performed ChIP-seq analyses of TRIM33 and H3K18ac in EB-AC and EB-SB and classified the top 10,000 ChIP-seq peaks by enrichment p value into three groups: group I are peaks enriched in EB-SB; group II are peaks enriched in EB-AC; and group III are peaks present in both (Figure 2D). We found 6,015 co-occupied sites in EB-AC (Figure S2A). Among these, promoter regions were highly over-represented (Figure 2E). The corresponding genes with TRIM33 and H3K18ac overlapping peaks at their promoter (i.e., the top 1,000 genes selected by TRIM33 signal intensity from a total of 2,122 genes; Figure 2F and Table S3) were enriched for nodal target genes, Wnt target genes, and genes involved in early embryonic development, including most of the GDNT gene set. In contrast, a set of genes (4,525 genes), with both TRIM33 and H3K18ac bound at their promoters specifically in the nodal-inhibited EB-SB, were not involved in development or TGF-β signaling (Figures S2B and S2C; Table S4). Furthermore, we identified 728 genes, including 19 of the 21 GDNT genes, in EB-AC that had TRIM33, Smad2/3, and H3K18ac cobinding (Table S5). Thus, these results suggest that, upon nodal signaling, TRIM33 and H3K18ac colocalize at nodal target genes that are important for early development.
Figure 2.
TRIM33 Colocalizes with Histone H3K18ac at the Promoters of Mesendodermal Genes
(A–C) Distribution of H3K18ac in E7.5 mouse embryos. (A) Immunofluorescence analysis of mouse whole embryo (50× magnification) sections with antibody against H3K18ac. The framed regions are node region and primitive streak. (B) Node region (200×), and (C) primitive streak region (200×).
(D) Heatmaps of ChIP-seq signal densities on H3K18ac and TRIM33 peaks genome wide (within ±1× peak length). Top 10,000 significant peaks selected by p value for H3K18ac or TRIM33 peaks in EB-AC. Heatmaps of ChIP-seq signals for H3K18ac (left panel) or TRIM33 (right panel) at these selected regions in EB-AC and EB-SB are shown. These peaks are divided into three groups based on their ChIP signal intensity in EB-AC or EB-SB: group I, SB-specific peaks; group II, AC-specific peaks; group III, shared peaks. Color bar represents ChIP signal intensity (normalized reads per kilobase per million [RPKM]) distribution.
(E) Pie chart shows the genome distribution of TRIM33 and H3K18ac overlapped peaks. The random peaks were set up for control. Thirty-five percent of the overlapped peaks are located at promoter regions (transcription start site [TSS] ±2.5kb).
(F) Gene ontology (GO) analysis of genes with overlapped TRIM33 and H3K18ac peaks at their promoter regions (TSS ±2.5kb). We selected the top 1,000 genes by TRIM33 signal intensity for GO analysis.
(G) The UCSC browser view shows ChIP-seq signals of Smad2/3, TRIM33, and H3K18ac at the indicated loci, and conditions. Regions with increased Smad2/3 and TRIM33 binding significantly increased in EBs compared with ESCs are shaded blue; control gene Smad7 with similar levels of Smad2/3 and TRIM33 binding in these two conditions are shaded gray. Different loci are separated by dashed lines. Cx3cr1 in gray is the gene in the control region. Target regions are shaded blue, control regions are shaded gray. Genome location in kilobases.
See also Figure S2.
We then examined H3K18ac marks at promoter regions of mesendodermal genes in both ESCs and EBs. Although the total levels of H3K18ac in ESCs and EBs remained largely the same (Figure S2D), their distribution pattern at mesendodermal gene promoters changed significantly from ESCs to EBs (Figures 2G and S2E). H3K18ac was present as small foci at promoters of the mesendodermal genes Eomes and Fgf8 in ESCs, but became more widely distributed in EB-AC (Figures 2G and S2E). Furthermore, this switching from small foci to the expanded pattern of H3K18ac correlated with TRIM33 and Smad2/3 recruitment to their promoters in EBs, and also correlated with corresponding gene expression. As a control, this was not observed at the promoter of a canonical TGF-β target gene, Smad7, which is a known TRIM33-independent gene (Xi et al., 2011) (Figures 2G and S2E). Interestingly, H3K18ac and TRIM33 ChIP analysis at the Mixl1 promoter region in ESCs and day-2.5 EBs with or without SB431542 treatment revealed higher H3K18ac levels and TRIM33 binding in day-2.5 EBs compared with ESCs (Figure S2F). Increasing the length of SB431542 treatments from 2 to 4 h in EBs gradually impaired TRIM33 binding to the chromatin, which also correlated with decreased H3K18ac modification at the same region and decreased Mixl1 gene expression (Figure S2F). These results demonstrate that TRIM33 and H3K18ac co-occupy the promoters of mesendodermal genes upon nodal signaling.
Histone Acetylation of H3K18ac Facilitates TRIM33 Recruitment to Promoters of Mesendodermal Genes
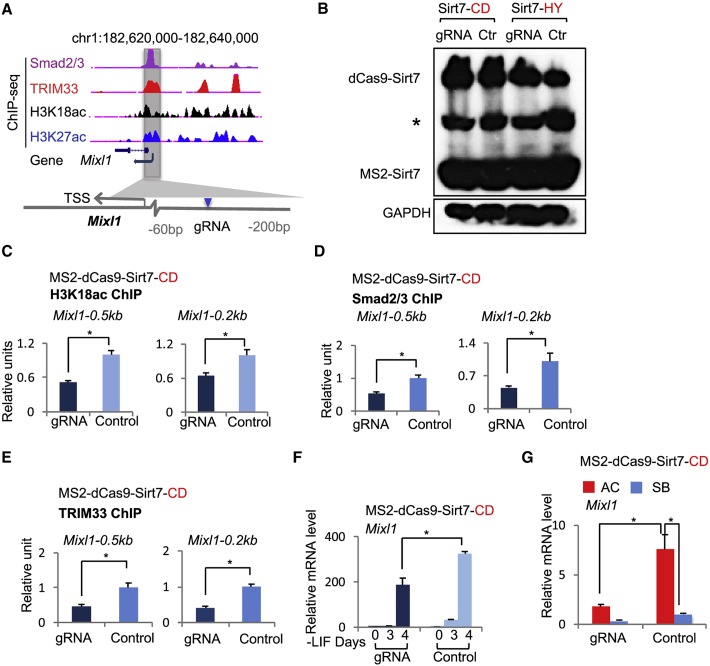
We next investigated whether the specific histone mark H3K18ac is important for the expression of mesendodermal genes. The histone deacetylase Sirt7 has been shown to specifically remove the acetyl group from H3K18ac, and its targeted recruitment to a promoter causes a local loss of H3K18ac (Barber et al., 2012). Therefore, we constructed a CRISPR-dCas9-Sirt7CD (Sirt7 Catalytic Domain) & MS2-Sirt7CD effector system (Konermann et al., 2015), and used it with specific guide RNAs (gRNAs) to recruit Sirt7 to the promoter region of Mixl1 (Figures 3A and 3B). We also constructed a CRISPR-dCas9-Sirt7HY & MS2-Sirt7HY effector system that expresses a deacetylase dead mutant Sirt7 H187Y (Barber et al., 2012) as a negative control. We found that recruited dCas9-Sirt7CD reduced levels of H3K18ac by 48% and 35% at the −0.5-kb and −0.2-kb promoter regions of Mixl1, respectively (Figure 3C), whereas Sirt7HY had no such effect (Figure S3A). Consistent with Sirt7 being an H3K18ac-specific HDAC, targeted recruitment of either Sirt7CD or Sirt7HY to the Mixl1 promoter had no effect on H3K14ac (Figures S3B and S3C). Furthermore, the decrease of H3K18ac, presumably caused by Sirt7, led to a marked reduction of TRIM33 and Smad2/3 at the Mixl1 promoter (Figures 3D, 3E, S3D, and S3E). Most importantly, Mixl1 induction by endogenous signals or exogenous activin was impaired (Figures 3F and 3G), whereas recruitment of Sirt7HY had no effect on Mixl1 expression (Figures S3F and S3G). Also, tethering Sirt7CD to the Mixl1 promoter did not affect expression of Smad7 (Figure S3H). These results indicate an important role for the specific histone modification H3K18ac in transcriptional activation of TRIM33-dependent nodal target genes such as Mixl1.
