Abstract
Introduction
Tuberculosis (TB) still remains an important public health problem in Iran. The genotyping of Mycobacterium tuberculosis isolates is expected to lead to a better understanding of M. tuberculosis transmission in Tehran, the most populated city of Iran.
Materials and Methods
A total of 2300 clinical specimens were obtained from TB suspected patients who were referred to a TB center in Tehran from Jan 2014 to Dec 2016. Identification was performed using both conventional and molecular methods. The presence of resistance to rifampicin was examined by the GeneXpert MTB/RIF. The standard 15-locus mycobacterial interspersed repetitive units-variable number of tandem repeats (MIRU-VNTR) typing method was applied to genotype of clinical isolates.
Results
Of 2300 specimens, 80 isolates were identified as M. tuberculosis by using biochemical and molecular tests. Of 80 M. tuberculosis isolates, 76 (95%) had unique genotypic profiles and 4 (5%) shared a profile with one or more other strains. Based on single loci variation (SLV) 4 clonal complexes were observed. NEW-1 was found to be the most predominant lineage (22.5%) followed by West African (1.25%), Central Asian (CAS)/Delhi (1.25%), Bovis (1.25%), H37Rv (1.25%) and multiple matches (1.25%). Loci MIRU10, MIRU26, MTUB21 and QUB26 were found as highly discriminative. No mutation was detected in the hotspot region of rifampicin by using GeneXpert MTB/RIF.
Conclusions
Our study findings show that there was considerable genotypic diversity among M. tuberculosis isolates in Tehran. The 15-locus MIRU-VNTR showed high HGDI and could be used as a first-line genotyping method for epidemiological studies.
Keywords: Mycobacterium tuberculosis, Genotyping, MIRU-VNTR, Tehran, Iran
Introduction
Tuberculosis (TB) caused by Mycobacterium tuberculosis is the first leading cause of death from infectious disease worldwide [1]. Despite the existence of effective treatment regimens, TB continues to be a major global health challenge with an estimated 6.3 million new cases and 1.8 million TB deaths in 2016 [1]. In Iran, according to the World Health Organization (WHO) report in 2016, the incidence of TB was 16 (12–20) per 100,000 [1]. This rate in countries around Iran (Afghanistan, Pakistan, Turkey, Armenia, Azerbaijan and Iraq) is 2–3 times greater than this amount [1], [2]. Tehran, the capital of Iran, is one of the most populated cities in the world with average of 12 million people and is considered a low TB incidence area [3]. However, due to the large number of migrants from countries with high rates of TB, monitoring of suspected TB cases and understanding of TB transmission dynamics is vitally important to avoid the spread of disease locally [4], [5], [6], [7], [8], [9]. However, the information available on the population structure of M. tuberculosis strains and transmission dynamics in Tehran is limited. Many genotyping methods have been used to evaluate the transmission dynamic of TB, among which the MIRU-VNTR is a widely used typing method [10], [11], [12], [13], [14], [15], [16]. In the current study, 15-locus MIRU-VNTR typing methods were applied for fingerprinting and following the transmission dynamics of M. tuberculosis strains isolated from TB patients in Tehran.
Materials and methods
Setting and study population
This cross-sectional study evaluated clinical specimens obtained from suspected TB patients referred to one of the main TB centers of Tehran, Iran, from Jan 2014 to Dec 2016. This center with drug susceptibility testing (DST) capability, acts as local center for diagnosis and treatment of infectious diseases. A total of 2300 clinical specimens (sputum) were obtained from TB suspected patients. Patients were from across the age-range (20–65 years), and had clinical signs and symptoms of TB, and undergoing examination for possible active TB. The Ethics Committee of Shahid Beheshti University of Medical Sciences approved the study and all the patients have signed informed consent.
Culture and identification
Samples from each patient were decontaminated by Petroff's method and were inoculated into Lowenstein-Jensen media [17], [18]. The inoculated media was incubated at 37 °C. Cultures with no growth until the eighth weeks were recorded as negative. Bacterial isolates were identified as M. tuberculosis complex using standard biochemical tests, including production of niacin, nitrate reduction, catalase and inhibition by thiophene-2-carboxylic acid hydrazide [17].
