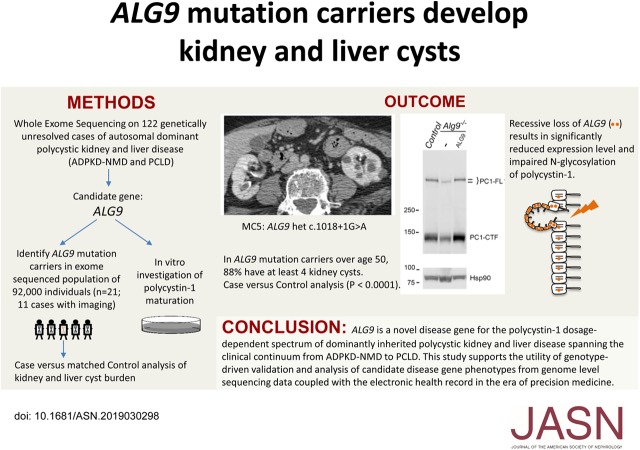
Significance Statement
Dominantly inherited polycystic kidney and liver phenotypes occur when epithelial cells in these organs have reduction of polycystin-1 functional dosage. In a cohort of genetically unresolved polycystic liver and kidney disease, the authors identified heterozygous loss of function mutations in ALG9. ALG9 encodes an endoplasmic reticulum enzyme that builds N-glycans, and the authors show that Alg9 inactivation results in impaired polycystin-1 maturation. Using a novel ‘genotype-first’ approach to ascertain individuals based strictly on their ALG9 genotype from a large cohort of exome-sequenced individuals, the authors find that 7/8 (88%) of ALG9 mutation carriers over age 50 have multiple kidney cysts. Together, these findings identify ALG9 as a novel human polycystic kidney and liver disease gene and support the utility of a genotype-driven approach to candidate disease gene validation and genotype-phenotype correlation.
Keywords: ADPKD, chronic renal disease, cystic kidney, human genetics, kidney stones, polycystic kidney disease
Visual Abstract
Abstract
Background
Mutations in PKD1 or PKD2 cause typical autosomal dominant polycystic kidney disease (ADPKD), the most common monogenic kidney disease. Dominantly inherited polycystic kidney and liver diseases on the ADPKD spectrum are also caused by mutations in at least six other genes required for protein biogenesis in the endoplasmic reticulum, the loss of which results in defective production of the PKD1 gene product, the membrane protein polycystin-1 (PC1).
Methods
We used whole-exome sequencing in a cohort of 122 patients with genetically unresolved clinical diagnosis of ADPKD or polycystic liver disease to identify a candidate gene, ALG9, and in vitro cell-based assays of PC1 protein maturation to functionally validate it. For further validation, we identified carriers of ALG9 loss-of-function mutations and noncarrier matched controls in a large exome-sequenced population-based cohort and evaluated the occurrence of polycystic phenotypes in both groups.
Results
Two patients in the clinically defined cohort had rare loss-of-function variants in ALG9, which encodes a protein required for addition of specific mannose molecules to the assembling N-glycan precursors in the endoplasmic reticulum lumen. In vitro assays showed that inactivation of Alg9 results in impaired maturation and defective glycosylation of PC1. Seven of the eight (88%) cases selected from the population-based cohort based on ALG9 mutation carrier state who had abdominal imaging after age 50; seven (88%) had at least four kidney cysts, compared with none in matched controls without ALG9 mutations.
Conclusions
ALG9 is a novel disease gene in the genetically heterogeneous ADPKD spectrum. This study supports the utility of phenotype characterization in genetically-defined cohorts to validate novel disease genes, and provide much-needed genotype-phenotype correlations.
Individuals with autosomal dominant polycystic kidney disease (ADPKD) develop fluid-filled cysts that originate from renal tubules and enlarge over time, eventually leading to renal failure in at least half of affected individuals by the sixth decade. In addition to kidney cysts, patients with ADPKD frequently develop liver cysts originating from the bile ducts.1 ADPKD most commonly results from mutations in PKD1 (77%) or PKD2 (15%), but approximately 8% of cases have no mutation detected (NMD) in either gene (ADPKD-NMD).2 Several genes which encode proteins in the endoplasmic reticulum (ER) that function in the biogenesis and quality control of integral membrane proteins have been implicated as disease genes for dominant polycystic kidney and liver phenotypes clinically characterized as isolated polycystic liver disease (PCLD) or ADPKD-NMD.2–8 PCLD, also known as autosomal dominant polycystic liver disease, presents with liver findings that are histologically and radiographically indistinguishable from those in ADPKD, but with few or absent kidney cysts.9 Although the ADPKD-PCLD diseases are inherited as dominant traits, accrual of somatic mutations resulting in biallelic loss of the respective disease genes in renal tubular and biliary epithelial cells is required for cyst initiation and accounts for the focal nature of the resulting polycystic phenotypes.10–14 Although there are a large number of client proteins that may be affected by loss of these ER-associated gene products, studies have shown that it is specifically the impaired post-translational maturation of the PKD1 gene product polycystin-1 (PC1) that produces the polycystic liver and kidney phenotypes.15 The factors that determine the relative occurrence of liver cysts and kidney cysts are not well understood but the ESKD that occurs in ADPKD due to PKD1 and PKD2 has not been attributed to mutations in any of the disease genes in the PCLD–ADPKD-NMD spectrum.2,6–8,15
In this study, we identified ALG9 as a novel candidate gene in our unsolved cases for the PCLD–ADPKD-NMD phenotypic spectrum. ALG9 encodes α-1,2-mannosyltransferase, which catalyzes the addition of the seventh and ninth mannose molecules to a growing N-glycan precursor in the ER lumen.16,17 Autosomal recessive loss of ALG9 results in a severe congenital disorder of glycosylation (CDG) with a multiorgan phenotype that includes kidney cysts.16,18,19 We validated ALG9 as a disease gene by in vitro functional bioassays showing both quantitative and qualitative defects in PC1 maturation after inactivation of Alg9. Amongst ALG9 loss-of-function mutation carriers identified in a large population-based cohort that had undergone whole exome sequencing (WES), we identified significant enrichment of kidney cysts.
Methods
PCLD–ADPKD-NMD Cohort and Exome Analysis
This study is approved by the Institutional Review Board at Yale University. The studied cohort consists of 122 unrelated cases of PCLD or ADPKD-NMD enrolled at Yale or received de-identified from collaborators, as well as selected mild–moderate severity ADPKD-NMD cases from the Consortium of Radiologic Imaging Study of PKD20 and HALT21 clinical cohorts obtained from the National Institute of Diabetes and Digestive and Kidney Diseases Central Repository. Cases solved by pathogenic variants in established disease genes (PKD1, PKD2, PRKCSH, SEC6, GANAB, ALG8, SEC61B, PKHD1, and DNAJB11) have been excluded. Extraction of genomic DNA, WES, variant calling, and analysis for this cohort have been described previously.7 New samples were captured by the xGen Exome Research Panel reagent from Integrated DNA Technologies and sequenced on the Illumina HiSeq4000 platform. Variants were filtered to consider only rare heterozygous likely deleterious variants (stop gain, frameshift, canonical splice, missense with Combined Annotation Dependent Depletion [CADD]22 score >20) of high sequencing quality. Rare variants were considered to be those with minor allele frequency (MAF) of <0.001 in the gnomAD Browser.23 False positive variant calls as defined by PLDiff/depth <724 were excluded from consideration in all analyses. As no single gene was enriched with variants among cohort cases in statistical analyses at a genome-wide level, we ranked all genes represented by at least one loss-of-function variant based on the number of individuals carrying potentially deleterious variants in the gene. ALG9 was then selected from among top candidates by review of available literature and resources, including data on protein function and localization, and mouse or human phenotypes. ALG9 variants investigated as part of this study are deposited in ClinVar (SCV000914195, SCV000914194, SCV000930618-SCV000930621).
