Short abstract
NK cells are innate lymphoid cells that exert a key role in immune surveillance through the recognition and elimination of transformed cells and viral, bacterial, and protozoan pathogen-infected cells without prior sensitization. Elucidating when and how NK cell-induced intracellular microbial cell death functions in the resolution of infection and host inflammation has been an important topic of investigation. NK cell activation requires the engagement of specific activating, co-stimulatory, and inhibitory receptors which control positively and negatively their differentiation, memory, and exhaustion. NK cells secrete diverse cytokines, including IFN-γ, TNF-α/β, CD95/FasL, and TRAIL, as well as cytoplasmic cytotoxic granules containing perforin, granulysin, and granzymes A and B. Paradoxically, NK cells also kill other immune cells like macrophages, dendritic cells, and hyper-activated T cells, thus turning off self-immune reactions. Here we first provide an overview of NK cell biology, and then we describe and discuss the life–death signals that connect the microbial pathogen sensors to the inflammasomes and finally to cell death signaling pathways. We focus on caspase-mediated cell death by apoptosis and pro-inflammatory and non-caspase-mediated cell death by necroptosis, as well as inflammasome- and caspase-mediated pyroptosis.
Keywords: Apoptosis, inflammasomes, NK cells, necroptosis, pyroptosis
Introduction
NK cells, which belong to group 1 of the innate lymphoid cell (ILC) family, are identified by their ability to produce “type I” cytokines, mainly IFN-γ.1 Contrasting with NKT, γδ T cells, and CD8+ T cells, NK cells do not express T cell receptors. NK cells recognize transformed and infected cells through non-specific germ-line encoding activating receptors. They then inhibit or terminate their activation responses via inhibitory receptors using MHC-1 molecules and class I-like molecules receptors. NK cells constitute only 5–15% of all circulating lymphocytes in human blood, but a large number of distinct NK cell subsets are found in many organs, including the uterus, liver, lungs, and placenta,2 where they play diverse roles in tissue homeostasis and immune surveillance.3,4 These NK cell populations contribute to the full development of innate and adaptive responses through the production of cytokines, including IFN-γ (type II IFN), TNF-β, IL-10, IL-13, and GM-CSF (granulocyte-macrophage-colony stimulating factor), which increase or dampen immune responses.5 IFN-γ stimulates the macrophage cytotoxic capacities and favors Th1 polarization of CD4 T cells. In addition, the direct cell-to-cell contact of NK cells with accessory cells such as dendritic cells (DCs), neutrophils, macrophages, mast cells, and epithelial cells, as well as B and T lymphocytes, is fundamental to their immune function.5 In this context, NK cells and DCs directly influence each other through the production of cytokines such as IFN-γ and/or type I IFNs. DC maturation and subsequent IL-12 production lead to the generation of highly protective CD8 T cells. On the other hand, DC activation and production of IL-18 can prime NK cells and increase their cytolytic effector function. However, excessive amplification or inhibition of NK cells immune response may have detrimental effects to the host.6 Indeed, the NK, accessory, and adaptive functions of NK cells in some specific microbial and pathological conditions are not entirely understood.1,5
Recent studies have demonstrated how dead cells and their constituents are recognized and processed by immune cells, such as phagocytes and DCs, leading to innate and adaptive responses.7 NK cells trigger multiple interconnected molecular mechanisms for the killing and elimination of pathogens and infected-host cells. A variety of pathogen-associated factors called PAMPs and infected cell-endogenous factors called DAMPs (damaged-associated molecular patterns), released by dying cells, activate intracellular innate immune receptors called PRRs. Depending on the immunogenicity of dying cells and their products, the host responds to a weak or strong inflammation and/or immunization.7 Here we attempt to describe in more detail how and when NK cells could potentially regulate cell death pathways and differentially influence, positively and/or negatively, the host’s immune defense.
We will first provide a short update on NK cell biology. Next, we will describe the similarities and differences of the molecular mechanisms used for killing intracellular virus, bacteria and parasites in host infected cells. In the second part, we will describe the interconnected signaling pathways that control programmed cell death via apoptosis, necroptosis and pyroptosis along with how and when NK cells can contribute to these processes. Finally, we will describe some examples of how pathogen virulence factors manipulate inflammasomes and cell death signaling pathways to avoid immediate innate and late adaptive host immune responses.
NK subtypes and functional properties
NK cells develop and begin maturation in the bone marrow, then migrate and continue to mature in peripheral tissues.8,9 NK precursor cells are also found in the liver, thymus and lymph nodes. The transcription factors runt-related transcription factor (RUNX3), T-bet, Eomes, and Blimp-1 play specific roles at distinct stages of NK cell development and maturation, whereas STAT1, STAT4, Zbtb32, and AhR are examples of transcription factors that regulate the different effector functions of NK cells.1,10,11 NK cell precursors, upon stimulation with IL-15, differentiate towards immature NK (iNK) and mature NK (mNK) cells.12 Three distinct subsets of NK cells, with respect to expression levels of CD56 glycoprotein and CD16 (the Fc receptor that binds the Fc portion of Abs), have been identified (Table 1). Around 10% of the NK cell population exhibit high levels of CD56 and low or no expression of CD16. The circulating CD56bright, CD16negative, CD62L+ NK cell subset is predominantly present in lymphoid nodes and sites of infection and inflammation. In addition, it is a potent producer of cytokines (mainly IFN-γ). The canonical CD56dimmer, CD16positive NK cell subset is highly cytotoxic and comprises around 90% of the NK cell population. Tissue-resident NK cells predominately consist of CD56bright CD16negative cells. NK cells expressing CD56negative/CD16positive are normally present in a low percentage of healthy individuals, but in an increasing percentage in elderly people and individuals infected by HIV and the human cytomegalovirus (HCV).4,12,13 Continuous maturation of peripheral NK cells enables the formation of these heterogeneous subsets with slight differences in the expression of germ-line encoded activating and inhibitory receptors, adhesion molecules, chemokines, cytokines, and growth factor receptors in addition to key functional capacities, as listed in Table 1.12,14
Table 1.
Relative expression patterns of activating and inhibiting receptors, differentiation markers, cytokines, chemokines, and functional capabilities in NK cell subsets.
| Receptors/markers/cytokinesa | CD56bright | CD56dim | CD56neg |
|---|---|---|---|
| Adhesion receptors | |||
| CD56/N-CAM | +++ | ++ | − |
| CD57 | − | +++ | + |
| LFA-1, -2, -3 | − | +++ | + |
| L-Selectin | − | +++ | + |
| Activating receptors | |||
| CD16/FcγRIIIa | − | +++ | + |
| NKG2A | +++ | ++ | + |
| NKG2C | +++ | ++ | + |
| NKG2D | ++ | ++ | ++ |
| NKp30 | +++ | +++ | ++ |
| NKp44 | − | +++ | − |
| NKp46 | +++ | ++ | + |
| Inhibitory receptors | |||
| KIRs | − | −/+ | +++ |
| LILRB1 | + | ++ | +++ |
| Siglec-7 | +++ | +++ | − |
| LAMP-1 | +++ | ++ | −/+ |
| Pore-forming proteins and proteases | |||
| Perforin | −/+ | +++ | + |
| Granulysin | − | +++ | + |
| Granzymes | −/+ | +++ | + |
| Cathepsins | − | +++ | − |
| Cytokine/chemokine/receptors | |||
| IL-2Rα | +++ | + | − |
| IL-7Rα | +++ | − | − |
| CXCR1 | − | +++ | ND |
| CXCR3 | +++ | +++ | ND |
| CXCR4 | +++ | +++ | ND |
| CXCL8 | +++ | ++ | ND |
| GM-GSF | +++ | + | + |
| TNF- | +++ | + | + |
| TNF- | +++ | + | ND |
| IFN- | +++ | ++ | + |
| IL-6 | ++ | + | ND |
| IL-10 | +++ | + | ND |
| IL-13 | +++ | + | ND |
| CCL3 | + | + | + |
| CCL4 | + | ++ | + |
| CCL5 | + | ++ | + |
| MIP-1α | + | ++ | + |
| Biological activities | |||
| Cytotoxicity | −/+ | +++ | −/+ |
| Proliferation | +++ | ++ | + |
| Degranulation | + | +++ | + |
| ADCC | + | +++ | + |
| LAK | + | +++ | |
aRelative expression levels and activities (graded as −/+, +, ++, or +++) were obtained from a compilation of results found in the literature. ADCC: Ab-dependent cellular cytotoxicity, LAK: lymphokine-activated killer.
