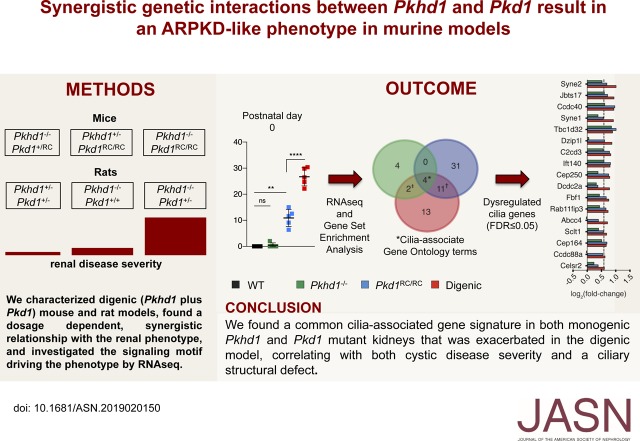
Significance Statement
The lack of rapidly progressive murine models reflecting the more severe end of the spectrum of autosomal recessive polycystic kidney disease (ARPKD) inhibits progress to understanding ARPKD pathogenesis. Defects in primary cilia have been implicated in polycystic kidney disease, but their potential role is poorly understood. The authors generated and characterized new mouse and rat models of ARPKD and autosomal dominant polycystic kidney disease (ADPKD) and investigated the interaction between causative genes for these two conditions. Their digenic models demonstrated a synergistic interaction that better reflects the early-onset disease characteristic of ARPKD. Analysis of mRNA expression in the models highlighted different disrupted pathways, but with a commonality of dysregulated mechanisms associated with primary cilia. These models may improve understanding of ARPKD and preclinical testing for this disease.
Keywords: ADPKD, polycystic kidney disease, genetics and development
Visual Abstract
Abstract
Background
Autosomal recessive polycystic kidney disease (ARPKD) and autosomal dominant polycystic kidney disease (ADPKD) are genetically distinct, with ADPKD usually caused by the genes PKD1 or PKD2 (encoding polycystin-1 and polycystin-2, respectively) and ARPKD caused by PKHD1 (encoding fibrocystin/polyductin [FPC]). Primary cilia have been considered central to PKD pathogenesis due to protein localization and common cystic phenotypes in syndromic ciliopathies, but their relevance is questioned in the simple PKDs. ARPKD’s mild phenotype in murine models versus in humans has hampered investigating its pathogenesis.
Methods
To study the interaction between Pkhd1 and Pkd1, including dosage effects on the phenotype, we generated digenic mouse and rat models and characterized and compared digenic, monogenic, and wild-type phenotypes.
Results
The genetic interaction was synergistic in both species, with digenic animals exhibiting phenotypes of rapidly progressive PKD and early lethality resembling classic ARPKD. Genetic interaction between Pkhd1 and Pkd1 depended on dosage in the digenic murine models, with no significant enhancement of the monogenic phenotype until a threshold of reduced expression at the second locus was breached. Pkhd1 loss did not alter expression, maturation, or localization of the ADPKD polycystin proteins, with no interaction detected between the ARPKD FPC protein and polycystins. RNA-seq analysis in the digenic and monogenic mouse models highlighted the ciliary compartment as a common dysregulated target, with enhanced ciliary expression and length changes in the digenic models.
Conclusions
These data indicate that FPC and the polycystins work independently, with separate disease-causing thresholds; however, a combined protein threshold triggers the synergistic, cystogenic response because of enhanced dysregulation of primary cilia. These insights into pathogenesis highlight possible common therapeutic targets.
Autosomal dominant polycystic kidney disease (ADPKD) and autosomal recessive polycystic kidney disease (ARPKD) are common monogenic causes of ESKD.1 These diseases are genetically distinct, with PKD1 or PKD2 being the major ADPKD loci (approximately 78% and 15% of families, respectively), whereas PKHD1 is causative for ARPKD.2–8 ADPKD is characterized by progressive renal cystogenesis often resulting in ESKD in the fifth to seventh decade of life, whereas the classic ARPKD presentation is in utero or perinatal with enlarged, echogenic kidneys that can be lethal or result in childhood ESKD.1,9–12 The liver disease also differs, with polycystic liver disease common in ADPKD, whereas congenital hepatic fibrosis is the typical lesion in ARPKD.9,13 Both diseases have a wide range of severity, partly due to genic and allelic effects. Biallelic PKD1 can be mistaken for ARPKD and weak PKHD1 allelic combinations, or even single alleles, can have adult-onset disease with kidneys similar to ADPKD or mild polycystic liver disease.14–18 Although the timing, severity, and apparent pathogenesis of the diseases are usually dissimilar, there is evidence of ADPKD and ARPKD commonalities. Heterozygous loss of Pkd1 in a Pkhd1−/− background results in aggravated cystic kidney disease,19,20 and inhibition of the V2 vasopressin receptor to block downstream cAMP signaling has proven beneficial in Pkd1, Pkd2, and Pkhd1 murine models, and in human ADPKD.21–26 Yet, the intersection of cellular pathways affected by the loss/reduction of Pkd1/Pkd2 or Pkhd1 has yet to be elucidated.
PKD1 and PKD2 encode polycystin-1 (PC1) and polycystin-2 (PC2), respectively, which form a heteromeric complex thought to function as a receptor channel. PKHD1 encodes a large, single-pass, transmembrane protein, fibrocystin/polyductin (FPC).5,27–31 There is evidence that all three proteins localize to the primary cilia—signaling antenna found on most cell types that, in kidney tubules, protrude into the luminal space to sense flow and respond to extracellular signals.32–40 Defects in the function of primary cilia have been implicated in PKD animal models, and a PKD phenotype is found in many syndromic ciliopathies—genetic diseases disrupting ciliary function.36–39,41–49 However, signaling pathways most clearly linked to simple forms of PKD, such as cAMP, are only tenuously linked to cilia.21,50 Further complexity is shown by ciliary deletion, in the context of induced Pkd1 loss, reducing cystic disease severity.51
Complete loss of PC1 or PC2 in mice or humans is associated with embryonic lethality.52–55 However, serviceable preclinical models have been developed which use conditional loss with various Cre drivers or viable biallelic hypomorphic/hypermutable models that generate some functional product.41,52,56–60 In particular, the hypomorphic Pkd1RC/RC model, with an approximately 60% reduction in mature PC1, develops as a slowly progressive disease similar to human ADPKD.41 For Pkhd1, available models are less satisfactory. The homozygous rat, PCK, generates slowly progressive cystic kidney and liver disease akin to ADPKD, and null mice develop only mild kidney tubule dilations in aged mice, although the liver disease is similar to human ARPKD.5,19,42,61–64
Here, we developed two digenic, Pkhd1-Pkd1, murine models with the hypothesis of a commonality in the causes of cystic disease in ADPKD and ARPKD. The results we obtained support our hypothesis because the digenic models show a synergistic interaction, accurately recapitulating the early-onset disease characteristic of human ARPKD.63 Careful analysis of the mouse model reveals processes important to cystogenesis and highlights a pivotal role of primary cilia in both diseases.
Methods
Animal Breeding and Phenotypic Analysis
The following mouse lines were used: Pkhd1LSL/LSL (termed Pkhd1−/−, which has a STOP cassette in exon 3 to ablate the full-length transcript that develops liver but only mild kidney disease),63 Pkd1RC/RC (a homozygous Pkd1 model with the hypomorphic, human missense mutation p.Arg3277Cys that develops slowly progressive disease),41 and Pkhd1SV5-Pk (termed Pkhd1V5, with an N-terminal endogenous tag after the signal peptide, allowing study of the FPC protein),63 each fully inbred into the C57BL/6 strain. Pkhd1-Pkd1 compound mutant mice were first generated by interbreeding Pkhd1−/− and Pkd1RC/RC animals to produce Pkhd1−/−;Pkd1+/RC and Pkhd1+/−;Pkd1RC/RC mice. Compound mutant animals were then used to generate homozygous digenic (Pkhd1−/−;Pkd1RC/RC) and compound mutant animals (Pkhd1+/−;Pkd1RC/RC, Pkhd1−/−;Pkd1+/RC). An equal number of male and female mice were used for analysis at each time point (perinatal day [P] 0, P12, P90, P180, and P270: n=5 per sex; for P3, five animals in total were analyzed). The sex of the animals at perinatal time points (P0, P3, and P12) was determined by PCR amplification of intron 9 from the Smcx/Smcy locus and size resolution of a 331- and 302-bp product by gel electrophoresis as previously described.65 Anatomic features were used to identify the sex of adult animals.
For the rat, digenic (Pkhd1−/− [PCK];Pkd1+/−), compound (Pkhd1+/−;Pkd1+/−), and single homozygous (Pkhd1−/−) animals were generated for phenotypic studies. See figure legends for the number of animals used at each time point. The Pkhd1−/− (PCK) rats are on the Sprague-Dawley background and the Pkd1+/− rats on the Lewis background, and so the digenic animals are on a mixed Sprague-Dawley/Lewis background (Supplemental Methods, Supplemental References).
