Abstract
Oligodendrocytes differentiate from oligodendrocyte progenitor cells (OPCs) in response to distinct extracellular signals. This process requires changes in gene expression resulting from the interplay between transcription factors and epigenetic modulators. Extracellular signals include chemical and physical stimuli. This review focuses on the signaling mechanisms activated in oligodendrocyte progenitors in response to mechanical forces. Of particular interest is a better understanding on how these forces are transduced into the OPC nuclei and subsequently reshape their epigenetic landscape. Here we will introduce the concept of epigenetic regulation of gene expression, first in general and then focusing on the oligodendrocyte lineage. We will then review the current literature on mechano-transduction in distinct cell types, followed by pathways identified in myelinating oligodendrocytes and their progenitors. Overall, the reader will be provided with a comprehensive review of the signaling pathways which allow oligodendrocyte progenitors to “sense” physical forces and transduce them into patterns of gene expression.
Keywords: brain, cytoskeleton, epigenetics, glia, myelin
1 ∣. INTRODUCTION
Myelination in the central nervous system (CNS) is carried out by oligodendrocytes, which are derived from oligodendrocyte progenitor cells (OPCs), that migrate from germinal zones throughout the developing nervous system (Baumann & Pham-Dinh, 2001). The progression from undifferentiated progenitors to functionally mature oligodendrocytes mostly coincides with the postnatal developmental period, although myelination can also occur in the adult brain (Dawson, Polito, Levine, & Reynolds, 2003; Hughes, Kang, Fukaya, & Bergles, 2013). This process is tightly regulated by the interplay of intrinsic and extrinsic factors. Intracellular components include epigenetic modulators and transcription factors. Extracellular signals include hormones and growth factors, as well as physical forces, such as tension or compression and rigidity or elasticity. This review will briefly discuss the concept of epigenetic regulation of gene expression in the oligodendrocyte lineage and then focus on the molecular mechanisms that are responsible for transducing physical forces into a functional response. In order to improve legibility, this review will be focused on mechanotransduction and the reader is referred to previous reviews published in Glia (Copray, Huynh, Sher, Casaccia-Bonnefil, & Boddeke, 2009; Hernandez & Casaccia, 2015) and other journals (Emery, 2010; Emery & Lu, 2015; Liu, Moyon, Hernandez, & Casaccia, 2016) on the topic of epigenetic regulation of oligodendrocyte differentiation.
2 ∣. EPIGENETIC REGULATION OF GENE EXPRESSION
Epigenetics refers to all the mechanisms modulating gene expression which do not involve changes in the DNA sequence. They include post-translational modification of nucleosomal histones, DNA methylation, chromatin remodeling and microRNAs (miRNA). Nucleosomes are the basic unit of chromatin and are comprised of 147 base pairs of DNA wrapped around octamers of the core histone proteins H2A, H2B, H3, and H4. Chromatin can be detected in a dispersed “relaxed” state, which is transcriptionally active (euchromatin) and accessible to the transcriptional machinery, or in a compacted state (heterochromatin), which is transcriptionally inactive, as the tightly packed histones prevent DNA accessibility to transcription factors and RNA polymerases. While all cells contain the same genome, the epigenetic landscapes between different cell types and cells at different developmental stages result in the activation and repression of different sets of genes that are critical for determining cell identity and behavior. Gene expression is therefore modulated by the accessibility of specific gene loci to the transcriptional machinery. Understanding which genomic regions are characterized by heterochromatin and euchromatin within each cell, allows for the definition of cell-specific patterns of gene expression, which are ultimately mediated by epigenetic events. These epigenetic modifications are characteristic for each cell-type and each developmental stage. Intriguingly, in the oligodendrocyte lineage, progenitor nuclei are characterized by euchromatin while myelinated oligodendrocyte nuclei are mostly heterochromatic (Mori & Leblond, 1970; Shen, Liu, Li, Wolubah, & Casaccia-Bonnefil, 2008), thereby suggesting that differentiation of progenitors into oligodendrocytes requires epigenetic repression of gene expression, a concept which has been covered also in other reviews (Hernandez & Casaccia, 2015; Liu et al., 2016).
Transcriptional activation within euchromatin is possible because post-translational modifications of specific lysine and arginine residues in the tail of nucleosomal histones modify the protein-DNA association. For a more thorough review of these changes and the interplay between transcription factors and epigenetic modulators in the oligodendrocyte lineage we refer the reader to previous work published in Glia (Copray et al., 2009; Hernandez & Casaccia, 2015). Here we highlight two well-studied mechanisms of gene activation: (1) histone acetylation (Table 1) and (2) chromatin remodeling. Acetylation of histones is catalyzed by histone acetyltransferases (HATs/KATs), which utilize acetyl-CoA to transfer acetyl groups onto lysine residues of N-terminal histone tails. The acetylation of histone H3 lysine 9 (H3K9ac) marks active promoters and H3K27ac marks active enhancers (Creyghton et al., 2010; Guillemette et al., 2011; Karmidoya, Krebs, Oulad-Abdelghani, Kimura, & Tora, 2012; Pokholok et al., 2005; Tie et al., 2009; Wolffe & Pruss, 1996). Acetylated lysine residues are then bound by α-helical structural motifs known as “bromodomains”, which act to promote gene activation through the recruitment of transcription factors, coactivators and components of the transcription initiation complex.
TABLE 1.
Transcriptional consequences of histone lysine acetylation and methylation
| Modification | Function | |
|---|---|---|
| Acetylation | H3K9ac | Transcriptional activation (promoters) |
| H3K14ac | Transcriptional activation (promoters) | |
| H3K18ac | Transcriptional activation (promoters) | |
| H3K27ac | Transcriptional activation (enhancers) | |
| H3K36ac | Transcriptional activation (promoters) | |
| H4K5ac | Transcriptional activation (promoters) | |
| H4K8ac | Transcriptional activation (promoters) | |
| H4K12ac | Transcriptional activation (promoters) | |
| H4K16ac | Transcriptional activation (promoters) | |
| H4K91ac | Transcriptional activation (promoters) | |
| Methylation | H3K4me1 | Transcriptional activation (enhancers) |
| H3K4me3 | Transcriptional activation (promoters) | |
| H3K9me3 | Transcriptional repression | |
| H3K27me3 | Transcriptional repression (promoters) | |
| H3K36me3 | Transcriptional activation | |
| H3K79me3 | Transcriptional activation (promoters) | |
| H4K20me1 | Transcriptional activation | |
| H4K20me3 | Transcriptional repression |
Histone lysine acetylation at both promoter and enhancer regions is associated with transcriptional activation. Histone lysine methylation is typically associated with transcriptional repression, but in some cases can also be associated with transcriptional activation.