Activation of NK cells is tightly regulated by the expression of specific activating and inhibitory receptors containing ITIM (immunoreceptor tyrosine-based inhibition motif) and ITAM (immunoreceptor tyrosine-based activation motif) molecules, which serve as intracellular docking sites for members of the spleen tyrosine kinase (SYK) and ζ-chain-associated protein kinase of 70 kDa (ZAP-70) family. The NK receptor families include the natural cytotoxicity receptors (NCRs), which are integral type 1 transmembrane proteins pertaining to the immunoglobulin-like superfamily receptors (KIRs). These KIRs bind to MHC class I molecules (HLAs) in humans and the Ly49 (also known as KLRA) receptors in mice, encoded by KLR genes located in the NK–gene complex (NKC) locus on chromosome 6 in mice and on chromosome 12 in humans. Inhibitory KIR receptors such as KIR2DL, KIR3DL, and CD94/NKG2A, among others, function to detect self-MHC class I ligands (HLA-A, HLA-B, HLA-C) in healthy cells.12,15 CD94/NKG2 (heterodimers), a C-type lectin family receptor, is conserved in both rodents and primates and recognizes non-classical MHC I molecules such as HLA-E. When inhibitory phosphorylated receptors interact via ITIM molecules with cytosolic SHP-1 and SHP-2 tyrosine phosphatases, the downstream signaling pathway is inhibited. KIRs and their cognate MHC class I ligands are critical for the educating or licensing processes by which NK cells achieve self-tolerance.8 Thus, they exert dominant inhibitory effects on NK cells and, in their absence, NK cells promote the killing of target cells.
NKG2D, DNAM-1, NKp46/NCR1 (mouse/human), NKp44/NCR2 (human), and NKp30/NCR3 (human) are members of the NCR family of germ-line encoded genes. NCRs interact, via ITAM molecules, with ZAP-70 and SYK, or other protein kinases, to initiate downstream signaling.12,15 Molecules associated with infection, transformation, or DNA damage can bind and activate these receptors NKG2D is a NCR receptor expressed on the surface of all NK cells as well as cytotoxic CD8+ T and γδ T cells. MICA/MICB (humans), RAE1 family (rodents), ULBP-1, -2 and -3 (only in humans) are proteins expressed on the surface of stressed, malignant, transformed, and infected cells that bind to the NKG2D receptor and activate NK cells.12,15 NKp44/NCR2 is induced upon activation of resting NK cells with IL-2 and can bind to viral hemagglutinins. NK cells interact with DCs via NKp30/NCR3 and this may result in down-regulation of NK activating receptors upon their death.5
NK cell-mediated immune responses are also controlled by immune checkpoint molecules, which are also present in cytotoxic T cells.16,17 The expression of the cytotoxic T lymphocyte associated antigen 4 (CTLA4), programmed cell death protein receptor (PDCD1 or PD-1), TIM-3, lymphocyte-activation gene 3 (LAG3), TIGIT, and B and T lymphocyte attenuator (BTLA/CD272) receptors and their cognate ligands triggers immunosuppressive signaling pathways to prevent excessive activation of T cell cytotoxicity activity.16,17 Various mAbs that block these receptors or their ligands have been successfully used to restore T cell cytotoxic activity and promote a potent immune response in patients with advanced cancers.17 Inhibition of these receptors may enhance the efficacy of antiviral and cancer therapies.18,19 With this idea, one clinical trial is underway to evaluate if pembrozilumab, an inhibitor of the PD-1 pathway, can restore cytotoxicity to exhausted PD-1 expressing NK cells.20
NK cell exhaustion is caused by different factors including chronic exposure to endogenous or foreign antigens; this can lead to cell death, hypoxia and metabolic deprivation of essential amino acids.16,21,22 The expression of different, multiple combinations of KIR receptors has also been associated with NK dysfunction or exhaustion.21,22 Clinical studies in patients with acute myeloid leukemia have suggested that the lack of response due to down-regulation in the expression of activating receptors NKp46/NCR1, NKp30/NCR3, CD226, 2B4, CD94/NKG2C, and NKG2D was caused by NK cell exhaustion.23 This dysfunction could not be overcome with an up-regulation in their ligands MICA and MICB.24 Posttranslational alterations can also decrease the affinity of MICA for NKGD2, blocking the activation signaling triggered by this receptor.25 In addition, CD4+CD25+Foxp3+ regulatory T cells (Treg) and myeloid-derived suppressor cells (MDSCs) as well as their products TGF-β, prostaglandin E2 (PGE2), or indoleamine 2,3-dioxygenase (IDO) contribute to maintaining a highly suppressive environment that limits NK cell activities. Treg cells are able to inhibit NKG2D expression leading to NK cell activation failure.26 MDSCs induced by the hepatitis C virus inhibit the release of IFN-γ by NK cells via the arginase-1 dependent mechanism.27
Repeated TCR stimulation leads to activation-induced cell death (AICD), a well-known mechanism for the regulation of T cell homeostasis.28 Recent studies have demonstrated that activated T cells are eliminated by necroptosis and not apoptosis.28 Exhausted NK cells may undergo cell death after repeated Ag stimulation, but this has not been established yet.22 Another mechanism that can lead to the inhibition of NK cell-mediated cytotoxicity is split anergy, defined as the cellular responses mediated by NK cells after engagement with target cells within a tumor or inflammatory microenvironment.29 These anergized or regulatory NK (NKreg) cells are characterized by the CD16−/dimCD56dim/+CD69+ phenotype, and, despite the low cytotoxicity activity, NKreg cells have increased cytokine production and promote the differentiation of stem cells and cancer stem cells.30
NK cytotoxicity can be triggered by IgG1 Abs that recognize with high-affinity cell surface Ags in tumor and infected cells via a mechanism known as Ab-dependent cellular cytotoxicity (ADCC).31 Three classes of receptors that recognize the Fc portion of the IgG Ab (FcγR) have been identified.32 They can activate or inhibit the effector function of the immune cells.32 Human NK cells can express FcγRIIc (CD32c) and FcγRIIIa (CD16a), which is highly expressed in the CD56dim/CD16+ NK subset.32 FcγRIIIa usually associates with FcϵRI-γ chains or CD3-ζ chains to activate distinct cellular signaling pathways, including NF-κB, PI3K, and ERK via a tyrosine based activating motif (ITAM) phosphorylation, leading to NK degranulation, cytokine production and cell death.31 In HIV positive patients, Chung et al. have demonstrated that ADCC responses are present in NK cells after exposure to gp140 Env or HIV peptide pool proteins, although no correlation with disease progression was observed.33 In cancers, ADCC has been used as a part of the antitumor immune response with mAbs, improving their efficacy by blocking ligand binding and cell signaling inhibition.23,31
Considerable efforts have been made toward identifying the phenotypic and functional features of memory NK cells.34,35 A subset of NK cells expressing the activating Ly49H receptor have been found to acquire memory-like or adaptive immune features that mediate a recall response following cytomegalovirus (CMV) infection in mice.36 Infection with CMV in humans is associated with expansions in potential memory NK cell subsets expressing NKG2C or self-specific inhibitory KIRs.37 NK cell memory for CMV infection is induced by IL-12, and requires microRNA-155 expression.36,38,39 Exposure of human NK cells to IL-12, IL-15, and IL-18 causes stable demethylation of the IFNG locus regulatory region suggesting that epigenetic imprinting and not gene rearrangement is responsible for NK cell memory.34,35
NK cells activate multiple molecular pathways to control intracellular viral, bacterial, and protozoal infections
NK cells can be activated directly by pathogens, including influenza virus, immunodeficiency virus, bacteria and protozoan after binding to specific surface receptors or cytokine receptors (Figure 1).5,14,40 NK cell activation depends on direct contact with accessory cells, including DCs, monocytes and macrophages. These cells provide cytokines such as IL-1, IL-2, IL-12, IL-15, IL-18, IL-21, and type I IFNs, which are key factors necessary for their proliferation, complete activation, and effector function.5,14,41 Activated NK cells initiate the production of IFN-γ, cytotoxic cytokines TNF-α/β, CD95/FasL, and TRAIL/Apo2L (TNF-related apoptosis-inducing ligand) and small cytotoxic granules in their cytoplasm.