In Vitro Gene Knockout
The parental cell line noted as “control” in this study is a mouse renal tubule epithelial cell line which contains a three-copy Pkd1F/H bacterial artificial chromosome (BAC) with amino-terminal (N-terminal) FLAGx3 and carboxy-terminal (C-terminal) HAx3 epitope tags.15 The cell line has stable expression of Cas9 introduced by lentiviral transduction using lentiCas9-Blast (Addgene 52962). Guide-RNA sequence targeting the reverse strand of Alg9 exon 6 (5′-TCCCGTCATGGCGATCAGCG-3′) was introduced transiently using electroporation of a modified pGL3-U6-sgRNA-PGK-puromycin plasmid (Addgene 51133) lacking the puromycin cassette to direct the Cas9 protein to make a double-stranded cut at this site. Transfected cells were clonally diluted after 72 hours and grown until sufficient quantity to passage. Cell line clones were screened by PCR and Sanger sequencing for frameshift mutations (Supplemental Figure 1).
Immunoblotting and Glycosylation Analysis
Immunoblotting was performed on whole cell lysate prepared using RIPA buffer as reported previously.7 For quantitative comparisons, cell lysate concentration was measured by Lowry protein assay using standard reagents. For Ponceau staining, membrane was washed in Tris-buffered saline/Tween 20 (TBST), submerged in Ponceau S Solution (Sigma) for 10 minutes, rinsed briefly in water to remove background staining, and dried on tissue. Membrane was rehydrated in 100% methanol before proceeding with blocking in 5% nonfat milk in TBST. The following primary antibodies were used at 1:3000 dilution in 5% milk in TBST unless otherwise noted: anti-hemagglutinin (anti-HA) (3F10; Roche), anti–PC1-LRR (7e12, dilution 1:1000; Santa Cruz Biotechnology), chicken anti-green fluorescent protein (anti-GFP) antibody (GFP-1020; Aves Labs), and rabbit anti-Hsp90 antibody (Cell Signaling). For glycosylation studies, cell lysate obtained in RIPA buffer was treated with Endoglycosidase H (EndoH) per nondenaturing protocol (NEB) for 3 hours at 37°C.
Missense Variant Analysis
In silico analysis of missense variants was performed using prediction algorithms including CADD,22 MetaSVM and MetaLR,25 REVEL,26 and FATHMM-MKL.27 Biochemical validation studies were conducted on stable cell lines generated by lentiviral transduction of C-terminal GFP fusion with human ALG9 cDNA (ENST00000616540.4; 618 amino acid transcript) into the Alg9−/− cell line. Human ALG9 cDNA sequence was amplified from total RNA of human embryonic kidney 293 T cells using a two-step PCR strategy. The 5′ untranslated region to the middle of the transcript was amplified separately from overlapping region spanning from the middle of the transcript to the 3′ untranslated region, and these products were then used as a template for a second PCR reaction, using primers designed to add restriction sites to each end. This product was cloned into Zero Blunt TOPO vector using a cloning kit from ThermoFisher, and the correct clone was confirmed by Sanger sequencing. This ALG9 sequence in the TOPO vector was modified using a site-directed mutagenesis protocol.28 Briefly, Pfu polymerase (Promega) amplified a modified plasmid primed by complimentary forward and reverse primers containing the sequence edit (Supplemental Methods). The resultant product was transformed into DH5α competent cells after digestion of parental template plasmid with DpnI (NEB). A single modified clone for each missense variant was confirmed by sequencing, cut by restriction digest, ligated into pLVX-IRES-Puro (Clontech), and again each was confirmed with Sanger sequencing. GFP expression and localization for each cell line was examined by immunoblotting and fluorescence microscopy, respectively.
MyCode Cohort Exome Analysis
Whole-exome sequencing and variant calling were performed before this study on 92,455 MyCode Community Health Initiative participants29 as part of the Geisinger-Regeneron DiscovEHR collaboration. Methods for whole-exome capture and variant calling in this cohort have been previously described. Briefly, NimbleGen (SeqCap VCRome) probes were used for exome capture of the first approximately 61,000 samples and a modified version of Integrated DNA Technologies xGen probes was used for the remaining approximately 31,000 samples.30,31 Variants were annotated using the Ensembl Variant Effect Predictor with Ensembl 90 definitions.32 PLdiff/depth and gnomAD were used as described above to filter out common and false positive variants. Carriers of ALG9 loss-of-function variants were identified from this cohort and confirmed with Sanger sequencing. We accessed pedigree reconstruction data assembled from exome-based identity-by-descent analysis to determine the relatedness between individuals to identify any first or second degree relatives for the ALG9 carriers in the cohort.31
MyCode Cohort Case versus Control Study
The study of the MyCode cohort was approved by the Institutional Review Board at Geisinger. Patients carrying the ALG9 mutation who were eligible for matching were defined as those with abdominal imaging studies available for digital review with a definable native kidney eGFR—i.e., not on dialysis or with kidney transplant. One imaging exam was selected for each case by a predefined algorithm: magnetic resonance imaging (MRI) > computed tomography (CT) > ultrasound, and exams with contrast > those without. Clinical information for carriers of ALG9 mutations was obtained from clinical databases and used to select random matched controls in a 2:1 ratio (see power calculation below) based on six parameters (see Results). A radiologist with expertise in abdominal imaging (W.J.T.), blinded to patient genotypes and clinical information, provided interpretation of kidney and liver findings in the selected imaging studies using clinical radiologic diagnostic criteria. Peripelvic cysts, although infrequent (data not shown), were excluded from the analysis given their lymphatic origin and inability to reliably delineate discrete cysts from the collecting system. For CT scans, a cyst was defined as a homogeneous mass of water attenuation (−10 to 20 HU) with an imperceptible wall and no enhancement after administration of contrast or, in the case of hemorrhagic or proteinaceous cysts, as a homogeneous, nonenhancing lesion with precontrast density >20 HU. Size thresholds were set based on the heterogeneity of the CT data, discussed below (see Results). For MRI, a cyst was defined as a lesion distinct from the surrounding parenchyma with either homogenous internal T2-weighted signal intensity consistent with simple fluid, or a lesion with T1 and T2 signal characteristics consistent with a hemorrhagic or proteinaceous cyst. In cases where postcontrast imaging was available, the lesions showed no enhancement. For ultrasound imaging, a cyst was defined as an anechoic structure with a distinct posterior wall and posterior enhancement.
Statistical Analyses
A two-tailed test was used to compare means of biologic repeats on Western blots. Sample size calculations for case versus control analysis were performed using Power Analysis Sample Size (PASS) with the goal of achieving >90% power in detecting a difference if cases and controls had an 80% and very conservatively selected 20% incidence of kidney cysts, respectively. Case versus control comparisons were performed using the Fisher exact test.
