Abstract
Immune evasion in the tumor microenvironment (TME) is a crucial barrier for effective cancer therapy and plasticity of innate immune cells may contribute to failures of targeted immuno-therapies. Here, we show that rivaroxaban, a direct inhibitor of activated coagulation factor X (FX) promotes antitumor immunity by enhancing infiltration of dendritic cells and cytotoxic T cells at the tumor site. Profiling FX expression in the TME identifies monocytes and macrophages as crucial sources of extravascular FX. By generating mice with immune cells lacking the ability to produce FX, we show that myeloid cell-derived FX plays a pivotal role in promoting tumor immune evasion. In mouse models of cancer, we report that the efficacy of rivaroxaban is comparable to anti-PD-L1 therapy and that rivaroxaban synergizes with anti-PD-L1 in improving anti-tumor immunity. Mechanistically, we demonstrate that FXa promotes immune evasion by signaling through protease activated receptor 2 and that rivaroxaban specifically targets this cell-autonomous signaling pathway to reprogram tumor-associated macrophages. Collectively, our results have uncovered the importance of coagulation factor X produced in the TME as a regulator of immune cell activation and suggest translational potential of direct oral anticoagulants to remove persisting roadblocks for immunotherapy and provide extravascular benefits in other diseases.
One Sentence Summary:
Rivaroxaban, an inhibitor of coagulation Factor Xa promotes antitumor immunity in mice.
Introduction
The coagulation system is a major innate defense pathway that cooperates with the complement cascade to limit infections and supports immunity during restoration of tissue integrity after injury. Many aspects of these host protective pathways can be exploited by tumor cells to shape the tumor microenvironment (TME) and to promote metastasis. Cancer cell expression of the coagulation initiator tissue factor (TF) (1), activation of platelets and platelet-leukocyte interactions facilitate tumor cell survival in the blood and distant metastasis (2). In contrast, cancer cell TF-FVIIa activating protease activated receptor (PAR) 2 directly promotes tumor progression independent of the intravascular blood clotting cascade (3). However, many of these studies were based on human xenografts in immune-deficient mice. These models have major limitations in evaluating full responses of the immune system as a driver of tumor progression as well as a gatekeeper and effector of anti-tumor immunity.
The innate immune and coagulation systems have many evolutionary ties and are connected in several hematopoietic and myeloid cell signaling pathways. Coagulation protease-mediated signaling via PARs not only regulate hematopoiesis (4) and viral infection (5), but also converge with innate immune sensing toll like receptor (TLR) 4 signaling controlling dendritic cell and macrophage phenotypes (6–11). Signaling of the TF-FVIIa-FXa coagulation initiation complex in particular regulates TLR4-dependent interferon responses and tumor immune-evasive chemokine Ccl22 expression (7), but how these intrinsic wirings between coagulation and immunity influence the TME is unknown.
Intriguingly, tumor-associated macrophages (TAM) synthesize the TF ligands FVII and FX (12, 13). TAM are associated with poor prognosis in a broad variety of tumor entities (14), display high plasticity (15–17), and shape the TME, supporting angiogenesis, metastasis and immune evasion (18–20). Targeting cell-intrinsic macrophage signaling may therefore improve anti-tumor immunity, but the potential of targeting macrophage signaling in the context of relevant checkpoint inhibitor and cancer immuno-therapies has not been explored extensively (14, 21, 22). Here, we identify FX synthesized by monocytes and macrophages as the activator of PAR2 and a crucial driver of such an innate immune signaling pathway. FXa-PAR2 signaling impedes anti-tumor immunity in the TME and targeting this pathway by clinically used oral FXa inhibitors provides synergistic benefit with checkpoint inhibitor therapy.
Results
Effects of direct small molecule FXa inhibitors on anti-tumor immunity
Target-specific oral FXa inhibitors are widely used for cardiovascular indications, but their clinical applications in the context of cancer therapy was limited prior to randomized trial validating their efficacy in comparison to standard of care for thrombosis prophylaxis, i.e. low molecular weight heparin (LMWH) (23, 24). LMWH requires antithrombin for inactivation of coagulation proteases, including FXa, and therefore preferentially inhibits coagulation in the intravascular and possibly perivascular space where antithrombin is detectable (25). Because small molecule FXa inhibitors have broader tissue distribution (26), we evaluated potential effects of these clinically relevant anticoagulants on tumor growth and metastasis of an aggressive preclinical tumor model in immune competent mice.
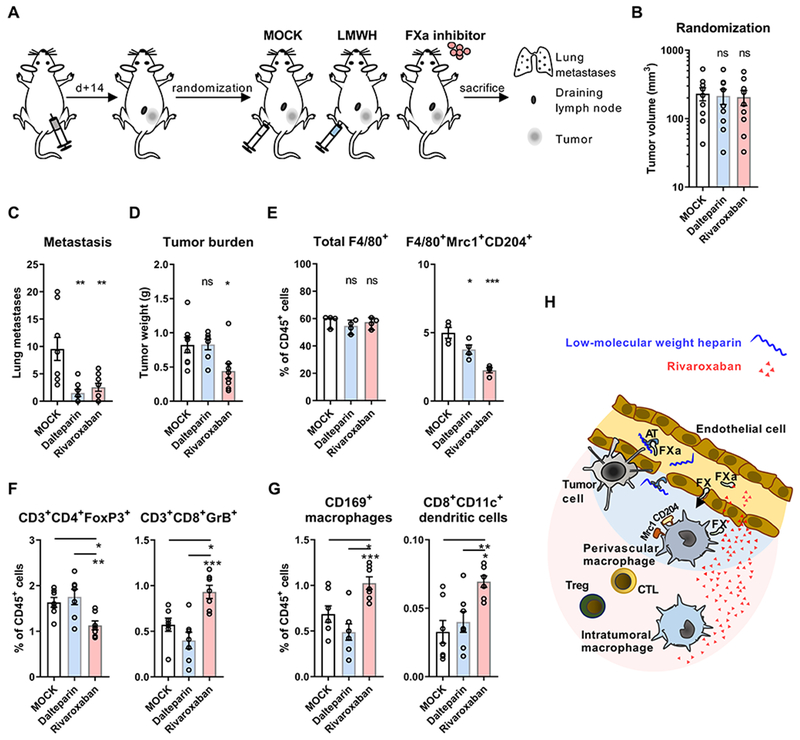
Mice injected with T241 fibrosarcoma cells were randomized after 14 days (Fig. 1A) to cohorts with similar tumor volumes (Fig. 1B) receiving therapy with the LMWH dalteparin, which has been used in prior cancer trials, or with the direct FXa inhibitor rivaroxaban. Both, heparin and rivaroxaban significantly reduced spontaneous metastasis (Fig. 1C) that is known to require intravascular coagulation. In contrast, only rivaroxaban attenuated tumor growth relative to the control group at the end of the experiment (Fig. 1D).
Figure 1: Direct FXa inhibitors improve anti-tumor immunity.
(A) Schematic illustration of treatment scheme. WT mice injected with 5×104 T241 cells were randomized at day 14 to treatment with saline injection (MOCK), LMWH injection (dalteparin 200 mg/kg body weight), or oral anticoagulation with FXa inhibitor (rivaroxaban 0.4 mg/g chow). (B) Tumor volumes of treatment groups (n=9) at day of randomization. (C) Macroscopic lung metastasis in treatment groups (day 22; n=9/8/9). (D) Tumor weights at the end of the experiment (day 22; n=8/7/8). (E) Quantification of total and Mrc1+CD204+ macrophages in the TME (n=4). (F) Regulatory and cytotoxic T cell frequencies in the TME (n=7). (G) Quantification of CD169+CD11c+ MHC II+ macrophages and CD8+CD11c+ dendritic cells in draining lymph nodes of T241 tumors (n=7); for all: one-way ANOVA, mean ± SD; *p< 0.05, **p <0.005, ***p <0.001, ns: not significant. (H) Illustration of predicted distribution of rivaroxaban and LMWH/antithrombin in the TME.
To better understand the surprising marked reduction in tumor growth seen specifically in rivaroxaban-treated mice, we analyzed the abundance of immune cell populations regulating anti-tumor immunity. Macrophage numbers were indistinguishable between treatment groups, but rivaroxaban markedly suppressed marker expression (Mrc1, CD204) characterizing TAM involved in immune evasion, angiogenesis and metastasis (Fig. 1E). Strikingly, treatment with rivaroxaban, but not heparin, significantly decreased immune-suppressive regulatory FoxP3+CD4+ T cell numbers and increased tumor-killing granzyme B+ CD8+ T cells in the TME (Fig. 1F).
Priming of CD8+ T cell anti-tumor immunity depends on tumor antigen cross-presentation by CD169+ macrophages and CD8+ dendritic cells (DC) in the tumor draining lymph nodes (27–29). Rivaroxaban, but not heparin, remarkably caused an expansion of these immune cell populations in tumor-draining lymph nodes relative to control mice (Fig. 1G). Thus, attenuating intra- and possibly perivascular cancer-associated hypercoagulability with heparin was insufficient for reprogramming innate immune cells in the TME and only the tissue-penetrating FXa inhibitor rivaroxaban improved antitumor immunity (Fig. 1H).
Factor X expression by tumor-associated myeloid cells
These data indicated that FXa had tumor evasive functions in the TME. Although coagulation factors are known to be expressed by human tumor cells (30–32), syngeneic tumor models, including the polyoma middle T (PyMT) breast cancer model exhibiting TF-dependent tumor development (33, 34), expressed neither FX nor its activator FVIIa (Fig. S1A), but other components of the TF pathway, i.e. TF pathway inhibitor (TFPI) (35) and the FX receptor endothelial protein C receptor (EPCR, Procr) (36). In contrast, peritoneal macrophages, known to express FVII (37, 38), also upregulated TF and FX upon stimulation with e.g. interferon γ (IFNγ) and lipopolysaccharide (LPS) (Fig. S1B). F7 and F10 mRNA levels in peritoneal macrophages were also comparable to levels in liver cells, the physiological site of plasma coagulation factor synthesis. Stimulation of macrophages resulted in vitamin K-dependent generation of FXa activity measurable in the cell supernatant (Fig. S1C) and synthesis of FX protein detectable intracellularly (Fig. S1D). Thus, innate immune cells express a cell autonomous extrinsic coagulation pathway capable of generating FXa.
