Abstract
The evolution of antibiotic resistance is influenced by a variety of factors, including the availability of resistance mutations, and the pleiotropic effects of such mutations. Here, we isolate and characterize chromosomal quinolone resistance mutations in E. coli, in order to gain a systematic understanding of the rate and consequences of resistance to this important class of drugs. We isolated over fifty spontaneous resistance mutants on nalidixic acid, ciprofloxacin, and levofloxacin. This set of mutants includes known resistance mutations in gyrA, gyrB, and marR, as well as two novel gyrB mutations. We find that, for most mutations, resistance tends to be higher to nalidixic acid than relative to the other two drugs. Resistance mutations had deleterious impacts on one or more growth parameters, suggesting that quinolone resistance mutations are generally costly. Our findings suggest that the prevalence of specific gyrA alleles amongst clinical isolates are driven by high levels of resistance, at no more cost than other resistance alleles.
Introduction
The increasing prevalence of antimicrobial resistance (AMR) has become an urgent public health problem worldwide. For example, resistance to ciprofloxacin, the most commonly purchased antimicrobial by hospitals in Canada between 2008–2014 [1], in Escherichia coli rose to 26.7% in 2015 from 21.6% in 2009 [2]. The present AMR crisis has been attributed to the misuse and overuse of antibiotics, as well as the scarcity of novel drug development [3–7]. Given the rapid increase in the prevalence of resistance, an understanding of the principles underlying resistance evolution is vital.
Adaptation, of which the evolution of AMR is a prime example, is driven by the interplay between mutation, selection, and demographic processes like drift. Mutation determines the rate at which beneficial variants are introduced into a population, while selection and demography govern the fates of these variants. Thus, in understanding the evolution of AMR, we are interested in both mutation and selection. We expect, for example, that higher mutation rates will generally lead to a more rapid evolution of resistance. The spread of a given mutation will then be influenced by its selective consequences, including its effect on resistance, and on its pleiotropic effects, such as fitness in the absence of antibiotic, and collateral sensitivity or cross resistance to other antibiotics.
The rate of mutation to resistance is given by the product of population size (N), overall mutation rate (μ), and the fraction (f) of mutations that are beneficial (i.e., those that grant resistance). Thus, resistance mutations will appear more frequently for larger populations, or for populations with higher mutation rates. Up to a point, this dependence on Nμ leads to an increase in the rate of adaptation; however, as Nμ approaches 1, a population is no longer limited by mutational input, but instead by competition between competing mutations (e.g., [8,9]).
Relevantly here, the fraction of mutations granting resistance almost certainly differs between different antibiotics; for some antibiotics, there will be a greater availability of resistance mutations. Variation in the availability of resistance mutations may reflect differences in the number of genetic loci that can contribute to resistance. For example, resistance to trimethoprim is largely conferred by mutations in a single gene encoding dihydrofolate reductase (DHFR) [10], while resistance to chloramphenicol and streptomycin can be conferred by mutations in a number of different genes [10,11]. Moreover, the number of individual mutations conferring resistance may differ between genes: while a wide range of loss-of-function lesions in the transcriptional regulator marR will grant multi-drug resistance [12], only a handful of mutations in gyrA confer quinolone resistance [13,14]. Thus, even given equal population size and overall mutation rates, we expect different rates of evolution to resistance to different antibiotics due to differences in the mutational target size.
Once a resistance mutation has arisen, its persistence and spread may be affected by its pleiotropic effects, including its fitness costs and effects on resistance to other drugs [15–18]. In the presence of antibiotic, a resistant bacterium has a clear advantage compared to susceptible genotypes. However, in an antibiotic-free environment, a resistance mutation may impose a burden, for example through reduced growth rates relative to sensitive strains [19–23]. However, while resistance mutations are often costly, not all resistance mutations bear a cost, and such cost-free mutations are likely to persist [24–27]. Melnyk et al. (2015) [28] conducted a meta-analysis including 179 single chromosomal mutations conferring resistance to 16 antibiotics from 8 bacterial species. They reported 8 no-cost mutations, with variable costs of resistance depending on antibiotic class and the species assayed.
Increased resistance to one antibiotic may be accompanied by increased cross-resistance to other antibiotics. Cross-resistance is often observed between members of the same class of antibiotic. For example, all quinolones target DNA gyrase and Topoisomerase IV, whose subunits are encoded by the gyrA/B and parC/E genes, respectively. Resistance to quinolones can be conferred by point mutations affecting specific portions of GyrA and ParC, known as the quinolone resistance-determining regions (QRDR) [13,29–31]. Changes at amino acid positions 83 and 87 of gyrA lead to a significant loss in quinolone susceptibility [32–34]. Moreover, known resistance mutations in gyrB mutations affect amino acid positions 426 and 464, sites that interact with the bound quinolone molecule close to the QRDR of GyrA [35–37].
Cross-resistance can also occur between drug classes—for example, marR mutations selected by quinolones also confer resistance to phenicols, tetracyclines, and rifampicin [31,38,39]. Mutations in marR, which encodes a negative regulator of the marRAB operon, render the repressor function inactive, resulting in increased efflux and reduced permeability [14,40–43]. By contrast, in collateral sensitivity or negative cross-resistance, acquisition of resistance to one antibiotic may grant sensitivity to other antibiotics. For example, resistance to aminoglycosides in E. coli can be conferred by electron transport chain mutations that reduce proton-motive force (PMF). This decrease in PMF negatively affects the activity of multi-drug efflux pumps, such as AcrAB-TolC, granting hypersensitivity to many other antibiotics [44,45].
Here, we assess mutation rates, levels of resistance, and pleiotropic effects for chromosomal mutations conferring resistance to different quinolones, in an effort to understand the full set of parameters contributing to the origin, spread, and persistence of quinolone resistance. Quinolones were first used clinically in the 1960’s, and have undergone multiple rounds of development. The first-generation quinolone, nalidixic acid (nal), possesses a limited spectrum of activity, but fluorination of the core structure generated the so-called 2nd -generation quinolones like ciprofloxacin (cip). Further overall structural developments resulted in 3rd-generation drugs such as levofloxacin (levo). We predict that, while broad mechanisms of resistance will be shared between quinolones, resistance mutations will differ in their effects towards different antibiotics owing to differences in antibiotic penetration and/or structural configuration.
Materials and methods
Bacterial strains and media
The E. coli laboratory strain K-12 (MG1655) was used for all experiments. Lysogeny broth (LB) (10 g/l tryptone, 5 g/l yeast extract, 10 g/l NaCl per litre; Bishop) was used for agar and broth cultures.
Quinolone susceptibility assays
Minimum inhibitory concentration (MIC) values for the ancestral strain and for antibiotic-resistant mutants were determined for nalidixic acid, ciprofloxacin and levofloxacin (Sigma-Aldrich) using a 96 well plate assay. Antibiotic concentrations started at 10μg/ml, 1μg/ml, and 8μg/ml for nal, cip, and levo, respectively, and were diluted in a two-fold series and dispensed with 125μl/well of LB into 96-well plates. The 96 well plates were incubated overnight at 30 C, with shaking at 150 rpm. The MIC was defined as the lowest concentration of antibiotic for which 90% growth inhibition was visibly observed after overnight culture.
Fluctuation analysis and estimation of mutation rates
Mutation rates to resistance were determined using fluctuation assays [46,47]. A single colony of E. coli MG1655 was grown overnight in liquid LB at 37 C, with shaking at 150 rpm. Fresh 200μl cultures were inoculated with ~100 cells each. Each independent culture was then grown to saturation overnight. The final number of cells, Nt, was estimated from plate counts on LB without antibiotic. Selective plates were supplemented with antibiotics (nal, cip or levo) at a concentration of 1xMIC or 2xMIC. 30 replicate populations were plated for each antibiotic at each concentration. The observed number of mutants, r, was then counted for each replicate.