Molecular characterization of the isolates
To confirm the identification of M. tuberculosis and the differentiation of M. tuberculosis and nontuberculous mycobacteria (NTM) from positive cultures, two methods were used; IS6110 based Polymerase chain reactions (PCR) assay and GeneXpert MTB/RIF assay. GeneXpert MTB/RIF was also used to evaluate the presence of resistance to rifampicin. Genomic DNA, for IS6110 based PCR assay, was extracted using boiling method. A 123 bp fragment of insertion element IS6110 of the M. tuberculosis complex was used as a target and amplified using previously described PCR primers [19]. Genomic DNA of M. tuberculosis H37Rv (ATCC27294) and M. fortuitum (ATCC 49404) were used as positive and negative controls, respectively. GeneXpert MTB/RIF assay was performed as described previously [20].
MIRU-VNTR typing
The standard 15-locus MIRU-VNTR typing method was performed as described by Supply et al. [14].
Analysis of VNTR allelic diversity and genetic relationships
All results were entered into Excel® spreadsheets, and analyzed by MIRU-VNTRplus software [21]. The allelic diversity of each VNTR loci was calculated by the Hunter–Gaston discriminatory index (HGDI), and Genetic relationships among the isolates were estimated using the categorical coefficient and UPGMA [21]. Cluster definition was based on a distance cut-off of 0 and similar patterns in 15-locus [22].
Database comparison
The 15-locus MIRU-VNTR patterns were compared with the patterns from the MIRU-VNTRplus database to determine M. tuberculosis strain lineages and relatedness [23]. Distribution of identified lineage was done by similarity search with reference strains. So that strains with distance lower than 0.3 with reference strains were determined. Minimum spanning tree was calculated using 15-locusMIRU-VNTR and clonal complexes were described in the means that Maximum loci difference within a clonal complex was single loci variation (SLV).
Results
Clinical isolates of M. tuberculosis
Of 2300 specimens, 104 were positive for culture. By using biochemical and molecular tests, 80 isolates were identified as M. tuberculosis and the others were NTM. Then we used all of the samples that confirmed for M. tuberculosis. Each isolates corresponding to a unique TB patient. All the isolates were obtained by culture of respiratory samples (sputum).
Confirmation of the isolates
By using IS6110 and GeneXpert, all 80 isolates was confirmed as M. tuberculosis. No mutation was detected in the hotspot region of rifampicin by using GeneXpert in investigated isolates.
MIRU-VNTR
Using 15-locus MIRU-VNTR genotyping, 78 different patterns were detected among the 80 isolates. Of these, 76 (95%) had unique genotypic profiles and 4 (5%) shared a profile with one or more other strains (Table 1). A comparison of the 15-locus MIRU patter obtained with the international MIRU-VNTR plus database (http://www.miru-vntrplus.org) showed that 2 of the strains matches in the most of loci (14 loci) with references strains that represent NEW-1 lineage. Distribution of other identified lineage was determined and the result showed that 71.25% of isolates were unknown based on similarity search with reference strains (with distance < 0.3) in MIRU-VNTR plus database. The most identified linage was NEW-1 with 22.5% and the others were West African with 1.25%, Central Asian (CAS)/Delhi with 1.25%, Bovis with 1.25%, H37Rv with 1.25% and multiple matches (Cameroon-NEW-1) with 1.25%. The discriminatory power of MIRU-VNTR typing for all isolates was high (HGDI = 0.99).
Table 1.
78 different profiles in dataset (displayed in descending order of frequency).
