Abstract
Background
To date, the impact and potential molecular mechanisms of CLDN12 and its association with malignancy in osteosarcoma have not been determined.
Materials and methods
In the present study, the expression profiles of CLDN12 in osteosarcoma cell lines and tissues were explored by immunohistochemistry. A fetal osteoblast cell line was transfected with a eukaryotic expression plasmid, and endogenous CLDN12 in osteosarcoma cells were silenced through an RNA interference (RNAi) method. These transfections were verified, and the activation state of Thr308 site in protein kinase B (Akt) was explored by Western blotting. Moreover, the malignant phenotype of osteosarcoma cells was evaluated by cell counting kit-8 (CCK-8), colony formation, Transwell, and wound-healing assays. Furthermore, osteoblast cells were treated with the phosphatidylinositol-3-kinase (PI3K) inhibitor LY294002 to determine the impact of the PI3K/Akt signaling pathway on cell migration ability.
Results
The results revealed that CLDN12 was overexpressed and localized in the cytoplasm of osteosarcoma cells, and its overexpression was associated with an unfavorable prognosis, irrespective of tumor node metastasis stage. In addition, the knockdown of CLDN12 in cultured osteosarcoma cells markedly attenuated cell proliferation and migration, as indicated by the Cell Counting Kit-8 assay, colony formation assay, scratch wound healing assay and Transwell migration assay. The results also demonstrated that the overexpression of CLDN12 increased the activation of Thr308 site in Akt in fetal osteoblast cells, and the PI3K inhibitor LY294002 partially decreased CLDN12-promoted proliferation and metastasis.
Conclusion
In conclusion, the results of the present study indicated that CLDN12 promoted cell proliferation and migration through the PI3K/Akt signaling pathway in osteosarcoma cells, suggesting that CLDN12 may be a potential agent in the treatment of patients with osteosarcoma.
Keywords: tight junction, claudin-12, osteosarcoma, metastasis, protein kinase B
Introduction
The poor prognosis of osteosarcoma is chiefly attributed to metastasis and recurrence, and the overall 5-year survival rate is less <20%.1,2 Previously, constraining metastasis and the recurrence of osteosarcoma has suggested to be foundation for the therapy of osteosarcoma.3 Therefore, the underlying molecular mechanism governing metastasis in osteosarcoma is of vital research significance. An increasing amount of evidence has suggested that the abnormal expression and location of tight junction (TJ) proteins and claudin (CLDN) serve a crucial role in the tumorigenesis of various types of human tumors via affecting the function of TJs and associated cellular signaling pathways.4,5 CLDNs are the principal components that constitute TJs, which are located at the peak of cell junctions and participate in maintaining cell polarity and permeability.6 The presence of CLDNs in TJs can limit the availability of growth factors and nutrients necessary for the progression of tumor cells, and the abnormal expression level and location of CLDNs contribute to the functional loss of the intercellular connection structure, which is a vital reason for the peripheral infiltration and distant metastasis of tumor cells.7 For instance, studies have demonstrated that the mRNA and protein expression levels of CLDN1 and CLDN7 in certain breast carcinoma cell lines and tissues were notably reduced, and were involved with the recurrence and metastasis of breast carcinoma.8,9 The traditional view is that CLDNs expression are down-regulated in human tumors. However, numerous reports have stated that the expression level of CLDNs in certain human tumors was notably higher than that in the surrounding normal tissues.10–12 For instance, studies investigating expression via gene sequence analysis demonstrated that CLDN3 and CLDN4 were up-regulated in various human tumors such as breast, prostate and pancreatic carcinoma.4,13 These studies suggest that the expression of the CLDN molecule is different among tumors of different tissue sources, and the expression and function of CLDN may exhibit tissue specificity during tumorigenesis in certain tumor types.14–16 The specific expression patterns of CLDNs in tumors suggests that CLDNs can be used as molecular markers for specific human tumor types.
Generally, CLDNs are integral membrane proteins localized at tight junctions, which are responsible for establishing and maintaining epithelial cell polarity and it is accepted that the CLDNs are low expressed in the mesenchymal cells.17–19 At present, there are also several literatures on CLDNs in sarcomas. For instance, the expression of CLDN4, −7 and 10 were found expressed in biphasic synovial sarcomas, and CLDN1 expression was also identified in 63% of Ewing’s sarcoma family tumors.20 Besides, CLDN1 is also found to be significantly expressed in low-grade fibromyxoid sarcomas.21 Moreover, CLDN4 expression has been reported in the glandular component of biphasic synovial sarcoma but has not been systematically evaluated in other sarcoma types.22 Furthermore, it is implied that CLDN1 was initially localized at cell junctions of normal osteoblasts, but substantially delocalized to the nucleus of metastatic osteosarcoma cells, the nucleus localization of CLDN1 contributed to metastatic capacity of osteosarcoma cells.23 At present, the expression of CLDNs in osteosarcoma remains to be further studied. In our work, CLDN12 was demonstrated to be highly expressed in osteosarcoma cell lines and lowly expressed in osteoblastic cell lines, suggesting that increased CLDN12 expression may have an impact on the occurrence and progression of osteosarcoma. To the best of our knowledge, the influence and mechanism of CLDN12 in osteosarcoma tumorigenesis has not yet been reported, and therefore, the present study aimed to investigate the effect of CLDN12 on human osteosarcoma cell migration with molecular biology technology, in an attempt to identify a novel target for the control of early metastasis and treatment in osteosarcoma.
Materials And Methods
Patients And Specimens
A total of 32 human osteosarcoma samples and 34 cases of noncancerous bone tissues were obtained during surgery at Shandong Qilu Hospital (Jinan, Shandong, China) from May 2010 to July 2015. Histologically non-cancerous bone tissues were gained from 34 knee arthritis patients who were treated at Shandong Qilu Hospital between June 2008 and October 2014, including 20 men and 14 women with an average age of 61 years. Snap-frozen samples were maintained in liquid nitrogen and stored at −80°C until subsequent use. Paraffin-embedded tissues were retrieved from the Tissue Bank of Shandong Qilu Hospital, and 4-μm tissue sections were prepared by the Department of Pathology at the same institution. The clinicopathological features of these patients are summarized in Table 1. The present study was approved by the institutional ethics committee at Shandong Qilu Hospital (Ethical Approval Form Number: 2016–002) and adhered to the principles of the Declaration of Helsinki. Written informed consents from all patients were obtained prior to tissue collection.