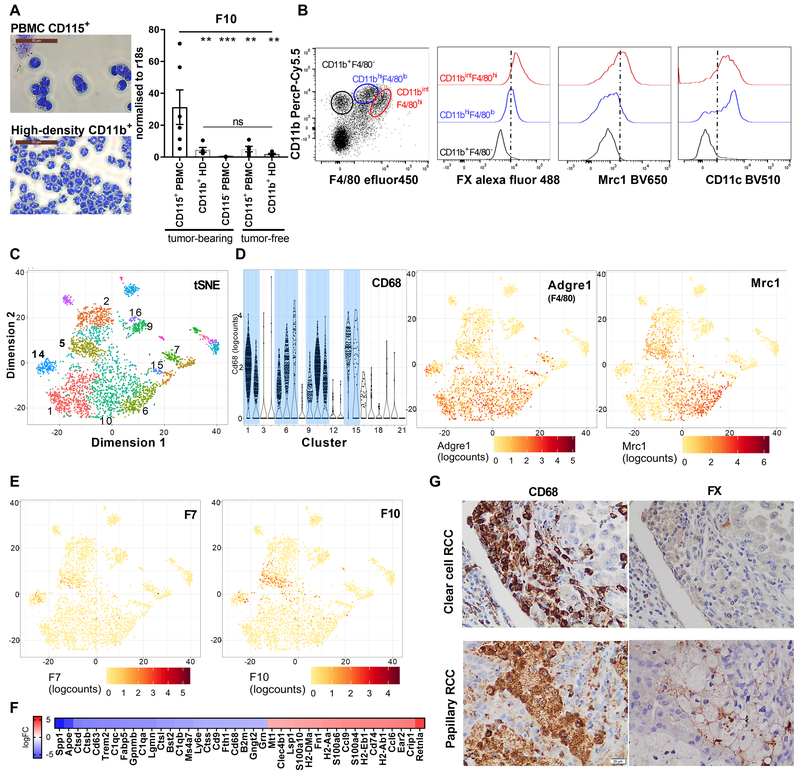
We next evaluated myeloid cell FX expression in PyMT mice with spontaneous breast cancer development. CD115+ blood monocytes isolated from tumor-bearing, but not tumor-free mice expressed F10 mRNA. However, F10 mRNA was not upregulated in blood neutrophils (Fig. 2A). Staining for FX protein in innate immune cells isolated from the TME of PyMT tumor bearing mice furthermore showed that FX was not detectable in CD11b+/F4/80− neutrophils but expressed by CD11b+/F4/80+ macrophages with heterogenous expression of Mrc1 and CD11c (Fig. 2B).
Figure 2: Expression of FX by tumor-associated immune cells.
(A) Peripheral blood monocytes and neutrophils from PyMT or tumor-free mice were isolated by density gradient centrifugation and additional CD115+ or CD11b+ bead selection. Shown are representative cytospins and F10 mRNA expression relative to r18s (n=6/5/7/5/6); one-way Anova, mean ± SEM, **p<0.005, ***p<0.001, ns: not significant. (B) Flow cytometric analysis of FX (intracellular), Mrc1 and CD11c expression in CD11b+/F4/80− (neutrophils) and CD11b+/F4/80+ (TAM) of PyMT mice. (C) Clustering of Gel bead-in-EMulsion (GEM) single cell sequenced CD11c-selected cells from early-stage PyMT WT tumors by t-distributed Stochastic Neighbor Embedding (t-SNE) analysis. (D) Violin and tSNE plots revealing F4/80 (Adgre1) and Mrc1 expressing myeloid cell clusters. (E) Expression of F7 and F10 in CD68+ and F4/80+ myeloid cells. (F) Differential transcript abundance in F10+ clusters 5 vs 14 (top 40 logFC). (G) FX expression by CD68+ TAM on serial sections from human clear cell (representative for 3/15 tumors) and papillary (representative for 8/10 tumors) renal cell cancer (RCC).
Gel bead-in-EMulsion (GEM) single cell sequencing of CD11c-selected TAM from two independent PyMT tumors further defined the expression of coagulation factors in the TME. The enriched 3’ sequences from 3647 viable cells of the combined data set yielded distinct clusters defined by the t-distributed Stochastic Neighbor Embedding (t-SNE) algorithm (Fig. 2C). Expression of macrophage markers CD68 and F4/80 in the depicted 11 clusters (Fig. 2D) showed that a large proportion of cells was myeloid cell-related, reflecting TAM heterogeneity previously demonstrated for human tumors (16), with remaining clusters consisting of small numbers of contaminating tumor and T cells.
No mRNA expression of Fga, Fgb, Fgg (fibrinogen), F2 (prothrombin), F5 (FV), F8 (FVIII), F9 (FIX), F11 (FXI), and F12 (FXII) was detected in macrophages. In contrast, F10 (FX) and F7 (FVII) were co-expressed predominantly in clusters 5 and 14. F10 was also more sparsely expressed in cells assigned to clusters 10 and 11 (Fig. 2E) which were positive for the TAM marker Mrc1 (Fig. 2D). Clusters 14 and 5 co-expressing F7 and F10 differed in their RNA profile (Fig. 2F). Expression of Spp1 (osteopontin), Apoe (apolipoprotein E) and Ctsb (cathepsin B) indicated that cells of cluster 14 were recruited monocytes in transition to macrophages, whereas cells of cluster 5 expressed higher levels of markers for alternatively activated macrophages, e.g. Retnla (Fizz1) (39). These data and protein expression (Fig. 2B) indicated that upstream coagulation protease regulate innate immune functions and/or phenotypic maturation of monocyte and macrophages.
We next confirmed macrophage FX expression in human tumors. Tumor transcriptome analysis in the Pathology Atlas showed that TF (F3), EPCR (Procr), and F10 transcript levels were correlated with poor prognosis in clear cell and papillary renal cell carcinoma (RCC), but F10 was not an independent prognostic marker in other tumor entities (40). Immunohistochemistry of human tumor samples demonstrated expression of FX in a subset of CD68+ TAM, but not in tumor or stromal cells, of both RCC types (Fig. 2G). Thus, TAM are the major cell type in human tumors with F10 expression being correlated with poor outcome in RCC (40).
Regulation of anti-tumor immunity by monocyte/macrophage-autonomous FX synthesis
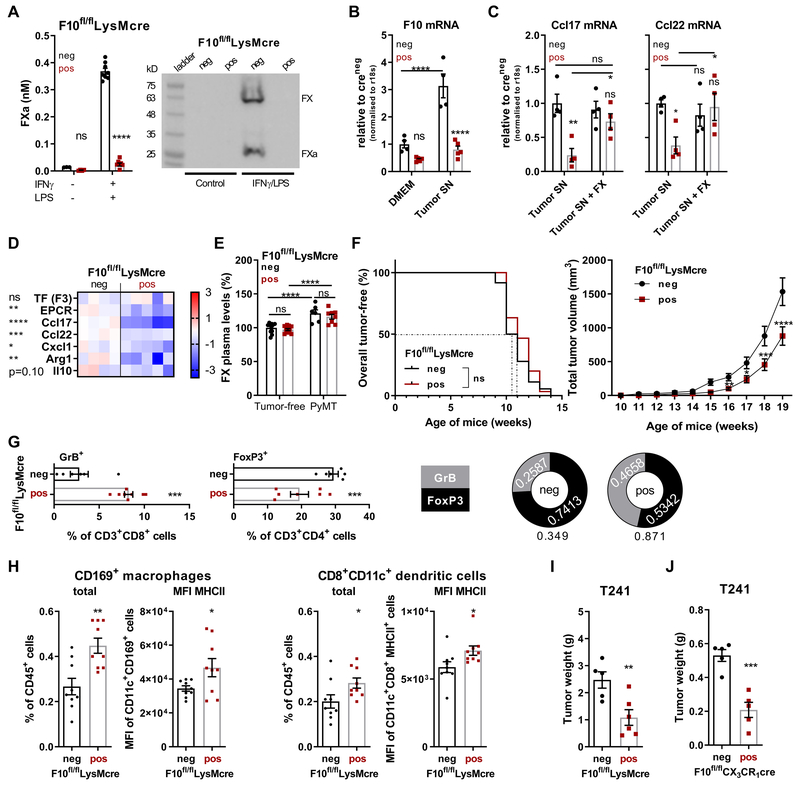
In order to directly demonstrate a role for monocyte/macrophage synthesized FX in immune evasion, we generated F10fl/flLysMcre mice deficient in macrophage FX synthesis and activation (Fig. 3A). Exposure of peritoneal macrophages to PyMT tumor cell supernatant supplemented with IFNγ compared to DMEM/IFNγ caused upregulation of F10 mRNA in vitro (Fig. 3B). In addition, this stimulation induced chemokines involved in the recruitment of suppressive immune cells (Ccl17, Ccl22), but the response was blunted in F10fl/flLysMcre macrophages and restored to WT levels upon addition of purified human FX (Fig. 3C). Other markers of immune-evasive macrophage polarization (Cxcl1, Arg1, Il10) (41) were similarly reduced in tumor supernatant-stimulated FX-deficient macrophages (Fig. 3D), demonstrating that endogenously synthesized FX promotes macrophage polarization.
Figure 3: Monocyte/macrophage-autonomous FX synthesis promotes immune evasion.
