Abstract
The tumor suppressor TP53 is mutated in the majority of human cancers and over 70% of pancreatic ductal adenocarcinoma (PDAC)1,2. Wild-type p53 accumulates in response to cellular stress and regulates gene expression to alter cell fate and prevent tumor development2. Wild-type p53 also modulates cellular metabolic pathways3, though p53-dependent metabolic alterations that constrain cancer progression remain poorly understood. Here, we find that p53 remodels cancer cell metabolism to enforce changes in chromatin and gene expression that favor premalignant cell fate. Restoring p53 function in cancer cells derived from Kras mutant murine PDAC models leads to the accumulation of α-ketoglutarate (αKG), a metabolite that also serves as an obligate substrate for a subset of chromatin modifying enzymes. p53 induces transcriptional programs characteristic of premalignant differentiation, an effect that can be partially recapitulated by addition of cell-permeable αKG. Increased levels of the αKG-dependent chromatin modification 5-hydroxymethylcytosine (5hmC) accompany tumor cell differentiation triggered by p53, while decreased 5hmC characterizes the transition from premalignant to de-differentiated malignant lesions associated with mutations in p53. Enforcing αKG accumulation in p53-deficient PDAC cells through the inhibition of the tricarboxylic acid (TCA) cycle enzyme oxoglutarate (αKG) dehydrogenase (Ogdh) specifically results in enhanced 5hmC, tumor cell differentiation, and decreased tumor cell fitness. Conversely, elevating intracellular levels of succinate, a competitive inhibitor of αKG-dependent dioxygenases, blunts p53-driven tumor suppression. Together these data nominate αKG as an effector of p53-mediated tumor suppression whose accumulation in p53-deficient tumors can drive tumor cell differentiation and antagonize malignant progression.
PDAC is a lethal cancer dominated by co-occurring oncogenic Kras mutations and inactivating TP53 mutations1. Mutant Kras is sufficient to trigger a spectrum of well-differentiated premalignant PDAC precursor lesions, including pancreatic intraepithelial neoplasia (PanIN)4. Malignant progression often coincides with the acquisition of TP53 mutations, which are associated with more poorly differentiated disease5. Whereas studies have established key roles for KRAS in the metabolic rewiring of PDAC cells6,7, it remains to be determined whether p53-regulated metabolism limits PDAC development or if the metabolic consequences of TP53 mutation sustain malignant disease.
To study this, we developed a modified version of the “KPC” mouse model that recapitulates many aspects of human PDAC progression and histopathology8,9 via pancreas-specific expression of oncogenic Kras (LSL-KrasG12D) and inactivation of Trp53 (hereafter, p53) through either conditional p53 deletion or mutation. In our system, loss of p53 function is achieved by a doxycycline (dox)-regulated p53 shRNA and an mKate2-linked rtTA tet-on allele. In this “KPsh”configuration, expression of mutant KrasG12D by pancreas-specific Cre recombination is followed by suppression of p53 expression upon dox administration, thus permitting reactivation of endogenous p53 following tumor formation. Like p53 mutations, shRNA-mediated p53 suppression together with Kras activation accelerates the onset of aggressive, moderately to poorly differentiated disease10.
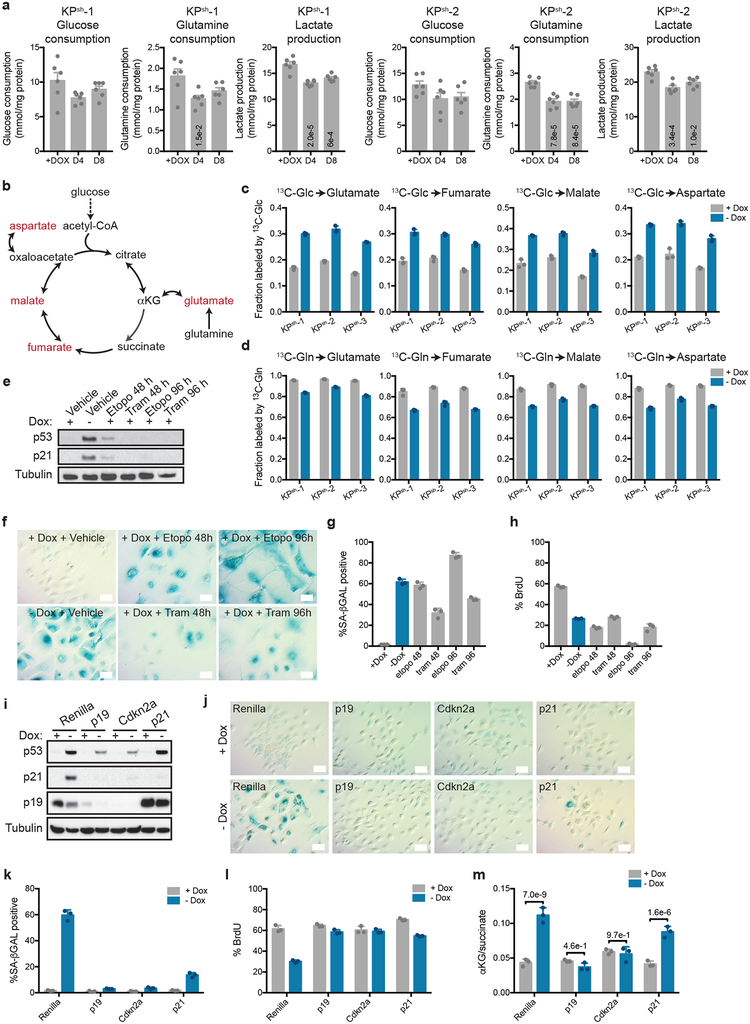
In PDAC cell lines derived from tumors arising in three independent dox fed mice (designated KPsh-1–3, Extended Data Fig. 1a)10, dox withdrawal resulted in the induction of p53 protein, its targets Cdkn1a/p21 and Mdm2, and cellular senescence (Fig. 1a and Extended Data Fig. 1b–f). Similarly, dox withdrawal from mice harboring orthopic tumors produced from KPsh cells extinguished shp53 expression, leading to Cdkn1a/p21 induction, decreased tumor growth and enhanced animal survival (Extended Data Fig. 1g–k). Of note, the levels of Cdkn1a/p21 that were induced in tumors upon p53 activation were higher than those observed in surrounding normal cells (Extended Data Fig. 1h), suggesting that Kras potentiates the p53 response11. Thus, as has been observed in other contexts11–14, p53 inactivation is required to sustain tumorigenesis.
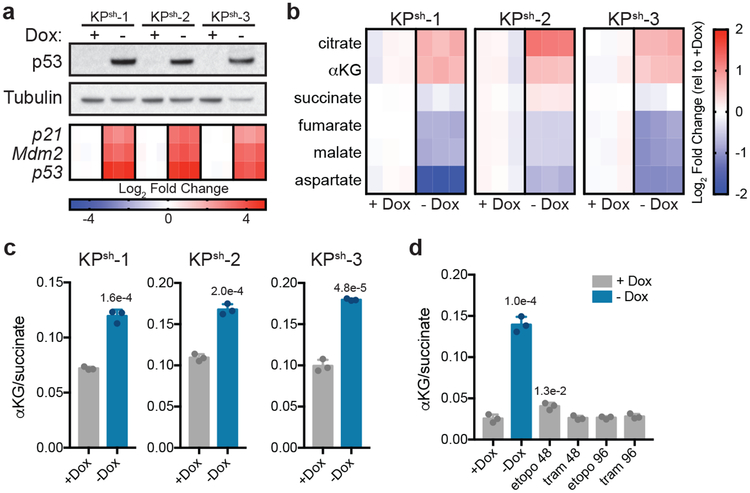
Figure 1. p53 restoration increases the cellular αKG/succinate ratio independently of changes in proliferation.
a, Western blot (top) and qRT-PCR (bottom) of KPsh-1–3 lines cultured with or without doxycycline (dox) for six days. Gene expression is represented as the log2 fold change relative to +dox controls for each line. b,c Steady-state levels of TCA cycle metabolites (b) or αKG/succinate ratio (c) in cells cultured with or without dox for eight days. d, αKG/succinate ratio in cells cultured on dox with 25 nM trametinib or 3 μM etoposide for 48 or 96 has shown. Cells cultured without dox for six days are included as a control. a-d were repeated twice with similar results. Data are presented as either a representative independently treated well (a, top), as individual data points (a bottom, b), or as mean ± SD of n=3 independently treated wells with individual data points shown (c,d). For gel source data (a), see Supplementary Figure 1. Significance assessed by two-tailed Student’s t-test (c) or 1-way ANOVA with Tukey’s post-test (d) in comparison with vehicle treated cells grown with dox.
Taking advantage of the cell lines described above, we studied the impact of wild-type p53 on PDAC metabolism. PDAC cells driven by oncogenic Kras and p53 disruption consume high amounts of glucose and glutamine, which provide the primary substrates that support anabolic proliferation in cultured cancer cells6,7. Surprisingly, p53 restoration did not result in major changes in glucose and glutamine consumption or lactate production, despite the induction of cell cycle arrest (Extended Data Fig. 1b,d,f, 2a). However, monitoring glucose and glutamine utilization via the mitochondrial tricarboxylic acid (TCA) cycle indicated that p53 triggered a metabolic switch marked by enhanced incorporation of glucose-derived carbons and reduced contribution of glutamine-derived carbons into TCA cycle intermediates (Extended Data Fig. 2b–d and Supplementary Table 1). Accordingly, citrate and αKG accumulated in response to p53, whereas metabolites derived from glutamine oxidation including succinate, malate and aspartate progressively decreased (Fig. 1b, and Supplementary Table 1). This metabolic shift produced an increase in the αKG/succinate ratio (Fig. 1c), which is implicated in cell fate decisions in some contexts15–17.
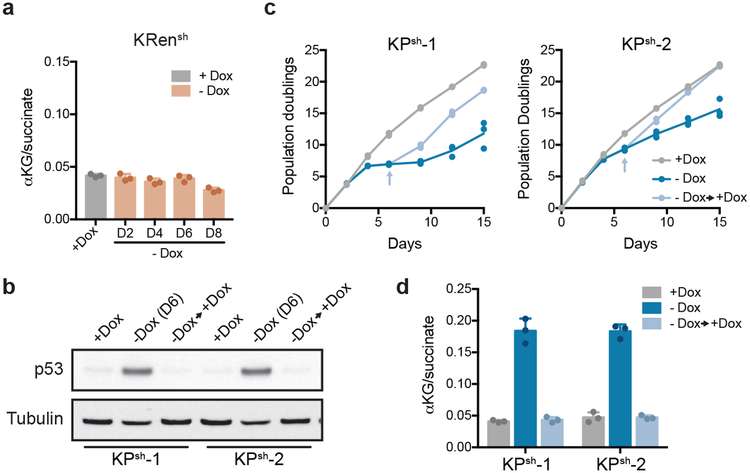
The elevated αKG/succinate ratio observed in cells responding to p53 was not a secondary consequence of decreased proliferation and could be uncoupled from p53-driven cell cycle arrest. Hence, treatment of p53-silenced cells with the chemotherapeutic drug etoposide or the MEK inhibitor trametinib induced cycle arrest and senescence but did not alter the αKG/succinate ratio (Extended Data Fig. 2e–h; Fig. 1d). Furthermore, KPsh cells transduced with constitutive shRNAs targeting either Arf or Cdkn1a/p21, which can attenuate p53 accumulation or act downstream to circumvent cell cycle arrest, respectively (Extended Data Fig. 2i)2,11, did not arrest to the same extent as controls following dox withdrawal (Extended Data Fig. 2j–l). However, while Arf-silenced cells failed to efficiently induce p53 and showed no increase in the αKG/succinate ratio (Extended Data Fig. 2i,m), Cdkn1a/p21-silenced cells still accumulated p53 and increased the αKG/succinate ratio (Extended Data Fig. 2i,m). Moreover, the increase in the αKG/succinate ratio was not due to withdrawal of dox after long term exposure (Extended Data Fig. 3a) and was reversible upon p53 suppression following dox re-addition (Extended Data Fig. 3b–d).
