Abstract
Pancreatic ductal adenocarcinoma (PDAC) remains one of the most lethal solid malignancies with very few therapeutic options to treat advanced or metastatic disease. The utilization of genomic sequencing has identified therapeutically relevant alterations in ~25% of PDAC patients, most notably in the DNA damage response (DDR) genes rendering cancer cells more sensitive to DNA damaging agents, and DNA damage response inhibitors, such as PARP inhibitors. ATM is one of the most commonly mutated DDR genes, with somatic mutations identified in 2 – 18% of PDACs and germline mutations identified in 1 – 34% of patients with PDAC. ATM plays a complex role as a cell cycle checkpoint kinase, regulator of a wide array of downstream proteins, and responder to DNA damage for genome stability. The disruption of ATM signaling leads to downstream reliance on ATR and CHK1 among other DNA repair mechanisms, which may enable exploiting the inhibition of downstream proteins as therapeutic targets in ATM-mutated PDACs. In this review we will detail the function of ATM, review the current data on ATM-deficiency in PDAC, examine the therapeutic implications of ATM alterations, and explore the current clinical trials surrounding the ATM pathway.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) remains one of the most lethal solid malignancies with fewer than 10% of patients surviving 5 years. Disease incidence is increasing and PDAC is projected to be the second most common cause of cancer-related death by 2030 (1,2). The high mortality is due to the majority of patients presenting with locally advanced or metastatic disease at the time of diagnosis. Unfortunately, despite recent improvements in outcomes with newer chemotherapy regimens, the median survival remains less than one year (3,4). Comprehensive genetic analysis is being pursued to identify mutational pathways for potential treatment options, and a recent study utilizing genomic sequencing identified therapeutically relevant (highly actionable) alterations in 27% of PDAC samples (5). These findings were consistent with several other publications, all of which have demonstrated findings of actionable targets in 17– 48% of PDAC samples (6–11). One commonality to these large-scale next generation sequencing efforts is the identification of mutational defects in the genes that regulate the DNA damage response and repair (DDR) system, found in 17 – 25% of PDACs.
During the cell cycle, there is a replication of over six billion base pairs of DNA. Such genomic replication is subject to numerous insults and replication stressors, which rely on essential response and repair mechanisms to ensure DNA’s integrity. Furthermore, the chemotherapies used to treat cancers, particularly pancreatic cancer, result in specific types of DNA damage. For example, alkylating agents such as platinums, and topoisomerase inhibitors such as irinotecan cause double-strand DNA breaks (DSBs); while anti-metabolites such as 5-fluorouracil and gemcitabine cause singe base pair damage that can lead to single strand DNA breaks (12). Deficiencies in DDR mechanisms have revealed targets for therapy and research has led to FDA approved treatments targeting cancers that harbor such deficiencies. For example, BRCA1 and BRCA2 play an integral role in the maintenance of genomic integrity, and germline mutations in either gene leads to increased risks for breast, ovarian, pancreatic, and prostate cancers(13). However, the presence of a BRCA1/2 mutation also predicts for an improved response and improved overall survival with platinum-based chemotherapy in both triple negative breast cancers as well as PDAC (14,15). Exploiting this DNA repair defect not only improves sensitivity to chemotherapy, but also allows targetable therapy through the inhibition of the poly (adenosine diphosphate [ADP]–ribose) polymerase (PARP), leading to the accumulation of single strand breaks which compromise DNA double strand integrity at the replication fork. PARP inhibitors increase progression free survival in advanced BRCA1/2 mutated ovarian and breast cancer (16–18), and are now FDA approved for these diseases. Additionally, responses to PARP inhibitors are also frequently seen in BRCA mutated castrate resistant prostate cancer with, for example, a response rate of 88% in a 50 patient trial; and in BRCA mutated pancreatic cancer, with responses of 16 to 22% in small trials of 19 and 23 patients, respectively (16,19–21).
ATM also plays a critical role in DDR. The ataxia telangiectasia mutated (ATM) gene, located on chromosome 11q 22–23, was first identified in 1995 during the evaluation of the ataxia telangiectasia syndrome. Germ-line mutations of ATM result in a well-characterized syndrome, as well as an increased predisposition for breast, pancreatic, and prostate cancers (22–28). Relevant here, mutations in the ATM gene, whether germline or somatic are found in up to ~6% of PDACs (further details below), and thus may represent a more prevalent DDR mutation than BRCA1/2 (7). In this review, we will detail the function of ATM, review the current data on ATM-deficiency in PDACs, and examine the therapeutic implications of ATM alterations.
Function of ATM
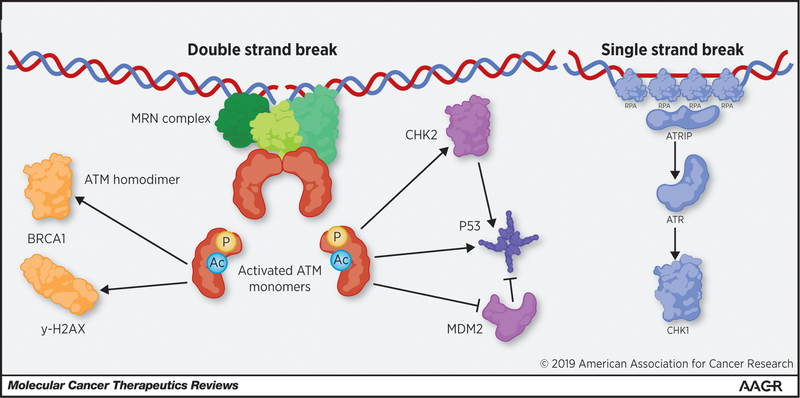
The ATM gene consists of 66 exons that encode a PI3K-related serine/threonine protein kinase that plays a central role in the response to, and ultimately the repair of DNA double-strand breaks (DSB). Structurally, this large protein (350kDa) contains serine or threonine residues susceptible for phosphorylation, followed by a glutamine amino acid located near its hydrophobic target region. Similar sites for post-translational modifications (PTMs) are found in the ataxia telangiectasia and RAD 3-related (ATR) kinase and in the DNA protein kinase (DNA-PK) proteins (29). ATM has important functions in the cell including the maintenance of: i) telomere length (30,31); and ii) the mitotic spindle structure during mitosis (32). However, this review will solely focus on the central role of ATM in the process of DDR, including as it relates to targeted therapies in cancer. As depicted in Figure 1, in order to repair damaged DNA, the MRE11-RAD50-NBS1 (MRN) complex acts as the primary sensor for DSBs and creates a physical bridge between the two broken ends (33). ATM can then interact directly with NBS1 (part of the MRN complex) through the direct binding of the C-terminus of NBS1 to several of the HEAT repeats that reside in ATM (34). It is believed that several PTMs are required for subsequent ATM activation. For instance, ATM has been shown to be activated through acetylation of K3016 by TIP60, a histone acetyltransferase that binds to ATM through recognition of the C-terminal FATC domain (35). ATM also requires autophosphorylation at S1981, which allows the kinase domain to dissociate from the FAT domain, enabling, in turn, the kinase to become active (36). These modifications allow ATM to transition from an inactive homodimer into an active monomer in response to DNA damage (36). This mechanism has been supported in the literature, but also questioned by others, demonstrating the need for further work in the field to clearly identify the role of S1981 and other ATM autophosphorylation events (29,36,37). Once activated, ATM phosphorylates multiple substrates, protein kinases, and sensor proteins in order to carry out DSB repair and also regulate normal cell cycle processes, such as apoptosis and checkpoint activation (36,38,39).
Figure 1: ATM functions and other related pathways for DNA repair.