Repressive heterochromatin can be further distinguished between facultative heterochromatin—referring to focal regions of the genome that are repressed by deacetylation and trimethylation of H3K27—and can be reactivated (Trojer & Reinberg, 2007), and constitutive heterochromatin—referring to stably repressed genomic regions, often characterized by deacetylation and H3K9 trimethylation of histones and DNA methylation—identified in centromeres, telomeres, high copy number of DNA tandem repeats, and transposons but also includes the heterochromatic regions identified at the nuclear periphery. Epigenetic mechanisms of gene repression include deacetylation and methylation of specific lysine residues in nucleosomal histone tails (Table 1), DNA methylation and microRNA expression. Trimethylation of lysines 9 and 27 on histone H3 (H3K9me3, H3K27me3) is associated with heterochromatin formation and transcriptional repression (Barski et al., 2007; Boggs et al., 2002; Kim & Kim, 2012). Repressive histone methylation is catalyzed by a family of histone methyltransferases (HMTs) with high specificity for residues K9 or K27 on histone H3. These enzymes catalyze the transfer of methyl groups from S-Adenosylmethionine onto the specific lysine residue in the histone tails. Methylated lysine residues can be recognized by specific domains of proteins called “chromodomains” (Jacobs & Khorasanizadeh, 2002). Transcriptional repression is also mediated by DNA methylation. This involves the methylation of cytosine residues within CpG dinucleotide sequences by DNA methyltransferases (DNMTs) which precludes the recognition of transcription factor binding sites (if it occurs at the promoter region) or prevents the elongation of transcripts (if it occurs at the gene body region). In addition to methylation, gene repression can also be mediated by miRNAs, which are short non-coding RNAs that target complementary mRNA transcripts for degradation. Chromatin remodelers may induce epigenetic activation or repression of gene expression, using energy from ATP hydrolysis to facilitate access of the transcriptional machinery to chromatin (Ho & Crabtree, 2010). These complexes include several proteins with distinct functions, such as binding to acetylated (via bromodomains) or to methylated (via chromodomains) lysine residues in the nucleosomal histones and catalyzing the ATP-dependent repositioning or ejecting of histones, to favor transcriptional activation or repression.
2.1 ∣. Epigenetic regulation in the oligodendrocyte lineage
Histone modifications, DNA methylation, and chromatin remodeling all play critical roles in regulating OPC differentiation. As OPCs transition into myelinating oligodendrocytes, the previously described morphological changes of nuclear chromatin (Mori & Leblond, 1970; Shen et al., 2008) are induced by the accrual of repressive epigenetic events, which result in global changes in gene expression, including downregulation of genes regulating proliferation and transcriptional inhibition of differentiation (Hernandez & Casaccia, 2015; Liu et al., 2016). The downregulation of these gene categories precedes myelin gene expression (Swiss et al., 2011) and is reflected in the progressive formation of heterochromatin (Liu et al., 2012; Magri et al., 2014; Shen, Li, & Casaccia-Bonnefil, 2005; Wu et al., 2012).
The transcriptional repression that drives early OPC differentiation is dependent on the removal of activating acetylated histone marks, catalyzed by histone deacetylase (HDAC) proteins. Pharmacological HDAC inhibition and conditional knockout models have demonstrated that progressive deacetylation of histone H3 by class I HDACs is required to promote oligodendrocyte differentiation (Cunliffe & Casaccia-Bonnefil, 2006; He & Casaccia-Bonnefil, 2008; Liu, Muggironi, Marin-Husstege, & Casaccia-Bonnefil, 2003; Marin-Husstege, Muggironi, Liu, & Casaccia-Bonnefil, 2002; Shen et al., 2005; Ye et al., 2009). Ablation of Hdac1 and Hdac2 or impairment of their recruitment to the genome of oligodendrocyte progenitors led to activation of the Wnt signaling pathway, resulting in upregulation of differentiation inhibitors Id2/4 and repression of oligodendrocyte specific genes, such as Olig2 and Mbp (He & Casaccia-Bonnefil, 2008; Ye et al., 2009).
Increasing levels of H3K9me3 and H3K27me3 were detected in white matter tracts over the course of developmental myelination, with H3K27me3 marks already present at the OPC stage and H3K9me3 accumulating during the transition to differentiated oligodendrocytes (Liu et al., 2015). These data were in agreement with the finding that EZH2 (Enhancer of Zeste Homolog 2), the enzyme responsible for the deposition of the H3K27me3 mark, was identified as an important mediator of OPC specification in neural stem cells (NSC), where it likely served the role of repressing genes regulating pluripotency as well as neuronal and astrocytic genes (Sher, Boddeke, Olah, & Copray, 2012; Sher et al., 2008). The genomic deposition of H3K9me3 marks and of DNA methylation marks, in contrast, increased during the transition from progenitors into oligodendrocytes (Liu et al., 2015; Moyon et al., 2016) and further contributed to stable repression of genes related to alternative lineages and regulation of proliferation. Oligodendrocyte lineage-specific ablation of the DNA methyltransferase Dnmt1 in mice severely impaired developmental myelination and resulted in aberrant alternative splicing and loss of immature oligodendrocytes, due to activation of an endoplasmic reticulum (ER) stress response (Moyon et al., 2016).
While repressive modifications contribute to the establishment of lineage specific formation of constitutive heterochromatin, the progression toward a fully differentiated, myelinating phenotype also requires transcriptional activation of pro-differentiation and myelin genes. Transcriptional activation during OPC differentiation has been proposed to be mediated by the ATP-dependent SWI/SNF (switch/sucrose non-fermentable) chromatin remodeling complex containing the enzyme BRG1 (Brahma-related gene-1). However the functional consequence of Brg1 ablation depended on timing and differentiation stage and suggested the existence of other mechanisms regulating the late transition. A severe phenotype was detected when Brg1 was ablated during OPC specification as it prevented oligodendrocyte differentiation by impairing the recruitment of RNA polymerase II and OLIG2 to the genome (Yu et al., 2013). A much milder phenotype was detected when ablation of Brg1 occurred at the early stages of oligodendrocytes (Bischof, Weider, Kuspert, Nave, & Wegner, 2015). This evidence suggested that the chromatin remodeling complex containing BRG1 plays an important role in driving the initial differentiation of OPCs, but is not essential for terminal differentiation, thereby supporting the need for further research into the chromatin remodeling complexes modulating the later stages.