Figure 3.
Histone Acetylation H3K18ac Is Essential for TRIM33 Recruitment to Promoters of Mesendodermal Genes
(A) Scheme of genomic organization at the Mixl1 locus with the indicated gRNA-targeting site used for targeted recruitment of Sirt7. The UCSC browser view (top) shows ChIP-seq signals of Smad2/3, TRIM33, H3K18ac, and H3K27ac at Mixl1 locus in EB-AC.
(B) Western blotting shows similar expression levels of dCas9-Sirt7-CD and MS2-Sirt7-HY fusion protein in ESCs expressing the specific or control gRNA. GAPDH was used as loading control. The asterisk indicates a non-specific band.
(C) H3K18ac ChIP analysis of the Mixl1 promoter (−0.2 kb and −0.5 kb) in ESCs expressing dCas9-MS2-Sirt7 (CD) and either specific or control gRNAs.
(D and E) Effect of targeted recruitment of Sirt7 on Smad2/3 (D) or TRIM33 (E) binding at the Mixl1 promoter during EB formation. Smad2/3 or TRIM33 binding at the Mixl1 promoter (−0.2 kb and −0.5 kb) in day-2.5 EBs were analyzed using ChIP assays.
(F) Effect of targeted recruitment of Sirt7 on Mixl1 mRNA levels during EB formation. ESCs expressing dCas9-MS2-Sirt7 (CD) and either specific or control gRNAs were set for EB formation, and Mixl1 mRNA levels were analyzed by qRT-PCR at indicated time points.
(G) Effect of targeted recruitment of Sirt7CD on activin response of Mixl1. ESCs expressing dCas9-MS2-Sirt7 (CD) and either specific or control gRNAs were set for EB formation for 2.5 days and then treated with AC or SB for 2 h. Mixl1 mRNA levels were analyzed by qRT-PCR.
In (C) to (G) the results are shown as mean ± SEM by unpaired, two-tailed Student's t test (n = 12, including 3 biological replicates and 4 technical replicates). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S3.
HDAC1 Associates with Transcriptionally Active Genes and Negatively Regulates Mesendodermal Gene Expression
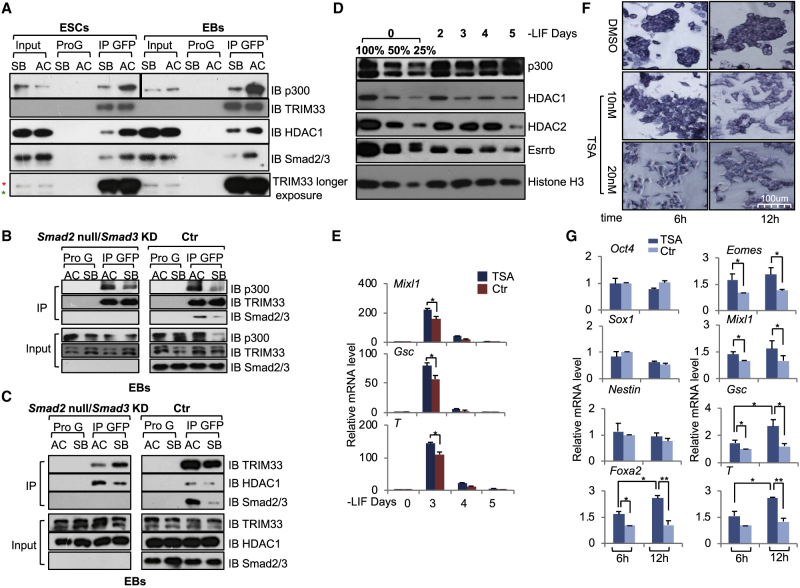
To search for factors that affect TRIM33 recruitment to chromatin, we took a proteomics approach using GFP-TRIM33 as bait for affinity purification followed by mass-spectrometry analysis to identify novel TRIM33 binding partners in ESCs and EBs (our unpublished data). Among the identified TRIM33-interacting factors, we focused on histone modifiers such as the histone deacetylase HDAC1/2 and the histone acetyltransferase p300 that serves as a transcription coactivator. We performed coimmunoprecipitation (coIP) assays to validate the interaction between TRIM33 and HDAC1/2 or p300 in vivo by coexpressing FLAG-TRIM33 and HA-HDAC1/2 or p300 ectopically in HEK293T cells (Figures S4A–S4C). We further showed that TRIM33 interacts with HDAC1 and p300 in both ESCs and EBs, and that both interactions are dependent on activin treatment (Figure 4A). Depletion of Smad2/3 did not interfere with the interaction between TRIM33 and p300 (Figure 4B), suggesting that it is Smad2/3 independent. Rather, the TRIM33-HDAC1 interaction appeared to be enhanced in Smad2/3-depleted EBs (Figure 4C), which suggests that TRIM33-Smad2/3 complex formation interferes with the TRIM33-HDAC1 interaction.
Figure 4.