ATM is recruited to DSBs by the MRN complex through direct interaction of NSB1 with ATM’s HEAT repeats. ATM is then activated through autophosphorylation, and acetylation by TIP60, this activation allows ATM to dissociate to the active monomeric state. ATM monomers can then signal for DNA repair through BRCA1 and γ-H2AX. ATM can also signal for cell cycle arrest and/or apoptosis through the activation of p53 through direct phosphorylation and indirect activation through CHK2 and MDM2. In parallel, ATR is recruited to long stretches of single strand DNA caused by single strand breaks, the resection of DSBs, or replication stress. PARP1 is another factor that is critical for the repair of single strand breaks.
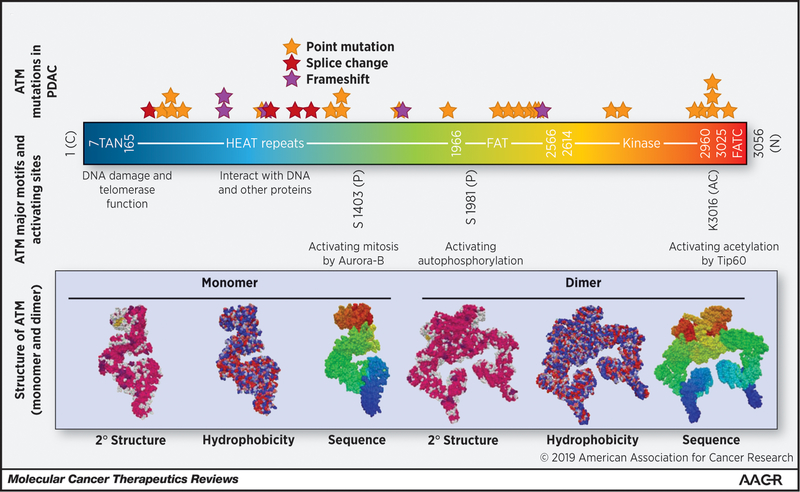
ATM plays a role in the signaling required to initiate DNA repair, and thus, ATM defects can lead to genomic instability and malignancy. Hereditary and sporadic ATM mutations span the functional domains of the entire ATM gene (Figure 2). These mutations occur mostly in the C-terminal end, which interacts with the PI3 kinase domain. This domain is involved with acetylation and activation of ATM (40). DDR is impaired when the ATM protein is dysfunctional, and loss of this DDR mechanism designed for DSBs can possibly lead, over time, to the accumulation of mutations, which can, in theory initiate the process of tumorigenesis. For instance, germline point mutations in ATM result in increased risks of breast cancers, specifically those associated with the S49C and S707P mutations. Melanoma, prostate, and oropharyngeal cancers are specifically associated with the S49C mutation. While thyroid or endocrine cancers are generally associated with the S707P mutation (41,42). Next-generation sequencing has also revealed somatic ATM mutations in many tumor types, including PDAC (Figure 2) (40).
Figure 2: ATM Structure-function domains and frequent mutations in PDAC.
ATM has several important domains that are critical for ATM function as either a monomer, dimer or both. The TAN domain is critical for telomerase function and recruitment to DSBs. This recruitment is also dependent on interactions between the ATM HEAT repeats and NBS1 (part of the MRN complex)(31,34). The FAT domain normally inhibits the kinase activity as a dimer, but after DNA damage induced autophosphorylation at S1981 and subsequent dissociation of the dimer the kinase domain becomes active (36). The FATC domain is critical for interaction with TIP60, TIP60 acetylation of ATM at K3016 is necessary for ATM activation. Mutational analysis of PDAC patients with ATM mutations from Cbio-portal (date of accession 01/21/2019) did not show significant clustering or hotspot mutations in ATM, but the number of patients was low (N=34).
Inactivating ATM Variants in Pancreatic Cancer
Multiple studies have reaffirmed the importance of TP53, KRAS, CDKN2A and SMAD4 mutations in PDAC (7,11,43,44). Inherited risks of PDAC are also well established to be in large part due to germline mutations in BRCA1/BRCA2 and CDKN2A, identified in 7.4% of familial pancreatic cancers (N=727(45)), as well as individuals affected by Lynch syndrome (mutations in the genes: MLH1, MSH2, PMS2 and MSH6) (46). Additionally, a study of familial PDAC patients showed that deleterious ATM mutations were significantly higher than the control group suggesting ATM’s role in malignancy (25). This knowledge has led to germline and somatic mutations in ATM as being identified and added to the catalog of predisposing gene mutations.
A genomic characterization of PDAC revealed 37% of the 71 samples carried alterations in DNA repair genes (6). This significant discovery has also been confirmed in a recent large PDAC profiling study that identified targetable alterations in 50% of 640 PDAC patients, of which 8.4% expressed BRCA1/2 or ATM mutations (5). A review of the International Cancer Genome Consortium, a large database of sporadic PDACs, in 2015 identified ATM mutations in 9 – 18% of PDACs, with an average of 12% of the 591 samples (47). An additional large population study which identified the prevalence of homologous recombination related gene mutations in 15.4% of PDACs (48). This study also revealed that ATM, ATRX, and CHEK2 mutations are present in 1.3% out of fifty thousand tumors samples. These mutations were most prevalently identified in PDAC (48). Further genomic studies support the high prevalence of ATM, CHEK2 and ATR mutations in PDAC (8). Our literature search, capturing 5,234 pancreatic cancer patients overall shows that the total prevalence of ATM mutations (germline or somatic) in PDAC is 6.4% (range 1 – 34%) (5,6,8,9,11,25,47–57). Importantly, in one study that showed that nearly 10% of PDAC patients carried a germline ATM mutation, in 44% of these patients a somatic second hit was identified (58). While this loss of heterozygosity (LOH) seems to occur frequently in tumors arising from patients with germline ATM mutations (e.g. as seen in breast and pancreatic cancers (59,60)), the necessity of LOH to confer therapeutic sensitivity (i.e. to platinums and DDR inhibitors, as discussed below) is uncertain.
ATM Deficiency: A Therapeutic Opportunity?
It is now well established that tumor cells with underlying defects in DDR are exquisitely sensitive to DNA-damaging agents, notably platinums, and more recently to PARP inhibitors in a phenomenon known as synthetic lethality (61). Similarly, inactivating mutations in ATM can also set up synthetic lethality in the presence of DNA damaging agents (62). Historically, ataxia telangiectasia patients were found to be profoundly radiosensitive at the chromosomal level suggesting the link between ATM and DNA repair (63). Pre-clinical data have demonstrated that knockdown of ATM results in radio-sensitization (64,65). Additional studies in the laboratory have demonstrated that ATM alterations in malignant cells can sensitize cells to platinum drugs (44,66), and outside of platinum therapy, pre-clinical studies have also demonstrated ATM suppression in p53 deficient mouse fibroblasts sensitives them to doxorubicin (67). Previous work has also demonstrated that ATM deficiency in p53 deficient cell lines causes a modest increase in 5-FU sensitivity (68). Lastly, one study took advantage of ATM-mutated PDAC cells in a mouse model and showed that treatment with the PARP inhibitor, olaparib or the ATR inhibitor, VE-822 led to dramatic accumulation of DSBs and reduced tumor cell viability in vitro and in vivo (62). The authors noted the compensation of alternate signaling routes to bypass ATM deficiency, including ATR in the replicative stress response. Thus ATR inhibition was efficient in promoting intolerable mitotic damage, an effect that was enhanced when combined with gemcitabine (62).