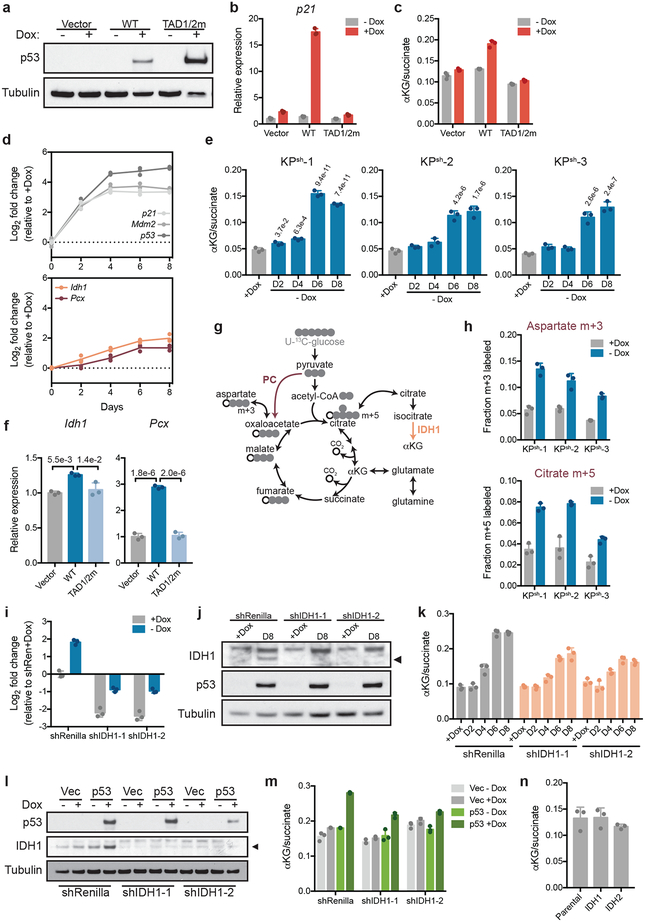
The tumor suppressor activity of p53 is closely associated with its ability to transcriptionally activate downstream target genes2. Accordingly, enforced expression of wild-type p53—but not a well-characterized transactivation-defective p53 mutant18—was able to induce Cdkn1a/p21 and increase the αKG/succinate ratio in p53 null PDAC cells (Extended Data Fig. 4a–c). Transcriptional profiling following p53 restoration in KPsh cells revealed a significant increase in the TCA cycle enzymes pyruvate carboxylase (Pcx, PC) and isocitrate dehydrogenase 1 (Idh1, IDH1) with kinetics similar to those of the increase in the αKG/succinate ratio (Extended Data Fig. 4d,e), and both were induced by ectopic p53 expression in a transactivation domain-dependent manner (Extended Data Fig. 4f).
PC enables glucose-derived anaplerosis that can relieve the requirement for consumption of glutamine-derived αKG by the TCA cycle19, while IDH1 generates cytosolic αKG from citrate, and so increased levels of either enzyme (or both) could facilitate αKG accumulation (Extended Data Fig. 4g). Indeed, p53 activation triggered an increase in glucose anaplerosis that is associated with PC activity (Extended Data Fig. 4h) and inhibition of p53-driven Idh1 expression blunted the p53-mediated increase in the αKG/succinate ratio (Extended Data Fig. 4i–m). Still, IDH1 overexpression is not sufficient to increase the αKG/succinate ratio in the absence of p53 (Extended Data Fig. 4n). Moreover, both the Pcx and Idh1 loci harbor predicted p53 targets sites that bind p53 in some cell types (Extended Data Fig. 5a,b). Therefore, although the precise mechanism whereby p53 increases the αKG/succinate ratio remains to be determined, our data collectively suggest that this process involves changes in multiple TCA enzyme levels arising as a part of canonical p53 transcriptional functions.
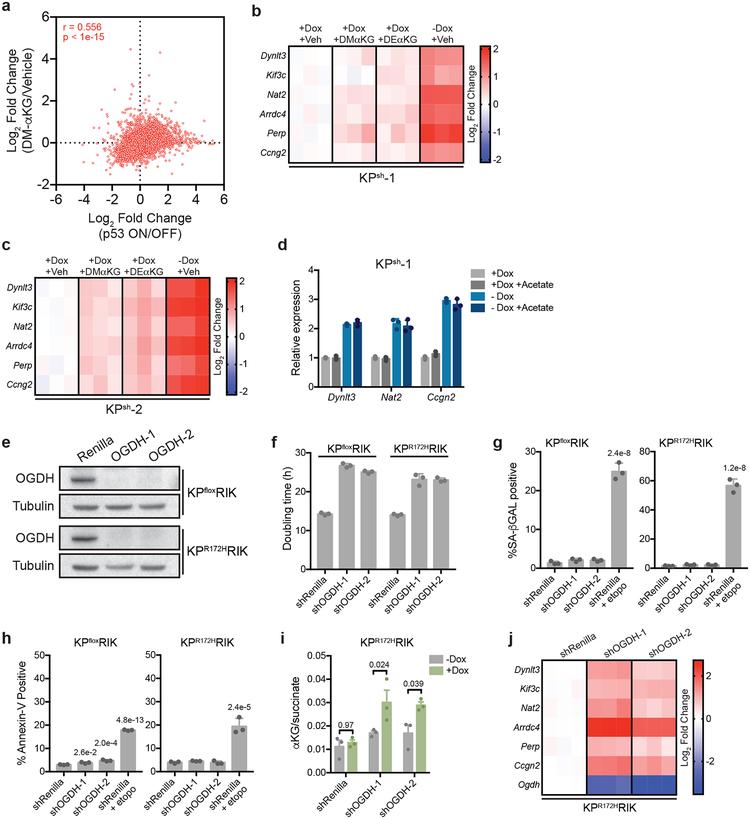
Alterations in the intracellular αKG/succinate ratio are associated with changes in the activity of αKG-dependent chromatin modifying enzymes that can alter transcription15–17. To determine the extent to which αKG alone recapitulates the effects of p53 on global chromatin states and gene expression, we performed ATAC-Seq (which provides a global view of chromatin accessibility that can reflect changes in αKG-dependent enzyme function20,21) and RNA-Seq on KPsh cells subjected either to p53 reactivation or treatment with cell-permeable αKG. Both p53 and αKG increased chromatin accessibility, with a strong correlation between those regions affected by p53 and αKG (r = 0.605, p < 2.2e-16) (Fig. 2a). Moreover, there was a significant correlation between transcriptional profiles produced by p53 and αKG treatment (r = 0.556; p < 1e-15; Extended Data Fig. 6a), with both treatments inducing gene expression signatures previously linked to p53 action (Fig. 2b, Supplementary Table 2), and cell-permeable αKG recapitulating trends observed in the transcriptional response produced by p53 restoration in KPsh cells (Fig. 2c).
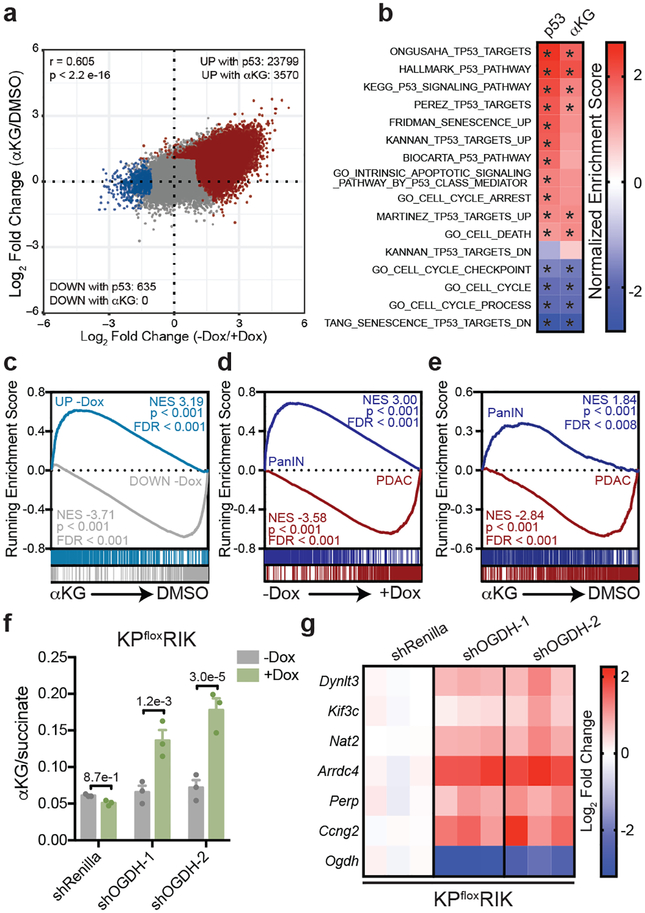
Figure 2. αKG recapitulates gene expression changes induced by p53 restoration.
a, Mean log2 fold change of all ATAC-Seq peaks following p53 reactivation or treatment with cell-permeable αKG in n=2, independently treated wells of KPsh-1 cells. All samples contained equivalent amounts of vehicle (DMSO). Pearson correlation r = 0.605, p < 2.2e-16. Number of peaks increased (red) or decreased (blue) at least two-fold with a false discovery rate < 0.1 in each condition are indicated. b, Normalized gene enrichment score of published p53-associated gene sets in KPsh-1 cells following p53 reactivation or αKG treatment. Gene sets significantly (p < 0.05) enriched (red) or depleted (blue) are marked with an asterisk. Genes are listed in Supplementary Table 2. c, GSEA analyses of RNA-Seq data of KPsh-1 cells treated with cell-permeable αKG showing enrichment of genes upregulated (genes UP) or downregulated (genes DOWN) at least 2-fold, padj < 0.05 following p53 restoration. d,e, GSEA of PanIN-cell and PDAC-cell associated genes associated genes following p53 restoration or αKG treatment (d,e). Gene sets derived from (Boj et al29) and listed in Supplementary Table 2. RNA-Seq in b-e performed on n=2, independently treated wells. f, αKG/succinate ratio of p53 null (KPfloxRIK) cells expressing dox-inducible shRNAs targeting Ogdh or Renilla luciferase (control) grown 4 days with or without dox. g, qRT-PCR of genes upregulated by both p53 restoration and αKG treatment in KPsh-1 cells by RNA-Seq. Gene expression is represented as the log2 fold change relative to shRenilla expressing cells. f,g were repeated twice with similar results. Data are presented as mean ± SEM of n=3, independently treated wells with individual data points shown (f), or as individual data points (g). Significance assessed in indicated comparisons by 2-way ANOVA with Sidak’s multiple comparisons post-test.
As p53 inactivation is associated with malignant progression during pancreatic tumorigenesis5,22, we asked whether gene expression programs regulated by p53 and αKG reflect defined stages of pancreatic cancer. Both p53 and cell-permeable αKG increased expression of genes selectively expressed in cells derived from premalignant PanIN lesions and decreased expression of genes enriched in malignant PDAC cells (Fig. 2d,e). Similar results were observed in cells treated with distinct forms of cell-permeable αKG, but not with acetate, a metabolite downstream of citrate that can regulate gene expression by contributing to histone acetylation23 (Extended Data Fig. 6b–d).
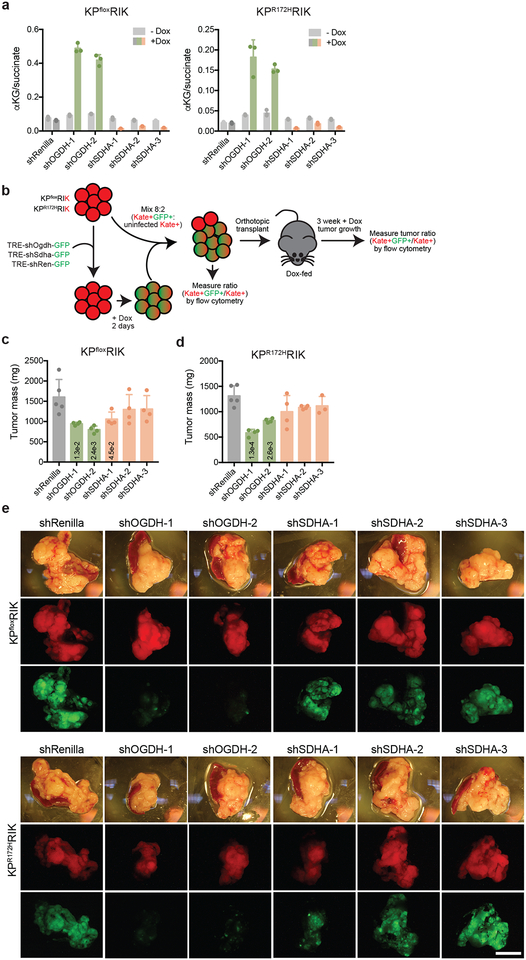
We also tested whether genetic manipulation of endogenous metabolic pathways predicted to increase the αKG/succinate ratio could recapitulate aspects of p53-dependent gene expression in the absence of wild-type p53. rtTA-expressing cell lines derived from p53 null (KPfloxRIK) or p53 mutant (KPR172HRIK) PDAC were engineered to express dox-inducible hairpins against oxoglutarate dehydrogenase (Ogdh), a subunit of the oxoglutarate dehydrogenase complex that converts αKG to succinyl-CoA in the TCA cycle (Extended Data Fig. 6e). While Ogdh knockdown slowed cell proliferation, no notable senescence or apoptosis was observed (Extended Data Fig. 6f–h). Still, knockdown of Ogdh elevated the αKG/succinate ratio to a similar degree as p53 and increased the expression of genes that were also induced by both p53 re-expression and αKG addition (Fig. 2f,g and Extended Data Fig. 6i,j).