Results
ALG9 Is a Candidate Gene for PCLD–ADPKD-NMD
We performed WES on 122 unrelated patients with ADPKD-NMD or PCLD without pathogenic mutations in previously established nor proposed disease genes. We identified one case with a heterozygous loss-of-function mutation in exon 6 of ALG9, p.W227X, and five other cases with rare (MAF<1×10−3) heterozygous missense variants of unknown consequence in ALG9 (Table 1). Each of the five missense variants was predicted to be in the top 0.3% most likely deleterious of all potential genome single nucleotide variants (CADD-phred ≥25) but had differing predictions with other in silico algorithms (Table 1).33 In vitro inactivation of the previously reported PCLD–ADPKD-NMD disease genes that encode ER proteins have shown significant quantitative reduction and qualitative alteration of mature PC1 protein.6–8,15 In vitro biallelic inactivation models the cellular recessive genotype resulting from somatic second hits in cyst-forming epithelium. We therefore investigated the effects of inactivation of ALG9 on PC1 in a cell culture system. The full-length PC1 protein (PC1-FL) is normally cleaved into an N-terminal fragment and an 11-transmembrane-spanning C-terminal fragment (PC1-CTF) before leaving the ER and trafficking to functional destinations on the cell surface. We used CRISPR/Cas9 to introduce a frameshift in exon 6 of Alg9 to inactivate it in a mouse kidney tubular epithelial cell line. The isogenic unmodified parental cell expressing Cas9 was used as control. The parental cell line expresses three copies of a BAC transgene, Pkd1F/H-BAC, containing the full genomic sequence of mouse Pkd1 modified with addition of N-terminal FLAGx3 and C-terminal HAx3 epitope tags.7,15 Sanger sequencing of the genomic region of Alg9 targeted by the guide RNA in a clonally selected candidate Alg9−/− cell line confirmed the presence of a homozygous frameshifting mutation (Supplemental Figure 1). Alg9−/− cells had no apparent morphologic changes on light microscopy compared with the parental cell line.
Table 1.
ALG9 heterozygous mutation carriers in 122 genetically unresolved patients with PCLD–ADPKD-NMD
| Identifier | Nucleotide Changea | Amino Acid Change | MAF gnomAD | CADDb | MetaSVMb | MetaLRb | REVELc | FATHMM-MKLb | Age/Gender | Kidney Cystsd | Liver Cysts |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases with pathogenic ALG9 mutations | |||||||||||
| G8261813e | c.681G>A | p.W227X | Novel | N/A | N/A | N/A | N/A | D | 60/M | Innumerable | N/A |
| YU202f | c.1109G>A | p.R370K | 4×10−6 | 33 | D | D | 0.912 | D | 56/M | 7:11 | Innumerable |
| Cases not explained by ALG9: benign missense variants only | |||||||||||
| YU22g | c.694G>C | p.A232P | 3×10−4 | 25 | D | T | 0.395 | D | 67/F | N/A | Multiple |
| YU36h | c.839C>T | p.A280V | 7×10−6 | 31 | T | T | 0.324 | D | 66/F | — | Multiple |
| 453976i | c.944A>G | p.N315S | 6×10−5 | 25 | T | T | 0.574 | D | 42/F | Multiple | Innumerable |
| YU394 | c.1550G>T | p.R517L | 4×10−6 | 27 | T | T | 0.462 | D | 63/F | 1:0 | Multiple |
Yale case cohort comprised of PCLD and mild-moderate ADPKD-NMD cases from Consortium of Radiologic Imaging Study of PKD (CRISP)/HALT Progression of Polycystic Kidney Disease (HALT PKD) cohorts. N/A, not applicable; D, deleterious; T, tolerated.
ENST00000616540.4 (CCDS73380).
Variant prediction (see Methods).
REVEL scores range from zero to one, with one representing all simulations suggest pathogenicity.
Cyst counts (right:left) or description from available imaging.
HALT identifier.
Mayo Clinic Family T57.
Mayo Clinic Family T39.
Mayo Clinic Family T17.
CRISP identifier.
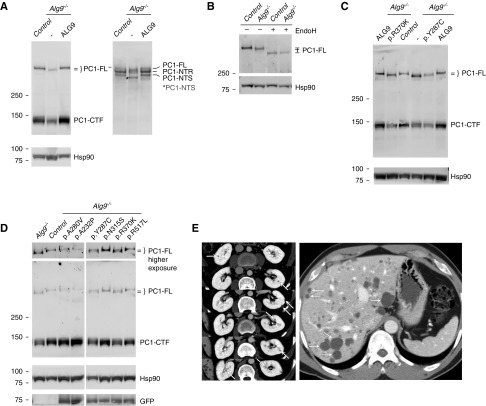
Alg9−/− cells showed a significant decrease in the steady-state level of PC1-CTF to approximately 70% of the control intensity, quantified in four biologic samples per genotype by anti-HA (Figure 1A, left panel; Supplemental Figure 2). A total of 57 of the 60 predicted N-glycosylation sites on PC1 are in the N-terminal fragment. After exit from the ER, enzymes in the Golgi modify the N-glycans conferring resistance to EndoH which is associated with slower migration on protein gel electrophoresis. This allows experimental distinction of the immature EndoH-sensitive PC1 N-terminal fragment (PC1-NTS) that has not traversed the Golgi from the EndoH-resistant fragment (PC1-NTR) that reaches the cell surface. Immunoblot analysis with antibody directed to the PC1 N-terminal resolves three fragments: PC1-FL, PC1-NTR, and PC1-NTS (Figure 1A, right panel). Alg9−/− cells show quantitatively decreased PC1-NTR, indicating reduced mature, cell surface–expressed PC1. In addition to quantitative differences, PC1-FL and PC1-NTS from Alg9−/− cells have faster migration in denaturing gel electrophoresis compared with control cells (Figure 1A, Supplemental Figure 2). This may also be true of PC1-NTR, but the overlapping migration of PC1-FL and PC1-NTR from Alg9−/− cells prevents definitive conclusion. The genotype-dependent differences in migration of both cleaved and uncleaved PC1 suggest that, in the absence of Alg9, there is altered glycosylation of PC1. Enzymatic removal of N-glycans, which on PC1-FL are all EndoH sensitive, results in identical migration between genotypes (Figure 1B), confirming that the faster migration of PC1-FL from Alg9−/− cells is due to altered N-glycosylation. Finally, both the altered protein expression levels and altered glycosylation of PC1 in Alg9−/− cells showed expression indistinguishable from control cells when human ALG9-GFP is re-expressed in Alg9−/− cells (Figure 1A). This confirms that the effects on PC1 maturation are a specific consequence of loss of Alg9 and provides strong support for the conclusion that ALG9 is a human polycystic disease gene.
Figure 1.