TRIM33 Associates with Histone Acetylation Modifiers upon Nodal Signaling and the HDAC Activity Negatively Affects Mesendodermal Differentiation
(A) Activin-dependent interaction between TRIM33 and HDAC1 or p300. GFP-TRIM33 overexpressing ESCs or day-2.5 EBs were treated with AC or SB for 2 h. Cell lysates were immunoprecipitated with anti-GFP-Trap affinity beads, and immune complexes were analyzed by immunoblotting using antibodies against the indicated proteins. The red asterisk indicates a band for GFP-TRIM33 and the green asterisk for the endogenous TRIM33.
(B) The interaction between TRIM33 and p300 in EBs is Smad2/3 independent. GFP-TRIM33 overexpressed in Smad2/3 depleted or control cell lines in day-2.5 EBs were treated with AC or SB for 2 h. Cell lysates were immunoprecipitated with anti-GFP-Trap affinity beads, and immune complexes were analyzed by immunoblotting using antibodies against the indicated proteins.
(C) TRIM33-HDAC1 interaction is enhanced in Smad2/3-depleted EBs. GFP-TRIM33 overexpressing in Smad2/3 depleted or control cell lines in day-2.5 EBs were treated with AC or SB for 2 h. Cell lysates were immunoprecipitated with anti-GFP-Trap affinity beads, and immune complexes were analyzed by immunoblotting using antibodies against the indicated proteins.
(D) HDAC1 protein level declined during differentiation. Protein samples were collected at indicated differentiation days and the protein level was measured by immunoblotting using antibodies against the indicated proteins. Histone H3 was used as loading control.
(E) Effect of trichostatin A (TSA) on mesendodermal differentiation. E14 ESCs were set up for EB formation, DMSO or TSA (20 nM) was added at day 2 for 48 h, and mRNA levels of indicated genes were quantified by qRT-PCR analysis.
(F) Effect of TSA on alkaline phosphatase (AP) staining of mESCs. mESCs were treated with TSA (10 nM or 20 nM) or DMSO for 6 h and 12 h and then stained for AP.
(G) Effect of TSA on gene expression in mESCs. mESCs were treated with TSA (20 nM) for indicated times, and mRNA levels of indicated genes were quantified by qRT-PCR analysis.
In (E) and (G) the results are shown as mean ± SEM by unpaired, two-tailed Student's t test (n = 12, including 3 biological replicates and 4 technical replicates). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S4.
HDAC1/2 are the most prevalently expressed HDACs in mESCs (Table S6). During mESC differentiation, we observed slightly decreased protein levels of HDAC1 (at day 3) and HDAC2 (at day 5) during EB differentiation (Figure 4D). Similarly, declining HDAC1 levels were also observed in human ESC mesendodermal differentiation (Figure S4D). These results suggest that HDAC1/2 could be one of the differentiation barriers that help maintain the ESC state. To probe the effect of HDAC1/2 on mesendodermal differentiation, we treated EBs at day 2 with a broad HDAC inhibitor, trichostatin A (TSA), to mimic the lowered HDAC activities caused by decreased protein levels of HDACs during EB differentiation (Figure 4D). As a result, the expression of Mixl1, Gsc, and T increased compared with non-treated control EBs (Figure 4E). This treatment did not affect ectoderm gene expression (Figure S4E). The effects of TSA on ESCs have been reported (Karantzali et al., 2008, McCool et al., 2007). Similarly, TSA treatment of ESCs (Kouzarides, 1999) also led to the increased expression of TRIM33-regulated mesendodermal genes Eomes, Mixl1, Gsc, and Foxa2, in addition to causing a loss of alkaline phosphatase staining (Figures 4F and 4G). Therefore, HDAC activity negatively affects mesendodermal gene activation (see also Brunmeir et al., 2009, Dovey et al., 2010).
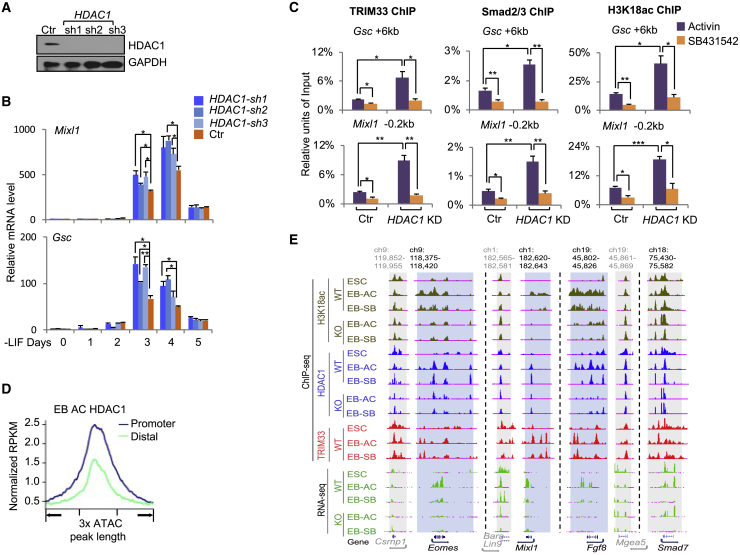
To determine which HDAC negatively affects mesendodermal differentiation, we depleted separately HDAC1 or HDAC2 with short hairpin RNAs (shRNAs) (Figures 5A and S5C) and determined their effects on mesendodermal gene expression. Depletion of HDAC1, but not HDAC2, led to enhanced expression of mesendodermal genes compared with control cells (Figures 5B and S5C). We further found that depletion of HDAC1, but not HDAC2, increased activin responses in EBs compared with control cells (Figures S5A and S5D). The increased activin responses of Gsc and Mixl1 were correlated with enhanced recruitment of TRIM33 and Smad2/3 to their regulatory element regions, and consequently more RNA polymerase II was recruited to the TSS (Figures 5C and S5B). The enhanced recruitment of TRIM33 and Smad2/3 was correlated with increased levels of H3K18ac at these loci in the HDAC1-depleted EBs (Figure 5C). Furthermore, the repressive effect of HDAC1 was specific to the TRIM33/Smad2/3 target genes Gsc and Mixl1, whereas the induction of Smad7, a Smad2/3-Smad4 target gene, was not affected by HDAC1 depletion (Figure S5A).
Figure 5.
HDAC1 Associates with Transcriptional Active Genes and Negatively Regulates Mesendodermal Gene Expression
(A) Western blotting shows efficient knockdown of HDAC1 by shRNAs in mESCs.
(B) qRT-PCR analysis of mRNA levels of Mixl1 and Gsc in control or HDAC1-depleted EBs at indicated time points.
(C) ChIP analysis of TRIM33, Smad2/3, and H3K18ac binding at indicated regions of Mixl1 and Gsc in control or HDAC1-depleted EBs at day 2.5 that were treated with AC or SB for 2 h.
(D) Line chart showing the signal intensity of HDAC1 on promoter and distal ATAC-seq peaks in EB-AC.