All animals were euthanized at the indicated time point, and tissue collections and measurements were completed as previously described.41 Cyst areas were determined from a single cross section in perinatal mice using the NIS software, whereas for adult mice and rats, the average of three cross sections per kidney was used (see figure legend for number of animals used for each analysis). A threshold was set at 500 μm2 for cyst identification in young mice (≤P12), 2000 μm2 for adult mice, and 10,000 μm2 for rats.
Magnetic Resonance Imaging and Analysis
Magnetic resonance imaging scans in rats ≤1 month were performed in a Bruker Advance 700 Mhz (16.4T) vertical bore nuclear magnetic resonance spectrometer, whereas adult rats were scanned using the GE clinical 3T scanner, following procedures approved by the Institutional Animal Care and Use Committee (IACUC). Total kidney volume was calculated from axial slices of the kidney using Analyze 12.0 software.
PC2 Cilia Localization
Kidneys were collected from Pkhd1V5/V5 and Pkhd1−/− animals euthanized at P12 and immediately dissociated using the Multi Tissue Dissociation Kit 2 from Miltenyi Biotec. The cell suspension was quenched with ice-cold 5% BSA solution containing 2 mM EDTA and passed through a 40-μm filter before centrifugation. The cell pellet was resuspended in Advanced DMEM with 5% FBS, GlutaMax, and penicillin/streptomycin and seeded onto collagen-coated plates/slides and cultured to confluence, followed by 24 hours of serum starvation before experimentation. Cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked with 3% BSA overnight at 4°C, followed by overnight incubation with primary antibodies at 4°C (acetylated tubulin [32-2700; Invitrogen], PC2 [PA5-77328; Invitrogen]).
Western Analysis
Kidneys were collected from P0 and P12 animals of both the Pkhd1V5/V5 and Pkhd1−/− genotype and crude membrane lysates prepared as previously described.40 For immunoprecipitation (IP), antibodies to the V5 epitope were incubated with 200 μg lysate overnight and incubated with 20 μl of packed, washed A/G agarose for 2 hours at 4°C. The agarose was washed three times in IP buffer and the protein eluted in 1× LDS plus 50 mM tris(2-carboxyethyl) phosphine hydrochloride. A total of 25 μg of input and 100% of the IP were analyzed by Western blotting. Samples were resolved on SDS-PAGE (3%–8% Tris-acetate), transferred to polyvinylidene difluoride membrane using wet transfer conditions, and blocked with 1% nonfat milk. The following antibodies were used for immunoblotting: V5, mouse IgG2a (catalog number MCA-1360; BioRad) or rabbit polyclonal (catalog number AB3792; Millipore); FPC (human), mouse IgG1 (clone ab11)66; PC1, mouse IgG1 (clone 7e12); PC2, mouse IgG2b (catalog number sc-28331; Santa Cruz); E-cadherin, mouse IgG1 (catalog number 14472; Cell Signaling Technology); Vinculin, rabbit polyclonal (catalog number 4650; Cell Signaling Technology).
RNA Sequencing
Right kidneys were extracted from single homozygous, digenic, and wild-type (WT) control animals at P0, P3, or P12 (n=3 per genotype per time point, nonlittermates). All animals were born between the hours of 10 pm and 6 am and euthanized at noon±2 hours. RNA was extracted from homogenized whole kidney tissue using RNAeasy Mini Kit (catalog number 74104; Qiagen) according to the manufacturer’s protocol and met RNA quality thresholds before submission. Briefly, RNA samples were processed through Mayo Clinic’s MAP-RSeq (version 2.1.0)67 pipeline after sequencing on the Illumina HiSeq 4000 platform (150 bp paired-end reads). Within MAP-RSeq, TopHat268 with the Bowtie169 option was used to align the reads to the GRCm38 reference genome. The gene counts were generated by FeatureCounts70 using the Ensemble GRCm38 gene definition file. RSeqQC71 was used to create quality control metrics before differential expression analysis by edgeR.72 Genes with an average of <25 read counts in WT controls were removed, resulting in the inclusion of 18,049 genes for subsequent analysis. Expression data have been deposited in Gene Expression Omnibus under accession number GSE131277.
Gene Set Enrichment Analysis
Gene ontology (GO) term enrichments were determined by the GOrilla platform (http://cbl-gorilla.cs.technion.ac.il/) using the minimum hypergeometric (mHG) analysis statistical tool.73,74 Genes were first ranked by the product of their log2 fold change (log2FC) and −log10(P value) determined by differential expression analysis before mHG statistical analysis of ranked gene lists. All enriched GO terms met a P value threshold of ≤10−5. A cilia-associated gene list was generated by use of the GO Consortium website (www.amino.geneontology.org/amigo), using the indicated GO identifiers and filtering the list by the organism Mus musculus (Supplemental Table 1).75,76
Scanning Electron Microscopy
Imaging was performed on left kidney excised from P0 mice, as previously described.42 Scanning electron microscopy (SEM) was performed with a Hitachi S-4700 microscope and cilium imaging was performed in morphologically similar tubules (brush-border positive).
Statistical Analysis
Prism 7 was used for all statistical measures, with the exception of differentially expressed gene (DEG) and gene set enrichment analysis (GSEA). See figure legends for specific statistical tests applied to experimental data sets.
Study Approval
These animal experiments were conducted under the following IACUC protocols: A00003539, A00001915, and A00003800. They were reviewed and approved by the IACUC of the Mayo Clinic, in accordance with National Institutes of Health, United States Department of Agriculture, and Association for Assessment and Accreditation of Laboratory Animal Care guidelines.
Results
Characterization of the Early-Onset Cystic Disease in Digenic Mice
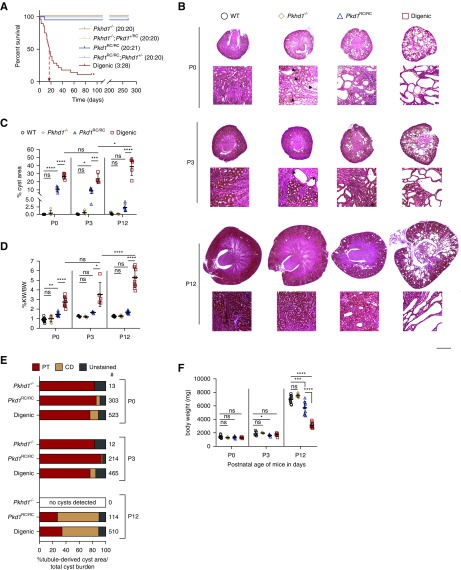
We interbred hypomorphic Pkd1RC/RC and null Pkhd1−/− mice to better understand the interaction between ADPKD and ARPKD. Pkd1RC/RC mice develop a mild, progressive renal cystic disease and Pkhd1−/− mice develop only mild renal tubular dilations by 6 months. However, when both the Pkhd1 and Pkd1 alleles were homozygous (Pkhd1−/−;Pkd1RC/RC, digenic), a synergistic renal phenotype resulted in early lethality (Figure 1A, Supplemental Figure 1, A and B). Because there was no indication of in utero lethality (Supplemental Figure 1C), we characterized the progression of cystic disease perinatally of monogenic (Pkhd1−/− and Pkd1RC/RC) and digenic animals, plus WTs. Consistent with previous findings,41 Pkd1RC/RC mice had multiple cysts by P0 that tended to regress by P12 (Figure 1, B and C). We found that Pkhd1−/− mice developed a few tubular dilations/cysts (P0 and P3), although neither the kidney weight/body weight (KW/BW) or cystic area (CA) significantly differed from WT (Figure 1, B–D), and these cysts regressed by P12. In contrast, the digenic kidneys had significant cystic disease, evident by KW/BW, CA, and number and size of cysts compared with the monogenics by P0 (Figure 1, B–D, Supplemental Figure 1, D and E). The origin of cysts was tested by immunofluorescence, using lotus tetragonolobus agglutinin (stains proximal tubules [PTs] red) and an aquaporin-2 antibody (stains collecting ducts [CDs] green; Figure 1E, Supplemental Figure 1F). At P0 and P3 in the monogenic and digenic models, cysts were largely derived from PTs, but by P12 the renal CA in Pkd1RC/RC and digenic kidneys was predominantly CD derived.41 As well as the rapidly progressive renal cystic disease, other ARPKD-like characteristics of the digenic mice include growth retardation, early lethality, and indications of biliary dysgenesis by P12 (Figure 1, A and F, Supplemental Figure 1G).
Figure 1.