The clinical experience in PDAC patients with confirmed ATM, ATR or CHEK2 mutations is very limited, and focused on the efficacy of oxaliplatin-based chemotherapy. One case series (N=71) utilizing real-time whole exome sequencing demonstrated that a majority of patients with such mutations experienced a partial response or stable disease with oxaliplatin-based chemotherapy (6). In this case series, 80% of those with ATM, ATR or CHEK2 mutations were treated with an oxaliplatin-based chemotherapy and 62.5% demonstrated partial response or stable disease on first follow-up scans (6). Another small study (N=13) demonstrated a 37.5% response rate to oxaliplatin-based chemotherapy regimens in patients with DDR mutated tumors (69). There was also a significantly longer progression free survival compared to those patients whose tumors were DDR-non-mutated (20.8 months vs 1.7 months respectively p=0.049) (69). Specifically, four of thirty patients had known pathogenic ATM mutations, with at least one patient experiencing a prolonged partial response of nearly 40 months on 5-FU, irinotecan, and oxaliplatin (FOLFIRINOX) (69). There are currently multiple ongoing trials targeting ATM-deficient tumors, including and especially with PARP inhibitors (Table 1).
Table 1: ATM-relevant trials.
The table summarizes currently open clinical trials that directly or indirectly target ATM-deficient tumors. The trials were captured from a search on clinicaltrials.gov on 03/02/2019. Clinical trials that potentially accept pancreatic cancer patients are in bold type.
| DDR Target | Drug name | NCT Identifier | Phase | Study Size | ATM Status Considered | Study Title (Pancreatic cancer eligible trials are in bold) |
|---|---|---|---|---|---|---|
| ATM | AZD0156 | I | 83 | No | Study to Assess the Safety and Preliminary Efficacy of AZD0156 at Increasing Doses Alone or in Combination With Other Anti-cancer Treatment in Patients With Advanced Cancer. (AToM) | |
| AZD1390 | I | 132 | No | A Study to Assess the Safety and Tolerability of AZD1390 Given With Radiation Therapy in Patients With Brain Cancer | ||
| ATR | M6620 (VX-970) | I | 60 | No | Veliparib (ABT-888), an Oral PARP Inhibitor, and VX-970, an ATR Inhibitor, in Combination With Cisplatin in People With Refractory Solid Tumors | |
| M6620 (VX-970) | I | 51 | No | VX-970 and Irinotecan Hydrochloride in Treating Patients With Solid Tumors That Are Metastatic or Cannot Be Removed by Surgery | ||
| M6620 (VX-970) | II | 28 | Exploratory Objective | ATR Kinase Inhibitor M6620 and Irinotecan in Treating Patients With Progressive, Metastatic, or Unresectable TP53 Mutant Gastric or Gastroesophageal Junction Cancer | ||
| M6620 (VX-970) | II | 130 | No | ATR Kinase Inhibitor VX-970 and Carboplatin With or Without Docetaxel in Treating Participants With Metastatic Castration-Resistant Prostate Cancer | ||
| M6620 (VX-970) | II | 90 | No | Cisplatin and Gemcitabine Hydrochloride With or Without ATR Kinase Inhibitor M6620 in Treating Patients With Metastatic Urothelial Cancer | ||
| M6620 (VX-970) | II | 70 | No | Gemcitabine Hydrochloride Alone or With M6620 in Treating Patients With Recurrent Ovarian, Primary Peritoneal, or Fallopian Tube Cancer | ||
| M6620 (VX-970) | I | 111 | No | Carboplatin and Gemcitabine Hydrochloride With or Without ATR Kinase Inhibitor VX-970 in Treating Patients With Recurrent and Metastatic Ovarian, Primary Peritoneal, or Fallopian Tube Cancer | ||
| M6620 (VX-970) | I/II | 70 | No | Trial of Topotecan With VX-970, an ATR Kinase Inhibitor, in Small Cell Cancers Amd Extrapulmonary Small Cell Cancers | ||
| M6620 (VX-970) | I | 65 | No | M6620 Plus Standard Treatment in Oesophageal and Other Cancer (CHARIOT) | ||
| M6620 (VX-970) | I | 45 | No | M6620, Cisplatin, and Radiation Therapy in Treating Patients With Locally Advanced HPV-Negative Head and Neck Squamous Cell Carcinoma | ||
| M6620 (VX-970) | I | 46 | No | VX-970 and Whole Brain Radiation Therapy in Treating Patients With Brain Metastases From Non-small Cell Lung Cancer, Small Cell Lung Cancer, or Neuroendocrine Tumors | ||
| M6620 (VX-970) | II | 223 | Yes | M6620 (VX-970) in Selected Solid Tumors | ||
| AZD6738 | II | 68 | No | Phase II Trial of AZD6738 Alone and in Combination With Olaparib | ||
| AZD6738 | II | 86 | No | Combination ATR and PARP Inhibitor (CAPRI) Trial With AZD6738 and Olaparib in Recurrent Ovarian Cancer (CAPRI) | ||
| AZD6738 | II | 47 | No | Targeting Resistant Prostate Cancer With ATR and PARP Inhibition (TRAP Trial) | ||
| AZD6738 | I/II | 62 | No | A Study of AZD6738 and Acalabrutinib in Subjects With Relapsed or Refractory Chronic Lymphocytic Leukemia (CLL) | ||
| AZD6738 | I | 50 | No | AZD6738 & Gemcitabine as Combination Therapy (ATRiUM) | ||
| AZD6738 | Ib | 52 | No | AZD6738 for Patients With Progressive MDS or CMML | ||
| AZD6738 | I | 250 | Yes | Ascending Doses of AZD6738 in Combination With Chemotherapy and/or Novel Anti Cancer Agents | ||
| BAY1895344 | I | 219 | Yes | First-in-human Study of ATR Inhibitor BAY1895344 in Patients With Advanced Solid Tumors and Lymphomas | ||
| CHK1/2 | Prexasertib | II | 50 | Yes | A Study of LY2606368 (Prexasertib) in Patients With Solid Tumors With Replicative Stress or Homologous Repair Deficiency | |
| Prexasertib | I | 24 | No | Combination Study of Prexasertib and Olaparib in Patients With Advanced Solid Tumors | ||
| Prexasertib | I | 28 | No | A Study of Prexasertib (LY2606368), CHK1 Inhibitor, and LY3300054, PD-L1 Inhibitor, in Patients With Advanced Solid Tumors | ||
| Prexasertib | II | 153 | No | A Phase II Single Arm Pilot Study of the Chk1/2 Inhibitor (LY2606368) in BRCA1/2 Mutation Associated Breast or Ovarian Cancer, Triple Negative Breast Cancer, High Grade Serous Ovarian Cancer, and Metastatic Castrate-Resistant Prostate Cancer | ||
| Prexasertib | I | 65 | No | Prexasertib in Treating Pediatric Patients With Recurrent or Refractory Solid Tumors | ||
| Prexasertib | I | 28 | No | Prexasertib in Combination With MEC in Relapsed/Refractory AML and High Risk MDS - a Phase I Trial | ||
| SRA737 | I/II | 170 | Chk1 or ATR or other related gene | A Phase 1/2 Trial of SRA737 in Subjects With Advanced Cancer | ||
| SRA737 | I/II | 140 | Yes | A Phase 1/2 Trial of SRA737 in Combination With Gemcitabine Plus Cisplatin or Gemcitabine Alone in Subjects With Advanced Cancer | ||
| Additional Trials Targeting ATM-Deficient Tumors | ||||||
| PARP | Olaparib | II | 64 | No | OLAParib COmbinations (OLAPCO) | |
| Olaparib | Ib | 102 | Yes | Copanlisib, Olaparib, and Durvalumab in Treating Patients With Metastatic or Unresectable Solid Tumors | ||
| Olaprib | II | 28 | Yes | Olaparib Monotherapy in Relapsed Small Cell Lung Cancer Patients With HR Pathway Gene Mutations Not Limited to BRCA 1/2 Mutations, ATM Deficiency or MRE11A Mutations (SUKSES-B) | ||
| Olaparib | II | 70 | Yes | Abiraterone/Prednisone, Olaparib, or Abiraterone/Prednisone + Olaparib in Patients With Metastatic Castration-Resistant Prostate Cancer With DNA Repair Defects | ||
| Olaparib | II | 20 | Yes | Study of Olaparib in Metastatic Renal Cell Carcinoma Patients With DNA Repair Gene Mutations (ORCHID) | ||
| Olaparib | Pilot | 15 | Yes | Olaparib Before Surgery in Treating Participants With Localized Prostate Cancer | ||
| Olaparib | II | 60 | Yes | Olaparib in Treating Patients With Metastatic or Advanced Urothelial Cancer With DNA-Repair Defects | ||