(E) The UCSC browser view shows H3K18ac, HDAC1, TRIM33 binding, and mRNA expression at Eomes, Mixl1, and Fgf8 in the indicated cell lines and indicated conditions. WT, Trim33 wild type; KO, Trim33 null. Smad7 was used as a control gene. Csrnp1, Bara, Lin9, and Mgea5 (in gray) are the nearby genes that are not nodal signaling target genes. Target regions are shaded blue, control regions are shaded gray. Genome location in kilobases.
In (B) and (C) the results are shown as mean ± SEM by unpaired, two-tailed Student's t test (n = 12, including 3 biological replicates and 4 technical replicates). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S5.
HDAC1 ChIP-seq and ATAC-seq (assay for transposase-accessible chromatin using sequencing) in EBs showed that HDAC1 is enriched at accessible gene promoters and enhancers, which is consistent with a previous report (Wang et al., 2009) (Figure 5D). Genome wide, there were 7,303 genes in EB-AC that had H3K18ac and HDAC1 co-occupancy at their promoter regions (TSS ±2.5 kb) including GDNT genes. We show the representative overlapping enrichment of HDAC1 and H3K18ac at GDNT genes Eomes, Mixl1, and Fgf8 in Figure 5E. This enrichment of HDAC1 and H3K18ac dramatically decreased in Trim33 null EBs (Figure 5E), suggesting that HDAC1 is recruited by TRIM33 to the regulatory regions of these genes. Maintenance of H3K18ac marks at TRIM33 target genes was dependent on TRIM33 because the H3K18ac peaks dropped significantly in Trim33 null EBs (Figure 5E). Thus, we surmise that HDAC1 reduces levels of histone acetylation at these regulatory regions, which would therefore necessitate constant recruitment of histone acetyltransferase activity to maintain the proper histone acetylation landscape at these genes.
The Coactivator p300 Enhances TRIM33 Recruitment to Mesendodermal Genes by Depositing H3K18ac and Promoting TRIM33-Chromatin Interaction
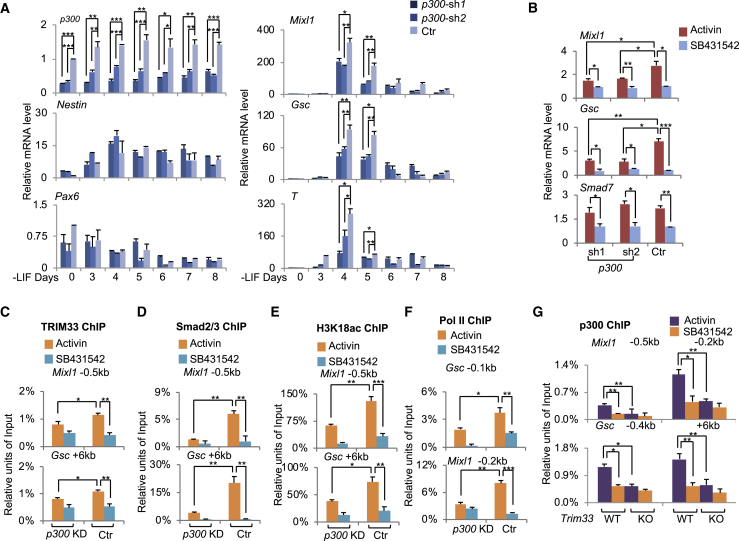
As mentioned earlier, we identified p300 as a TRIM33-interacting partner. To investigate the role of TRIM33-interacting p300 in activation of TRIM33-regulated mesendodermal genes, we depleted p300 by shRNA in ESCs and induced differentiation. Since p300 is responsible for both H3K18ac and H3K27ac modifications (Tang et al., 2013), depletion of p300 consequently decreased overall H3K18ac and H3K27ac, but not H3K14ac (Figure S6A). Depletion of p300 caused a drastic decrease in expression of mesendodermal genes (T, Mixl1, Gsc, and Foxa2), but had no such effect on the expression of the ectoderm marker genes Pax6 and Nestin (Figure 6A). Similarly, p300 knockdown impaired activin responses of Mixl1 and Gsc in EBs, but not that of Smad7 (Figure 6B). In addition, we did not observe any effect of CBP depletion on mesendodermal differentiation (Figures S6B–S6D), indicating a specific role of p300 in activation of TRIM33-dependent nodal responses of mesendodermal genes.
Figure 6.
The Coactivator p300 Enhances TRIM33 Recruitment to Mesendodermal Genes by Depositing H3K18ac and Promoting TRIM33-Chromatin Interaction
(A) Control or p300 knockdown ESCs were set for EB formation, and total RNA at indicated time points was analyzed by qRT-PCR using primers for the indicated genes.
(B) qRT-PCR analysis of mRNA levels of Mixl1, Gsc, and Smad7 in control and p300-depleted cells under EB suspension at day 2.5 that were treated with AC or SB.
(C–F) ChIP analysis of TRIM33 (C), Smad2/3 (D), and H3K18ac (E), and Pol II (F) at indicated regions of Mixl1 and Gsc in control or p300-depleted day-2.5 EBs that were treated with AC or SB.
(G) ChIP analysis of p300 at indicated regions of Mixl1 and Gsc in wild-type (WT) or Trim33 null (KO) day-2.5 EBs that were treated with AC or SB.
The results are shown as mean ± SEM by unpaired, two-tailed Student's t test (n = 12, including 3 biological replicates and 4 technical replicates). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S6.
We further investigated whether p300 facilitates TRIM33 recruitment to mesendodermal genes. ChIP analysis showed that depletion of p300 caused a significant reduction of TRIM33 and Smad2/3 binding at Mixl1 −0.5-kb region and Gsc +6.0-kb region in EBs (Figures 6C and 6D). Gsc +6.0-kb region contains an enhancer element required for efficient expression of Gsc in EBs (Wang et al., 2017b). RNA polymerase II recruitment was significantly decreased in p300-depleted EBs (Figure 6F). We also observed a marked reduction of H3K18ac at both regions in p300-depleted cells (Figure 6E), supporting the notion that p300 deposits H3K18ac at these regions (Jin et al., 2011). Our Sirt7 tethering experiment indicated that specific removal of H3K18ac modification is enough to influence TRIM33 recruitment (Figure 3). Moreover, Trim33 knockout led to a significant reduction in p300 recruitment (Figure 6G). Altogether, these data, i.e., p300 depletion reduces TRIM33 recruitment and H3K18ac modification (Figures 6C–6F), and Trim33 knockout reduces both p300 recruitment (Figure 6G) and H3K18ac modification (Xi et al., 2011), collectively suggest a scenario whereby p300 deposits the histone mark H3K18ac, H3K18ac facilitates TRIM33 recruitment to the chromatin, and TRIM33 further interacts with p300 to maintain the H3K18ac modification, thereby reinforcing each other's recruitment to target promoter regions.