Alg9 loss causes abnormal biogenesis of PC1. (A) Immunoblots of cell lysate with anti-HA (left panel) and anti-LRR PC1–N-terminal antibody (7e12) (right panel) show quantitative decrease in PC1–C-terminal fragment (PC1-CTF) and the mature EndoH-resistant fraction of PC1–N-terminal fragment (PC1-NTR), as well as faster migration of PC1 full-length (PC1-FL) and the immature EndoH-sensitive PC1–N-terminal fragment (PC1-NTS) in Alg9−/− cells compared with controls. Re-expression of human ALG9-GFP in Alg9−/− cells rescues both the quantitative and migration differences (third lane). (B) Cell lysate was treated with EndoH and blotted with anti-HA. The migration difference of PC1-FL between control and Alg9−/− cell lysates is eliminated after EndoH treatment, showing that there is altered glycosylation of PC1 in the absence of Alg9. (C and D) Anti-HA immunoblots of cell lysate from Alg9−/− cells with or without stable re-expression of human ALG9-GFP with either wild-type (ALG9) or the indicated missense variants. The known pathogenic missense mutation, p.Y287C, and the experimental missense variant, p.R370K, do not rescue the PC1 phenotype in Alg9−/− cells. (D) The other experimental missense variants rescue the PC1 phenotypes. This bioassay identifies p.R370K as a deleterious missense mutation and p.A232P, p.A280V, pN315S, and p.R517L as benign variants. (E) Imaging for YU202 with ALG9-p.R370K. Serial CT scan sections show multiple small kidney cysts (arrows) many of which are likely hemorrhagic/proteinaceous (left panel) and innumerable liver cysts (right panel).
Assessment of ALG9 Missense Variants Found in Patients with PCLD–ADPKD-NMD
We next used this system to evaluate the functional consequences of the ALG9 missense variants identified in our patients (Table 1). We expected that re-expression of benign human ALG9 missense variants would rescue PC1 properties in Alg9−/− cells, whereas deleterious missense variants would fail to do so. We modified the human wild-type ALG9-GFP cDNA sequence to introduce each of the five missense mutations (Table 1). In addition, we produced a clone with a known deleterious missense variant ALG9-p.Y287C16 as a positive control. Each cDNA was introduced into the Alg9−/− cell line by lentiviral transduction and stably expressing cell lines were selected. Expression of each clone was verified by GFP epifluorescence in cell culture (data not shown). Alg9−/− cells expressing the positive control ALG9-p.Y287C, as well as the line expressing the patient-derived variant ALG9-p.R370K, showed quantitatively reduced PC1-CTF and slower migrating PC1-FL indistinguishable from that seen in the Alg9−/− (Figure 1C). The Alg9−/− cells expressing ALG9-p.A232P, ALG9-p.A280V, ALG9-p.N315S, or ALG9-p.R517L each showed complete rescue of PC1 quantitative and qualitative maturation defects (Figure 1D). The data indicate that, among the rare missense mutations found in the patients, only ALG9-p.R370K resulted in ALG9 loss of function (Table 1). The demonstration of significant impairment of PC1 maturation resulting from ALG9 exon 6 truncation or ALG9-p.R370K, and the occurrence of these variants in patients with the ADPKD-PCLD phenotype support these as the causative variants for these two patients in our cohort (Table 1). YU202, with pathogenic missense variant ALG9-p.R370K, is a 56-year-old male with clinically diagnosed PCLD (Figure 1E). His father also had PCLD, but no genetic diagnosis. G8261813, with ALG9-p.W227X, was diagnosed with ADPKD-NMD as an enrolled participant in the HALT PKD study (imaging not available). These clinical data indicate that ALG9-related disease can span the spectrum from PCLD to ADPKD.
Genotype-First Approach To Characterize ALG9 Mutation Carriers
To evaluate these findings further, we determined the burden of polycystic kidney and liver phenotypes in cases selected based solely on the presence of ALG9 loss-of-function mutations from a large population-based cohort in which WES data are linked to electronic health record data.30 Among the 92,455 exomes queried, we identified 21 carriers of rare (MAF<1×10−3) heterozygous ALG9 loss-of-function mutations (nonsense, frameshift, canonical splice, or start loss) meeting sequence quality control thresholds (see Methods). There were no instances of individuals with homozygous or compound heterozygous ALG9 loss-of-function mutations. Principle component analysis and relatedness analysis of exome data indicated that all 21 of these patients were of European descent and no close familial relations were present between them. Of the 21 patients, 14 (67%) had at least one abdominal imaging report available for evaluation. The ALG9 variants (Table 2) in these 14 individuals were confirmed by Sanger sequencing. None carried loss-of-function mutations in established cystic disease genes, and missense variants in these genes were benign by in silico prediction (Supplemental Table 1). Clinical radiology reports described kidney cysts (n=7) or hypodensities suggestive of cysts (n=4) in these 14 patients, and liver cysts in only one. To characterize and quantify these findings and determine the specificity of these phenotypes to the ALG9 genotype in light of the sporadic occurrence of kidney and liver cysts in some individuals with increasing age, we designed a case versus control analysis in which a radiologist blinded to case-control status evaluated and quantitated cyst burden.
Table 2.
Alg9 loss-of-function mutations carriers with abdominal imaging among 92,455 patients in a health system
| Identifier/Gender | Nucleotide Changea | Amino Acid Change | MAF gnomAD | Imaging Type (age)b | Kidney Cystsc | TSTCc | Nephrolithiasisd | eGFR (age)e |
|---|---|---|---|---|---|---|---|---|
| Cases in case-control analysis | ||||||||
| Kidney cysts | ||||||||
| MC2/F | c.427C>T | p.R143X | Novel | MRI (65) | 10 | 18 | Y | 60 (65) |
| MC3/M | c.1295C>A | p.S432X | Novel | US (73) | Innumerable | — | + | 26 (75) |
| MC5/F | c.1018+1G>A | Splice variant | Novel | CT+ (86) | 39f | 51 | Yg | 38 (96) |
| MC7/M | c.1472delA | p.N491IfsTer33 | Novel | CT+ (69) | 6 (9) | 22 | Y | 74 (73) |
| MC9/F | c.506dupT | p.S170EfsTer24 | Novel | US (66) | 5 | — | + | 90 (66) |
| MC10/M | c.883_885delCCT_insGTAAA | p.P295VfsTer13 | Novel | CT+ (68) | 9 | 21 | Y | 52 (75) |
| MC12/F | c.511C>T | p.R171X | 8.13E−06 | CT+ (61) | 4 (9) | 20 | — | 46 (63) |
| One or fewer cysts | ||||||||
| MC1/M | c.1018+1G>A | Splice variant | Novel | CT+ (42) | — | 1 | — | 50 (51) |
| MC4/F | c.1088_1091delTCCA | p.I363SfsTer52 | 4.06E−06 | CT+ (82) | — | — | — | 47 (85) |
| MC8/F | c.3G>A | Start loss | Novel | CT+ (37) | — | 1 | — | 72 (44) |
| MC14/F | c.3G>A | Start loss | Novel | MRI+ (37) | 1 | 3 | + | 90 (44) |
| Cases not matched for case-control analysis | ||||||||
| MC6/M | c.1018+1G>A | Splice variant | Novel | CT+ (76) | Yh | — | 47 (78) | |
| MC11/F | c.1363C>T | p.R455X | 4.06E−06 | CT (70) | Yh,i | — | ESKD (66) | |
| MC13/F | c.566-1G>A | Splice variant | 1.22E−05 | CT+ (49) | Yh | — | 97 (50) | |
US, ultrasound.
ENST00000616540.4 (CCDS73380).
+, with contrast.
