Short abstract
Genetic factors play a role in periodontitis. Here we examined whether the risk haplotype of MHC class III region BAT1-NFKBIL1-LTA and lymphotoxin-α polymorphisms associate with salivary biomarkers of periodontal disease. A total of 455 individuals with detailed clinical and radiographic periodontal health data were included in the study. A 610 K genotyping chip and a Sequenom platform were used in genotyping analyses. Phospholipid transfer protein activity, concentrations of lymphotoxin-α, IL-8 and myeloperoxidase, and a cumulative risk score (combining Porphyromonas gingivalis, IL-1β and matrix metalloproteinase-8) were examined in saliva samples. Elevated IL-8 and myeloperoxidase concentrations and cumulative risk scores associated with advanced tooth loss, deepened periodontal pockets and signs of periodontal inflammation. In multiple logistic regression models adjusted for periodontal parameters and risk factors, myeloperoxidase concentration (odds ratio (OR); 1.37, P = 0.007) associated with increased odds for having the risk haplotype and lymphotoxin-α concentration with its genetic variants rs2857708, rs2009658 and rs2844482. In conclusion, salivary levels of IL-8, myeloperoxidase and cumulative risk scores associate with periodontal inflammation and tissue destruction, while those of myeloperoxidase and lymphotoxin-α associate with genetic factors as well.
Keywords: Periodontal disease, lymphotoxin-alpha, phospholipid transfer protein, haplotypes, major histocompatibility complex, interleukin-8, myeloperoxidase
Introduction
The human MHC region carries genes involved in infection, innate and adaptive immune response, and chronic inflammation, including periodontitis.1–3 Genome-wide association studies have given an estimate that one locus in the MHC region (6p21.3) may associate with ‘high periodontal pathogen colonization’ traits.4 The MHC class III region BAT1-NFKBIL1-LTA risk haplotype (AGCGAC) has proven to be a significant contributor to the risk of periodontitis and the strongest association with this risk haplotype was observed with bleeding on probing (BOP), deepened periodontal pockets of ≥ 6 mm and severe periodontitis.3 The telomeric end of the MHC class III region, which joins the HLA class I loci, carries genes of several inflammation-related proteins, e.g. heat shock proteins, tumor necrosis factors (TNF) and lymphotoxins.5 Lymphotoxins are mostly produced by lymphocytes and NK cells,6 and high serum concentrations of lymphotoxin-α (LTA, formerly known as TNF-β) have been associated with periodontal risk alleles of the LTA.3
Binding and neutralization of LPS represents an important mechanism against infection and is regulated by several proteins including antimicrobial peptides7 and plasma phospholipid transfer protein (PLTP).8,9 The initial response against bacteria is regulated by resident leukocytes, which are mainly neutrophils. IL-8 and LTA are pro-inflammatory cytokines that can induce neutrophil chemotaxis and myeloperoxidase (MPO) secretion, respectively.10,11 MPO is involved in the activation of matrix metalloproteinases (MMPs), especially MMP-8 and -9, and thus, degradation of soft and hard tissues around affected teeth.12,13
It would be a great step forward for physicians, dentists and even the public to detect periodontitis at an early stage. Several salivary proteins, including IL-8, MPO and MMPs, have been suggested as diagnostic biomarkers of periodontitis; however, their clinical use has been challenged by the multifactorial characteristics of the disease. Moreover, salivary levels of some biomarkers are genetically regulated.14 To overcome these limitations, a novel diagnostic approach, namely the cumulative risk score (CRS), which statistically combines the levels of three biomarkers (Porphyromonas gingivalis, IL-1β and MMP-8), was developed.15–17 Yet there is still a lack of data on the salivary levels of many proteins that may play roles in pathogenesis of periodontitis, thus being potentially useful as salivary biomarkers of the disease. For example, an increased plasma PLTP activity has been observed in periodontitis patients,18 whereas no data exist on its levels in saliva. Also, data on salivary LTA are scarce, including one study with a small number of individuals with chronic periodontitis.19
Genomic approaches allow researchers to characterize the behaviour of salivary biomarkers in individuals with different genetic backgrounds.20 It has been previously demonstrated that LTA gene polymorphisms may affect the expression of inflammatory biomarkers.21,22 In the present study, the aim was to test the diagnostic ability of salivary PLTP activity, LTA, IL-8 and MPO concentrations, and CRS as salivary markers of periodontitis and to examine whether there are independent associations between these markers and the MHC class III region BAT1-NFKBIL1-LTA risk haplotype or LTA gene polymorphisms.
Materials and methods
Study population
The Corogene study is a prospective cohort including consecutive patients with an indication to coronary artery angiography.23 Their clinical and radiographic oral health examinations were performed in a substudy, the Parogene, as described in detail elsewhere.3,24 All study participants signed an informed consent and the study was approved by the Helsinki University Central Hospital ethics committee. The present study population consists of the Parogene study participants based on the availability of both saliva samples and genetic data (n = 455, 89.6% of the Parogene cohort). Briefly, the clinical examinations were performed by two calibrated periodontists. Probing pocket depths (PPD) were recorded from six sites and BOP from four sites of each tooth. The extent of alveolar bone loss (ABL) was evaluated from digital panoramic radiographs by an oral radiologist and then registered according to the most severe bone loss in each dentate sextant. The study participants filled in a questionnaire about their dental hygiene and smoking habits. In the present study, the participants were grouped according to periodontal parameters as follows: BOP 0–25%, 26–50%, or 51–100%; presence of PPD < 4 mm, 4–5 mm, or ≥ 6 mm; and no ABL, ABL <1/3 or ≥1/3 of the root length.3,24 The present study includes 23 edentulous individuals.