We next tested whether Ogdh inhibition could recapitulate phenotypes associated with p53-driven tumor suppression in the absence of p53 in vivo. Consistent with the ability of p53 to produce a premalignant gene expression profile in vitro, tumors responding to p53 activation exhibited more differentiated histopathology, defined by the emergence of glandular structures with cuboidal and columnar cellular morphology that were strongly positive for the epithelial cytokeratin CK19 (Fig. 3a and Extended Data Fig. 7a). Orthotopic tumors arising from p53 null cells expressing constitutive GFP-linked Ogdh shRNAs were smaller than those produced from cells expressing control shRNAs and had a notable decrease in the contribution of shRNA-expressing cells to tumor areas (Extended Data Fig. 7b). Similarly, induction of dox-inducible Ogdh shRNAs in established p53 null tumors also reduced tumor growth (Extended Data Fig. 7c). In both settings, tumors harboring Ogdh shRNAs were characterized by abundant, well-differentiated, glandular structures with cuboidal/columnar cellular morphology and increased CK19 staining (Fig. 3b, Extended Data Fig. 7d–f).
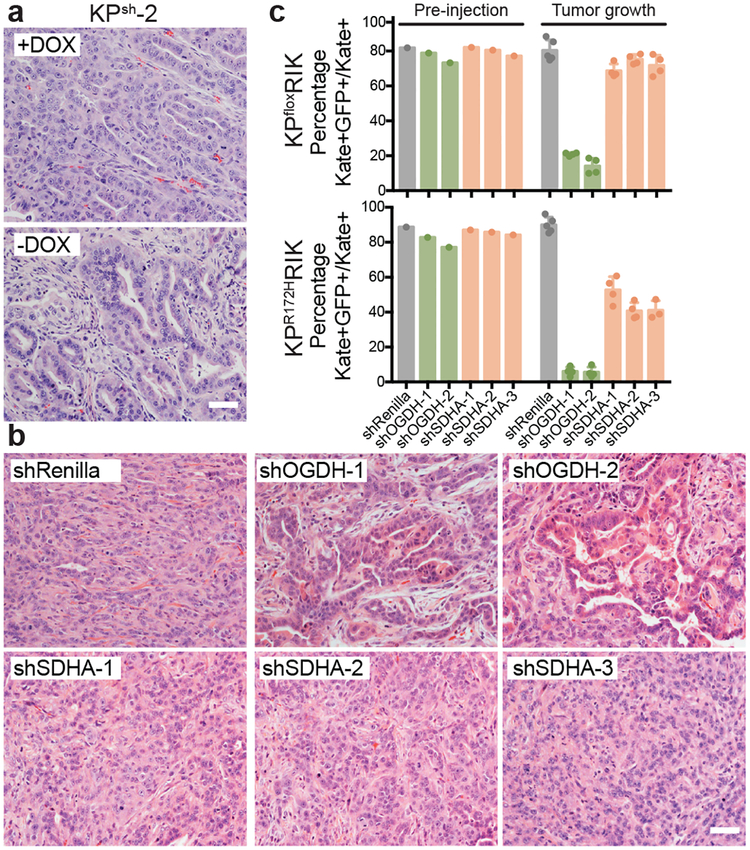
Figure 3. Both p53 restoration and Ogdh inhibition promote tumor cell differentiation and tumor suppression.
a, Representative hematoxylin and eosin (H&E) staining of orthotopic tumors derived from KPsh-2 cells grown in mice on dox-diet (n=3) or ten days after dox withdrawal (n=3). b, Representative H&E staining of orthotopic tumors derived from KPflox cells expressing dox-inducible shRenilla, shOgdh, or shSdha two weeks after injection in mice maintained on dox. n= 5 mice shRenilla, n=4 mice (shOgdh, shSdha). c, In vivo competition assay tracking frequency of KPfloxRIK (top) or KPR172KRIK (bottom) cells expressing shRenilla, shOgdh, or shSdha (GFP+) after three weeks of tumor growth in dox-fed mice. Data are presented as mean ± SD of individual tumors. KPfloxRIK: n= 5 mice shRenilla, n=4 mice (shOgdh, shSdha). KPR172KRIK : n= 5 mice shRenilla, (shOgdh, shSdha-1,2), n=3 mice shSdha-3. Scale bar 50 μm.
To rule out the possibility that the anti-tumor and pro-differentiation effects of Ogdh inhibition were a generic consequence of disrupting the TCA cycle, we performed a parallel series of experiments using potent shRNAs targeting succinate dehydrogenase subunit a (Sdha) to perturb the TCA cycle without increasing the αKG/succinate ratio (Extended Data Fig 7g). shSdha-expressing cells more uniformly contributed to tumor areas (Extended Data Fig. 7b) and did not display the same degree of morphological features of differentiation or CK19 expression as observed in shOgdh expressing cells (Fig. 3b, Extended Data Fig. 7d–f). Furthermore, Sdha inhibition in established tumors only modestly inhibited tumor growth and did not trigger notable differentiation (Extended Data Fig. 7c–f). shOgdh-specific anti-tumor effects on in vivo competitive fitness extend to both p53 null and p53 mutant PDAC cells (Fig. 3c and Extended Data Fig. 8a–e). Apparently, the p53-triggered increase in the αKG/succinate ratio results in specific tumor suppressive effects that are not a generalized response to perturbation of the TCA cycle function.
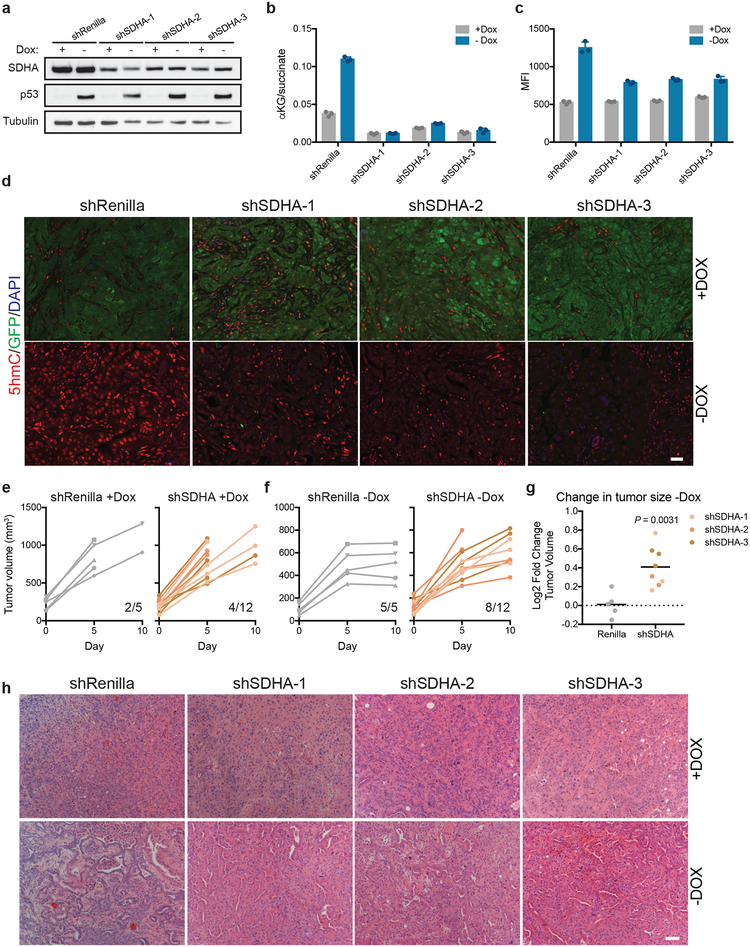
To assess whether the activity of αKG-dependent enzymes linked to gene regulation reflects p53 status in vivo, we performed histological analysis of 5-hydroxymethycytosine (5hmC), the product of the strictly αKG-dependent ten eleven translocation (Tet) enzyme family24, across different stages of mouse and human PDAC development. In the KPC mouse model, which is initially heterozygous for a missense p53R172H allele, loss of wild-type p53 precedes stabilization of the mutant p53 protein such that cells harboring high p53 staining are those with inactive p538. Normal pancreatic epithelial cells and stromal cells25, as well as premalignant cells with PanIN features, displayed low p53 (p53 wild-type) and high 5hmC staining (Fig. 4a, Supplementary Figure 3). In contrast, malignant cells displaying high p53 levels (p53 mutant) showed very low 5hmC staining (Fig. 4a, Supplementary Figure 3). Consistent with other reports suggesting that 5hmC is frequently lost in advanced cancers24, decreased 5hmC levels were observed during human PDAC progression, with the the reduction in 5hmC coinciding with the transition from benign to malignant disease that is frequently characterized by acquisition of TP53 mutations (Fig. 4b,c, Supplementary Figure 3).
Figure 4. p53 status and cellular αKG/succinate ratio dictates 5hmC levels in mouse models of PDAC.
a, Representative 5hmC and p53 staining in PDAC arising in KPC mice (n=3). High p53 staining denotes malignant cells. b, Representative 5hmC staining in human PanIN 1–3 and PDAC samples. b-catenin marks tumor epithelium. c, Fraction 5hmC-positive nuclei (binned into quartiles) in indicated numbers of human tumors. d, Representative 5hmC staining in orthotopic tumors derived from KPsh-2 cells in mice maintained on dox (n=3) or 10 days off dox (n=3). GFP denotes cells expressing shp53. e, Nuclear 5hmC intensity in lineage-traced (i.e., GFP high, +DOX; GFP low −DOX) tumor cells from three images each from n=3 independent tumors from d. n= 1074, 1571, 1359, 1569, 1253, 781 nuclei quanitified per mouse. f, Nuclear 5hmC intensity in lineage-traced (i.e., GFP+) tumor cells from three images each from n=3 independent tumors derived by orthotopic injection of KPflox cells expressing dox-inducible shRenilla, shOgdh, or shSdha two weeks after injection in mice maintained on dox. n =1609, 1796, 1947, 1581, 1751, 1619, 1636, 1786, 1907, 1801, 1892, 1758, 1829, 2001, 1898, 1982, 1839, 1926 nuclei quanitified per mouse. GFP denotes shRNA-expressing cells. g, Nuclear 5hmC intensity in lineage-traced (i.e., GFP high, +Dox; GFP low −Dox) tumor cells from three images each from n=3 independent tumors derived by orthotopic injection of KPsh-2 cells expressing shRenilla or shSdha maintained on dox or 10 days off dox. n=1810, 1720, 1980, 1837, 1739, 1670, 1695, 1592, 1583, 1690, 1428, 1734, 1351, 1434, 1708, 1543, 1308, 1413, 1641, 1893, 1476, 1586, 1597, 1547 nuclei quanitified per mouse. e-g, Population medians were taken for each mouse and points represent total 5hmC levels of individual nuclei normalized to DAPI. e-g, Significance assessed in indicated comparisons using two tailed Students t-test. Scale bar 50 μm.
We also asked whether 5hmC levels are sensitive to changes in p53 or αKG manipulation. Despite modest changes in expression of Tet enzymes, p53 reactivation induced 5hmC in KPsh cells in a Tet-dependent manner (Extended Data Fig. 9a–d). Furthermore, cell-permeable αKG induced cellular 5hmC in dox-treated KPsh cells and p53 mutant human PDAC cells (Extended Data Fig. 9e,f), while Ogdh inhibition increased 5hmC in p53 null and mutant mouse PDAC cells (Extended Data Fig. 9g). Remarkably, both p53 restoration and Ogdh inhibition during tumor initiation or in established tumors led to increases in 5hmC in vivo that coincided with areas of glandular differentiation rarely observed with Sdha suppression (Fig. 4d–f, Extended Data Fig. 9h,i, Supplementary Figure 3). Therefore, the decline in 5hmC associated with p53 loss during PDAC progression is specifically reversible by interventions that increase the αKG/succinate ratio and is associated with the reversion to a more premalignant like cell fate.
To test whether an elevated αKG/succinate ratio was required for p53-mediated increases in 5hmC, we took advantage of the fact that Sdha silencing induces high levels of succinate, a competitive inhibitor of αKG-dependent dioxygenases26. Constitutive suppression of Sdha in KPsh cells blocked p53-mediated induction of the αKG/succinate ratio despite abundant accumulation of p53 following dox withdrawal (Extended Data Fig. 10a,b) and significantly reduced 5hmC levels in vitro and in vivo (Extended Data Fig. 10c,d, Fig. 4g, Supplementary Figure 3). Furthermore, Sdha silencing resulted in sustained tumor growth and reduced survival following prolonged dox withdrawal (Extended Data Fig. 10e–g), corresponding with a intermediate histological phenotype with reduced frequency of clearly articulated glandular structures (Extended Data Fig. 10h). Therefore, an elevated αKG/succinate ratio appears necessary for the induction of chromatin marks characteristic of premalignant cell fate and functionally contributes to p53-driven tumor suppression. While several αKG-dependent activities27 are undoubtedly affected by p53-driven metabolic reprogramming, these data suggest that 5hmC serves as a biomarker of p53-triggered, tumor suppressive changes in the αKG/succinate ratio.
Our results identify a metabolic link between p53 function, chromatin regulation, and tumor cell fate. When responding to oncogenic signaling, p53 rewires glucose and glutamine metabolism to favor accumulation of αKG at the expense of succinate, thereby reinforcing the activity of αKG-dependent effectors and promoting premalignant patterns of gene expression. Loss of p53 prevents these metabolic effects and enables transition to more aggressive and less differentiated carcinomas that display hallmarks of reduced αKG-dependent activity. Increases in αKG levels, for example as produced by OGDH suppression, are sufficient to impose a p53-like chromatin and transcriptional profile in tumor cells lacking p53, thereby enabling cells to reacquire a premalignant identity. These data illustrate how metabolic consequences of p53 action contribute to p53-driven tumor suppression and further suggest that αKG can play an active role in tumor suppression, a hypothesis that is strengthened by the observation that mutations in isocitrate dehydrogenase that produce 2-hydroxyglutarate—an antagonist of αKG-dependent enzymes—block differentiation and promote tumorigenesis28. These results also nominate therapeutic strategies to increase αKG levels as a mechanism to engage latent tumor suppressive pathways in p53-deficient tumors.