| Olaparib | I/II | 427 | No | A Phase I/II Study of MEDI4736 in Combination With Olaparib in Patients With Advanced Solid Tumors. (MEDIOLA) | ||
| Talozaparib | II | 200 | Yes | Javelin BRCA/ATM: Avelumab Plus Talazoparib in Patients With BRCA or ATM Mutant Solid Tumors | ||
| Talozaparib | II | 150 | Yes | Study of the PARP Inhibitor BMN 673 in Advanced Cancer Patients With Somatic Alterations in BRCA1/2, Mutations/Deletions in PTEN or PTEN Loss, a Homologous Recombination Defect, Mutations/Deletions in Other BRCA Pathway Genes and Germline Mutation in BRCA1/2 (Not Breast or Ovarian Cancer) | ||
| Talozaparib | Ib/2 | 242 | No | Javelin Parp Medley: Avelumab Plus Talazoparib In Locally Advanced Or Metastatic Solid Tumors | ||
| Talozaparib | II | 64 | Yes | Lung-MAP: Talazoparib in Treating Patients With HRRD Positive Recurrent Stage IV Squamous Cell Lung Cancer | ||
| Talozparib | II | 58 | Yes | Phase II Talazoparib in BRCA1 +BRCA2 Wild-Type &Triple-Neg /HER2-Negative Breast Cancer /SolidTumors | ||
| Niraparib | I | 146 | Yes | Niraparib Plus Carboplatin in Patients With Homologous Recombination Deficient Advanced Solid Tumor Malignancies | ||
| Niraparib | II | 47 | Yes | A Trial of Niraparib in BAP1 and Other DNA Damage Response (DDR) Deficient Neoplasms (UF-STO-ETI-001) | ||
| Rucaparib | II | 360 | Yes | A Study of Rucaparib in Patients With Metastatic Castration-resistant Prostate Cancer and Homologous Recombination Gene Deficiency (TRITON2) | ||
Since ATM, ATR and CHK1 are all important for resolving DNA damage (Figure 1), utilizing an underlying DDR defect and inducing synthetic lethality by inhibiting an additional kinase is an innovative way to induce cancer cell death. The use of small molecule inhibitors of ATM, ATR and CHK1 are promising avenues of cancer treatment due to the malignant cells’ rapid and unregulated cell division. There are currently phase I and II clinical trials utilizing ATM or ATR inhibitors as monotherapy as well as in combination with chemotherapy (70,71).
ATM Inhibitors
The first compound described to inhibit ATM was wortmannin (72), however, there are now a host of newer, more potent compounds that inhibit ATM. One of the newer generation of ATM inhibitors published in 2004 was KU55933. This compound was shown to inhibit downstream ATM phosphorylation after radiation, and it also enhanced responses to the topoisomerase inhibitors etoposide, camptothecins, and doxorubicin (73). A similar sensitization to topoisomerase inhibitors was later demonstrated with the ATM inhibitor, AZ31 which was shown to increase the efficacy of Irinotecan in resistant tumors in PDX models (74). KU60019 is another compound that was introduced in 2009 as an improved analogue of KU55933 (75), and early work demonstrated that KU60019 is a potent radiosensitizer (75). This molecule is currently being studied in in clear cell renal cell carcinoma in combination with another known sensitizing agent, CX4945, which is an inhibitor of the protein kinase CK2 (82). The rational for this combination comes from a compound screen where CK2 and ATM inhibitors were found to be highly synergistic in renal cancer. Interestingly when CK2 inhibitors were tested in isogenic ATM proficient and deficient mouse cell lines there was little difference in downstream effectors of DNA repair such as AKT1 and BID although overall viability in the ATM proficient and deficient cell lines treated with CK2 inhibitors was not assessed (76). This brings to light an important point when comparing the efficacy of interventions performed in combination with ATM inhibitors, as compared to these same interventions performed in patients with ATM deficient tumors. The results may potentially be divergent as tumors with constitutive deficiency in ATM may have adapted to chronic loss of ATM function as opposed to acute loss as induced by ATM inhibitors. Conversely, other interventions may be both synergistic with ATM inhibitors and more potent in ATM deficient patients.
Beyond the pre-clinical explorations of ATM inhibitors, there are currently two ATM inhibitors in clinical trials which are being investigated in combination with other therapies (Table 1). AZD0156, an oral ATM inhibitor is currently in clinical trials in combination with olaparib or FOLFIRI (84). These combinations are rational since, as previously mentioned, ATM inhibitors have been shown to sensitize cells to PARP inhibitors, and also to both 5-FU and irinotecan (61,62,68). AZD1390, another oral ATM inhibitor that penetrates the blood-brain barrier is currently being tested in combination with radiation, given that radiation has been demonstrated to be more effective in ATM deficient cancers (65,77,78). Importantly in considering the potential adverse events for this trial, knockout of ATM in healthy tissue as compared to cancerous tissue was shown to induce less radiation sensitivity (78). This work demonstrated that increased sensitivity to radiation through ATM inhibition was primarily seen in cells that were rapidly replicating. As ATM inhibitors are further explored in the clinic, it will, of course, be important to monitor the side effects of ATM inhibitors particularly in combination with other therapies.
Increased sensitivity of ATM-deficient tumors to PARP inhibitors has been previously shown (64,75,79), and in a clinical trial of 124 patients it has been demonstrated that dosing with olaparib and paclitaxel was more effective at increasing overall survival in patients with less ATM activity (HR, 0.35; 80% CI, 0.22 to 0.56; P = .002; median OS, not reached v 8.2 months) (80). Unfortunately, the subsequent Phase III trial with 525 patients did not enrich for patients with ATM-deficient tumors and was a negative study (81). Nevertheless, there are multiple ongoing trials of PARP inhibitor-based therapy targeting patients, at least in part, with ATM-deficient tumors (Table 1).
ATR Inhibitors
ATR is a phosphoinositide 3-kinase-related protein kinase that primarily responds to and repairs single-strand DNA breaks. It also shares functional sequences with ATM and DNA-PK, which respond to DSBs (29,82). Upstream protein phosphorylation by ATM and autophosphorylation at the T1989 site stimulates ATR activity as well as TopBP1 which contains an ATR-activation domain to stimulate the kinase’s activity (29,83). The ATR kinase responds to a wide array of cell stressors, maintains DNA’s integrity during replication, and is essential for proliferating cell survival. In the rapidly dividing cancer cell, there exists a high degree of replicative stress, creating an environment in which, as preclinical research has shown, suppression of ATR activity further increases replication stress leading to cell death (84). Furthermore, while normal dividing cells utilize ATM dependent pathways for assistance in DNA repair, cancer cells, which are often deficient in ATM/p53 signaling, may rely solely on the ATR pathway for survival (67,85,86). This was demonstrated in genetically engineered mouse models of cancer, in which 90% genetic reduction of ATR expression suppressed the development of fibrosarcomas and acute myeloid leukemias with minimal side effects in normal tissues. This work affirmed the tumor selectivity of ATR inhibition (84). Moreover, inhibition of ATR selectively sensitizes tumor cells, but not normal cells, to radiation and chemotherapy (87).