| MIRU pattern | Frequency | MIRU pattern | Frequency |
|---|---|---|---|
| 3, 4, 2, 3, 2, 3, 6, 3, 2, 7, 3, 3, 1, 5, 1 | 2 | 4, 2, 1, 4, 5, 3, 7, 3, 2, 8, 2, 3, 2, 4, 2 | 1 |
| 3, 4, 2, 2, 2, 2, 6, 1, 2, 6, 2, 3, 1, 5, 2 | 2 | 3, 4, 2, 3, 5, 1, 1, 2, 2, 7, 3, 2, 2, 2, 2 | 1 |
| 4, 7, 3, 7, 9, 2, 1, 2, 2, 8, 6, 8, 1, 6, 1 | 1 | 2, 2, 2, 4, 7, 3, 9, 3, 2, 4, 3, 3, 3, 4, 2 | 1 |
| 2, 4, 3, 4, 5, 4, 7, 3, 3, 7, 2, 3, 8, 6, 4 | 1 | 3, 4, 2, 3, 2, 3, 5, 3, 2, 8, 2, 3, 2, 5, 2 | 1 |
| 4, 2, 2, 3, 2, 2, 6, 1, 2, 8, 4, 3, 2, 4, 2 | 1 | 4, 2, 2, 4, 5, 3, 7, 4, 2, 8, 2, 0, 3, 4, 2 | 1 |
| 3, 3, 2, 3, 3, 2, 3, 4, 4, 8, 4, 3, 2, 3, 2 | 1 | 3, 4, 2, 4, 2, 3, 6, 3, 2, 5, 2, 0, 2, 5, 2 | 1 |
| 3, 4, 2, 3, 2, 2, 6, 3, 2, 8, 2, 3, 2, 5, 4 | 1 | 3, 4, 2, 3, 4, 1, 1, 2, 2, 4, 3, 2, 2, 2, 2 | 1 |
| 4, 2, 2, 5, 6, 2, 6, 3, 2, 5, 2, 3, 2, 4, 4 | 1 | 3, 4, 2, 3, 3, 3, 5, 1, 4, 5, 2, 4, 2, 2, 2 | 1 |
| 3, 4, 2, 3, 2, 2, 6, 3, 2, 3, 2, 3, 2, 5, 3 | 1 | 4, 4, 2, 4, 3, 3, 5, 3, 6, 8, 3, 4, 3, 4, 2 | 1 |
| 3, 4, 2, 3, 2, 2, 6, 3, 2, 6, 2, 3, 2, 5, 1 | 1 | 3, 4, 2, 3, 6, 3, 5, 1, 3, 4, 2, 4, 0, 3, 1 | 1 |
| 3, 2, 2, 4, 2, 2, 4, 3, 5, 5, 2, 2, 2, 7, 0 | 1 | 2, 4, 2, 3, 2, 1, 1, 2, 2, 4, 4, 2, 2, 2, 1 | 1 |
| 3, 4, 2, 3, 2, 3, 4, 3, 2, 7, 2, 3, 2, 5, 8 | 1 | 2, 4, 2, 3, 6, 3, 5, 3, 2, 7, 2, 3, 1, 5, 1 | 1 |
| 4, 2, 2, 4, 6, 3, 6, 3, 2, 8, 2, 3, 2, 4, 2 | 1 | 2, 4, 2, 2, 2, 3, 5, 2, 3, 5, 2, 3, 1, 2, 1 | 1 |
| 4, 2, 2, 3, 4, 3, 5, 3, 2, 3, 4, 3, 2, 4, 2 | 1 | 2, 2, 2, 4, 2, 3, 7, 3, 2, 4, 4, 3, 2, 4, 1 | 1 |
| 3, 4, 2, 3, 2, 3, 4, 3, 2, 3, 2, 3, 2, 5, 3 | 1 | 2, 7, 2, 3, 5, 1, 5, 3, 2, 8, 2, 3, 1, 3, 1 | 1 |
| 3, 2, 3, 3, 2, 3, 5,0, 2, 5, 2, 0, 2, 5, 3 | 1 | 4, 5, 2, 3, 3, 1, 1, 3, 2, 6, 4, 3, 2, 3, 1 | 1 |
| 3, 4, 2, 3, 2, 3, 5, 3, 2, 2, 2, 3, 2, 5, 3 | 1 | 2, 4, 2, 2, 1, 3, 4, 3, 2, 7, 2, 3, 1, 3, 2 | 1 |
| 4, 4, 2, 3, 2, 3, 4, 1, 3, 2, 2, 2, 2, 3, 3 | 1 | 2, 2, 2, 4, 3, 4, 6, 3, 2, 6, 3, 3, 2, 3, 3 | 1 |
| 3, 4, 2, 3, 3, 2, 3, 0, 5, 5, 3, 5, 2, 2, 1 | 1 | 4, 4, 2, 2, 1, 3, 5, 3, 2, 7, 2, 4, 1, 3, 2 | 1 |
| 3, 5, 2, 3, 2, 3, 5, 0, 2, 7, 3, 3, 2, 3, 1 | 1 | 3, 4, 2, 2, 1, 3, 6, 3, 2, 6, 2, 3, 1, 4, 2 | 1 |