For estimating the number of mutational events m, the MSS maximum-likelihood method was used [48]. This method is based on a recursive algorithm for estimating the Luria-Delbruck distribution for a given number of mutational events [46]. This method is valid over the entire range of values of m [49,50]. The mutation rate per cell per generation, M, is calculated as m divided by the total number of bacteria plated on selective plates (Nt) [47,51,52].
PCR amplification and sequencing of candidate genes
Targeted sequencing of the known resistance loci gyrA and marR was carried out in order to identify potential resistance mutations. To ensure independence, a single mutant colony was picked from every plate from the fluctuation assay on which there was growth, and inoculated overnight in LB broth without antibiotic. DNA was then extracted using the EZ-10 spin column bacterial DNA miniprep kit (Bio Basic) and PCR was used to amplify the QRDR of gyrA (Gyrase forward—5’GTAAAACGACGGCCAGTGATGAGCGAC3’, Gyrase reverse—5’CGGTACGGTAAGCTTCTTC3’) and the entire marR gene (MarR-forward 5’GTAAAACGACGGCCAGTGGTCAATTCA3’, MarR reverse—5’TCTGGACATCGTCATACCTC3’). PCR amplicons were sent to Genome Quebec for Sanger sequencing. Mutations in these particular regions were compared with the wild type MG1655 strain of E. coli.
Whole-genome sequencing
Whole-genome sequencing was carried out to identify potential resistance mutations in clones for which mutations were observed in neither gyrA nor marR. Sequencing libraries were prepared using the Nextera XT kit (Illumina), and sequencing was carried out on the MiSeq platform using paired-end 300bp reads. Raw reads were processed using Trimmomatic-0.35 [53], allowing for a minimum Phred-scaled quality score of 20 for leading and trailing bases, truncating reads once average quality dropped below 20 in a 4bp sliding window, and dropping all reads of length less than 36. Read quality was assessed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
Reference-based assembly was carried out using Bowtie-2 [54], with E. coli K-12 (MG1655; NC_00913.2) as the reference genome. SNPs were called using Samtools [55] and SNP effects were inferred using SNPeff [56].
24-hour growth curve analysis
Fitness in the absence of antibiotic was estimated using 24-hour growth curves for each single colony isolate. Growth curves were obtained in triplicate in 96-well plates, inoculated at a 1:100 dilution from overnight cultures. OD600 was measured on a Bioteck ELx808 plate reader every 37 minutes for 24 hours, incubating at 37 C with 30 s of shaking every 5 minutes. Three growth parameters, lag time, maximum growth rate and optical density at stationary phase after 24 hours, were estimated using the program GrowthRates [20].
Results and discussion
Estimation of mutation rates
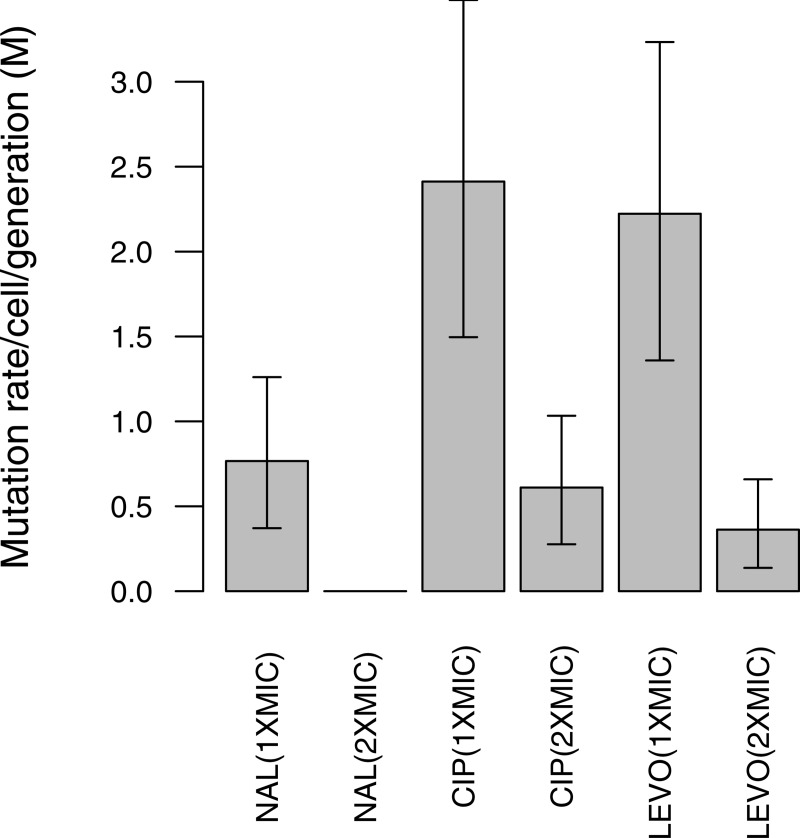
We estimated mutation rates to resistance for each of the three quinolones nal, cip, and levo using fluctuation assays [46]. The probability of a mutational event per cell per generation was estimated at 1x and 2x MIC where MIC values were 10 μg/ml, 15 ng/ml, and 31 ng/ml for nal, cip, and levo, respectively. The number of observed mutants (r) on each of 30 selective plates was used to estimate mutation rates to nalR, cipR, and levoR, using the MSS maximum likelihood method [50]. At 1x and 2x MIC, mutation rates differed between antibiotics, with lower rates to nalR than to cipR or levoR (cip>levo>nal), presumably because fewer resistance mutations are available for nal that can grant sufficiently high levels of resistance (Fig 1, Table 1).
Fig 1. Spontaneous mutation rate per 108 cells to quinolone resistance among E. coli K-12 (MG1655).
Mutation rates were estimated from 30 independent cultures at 1x and 2x MIC. Error bars represent 95% confidence intervals. Note that no colonies were obtained at 2xMIC for Nal.
Table 1. Spontaneous mutation rates to quinolone resistance in E. coli K-12 (MG1655).