Digenic mice develop early-onset and rapidly progressive renal cystic disease. (A) Kaplan–Meier survival curve of the different genotypic groups, as shown. For each genotype, the number of mice surviving to 270 days and total mice are indicated in parentheses. Digenic mice show significantly decreased survival (median age, P=17 days [arrow; P<0.001]), unlike the other genotypic combinations (one of 21 Pkd1RC/RC mice died at P10 likely due to an unrelated cause). *Euthanasia of the three digenic animals surviving to 3 months. (B) Representative Masson trichrome sections of Pkhd1−/− (orange diamond), Pkd1RC/RC (blue triangle), digenic (red square), and WT (black circle) kidneys from P0, P3, and P12 mice. Cystic disease was exacerbated in digenic kidneys as early as P0 compared with single homozygous kidneys and progressively worsened during postnatal development. Scale bar, 500 μm (kidney cross section) and 100 μm (magnified region). Arrowheads in magnified regions denote representative cysts or dilated tubules (≥500 μm2) in Pkhd1−/− kidneys. (C and D) Graphical representation of percentage cyst area and percentage KW/BW from P0, P3, and P12 animals. (C) Cyst area was significantly increased in digenic kidneys compared Pkd1RC/RC kidneys at P0, P3, and P12 (n=5 animals per genotype per time point). (D) %KW/BW was significantly increased in Pkd1RC/RC (57%) compared with WT only at P0, whereas the KW/BW in digenic kidneys was significantly increased at P0 (204%), P3 (181%), and P12 (319%) compared with Pkd1RC/RC (P0 and P12, n=10 animals per genotype per time point; P3, n=5 animals per genotype per time point). (E) Quantification of cyst origin as a percentage of total cyst area. Cysts were identified (≥500 μm) and binned as lotus tetragonolobus agglutinin (LTA)+ (PT), aquaporin-2 (AQP2)+ (CD), or LTA−, AQP2− (unstained). Cystogenesis primarily occurred in PTs at P0: Pkhd1−/−, 77%; Pkd1RC/RC, 91.9%; digenic, 79.2%, with minimal CD-derived (AQP2) cystic burden in both digenic and Pkd1RC/RC kidneys (2.1% and 2.6%, respectively). By P12, there was regression of the P0/P3-PT–derived cysts and initiation and expansion of CD-derived cysts that occurred in both Pkd1RC/RC and digenic kidneys (See Supplemental Figure 1G for representative immunofluorescence image). #Total cysts counted in analysis (n=3 mice per genotype per time point with one axial slice used for counting). (F) Graphical analysis of BW measurements from P0, P3, and P12 animals (P0 and P12, n=10 animals per genotype per time point; P3, n=5 animals per genotype per time point). Digenic mice showed significant growth retardation by P12. Statistical values were obtained by one-way ANOVA, followed by Tukey post-hoc analysis. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001; error bars indicate ±SD.
The genetic interaction between Pkhd1 and Pkd1 was dosage dependent; addition of a single allele to homozygosity at the second locus (Pkhd1−/−;Pkd1+/RC or Pkhd1+/−;Pkd1RC/RC) or, as expected, monogenic mice showed no decrease in survival to 9 months (Figure 1A). Consistent with this, KW normalized to ex-hepatic BW (KW/ex-hepatic BW) did not differ in Pkhd1−/−;Pkd1+/RC and Pkhd1+/−;Pkd1RC/RC animals compared with their monogenic counterparts at 3, 6, or 9 months, in contrast to the three surviving 3-mo digenic animals that had extreme cystic disease (Supplemental Figure 2, A–D). Furthermore, we found no significant difference in disease severity between males and females, with the exception of a small but statistically significant difference in the Pkhd1−/− model at P12 (mean percentage KW/BW: 1.31 females versus 1.18 males, P=0.02), and only when KWs were normalized to BWs (Supplemental Table 2). This discordance with previously published data,63 where only female Pkhd1−/− mice developed significant renal disease in animals of at least 6 months, is likely due to the much earlier time points analyzed in this study. Analysis of the liver pathology at 3 months in these different genotypes showed only a mild increase in severity in the digenic mice (Supplemental Figure 2, E and F).
This analysis shows a synergistic interaction between the level of Pkd1 and presence of Pkhd1 in mice, but we wanted to see if this interaction is more generally true.
Heterozygous Loss of Pkd1 Exacerbates the Renal Phenotype in Pkhd1−/− (PCK) Rats
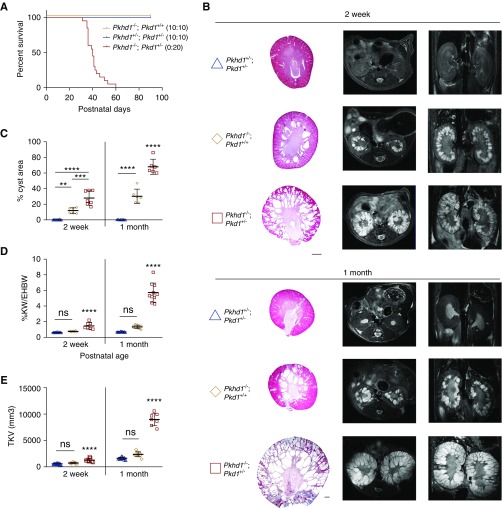
Pkd1 mutant rats were generated by use of transcription activator–like effector nucleases, resulting in an 8-bp frameshifting deletion in exon 29 (c.9793_9800del; p.Arg3274fs). Although Pkd1−/− rats were embryonically lethal (Supplemental Figure 3A), heterozygotes developed just a few cysts with a slight increase in KW/ex-hepatic BW by 12 months, and had no postnatal lethality (Figure 2A, Supplemental Figure 3B). Again, a strong genetic interaction was observed in Pkhd1−/−;Pkd1+/− (digenic) rats, with animals experiencing early postnatal lethality and rapidly progressive renal cystic disease (Figure 2). Similar to mice, there was a substantial PT-derived CA at P0 (approximately 50%), but by P14 almost all cysts were CD derived in the digenic and Pkd1−/− rats (Supplemental Figure 3D). Although digenic rats did not exhibit significant growth retardation compared with Pkhd1−/− or digenic heterozygotes, there was a significant increase in BUN levels at 1 month indicating uremia (Supplemental Figure 3, E and F). The digenic liver phenotype showed slightly increased (but NS) severity (Supplemental Figure 3, G and H).
Figure 2.
Heterozygous loss of Pkd1 in the Pkhd1−/− (PCK) rat model causes a synergistic renal phenotype. (A) Kaplan–Meier survival curve of the different genotypic groups, as shown. For each genotype, the number of rats surviving to 90 days and total rats are indicated in parentheses. Pkhd1−/−;Pkd1+/− rats show significantly decreased survival (median age=40 days). (B) Representative Masson trichrome sections and magnetic resonance imaging (left, axial; right, coronal) of Pkhd1+/−;Pkd1+/− (blue triangle), Pkhd1−/−;Pkd1+/+ (orange diamond), and Pkhd1−/−;Pkd1+/− (red square) kidneys from rats aged 2 weeks (w) and 1 month (m). The cystic phenotype was more severe in Pkhd1−/−;Pkd1+/− by 2 weeks compared with other genotypic combinations. Scale bar, 1000 μm. (C–E) Graphical analysis of percentage cyst area, percentage KW/ex-hepatic BW (%KW/EHBW), and total kidney volume (TKV) measurements, with each measurement showing significantly increased disease in Pkhd1−/−; Pkd1+/− animals at both the 2 weeks and 1 month time point compared with other genetic cohorts. Values in (C–E) were obtained from n=6–10 animals per genotype per time point. Statistical values were obtained by one-way ANOVA, followed by Tukey post-hoc analysis. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001; error bars indicate ±SD.
As the synergistic relationship between Pkhd1-Pkd1 was similar in both rats and mice, we next explored the possible mechanism of this genetic interaction.
Biochemical Analyses Suggest Different Functions for FPC and the PC1/PC2 Protein Complex
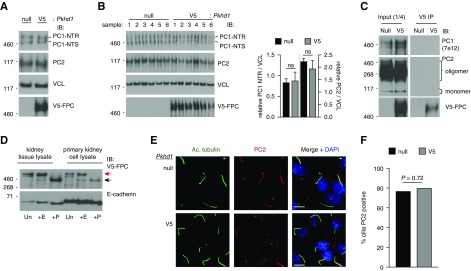
To determine if loss of FPC exacerbates the Pkd1 phenotype by affecting PC1/PC2 expression or localization, we used the Pkhd1V5 mouse model.63 As suggested,77,78 loss of FPC did not affect PC1 biogenesis or decrease PC2 expression (Figure 3, A and B), indicating that Pkhd1 loss does not modulate the PKD phenotype through a reduction in PC1/PC2 protein levels. Additionally, coimmunoprecipitation assays employing the V5 tag of FPC did not reveal an intracellular interaction between FPC and either PC1 or PC2 (Figure 3C).
Figure 3.