Thus, small molecule inhibitors of ATR may be particularly potent in PDACs with somatic mutations in ATM given that the lack of ATM’s function may lead to increased dependence on ATR, and ATR inhibition could thus significantly promote cancer cell death. The ATR inhibitor, VE-821, sensitizes cancer cells but not normal cells to chemotherapy (87), and these effects were synergistic in ATM deficient cells (87). Another ATR inhibitor, AZD6738, causes accumulation of DNA damage, S phase arrest, and apoptosis in ATM dysfunctional gastric cells while not affecting those with functional ATM (88). Similar pre-clinical studies also suggest synthetic lethality between ATR inhibition with VE-822 and ATM deficiency in PDAC as well as lung adenocarcinoma cell lines, reaffirming the actionable molecular dependencies on ATR (62,79). VE-822 has also been shown to potentially synergize with cisplatin in ATM-deficient esophageal squamous cells (89). This effect of potentiating the cytotoxicity of cisplatin and gemcitabine is also seen with AZD6738 in ATM deficient non-small cell lung cancer cells (90). Several ongoing trials of ATR inhibitor-based therapies are listed in Table 1.
CHK1 and CHK2 Inhibitors
Downstream to and activated by ATR is the checkpoint kinase 1 (CHK1) pathway. CHK1 promotes proteasomal degradation of CDC25A in response to genome stress (29). The combined activitry of ATR, CHK1, and CDC25A results in cell cycle arrest and stabilization of replication stress at DNA forks. The inhibition of this complex leads to a decreased rate of fork progression, massive fork collapse in S phase cells and ultimately cell death (82). Preclinical studies utilizing the CHK1 inhibitors, MK8776 and LY2603618, with gemcitabine-based chemoradiation showed synergistic effects to induce apoptosis of PDAC cells (91,92). The combination of gemcitabine, a CHK1 inhibitor, PF-477736, and Lutetium-177–labeled anti-EGFR antibody lead to extensive DNA damage, apoptosis, and tumor degeneration in patient-derived xenografts (93). Additional preclinical studies with a tumor stem cell marker Doublecortin-like kinase 1 (Dclk1) inhibitor LRRK2-IN-1 (LRRK) showed decreased expression of phosphorylated Chk1 (94). This same study demonstrated the combination of gemcitabine with LRRK significantly reduced cell survival compared treatment with gemcitabine alone (94). Thus, CHK1 and Dclk1 are both potential targets in ATM-deficient malignancies as they also play a large role in single-strand break DNA repair.
CHK1 activation is primarily dependent on ATR at stalled replication forks and single-strand DNA, whereas CHK2 is mainly activated by ATM induced by DNA DSBs. One preclinical study examined the antitumor effects of a CHK2 inhibitor, NSC109555, in combination with gemcitabine. This combination increased apoptosis in pancreatic cells (95). Clinical trials of CHK1/2 inhibitors are also listed in Table 1.
Conclusions
There are currently no FDA approved targeted therapies for patients with pancreatic cancer. However, genomic profiling of pancreatic adenocarcinomas is revealing therapeutically relevant alterations, and 17 – 25% of pancreatic cancers harbor mutations in the DDR pathway. Therapy targeted towards inhibiting the DNA damage response, including with PARP inhibitors is proving to be highly effective particularly in DDR deficient cancers, as has been established in BRCA1/2 mutated cancers. However, DNA damage response is a highly complicated process, involving several overlapping pathways. It is reasonable to hypothesize that, depending upon the specific DDR mutation, there may be different optimal therapies to be utilized. PARP inhibitors are showing early promise in PDACs that harbor BRCA1/2 or PALB2 mutations, but consistently the most common DDR gene mutated in PDAC is actually ATM. It will be critical in the coming years to explore what DDR-targeted therapies might work best in ATM-deficient tumors. As with any therapeutic breakthrough, the future exploration of the complexity of the DDR pathway also justifies the need for a better understanding of compensatory and resistance mechanisms that may arise in the setting of ATM/ATR/CHK1 targeted therapies.
ATM-deficiency may provide sensitivity for other elements of conventional therapies for PDAC including radiation (96) and oxaliplatin. However, emerging targeted strategies, including immunotherapeutic combination approaches (97) will likely provide even better matches for ATM-deficient tumors. For example, mechanistically, it seems reasonable to consider that PARP inhibitors may be effective in treating ATM-deficient tumors. But potentially more promising might be the combination of a PARP inhibitor with an ATR inhibitor in ATM deficient PDAC – essentially exploiting a new node of synthetic lethality in the DNA damage response pathway. Similarly, there is a mechanistic reason to explore the role of CHK1 inhibition in ATM-deficient tumors. In both cases, understanding the need for inducing DNA damage with DNA damaging chemotherapy will be critical as well. There are several ongoing clinical trials as discussed above, but clinical trials in PDAC in which there is such a high unmet need, and where ATM deficiency is common, would be ideal.
Additionally, recent genetic studies have revealed that specific ATM genotypes correlate to susceptibility to different diseases including cancer, which may provide valuable clinical information with regards to early detection, the subtyping of, and the treatment of PDACs (98). These genetic studies may complement and/or be evaluated in published genetically engineered mouse models (47,62,99) that have identified ATM’s various roles (i.e., EMT, genetic instability, and metastases) in the progression model of PDAC. Moving forward, the research community should evaluate novel agents and combination therapies discussed above in these isogenic, in vivo models with the ultimate aim of classifying each ATM pathogenic genotype observed in patients with an optimally tailored, matched targeted therapeutic strategy.
Acknowledgments
Funding Sources:
J.R. Brody and M.J. Pishvaian are supported by an NIH-NCI R01 CA212600 grant and J.R. Brody is also supported by a NIH-NCI P30CA056036 core grant given to K. Knudsen. Research supported by the 2015 Pancreatic Cancer Action Network-AACR Research Acceleration Network Grant, Grant Number 15–90-25-BROD to J.R. Brody and M.J. Pishvaian.
Footnotes
Authors’ disclosures of potential conflicts of interest:
Samantha Armstrong: None
Christopher Shultz: None
Ariana Azimi-Sadjadi: None
Jonathan R. Brody: Scientific Advisory Board for Perthera
Michael J. Pishvaian: None directly related to this work. Dr. Pishvaian does have the following conflicts to report: Speaker/Consultant: AstraZeneca/MedImmune, Caris Life Sciences, Celgene, Merrimack, Perthera, RenovoRx and Sirtex Medical. Travel, Accommodations and Expenses Support: AstraZeneca/MedImmune, Caris Life Sciences, Perthera and Sirtex Medical. Stock: Perthera. Research Funding to my Institution: ARMO BioSciences, Bavarian Nordic, Bayer, Bristol-Myers Squibb, Calithera Biosciences, Celgene, Celldex, Curegenix, Fibrogen, Genentech, Gilead Sciences, GlaxoSmithKline, Halozyme, Karyopharm Therapeutics, MedImmune, Merck, Novartis, Regeneron, Pfizer, Pharmacyclics and Tesaro.
