Abstract
Neonates at risk of childhood atopy and asthma exhibit perturbation of the gut microbiome, metabolic dysfunction and increased concentrations of 12,13-diHOME in their faeces. However, the mechanism, source and contribution of this lipid to allergic inflammation remain unknown. Here, we show that intra-abdominal treatment of mice with 12,13-diHOME increased pulmonary inflammation and decreased the number of regulatory T (Treg) cells in the lungs. Treatment of human dendritic cells with 12,13-diHOME altered expression of PPARy-regulated genes and reduced anti-inflammatory cytokine secretion and the number of Treg cells in vitro. Shotgun metagenomic sequencing of neonatal faeces indicated that bacterial epoxide hydrolase (EH) genes are more abundant in the gut microbiome of neonates who develop atopy and/or asthma during childhood. Three of these bacterial EH genes (3EH) specifically produce 12,13-diHOME, and treatment of mice with bacterial strains expressing 3EH caused a decrease in the number of lung Treg cells in an allergen challenge model. In two small birth cohorts, an increase in the copy number of 3EH or the concentration of 12,13-diHOME in the faeces of neonates was found to be associated with an increased probability of developing atopy, eczema or asthma during childhood. Our data indicate that elevated 12,13-diHOME concentrations impede immune tolerance and may be produced by bacterial EHs in the neonatal gut, offering a mechanistic link between perturbation of the gut microbiome during early life and atopy and asthma during childhood.
The development of severe asthma during childhood is often preceded by eczema and/or atopy—a heightened immune response to multiple food allergens and aeroallergens1. Although genetic risk factors have been identified, the increased prevalence of atopy, eczema and asthma, predominantly in developed nations, suggests that changes in lifestyle and environmental exposures play a key role in the development of these diseases. Indeed, a number of early-life exposures have been identified that increase childhood atopy and asthma risk, including formula feeding, birth by caesarian section, and prenatal and postnatal antibiotic use, whereas early-life exposure to livestock and furred pets confer protection2–6. Many of these exposures shape the composition and function of the human microbiome, which has now been established as a critical driver of immune function7–9.
Independent US and Canadian birth cohorts revealed a link between the early-life gut microbiome and the subsequent development of childhood atopy and asthma10–12. The gut microbiota of infants and neonates at increased risk of childhood atopy and/ or asthma are distinct, characteristically depleted of bacterial genera and exhibit fungal expansion and metabolic reprogramming10–12. Faecal metabolic alterations in high-risk infants include depletion of anti-inflammatory short-chain or polyunsaturated fatty acids and enrichment of monohydroxy fatty acids, including 12,13-diHOME10–12. Importantly, in mice, gut-microbiome-derived short-chain and polyunsaturated fatty acids protect against allergic inflammation of the airways13,14. By contrast, in vitro co-culture studies of human dendritic cells (DCs) and T cells revealed that pretreatment of DCs with 12,13-diHOME caused a dose-dependent decrease in the frequency of Treg cells11, a characteristic immunological feature of untreated children with asthma15.
12,13-diHOME is a relatively uncharacterized linoleic acid (LA) metabolite previously shown to be induced in the airways of adults with asthma following bronchial provocation with an allergen16. More recent in vivo studies have linked 12,13-diHOME with increased thermal hyperalgesia and fatty-acid uptake in adipose and skeletal muscle17–19, implicating it as an important lipid mediator in a range of physiological processes. However, the mechanism and source of this lipid in the faeces of neonates at risk of asthma remain unknown. Given our previous observations11, we hypothesized that elevated faecal concentrations of 12,13-diHOME promote allergic inflammation by inducing DC dysfunction, resulting in a subsequent reduction in the number of anti-inflammatory Treg cells. Furthermore, we hypothesized that this lipid is produced by gut microorganisms in high-risk neonates. Here we show that—at elevated concentrations—12,13-diHOME caused an increase in pulmonary inflammation, a reduction in the number of lung Treg cells in vivo and reprogrammed human DC activity in vitro. We identified three strain-specific bacterial genes within the neonatal gut microbiome that catalyse production of 12,13-diHOME and are sufficient, when introduced to the gut microbiome of mice, to recapitulate the effects of 12,13-diHOME on pulmonary Treg cells in vivo. Finally, we show that an increase in the faecal copy number of these three bacterial epoxide hydrolase genes (3EH) or the concentration of their lipid product in one-month-old neonates significantly increases the probability of developing childhood allergies, eczema and asthma.
Results
12,13-diHOME exacerbates lung inflammation in mice challenged with allergens.
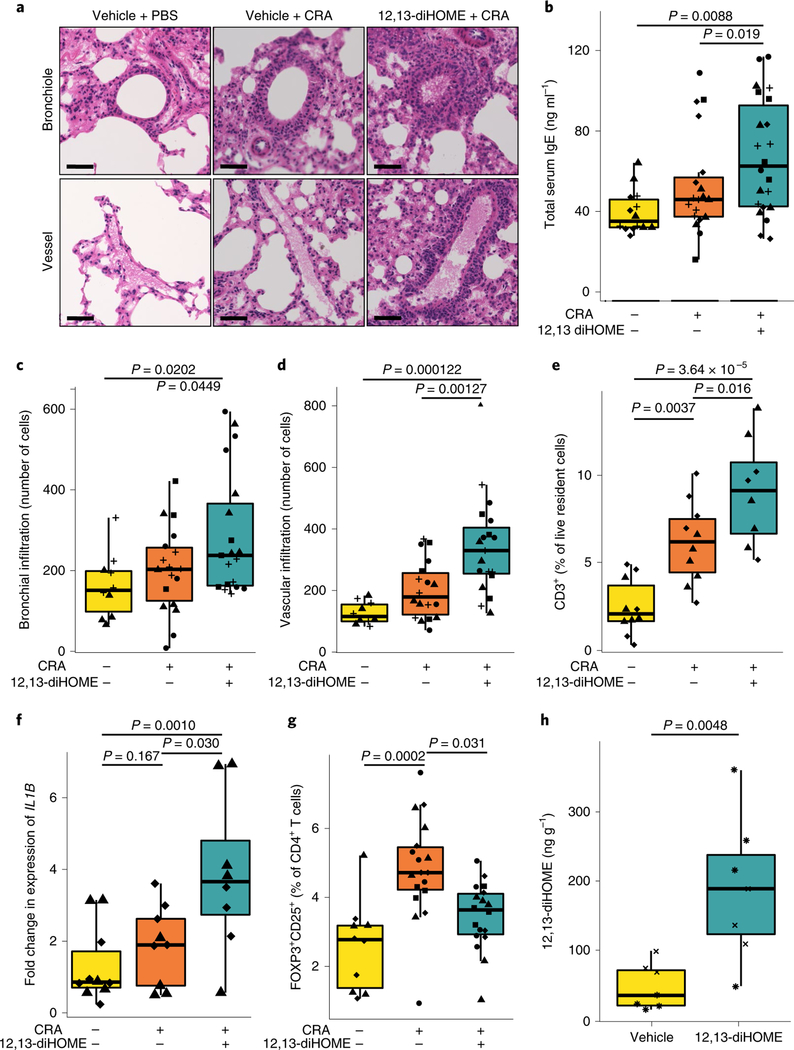
Initially, we examined whether 12,13-diHOME exacerbates allergic sensitization in vivo by treating mice with 12,13-diHOME (30 mg kg−1) or vehicle control (10% DMSO) by peritoneal injection before subjecting them to airway sensitization and challenge with cockroach antigen (CRA). Treated animals showed significantly increased peribronchial and perivascular inflammatory infiltrates and total serum IgE compared with those treated with vehicle alone (Fig. 1a–d, Supplementary Fig. 1a,b). Furthermore, 12,13-diHOME-treated animals also exhibited increases in lung-resident T cells, neutrophils and monocytes, and pulmonary expression of pro-inflammatory innate cytokines IL-1β, IL-1α and TNF, as well as a significant decrease in the number of lung Treg cells and a decrease in the number of alveolar macrophages in the lungs (Fig. 1e–g, Supplementary Fig. 1j–m).
Fig. 1 |. Peritoneal treatment with 12,13-diHOME exacerbates lung inflammation in mice challenged with CRA.
a, Haematoxylin and eosin-stained bronchioles and blood vessels from the lungs of mice treated with vehicle (10% DMSO) and challenged with PBS or CRA, or treated with 30 mgkg−1 12,13-diHOME (solubilized in 10% DMSO) and challenged with CRA. Scale bars, 50 μm. Representative of four experimental repeats. b, Peritoneal 12,13-diHOME treatment (n = 22 mice) increases serum IgE compared with vehicle-treated CRA-challenged mice (n = 19 mice; linear mixed effects (LME), P= 0.019) or vehicle-treated, PBS-challenged mice (n = 14 mice; LME, P= 0.0088). c,d, 12,13-diHOME treatment (n = 19 mice) increases the number of infiltrating cells surrounding the bronchioles (c) and veins (d) of CRA-challenged mice compared with mice treated with vehicle and challenged with PBS (n = 10 mice; LME, P = 0.0202 (bronchial), P = 0.000122 (venous)) or mice treated with vehicle and challenged with CRA (n = 18 mice; LME, P=0.0449 (bronchial), P = 0.00127 (venous)). e, Peritoneal 12,13-diHOME treatment increases the number of lung-resident T cells (CD3+) in CRA-challenged mice (n = 8 mice) compared with mice treated with vehicle and challenged with PBS (n = 10 mice; LME, P = 4.69×10−6) or mice treated with vehicle and challenged with CRA (n = 10 mice; LME, P = 0.016). f, 12,13-diHOME treatment of mice challenged with CRA (n = 8 mice) increases the expression of IL1B compared with expression in mice treated with vehicle and challenged with PBS (n = 9 mice; LME, P=0.0010) or mice treated with vehicle and challenged with CRA (n = 9 mice; LME, P = 0.030). g, 12,13-diHOME treatment of mice challenged with CRA (n = 18 mice) decreases the number of Treg cells compared with the number in mice treated with vehicle and challenged with CRA (n = 18 mice; LME, P= 0.031). h, Peritoneal treatment of mice with 12,13-diHOME significantly increases the concentration of 12,13-diHOME in the lungs (n = 7 mice; LME, P = 0.0048) at 3h after delivery. Unique symbols (triangles, squares, diamonds, circles, pluses, asterisks and crosses) represent mice from independent repeats. All data were plotted using boxplots from ggplot2, which displays the median, the 25th and 75th percentiles, and the smallest and largest values within 1.5× the interquartile range (whiskers). See also Supplementary Fig. 1.