Table 1.
Expression Of CLDN12 And Clinicopathological Characteristics In OS Patients
| Item | N | CLDN12 Positive | CLDN12 Negative | P |
|---|---|---|---|---|
| Tumor tissue | 32 | 21 | 11 | <0.05a |
| Noncancerous bone tissue | 34 | 12 | 22 | |
| Age (years) | ||||
| ≤19 | 15 | 11 | 4 | 0.347a |
| >19 | 17 | 10 | 7 | |
| Gender | ||||
| Male | 17 | 12 | 5 | 0.648a |
| Female | 15 | 9 | 6 | |
| Stage | ||||
| IA- IIA | 14 | 5 | 9 | <0.001a |
| IIB- III | 18 | 16 | 2 | |
| Response to chemotherapy | ||||
| Poor | 12 | 7 | 5 | 0.748a |
| Good | 6 | 4 | 2 | |
| NA (n =12) | 14 | 10 | 4 | |
| Pulmonary metastasis | ||||
| + | 18 | 15 | 3 | <0.001a |
| - | 14 | 6 | 8 | |
| p-Akt | ||||
| + | 17 | 14 | 3 | <0.001a |
| - | 15 | 7 | 8 |
Note: aStatistical significance was determined with the χ2 test/χ2 Goodness-of-Fit Test.
Immunohistochemistry (IHC)
The 4-μm thick sections were deparaffinized in xylene, rehydrated through a graded ethanol, series and coated in 3% hydrogen peroxide for 10 min at room temperature (RT) in order to block endogenous peroxidase activity, according to the protocol outlined in a previous study.24 The sections were then stained with an anti-CLDN12 at a 1:350 dilution (cat. no. ab53032, Abcam, Cambridge, UK) and anti-phosp-Akt (Thr308) polyclonal antibody at a 1:300 dilution (cat. no. #13038, Cell Signaling Technology, Inc., Danvers, MA, USA) at 4°C overnight using IHC in the Department of Pathology at Shandong Qilu Hospital. Normal rat IgG (1:350 dilution; D110504, Sangon Biotech, Co., Ltd., Shanghai, China) in place of the primary antibody was used as a negative control. Following washing with PBS, the sections were incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody (1:2,000; goat anti-rat cat. no. A0192, Beyotime, Institute of Biotechnology Jiangsu, China) for 1 h at RT. These sections were subsequently stained with 3.39-diaminobenzidine (DAB) (GK500705, Gene Tech, Co., Ltd., Shanghai, China) and were counterstained with hematoxylin. A modified H score system was used to semi-quantitate CLDN12 expression, as described previously25 The expression of CLDN12 in cytoplasm was taken as positive. Briefly, the maximal intensity of staining (0, negative; 1, weak; 2, moderate; and 3, strong) was multiplied by the percentage of positive tumor cells (0–100%) to generate the modified H score (range: 0–300). H-score was calculated using the following formula: H-score= (percentage of cells of weak ×1) + (percentage of cells of moderate×2) + (percentage of cells of strong×3). The maximum H-score would be 300, corresponding to 100% of cells with strong intensity (3+).
Follow-Up
Patients underwent follow-up from the time of diagnosis until 60 months to assess the presence of distant metastasis and to determine the overall survival times. Survival time was calculated as from the date of diagnosis until mortality, or loss to follow-up. By the end of June 2018, all patients had received follow-up via outpatient basis or via a telephone interview to confirm the living status of each patient.
RNA Extraction And Reverse Transcription-Quantitative PCR (RT-qPCR)
Total RNA was isolated from cultured cells using TRIzol® reagent (Thermo Fisher Scientific, Inc.). First-strand cDNA for CLDN12 was synthesized using a High Capacity Reverse Transcription System Kit (Takara). Subsequently, qPCR was performed using a Universal SYBR Green PCR Kit (Takara) and the samples were tested on an ABI 7500 Real-Time PCR System (Applied Biosystems). The results were assessed with the 2-ΔΔCt method using GAPDH or U6 small RNA as the housekeeping genes for the normalization of CLDN12 expression. The RT-qPCR primers included: for CLDN12 forward, 5ʹ- TGCCATGGGCTGTCGGGATGT −3ʹ, and reverse, 5ʹ- TCACTGCTCCCGTCATAC −3ʹ; and β-actin forward, 5ʹ-GCACCACACCTTCTACAATGAG-3ʹ, and reverse, 5ʹ-ACAGCCTGGATGGCTACGT-3ʹ.
Subcellular Fractionation
The cytoplasmic proteins in non-cancerous and osteosarcoma tissues were obtained using NE-PER Nuclear and Cytoplasmic Extraction Reagent kit (Pierce, Rockford, IL, USA). The extraction procedure was conducted based on the manufacturer’s instructions. Thereafter, expression of CLDN12 in non-cancerous and osteosarcoma tissues was detected followed by Western blot analysis.
Western Blotting Analysis
The cytoplasmic proteins in non-cancerous and osteosarcoma tissues were obtained using NE-PER Nuclear and Cytoplasmic Extraction Reagent kit (Pierce, Rockford, IL, USA). Total protein was extracted from cell cultures using an immunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology) containing proteinase promoter cocktail solution (Roche Diagnostics, Basel, Switzerland), and quantified using a bicinchoninic acid protein (BCA) assay (Beyotime Institute of Biotechnology) as according to the manufacturer’s protocol. Western blotting was performed according to standard protocols as previously described.26 Briefly, proteins (30 µg) were separated by 10% SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) membrane. Subsequently, the membrane was blocked in 5% fat-free dry milk at RT for 1 h, and incubated with a rabbit anti-CLDN12 polyclonal antibody (1:1,000 dilution; cat. no. #ab107061, Abcam), rabbit monoclonal antibodies anti-PI3K-p110α (cat. no.# 4255, Cell Signaling Technology, Inc.), anti-Akt (cat. no. #2920, Cell Signaling Technology, Inc.), anti-pAkt (Thr308) (cat. no. #13038, Cell Signaling Technology, Inc.) and mouse anti-human β-actin (cat. no. #ab8227, Abcam) overnight at 4°C. The membranes were washed again with TBST and incubated with respective horseradish peroxidase (HRP)-conjugated secondary antibodies (1:2,000; goat anti-mouse cat. no. A0216 or goat anti-rabbit cat. no. A0239, Beyotime Institute of Biotechnology) at RT for 1 h. The proteins were finally examined by an enhanced chemiluminescence system (ECL) (P0018AS, Beyotime Institute of Biotechnology). The grayscale values of protein bands were analyzed using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Cell Culture And Transfection
The human osteosarcoma cell lines Saos2, 143B, MG63, and U2OS and a human fetal osteoblast cell line hFOB.1.19 were purchased from the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences (Beijing, China). All cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS, Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a humidified atmosphere with 5% CO2. Full-length fragments of CLDN12 were inserted into the pNSE-IRES2-EGFP-C1 vector to generate the CLDN12-overexpression plasmid (constructed and amplified by Nanjing KeyGEN BioTECH Co., Ltd., Nanjing, China), and the pNSE-IRES2-EGFP-C1 vector was the negative control (vector). hFOB.1.19 cells were seeded into 6-well plates at a density of 1×104 cells per cm2, followed by transfection of the above products with Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and G418 (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) was used to expand G418-resistant clone in culture as a monoclonal population.