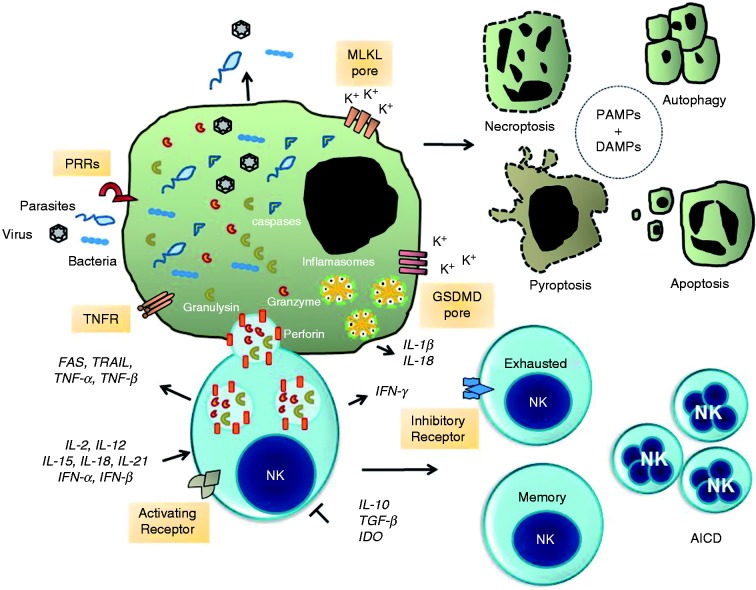
Figure 1.
NK cell-mediated signaling pathways to intracellular pathogen and infected cell death. The formation of immunological synapse between a NK cell and the target cell occurs upon identification of specific self-deficiencies by an array of either paired activating (AR) or inhibitory membrane receptors (IR) and their ligands. NK cells release cytotoxic granules containing the pore-forming protein perforin, granulysin and serine proteases known as granzymes. Granzymes promote the cleavage and activation of a family of protease known as caspases. Caspases promote proteolytic cleavage of cellular substrates leading to apoptosis. Alternatively, the assembly of the channels and pores formed by MLKL and GSDMD promote the exchange of Ca2+, Na+, and K+ ions that contribute to osmotic perturbation and ultimately cell death by necroptosis or pyroptosis, respectively. Infected cells may die by autophagy, which is a process that can inhibit or activate programmed cell death. Finally, sustained expression and activation of IRs promotes NK cell exhaustion whereas over-activation of ARs leads to activation-induced cell death (AICD). Concomitantly, survival NK cell clones expand to make a memory repertoire of NK cells for a specific pathogen. Extracellular and intracellular pathogens or their products (PAMPs and DAMPs) activate the inflammasome signaling pathways and production of pro-inflammatory cytokine IL-1β and IL-18. Production of the immune suppressive cytokines TGF-β and IL-10 and indoleamine 2,3-dioxygenase (IDO) abolish NK cell activation and tissue damage. Abbreviations: GM-CSF, granulocyte-macrophage CSF; CD95/FasL (Fas ligand); TRAIL, TNF-related apoptosis-inducing ligand; TNFR, TNF receptor; DAMPs, danger-associated molecular patterns; MLKL, mixed lineage kinase domain-like; GSDMD, gasdermin D.
NK cell recognition of host infected cells occurs through immunological synapse formation which is mediated by mobilization, fusion and delivery of exocytic granules (Figure 1).42,43 Cytotoxic granules are acidic secretory lysosomes (pH ≈ 5) that contain the death effector proteins perforin (PFN), granzymes (Gzms), and granulysin (GNLY), in addition to lysosomal associated membrane glycoproteins, such as LAMP-1 (CD107a), LAMP-2, and LAMP-3, among other membrane proteins.43 PFN belongs to a family of pore-forming proteins that share a domain with the C9 complement protein.44,45 PFN monomers undergo homopolymerization in a calcium-dependent manner moving into the plasma membrane to produce pores.44,46,47 Gzms are serine proteases delivered to the target cell by PFN and contain a conserved His-Asp-Ser catalytic triad in their active site. Gzms are synthesized as pro-enzymes and become active after processed by cathepsin C. There are five isoforms in humans and the Gzm family members A, B, H, K and M. They recognize and cleave hundreds of extracellular and intracellular proteins.43,47,48 GNLY is present in cytotoxic granules together with PFN and Gzms. GNLY is produced as an inactive 15 kDa pro-peptide that is processed in killer cell cytotoxic granules to a 9 kDa peptide.49 GNLY itself possesses antiviral and antimicrobial effects against a variety of pathogens including bacteria, fungi, and parasites.
The role of NK cells in controlling viral infections was first recognized in 1999.50 Many viruses modify the surface membrane of their host cells, leading to down-regulation of MHC class I molecules or up-regulation of host-derived, stress-related molecules that serve as a target for infected cells by NK cells. For example, expression of viral envelope proteins such as the influenza virus hemagglutinins (H1-7) are recognized by NKp30, NKp44, and NKp46 receptors which then induce NK cell cytotoxicity. Co-stimulatory accessory cell-derived signals contribute to NK activation and control of viral infections.5 Secretion of IFN-γ by NK cells increases the production of IFN-inducible anti-viral proteins including dsRNA-dependent protein kinase (PKR) and IFN regulatory factors (IRF-1 to -9). PKR induces apoptosis of viral infected cells mediated by the eIF2alpha NF-κB and p53 pathways.51,52 The transcription factor p53 induces the expression of hundreds of genes simultaneously, including ATM (ataxia-telangiectasia-mutated) and ATR (ataxia, and rad3 related), which mediate DNA repair, cell-cycle arrest, senescence, and apoptosis of target cells. Nonetheless, many viruses have evolved strategies to evade p53-mediated apoptosis in order to increase progeny virus production in infected cells.53
Recent in vitro and in vivo studies have shown that NK cells can directly promote the killing of the intracellular bacteria Listeria monocytogenes and Mycobacterium tuberculosis,54 as well as the protozoan parasites Trypanosoma cruzi, Toxoplasma gondii, and Leishmania major.55,56 Walch and collaborators have shown that GNLY delivers Gzms A and B into cytosol of infected cell.54 These proteolytic enzymes generate reactive oxygen species (ROS) to trigger rapid death of aerobic Escherichia coli.54 Gzm A cleaves various intracellular proteins, including electron transport chain (ETC) complex I proteins, which leads to the generation of superoxide anions and peroxide.57 In addition, Gzm B promotes the degradation of multiple ribosomal proteins, including aminoacyl tRNA synthetases, ribosome-associated chaperone trigger factor, heat shock protein DnaK, and chaperonin GroEL.55 Thus, NK cells delivery of Gzms leads to the inactivation of multiple proteins involved in the cellular oxidative stress defense as well as biosynthetic and metabolic pathways of the intracellular pathogens.54,56
Gzm B is known to activate the classical caspase-dependent apoptotic pathway in mammalian cells.48 HeLa cells infected with intracellular L. monocytogenes are killed by the cooperative actions of Gzm A and B, GNLY and PFN; however, death occurs independently of caspase activation.54 Huang et al. generated a human GNLY expressing transgenic mouse strain to evaluate the role of NK and CD8+T cells in tumor rejection.58 The authors demonstrated that GNLY mice showed improved survival rates in an experimental lymphoma model in relation to wild type controls.58 As expected, Walch et al. showed that GNLY transgenic mice had a reduced intracellular L. monocytogenes load when compared to wild type mice.54 The in vivo cytotoxic effect of GNLY was not observed in transgenic mice deficient of Prf1 gene.
NK cells also kill T. cruzi, T. gondii, and L. major which are obligate intracellular parasites.55 PFN is necessary for GNLY and Gzm mediated protozoan death.55 Dying cells displayed apoptosis-like features, including increased production of mitochondrial superoxide radicals, DNA damage, mitochondrial outer membrane breaks, blebbing, as well as phosphatidylserine exposure and chromatin condensation in parasites and dead cells. Again, all these events were independent of intracellular caspase activation.55 Mice genetically engineered to express human GNLY gene in NK cells and CD8+ T cells58 were also partially protected from lethal infection by T. cruzi and T. gondii.55
NK cells also play a relevant role in combating fungal infections in vivo.59 Different fungal species can be recognized and eliminated by NK cells, although the specific receptors involved in this recognition remains poorly understood. Li et al. have demonstrated that the NKp30 receptor is essential for identification and elimination of C. neoformans. In addition, the decreased NK cell cytotoxicity in HIV infected patients against fungi is dependent on the NKp30 receptor and can be restored with IL-12. NKp46 (human) or NCR1 (mouse) recognize the C. glabrata virulence protein as Epa1, Epa6, and Epa7 in order to clear fungal infections in vivo.60
NK cells modulate cell death pathways and inflammasome activation in microbial infected cells
Host cells have a variety of ways to deal with a wide range of pathogens that cause intracellular infection, including bacteria, viruses, fungi and protozoa. The death of professional phagocytes, like macrophage, monocytes and DCs and neutrophils, are considered a defense mechanism against infection.61 Dying cells stimulate an inflammatory response (sterile or non-sterile) with broad biological impact on the host’s pathophysiological response.61,62 Apoptosis, necroptosis, pyroptosis, netosis, efferocytosis, and autophagic cell death are distinct cell death processes that cooperate in the presentation and clearance of various microbial pathogens.61,63–65
We will now examine how and when NK cell-induced microbial and infected cell death could help innate and adaptive immune cell responses.