| Antibiotic | Antibiotic concentration (μg/ml) |
Mutation rate per culture ‘m’ | Mutation rate (M) per 108 cells | Upper 95% Confidence interval | Lower 95% Confidence interval |
|---|---|---|---|---|---|
| Nalidixic acid (1xMIC) | 11.11 | 0.368 | 0.76 | 0.49 | 0.39 |
| Ciprofloxacin (1xMIC) |
0.015 | 1.158 | 2.41 | 1.06 | 0.91 |
| Ciprofloxacin (2xMIC) |
0.030 | 0.293 | 0.61 | 0.42 | 0.33 |
| Levofloxacin (1xMIC) |
0.0312 | 1.067 | 2.22 | 1.01 | 0.86 |
| Levofloxacin (2xMIC) |
0.0625 | 0.174 | 0.36 | 0.29 | 0.22 |
Identification of resistance mutations
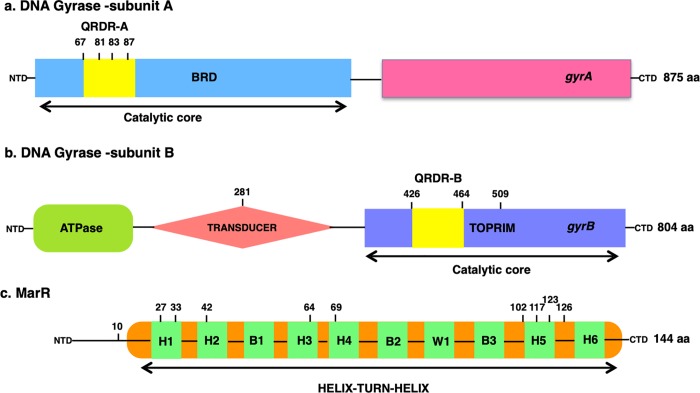
We obtained a total of 56 spontaneous quinolone resistant mutants from the fluctuation assays (one from each plate), and identified putative resistance mutations in 50 of these (Table 2). Targeted sequencing identified 36 gyrA mutants, 9 marR mutants, and 1 double gyrA, marR mutant. Whole-genome sequencing identified an additional 4 gyrB mutants. We note that gyrA/gyrB or marR/gyrB double mutants will be undetected by our approach. However, given that only one gyrA/marR double mutant was detected, the frequency of double mutants is probably low. In gyrA, mutations were observed at nucleotides encoding amino acid positions 67, 81, 83, and 87 (Fig 2A), consistent with previous findings that the GyrA QRDR spans amino acid 67 to 107 [29,57–59]. The most common alterations in the gyrA QRDR region were S83L (n = 18) and D87G/N/Y (n = 11). These mutations were found in mutants isolated against all three drugs, presumably because these positions are located within the positively charged region close to the DNA-enzyme binding site [31]. Amino acids 83 and 87 are located near the active site of DNA gyrase, along with the tyrosine-122 residue that interacts with the broken DNA strand following cleavage [59–66]. The α helix-4 region is particularly essential to quinolone binding and the substitution to leucine at position 83 makes the vicinity of α helix-4 of the gyrase less electron rich, crippling gyrase-quinolone binding [33,61,67]. Mutations at position 87 and 81 also perturb the alignment of the α helix-4 structure.
Table 2. MIC fold-increase and mutations in gyrA, gyrB and marR among nalR, cipR, and levoR mutants of E. coli K-12 (MG1655).
| S.No. | Antibiotic | MIC WT (ug/ml) | MIC fold increase of mutants | Resistance mutation | Total number of nalR, cipR and levoR mutants | Number of gyrA, gyrB and marR mutants | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Nal | Cip | Levo | gyrA | gyrB | marR | |||||
| Nalidixic acid | 10 | 11 | ||||||||
| 1 | 64 | A67S | 2 | |||||||
| 2 | 256 | S83L | 6 | |||||||
| 3 | 128 | D87G | 2 | |||||||
| 4 | 64 | D426N | 1 | |||||||
| Ciprofloxacin | 0.015 | 24 | ||||||||
| 1 | 64 | G81C/D | 3 | |||||||
| 2 | 128 | S83L | 4 | |||||||
| 3 | 32 | S83W | 2 | |||||||
| 4 | 64 | D87Y/G/N | 7 | |||||||
| 5 | 32 | H281L | 1 | |||||||
| 6 | 32 | S464Y | 1 | |||||||
| 7 | 16 | L509G | 1 | |||||||
| Levofloxacin | 0.031 | 21 | ||||||||
| 1 | 8 | G81C | 1 | |||||||
| 2 | 16 | S83L | 8 | |||||||
| 4 | 8 | D87G | 1 | |||||||
| 3 | 16 | D87G | R27P | 1 | ||||||
| 5 | 16 | E10stop | 1 | |||||||
| 6 | 16 | L33R | 1 | |||||||
| 7 | 8 | Q42E | 1 | |||||||
| 8 | 4 | L64fs | 1 | |||||||
| 9 | 16 | G69E | 1 | |||||||
| 10 | 8 | T102S | 1 | |||||||
| 11 | 8 | Q117stop | 1 | |||||||
| 12 | 4 | L123S | 1 | |||||||
| 13 | 8 | N126fs | 1 | |||||||
Fig 2. The domain structures of Gyrase A, B, and MarR.
Mutations obtained in this study are indicated in bold above. Panel A: Arrangement of GyrA. This subunit of DNA gyrase consists of the breakage-reunion (BRD) domain, and the quinolone resistance determining region (QRDR-A) site. Panel B: Arrangement of GyrB, with the ATPase, Transducer (221–392), and Toprim (418–533) regions. The QRDR-B is shown within the Toprim domain of GyrB [73]. Panel C. MarR domain structure, comprising four helices (H) and three ß-sheets (B). H3 and H4 (57–80) are the recognition and DNA binding motifs containing H-T-H motifs and the ß-sheet winged structure. H1, H5, and H6 are associated with dimerization [42,74–76].
marR mutations were obtained only in levoR isolates. These point, frameshift, and missense mutations were dispersed throughout the gene, as expected given that the Mar phenotype can arise from any loss-of-function mutation (Fig 2C). MarR consists of two domains, one N-C terminal domain and a helix-turn-helix (HTH) DNA binding domain. The closely packed hydrophobic core and intermolecular hydrogen bonds stabilize the N-terminus (residues 10–21) of one subunit and the C-terminus (residues 123–144) of the second subunit, holding the dimer together. Some of the mutations reported in this study belong to the oligomerization dimer domain of MarR, such as those at positions 10, 27, and 33 of one terminal subunit, and positions 123 and 126 of the other equivalent subunit. The rest of the MarR protein is linked via antiparallel helices emerging out from each of the subunits, encompassing the DNA binding domain (residues 55–100) [12,42]. Mutations at positions 64 and 69 fall in the HTH DNA binding motif. Other mutations reported here at position 42 and 102 residue at α and β sheets of MarR, which are essential for the interaction between the two antiparallel strands of HTH DNA binding domain [42].
WGS revealed gyrB mutations in four isolates for which no gyrA or marR mutations were detected (Fig 2B). H281L, S464F, and L509Q mutations were found in strains isolated on cip, and a D426N mutation was found in a strain isolated on nal. The GyrB enzyme consists of two domains, an N-terminal domain (amino acid 2–393) that incorporates the ATPase catalytic site, and a C-terminal domain (amino acid 394–804) that interacts with GyrA [68,69]. D426N and S464F have been previously reported to confer quinolone resistance [35,59,64,70,71]. Both of these mutations are part of the QRDR of GyrB and cause conformational changes in the structure of the gyrase subunits [37,59]. The gyrB D426N mutation has been reported before along with a mutation at position L447, both of which provide a neutral vicinity, owing to their respective opposite charges. Both of these residues are suggested to be part of a quinolone-binding pocket [31,35,36,70,72].
Interestingly, the H281L and L509G mutations have not been previously reported to confer quinolone resistance. These two novel mutations are located outside the GyrB QRDR. Position 281 is located in the transducer region of GyrB, which forms a cavity just large enough to facilitate the transfer of the trapped double-stranded DNA through the DNA gate in the presence of ATP [73,77–79]. Position 509 is within the TOPRIM domain of GyrB, part of the catalytic DNA cleavage-rejoining complex along with the GyrA winged helix domain [79,80].