Saliva collection
Prior to the oral examination, the study participants chewed a piece of paraffin for 5 min and at least 2 ml of stimulated whole saliva was collected. Saliva samples were stored at −70°C until further use. After thawing, the samples were centrifuged at 9.300 g for 5 min and the supernatants were used for the analyses. In the laboratory, the samples were analysed blindly.
Genotyping protocol and performing genetic risk profiles
A standard salt precipitation protocol was used to isolate genomic DNA from blood. A subpopulation with acute coronary symptoms (n = 169) was genotyped for single nucleotide polymorphisms (SNPs) with a 610 K genotyping chip (HumanHap 610-Quad SNP array Illumina, San Diego, CA, USA) at the Wellcome Trust Sanger Institute (Hinxton, Cambridge, United Kingdom). Information of the genotyping protocol, imputation, quality controls and constructing the risk haplotype is given in detail elsewhere.3 Briefly, we analysed 13,245 SNPs in the MHC region covering the area between positions 25749179 (rs932316) and 33473618 (rs211457). A total of 18 SNPs were found to associate with periodontal parameters in univariate analyses. These SNPs were selected for the replication analyses, where the rest of the patients in the Parogene population (n = 339) were genotyped with a Sequenom platform (iPlex MassARRAY, San Diego, CA, USA) at the Institute for Molecular Medicine (Helsinki, Finland). A risk haplotype associating with periodontal parameters was constructed from SNPs with a high linkage disequilibrium (r2 > 0.9). Six SNPs, rs11796, rs3130059, rs2239527, rs2071591, rs909253 and rs1041981, constituted a genetic risk profile for periodontitis, AGCGAC, respectively.3 These SNPs were located in BAT1 intron (rs11796 and rs3130059), BAT5’ untranslated region (rs2239527), NFKBIL1 intron (rs2071591) and LTA intron (rs909253) and coding exon (rs1041981). In the present study, the individuals were further divided as heterozygous or homozygous for the risk haplotype, or without the risk haplotype. In addition, genotype information of 291 patients was available to analyse the association between saliva biomarkers and genetic markers in the LTA gene, that were in > 0.7 linkage disequilibrium with the lead-SNP in the coding region of the LTA gene, rs1041981, included in the risk haplotype. The three selected SNPs were rs2857708 (LTA promoter), rs2009658 (LTA promoter) and rs2844482 (LTA intron).
Biomarker measurements from saliva
PLTP-facilitated phospholipid transfer activity was measured with a lipoprotein-independent assay as previously described.25 Briefly, the assay measures the transfer of radioactively labelled phosphatidylcholine from liposomes to acceptors, which are human high-density lipoprotein (HDL3) particles. Each series contained assays without sample (spontaneous transfer) and control plasma pool diluted in 1:10. After incubation, the reaction was stopped by adding 536 mM NaCl, 363 mM MnCl2 and 52 units of heparin. After 10 min incubation, vortexing and 10 min centrifugation (13.000 g), the radioactivity was counted in the supernatant.
LTA concentrations were detected by the magnetic bead flow cytometry (Luminex® xMAP™ technique, Luminex Corporation, Austin, TX, USA) with the commercially optimized assay (Bio-Plex Pro Human Cytokine Assay Panel, 171-B6020M TNF-β Set, Bio-Rad Laboratories Inc., Hercules, California, USA) according to the manufacturers’ instructions. The detection limit of the assay was 0.3 pg/ml and the mean assay precision of coefficient of variation (CV) was 4.0%.
IL-8 concentrations were detected by the conventional bead flow cytometry (Luminex® xMAP™ technique, Luminex Corporation, Austin, TX, USA) with the commercially optimized assay (MILLIPLEX® Map Kit, Human Cytokine/Chemokine panel MPXHCYTO-60K, Millipore, Billerica, MA, USA) according to the manufacturers’ instructions. The detection limit of the assay was 0.2 pg/ml for IL-8 and the mean assay precision of CV was 11.6%.
Salivary MPO concentrations were determined by a commercial ELISA kit (Immundiagnostik, Bensheim, Germany) according to the manufacturer’s instructions. The mean assay precision of CV for the MPO assay was 7.3% and the detection limit was 0.2 ng/ml.