Apoptosis is the major cell death that depends on the activity of the cysteine aspartyl proteases of the caspase family.66 Apoptosis is triggered by two distinct signaling pathways, the extrinsic pathway and the intrinsic pathway.66 The intrinsic pathway is mediated by cytotoxic cytokines TNF-α, TRAIL, and CD95/FasL after their interaction with specific membrane receptors. It is followed by the formation of a large structure called DISC (death-inducing signaling complex) containing TNFR-associated death domain (TRADD), RIPK1 (the receptor-interacting protein kinase 1), cellular inhibitor of apoptosis 1 (cIAP1), cIAP2, FLIP (Flice-like inhibitory protein), TNFR-associated factor 2 (TRAF2) and TRAF5, and an apical, either procaspase-8 or -10.66 Caspase-8 cleaves and activates the executioner procaspase-3, -6, and -7, which in turn cleave and activate or inactivate hundreds of cellular proteins, leading to the morphological and biochemical features of the apoptotic process.63,66,67
The intrinsic pathway of apoptosis is controlled by a relative abundance of BCL-2 (B cell lymphoma) family proteins in which anti-apoptotic member BCL-2, BCL-XL, MCL-1, and other members prevent the release and/or activation of the pro-apoptotic proteins BAX (BCL2 Associated X) and BAK (BCL2-antagonist/killer 1). Once released, BAX and BAK take part in the molecular composition of multi-complex membrane pores leading to inner and outer mitochondrial permeabilization and the release of cytochrome c which catalyzes the cleavage and activation of initiator procaspase 9/apaf-1 complex.68 Caspase 9 promotes the cleavage and activation of executioner procaspase-3, -6, and -7, which act together to promote apoptosis.63,66,67
Necroptotic cell death is triggered by ligands of the TNF family of receptors, TLRs, TCR, DAI/ZBP1/DML-1 (DNA-dependent activator of IFN-regulatory factors), and IFN receptors. Necroptosis does not depend directly on caspases and is negatively regulated by caspase-8, FLIP, and inhibitors of apoptosis proteins (IAPs).63,69,70 Necroptosis was originally characterized in cells treated with the pan-caspase inhibitor Z-VAD-fmk.71 Caspase-8 inhibits necroptosis via cleavage of the RIPK1 at Asp324. However, when caspase-8 is inhibited or eliminated, un-cleaved RIPK1 interacts and oligomerizes with RIPK3 through their respective RIP homotypic interaction motifs (RHIMs). This leads to the assembly of a large, signal-induced multiprotein complex named necrosome.72 RIPK3 promotes the phosphorylation and activation of mixed lineage kinase domain-like protein (MLKL) at Thr357 and Ser358.73 MLKL binds the inner membrane phospholipids, particularly phosphoinositides and cardiolipin. The N-terminal coiled-coil domain of MLKL folds into four-helix bundles, and these bundles form transmembrane pores allow the exchange of Na+, Cl−, and K+.73 Increases in the osmotic concentration and cell swelling cause them to burst and die.
Pyroptosis is a programmed inflammatory cell death that occurs in macrophages and DCs infected with intracellular bacterial pathogens such as Salmonella typhimurium, Francisella tularensis, and Bacillus anthracis.74–76 A genetic screen with ethyl-N-nitrosourea (ENU) mutant mice revealed that the gasdermin D genes (GSDMD) act as critical mediators of host response to excessive LPS and Gram-negative bacterial infection.77 GSDMD is a member of the gasdermin family of conversed proteins which include gasdermin A, B, C, D, E (DFNA5) and DFNB59.78 GSDMD was recently discovered as one substrate of pro-inflammatory caspases-1, -4, -5, and -11.75,76,79–81 The cleavage of GSDMD releases its N-terminal domain from the inhibitory C terminal domain.82 GSDMD-N-terminal domain displays an α-helix structure that interacts with cardiolipin or phosphorylated head groups (phosphatidylinositol phosphates PIP1 and PIP2) of inner leaflet of plasma membrane of infected cells, forming non-selective permeable pores.82 Permeabilization of the endoplasmic reticulum allows Ca2+ release and activation of endonucleases leading to nuclear condensation and DNA fragmentation.76,80,83 GSDME (also known as DFNA5, deafness, autosomal dominant 5) is cleaved and activated specifically by caspase-3, but, in this case, the cells die by pyroptosis.84 More importantly, overexpression of the N domain of all members of the gasdermin family is capable of triggering pyroptosis in many types of mammalian cells.80,84 It is interesting that GSDMD itself is an antimicrobial peptide and can kill bacteria within the cytosol as well as free-bacteria outside of cells.82
There is no clear evidence that pyroptosis kills intracellular bacteria in vivo, thus suggesting that other mechanisms are required for intracellular clearance of infections.61,74 Previous studies have shown that pyroptosis causes intracellular bacteria to escape to the extracellular environment, thereby allowing their destruction via neutrophils, antimicrobial peptides, Ab immune complexes and complement system.74,75,85 Neutrophils kill free pathogens through the release of extracellular traps (NETs) and pore-induced intracellular traps (PITs), which are formed by a meshwork of chromatin fibers, proteases, and antimicrobial peptides.74,85
NK cells induce apoptosis of infected cells by either the extracellular release of cytotoxic cytokines or the intracellular release of cytotoxic granules containing Gzm A and B, GNLY, and PFN.45,78,86 PFN pores allow the translocation of Gzms A and B into targeted cells.87,88 Gzms A and B promote direct cleavage and activation of caspase-3, -7, -8, and -10. Gzms A and B also promote the cleavage of BID, leading to the release of cytochrome c from mitochondria, initiating the intrinsic pathway of apoptosis. The combination of recombinant Gzm A with PFN induces a new form of cell death, which has many of the features of apoptosis, but is independent of caspase activation.88 Gzms A and B can also cleave many intracellular and extracellular proteins, including cell surface receptors and extracellular matrix (ECM) components.86,89 Mutations in PFN locus at 10q22 disrupt normal PFN levels contributing to the development of the fatal human hyper-inflammatory disease designated familial hemophagocytic lymphohistiocystosis.43 Gzm B-deficient mice are as resistant to infection by the poxvirus and herpes simplex virus as wild type mice, whereas Gzm A-deficient mice are highly susceptible to these viruses.45,87,88
GNLY is a membrane-perturbing saposin-like protein which causes lipid degradation.49,90 Human CTLs and NK cells produce the largest GNLY precursor of 15 kDa and a shorter form of 9 kDa. Both molecules show the highest amount of activity against cancer cells.91,92 GNLY activates an apoptosis cell death pathway that is distinct from those induced by CD95/FasL or Gzms.93 Caspase 3, but not caspase 9, is processed in GNLY treated cells. NK cells expressing GNLY (whole protein) induce endoplasmatic reticulum (ER) damage and activation of caspase-7, which in turn activates caspase-12.92 On the other hand, the recombinant 9 kDa form of GNLY promotes mitochondrial damage through activation of caspase-3, -7, and -9.92 Diversely, the GNLY 15 kDa form is able to activate monocytes and cause their differentiation into immature DCs.90,94 It has been demonstrated that pores formed by GNLY molecules promote Ca2+ and K+ ion translocation which activates the intracellular signal pathway for DC activation and maturation.90,94 Transgenic mice expressing human GNLY in T and NK cells respond normally to infection.58 However, they are much more resistant to lethal doses of bacteria, fungi and parasite infection, including M. tuberculosis, when compared to wild type mice.90 Thus, these two GNLY isoforms have major implications in the control of intracellular pathogen infection. It is important to mention here that the role of NK cells in pyroptosis is very limited and require further investigation.