| 2, 4, 2, 3, 3, 2, 5, 2, 1, 4, 3, 3, 2, 2, 1 | 1 | 3, 4, 2, 2, 1, 3, 5, 3, 2, 7, 2, 3, 1, 5, 2 | 1 |
| 2, 4, 2, 3, 3, 2, 5, 3, 3, 5, 3, 4, 2, 2, 1 | 1 | 4, 4, 2, 2, 2, 3, 5, 2, 3, 4, 2, 4, 1, 3, 2 | 1 |
| 2, 2, 2, 4, 6, 3, 9, 3, 2, 4, 4, 3, 3, 4, 1 | 1 | 3, 4, 2, 2, 1, 3, 5, 3, 2, 7, 2, 3, 1, 3, 2 | 1 |
| 3, 5, 2, 3, 3, 3, 5, 2, 3, 7, 3, 4, 1, 3, 1 | 1 | 4, 2, 2, 4, 3, 3, 7, 3, 2, 6, 2, 3, 3, 4, 1 | 1 |
| 3, 4, 2, 3, 3, 3, 4, 3, 5, 5, 3, 3, 1, 2, 1 | 1 | 2, 2, 2, 4, 6, 3, 9, 3, 2, 4, 4, 3, 3, 4, 3 | 1 |
| 3, 4, 2, 3, 2, 3, 5, 3, 2, 8, 3, 3, 1, 5, 1 | 1 | 3, 4, 2, 2, 1, 3, 5, 3, 2, 6, 2, 3, 0, 5, 2 | 1 |
| 2, 4, 2, 3, 3, 3, 5, 2, 2, 4, 3, 3, 1, 2, 1 | 1 | 2, 4, 2, 2, 1, 3, 5, 0, 2, 7, 2, 3, 1, 5, 2 | 1 |
| 4, 4, 3, 3, 3, 3, 5, 3, 3, 5, 3, 1, 2, 1, 1 | 1 | 2, 4, 2, 3, 3, 3, 7, 3, 3, 5, 3, 9, 3, 2, 2 | 1 |
| 3, 4, 2, 3, 2, 3, 6, 1, 2, 7, 3, 3, 1, 5, 1 | 1 | 3, 4, 2, 2, 2, 1, 5, 3, 4, 5, 2, 8, 1, 2, 2 | 1 |
| 3, 4, 2, 3, 0, 3, 5, 3, 2, 2, 3, 2, 1, 4, 1 | 1 | 3, 4, 2, 2, 1, 3, 5, 2, 2, 6, 2, 3, 1, 5, 1 | 1 |
| 4, 3, 2, 3, 2, 3, 4, 2, 3, 4, 3, 2, 1, 3, 1 | 1 | 4, 2, 2, 3, 6, 3, 7, 2, 2, 7, 2, 3, 1, 4, 3 | 1 |
| 3, 4, 2, 3, 2, 3, 6, 3, 2, 8, 3, 3, 1, 5, 2 | 1 | 2, 4, 2, 2, 3, 1, 1, 0, 2, 7, 4, 3, 1, 3, 1 | 1 |
| 3, 4, 2, 3, 5, 2, 1, 1, 2, 7, 3, 3, 2, 2, 3 | 1 | 4, 2, 2, 4, 4, 3, 7, 3, 2, 3, 4, 3, 2, 4, 3 | 1 |
| 4, 2, 2, 3, 5, 3, 6, 3, 2, 7, 3, 4, 2, 4, 3 | 1 | 3, 4, 2, 2, 2, 2, 6, 3, 2, 6, 2, 3, 1, 5, 2 | 1 |
| 3, 2, 2, 4, 5, 2, 6, 3, 2, 7, 2, 1, 2, 4, 2 | 1 | 3, 4, 2, 2, 2, 3, 6, 3, 2, 6, 2, 3, 1, 5, 2 | 1 |
| 3, 4, 2, 3, 2, 2, 5, 3, 4, 5, 2, 3, 2, 2, 2 | 1 | 3, 2, 2, 4, 5, 1, 5, 3, 2, 6, 2, 3, 1, 6, 3 | 1 |
| 3, 5, 2, 3, 2, 3, 5, 3, 2, 7, 2, 6, 2, 5, 2 | 1 | 3, 0, 2, 4, 2, 1, 5, 0, 2, 6, 2, 3, 1, 6, 1 | 1 |
| 3, 4, 2, 3, 2, 3, 4, 3, 3, 8, 2, 3, 2, 3, 2 | 1 | 3, 2, 2, 4, 5, 2, 5, 3, 2, 6, 2, 3, 1, 6, 4 | 1 |
| 3, 4, 2, 3, 2, 3, 6, 3, 2, 7, 2, 3, 2, 5, 2 | 1 | 4, 2, 2, 4, 5, 2, 6, 3, 2, 3, 4, 3, 2, 4, 4 | 1 |
MIRU Pattern: ETR A, ETR C, MIRU 04, ETR E, MIRU 10, MIRU 16, MIRU 26, MIRU 40, QUB11b, QUB 26, MTUB 30, MTUB 39, MTUB 04, MTUB 21, QUB4156.