(A) FXa activity and FX protein in serum-free supernatant of F10fl/flLysMcre peritoneal macrophages after stimulation as indicated for 18 hours (n=3/4/8/8); mean ± SEM, two-way ANOVA, Sidak`s multiple comparisons test. (B) Expression of F10 mRNA of peritoneal macrophages exposed to DMEM + IFNγ or PyMT cell culture supernatant supplemented with IFNγ (n=4/5/4/5). (C) Ccl17 and Ccl22 mRNA expression of peritoneal macrophages exposed to PyMT cell culture supernatant with or without purified human FX (n=4); mean ± SEM, two-way ANOVA, Sidak`s multiple comparisons test. (D) Effect of FX deletion in F10fl/flLysMcre peritoneal macrophages on polarization marker expression after 18 hours stimulation in PyMT tumor cell-conditioned medium with IFNγ; Z-score, individual replicates (n=4/5) are shown, unpaired, two-tailed t-test. (E) FX plasma levels in 19 weeks-old female mice with or without PyMT tumors (n=12/12(6/8); mean ± SEM, two-way ANOVA, Sidak`s multiple comparisons test. (F) Tumor-free survival (n=30) and tumor growth (n=33) of PyMT F10fl/flLysMcre and PyMT F10fl/fl littermate controls (n=36; n=37) mice; log-rank (Mantel-Cox) test (tumor-free survival), mean ± SEM, two-way ANOVA, Sidak`s multiple comparisons test (tumor growth). (G) Quantification of GrB+CD8+ T cell (n=6/8) and FoxP3+CD4+ T cells (n=6/7) in the TME and (H) of CD169+ macrophages and CD8+ dendritic cells in the draining lymph nodes of late-stage tumors of 20 weeks-old PyMT F10fl/flLysMcre or PyMT F10fl/fl littermate control mice (n=9/9). Tumor weight at sacrifice (d22) of (I) F10fl/flLysMcre (n=5/6) and (J) F10fl/flCX3CR1cre (n=5/5) mice injected with 5×104 T241 fibrosarcoma cells; mean ± SEM, unpaired, two-tailed t-test. *p< 0.05, **p <0.005, ***p <0.0005, ****p <0.0001, ns: not significant.
To evaluate roles of FX in tumor growth, we crossed F10fl/flLysMcre mice with mice spontaneously developing PyMT breast cancer. Although TAM-expressed macrophage scavenger receptor 1 (Msr1, CD204) had been implicated in the regulation of FX plasma levels (42), macrophage FX deficiency had no effect on plasma FX levels in tumor-free or tumor-bearing littermate mice (Fig. 3E). Importantly, myeloid cell-specific FX ablation diminished tumor growth without delaying tumor initiation in the PyMT model of spontaneous tumor development (Fig. 3F). In line with reduced tumor growth, the balance of immune-suppressive regulatory CD4+FoxP3+ T cells was shifted in favor of tumor-eliminating granzyme B+ CD8+ T cells in FX-deleted versus control mice (Fig. 3G). Moreover, in draining lymph nodes of PyMT tumors, macrophage FX deletion increased the abundance of tumor antigen-processing CD169+ macrophages (27, 28, 43) and CD8+ cross-presenting DC (29) that appeared to be activated based on increased MHC class II expression (Fig. 3H). Although we did not detect FX in neutrophils (Fig. 2A, B), we compared transplanted fibrosarcoma growth in F10fl/flLysMcre mice, with deletion in myeloid lineages including neutrophils (Fig. 3I), and F10fl/flCX3CR1cre mice, with more specific monocyte F10 deletion (Fig. 3J). Deletion with both cre driver lines similarly reduced tumor growth relative to littermate F10fl/fl controls. In addition, lung metastasis was also reduced in mice lacking FX synthesis or PAR2 signaling in the mono-myeloid compartment (Fig. S2). Thus, monocyte/macrophage FX synthesis promotes tumor immune evasion independent of the coagulation cascade in the circulating blood.
Targeting of macrophage FXa by direct FXa inhibitors
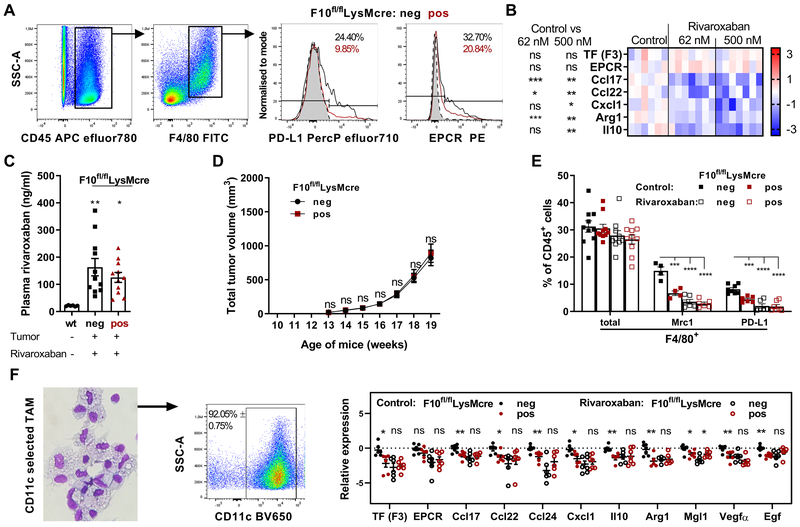
As seen in rivaroxaban-treated mice (Fig. 1E), FX-deficiency caused phenotypic changes in TAM, as evidenced by diminished expression of the immune suppressor PD-L1 and the FX receptor EPCR (Fig. 4A). Akin to FX-deficient macrophages, therapeutic concentrations of the direct FXa inhibitor rivaroxaban reduced macrophage expression of immune-suppressive mediators following exposure to PyMT tumor supernatant in vitro (Fig. 4B). These data raised the question whether myeloid cell FXa was the therapeutic target for tissue penetrating small molecule FXa inhibitors in vivo. Oral administration of rivaroxaban to tumor-bearing mice achieved comparable therapeutic plasma levels irrespective of FX expression in myeloid cells (Fig. 4C). Importantly, the breast cancer growth difference between macrophage FX-deficient and FX-expressing PyMT mice (Fig. 3F) was no longer seen in rivaroxaban-treated mice (Fig. 4D) and rivaroxaban inhibited spontaneous metastasis (Fig. S2).
Figure 4: Pharmacological targeting of macrophage FXa-PAR2 signaling by direct FXa inhibitors.
(A) PD-L1 and EPCR Expression in TAM from late-stage tumors of PyMT F10fl/flLysMcre mice compared to PyMT F10fl/fl littermate controls. (B) Effects of rivaroxaban on TAM polarization marker expression in WT peritoneal macrophages after 18 hours in PyMT tumor cell-conditioned medium with hirudin/vitamin K and IFNγ; heatmap (control: n=6; 62 nM rivaroxaban: n=7; 500 nM rivaroxaban: n=7); log2 scale; values relative to mean of control; Z-score, unpaired, two-tailed Student’s t-test. (C) Rivaroxaban plasma levels of untreated tumor-free WT mice and PyMT F10fl/flLysMcre and PyMT F10fl/fl littermate controls receiving rivaroxaban in the diet (n=6/11/11); means ± SEM, one-way ANOVA with Dunnett’s multiple comparisons test. (D) Tumor growth of PyMT F10fl/flLysMcre (n=28) and PyMT F10fl/fl littermate control (n=36) mice with oral rivaroxaban administration; means ± SEM; two-way ANOVA, Sidak’s multiple comparisons test. (E) Quantification of CD11b+CD11c+F4/80+ TAM (n=10) and Mrc1+ (n=4/4/6/6) and PD-L1+ (n=8/6/6/7) subpopulations in late-stage tumor TME of 20 weeks-old PyMT mice with or without oral rivaroxaban therapy; means ± SEM; two-way ANOVA, Dunnett’s multiple comparisons test. (F) Expression profiles of CD11c-selected TAM from the TME of late-stage tumors in 20 weeks-old PyMT mice (n=7/6/7/7); mean ± SEM, unpaired, two-tailed Student’s t-test. *p< 0.05, **p <0.005, ****p <0.0001, ns: not significant.
Comparison of macrophage phenotypes showed that myeloid cell FX deficiency reduced the abundance of Mrc1+ and PD-L1+ TAM. This reduction was also seen in rivaroxaban-treated mice (Fig. 4E). In addition, we profiled CD11c-selected macrophages (Fig. 4F) from these groups and found reduced expression of immune-evasive Ccl17, Ccl22, Cxcl1, Arg1, Il10, pro-angiogenic Vegfa and pro-metastatic Egf (20) in FX-deficient mice and rivaroxaban-treated mice (Fig. 4G). Importantly, there was no additional reduction in the expression of these markers, when macrophage FX-deficient mice were treated with rivaroxaban, demonstrating that the oral FXa inhibitor primarily targeted immune-evasive functions of macrophage-synthesized FXa.
Synergy of therapeutic FXa blockade with checkpoint inhibitor therapy
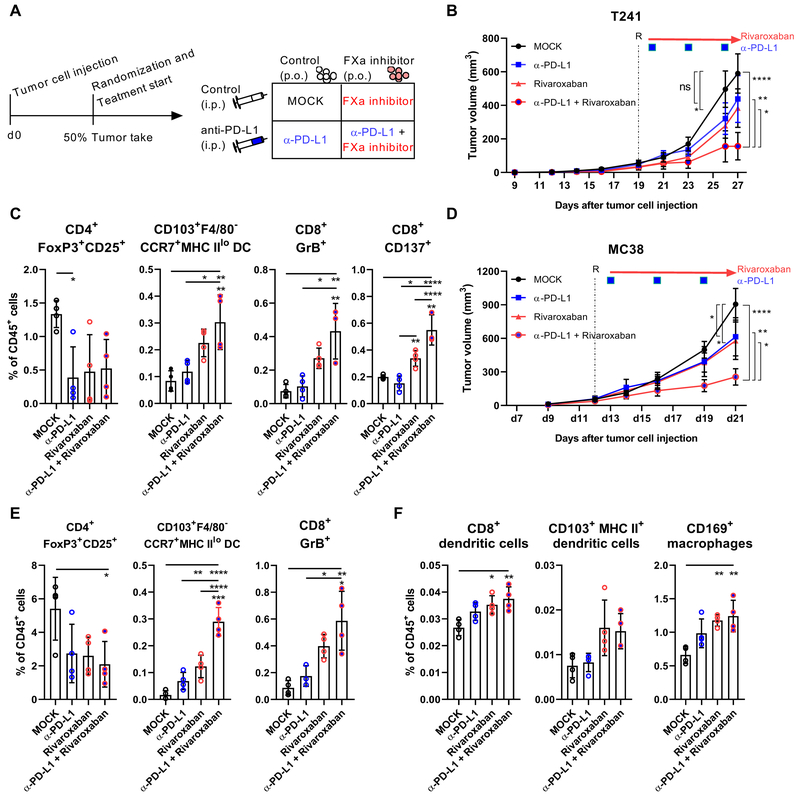
The finding that rivaroxaban inhibited expansion of immune-suppressive PD-L1+ TAM (44), raised the question whether therapeutic blockade of FXa could aid immuno-therapeutic regimens. We therefore randomized tumor-bearing mice to single or combination therapy with rivaroxaban and anti-PD-L1 antibody (Fig. 5A). As monotherapy, rivaroxaban was comparable to anti-PD-L1 in attenuating tumor growth of T241 fibrosarcoma, but the combination produced a synergistic effect in tumor suppression (Fig. 5B). While both agents similarly reduced Treg frequencies, only the TME of mice treated with rivaroxaban showed an increase in CD103+F4/80−CCR7+ MHC IIlo DC and GrB+ cytotoxic CD8+ T cells (Fig. 5C). Remarkably, rivaroxaban and anti-PD-L1 synergized to increase antigen-activated CD137+ cytotoxic CD8+ T cells (Fig. 5C). In the colon cancer model MC38, rivaroxaban and anti-PD-L1 also attenuated tumor growth as single therapy, but in combination they were more effective (Fig. 5D). While both agents suppressed Treg abundance, rivaroxaban again had a predominant effect on expanding CD103+F4/80−CCR7+ MHC IIlo DC and GrB+ CD8+ T cells in the TME (Fig. 5E) and of cross-presenting CD8+ DC in draining lymph nodes (Fig. 5F), providing a mechanism for enhancing anti-tumor immunity.