Methods
Mouse strains.
All animal experiments were performed in accordance with a protocol approved by the Memorial Sloan-Kettering Institutional Animal Care and Use Committee. All mouse strains have been previously described. Pdx1-Cre30, p48-Cre31, p48-CreER32, LSLKrasG12D33, LSL-p53R172H34, p53flox 35, CHC-TRE-shp53-shRenilla and CHC-shRenilla10,36, and CAGs-LSL-RIK strains10,37 were interbred and maintained on mixed Bl6/129J backgrounds. Athymic nude mice (Envigo) were used for all transplant experiments.
Cell lines and cell culture treatments.
KPsh-1–3 cells were derived from PDAC that developed in 3 separate Pdx1-Cre; LSL-KrasG12D; Col1a1-TRE-shp53-shRenilla; Rosa26-CAGs-LSL-rtTA-IRES-mKate2 mice generated by blastocyst injection and maintained on dox chow (625 mg/kg, Harlan Laboratories) 5 days from birth until sacrifice as previously described10. KRsh cells were generated from PDAC that developed in one Pdx1-Cre; LSL-KrasG12D; Col1a1-TRE-shRenilla; Rosa26-CAGs-LSL-rtTA-IRES-mKate2 mouse generated by blastocyst injection and maintained on dox chow 5 days from birth until sacrifice as previously described10. Guide strands for Col1a1 targeted tandem shRNAs are: p53, TTACACATGTACTTGTAGTGG and Renilla, TAGATAAGCATTATAATTCCT. KPfloxRIK cells were derived from PDAC that developed in a p48-Cre; LSL-KrasG12D; p53flox/+; Rosa26-CAGs-LSL-rtTA-IRES-mKate2 mouse generated by blastocyst injection. KPR172HRIK cells were derived from tumors that developed in a p48CreER; LSL-KrasG12D; LSL-p53R172H/+; Rosa26-CAGs-LSL-rtTA-IRES-mKate2 mouse generated by blastocyst injection. This animal was treated with tamoxifen (1 mg dissolved in corn oil, 5 consecutive daily i.p. injections) at 4 weeks of age to induce Cre recombination. Tumors were diced with scissors and resulting pieces sequentially digested with1 mg/ml collagenase V (Sigma) diluted in Hanks Buffered saline solution followed by 0.25% Trypsin. Digested suspensions were washed with complete DMEM (DMEM, 10%FBS (GIBCO), 1× Penicillin/Streptomycin) and propagated in complete DMEM on collagen-coated plates (PurCol, Advanced Biomatrix, 0.1 mg/ml). KPsh cells were grown in complete DMEM supplemented with 1 μg/ml doxycycline. KPflox cells38 were grown in complete DMEM on non-coated, tissue culture treated vessels. 1μg/ml doxycycline was added to complete media to induce shRNA expression in KPflox, KPfloxRIK, and KPR172HRIK cells infected with lentiviral or retroviral vectors encoding TRE-linked shRNAs. Where indicated, cells were treated with H2O (vehicle), DMSO (vehicle), 3 μM Etoposide (Sigma), 25 nM Trametinib (Selleck Chemicals), 4 mM sodium acetate (Sigma), 4 mM dimethyl-αKG (Sigma), or 4 mM diethyl-αKG (Sigma).
Lentiviral and retroviral production and infection.
Lentivirus was generated by co-transfection of shRNA expressing viral vectors with packaging plasmids psPAX2 and pMD2.G (Addgene) into 293T cells. Retroviruses were generated by co-transfection of shRNA expressing viral vectors with pCMV-VSVG (Addgene) into 293GP cells. Viral containing supernatants were cleared of cellular debris by 0.45 μM filtration and mixed with 8 μg/ml polybrene. Target cells were exposed to viral supernatants for two 12 hour periods before being washed, grown for 24 h in fresh media, then subjected to antibiotic selection. Cells were maintained in antibiotic selection until used for experiments.
Lentiviral and retroviral shRNA and cDNA vectors and sequences.
Renilla (TAGATAAGCATTATAATTCCT); p19 (Arf) (ATGTTCACGAAAGCCAGAGCG); p16/p19 (Cdkn2a) (AACACAAAGAGCACCCAGCGG); p21 (Cdkn1a) (TTTAAGTTTGGAGACTGGGAG); Sdha-1 (TTAATTGAAGGAACTTTATCTC); Sdha-2 (TTCATAACCGATTCTTCTCCAG); Sdha-3 (TCTGATGTTCTTATACTTCCAT); Idh1–1 (TTCAATTGACTTATCCTGGTTG), and Idh1–2 (TTGTATTTCTTTATAGCCTCTG) shRNAs (guide strand sequences) were constitutively expressed in KPsh cells with retroviral LMNe-BFP, a modified version of LMP-GFP39 in which the mir30 context has been replaced by the optimized “mirE” context40, the puromycin resistance gene has been replaced by a neomycin resistance cassette, and GFP replaced by BFP, respectively. Cells infected with LMNe-BFP were selected in 1 mg/ml neomycin. Renilla; Ogdh-1 (TAAATGAAACATTTTGTCCTG); Ogdh-2 (TAGCAATTCTGCATACTTCTG); Sdha-1; Sdha-2, and Sdha-3 shRNAs were introduced into KPflox cells to enable dox inducible expression using lentiviral LT3GEPIR40. Cells infected with LT3GEPIR were selected with 1 μg/ml puromycin. Renilla, Ogdh-1; Ogdh-2; Sdha-1; Sdha-2, and Sdha-3 shRNAs were introduced into KPfloxRIK and KPR172HRIK cells to enable dox inducible expression using retroviral RT3GEN40. Cells infected with RT3GEN were selected with 1 mg/ml neomycin. Wildtype (WT) p53 cDNA was obtained from Dharmacon and exchanged with the dsRED-shRNA cassette of RT3REVIN40. Site directed mutagenesis was performed according to manufacturer’s instructions to sequentially introduce mutations resulting in amino acid substitutions in the TAD1 and TAD2 regions of WT p53 (L25Q;W26S;F53Q;F54S) using a Q5 Site-Directed Mutagenesis kit (NEB, E0554) and a QuikChange II XL Site-Directed Mutagenesis Kit (Agilent 200522), respectively. KPfloxRIK cells were infected with virus generated from transfection of RT3-REVIN without cDNA (vector), RT3-REVIN-p53WT, RT3-REVIN p53TAD1/2M as described above and selected with 1 mg/ml neomycin. Human IDH1 and IDH2 cDNA were obtained from pMIG-Idh1 and pMIG-Idh2 vectors41. Lenti-EFSIDH1-PGK-Neo and Lenti-EFS-IDH2-PGK-Neo were generated through Gibson assembly by combining the following four DNA fragments: (i) PCR-amplified EF1s promoter, (ii) PCR-amplified IDH1 cDNA or IDH2 cDNA, (iii) PCR-amplified PGK-Neo cassette, and (iv) BsrGI/PmeI-digested pLL3-based lentiviral backbone. KPsh-2 cells were infected with Lenti-EFS-IDH1-PGK-Neo and Lenti-EFS-IDH2-PGK-Neo and selected with neomycin as described above. Cas9 was constitutively expressed in KPsh-2 cells via infection with lentiviral lentiCas9-Blast42 (generous gift from Feng Zhang (Addgene plasmid #52962; http://n2t.net/addgene:52962; RRID:Addgene_52962)). LentiCas9-Blast cells were selected with 10 μg/ml blastocidin. sgRNAs targeting Tet1 (CCTACGGGAAGCGACCATAA; GACACCGGCGCCGAGTTTT); Tet2 (GGGAGAAAGCAATATCTTCG; TGCGACGGCGGTGGACTGCG), and Tet3 (CTGGGATCAAGACCAGTGTC; CCGGGCCCCCTCATGGCCTG) were constitutively introduced into stable KPsh-2-Cas9 cells using a modified version of pUSEPB, a modified version of pUSEPR43 in which the RFP cassette has been replaced with BFP. pUSEPB infected cells were selected with 4 μg/ml puromycin.
Growth curves.
Population doubling curves were generated from KPsh cells as follows. Cells were washed with PBS, trypsinized, and 50,000 cells were plated in triplicate in 6 well dishes off and on dox. Every 48 h cell number was recorded and 1/8 of the total cell number was replated. Population doublings for each 48 h period were calculated with the formula 3.32*(log(final cell number)-log(initial cell number)). Growth curve analysis for shOGDH expressing KPfloxRIK and KPR172HRIK cells were performed by plating 10,000 cells on dox into 12 well dishes and counting the number of cells in triplicate every 24 h from day 1 to 4.
Senescence associated Beta-galactosidase (SA-βgal) assay.
SA-β-gal activity was detected as described previously44. Briefly, cells were washed twice with PBS, fixed with 0.5% gluteraldehyde in phosphate-buffered saline (PBS) for 15 min, washed with PBS supplemented with 1 mM MgCl2, pH 5.5, and incubated overnight at 37°C in PBS containing 1 mM MgCl2, 1 mg/mL X-Gal (Roche), and 5 mM each potassium ferricyanide (Sigma) and potassium ferrocyanide (Sigma), pH 5.5. At least 200 cells were analyzed from triplicate wells at all conditions. Cells were plated for analysis 2 days prior to staining.
BrdU, Annexin-V, 5hmC flow cytometry assays.
BrdU (BD Pharmigen) and Annexin V (BD Pharmigen) analysis was performed in triplicate in cells plated in 6 well dishes under indicated conditions via manufacturers’ protocols. 5hmC flow was performed in triplicate in cells plated in 6 well dishes. Briefly, adherent cells were washed with PBS and collected by trypsinization, fixed on ice for 15 min in fixation/permeabilization solution (BD Cat#554714), washed with 1X perm/wash buffer (BD Cat#554714) supplemented with 0.1% Triton X, treated with 1X perm/wash buffer supplemented with 0.5% Triton X and 0.5% Nonidet P-40 substitute for 30 min, washed with 1X perm/wash buffer supplemented with 0.1% Triton X, incubated overnight at 4°C with rabbit anti 5hmC antibody diluted 1:100 in 1X perm/wash buffer supplemented with 0.1% Triton X, washed with 1X perm/wash buffer supplemented with 0.1% Triton X, incubated for 1 h at room temperature with goat anti-rabbit Alexa Fluor 700 diluted 1:200 in 1X perm/wash buffer supplemented with 0.1% Triton X, washed with 1X perm/wash buffer supplemented with 0.1% Triton X, resuspended in 1X perm/wash buffer and analyzed. Example gating strategies for all 3 assays are shown in Supplementary Figure 2.
Orthotopic tumors.
For in vivo p53 restoration experiments parental KPsh cells, or KPsh cells expressing the indicated constitutively expressed shRNA, maintained on dox were washed with PBS, trypsinized, and counted. 5×105 cells in serum-free DMEM were mixed 1:1 with growth factor reduced matrigel (Corning) and injected into the exposed pancreas of athymic nude mice using a Hamilton syringe fitted with a 26 gauge needle. Recipient mice were enrolled on dox chow (625 mg/kg, Harlan Laboratories) 2 days before surgery and maintained on dox chow until mice were randomized for analysis. Tumor volume was measured by small animal ultrasound (Vevo 2100) 2 weeks post-transplant at which point 3 mice were maintained on dox chow and 6 mice switched to dox-free chow. Tumor volume was measured at either 5, 10, and 18 days, or 5 and 10 days after randomization unless mice were sacrificed due to reaching IACUC approved humane endpoints for tumor burden and mouse body condition (see below). For analysis of in vivo p53 restoration in tumors derived from parental KPsh cells, 3 mice were randomly censored for sacrifice and tissue analysis 10 days after being enrolled off dox. At sacrifice, epifluorescence for mKate2 and GFP were recorded with a fluorescent dissection scope (Nikon SMZ1500) before tissues were fixed for histological analysis. For KPflox orthotopic tumor growth assays, cells were infected with LT3GEPIR lentiviral vectors expressing GFP linked shRNAs targeting Renilla luciferase or mouse Ogdh and subjected to antibiotic selection. Cells were treated with doxycycline 2 days before injection and 5×105 cells transplanted into the pancreas of dox fed mice as described above. Pancreata were removed 2 weeks after transplant and GFP epifluorescence was recorded before tissues were fixed for histological analysis. For evaluation of effects of shRNAs in established tumors derived from KPflox cells, cells infected with LT3GEPIR-shRNA viruses were grown and selected in dox free medium and 5×105 cells were injected into mice maintained on normal chow. 2 weeks after injection, tumor volume was measured by small animal ultrasound and mice were enrolled on dox chow. Tumor volume was measured 3, 6 and 9 days after enrolling on dox chow. 3–5 independently derived tumors were analyzed per group as approved by MSKCC IACUC protocol 11-06-018. As specified in this protocol all mice bearing orthotopic tumors were evaluated daily for signs of distress or endpoint criteria. Specifically, mice were immediately euthanized if they showed signs of discomfort, cachexia, weight loss >20% of initial weight, blood loss, breathing difficulties, severe infections, or developed tumors 15 mM in diameter. No tumors exceeded this limit.