Transfection Assay For RNAi
The Frozen glycerol bacterial stocks comprising pGCSIL-eGFP-CLDN12-RNAi and pGCSIL-eGFP- vector were obtained from Nanjing KeyGen Biotech Co., Ltd. Transfection of plasmids was achieved using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer’s protocol. Briefly, Saos2 cells (5.6×104 per cm2) were plated into 6-well plates and cultured for 24 h to 75–80% convergence prior to transfection. The plasmid and Lipofectamine 2000 were then combined at a ratio of 3:5 (3 µg of plasmid and 5 µL of Lipofectamine 2000) and were mixed gently and incubated in 500 µL serum-free Opti-MEM (Gibco; Thermo Fisher Scientific, Inc.) for 30 min at RT. The total of the combination (volume 500 uL) was supplemented with RPMI-1640 medium to a final volume of 2mL per well, and incubated for 48 h. Prior to the following experiments being conducted, the flow cytometry was used in the selection of RNAi-transfected cells with eGFP expression.
Cell Proliferation Assay
The Saos2 or hFOB.1.19 cells were seeded at a concentration of 3.6x104 cells per cm2 into 96-well plates in triplicate and cultured in an incubator at 37°C overnight. A growth curve was drawn based on the growth every 12 h over 4 days as analyzed via a colorimetric water-soluble tetrazolium salt kit (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) according to the manufacturer’s protocol. The doubling time was calculated in the exponential phase of growth to describe the proliferation rate.
Cell Colony Formation Assay
A plate colony formation assay was conducted as described previously.27 Briefly, The Saos2 or hFOB.1.19 cells (51 cells per cm2) were seeded in six-well plates and cultivated in RPMI-1640 complete medium at 37°C for 14 days. The cell colonies were washed twice with phosphate-buffered saline (PBS), fixed with methanol for 20 min, and stained with 0.1% crystal violet in PBS (Beyotime Institute of Biotechnology, Haimen, China) for 15 min. The colonies containing >50 cells were counted and the experiments were performed in triplicate.
Cell Migration Assay
A Transwell® migration assay was performed as according to the protocol (Cat. No. 3422, Corning, Tewksbury, MA, USA.) outlined. In brief, The Saos2 or hFOB.1.19 cells (4×105 cells/mL) were plated in serum-free RPMI1640 medium in the top chamber of a Transwell® insert. The corresponding medium supplemented with 15% FBS was placed in the lower chamber and used as a chemoattractant. Then, the cells on the top side of the membrane were removed gently with a cotton swab following a 12 h incubation at 37°C, and cells that had passed through the membrane were fixed with methanol for 15 min and then stained with crystal violet (diluted in 0.1% in PBS) for 10 min. The migrating cells were counted in six randomly selected fields per well, were photographed and the numbers of migrated cells were counted using a light microscope (magnification x200, Olympus Corporation, Tokyo, Japan).
Scratch Wound Healing Assay
A monolayer scratch wound assay was employed as previously described.28 Briefly, cells (4×104 cells per cm2) were plated in 12-well plates and cultured to nearly 100% confluence. A scratch wound was generated with a 200 μl pipette tip. A light microscope (Olympus, Tokyo, Japan) was used to photograph the wound closure at 0, 24 h and 48 h.
Immunofluorescence Method
A cell immunofluorescence assay was conducted as previously described.24 In brief, after washing three times with PBS and fixed with 4% paraformaldehyde for 10 min at RT, the cells were permeabilized with 0.1% Triton X-100 (cat. no. #9002-93-1; Sigma-Aldrich; Merck KGaA). The cells were subsequently blocked with 2% bovine serum albumin (BSA) diluted in PBS (Changchun Changsheng Biological Technology, Co., Ltd., Changchun, China) for 1 h at RT and probed with the anti-human CLDN12 primary antibody diluted in blocking solution (1:1,000) for 30 min at RT. For observation of the nuclei by microscopy the cells were stained with 2% Hoechst 33342 (CAS No. 23491-52-3, Sigma Chemical Co.) solution. Next, cells were stained with Alexa Fluor 647-conjugated anti-rabbit IgG antibodies (cat. no. #ab150093; Santa Cruz Biotechnology, Inc.; 1:1,000) and photographed using a fluorescence microscope (Nikon Eclipse TE2000-S, Nikon Corporation; magnification, ×400).
LY294002 Treatment
The PI3K/Akt inhibitor LY294002 (cat. no.#9901) was obtained from Cell Signaling Technology, Inc. The PI3K/Akt inhibitor LY294002 (5 mg) was dissolved in DMSO to a concentration of 50 mM and stored at −20°C. LY294002 was diluted to a final concentration of 10 nM, and cells were treated for 24 h at this concentration to achieve the full effect of the inhibitor. Cells treated with DMSO at the same volume served as the control group.