TRIM33 Maintains Chromatin Accessibility at Mesendodermal Genes and Promotes Their Activation during mESC Differentiation
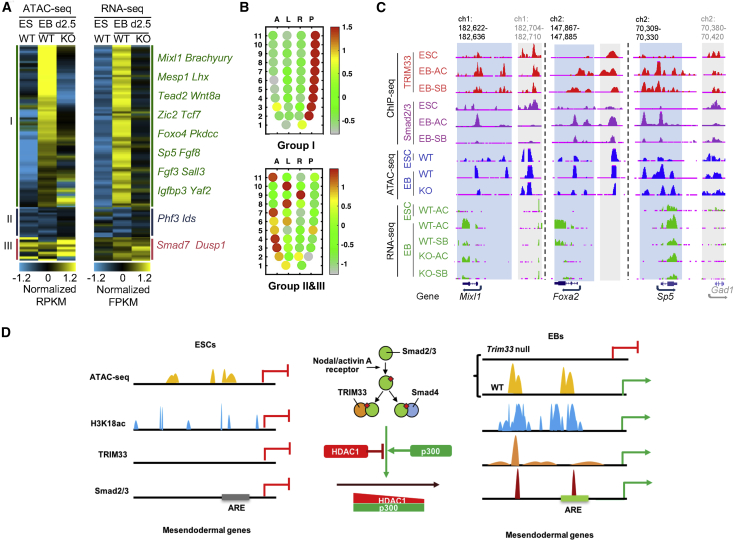
Next, we asked whether TRIM33 promotes mesendodermal gene activation by promoting chromatin accessibly at their regulatory regions. To this end, we performed an ATAC-seq assay in ESCs and in EBs, both wild-type and knockout, for Trim33 (Figure S7A). By analyzing the ATAC-seq data, we identified about the same number of genes with open chromatin regions (i.e., ATAC-seq peaks) at their promoters in wild type as in Trim33 knockout EBs. A total of 1,730 ATAC-seq peaks were only present in wild-type EBs, but not in Trim33 knockout EBs (Figure S7B). Among these genes with open chromatin regions only in wild-type EBs, two biological processes, early development and TGF-β signaling, were significantly enriched (Figure S7C), indicating an important role of TRIM33 in gene activation in both processes. Indeed, 40% of the open chromatin regions identified by ATAC-seq contained TRIM33 binding sites in EBs (Figure S7D). Furthermore, many genes (n = 67) become more accessible at their promoters (up to 5 kb upstream) when ESCs were differentiated to EBs and the increased accessibility depended on TRIM33 (group I genes). Consequently, most of these genes were highly induced in a TRIM33-dependent manner at early stages of EB formation (Figure 7A). Additionally, a significant portion of these genes are involved in mesendodermal differentiation according to gene ontology analysis (Figure S7E). Consistent with these findings, TRIM33-dependent group I genes were strongly enriched at the posterior section of E7.0, where most of the mesendodermal genes are induced (Figure 7B) (Peng et al., 2016). Thus, TRIM33 is specifically involved in activation of mesendodermal genes in EBs by promoting chromatin accessibility at their promoters. Both TRIM33 and Smad2/3 are recruited to three mesendodermal genes, Mixl1, Foxa2, and Sp5, in activin-treated EBs but not in ESCs. Moreover, chromatin opening of TRIM33/Smad2/3 binding regions at these genes depended on TRIM33, and depletion of Trim33 caused a drastic decrease in their expression in EBs (Figures 7A and 7C). Therefore, TRIM33, like Smad2/3, is indispensable for activation of these mesendodermal genes.
Figure 7.
TRIM33 Maintains Chromatin Accessibility at Mesendodermal Genes and Promotes their Activation during mESC Differentiation
(A) Effect of TRIM33 on chromatin accessibility and gene expression during EB formation. Left panel: heatmap of ATAC-seq signals (normalized RPKM) at promoters (TSS ±2.5 kb) for genes whose signals significantly increased in EBs compared with mESCs (group I genes) or not changed (groups II and III genes) (see Supplemental Experimental Procedures for details). Right panel: heatmap representation of expression levels (normalized FPKM), revealed by RNA-seq analysis, of the same genes as in left panel. Representative genes of each group are also listed.
(B) The spatial expression pattern of genes from (A) at E7.0. The corn plots represent spatial expression patterns of group I genes (left panel) and groups II and III genes (right panel) (Peng et al., 2016). Each dot in the plot represents the cell sample at a specific position—anterior (A), posterior (P), left lateral (L), or right lateral (R)—of each sample section (1–11) in the embryo, and the color indicates the enrichment level of gene expression.
(C) The UCSC browser view shows TRIM33 and Smad2/3 binding, ATAC-seq signals, and RNA-seq signals at Mixl1, Foxa2, and Brachyury (T) in the indicated cell lines and conditions. TRIM33-dependent open chromatin regions are shaded blue, control regions are shaded gray. GAD1 (in gray) is the gene in the control region. Genome location in kilobases.
(D) Working model for TRIM33-mediated mesendodermal differentiation of mouse ESCs.
See also Figure S7.
In sum, during mesendodermal differentiation, TRIM33 binds to H3K18ac at promoter regions of mesendodermal genes and promotes chromatin accessibility. HDAC1 exerts repressive effects specifically on the expression of mesendodermal genes because it interferes with TRIM33 recruitment to these genes by reducing H3K18ac levels. On the other hand, TRIM33-interacting p300 deposits H3K18ac on the chromatin, reinforcing TRIM33 recruitment. Thus, p300 and TRIM33 together maintain chromatin accessibility at mesendodermal genes by sustaining the high levels of H3K18ac at their promoters to promote mesendodermal differentiation (Figure 7D).
Discussion
H3K18ac Primes Mesendodermal Differentiation upon Nodal Signaling
A nodal-dependent interaction between TRIM33 and Smad2/3 facilitates recruitment of the Smad2/3-Smad4 complex to chromatin. TRIM33 binds to the chromatin through its PHD-Bromo cassette by recognizing dual histone marks H3K9me3 and H3K18ac, and bromodomain deletion of TRIM33 fails to recognize acetylated histone peptide in vitro, suggesting an essential role of histone acetylation in recruiting TRIM33 (Xi et al., 2011). In this report, we present several lines of strong evidence suggesting that TRIM33 serves as a mediator for nodal signaling crosstalk with the epigenome through its binding to H3K18ac during mesendodermal differentiation, and that H3K18ac is the key epigenetic mark that modulates mesendodermal differentiation. First, we show that the epigenetic landscape of H3K18ac expands significantly at promoter regions of mesendodermal genes when ESCs differentiate to EBs. Second, TRIM33 and H3K18ac binding regions genome wide are significantly overlapped. Third, many of these overlapped binding regions in EB-AC are located at promoters of genes that are involved in mesendodermal differentiation. Lastly, local removal of the H3K18ac mark by artificial recruitment of Sirt7 to the promoter of Mixl1 impairs recruitment of TRIM33 and Smad2/3, and consequently causes a significant reduction of Mixl1 expression. Therefore, the H3K18ac-TRIM33 interaction is central to the appropriate nodal response in mesendodermal differentiation. We speculate that this mechanism, i.e., a particular epigenetic mark interacting with its reader, could be utilized in other lineage differentiations. More work will be needed to explore this possibility.