Kidney cysts (>8 mm) and lesions TSTC (4–8 mm) as described in Methods and Results. When additional imaging allowed for recharacterization of indeterminate masses as cysts, cyst count inclusive of these is noted in parentheses after count from originally selected image.
Noted during blinded analysis. Y, yes; +, nephrolithiasis noted on additional CT scan if available.
Most recent outpatient eGFR.
Among cases with liver imaging, this was the only case with liver cysts noted.
Pathology of bilateral nephrolithiasis demonstrated uric acid composition.
Based on clinical radiology report.
Native kidneys by CT 2/4 yr after transplant/ESKD.
Case versus Matched Control Analysis
We included 11 of the 14 ALG9 cases for the case-control study. The excluded patients include two for which only a scanned radiology report but no digital imaging was available for re-evaluation (MC6, MC13; Table 2). The third patient had reached ESKD before first available imaging (MC11; Table 2). We selected two random controls for each of the 11 remaining cases from exome-sequenced individuals lacking any coding region variants in PKD1 or PKD2 (MAF<2×10−3) or other established PCLD gene (MAF<1×10−2); they were matched for six parameters: age at time of imaging, type of imaging study including use of contrast, year of imaging, CKD stage at time of imaging, gender, and ethnicity. We defined four or more cysts in patients aged 50 years or more as a threshold for positive evidence of cystic disease. This criterion was informed by Rule et al.34 who report one, two, or three cysts of ≥5 mm seen in 26%, 9.8%, and 4.3%, respectively, in healthy organ donors aged 50 or more. Four or more cysts only occurred 1.2% of those healthy individuals.34 We assessed kidney and liver lesions in a blinded fashion using prespecified radiographic criteria (see Methods) for all 33 individuals (11 cases, 22 matched controls; Supplemental Table 2; Table 2). Although matched between cases and controls, the 33 imaging studies included different imaging modalities with variable technical parameters inherent in a cohort spanning multiple decades. In aggregate, the CT exams, which were the majority of the data, dictated that a size threshold of ≥8 mm allowed for a definitive application of the specified criteria for a cyst, and a minimum size threshold of ≥4 mm allowed for lesions to be detectable across all exams. As such, lesions ≥8 mm were classified as either “cyst” or “indeterminate mass,” and lesions <8 mm but ≥4 mm were reported as “too small to characterize” (TSTC) per common nomenclature. To facilitate direct comparison to the CT data, the same TSTC thresholds were applied to the MRI data. In contrast to CT and MRI, the concept of TSTC does not apply to the ultrasound data, as such findings would not be reliably detected by that modality.
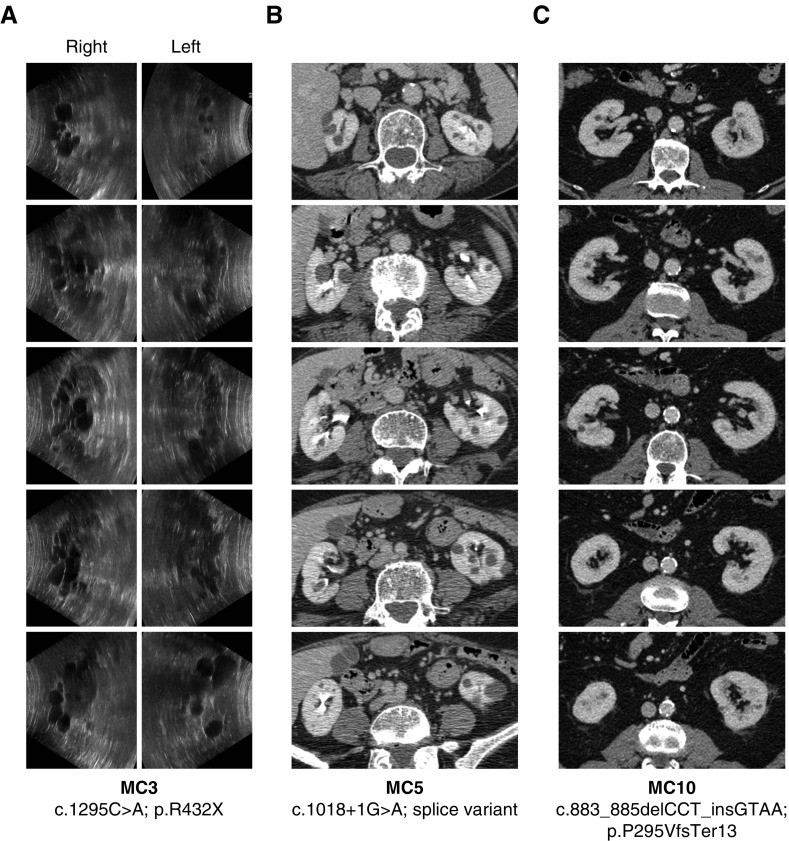
Among the 11 ALG9 mutation carriers (cases) analyzed in the case-control analysis, seven (64%) had four or more kidney cysts (Figure 2, Table 2). None of the 22 controls met this threshold (P<0.001; Tables 2 and 3; see Methods). The four ALG9 cases with less than four kidney cysts included the three youngest patients, aged 37, 37, and 42, each of whom also had at least one TSTC in addition to the observed cysts. The fourth case was an 82-year-old female with no cysts. Considering only the cases and controls ≥50 years of age, seven of eight cases (88%) versus none of 16 controls had four or more kidney cysts (P<0.001; Table 3). The background occurrence of a single cyst or TSTC was relatively common in controls (ten of 22; 45%), but significantly less than in cases (ten of 11; 91%; P=0.02). The case versus control cyst count comparison for the selected threshold of four or more kidney cysts remained robustly significant (P<0.01) even when criteria were modified to include TSTCs or any number of bilateral cysts (Table 3).
Figure 2.
Cystic kidney phenotypes in carriers of ALG9 heterozygous loss-of-function variants are of varying severity. (A) Ultrasound long-axis views from anteroposterior sweep, and (B and C) axial CT scan sections. Quantification of lesions in all cases is presented in Table 2.
Table 3.
Case versus matched control analysis
| Cyst count | Cases | Controls | P Value |
|---|---|---|---|
| Case-control cohort | n=11 | n=22 | |
| Cyst count ≥4 | 7 | 0 | <0.001 |
| Cysts plus TSTC combined count ≥4 | 8 | 4 | 0.0055 |
| Bilateral cystsa | 7 | 2 | 0.0021 |
| Subset of age over 50 | n=8 | n=16 | |
| Cyst count ≥4 | 7 | 0 | <0.001 |
| Cysts plus TSTC combined count ≥4 | 7 | 4 | 0.0087 |
| Bilateral cystsa | 7 | 2 | 7.5×10−4 |
Bilateral cysts: at least one cyst in each kidney.
The severity of the cystic phenotype in cases is highly variable between individuals. Cyst numbers ranged from zero to innumerable kidney cysts (Figure 2, Table 2). In contrast, no controls had more than two cysts. Cysts were bilateral in all seven affected cases, and were mentioned in the clinical radiology reports for all seven cases. Five of the six cases with a finite number of cysts had 18 or more lesions of 4–8 mm diameter, suspected to be early cysts but noted as TSTCs (Figure 2, Table 2). The largest kidney cysts in cases ranged from 1.2 to 4.8 cm. For two cases with four and six cysts respectively, specific additional kidney lesions counted as indeterminate masses could be categorically defined to be cysts upon review of additional imaging studies (Table 2). The liver was not imaged in the two ALG9 carriers who only had renal ultrasound available; only one of the nine cases with liver imaging (MC5) had liver cysts. MC5’s only partially imaged liver contained two cysts, up to 1.1 cm in size, and four TSTCs.