Statistical Analysis
The data for histogram was presented as means ± SD of three independent experiments. Data analyses were conducted using GraphPad Prism7 (GraphPad Software, Inc., La Jolla, CA, USA) and SPSS statistical software for Windows, version 22 (SPSS, Chicago, IL, USA). Paired or unpaired Student’s t-tests were performed for continuous variables and Pearson’s χ2 test and Fisher’s exact test were conducted for categorical comparisons. The Kaplan-Meier method was conducted for survival analyses and the log rank test was used to examine the differences in survival. All statistical tests were two-sided and P<0.05 was considered to indicate a statistically significant difference.
Results
Expression Of CLDN12 In Osteosarcoma Cells And Osteoblast Cells
The expression patterns of CLDN12 in the Saos2, MG63,143B, and U2OS osteosarcoma cell lines and the human fetal osteoblast cell line (hFOB.1.19) were explored via RT-qPCR or Western blotting assays. The data revealed that CLDN12 mRNA and protein were highly expressed in the osteosarcoma cell lines Saos2, 143B, MG63 and U2OS yet were expressed at low levels in the human fetal osteoblast cell line hFOB.1.19 (Figure 1A–C).
Figure 1.
The mRNA and protein expression levels of CLDN12 in human fetal-osteoblast line and osteosarcoma cell lines was explored via RT-PCR and Western blot. (A) The relative mRNA expression of CLDN12 in fetal-osteoblast line and osteosarcoma cell lines. (B) The relative protein expression of CLDN12 in fetal-osteoblast line and osteosarcoma cell lines. (C) The corresponding statistical analysis of CLDN12 protein expression. **P<0.01, compared with the fetal-osteoblast line.
Abbreviation: CLDN12, claudin-12.
Expression Of CLDN12 In Human Osteosarcoma Tissues Was Upregulated And Predicts An Unfavorable Prognosis
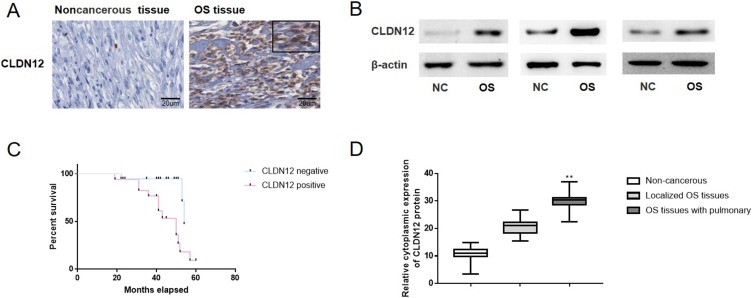
CLDN12 expression was detected in the 32 osteosarcoma tissues specimens and 34 sections of noncancerous bone tissue (Figure 2A). As presented in Table 1, high-level cytoplasmic expression of CLDN12 was observed in 65.6% (21/32) of the osteosarcoma tissues and in 35.3% (12/34) of the noncancerous bone tissues (χ2 test, P=0.032). The CLDN12 was mainly substantially localized to the cytoplasm of metastatic osteosarcoma cells. Moreover, the expression of CLDN12 was not significantly associated with clinical indictors, such as patient age (P=0.347), sex (P=0.648) and response to chemotherapy (P=0.748); however, was significantly associated with TNM stage (P<0.001), phosphate-Akt expression (P<0.001) and pulmonary metastasis (P<0.001; Table 1). These data revealed that the expression of CLDN12 was increased in osteosarcoma tissues and was associated with lymph node phosphate-Akt expression metastasis and pulmonary metastasis.
Figure 2.
The expression of CLDN12 in patients with osteosarcoma and noncancerous tissues were explored via IHC and western-blotting. (A) Detection of CLDN12 in primary osteosarcoma tissues and noncancerous tissues. (B) Protein expression of CLDN12 in human primary osteosarcoma tissues and noncancerous tissues. (C) Kaplan-Meier Survival Curves and the log rank test were used in the analysis of the association between CLDN12 and survival time. (D) The corresponding statistical analysis of CLDN12 protein expression in human primary osteosarcoma tissues and noncancerous tissues. **P<0.01.
Abbreviations: CLDN12, claudin-12; NC, Noncancerous tissues; OS, osteosarcoma tissues.
The cytoplasmic proteins in non-cancerous and osteosarcoma tissues were extracted and Western blotting analysis was also performed with the patient tissues in order to analyze the variations in cytoplasmic expression of CLDN12 between noncancerous tissues, localized osteosarcoma tissues and osteosarcoma tissues with pulmonary metastasis. As it showed in Figure 2B and D, the cytoplasmic expression of CLDN12 was notably upregulated in the osteosarcoma tissues with lung metastasis vs the noncancerous bone tissues (P=0.0007) or localized osteosarcoma tissues (P=0.0011).
The association between CLDN12 and survival time were analyzed using the Kaplan-Meier survival curves and the log rank test. As illustrated in Figure 2C, the patients with high level of CLDN12 expression (median survival, 32.85 months) had a notably shorter survival time than those whose with low expression level of CLDN12 protein (median survival, 45.35 months; P=0.0062).
Knockdown Of CLDN12 Decreased The Migration Ability Of Osteosarcoma Cells
Given that CLDN12 overexpression was observed in osteosarcoma cells, the present study then investigated the impacts of CLDN12 knockdown on osteosarcoma malignancy. As illustrated in Figure 3A, the activation state of the Akt signal pathway in osteosarcoma cell line Saos2 was explored via Western blotting, as displayed in Figure 3A, the ratios of phosphorylated Akt (P=0.0012) were notably decreased in the Saos2 cells with silenced CLDN12. Immunofluorescence was used to detect the expression and localization of CLDN12 in Saos2 cells. The results suggested that CLDN12 was primarily localized on cell cytoplasm (Figure 3B).
Figure 3.
Loss of CLDN12 decreased the migration ability of osteosarcoma cells. (A) Western blotting was used to examine the effects of silencing CLDN12 and the activation of the Akt signaling pathway in the Saos2 cell line. (B) The expression location of CLDN12 was detected via immunofluorescence. The CLDN12 protein was stained with red color and the nuclear was stained with blue color. (C) Growth curve of Saos2 cells detected by the CCK-8 assay. (D) The abilities of Saos2 cells to form colonies under 2D culture conditions were determined via a colony formation assay. (E) The wound-healing assay was utilized to explore the migration ability of Saos2 cells in vitro. (F) Transwell chambers were utilized to explore the impact of CLDN12 silence on the migratory ability of Saos2 cells in vitro. **P<0.01, compared with the scramble group.