3 ∣. RESPONSE OF CELLS TO PHYSICAL STIMULI
During development, highly plastic stem cells need to decide their fate and differentiate into distinct cell types. This decision is in part dependent on the distinct mechanical properties of different tissues (Engler, Sen, Sweeney, & Discher, 2006). In other words, stem cell lineage commitment is modulated by the mechanical properties of their microenvironment. Mesenchymal stem cells (MSCs), for instance, can be directed toward distinct lineages, by varying the mechanical properties of the substrate on which they are cultured. MSCs grown on soft acrylamide gels with a stiffness similar to brain tissue (0.1–1 kPa) will exhibit neurogenic transcript profiles, while MSCs grown on gels with muscle-like stiffness (11 kPa) or bone-like stiffness (34 kPa) will display myogenic or osteogenic transcriptional profiles, respectively (Engler et al., 2006).
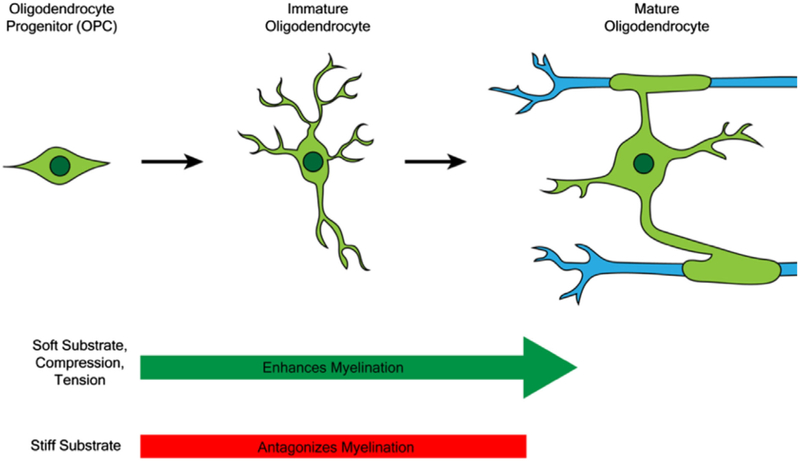
The specification and development of the diverse cell types within the CNS is also modulated by mechanostimuli. For instance, the decision of NSCs to differentiate into either neurons, oligodendrocytes, or astrocytes, is influenced by substrate stiffness (Georges, Miller, Meaney, Sawyer, & Janmey, 2006; Leipzig & Shoichet, 2009; Saha et al., 2008). When cultured on soft gels (0.1–0.5 kPa), NSCs were directed toward the neuronal lineage, while on slightly stiffer gels (1–10 kPa) they were directed toward glial lineages (Georges et al., 2006; Leipzig & Shoichet, 2009; Saha et al., 2008). OPC differentiation is also modulated by surface rigidity, as shown by their response to culture on polyacrylamide gels of varying stiffness and on glass surfaces. OPC migration and proliferation was enhanced by growth on soft substrates, in the range of 0.4–1 kPa (Jagielska et al., 2012). Their differentiation was potentiated on substrates with stiffness between 1.5 and 6.5 kPa, comparable to healthy brain tissue (Jagielska et al., 2012; Lourenço et al., 2016; Urbanski et al., 2016), further supporting OPCs as mechano-sensitive cells (Figure 1).
FIGURE 1.
Mechanical stimuli modulate oligodendrocyte differentiation and myelination. The growth of OPCs on soft substrates and the exposure of OPCs to tension and compression promote myelination in co-culture while growth of OPCs on stiff substrates antagonizes myelination
OPCs are also capable of functionally responding to spatial constraints, modeled in vitro through high density plating, co-culture with inert microspheres and direct mechano-stimulation (Hernandez et al., 2016; Rosenberg, Kelland, Tokar, De la Torre, & Chan, 2008). OPCs also responded to tensile strains, mimicked by laminin-coated elastomeric culture surfaces, which affected the proliferative and migratory properties of OPCs and induced their differentiation (Jagielska et al., 2017; Makhija et al., 2018) in a lineage-specific fashion, as the same tensile forces impaired the differentiation from neural stem cells (Arulmoli et al., 2015).
4 ∣. MECHANO-TRANSDUCTION IN THE OLIGODENDROCYTE LINEAGE
It is clear that while all cells are exposed to varying forms of mechanical cues, the intracellular response is dependent not only on the stimulus, but also on the cell type and stage of differentiation, which are characterized by distinctive epigenetic landscapes and gene expression patterns. Exposure of OPCs to mechanical strain and high density induced by microspheres, for instance, has been recently reported to modulate chromatin structure as it promoted the formation of heterochromatin and of the associated H3K9me3 histone marks (Hernandez et al., 2016). Significantly reduced fluctuations in nuclear shape (a characteristic feature of differentiated cells) were also reported in OPCs exposed to constant mechanical strain compared with unstrained cells (Makhija et al., 2018). Strain-induced OPC differentiation required histone deacetylation as pharmacological inhibitors of HDACs retained cells at an immature state (Jagielska et al., 2017). In addition to changes in chromatin structure through modulation of levels of epigenetic modifiers, mechano-stimulation has also been shown to modulate the expression levels of transcription factors in OPCs. RNA-sequencing of OPCs exposed to tensile strain, for instance, revealed upregulation of Tcf7l2, Sox10, Nkx2–2, Zfp191, and Olig2 (Jagielska et al., 2017). OPCs grown on soft substrates show cytoplasmic localization of OLIG1 (Urbanski et al., 2016), which is a characteristic feature of differentiated oligodendrocytes (Arnett et al., 2004). It remains to be explored whether distinct forms of mechano-stimulation (tension, compression, and substrate rigidity) differentially modulate the expression levels and subcellular localization of transcription factors, or they converge onto the same intracellular signal. Together these studies underscored the importance of mechanical stimuli in inducing both epigenetic and transcriptional changes. However, the molecular intermediates linking external force to nuclear transcriptional changes are only partially understood. Three potential molecular pathways have been suggested to modulate OPC differentiation and will be sequentially reviewed here: (1) the linker of nucleoskeleton and cytoskeleton (LINC), which bridges the cytoskeleton and chromatin; (2) the cytosolic transcriptional coactivators yes-associated protein (YAP) and WW domain containing transcription regulator 1 (TAZ/WWTR1) that shuttle into the nucleus in response to mechano-stimulation; and (3) nuclear actin.