Genetic deletion of Pkhd1 does not perturb the maturation and localization of the PC1/PC2 protein complex. (A) Immunoblotting (IB) of PC1 (NTR/NTS [NT, N-terminal; R, EndoH resistant; S, EndoH sensitive]) glycoforms and PC2 in Pkhd1−/− (null) kidney tissue crude membrane preparations compared with Pkhd1V5/V5 (V5), each representing a pool of multiple kidneys extracted from P12 mice, demonstrates no change in PC1/PC2 expression. (B) IB of PC1 and PC2 in Pkhd1 null kidney tissue crude membrane preparations compared with Pkhd1V5/V5 (V5), each sample prepared from individual animals at P0. Relative quantification of PC1 NTR (left axis) and PC2 densitometry (right axis), both normalized to vinculin (VCL) densitometry, reveal no significant change in expression. (C) IB of PC1 and PC2 from V5 immunoprecipitation (IP) eluates from Pkhd1 null and Pkhd1V5/V5 (V5) kidney tissue. No interaction was observed between FPC and the PC1/PC2 proteins. Input samples (25 μg) represent a quarter of total protein used in IP reaction. Each Western blot for input and V5 IP samples are from the same exposure. (D) Deglycosylation analysis of kidney tissue or cultured primary cells from the Pkhd1V5/V5 mouse model. Lysates were treated with EndoH (+E) or PNGaseF (+P), or untreated (Un), to reveal glycoforms of endogenous FPC. FPC exists as a mature, post-Golgi glycoform (EndoH resistant, red arrow), and an immature, ER glycoform (EndoH sensitive, black arrow). E-cadherin, positive control. (E) Immunofluorescence detection of cilia (acetylated α-tubulin, Ac. tubulin) and PC2 in Pkhd1 null and Pkhd1V5/V5 (V5) primary cells, counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Scale bar, 100 μm. (F) Quantification of PC2-positive cilia. PC2 localization to cilia was observed in 314 of 411 (76.4%) and 386 of 486 (79.4%) in null and Pkhd1V5/V5 (V5) cells, respectively. No significant difference was observed, two-tailed Fisher exact test.
To test whether loss of FPC affects PC2 cilia localization, we optimized the culture of primary cells from enzymatically digested Pkhd1V5/V5 and Pkhd1−/− kidneys, because we found established human renal cell lines (using a previously validated antibody to FPC66), and even immortalized cell lines generated from the Pkhd1V5 mouse, did not maintain FPC expression, and exogenously expressed FPC did not mature (Supplemental Figure 4, A–C). In contrast, cultured primary Pkhd1V5 cells maintained expression of appropriately matured FPC, based on deglycosylation analysis (Figure 3D, Supplemental Figure 4D). Using these primary cell cultures, we did not find a significant change in PC2 cilia localization in Pkhd1−/− cells compared with WT (Pkhd1V5) (Figure 3, E and F). Although we were unable to test for FPC localization to cilia due to V5 antibody nonspecificity (data not shown), there was no indication of an effect on ciliation between WT and Pkhd1−/− cells (Supplemental Figure 4E).
Taken together, these data indicate that the synergistic interaction observed in the digenic models did not occur through disruption of an FPC plus PC1/PC2 protein complex nor a reduction in PC1/PC2 expression, maturation, or localization. Next, we analyzed relationships between expression differences in the three models.
Kidney Transcriptome Analysis of WT, Monogenic, and Digenic Mice
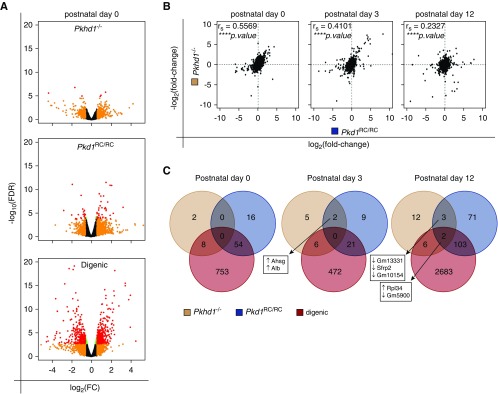
To determine if there exists a commonality in ARPKD and ADPKD signaling pathways and if further dysregulation of a specific signaling motif explains the exacerbated cystic disease in digenic animals, we used RNA sequencing (RNA-seq) on digenic, monogenic, and WT kidneys from P0, P3, and P12 mice (Supplemental Table 3). Transcriptome profiles from monogenic and digenic mice (versus WT) were plotted as volcano plots, with the number of DEGs correlating with cystic disease severity, digenic>Pkd1RC/RC>Pkhd1−/−, and time of analysis, P12>P3>P0 (Figures 1B and 4A, Supplemental Figure 5, A and B, Supplemental Table 4). As expected, we only observed loss of Pkhd1 expression in Pkhd1−/− and digenic mutant data sets (Supplemental Figure 5C), and Pkd1 mRNA expression was not decreased in Pkd1RC/RC kidneys.41 We also did not observe a significant change in Pkhd1 expression in Pkd1RC/RC mutant kidneys, nor an expression change of either Pkd1 or Pkd2 in any of the mutant data sets.
Figure 4.
Transcriptome profiles in single homozygous and digenic kidneys indicate common gene networks. (A) Volcano plots of analyzed genes to identify significantly DEGs in Pkhd1−/−, Pkd1RC/RC, and digenic kidneys at P0 compared with the WT control. Red points denote log2FC ≥0.585 and −log10FDR ≥1.3; orange, log2FC ≥0.585 only; green, −log10FDR ≥1.3 only. The number of DEGs reaching significance (−log10FDR≥1.3) increases with disease severity. (B) Scatterplots comparing Pkhd1−/− gene expression to Pkd1RC/RC at P0, P3, and P12. Spearman rank coefficients (rs) and P values are noted in top left corner of each plot. There is a positive correlation, strongest at P0, indicating that there are shared gene regulatory networks between Pkhd1 and Pkd1. Spearman rank coefficients (rs) and associated P values were calculated using Prism 7 (****P<0.0001). (C) Venn diagram showing the number of DEGs significantly different from WT (−log10FDR≥1.3, red and orange data points from volcano plots), shared, and unique for each genotype at P0, P3, and P12. Genes found at the intersection of the Pkhd1 and Pkd1 data sets are noted, along with their direction of change in expression compared with WT controls.
Testing the hypothesis that ARPKD and ADPKD share similar transcriptional networks, we first performed a Spearman rank correlation analysis of all genes (Figure 4B, Supplemental Figure 5D). Here, we found a consistent positive correlation between the mutant DEG profiles, indicating that loss of Pkhd1 or Pkd1 reduction induced dysregulation of common gene networks. The correlation coefficients were strongest at the P0 time point, likely due to later secondary cystogenic signatures in both Pkd1RC/RC and digenic kidneys (i.e., inflammatory and oxidative stress). However, when we applied a P value threshold to compare all significant individual DEGs relative to WT, we found no common ones between Pkhd1 and Pkd1 at P0, and only two at P3 and five at P12 (Figure 4C). Because just a small number of common genes with dissimilar functions and likely unrelated to cystic disease were identified, it is unlikely that FPC and PC1 directly coregulate these genes. Because Pkhd1−/− mice have a very mild kidney phenotype, a reasonable conclusion is that the underlying transcriptional dysregulation due to Pkhd1 loss does not meet a P value–based threshold cutoff.
Although a correlation between dysregulated gene networks was seen in the three models, we did not find commonly dysregulated genes. We, therefore, concluded other statistical measures are needed to uncover related dysregulated gene networks.
Transcriptome Analysis Reveals a Dysregulated Ciliary Compartment at the Intersection of the Pkhd1-Pkd1 Genetic Interaction
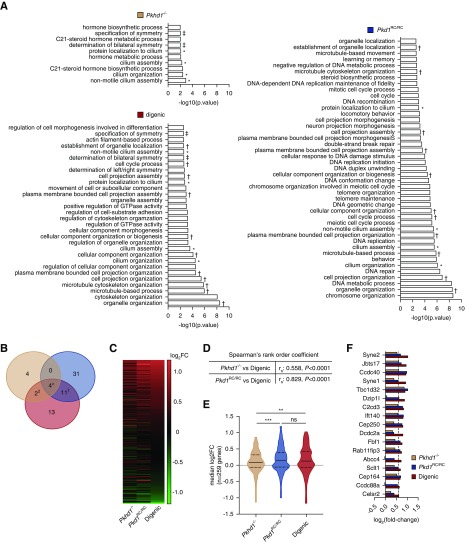
As we found no clear evidence of direct coregulation of genes by FPC and PC1, although there was a positive correlation between the transcriptome profiles, we hypothesized that loss of FPC or reduction of functional PC1 alone was insufficient to cause a robust DEG response. Thus, we performed mHG analysis73,74 from each of the monogenic and digenic P0 data sets, compared with WT, to determine if there were GO terms associated with biologic processes significantly over-represented at the top of the ranked gene list across the entire transcriptome (ranking based on the product of their log2FC and −log10[P value]). Here, we found a number of GO terms significantly enriched in one or more genotypic group (Figure 5, A and B, Supplemental Table 5). Of the ten GO terms enriched in the Pkhd1−/− group, four were unique to Pkhd1−/− with two also enriched in the digenic group (Figure 5, A and B). As expected, we had more GO terms enriched in the Pkd1RC/RC46 or digenic30 groups, including several semantically similar terms. Most prominently we found GO terms associated with cell proliferation: DNA metabolism/replication, organelle organization, and cell cycle. The increase in expression of genes annotated to these processes is likely due to hyperproliferation of cystic cells in both Pkd1RC/RC and digenic kidneys. Although DNA repair pathways were uniquely enriched in the Pkd1RC/RC group, pathways associated with increased GTPase activity and cell morphogenesis were more closely aligned with the digenic transcriptome. The dysregulation of these cellular pathways may be an indicator of cystic disease severity.