Apoptosis is characterized by nuclear shrinkage, plasma membrane blebbing and formation of apoptotic bodies.95–97 A hallmark of apoptotic cells is their engulfment by phagocytic cells. Apoptotic cells release soluble “find me”, “catch me,” and “eat me” molecules, which are distinct molecules that attract phagocytes that help with their fast removal.97 Apoptotic bodies containing microbial debris are engulfed by monocytes and macrophages, leading to rapid degradation of both the apoptotic cell and the intracellular pathogen, a process referred to as efferocytosis.95,96 The process is highly regulated and begins with the exposure to phosphatidylserine (find me) and terminates with macrophage engulfment (eat me) of the dying cell.96,97 Macrophages increase the synthesis of IL-10 and TGF-β, which participate in the resolution of inflammation. However, apoptotic cells that were not rapidly efferocytosed acquire a cell phenotype related to that of necrosis which can lead to a pro-inflammatory response.96
Fundamental differences exist in the plasma membrane permeability of pyroptotic and necroptotic cells.63,72,97,99 Pyroptotic and necroptotic cells elicit an inflammatory response thought the release PAMPs and DAMPs and other endogenous danger factors through plasma membrane pores. A variety of DAMPs have been identified with potential pro-inflammatory activity. The best studied are ATP, HMGB1, heat-shock proteins, uric acid, single-stranded RNA, genomic DNA, and IL-1α. These molecules are recognized by PRRs, including TLRs, NLRs, RIG-I-like receptors (RLRs), and AIM2-like receptors (ALRs). These distinct families of sensors are found on the plasma membrane, cytoplasm and endosomes of epithelial cells in tissues with mucosal surfaces and in immune cells of the myeloid lineage.100,101
Extracellular and intracellular PAMPs and DAMPs are also recognized by the inflammasome-associated molecules which act as specific sensors and effectors that activate immune responses.100–102 The inflammasome multiprotein complexes are organized under the scaffold of NLR family proteins, including ALR proteins, pyrin domain-containing 3 (NLRP3), CARD-containing 4 (NLRC4), and pyrin and the IFN-inducible p200-protein IFI16, which share a protein–protein interaction pyrin domain (PYD) in the N-terminus region.61,100 Pyrin is able to find bacterial modifications induced by Rho GTPases, mainly glucosylation, adenylylation, ADP-ribosylation, and deamidation.103 Specifically, these complexes detect the contamination in the cytosol by foreign DNA, bacterial flagellin, type 3 secretion system (T3SS) needle and rod subunits, crystalline structures, toxins and cellular perturbations. RIG-I and MDA-5 helicases and AIM2 and IFI16 are RNA and DNA sensors that initiate transcription of type I IFNs via STING (stimulator of IFN genes). IFI16 detects intracellular HIV viral nucleic acids in CD4+ cells.103 Certain inflammasome are activated directly by oxidative free radicals and K+ efflux either through potassium channels (P2Xs 1-7), hemichannels pannexin-1 or various bacterial pore-forming toxins, including T3SS.100,103
Macrophages and DCs require at least two signals for activation; the first drives the synthesis of both pro-IL-1-β and pro-IL-18, and the second drives the assembly of one specific inflammasome.104,105 The most intensely studied complex of the inflammasome is formed by the NLR family member NLRP3 and the adapter protein ASC (apoptosis-associated speck-like protein). ASC contains a CARD domain and an N-terminal pyrin domain (PYD) that allows its interaction with one NLR or ALR member and the pro-caspase-1 protein. This interaction drives the intermolecular cleavage and enzymatic activation of caspase-1. Caspase-1 promotes the cleavage of pro-inflammatory cytokines IL-1β and IL-18, which are then released from secretory lysosomes or through cellular membrane leakage.100,103,106 Murine caspase-11 and its human homologs’ caspase-4 and -5 are activated directly by the binding of bacterial LPS that enter the cytosol of the host cells.75,80,107 Macrophages from caspase-11 knockout mice normally secrete IL-1β in response to ATP and monosodium urate, indicating that caspase-11 activates a non-canonical inflammasome pathway.107 Caspase-11 is critical for the host’s systemic lethal sepsis. Transgenic mice deficient of caspase-1, -11, and -12 are all resistant to LPS-induced shock.
NK activation also enhances the production of pro-inflammatory cytokines through the release of Gzms.89 Gzm A released in the extracellular space can re-enter and promote the activation of macrophages and production of IL-1β.86,108 Gzm B can cleave both human and mouse IL-1α.109 However, Gzm B itself does not cleave IL-1β or IL-18.109 Wild type mice or mice deficient of the Gzm B gene are resistant to viruses, but mice deficient of Gzm A are highly susceptible to poxviruses and herpes simplex virus infection.110
The different inflammasome platforms have redundant roles during infection and their activation can be modulated specifically by many bacterial and virus virulence factors.100 For example, Yersinia enterocolitica YopE, YopK, and YopT proteins inhibit caspase-1 activation and prevent secretion of mature IL-1β or IL-18 and pyroptosis.111 However, YopJ induces rapid macrophage cell death through regulation of the caspase-8, RIPK1, and RIPK3 signaling pathways.112 Pseudomonas aeruginosa, through the virulence factor called exoenzyme (ExoU) and phospholipase, can block inflammasome-mediated caspase-1 activation. Similarly, viruses can either accelerate or inhibit apoptosis cell death pathways. The cowpox virus protein cytokine response modifier A (CrmA) is one of many viral coding natural caspase inhibitors that acts as a pseudo substrate and inhibits caspase-1, -6, and -8.100 Vaccinia virus also expresses potent apoptosis inhibitors that block caspases as well as the BCL-2 protein family.113
Apoptosis and necroptosis cell death pathways are interconnected by the axis formed by pro-caspase-8, RIPK-1, and RIPK-3.63,100,114,115 The specific role of necroptosis in resolving bacterial infection is not well established, but its participation in the control of viral infection is well defined.72,97,116 RIP1 is the molecular target for certain viruses to impair the necroptosis cell death defense initiated by dsRNA through TLR3.81 The murine cytomegalovirus M45 protein interacts directly with RIPK-1 and RIPK-3 via RHIM to suppress cell death. HSV can also manipulate caspase-8-induced apoptosis and RIP3-induced necroptosis.116,117 HSV and many other pathogenic viruses suppress apoptotic response via the production of the anti-apoptotic viral proteins gD, gJ, Us3, and latency-associated transcript (LAT). Nonetheless many viruses use apoptotic caspase activity to facilitate their own proliferation.116
Pyroptosis and apoptosis pathways are linked via a direct interplay between the inflammatory and apoptotic caspases. GSDMD cleavage by caspase-1, -4/-5, and/or caspase-11 at D276 in a linker between the N-terminal and C-terminal domains of GSDMD is essential for the induction of pyroptosis.80 Two recent studies have shown that the cleavage site for caspase-3 and 7 in GSDMD is also located between the N-and C-terminal domains.118,119 In these studies, the authors used DPP8 and DPP9 (DPP8/9), two non-selective inhibitors of the S9 cytosolic serine peptides, and potent immune and anti-cancer agents in vivo.118 As shown in this research, treatment with DPP8/9 inhibitors increases the production of caspase-1 that cleaves GSDMD thereby inducing pyroptotic cell death in murine and human monocytes and macrophages.118,119 In addition, the authors showed that caspase-1 directly cleaves caspase-3 and -7 thus delaying the onset of apoptotic cell death. Notably, GSDMD molecules were cleaved at the Asp275 site by caspase-1, and at Asp87 by caspase-3 and-7, as recognized by the presence of p30 and p22 fragments, and a p43 fragment, respectively. Therefore, the authors proposed that p43 fragments promote the blockade of pyroptosis which is induced by caspase-1 activation, thereby preventing an inflammatory response.118,119
In accordance with these studies, Lamkanfi et al. have shown that the stimulation of macrophages with LPS plus nigericin or their infection with S. typhimurium and L. pneumophila can activate caspase-3 and -7.120 They had also previously demonstrated that caspase-3 and -7 induce apoptosis in macrophages deficient of the Gsdmd gene.81 Therefore, these two cleavage sites in the GSDMD protein are now considered checkpoints for controlling whether infected cells will undergo pyroptosis or apoptosis (Figure 2). Thus, it remains to be proven if these checkpoints ensure complete and efficient elimination of microbes and infected cells during pathological conditions.
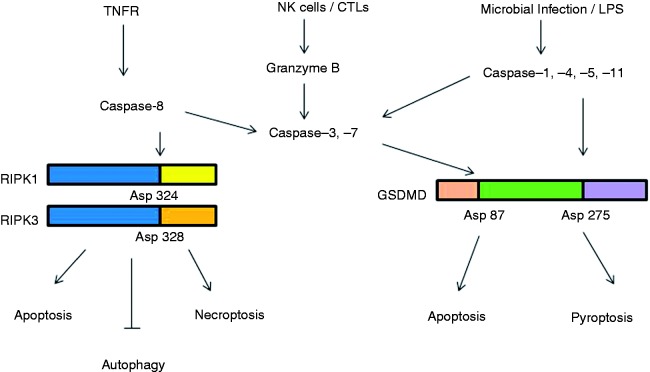
Figure 2.