Figure 5: FXa inhibitor synergizes with check point inhibitor therapy.
(A) Treatment scheme. (B) Tumor growth of 5×104 T241 cells injected into WT mice with or without treatment of rivaroxaban and/or α-PD-L1 (n=9/9/10/8); mean ± SEM, two-way ANOVA, Tukey’s multiple comparisons test. (C) Quantification of CD103+ CCR7+ MHCIIlo DCs, GrB+ CTL, activated CD137+ CD8+ T cells and Treg (n=4) in the TME at sacrifice 27 days after tumor cell injection (n=4); mean ± SEM, one-way ANOVA, Tukey’s multiple comparison test. (D) Tumor growth of 5×104 MC38 cells injected into WT mice with or without treatment of rivaroxaban and/or α-PD-L1 (n=9/11/11/11); mean ± SEM, two-way ANOVA, Tukey’s multiple comparisons test. Quantification of (E) CD103+ CCR7+ MHCIIlo DCs, GrB+ CTL and Treg (n=4) in the TME and of (F) CD8+ DCs, CD103+ DCs and CD169+ macrophages (n=4) at sacrifice 21 days after tumor injection; *p< 0.05, **p <0.005, ***p <0.0005, ****p <0.0001, ns: not significant.
Inhibition of tumor immune-evasive macrophage FXa-PAR2 signaling by FXa inhibitors
We hypothesized that extravascular coagulation primarily regulated anti-tumor immunity through proteolytic signaling. To pinpoint the specific signaling functions of FXa, we used mouse strains with point mutations in PAR2 causing resistance to cleavage by specific proteases (Fig. 6A). We analyzed cancer growth in PAR2-G37I mice (45) carrying a mutation preventing cleavage by the TF-FVIIa-FXa signaling complex, which is dependent on the FX receptor EPCR (Procr) (36, 46) (Fig. 6B). In addition, we used PAR2-R38E mice with abolished canonical proteolysis by all proteases (7, 47). As expected from cancer cell TF-FVIIa-PAR2 proangiogenic signaling (3) and delayed breast cancer development in PAR2-deficient PyMT mice (33, 34), tumors appeared later (Fig. 6C) and progressed slower (Fig. 6D) in completely cleavage-resistant PAR2-R38E relative to wild-type (WT) mice. In support of FXa-PAR2 signaling specifically causing immune evasion, PyMT tumor initiation in FXa-resistant PAR2-G37I mice was unaltered, but tumor growth was attenuated compared to WT.
Figure 6: Attenuated tumor growth in FXa-resistant PAR2 mutant mice.
(A) Mature residue numbering, coding and amino-acid sequence of PAR2-WT, G37I, and R38E mice and sensitivity to coagulation proteases. (B) Confirmation of FXa-resistance of mouse PAR2-G37I to cleavage by TF-FVIIa-FXa on CHO-cells expressing mouse EPCR (n=4); mean ± SEM, two-tailed, unpaired Student`s t-test. (C) Tumor-free survival (n=29, 34, 28); log-rank (Mantel-Cox) test, and (D) total tumor volume and weights of the largest tumors at sacrifice (n=30, 34, 30) of PyMT mice; means ± SEM, two-way ANOVA, Tukey’s multiple comparisons test. (E) Quantification of CD11b+CD11c+CD68+ TAM (n=9/9/10) and Mrc1+ (n=7) and EPCR+ (n=6/5/4) subpopulations in late-stage TME of 20 weeks old PyMT PAR2-WT, G37I, or R38E mice; means ± SEM, one-way ANOVA. (F) CD3+CD8+ T-cell (n=14/14/13) frequencies in late-stage tumors of 20 weeks old PyMT mice; means ± SEM, one-way ANOVA. (G) Tumor weights 22 days after s.c. injection of B16F10 (n=12/12/10) or T241 (n=9) cells into PAR2-WT, G37I or R38E mice; pooled data from two independent experiments; means ± SEM, one-way ANOVA. (H) Tumor weights 22 days after s.c. injection of T241 in PAR2fl/flLysMcre or littermate control mice (n=5/4); means ± SEM, unpaired, two tailed t test; (I) Frequencies of Granzyme B+ (GrB+) CD8+ T cells in TME of F10fl/flLysMcre mice (n=4) or (F) PAR2-G37I mice and controls with or without rivaroxaban therapy (n=4) evaluated 21 days after T241 injection. T cells were co-cultured with T241 cells for quantification of anti-tumor CD8+ T cell responses by ELISpot assay shown by representative spots; means ± SEM, unpaired, two tailed t test; *p< 0.05, **p <0.005, ***p <0.0005, ****p <0.0001, ns: not significant.
Consistent with the data obtained in myeloid cell FX-deficient and rivaroxaban-treated mice, CD11b+CD11c+CD68+ macrophage numbers in the TME were unchanged upon disabling PAR2 signaling, but TAM phenotypes differed. Specifically, TAM expressing Mrc1 (CD206) (16, 21, 48) and the TF-FVIIa-FXa signaling co-receptor EPCR were reduced (Fig. 6E). Conversely, total CD8+, but not CD4+ T cell numbers were increased in PAR2 signaling-deficient mice (Fig. 6F). A similar reduction in the growth of syngeneic tumors transplanted into mutant PAR2-R38E or PAR2-G37I mice (Fig. 6G) confirmed that impaired PAR2 signaling by non-tumor cells in the TME limited tumor expansion due to FXa cleavage resistance. Furthermore, reduced tumor growth of T241 fibrosarcoma transplanted into myeloid cell PAR2-deficient PAR2fl/flLysMcre mice (Fig. 6H) provided evidence that FXa induced cell autonomous PAR2 signaling in macrophages.
In the aggressive T241 model, we furthermore documented that CD8+ T cells with anti-tumor activity were expanded following genetic or pharmacological FXa-PAR2 signaling blockade. ELISpot assays showed increased numbers of tumor-specific cytotoxic granzyme B+ CD8+ T cells in the TME of F10fl/flLysMcre mice versus littermate controls (Fig. 6I). Treatment with the FXa inhibitor rivaroxaban also increased CD8+ T cell anti-tumor responses, but rivaroxaban had no additional effect on already increased tumor-specific granzyme B+ CD8+ T cell levels in FXa signaling-deficient PAR2-G37I mice (Fig. 6J). Thus, rivaroxaban improves anti-tumor immunity specifically by targeting myeloid cell FXa-PAR2 signaling.
Tumor stage-independent suppression of anti-tumor immunity by FXa-PAR2 signaling
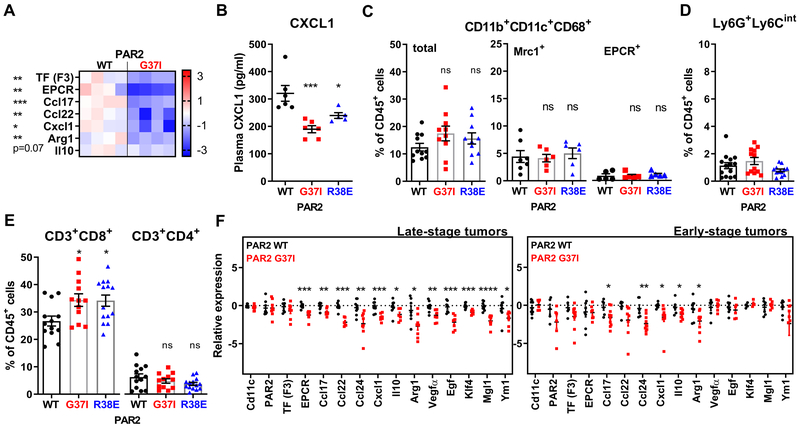
TAM phenotypes are determined by a complex interplay of polarizing cytokines, vascularization, hypoxia and tumor cell metabolism. However, simple exposure to PyMT tumor cell-conditioned medium independent of complex immune cell alterations caused macrophage polarization, which was also prevented in PAR2-G37I relative to WT macrophages (Fig. 7A), in line with data obtained with FX-deficient (Fig. 3D) or rivaroxaban-treated (Fig. 4B) cells. In addition, CXCL1 plasma levels were already reduced during early stages of PyMT breast cancer development in PAR2 mutant mice (Fig. 7B), indicating that macrophage PAR2 signaling played an immune-modulatory role throughout tumor progression.
Figure 7: Tumor stage-independent reprogramming of TAM immune-evasive phenotypes in PAR2 FXa-resistant mice.