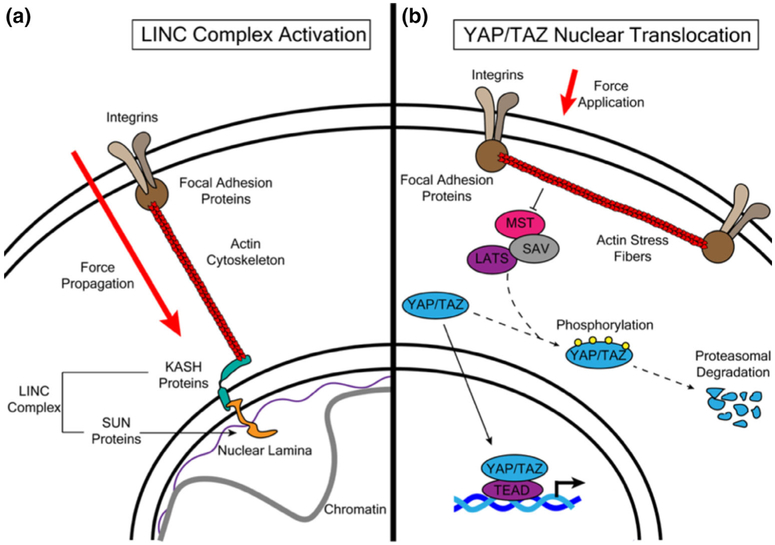
The protein complex called LINC acts as a direct physical connection between the plasma membrane and nuclear chromatin (Figure 2a). It is composed of spectrin repeat containing nuclear envelope (SYNE) proteins that bind to the cytoskeleton and Sad1 and UNC-84 (SUN) proteins that bind to nuclear lamins and chromatin fibers (Martins, Finan, Farshid, & Lee, 2012). The LINC complex has been demonstrated to be integral in the propagation of force from the cytoskeleton to the nucleus (Lombardi et al., 2011). Isolated cell nuclei have been shown to respond to tension and increase their stiffness through a mechanism involving LINC complex-dependent phosphorylation and activation of Emerin, an inner nuclear membrane protein, by Src family kinases (Guilluy et al., 2014). The same complex has also been shown to mediate global changes in gene expression in response to shifts in substrate rigidity (Alam et al., 2016). The levels of the LINC complex components SUN1 and SYNE4 in oligodendrocytes are responsive to tensile strain (Jagielska et al., 2017). In this lineage, the SYNE protein SYNE1 was shown to be required for heterochromatin formation in OPCs and for compression-induced enhancement of myelination in co-culture (Hernandez et al., 2016).
FIGURE 2.
The LINC complex and the YAP/TAZ pathway are canonical signaling pathways that transduce mechanical forces to the nucleus. The LINC complex interacts with the actin cytoskeleton (a). Forces applied to the cellular membrane are propagated through focal adhesions and actin fibers, activating the LINC complex and inducing changes in chromosomal structure and gene expression. In addition to LINC complex activation, force application also induces activation of the transcriptional co-activators YAP/TAZ and of the transcription factor TEAD (b). The application of physical forces induces actin stress fiber formation, which inhibits phosphorylation and subsequent degradation of YAP/TAZ. YAP/TAZ then translocate to the nucleus to bind TEAD and activate gene expression
Additional possible mediators of transcriptional changes in response to external forces are members of the YAP/TAZ family (Figure 2b) of mechano-responsive transcriptional coactivators that shuttle from the cytoplasm to the nucleus upon activation and interact with TEA domain (TEAD) transcription factors (Dupont et al., 2011; Low et al., 2014; Vassilev, Kaneko, Shu, Zhao, & Depamphilis, 2001). The biological effect of YAP/TAZ activation by mechanical cues is highly cell-type specific. For example, YAP activation in embryonic stem cells promotes self-renewal (Lian et al., 2010; Ohgushi, Minaguchi, & Sasai, 2015), while in MSCs and cardiac progenitors YAP alone (Zhong et al., 2013), TAZ alone (Kyung et al., 2014), or YAP or TAZ promote differentiation (Dupont et al., 2011; Mosqueira et al., 2014). In oligodendrocytes, YAP was shown to translocate to the nucleus in response to both tensile strain (Shimizu et al., 2016) and rigid substrates (Urbanski et al., 2016). YAP was also upregulated in OPCs exposed to tensile strain (Jagielska et al., 2017). Therefore YAP is clearly responsive to mechanical stimulation in OPCs, but the exact molecular mechanism through which it modulates oligodendroglial gene expression is still not well understood. It is likely that YAP and the LINC complex might exert their effects through overlapping mechanisms. YAP is seen to translocate from the cytoplasm to the nucleus in response to both tension and rigid substrates (Driscoll, Cosgrove, Heo, Shurden, & Mauck, 2015; Elosegui-Artola et al., 2017; Guilluy et al., 2014). Ablation of LINC complex components in both fibroblasts (Guilluy et al., 2014) and MSC (Driscoll et al., 2015) reduced nuclear localization of YAP/TAZ in response to stretch. Similarly, ablation of LINC complex components in fibroblasts inhibited YAP nuclear shuttling (Elosegui-Artola et al., 2017). Interestingly, it was demonstrated in fibroblasts that translocation of YAP into the nucleus was mediated by an increase in nuclear pore permeability in response to the mechanical force exerted by stiffer substrates (Elosegui-Artola et al., 2017).