The CRS groups of the present study population have been described previously.16 Briefly, the concentrations of IL-1β and MMP-8 were determined from the saliva supernatants and the quantity of P. gingivalis from the saliva pellets. IL-1β was determined by the flow cytometry-based Luminex technology using a commercially available kit (Milliplex Map Kit; MPXHCYTO-60k, Millipore, Billerica, MA, USA). MMP-8 concentrations were determined with a time-resolved immunofluorometric assay as published earlier.26 Quantitative real-time PCR was used to determine P. gingivalis concentrations using a method described previously.27 The bacterial levels were determined using standard curves deriving from serial dilutions of the bacterium reference strain (W50) DNA and P. gingivalis genome size was used in the calculations of the concentrations. For the CRS calculations, the concentrations of MMP-8, IL-1β and P. gingivalis were divided into tertiles, which were multiplied with each other and used to categorize the patients into groups with low (subscore 1 or 2), moderate (subscore 3, 4, 6 or 8) or high (subscore 9, 12, 18 or 27) risk of having periodontitis.15
Isolation and visualization of PLTP
Next, 1 ml of combined saliva supernatant from six apparently healthy volunteers was applied to a Heparin-Sepharose CL-6B-GE column (Sigma-Aldrich, St. Louis, MO, USA), which was equilibrated with 25 mM Tris-HCl, pH 7.4, containing 1 mM EDTA, at a flow-rate of 1.0 ml/min/fraction. After washing, the bound material was eluted with 0.5 M NaCl, followed by 1 M and 2 M NaCl in the equilibration buffer. The elution of PLTP was followed by the absorbance at 280 nm and the activity assay described above.25 The fractions containing PLTP activity were combined and applied to a 12.5% SDS-PAGE under either reducing or non-reducing conditions. The proteins were transferred to a nitrocellulose membrane and visualized using the monoclonal anti-PLTP Ab, Mab59,28 followed by an HRP-labelled goat-anti-mouse Ab.
Statistical analyses
The statistical analyses were performed with the statistical programme (SPSS version 21.0; IBM, Armonk, NY, USA). The LTA values under the detection limit were substituted with 0.01 pg/ml for the statistical analyses.29 The concentrations of salivary biomarkers were logarithmically transformed before the analysis, as they had a skewed distribution. ANOVA and t-tests were used when examining the association of single salivary biomarkers and CRS with the indicated periodontal parameters, number of teeth and the risk haplotype. Associations between the risk haplotype and single and combined salivary biomarkers were analysed by logistic regression models adjusted for number of teeth (model 1) and further for age, gender, smoking (ever/never) and diabetes (Model 2), and additionally for PPD (model 3), BOP (model 4), or ABL (model 5). Since five biomarkers were analysed, a P value below 0.01 was considered significant. Linear regression models adjusted for age were used to examine the association between salivary biomarkers, including CRS, and LTA-associated polymorphisms.
Results
The biomarker levels in saliva were above the detection limit except for 48.4% of the LTA concentrations and 1.8% of PLTP activity. Table 1 presents the mean biomarker concentrations and CRS values according to clinical (BOP%, PPD) and radiographic (ABL) periodontal parameters and the number of teeth. PLTP activity and LTA concentration did not differ according to any periodontal parameter. Elevated IL-8 concentrations were found in individuals with BOP >25%, deepened pockets and more teeth, while elevated MPO concentrations associated also with ABL, as did CRS.
Table 1.
Single and combined (CRS) salivary biomarkers in association with clinical and radiographic periodontal parameters and the number of teeth.
|
Saliva concentration, mean (SD)a |
CRS, n (%)b |
||||||
|---|---|---|---|---|---|---|---|
| PLTP (nmol/h/ml) | LTA (pg/ml) | IL-8 (pg/ml) | MPO (ng/ml) | I | II | III | |
| Edentulous | 595.8 (454.1) | 0.345 (0.605) | 1299 (1017) | 755 (980) | 14 (60.9) | 7 (30.4) | 2 (8.7) |
| BOP% | |||||||
| 0–25 | 363.4 (386.0) | 0.263 (0.335) | 1225 (1077) | 3563 (4940) | 40 (33.9) | 43 (36.4) | 35 (29.7) |
| 26–50 | 475.3 (456.2) | 0.262 (0.515) | 1520 (1316) | 4586 (6100) | 38 (21.5) | 74 (41.8) | 65 (36.7) |
| 50–100 | 481.0 (477.0) | 0.222 (0.588) | 1416 (1173) | 6363 (8035) | 18 (17.6) | 30 (29.4) | 54 (52.9) |
| P valuec | 0.081 | 0.369 | 0.006 | < 0.001 | < 0.001 | ||
| PPD | |||||||
| < 4mm | 429.6 (400.8) | 0.138 (0.242) | 957 (1131) | 2037 (3656) | 21 (52.5) | 14 (35.0) | 5 (12.5) |
| 4–5mm | 460.0 (461.7) | 0.287 (0.508) | 1403 (1258) | 3705 (4794) | 47 (28.0) | 67 (39.9) | 54 (32.1) |
| ≥ 6mm | 432.8 (439.1) | 0.244 (0.509) | 1503 (1179) | 6278 (7736) | 28 (14.8) | 66 (34.9) | 95 (50.3) |
| P valuec | 0.416 | 0.088 | 0.001 | < 0.001 | < 0.001 | ||
| ABL | |||||||
| No | 438.9 (420.7) | 0.189 (0.267) | 1420 (1125) | 3642 (4808) | 26 (27.4) | 41 (43.2) | 28 (29.5) |
| 1/3 | 484.5 (476.1) | 0.249 (0.497) | 1396 (1297) | 4382 (6132) | 47 (27.0) | 67 (28.5) | 60 (34.5) |
| ≥ 2/3 | 391.8 (411.0) | 0.305 (0.601) | 1418 (1166) | 6140 (7650) | 23 (18.0) | 39 (30.5) | 66 (51.6) |
| P valuec | 0.285 | 0.467 | 0.164 | < 0.001 | < 0.001 | ||
| Number of teeth | |||||||
| 1–10 | 356.0 (344.8) | 0.209 (0.318) | 1110 (1502) | 2381 (3823) | 27 (45.0) | 18 (30.0) | 15 (25.0) |
| 11–20 | 373.4 (393.4) | 0.280 (0.320) | 1300 (1112) | 4367 (5988) | 12 (16.9) | 28 (39.4) | 31 (43.7) |
| 21–32 | 482.3 (472.2) | 0.254 (0.554) | 1498 (1165) | 5356 (6876) | 57 (21.4) | 101 (38.0) | 108 (40.6) |
| P valuec | 0.049 | 0.151 | < 0.001 | < 0.001 | < 0.001 | ||
aLog, logarithmic transformation preceding the ANOVA test.
bThe Chi-square test used to produce the P values.
cP value for the whole population including the edentulous patients.