References
- 1.Quante AS, Ming C, Rottmann M, Engel J, Boeck S, Heinemann V, et al. Projections of cancer incidence and cancer-related deaths in Germany by 2020 and 2030. Cancer Med 2016;5(9):2649–56 doi 10.1002/cam4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014;74(11):2913–21 doi 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 3.Conroy T, Desseigne F, Ychou M, Bouche O, Guimbaud R, Becouarn Y, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med 2011;364(19):1817–25 doi 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 4.Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med 2013;369(18):1691–703 doi 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pishvaian MJ, Bender RJ, Halverson D, Rahib L, Hendifar AE, Mikhail S, et al. Molecular Profiling of Patients with Pancreatic Cancer: Initial Results from the Know Your Tumor Initiative. Clin Cancer Res 2018;24(20):5018–27 doi 10.1158/1078-0432.CCR-18-0531. [DOI] [PubMed] [Google Scholar]
- 6.Aguirre AJ, Nowak JA, Camarda ND, Moffitt RA, Ghazani AA, Hazar-Rethinam M, et al. Real-time Genomic Characterization of Advanced Pancreatic Cancer to Enable Precision Medicine. Cancer Discov 2018;8(9):1096–111 doi 10.1158/2159-8290.CD-18-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531(7592):47–52 doi 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 8.Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 2015;6:6744 doi 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lowery MA, Jordan EJ, Basturk O, Ptashkin RN, Zehir A, Berger MF, et al. Real-Time Genomic Profiling of Pancreatic Ductal Adenocarcinoma: Potential Actionability and Correlation with Clinical Phenotype. Clin Cancer Res 2017;23(20):6094–100 doi 10.1158/1078-0432.CCR-17-0899. [DOI] [PubMed] [Google Scholar]
- 10.Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet 2015;47(10):1168–78 doi 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012;491(7424):399–405 doi 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer 2008;8(3):193–204 doi 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 13.Chen CC, Feng W, Lim PX, Kass EM, Jasin M. Homology-Directed Repair and the Role of BRCA1, BRCA2, and Related Proteins in Genome Integrity and Cancer. Annu Rev Cancer Biol 2018;2:313–36 doi 10.1146/annurev-cancerbio-030617-050502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tutt A, Tovey H, Cheang MCU, Kernaghan S, Kilburn L, Gazinska P, et al. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TNT Trial. Nat Med 2018;24(5):628–37 doi 10.1038/s41591-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golan T, Kanji ZS, Epelbaum R, Devaud N, Dagan E, Holter S, et al. Overall survival and clinical characteristics of pancreatic cancer in BRCA mutation carriers. Br J Cancer 2014;111(6):1132–8 doi 10.1038/bjc.2014.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaufman B, Shapira-Frommer R, Schmutzler RK, Audeh MW, Friedlander M, Balmana J, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol 2015;33(3):244–50 doi 10.1200/JCO.2014.56.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N Engl J Med 2017;377(6):523–33 doi 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 18.Litton JK, Rugo HS, Ettl J, Hurvitz SA, Goncalves A, Lee KH, et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N Engl J Med 2018;379(8):753–63 doi 10.1056/NEJMoa1802905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med 2015;373(18):1697–708 doi 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domchek SM, Aghajanian C, Shapira-Frommer R, Schmutzler RK, Audeh MW, Friedlander M, et al. Efficacy and safety of olaparib monotherapy in germline BRCA1/2 mutation carriers with advanced ovarian cancer and three or more lines of prior therapy. Gynecol Oncol 2016;140(2):199–203 doi 10.1016/j.ygyno.2015.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shroff RT, Hendifar A, McWilliams RR, Geva R, Epelbaum R, Rolfe L, et al. Rucaparib Monotherapy in Patients With Pancreatic Cancer and a Known Deleterious BRCA Mutation. JCO Precis Oncol 2018;2018 doi 10.1200/PO.17.00316. [DOI] [PMC free article] [PubMed]
- 22.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 1995;268(5218):1749–53. [DOI] [PubMed] [Google Scholar]
- 23.Antoniou AC, Easton DF. Models of genetic susceptibility to breast cancer. Oncogene 2006;25(43):5898–905 doi 10.1038/sj.onc.1209879. [DOI] [PubMed] [Google Scholar]
- 24.Cavaciuti E, Lauge A, Janin N, Ossian K, Hall J, Stoppa-Lyonnet D, et al. Cancer risk according to type and location of ATM mutation in ataxia-telangiectasia families. Genes Chromosomes Cancer 2005;42(1):1–9 doi 10.1002/gcc.20101. [DOI] [PubMed] [Google Scholar]
- 25.Roberts NJ, Jiao Y, Yu J, Kopelovich L, Petersen GM, Bondy ML, et al. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov 2012;2(1):41–6 doi 10.1158/2159-8290.CD-11-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grant RC, Al-Sukhni W, Borgida AE, Holter S, Kanji ZS, McPherson T, et al. Exome sequencing identifies nonsegregating nonsense ATM and PALB2 variants in familial pancreatic cancer. Hum Genomics 2013;7:11 doi 10.1186/1479-7364-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N Engl J Med 2016;375(5):443–53 doi 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Angele S, Falconer A, Edwards SM, Dork T, Bremer M, Moullan N, et al. ATM polymorphisms as risk factors for prostate cancer development. Br J Cancer 2004;91(4):783–7 doi 10.1038/sj.bjc.6602007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol Cell 2017;66(6):801–17 doi 10.1016/j.molcel.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 30.Lee SS, Bohrson C, Pike AM, Wheelan SJ, Greider CW. ATM Kinase Is Required for Telomere Elongation in Mouse and Human Cells. Cell Rep 2015;13(8):1623–32 doi 10.1016/j.celrep.2015.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seidel JJ, Anderson CM, Blackburn EH. A novel Tel1/ATM N-terminal motif, TAN, is essential for telomere length maintenance and a DNA damage response. Mol Cell Biol 2008;28(18):5736–46 doi 10.1128/MCB.00326-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palazzo L, Della Monica R, Visconti R, Costanzo V, Grieco D. ATM controls proper mitotic spindle structure. Cell Cycle 2014;13(7):1091–100 doi 10.4161/cc.27945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR 3rd, et al. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 1998;93(3):477–86. [DOI] [PubMed] [Google Scholar]
- 34.You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol 2005;25(13):5363–79 doi 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Y, Xu Y, Roy K, Price BD. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol Cell Biol 2007;27(24):8502–9 doi 10.1128/MCB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 2003;421(6922):499–506 doi 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 37.Lau WC, Li Y, Liu Z, Gao Y, Zhang Q, Huen MS. Structure of the human dimeric ATM kinase. Cell Cycle 2016;15(8):1117–24 doi 10.1080/15384101.2016.1158362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lavin MF, Kozlov S. ATM activation and DNA damage response. Cell Cycle 2007;6(8):931–42 doi 10.4161/cc.6.8.4180. [DOI] [PubMed] [Google Scholar]
- 39.Mochan TA, Venere M, DiTullio RA Jr., Halazonetis TD. 53BP1 and NFBD1/MDC1-Nbs1 function in parallel interacting pathways activating ataxia-telangiectasia mutated (ATM) in response to DNA damage. Cancer Res 2003;63(24):8586–91. [PubMed] [Google Scholar]
- 40.Choi M, Kipps T, Kurzrock R. ATM Mutations in Cancer: Therapeutic Implications. Mol Cancer Ther 2016;15(8):1781–91 doi 10.1158/1535-7163.MCT-15-0945. [DOI] [PubMed] [Google Scholar]