(A) Macrophage polarization marker expression in peritoneal macrophages after 18 hours culture in PyMT tumor cell-conditioned, serum-free DMEM supplemented with hirudin/vitamin K and IFNγ (n=4); Z-score, unpaired, two-tailed t-test. (B) CXCL1 plasma levels of 13-weeks old PyMT PAR2-WT, G37I, or R38E mice (n=6/6/5); means ± SEM, one-way ANOVA. (C-E) Quantification of (C) CD11b+CD11c+CD68+ TAM (n=11/10/10) and Mrc1+ (n=7/6/6) and EPCR+ (n=5/5/5) subpopulations, (D) of neutrophils (n=14/12/12), and (E) of CD8+ and CD4+ T cells in early-stage TME of 20-week old PyMT PAR2-WT, G37I, or R38E mice (n=13/12/14); means ± SEM, one-way ANOVA. (F) Expression profiles of CD11c-selected TAM from late- (n=8/8) and early-stage (n=5-9/7-10) tumors of 20 weeks-old mice; mean ± SEM, unpaired, two-tailed t-test; *p< 0.05, **p <0.005, ***p <0.0005, ****p <0.0001, ns: not significant.
PyMT breast cancer is a multifocal disease model with sequential development of tumors in several mammary glands during the growth of typically one large tumor that determines termination of the experiment (Fig. S3A). At time of sacrifice, the early-stage tumors isolated from the same animals differed from late-stage tumors in TAM populations. Early-stage tumors had reduced numbers of macrophages with a CD11blowF4/80hi phenotype and showed attenuated expression of PD-L1, Mrc1 and VCAM1 by F4/80+ cells, which was typical for TAM of late-stage tumors (49) (Fig. S3B).
In line with diminished immune evasion in the TME, infiltration of late-stage tumors with neutrophils, immature Ly6Glow and low-density neutrophil populations (50, 51) was reduced in PAR2 signaling-deficient PyMT mice (Fig. S3C); and transcript levels of genes implicated in immune suppression by myeloid cells (Bv8, Il4ra, PD-L1) (51) were diminished in isolated CD11b+CD11c− cells from PAR2-G37I relative to WT mice (Fig. S3D). Neutrophil infiltration was also reduced in myeloid cell FX-deficient PyMT mice (Fig. S3E) and in transplanted T241 fibrosarcoma tumors of PAR2-G37I mice along with reduced numbers of Mrc1+PD-L1+ and EPCR+ TAM (Fig. S3F).
In contrast to late-stage tumors, early-stage PyMT tumors showed no differences in Mrc1+ and EPCR+ TAM between WT and PAR2 signaling-deficient mice (Fig. 7C) and neutrophil infiltration was overall very low (Fig. 7D). Remarkably, CD8+ T cells were already expanded at this stage without changes in CD4+ T cell numbers in PAR2 cleavage-resistant mice (Fig. 7E). These data indicated that FXa-PAR2 signaling not only regulated typical TAM populations in advanced tumors, but also CD11b+CD11c+F4/80+ macrophages in early tumor progression. We therefore compared the expression profile of CD11c+ cells isolated from these stages of tumor development. In CD11c+ cells isolated from late-stage tumors, TF and PAR2 transcript levels were similar between WT and PAR2-G37I mice, but mutant cells had lower expression of EPCR, immune modulatory chemokines (Ccl17, Ccl22, Ccl24, Cxcl1), immune-suppressive Il10 and Arg1, proangiogenic Vegfa and prometastatic Egf (20), and other markers of alternatively activated macrophages (Klf4, Mgl1, Ym1) (41) (Fig. 7F).
Similarly, loss of FXa-PAR2 signaling caused reduced expression of immune-modulatory chemokines and immune-suppressive IL10 and Arg1 in CD11c-selected cells from early-stage tumors (Fig. 7F). These TAM expression data are consistent with the observed lower plasma levels of CXCL1 in PAR2 mutant mice at early stages of PyMT tumor development (Fig. 7B). In line with low expression of certain TAM markers, including Mrc1 (CD206), FXa-PAR2 signaling-deficiency had no effect on the expression of other alternatively activated macrophage markers, i.e. Klf4, Mgl1, Ym1, as well as Procr, Vegfa, and Egf. Thus, while PAR2 signaling promoted the development of proangiogenic TAM in advanced tumors, remarkably, the immune-evasive phenotype of macrophages was already reprogrammed in FXa-PAR2 signaling-deficient mice at early stages of tumor progression.
Discussion
These experiments uncover tumor-evasive immune signaling roles of monocyte- and macrophage-synthesized FX that functions independently of the coagulation cascade in the circulating blood. Loss of this signaling pathway causes a reprogramming of TAM and a shift in TME immune cell composition to anti-tumor immunity already in early stages of tumor progression. Remarkably, macrophage FX synthesis also determines TAM phenotypes in late-stage tumors where vessel hyperpermeability might have provided coagulation factors for extravascular signaling. Limited vascular leakage of plasma coagulation inhibitors, which are restricted to the intravascular and perivascular space (25), or degradation of antithrombin in the TME (52) may contribute to the inefficient TAM reprogramming with LMWH, which activates antithrombin for FXa inhibition. In contrast, small molecule direct FXa inhibitors are highly efficacious and at clinically relevant concentrations achieve TAM reprogramming indistinguishable from genetic ablation of FXa-specific PAR2 signaling. Improving tissue penetrance of oral FXa inhibitors at the expense of intravascular distribution may represent a strategy to improve anti-tumor immunity while limiting bleeding side effects associated with inhibition of the blood clotting cascade.
Experiments with isolated macrophages in vitro confirmed FXa-PAR2 cell autonomous signaling as the driver for alternative macrophage activation. Extensive studies have delineated diverse pathways by which the hemostatic system and tumor cell coagulation signaling promote tumor initiation, invasion and metastasis (1). Because these studies have largely relied on xenograft models in immune-deficient mice, the crucial function of coagulation innate immune signaling in promoting immune evasion and the potency of FXa inhibitors to induce antigen-specific anti-tumor immunity has so far escaped detection. Although signaling by PAR2 has been implicated as a stimulator of DC migration to lymph nodes (53) and a suppressor of CD4+ T cell priming (54), the newly developed FXa-resistant mutant mouse clearly defined a protease-selective PAR2 signaling pathway central for alternative activation of TAM. This pathway ultimately proved to be the target for oral FXa-inhibitors to reprogram TAM phenotypes and to promote anti-tumor immunity. Targeting PAR2 with appropriate antagonists (55) may represent an alternative approach to prevent immune evasion.
The presented data indicate that FXa-PAR2 signaling is central to the synthesis of immune-evasive chemokines and mediators at all stages of tumor development, as well as TAM phenotypes promoting angiogenesis and metastasis in advanced tumors. Consequently, immune-suppressive neutrophil and regulatory T cell recruitment to the TME is attenuated upon genetic deletion or pharmacological disruption of FXa-PAR2 signaling. However, in early-stage tumors only minimal infiltration with neutrophils is seen, but loss of FXa-PAR2 signaling nevertheless leads to an expansion of cytotoxic T cells in the TME and activation of cross-presenting macrophages and dendritic cells in tumor-draining lymph nodes. Thus, our study indicates that intervention in the FXa-PAR2 signaling pathway primarily boosts anti-tumor immunity with additional benefits on immune-suppressive cell populations in the TME. The predicted extravascular and lymphatic accumulation of tissue-penetrating oral FXa inhibitors may be harnessed for modulation of immunity not only in the context of cancer immuno-therapy, but also cardio-metabolic diseases (10, 11).
The demonstrated FX synthesis by TAM subsets in the mouse and human TME adds to the expanding paradigm that the innate defense coagulation and complement pathways interact to regulate extravascular milieus. Coagulation proteases not only have important extravascular functions in wound repair, but also control internal milieus in the hematopoietic stem cell niche (4, 56) in conjunction with complement (57) and fibrinolytic (58) systems. As shown here, TAM-expressed FXa activates PAR2 for immune suppression in the internal TME. In addition, protein S (Pros1) which participates in the regulation of FXa-PAR2 signaling (6) can be derived from tumor cells to suppress TAM pro-inflammatory polarization through tyrosine kinase receptor MER signaling (59). The fibrinolytic system controls TAM phenotypes by urokinase-dependent plasmin generation that in turn activates complement signaling suppressing cytotoxic T cell responses under chemotherapy (60).
Our data show that pharmacological targeting of FX-PAR2 signaling with oral FXa inhibitors preferentially acts on professional antigen presenting cells and increases cytotoxic T cell activation and expansion and thereby synergizes with clinically effective check-point inhibitor therapy. While we demonstrate that rivaroxaban broadly attenuates the growth of different tumor models as mono or combination therapy, clinical trials will be required to determine whether these beneficial effects are generally applicable to human cancers with variable immunogenic profiles. Selection of patients for such anticoagulant trials is challenging and needs to take into consideration comorbidities, pretreatments, drug interactions and anatomical location of tumors that may predispose to unwanted bleeding under therapy with oral anticoagulants. It will be necessary to identify biomarker profiles of patients who benefit from intervention in FXa-PAR2 immune-evasive pathways to improve anti-tumor immunity. This approach will ultimately help decide on suitable anti-thrombotic therapy and may also lead to potential strategies for simultaneous prevention of cancer-associated thrombosis and immune-suppression in selective tumor types.
Materials and Methods
Study design
Adult mice of similar age (10–12 weeks) and same sex from the same breeding colony were used. Comparisons of PyMT tumor development involved cohorts in the same animal facility to avoid environmental variability. Tumor cell suspensions of transplantable tumors were randomly injected into different lines that originated from littermate founders or were cre-deleter strains of floxed alleles and littermate controls without cre-recombinase. Effects of host genetic mutations were independently confirmed in repeat experiments or different tumor models and pooled data were analyzed. For treatment experiments, groups were randomized by cage, assuring equal tumor volume at the beginning of treatment. Treatment and tumor monitoring were performed by the same investigators, but analysis of flow cytometry data was performed without knowledge of the treatment groups. Flow cytometry comparisons were based on biological replicates stained with the same fluorophore combinations and compensations analyzed on the same flow cytometer. Expression profiles were obtained from biological replicates randomly analyzed by quantitative PCR in parallel; technical failures were eliminated from the analysis.