Caspases-mediated cleavage and the switch in programmed cell death signaling pathways. In response to pathogen invasion, RIPK1 is activated either directly, by ligation of the TNF receptor, or indirectly, by ligation of TLRs (via TRIF) or IFN receptors. Cleavage of RIPK1 at Asp324 and RIPK3 at Asp328 by caspase-8 induces apoptosis. Inhibition of caspase-8 by chemical or viral inhibitors leads to the formation of RIPK-1/RIPK-3 complex (necrosome). RIP3 phosphorylates its substrate MLKL and this event drives MLKL oligomerization and translocation to the plasma membrane, leading to necroptosis. Silencing of RIPK1 under both normal and starvation conditions enhances autophagy. NK cells and cytotoxic lymphocytes (CTLs) deliver cytotoxic molecules, including perforin, granulysin and granzymes (Gzm), into the cytosol of infected cells. Gzm B mediates the cleavage and activation of caspase-3 and -7. Microbial pathogens and inflammatory agents or LPS via interaction cytosolic sensor proteins or with caspase -4, -5 and -11, can activate the canonical and non-canonical inflammasome pathways, respectively. Inflammasome complexes activate caspase-1, which promotes the cleavage and activation of IL-1β and IL-18. In parallel, caspase-1 cleaves the protein gasdermin-D (GSDMD) at Asp275 to induce pyroptosis cell death events. If GSDMD is cleaved at Asp87 by caspase-3 or -7 pyroptosis events are put on hold and cells switch to apoptosis.
Concluding remarks
In the last decade we have gained new insights into NK cell biology through definition of the roles of the multiple transcription factors responsible for surface membrane receptors and signaling pathways maturation, education, memory, exhaustion, and cytotoxicity activity. Many more studies are needed to gain a more complete understanding of how, when and which NK activating and inhibitor receptors act in the NK-mediated innate and long-lived (memory) adaptive responses to viral, bacterial, and protozoal antigens. This knowledge will help us to develop new strategies for immunization via vaccines.
NK cells trigger various cell death signaling pathways to kill pathogens and host infected cells through the release of cytokines, cytotoxic granules and inflammasome-induced caspase activation and cytokine synthesis (Figure 1). Apoptosis is a cell death program dependent on caspase-8 with low inflammatory response whereas necroptosis and pyroptosis are two pro-inflammatory cell death programs which are caspase-8-independent and caspase-1-dependent, respectively. Type of cell death might positively or negatively influence how the immune system cells will recognize and eliminate the intracellular pathogen during infection. Reciprocal interaction of inflammasome-dependent cell death and cytokines is likely to elicit protective inflammation and resolution of certain infections. In vitro and in vivo studies have revealed that many pathogens have evolved strategies to inhibit different types of cell death to stimulate their replication and cause diseases.
Caspases act as key regulators of multiple signaling nodes for cell death programs by apoptosis, necroptosis and pyroptosis. Cleavages of RIPK 1 and RIPK3 by caspase-8 works as a checkpoint control between apoptosis and necroptosis. Meanwhile cleavage of GSDMD by caspases -1, -11, -3, and -7 works as checkpoint control and decision point between the pyroptotic and apoptotic cell death pathways (Figure 2). On the other hand, inhibition of caspase-8 may direct cells towards autophagic cell death. Microbial factors may interfere in the regulation of these events in infected cells and this seems to contribute to remarkable differences in host cell death and the induction of different outcomes during one immune response.
Future studies aimed at gaining a further understanding of when and how NK induces host cell death and the killing of virus, bacteria, fungi and protozoan parasites may help us develop new strategies to increase or limit our body’s self-defense during overwhelming infection or sepsis.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: this work was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível (CAPES/PNPD proc 188-37/2011), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, proc 486048/2011 and 312206/2016-0), and Fundação de Amparo a Pesquisa do Estado de São Paulo (2014/12658-3, 2018/11053-1).
References
- 1.Sun JC, Lanier LL. Cutting edge: viral infection breaks NK cell tolerance to “missing self”. J Immunol 2008; 181: 7453–7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cella M, Miler H, Song C. Beyond NK cells: the expanding universe of innate lymphoid cells. Front Imunnol 2014; 5: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi FD, Ljunggren HG, La Cava A, et al. Organ-specific features of natural killer cells. Nat Rev Immunol 2011; 11: 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjorkstrom NK, Kekalainen K, Mjosberg J. Tissue-specific effector functions of innate lymphoid cells. Immunology 2013; 139: 416–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newman KC, Riley EM. Whatever turns you on: accessory-cell-dependent activation of NK cells by pathogens. Nat Rev Immunol 2007; 7: 279–291. [DOI] [PubMed] [Google Scholar]
- 6.Souza-Fonseca-Guimaraes F, Cavaillon JM, Adib-Conquy M. Bench-to-bedside review: natural killer cells in sepsis - guilty or not guilty? Crit Care 2013; 17: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garg AD, Martin S, Golab J, et al. Danger signalling during cancer cell death: origins, plasticity and regulation. Cell Death Differ 2014; 21: 26–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Höglund P, Brodin P. Current perspectives of natural killer cell education by MHC class I molecules. Nat Rev Immunol 2010; 10: 724–734. [DOI] [PubMed] [Google Scholar]
- 9.Shifrin N, Raulet DH, Ardolino M. NK cell self-tolerance, responsiveness and missing self-recognition. Semin Immunol 2014; 26: 138–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon SM, Chaix J, Rupp LJ, et al. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity 2012; 36: 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinette ML, Fuchs A, Cortez VS, et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol 2015; 16: 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morvan MG, Lanier LL. NK cells and cancer: you can teach innate cells new tricks. Nat Rev Cancer 2016; 16: 7–19. [DOI] [PubMed] [Google Scholar]
- 13.Björkström NK, Ljunggren HG, Sandberg JK. CD56 negative NK cells: origin, function, and role in chronic viral disease. Trends Immunol 2010; 31: 401–406. [DOI] [PubMed] [Google Scholar]
- 14.Horowitz A, Strauss-Albee DM, Leipold M, et al. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med 2013; 5: 208ra145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orr MT, Lanier LL. Natural killer cell education and tolerance. Cell 2010; 142: 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuertes-Marraco SAF, Neubert NJ, Verdeil G, et al. Inhibitory receptors beyond T cell exhaustion. Front Immunol 2015; 6: 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guillerey C, Huntington ND, Smyth M. Targeting natural killer cells in cancer immunotherapy. Nat Immunol 2016; 17: 1025–1036. [DOI] [PubMed] [Google Scholar]
- 18.Norris S, Coleman A, Kuri-Cervantes L, et al. PD-1 expression on natural killer cells and CD8(+) T cells during chronic HIV-1 infection. Viral Immunol 2012; 25: 329–332. [DOI] [PubMed] [Google Scholar]
- 19.Pesce S, Greppi M, Tabellini G, et al. Identification of a subset of human natural killer cells expressing high levels of programmed death 1: a phenotypic and functional characterization. J Allergy Clin Immunol 2017; 139: 335–346. [DOI] [PubMed] [Google Scholar]
- 20.Beldi-Ferchiou A, Lambert M, Dogniaux S, et al. PD-1 mediates functional exhaustion of activated NK cells in patients with Kaposi sarcoma. Oncotarget 2016; 7: 72961–72977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wherry J, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 2015; 15: 486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bi J, Tian Z. NK cell exhaustion. Front Immunol 2017; 8: 760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martín-Antonio B, Suñe G, Perez-Amill L, et al. Natural killer cells: angels and devils for immunotherapy. Int J Mol Sci 2017; 18: 868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez-Correa B, Morgado S, Gayoso I, et al. Human NK cells in acute myeloid leukaemia patients: analysis of NK cell-activating receptors and their ligands. Cancer Immunol Immunother 2011; 60: 1195–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuboi S, Sutoh M, Hatakeyama S, et al. A novel strategy for evasion of NK cell immunity by tumours expressing core2 O-glycans. EMBO J 2011; 30: 3173–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghiringhelli F, Ménard C, Terme M, et al. CD4+CD25+ regulatory T cells inhibit natural killer cell functions in a transforming growth factor-beta-dependent manner. J Exp Med 2005; 202: 1075–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goh CC, Roggerson KM, Lee HC, et al. Hepatitis C virus-induced myeloid-derived suppressor cells suppress NK cell IFN-γ production by altering cellular metabolism via arginase-1. J Immunol 2016; 196: 2283–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green DR, Droin N, Pinkoski M. Activation-induced cell death in T cells. Immunol Rev 2003; 193: 70–81. [DOI] [PubMed] [Google Scholar]