- 41.Stredrick DL, Garcia-Closas M, Pineda MA, Bhatti P, Alexander BH, Doody MM, et al. The ATM missense mutation p.Ser49Cys (c.146C>G) and the risk of breast cancer. Hum Mutat 2006;27(6):538–44 doi 10.1002/humu.20323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dombernowsky SL, Weischer M, Allin KH, Bojesen SE, Tybjaerg-Hansen A, Nordestgaard BG. Risk of cancer by ATM missense mutations in the general population. J Clin Oncol 2008;26(18):3057–62 doi 10.1200/JCO.2007.14.6613. [DOI] [PubMed] [Google Scholar]
- 43.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008;321(5897):1801–6 doi 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015;518(7540):495–501 doi 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhen DB, Rabe KG, Gallinger S, Syngal S, Schwartz AG, Goggins MG, et al. BRCA1, BRCA2, PALB2, and CDKN2A mutations in familial pancreatic cancer: a PACGENE study. Genet Med 2015;17(7):569–77 doi 10.1038/gim.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kastrinos F, Mukherjee B, Tayob N, Wang F, Sparr J, Raymond VM, et al. Risk of pancreatic cancer in families with Lynch syndrome. JAMA 2009;302(16):1790–5 doi 10.1001/jama.2009.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russell R, Perkhofer L, Liebau S, Lin Q, Lechel A, Feld FM, et al. Loss of ATM accelerates pancreatic cancer formation and epithelial-mesenchymal transition. Nat Commun 2015;6:7677 doi 10.1038/ncomms8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heeke AL, Pishvaian MJ, Lynce F, Xiu J, Brody JR, Chen WJ, et al. Prevalence of Homologous Recombination-Related Gene Mutations Across Multiple Cancer Types. JCO Precis Oncol 2018;2018 doi 10.1200/PO.17.00286. [DOI] [PMC free article] [PubMed]
- 49.Murphy SJ, Hart SN, Lima JF, Kipp BR, Klebig M, Winters JL, et al. Genetic alterations associated with progression from pancreatic intraepithelial neoplasia to invasive pancreatic tumor. Gastroenterology 2013;145(5):1098–109 e1 doi 10.1053/j.gastro.2013.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim H, Saka B, Knight S, Borges M, Childs E, Klein A, et al. Having pancreatic cancer with tumoral loss of ATM and normal TP53 protein expression is associated with a poorer prognosis. Clin Cancer Res 2014;20(7):1865–72 doi 10.1158/1078-0432.CCR-13-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grant RC, Selander I, Connor AA, Selvarajah S, Borgida A, Briollais L, et al. Prevalence of germline mutations in cancer predisposition genes in patients with pancreatic cancer. Gastroenterology 2015;148(3):556–64 doi 10.1053/j.gastro.2014.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Connor AA, Denroche RE, Jang GH, Timms L, Kalimuthu SN, Selander I, et al. Association of Distinct Mutational Signatures With Correlates of Increased Immune Activity in Pancreatic Ductal Adenocarcinoma. JAMA Oncol 2017;3(6):774–83 doi 10.1001/jamaoncol.2016.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shahda STK, Ibrahim A, et al. Homologous recombination deficiency (HRD) in patients with pancreatic cancer (PS) and response to chemotherapy American Society of Clinical Oncology Gastrointestinal Cancer Symposium, San Francisco, CA, January 21–23, 2016 (abstr 317) 2016. [Google Scholar]
- 54.Shindo K, Yu J, Suenaga M, Fesharakizadeh S, Cho C, Macgregor-Das A, et al. Deleterious Germline Mutations in Patients With Apparently Sporadic Pancreatic Adenocarcinoma. J Clin Oncol 2017;35(30):3382–90 doi 10.1200/JCO.2017.72.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu C, Hart SN, Polley EC, Gnanaolivu R, Shimelis H, Lee KY, et al. Association Between Inherited Germline Mutations in Cancer Predisposition Genes and Risk of Pancreatic Cancer. JAMA 2018;319(23):2401–9 doi 10.1001/jama.2018.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ohmoto A, Morizane C, Kubo E, Takai E, Hosoi H, Sakamoto Y, et al. Germline variants in pancreatic cancer patients with a personal or family history of cancer fulfilling the revised Bethesda guidelines. J Gastroenterol 2018;53(10):1159–67 doi 10.1007/s00535-018-1466-y. [DOI] [PubMed] [Google Scholar]
- 57.Chaffee KG, Oberg AL, McWilliams RR, Majithia N, Allen BA, Kidd J, et al. Prevalence of germ-line mutations in cancer genes among pancreatic cancer patients with a positive family history. Genet Med 2018;20(1):119–27 doi 10.1038/gim.2017.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yurgelun MB, Chittenden AB, Morales-Oyarvide V, Rubinson DA, Dunne RF, Kozak MM, et al. Germline cancer susceptibility gene variants, somatic second hits, and survival outcomes in patients with resected pancreatic cancer. Genet Med 2018. doi 10.1038/s41436-018-0009-5. [DOI] [PMC free article] [PubMed]
- 59.Weigelt B, Bi R, Kumar R, Blecua P, Mandelker DL, Geyer FC, et al. The Landscape of Somatic Genetic Alterations in Breast Cancers From ATM Germline Mutation Carriers. J Natl Cancer Inst 2018;110(9):1030–4 doi 10.1093/jnci/djy028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lowery MA, Wong W, Jordan EJ, Lee JW, Kemel Y, Vijai J, et al. Prospective Evaluation of Germline Alterations in Patients With Exocrine Pancreatic Neoplasms. J Natl Cancer Inst 2018;110(10):1067–74 doi 10.1093/jnci/djy024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017;355(6330):1152–8 doi 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perkhofer L, Schmitt A, Romero Carrasco MC, Ihle M, Hampp S, Ruess DA, et al. ATM Deficiency Generating Genomic Instability Sensitizes Pancreatic Ductal Adenocarcinoma Cells to Therapy-Induced DNA Damage. Cancer Res 2017;77(20):5576–90 doi 10.1158/0008-5472.CAN-17-0634. [DOI] [PubMed] [Google Scholar]
- 63.Taylor AM, Metcalfe JA, Oxford JM, Harnden DG. Is chromatid-type damage in ataxia telangiectasia after irradiation at G0 a consequence of defective repair? Nature 1976;260(5550):441–3. [DOI] [PubMed] [Google Scholar]
- 64.Wang L, et al. Abstract A93: Radiation-Induced Phosphorylation of ATDC via ATM/MAPKAP Kinase 2 Signaling Mediates Radioresistance of Pancreatic Cancer Cells. Cancer Research, vol 72, no 14 Supplement, 2012, doi:101158/1538–7445panca2012-a93 2012. [Google Scholar]
- 65.Ayars M, Eshleman J, Goggins M. Susceptibility of ATM-deficient pancreatic cancer cells to radiation. Cell Cycle 2017;16(10):991–8 doi 10.1080/15384101.2017.1312236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pennington KP, Walsh T, Harrell MI, Lee MK, Pennil CC, Rendi MH, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res 2014;20(3):764–75 doi 10.1158/1078-0432.CCR-13-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiang H, Reinhardt HC, Bartkova J, Tommiska J, Blomqvist C, Nevanlinna H, et al. The combined status of ATM and p53 link tumor development with therapeutic response. Genes Dev 2009;23(16):1895–909 doi 10.1101/gad.1815309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fedier A, Schlamminger M, Schwarz VA, Haller U, Howell SB, Fink D. Loss of atm sensitises p53-deficient cells to topoisomerase poisons and antimetabolites. Ann Oncol 2003;14(6):938–45. [DOI] [PubMed] [Google Scholar]
- 69.Kondo T, Kanai M, Kou T, Sakuma T, Mochizuki H, Kamada M, et al. Association between homologous recombination repair gene mutations and response to oxaliplatin in pancreatic cancer. Oncotarget 2018;9(28):19817–25 doi 10.18632/oncotarget.24865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O’Connor MJ. Targeting the DNA Damage Response in Cancer. Mol Cell 2015;60(4):547–60 doi 10.1016/j.molcel.2015.10.040. [DOI] [PubMed] [Google Scholar]