ABL: alveolar bone loss; BOP: bleeding on probing; CRS: cumulative risk score; LTA: lymphotoxin-α; MPO: myeloperoxidase; PPD: probing pocket depths; PLTP: phospholipid transfer protein.Significant P values are highlighted in bold. Analyses were performed in the Parogene population (n = 455) including patients with an indication to coronary artery angiography. All patients underwent a clinical and radiographic oral examination.
The characteristics of the population are presented according to the presence of the risk haplotype in Table 2. Individuals with the risk haplotype in either chromosome had fewer teeth. The means of single biomarker concentrations and CRS are presented in association with the dominant risk haplotype in Table 3. In dominant and recessive (data not shown) models of periodontal risk haplotype comparisons, none of the single biomarkers or CRS differed significantly between the genetic groups. This was further examined with multiple logistic regression models with the dominant risk haplotype as an independent variable (Table 4). Salivary MPO concentrations associated in all models (1–3) with increased odds for having the risk haplotype and reached a statistical significance (strongest in Model 3) with an OR of 1.372 (P = 0.007) per increase of logarithmically transformed unit. The association of CRS was of borderline significance with an OR of 1.793 (P = 0.052) (model 1) and OR of 1.831 (P = 0.054) (model 2) for the change from low to moderate/high risk of having periodontitis.
Table 2.
Characteristics of the population according to the presence of periodontitis risk haplotype.
| Whole population (n = 455) | Periodontitis risk haplotype in either chromosome (n = 387) | Other (n = 68) | ||
|---|---|---|---|---|
| Mean (SD) | P valuea | |||
| Age (yr) | 63.6 (9.2) | 63.5 (9.2) | 63.5 (8.6) | 0.996 |
| BMI (kg/m2) | 28.1 (5.1) | 28.1 (5.1) | 28.0 (4.8) | 0.793 |
| Number of teeth | 20.0 (8.8) | 19.5 (9.1) | 22.8 (6.2) | 0.001 |
| n (%) | P valueb | |||
| Sex (males) | 279 (61.3) | 236 (60.9) | 43 (63.2) | 0.597 |
| Ex or current smoker | 223 (49.0) | 193 (50.0) | 30 (44.1) | 0.559 |
| Dyslipidemia | 344 (75.6) | 295 (76.2) | 49 (72.0) | 0.495 |
| Diabetes | 106 (23.3) | 91 (23.5) | 15 (22.1) | 0.819 |
| Coronary artery disease | 317 (69.7) | 270 (70.0) | 47 (69.1) | 0.626 |
aT-Test for comparisons between the two groups.
bChi-square test. BMI, body mass index.Significant P values are highlighted in bold.
Analyses were performed in the Parogene population (n = 455) including patients with an indication to coronary artery angiography. The individuals were divided as being heterozygous for the periodontitis risk haplotype (dominant model, risk haplotype in either chromosome), or without the risk haplotype.3
Table 3.
Salivary biomarkers and CRS in association with risk haplotype.
| Periodontitis risk haplotype in either chromosome(n = 387) | Other(n = 68) | ||
|---|---|---|---|
|
Saliva concentration, mean (SD) |
P valuea | ||
| PLTP (nmol/h/ml) | 455.7 (443.7) | 420.2 (483.4) | 0.164 |
| LTA (pg/ml) | 0.261 (0.538) | 0.259 (0.334) | 0.401 |
| IL-8 (pg/ml) | 1409 (1182) | 1480 (1502) | 0.634 |
| MPO (ng/ml) | 4525 (6305) | 3894 (6237) | 0.240 |
| CRS, n (%) | P valueb | ||
| I | 91 (25.4) | 19 (30.6) | |
| II | 133 (37.2) | 21 (33.9) | 0.684 |
| III | 134 (37.4) | 22 (35.5) | |
aT-Test for comparisons between the two groups.
bChi-square test.
CRS: cumulative risk score; LTA: lymphotoxin-α; MPO: myeloperoxidase; PLTP: phospholipid transfer protein.Significant P values are highlighted in bold.
Analyses were performed in the Parogene population (n = 455) including patients with an indication to coronary artery angiography. The individuals were divided as being heterozygous for the periodontitis risk haplotype (dominant model, risk haplotype in either chromosome), or without the risk haplotype.3
Table 4.