To evaluate whether oxylipins injected into the peritoneum reach the circulation, we quantified the concentration of 12,13-diHOME in plasma using targeted liquid chromatography with mass spectrometry (LC-MS)20 3 h after peritoneal injection of 12,13-diHOME and observed a significant increase in plasma concentration compared with vehicle-treated animals (Fig. 1h). Furthermore, the 12,13-diHOME-treated animals exhibited a significantly increased concentration of 12,13-diHOME in their lungs (Supplementary Fig. 1n). Together, these data suggest that—following peritoneal injection—12,13-diHOME can enter the circulatory system and may interact directly with the lung mucosa to exacerbate allergic inflammation of the airways. This does not preclude local synthesis of 12,13-diHOME by pulmonary microbiota or by pulmonary tissue, but rather indicates that lower-airway allergic inflammation may be exacerbated by circulating metabolites originating in the gut.
12,13-diHOME reprograms lipid handling and IL-10 expression of DCs.
To determine a mechanism by which 12,13-diHOME exacerbates inflammatory responses, we considered its structural similarity to known ligands of peroxisome proliferator-activated receptor-γ (PPARγ)—a lipid-activated nuclear receptor that regulates adipogenesis, fatty-acid uptake, inflammation, glucose metabolism and intestinal microbiota21–23. In mice, loss of PPARγ in DCs leads to a loss of immune tolerance and exacerbation of pulmonary inflammation22. We therefore hypothesized that 12,13-diHOME induces pro-allergic immune dysfunction through PPARγ signalling in DCs. Human DCs treated with 12,13-diHOME exhibit decreased secretion of IL-10, an anti-inflammatory cytokine that protects against allergic inflammation24 (Fig. 2a). Co-culture of 12,13-diHOME-treated DCs and autologous T cells altered the distribution of CD4+ T cells, specifically decreasing the frequency of Treg cells without decreasing cell viability (Fig. 2b, Supplementary Fig. 2c–e), confirming that the observed reduction in IL-10 secretion by DCs has a functional consequence for Treg cell populations.
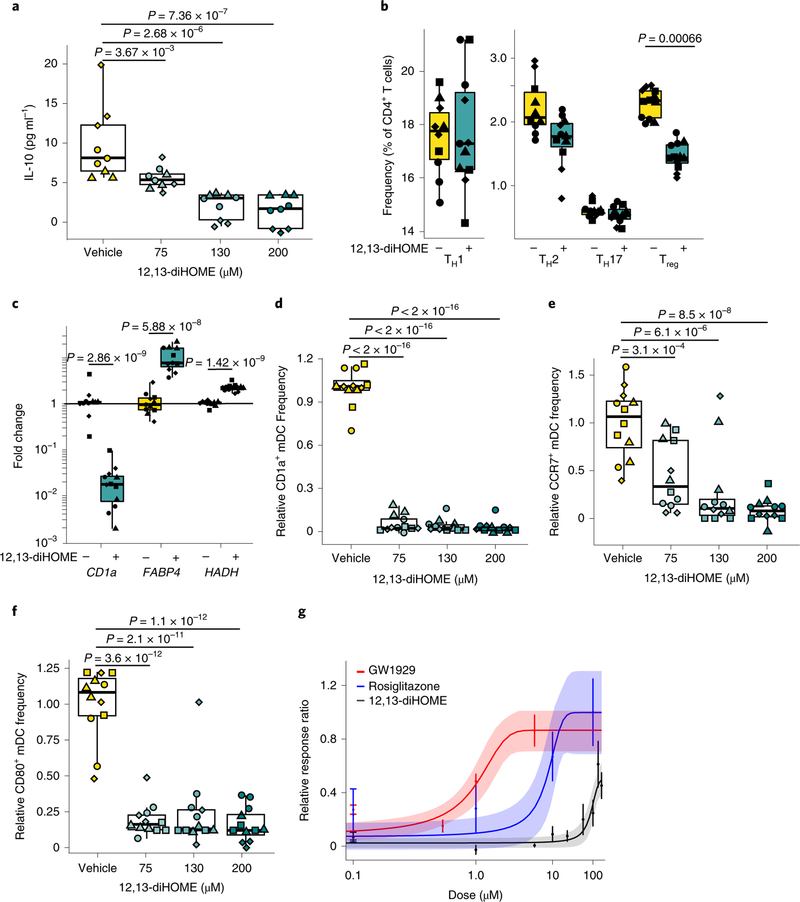
Fig. 2 |. 12,13-diHOME acts through PPARγ on DCs to decrease the number of Tregs.
a, Treatment with 12,13-diHOME causes a dose-dependent decrease in IL-10 secretion from human DCs (n = 3 human donors; LME, P = 3.67×10−3, P= 2.68 ×10−6 and P = 7.36×10−7 for concentrations of 75μM, 130μM and 200 μM, respectively). b, Treatment of human DCs with 130 μM 12,13-diHOME causes a specific decrease in the frequency of Treg cells (n = 4 human donors; CD3+CD4+CD25+FOXP3+; LME, P= 0.00066). c, Treatment of human DCs with 130μM 12,13-diHOME causes changes in gene expression consistent with activation of PPARγ (n = 4 human donors; LME, P = 2.86×10−9, P= 5.88×10−8 and P = 1.42×10−9 for CD1a, FABP4 and HADH, respectively). Fold changes (2−ΔΔCt) relative to vehicle control are presented on a log scale. d-f, 12,13-diHOME decreased the expression of CD1a (d), CCR7 (e) and CD80 (f) on human DCs (CD3−CD19−CD11c+; n = 4 human donors; LME, comparing vehicle with 75 μM 12,13-diHOME the P values were P<2.0×10−16, P< 3.1 ×10−4 and P<3.6×10−12 for CD1a, CCR7 and CD80, respectively). g, Raw264.7 cells transfected with a PPARγ-activated luciferase reporter and treated with 12,13-diHOME or known PPARγ agonists GW1929 (Kd = 1.4nM)56 and rosiglitazone (Kd = 40 nM)56 (n = 3 independent experiments; graph depicts median and s.e.m. for all datapoints). For a-f, data are presented as boxplots generated using ggplot2, which displays the median, the 25th and 75th percentiles, and the smallest and largest values within 1.5× the interquartile range (whiskers). For a-g, experiments were performed with n = 3–4 treatment replicates using cells isolated from three or four independent donors (biological replicates are represented by triangles, diamonds, circles and squares). See also Supplementary Fig. 2.
To evaluate whether 12,13-diHOME exerts this effect through PPARγ, we isolated RNA from human DCs treated with 12,13-diHOME in vitro and studied PPARγ-regulated gene expression. Treatment with 12,13-diHOME mimicked previously characterized effects of PPARγ activation, including decreased expression of CD1a—an immune marker involved in lipid-antigen presentation25—and increased expression of fatty acid binding protein 4 (FABP4) and hydroxyacyl-CoA-dehydrogenase (HADH)—genes involved in fatty-acid uptake and oxidation, respectively25 (Fig. 2c). Using flow cytometry, we confirmed that 12,13-diHOME activated PPARγ in human DCs in vitro, decreasing expression of CD1a, CD80 and CCR7—surface proteins involved in lipid presentation, antigen presentation and cell trafficking in human DCs25 (Fig. 2d–f). To test whether 12,13-diHOME acts on PPARγ directly, we used a modified PPARγ reporter assay26 and found that treatment with 12,13-diHOME in excess of 50 μM leads to PPARγ activation (Fig. 2g). These observations are consistent with recently reported increases in lipid uptake by adipose tissue treated with 12,13-diHOME17.
Although PPARγ activation in DCs is traditionally considered to be anti-inflammatory27, more-recent studies suggest that it may promote allergic inflammation28. Alternatively, 12,13-diHOME may act as a weak agonist of PPARγ and compete with endogenous PPARγ ligands25. Emerging data indicate that 12,13-diHOME influences a range of mammalian tissues17-19. Our data provide evidence that the effects of 12,13-diHOME extend to antigen-presenting cells and that elevated concentrations of this lipid cause a change in lipid handling and a reduction in IL-10 secretion by DCs. Given the emerging importance of 12,13-diHOME in a number of different tissues, it is plausible that it may also signal through other lipid receptors. Indeed, 12,13-diHOME was recently identified as an activator of transient receptor potential vanilloid 1 (TRPVI)29, the loss of which is associated with decreased asthma prevalence in humans and protection against airway inflammation in animal models30,31. Thus, 12,13-diHOME may have multiple cellular targets and its effects are probably not mediated exclusively by PPARγ.
12,13-diHOME is enriched in the faeces of neonates who are at risk of atopy and asthma.