Figure 5.
Transcriptome analyses reveal a dysregulated ciliary compartment at the intersection of the Pkhd1-Pkd1 genetic interaction. (A) GO terms significantly enriched (P≤10−5) at P0 in each ranked gene list (18,049 genes, ranked by the product of their log2FC and −log10[P value]) by mHG test analysis using the GOrilla software tool (http://cbl-gorilla.cs.technion.ac.il/). Note, gene ranking by FC only did not appreciably change the GSEA results, data not shown. Common GO terms shared between mutant mouse models are noted as follows: ‡, Pkhd1−/− and digenic; †, Pkd1RC/RC and digenic; *, all three genotypes. All four GO terms at the intersection of the Pkhd1-Pkd1-digenic GSEA are involved in cilia morphogenesis/function. (B) Venn diagram showing the number of enriched GO terms. (C–E) Graphical analyses of expression profiles of genes annotated to common cilia-associated GO terms (n=259 genes [see Supplemental Table 1), GO: 0044782, 0060271, 0061512, 1905515). (C) Heatmap of gene expression in Pkhd1−/−, Pkd1RC/RC, and digenic kidneys at P0. (D) Spearman rank order coefficient (rs) for the comparison of digenic expression profile to the Pkhd1−/− and Pkd1RC/RC expression profiles. (E) Violin plots showing distribution of gene expression: solid line, median; dashed line, interquartile. There is a significant increase in the median expression in Pkd1RC/RC and digenic kidneys compared with Pkhd1−/−. (F) Expression of genes annotated to cilia-associated GO terms that reach statistical significance (FDR≤0.05). Spearman rank coefficients were calculated using Prism 7. Statistical values were obtained by one-way ANOVA, followed by Tukey post-hoc analysis (**P<0.01, ***P<0.001).
Most strikingly, we found four biologic processes commonly enriched in each of the mutant models that were all associated with primary cilia (cilium organization [GO:0044782], cilium assembly [GO:0060271], protein localization to cilium [GO:0061512], and nonmotile cilium assembly [GO:1905515]). Given that loss of Pkhd1 does not induce a robust DEG response (see Figure 4A, volcano plot), we were concerned that mHG analysis may generate erroneous results. Therefore, we also performed GSEA on DEGs (log2FC≥1.5; P value≤0.05) in this gene set compared with the background gene set and confirmed the cilia-associated enrichment (nonmotile cilium assembly; false discovery rate [FDR] ≤0.0088, enrichment score 8.79). Furthermore, a heatmap of the expression of genes annotated to the commonly enriched cilia-associated GO terms revealed a similar change in expression for all three genotypes (Figure 5C, Supplemental Table 1). In addition, the Spearman rank correlation analysis of this gene subset showed a strongly positive relationship between the digenic and Pkd1RC/RC expression profiles and a moderately positive correlation between the digenic and Pkhd1−/− profiles (Figure 5D). The median log2FC of this gene set exhibited an overall increase in expression for all genotypes compared with WT controls, and a significant increase for both the Pkd1RC/RC and digenic compared with Pkhd1−/− (Figure 5E), emphasizing the relationship of the dysregulated ciliary compartment and cystic disease severity. To gain a better interpretation of the ciliary-associated genes dysregulated in these analyses, we plotted the FC of those genes reaching significance (FDR≤0.05) (Figure 5F). We found 17 genes reaching significance in the digenic model, with a similar expression trend in the other genotypes. Many of these upregulated genes localize to the basal body/transition zone and are implicated in regulating ciliogenesis or protein transport into/out of cilia. For example, Cep164, Fbf1, Sclt1, and C2cd3 each localize to the distal appendages of centrioles to promote membrane docking and subsequent initiation of ciliogenesis.79 We also observed dysregulation of several known ciliopathy genes, Jbts17,80 Cep164,81 Cep250,82 Sclt1,83 Ift140,84 and C2cd3.85,86 Additionally, Dzip1l, which is associated with an ARPKD-like phenotype in humans and mice by altering PC1/PC2 ciliary localization,87 showed a significant increase in gene expression.
Here, by ranking dysregulated biologic processes, primary cilia were highlighted as common in each model, suggesting a shared axis, and so cilia structure was analyzed.
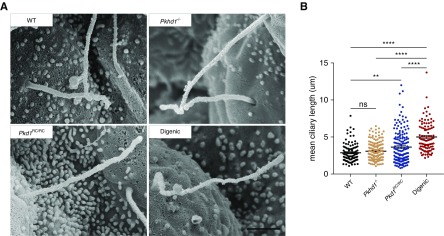
Ciliary Length Is Longer in Digenic Kidneys
Based on our findings of a dysregulated ciliary-associated transcriptome network, plus the previously observed ciliary defects associated with the Pkd1RC model,41 we analyzed cilia structure in the different genotypes. SEM of kidneys at P0 confirmed the average length of primary cilia was longer in the Pkd1RC/RC model compared with WT but not in the Pkhd1−/− (Figure 6, A and B).41,42 In addition, we found more extended cilia in the digenic compared with Pkd1RC/RC mice (4.99 versus 3.63 μm, respectively). We observed a wide spread of primary cilia lengths across all genotypes, however, there were noticeably fewer shorter cilia (1–2 μm) in digenic kidneys and more longer cilia (>6 μm) in both the digenic and Pkd1RC/RC models.
Figure 6.
Ciliary length defect is exacerbated in digenic kidneys. The length of tubular primary cilia in the kidney was measured in WT, single homozygous, and digenic animals at P0. (A) Representative SEM images of primary cilia from each genotype (WT; Pkhd1−/− Pkd1RC/RC; digenic). (B) Graphical representation of the length of each primary cilium measured (WT (black), n=104 [three animals]; Pkhd1−/− (orange), n=185 [three animals]; Pkd1RC/RC (blue), n=162 [three animals]; digenic (red), n=98 [two animals]). Average cilia length was significantly elongated in digenic kidneys compared with Pkd1RC/RC cilia. Scale bar, 1 μm. Statistical values were obtained by one-way ANOVA, followed by Tukey post-hoc analysis. *P<0.05, **P<0.01, ****P<0.0001; error bars indicate ±SD.
Therefore, structural changes in cilia also highlighted a role for this organelle in cystogenesis in these models.
Discussion
The pathogenic relationship of the two common, simple forms of PKD (ADPKD and ARPKD) has been much debated but not fully resolved.19,20,30,78,88,89 Similarities beyond the renal phenotype include possible protein localization to cilia, but differences include the mode of inheritance and liver disease.40,66,90,91 Study of ARPKD has been hampered by the much weaker murine renal phenotype, especially in mice, compared with complete loss of Pkhd1 in humans.11,19,42,61–63 Here, we more clearly illustrate than previously described19,20 a synergistic interaction between Pkd1 and Pkhd1, in two species. The resulting digenic phenotypes of rapidly progressive, severe renal cystic disease, including a phenotypic switch from predominant PT to CD cysts, plus early lethality, match the canonical human disorder.11,92,93 However, the genetic interaction in both murine models reveal distinct thresholds for both Pkhd1 and Pkd1: haploinsufficiency for Pkhd1 or a single Pkd1RC allele do not exacerbate the phenotype, but homozygosity of either allele in the digenic context results in severe disease. These results further highlight that ADPKD is a dosage-dependent disorder, which is also the case for both diseases in humans.11,41 The rapidly progressive PKD phenotype induced by a low level of functional PC1 (<20%)41,56 can be mimicked by a less marked reduction of PC1 (approximately 40% in mice, 50% in rats) in the context of FPC loss, indicating a related downstream effect of the PC complex and FPC. These results have possible implications for modification of the human ADPKD phenotype by coinheritance of a PKHD1 mutation, which is expected by the carrier rate in approximately one in 70 individuals.94 Whether this will be a phenotypic modifier depends of the effect of a 50% reduction of FPC in the setting of ADPKD gene haploinsufficiency, bearing in mind that in the human setting FPC loss is associated with much more severe kidney disease than in murine models.
The PC1/PC2 dosage dependence of cystic disease severity is related to the PC1 and PC2 interaction and their corequirement for appropriate maturation and localization.40,41,95 Our studies and related data indicate that FPC loss does not directly alter the level or localization of mature PC1/PC2, or that FPC and the PCs are in the same complex,77 but rather that the role of these proteins/complexes overlap. Although the liver phenotype in our digenic mice is also enhanced, the dosage dependence is not as dramatic as for the kidney and likely reflects the different Pkd1 and Pkhd1 liver phenotypes, with an additive rather than synergistic relationship.