A p300-Mediated Positive Feedback Mechanism Sustains the Function of TRIM33 upon Nodal Signaling in a Cell-Context-Dependent Manner
TRIM33 is ubiquitously expressed in all tissues (He et al., 2006). But how TRIM33 functions differently in different cell contexts in response to TGF-β is not well understood. In this study, we shed some light on how TRIM33 activity is sustained during ESC differentiation. We showed that recruitment of both TRIM33 and Smad2/3 to mesendodermal genes occurs only in EBs (i.e., not in ESCs). In search of any factors that regulate TRIM33-H3K18ac interaction, we found that histone modifiers HDAC1 and p300 both interact with TRIM33 and affect nodal responses of mesendodermal genes. HDAC1 associated with actively transcribed genes and plays a negative role in regulating TRIM33 function presumably by interfering with TRIM33 recruitment to the chromatin. Indeed, the TRIM33-Smad2/3 complex reduced the interaction between TRIM33 and HDAC1, but p300, a transcriptional coactivator with HAT activity, interacts with TRIM33 independent of Smad2/3 and enhances TRIM33 recruitment to mesendodermal genes during early differentiation. Thus, TRIM33-interacting p300 deposits both H3K18ac and H3K27ac on chromatin, and the increased level of H3K18ac in turn augments recruitment of TRIM33 and p300. The facts that p300 null mice die around E9–E11.5 (Yao et al., 1998), p300 has also been reported to play important roles in endoderm development (Shikama et al., 2003), p300 but not CBP is essential for proper hematopoietic differentiation (Rebel et al., 2002), and Hdac1 null mice died around E9.5 (Montgomery et al., 2007), together suggest that these factors are essential for early development. Our proposed model of TRIM33 regulation by p300 and HDAC1 is based on our finding that the TRIM33-H3K18ac interaction is critically important for activation of mesendodermal genes during ESC differentiation. Thus, the H3K18ac landscape maintained by these histone modifiers shapes the nodal response via its interaction with TRIM33 in mesendodermal differentiation.
TRIM33-Dependent Nodal Response and Chromatin Accessibility
During early development, the spatiotemporal expression of genes is tightly regulated. Chromatin regulation is intimately involved in this process to establish cell-type-specific transcription programs. Cell-type-specific gene expression is highly correlated with the status of chromatin accessibility at specific regulatory elements (Heintzman et al., 2009, Song et al., 2011). We showed here that TRIM33 maintains chromatin accessibility at mesendodermal genes and promotes their activation upon nodal signaling. First, TRIM33-dependent open chromatin regions in EBs are mostly located at nodal target genes that are activated during early development; second, TRIM33 binds to 40% of these open chromatin regions; third, depletion of TRIM33 reduces the enrichment of H3K18ac at certain promoter regions; fourth, mesendodermal genes undergo a TRIM33-dependent increase in chromatin accessibility at their promoters during EB formation and, more importantly, the expression of these genes is dependent on TRIM33.
Experimental Procedures
Cell Lines and Differentiation Assays
Mouse ESCs E14Tg2a.IV and Trim33 null ESCs (Xi et al., 2011) were maintained in feeder-layer free leukemia inhibitory factor-supplemented medium (Keller, 1995). Stable HDAC1, HDAC2, p300, or CBP knockdown in mESCs cells were done using LKO1 lentiviral constructs expressing shRNA against mouse HDAC1, HDAC2, p300, or CBP (for shRNA information, see Table S7). Lentiviral vector infections and plasmid transfection were performed as previously described (Sapkota et al., 2007, Stewart et al., 2003). mESCs were maintained on gelatin-coated plates. Embryoid body (EB) formation and differentiation were carried out according to the protocol provided by the supplier (ATCC).
HEK293T human embryonic kidney cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum.
Chromatin Immunoprecipitation
EBs at 2.5 days were incubated with human recombinant activin A (50 ng/mL, R&D Systems) or SB431542 (10 μM, Tocris) for 2 h. Cells were then crosslinked with 1% formaldehyde at 37°C for 10 min and quenched with 0.125 M glycine for 5 min at room temperature. ChIP was performed as described previously. The primer pairs used to amplify the unrelated-control or promoter regions of indicated genes were described previously (Wang et al., 2017b, Xi et al., 2011).
CRISPR-dCas9-Sirt7 & MS2-Sirt7 Effector System
Lentiviral vector of dCas9-Sirt7 (CD) or dCas9-Sirt7 (HY) were constructed by inserting Sirt7 (CD) and Sirt7 (HY) cDNAs digested with BamHI/BsrGI into BamHI/BsrGI cut lenti dCas9-VP64_Blast (Addgene plasmid #61425). The same strategy was used to construct the MS2-Sirt7 (CD or HY) with lentiviral vector, lenti MS2-P65-HSF1_Hygro (Addgene plasmid #61426).
Specific targeting gRNA vector was cloned as described previously (Konermann et al., 2015). In brief, gRNA oligos 5′-ACG AAC CAA GCC CCC AAG AG-3′ (forward), 5′-TGC TTG GTT CGG GGG TTC TC-3′ (reverse) were annealed and cloned into BsmBI enzyme site of lenti single guide RNA (MS2) zeocin backbone (Addgene plasmid #61427).
Statistical Analysis
All of the values of bar graphs are shown as mean ± SEM with a two-way ANOVA test. The significance between different groups was determined by Student's t test, denoted in figures as ∗p<0.05, ∗∗p< 0.01, and ∗∗∗p< 0.001.
Author Contributions
J.B., B.L., and P.Y. contributed equally. M.L. performed ATAC-seq; M.L. and Y.S. performed ChIP-seq experiments; B.L. and W.X. performed analyses for ATAC-seq and ChIP-seq; Y.Y. and Z.L. performed RNA-seq analysis; Z.S. and X.L. performed E7.0 spatial transcriptome distribution analysis; M.L. performed affinity purification assay; H.D. supervised mass-spectrometry analysis; J.M. supervised RNA-seq experiments; Q.X. performed RNA-seq experiments; M.L. performed immunofluorescence; J.B., H.S., and M.L. performed coIP experiments; F.Z., H.S., P.Y., and M.L. established all cell lines and performed differentiation assays and ChIP-qPCR; X.X. and Y.C. performed differentiation assays in human ESCs; Q.X. designed the work. All authors contributed ideas to the project. Q.X. supervised the project and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Conflicts of Interest
Dr Joan massagué (JM) is a scientific advisor and owns company stock in Scholar Rock.