Abbreviations: CLDN12, claudin-12; CCK-8, Cell Counting Kit-8.
To determine the impact of CLDN12 on the progression of osteosarcoma cell lines, the CCK8 method was conducted to generate the growth curve of the Saos2 cell line. As depicted in Figure 3C, the doubling time of CLDN12-siRNA transfected cells were notably lower than that of the scramble-transfected cells (P=0.001). The present study also determined the ability of CLDN12-silenced cells to form colonies in 2D monolayer cultures (Figure 3D). The number of colonies formed by CLDN12-siRNA cells were markedly lower than the number formed by the scramble-transfected cells (P=0.0027; P=0.0013, respectively). Upon performing wound-healing and Transwell assays (Figure 3E and F), the present study suggest that CLDN12 silencing inhibits cell migration in the human osteosarcoma cell line Saos2.
Overexpression Of CLDN12 Notably Promoted The Malignant Phenotype Of Human Fetal Osteoblast Cells
Given that low CLDN12 expression was observed in the human fetal osteoblast cell line hFOB.1.19, the present study attempted to explore the effects of CLDN12 overexpression on the metastatic phenotype of human fetal osteoblast cells. The hFOB.1.19 cells were transfected with the pNSE-IRES2-EGFP-C1/CLDN12 plasmid (termed the CLDN12 group) or with the pNSE-IRES2-EGFP-C1 vector as a negative control. The expression of CLDN12 and the changes in the activation state of the PI3K/Akt pathway in these cells were detected via Western blotting. As presented in Figure 4A, the proportions of phosphorylated Akt (P=0.0026) were notably increased in the hFOB.1.19 cells overexpressing CLDN12 (P=0.0001). Immunofluorescence was used to detect the expression and localization of CLDN12 in hFOB.1.19 cells, and the results suggested that CLDN12 was primarily localized on cell cytoplasm (Figure 4B).
Figure 4.
The overexpression of CLDN12 significantly promoted the malignant phenotype of fetal-osteoblast line cell. (A) Western blotting was utilized to examine the expression of CLDN12 and the activities level of the Akt protein. (B) The expression location of CLDN12 was detected via immunofluorescence. The CLDN12 protein was stained with red color and the nuclear was stained with blue color. (C) Growth curve of hFOB.1.19 cells detected by the CCK-8 assay. (D) The abilities of hFOB.1.19 cells to form colonies under 2D culture conditions were determined via the colony formation assay. (E) The wound-healing assay was utilized to explore the migration ability of hFOB.1.19 cells in vitro. (F) The Transwell chambers method was utilized to explore the impact of CLDN12 overexpression on the migratory ability of hFOB.1.19 cells in vitro. **P<0.01, compared with the vector group.
Abbreviations: CLDN12, claudin-12; CCK-8, Cell Counting Kit-8.
To determine the impacts of CLDN12 on the malignant phenotype of hFOB.1.19 cells, the CCK8 method was performed to generate a growth curve for the hFOB.1.19 cell line. As depicted in Figure 4C, the doubling time of hFOB.1.19-CLDN12 cells was notably higher (P=0.0012) than that of the vector plasmid transfected cells. The present study also determined the ability of CLDN12-overexpressing cells to form colonies in 2D monolayer cultures (Figure 4D). The number of colonies formed by CLDN12-overexpressing cells was notably higher than the number formed by the scramble-transfected cells (P=0.0012). Upon performing wound-healing assays (Figure 4E) and Transwell (Figure 4F), cell migration ability was observed to be enhanced following the overexpression of CLDN12 in hFOB.1.19 cells. Taken together, these results suggest that CLDN12 expression has a promotive effect on the malignant phenotype in fetal osteoblast cells.
CLDN12 Modulated Cell Migration Via PI3K/Akt Signaling Pathway In Fetal Osteoblast Cells
To explore the impacts of the PI3K/Akt signaling pathway on cell migration ability, osteoblast cells were treated with the PI3K tyrosinase inhibitor LY294002 (IC50 of 1.6 μM). Following treatment with 10 nM LY294002 for 24 h, the activation state of the PI3K/Akt pathway in osteoblast cells was explored via Western blotting. As depicated in Figure 5A, the ratios of PI3K-p110alpha (P=0.0029) and phosphorylated Akt (P= 0.0017) were significantly decreased in hFOB.1.19 fetal osteoblast cells that overexpressed CLDN12.
Figure 5.
The specific PI3K inhibitor LY294002 was used to inhibit the PI3K activity in hFOB.1.19 cells. (A) Western blotting was utilized to examine the expression of CLDN12 and the activity level of the PI3K and Akt protein. (B) Growth curve of hFOB.1.19 cells detected by the CCK-8 assay. (C) The abilities of hFOB.1.19 cells to form colonies under 2D culture condition were determined via a colony formation assay. (D) The Transwell chambers method was utilized to explore the impact of CLDN12 overexpression on the migratory ability of hFOB.1.19 cells in vitro. (E) The wound-healing assay was utilized to explore the migration ability of hFOB.1.19 cells in vitro. **P<0.01, compared with the DMSO group.
Abbreviations: CLDN12, claudin-12; CCK-8, Cell Counting Kit-8; PI3K, phosphoinositide 3-kinase.
The observations obtained from the CCK8 (Figure 5B) and colony formation (Figure 5C) assays revealed that the doubling time (P=0.0021) and the number of colonies in LY294002-treated cells (P=0.0003) were markedly decreased when compared with the DMSO group. Furthermore, the number of migratory hFOB.1.19 cells was markedly decreased (P=0.0016) following treatment with LY294002 (Figure 5D). In addition, the migration distances of LY294002-treated cells were notably decreased when compared with the DMSO-treated group at 12 and 24 h (P=0.0034 and P=0.0002, respectively; Figure 5E).