Association between risk haplotype and salivary biomarkers.
|
OR (95% CI) for periodontitis risk haplotype, P value |
|||
|---|---|---|---|
| Model 1a | Model 2a | Model 3a | |
| PLTP (nmol/h/ml)b | 1.194 (0.950–1.502), 0.129 | 1.195 (0.949–1.505), 0.129 | 1.197 (0.948–1.512), 0.130 |
| LTA (pg/ml)b | 0.962 (0.880–1.050), 0.383 | 0.967 (0.884–1.058), 0.467 | 0.974 (0.890–1.067), 0.572 |
| IL-8 (pg/ml)b | 1.155 (0.878–1.521), 0.303 | 1.209 (0.907–1.611), 0.195 | 1.228 (0.919–1.641), 0.165 |
| MPO (ng/ml)b | 1.264 (1.028–1.556), 0.027 | 1.321 (1.063–1.641), 0.012 | 1.372 (1.092–1.723), 0.007 |
| CRSc | 1.793 (0.988–3.400), 0.052 | 1.831 (0.981–3.416), 0.054 | 1.807 (0.958–3.406), 0.068 |
Logistic regression model for periodontal risk haplotype in either chromosome (dominant model).
aModel 1, adjusted for number of teeth; model 2, adjusted additionally for age, gender, diabetes, smoking (never/ever); model 3, adjusted additionally for PPD.
bOR (95% CI) per unit increase of logarithmically transformed values.
cOR (95% CI) per change from low risk to moderate/high risk of having periodontitis (CRS II-III vs. I).
CI: confidence interval; CRS: cumulative risk score; LTA: lymphotoxin-α; MPO: myeloperoxidase; OR: odds ratio; PLTP: phospholipid transfer protein.
Significant P values are highlighted in bold. Analyses were performed in the Parogene population (n = 455) including patients with an indication to coronary artery angiography.
Supplemental Table 1 presents the association of the salivary concentration of LTA with four different LTA gene-associated SNPs controlled by age. LTA concentrations associated with the variants rs2857708, rs2009658 and rs2844482, but not with rs1041981, which is included in the risk haplotype. The allele frequencies are presented in Supplemental Table 2.
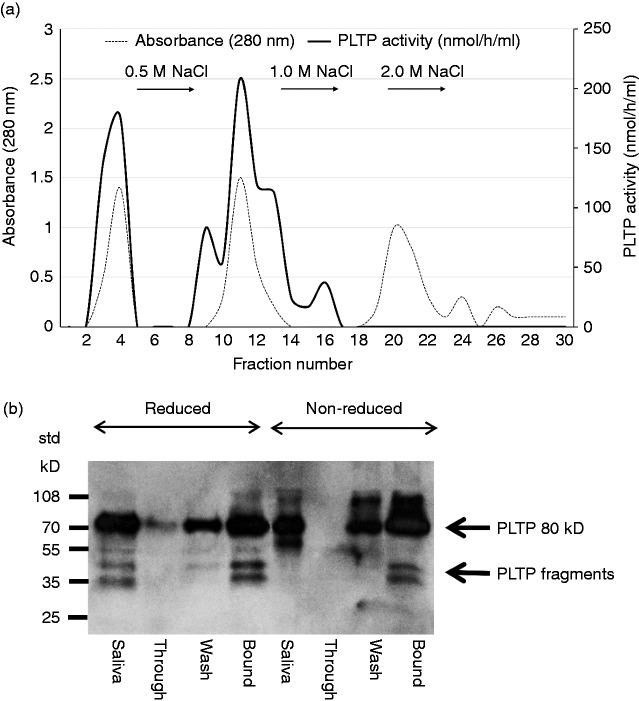
The elution profile of PLTP is presented in Figure 1a. From the total PLTP activity applied on the column, 68% was bound and could be eluted with 0.5 M NaCl. The Western blot of the fractions is presented in Figure 1b. The proteins visualized by the monoclonal anti-PLTP Ab revealed similar patterns as in the fresh saliva sample with the main band of 80 kD and some smaller-sized fragments. There were no obvious differences between the protein patterns detected in either under reducing or non-reducing conditions.
Figure 1.
Isolation of saliva plasma phospholipid transfer protein (PLTP) by heparin affinity column run with a HPLC technique. (a) Saliva was collected from six apparently healthy laboratory staff members and pooled. Then 1 ml of saliva was applied on Heparin-Sepharose CL-6B-GE column and the elution of PLTP was followed by absorbance (280 nm) and PLTP activity. Fractions 1-2, 3-4, and 11-13 were combined separately representing non-bound ‘through’, non-bound ‘wash’ and bound material, respectively. (b) Western blot of the combined fractions with PLTP activity after heparin affinity chromatography. The SDS-PAGE was run under reducing and non-reducing conditions. Volumes applied on the gel: saliva 5 µl, non-bound ‘through’ 10 µl, non-bound ‘wash’ 30 µl, and bound 40 µl. A representative elution curve and Western blot from three experiments using the same saliva pool is shown.
Discussion
To our knowledge, this is the first study to demonstrate independent associations between the MHC class III region BAT1-NFKBIL1-LTA risk haplotype and salivary MPO concentrations, as well as LTA gene polymorphisms and salivary concentrations of LTA. Moreover, salivary PLTP activity was also measured for the first time. Based on our results, elevated MPO concentrations and higher CRS scores associate significantly with clinical (bleeding on probing and deepened pockets) and radiographic (alveolar bone loss) indicators of periodontitis, whereas elevated IL-8 concentrations associate with clinical indicators only. Salivary PLTP activity was lowest in individuals with advanced tooth loss and LTA concentration in those with no deepened pockets (probing pocket depth < 4 mm). Although none of the single biomarkers or CRS differed between individuals with and without the risk haplotype in univariate analyses, in the multiple logistic regression models adjusted for periodontal parameters and risk factors, MPO concentration associated with increased odds for having the risk haplotype. The association of the same magnitude with CRS did not reach statistical significance. LTA concentration associated indirectly with the variants, rs2857708, rs2009658 and rs2844482, whereas other salivary biomarkers did not.