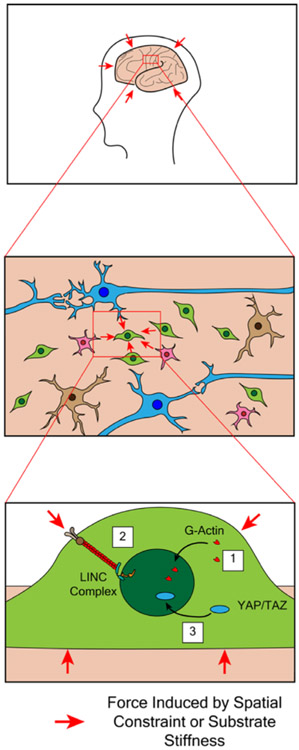
The transduction of mechanical signals from the plasma membrane to the nucleus is likely to be mediated through a combination of LINC complex activation and shuttling of YAP/TAZ between the cytoplasm and nucleus (Figure 3). The functional interaction between YAP/TAZ and the LINC complex has been well documented in other cell types (Driscoll et al., 2015; Elosegui-Artola et al., 2017; Guilluy et al., 2014), but this remains to be confirmed in OPCs in response to mechanical stimuli. In addition to YAP/TAZ and the LINC complex, a third and final proposed mechanism for the transduction of mechanical cues into the OPC nucleus is nuclear actin.
FIGURE 3.
Oligodendrocyte progenitors are exposed to mechanical signals during development. Development of the central nervous system is characterized by expansive growth within the spatial constraints of the skull, thereby exerting space-related sensing mechanisms over time. In addition, progenitors make contact with substrates of different stiffness, due to the distinct molecular composition of tissues. These mechanical stimuli are ultimately all sensed by OPCs and transduced into nuclear changes through actin remodeling (1), LINC complex activation (2), and YAP/TAZ activation (3)
5 ∣. NUCLEAR ACTIN AS AN EPIGENETIC MODULATOR OF GENE EXPRESSION
In the oligodendrocyte lineage, actin dynamics has been mostly studied in relation to the process of extension and wrapping of the oligodendrocyte membrane around the axons (Nawaz et al., 2015; Zuchero et al., 2015). Acto-myosin contractility mediated by non-muscle myosin II (NMII), in response to activated RhoA signaling (Lourenço & Grãos, 2016), has also been proposed to affect branching and myelination. Pharmacological inhibition of NMII increased oligodendrocyte branching complexity, membrane spreading and the number of myelinated segments in oligodendrocyte-dorsal root ganglion (DRG) neuron co-cultures (Kippert, Fitzner, Helenius, & Simons, 2009; Wang et al., 2012; Wang, Tewari, Einheber, Salzer, & Melendez-Vasquez, 2008). In addition, exposure of OPCs to mechanical stimuli (i.e., tension) was associated with upregulation of the two upstream activators of NMII RhoA and ROCK-II (Jagielska et al., 2017) and the in vivo ablation of this myosin removed the inhibitory effect of rigid substrates on branching complexity of oligodendrocytes (Urbanski et al., 2016).
Besides these roles of actin in the cytoplasm and membrane extensions, it is now well established that actin plays important functions in the cell nucleus. In HeLa cells, for instance, it has been shown that the subcellular localization of actin is regulated through active nuclear import (mediated by Importin 9) (Dopie, Skarp, Rajakylä, Tanhuanpää, & Vartiainen, 2012) and export (mediated by Exportin 6) (Stüven, Hartmann, & Görlich, 2003). In vitro studies have demonstrated that nuclear actin associates with all three RNA polymerases. Since RNA polymerase I is responsible for over 50% of the ribosomal RNA synthesis, RNA polymerase II is responsible for transcription of protein-encoding genes and microRNAs and RNA polymerase III is required for efficient transcription of tRNAs and small ribosomal subunit synthesis (Hofmann et al., 2004; Hu, Wu, & Hernandez, 2004; Kukalev, Nord, Palmberg, Bergman, & Percipalle, 2005; Philimonenko et al., 2004), we suggest that actin may play an important role in the communication between the cytoplasm and the nuclear expression of protein encoding genes as well as for components of ribosomal assembly. Nuclear actin has also been shown to complex with heterogeneous ribonucleoprotein U (hnRNP U), a protein that functions in pre-mRNA processing and transport (Kukalev et al., 2005). The actin-hnRNP U complex has been reported to interact with the lysine histone acetyltransferase KAT2B and disruption of the actin-hnRNP U complex has important functional consequence on the activity of this HAT, as it results in decreased levels of histone acetylation (Obrdlik et al., 2008). Nuclear actin also associates with and inhibits the activity of HDAC1 and HDAC2 (Serebryannyy, Cruz, & de Lanerolle, 2016), thereby supporting an overall function in regulation of histone acetylation levels. Finally, nuclear actin is required for the integrity and the activity of the BRG1-containing SWI/SNF chromatin remodeling complex (Kapoor & Shen, 2014; Wang et al., 1996; Zhao et al., 1998). Together, these studies in distinct cell types support a role for nuclear actin as regulator of histone acetylation and transcriptional activation.
In OPCs, localization of globular actin (G-actin) to the nucleus is induced by exposure of cells to spatial constraints, which also activates the LINC complex (Hernandez et al., 2016). The exact mechanism through which nuclear import occurs has not yet been investigated. Given that this effect is seen within 1 min of OPC stimulation, it could likely be consequent of the rapid depolymerization of filamentous actin (F-actin). The concept of rapid depolymerization of cytoskeletal F-actin in oligodendrocyte lineage cells has been previously reported within the context of process outgrowth and myelin wrapping (Liu et al., 2003; Nawaz et al., 2015; Zuchero et al., 2015). However depolymerization of F-actin in the cytoplasm may occur also in response to mechanical forces caused by physical constraints such as those induced by high cell density, or culturing in the presence of high-density microspheres (Hernandez et al., 2016; Rosenberg et al., 2008). Actin was shown to co-immunoprecipitate with both the LINC complex and the repressive H3K9me3 histone mark in protein extracts from white matter tracts (Hernandez et al., 2016). This was of particular interest, especially since ablation of actin in other cell types (i.e., fibroblasts) led to aberrant genome-wide localization of H3K9me3 (Xie et al., 2018). Therefore, these studies conducted in oligodendrocyte lineage cells suggest that nuclear actin in this glial lineage may play an important role in regulating the repressive histone marks that are necessary for heterochromatin formation and for OPC differentiation (Liu et al., 2015). In addition to establishing heterochromatin, it is worth exploring the role of nuclear actin in mediating gene activation in OPCs, as actin has been shown to associate with all three RNA polymerases (Hofmann et al., 2004; Hu et al., 2004; Kukalev et al., 2005; Philimonenko et al., 2004). Ablation of actin in fibroblasts results in a dramatic reduction of genome-wide BRG1 binding (Xie et al., 2018). Given the role of BRG1 in activating pro-differentiation genes during OPC differentiation (Yu et al., 2013), it is conceivable that actin might also regulate gene activation in the oligodendrocyte lineage through BRG1. This redistribution of globular actin to the nucleus may therefore be critical for mediating both epigenetic repression and activation of gene expression in response to mechanical cues during OPC differentiation.