Discussion
CLDNs are integral components of TJs, and its membrane expression maintain paracellular permeability and are critical for epithelial cell polarity.19 It is revealed that the change of expression level and localization of CLDNs was an important cause of tumor development.29,30 For example, a concurrent cytoplasmic subcellular location of CLDN1 expression is associated with malignant canine thyroid tumors and their vascular invasion thyroid tumors.31 Similarly, a recent study revealed that the delocalization of CLDN6 promoted a malignancy in myxofibrosarcomas, and could be used as a potential marker for myxofibrosarcomas.32 In lung adenocarcinoma (ACA), CLDN3 and CLDN4 were hypothesized to be biomarkers of carcinoma when discriminating lung ACA from mesothelioma. Additionally, Dhawan et al reported that the de-localization of CLDN1 from TJs to the cytoplasm occurred in tumors with unfavorable behavior in human colorectal cancer.33 The subcellular alteration of CLDN1 from its typical membranous location may be on account of structural changes in TJs that would contribute to significant modifications in paracellular permeability, and thereby lead to an abnormal fluidity of nutrients and signaling proteins, which is suggested to be vital for tumor progression.4,34 The results of the present study suggest that the cytoplasmic expression of CLDN12 was overexpressed in human osteosarcoma, and that this overexpression was involved with distant metastasis. To validate this hypothesis, a CLDN12-overexpressing fetal osteoblast cell line and a CLDN12-knockdown osteosarcoma cell line were created, and the results indicated that the loss of CLDN12 inhibited the ability of proliferation and migration in osteosarcoma cells.
Previously, numerous evidences revealed that the pAkt expression in osteosarcoma promotes the progression of osteosarcoma while considering the introduction of Akt inhibitors for osteosarcoma treatment.35,36 Additionally, at present study, the PI3K/Akt signaling pathway was revealed to be associated with the influence of CLDN12 on the metastatic ability of osteosarcoma cells. Previous studies have demonstrated that the carboxyl terminal in the cytoplasm of CLDNs is rich in the PDZ binding domain, which can bind with other TJ membrane-related proteins to participate in intercellular signal transduction.37,38 For example, CLDNs can interact with the zonula occludens to participate in cell barrier function, cell proliferation and apoptosis.39,40 The PI3K/Akt signaling pathway was observed to serve a vital role in tumorigenesis and regulates critical cellular functions including survival, proliferation and metabolism.41–43 Currently, a limited number of studies have demonstrated that PI3K/Akt signaling was associated with the role of CLDNs protein in human tumorigenesis. The results of the present study indicated that the upregulation of CLDN12 contributed to an enhancement of cell migration via the PI3K/Akt signaling pathway in osteosarcoma. There are serval literatures to support that a similar mechanism in sarcomas. For instance, the CLDN8 was reported to contribute to a malignant proliferation in human osteosarcoma U2OS cells.44 On the contrary, CLDN2 and afadin were found to be under-expressed in osteosarcoma tissues, and the overexpression of CLDN2 significantly inhibited the migration abilities of osteosarcoma cells.45 However, the exact molecular mechanisms of the signal transduction from cytoplasm-anchored CLDN12 to Akt has yet to be completely elucidated.
The presently available treatment for patients with advanced osteosarcoma remains unsatisfactory, therefore it is crucial to elucidate the detailed mechanisms that culminates in disease progression of osteosarcoma.3 To date, the majority of these studies have demonstrated that CLDN12 impacts epithelial permeability and cell junctions.46–49 However, whether CLDN12 is involved in oncogenesis or tumor progression remains unclear. In the present research, the pro-oncogenic role of CLDN12 on osteosarcoma was investigated in fetal osteoblast and osteosarcoma cells and clarified the possible mechanism by which CLDN12 promoted osteosarcoma cells proliferation and migration. In addition, the pro-cancer effects of CLDN12 is association with the PI3K/Akt signaling pathway were reported, which supports the results of a previous study that stated that the PI3K/Akt signaling pathway is associated with cell migration in osteosarcoma.50 The results of the present study revealed that PI3K inhibition partially reversed CLDN12-induced hFOB.1.19 cell proliferation and migration. This phenomenon indicates that CLDN12-induced PI3K/Akt signaling pathway is closely associated with the proliferation and migration of osteosarcoma. However, further studies should investigate the effect of CLDN12 on other signaling pathway in osteosarcoma cells.
Conclusion
In summary, the findings of the present study suggest that CLDN12 can promote proliferation and migration ability of osteosarcoma cells via the PI3K/Akt signaling pathway. The present study provides a foundation for further preclinical evaluations, which may serve as a potential anti-cancer agent in the therapy of patients with osteosarcoma.
Acknowledgements
We would like to thank American Journal Experts (AJE) and Spandidos Publications English Language Editing Service for help with this manuscript. No funding was received.
Abbreviations
TJ, Tight junction; CLDNs, claudins; CCK8, Cell Counting Kit-8; Akt, protein kinase B; PI3K, phosphatidylinositol-3-kinase.