Allele frequencies in the sample studied
Based on the discriminatory index, 4 MIRU loci were designated as highly discriminative (h > 0.6) in our sample (MIRU26, MIRU10, MTUB21 and QUB26), 9 were designated as moderately discriminative (0.3 ≤ h ≤ 0.6; MIRU40, MIRU 16, QUB11b, MTUB30, MTUB39, MTUB04, QUB4156, ETRA and ETRC), and among 15 loci, loci MIRU04 and ETRE had the lowest allelic diversity, and loci QUB26 had the highest allelic diversity. Results of the distribution of the MIRU alleles are shown in Table 2.
Table 2.
Allelic diversity for each locus.
| Allele | ETR A | ETR C | MIRU 04 | ETR E | MIRU 10 | MIRU 16 | MIRU 26 | MIRU 40 | QUB11b | QUB 26 | MTUB 30 | MTUB 39 | MTUB 04 | MTUB 21 | QUB4156 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | – | 1 | – | – | 1 | – | – | 6 | – | – | – | 3 | 2 | – | 1 |
| 1 | – | – | 1 | – | 8 | 9 | 7 | 8 | 1 | – | – | 2 | 32 | 1 | 28 |
| 2 | 17 | 21 | 75 | 16 | 33 | 19 | – | 12 | 60 | 3 | 45 | 7 | 38 | 14 | 33 |
| 3 | 44 | 2 | 4 | 43 | 15 | 50 | 2 | 52 | 11 | 5 | 23 | 55 | 7 | 15 | 12 |
| 4 | 19 | 50 | – | 19 | 3 | 2 | 8 | 2 | 4 | 11 | 11 | 8 | – | 19 | 5 |
| 5 | – | 4 | – | 1 | 11 | – | 31 | – | 3 | 13 | – | 1 | – | 25 | – |
| 6 | – | – | – | – | 7 | – | 21 | – | 1 | 14 | 1 | 1 | – | 5 | – |
| 7 | – | 2 | – | 1 | 1 | – | 8 | – | – | 21 | – | – | – | 1 | – |
| 8 | – | – | – | – | – | – | – | – | – | 13 | – | 2 | 1 | – | 1 |
| 9 | – | – | – | – | 1 | – | 3 | – | – | – | – | 1 | – | – | – |
| HGDI | 0.6 | 0.5 | 0.1 | 0.2 | 0.7 | 0.5 | 0.7 | 0.5 | 0.4 | 0.8 | 0.5 | 0.5 | 0.6 | 0.7 | 0.6 |
| Total | 3 | 6 | 3 | 5 | 9 | 4 | 7 | 5 | 6 | 7 | 4 | 9 | 5 | 7 | 6 |
HGDI total: 0.99 (HGDI, Hunter–Gaston discriminatory index.).