6 ∣. CONCLUSION
Over the course of CNS development, cells are exposed to both chemical and physical signals. The latter include signals of spatial constraint, tension and substrate elasticity or stiffness. These biophysical signals, in tandem with soluble chemical factors, are transduced to the nucleus of undifferentiated progenitor cells and ultimately impact cell fate and function. Different regions of the brain and spinal cord, particularly the white vs. gray matter, have distinct mechanical properties (Budday et al., 2015; Christ et al., 2010; Johnson et al., 2013), which can also be impacted by the process of inflammation or injury and scar formation. These variations in tissue stiffness ultimately create diverse cellular microenvironments that affect how oligodendrocyte progenitors in those specific regions develop and behave. Additionally, the mechanical properties of brain tissue have been shown to be altered in multiple sclerosis (Makhija et al., 2018; Ruscielewicz et al., 2014; Streitberger et al., 2012; Wuerfel et al., 2010) and in response to injury (Moeendarbary et al., 2017). A better understanding of pathways governing the mechanobiology of OPC differentiation is therefore also likely to be essential for promoting remyelination.
In recent years, much work has been done to identify the molecular players that are responsible for mediating cellular responses to mechanical stimuli. This review highlights the effect that external mechanical signals have on nuclear events modulating gene expression during oligodendrocyte progenitor differentiation. Since gene expression is driven by the interplay of histone marks, DNA methylation and chromatin remodeling complexes, much work still remains to resolve the exact pathways bridging mechanostimulation and epigenetic changes in this lineage.
ACKNOWLEDGMENT
PC is funded by NIH-NINDS (R37-NS42925 and R01NS52738).
Funding information
NIH-NINDS, Grant/Award Numbers: R01NS52738, R37-NS42925
GLOSSARY
- Bromodomain
α-helical structural domain that recognizes and binds acetylated lysine residues on histones
- BRG1 (Brahma-Related Gene-1)
major catalytic subunit of the SWI/SNF complex with both helicase and ATPase activities
- Chromodomain
structural domain that recognizes and binds methylated lysine residues on histones
- CNS (central nervous system)
nerve tissues comprising of the brain and spinal cord
- CpG
region of DNA that contains a cytosine followed by a guanine and is targeted by DNA methyltransferases
- DNMT (DNA Methyltransferase)
enzyme that catalyzes the methylation of cytosine residues in CpG dinucleotide sequences in DNA
- DNMT1 (DNA Methyltransferase 1)
DNA methyltransferase that is responsible for maintaining DNA methylation patterns following DNA replication in proliferating cells
- Euchromatin
regions of chromatin containing active genes where DNA is accessible to transcription factors and RNA Polymerases
- EZH2 (Enhancer of Zeste Homolog 2)
histone methyltransferase that catalyzes the transfer of 3 methyl groups onto H3K27, mediating transcriptional repression
- H3K9ac
acetylation of lysine residue 9 in histone H3, marking transcriptional activation (Table 1)
- H3K27ac
acetylation of lysine residue 27 in histone H3, marking transcriptional activation (Table 1)
- H3K9me3
trimethylation of lysine residue 9 in histone H3, marking transcriptional repression (Table 1)
- H3K27me3
trimethylation of lysine residue 27 in histone H3, marking transcriptional repression (Table 1)
- HAT/KAT (Histone Acetyltransferase)
enzyme that catalyzes the transfer of acetyl groups from acetyl-CoA to lysine residues on N-terminal tails of histones
- HDAC (Histone Deacetylase)
enzyme that removes acetyl groups from histone lysine residues
- HDAC1/2 (Histone Deacetylase 1/2)
members of the class I histone deacetylases that remove acetyl groups from lysine residues on all 4 core histones, mediating transcriptional repression
- Heterochromatin
regions of chromatin containing repressed genes where DNA is tightly wound around the histones and is inaccessible to the transcriptional machinery
- HMT (Histone Methyltransferase)
enzyme that catalyzes the transfer of methyl groups from S-Adenosylmethionine to lysine residues on N-terminal tails of histones
- hnRNP U (Heterogeneous Ribonucleoprotein U)
RNA binding protein that associates with pre-mRNAs and are involved in pre-mRNA processing, metabolism, and transport
- ID2/4 (Inhibitors of DNA Binding 2/4)
transcriptional regulators that dimerize with and inhibit the function of basic helix–loop–helix transcription factors
- LINC (Linker of Nucleoskeleton and Cytoskeleton) complex
nuclear membrane protein complex that interacts with components of the both the cytoplasm (microtubules and actin filaments) and the nucleus (nuclear lamins and chromatin)
- KAT2B (LysineAcetyltransferase 2B)
acetyltransferase that catalyzes the acetylation of lysine residues on histones H3 and H4, mediating transcriptional activation
- MBP (Myelin Basic Protein)
membrane protein that is a major component of the myelin sheath
- miRNA (micro RNA)
short non-coding RNA sequence that targets mRNA transcripts for degradation
- MSC (Mesenchymal Stem Cell)
multipotent adult stem cells that can differentiate into a variety of cell types, including osteoblasts, myocytes, and neural cells
- NKX2-2 (NK2 Homeobox2)
homeobox domain-containing transcription factor that is required for oligodendrocyte progenitor differentiation
- NMII (Non-Muscle Myosin II)
motor protein that binds to and mediates the contraction of actin filaments, which is critical for processes such as cell adhesion, migration, and division
- NSC (Neural Stem Cell)
self-renewing multipotent cells that are capable of differentiating into neurons, astrocytes, and oligodendrocytes
- OLIG2 (Oligodendrocyte Transcription Factor 2)
basic helix–loop–helix transcription factor that is required for oligodendrocyte specification
- OPC (Oligodendrocyte Progenitor Cell)
proliferative precursor cells that derive from neural stem cells and are able to differentiate into oligodendrocytes
- RhoA (Ras Homolog Family Member A)