Direct responses to selection
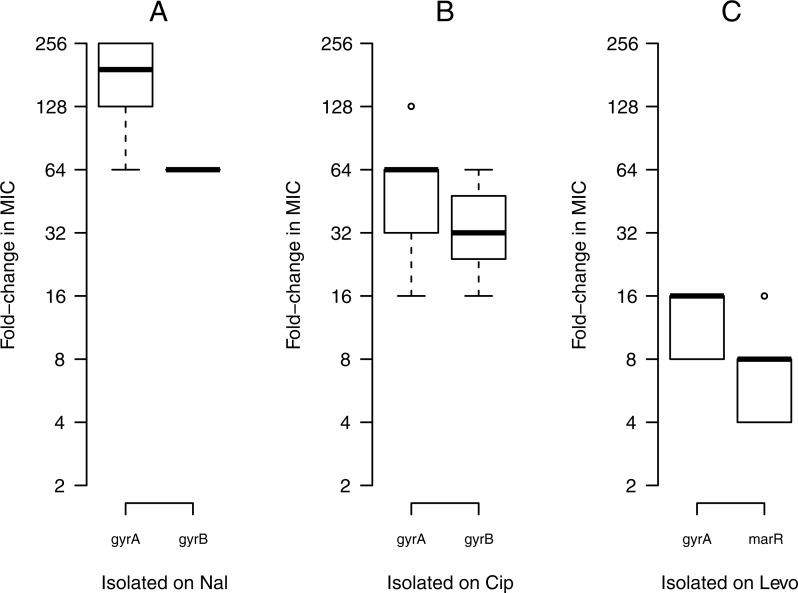
An increase in fitness in the selective environment is referred to as the direct response to selection; here, the direct response to selection is measured by an increase in MIC towards the drug on which a mutant was selected. We found substantial variation between drugs in the magnitude of the direct response. Mutants isolated on nal showed a stronger direct response to selection than did mutants isolated on cip or levo, with a mean increase of 256-fold MIC towards nal. Mutants isolated on cip and levo showed mean increases of 64-fold and 16-fold towards cip and levo, respectively (Fig 3).
Fig 3. Direct responses to selection.
Changes in MIC for resistant mutants towards the drug on which they were selected: nal (A), cip (B), and levo (C). The boxplot presents the median, first, and third quartiles, with whiskers showing either the maximum (minimum) value or 1.5 times the interquartile range of the data, whichever is smaller (larger).
Among resistance mutations, gyrA mutations consistently showed higher levels of resistance than gyrB or marR, regardless of the antibiotic they were isolated on, which impacts the variation in MIC values significantly (Table 3). Furthermore, within each gene, the level of resistance varied by mutation. In the case of gyrA, the S83L mutation conferred higher resistance among all the isolates compared to other mutational sites of gyrA (87, 81 or 67). This suggests that the widespread occurrence of the S83L mutation amongst clinical isolates is due to the high level of resistance (S1 Fig).
Table 3. Two-way analysis of variance (ANOVA) for the effects of antibiotic and gene on levels of resistance.
| Factor | F-value | P-value |
|---|---|---|
| Gene | 10.00 | 0.00025* |
| Antibiotic | 39.30 | 1.34e-10* |
| Gene*Antibiotic | 2.83 | 0.09 |
Cross resistance between quinolones
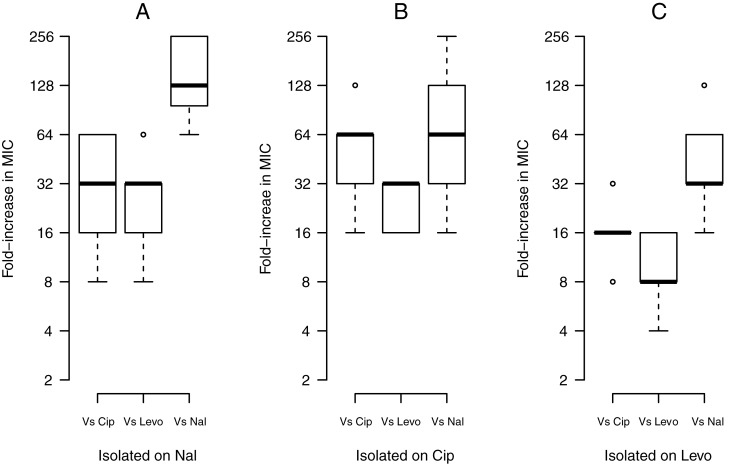
Cross-resistance between quinolones was widespread: all of the resistant mutants isolated on one quinolone displayed increased resistance, in varying degrees, to the other two quinolones. Nonetheless, different quinolones were not equally affected by the resistance mutations (Fig 4). Overall, mutants were more resistant towards nal than they were towards cip or levo. Among cip and levo, mutations showed smaller gains in resistance on levo. Nevertheless, significant correlations between levels of resistance for cip and levo (Pearson’s r = 0.64, t = 6.2, P = 8.433e-08), nal and cip (r = 0.47, t = 3.93, P = 0.0002), and nal and levo (r = 0.38, t = 3.07, P = 0.0032) suggest a closer relationship between the newer quinolones. This variation in resistance among quinolones can be explained by the intrinsic structural drug differences between older and newer quinolone classes. Nal is devoid of any cyclic derivatives whereas cip and levo have substituents at positions C-6, C-7 and C-8, that offer greater spectrum/potency of activity. Thus, the modified quinolone substituents likely reduce resistance levels by increasing the affinity for GyrA, and by stabilizing the quinolone-DNA complex [33,81–96].
Fig 4. Cross-resistance between antibiotics.
Fold-increase in MIC of resistant mutants isolated on nal (A), cip (B), and levo (C) against all three antibiotics. The boxplot presents the median, first, and third quartiles, with whiskers showing either the maximum (minimum) value or 1.5 times the interquartile range of the data, whichever is smaller (larger).
These trends are also evident for specific loci. gyrA mutants showed the highest gain in resistance (average 128xMIC) on nal in comparison with cip (32xMIC) or levo (16xMIC). gyrB mutants also displayed a higher increase in resistance to nal (64xMIC), but the same increase on cip and levo (16xMIC). The novel H281L and L509G mutations gained similar increases in resistance with cip and levo, at 32xMIC and 16xMIC respectively. On nal, H281L gained 64xMIC whereas L509G gained similar increase as with cip or levo, i.e. 16xMIC. Meanwhile, marR mutants did not show as high increases in MICs, with increases of 32x, 16x, and 8xMIC on nal, cip, and levo.