Given the evidence for 12,13-diHOME as a driver of allergic inflammation in mice and humans, we focused on determining the concentration and potential sources of 12,13-diHOME in neonatal faeces. Initially, using LC-MS, we quantified the concentration of 12,13-diHOME in a subset of one-month-old neonates from the Wayne County Health, Environment, Allergy and Asthma Longitudinal Study (WHEALS; Supplementary Tables 6 and 11; total n = 41; atopic n = 7; asthmatic n = 8; atopy and/or asthma n = 19)11. These included 26 stool samples from one-month-old neonates—microbiologically representative of the variance in the composition of the gut microbiome observed across a larger cohort of 130 one-month-old neonates11—and a further 15 randomly selected samples (with >50 mg of stool and >10 ng of extracted faecal DNA) from the same cohort of one-month-old neonates for whom childhood atopy and asthma outcomes were available. Targeted LC-MS demonstrated that 12,13-diHOME was present in all neonatal stool; however, significantly higher concentrations were detected in the stool of neonates who subsequently developed atopy and/or asthma, even after adjusting for potential confounding factors (Fig. 3a, Supplementary Table 8). By contrast, the concentration of 9,10-diHOME—an enantiomer of 12,13-diHOME—did not differ between the two groups (Supplementary Fig. 3a, Supplementary Table 8).
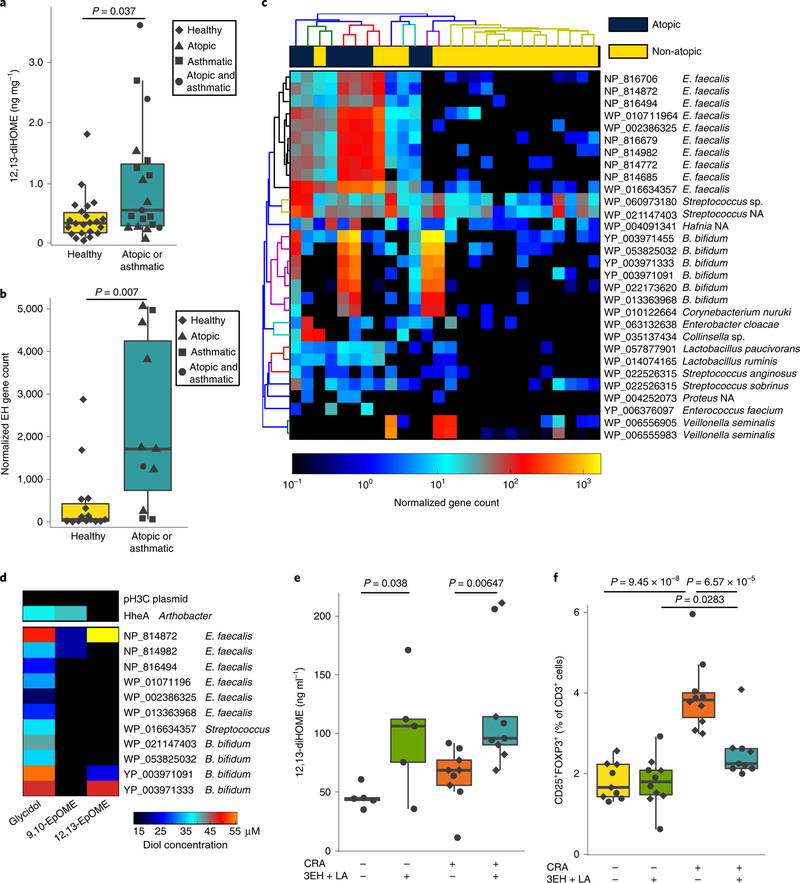
Fig. 3 |. Neonatal gut-microbiome-derived EH genes from B. bifidum and E. faecalis produce 12,13-diHOME and decrease the number of lung Treg cells in mice challenged with CRA.
a, Neonates who develop childhood atopy and/or asthma exhibit increased faecal concentrations of 12,13-diHOME (n = 41 neonates; LR adjusting for race, P= 0.037; see Supplementary Table 6 for a full summary of the statistical analysis). b, Neonates who develop atopy and/ or asthma during childhood have more bacterial EH genes in their stool (n = 26 neonates; LR adjusting for formula feeding; P = 0.007; Supplementary Table 7). c, The 30 most abundant EH genes identified in stool are enriched in neonates who develop atopy. Dendograms represent hierarchical clustering of bacteria and genes generated using HClust2. NA, not applicable. d, Three of the candidate EH genes, E. faecalis NP_814872, B. bifidum YP_003971091 and B. bifidum YP_003971333, convert 12,13-EpOME to its conjugate diol, 12,13-diHOME. e, Mice orally gavaged with LA and E. coli overexpressing NP_814872, YP_003971091 and YP_003971333 (3EH and LA) have significantly more 12,13-diHOME in their plasma than those supplemented with vehicle (LME; no CRA n= 5 mice, P= 0.038; CRA-challenged n = 9 mice, P = 0.0065). f, Mice treated with 3EH and LA and challenged intratracheally with CRA had significantly fewer lung Teg cells than mice treated with vehicle and challenged with CRA (LME; CRA-challenged n= 9 mice, P= 6.57×10−5). For a, b, e and f, data are presented as boxplots that were generated using ggplot2, which displays the median, the 25th and 75th percentiles, and the smallest and largest values within 1.5× the interquartile range (whiskers). See also Supplementary Fig. 3 and Supplementary Tables 6 and 7.
Specific bacterial EH genes produce 12,13-diHOME and decrease lung Treg cells in CRA-challenged mice.
12,13-diHOME is a terminal product of linoleic acid (LA) metabolism, which is initially converted to 12,13-EpOME either enzymatically via a cytochrome P450 epoxygenase32 or spontaneously through oxidation. Then, 12,13-EpOME is converted to 12,13-diHOME by an epoxide hydrolase (EH)—enzymes encoded by humans, bacteria and fungi33–35. To screen for potential sources of 12,13-diHOME in neonatal faeces, the aforementioned 26 neonatal stool samples were sequenced using shotgun metagenomic sequencing. A database of known bacterial (~73,000), fungal (~5,000) and human (~50) EH genes was assembled and analysed using ShortBred36 to probe the neonatal metagenomic data for sequence reads with EH homology. No fungal or human EH genes were detected in the neonatal shotgun sequence dataset; however, approximately 1,400 putative bacterial EH genes were identified. Bacterial EH genes were significantly more abundant in the stool of neonates who developed atopy and/or asthma during childhood (Fig. 3b, Supplementary Table 9). The 30 most abundant of which were primarily encoded by the strains Enterococcus faecalis, Streptococcus, Bifidobacterium bifidum and Lactobacillus (Fig. 3c), common early-life gut micro-biome members.
To evaluate whether these putative EH genes are capable of hydrolysing epoxides and producing 12,13-diHOME, we developed a cell-based functional assay. A subset (n = 11) of the most frequently detected putative bacterial EH genes with ≥75% of homologous EH marker regions identified in the metagenomic data were selected for functional screening (Supplementary Table 1). These genes were synthesized, cloned into Escherichia coli for expression and EH activity was measured using modifications of a previously described colorimetric assay37. All 11 genes screened were capable of hydrolysing glycidol, a generic epoxide, to its conjugate diol, glycerol, and three of them hydrolysed 9,10-EpOME to 9,10-diHOME. However, only NP_814872 (E. faecalis), YP_003971091 (B. bifidum) and YP_003971333 (B. bifidum) converted 12,13-EpOME to 12,13-diHOME (Fig. 3d), indicating that—although bacterial EH activity is common in the neonatal gut microbiome—bacterial capacity to produce 9,10-diHOME or 12,13-diHOME appears less common and strain specific.
Next, we examined whether the introduction of the 3EHs capable of synthesizing 12,13-diHOME could recapitulate the reduction in Treg cells observed in 12,13-diHOME-treated mice (Fig. 1). To test this, mice were orally supplemented with E. coli strains that overexpressed NP_814872 (E. faecalis), YP_003971091 (B. bifidum) and YP_003971333 (B. bifidum), and LA, a precursor to 12,13-diHOME. Mice were then subjected to the CRA model of allergic airway inflammation used in Fig. 1 (Supplementary Fig. 3e). Mice supplemented with the 3EH strains and LA showed significantly higher concentrations of 9,10-diHOME and 12,13-diHOME in their plasma compared with vehicle-treated controls (Fig. 3e, Supplementary Fig. 3f). Although the plasma concentrations of 12,13-diHOME in this experiment were almost an order of magnitude lower than those observed after peritoneal injection (compare Fig. 3e with Supplementary Fig. 1n), we still observed a significant decrease in the number of Treg cells in the lungs of CRA-challenged mice supplemented with the 3EH-expressing bacteria and LA (Fig. 3f). Together, these results indicate that oral supplementation with 12,13-diHOME-producing bacteria and LA is sufficient to recapitulate the decrease in mouse lung Treg cells, a cardinal immune feature of airway allergic inflammation.
An increase in neonatal faecal 3EH gene copy number and 12,13-diHOME concentration are associated with atopy, eczema and asthma during childhood.
Our data indicated that elevated concentrations of 12,13-diHOME promote allergic inflammation in vitro and in vivo, and that specific bacterial epoxide hydrolases identified in the gut microbiome of neonates at high risk of asthma both produce 12,13-diHOME and promote airway allergic inflammation in mouse models. We therefore investigated whether neonatal gut microbiome 3EH copy number or the concentration of 12,13-diHOME could be used to predict risk of atopy or asthma in childhood. Indeed, faecal 3EH copy number was significantly increased in one-month-old neonates who subsequently developed atopy and/or asthma during childhood (Fig. 4a, Supplementary Table 10; n = 41). Using our subset of the WHEALS cohort (n = 41; atopic/asthmatic, n = 19; Supplementary Tables 8 and 11) and logistic regression (LR), we examined the probability of children developing atopy at two years of age and/or asthma at four years of age on the basis of neonatal faecal 12,13-diHOME concentration, faecal 3EH copy number or known early-life risk factors selected a priori on the basis of previously published literature, including gender, race, maternal asthma, lack of pets, maternal smoking, delivery by caesarean section and formula feeding at one month of age38–41.