Microbial burden plays a key role in the initiation of periodontal inflammation when LPS of Gram-negative periodontal pathogens stimulates the immune response.30,31 According to current knowledge, PLTP has the ability to promote LPS disaggregation and its binding to HDL eliminates LPS from the body.8 Moreover, PLTP has other inflammation-independent roles, for example, in lipid metabolism.9 To the authors’ knowledge, PLTP activity is measured for the first time from saliva samples and its association with periodontal parameters analysed. The semi-isolation of saliva PLTP revealed that the largest portion of PLTP was in a catalytically active form.32 The observation that saliva PLTP activity was lowest in individuals with advanced tooth loss could suggest it associates with inflammation. However, no association was found with periodontal parameters, the risk haplotype, or LTA-related gene variants. This contrasts with studies where plasma PLTP activity was shown to be associated with periodontitis.18,33 The current levels of PLTP activity in saliva were 10 times lower than those previously measured from plasma samples.18 This difference in serum and saliva levels may reflect the importance of PLTP systemically and locally: it probably plays its main role elsewhere than in the oral cavity.
LTA is a pro-inflammatory cytokine involved in the recruitment of inflammatory cells as well as in alveolar bone loss in periodontal pathogenesis.34 Recently, it was shown that serum LTA concentrations associate with deepened periodontal pockets and the number of teeth,3 whereas LTA concentrations in saliva associated inversely with the percentage of periodontal pockets.19 Previous studies also suggest that polymorphism of LTA is connected to susceptibility of periodontitis1,35–37 and serum LTA concentrations.3 In patients with coronary artery disease, the LTA+496C variant was shown to associate significantly with alveolar bone loss.1 The present study demonstrates an association between reduced salivary LTA concentration and variants rs2857708, rs2009658 and rs2844482, which are in high linkage disequilibrium with each other and with the SNP in the LTA gene (rs1041981). Indeed, polymorphisms of the LTA-related variants seem to have an impact on LTA concentrations in saliva, although the SNP included in the genetic risk haplotype and located in the coding exon of the LTA gene (rs1041981) did not associate with the saliva concentrations. According to the results of this study, salivary LTA concentrations were lower in the absence of deepened pockets, which is in line with the previous literature. However, no associations between the risk haplotype, number of teeth, or other periodontal parameters and the salivary concentrations of LTA were found. This may be explained by relatively low concentrations of LTA in saliva and a considerable number of samples under the detection limit. In fact, the present LTA levels in saliva are approximately 1/100 compared to those measured previously from serum samples.3 For the suitability of LTA measurements in salivary diagnostics, the development of sensitive laboratory methods are required. Overall, however, the role of LTA in periodontitis warrants for further research.
IL-8 is an acute-phase protein and its profile is connected to chemotactic activity,11,38 thus differing from a typical profile of pro-inflammatory biomarkers. In the present study, IL-8 concentrations associated with bleeding on probing, probing pocket depth and the number of teeth, but not with alveolar bone loss, supporting its role at early phase of inflammation. No association with the risk haplotype or LTA-related SNPs was found. MPO is an oxidative activator of neutrophilic MMPs12 and LTA can induce the secretion of MPO.10 In the present study, elevated salivary MPO concentrations associated significantly with all periodontal parameters and the number of teeth. In further analyses taking into account both periodontitis and its risk factors, MPO associated significantly with increased odds for having the dominant risk haplotype. Because the adjustment for periodontal parameters strengthened the association, residual confounding may be present. Because no association between LTA-related SNPs and MPO was found, other components of the risk haplotype may be involved in MPO regulation. These findings enhance the usability of MPO as a potential biomarker of periodontitis in salivary diagnostics. Underlying mechanisms between MPO and the risk haplotype remain open, thus further research is needed to confirm this putative association. CRS, combining three salivary biomarkers (P. gingivalis, IL-1β and MMP-8), seems to be a promising tool for salivary diagnostics of periodontitis.17 In the present study, higher CRS values associated with increased bleeding on probing, probing pocket depths, alveolar bone loss and the number of teeth. In association analyses, a borderline significance for the change from the low CRS value to moderate/high values and having the risk haplotype was observed. There were no associations between the allele frequencies of the LTA-related SNPs and CRS.
Among the strengths of the present study are the relatively large sample size (n = 455) and verified background data on general and oral health of the study population. The population, however, consists of patients with symptomatic heart disease, including stable coronary artery disease, acute coronary syndrome, stable or atypical chest pains, valvular heart disease or cardiomyopathy. Thus, the results may not be fully applicable to a normal population. Also, the heart disease status may contribute to some salivary biomarker levels or the genotype distributions. A cross-sectional study design was a limitation, although the genetic information is not bound to any time point. Instead of long-term monitoring, the current study design made it possible to show the impact of genetics on the association between periodontal disease and levels of studied biomarkers.