There was limited availability of familial data in this study. At the time of YU202’s enrollment, he reported that his father had multiple liver cysts at age 83, and mother was living and well. For the cases in the MyCode cohort, we used the available pedigree reconstruction from the 92,455 individuals to supplement the absence of relevant family history data in clinical charts. Only MC1 and MC3 have relatives in the cohort, a mother and a brother, respectively, and each are noncarriers of the respective ALG9 mutations. MC3 (Figure 2A, Table 2) is the most significantly affected case in this cohort, and notably the brother of this individual did not share the ALG9 mutation and did not have kidney cysts on a CT scan performed at age 70.
The blinded review incidentally noted nephrolithiasis in four ALG9 mutation carrier cases (two CT scan, one ultrasound, and one MRI) on the single imaging study used in the study. This was only noted in one control (ultrasound exam). As MRI and ultrasound are inferior for identifying nephrolithiasis, and nephrolithiasis can be transient on imaging, we performed additional chart review to consider other available CT scans for the cases and controls. CT scan clinical reports confirmed the nephrolithiasis found in the four cases, and reported nephrolithiasis in an additional three cases for a total of seven of 11 (64%) cases with digital imaging (Table 2). Six of these seven were among the cases with multiple kidney cysts and the seventh had one kidney cyst and three TSTCs. The most recent CT scan clinical reports for the controls did not describe any additional cases of nephrolithiasis, however, we did not review every available scan for each control as this study was not designed to make a case versus control comparison for this finding. Of the remaining ten ALG9 mutation carriers who lacked digital imaging data, there was no notation of nephrolithiasis by International Classification of Diseases code, an insensitive measure, nor on the imaging report for MC6, MC11, or M13.
In aggregate, this genotype-first analysis shows a significantly greater incidence of multiple kidney cysts in ALG9 loss-of-function carriers compared with individuals without this genotype. Given the expanding availability of exome-sequenced cohorts tied to the live electronic health record, this genotype-first approach will be a valuable adjunct in validating the pathogenicity of candidate genes for rare Mendelian disease traits.
Discussion
Our study is the first to complement gene discovery in clinically ascertained polycystic kidney and liver disease with phenotypic characterization in genetically defined at-risk patients from population-based exome sequencing efforts. We show the significant benefits of combining these complementary strategies. Recent implication and analyses of disease genes for ADPKD and PCLD demonstrate that these clinically defined human phenotypes are found in individuals with heterozygous mutations in genes required for sufficient PC1 functional dosage. As such, the implication and study of novel disease genes for this genetically heterogeneous phenotype offers concrete entry points for novel biologic investigations into PC1 function, an entity of central importance to the design of targeted therapies for ADPKD and PCLD–ADPKD-NMD. These phenotypes exist both mechanistically and phenotypically on more of a continuum than was realized before these studies.6–8,15 This study adds the novel benefit of being able to detail findings in mutation carriers of a disease gene independent from the influence of the clinical polycystic cohort from which it was discovered.
The population-based ascertainment of ALG9 mutation carriers in the MyCode cohort provides several insights. Firstly, because human variation assures that no additional gene variants would be over-represented in the carriers of ten novel or ultra-rare ALG9 variants, it provides the strength of a randomized study investigating the effect of the ALG9 variant alone, not requiring a co-occurring variant. We found that the penetrance of four or more definitive kidney cysts in individuals over age 50 with ALG9 mutations is 88% (seven of eight). The size and number of cysts found in ALG9 carriers is generally milder than typical ADPKD, showing large variation in expressivity. ALG9 carriers show a kidney-predominant phenotype, differing from hypothesis based on four of the six PCLD–ADPKD-NMD disease genes encoding proteins in the ER which have been described in patients exclusively with PCLD. This raises the possibility that similar genotype-first analyses for other PCLD genes related to ER biogenesis may establish whether the stronger correlation with PCLD is a reflection of the disease mechanism or related to the criteria for ascertainment used in previous discovery cohorts.3–5,7 The generally mild phenotype in human ALG9 disease is likely determined by two factors: the relative frequency of somatic second hit mutations inactivating the normal copy of ALG9, and the level of damage to the PC1 functional dose that results from loss of ALG9.7
ALG9 joins two other genes, ALG8 and PMM2, implicated in kidney and liver cyst phenotypes whose recessive loss results in rare CDG.7,35 Kidney cystic disease attributed to PMM2 mutations is described as part of a hyperinsulinemic hypoglycemia and polycystic kidney disease syndrome attributed to recessively inherited biallelic nontruncating mutations, at least one of which is a unique promotor variant proposed to result in decreased expression of PMM2 specifically in the pancreas, kidney, and liver, avoiding the typical neurologic symptoms of PMM2-CDG.35 Due to a germline recessive genotype, the kidney cystic phenotype in hyperinsulinemic hypoglycemia and polycystic kidney disease does not require accrual of somatic second hit mutations, is diagnosed at a younger age, and affects the organ diffusely. Recessive loss of the α-1,2-mannosyltransferase encoded by ALG9 causes the rare ALG9-CDG. Unique from other CDGs, kidney cysts are present in all five of five cases in which kidney examination was included among the 12 reported cases of ALG9-CDG. The pathogenic ALG9 missense variant p.R370K described in this report, as well as the variants that cause ALG9-CDG (p.Y287C, p.N527K, p.E530K; ENST00000616540.4; CCDS73380), are all predicted to be on the ER luminal side of the ALG9 ER transmembrane protein.16,19,36 ALG9 functions to add specific mannose molecules to the assembling N-glycan precursors in the ER lumen.17 The presence of kidney cysts in ALG9-CDG at birth, in contrast to the age-dependent and incomplete penetrance in patients heterozygous for ALG9, is supportive of the hypothesis that a cellular recessive mechanism established for ADPKD and PCLD is required to initiate cysts due to ALG9 mutations. As the proteins encoded by ALG9, ALG8, and PMM2 are enzymes with nonredundant roles in the assembly of N-glycans, our findings suggest a requirement of N-glycosylation in renal tubular and biliary epithelial homeostasis.
ALG9 is now the third ER protein–encoding polycystic disease gene, after GANAB and DNAJB11, described to explain either kidney-predominant polycystic phenotypes or PCLD.6,8 We also reported a young female with a mutation in ALG8 who had eight bilateral kidney cysts without liver cysts.7 DNAJB11 is the only of these genes with reported cases of ESKD, but this was proposed to result from tubulointerstitial fibrosis rather than polycystic kidney disease.8 In this study, one ALG9 mutation carrier reached ESKD at age 66 with atrophic native kidneys containing bilateral renal cysts on imaging 4 years after onset of ESKD. Lack of records from earlier time points precludes drawing any conclusions in this case. In the two ALG9 cases with the largest number of kidney cysts (MC3, MC5), the eGFRs were 26 and 38, and the kidney size was mildly enlarged or normal at 14/15 and 10/12 cm, respectively. The patient we report from the HALT ADPKD cohort, for whom imaging and organ size measurements were not available, had an eGFR of approximately 30 ml/min at age 60. It remains possible that ALG9 falls in the group of ER-associated polycystic kidney disease genes that may on rare occasions be associated with CKD, although additional study will be required to validate this clinical association.