Immunofluorescence.
Tissues were fixed overnight at 4°C in 10% formalin prior to paraffin embedding. Five-micron sections were deparaffinized and rehydrated with a histoclear/alcohol series and subjected to antigen retrieval by boiling in citrate antigen retrieval buffer (Vector). Slides were blocked in PBS with 5% BSA and primary antibody staining was performed in blocking buffer overnight at 4°C. The following primary antibodies were used: chicken anti-GFP (1:500, Abcam 13970), rabbit anti-mKate2/Turbo RFP (1:1000, Evrogen, AB233), mouse anti-p21 (1:1000, 556431, BD), mouse anti-Ki67 (1:500, BD, 550609), rat anti-CK19 (Troma III) (1:1000, Developmental Studies Hybridoma Bank, AB_2133570), rabbit anti-p53 (1:500, NCLL-p53-CM5p, Leica Biosystems), rabbit anti-5hmC (1:500, Active Motif, 39769), mouse anti-5hmC (1:200, Abcam, 178771) and mouse anti-β-Catenin (1:200, BD, 610153). Primary antibodies were detected with the following fluorescently conjugated secondary antibodies: goat-anti-chicken AF488 (Life Technologies A-11039), goat anti-rabbit AF488 (Life Technologies A-32723), goat anti-rabbit AF594 (Life Technologies A-11037), goat anti-mouse AF488 (Life Technologies, A-32723), goat anti-mouse AF594 (Life Technologies, A-11032), goat anti-rat AF488 (Life Technologies, A-11006) and goat anti-rat 594 (Life Technologies, A-11007). All secondary antibodies were diluted in blocking buffer and incubated for 1 hour at room temperature. Subsequently, slides were washed and nuclei counterstained with PBS containing DAPI and mounted under cover slips with ProLong Gold (LifeTechnologies). Images were acquired with a Zeiss AxioImager microscope using Axiovision software.
In vivo competition assay.
KPfloxRIK and KPR172HRIK cells were infected with RT3GEN retroviral vectors expressing GFP linked shRNAs targeting Renilla luciferase or mouse Sdha and Ogdh and subjected to antibiotic selection. Cells were treated with doxycycline 2 days before injection, analyzed for GFP expression (Supplementary Figure 2, Extended Data Fig. 8), and transplanted into the pancreas of dox fed mice. Tumors were removed 3 weeks after transplant, epifluorescence for mKate2 and GFP was recorded, and tumors prepared for flow cytometry as described45. Briefly, tumors were minced with scissors and sequentially incubated with collagenase V (1 mg/ml (Fisher) in Hanks buffered saline solution), trypsin (0.05%), and dispase (2 U/ml, Invitrogen). Dnase1 (100 μg/ml, Sigma) was added during all enzyme incubations. Cells were washed with PBS between the collagenase and trypsin steps, and with FACs buffer (2% FBS, 10 mM EGTA, in PBS) between the trypsin and dispase steps. Suspensions were then filtered through 40 μM mesh and resuspended in FACs buffer with 300 nM DAPI for flow analysis (Supplementary Figure 2).
qRT-PCR.
Total RNA was extracted in triplicate wells from indicated conditions using the RNAeasy Mini Kit (Qiagen) or Trizol (Invitrogen) according to manufacturer protocols. cDNA was synthesized from 1 μg of RNA (AfinityScript QPCR cDNA synthesis kit, Agilent or iScript, BioRad) and QPCR amplification performed with SYBR green (Perfecta SYBR green fast mix, QuantaBio) using the following primer pairs on a ViiA 7 Real-Time PCR System (Life technologies) or QuantStudio 5 (Applied Biosystems byThermo Fisher Scientific). 36B4 was utilized as endogenous control.
Dynlt3: Left: TGGACTGCAAGCATAGTGGAA Right: GTGAAATCCATACGGGCTCCT
Kif3c: Left: CAGGCCGACCTGTATGACG Right: GTCCCCTGCATGGTGTAGG
Nat2: Left: ACACTCCAGCCAATAAGTACAGC Right: GGTAGGAACGTCCAAACCCA
Arrdc4: Left: CCCTGGTGCTAAAAGATTGATGC Right: TGAACTGGCTTGCGACACTG
Perp: Left: ATCGCCTTCGACATCATCGC Right: CCCCATGCGTACTCCATGAG
Ccng2: Left: AGGGGTTCAGCTTTTCGGATT Right: AGTGTTATCATTCTCCGGGGTAG
Cdkn1a: Left: CGGTGTCAGAGTCTAGGGGA Right: ATCACCAGGATTGGACATGG
tp53: Left: CTAGCATTCAGGCCCTCATC Right: TCCGACTGTGACTCCTCCAT
Mdm2: Left: TGTCTGTGTCTACCGAGGGTG Right: TCCAACGGACTTTAACAACTTCA
Idh1: Left: ATGCAAGGAGATGAAATGACACG Right: GCATCACGATTCTCTATGCCTAA
Pcx: Left: GGCCAAGGAAAATGGTGTAG Right: CTTCCACCTTGTCTCCCATC
Ogdh: Left: GGTGGAAGCACAACCTAACG Right: CATGGTGCCCTCGTATCTGA
Tet1: Left: GAAGCTGCACCCTGTGACTG Right: GACAGCAGCCACACTTGGTC
Tet2: Left: AAGCTGATGGAAAATGCAAGC Right: GCTGAAGGTGCCTCTGGAGT
Tet3: Left: TCACAGCCTGCATGGACTTC Right: ACGCAGCGATTGTCTTCCTT
36B4: Left: GCTCCAAGCAGATGCAGCA Right: CCGGATGTGAGGCAGCAG
Western Blotting.
Cell lysates were extracted using RIPA buffer and protein concentration was determined by BCA assay. Samples were boiled for 5 minutes and 20 to 30 μg of protein were separated by SDS-PAGE, transferred to nitrocellulose membranes, blocked with 3% milk prepared in 1X TBS-Tween and probed with the relevant primary antibody overnight at 4°C. Membranes were then incubated with horseradish peroxide (HRP)-conjugated secondary antibodies at room temperature and proteins were detected using Pierce ECL Western Blotting Substrate (34095, Thermo Fisher Scientific). Blots were imaged using HyBlot CL Autoradiography Film (E3018, Denville Scientific) and Konica Medical Film Processor (Model SRX-101A). Antibodies were diluted as follows: p53 (CM5) (1:500, NCL-L-p53-CM5p, Leica Biosystems), p21 (F-5) (1:500, sc-6246, Santa Cruz Biotechnology), p19 (M-167) (1:500, sc-1063, Santa Cruz Biotechnology), Ogdh (1:1000, 15212–1-AP, ProteinTech), Idh1 (1:1000, 12332–1-AP, ProteinTech), Sdha (1:1000, ab14715, Abcam) and Tubulin (1:10,000, T9026, Sigma-Aldrich).
Image analysis.
The tumors of three mice were stained for 5hmC and DAPI. These tumors were imaged at three randomly chosen 20X fields and these images were analyzed using custom-made Matlab® scripts. Briefly, a GFP mask was created to identify cells of interest. Within these regions we located and segmented cell nuclei using the DAPI staining. We then quantified nuclear 5hmC levels within GFP+ cells. To account for differences in nuclear area, we used median 5hmC values, but similar results were obtained if using mean or total 5hmC levels. To account for changes on DNA levels, we normalized per cell 5hmC values to corresponding DAPI levels. We also tested different values for the parameter than determined the stringency of the GFP mask and our results remained qualitatively the same. Raw images as well as analysis routines are available upon request.
sgRNA editing analysis.
Target locus analysis was performed and analyzed as previously described46. Briefly, genomic DNA was extracted as described from KPsh-2-Cas9 cells expressing sgRNAs targeting Tet1, Tet2, and Tet3 (listed above) and amplification of target regions was performed from 100 ng of genomic DNA using Herculase II Fusion DNA polymerase (Agilent 600675) per manufacturer instructions. Edited regions were amplified using the following primers:
Tet1 sg1: Left: CAAGCTGTCTGATCCTTCTCC Right: ACAGAGGTGGCATCCAGAAC
Tet1 sg2: Left: CCGGAAAACCGAAGCAATTA Right: TCGCCAGCTAAGAGAGGTTC
Tet2 sg1: Left: ACACCAAGTGGCAATCTTCC Right: GCTGCTTTTACCGTGGTTTC
Tet2 sg2: Left: GCAGAAGGAAGCAAGATGG Right: AAGGCCGAGAGAAAGAGAGG
Tet3 sg1: Left: GCCTCCTTCCCTACTTCCAC Right: CCTGGACCTGGATTTCTTGA
Tet3 sg2: Left: TTCAGGTCTCCCCAGTCCTA Right: CCCAATAGCTGCTCCAGTTC
PCR products were column purified (Qiagen) for MiSeq. DNA-library preparation and sequencing were performed at GENEWIZ. An NEB NextUltra DNA Library Preparation kit was used according to the manufacturer’s recommendations (Illumina). Adaptor-ligated DNA was indexed and limited-cycle PCR used for enrichment. DNA libraries were validated via TapeStation (Agilent) and measured with a Qubit 2.0 fluorometer. Libraries were further quantified through real-time PCR (Applied Biosystems) and loaded on an Illumina MiSeq instrument according to the manufacturer’s instructions (Illumina). Sequencing was performed with a 2 × 150 paired-end configuration. Image analysis andbase calling were conducted in MiSeq Control Software on a MiSeq instrument. Raw Fastq data was first trimmed to remove low quality data using sickle (https://github.com/najoshi/sickle). Pandaseq (https://github.com/neufeld/pandaseq) was then used to merge read1 and read2. The merged reads were mapped to the reference sequence using BWA (http://bio-bwa.sourceforge.net/). Lastly, a variant detection analysis was performed using GENEWIZ’s custom in-house developed scripts, which has been independently validated with a custom bioinformatic pipeline46.
RNA-Seq.
Total RNA was isolated from duplicate wells of indicated conditions. RNA integrity and concentration were assessed using a BioAnalyzer (Agilent). RNA sequencing libraries were generated using Illumina mRNA TruSeq kit with dual index barcoding. Multiplexed libraries were sequenced at the Cold Spring Harbor Labs core sequencing facility. Approximately 8 million paired-end 76 bp reads were sequenced per replicate on a HiSeq 2500 instrument on RAPID mode. After removing adaptor sequences with Trimmomatic47, RNA-Seq reads were aligned to GRCm38 – mm10 with STAR48. Genome wide transcript counting was performed by HTSeq or featureCounts to generate FPKM matrix49,50. Differential expression analysis was performed with DESeq2 package in R51. Genes with a real adjusted p value were used for downstream analyses.
Gene set enrichment analysis.
Genes were ranked by fold change and gene set enrichment analysis (GSEA)52 was performed using ranked lists. Gene lists associated with p53 activity are found in the MSigDB52 and all gene sets used in this study are detailed in Supplementary Table 2. GSEAPreranked version 4 was used with default parameters and data were exported and graphed in GraphPad Prism version 7.
Glucose, glutamine and lactate measurements.
Glucose, glutamine and lactate levels in culture medium were measured using a YSI 7100 multichannel biochemistry analyzer (YSI Life Sciences). 1×106 KPsh cells were washed with PBS and plated on and off dox 8 days (-D8) before YSI analysis. Cells maintained on dox and off dox were passaged every 48 h and additional cells were grown off dox starting at −D4. All cell groups (on dox, off dox 4 days, off dox 8 days) were split into 6 well plates in sextuplicate for collection on −D2 in fresh media. 24 h before collection media was refreshed. Media was harvested on D0. Changes in metabolite concentrations were determined relative to media maintained on 6-well plates without cells under similar conditions to control for evaporation. Values were further normalized to protein content of 6 replicate wells. These experiments were performed independently at least two times.
Metabolite profiling.