Molecular epidemiology relationships
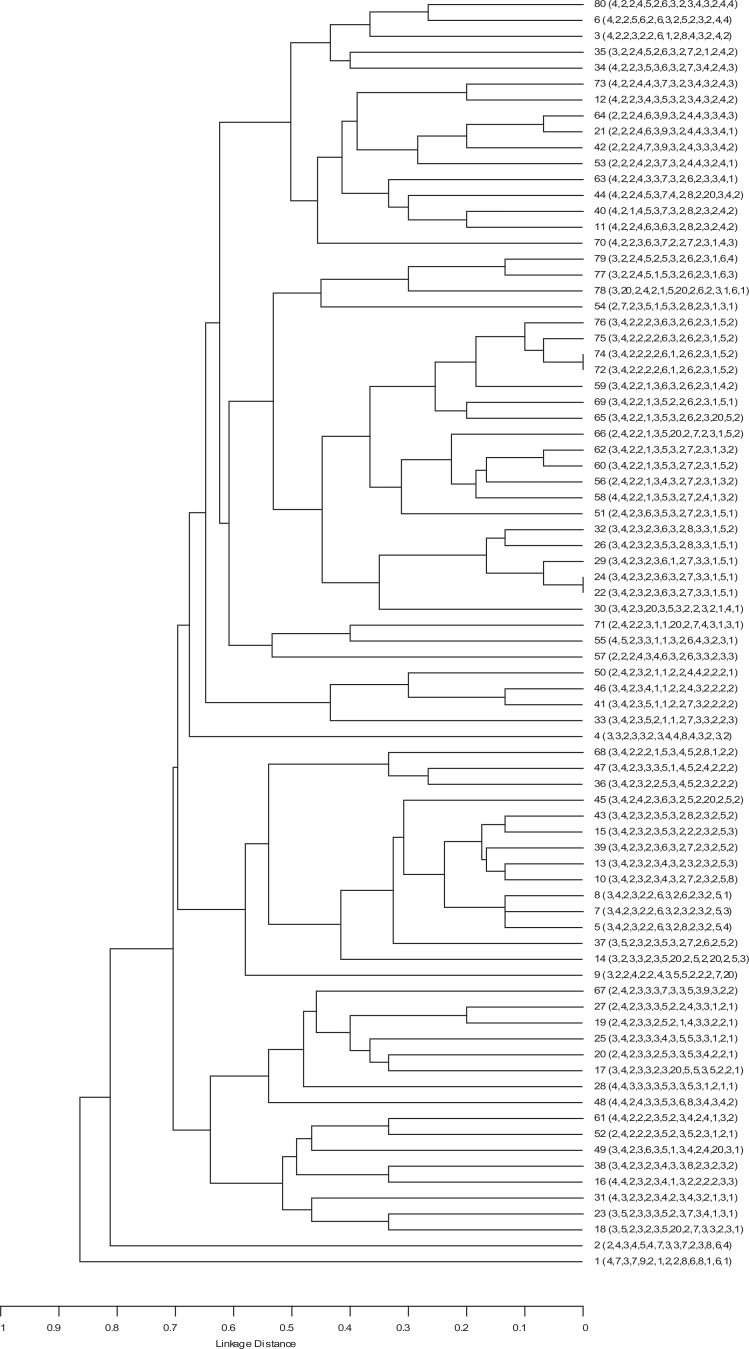
The dendrogram was generated by using the UPGMA algorithm based on the MIRU-VNTR data explaining the genetic relationships of included isolates (Fig. 1). The minimum spanning tree was showed that based on SLV, 4 clonal complexes including 9 isolates existed. Three of these clonal complexes include 2 strains and 1 of the clonal complexes had 3 isolates (Fig. 2). Lineage of all of these 9 isolates was NEW-1.
Fig. 1.
Genetic relationships of 80 isolates.
Fig. 2.
Minimum spanning tree showed the 4 clonal complex.
Discussion
Our study findings indicated that there was considerable genotypic diversity among M. tuberculosis isolates in Tehran, Iran. This finding was consistent with previous studies of M. tuberculosis genotypes in different provinces of Iran including Tehran, which also found a high level of strain diversity among M. tuberculosis isolates [22], [24], [25]. Furthermore, several major TB families/lineages found throughout the world were represented in our studied TB population, which reflects the epidemiology of M. tuberculosis in Tehran. According to the current study, NEW-1, West African and CAS/Delhi were the most frequent lineage. In other studies conducted in Tehran, Ural was found to be the most predominant lineage followed by CAS/Delhi, T and Latin American-Mediterranean (LAM) [26].
Over the last decades, Tehran has experienced rapid development in different areas, which permit movement of people from within the country. According to the statistical information, more than half of the Tehran's population is immigrant from all over the country. Consequently, this might have played an important role in the distributions of different M. tuberculosis strains in this region. These findings have important implications for TB control programs. If infections occur mostly from different sources and diversity in M. tuberculosis isolates are more common in community, efforts should focus on identifying the transmission chain of M. tuberculosis. Furthermore, factors such as high population density, increased prevalence of HIV/AIDS and frequent travel of immigrants should be taken into account when developing infection control strategies [27], [28], [29].
In the present study, MIRU10, MIRU26, MTUB21 and QUB26 were designated as highly discriminative. Previous studies have suggested that different VNTR loci show different discriminatory power for M. tuberculosis strains [30], [31], [32], [33], [34]. Similar to the present study, loci MIRU10, MTUB26, QUB11b and QUB26 were moderately to highly discriminative in studies conducted in Iran [3], [34].
Accordingly, we thought MIRU-VNTR was an easy, reliable, and reproducible method and has high discriminatory power for genetic analysis of epidemiological events such as transmission dynamics.