Animal studies
All animal experiments had approved protocols at Scripps Research (IACUC protocol 09–0111) or at Johannes-Gutenberg Medical Center Mainz (Landesuntersuchungsamt Koblenz, AZ 23 177–07/G 14–1-055 and 23 177–07/G 14–1-041). PAR2 G37I (F2rl1tm2.1Wmrf), PAR2 R38E (F2rl1tm1.1Wmrf), and PAR2fl/fl (F2rl1tm3.1Wmrf) mutant mice were generated by homologous recombination of sequence confirmed mutated F2rl1 exon 2 with flanking intron 1 and 2 into pBS-FRT-Neo-FRT (a gift of Dr. Uli Müller, Scripps Research) in C2 C57BL/6N embryonic stem cells (61) at the Scripps Transgenic Core Facility. A loxP site was also introduced by the targeting strategy in introns 1 and 2, respectively, for potential tissue specific deletion of mutant and WT alleles (Fig. S4A). Targeted ES cells were identified by PCR screening for recombination that introduced the 5’ and 3’ LoxP sites and for unique restriction sites created by silent mutations in the mutated exons (Fig. S4B). After germ line transmission and excision of the resistance cassette by crossing with a flippase-deleter strain, the mutants were bred to homozygosity and the entire coding sequence of the targeted exon confirmed by DNA sequencing (Fig. S4C–E). Mutants were crossed with C57BL/6N for generation of littermate progeny of mutant and WT mice. PAR2fl/fl mice were crossed with LysMcre mice on a C57BL/6N background and used as littermates.
F10tm1a(EUCOMM) mice (EMMA ID EM: 06365; HEPD0602_2_G07) were obtained from the European Mouse Mutant Archive (EMMA) and crossed with flippase-expressing C57BL/6 mice to generate a conditional F10 allele for cross breeding with CX3CR1cre or LysMcre mice on a C57BL/6N background and subsequently with PyMT C57BL/6N mice. The PAR2-R38E and G37I mutant strains were also crossed with PyMT C57BL/6N mice to establish cohorts for monitoring spontaneous breast cancer development, as previously described (33). Tumor-free survival was defined by appearance of the first palpable tumor confirmed by continuing local tumor growth. Tumor growth was quantified by measuring every tumor nodule with a digital caliper and calculating the total tumor volume based on the formula length x width2 × 0.51. Visible metastasis was quantified by counting lungs stained with Bouin’s solution.
For transplanted tumor studies, we injected 5×104 melanoma B16F10 cells (ATCC CRL6475), fibrosarcoma T241 (62) or PyMT tumor cells established as described (33). Cells were confirmed mycoplasma-free and cultured in RPMI /10% FCS/1% penicillin/streptomycin. Rivaroxaban, kindly provided by Dr. Stefan Heitmeier (Bayer Pharma AG), was formulated at a concentration of 0.4 mg/g of chow from SNIFF (Soest, Germany) and cohorts were fed the drug formulation or a control diet from the same manufacturer. LMWH-treated mice were injected s.c. once daily with dalteparin (200 IE/kg body weight) into the contra-lateral side of the tumor, control mice were received saline injections. Control experiments confirmed the efficacy of anticoagulation 3 hours after injection. The checkpoint inhibitor α-PD-L1 (10F.9G2) was given i.p. at a dose of 100 μg three times with three days intervals.
Cell isolation and flow cytometry
Peripheral blood monocytes and neutrophils were isolated by density gradient centrifugation using a 1.077/1.119 g/ml double layer. PBMC were collected from the top layer and neutrophils from the interface between 1.077/1.119 layers. Both fractions were washed three times and applied to either CD115+ bead selection for PBMCs or CD11b+ bead selection for neutrophil selection. Purity of cells was checked by cytospin and staining with May-Grünwald Giemsa. Cells were used for RNA isolation and downstream cDNA.
Single cell suspensions of tumors were prepared by mincing and digesting the tumor with 2 mg/ml collagenase A, 5 U/ml DNaseI in DMEM under gentle rotation at 200 rpm for 1–2 hours at 37°C. Cells passed through 40 μm cell strainers were further processed on a 30%−40%−78% or 44%−66% Percoll gradient for separation of dead cells and enrichment of tumor-infiltrating immune cells in layers at the interface. For endothelial cell staining, Percoll gradient was omitted. Single cells from draining lymph nodes were obtained by passing through 40 μm cell strainers. Recovered cells were washed after red blood cell lysis, resuspended in PBS for staining with fixable viability dye, and further processed in PBS/0.5% BSA or directly resuspended in PBS/1% FCS/0.5 mM EDTA for staining without fixable viability dye for flow cytometry. Staining of F10fl/flLysMcre cells used BSA instead of FCS to avoid FX carryover. Antibodies listed in Supplementary Table 2 were used at 1:200 dilution. Intracellular staining used the FoxP3 Fix/Perm Buffer Set (ThermoFischer). Flow cytometry analysis used LSR II flow cytometer (BD), FACS Canto II (BD) or an Attune NxT acoustic focusing cytometer (ThermoFischer). Data were analyzed with FloJo 10.
mRNA expression was analyzed in CD11c+ TAM isolated with anti-mouse CD11c UltraPure MicroBeads (Miltenyi) from digested tumors. The flow-through from the first selection was used to select CD11c−CD11b+ cells (neutrophils/monocytes) using anti-mouse CD11b MicroBeads (Miltenyi). Efficiency of selection was confirmed by flow cytometry and Cytospin (ThermoFisher) preparations stained with May-Grünwald Giemsa.
For ELISpot assays, CD8+ cells from the TME of T241 tumors were selected with anti-mouse CD8α (Ly-2) MicroBeads (Miltenyi). Purity of the selected cells was confirmed by flow cytometry. Anti-mouse Granzyme B ELISpot Kit (R&D) used 2×104 T241 cells plated at 37°C for at least 3 hours and CD8+ T cells were added for co-culture of 20 hours, followed by ELISpot analysis on an Immunospot S5 Versa Analyzer.
Real-time PCR (qRT-PCR)
Total RNA was extracted with Trizol, cDNA was synthesized from 100 ng total RNA with VILO cDNA synthesis kit. Relative expression levels were determined by RT-PCR on a BioRad Real-Time System (CFX Connect Real-Time System; CFX96 Real-Time System) using SYBRgreen or PowerUp SYBRgreen kit and the primer sequences listed in Supplementary Table 3. r18s was used for normalization and standard curves for each target gene were generated by pooled cDNA. Data are presented at a log2 scale as normalized expression to mean WT expression analyzed in parallel with mutant samples or as normalized expression of the respective transcripts in total RNA from mouse liver.
Characterization of thioglycollate-evoked peritoneal macrophages
Peritoneal macrophages were harvested four days after i.p. injection of 3 ml thioglycollate (19.25 g powder/500 ml H2O) by sterile flushing the abdominal cavity. Macrophages after red blood cell lysis were plated in 12-well plates at 1×106/well in serum-free DMEM. After 3 hours of adhesion and washing, cells were cultured in 0.5 ml DMEM, 1 U/ml hirudin, 80 ng/ml konakion or serum-free tumor cell-conditioned medium 1 U/ml hirudin/80 ng/ml konakion with or without 100 ng/ml IFNγ, 500 ng/ml LPS, 100 nM of the vitamin K antagonist (VKA) acenocoumarol. FXa generated by cell autonomous activation was quantified in cell-free supernatants with Spectrozyme FXa (Sekisui Diagnostics) in the presence or absence of the FXa inhibitor NAP5 and a calibration curve with purified FXa on a Spectramax i3 (Molecular Devices).
For FX staining of peritoneal macrophages used stimulated cells fixed with 4% formaldehyde in the culture medium, fixed again after wash in 4% histofix, and blocked with 1% BSA/0.3% glycine/0.1% Tween/10% donkey serum in PBS. Cells stained with rabbit anti-human FX antibody cross-reactive with mouse FX (R01915; 1:500 ) (63) in PBS/1% BSA over night at 4°C, were counterstained with anti-rabbit 594 and mounted with ProLong ® Gold antifade with DAPI for imaging on a Spinning Disk CSU-W1 with dual camera port.
FX plasma levels
Citrated plasma was collected from 19 weeks-old female controls and F10fl/flLysMcre mice with or without spontaneous PyMT breast cancer or from 13 weeks old female controls or F10fl/flLysMcre mice with or without tumors 22 days after s.c. injection of T241 cells. FX levels were determined using STA–deficient X plasma on a Stago STart coagulometer.
FX staining of human tumor macrophages
After heat-induced antigen retrieval using the target retrieval solution (K8005, DakoCytomation, Glostrup, Denmark) whole tissue slides were stained with a polyclonal anti-FX rabbit antibody (R01915; 1:2500) (63) or a ready-to-use monoclonal anti-CD68 mouse antibody (IR613, DakoCytomation). An automated staining system (Autostainer Plus, DakoCytomation) was used in accordance with the manufacturer’s instructions. Protocol was modified as follows: Dako Envision Polymer/AP was replaced by Dako Envision Polymer/HRP and chromogen incubation was increased to 30 min for DAB and 20 min for Green HRP, respectively.
CXCL1 plasma levels
Citrated plasma was collected from 13-week old female PyMT PAR2-WT, G37I and R38E mice. CXCL1 levels were determined with the mouse CXCL1/KC Quantikine ELISA Kit (R&D).
PAR2 cleavage assay
PAR2 cleavage was determined by transiently transfecting CHO-K1 cells expressing human TF and murine EPCR with amino-terminal FLAG-tagged murine PAR2 WT (mPAR2 WT) or mPAR2-G37I cloned into pcDNA3.1/Zeo(+) (Invitrogen), as described in previous publications (36, 46, 64).
Western blotting
Cell-free supernatants were precipitated with 30% trichloroacetic acid (TCA), washed with acetone and separated by SDS-PAGE for western blotting using rabbit anti-human FX antibody cross-reactive with mouse FX (R01915; 1:500 ) (63) and detection with anti-rabbit IgG HRP-linked (dilution 1:5000) for quantification of FX on a Fusion Fx Vilber Lourmat (Peqlab).
Single cell sequencing of TAM
CD11c-bead selected cells from early-stage PyMT tumors were frozen in 90%FCS/10%DMSO until single cell library preparation. Thawed cell suspensions were adjusted to 3000 viable cells per sample for capture in droplet emulsions using the GemCode Single-Cell Instrument (10x Genomics), and scRNA-seq libraries were constructed using GemCode Single-Cell 3′ Gel Bead and Library V2 Kit. Sequences from the microfluidic droplet platform were de-multiplexed and aligned using CellRanger version 2.0.1, available from 10x Genomics with default parameters.