- 71.Brown JS, O’Carrigan B, Jackson SP, Yap TA. Targeting DNA Repair in Cancer: Beyond PARP Inhibitors. Cancer Discov 2017;7(1):20–37 doi 10.1158/2159-8290.CD-16-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sarkaria JN, Tibbetts RS, Busby EC, Kennedy AP, Hill DE, Abraham RT. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res 1998;58(19):4375–82. [PubMed] [Google Scholar]
- 73.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res 2004;64(24):9152–9 doi 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 74.Greene J, Nguyen A, Bagby SM, Jones GN, Tai WM, Quackenbush KS, et al. The novel ATM inhibitor (AZ31) enhances antitumor activity in patient derived xenografts that are resistant to irinotecan monotherapy. Oncotarget 2017;8(67):110904–13 doi 10.18632/oncotarget.22920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Golding SE, Rosenberg E, Valerie N, Hussaini I, Frigerio M, Cockcroft XF, et al. Improved ATM kinase inhibitor KU-60019 radiosensitizes glioma cells, compromises insulin, AKT and ERK prosurvival signaling, and inhibits migration and invasion. Mol Cancer Ther 2009;8(10):2894–902 doi 10.1158/1535-7163.MCT-09-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Olsen BB, Fritz G, Issinger OG. Characterization of ATM and DNA-PK wild-type and mutant cell lines upon DSB induction in the presence and absence of CK2 inhibitors. Int J Oncol 2012;40(2):592–8 doi 10.3892/ijo.2011.1227. [DOI] [PubMed] [Google Scholar]
- 77.Neubauer S, Arutyunyan R, Stumm M, Dork T, Bendix R, Bremer M, et al. Radiosensitivity of ataxia telangiectasia and Nijmegen breakage syndrome homozygotes and heterozygotes as determined by three-color FISH chromosome painting. Radiat Res 2002;157(3):312–21. [DOI] [PubMed] [Google Scholar]
- 78.Moding EJ, Lee CL, Castle KD, Oh P, Mao L, Zha S, et al. Atm deletion with dual recombinase technology preferentially radiosensitizes tumor endothelium. J Clin Invest 2014;124(8):3325–38 doi 10.1172/JCI73932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schmitt A, Knittel G, Welcker D, Yang TP, George J, Nowak M, et al. ATM Deficiency Is Associated with Sensitivity to PARP1- and ATR Inhibitors in Lung Adenocarcinoma. Cancer Res 2017;77(11):3040–56 doi 10.1158/0008-5472.CAN-16-3398. [DOI] [PubMed] [Google Scholar]
- 80.Bang YJ, Im SA, Lee KW, Cho JY, Song EK, Lee KH, et al. Randomized, Double-Blind Phase II Trial With Prospective Classification by ATM Protein Level to Evaluate the Efficacy and Tolerability of Olaparib Plus Paclitaxel in Patients With Recurrent or Metastatic Gastric Cancer. J Clin Oncol 2015;33(33):3858–65 doi 10.1200/JCO.2014.60.0320. [DOI] [PubMed] [Google Scholar]
- 81.Bang YJ, Xu RH, Chin K, Lee KW, Park SH, Rha SY, et al. Olaparib in combination with paclitaxel in patients with advanced gastric cancer who have progressed following first-line therapy (GOLD): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol 2017;18(12):1637–51 doi 10.1016/S1470-2045(17)30682-4. [DOI] [PubMed] [Google Scholar]
- 82.Saldivar JC, Cortez D, Cimprich KA. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat Rev Mol Cell Biol 2017;18(10):622–36 doi 10.1038/nrm.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yoo HY, Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. Ataxia-telangiectasia mutated (ATM)-dependent activation of ATR occurs through phosphorylation of TopBP1 by ATM. J Biol Chem 2007;282(24):17501–6 doi 10.1074/jbc.M701770200. [DOI] [PubMed] [Google Scholar]
- 84.Schoppy DW, Ragland RL, Gilad O, Shastri N, Peters AA, Murga M, et al. Oncogenic stress sensitizes murine cancers to hypomorphic suppression of ATR. J Clin Invest 2012;122(1):241–52 doi 10.1172/JCI58928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009;461(7267):1071–8 doi 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bolt J, Vo QN, Kim WJ, McWhorter AJ, Thomson J, Hagensee ME, et al. The ATM/p53 pathway is commonly targeted for inactivation in squamous cell carcinoma of the head and neck (SCCHN) by multiple molecular mechanisms. Oral Oncol 2005;41(10):1013–20 doi 10.1016/j.oraloncology.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 87.Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA, et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol 2011;7(7):428–30 doi 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 88.Min A, Im SA, Jang H, Kim S, Lee M, Kim DK, et al. AZD6738, A Novel Oral Inhibitor of ATR, Induces Synthetic Lethality with ATM Deficiency in Gastric Cancer Cells. Mol Cancer Ther 2017;16(4):566–77 doi 10.1158/1535-7163.MCT-16-0378. [DOI] [PubMed] [Google Scholar]
- 89.Shi Q, Shen LY, Dong B, Fu H, Kang XZ, Yang YB, et al. The identification of the ATR inhibitor VE-822 as a therapeutic strategy for enhancing cisplatin chemosensitivity in esophageal squamous cell carcinoma. Cancer Lett 2018;432:56–68 doi 10.1016/j.canlet.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 90.Vendetti FP, Lau A, Schamus S, Conrads TP, O’Connor MJ, Bakkenist CJ. The orally active and bioavailable ATR kinase inhibitor AZD6738 potentiates the anti-tumor effects of cisplatin to resolve ATM-deficient non-small cell lung cancer in vivo. Oncotarget 2015;6(42):44289–305 doi 10.18632/oncotarget.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Engelke CG, Parsels LA, Qian Y, Zhang Q, Karnak D, Robertson JR, et al. Sensitization of pancreatic cancer to chemoradiation by the Chk1 inhibitor MK8776. Clin Cancer Res 2013;19(16):4412–21 doi 10.1158/1078-0432.CCR-12-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liang M, Zhao T, Ma L, Guo Y. CHK1 inhibition sensitizes pancreatic cancer cells to gemcitabine via promoting CDK-dependent DNA damage and ribonucleotide reductase downregulation. Oncol Rep 2018;39(3):1322–30 doi 10.3892/or.2017.6168. [DOI] [PubMed] [Google Scholar]
- 93.Al-Ejeh F, Pajic M, Shi W, Kalimutho M, Miranda M, Nagrial AM, et al. Gemcitabine and CHK1 inhibition potentiate EGFR-directed radioimmunotherapy against pancreatic ductal adenocarcinoma. Clin Cancer Res 2014;20(12):3187–97 doi 10.1158/1078-0432.CCR-14-0048. [DOI] [PubMed] [Google Scholar]
- 94.Kawamura D, Takemoto Y, Nishimoto A, Ueno K, Hosoyama T, Shirasawa B, et al. Enhancement of cytotoxic effects of gemcitabine by Dclk1 inhibition through suppression of Chk1 phosphorylation in human pancreatic cancer cells. Oncol Rep 2017;38(5):3238–44 doi 10.3892/or.2017.5974. [DOI] [PubMed] [Google Scholar]
- 95.Duong HQ, Hong YB, Kim JS, Lee HS, Yi YW, Kim YJ, et al. Inhibition of checkpoint kinase 2 (CHK2) enhances sensitivity of pancreatic adenocarcinoma cells to gemcitabine. J Cell Mol Med 2013;17(10):1261–70 doi 10.1111/jcmm.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hallahan D ATM Inhibition Sensitizes Tumors to High-Dose Irradiation. Cancer Res 2019;79(4):704–5 doi 10.1158/0008-5472.CAN-18-4072. [DOI] [PubMed] [Google Scholar]
- 97.Sen T, Rodriguez BL, Chen L, Della Corte C, Morikawa N, Fujimoto J, et al. Targeting DNA damage response promotes anti-tumor immunity through STING-mediated T-cell activation in small cell lung cancer. Cancer Discov 2019. doi 10.1158/2159-8290.CD-18-1020. [DOI] [PMC free article] [PubMed]
- 98.van Os NJH, Chessa L, Weemaes CMR, van Deuren M, Fievet A, van Gaalen J, et al. Genotype-phenotype correlations in ataxia telangiectasia patients with ATM c.3576G>A and c.8147T>C mutations. J Med Genet 2019. doi 10.1136/jmedgenet-2018-105635. [DOI] [PubMed]
- 99.Drosos Y, Escobar D, Chiang MY, Roys K, Valentine V, Valentine MB, et al. ATM-deficiency increases genomic instability and metastatic potential in a mouse model of pancreatic cancer. Sci Rep 2017;7(1):11144 doi 10.1038/s41598-017-11661-8. [DOI] [PMC free article] [PubMed] [Google Scholar]