Availability Of Data And Materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Wang W, Yang J, Wang Y, et al. Survival and prognostic factors in Chinese patients with osteosarcoma: 13-year experience in 365 patients treated at a single institution. Pathol Res Pract. 2017;213(2):119–125. doi: 10.1016/j.prp.2016.11.009 [DOI] [PubMed] [Google Scholar]
- 2.Anderson ME. Update on Survival in Osteosarcoma. Orthop Clin North Am. 2016;47(1):283–292. doi: 10.1016/j.ocl.2015.08.022 [DOI] [PubMed] [Google Scholar]
- 3.Salunke AA, Shah J, Gupta N, Pandit J. Pathologic fracture in osteosarcoma: association with poorer overall survival. Eur J Surg Oncol. 2016;42(6):889–890. doi: 10.1016/j.ejso.2016.02.255 [DOI] [PubMed] [Google Scholar]
- 4.Osanai M, Takasawa A, Murata M, Sawada N. Claudins in cancer: bench to bedside. Pflugers Archiv. 2017;469(1):55–67. doi: 10.1007/s00424-016-1877-7 [DOI] [PubMed] [Google Scholar]
- 5.Kwon MJ. Emerging roles of claudins in human cancer. Int J Mol Sci. 2013;14(9):18148–18180. doi: 10.3390/ijms140918148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balda MS, Matter K. Tight junctions at a glance. J Cell Sci. 2008;121(22):3677–3682. doi: 10.1242/jcs.023887 [DOI] [PubMed] [Google Scholar]
- 7.Zorn-Kruppa M, Vidal YSS, Houdek P, et al. Tight Junction barriers in human hair follicles - role of claudin-1. Sci Rep. 2018;8(1):12800. doi: 10.1038/s41598-018-30341-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katayama A, Handa T, Komatsu K, et al. Expression patterns of claudins in patients with triple-negative breast cancer are associated with nodal metastasis and worse outcome. Pathol Int. 2017;67(8):404–413. doi: 10.1111/pin.v67.8 [DOI] [PubMed] [Google Scholar]
- 9.Upmanyu N, Bulldan A, Papadopoulos D, Dietze R, Malviya VN, Scheiner-Bobis G. Impairment of the Gnalpha11-controlled expression of claudin-1 and MMP-9 and collective migration of human breast cancer MCF-7 cells by DHEAS. J Steroid Biochem Mol Biol. 2018;182:50–61. doi: 10.1016/j.jsbmb.2018.04.010 [DOI] [PubMed] [Google Scholar]
- 10.Blanchard AA, Ma X, Wang N, et al. Claudin 1 is highly upregulated by PKC in MCF7 human breast cancer cells and correlates positively with PKCepsil on in patient biopsies. Transl Oncol. 2019;12(3):561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaaskelainen A, Soini Y, Jukkola-Vuorinen A, Auvinen P, Haapasaari KM, Karihtala P. High-level cytoplasmic claudin 3 expression is an independent predictor of poor survival in triple-negative breast cancer. BMC Cancer. 2018;18(1):223. doi: 10.1186/s12885-018-4141-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Z, Shi J, Song Y, et al. Claudin-7 (CLDN7) is overexpressed in gastric cancer and promotes gastric cancer cell proliferation, invasion and maintains mesenchymal state. Neoplasma. 2018;65(3):349–359. doi: 10.4149/neo_2018_170320N200 [DOI] [PubMed] [Google Scholar]
- 13.Tabaries S, Siegel PM. The role of claudins in cancer metastasis. Oncogene. 2017;36(9):1176–1190. doi: 10.1038/onc.2016.289 [DOI] [PubMed] [Google Scholar]
- 14.Okui N, Kamata Y, Sagawa Y, et al. Claudin 7 as a possible novel molecular target for the treatment of pancreatic cancer. Pancreatology. 2019;19(1):88–96. doi: 10.1016/j.pan.2018.10.009 [DOI] [PubMed] [Google Scholar]
- 15.Martin de la Fuente L, Malander S, Hartman L, et al. Claudin-4 expression is associated with survival in ovarian cancer but not with chemotherapy response. Int J Gynecoll Pathol. 2018;37(2):101–109. doi: 10.1097/PGP.0000000000000394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K, Xu C, Li W, Ding L. Emerging clinical significance of claudin-7 in colorectal cancer: a review. Cancer Manag Res. 2018;10:3741–3752. doi: 10.2147/CMAR.S175383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berndt P, Winkler L, Cording J, et al. Tight junction proteins at the blood-brain barrier: far more than claudin-5. Cell Mol Life Sci. 2019;76:1987–2002. doi: 10.1007/s00018-019-03030-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao J, Krystofiak ES, Ballesteros A, et al. Multiple claudin-claudin cis interfaces are required for tight junction strand formation and inherent flexibility. Commun Biol. 2018;1:50. doi: 10.1038/s42003-018-0051-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vermette D, Hu P, Canarie MF, Funaro M, Glover J, Pierce RW. Tight junction structure, function, and assessment in the critically ill: a systematic review. Intensive Care Med Exp. 2018;6(1):37. doi: 10.1186/s40635-018-0203-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ouban A, Ahmed A. Claudins in human cancer, a review. Histol Histopathol. 2010. [DOI] [PubMed] [Google Scholar]
- 21.Thway K, Fisher C, Debiec-Rychter M, Calonje E. Claudin-1 is expressed in perineurioma-like low-grade fibromyxoid sarcoma. Hum Pathol. 2009;40(11):1586–1590. doi: 10.1016/j.humpath.2009.04.003 [DOI] [PubMed] [Google Scholar]
- 22.Schaefer I-M, Agaimy A, Fletcher CD, Hornick JL. Claudin-4 expression distinguishes SWI/SNF complex-deficient undifferentiated carcinomas from sarcomas. Mod Pathol. 2017;30(4):539. doi: 10.1038/modpathol.2016.230 [DOI] [PubMed] [Google Scholar]
- 23.Jian Y, Chen C, Li B, Tian X. Delocalized Claudin-1 promotes metastasis of human osteosarcoma cells. Biochem Biophys Res Commun. 2015;466(3):356–361. doi: 10.1016/j.bbrc.2015.09.028 [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Ruan Y, Li Y, Lin D, Quan C. Tight junction protein claudin-6 inhibits growth and induces the apoptosis of cervical carcinoma cells in vitro and in vivo. Med Oncol. 2015;32(5):148. doi: 10.1007/s12032-015-0600-4 [DOI] [PubMed] [Google Scholar]
- 25.Howitt BE, Sun HH, Roemer MG, et al. Genetic basis for PD-L1 expression in squamous cell carcinomas of the cervix and vulva. JAMA Oncol. 2016;2(4):518–522. doi: 10.7150/ijms.22927 [DOI] [PubMed] [Google Scholar]
- 26.Zhao S, Zhou L, Niu G, Li Y, Zhao D, Zeng H. Differential regulation of orphan nuclear receptor TR3 transcript variants by novel vascular growth factor signaling pathways. FASEB J. 2014;28(10):4524–4533. doi: 10.1096/fj.13-248401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu HM, Chen Y, Liu L, et al. C1orf61 acts as a tumor activator in human hepatocellular carcinoma and is associated with tumorigenesis and metastasis. FASEB J. 2013;27(1):163–173. doi: 10.1096/fj.12-216622 [DOI] [PubMed] [Google Scholar]