As a conclusion, it can be stated that salivary biomarkers IL-8, MPO and CRS associate with periodontal inflammation and tissue destruction, whereas MPO and LTA associate independently with genetic factors. Further research is needed to increase understanding and find means to utilize genetic information in practice.
Supplemental Material
Supplemental material for Salivary biomarkers in association with periodontal parameters and the periodontitis risk haplotype by Joonas Liukkonen, Ulvi K Gürsoy, Eija Könönen, Mervi Gürsoy, Jari Metso, Aino Salminen, Elisa Kopra, Matti Jauhiainen, Päivi Mäntylä, Kåre Buhlin, Susanna Paju, Timo Sorsa, Markku S Nieminen, Marja-Liisa Lokki, Juha Sinisalo and Pirkko J Pussinen in Innate Immunity
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from the Academy of Finland (1266053 to PJP), the Sigrid Juselius Foundation (PJP and TYH2012209 and TKK2012005 to JS), the Finnish Medical Society's Swedish Funds (Einar and Karin Stroems Foundation to KB), the Finnish Dental Society Apollonia (JL and PJP), the Paulo foundation (PJP) and University of Turku joint research grant fund (JL).
References
- 1.Palikhe A, Lokki ML, Pussinen PJ, et al. Lymphotoxin alpha LTA+496C allele is a risk factor for periodontitis in patients with coronary artery disease. Tissue Antigens 2008; 71: 530–537. [DOI] [PubMed] [Google Scholar]
- 2.Folwaczny M, Henninger M, Glas J. Impact of MICA-TM, MICB-C1_2_A and C1_4_1 microsatellite polymorphisms on the susceptibility to chronic periodontitis in Germany. Tissue Antigens 2011; 77: 298–304. [DOI] [PubMed] [Google Scholar]
- 3.Kallio KA, Marchesani M, Vlachopoulou E, et al. Genetic variation on the BAT1-NFKBIL1-LTA region of major histocompatibility complex class III associates with periodontitis. Infect Immun 2014; 82: 1939–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Divaris K, Monda KL, North KE, et al. Genome-wide association study of periodontal pathogen colonization. J Dent Res 2012; 91: 21S–28S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yung Yu C, Yang Z, Blanchong CA, et al. The human and mouse MHC class III region: A parade of 21 genes at the centromeric segment. Immunol Today 2000; 21: 320–328. [DOI] [PubMed] [Google Scholar]
- 6.von Boehmer H. Lymphotoxins: From cytotoxicity to lymphoid organogenesis. Proc Natl Acad Sci USA 1997; 94: 8926–8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gürsoy UK, Könönen E. Understanding the roles of gingival beta-defensins. J Oral Microbiol 2012; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gautier T, Lagrost L. Plasma PLTP (phospholipid-transfer protein): An emerging role in 'reverse lipopolysaccharide transport' and innate immunity. Biochem Soc Trans 2011; 39: 984–988. [DOI] [PubMed] [Google Scholar]
- 9.Yu Y, Cui Y, Zhao Y, et al. The binding capability of plasma phospholipid transfer protein, but not HDL pool size, is critical to repress LPS induced inflammation. Sci Rep 2016; 6: 20845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richter J. Lymphotoxin induces secretion of granule proteins from adherent neutrophils: Possible role of intracellular free calcium. J Leukoc Biol 1990; 47: 506–513. [DOI] [PubMed] [Google Scholar]
- 11.Groeger SE, Meyle J. Epithelial barrier and oral bacterial infection. Periodontol 2000 2015; 69: 46–67. [DOI] [PubMed] [Google Scholar]
- 12.Spallarossa P, Garibaldi S, Barisione C, et al. 2008. Postprandial serum induces apoptosis in endothelial cells: Role of polymorphonuclear-derived myeloperoxidase and metalloproteinase-9 activity. Atherosclerosis 2008; 198: 458–467. [DOI] [PubMed] [Google Scholar]
- 13.Hernández M, Dutzan N, García-Sesnich J, et al. Host-pathogen interactions in progressive chronic periodontitis. J Dent Res 2011; 90: 1164–1170. [DOI] [PubMed] [Google Scholar]
- 14.Gürsoy UK, Könönen E. Editorial: Use of saliva in diagnosis of periodontitis: Cumulative use of bacterial and host-derived biomarkers. Front Cell Infect Microbiol 2016; 6: 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gürsoy UK, Könönen E, Pussinen PJ, et al. Use of host- and bacteria-derived salivary markers in detection of periodontitis: A cumulative approach. Dis Markers 2011; 30: 299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salminen A, Gürsoy UK, Paju S, et al. Salivary biomarkers of bacterial burden, inflammatory response, and tissue destruction in periodontitis. J Clin Periodontol 2014; 41: 442–450. [DOI] [PubMed] [Google Scholar]
- 17.Gürsoy UK, Pussinen PJ, Salomaa V, et al. Cumulative use of salivary markers with an adaptive design improves detection of periodontal disease over fixed biomarker thresholds. Acta Odontol Scand Epub ahead of print 2018: 1–4. [DOI] [PubMed] [Google Scholar]