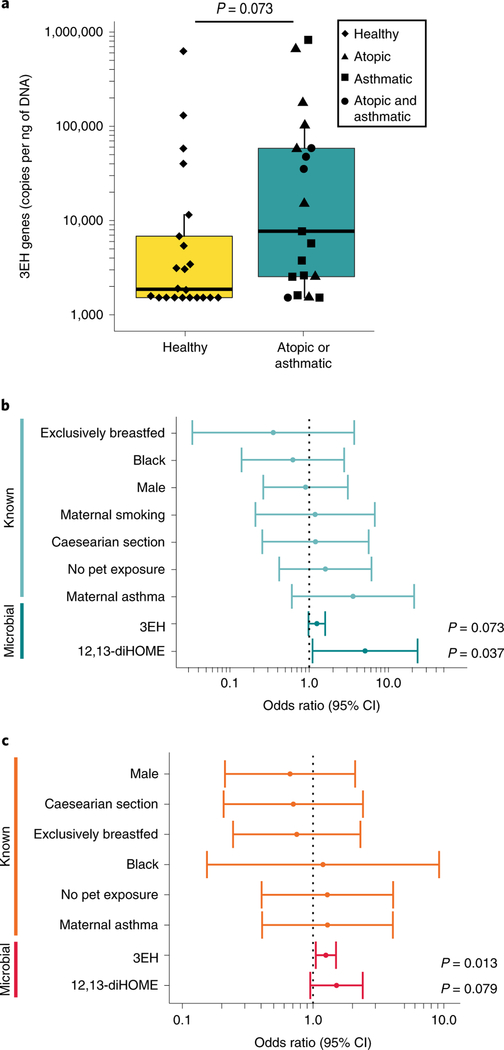
Fig. 4 |. Increased concentrations of 12,13-diHOME and the 3EH genes in neonatal stool are associated with the development of childhood atopy, eczema and/or asthma in two US cohorts.
a, Increased bacterial 3EH copy number is observed in neonates who developed atopy and/or asthma, and corresponds with the significant increase in overall faecal EH genes identified by ShortBred analysis (n = 41 neonates; LR, P = 0.0387; Fig. 3b, Supplementary Table 10). Data are presented as a boxplot that was generated using ggplot2, which displays the median, the 25th and 75th percentiles, and the smallest and largest values within 1.5× the interquartile range (whiskers). b, In a subset of the WHEALS cohort, an increase in 12,13-diHOME concentration in neonatal stool significantly increased the relative probability of developing atopy at two years of age and/or asthma at four years of age (n = 41 neonates; LR, 3EH+: OR = 1.25, 95% CI = 0.98–1.59, P = 0.073; 12,13-diHOME adjusted for race: OR = 5.07, 95% CI =1.10–23.3, P = 0.037; see Supplementary Table 12 for a full summary of the statistical analysis). For 3EH, analysis was performed using a log2 transformation of the 3EH copy number. c, In a subset of the TIPS birth cohort, an increase in faecal 3EH copy number was associated with a significantly increased relative probability of developing eczema in two-year-old children and/or asthma in four-year-old children (n = 50 neonates; LR, 3EH+: OR = 1.25, 95% CI = 1.05–1.50, P = 0.013; 12,13-diHOME: OR = 1.51, 95% CI = 0.95–2.40, P = 0.079; see Supplementary Table 14 for a full summary of the statistical analysis). For 3EH, analysis was performed using a log2 transformation of the 3EH copy number. The OR and CI for each risk factor are shown in b and c. See also Supplementary Fig. 3 and Supplementary Tables 6–14.
In our subset of the WHEALS cohort, we found that increased 12,13-diHOME concentration in neonatal stool was associated with a significantly increased probability of developing atopy and/ or asthma during childhood (Fig. 4b, Supplementary Table 12). Logistic regression (LR) was used to model this relationship and test for potential confounding by known risk factors. Multivariate LR identified race as a potential confounding factor of the relationship between faecal 12,13-diHOME concentration and disease development (see Supplementary Table 8 for the full analysis). Adjusting for race, we found that each additional ng of 12,13-diHOME per mg of neonatal faeces increased the probability of developing childhood atopy and/or asthma by a factor of 5 (adjusted odds ratio (OR) = 5.07, 95% confidence interval (CI) = 1.10–23.3, P = 0.016; Supplementary Table 12).
Given our small sample size, we sought to validate these findings in an independent neonatal cohort. Accordingly, we quantified 12,13-diHOME concentration and 3EH copy number in one-month stool samples from the placebo arm of the Trial of Infant Probiotic Supplementation (TIPS) study42. This trial—performed in a predominantly white birth cohort in San Francisco, California—examined the effect of probiotic supplementation on the development of eczema and asthma. Those in the placebo arm with >300 mg of stool collected at one month of age, eczema outcomes collected at two years of age and asthma outcomes collected at 4 years of age were included in our analysis (Supplementary Tables 7 and 13; total n = 50, eczema n = 22, asthma n = 6, eczema and/or asthma n = 23).
In our subset of the TIPS cohort, increased faecal 3EH copy number was associated with the development of eczema at two years of age and/or asthma at four years of age (OR = 1.25, 95% CI = 1.05–1.50, P = 0.013; Supplementary Table 14), whereas increased faecal 12,13-diHOME concentration did not reach our threshold for significance (12,13-diHOME, OR = 1.51, 95% CI = 0.95–2.40, P = 0.079; Fig. 4c, Supplementary Table 14). In this predominantly white cohort, the associations between one-month-old faecal 3EH copy number or 12,13-diHOME and the development of eczema and/or asthma during childhood did not show strong evidence of confounding by known risk factors (all change in OR < 10%; Supplementary Table 14). Despite the differences in geography and study design between the WHEALS and TIPS cohorts (see Methods), neonates who went on to develop eczema and/or asthma during childhood exhibited a significant increase in the faecal abundance of 3EH genes. Although these microbial risk genes require validation in additional larger cohorts, they represent exciting targets for future study and potential intervention.
Discussion
Our previous study determined that one-month-old neonates who subsequently developed childhood atopy and/or asthma exhibited a perturbed gut microbiome and increased concentrations of 12,13-diHOME11. In this study, we aimed to determine whether this oxylipin promotes proallergic immune dysfunction in vitro and in vivo, in an attempt to establish a mechanistic link between metabolic dysfunction of the gut microbiome during early life and the development of childhood disease. Consistent with this hypothesis, intra-abdominal delivery of 12,13-diHOME exacerbated airway inflammation, reduced the frequency of lung Treg cells and increased the concentration of serum IgE in mice (Fig. 1). In vitro, we found that 12,13-diHOME interacts directly and agonistically with PPARγ on DCs, altering expression of PPARγ-regulated genes involved in fatty acid uptake, metabolism and presentation (Fig. 2c). We observed that the profile of DC gene expression is consistent with that shown by Lynes et. al., who demonstrated that 12,13-diHOME leads to an increase in fatty-acid uptake in skeletal muscle and brown adipose tissue17,18. Here we showed that 12,13-diHOME upregulated FABP4 and HADH—PPARγ-regulated genes involved in fatty acid uptake and metabolism—indicating that common cellular mechanisms may underlie the effect of this molecule on distinct mammalian cell populations and tissues.
Given that 12,13-diHOME promoted allergic inflammation in vitro and in vivo, we aimed to identify potential sources of 12,13-diHOME in the neonatal gut microbiome. Metagenomic screening identified a large number of putative bacterial EHs and indicated that the faeces of high-risk neonates are enriched with these bacterial genes (Fig. 3c). Of the most frequently detected putative EH genes, several distinct genes were encoded by B. bifidum and E. faecalis, indicating that strain-specific variance in EH gene content and function exists in the neonatal gut microbiome. Indeed of the 11 EHs screened, only 3 were capable of producing 12,13-diHOME (Fig. 3d). The copy number of these three bacterial genes was both significantly enriched in the stool of neonates at high risk for childhood atopy and/or asthma and associated with an increased probability of allergic disease in two small, but racially, geographically and socioeconomically distinct birth cohorts. Notably, the TIPS and WHEALS cohorts were collected in the 2000s, in different cities, several years apart. Previous studies have demonstrated that collection and storage conditions alter the microbial and metabolic profiling of stool43,44. Although this represents a limitation, the associations between 12,13-diHOME, the 3EH genes and development of allergic disease remained consistent between these distinct cohorts. Although further testing in larger cohorts is required, this suggests that the observed associations may be generalizable to a broader population.
Our data indicate that an increased copy number of specific bacterial EH genes and elevated concentration of their product—12,13-diHOME—in the gut microbiome during early life are related to clinical outcomes of atopy, eczema and asthma during childhood. Humans have the capacity to produce 12,13-diHOME35, and all of the infants in our study showed baseline levels of faecal 12,13-diHOME. However, neonates whose gut microbiomes encode bacterial EH genes with the confirmed ability to produce this lipid exhibited increased concentrations of 12,13-diHOME, highlighting the importance of bacterial production of 12,13-diHOME in highrisk neonates. Our data support a role for 12,13-diHOME in promoting allergic inflammation both in vitro and in vivo. The observation that 12,13-diHOME induces altered lipid uptake, metabolism and presentation, and reduced IL-10 secretion by human DCs suggests that it may mediate its proallergic effect in part by altering immune cell metabolism. We acknowledge that other microbial-derived products likely contribute to early-life immune dysfunction associated with childhood atopy and asthma. This study exemplifies just one component of a complex microbiome-immune interaction in high-risk neonates that promotes allergic inflammation and offers a mechanism by which metabolic products of neonatal gut bacteria may increase susceptibility to allergies and asthma during childhood.