For all metabolite experiments, cells were seeded 2 days before collection in 6-well plates such that cell density was ~75% confluent at time of analysis. Media was refreshed 16–18 h before metabolite collection. Metabolites were extracted with 1 mL ice-cold 80% methanol supplemented with 2 μM deuterated 2-hydroxyglutarate (D-2-hydroxyglutaric-2,3,3,4,4-d5 acid, d5–2HG) as an internal standard. Lysates were incubated overnight incubation at −80°C and then harvested and centrifuged at 21,000g for 20 min. Metabolite extracts were dried in an evaporator (Genevac EZ-2 Elite) and resuspended in 50 uL of 40 mg/mL methoxyamine hydrochloride in pyridine with incubation at 30°C for 2 h. Metabolites were further derivatized by adding 80 μL of MSTFA + 1% TCMS (Thermo Scientific) and 70 μl ethyl acetate (Sigma) with incubation at 37°C for 30 min. Samples were analyzed using an Agilent 7890A GC coupled to Agilent 5975C mass selective detector. The GC was operated in splitless mode with helium gas flow at 1 mL/min. 1 μl of sample was injected onto an HP-5MScolumn and the GC oven temperature ramped from 60°C to 290°C over 25 min. Peaks representing compounds of interest were extracted and integrated using MassHunter software (Agilent Technologies) and peak area was normalized to the internal standard (d5–2HG) peak area and protein content of duplicate samples as determined by BCA protein assay (Thermo Scientific). Ions used for quantification of metabolite levels are as follows: d5–2HG m/z 354; citrate, m/z 465; αKG, m/z 304; aspartate, m/z 334; fumarate, m/z 245; malate, m/z 335 and succinate, m/z 247. All peaks were manually inspected and verified relative to known spectra for each metabolite. For isotope tracing studies, experiments were set up as described above. 4 h before metabolite collection, cells were washed and incubated with glucose- and glutamine-free DMEM media base supplemented with 12C-glucose (Sigma) and 12C-glutamine (Gibco) or the 13C versions of each metabolite, [U-13C]glucose or [U-13C]glutamine (Cambridge Isotope Labs). Enrichment of 13C was determined by quantifying the abundance of the following ions: citrate, 465–482; αKG, 304–315, aspartate, m/z 334–346; fumarate, m/z 245–254; glutamate, m/z 363–377 and malate, m/z 335–347. Correction for natural isotope abundance was performed using IsoCor software53. All experiments were performed independently at least twice and a representative experiment is shown. Peak areas for all individual metabolites as well as the technical normalizations that were performed are found in Supplementary Table 1.
Epigenomic analysis.
1×106 KPsh cells were washed with PBS and plated on and off dox 8 days (-D8) before collecting cells for ATAC-Seq analysis. Cells maintained on dox and off dox were passaged every 48 h. Three days prior to harvest, dimethyl-α-ketoglutarate (DM-αKG) was added to cells grown on dox, while DMSO was added to cells maintained on dox and cells maintained off dox. All cell groups (On dox, DMSO; On dox, DM-αKG; Off dox, DMSO) were split into 6 well plates in duplicate in fresh media containing dox, DM-αKG, and DMSO as indicated two days prior to harvest. This media was refreshed 24 h before collection. Upon harvest, were washed with PBS, trypsinized, and collected as a single cell suspension in complete DMEM. 50,000 cells of each group were sorted using a BD-FACS-ARIA into complete DMEM and washed with PBS. Sorted cells were processed for ATAC-Seq54. Nuclei from washed pellets were extracted using lysis buffer (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630) and transposition performed at 37 degrees for 30 min using the Nextera DNA Library Prep Kit (Illumina). Transposed DNA was purified using the QIAgen MinElute PCR Purification kit and amplified for 12 cycles using the barcoded primers below. Libraries were purified and library assessment was performed using a spectrophotometer (Nanodrop) and automated capillary electrophoresis (Agilent Bioanalyzer). Barcoded libraries were pooled (2–4 samples/lane) and run on an Illumina HiSeq 2500 sequencer using 50 bp paired-end reads.
Primer sequences used for ATAC-Seq sample barcoding (Buenrostro et al-24097267)
Ad1_noMX
AATGATACGGCGACCACCGAGATCTACACTCGTCGGCAGCGTCAGATGTG
Ad2.1_TAAGGCGA
CAAGCAGAAGACGGCATACGAGATTCGCCTTAGTCTCGTGGGCTCGGAGATGT
Ad2.2_CGTACTAG
CAAGCAGAAGACGGCATACGAGATCTAGTACGGTCTCGTGGGCTCGGAGATGT
Ad2.3_AGGCAGAA
CAAGCAGAAGACGGCATACGAGATTTCTGCCTGTCTCGTGGGCTCGGAGATGT
Ad2.4_TCCTGAGC
CAAGCAGAAGACGGCATACGAGATGCTCAGGAGTCTCGTGGGCTCGGAGATGT
Ad2.5_GGACTCCT
CAAGCAGAAGACGGCATACGAGATAGGAGTCCGTCTCGTGGGCTCGGAGATGT
Ad2.6_TAGGCATG
CAAGCAGAAGACGGCATACGAGATCATGCCTAGTCTCGTGGGCTCGGAGATGT
Ad2.7_CTCTCTAC
CAAGCAGAAGACGGCATACGAGATGTAGAGAGGTCTCGTGGGCTCGGAGATGT
Ad2.8_CAGAGAGG
CAAGCAGAAGACGGCATACGAGATCCTCTCTGGTCTCGTGGGCTCGGAGATGT
Ad2.9_GCTACGCT
CAAGCAGAAGACGGCATACGAGATAGCGTAGCGTCTCGTGGGCTCGGAGATGT
Ad2.10_CGAGGCTG
CAAGCAGAAGACGGCATACGAGATCAGCCTCGGTCTCGTGGGCTCGGAGATGT
Ad2.11_AAGAGGCA
CAAGCAGAAGACGGCATACGAGATTGCCTCTTGTCTCGTGGGCTCGGAGATGT
Ad2.12_GTAGAGGA
CAAGCAGAAGACGGCATACGAGATTCCTCTACGTCTCGTGGGCTCGGAGATGT
Ad2.13_GTCGTGAT
CAAGCAGAAGACGGCATACGAGATATCACGACGTCTCGTGGGCTCGGAGATGT
Ad2.14_ACCACTGT
CAAGCAGAAGACGGCATACGAGATACAGTGGTGTCTCGTGGGCTCGGAGATGT
Ad2.15_TGGATCTG
CAAGCAGAAGACGGCATACGAGATCAGATCCAGTCTCGTGGGCTCGGAGATGT
Ad2.16_CCGTTTGT
CAAGCAGAAGACGGCATACGAGATACAAACGGGTCTCGTGGGCTCGGAGATGT
Ad2.17_TGCTGGGT
CAAGCAGAAGACGGCATACGAGATACCCAGCAGTCTCGTGGGCTCGGAGATGT
Ad2.18_GAGGGGTT
CAAGCAGAAGACGGCATACGAGATAACCCCTCGTCTCGTGGGCTCGGAGATGT
Ad2.19_AGGTTGGG
CAAGCAGAAGACGGCATACGAGATCCCAACCTGTCTCGTGGGCTCGGAGATGT
Ad2.20_GTGTGGTG
CAAGCAGAAGACGGCATACGAGATCACCACACGTCTCGTGGGCTCGGAGATGT
Ad2.21_TGGGTTTC
CAAGCAGAAGACGGCATACGAGATGAAACCCAGTCTCGTGGGCTCGGAGATGT
Ad2.22_TGGTCACA
CAAGCAGAAGACGGCATACGAGATTGTGACCAGTCTCGTGGGCTCGGAGATGT
Ad2.23_TTGACCCT
CAAGCAGAAGACGGCATACGAGATAGGGTCAAGTCTCGTGGGCTCGGAGATGT
Ad2.24_CCACTCCT
CAAGCAGAAGACGGCATACGAGATAGGAGTGGGTCTCGTGGGCTCGGAGATGT
For data analysis, quality and adapter filtering was applied to raw reads using ‘trim_galore’ before aligning to mouse assembly mm9 with bowtie2 using the default parameters. The Picard tool MarkDuplicates (http://broadinstitute.github.io/picard/) was used to remove reads with the same start site and orientation. The BEDTools suite (http://bedtools.readthedocs.io) was used to create read density profiles. Enriched regions were discovered using MACS2 and scored against matched input libraries (fold change > 2 and FDR-adjusted p-value < 0.1). A consensus peak atlas was then created by filtering out blacklisted regions (http://mitra.stanford.edu/kundaje/akundaje/release/blacklists/mm9-mouse/mm9-blacklist.bed.gz) and then merging all peaks within 500 bp. A raw count matrix was computed over this atlas using featureCounts (http://subread.sourceforge.net/) with the ‘-p’ option for fragment counting. The count matrix and all genome browser tracks were normalized to a sequencing depth of ten million mapped fragments. DESeq2 was used to classify differential peaks between two conditions using fold change > 2 and FDR-adjusted p-value < 0.1. Peak-gene associations were made using linear genomic distance to the nearest transcription start site with Homer (http://homer.ucsd.edu). ChIP-Seq data55 were processed in the same way as ATAC-seq data, except read density profiles for ChIP included a read extension equivalent to the average library fragment size. Motif signatures for p53 were discovered using Homer’s annotatePeaks.pl script using the default settings.
Statistics and reproducibility.
GraphPad PRISM 7 and R software was used for statistical analyses. Sample size, error bars and statistical tests are reported in the figure legends. Exact p-values are shown where possible. P value ranges reported when the number of significant digits exceeds the limits calculated by the statistical package. Statistical tests include unpaired two-tailed Student’s t-test, one-way analysis of variance (ANOVA), two-way ANOVA and Fisher’s exact test. All sequencing experiments were done once. The number of times experiments were performed with similar results is indicated in each legend.
Data availability Statement.
RNA-Seq and ATAC-Seq data that support the findings of this study have been deposited in the Gene Expression Omnibus under the accession codes GSE114263 and GSE114342. ChIP-Seq data reanalyzed for this study is available under the accession code GSE46240. All other data supporting the findings of this study are available from the corresponding authors upon reasonable request.
Extended Data
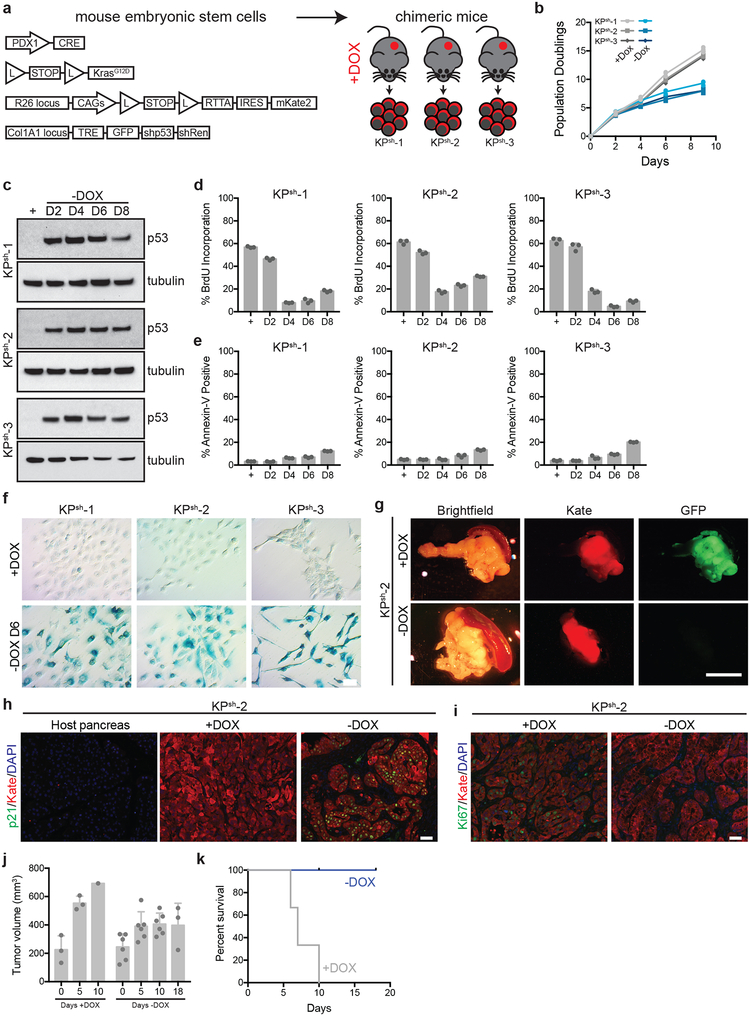
Extended Data Figure 1. KPsh ESC-GEMM PDAC model driven by mutant Kras and inducible and reversible p53 silencing.
a, KPsh embryonic stem cell-based genetically engineered mouse model (ESC-GEMM) of pancreatic ductal adenocarcinoma (PDAC). Embryonic stem cells express Pdx1-Cre (transgenic expression of Cre in pancreatic progenitors); LSL-KrasG12D (knock-in, conditional heterozygous expression of mutant Kras); RIK (knock in, conditional heterozygous expression of rtTA and fluorescent mKate2 from the Rosa26 locus); Col1a1-TRE-GFP-shp53-shRenilla (Col1a1 homing cassette (CHC) targeted with doxycycline inducible tandem shRNA expressing shp53 and shRenilla linked to GFP). KPsh mice were generated by blastocyst injection and mothers enrolled on dox chow at day 5. Cell lines were derived and maintained in dox-containing media from tumors arising in dox fed mice. All KPsh cells constitutively express mKate2 (Kate) and rtTA. b, Population doublings of KPsh-1–3 lines grown on/off dox. c-f, Characterization of p53 levels (c), BrdU incorporation (d), Annexin-V staining (e) and senescence-associated β-galactosidase (SA-β GAL) staining (f) in three independent KPsh lines grown on/off dox (D, day). g, Representative gross pathology and epifluorescence images of pancreatic tumors resulting from orthotopic transplant of KPsh-2 cells into dox fed mice maintained on dox chow (top, n=3 mice) or withdrawn from dox chow for 10 days (bottom, n=3 mice). KPsh cells uniformly express Kate, while GFP expression indicates cell actively expressing the p53 shRNA. h, Representative Cdkn1a/p21 immunostainig in matched normal host pancreas, or in orthotopic KPsh-2 tumors maintained on dox (n=3) or 10 days following dox withdrawal (n=3). Kate indicates injected KPsh-2 cells. i, Representative Ki67 immunostaining in orthotopic KPsh-2 tumors maintained on dox (n= 3 mice) or 10 days following dox withdrawal (n=3 mice). Kate indicates injected KPsh-2 cells. j, Small animal ultrasound measurement of tumor volume. KPsh-2 cells were injected into dox-fed mice and mice were maintained on dox diet for 2 weeks. After two weeks (D0), tumor size was measured and mice were randomized into off (n=6 mice) and on dox chow groups (n=3 mice). Subsequent tumor size was measured at the indicated time points. n=3 mice on dox were collected for analysis upon sacrifice, n=3 mice were analyzed after 5 and 10 days of dox withdrawal, respectively. k, Survival of mice shown in i after randomization into groups maintained on dox food (n=3 mice) or following dox withdrawal (n= 6 mice). b-f were repeated twice with similar results. Data are presented as either representative independently treated wells (c,f) or mean ± SD of n=3, independently treated wells with individual data points shown (b,d,e). For gel source data (c), see Supplementary Figure 1. Scale bar for immunostaining 50 μM. Scale bar for pathology 1 cm.