Costs of resistance
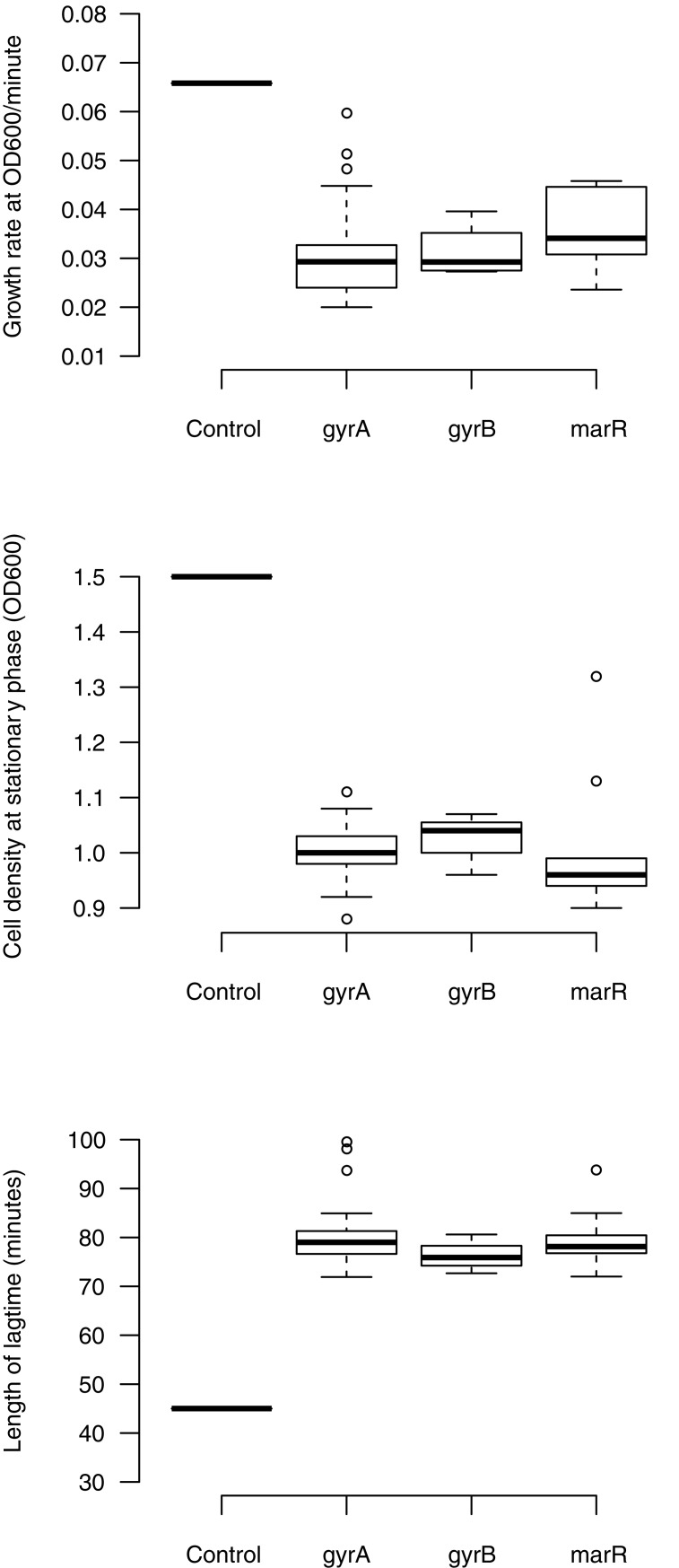
The persistence of resistance in the absence of antibiotic is determined in part by the fitness costs associated with resistance mutations [97–99]. No-cost mutations may contribute to the persistence of resistance mutations in the absence of antibiotic. We measured three fitness components in the absence of antibiotic for our set of quinolone resistant mutants: maximum growth rate (Vmax), density at stationary phase (Max OD), and length of lag phase. Resistant mutants were consistently found to be costly, exhibiting significant differences in Vmax, Max OD, and lag time compared to their drug-susceptible ancestor MG1655 (Fig 5, Table 4) (S2 Fig). Thus, overall we observe significant costs of resistance for quinolone resistance mutations, consistent with previous studies [92,100–102].
Fig 5. Costs of resistance of quinolone resistant mutants.
The fitness components measured are growth rate, cell density, and lag time between gyrA, gyrB, and marR resistance mutations. All the fitness components are compared to control E. coli K-12 (MG1655). The boxplot presents the median, first, and third quartiles, with whiskers showing either the maximum (minimum) value or 1.5 times the interquartile range of the data, whichever is smaller (larger).
Table 4. Effects of resistance mutations on growth parameters.
| Factor | Post hoc (Tukey HSD) comparisons with E. coli K-12 MG1655 | ||
|---|---|---|---|
| Growth rate Length of Cell density at (OD600/minute) lag time (minutes) stationary phase (OD600) | |||
| Mean P-value | Mean P-value | Mean P-value | |
| gyrA-MG1655 | -0.032 0.0000011 | 33.4 <2.0e-16 | -0.55 <2.0e-16 |
| gyrB-MG1655 | -0.033 0.0000191 | 28.9 <2.0e-16 | -0.52 <2.0e-16 |
| marR-MG1655 | -0.029 0.0000173 | 32.3 <2.0e-16 | -0.53 <2.0e-16 |
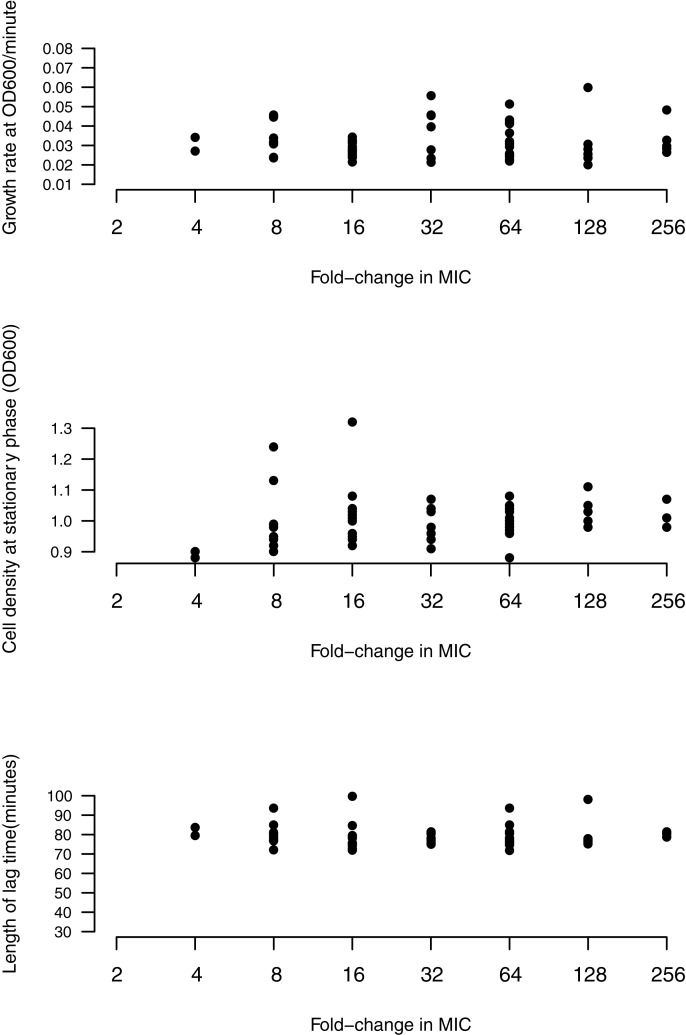
Some studies have reported that mutations granting higher levels of resistance impose higher costs [28]. However, we fail to find a significant relationship between MIC and any fitness component. No correlations were found between MIC and growth rate (P = 0.42, Kendall’s tau = 0.08), length of lag phase (P = 0.82, tau = 0.02) or cell density (P = 0.12, tau = 0.15) (Fig 6). We note that gyrA mutations confer no greater costs than other resistance mutations (gyrB, marR). Moreover, amongst the handful of mutations in gyrA (S83, D87, G81) that can confer high level resistance [58,63], a few prominent alleles of gyrA tend to be found in E. coli clinical isolates [24,62,103,104]. That these mutations confer high levels of resistance, but are no more costly than other gyrA mutations (Fig 6), could help to explain the prevalence of specific gyrA mutations amongst clinical isolates.
Fig 6. No correlation between level of resistance (fold-increase in MIC) and growth rate, cell density or length of lag phase for all mutants.
Conclusions and perspectives
Quinolones target DNA gyrase and topoisomerase IV. Resistance against these drugs can be achieved by target alteration and/or through efflux and permeability associated mutations. We find that all 50 spontaneous mutants obtained through fluctuation assays were resistant through mutations in the known resistance conferring genes gyrA, gyrB, and marR. This finding suggests that there are few other quinolone resistance mutations available in E. coli K-12; this is somewhat surprising, given that selection experiments in P. aeruginosa have identified novel resistance mutations [105]. We find significant costs of resistance, and differences in the mutational supply rate among resistant isolates. Notably, we find gyrA mutations conferred higher resistance, without greater fitness cost, than other mutations. This finding may explain the prevalence of gyrA mutations in clinical samples. We also find variation in cross-resistance amongst quinolone resistant isolates, implying that different resistance mutations respond differently to quinolone variants. Thus, antibiotic variants may have different implications for the evolution of resistance. Optimally, we should choose an antibiotic for which resistance is costly, and where single mutations have relatively small effect, as was the case here for levofloxacin.
Supporting information
Resistance mutations isolated as a direct selection on nal (A), cip (B), and levo (C).