The unbiased genome-wide gene expression analysis in whole kidneys from monogenic and digenic animals allowed us to ascertain early markers of cystogenesis in perinatal kidneys before the involvement of strong secondary effects as cystic remodeling takes hold, including from infiltrating immune cells.96 GSEA analysis of the RNA-seq data highlighted a dysregulated transcriptional network implicated in ciliogenesis/cilia function in both monogenic and digenic kidneys, with the degree of dysregulation correlating with cystic disease severity. Interestingly, this transcriptome response occurred in the absence of significant cystic disease in the Pkhd1−/− kidneys, suggesting the transcriptional changes precede cystogenesis. This transcriptome data, plus observations of enhanced ciliary lengths in the digenic compared with the Pkd1RC/RC model, further implicate the dysregulation of the primary ciliary compartment in both ARPKD and ADPKD.41,97 However, the exact role of this organelle in both disorders is not yet fully resolved. The cilia-related expression and length differences may be associated with the basic defect or are a response to reduced PC-complex/FPC signaling.
Our novel, digenic murine models will aid understanding of early-onset PKD and act as ARPKD surrogate animal models, where loss of Pkhd1 alone does not fully recapitulate the human disease. In this age of PKD therapeutics,24,26 there is an urgent need of a severe ARPKD-related model for preclinical testing and our unique cellular system will also aid in resolving pathogenesis. Our data, supportive of a common cilia-dependent function dysregulated in both ARPKD and ADPKD, suggest ADPKD therapeutics, particularly those targeting ciliary dysregulation, may also be beneficial in ARPKD.
Disclosures
Dr. Geurts reports grants from NIH, during the conduct of the study. Dr. Torres reports grants from NIH, grants and other from Otsuka Pharmaceuticals, grants and other from Palladio Biosciences, other from Mironid, grants and other from Sanofi Genzyme, other from Vertex, grants from Acceleron Pharma Inc., grants from Regulus Therapeutics, grants from Palladio Biosciences, and grants from Blueprint Medicines, outside the submitted work. Dr. Harris reports grants from NIH, grants and other from Otsuka Pharmaceuticals, other from Mitobridge, other from Regulus Therapeutics, other from Vertex Pharmeceuticals, other from Goldfinch Bio, and other from Chinook, outside the submitted work. In addition, the Pkd1RC mouse is a licensed reagent (Dr. Harris and Hopp), with royalties paid by Amgen Inc., Bayer AG, GlaxoSmithKline and Sanofi.
Funding
This work was supported by the Mayo Graduate School of Biomedical Sciences Initiative for Maximizing Student Development and an F31 predoctoral fellowship, DK109597 (to Olson); National Institute of Diabetes and Digestive and Kidney Diseases grants DK058816 and DK59597 (to Dr. Harris); the Mayo Translational PKD Center (DK090728); and an endowment from Robert M. and Billie J. Pirnie for Kidney Research.
Supplementary Material
Acknowledgments
We thank the Mayo Gene Expression Core for performing the RNA-seq and differential expression analysis; the Mayo Clinic Electron Microscopy Core, especially Scott Gamb for training Dr. Olson to use SEM on kidney tissue; and Dr. Vladimir Gainullin for training Dr. Olson in the world of PC/FPC Western blotting.
Drs. Olson, Hopp, and Harris conceived the study. Drs. Olson, Hopp, and Mr. Wells, Ms. Smith, Furtado, Constans, and Escobar performed the experiments. Dr. Geurts generated the Pkd1 rat model. Drs. Olson and Hopp, Mr. Wells, and Ms. Smith analyzed the data. Drs. Olson, Hopp, and Harris wrote and edited the paper. All authors reviewed the results and approved the final version of the manuscript.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2019020150/-/DCSupplemental.
Supplemental Figure 1. Additional phenotypic characterization of Pkhd1-Pkd1 mutant mice.
Supplemental Figure 2. The Pkhd1-Pkd1 genetic interaction is dosage dependent.
Supplemental Figure 3. Characterization of Pkd1 mutant rats.
Supplemental Figure 4. FPC expression and maturation is tightly regulated.
Supplemental Figure 5. Additional analysis of DEG profiles.
Supplemental Table 1. Merged cilia-associated annotated gene list from Gene Ontology Consortium.
Supplemental Table 2. Sexual dimorphism in renal phenotype.
Supplemental Table 3. Animals used in RNAseq analysis.
Supplemental Table 4. Differential expressed genes in Pkhd1−/−, Pkd1RC/RC, and digenic kidneys.
Supplemental Table 5. Pkhd1−/−, Pkd1RC/RC, and digenic gene set enrichment analysis.
References
- 1.Harris PC, Torres VE: Polycystic kidney disease. Annu Rev Med 60: 321–337, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The European Polycystic Kidney Disease Consortium : The polycystic kidney disease 1 gene encodes a 14 kb transcript and lies within a duplicated region on chromosome 16. Cell 77: 881–894, 1994 [DOI] [PubMed] [Google Scholar]
- 3.Hughes J, Ward CJ, Peral B, Aspinwall R, Clark K, San Millán JL, et al.: The polycystic kidney disease 1 (PKD1) gene encodes a novel protein with multiple cell recognition domains. Nat Genet 10: 151–160, 1995 [DOI] [PubMed] [Google Scholar]
- 4.Mochizuki T, Wu G, Hayashi T, Xenophontos SL, Veldhuisen B, Saris JJ, et al.: PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science 272: 1339–1342, 1996 [DOI] [PubMed] [Google Scholar]
- 5.Ward CJ, Hogan MC, Rossetti S, Walker D, Sneddon T, Wang X, et al.: The gene mutated in autosomal recessive polycystic kidney disease encodes a large, receptor-like protein. Nat Genet 30: 259–269, 2002 [DOI] [PubMed] [Google Scholar]
- 6.Onuchic LF, Furu L, Nagasawa Y, Hou X, Eggermann T, Ren Z, et al.: PKHD1, the polycystic kidney and hepatic disease 1 gene, encodes a novel large protein containing multiple immunoglobulin-like plexin-transcription-factor domains and parallel beta-helix 1 repeats. Am J Hum Genet 70: 1305–1317, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornec-Le Gall E, Audrézet MP, Chen JM, Hourmant M, Morin MP, Perrichot R, et al.: Type of PKD1 mutation influences renal outcome in ADPKD. J Am Soc Nephrol 24: 1006–1013, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heyer CM, Sundsbak JL, Abebe KZ, Chapman AB, Torres VE, Grantham JJ, et al. HALT PKD and CRISP Investigators : Predicted mutation strength of nontruncating PKD1 mutations aids genotype-phenotype correlations in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 27: 2872–2884, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris PC, Torres VE: Polycystic Kidney Disease, Autosomal Dominant. In GeneReviews, edited by Adam MP, Ardinger HH, Pagon RA, et al., Seattle, WA, University of Washington, 2018 [Google Scholar]
- 10.Torres VE, Harris PC, Pirson Y: Autosomal dominant polycystic kidney disease. Lancet 369: 1287–1301, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Bergmann C, Senderek J, Windelen E, Küpper F, Middeldorf I, Schneider F, et al. APN (Arbeitsgemeinschaft für Pädiatrische Nephrologie) : Clinical consequences of PKHD1 mutations in 164 patients with autosomal-recessive polycystic kidney disease (ARPKD). Kidney Int 67: 829–848, 2005 [DOI] [PubMed] [Google Scholar]
- 12.Guay-Woodford LM: Autosomal recessive polycystic kidney disease: The prototype of the hepato-renal fibrocystic diseases. J Pediatr Genet 3: 89–101, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guay-Woodford LM, Desmond RA: Autosomal recessive polycystic kidney disease: The clinical experience in North America. Pediatrics 111: 1072–1080, 2003 [DOI] [PubMed] [Google Scholar]
- 14.Rossetti S, Kubly VJ, Consugar MB, Hopp K, Roy S, Horsley SW, et al.: Incompletely penetrant PKD1 alleles suggest a role for gene dosage in cyst initiation in polycystic kidney disease. Kidney Int 75: 848–855, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vujic M, Heyer CM, Ars E, Hopp K, Markoff A, Orndal C, et al.: Incompletely penetrant PKD1 alleles mimic the renal manifestations of ARPKD. J Am Soc Nephrol 21: 1097–1102, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adeva M, El-Youssef M, Rossetti S, Kamath PS, Kubly V, Consugar MB, et al.: Clinical and molecular characterization defines a broadened spectrum of autosomal recessive polycystic kidney disease (ARPKD). Medicine (Baltimore) 85: 1–21, 2006 [DOI] [PubMed] [Google Scholar]