Methods
Animal models.
Six-week-old female C57BL/6 mice were obtained from Jackson Laboratories and randomized to treatment groups. All experiments were performed with 3–5 mice per cage; mice were cohoused for 2–3 weeks before treatment to minimize the effect of cages. Each treatment group was housed independently to mitigate potential effects of cohousing and coprophagy. Housing from the same assay were stored in the same rack and handled identically to minimize microbial variation45. To determine sample size, a pilot study was performed to estimate the effect size (d = 5.47) of 12,13-diHOME on Treg cell frequency in mouse lungs. This estimated effect size was used to determine the minimal sample size (n = 3) given an α = 0.05 and β = 0.80.
For the studies presented in Fig. 1a–g and Supplementary Fig. 1a–k, mice were treated on days 1, 2, 3, 14 and 21 with 30 mg kg−1 12,13-diHOME solubilized in 10% dimethylsulfoxide (DMSO) or vehicle (10% DMSO) by peritoneal injection. Three hours after injection, mice were challenged intratracheally with either PBS or CRA (20,000 PNU ml−1; Greer). Twenty-four hours after the final challenge, mice were anaesthetised, injected retro-orbitally with 100 μl CD45-APC (1:10), allowed to recover from anaesthesia and subsequently euthanized. Serum and lung tissue were collected from all mice subjected to allergen challenge. Serum concentrations of IgE were determined for all animals from which more than 20 μl of serum was obtained by cardiac puncture (n = 57). After euthanasia, the right and left lungs were separated. In the majority of repeats, the inferior lobes of the left lungs were processed for histological analysis (n = 47; Fig. 1c,d, Supplementary Fig. 1a,b). The remaining lobes of the left lung were processed for flow cytometry and stained for Treg cells (n = 47; Fig. 1g, Supplementary Fig. 1d), whereas the right lung was either processed for qPCR (n = 27) or flow cytometry (n = 28). There were two exceptions to this: in repeat P1A6 (represented by plus symbols in Fig. 1) the left lung was not processed or stained for Treg cells; in repeat P1A5 (represented by diamond symbols in Fig. 1) histological analysis was not performed.
Serum was isolated, and serum IgE levels were measured using a mouse IgE ELISA Max kit (Biolegend). Lungs were sent to the Mouse Pathology Core at University of California, San Francisco (UCSF) for haematoxylin and eosin staining of paraffin-embedded tissue sections. Two bronchioles and two vessels from each stained tissue section were scored on a 0–4 scale in which 0 represents structures with no inflammatory infiltrates; 1 represents few inflammatory cells; 2 represents a ring of inflammatory cells that is 1 cell layer wide; 3 represents a ring of inflammatory cells that is 2–4 cells wide; and 4 represents a ring of inflammatory cells more than 4 cells wide. Each structure was scored by two blinded individuals, and scores were averaged for each animal. Cell counts were determined for each structure by a blinded individual. In brief, the ImageJ freehand selection tool was used to trace the perimeter of each bronchiole and vessel. The area extending beyond the perimeter of the vessel was cleared and the colour threshold of the image was adjusted using the default method with the following parameters: hue = 0–255, saturation = 0–255, brightness = 130–255, threshold colour = white, background = dark, colour space = HSB. The image was converted to an 8-bit greyscale and the threshold was adjusted using the B&W defaults and a range of 0–150. Counts were outlined and summarized using the analyse particles window (size = 0–infinity, circularity = 0.0–1.00).
Lung cell subsets were assessed using flow cytometry. Lung tissue was manually dissected, digested with 5 mg per sample collagenase (Sigma-Aldrich) and passed through a 40 μm filter to generate single-cell suspensions. CD45- lung cells were considered to be resident cells and further classified as T cells (CD3+), neutrophils (Ly6G+CD11b+), monocytes (F4/80+Ly6C+Ly6G−CD11b+) and alveolar macrophages (Siglec-F+F4/80+CD11c+CD11b−). The gating strategy can be found in Supplementary Fig. 1c. Treg cells were identified as CD3+CD4+CD25+FOXP3+ cells using the human T cell gating strategy. A representative flow plot can be found in Supplementary Fig. 1d. Investigators were blinded to treatment groups while collecting and gating flow cytometry data.
Lung tissue was preserved in RNAlater (Ambion). Preserved lung tissue was homogenized in Lysing Matrix E Tubes (MP Biomedicals) using a FastPrep 24 homogenizer (MP Biomedicals) and extracted using an RNAeasy Mini Kit (Qiagen). An RT2 First Strand Kit (Qiagen) was used to synthesize cDNA, and expression of ILIA, IL1B and TNF relative to GAPDH was measured using a Power SYBR Green PCR Master Mix (ThermoFisher Scientific) and a QuantStudio 6 Real-Time PCR system (ThermoFisher Scientific). Primers are summarized in Supplementary Table 3.
For the quantification of 12,13-diHOME in mouse lung and plasma samples (Fig. 1h, Supplementary Fig. 1n), six-week-old female C57BL/6 mice were purchased from Jackson Laboratories and treated with 30 mg kg−1 12,13-diHOME solubilized in 10% DMSO or vehicle (10% DMSO) by peritoneal injection. Three hours after a single injection, mice were euthanized and lung tissue and plasma were collected and frozen immediately in liquid nitrogen. Then, 12,13-diHOME was extracted from the tissue and plasma using an established solid-phase extraction protocol46. In brief, flash-frozen tissue was massed and added to a Lysing Matrix E tube containing 1 ml of methanol, 10 μl 0.2 mg ml−1 BHT/EDTA and 1.25 ng 12,13-diHOME-D4 (internal standard, Cayman Chemical) then homogenized as described above. Tissue samples were centrifuged for 10 min at 2,125g, and the supernatant was transferred to a Falcon tube containing 19 ml deionized water to generate a 5% methanol solution. Plasma samples were thawed on ice and 10 μl 0.2 mg ml−1 BHT/EDTA and 1.25 ng 12,13-diHOME-D4 were added to 250 μl of thawed plasma. All samples were extracted using Waters Oasis HLB Cartridges (60 mg of sorbent, 30 μM particle size) as previously described20.
Extracted samples were resuspended in methanol, and LC-MS was performed using a QTRAP 6500 mass spectrometer (AB Sciex) and a Nextera X2 HPLC (Shimadzu Scientific Instruments) as previously described20. Linear standard curves were generated using six injections of 12,13-diHOME, 9,10-diHOME and 12,13-diHOME-D4 (internal standard). Peaks were manually integrated, and recovery of the internal standard was used to correct for extraction efficiency.
For the mouse studies shown in Fig. 3e,f and Supplementary Fig. 3e–h, six-week-old female C57BL/6 mice were obtained from Jackson Laboratories and orally gavaged with the three E. coli strains that overexpress NP_814872, YP_003971091 and YP_003971333, and LA before intratracheal challenge with CRA. In brief, BL21 cells containing recombinant plasmids (described below in the ‘Colorimetric detection of EH Activty’ section) were streaked onto Lauria-Bertani (LB) agar plates supplemented with 50 μg ml−1 kanamycin. Isolated colonies were selected and used to inoculate 10 μl LB supplemented with 50 μg ml−1 kanamycin. Starter cultures were grown for 18 h at 37 °C and 200 r.p.m. and used to inoculate 120 ml LB supplemented with kanamycin. These cultures were grown until the optical density at 660 nm (OD660) reached 0.6, at which point expression was induced with 1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG; Sigma-Aldrich). The temperature was reduced to 25 °C and incubation continued for 18 h at 200 r.p.m. Cultures were then centrifuged for 15 min at 3,000g, and the OD660 was adjusted to 6. The three strains were combined to make a 25% glycerol stock solution with approximately 1 × 109 colony forming units per strain and frozen in 500 μl aliquots.
Mice were treated by oral gavage and subjected to intratracheal challenge on days 1, 2, 3, 14, and 21 (Supplementary Fig. 3e). Each challenge was preceded by 4 d of oral gavage with either 75 μl 3EH and LA or 25% glycerol. Immediately before treatment, 250 μl filter-sterilized LA (Sigma-Aldrich) was added to each 500 μl 3EH glycerol stock. Mice were challenged intratracheally with either 50 μl CRA or PBS. On day 22, mice were euthanized and their lungs and plasma were collected. The concentration of 12,13-diHOME and 9,10-diHOME in the plasma and the frequency of lung Treg cells were determined as described above. All mouse studies were reviewed and approved by the University of California San Francisco’s Institutional Care and Use Program. Investigators were blinded to treatment groups while collecting and scoring all flow cytometry and mass spectrometry data.
Human immune assays.
12,13-diHOME purchased from Cayman Chemical was stored at −20 °C in methyl acetate until use. Although 12,13-diHOME may be stably stored in organic solvents for several years, it is relatively unstable in aqueous solutions; this was reflected in the need for high concentrations of this lipid in our in vitro assays. We attempted to mitigate these limitations by solubilizing 12,13-diHOME in DMSO and preparing aqueous solutions immediately before use; however, the concentrations used in our in vitro assays are unlikely to directly reflect those found in tissue.
To prevent degradation, fresh solutions of 12,13-diHOME solubilized in DMSO were prepared immediately before use (<2 h). For all experiments, methyl acetate was removed under a nitrogen stream, and the compound was resuspended in DMSO (maximum solubility 20 mg ml−1) before addition of PBS or media. Fresh media containing 12,13-diHOME was added every 48 h to in vitro assays. This strategy of solvent evaporation and subsequent resuspension in DMSO was used for several related oxylipins sold by Cayman Chemical, including 9,10-diHOME, 12,13-EpOME, 9-HODE and 13-HODE, none of which had a significant effect on the frequency of Treg cells (Supplementary Fig. 2b).