Acknowledgments
We would like to thank R. Roeder, J. Guan, and M. Chen for reagents; C.J. David and Q. Zhang for helpful discussions; A. Viale for valuable help with RNA-seq; K. Liu for technical help; and W. Zhang for assistance with mass spectrometry analysis. We thank Life Science Editors for editorial assistance. J.M. was supported by NIH Grant CA34610 and NIH grant P30 CA008748 - Thompson- MSKCC Core grant. W.X. was supported by the National Key R&D Program of China (2016YFC0900300 to W.X.), and the National Basic Research Program of China (2015CB856201 to W.X.). W.X. is a recipient of an HHMI International Research Scholarship. Q.X. is supported by the National Natural Science Foundation of China (31471229, 31771622, and 91540108 to Q.X.) and by the Ministry of Science and Technology of China (2018YFA0107702).
Published: September 26, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.08.016.
Accession Numbers
The ATAC-seq, RNA-seq, and ChIP-seq datasets have been deposited in the GEO under accession number GEO: GSE115169.
Supplemental Information
(See Excel File Table S2).
(See Excel File Table S3).
(See Excel File Table S4).
(See Excel File Table S5).
References
- Agricola E., Randall R.A., Gaarenstroom T., Dupont S., Hill C.S. Recruitment of TIF1gamma to chromatin via its PHD finger-bromodomain activates its ubiquitin ligase and transcriptional repressor activities. Mol. Cell. 2011;43:85–96. doi: 10.1016/j.molcel.2011.05.020. [DOI] [PubMed] [Google Scholar]
- Arnold S.J., Robertson E.J. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 2009;10:91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- Badeaux A.I., Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nat. Rev. Mol. Cell Biol. 2013;14:211–224. [PubMed] [Google Scholar]
- Bai J., Xi Q. Crosstalk between TGF-beta signaling and epigenome. Acta Biochim. Biophys. Sin. (Shanghai) 2018;50:60–67. doi: 10.1093/abbs/gmx122. [DOI] [PubMed] [Google Scholar]
- Bai X., Kim J., Yang Z., Jurynec M.J., Akie T.E., Lee J., LeBlanc J., Sessa A., Jiang H., DiBiase A. TIF1gamma controls erythroid cell fate by regulating transcription elongation. Cell. 2010;142:133–143. doi: 10.1016/j.cell.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber M.F., Michishita-Kioi E., Xi Y., Tasselli L., Kioi M., Moqtaderi Z., Tennen R.I., Paredes S., Young N.L., Chen K. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487:114–118. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunmeir R., Lagger S., Seiser C. Histone deacetylase HDAC1/HDAC2-controlled embryonic development and cell differentiation. Int. J. Dev. Biol. 2009;53:275–289. doi: 10.1387/ijdb.082649rb. [DOI] [PubMed] [Google Scholar]
- Chen W., Ten Dijke P. Immunoregulation by members of the TGFbeta superfamily. Nat. Rev. Immunol. 2016;16:723–740. doi: 10.1038/nri.2016.112. [DOI] [PubMed] [Google Scholar]
- David C.J., Massague J. Contextual determinants of TGFbeta action in development, immunity and cancer. Nat. Rev. Mol. Cell Biol. 2018;19:419–435. doi: 10.1038/s41580-018-0007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R., Akhurst R.J. Differentiation plasticity regulated by TGF-beta family proteins in development and disease. Nat. Cell Biol. 2007;9:1000–1004. doi: 10.1038/ncb434. [DOI] [PubMed] [Google Scholar]
- Dovey O.M., Foster C.T., Cowley S.M. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc. Natl. Acad. Sci. U S A. 2010;107:8242–8247. doi: 10.1073/pnas.1000478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S., Zacchigna L., Cordenonsi M., Soligo S., Adorno M., Rugge M., Piccolo S. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell. 2005;121:87–99. doi: 10.1016/j.cell.2005.01.033. [DOI] [PubMed] [Google Scholar]
- Estaras C., Benner C., Jones K.A. SMADs and YAP compete to control elongation of beta-catenin:LEF-1-recruited RNAPII during hESC differentiation. Mol. Cell. 2015;58:780–793. doi: 10.1016/j.molcel.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell R.A., Sanjabi S., Wrzesinski S.H., Licona-Limon P. The polarization of immune cells in the tumour environment by TGFbeta. Nat. Rev. Immunol. 2010;10:554–567. doi: 10.1038/nri2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaarenstroom T., Hill C.S. TGF-beta signaling to chromatin: how Smads regulate transcription during self-renewal and differentiation. Semin. Cell Dev. Biol. 2014;32:107–118. doi: 10.1016/j.semcdb.2014.01.009. [DOI] [PubMed] [Google Scholar]
- He W., Dorn D.C., Erdjument-Bromage H., Tempst P., Moore M.A., Massague J. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell. 2006;125:929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- Heintzman N.D., Hon G.C., Hawkins R.D., Kheradpour P., Stark A., Harp L.F., Ye Z., Lee L.K., Stuart R.K., Ching C.W. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C.S. Spatial and temporal control of NODAL signaling. Curr. Opin. Cell Biol. 2017;51:50–57. doi: 10.1016/j.ceb.2017.10.005. [DOI] [PubMed] [Google Scholar]
- Inman G.J., Nicolas F.J., Callahan J.F., Harling J.D., Gaster L.M., Reith A.D., Laping N.J., Hill C.S. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- Jin Q., Yu L.R., Wang L., Zhang Z., Kasper L.H., Lee J.E., Wang C., Brindle P.K., Dent S.Y., Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantzali E., Schulz H., Hummel O., Hubner N., Hatzopoulos A., Kretsovali A. Histone deacetylase inhibition accelerates the early events of stem cell differentiation: transcriptomic and epigenetic analysis. Genome Biol. 2008;9:R65. doi: 10.1186/gb-2008-9-4-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G.M. In vitro differentiation of embryonic stem cells. Curr. Opin. Cell Biol. 1995;7:862–869. doi: 10.1016/0955-0674(95)80071-9. [DOI] [PubMed] [Google Scholar]
- Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Histone acetylases and deacetylases in cell proliferation. Curr. Opin. Genet. Dev. 1999;9:40–48. doi: 10.1016/s0959-437x(99)80006-9. [DOI] [PubMed] [Google Scholar]