(EPS)
Variation in costs of resistance—Growth rate, cell density, and lag phase between gyrA (A), gyrB (B), and marR (C) resistance mutations. All the fitness components are compared to control E. coli K-12 (MG1655). The boxplots present the median, first, and third quartiles, with whiskers showing either the maximum (minimum) value or 1.5 times the interquartile range of the data, whichever is smaller (larger).
(EPS)
(XLSX)
Acknowledgments
We thank two anonymous reviewers for helpful comments and discussion of this work.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by a Natural Sciences and Engineering Research Council (NSERC) Discovery Grant 435248 to AW, a Canadian Institutes for Health Research (CIHR) New Investigator Award 141995 to AW, and by an Ontario Graduate Scholarship (OGS) to KB. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Public health agency of Canada (PHAC). (2015–2016). Available from: http://www.phac-aspc.gc.ca/rpp/index-eng.php.
- 2.Canadian antimicrobial resistance alliance (CARA). CANWARD susceptibility report. 2015. Available from:www.can-r.com. [Google Scholar]
- 3.Piddock LJ. The crisis of no new antibiotics—what is the way forward? Lancet Infect Dis. 2012;12(3):249–253. 10.1016/S1473-3099(11)70316-4 [DOI] [PubMed] [Google Scholar]
- 4.Bartlett JG, Gilbert DN, Spellberg B. Seven ways to preserve the miracle of antibiotics. Clin Infect Dis. 2013;56(10):1445–1450. 10.1093/cid/cit070 [DOI] [PubMed] [Google Scholar]
- 5.Gross M. Antibiotics in crisis. Curr Biol. 2013;23(24):1063–1065. [DOI] [PubMed] [Google Scholar]
- 6.Gould IM, Bal AM. New antibiotic agents in the pipeline and how they can overcome microbial resistance. Virulence. 2013;4(2):185–191. 10.4161/viru.22507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Viswanathan VK. Off-label abuse of antibiotics by bacteria. Gut Microbes. 2014;5(1):3–4. 10.4161/gmic.28027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerrish PJ, Lenski RE. The fate of competing beneficial mutations in an asexual population. Genetica. 1998;102–103(1–6):127–144. [PubMed] [Google Scholar]
- 9.Wilke CO. The speed of adaptation in large asexual populations. Genetics. 2004;167(4):2045–2053. 10.1534/genetics.104.027136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toprak E, Veres A, Michel J, Chait R, Hartl D, Kishony R. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet. 2011;44(1):101–105. 10.1038/ng.1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winkler J, Kao KC. Harnessing recombination to speed adaptive evolution in Escherichia coli. Metab Eng. 2012;14(5):487–495. 10.1016/j.ymben.2012.07.004 [DOI] [PubMed] [Google Scholar]
- 12.Alekshun MN, Kim YS, Levy SB. Mutational analysis of MarR, the negative regulator of marRAB expression in Escherichia coli, suggests the presence of two regions required for DNA binding. Mol Microbiol. 2000;35(6):1394–1404. 10.1046/j.1365-2958.2000.01802.x [DOI] [PubMed] [Google Scholar]
- 13.Ruiz J. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother. 2003;51(5):1109–1117. 10.1093/jac/dkg222 [DOI] [PubMed] [Google Scholar]
- 14.Hooper DC. Bacterial resistance to fluoroquinolones: mechanisms and patterns. Adv Exp Med Biol. 1995;390:49–57. 10.1007/978-1-4757-9203-4_4 [DOI] [PubMed] [Google Scholar]
- 15.Andersson DI, Levin BR. The biological cost of antibiotic resistance. Curr Opin Microbiol. 1999;2(5):489–493. [DOI] [PubMed] [Google Scholar]
- 16.Andersson MI, MacGowan AP. Development of the quinolones. J Antimicrob Chemother. 2003;51(suppl 1):1–11. [DOI] [PubMed] [Google Scholar]
- 17.Pál C, Papp B, Lázár V. Collateral sensitivity of antibiotic-resistant microbes. Trends Microbiol. 2015;23(7):401–407. 10.1016/j.tim.2015.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogwill T, MacLean RC. The genetic basis of the fitness costs of antimicrobial resistance: a meta-analysis approach. Evol Appl. 2015;8(3):284–295. 10.1111/eva.12202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall AR, Iles JC, MacLean RC. The fitness cost of rifampicin resistance in Pseudomonas aeruginosa depends on demand for RNA polymerase. Genetics. 2011;187(3):817–822. 10.1534/genetics.110.124628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall BG, Acar H, Nandipati A, Barlow M. Growth rates made easy. Mol Biol Evol. 2014;31(1):232–238. 10.1093/molbev/mst187 [DOI] [PubMed] [Google Scholar]
- 21.Tenover FC. Mechanisms of antimicrobial resistance in bacteria. Am J Infect Control. 2006;34(5 Suppl 1):S3–S10. [DOI] [PubMed] [Google Scholar]
- 22.Reynolds MG. Compensatory evolution in rifampin-resistant Escherichia coli. Genetics. 2000;156(4):1471–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levin BR, Perrot V, Walker N. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics. 2000;154(3):985–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindgren PK, Karlsson A, Hughes D. Mutation rate and evolution of fluoroquinolone resistance in Escherichia coli isolates from patients with urinary tract infections. Antimicrob Agents Chemother. 2003;47(10):3222–3232. 10.1128/AAC.47.10.3222-3232.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo N, Pereira S, Sahin O, Lin J, Huang S, Michel L, Zhang Q. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc Nat Acad Sci of the U S A. 2005;102(3):541–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rozen DE, McGee L, Levin BR, Klugman KP. Fitness costs of fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 2007;51(2):412–416. 10.1128/AAC.01161-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Q, Sahin O, McDermott PF, Payot S. Fitness of antimicrobial-resistant Campylobacter and Salmonella. Microbes Infect. 2006;8(7):1972–1978. 10.1016/j.micinf.2005.12.031 [DOI] [PubMed] [Google Scholar]
- 28.Melnyk AH, Wong A, Kassen R. The fitness costs of antibiotic resistance mutations. Evol Appl. 2015;8(3):273–283. 10.1111/eva.12196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horowitz DS, Wang JC. Mapping the active site tyrosine of Escherichia coli DNA gyrase. J Biol Chem. 1987;262(11):5339–5344. [PubMed] [Google Scholar]
- 30.Yoshida H, Bogaki M, Nakamura S, Ubukata K, Konno M. Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J Bacteriol. 1990;172(12):6942–6949. 10.1128/jb.172.12.6942-6949.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hooper DC. Mechanisms of quinolone resistance. Drug Resist Updat. 1999;2(1):38–55. 10.1054/drup.1998.0068 [DOI] [PubMed] [Google Scholar]
- 32.Sullivan EA, Kreiswirth BN, Palumbo L, Kapur V, Musser JM, Ebrahimzadeh A, et al. Emergence of fluoroquinolone- resistant tuberculosis in New York City. Lancet. 1995;345(8958):1148–1150. 10.1016/s0140-6736(95)90980-x [DOI] [PubMed] [Google Scholar]