Extended Data Figure 2. p53 restoration increases the αKG/succinate ratio independently of changes in proliferation or senescence.
a, Glucose/glutamine consumption, lactate production in KPsh-1,2 cultured on/off dox for 4 or 8 days (D, days). b, TCA cycle schematic indicating entry points for glucose- and glutamine-derived carbons. Metabolites in red were assessed by isotope tracing experiments. c,d, Metabolite fraction containing 13C derived from [U-13C]glucose (13C-Glc) (c) or derived from [U-13C]glutamine (13C-Gln) (d) after four h of labeling off/on dox for six days. e, f p53 immunoblot (e) or Senescence-associated β-galactosidase (SA-β GAL) staining (f) in on dox KPsh-2 treated with 3 μM etoposide (Etopo) or 25 nM trametinib (Tram) for 48 or 96 h. Cells grown in the absence of dox (-dox) for six days are included as a positive control. g,h, SA-βGAL (g) or BrdU positive (h) cells treated as described in (e). i, Western blot of cells expressing shRenilla, shp19, shp16/p19, or shCdkn1a/p21 on/off dox for six days. j, SA-βGAL staining of cells described in (i). k,l, SA-βGAL (k) or BrdU positive (l) cells treated as described in (i). m, αKG/succinate ratio in cells expressing shRenilla, shp19, p16/p19, or shCdkn1a/p21 on/off dox for six days. a,c,d,f-m were repeated twice with similar results and e was performed once. Data are presented as mean ± SEM of n=6, independently treated wells (a), mean ± SD of n=3, independently treated wells from a representative experiment with individual data points shown (c,d,f,g,h,k,l,m), or representative of 1 independently treated well (e,f,i,j). For gel source data (e,i), see Supplementary Figure 1. Significance assessed in comparison to cells grown with dox by 1-way ANOVA with Tukey’s multiple comparison post-test (a) or in the indicated comparisons by 2-way ANOVA with Sidak’s post-test (m). Scale bar 50 μM.
Extended Data Figure 3. Characterization of reversibility of p53 dependent effects in KPsh cells.
a, αKG/succinate ratio in KrasG12D; TRE-shRenilla (KRsh) PDAC cells cultured with or without dox for indicated number of days. b-d, Western blot for p53 (b), population doublings (c) and αKG/succinate ratio (d) in 2 KPsh lines cultured with (+Dox) or without dox (-Dox) for 6 days days or cultured without dox for 6 days, followed by 6 days of culture with dox (-Dox→+Dox, arrow indicates when dox was re-introduced). a, c were performed twice with similar results and b,d were performed once. For gel source data (b), see Supplementary Figure 1. Data are presented as mean ± SD of n=3, independently treated wells of a representative experiment with individual data points shown (a,c,d) or or representative of 1 independently treated well (b).
Extended Data Figure 4. Functional p53 transactivation is required to increase the cellular αKG/succinate ratio.
a-c, p53 immunoblot (a), Cdkn1a/p21 qRT-PCR (b), and αKG/succinate ratio (c) in KPfloxRIK-TRE-Empty, KPfloxRIK-TRE-p53WT (dox inducible expression, wt p53), and KPfloxRIK-TRE-p53TAD1/2M (dox inducible expression, p53 with mutations in both transactivation domain 1 and 2) cells 2 days off/on dox. d Cdkn1a/p21, Mdm2, p53 (top), Idh1 and Pcx (bottom) qRT-PCR in KPsh-2 off/on dox. Day 0 and day 6 Cdkn1a/p21, Mdm2, and p53 values also shown in Fig. 1a. e, αKG/succinate ratio in KPsh1–3 grown on/off dox. f, Idh1 and Pcx qRT-PCR in KPfloxRIK-TRE-Empty, KPfloxRIK-TRE-p53WT, and KPfloxRIK-TRE-p53TAD1/2M off/on dox for 2 days. g, PC activity associated glucose labeling patterns. IDH1 and PC dependent reactions labeled. h, Fractional m+3 (top) or m+5 (bottom) labeling of aspartate and citrate in KPsh1–3 on/off dox for six days after four hours with [U-13C]glucose. i, Idh1 qRTPCR in KPsh-2 expressing shRenilla or shIdh1 on/off dox for 8 days. j, Idh1 and p53 levels in KPsh-2 expressing shRenilla or shIdh1 grown on/off dox for 8 days. Arrowhead: specific Idh1 band. k, αKG/succinate ratio in KPsh-2 expressing shRenilla or shIdh1 grown on/off dox. l,m. p53 and Idh1 immunoblot (l) and αKG/succinate ratio (m) in KPfloxRIK-TRE-Empty, KPfloxRIKTRE-p53WT, and KPfloxRIK-TRE-p53TAD1/2M expressing shRenilla or shIdh1 grown with dox for 2 days. n, αKG/succinate ratio in parental KPsh-2 versus KPsh-2 expressing IDH1 or IDH2 cDNA grown on dox. a-e,h-n were repeated twice with similar results. Data are presented as representative of 1 independently treated well (a,j,l) or as mean ± SD of n=3 independently treated wells of a representative experiment with individual data points shown (b-f, h,i,k,m,n). For gel source data (a,j,l), see Supplementary Figure 1. Significance assessed compared to cells grown on dox by 1-way ANOVA with Sidak’s multiple comparisons post-test (e) or indicated comparisons (f).
Extended Data Figure 5. p53 binding at Pcx and Idh1.
a. Analysis of ChIP-Seq signal at the Pcx locus in primary p53WT and p53null (KO) mouse embryonic fibroblasts after treatment with doxorubicin. b. Analysis of ChIP-Seq signal at the Idh1 locus in primary p53WT and p53null (KO) mouse embryonic fibroblasts after treatment with doxorubicin. p53 binding sites predicted by MACs comparison of immunoprecipitation samples with input. Response elements predicted by Homer analysis as described in methods. ChIP-Seq data from ref55.
Extended Data Figure 6. Elevating intracellular αKG levels phenocopies the effect of p53 reactivation on gene expression.
a, Mean log2 fold change of all genes following p53 reactivation or cell-permeable αKG in n=2, independently treated wells of KPsh-1 cells. All samples treated with equal amounts of DMSO (vehicle). Spearman correlation r = 0.556, p < 1e-15. b, c qRT-PCR of genes upregulated with both p53 restoration and αKG in KPsh-1 (b) and KPsh-2 (c) cells treated for 72 hours with vehicle, dimethyl-αKG (DM-αKG), diethyl-αKG (DE-αKG) or following p53 restoration (-dox 8 days). d, qRT-PCR of PanIN-cell associated genes in KPsh-1 cells grown on/off dox and treated with 4 mM sodium acetate for 72 h. e, Ogdh immunoblot in p53 null KPfloxRIK or p53 mutant KPR172HRIK cells expressing shOgdh for 4 days. f, Doubling time, day 1–4, of KPfloxRIK and KPR172HRIK cells expressing dox-inducible shOgdh or shRenilla. g,h, Percentage SA-βGAL positive (g) or Annexin-V positive (h) shRNA expressing KPfloxRIK and KPR172HRIK cells off/on dox for 4 days. Etoposide (etopo, 96hrs, 3 μM) included as a positive control. i, αKG/succinate ratio of KPCR172HRIK cells expressing dox-inducible shOgdh or shRenilla grown 4 days off/on dox. j, qRT-PCR of p53/αKG co-regulated genes in shOgdh KPR172HRIK cells compared to shRenilla controls. b-d,i,j were repeated twice with similar results and e was performed once. f-h were repeated in 2 additional lines. For gel source data (e), see Supplementary Figure 1. Data presented as individual data points (b,c,j). mean ± SD (d,f,g,h), mean ± SEM (i), or as a representative image (e) of n=3, independently treated wells of a representative experiment with individual data points shown. Significance assessed in the indicated comparisons by 2-way ANOVA with Sidak’s multiple comparison post test (i) or compared to shRenilla expressing cells by 1-way ANOVA with Tukey’s multiple comparison post-test (g,h).
Extended Data Figure 7. Ogdh depletion induces differentiation and decreases tumor growth.
a, Representative CK19 staining of KPsh-2 derived orthotopic tumors injected in mice on dox (n=3 mice) or ten days after dox withdrawal (n=3 mice). Kate marks injected KPsh-2 cells. b, Representative gross images of KPflox derived orthotopic tumors 12 days following injection of cells expressing shRenilla (n=5), shOgdh (n=4 mice), or shSdha (n=4 mice) into dox fed mice. GFP indicates shRNA expressing cells. c, Small animal ultrasound measurement of tumors derived from KPflox cells dox inducuble shRenilla (n=4 mice), shOgdh (Ogdh-1, n=4 mice, Ogdh-2, n=3 mice), or shSdha (n=4) after enrolling on dox chow. d, Representative H&E staining of KPflox derived orthotopic tumors 9 days after expression of dox-inducible shRenilla, shOgdh, or shSdha in established tumors from c. GFP marks injected KPflox cells expressing indicated shRNA. e, Representative CK19 staining of KPflox derived orthotopic tumors expressing dox-inducible shRenilla, shOgdh, or shSdha from Figure 3b. GFP marks injected KPflox cells expressing indicated shRNA. f, Representative CK19 staining of KPflox derived orthotopic tumors 9 days after expression of dox-inducible shRenilla, shOgdh, or shSdha in established tumors from c. GFP marks cells expressing indicated shRNA. g, Western blot (top) of Sdha in KPflox cells expressing dox-inducible shRenilla or shSdha grown with dox for 4 days. αKG/succinate ratio (bottom) in in KPflox cells expressing dox-inducible shRenilla or shSdha grown with dox for 4 days. For c, data are presented as mean ± SD and individual, longitudinally tracked tumor volumes are presented in the source data. For gel source data (g), see Supplementary Figure 1. g was performed once and GCMS is presented as mean ± SD of n=3, independently treated wells with individual data points shown. Scale bar for immunostaining 50 μM. Scale bar for pathology 1 cm.
Extended Data Figure 8. Ogdh inhibition reduces tumor cell competitive fitness in vivo.
a, αKG/succinate ratio of KPCfloxRIK and KPCR172HRIK cells expressing dox-inducible shRNAs targeting Renilla, Ogdh, and Sdha grown 4 days with or without dox. b, Schematic of in vivo competition assay. Kate positive KPfloxRIK and KPR172HRIK cells were infected with retroviruses encoding dox inducible, GFP linked shRNAs targeting Renilla, Ogdh, or Sdha. Cells were selected for viral integration, induced with dox for 2 days, mixed with uninfected parental cellsat a ratio of 8:2 and analyzed by flow cytometry to determine initial ratio of shRNA expressing cells to uninfected cells. This cell mixture was injected orthotopically into dox fed recipient mice. After 3 weeks of tumor growth, pancreatic tumors were removed, weighed, dissociated, and analyzed by flow cytometry to measure final ratio of shRNA expressing to uninfected cells. Data are presented in Fig. 3c. c,d, Tumor mass of orthotopically injected KPfloxRIK (c) or KPR172HRIK (d) cells expressing a mixture of shRNAs targeting Renilla, Ogdh, or Sdha from Fig. 3c. KPfloxRIK: n= 5 mice shRenilla, n=4 mice shOgdh, shSdha. KPR172KRIK : n= 5 mice shRenilla, n=4 mice shOgdh, shSdha-1,2, n=3 mice shSdha-3. e, Representative gross images of pancreatic tumors arising in dox fed mice 3 weeks days following orthotopic transplant of KPCfloxRIK (top) and KPCR172HRIK (bottom) cells expressing dox-inducible shRNAs targeting Renilla, Ogdh, and Sdha mixed 8:2 from Fig. 3c. a was performed once. Data are presented as mean ± SD of n=3, independently treated wells of a representative experiment with individual data points shown. Significance assessed compared to shRenilla controls by 1-way ANOVA with Dunnet’s multiple comparison post-test (c). Scale bar 1 cm.