Two independent biological replicates of single cell libraries were sequenced on an Illumina sequencer. Single cell sequences were identified by detecting significant deviations from the expression profile of the ambient solution (65). The dataset was annotated to gene level information based on ENSEMBL v92. Quality control was performed on each data set independently to remove poor quality cells, using the scater package (v1.11.1) (66). The proportion of mitochondrial gene content was used as a proxy for damaged cells, using three median absolute deviations (MADs) as a threshold. Normalization of cell specific biases was performed on the remaining sets of 2285 and 1362 cells using the deconvolution method of Lun et al. (65). Counts were divided by size factors to obtain normalized expression values that were log-transformed after adding a pseudo-count of one. Highly variable genes were identified on the pooled set of 3647 cells, after modeling the technical noise as Poisson distributed. Genes with large biological components were provided as input to the denoisePCA function in the scran package (v1.11.1). The results of this PCA were then provided as initialization to the t-SNE algorithm (http://www.jmlr.org/papers/v9/vandermaaten08a.html) to obtain a reduced dimensionality representation of the data. To identify clusters of cells, we grouped them with unbiased, the shared nearest neighbor graph method (67) was applied, and we used the Walktrap algorithm to identify clusters. We identified marker genes for each cluster using the function findMarkers from the scran package, which performs Welch t-tests on the log-expression values for every gene and between every pair of clusters. All visualizations were generated with the iSEE package (v1.3.1, Rue-Albrecht et al., 2018 - https://f1000research.com/articles/7–741/v1).
Statistics
Statistical analyses used GraphPad Prism 6.07 or 7.0 as indicated in the figures.
Supplementary Material
Figure S1: Expression of TF signaling components in macrophages and tumor cells.
Figure S2: Regulation of metastasis by macrophage FXa-PAR2 signaling.
Figure S3: Tumor progression alters myeloid cell composition of the TME.
Figure S4: Generation of PAR2 mutant mice.
Table S2: Sources of materials used
Table S3: Primer list for RT-PCR
Table S1: Raw data file
Acknowledgments:
We thank Cindi Biazak, Jennifer Royce, Sebastian Schubert, Mona Saffarzadeh, Son Thanh Nguyen, Alexandra Grill for technical assistance and advice; Martin Suchan, Irina Eichelbroenner, and Barbara Schrörs (TrOn) for mRNA profiling, Jennifer Disse for PAR2 cleavage assay, Henrik Østergaard for recombinant mouse FVIIa, and Stefan Heitmeier for rivaroxaban.
Funding: This study was supported by grants from the National Institutes of Health (NHLBI, HL60742), the Humboldt Foundation of Germany (Humboldt Professorship Ruf), the Federal Ministry of Education and Research Germany (BMBF 01EO1003 and 01EO1503) and the German Research Foundation (DFG SFB 1292).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Single cell sequencing data are available through the NCBI Gene expression omnibus, accession number GSE134904. All materials and mouse strains are available through a simple academic MTA, where necessary for institutional transfer.
References
- 1.Ruf W, Disse J, Carneiro-Lobo TC, Yokota N, Schaffner F, Tissue factor and cell signalling in cancer progression and thrombosis. J. Thromb. Haemost 9 Suppl 1, 306–315 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haemmerle M, Stone RL, Menter DG, Afshar-Kharghan V, Sood AK, The Platelet Lifeline to Cancer: Challenges and Opportunities. Cancer Cell 33, 965–983 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Versteeg HH, Schaffner F, Kerver M, Petersen HH, Ahamed J, Felding-Habermann B, Takada Y, Mueller BM, Ruf W, Inhibition of tissue factor signaling suppresses tumor growth. Blood 111, 190–199 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gur-Cohen S, Itkin T, Chakrabarty S, Graf C, Kollet O, Ludin A, Golan K, Kalinkovich A, Ledergor G, Wong E, Niemeyer E, Porat Z, Erez A, Sagi I, Esmon CT, Ruf W, Lapidot T, PAR1 signaling regulates the retention and recruitment of EPCR-expressing bone marrow hematopoietic stem cells. Nature Medicine 21, 1307–1317 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antoniak S, Mackman N, Multiple roles of the coagulation protease cascade during virus infection. Blood 123, 2605–2613 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang HP, Kerschen EJ, Basu S, Hernandez I, Zogg M, Jia S, Hessner MJ, Toso R, Rezaie AR, Fernandez JA, Camire RM, Ruf W, Griffin JH, Weiler H, Coagulation factor V mediates inhibition of tissue factor signaling by activated protein C in mice. Blood 126, 2415–2423 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang HP, Kerschen EJ, Hernandez I, Basu S, Zogg M, Botros F, Jia S, Hessner MJ, Griffin JH, Ruf W, Weiler H, EPCR-dependent PAR2 activation by the blood coagulation initiation complex regulates LPS-triggered interferon responses in mice. Blood 125, 2845–2854 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nhu QM, Shirey K, Teijaro JR, Farber DL, Netzel-Arnett S, Antalis TM, Fasano A, Vogel SN, Novel signaling interactions between proteinase-activated receptor 2 and Toll-like receptors in vitro and in vivo. Mucosal. Immunol 3, 29–39 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rallabhandi P, Nhu QM, Toshchakov VY, Piao W, Medvedev AE, Hollenberg MD, Fasano A, Vogel SN, Analysis of proteinase-activated receptor 2 and TLR4 signal transduction: a novel paradigm for receptor cooperativity. J Biol. Chem 283, 24314–24325 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Badeanlou L, Furlan-Freguia C, Yang G, Ruf W, Samad F, Tissue factor-PAR2 signaling promotes diet-induced obesity and adipose inflammation. Nat. Med 17, 1490–1497 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghorpade DS, Ozcan L, Zheng Z, Nicoloro SM, Shen Y, Chen E, Bluher M, Czech MP, Tabas I, Hepatocyte-secreted DPP4 in obesity promotes adipose inflammation and insulin resistance. Nature 555, 673–677 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biswas SK, Gangi L, Paul S, Schioppa T, Saccani A, Sironi M, Bottazzi B, Doni A, Vincenzo B, Pasqualini F, Vago L, Nebuloni M, Mantovani A, Sica A, A distinct and unique transcriptional program expressed by tumor-associated macrophages (defective NF-kappaB and enhanced IRF-3/STAT1 activation). Blood 107, 2112–2122 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Schaffner F, Yokota N, Carneiro-Lobo TC, Kitano M, Schaffer M, Anderson GM, Mueller BM, Esmon CT, Ruf W, Endothelial Protein C Receptor Function in Murine and Human Breast Cancer Development. PLoS. ONE 8, e61071-. (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noy R, Pollard JW, Tumor-associated macrophages: from mechanisms to therapy. Immunity 41, 49–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perry CJ, Munoz-Rojas AR, Meeth KM, Kellman LN, Amezquita RA, Thakral D, Du VY, Wang JX, Damsky W, Kuhlmann AL, Sher JW, Bosenberg M, Miller-Jensen K, Kaech SM, Myeloid-targeted immunotherapies act in synergy to induce inflammation and antitumor immunity. J Exp Med 215, 877–893 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chevrier S, Levine JH, Zanotelli VRT, Silina K, Schulz D, Bacac M, Ries CH, Ailles L, Jewett MAS, Moch H, van den Broek M, Beisel C, Stadler MB, Gedye C, Reis B, Pe’er D, Bodenmiller B, An Immune Atlas of Clear Cell Renal Cell Carcinoma. Cell 169, 736–749 e718 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ginhoux F, Schultze JL, Murray PJ, Ochando J, Biswas SK, New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat Immunol 17, 34–40 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Colegio OR, Chu NQ, Szabo AL, Chu T, Rhebergen AM, Jairam V, Cyrus N, Brokowski CE, Eisenbarth SC, Phillips GM, Cline GW, Phillips AJ, Medzhitov R, Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 513, 559–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis CE, Harney AS, Pollard JW, The Multifaceted Role of Perivascular Macrophages in Tumors. Cancer Cell 30, 18–25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qian BZ, Pollard JW, Macrophage diversity enhances tumor progression and metastasis. Cell 141, 39–51 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P, Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol 14, 399–416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Y, Knolhoff BL, Meyer MA, Nywening TM, West BL, Luo J, Wang-Gillam A, Goedegebuure SP, Linehan DC, DeNardo DG, CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res 74, 5057–5069 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carrier M, Abou-Nassar K, Mallick R, Tagalakis V, Shivakumar S, Schattner A, Kuruvilla P, Hill D, Spadafora S, Marquis K, Trinkaus M, Tomiak A, Lee AYY, Gross PL, Lazo-Langner A, El-Maraghi R, Goss G, Le Gal G, Stewart D, Ramsay T, Rodger M, Witham D, Wells PS, Investigators A, Apixaban to Prevent Venous Thromboembolism in Patients with Cancer. N Engl J Med 380, 711–719 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Raskob GE, van Es N, Verhamme P, Carrier M, Di Nisio M, Garcia D, Grosso MA, Kakkar AK, Kovacs MJ, Mercuri MF, Meyer G, Segers A, Shi M, Wang TF, Yeo E, Zhang G, Zwicker JI, Weitz JI, Buller HR, Hokusai VTECI, Edoxaban for the Treatment of Cancer-Associated Venous Thromboembolism. N Engl J Med 378, 615–624 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Carlson TH, Simon TL, Atencio AC, In vivo behavior of human radioiodinated antithrombin III: distribution among three physiologic pools. Blood 66, 13–19 (1985). [PubMed] [Google Scholar]