Some limitations of this study should be considered for results interpretation. First, as TB genotyping currently requires isolation of M. tuberculosis by culture, the major limitation of this study is that it only considered culture-positive patients. We cannot therefore assess the full extent of TB transmission in Tehran. Second, the potential influence of age, sex, HIV/AIDS status and previous treatment could not be analyzed because of the limited information obtained from the patients. Third, due to the limited information, the epidemiological links between patients with similar strains could not be determined.
In conclusion, the findings of this study suggest that there was considerable genotypic diversity among M. tuberculosis isolates in Tehran. The 15-locus MIRU-VNTR showed a high HGDI and could be used as a first-line genotyping method for epidemiological studies. A study with a large sample size for the whole country, in order to provide a detailed population structure of M. tuberculosis circulating in Iran, are strongly recommended.
Acknowledgments
Acknowledgment
This study was supported by Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Conflict of interest
None.
Founding
None.
References
- 1.World Health Organization . 2016. Global tuberculosis report 2016. [Google Scholar]
- 2.Nasiri M.J. Prevalence of drug-resistant tuberculosis in Iran: systematic review and meta-analysis. Am J Infect Control. 2014;42(11):1212–1218. doi: 10.1016/j.ajic.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 3.Zamani S. MIRU-VNTR analysis of the Mycobacterium tuberculosis isolates from three provinces of Iran. Scand J Infect Dis. 2013;45(2):124–130. doi: 10.3109/00365548.2012.717233. [DOI] [PubMed] [Google Scholar]
- 4.Barnes P.F., Cave M.D. Molecular epidemiology of tuberculosis. N Engl J Med. 2003;349(12):1149–1156. doi: 10.1056/NEJMra021964. [DOI] [PubMed] [Google Scholar]
- 5.Hoza A.S. Molecular characterization of Mycobacterium tuberculosis isolates from Tanga, Tanzania: first insight of MIRU-VNTR and microarray-based spoligotyping in a high burden country. Tuberculosis. 2016;98:116–124. doi: 10.1016/j.tube.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Jagielski, T. Methodological and clinical aspects of the molecular epidemiology of Mycobacterium tuberculosis and other Mycobacteria. Clin Microbiol Rev. 2016;29(2):239–290. doi: 10.1128/CMR.00055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamblion E.L. Recent TB transmission, clustering and predictors of large clusters in London, 2010–2012: results from first 3 years of universal MIRU-VNTR strain typing. Thorax. 2016;71(8):749–756. doi: 10.1136/thoraxjnl-2014-206608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Black A.T. Tracking and responding to an outbreak of tuberculosis using MIRU-VNTR genotyping and whole genome sequencing as epidemiological tools. J Public Health. 2017:1–8. doi: 10.1093/pubmed/fdx075. [DOI] [PubMed] [Google Scholar]
- 9.Heidary M., Nasiri M.J. Why has MDR-TB prevalence increased in Iran? J. Clin Tuberculosis Mycobacterial Dis. 2016;5:9. doi: 10.1016/j.jctube.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouklata N. Molecular typing of Mycobacterium Tuberculosis complex by 24-locus based MIRU-VNTR typing in conjunction with spoligotyping to assess genetic diversity of strains circulating in Morocco. PloS One. 2015;10(8) doi: 10.1371/journal.pone.0135695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crudu V. Nosocomial transmission of multidrug-resistant tuberculosis. Int J Tuberculosis Lung Dis. 2015;19(12):1520–1523. doi: 10.5588/ijtld.15.0327. [DOI] [PubMed] [Google Scholar]
- 12.Globan M. Molecular epidemiology of tuberculosis in Victoria, Australia, reveals low level of transmission. Int J Tuberculosis Lung Dis. 2016;20(5):652–658. doi: 10.5588/ijtld.15.0437. [DOI] [PubMed] [Google Scholar]
- 13.Stucki D. Standard genotyping overestimates transmission of Mycobacterium tuberculosis among immigrants in a low-incidence country. J Clin Microbiol. 2016;54(7):1862–1870. doi: 10.1128/JCM.00126-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Supply P. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006;44(12):4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker T.M. A cluster of multidrug-resistant Mycobacterium tuberculosis among patients arriving in Europe from the Horn of Africa: a molecular epidemiological study. Lancet Infect Dis. 2018 [Google Scholar]