small GTPase that acts upstream of serine/threonine kinases that phosphorylate and activate non-muscle myosin II
- ROCK-II (Rho Associated Coiled-Coil Containing Protein Kinase 2)
serine/threonine kinase that phosphorylates and activates non-muscle myosin II
- SOX10 (Sex Determining Region Y Box10)
transcription factor that activates myelin genes during oligodendrocyte progenitor differentiation
- SUN (Sad1 and UNC-84) domain protein
component of the LINC complex that crosses the inner nuclear membrane and interacts with the nuclear lamina and chromatin
- SWI/SNF (Switch/Sucrose Non-Fermentable) chromatin remodeling complex
large multi-subunit complex that uses ATP hydrolysis to alter the positioning of nucleosomes
- SYNE (Spectrin Repeat Containing Nuclear Envelope) domain protein
component of the LINC complex that crosses the outer nuclear membrane and interacts with microtubule and actin filaments. Also referred to as KASH (Klarsicht, ANC-1, Syne Homology) domain proteins
- TAZ/WWTR1 (WW Domain Containing Transcription Regulator 1)
transcriptional coactivator that responds to mechanical stimuli by shuttling into the nucleus and complexing with TEAD transcription factors
- TCF7L2 (Transcription Factor 7Like 2)
transcription factor that is critical for oligodendrocyte progenitor differentiation
- TEAD transcription factors
transcription factors containing TEA/ATTS DNA binding domains that require interaction with transcriptional coactivators to activate transcription
- tRNA (Transfer RNA)
short RNA molecule that acts to transfer amino acids to the ribosome during translation
- YAP (Yes-Associated Protein)
transcriptional coactivator that responds to mechanical stimuli by shuttling into the nucleus and complexing with TEAD transcription factors
REFERENCES
- Alam SG, Zhang Q, Prasad N, Li Y, Chamala S, Kuchibhotla R, … Lele TP (2016). The mammalian LINC complex regulates genome transcriptional responses to substrate rigidity. Scientific Reports, 6, 38063 10.1038/srep38063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnett HA, Fancy SPJ, Alberta JA, Zhao C, Plant SR, Kaing S, … Stiles CD (2004). bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science, 306(5704), 2111–2116. 10.1126/science.1103709 [DOI] [PubMed] [Google Scholar]
- Arulmoli J, Pathak MM, McDonnell LP, Nourse JL, Tombola F, Earthman JC, & Flanagan LA (2015). Static stretch affects neural stem cell differentiation in an extra cellular matrix-dependent manner. Scientific Reports, 5, 8499 10.1038/srep08499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann N, & Pham-Dinh D (2001). Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiological Reviews, 81(2), 871–927. 10.1152/physrev.2001.81.2.871 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, … Zhao K (2007). High-resolution profiling of histone methylations in the human genome. Cell, 129(4), 823–837. 10.1016/j.cell.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, & Allis CD (2002). Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nature Genetics, 30(1), 73–76. 10.1038/ng787 [DOI] [PubMed] [Google Scholar]
- Bischof M, Weider M, Kuspert M, Nave KA, & Wegner M (2015). Brg1-dependent chromatin remodelling is not essentially required during oligodendroglial differentiation. Journal of Neuroscience, 35(1), 21–35. 10.1523/JNEUROSCI.1468-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budday S, Nay R, de Rooji R, Steinmann P, Wyrobek T, Ovaert TC, & Kuhl E (2015). Mechanical properties of gray and white matter brain tissue by indentation. Journal of the Mechanical Behavior of Biomedical Materials, 46, 318–330. 10.1016/j.jmbbm.2015.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ AF, Franze K, Gautier H, Moshayedi P, Fawcett J, Franklin RJM, … Guck J (2010). Mechanical difference between white and gray matter in the rat cerebellum measured by scanning force microscopy. Journal of Biomechanics, 43(15), 2986–2992. 10.1016/j.jbiomech.2010.07.002 [DOI] [PubMed] [Google Scholar]
- Copray S, Huynh JL, Sher F, Casaccia-Bonnefil P, & Boddeke E (2009). Epigenetic mechanisms facilitating oligodendrocyte development, maturation, and aging. Glia, 57(15), 1579–1587. 10.1002/glia.20881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, … Jaenisch R (2010). Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America, 107(50), 21931–21936. 10.1073/pnas.1016071107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe VT, & Casaccia-Bonnefil P (2006). Histone deacetylase 1 is essential for oligodendrocyte specification in the zebrafish CNS. Mechanisms of Development, 123(1), 24–30. 10.1016/j.mod.2005.10.005 [DOI] [PubMed] [Google Scholar]
- Dawson MR, Polito A, Levine JM, & Reynolds R (2003). NG2-expressing glial progenitor cells: An abundant and widespread population of cycling cells in the adult rat CNS. Molecular and Cellular Neurosciences, 24(2), 476–488. 10.1016/S1044-7431(03)00210-0 [DOI] [PubMed] [Google Scholar]
- Dopie J, Skarp K-P, Rajakylä EK, Tanhuanpää K, & Vartiainen MK (2012). Active maintenance of nuclear actin by importin 9 supports transcription. Proceedings of the National Academy of Sciences of the United States of America, 109(9), E544–E552. 10.1073/pnas.1118880109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll TP, Cosgrove BD, Heo S-J, Shurden ZE, & Mauck RL (2015). Cytoskeletal to nuclear strain transfer regulates YAP signaling in mesenchymal stem cells. Biophysical Journal, 108(12), 2783–2793. 10.1016/j.bpj.2015.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, … Piccolo S (2011). Role of YAP/TAZ in mechanotransduction. Nature, 474(7350), 179–183. 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- Elosegui-Artola A, Andreu I, Beedle AEM, Lezamiz A, Uroz M, Kosmalska AJ, … Roca-Cusachs P (2017). Force triggers YAP nuclear entry by regulating transport across nuclear pores article force triggers YAP nuclear entry by regulating transport across nuclear pores. Cell, 171(6), 1397–1410. 10.1016/j.cell.2017.10.008 [DOI] [PubMed] [Google Scholar]
- Emery B (2010). Transcriptional and post-transcriptional control of CNS myelination. Current Opinion in Neurobiology, 20, 601–607. 10.1016/j.conb.2010.05.005 [DOI] [PubMed] [Google Scholar]