The finding of nephrolithiasis in the clinical imaging reports of seven of 11 ALG9 carrier cases is of interest but should be interpreted with caution. Clinical indication for CT scanning was not noted in our analyses, therefore indication bias enriching nephrolithiasis in the patients who had imaging is possible. Nonetheless, considering the unbiased denominator of 21 ALG9 mutation carriers, we can report a lifetime prevalence of at least 33% for nephrolithiasis in ALG9 loss-of-function mutation carriers. This is comparable to the 28% found in patients with ADPKD by CT scan imaging.37 Nephrolithiasis prevalence in the United States is reported at approximately 9% when self-reported by patients,38,39 and similarly when 1220 veterans were screened with ultrasound,40 but comparable CT scan studies have not been done. The available data thus suggest that nephrolithiasis in ALG9 carriers is of similar prevalence to that in typical ADPKD, which is considered enriched, but that comparisons cannot be drawn to the general population.
Our novel use of the genotype-first approach to disease gene validation and phenotype characterization was able to uncover a kidney-predominant cystic phenotype which would not have been possible from our phenotypically defined cohort of predominantly PCLD. It also allows for quantification of additional imaging findings such as nephrolithiasis. This approach required the initial implication of ALG9 in phenotype-based cohorts. Although our data describe polycystic kidneys and suggest that a liver-specific (PCLD) phenotype may occur in fewer than one in 14 ALG9 mutation carriers, one of our two clinically ascertained patients had PCLD, thus some patients may manifest this way. As is the case for GANAB and DNAJB11, and potentially other genes on the PCLD–ADPKD-NMD spectrum, ALG9 mutations will likely explain only a very small fraction of cases with clinically diagnosed ADPKD. Nonetheless, ALG9 may be considered when mutations in PKD1 or PKD2 are not found in mild–moderate cases of ADPKD-NMD, as well as for PCLD and phenotypes spanning these clinical distinctions. We suggest the genotype-based ascertainment approach be applied to other diseases with extensive genetic heterogeneity to help validate candidate genes and better inform genotype-phenotype correlations and prognostic advice in the era of precision medicine.
Disclosures
Dr. Torres reports grants and honoraria from Otsuka Pharmaceuticals, grants from Palladio Biosciences, Mironid, Sanofi Genzyme, Acceleron Pharma Inc., Regulus Therapeutics, and Blueprint Medicines, all outside the submitted work. Dr. Somlo reports personal fees from Goldfinch Bio, outside the submitted work. Dr. Mirshahi reports grants from National Institutes of Health (NIH) National Institute of General Medical Sciences and grants from NIH National Human Genome Research Institute, during the conduct of the study.
Funding
This work was supported by PKD Foundation Research Grant and Fellowships to Dr. Besse (217G18a and 190F15a) and Dr. Gulati (207F17a); NIH grants DK100592 and DK051041 to Dr. Somlo, DK090728 to Dr. Torres, GM111913 to Dr. Mirshahi, and DK106515-01 to Dr. Chang; the George M. O’Brien Kidney Center at Yale (P30 DK079310); the Mayo Clinic Robert M. and Billie Kelley Pirnie Translational PKD Center; and the Yale CMG (NIH M#UM1HG006504-05).
Supplementary Material
Acknowledgments
We thank Dr. F. Perry Wilson and Dr. Brandon K. Fornwalt for advice on study design.
Dr. Besse performed experiments and cowrote the manuscript. Dr. Chang, Mr. Luo, Dr. Triffo, Ms. Moore, Dr. Gulati, and Mr. Hartzel analyzed data. Dr. Mane provided exome sequencing. Regeneron Genetics Center (RGC) provided access to exome sequencing data. Dr. Torres contributed patient samples and data. Dr. Besse, Dr. Chang, Mr. Luo, Dr. Triffo, Dr. Somlo, and Dr. Mirshahi designed the study. Dr. Somlo and Dr. Mirshahi cowrote the manuscript.
The list of contributors from RGC and Yale Center for Mendelian Genomics (CMG) is provided in the Supplemental Acknowledgments.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related editorial, “New Ways of Finding New Genes for Old Diseases,” on pages 2037–2039.
Contributor Information
Collaborators: Goncalo Abecasis, Aris Baras, Michael Cantor, Giovanni Coppola, Aris Economides, John D. Overton, Jeffrey G. Reid, Alan Shuldiner, Christina Beechert, Caitlin Forsythe, Erin D. Fuller, Zhenhua Gu, Michael Lattari, Alexander Lopez, Thomas D. Schleicher, Maria Sotiropoulos Padilla, Karina Toledo, Louis Widom, Sarah E. Wolf, Manasi Pradhan, Kia Manoochehri, Ricardo H. Ulloa, Xiaodong Bai, Suganthi Balasubramanian, Leland Barnard, Andrew Blumenfeld, Yating Chai, Gisu Eom, Lukas Habegger, Young Hahn, Alicia Hawes, Shareef Khalid, Evan K. Maxwell, John Penn, Jeffrey C. Staples, Ashish Yadav, Paloma M. Guzzardo, Marcus B. Jones, and Lyndon J. Mitnaul
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2019030298/-/DCSupplemental.
Supplemental Table 1. Additional coding region genetic variants with MAF <1% by whole exome sequencing in published ADPKD-PCLD genes in ALG9 loss-of-function carriers.
Supplemental Table 2. Matched Control Phenotype Data.
Supplemental Figure 1. Sanger sequencing of Alg9−/− cell line.
Supplemental Figure 2. Quantification of relative PC1 protein expression level in Alg9−/− cells.