- 33.Lu T, Zhao X, Drlica K. Gatifloxacin activity against quinolone-resistant gyrase: allele-specific enhancement of bacteriostatic and bactericidal activity by the C-8-methoxy group. Antimicrob Agents Chemother. 1999;43(12):2969–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedman SM, Lu T, Drlica K. Mutation in the DNA gyrase A gene of Escherichia coli that expands the quinolone resistance-determining region. Antimicrob Agents Chemother. 2001;45(8):2378–2380. 10.1128/AAC.45.8.2378-2380.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida H, Bogaki M, Nakamura M, Yamanaka LM, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob Agents Chemother. 1991;35(8):1647–1650. 10.1128/aac.35.8.1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heddle J, Maxwell A. Quinolone-binding pocket of DNA gyrase: Role of GyrB. Antimicrob Agents Chemother. 2002;46(6):1805–1815. 10.1128/AAC.46.6.1805-1815.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weigel LM, Anderson GJ, Tenover FC. DNA gyrase and topoisomerase IV mutations associated with fluoroquinolone resistance in Proteus mirabilis. Antimicrob Agents Chemother. 2002;46(8):2582–2587. 10.1128/AAC.46.8.2582-2587.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alekshun MN, Levy SB. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli mar locus, by multiple chemicals in vitro. J Bacteriol. 1999a;181(15):4669–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alekshun MN, Levy SB. Characterization of MarR superrepressor mutants. J Bacteriol. 1999b;181(10):3303–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen SP, Hächler H, Levy SB. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol. 1993;175(5):1484–1492. 10.1128/jb.175.5.1484-1492.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alekshun MN, Levy SB. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother. 1997;41(10):2067–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alekshun MN, Levy SB, Mealy TR, Seaton BA, Head JF. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat Struct Biol. 2001;8(8):710–714. 10.1038/90429 [DOI] [PubMed] [Google Scholar]
- 43.Randall LP, Woodward MJ. The multiple antibiotic resistance (mar) locus and its significance. Res Vet Sci. 2002;72(2):87–93. 10.1053/rvsc.2001.0537 [DOI] [PubMed] [Google Scholar]
- 44.Imamovic L, Sommer MO. Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci Transl Med. 2013;5(204):204ra132 [DOI] [PubMed] [Google Scholar]
- 45.Lázár V, Pal Singh G, Spohn R, et al. Bacterial evolution of antibiotic hypersensitivity. Mol Syst Biol. 2013;9:700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luria SE, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28(6):491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foster PL. Methods for determining spontaneous mutation rates. Methods Enzymol. 2006;409:195–213. 10.1016/S0076-6879(05)09012-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma WT, Sandri GH, Sarkar S. Analysis of the Luria-Delbrück distribution using discrete convolution powers. J Appl Probab. 1992;29(2):255–267. [Google Scholar]
- 49.Sarkar S, Ma WT, Sandri GH. On fluctuation analysis: a new, simple and efficient method for computing the expected number of mutants. Genetica. 1992;85(2):173–179. 10.1007/bf00120324 [DOI] [PubMed] [Google Scholar]
- 50.Hall BM, Ma CX, Liang P, Singh KK. Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria- Delbrück fluctuation analysis. Bioinformatics. 2009;25(12):1564–1565. 10.1093/bioinformatics/btp253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosche WA, Foster PL. Determining mutation rates in bacterial populations. Methods. 2000;20(1):4–17. 10.1006/meth.1999.0901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng Q. Progress of a half century in the study of the Luria–Delbrück distribution. Math Biosci. 1999;162(1–2):1–32. 10.1016/s0025-5564(99)00045-0 [DOI] [PubMed] [Google Scholar]
- 53.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25–R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cingolani P, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6(2):80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990;34(6):1271–1272. 10.1128/aac.34.6.1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heisig P, Tschorny R. Characterization of fluoroquinolone-resistant mutants of Escherichia coli selected in vitro. Antimicrob Agents Chemother. 1994;38(6):1284–1291. 10.1128/aac.38.6.1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hopkins KL, Davies RH, Threlfall EJ. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int J Antimicrob Agents. 2005;25(5):358–373. 10.1016/j.ijantimicag.2005.02.006 [DOI] [PubMed] [Google Scholar]
- 60.Oram M, Fisher LM. 4-Quinolone resistance mutations in the DNA gyrase of Escherichia coli clinical isolates identified by using the polymerase chain reaction. Antimicrob Agents Chemother. 1991;35(2):387–389. 10.1128/aac.35.2.387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maxwell A. The molecular basis of quinolone action. J Antimicrob Chemother. 1992;30(4):409–414. 10.1093/jac/30.4.409 [DOI] [PubMed] [Google Scholar]
- 62.Everett MJ, Jin YF, Ricci V, Piddock LJ. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob Agents Chemother. 1996;40(10):2380–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heisig P. Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 1996;40(4):879–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lindgren PK, Marcusson LL, Sandvang D, Frimodt-Møller N, Hughes D. Biological cost of single and multiple norfloxacin resistance mutations in Escherichia coli implicated in urinary tract infections. Antimicrob Agents Chemother. 2005;49(6):2343–2351. 10.1128/AAC.49.6.2343-2351.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moon DC, Seol SY, Gurung M, Jin JS, Choi CH, Kim J, et al. Emergence of a new mutation and its accumulation in the topoisomerase IV gene confers high levels of resistance to fluoroquinolones in Escherichia coli isolates, Int J of Antimicrob Agents. 2010;35(1):76–79. [DOI] [PubMed] [Google Scholar]
- 66.Marcusson LL, Frimodt-Møller N, Hughes D. Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog. 2009;5(8):e1000541 10.1371/journal.ppat.1000541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hallett P, Maxwell A. Novel quinolone resistance mutations of the Escherichia coli DNA gyrase A protein: enzymatic analysis of the mutant proteins. Antimicrob Agents Chemother. 1991;35(2):335–340. 10.1128/aac.35.2.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reece R, Maxwell A. DNA gyrase: structure and function. Crit Rev Biochem Mol Biol. 1991;26(3–4):335–375. 10.3109/10409239109114072 [DOI] [PubMed] [Google Scholar]
- 69.Brino L, Urzhumtsev A, Mousli M, Bronner C, Mitschler A, Oudet P, et al. Dimerization of Escherichia coli DNA-gyrase B provides a structural mechanism for activating the ATPase catalytic center. J Biol Chem. 2000;275(13):9468–9475. 10.1074/jbc.275.13.9468 [DOI] [PubMed] [Google Scholar]
- 70.Nakamura S, Nakamura M, Kojima T, Yoshida H. gyrA and gyrB mutations in quinolone-resistant strains of Escherichia coli. Antimicrob Agents Chemother. 1989;33(2):254–255. 10.1128/aac.33.2.254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vila J, Ruiz J, Goñi P, De Anta MT. Detection of mutations in parC in quinolone-resistant clinical isolates of Escherichia coli. Antimicrob Agents Chemother. 1996;40(2):491–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gensberg K, Jin YF, Piddock LJ. A novel gyrB mutation in a fluoroquinolone-resistant clinical isolate of Salmonella typhimurium. FEMS Microbiol Lett. 1995;132(1–2):57–60. 10.1111/j.1574-6968.1995.tb07810.x [DOI] [PubMed] [Google Scholar]
- 73.Stanger FV, Dehio C, Schirmer T. Structure of the N-terminal gyrase B fragment in complex with ADPPi reveals rigid-body motion induced by ATP hydrolysis. PLoS One. 2014;9(9):e107289 10.1371/journal.pone.0107289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sulavik MC, Gambino LF, Miller PF. The MarR repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli: prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol Med. 1995;1(4):436–446. [PMC free article] [PubMed] [Google Scholar]