IL-10 secretion was measured using a cytometric bead array (BD Biosciences). Human DCs were isolated from the peripheral blood mononuclear cells (PBMCs) of two healthy human donors as previously described11 and treated with increasing concentrations of 12,13-diHOME (Cayman Chemical) solubilized in 0.2% DMSO. Human PBMCs were isolated from human plasma that was purchased from the Blood Centers of the Pacific and were not subject to UCSF institutional review board (IRB) approval. Supernatant was collected after 24 h. IL-10 concentrations were determined according to the manufacturer’s instructions.
Co-culture of human DCs and T cells in the presence of 12,13-diHOME was performed as previously described11. In brief, DCs were isolated from the PBMCs of two healthy human donors and treated for 5 d with 130 μM 12,13-diHOME solubilized in 0.2% DMSO or vehicle (0.2% DMSO) in the presence of 20 ng ml−1 IL-4 (R&D Systems) and 10 ng ml−1 GM-CSF (R&D Systems). Fresh treatment media was added every 48 h throughout the course of the study. Then, 18 h before co-culture, DCs were stimulated with 10 ng ml−1 TNF, IL-1 β and IL-6 (Peprotech) and 1 mM prostaglandin E2 (Stemcell Technologies). DCs were subsequently washed and co-cultured with autologous T cells in the presence of 10 ng ml−1 anti-CD28 and anti-CD49d antibodies (BD Bioscience). Flow cytometry was used to authenticate human DCs and T cells before co-culture, and primary cells were not tested for mycoplasma.
After 5 d of co-culture, T cell subsets were analysed by flow cytometry. To assess cytokine secretion, cells were stimulated for 16 h with phorbol myristate acetate and ionomycin (ACROS) and GolgiPlug (Gplug; BD Biosciences). Antibodies used for staining are summarized in Supplementary Table 2. Flow cytometry data were collected on a BD LSR II flow cytometer. Helper T cell subsets were defined as follows: TH1, CD3+CD4+IFNγ+; TH2, CD3+CD4+IL-4+; TH17, CD3+CD4+IL-17+; Treg cells, CD3+CD4+CD25+FOXP3+. The human T cell gating strategy can be found in Supplementary Fig. 1e.
Human DC maturation was assessed by flow cytometry. PBMCs were isolated from two healthy human donors using Ficoll-Hypaque gradient centrifugation as previously described11 and cultured for 5 d in the presence of 20 ng ml−1 IL-4 and 10 ng ml−1 GM-CSF. Monocytic DCs were defined as CD3−CD19−CD11c+. The DC gating strategy can be found in Supplementary Fig. 1h.
Gene expression was examined in human DCs. DCs were isolated as described above and treated for 2 d with 130 μM 12,13-diHOME. Cells were washed twice in PBS and RNA was extracted using the RNAqueous Micro Kit (Ambion). An RT2 First Strand Kit (Qiagen) was used to synthesize cDNA, and expression of CD36, CD1A, FABP4 and HADH relative to ACTB was measured using Power SYBR Green PCR Master Mix (ThermoFisher Scientific) and a QuantStudio 6 Real-Time PCR system (ThermoFisher Scientific). Fold changes (2−ΔΔCt) relative to vehicle treatment were plotted on a log scale in Fig. 1d,e. Primers are summarized in Supplementary Table 3.
Luciferase assay.
A modified PPARγ luciferase assay was performed as described in Ye et. al.45. A PPRE-luciferase reporter plasmid, PPRE X3-TK-luc from B. Spiegelman (1015; Addgene), and a PPARγ overexpression plasmid, pGST-PPARγ from B. Vogelstein (16549; Addgene), were purified using a Plasmid Plus Maxi Kit (Qiagen). Raw264.7 cells were obtained from and authenticated by the UCSF Cell Culture Facility and were not tested for mycoplasma contamination. Cells were grown in R10 medium (Roswell Park Memorial Institute (RPMI) medium 1640 supplemented with 10% heat-inactivated fetal bovine serum, 2 mM L-glutamine and 100 U ml−1 penicillin-streptomycin) were transfected with PPRE reporter:PPARγ:Renilla luciferase plasmid DNA at a ratio of 1:20:40 using 50 ng per well of reporter plasmid DNA. Fugene HD transfection reagent (Promega) was combined with plasmid DNA at a ratio of 4:1 and the total volume was brought to 5 μl per well with RPMI. The transfection mixture was gently combined with diluted Raw264.7 cells and plated in black clear-bottom 96-well plates at a density of 50,000 cells per well. Twenty-four hours after transfection, cells were treated with 12,13-diHOME, rosiglitazone or GW1929 (Cayman Chemical) solubilized in 0.1% DMSO. Twenty-four hours after treatment, Firefly luciferase and Renilla luminescence were measured using the Dual-Glo Luciferase Assay kit (Promega) on a Cytation 3 plate reader (BioTek Instruments). Negative controls included the following: Raw264.7 cells transfected with the reporter plasmids (but no PPARγ plasmid) and treated 12,13-diHOME; untransfected untreated cells; and transfected untreated cells (Supplementary Fig. 1i).
Study population and definitions for the WHEALS cohort.
WHEALS recruited pregnant women (n = 1,258) between the ages of 21 and 49 between August 2003 and November 2007 in southeast Michigan47. Women were considered to be eligible if they lived in a predefined cluster of contiguous zip codes near Detroit, Michigan, had no intention of moving out of the area and provided informed written consent. Follow-up interviews were conducted at 1, 6, 12, 24 and 48 months after birth. The appointment at 24 months occurred at a standardized study clinic, in which the child underwent evaluation by a board-certified allergist. Stool samples from children were collected during home visits at one month and used in this study.
Samples were shipped to the UCSF on dry ice, where they were stored at −80 °C until processing. Faecal DNA extracted using the modified cetyl trimethylammonium bromide (CTAB) method9 and previously used for fungal and bacterial profiling was stored at −20 °C until analysed in this study. Atopy was defined as per Havstad et al.1. Children with asthma were identified by parent-reported doctor diagnosis of asthma during interview at 48 months. All research on human participants was reviewed and approved by the Henry Ford Hospital Institutional Review Board.
Characteristics of the individuals in our subset of the WHEALS birth cohort.
A subset of 41 neonates were selected from the WHEALS cohort (median age = 35 d when stool samples were taken) who had previously undergone faecal fungal and bacterial microbiota profiling11 and had >50 mg of stool and >10 ng of extracted faecal DNA remaining from the samples collected during home visits at one month of age (Supplementary Table 6; total n = 41, atopic n = 7, asthmatic n = 8, atopic asthmatic n = 4). These included 26 stool samples, microbiologically representative of the variance in gut microbiota observed in this larger (n = 130) cohort, that had previously undergone untargeted metabolomic profiling11 and 15 randomly selected faecal samples with >50 mg of stool and >10 ng of DNA, all of which had available childhood atopy and/or asthma outcomes. Power calculations were performed in G*Power v.3.1. Effect size was estimated on the basis of the relative concentration of 12,13-diHOME in high-and low-risk neonates, shown in supplementary figure 9 of Fujimura et. al. 201611. The estimated effect size (d = 1.228), α = 0.05 and power = 0.95 were used to estimate the required sample size (n = 38, assuming allocation of N2/N1 = 1). Of the 41 samples in our subset, 19 (46%) had atopy and/or asthma, 20 (48.8%) were male and 32 (78%) were black, based on maternal self-reports. Furthermore, 8 (19.5%) were born by caesarean section, 4 (9.75%) were exclusively breastfed until one month of age, 7 (17%) reported diagnosis by a doctor of maternal asthma, 6 (14.6%) reported maternal smoking during pregnancy and 13 (31.7%) lived with an indoor or outdoor dog or cat before delivery. The results presented in this study require replication in a larger neonatal population.
Study population and definitions for the TIPS birth cohort.
The TIPS study recruited infants in the San Francisco Bay area who had at least one parent with a history of asthma and parents who were willing to supplement breast milk or formula with a probiotic supplement or placebo for at least one feed each day48. Eligible infants were randomized to either the intervention arm, in which they received probiotic supplementation, or the placebo arm, in which they were supplemented daily for six months with 325 mg daily inulin42—a fermentable prebiotic shown to increase the frequency of Bifidobacterium species in paediatric and adult populations42. Stool was collected throughout the study. Demographics data were collected after the birth of an infant and obstetric medical records were used to confirm the method of delivery. Physical examinations were conducted by study clinicians at 1, 3, 6, 12 and 24 months of age, and interviews with the family were conducted monthly for the first year of life and semi-annually in all subsequent years42. The TIPS study was reviewed and approved by the UCSF IRB (10–02968).
Stool samples collected from one-month-old participants in the placebo arm of randomized control trial were used in this study. Stool was collected from diapers, mailed overnight to the study team and immediately stored at −80 °C until processing. Faecal DNA was extracted using the previously described modified CTAB method49 and stored at −20 °C until analysis.
Characteristics of the individuals in our subset of the TIPS cohort.
A random subset of 50 neonates from the placebo arm of the TIPS cohort (median infant age = 32 d when stool samples were collected) that had >300 mg stool remaining from collection at one month of age were selected (total n = 50, eczema n = 22, asthma n = 6, eczema and asthma n = 23; Supplementary Table 7). Of the 50 neonates, 20 (40%) were male and 4 (8%) were black, based on a family self-report. Furthermore, 15 (30%) were born by caesarean section, 28 (56%) were exclusively breastfed until one month of age, 31 (62%) reported doctor-diagnosis of maternal asthma, 1 (2%) reported maternal smoking during pregnancy and 18 (36%) did not live with a cat or dog.