- 29.Jewett A, Man YG, Cacalano N, et al. Natural killer cells as effectors of selection and differentiation of stem cells: role in resolution of inflammation. J Immunotoxicol 2014; 11: 297–307. [DOI] [PubMed] [Google Scholar]
- 30.Kaur K, Nanut MP, Ko MW, et al. Natural killer cells target and differentiate cancer stem-like cells/undifferentiated tumors: strategies to optimize their growth and expansion for effective cancer immunotherapy. Curr Opin Immunol 2018; 51: 170–180. [DOI] [PubMed] [Google Scholar]
- 31.Ochoa MC, Minute L, Rodriguez I, et al. Antibody-dependent cell cytotoxicity: immunotherapy strategies enhancing effector NK cells. Immunol Cell Biol 2017; 95: 347–355. [DOI] [PubMed] [Google Scholar]
- 32.Seidel UJ, Schlegel P, Lang P. Natural killer cell mediated antibody-dependent cellular cytotoxicity in tumor immunotherapy with therapeutic antibodies. Front Immunol 2013; 4: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chung AW, Navis M, Isitman G, et al. Activation of NK cells by ADCC antibodies and HIV disease progression. J Acquir Immune Defic Syndr 2011; 58: 127–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reeves RK, Li H, Jost S, et al. Antigen-specific NK cell memory in rhesus macaques. Nat Immunol 2015; 16: 927–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gabrielli S, Ortolani C, Zotto GD, et al. The memories of NK cells: innate-adaptive immune intrinsic crosstalk. J Immunol Res. 2016: 1376595. [DOI] [PMC free article] [PubMed]
- 36.Sun JC, Madera S, Bezman NA, et al. Proinflammatory cytokine signaling required for the generation of natural killer cell memory. J Exp Med 2012; 209: 947–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beziat V, Liu LL, Malmberg JA, et al. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood 2013; 121: 2678–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Leary JG, Goodarzim M, Drayton DL, et al. T cell-and B cell-independent adaptive immunity mediated by natural killer cells. Nat Immunol 2006; 7: 507–516. [DOI] [PubMed] [Google Scholar]
- 39.O’Sullivan TE, Sun JC, Lanier LL. Natural killer cell memory. Immunity 2015; 43: 634–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arase H, Mocarski ES, Campbell AE, et al. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science 2002; 296: 1323–1326. [DOI] [PubMed] [Google Scholar]
- 41.Chalifour A, Jeannin P, Gauchat JF, et al. Direct bacterial protein PAMP recognition by human NK cells involves TLRs and triggers alpha-defensin production. Blood 2004; 104: 1778–1783. [DOI] [PubMed] [Google Scholar]
- 42.Davis DM, Dustin ML. What is the importance of the immunological synapse? Trends Immunol 2004; 25: 323–327. [DOI] [PubMed] [Google Scholar]
- 43.Voskoboinik I, Whisstock JC, Trapani JA. Perforin and granzymes: function, dysfunction and human pathology. Nat Rev Immunol 2015; 15: 388–400. [DOI] [PubMed] [Google Scholar]
- 44.Pipkin ME, Liberman J. Delivering the kiss of death: progress on understanding how perforin works. Curr Op Immunol 2007; 19: 301–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thiery J, Lieberman J. Perforin: a key pore-forming protein for immune control of viruses and cancer. Subcell Biochem 2014; 80: 197–220. [DOI] [PubMed] [Google Scholar]
- 46.Kagi D, Ledermann B, Burki K, et al. Cytotoxicity mediated by T cells and natural killer cells is greatly impaired in perforin-deficient mice. Nature 1994; 369: 31–37. [DOI] [PubMed] [Google Scholar]
- 47.Krzewski K, Coligan JE. Human NK cell lytic granules and regulation of their exocytosis. Front Immunol 2012; 3: 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chowdhury D, Lieberman J. Death by a thousand cuts: granzyme pathways of programmed cell death. Annu Rev Immunol 2008; 26: 389–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krensky AM, Clayberger C. Biology and clinical relevance of granulysin. Tissue Antigens 2009; 73: 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biron CA, Nguyen KB, Pien GC, et al. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol 1999; 17: 189–220. [DOI] [PubMed] [Google Scholar]
- 51.Balachandran S, Roberts PC, Kipperman T, et al. Alpha/beta interferons potentiate virus-induced apoptosis through activation of the FADD/caspase-8 death signaling pathway. J Virol 2000; 74: 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barber GN. Host defense, viruses and apoptosis. Cell Death Differ 2001; 8: 113–126. [DOI] [PubMed] [Google Scholar]
- 53.Sato Y, Tsurumi T. Genome guardian p53 and viral infections. Rev Med Virol 2012; 23: 213–220. [DOI] [PubMed] [Google Scholar]
- 54.Walch M, Dotiwala F, Mulik S, et al. Cytotoxic cells kill intracellular bacteria through granulysin-mediated delivery of granzymes. Cell 2014; 157: 1309–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dotiwala F, Mulik S, Polidoro RB. Killer lymphocytes use granulysin, perforin and granzyme to kill intracellular parasites. Nat Med 2016; 22: 210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dotiwala F, Santara SS, Binker-Cosen AA, et al. Granzyme B disrupts central metabolism and protein synthesis in bacteria to promote an immune cell death program. Cell 2017; 171: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martinvalet D, Dykxhoorn DM, Ferrini R, et al. Granzyme A cleaves a mitochondrial complex I protein to initiate caspase-independent cell death. Cell 2008; 133: 681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang LP, Lyu SC, Clayberger C, et al. Granulysin-mediated tumor rejection in transgenic mice. J Immunol 2007; 178: 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bär E, Whitney PG, Moor K, et al. IL-17 regulates systemic fungal immunity by controlling the functional competence of NK cells. Immunity 2014; 40: 117–127. [DOI] [PubMed] [Google Scholar]
- 60.Vitenshtein A, Charpak-Amikam Y, Yamin R, et al. NK cell recognition of candida glabrata through binding of NKp46 and NCR1 to fungal ligands Epa1, Epa6, and Epa7. Cell Host Microbe 2016; 20: 527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jorgensen I, Rayamajhi M, Miao EA. Programmed cell death as a defence against infection. Nat Rev Immunol 2017; 17: 151–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin SJ, Henry CM, Cullen SP. A perspective on mammalian caspases as positive and negative regulators of inflammation. Mol Cell 2012; 46: 387–397. [DOI] [PubMed] [Google Scholar]
- 63.Berghe TV, Linkermann, Jouan-Lanhouet S, et al. Regulated necrosis: the expanding network of non-apoptoic cell death pathways. Nat Rev 2014; 15: 135–147. [DOI] [PubMed] [Google Scholar]
- 64.Ohsumi Y. Historical landmarks of autophagy research. Cell Res 2014; 24: 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chan F K-M, Luz NF, Moriwaki K. Programmed necrosis in the cross talk of cell death and inflammation. Annu Rev Immunol 2015; 33: 79–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J 2004; 384: 201–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Belizario JE, Vieira-Cordeiro LA, Enns SC. Necrotic cell death signaling and execution pathway: lessons from knockout mice. Mediat Inflamm 2015; 2015: 128076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Belizário JE, Alves J, Occhiucci JM, et al. A mechanistic view of mitochondrial death decision pores. Braz J Med Biol Res 2007; 40: 1011–1024. [DOI] [PubMed] [Google Scholar]
- 69.Silke J, Richard JA, Gerlic M. The diverse role of RIP kinases in necroptosis and inflammation. Nat Immunol 2015; 16: 689–691. [DOI] [PubMed] [Google Scholar]
- 70.Murphy JM, Vince JE. Post-translational control of RIPK3 and MLKL mediated necroptotic cell death. F1000Research 2015, 4: 1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vercammen D, Beyaert R, Denecker G, et al. Inhibition of caspases increases the sensitivity of L929 cells to necrosis mediated by tumor necrosis factor. J Exp Med 1998; 187: 1477–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hitomi J, Chrostofferson DE, Ng A, et al. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell 2008; 135: 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dondelinger Y, Declercq W, Montessuit S, et al. MLKL compromises plasma membrane integrity by binding to phosphatidylinositol phosphates. Cell Rep 2014; 7: 971–981. [DOI] [PubMed] [Google Scholar]