- 18.Pussinen PJ, Jauhiainen M, Vilkuna-Rautiainen T, et al. Periodontitis decreases the antiatherogenic potency of high density lipoprotein. J Lipid Res 2004; 45: 139–147. [DOI] [PubMed] [Google Scholar]
- 19.Shyu KG, Choy CS, Wang DC, et al. Change of scaling-induced proinflammatory cytokine on the clinical efficacy of periodontitis treatment. Sci World J 2015; 2015: 289647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeidán-Chuliá F, Gürsoy M, Neves de Oliveira BH, et al. A systems biology approach to reveal putative host-derived biomarkers of periodontitis by network topology characterization of MMP-REDOX/NO and apoptosis integrated pathways. Front Cell Infect Microbiol 2016; 5: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asselbergs FW, Pai JK, Rexrode KM, et al. Effects of lymphotoxin-alpha gene and galectin-2 gene polymorphisms on inflammatory biomarkers, cellular adhesion molecules and risk of coronary heart disease. Clin Sci (Lond) 2007; 112: 291–298. [DOI] [PubMed] [Google Scholar]
- 22.Calmon-Hamaty F, Combe B, Hahne M, et al. Lymphotoxin α stimulates proliferation and pro-inflammatory cytokine secretion of rheumatoid arthritis synovial fibroblasts. Cytokine 2011; 53: 207–214. [DOI] [PubMed] [Google Scholar]
- 23.Vaara S, Nieminen MS, Lokki ML, et al. Cohort Profile: The Corogene study. Int J Epidemiol 2012; 41: 1265–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buhlin K, Mäntylä P, Paju S, et al. Periodontitis is associated with angiographically verified coronary artery disease. J Clin Periodontol 2011; 38: 1007–1014. [DOI] [PubMed] [Google Scholar]
- 25.Jauhiainen M, Ehnholm C. Determination of human plasma phospholipid transfer protein mass and activity. Methods 2005; 36: 97–101. [DOI] [PubMed] [Google Scholar]
- 26.Gürsoy UK, Könönen E, Pradhan-Palikhe P, et al. Salivary MMP-8, TIMP-1, and ICTP as markers of advanced periodontitis. J Clin Periodontol 2010; 37: 487–493. [DOI] [PubMed] [Google Scholar]
- 27.Hyvärinen K, Mäntylä P, Buhlin K, et al. A common periodontal pathogen has an adverse association with both acute and stable coronary artery disease. Atherosclerosis 2012; 223: 478–484. [DOI] [PubMed] [Google Scholar]
- 28.Huuskonen J, Jauhiainen M, Ehnholm C, et al. Biosynthesis and secretion of human plasma phospholipid transfer protein. J Lipid Res 1998; 39: 2021–2030. [PubMed] [Google Scholar]
- 29.Whitcomb BW, Schisterman EF. Assays with lower detection limits: Implications for epidemiological investigations. Paediatr Perinat Epidemiol 2008; 22: 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol 2010; 8: 481–490. [DOI] [PubMed] [Google Scholar]
- 31.Kato H, Tagushi Y, Tominaga K, et al. Porphyromonas gingivalis LPS inhibits osteoblastic differentiation and promotes pro-inflammatory cytokine production in human periodontal ligament stem cells. Arch Oral Biol 2014; 59: 167–175. [DOI] [PubMed] [Google Scholar]
- 32.Jänis MT, Siggins S, Tahvanainen E, et al. Active and low-active forms of serum phospholipid transfer protein in a normal Finnish population sample. J Lipid Res 2004; 45: 2303–2309. [DOI] [PubMed] [Google Scholar]
- 33.Vuletic S, Taylor BA, Tofler GH, et al. SAA and PLTP activity in plasma of periodontal patients before and after full-mouth tooth extraction. Oral Dis 2008; 14: 514–519. [DOI] [PubMed] [Google Scholar]
- 34.Assuma R, Oates T, Cochran D, et al. 1998. IL-1 and TNF antagonists inhibit the inflammatory response and bone loss in experimental periodontitis. J Immunol 1998; 160: 403–409. [PubMed]
- 35.Hollá LI, Fassmann A, Vask ů A, et al. Interactions of lymphotoxin alpha (TNF-beta), angiotensin-converting enzyme (ACE), and endothelin-1 (ET-1) gene polymorphisms in adult periodontitis. J Periodontol 2001; 72: 85–89. [DOI] [PubMed] [Google Scholar]
- 36.Fassmann A, Holla LI, Buckova D, et al. Polymorphisms in the +252(A/G) lymphotoxin-alpha and the -308(A/G) tumor necrosis factor-alpha genes and susceptibility to chronic periodontitis in a Czech population. J Periodontal Res 2003; 38: 394–399. [DOI] [PubMed] [Google Scholar]
- 37.Vasconcelos DF, da Silva MA, Marques MR, et al. Lymphotoxin-alpha gene polymorphism +252A/G (rs909253, A/G) is associated with susceptibility to chronic periodontitis: A pilot study. ISRN Dent 2012; 2012: 617245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ebersole JL, Dawson DR, 3rd, Morford LA, et al. Periodontal disease immunology: 'Double indemnity' in protecting the host. Periodontol 2000 2013; 62: 163–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material for Salivary biomarkers in association with periodontal parameters and the periodontitis risk haplotype by Joonas Liukkonen, Ulvi K Gürsoy, Eija Könönen, Mervi Gürsoy, Jari Metso, Aino Salminen, Elisa Kopra, Matti Jauhiainen, Päivi Mäntylä, Kåre Buhlin, Susanna Paju, Timo Sorsa, Markku S Nieminen, Marja-Liisa Lokki, Juha Sinisalo and Pirkko J Pussinen in Innate Immunity