- 26.Mueck W, Stampfuss J, Kubitza D, Becka M, Clinical pharmacokinetic and pharmacodynamic profile of rivaroxaban. Clin Pharmacokinet 53, 1–16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asano K, Nabeyama A, Miyake Y, Qiu CH, Kurita A, Tomura M, Kanagawa O, Fujii S, Tanaka M, CD169-positive macrophages dominate antitumor immunity by crosspresenting dead cell-associated antigens. Immunity 34, 85–95 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Bernhard CA, Ried C, Kochanek S, Brocker T, CD169+ macrophages are sufficient for priming of CTLs with specificities left out by cross-priming dendritic cells. Proc Natl Acad Sci U S A 112, 5461–5466 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shortman K, Heath WR, The CD8+ dendritic cell subset. Immunol. Rev 234, 18–31 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Magnus N, Garnier D, Rak J, Oncogenic epidermal growth factor receptor up-regulates multiple elements of the tissue factor signaling pathway in human glioma cells. Blood 116, 815–818 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Koizume S, Jin M-S, Miyagi E, Hirahara F, Nakamura Y, Piao J-H, Asai A, Yoshida A, Tsuchiya E, Ruf W, Miyagi Y, Activation of cancer cell migration and invasion by ectopic synthesis of coagulation factor VII. Cancer Res 66, 9453–9460 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Danckwardt S, Gantzert AS, Macher-Goeppinger S, Probst HC, Gentzel M, Wilm M, Grone HJ, Schirmacher P, Hentze MW, Kulozik AE, p38 MAPK controls prothrombin expression by regulated RNA 3’ end processing. Mol Cell 41, 298–310 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Schaffner F, Versteeg HH, Schillert A, Yokota N, Petersen LC, Mueller BM, Ruf W, Cooperation of tissue factor cytoplasmic domain and PAR2 signaling in breast cancer development. Blood 116, 6106–6113 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryden L, Grabau D, Schaffner F, Jonsson PE, Ruf W, Belting M, Evidence for tissue factor phosphorylation and its correlation with protease activated receptor expression and the prognosis of primary breast cancer. Int. J Cancer 126, 2330–2340 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahamed J, Belting M, Ruf W, Regulation of tissue factor-induced signaling by endogenous and recombinant tissue factor pathway inhibitor 1. Blood 105, 2384–2391 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Disse J, Petersen HH, Larsen KS, Persson E, Esmon N, Esmon CT, Teyton L, Petersen LC, Ruf W, The Endothelial Protein C Receptor Supports Tissue Factor Ternary Coagulation Initiation Complex Signaling through Protease-activated Receptors. J Biol. Chem 286, 5756–5767 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, Mazloom AR, Ma’ayan A, Chua WJ, Hansen TH, Turley SJ, Merad M, Randolph GJ, Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol 13, 1118–1128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang N, Czepielewski RS, Jarjour NN, Erlich EC, Esaulova E, Saunders BT, Grover SP, Cleuren AC, Broze GJ, Edelson BT, Mackman N, Zinselmeyer BH, Randolph GJ, Expression of factor V by resident macrophages boosts host defense in the peritoneal cavity. J Exp Med 216, 1291–1300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gundra UM, Girgis NM, Ruckerl D, Jenkins S, Ward LN, Kurtz ZD, Wiens KE, Tang MS, Basu-Roy U, Mansukhani A, Allen JE, Loke P, Alternatively activated macrophages derived from monocytes and tissue macrophages are phenotypically and functionally distinct. Blood 123, e110–e122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uhlen M, Zhang C, Lee S, Sjostedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, Sanli K, von Feilitzen K, Oksvold P, Lundberg E, Hober S, Nilsson P, Mattsson J, Schwenk JM, Brunnstrom H, Glimelius B, Sjoblom T, Edqvist PH, Djureinovic D, Micke P, Lindskog C, Mardinoglu A, Ponten F, A pathology atlas of the human cancer transcriptome. Science 357, (2017). [DOI] [PubMed] [Google Scholar]
- 41.Biswas SK, Mantovani A, Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat. Immunol 11, 889–896 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Muczynski V, Bazaa A, Loubiere C, Harel A, Cherel G, Denis CV, Lenting PJ, Christophe OD, Macrophage receptor SR-AI is crucial to maintain normal plasma levels of coagulation factor X. Blood 127, 778–786 (2016). [DOI] [PubMed] [Google Scholar]
- 43.van Dinther D, Lopez Venegas M, Veninga H, Olesek K, Hoogterp L, Revet M, Ambrosini M, Kalay H, Stockl J, van Kooyk Y, den Haan JMM, Activation of CD8(+) T Cell Responses after Melanoma Antigen Targeting to CD169(+) Antigen Presenting Cells in Mice and Humans. Cancers (Basel) 11, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang H, Liang Y, Anders RA, Taube JM, Qiu X, Mulgaonkar A, Liu X, Harrington SM, Guo J, Xin Y, Xiong Y, Nham K, Silvers W, Hao G, Sun X, Chen M, Hannan R, Qiao J, Dong H, Peng H, Fu YX, PD-L1 on host cells is essential for PD-L1 blockade-mediated tumor regression. J Clin Invest 128, 580–588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ebert J, Wilgenbus P, Teiber JF, Jurk K, Schwierczek K, Dohrmann M, Xia N, Li H, Spiecker L, Ruf W, Horke S, Paraoxonase-2 regulates coagulation activation through endothelial tissue factor. Blood 131, 2161–2172 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Larsen KS, Ostergaard H, Olsen OH, Bjelke JR, Ruf W, Petersen LC, Engineering of substrate selectivity for tissue factor-factor VIIa complex signaling through protease activated receptor 2. J Biol. Chem 285, 19959–19966 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rothmeier AS, Liu E, Chakrabarty S, Disse J, Mueller BM, Ostergaard H, Ruf W, Identification of the integrin-binding site on coagulation factor VIIa required for proangiogenic PAR2 signaling. Blood 131, 674–685 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carmona-Fontaine C, Deforet M, Akkari L, Thompson CB, Joyce JA, Xavier JB, Metabolic origins of spatial organization in the tumor microenvironment. Proc Natl Acad Sci U S A 114, 2934–2939 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Franklin RA, Liao W, Sarkar A, Kim MV, Bivona MR, Liu K, Pamer EG, Li MO, The cellular and molecular origin of tumor-associated macrophages. Science 344, 921–925 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coffelt SB, Wellenstein MD, de Visser KE, Neutrophils in cancer: neutral no more. Nat Rev Cancer 16, 431–446 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Marvel D, Gabrilovich DI, Myeloid-derived suppressor cells in the tumor microenvironment: expect the unexpected. J. Clin. Invest 125, 3356–3364 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Reilly MS, Pirie-Shepherd S, Lane WS, Folkman J, Antiangiogenic activity of the cleaved conformation of the serpin antithrombin. Science 285, 1926–1928 (1999). [DOI] [PubMed] [Google Scholar]
- 53.Ramelli G, Fuertes S, Narayan S, Busso N, Acha-Orbea H, So A, Protease-activated receptor 2 signalling promotes dendritic cell antigen transport and T-cell activation in vivo. Immunology 129, 20–27 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shrivastava S, Ma L, Tham e., McVey H, Chen D, Dorling A, Protease-activated receptor-2 signalling by tissue factor on dendritic cells suppresses antigen-specific CD4+ T-cell priming. Immunology 139, 219–226 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng RKY, Fiez-Vandal C, Schlenker O, Edman K, Aggeler B, Brown DG, Brown GA, Cooke RM, Dumelin CE, Dore AS, Geschwindner S, Grebner C, Hermansson NO, Jazayeri A, Johansson P, Leong L, Prihandoko R, Rappas M, Soutter H, Snijder A, Sundstrom L, Tehan B, Thornton P, Troast D, Wiggin G, Zhukov A, Marshall FH, Dekker N, Structural insight into allosteric modulation of protease-activated receptor 2. Nature 545, 112–115 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Geiger H, Pawar SA, Kerschen EJ, Nattamai KJ, Hernandez I, Liang HP, Fernandez JA, Cancelas JA, Ryan MA, Kustikova O, Schambach A, Fu Q, Wang J, Fink LM, Petersen KU, Zhou D, Griffin JH, Baum C, Weiler H, Hauer-Jensen M, Pharmacological targeting of the thrombomodulin-activated protein C pathway mitigates radiation toxicity. Nat. Med 18, 1123–1129 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adamiak M, Ratajczak MZ, Innate Immunity and Mobilization of Hematopoietic Stem Cells. Curr. Stem Cell Rep 3, 172–180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heissig B, Lund LR, Akiyama H, Ohki M, Morita Y, Romer J, Nakauchi H, Okumura K, Ogawa H, Werb Z, Dano K, Hattori K, The plasminogen fibrinolytic pathway is required for hematopoietic regeneration. Cell Stem Cell 1, 658–670 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ubil E, Caskey L, Holtzhausen A, Hunter D, Story C, Earp HS, Tumor-secreted Pros1 inhibits macrophage M1 polarization to reduce antitumor immune response. J Clin Invest 128, 2356–2369 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Medler TR, Murugan D, Horton W, Kumar S, Cotechini T, Forsyth AM, Leyshock P, Leitenberger JJ, Kulesz-Martin M, Margolin AA, Werb Z, Coussens LM, Complement C5a Fosters Squamous Carcinogenesis and Limits T Cell Response to Chemotherapy. Cancer Cell 34, 561–578 e566 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gertsenstein M, Nutter LM, Reid T, Pereira M, Stanford WL, Rossant J, Nagy A, Efficient generation of germ line transmitting chimeras from C57BL/6N ES cells by aggregation with outbred host embryos. PLoS One 5, e11260 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sugiura K, Stock CC, Studies in a tumor spectrum. I. Comparison of the action of methylbis (2-chloroethyl)amine and 3-bis(2-chloroethyl)aminomethyl-4-methoxymethyl −5-hydroxy-6-methylpyridine on the growth of a variety of mouse and rat tumors. Cancer 5, 382–402 (1952). [DOI] [PubMed] [Google Scholar]
- 63.Fischer EG, Riewald M, Huang HY, Miyagi Y, Kubota Y, Mueller BM, Ruf W, Tumor cell adhesion and migration supported by interaction of a receptor-protease complex with its inhibitor. J. Clin. Invest 104, 1213–1221 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Disse J, Ruf W, Endothelial protein C receptor is required for tissue factor ternary complex signaling in the mouse. J Thromb Haemost 9, 2516–2518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lun AT, Bach K, Marioni JC, Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol 17, 75 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCarthy DJ, Campbell KR, Lun AT, Wills QF, Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu C, Su Z, Identification of cell types from single-cell transcriptomes using a novel clustering method. Bioinformatics 31, 1974–1980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Expression of TF signaling components in macrophages and tumor cells.
Figure S2: Regulation of metastasis by macrophage FXa-PAR2 signaling.
Figure S3: Tumor progression alters myeloid cell composition of the TME.
Figure S4: Generation of PAR2 mutant mice.
Table S2: Sources of materials used
Table S3: Primer list for RT-PCR
Table S1: Raw data file