- 74.Miao EA, Leaf IA, Treuting PM, et al. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol 2010; 11: 1136–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aachoui Y, Kajiwara Y, Leaf IA, et al. Canonical inflammasomes drive IFN-γ to prime caspase-11 in defense against a cytosol-invasive bacterium. Cell Host Microbe 2015; 8: 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kayagaki N, Stowe IB, Lee BL, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 2015; 526: 666–671. [DOI] [PubMed] [Google Scholar]
- 77.Nelms KA, Goodnow CC. Genome-wide ENU mutagenesis to reveal immune regulators. Immunity 2001; 15: 409–418. [DOI] [PubMed] [Google Scholar]
- 78.Kovacs SB, Miao EA. Gasdermins: effectors of pyroptosis. Trends Cell Biol 2017; 27: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shi J, Zhao Y, Wang Y, et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 2014; 514: 187–192. [DOI] [PubMed] [Google Scholar]
- 80.Shi J, Zhao Y, Wang K, et al. Cleavage of GSDMD by inflammatory caspase determines pyroptotic cell death. Nature 2015; 526: 660–665. [DOI] [PubMed] [Google Scholar]
- 81.He WT, Wan H, Hu L, et al. Gasdermin D is an executor of pyroptosis and required for interleukin-1beta secretion. Cell Res 2015; 25: 1285–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu X, Zhang Z, Ruan J, et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 2016; 535: 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aglietti RA, Dueber EC. Recent insights into the molecular mechanisms underlying pyroptosis and gasdermin family functions. Trends Immunol 2017; 38: 261–271. [DOI] [PubMed] [Google Scholar]
- 84.Wang Y, Gao W, Shi X, et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature 2017; 547: 99–105. [DOI] [PubMed] [Google Scholar]
- 85.Jorgensen I, Miao EA. Pyroptotic cell death defends against intracellular pathogens. Immunol Rev 2015; 265: 130–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wensink AC, Hack CE, Bovenschen N. Granzymes regulate proinflammatory cytokine responses. J Immunol 2015; 194: 491–497. [DOI] [PubMed] [Google Scholar]
- 87.Simon MM, Hausmann M, Tran T, et al. In vitro- and ex vivo-derived cytolytic leukocytes from granzyme A × B double knockout mice are defective in granule-mediated apoptosis but not lysis of target cells. J Exp Med 1997; 186: 1781–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pardo J, Bosque A, Brehm R, et al. Apoptotic pathways are selectively activated by granzyme A and/or granzyme B in CTL-mediated target cell lysis. J Cell Biol 2004; 167: 457–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cullen SP, Brunet M, Martin SJ. Granzymes in cancer and immunity. Cell Death Differ 2010; 17: 616–623. [DOI] [PubMed] [Google Scholar]
- 90.Wei H-M, Lin L-C, Wang C-F, et al. Antimicrobial properties of an immunomodulator - 15 kDa human granulysin. PLoS One 2016; 11: e0156321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Anderson DH, Sawaya MR, Cascio D, et al. Granulysin crystal structure and a structure-derived lytic mechanism. J Mol Biol 2003; 325: 355–365. [DOI] [PubMed] [Google Scholar]
- 92.Saini RV, Wilson C, Finn MW, et al. Granulysin delivered by cytotoxic cells damages endoplasmic reticulum and activates caspase-7 in target cells. J Immunol 2011; 186: 3497–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kaspar AA, Okada S, Kumar J, et al. A distinct pathway of cell-mediated apoptosis initiated by granulysin. J Immunol 2001; 167: 350–356. [DOI] [PubMed] [Google Scholar]
- 94.Tewary P, Yang D, de la Rosa G, et al. Granulysin activates antigen-presenting cells through TLR4 and acts as an immune alarmin. Blood 2010; 116: 3465–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Poon IKH, Lucas CD, Rossi AG, et al. Apoptotic cell clearance: basic biology and therapeutic potential. Nat Rev Immunol 2014; 14: 166–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martin CJ, Peters KN, Behar SM. Macrophages clean up: efferocytosis and microbial control. Curr Opin Microbiol 2014; 17: 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Green DR, Oguin TH, Martinez J. The clearance of dying cells: table for two. Cell Death Differ 2016; 23: 915–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cho YS, Challa S, Moquin D, et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 2009; 137: 1112–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen X, He WT, Hu L, et al. Pyroptosis is driven by non-selective gasdermin-D pore and its morphology is different from MLKL channel-mediated necroptosis. Cell Res 2016; 26: 1007–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lamkanfi M, Dixit VM. Modulation of inflammasome pathways by bacterial and viral pathogens. J Immunol 2011; 187: 597–602. [DOI] [PubMed] [Google Scholar]
- 101.Sangiuliano B, Perez M, Moreira D, et al. Cell death-associated molecular-pattern molecules: inflammatory signaling and control. Mediat Inflamm 2014; 2014: 821043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Franchi L, Munoz-Planillo R, Reimer T, et al. Inflammasomes as microbial sensors. Eur J Immunol 2010; 40: 595–653. [DOI] [PubMed] [Google Scholar]
- 103.Rathinam VAK, Fitzgerald KA. Inflammasome complexes: emerging mechanisms and effector functions. Cell 2016; 165: 792–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Netea MG, Nold-Petry CA, Nold MF, et al. Differential requirement for the activation of the inflammasome for processing and release of IL-1beta in monocytes and macrophages. Blood 2009; 113: 2324–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Van de Veerdonk FL, Netea MG, Dinarello CA, et al. Inflammasome activation and IL-1β and IL-18 processing during infection. Trends Immunol 2011; 32: 110–106. [DOI] [PubMed] [Google Scholar]
- 106.Garlanda C, Dinarello CA, Mantovani A. The interleukin family: back to the future. Immunity 2013; 39: 1003–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kayagaki N, Warming S, Lamkanfi M. Non-canonical inflammasome activation targets caspase-11. Nature 2011; 479: 117–121. [DOI] [PubMed] [Google Scholar]
- 108.Metkar SS, Menaa C, Pardo J, et al. Human and mouse granzyme A induce a proinflammatory cytokine response. Immunity 2008; 29: 720–733. [DOI] [PubMed] [Google Scholar]
- 109.Afonina IA, Tynan GA, Logue SE, et al. Granzyme B-dependent proteolysis acts as a switch to enhance the proinflammatory activity of IL-1α. Mol Cell 2011; 44: 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pereira RA, Simon MM, Simmons A. Granzyme A, a noncytolytic component of CD8+ cell granules, restricts the spread of herpes simplex virus in the peripheral nervous systems of experimentally infected mice. J Virol 2000; 74: 1029–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bergsbaken T, Cookson BT. Macrophage activation redirects yersinia-infected host cell death from apoptosis to caspase-1-dependent pyroptosis. PLoS Pathog 2007; 3: e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Weng D, Marty-Roix R, Ganesan S, et al. Caspase-8 and RIP kinases regulate bacteria-induced innate immune responses and cell death. Proc Natl Acad Sci 2014; 111: 7391–7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Aoyagi M, Zhai D, Jin C, et al. Vaccinia virus N1L protein resembles a B cell lymphoma-2 (Bcl-2) family protein. Protein Sci 2007; 16: 118–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kang TB, Yang SH, Toth B, et al. Caspase-8 blocks kinase RIPK3-mediated activation of the NLRP3 inflammasome. Immunity 2013; 38: 27–40. [DOI] [PubMed] [Google Scholar]
- 115.Conos SA, Chen KW, De Nardo D, et al. Active MLKL triggers the NLRP3 inflammasome in a cell-intrinsic manner. Proc Natl Acad Sci 2013; 114: E961–E969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yu X, He S. The interplay between human herpes simplex virus infections and the apoptosis and necroptosis cell death pathways. Virol J 2016; 13: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Guo H, Omoto S, Harris PA, et al. Herpes simplex virus suppresses necroptosis in human cells. Cell Host Microbe 2015; 17: 243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Okondo MC, Johnson DC, Sridharan R, et al. DPP8 and DPP9 inhibition induces pro-caspase-1-dependent monocyte and macrophage pyroptosis. Nat Chem Biol 2017; 13: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Taabazuing CY, Okondo MC, Bachovcin DA. Pyroptosis and apoptosis pathways engage in bidirectional crosstalk in monocytes and macrophages. Cell Chem Biol 2017; 24: 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lamkanfi M, Kanneganti TD, Van Damme P, et al. Targeted peptide-centric proteomics reveals caspase-7 as a substrate of the caspase-1 inflammasomes. Mol Cell Proteomics 2008; 7: 2350–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]