- Massague J. TGFbeta in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. TGFbeta signalling in context. Nat. Rev. Mol. Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J., Seoane J., Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- McCool K.W., Xu X., Singer D.B., Murdoch F.E., Fritsch M.K. The role of histone acetylation in regulating early gene expression patterns during early embryonic stem cell differentiation. J. Biol. Chem. 2007;282:6696–6706. doi: 10.1074/jbc.M609519200. [DOI] [PubMed] [Google Scholar]
- Montgomery R.L., Davis C.A., Potthoff M.J., Haberland M., Fielitz J., Qi X., Hill J.A., Richardson J.A., Olson E.N. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen A.C., Orlando D.A., Newman J.J., Loven J., Kumar R.M., Bilodeau S., Reddy J., Guenther M.G., DeKoter R.P., Young R.A. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen A.C., Wrana J.L. TGF-beta family signaling in embryonic and somatic stem-cell renewal and differentiation. Cold Spring Harb. Perspect. Biol. 2017;9 doi: 10.1101/cshperspect.a022186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshimori N., Fuchs E. The harmonies played by TGF-beta in stem cell biology. Cell Stem Cell. 2012;11:751–764. doi: 10.1016/j.stem.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G., Suo S., Chen J., Chen W., Liu C., Yu F., Wang R., Chen S., Sun N., Cui G. Spatial transcriptome for the molecular annotation of lineage fates and cell identity in mid-gastrula mouse embryo. Dev. Cell. 2016;36:681–697. doi: 10.1016/j.devcel.2016.02.020. [DOI] [PubMed] [Google Scholar]
- Rebel V.I., Kung A.L., Tanner E.A., Yang H., Bronson R.T., Livingston D.M. Distinct roles for CREB-binding protein and p300 in hematopoietic stem cell self-renewal. Proc. Natl. Acad. Sci. U S A. 2002;99:14789–14794. doi: 10.1073/pnas.232568499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson E.J. Dose-dependent Nodal/Smad signals pattern the early mouse embryo. Semin. Cell Dev. Biol. 2014;32:73–79. doi: 10.1016/j.semcdb.2014.03.028. [DOI] [PubMed] [Google Scholar]
- Sanjabi S., Oh S.A., Li M.O. Regulation of the Immune response by TGF-beta: from conception to autoimmunity and infection. Cold Spring Harb. Perspect. Biol. 2017;9 doi: 10.1101/cshperspect.a022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota G., Alarcon C., Spagnoli F.M., Brivanlou A.H., Massague J. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol. Cell. 2007;25:441–454. doi: 10.1016/j.molcel.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Shen M.M. Nodal signaling: developmental roles and regulation. Development. 2007;134:1023–1034. doi: 10.1242/dev.000166. [DOI] [PubMed] [Google Scholar]
- Shikama N., Lutz W., Kretzschmar R., Sauter N., Roth J.F., Marino S., Wittwer J., Scheidweiler A., Eckner R. Essential function of p300 acetyltransferase activity in heart, lung and small intestine formation. EMBO J. 2003;22:5175–5185. doi: 10.1093/emboj/cdg502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Zhang Z., Grasfeder L.L., Boyle A.P., Giresi P.G., Lee B.K., Sheffield N.C., Graf S., Huss M., Keefe D. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S.A., Dykxhoorn D.M., Palliser D., Mizuno H., Yu E.Y., An D.S., Sabatini D.M., Chen I.S., Hahn W.C., Sharp P.A. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Jiang Y., Martinez G.J., Tanaka K., Yan X., Kurosaki T., Kaartinen V., Feng X.H., Tian Q., Wang X. Trim33 mediates the proinflammatory function of Th17 cells. J. Exp. Med. 2018;215:1853–1868. doi: 10.1084/jem.20170779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Chen W.Y., Shimada M., Nguyen U.T., Kim J., Sun X.J., Sengoku T., McGinty R.K., Fernandez J.P., Muir T.W. SET1 and p300 act synergistically, through coupled histone modifications, in transcriptional activation by p53. Cell. 2013;154:297–310. doi: 10.1016/j.cell.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kraan P.M. The changing role of TGFbeta in healthy, ageing and osteoarthritic joints. Nat. Rev. Rheumatol. 2017;13:155–163. doi: 10.1038/nrrheum.2016.219. [DOI] [PubMed] [Google Scholar]
- Waddington C.H. Allen & Unwin; 1957. The Strategy of the Genes; a Discussion of Some Aspects of Theoretical Biology. [Google Scholar]
- Wang E., Kawaoka S., Roe J.S., Shi J., Hohmann A.F., Xu Y., Bhagwat A.S., Suzuki Y., Kinney J.B., Vakoc C.R. The transcriptional cofactor TRIM33 prevents apoptosis in B lymphoblastic leukemia by deactivating a single enhancer. Elife. 2015;4:e06377. doi: 10.7554/eLife.06377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Xu X., Cao Y., Li Z., Cheng H., Zhu G., Duan F., Na J., Han J.J., Chen Y.G. Activin/Smad2-induced histone H3 Lys-27 trimethylation (H3K27me3) reduction is crucial to initiate mesendoderm differentiation of human embryonic stem cells. J. Biol. Chem. 2017;292:1339–1350. doi: 10.1074/jbc.M116.766949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zou Y., Nowotschin S., Kim S.Y., Li Q.V., Soh C.L., Su J., Zhang C., Shu W., Xi Q. The p53 family coordinates Wnt and nodal inputs in mesendodermal differentiation of embryonic stem cells. Cell Stem Cell. 2017;20:70–86. doi: 10.1016/j.stem.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.B., Zang C.Z., Cui K.R., Schones D.E., Barski A., Peng W.Q., Zhao K.J. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe T., Miyazono K. Roles of TGF-beta family signaling in stem cell renewal and differentiation. Cell Res. 2009;19:103–115. doi: 10.1038/cr.2008.323. [DOI] [PubMed] [Google Scholar]
- Wei S., Wang Q. Molecular regulation of Nodal signaling during mesendoderm formation. Acta Biochim. Biophys. Sin. (Shanghai) 2017;50:74–81. doi: 10.1093/abbs/gmx128. [DOI] [PubMed] [Google Scholar]
- Xi Q., Wang Z., Zaromytidou A.I., Zhang X.H., Chow-Tsang L.F., Liu J.X., Kim H., Barlas A., Manova-Todorova K., Kaartinen V. A poised chromatin platform for TGF-beta access to master regulators. Cell. 2011;147:1511–1524. doi: 10.1016/j.cell.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T.P., Oh S.P., Fuchs M., Zhou N.D., Ch'ng L.E., Newsome D., Bronson R.T., Li E., Livingston D.M., Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(See Excel File Table S2).
(See Excel File Table S3).
(See Excel File Table S4).
(See Excel File Table S5).