Extended Data Figure 9. p53 reactivation and Ogdh inhibition induce 5hmC accumulation in PDAC cells.
a, Median fluorescence intensity of 5hmC in KPsh-2 cells grown with or without dox for 8 days. b, qRT-PCR of Tet1, Tet2, and Tet3 expression in KPsh-2 cells grown with or without dox for indicated number of days. c, Sequence analysis of CRISPR/Cas9 editing. Percentage of amplicons flanking sgRNA target sequence with indicated genotype amplified from KPsh-2 cells expressing sgRNAs targeting Tet1, Tet2, and Tet3. d, 5hmC Median fluorescence intensity (MFI) in KPsh-2 cells expressing sgRNAs targeting Tet1, Tet2, and Tet3 grown with or without dox for 8 days. e, 5hmC MFI in KPsh-2 cells grown with 4 mM DM-αKG for 72 h. f, 5hmC MFI in 8988 and Panc1 cells grown with 4 mM DM-αKG for 72 h. g, 5hmC MFI in KPCfloxRIK and KPCR172HRIK cells expressing dox-inducible shRNAs targeting Renilla or Ogdh grown 4 days with or without dox. h, Representative 5hmC staining in orthotopic tumors derived from KPflox cells expressing dox-inducible hairpins targeting Renilla (n=5 mice), Ogdh (n=4 mice), or Sdha (n=4 mice) two weeks after injection in mice maintained on dox. i, Representative 5hmC staining of orthotopic tumors derived from KPflox cells 9 days after activation of dox-inducible hairpins targeting Renilla (n=4 mice), Ogdh (Ogdh-1, n=4 mice, Ogdh-2, n=3 mice), or Sdha-1–3 (n=4) in established tumors. GFP marks cells expressing indicated shRNA. a,d-g were repeated twice with similar results, b was repeated in an additional line. Data are presented as mean ± SD of n=3, independently treated wells of a representative experiment with individual data points shown. Significance assessed by two-tailed Student’s t-test (a,e,f) or compared to shRenilla controls by 1-way ANOVA with Tukey’s multiple comparison post-test (g). Scale bar 50 μM
Extended Data Figure 10. Increase in the cellular αKG/succinate ratio enforces p53 driven tumor suppression.
a-c, Sdha and p53 immunoblot (a), αKG/succinate ratio (b) and 5hmC MFI (c) in KPsh-2 cells expressing constitutive shRNAs targeting Sdha or Renilla grown with or without dox for 8 days. d, Representative 5hmC staining in orthotopic tumors derived from KPsh-2 cells expressing constitutive shRNAs targeting Sdha or Renilla in mice maintained on dox (n=3 mice) or ten days following dox withdrawal (n=3 mice). GFP denotes cells expressing hairpin targeting p53. 5hmC staining per nucleus is quantified in Fig. 4g. e,f, Small animal ultrasound measurement of tumors derived from KPsh-2 cells expressing constitutive shRNAs targeting Renilla (left) or Sdha (right) orthotopically injected into dox-fed mice maintained on dox diet for 2 weeks. After two weeks (D0), tumor size was measured and mice were randomized into off and on dox chow groups (shRenilla n=5 mice, shSdha n=4 mice). Subsequent tumor size and mouse survival was monitored up to 10 days after dox withdrawal. g, Fold change in tumor size from day 5 to day 10 following withdrawal of dox chow (D0) from mice bearing orthotopic tumors derived from KPsh-2 cells expressing constitutive shRNAs targeting Sdha or Renilla. h, Representative H&E staining of orthotopic tumors derived from KPsh-2 cells expressing constitutive shRNAs targeting Sdha or Renilla maintained on dox or 10 days following dox withdrawal. a and b were performed once, c was repeated twice with similar results. For gel source data (a), see Supplementary Figure 1. Data are presented as either a representative independently treated well (a), or as mean ± SD of n=3 independently treated wells with individual data points shown (b,c). Significance assessed by two-tailed, unpaired t-test (g). Scale bar 50 μM.
Supplementary Material
Acknowledgements.
We thank members of the Lowe and Finley laboratory. PanIN and PDAC tissue microarrays were generated by Gokce Askan and Olca Basturk and generously provided as a resource from the David M Rubenstein Center for Pancreatic Cancer Research at Memorial Sloan Kettering Cancer Center. J.P.M IV was supported by an American Cancer Society Postdoctoral Fellowship (126337-PF-14-066-01-TBE). J.Y. is supported by a T32 training grant from the NICHD (T32HD060600). F.J.S.-R. is an HHMI Hanna Gray Fellow and was partially supported by an MSKCC Translational Research Oncology Training Fellowship (NIH T32-CA160001). L.W.S.F is a Dale F. Frey-William Raveis Charitable Fund Scientist supported by the Damon Runyon Cancer Research Foundation (DFS-23–17) and a Searle Scholar. S.W.L.is an investigator in the Howard Hughes Medical Institute. This work was additionally supported by a Lustgarten Research Investigator Award (S.W.L.), a Program Program Project grant from the National Cancer Institute (S.W.L.), research grants from the Emerson Collective (S.W.L.), the Starr Cancer Consortium (I11–039 L.W.S.F.), the Concern Foundation (L.W.S.F.) the Anna Fuller Fund (L.W.S.F.), and the Memorial Sloan Kettering Cancer Center Support Grant P30 CA008748.
Footnotes
Competing interests. S.W.L. is a founder and scientific advisory board member of Blueprint Medicines, Mirimus Inc., and ORIC pharmaceuticals, and on the scientific advisory board of Constellation Pharmaceuticals, Petra Pharmaceuticals, and PMV Pharmaceuticals. C.B.T. is a founder of Agios Pharmaceuticals and a member of its scientific advisory board. He also previously served on the board of directors of Merck and Charles River Laboratories.
References.
- 1.Waddell N et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518, 495–501, doi: 10.1038/nature14169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kastenhuber ER & Lowe SW Putting p53 in Context. Cell 170, 1062–1078, doi: 10.1016/j.cell.2017.08.028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kruiswijk F, Labuschagne CF & Vousden KH p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nature reviews. Molecular cell biology 16, 393–405, doi: 10.1038/nrm4007 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Morris J. P. t., Wang SC & Hebrok M KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nature reviews. Cancer 10, 683–695, doi: 10.1038/nrc2899 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey P et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52, doi: 10.1038/nature16965 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Ying H et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670, doi: 10.1016/j.cell.2012.01.058 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Son J et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496, 101–105, doi: 10.1038/nature12040 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hingorani SR et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer cell 7, 469–483, doi: 10.1016/j.ccr.2005.04.023 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Rhim AD et al. EMT and dissemination precede pancreatic tumor formation. Cell 148, 349–361, doi: 10.1016/j.cell.2011.11.025 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saborowski M et al. A modular and flexible ESC-based mouse model of pancreatic cancer. Genes & development 28, 85–97, doi: 10.1101/gad.232082.113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldser DM et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature 468, 572–575, doi: 10.1038/nature09535 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Junttila MR et al. Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature 468, 567–571, doi: 10.1038/nature09526 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martins CP, Brown-Swigart L & Evan GI Modeling the therapeutic efficacy of p53 restoration in tumors. Cell 127, 1323–1334, doi: 10.1016/j.cell.2006.12.007 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Xue W et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445, 656–660, doi: 10.1038/nature05529 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carey BW, Finley LW, Cross JR, Allis CD & Thompson CB Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518, 413–416, doi: 10.1038/nature13981 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raffel S et al. BCAT1 restricts alphaKG levels in AML stem cells leading to IDHmut-like DNA hypermethylation. Nature 551, 384–388, doi: 10.1038/nature24294 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Liu PS et al. alpha-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat Immunol 18, 985–994, doi: 10.1038/ni.3796 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Brady CA et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 145, 571–583, doi: 10.1016/j.cell.2011.03.035 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng T et al. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proceedings of the National Academy of Sciences of the United States of America 108, 8674–8679, doi: 10.1073/pnas.1016627108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–1218, doi: 10.1038/nmeth.2688 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davie K et al. Discovery of transcription factors and regulatory regions driving in vivo tumor development by ATAC-seq and FAIRE-seq open chromatin profiling. PLoS Genet 11, e1004994, doi: 10.1371/journal.pgen.1004994 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hosoda W et al. Genetic analyses of isolated high-grade pancreatic intraepithelial neoplasia (HG-PanIN) reveal paucity of alterations in TP53 and SMAD4. The Journal of pathology 242, 16–23, doi: 10.1002/path.4884 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wellen KE et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080, doi: 10.1126/science.1164097 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cimmino L & Aifantis I Alternative roles for oxidized mCs and TETs. Curr Opin Genet Dev 42, 1–7, doi: 10.1016/j.gde.2016.11.003 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang H et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene 32, 663–669, doi: 10.1038/onc.2012.67 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao M et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes & development 26, 1326–1338, doi: 10.1101/gad.191056.112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaelin WG Jr. & McKnight SL Influence of metabolism on epigenetics and disease. Cell 153, 56–69, doi: 10.1016/j.cell.2013.03.004 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schvartzman JM, Thompson CB & Finley LWS Metabolic regulation of chromatin modifications and gene expression. The Journal of cell biology 217, 2247–2259, doi: 10.1083/jcb.201803061 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boj SF et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338, doi: 10.1016/j.cell.2014.12.021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hingorani SR et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer cell 4, 437–450 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Kawaguchi Y et al. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nature genetics 32, 128–134, doi: 10.1038/ng959 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Pan FC et al. Spatiotemporal patterns of multipotentiality in Ptf1a-expressing cells during pancreas organogenesis and injury-induced facultative restoration. Development 140, 751–764, doi: 10.1242/dev.090159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson EL et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes & development 15, 3243–3248, doi: 10.1101/gad.943001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olive KP et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 119, 847–860, doi: 10.1016/j.cell.2004.11.004 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Marino S, Vooijs M, van Der Gulden H, Jonkers J & Berns A Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes & development 14, 994–1004 (2000). [PMC free article] [PubMed] [Google Scholar]
- 36.Beard C, Hochedlinger K, Plath K, Wutz A & Jaenisch R Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis 44, 23–28, doi: 10.1002/gene.20180 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Dow LE et al. Conditional reverse tet-transactivator mouse strains for the efficient induction of TRE-regulated transgenes in mice. PloS one 9, e95236, doi: 10.1371/journal.pone.0095236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weissmueller S et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor beta signaling. Cell 157, 382–394, doi: 10.1016/j.cell.2014.01.066 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dickins RA et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nature genetics 37, 1289–1295, doi: 10.1038/ng1651 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Fellmann C et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep 5, 1704–1713, doi: 10.1016/j.celrep.2013.11.020 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Chen C et al. Cancer-associated IDH2 mutants drive an acute myeloid leukemia that is susceptible to Brd4 inhibition. Genes & development 27, 1974–1985, doi: 10.1101/gad.226613.113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanjana NE, Shalem O & Zhang F Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784, doi: 10.1038/nmeth.3047 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruscetti M et al. NK cell-mediated cytotoxicity contributes to tumor control by a cytostatic drug combination. Science 362, 1416–1422, doi: 10.1126/science.aas9090 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aksoy O et al. The atypical E2F family member E2F7 couples the p53 and RB pathways during cellular senescence. Genes & development 26, 1546–1557, doi: 10.1101/gad.196238.112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morris J. P. t. et al. Dicer regulates differentiation and viability during mouse pancreatic cancer initiation. PloS one 9, e95486, doi: 10.1371/journal.pone.0095486 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zafra MP et al. Optimized base editors enable efficient editing in cells, organoids and mice. Nat Biotechnol 36, 888–893, doi: 10.1038/nbt.4194 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolger AM, Lohse M & Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120, doi: 10.1093/bioinformatics/btu170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, doi: 10.1093/bioinformatics/bts635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao Y, Smyth GK & Shi W featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930, doi: 10.1093/bioinformatics/btt656 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Anders S, Pyl PT & Huber W HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169, doi: 10.1093/bioinformatics/btu638 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Millard P, Letisse F, Sokol S & Portais JC IsoCor: correcting MS data in isotope labeling experiments. Bioinformatics 28, 1294–1296, doi: 10.1093/bioinformatics/bts127 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Buenrostro JD, Wu B, Chang HY & Greenleaf WJ ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21 29 21–29, doi: 10.1002/0471142727.mb2129s109 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kenzelmann Broz D et al. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes & development 27, 1016–1031, doi: 10.1101/gad.212282.112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq and ATAC-Seq data that support the findings of this study have been deposited in the Gene Expression Omnibus under the accession codes GSE114263 and GSE114342. ChIP-Seq data reanalyzed for this study is available under the accession code GSE46240. All other data supporting the findings of this study are available from the corresponding authors upon reasonable request.