Mass spectrometry in human samples.
Faecal oxylipin (9,10-diHOME and 12,13-diHOME) concentrations were assessed in our subset of the WHEALS cohort (n = 41). Oxylipins were extracted from approximately 50 mg of neonatal stool using the protocol described above. LC-MS was performed on a Thermo LTQ-Orbitrap-XL mass spectrometer equipped with an electrospray ionization source (ThermoFisher Scientific). The concentrations of 12,13-diHOME used in our ex vivo cell assays (~40 μg g−1) and in vivo animal models (~0.2 μg g−1) were within an order of magnitude of the concentrations measured in stool from high-risk neonates (0.2–4 μg g−1). The stability of oxylipins in faeces is unknown. Although 12,13-diHOME is relatively stable in organic solvent, we noticed that the median concentration of 12,13-diHOME was higher in the TIPS samples compared with the older WHEALS samples (P = 0.0143; Supplementary Fig. 3i), indicating the potential for lipid-signal degradation in long-term-stored samples.
Metagenomic data analysis.
DNA from the 26 stool samples from the WHEALS cohort that had previously undergone untargeted LC-MS11 and targeted oxylipin quantification (described above) was extracted using the modified CTAB method and sent to the Vincent J. Coates Genomic Sequencing Laboratory at the California Institute for Quantitative Biosciences for 150-bp paired-end sequencing on an Illumina HiSeq 4000 (www.qb3.berkeley.edu/gsl). Recognizing that the duration of sample storage affects concentration and integrity of DNA43,50,51 and that the WHEALS stool samples were collected in the early 2000s, metagenomic sequencing was only performed on samples that had sufficient concentrations of DNA with sufficient integrity to produce high-quality sequencing data for our metagenomic screen (n = 26). Sequencing efforts resulted in an average of 4.41 Gb (range of 3.05–7.68 Gb) of data per sample with an average coverage of 66.5 reads per contig (range of 21.8–148.8) as determined by Megahit52. Sequencing reads were quality-trimmed to Q17 with BBDuk (https://sourceforge.net/projects/bbmap/). A database of approximately 78,000 known bacterial (~73,000), fungal (~5,000) and human (~50) EH genes was generated using the NCBI protein database. All genes that had been tagged as ‘epoxide hydrolases’ were included in the database. The EH database and the UniRef50 database53 were input into the ShortBred identify pipeline36 and used to generate EH-specific markers. These markers were inputted into ShortBred quantify and were used to probe the quality-trimmed metagenomes for EH markers. The normalized marker abundance for each gene was summed and used to generate the normalized EH gene counts in Fig. 3b. The 30 most abundant EH genes were visualized using a heatmap generated by HClust2 (https://bitbucket.org/nsegata/hclust2) and Canberra distance matrices (Fig. 3c).
Colorimetric detection of EH activity.
A subset of 13 of the most-abundant EH genes were selected for functional investigation on the basis of the number of EH markers per gene identified in the metagenomic analysis (Supplementary Table 1). Genes with fewer than 75% of markers were excluded from further analysis. The 13 genes were structurally aligned with EH genes with known crystal structures from Mycobacterium tuberculosis (PDB ID: 2BNG), Pseudomonas aeruginosa (PDB ID: 4DLN) and Rhodococcus erythopolis (PDB ID: 1NWW) using the Promals3D server (http://prodata.swmed.edu/promals3d/; Supplementary Fig. 4). Amino acids extending beyond the EH domain were trimmed. Amino acid sequences were converted into DNA sequences using the default codon usage table for E. coli (http://www.bioinformatics.org/sms2/rev_trans.html), and restriction enzyme sites for EcoRI and Sall were added to the 5’ and 3’ ends of the DNA fragment, respectively. Genes were synthesized as gBlock gene fragments by IDT (https://www.idtdna.com/). Sequences of the synthetic constructs are summarized in Supplementary Table 4.
Synthetic genes were sub-cloned into the EcoRI and SalI sites of a pH3C plasmid—provided by O. Rosenberg—to generate proteins with an N-terminal 8xHis fusion. Constructs were verified by forward and reverse sequencing of the T7 promoter (5′-TAATACGACTCACTATAGGG-3’) and terminator (5′-GCTAGTTATTGCTCAGCGG-3′) performed by Quintara Bio. Subsequently, 11 of the 13 plasmids (NP_814872, NP_814982, NP_816494, WP_01071196, WP_002386325, WP_013363968, WP_01663157, WP_021147403, WP_053825032, YP_003971091 and YP_003971333) were validated and transformed into BL21(DE3) competent E. coli (New England Biolabs) for expression. BL21 cells containing recombinant plasmids were streaked onto LB agar plates supplemented with 50 μg ml−1 kanamycin and grown overnight. Isolated colonies were selected and used to inoculate 200 μl LB supplemented with 50 μg ml−1 kanamycin and 1 mM IPTG (Sigma-Aldrich).
Inoculates in 96-well V-bottom plates were incubated for 18 h at 37 °C at 200 r.p.m. After incubation, plates were centrifuged for 10 min at 3,000 r.p.m. and washed with 20 mM HEPES pH 7.5. Cells were resuspended and 40 μl of resuspended cells per well were added to 96-well flat-bottom plates. The OD600 was measured to estimate the protein concentration, and 40 μl of 13 mM epoxide was added to each well. Known concentrations of the conjugate diol plated in triplicate were included on each plate and used to generate standard curves (Supplementary Fig. 3b–d). To each well, 20 μl of sodium nitrite was added (final concentration 20 mM). Plates were incubated for 1 h at 37 °C at 200 r.p.m. The reaction was stopped by centrifugation for 10 min at 4 °C and 3,000g. Then, 85 μl of supernatant was added to a new 96-well flat bottom plate containing 50° μl 8 mM sodium periodate (final concentration of 2 mM). Plates were incubated for 30 min at room temperature with shaking. Background absorbance at 490 nm was recorded before addition of 50 μl 12 mM adrenaline (final concentration of 3 mM; Sigma-Aldrich). Plates were incubated for 5 min at room temperature with shaking, and the absorbance at 490 nm was recorded. BL21 cells containing the pH3C plasmid or expressing HheA—a previously characterized EH54—were included on each plate as controls. Enzymatic activity was assessed in triplicate for three epoxides, glycidol (Sigma-Aldrich), 9,10-Ep0ME and 12,13-EpOME (Cayman Chemical), and their conjugate diols, glycerol, 9,10-diH0ME and 12,13-diHOME.
Quantification of 3EH copy number by qPCR.
The abundance of the three active EH genes—NP_814872, YP_003971091 and YP_003971333—was quantified in faecal DNA that was extracted using the modified CTAB method49. Gene fragments containing the target sequences (Supplementary Table 5) were ordered from IDT (https://www.idtdna.com/) and used to generate a standard curve. In brief, gene fragments were amplified by PCR, normalized to 2 × 108 copies μl−1 and eight 1:10 serial dilutions were made for use as standards. qPCR was performed using the TaqMan Universal PCR Master Mix (Applied Biosystems) and a QuantStudio 6 Real-Time PCR system (ThermoFisher Scientific). The 3EH copy number represents the total number of copies of all three genes per ng of faecal DNA. Sequences of gene fragments, primers and probes are summarized in Supplementary Tables 3 and 5.
Statistical analysis.
All analyses were conducted using the R statistical programming environment (v3.2.3). All comparisons made in mouse models (Fig. 1, Supplementary Fig. 1) were made using a LME (R package lmerTest) with the random effect attributed to the experimental repeat and visualized using Visualization of Regression Models (visreg; R package visreg) and boxplots generated using ggplot2 (R package ggplot2). All human cytokine, T cell, DC and gene expression differences (Fig. 2, Supplementary Fig. 2) were tested using a LME with the random effect attributed to donor and visualized using visreg. Boxplots were generated using ggplot2. Univariate LR was used to determine the statistical significance of faecal concentrations of 12,13-diHOME and 9,10-diHOME—normalized EH gene count—and 3EH copy number differences (LR, R; Figs. 3a,b, 4a, Supplementary Fig. 3a, Supplementary Tables 8–10). Multivariate LR was used to identify potential confounding factors of associations between faecal factors and the development of atopy and/or asthma during childhood. Potential confounding factors were defined55 as risk factors that changed the adjusted OR by >10%. Risk factors were selected a priori and included race, gender, maternal asthma, maternal smoking before delivery, lack of pets before delivery, caesarean delivery and formula feeding at one month38–41.
The characteristics of cohorts were compared using Fisher exact tests, with the exception of the age comparisons, which used a two-sided Student’s t-test (Supplementary Tables 1, 2, 11 and 13). Univariate LR was used to model the relationship between known risk factors (selected a priori), log2-transformed 3EH copy number or 12,13-diHOME concentration (ng ml−1) and the development of atopy and/or asthma during childhood (Supplementary Table 12) or eczema and/ or asthma during childhood (Supplementary Table 14). Multivariate LR was used to identify potential confounders of significant associations, as described above and summarized in Supplementary Tables 12 and 14. Forest plots were created using OR-and CI-determined regression models with adjustment for potential confounding factors (Fig. 4b,c). All comparisons presented in this manuscript were analysed for effects of potential outliers. We identified potential outliers as points outside of the whiskers. Removal of these datapoints from the analysis did not change the significance of any comparison. The comparisons presented in the manuscript include all data; however, this additional outlier analysis is available in our GitHub repository (https://github.com/srlevan).