- 16.Guthrie J.L. Molecular epidemiology of tuberculosis in British Columbia, Canada–a 10-year retrospective study. Clin Infect Dis. 2017 doi: 10.1093/cid/cix906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nasiri M.J. Drug resistance pattern of Mycobacterium tuberculosis isolates from patients of five provinces of Iran. Asian Pacific J Tropical Med. 2014;7(3):193–196. doi: 10.1016/S1995-7645(14)60019-5. [DOI] [PubMed] [Google Scholar]
- 18.Caulfield A.J., Wengenack N.L. Diagnosis of active tuberculosis disease: from microscopy to molecular techniques. J Clin Tuberculosis Mycobacterial Dis. 2016;4:33–43. doi: 10.1016/j.jctube.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisenach K.D. Polymerase chain reaction amplification of a repetitive DNA sequence specific for Mycobacterium tuberculosis. J Infect Dis. 1990;161(5):977–981. doi: 10.1093/infdis/161.5.977. [DOI] [PubMed] [Google Scholar]
- 20.Boehme C.C. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363(11):1005–1015. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allix-Béguec C. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J Clin Microbiol. 2008;46(8):2692–2699. doi: 10.1128/JCM.00540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zamani S. Genotyping of Mycobacterium tuberculosis Isolates from Hormozgan Province of Iran Based on 15-Locus MIRU-VNTR and Spoligotyping. Int J Bacteriol. 2016:2016. doi: 10.1155/2016/7146470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weniger T. MIRU-VNTR plus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucl Acids Res. 2010;38(suppl_2):W326–W331. doi: 10.1093/nar/gkq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ravansalar H. Genetic Diversity of Mycobacterium tuberculosis Complex Isolated from Patients in the Northeast of Iran by MIRU-VNTR and Spoligotyping. Jundishapur J Microbiol. 2017;10(4) [Google Scholar]
- 25.Boroujeni A.D.K. Genetic diversity of multidrug resistant Mycobacterium tuberculosis strains isolated from tuberculosis patients in Iran, using MIRU-VNTR technique. Eur Respiratory Soc. 2016 [Google Scholar]
- 26.Haeili M. Spoligotyping and drug resistance patterns of Mycobacterium tuberculosis isolates from five provinces of Iran. MicrobiologyOpen. 2013;2(6):988–996. doi: 10.1002/mbo3.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heidary M., Nasiri M.J. Why has HIV/AIDS prevalence increased in Iran? Clin Infect Dis. 2016:ciw361. doi: 10.1093/cid/ciw361. [DOI] [PubMed] [Google Scholar]
- 28.Mirsaeidi S.M. Treatment of multiple drug-resistant tuberculosis (MDR-TB) in Iran. Int J Infect Dis. 2005;9(6):317–322. doi: 10.1016/j.ijid.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 29.Moosazadeh M. Does tuberculosis have a seasonal pattern among migrant population entering Iran? Int J Health Policy Manage. 2014;2(4):181. doi: 10.15171/ijhpm.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L. Highly polymorphic variable-number tandem repeats loci for differentiating Beijing genotype strains of Mycobacterium tuberculosis in Shanghai, China. FEMS Microbiol Lett. 2008;282(1):22–31. doi: 10.1111/j.1574-6968.2008.01081.x. [DOI] [PubMed] [Google Scholar]
- 31.Stavrum R. High diversity of Mycobacterium tuberculosis genotypes in South Africa and preponderance of mixed infections among ST53 isolates. J Clin Microbiol. 2009;47(6):1848–1856. doi: 10.1128/JCM.02167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Y.-J. Use of mycobacterial interspersed repetitive unit-variable-number tandem repeat typing to examine genetic diversity of Mycobacterium tuberculosis in Singapore. J Clin Microbiol. 2004;42(5):1986–1993. doi: 10.1128/JCM.42.5.1986-1993.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou A. Molecular genotyping of Mycobacterium tuberculosis in Xi'an, China, using MIRU-VNTR typing. Int J Tuberculosis Lung Dis. 2011;15(4):517–522. doi: 10.5588/ijtld.10.0495. [DOI] [PubMed] [Google Scholar]
- 34.Asgharzadeh M. Tuberculosis transmission in Northwest of Iran: using MIRU-VNTR, ETR-VNTR and IS6110-RFLP methods. Infect Genet Evol. 2011;11(1):124–131. doi: 10.1016/j.meegid.2010.09.013. [DOI] [PubMed] [Google Scholar]