- 28.Niu G, Ye T, Qin L, et al. Orphan nuclear receptor TR3/Nur77 improves wound healing by upregulating the expression of integrin beta4. FASEB J. 2015;29(1):131–140. doi: 10.1096/fj.14-257550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ouban A. Claudin-1 role in colon cancer: an update and a review. Histol Histopathol. 2018;33:11980. [DOI] [PubMed] [Google Scholar]
- 30.Che J, Yue D, Zhang B, et al. Claudin-3 inhibits lung squamous cell carcinoma cell epithelial-mesenchymal Transition and invasion via suppression of the Wnt/beta-catenin signaling pathway. Int J Med Sci. 2018;15(4):339–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suren D, Yildirim M, Sayiner A, et al. Expression of claudin 1, 4 and 7 in thyroid neoplasms. Oncol Lett. 2017;13(5):3722–3726. doi: 10.3892/ol.2017.5916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bekki H, Yamamoto H, Takizawa K, et al. Claudin 6 expression is useful to distinguish myxofibrosarcomas from other myxoid soft tissue tumors. Pathol Res Pract. 2017;213(6):674–679. doi: 10.1016/j.prp.2016.12.001 [DOI] [PubMed] [Google Scholar]
- 33.Jiang L, Yang L, Huang H, Liu BY, Zu G. Prognostic and clinical significance of claudin-1 in colorectal cancer: a systemic review and meta-analysis. Int J Surg. 2017;39:214–220. doi: 10.1016/j.ijsu.2017.02.005 [DOI] [PubMed] [Google Scholar]
- 34.Takasawa K, Takasawa A, Osanai M, et al. Claudin-18 coupled with EGFR/ERK signaling contributes to the malignant potentials of bile duct cancer. Cancer Lett. 2017;403:66–73. doi: 10.1016/j.canlet.2017.05.033 [DOI] [PubMed] [Google Scholar]
- 35.Van De Luijtgaarden AC, Roeffen MH, Leus MA, et al. IGF signaling pathway analysis of osteosarcomas reveals the prognostic value of pAKT localization. Future Oncol. 2013;9(11):1733–1740. doi: 10.2217/fon.13.118 [DOI] [PubMed] [Google Scholar]
- 36.Zhang G, Li M, Zhu X, Bai Y, Yang C. Knockdown of Akt sensitizes osteosarcoma cells to apoptosis induced by cisplatin treatment. Int J Mol Sci. 2011;12(5):2994–3005. doi: 10.3390/ijms12052994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marunaka K, Furukawa C, Fujii N, et al. The RING finger- and PDZ domain-containing protein PDZRN3 controls localization of the Mg(2+) regulator claudin-16 in renal tube epithelial cells. J Biol Chem. 2017;292(31):13034–13044. doi: 10.1074/jbc.M117.779405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeansonne B, Lu Q, Goodenough DA, Chen YH. Claudin-8 interacts with multi-PDZ domain protein 1 (MUPP1) and reduces paracellular conductance in epithelial cells. Cell Mol Biol. 2003;49(1):13–21. [PubMed] [Google Scholar]
- 39.Merino-Gracia J, Costas-Insua C, Canales MA, Rodriguez-Crespo I. Insights into the C-terminal Peptide Binding Specificity of the PDZ Domain of Neuronal Nitric-oxide Synthase: characterization of the interaction with the tight junction protein claudin-3. J Biol Chem. 2016;291(22):11581–11595. doi: 10.1074/jbc.M116.724427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monteiro AC, Sumagin R, Rankin CR, et al. JAM-A associates with ZO-2, afadin, and PDZ-GEF1 to activate Rap2c and regulate epithelial barrier function. Mol Biol Cell. 2013;24(18):2849–2860. doi: 10.1091/mbc.e13-06-0298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamilton BJ, Tse D, Stan RV. Phorbol esters induce PLVAP expression via VEGF and additional secreted molecules in MEK1-dependent and p38, JNK and PI3K/Akt-independent manner. J Cell Mol Med. 2019;23(2):920–933. doi: 10.1111/jcmm.2019.23.issue-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lv Y, Liu W, Ruan Z, Xu Z, Fu L. Myosin IIA regulated tight junction in oxygen glucose-deprived brain endothelial cells via activation of TLR4/PI3K/Akt/JNK1/2/14-3-3epsilon/NF-kappaB/MMP9 signal transduction pathway. Cell Mol Neurobiol. 2019;39(2):301–319. doi: 10.1007/s10571-019-00654-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manthari RK, Tikka C, Ommati MM, et al. Arsenic induces autophagy in developmental mouse cerebral cortex and hippocampus by inhibiting PI3K/Akt/mTOR signaling pathway: involvement of blood-brain barrier’s tight junction proteins. Arch Toxicol. 2018;92(11):3255–3275. doi: 10.1007/s00204-018-2304-y [DOI] [PubMed] [Google Scholar]
- 44.Xu J, Yang Y, Hao P, Ding X. Claudin 8 contributes to malignant proliferation in human osteosarcoma U2OS cells. Cancer Biother Radiopharm. 2015;30(9):400–404. doi: 10.1089/cbr.2015.1815 [DOI] [PubMed] [Google Scholar]
- 45.Zhang X, Wang H, Li Q, Li T. CLDN2 inhibits the metastasis of osteosarcoma cells via down-regulating the afadin/ERK signaling pathway. Cancer Cell Int. 2018;18(1):160. doi: 10.1186/s12935-018-0662-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cetinkaya A, Taskiran E, Soyer T, et al. Dermal fibroblast transcriptome indicates contribution of WNT signaling pathways in the pathogenesis of Apert syndrome. Turk J Pediatr. 2017;59(6):619–624. doi: 10.24953/turkjped.2017.06.001 [DOI] [PubMed] [Google Scholar]
- 47.Sabri A, Lai D, D’Silva A, et al. Differential placental gene expression in term pregnancies affected by fetal growth restriction and macrosomia. Fetal Diagn Ther. 2014;36(2):173–180. doi: 10.1159/000360535 [DOI] [PubMed] [Google Scholar]
- 48.Chen YC, Hsiao CJ, Jung CC, et al. Performance metrics for selecting single nucleotide polymorphisms in late-onset Alzheimer’s disease. Sci Rep. 2016;6:36155. doi: 10.1038/srep36155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hashimoto R, Ikeda M, Ohi K, et al. Genome-wide association study of cognitive decline in schizophrenia. Am J Psychiatry. 2013;170(6):683–684. doi: 10.1176/appi.ajp.2013.12091228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hao NB, Tang B, Wang GZ, et al. Hepatocyte growth factor (HGF) upregulates heparanase expression via the PI3K/Akt/NF-kappaB signaling pathway for gastric cancer metastasis. Cancer Lett. 2015;361(1):57–66. doi: 10.1016/j.canlet.2015.02.043 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.