- 17.Audrézet MP, Corbiere C, Lebbah S, Morinière V, Broux F, Louillet F, et al.: Comprehensive PKD1 and PKD2 mutation analysis in prenatal autosomal dominant polycystic kidney disease. J Am Soc Nephrol 27: 722–729, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunay-Aygun M, Turkbey BI, Bryant J, Daryanani KT, Gerstein MT, Piwnica-Worms K, et al.: Hepatorenal findings in obligate heterozygotes for autosomal recessive polycystic kidney disease. Mol Genet Metab 104: 677–681, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Gonzalez MA, Menezes LF, Piontek KB, Kaimori J, Huso DL, Watnick T, et al.: Genetic interaction studies link autosomal dominant and recessive polycystic kidney disease in a common pathway. Hum Mol Genet 16: 1940–1950, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fedeles SV, Tian X, Gallagher AR, Mitobe M, Nishio S, Lee SH, et al.: A genetic interaction network of five genes for human polycystic kidney and liver diseases defines polycystin-1 as the central determinant of cyst formation. Nat Genet 43: 639–647, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gattone VH 2nd, Wang X, Harris PC, Torres VE: Inhibition of renal cystic disease development and progression by a vasopressin V2 receptor antagonist. Nat Med 9: 1323–1326, 2003 [DOI] [PubMed] [Google Scholar]
- 22.Torres VE, Wang X, Qian Q, Somlo S, Harris PC, Gattone VH 2nd: Effective treatment of an orthologous model of autosomal dominant polycystic kidney disease. Nat Med 10: 363–364, 2004 [DOI] [PubMed] [Google Scholar]
- 23.Torres VE: Vasopressin antagonists in polycystic kidney disease. Semin Nephrol 28: 306–317, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torres VE, Chapman AB, Devuyst O, Gansevoort RT, Grantham JJ, Higashihara E, et al. TEMPO 3:4 Trial Investigators : Tolvaptan in patients with autosomal dominant polycystic kidney disease. N Engl J Med 367: 2407–2418, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hopp K, Hommerding CJ, Wang X, Ye H, Harris PC, Torres VE: Tolvaptan plus pasireotide shows enhanced efficacy in a PKD1 model. J Am Soc Nephrol 26: 39–47, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torres VE, Chapman AB, Devuyst O, Gansevoort RT, Perrone RD, Koch G, et al. REPRISE Trial Investigators : Tolvaptan in later-stage autosomal dominant polycystic kidney disease. N Engl J Med 377: 1930–1942, 2017 [DOI] [PubMed] [Google Scholar]
- 27.Hanaoka K, Qian F, Boletta A, Bhunia AK, Piontek K, Tsiokas L, et al.: Co-assembly of polycystin-1 and -2 produces unique cation-permeable currents. Nature 408: 990–994, 2000 [DOI] [PubMed] [Google Scholar]
- 28.Newby LJ, Streets AJ, Zhao Y, Harris PC, Ward CJ, Ong AC: Identification, characterization, and localization of a novel kidney polycystin-1-polycystin-2 complex. J Biol Chem 277: 20763–20773, 2002 [DOI] [PubMed] [Google Scholar]
- 29.Ong AC, Harris PC: A polycystin-centric view of cyst formation and disease: The polycystins revisited. Kidney Int 88: 699–710, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fedeles SV, Gallagher AR, Somlo S: Polycystin-1: A master regulator of intersecting cystic pathways. Trends Mol Med 20: 251–260, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su Q, Hu F, Ge X, Lei J, Yu S, Wang T, et al.: Structure of the human PKD1-PKD2 complex. Science 361: eaat9819, 2018 [DOI] [PubMed] [Google Scholar]
- 32.Bloodgood RA: From central to rudimentary to primary: The history of an underappreciated organelle whose time has come. The primary cilium. Methods Cell Biol 94: 3–52, 2009 [DOI] [PubMed] [Google Scholar]
- 33.Praetorius HA, Spring KR: Bending the MDCK cell primary cilium increases intracellular calcium. J Membr Biol 184: 71–79, 2001 [DOI] [PubMed] [Google Scholar]
- 34.Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF: Vertebrate Smoothened functions at the primary cilium. Nature 437: 1018–1021, 2005 [DOI] [PubMed] [Google Scholar]
- 35.Delling M, DeCaen PG, Doerner JF, Febvay S, Clapham DE: Primary cilia are specialized calcium signalling organelles. Nature 504: 311–314, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masyuk TV, Huang BQ, Ward CJ, Masyuk AI, Yuan D, Splinter PL, et al.: Defects in cholangiocyte fibrocystin expression and ciliary structure in the PCK rat. Gastroenterology 125: 1303–1310, 2003 [DOI] [PubMed] [Google Scholar]
- 37.Ward CJ, Yuan D, Masyuk TV, Wang X, Punyashthiti R, Whelan S, et al.: Cellular and subcellular localization of the ARPKD protein; fibrocystin is expressed on primary cilia. Hum Mol Genet 12: 2703–2710, 2003 [DOI] [PubMed] [Google Scholar]
- 38.Wang S, Luo Y, Wilson PD, Witman GB, Zhou J: The autosomal recessive polycystic kidney disease protein is localized to primary cilia, with concentration in the basal body area. J Am Soc Nephrol 15: 592–602, 2004 [DOI] [PubMed] [Google Scholar]
- 39.Yoder BK, Hou X, Guay-Woodford LM: The polycystic kidney disease proteins, polycystin-1, polycystin-2, polaris, and cystin, are co-localized in renal cilia. J Am Soc Nephrol 13: 2508–2516, 2002 [DOI] [PubMed] [Google Scholar]
- 40.Gainullin VG, Hopp K, Ward CJ, Hommerding CJ, Harris PC: Polycystin-1 maturation requires polycystin-2 in a dose-dependent manner. J Clin Invest 125: 607–620, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hopp K, Ward CJ, Hommerding CJ, Nasr SH, Tuan HF, Gainullin VG, et al.: Functional polycystin-1 dosage governs autosomal dominant polycystic kidney disease severity. J Clin Invest 122: 4257–4273, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woollard JR, Punyashtiti R, Richardson S, Masyuk TV, Whelan S, Huang BQ, et al.: A mouse model of autosomal recessive polycystic kidney disease with biliary duct and proximal tubule dilatation. Kidney Int 72: 328–336, 2007 [DOI] [PubMed] [Google Scholar]
- 43.Menezes LF, Cai Y, Nagasawa Y, Silva AM, Watkins ML, Da Silva AM, et al.: Polyductin, the PKHD1 gene product, comprises isoforms expressed in plasma membrane, primary cilium, and cytoplasm. Kidney Int 66: 1345–1355, 2004 [DOI] [PubMed] [Google Scholar]
- 44.Pazour GJ, San Agustin JT, Follit JA, Rosenbaum JL, Witman GB: Polycystin-2 localizes to kidney cilia and the ciliary level is elevated in orpk mice with polycystic kidney disease. Curr Biol 12: R378–R380, 2002 [DOI] [PubMed] [Google Scholar]
- 45.Badano JL, Mitsuma N, Beales PL, Katsanis N: The ciliopathies: An emerging class of human genetic disorders. Annu Rev Genomics Hum Genet 7: 125–148, 2006 [DOI] [PubMed] [Google Scholar]
- 46.Ko JY, Park JH: Mouse models of polycystic kidney disease induced by defects of ciliary proteins. BMB Rep 46: 73–79, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barr MM, Sternberg PW: A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature 401: 386–389, 1999 [DOI] [PubMed] [Google Scholar]
- 48.Pazour GJ, Dickert BL, Vucica Y, Seeley ES, Rosenbaum JL, Witman GB, et al.: Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J Cell Biol 151: 709–718, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin F, Hiesberger T, Cordes K, Sinclair AM, Goldstein LS, Somlo S, et al.: Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease. Proc Natl Acad Sci U S A 100: 5286–5291, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delling M, Indzhykulian AA, Liu X, Li Y, Xie T, Corey DP, et al.: Primary cilia are not calcium-responsive mechanosensors. Nature 531: 656–660, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma M, Tian X, Igarashi P, Pazour GJ, Somlo S: Loss of cilia suppresses cyst growth in genetic models of autosomal dominant polycystic kidney disease. Nat Genet 45: 1004–1012, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson PD: Mouse models of polycystic kidney disease. Curr Top Dev Biol 84: 311–350, 2008 [DOI] [PubMed] [Google Scholar]
- 53.Lu W, Peissel B, Babakhanlou H, Pavlova A, Geng L, Fan X, et al.: Perinatal lethality with kidney and pancreas defects in mice with a targetted Pkd1 mutation. Nat Genet 17: 179–181, 1997 [DOI] [PubMed] [Google Scholar]
- 54.Paterson AD, Wang KR, Lupea D, St George-Hyslop P, Pei Y: Recurrent fetal loss associated with bilineal inheritance of type 1 autosomal dominant polycystic kidney disease. Am J Kidney Dis 40: 16–20, 2002 [DOI] [PubMed] [Google Scholar]
- 55.Wu G, Markowitz GS, Li L, D’Agati VD, Factor SM, Geng L, et al.: Cardiac defects and renal failure in mice with targeted mutations in Pkd2. Nat Genet 24: 75–78, 2000 [DOI] [PubMed] [Google Scholar]
- 56.Lantinga-van Leeuwen IS, Dauwerse JG, Baelde HJ, Leonhard WN, van de Wal A, Ward CJ, et al.: Lowering of Pkd1 expression is sufficient to cause polycystic kidney disease. Hum Mol Genet 13: 3069–3077, 2004 [DOI] [PubMed] [Google Scholar]