References
- 1.Torres VE, Harris PC, Pirson Y: Autosomal dominant polycystic kidney disease. Lancet 369: 1287–1301, 2007 [DOI] [PubMed] [Google Scholar]
- 2.Cornec-Le Gall E, Torres VE, Harris PC: Genetic complexity of autosomal dominant polycystic kidney and liver diseases. J Am Soc Nephrol 29: 13–23, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li A, Davila S, Furu L, Qian Q, Tian X, Kamath PS, et al.: Mutations in PRKCSH cause isolated autosomal dominant polycystic liver disease. Am J Hum Genet 72: 691–703, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drenth JP, te Morsche RH, Smink R, Bonifacino JS, Jansen JB: Germline mutations in PRKCSH are associated with autosomal dominant polycystic liver disease. Nat Genet 33: 345–347, 2003 [DOI] [PubMed] [Google Scholar]
- 5.Davila S, Furu L, Gharavi AG, Tian X, Onoe T, Qian Q, et al.: Mutations in SEC63 cause autosomal dominant polycystic liver disease. Nat Genet 36: 575–577, 2004 [DOI] [PubMed] [Google Scholar]
- 6.Porath B, Gainullin VG, Cornec-Le Gall E, Dillinger EK, Heyer CM, Hopp K, et al.: Genkyst Study Group; HALT Progression of Polycystic Kidney Disease Group; Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease : Mutations in GANAB, encoding the glucosidase IIα subunit, cause autosomal-dominant polycystic kidney and liver disease. Am J Hum Genet 98: 1193–1207, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Besse W, Dong K, Choi J, Punia S, Fedeles SV, Choi M, et al.: Isolated polycystic liver disease genes define effectors of polycystin-1 function. J Clin Invest 127: 3558, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornec-Le Gall E, Olson RJ, Besse W, Heyer CM, Gainullin VG, Smith JM, et al.: Genkyst Study Group; HALT Progression of Polycystic Kidney Disease Group; Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease : Monoallelic mutations to DNAJB11 cause atypical autosomal-dominant polycystic kidney disease. Am J Hum Genet 102: 832–844, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qian Q, Li A, King BF, Kamath PS, Lager DJ, Huston J 3rd, et al.: Clinical profile of autosomal dominant polycystic liver disease. Hepatology 37: 164–171, 2003 [DOI] [PubMed] [Google Scholar]
- 10.Qian F, Watnick TJ, Onuchic LF, Germino GG: The molecular basis of focal cyst formation in human autosomal dominant polycystic kidney disease type I. Cell 87: 979–987, 1996 [DOI] [PubMed] [Google Scholar]
- 11.Wu G, D’Agati V, Cai Y, Markowitz G, Park JH, Reynolds DM, et al.: Somatic inactivation of Pkd2 results in polycystic kidney disease. Cell 93: 177–188, 1998 [DOI] [PubMed] [Google Scholar]
- 12.Watnick TJ, Torres VE, Gandolph MA, Qian F, Onuchic LF, Klinger KW, et al.: Somatic mutation in individual liver cysts supports a two-hit model of cystogenesis in autosomal dominant polycystic kidney disease. Mol Cell 2: 247–251, 1998 [DOI] [PubMed] [Google Scholar]
- 13.Janssen MJ, Salomon J, Cnossen WR, Bergmann C, Pfundt R, Drenth JP: Somatic loss of polycystic disease genes contributes to the formation of isolated and polycystic liver cysts. Gut 64: 688–690, 2015 [DOI] [PubMed] [Google Scholar]
- 14.Tan AY, Zhang T, Michaeel A, Blumenfeld J, Liu G, Zhang W, et al.: Somatic mutations in renal cyst epithelium in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 29: 2139–2156, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fedeles SV, Tian X, Gallagher AR, Mitobe M, Nishio S, Lee SH, et al.: A genetic interaction network of five genes for human polycystic kidney and liver diseases defines polycystin-1 as the central determinant of cyst formation. Nat Genet 43: 639–647, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis K, Webster D, Smith C, Jackson S, Sinasac D, Seargeant L, et al.: ALG9-CDG: New clinical case and review of the literature. Mol Genet Metab Rep 13: 55–63, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freeze HH: Genetic defects in the human glycome. Nat Rev Genet 7: 537–551, 2006 [DOI] [PubMed] [Google Scholar]
- 18.Tham E, Eklund EA, Hammarsjö A, Bengtson P, Geiberger S, Lagerstedt-Robinson K, et al. : A novel phenotype in N-glycosylation disorders: Gillessen-Kaesbach-Nishimura skeletal dysplasia due to pathogenic variants in ALG9. Eur J Hum Genet 24: 198–207, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arora V, Shah N, Khatter S, Puri RD, Kumar R, Gupta P, et al. : ALG9 associated gillessen-kaesbach–nishimura syndrome (GIKANIS): An uncommon aetiology of enlarged foetal kidneys. J Foetal Med 5: 237–239, 2018 [Google Scholar]
- 20.Chapman AB, Guay-Woodford LM, Grantham JJ, Torres VE, Bae KT, Baumgarten DA, et al.: Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease cohort : Renal structure in early autosomal-dominant polycystic kidney disease (ADPKD): The Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease (CRISP) cohort. Kidney Int 64: 1035–1045, 2003 [DOI] [PubMed] [Google Scholar]
- 21.Torres VE, Abebe KZ, Chapman AB, Schrier RW, Braun WE, Steinman TI, et al.: HALT-PKD Trial Investigators : Angiotensin blockade in late autosomal dominant polycystic kidney disease. N Engl J Med 371: 2267–2276, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J: A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46: 310–315, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al.: Exome Aggregation Consortium : Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zook JM, Chapman B, Wang J, Mittelman D, Hofmann O, Hide W, et al.: Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotechnol 32: 246–251, 2014 [DOI] [PubMed] [Google Scholar]
- 25.Dong C, Wei P, Jian X, Gibbs R, Boerwinkle E, Wang K, et al.: Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet 24: 2125–2137, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, et al.: REVEL: An ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet 99: 877–885, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shihab HA, Rogers MF, Gough J, Mort M, Cooper DN, Day IN, et al.: An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics 31: 1536–1543, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maniatis T, Fritsch EF, Sambrook J: Molecular cloning: A laboratory manual, New York, Cold Spring Harbor Laboratory, 1982 [Google Scholar]
- 29.Carey DJ, Fetterolf SN, Davis FD, Faucett WA, Kirchner HL, Mirshahi U, et al. : The Geisinger MyCode community health initiative: An electronic health record-linked biobank for precision medicine research. Genet med 18: 906–913, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dewey FE, Murray MF, Overton JD, Habegger L, Leader JB, Fetterolf SN, et al.: Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science 354: aaf6814, 2016 [DOI] [PubMed] [Google Scholar]
- 31.Staples J, Maxwell EK, Gosalia N, Gonzaga-Jauregui C, Snyder C, Hawes A, et al.: Profiling and leveraging relatedness in a precision medicine cohort of 92,455 exomes. Am J Hum Genet 102: 874–889, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al.: The ensembl variant effect predictor. Genome Biol 17: 122, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghosh R, Oak N, Plon SE: Evaluation of in silico algorithms for use with ACMG/AMP clinical variant interpretation guidelines. Genome Biol 18: 225, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rule AD, Sasiwimonphan K, Lieske JC, Keddis MT, Torres VE, Vrtiska TJ: Characteristics of renal cystic and solid lesions based on contrast-enhanced computed tomography of potential kidney donors. Am J Kidney Dis 59: 611–618, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cabezas OR, Flanagan SE, Stanescu H, García-Martínez E, Caswell R, Lango-Allen H, et al.: Polycystic kidney disease with hyperinsulinemic hypoglycemia caused by a promoter mutation in phosphomannomutase 2. J Am Soc Nephrol 28: 2529–2539, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The UniProt Consortium : UniProt: The universal protein knowledgebase. Nucleic Acids Res 45[D1]: D158–D169, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishiura JL, Neves RF, Eloi SR, Cintra SM, Ajzen SA, Heilberg IP: Evaluation of nephrolithiasis in autosomal dominant polycystic kidney disease patients. Clin J Am Soc Nephrol 4: 838–844, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfau A, Knauf F: Update on nephrolithiasis: Core curriculum 2016. Am J Kidney Dis 68: 973–985, 2016 [DOI] [PubMed] [Google Scholar]
- 39.Scales CD Jr, Smith AC, Hanley JM, Saigal CS; Urologic Diseases in America Project : Prevalence of kidney stones in the United States. Eur Urol 62: 160–165, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bansal AD, Hui J, Goldfarb DS: Asymptomatic nephrolithiasis detected by ultrasound. Clin J Am Soc Nephrol 4: 680–684, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.