Supplementary Material
Acknowledgements
We thank the WHEALS and TIPS study participants; A. Iavarone and the mass spectrometry facility and genomic sequencing laboratory at QB3 Berkeley (http://qb3.berkeley.edu/), O. Rosenberg, B. Vogelstein and B. Spiegelman for plasmid donations and N. Lukacs for assessment of the manuscript. This research was funded by NIH/NIAID award AI089473.
Footnotes
Competing interests
S.V.L. is co-founder of Siolta Therapeutics Inc., and serves as both a consultant and a member of its Board of Directors. Furthermore, the Regents of the University of California, UCSF have filed a provisional patent application (Application number 62/637,175) on behalf of S.V.L. and S.R.L. relating to the methods and compositions of EH genes.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41564–019-0498–2.
Reprints and permissions information is available at www.nature.com/reprints.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reporting Summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Metagenomic data generated in this study are available in the EMBLI repository as PRJEB24006 (https://www.ebi.ac.uk/ena/). Further datasets and materials are available from the corresponding author on reasonable request.
Code availability
Datasets and R scripts used for statistical analysis and figures are available on GitHub (https://github.com/srlevan/).
References
- 1.Havstad S. et al. Atopic phenotypes identified with latent class analyses at age 2 years. J. Allergy Clin. Immunol 134, 722–727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamamoto-Hanada K., Yang L., Narita M., Saito H. & Ohya Y. Influence of antibiotic use in early childhood on asthma and allergic diseases at age 5. Ann. Allergy Asthma Immunol 119, 54–58 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Chu S. et al. Cesarean section without medical indication and risks of childhood allergic disorder, attenuated by breastfeeding. Sci. Rep 7, 9762 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silvers KM et al. Breastfeeding protects against current asthma up to 6 years of age. J. Pediatr 160, 991–996 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Fall T. et al. Early exposure to dogs and farm animals and the risk of childhood asthma. JAMA Pediatr 169, e153219 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Genuneit J. Exposure to farming environments in childhood and asthma and wheeze in rural populations: a systematic review with meta-analysis. Pediatr. Allergy Immunol 23, 509–518 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez-Perez G. et al. Maternal antibiotic treatment impacts development of the neonatal intestinal microbiome and antiviral immunity. J. Immunol 196, 3768–3779 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Dominguez-Bello MG et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl Acad. Sci. USA 107, 11971–11975 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujimura KE et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc. Natl Acad. Sci. USA 111, 805–810 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arrieta M-C et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med 7, 307ra152 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Fujimura KE et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med 22, 1187–1191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durack J. et al. Delayed gut microbiota development in high-risk for asthma infants is temporarily modifiable by Lactobacillus supplementation. Nat. Commun 9, 707 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trompette A. et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med 20, 159–166 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Fonseca W. et al. Lactobacillus johnsonii supplementation attenuates respiratory viral infection via metabolic reprogramming and immune cell modulation. Mucosal Immunol 10, 1569–1580 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartl D. et al. Quantitative and functional impairment of pulmonary CD4+ CD25hi regulatory T cells in pediatric asthma. J. Allergy Clin. Immunol 119, 1258–1266 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Lundstrom SL et al. Allergic asthmatics show divergent lipid mediator profiles from healthy controls both at baseline and following birch pollen provocation. PLoS ONE 7, e33780 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynes MD et al. The cold-induced lipokine 12,13-diHOME promotes fatty acid transport into brown adipose tissue. Nat. Med 37, 631–637 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stanford KI et al. 12,13-diHOME: an exercise-induced lipokine that increases skeletal muscle fatty acid uptake. Cell Metab. 27, 1111–1120 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zimmer B. et al. The oxidized linoleic acid metabolite 12,13-DiHOME mediates thermal hyperalgesia during inflammatory pain. Biochim. Biophys. Acta 1863, 669–678 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Gouveia-Figueira S., Spath J., Zivkovic AM & Nording ML Profiling the oxylipin and endocannabinoid metabolome by UPLC-ESI-MS/MS in human plasma to monitor postprandial inflammation. PLoS ONE 10, e0132042 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Byndloss MX et al. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357, 570–575 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khare A., Chakraborty K., Raundhal M., Ray P. & Ray A. Cutting edge: dual function of PPARγ in CD11c+ cells ensures immune tolerance in the airways. J. Immunol 195, 431–435 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wahli W. & Michalik L. PPARs at the crossroads of lipid signaling and inflammation. Trends Endocrinol. Metab. 23, 351–363 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Iyer SS & Cheng G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit. Rev. Immunol 32, 23–63 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szatmari I. et al. PPAR regulates the function of human dendritic cells primarily by altering lipid metabolism. Blood 110, 3271–3280 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Choo J. et al. A novel peroxisome proliferator-activated receptor (PPAR)γ agonist 2-hydroxyethyl 5-chloro-4,5-didehydrojasmonate exerts anti-inflammatory effects in colitis. J. Biol. Chem 290, 25609–25619 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woerly G. et al. Peroxisome proliferator-activated receptors α and γ down-regulate allergic inflammation and eosinophil activation. J. Exp. Med 198, 411–421 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nobs SP et al. PPARγ in dendritic cells and T cells drives pathogenic type-2 effector responses in lung inflammation. J. Exp. Med 8, 3015 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Green D. et al. Central activation of TRPV1 and TRPA1 by novel endogenous agonists contributes to mechanical allodynia and thermal hyperalgesia after burn injury. Mol. Pain 12, 1–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q. et al. [TRPV1 UTR-3 polymorphism and susceptibility of childhood asthma of the Han nationality in Beijing]. Wei Sheng Yan Jiu 38, 516–521 (2009). [PubMed] [Google Scholar]
- 31.Baker K. et al. Role of the ion channel, transient receptor potential cation channel subfamily V member 1 (TRPV1), in allergic asthma. Respir. Res 17, 67 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ha J., Dobretsov M., Kurten RC, Grant DF & Stimers JR Effect of linoleic acid metabolites on Na+/K+ pump current in N20.1 oligodendrocytes: role of membrane fluidity. Toxicol. Appl. Pharmacol 182, 76–83 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Morisseau C. Role of epoxide hydrolases in lipid metabolism. Biochimie 95, 91–95 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biswal BK et al. The molecular structure of epoxide hydrolase B from Mycobacterium tuberculosis and its complex with a urea-based inhibitor. J. Mol. Biol 381, 897–912 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Decker M., Arand M. & Cronin A. Mammalian epoxide hydrolases in xenobiotic metabolism and signalling. Arch. Toxicol 83, 297–318 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Kaminski J. et al. High-specificity targeted functional profiling in microbial communities with ShortBRED. PLoS Comput. Biol 11, e1004557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cedrone F., Bhatnagar T. & Baratti JC Colorimetric assays for quantitative analysis and screening of epoxide hydrolase activity. Biotechnol. Lett 27, 1921–1927 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Wegienka G. et al. Combined effects of prenatal medication use and delivery type are associated with eczema at age 2 years. Clin. Exp. Allergy 45, 660–668 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Havstad S. et al. Effect of prenatal indoor pet exposure on the trajectory of total IgE levels in early childhood. J. Allergy Clin. Immunol 128, 880–885 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burke H. et al. Prenatal and passive smoke exposure and incidence of asthma and wheeze: systematic review and meta-analysis. Pediatrics 129, 735–744 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Bao Y. et al. Risk factors in preschool children for predicting asthma during the preschool age and the early school age: a systematic review and meta-analysis. Curr. Allergy Asthma Rep 17, 85 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Cabana MD et al. Early probiotic supplementation for eczema and asthma prevention: a randomized controlled trial. Pediatrics 140, e20163000 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gratton J. et al. Optimized sample handling strategy for metabolic profiling of human feces. Anal. Chem 88, 4661–4668 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Tedjo DI et al. The effect of sampling and storage on the fecal microbiota composition in healthy and diseased subjects. PLoS ONE 10, e0126685 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye F. et al. The dipeptide H-Trp-Glu-OH shows highly antagonistic activity against PPARγ: bioassay with molecular modeling simulation. Chembiochem 7, 74–82 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Laukens D., Brinkman BM, Raes J., De Vos M. & Vandenabeele P. Heterogeneity of the gut microbiome in mice: guidelines for optimizing experimental design. FEMS Microbiol. Rev 40, 117–132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aichbhaumik N. et al. Prenatal exposure to household pets influences fetal immunoglobulin E production. Clin. Exp. Allergy 38, 1787–1794 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cabana MD, McKean M., Wong AR, Chao C. & Caughey AB Examining the hygiene hypothesis: the Trial of Infant Probiotic Supplementation. Paediatr. Perinat. Epidemiol 21, 23–28 (2007). [DOI] [PubMed] [Google Scholar]
- 49.DeAngelis KM et al. Selective progressive response of soil microbial community to wild oat roots. ISME J. 3, 168–178 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Han M. et al. A novel affordable reagent for room temperature storage and transport of fecal samples for metagenomic analyses. Microbiome 6, 43 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rounge TB et al. Evaluating gut microbiota profiles from archived fecal samples. BMC Gastroenterol 18, 171 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li D., Liu C-M, Luo R., Sadakane K. & Lam T-W MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Suzek BE et al. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31, 926–932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang L. et al. A high-throughput adrenaline test for the exploration of the catalytic potential of halohydrin dehalogenases in epoxide ring-opening reactions. Biotechnol. Appl. BioChem 62, 451–457 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Maldonado G. & Greenland S. Simulation study of confounder-selection strategies. Am. J. Epidemiol 138, 923–936 (1993). [DOI] [PubMed] [Google Scholar]
- 56.Henke BradR. et al. N-(2-Benzoylphenyl)-L-tyrosine PPARy agonists. 1. Discovery of a novel series of potent antihyperglycemic and antihyperlipidemic agents. J. Med. Chem 41, 5020–5036 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.