- 57.Lantinga-van Leeuwen IS, Leonhard WN, van der Wal A, Breuning MH, de Heer E, Peters DJ: Kidney-specific inactivation of the Pkd1 gene induces rapid cyst formation in developing kidneys and a slow onset of disease in adult mice. Hum Mol Genet 16: 3188–3196, 2007 [DOI] [PubMed] [Google Scholar]
- 58.Piontek K, Menezes LF, Garcia-Gonzalez MA, Huso DL, Germino GG: A critical developmental switch defines the kinetics of kidney cyst formation after loss of Pkd1. Nat Med 13: 1490–1495, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harris PC, Torres VE: Genetic mechanisms and signaling pathways in autosomal dominant polycystic kidney disease. J Clin Invest 124: 2315–2324, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu G, D’Agati V, Cai Y, Markowitz G, Park JH, Reynolds DM, et al.: Somatic inactivation of Pkd2 results in polycystic kidney disease. Cell 93: 177–188, 1998 [DOI] [PubMed] [Google Scholar]
- 61.Lager DJ, Qian Q, Bengal RJ, Ishibashi M, Torres VE: The pck rat: A new model that resembles human autosomal dominant polycystic kidney and liver disease. Kidney Int 59: 126–136, 2001 [DOI] [PubMed] [Google Scholar]
- 62.Gallagher AR, Esquivel EL, Briere TS, Tian X, Mitobe M, Menezes LF, et al.: Biliary and pancreatic dysgenesis in mice harboring a mutation in Pkhd1. Am J Pathol 172: 417–429, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bakeberg JL, Tammachote R, Woollard JR, Hogan MC, Tuan HF, Li M, et al.: Epitope-tagged Pkhd1 tracks the processing, secretion, and localization of fibrocystin. J Am Soc Nephrol 22: 2266–2277, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Williams SS, Cobo-Stark P, James LR, Somlo S, Igarashi P: Kidney cysts, pancreatic cysts, and biliary disease in a mouse model of autosomal recessive polycystic kidney disease. Pediatr Nephrol 23: 733–741, 2008 [DOI] [PubMed] [Google Scholar]
- 65.Clapcote SJ, Roder JC: Simplex PCR assay for sex determination in mice. Biotechniques 38: 702, 704, 706, 2005 [DOI] [PubMed] [Google Scholar]
- 66.Hogan MC, Manganelli L, Woollard JR, Masyuk AI, Masyuk TV, Tammachote R, et al.: Characterization of PKD protein-positive exosome-like vesicles. J Am Soc Nephrol 20: 278–288, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kalari KR, Nair AA, Bhavsar JD, O’Brien DR, Davila JI, Bockol MA, et al.: MAP-RSeq: Mayo analysis pipeline for RNA sequencing. BMC Bioinformatics 15: 224, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL: TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H, Durbin R: Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liao Y, Smyth GK, Shi W: featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930, 2014 [DOI] [PubMed] [Google Scholar]
- 71.Wang L, Wang S, Li W: RSeQC: Quality control of RNA-seq experiments. Bioinformatics 28: 2184–2185, 2012 [DOI] [PubMed] [Google Scholar]
- 72.Robinson MD, McCarthy DJ, Smyth GK: edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z: GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eden E, Lipson D, Yogev S, Yakhini Z: Discovering motifs in ranked lists of DNA sequences. PLOS Comput Biol 3: e39, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.The Gene Ontology Consortium : The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res 47[D1]: D330–D338, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. The Gene Ontology Consortium : Gene ontology: Tool for the unification of biology. Nat Genet 25: 25–29, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Outeda P, Menezes L, Hartung EA, Bridges S, Zhou F, Zhu X, et al.: A novel model of autosomal recessive polycystic kidney questions the role of the fibrocystin C-terminus in disease mechanism. Kidney Int 92: 1130–1144, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Besse W, Dong K, Choi J, Punia S, Fedeles SV, Choi M, et al.: Isolated polycystic liver disease genes define effectors of polycystin-1 function [published correction appears in J Clin Invest 127: 3558, 2017]. J Clin Invest 127: 1772–1785, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Garcia-Gonzalo FR, Reiter JF: Open sesame: How transition fibers and the transition zone control ciliary composition. Cold Spring Harb Perspect Biol 9: a0281349, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Srour M, Schwartzentruber J, Hamdan FF, Ospina LH, Patry L, Labuda D, et al. FORGE Canada Consortium : Mutations in C5ORF42 cause Joubert syndrome in the French Canadian population. Am J Hum Genet 90: 693–700, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chaki M, Airik R, Ghosh AK, Giles RH, Chen R, Slaats GG, et al.: Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell 150: 533–548, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Castro-Miró M, Tonda R, Escudero-Ferruz P, Andrés R, Mayor-Lorenzo A, Castro J, et al.: Novel candidate genes and a wide spectrum of structural and point mutations responsible for inherited retinal dystrophies revealed by exome sequencing. PLoS One 11: e0168966, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Adly N, Alhashem A, Ammari A, Alkuraya FS: Ciliary genes TBC1D32/C6orf170 and SCLT1 are mutated in patients with OFD type IX. Hum Mutat 35: 36–40, 2014 [DOI] [PubMed] [Google Scholar]
- 84.Perrault I, Saunier S, Hanein S, Filhol E, Bizet AA, Collins F, et al.: Mainzer-Saldino syndrome is a ciliopathy caused by IFT140 mutations. Am J Hum Genet 90: 864–870, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boczek NJ, Hopp K, Benoit L, Kraft D, Cousin MA, Blackburn PR, et al.: Characterization of three ciliopathy pedigrees expands the phenotype associated with biallelic C2CD3 variants. Eur J Hum Genet 26: 1797–1809, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thauvin-Robinet C, Lee JS, Lopez E, Herranz-Pérez V, Shida T, Franco B, et al.: The oral-facial-digital syndrome gene C2CD3 encodes a positive regulator of centriole elongation. Nat Genet 46: 905–911, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu H, Galeano MCR, Ott E, Kaeslin G, Kausalya PJ, Kramer C, et al.: Mutations in DZIP1L, which encodes a ciliary-transition-zone protein, cause autosomal recessive polycystic kidney disease. Nat Genet 49: 1025–1034, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kaimori JY, Germino GG: ARPKD and ADPKD: First cousins or more distant relatives? J Am Soc Nephrol 19: 416–418, 2008 [DOI] [PubMed] [Google Scholar]
- 89.Wang S, Zhang J, Nauli SM, Li X, Starremans PG, Luo Y, et al.: Fibrocystin/polyductin, found in the same protein complex with polycystin-2, regulates calcium responses in kidney epithelia. Mol Cell Biol 27: 3241–3252, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kaimori JY, Nagasawa Y, Menezes LF, Garcia-Gonzalez MA, Deng J, Imai E, et al.: Polyductin undergoes notch-like processing and regulated release from primary cilia. Hum Mol Genet 16: 942–956, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bergmann C: ARPKD and early manifestations of ADPKD: The original polycystic kidney disease and phenocopies. Pediatr Nephrol 30: 15–30, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nakanishi K, Sweeney WE Jr, Zerres K, Guay-Woodford LM, Avner ED: Proximal tubular cysts in fetal human autosomal recessive polycystic kidney disease. J Am Soc Nephrol 11: 760–763, 2000 [DOI] [PubMed] [Google Scholar]
- 93.Konrad M, Zerres K, Wühl E, Rudnik-Schöneborn S, Holtkamp U, Schärer K; Arbeitsgemeinschaft für Pädiatrische Nephrologie : Body growth in children with polycystic kidney disease. Acta Paediatr 84: 1227–1232, 1995 [DOI] [PubMed] [Google Scholar]
- 94.Zerres K, Rudnik-Schöneborn S, Steinkamm C, Becker J, Mücher G: Autosomal recessive polycystic kidney disease. J Mol Med (Berl) 76: 303–309, 1998 [DOI] [PubMed] [Google Scholar]
- 95.Chapin HC, Rajendran V, Caplan MJ: Polycystin-1 surface localization is stimulated by polycystin-2 and cleavage at the G protein-coupled receptor proteolytic site. Mol Biol Cell 21: 4338–4348, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kleczko EK, Marsh KH, Tyler LC, Furgeson SB, Bullock BL, Altmann CJ, et al.: CD8+ T cells modulate autosomal dominant polycystic kidney disease progression. Kidney Int 94: 1127–1140, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Masyuk TV, Huang BQ, Masyuk AI, Ritman EL, Torres VE, Wang X, et al.: Biliary dysgenesis in the PCK rat, an orthologous model of autosomal recessive polycystic kidney disease. Am J Pathol 165: 1719–1730, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.