- 75.Finn RD, Coggill P, Eberhardt RY, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–D285. 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.UniProt Consortium, T. UniProt: the universal protein knowledgebase. Nuc Acids Res. 2018;45(D1):D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kampranis SC, Bates AD, Maxwell A. A model for the mechanism of strand passage by DNA gyrase. Proc Natl Acad Sci U S A. 1999;96(15):8414–8419. 10.1073/pnas.96.15.8414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Champoux JJ. DNA topoisomerases: Structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. 10.1146/annurev.biochem.70.1.369 [DOI] [PubMed] [Google Scholar]
- 79.Sissi C, Palumbo M. In front of and behind the replication fork: bacterial type IIA topoisomerases. Cell Mol Life Sci. 2010;67(12):2001–2024. 10.1007/s00018-010-0299-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gubaev A, Klostermeier D. The mechanism of negative DNA supercoiling: a cascade of DNA-induced conformational changes prepares gyrase for strand passage. DNA Repair. 2014;16:23–34. 10.1016/j.dnarep.2014.01.011 [DOI] [PubMed] [Google Scholar]
- 81.Domagala JM. Structure-activity and structure side effect relationships for the quinolone antibacterials. J Antimicrob Chemother. 1994;33(4):685–706. 10.1093/jac/33.4.685 [DOI] [PubMed] [Google Scholar]
- 82.Tillotson GS. Quinolones: structure-activity relationships and future predictions. J Med Microbiol. 1996;44(5):320–324. 10.1099/00222615-44-5-320 [DOI] [PubMed] [Google Scholar]
- 83.Zhao X, Wang JY, Xu C, Dong Y, Zhou J, Domagala J, Drlica K. Killing of Staphylococcus aureus by C-8-methoxy fluoroquinolones. Antimicrob Agents Chemother. 1998;42(4):956–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao X, Drlica K. Restricting the selection of antibiotic-resistant mutants: a general strategy derived from fluoroquinolone studies. Clin Infect Dis. 2001;33(Suppl 3):S147–S156. [DOI] [PubMed] [Google Scholar]
- 85.Fukuda H, Hiramatsu K. Primary targets of fluoroquinolones in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1999;43(2):410–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jorgensen JH, Weigel LM, Ferraro MJ, Swenson JM, Tenover FC. Activities of newer fluoroquinolones against Streptococcus pneumoniae clinical isolates including those with mutations in the gyrA, parC, and parE loci. Antimicrob Agents Chemother. 1999;43(2):329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dong Y, Zhao X, Domagala J, Drlica K. Effect of fluoroquinolone concentration on selection of resistant mutants of Mycobacterium bovis BCG and Staphylococcus aureus. Antimicrob Agents Chemother. 1999;43(7):1756–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pestova E, Millichap JJ, Noskin GA, Peterson LR. Intracellular targets of moxifloxacin: a comparison with other fluoroquinolones. J Antimicrob Chemother. 2000;45(5):583–590. 10.1093/jac/45.5.583 [DOI] [PubMed] [Google Scholar]
- 89.Sanders CC. Mechanisms responsible for cross-resistance and dichotomous resistance among the quinolones. Clin Infect Dis. 2001;32(Suppl 1):S1–S8. [DOI] [PubMed] [Google Scholar]
- 90.Peterson LR. Quinolone molecular structure-activity relationships: what we have learned about improving antimicrobial activity. Clin Infect Dis. 2001;33(Suppl 3):S180–S186. [DOI] [PubMed] [Google Scholar]
- 91.Michot JM, Seral C, Van Bambeke F, Mingeot-Leclercq MP, Tulkens PM. Influence of efflux transporters on the accumulation and efflux of four quinolones (ciprofloxacin, levofloxacin, garenoxacin, and moxifloxacin) in J774 macrophages. Antimicrob Agents Chemother. 2005;49(6):2429–2437. 10.1128/AAC.49.6.2429-2437.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Barnard FM, Maxwell A. Interaction between DNA gyrase and quinolones: Effects of alanine mutations at GyrA subunit residues Ser83 and Asp87. Antimicrob Agents Chemother. 2001;45(7):1994–2000. 10.1128/AAC.45.7.1994-2000.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Morgan-Linnell SK, Zechiedrich L. Contributions of the combined effects of topoisomerase mutations toward fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 2007;51(11):4205–4208. 10.1128/AAC.00647-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Morgan-Linnell SK, Becnel Boyd L, Steffen D, Zechiedrich L. Mechanisms accounting for fluoroquinolone resistance in Escherichia coli clinical isolates. Antimicrob Agents Chemother. 2008;53(1):235–241. 10.1128/AAC.00665-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Becnel Boyd L, Maynard MJ, Morgan-Linnell SK, et al. Relationships among ciprofloxacin, gatifloxacin, levofloxacin, and norfloxacin MICs for fluoroquinolone-resistant Escherichia coli clinical isolates. Antimicrob Agents Chemother. 2009;53(1):229–234. 10.1128/AAC.00722-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Azéma J, Guidetti B, Korolyov A, Kiss R, Roques C, Constant P, et al. Synthesis of lipophilic dimeric -7/-7-linked ciprofloxacin and -6/-6-linked levofloxacin derivatives. Versatile biological evaluations of monomeric and dimeric fluoroquinolone derivatives as potential antitumor, antibacterial or antimycobacterial agents. Euro J of Medicinal Chem. 2011;46(12):6025–6038. [DOI] [PubMed] [Google Scholar]
- 97.Björkman J, Nagaev I, Berg OG, Hughes D, Andersson DI. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science. 2000;287(5457):1479–1482. 10.1126/science.287.5457.1479 [DOI] [PubMed] [Google Scholar]
- 98.Gagneux S, Long CD, Small PM, Van T, Schoolnik GK, Bohannan BJ. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science. 2006;312(5782):1944–1946. 10.1126/science.1124410 [DOI] [PubMed] [Google Scholar]
- 99.Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Micro. 2010;8(4):260–271. [DOI] [PubMed] [Google Scholar]
- 100.Bagel S, Hüllen V, Wiedemann B, Heisig P. Impact of gyrA and parC mutations on quinolone resistance, doubling time, and supercoiling degree of Escherichia coli. Antimicrob Agents Chemother. 1999;43(4):868–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kugelberg E, Löfmark S, Wretlind B, Andersson DI. Reduction of the fitness burden of quinolone resistance in Pseudomonas aeruginosa. J Antimicrob Chemother. 2005;55(1):22–30. 10.1093/jac/dkh505 [DOI] [PubMed] [Google Scholar]
- 102.MacLean RC, Vogwill T. Limits to compensatory adaptation and the persistence of antibiotic resistance in pathogenic bacteria. Evol Med Public Health. 2014;2015(1):4–12. 10.1093/emph/eou032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Betitra Y, Teresa V, Miguel V, Abdelaziz T. Determinants of quinolone resistance in Escherichia coli causing community-acquired urinary tract infection in Bejaia, Algeria. Asian Pac J Trop Med. (2014);7(6):462–467. 10.1016/S1995-7645(14)60075-4 [DOI] [PubMed] [Google Scholar]
- 104.Basra P, Alsaadi A, Bernal-Astrain G, et al. Fitness tradeoffs of antibiotic resistance in extraintestinal pathogenic Escherichia coli. Genome Biol Evol. 2018;10(2):667–679. 10.1093/gbe/evy030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wong A, Rodrigue N, Kassen R. Genomics of adaptation during experimental evolution of the opportunistic pathogen Pseudomonas aeruginosa. PLoS Genet. 2012;8(9):e1002928 10.1371/journal.pgen.1002928 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Resistance mutations isolated as a direct selection on nal (A), cip (B), and levo (C).
(EPS)
Variation in costs of resistance—Growth rate, cell density, and lag phase between gyrA (A), gyrB (B), and marR (C) resistance mutations. All the fitness components are compared to control E. coli K-12 (MG1655). The boxplots present the median, first, and third quartiles, with whiskers showing either the maximum (minimum) value or 1.5 times the interquartile range of the data, whichever is smaller (larger).
(EPS)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.