- Emery B, & Lu QR (2015). Transcriptional and epigenetic regulation of oligodendrocyte development and myelination in the central nervous system. Cold Spring Harbor Perspectives in Biology, 7(9), a020461 10.1101/cshperspect.a020461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, & Discher DE (2006). Matrix elasticity directs stem cell lineage specification. Cell, 126(4), 677–689. 10.1016/j.cell.2006.06.044 [DOI] [PubMed] [Google Scholar]
- Georges PC, Miller WJ, Meaney DF, Sawyer ES, & Janmey PA (2006). Matrices with compliance comparable to that of brain tissue select neuronal over glial growth in mixed cortical cultures. Biophysical Journal, 90(8), 3012–3018. 10.1529/biophysj.105.073114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette B, Drogaris P, Lin HH, Armstrong H, Hiragami-Hamada K, Imhof A, … Festenstein RJ (2011). H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. PLoS Genetics, 7(3), e1001354 10.1371/journal.pgen.1001354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilluy C, Osborne LD, Van LL, Sharek L, Superfine R, Garcia-Mata R, & Burridge K (2014). Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nature Cell Biology, 16, 376–381. 10.1038/ncb2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, & Casaccia-Bonnefil P (2008). The Yin and Yang of YY1 in the nervous system. Journal of Neurochemistry, 106(4), 1493–1502. 10.1111/j.1471-4159.2008.05486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez M, & Casaccia P (2015). Interplay between transcriptional control and chromatin regulation in the oligodendrocyte lineage. Glia, 63(8), 1357–1375. 10.1002/glia.22818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez M, Patzig J, Mayoral SR, Costa KD, Chan JR, & Casaccia P (2016). Mechanostimulation promotes nuclear and epigenetic changes in oligodendrocytes. Journal of Neuroscience, 36(3), 806–813. 10.1523/JNEUROSCI.2873-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, & Crabtree GR (2010). Chromatin remodelling during development. Nature, 463(7280), 474–484. 10.1038/nature08911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann WA, Stojiljkovic L, Fuchsova B, Vargas GM, Mavrommatis E, Philimonenko V, … de Lanerolle P (2004). Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nature Cell Biology, 6(11), 1094–1101. 10.1038/ncb1182 [DOI] [PubMed] [Google Scholar]
- Hu P, Wu S, & Hernandez N (2004). A role for β-actin in RNA polymerase III transcription. Genes & Development, 18, 3010–3015. 10.1101/gad.1250804.an [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes EG, Kang SH, Fukaya M, & Bergles DE (2013). Oligodendrocyte progenitors balance growth with self-repulsion to achieve homeostasis in the adult brain. Nature Neuroscience, 16, 668–676. 10.1038/nn.3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, & Khorasanizadeh S (2002). Structure of HP1 chromodomain bound to a lysin 9-methylated histone H3 tail. Science, 295(5562), 2080–2083. 10.1126/science.1069473 [DOI] [PubMed] [Google Scholar]
- Jagielska A, Lowe AL, Makhija E, Wroblewska L, Guck J, Franklin RJM, … Van Vliet KJ (2017). Mechanical strain promotes oligodendrocyte differentiation by global changes of gene expression. Frontiers in Cellular Neuroscience, 11, 93 10.3389/fncel.2017.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagielska A, Norman AL, Whyte G, van Vliet KJ, Guck J, & Franklin RJM (2012). Mechanical environment modulates biological properties of oligodendrocyte progenitor cells. Stem Cells and Development, 21(16), 2905–2914. 10.1089/scd.2012.0189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CL, McGarry MDJ, Gharibans AA, Weaver JB, Paulsen KD, Wang H, … Georgiadis JG (2013). Local mechanical properties of white matter structures in the human brain. NeuroImage, 79(1), 145–152. 10.1016/j.neuroimage.2013.04.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor P, & Shen X (2014). Mechanisms of nuclear actin in chromatin-remodeling complexes. Trends in Cell Biology, 24(4), 238–246. 10.1016/j.tcb.2013.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmidoya K, Krebs AR, Oulad-Abdelghani M, Kimura H, & Tora L (2012). H3K9 and H3K14 acetylation co-occur at many gene regulator elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics, 13, 424 10.1186/1471-2164-13-424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, & Kim H (2012). Recruitment and biological consequences of histone modification of H3K27me3 and H3K9me3. ILAR Journal, 53, 232–239. 10.1093/ilar.53.3-4.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippert A, Fitzner D, Helenius J, & Simons M (2009). Actomyosin contractility controls cell surface area of oligodendrocytes. BMC Cell Biology, 10(1), 71 10.1186/1471-2121-10-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukalev A, Nord Y, Palmberg C, Bergman T, & Percipalle P (2005). Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nature Structural & Molecular Biology, 12(3), 238–244. 10.1038/nsmb904 [DOI] [PubMed] [Google Scholar]
- Kyung MK, Yoon JC, Hwang J, Kim AR, Cho HJ, Hwang ES, … Hong J (2014). Shear stress induced by an interstitial level of slow flow increases the osteogenic differentiation of mesenchymal stem cells through TAZ activation. PLoS One, 9, e92427 10.1371/journal.pone.0092427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipzig ND, & Shoichet MS (2009). The effect of substrate stiffness on adult neural stem cell behavior. Biomaterials, 30(36), 6867–6878. [DOI] [PubMed] [Google Scholar]
- Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, … Guan K (2010). The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes & Development, 24, 1106–1118. 10.1101/gad.1903310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Muggironi M, Marin-Husstege M, & Casaccia-Bonnefil P (2003). Oligodendrocyte process outgrowth in vitro is modulated by epigenetic regulation of cytoskeletal severing proteins. Glia, 44(3), 264–274. 10.1002/glia.10290 [DOI] [PubMed] [Google Scholar]
- Liu J, Dietz K, DeLoyht JM, Pedre X, Kelkar D, Kaur J, … Casaccia P (2012). Impaired adult myelination in the prefrontal cortex of socially isolated mice. Nature Neuroscience, 15(12), 1621–1623. 10.1038/nn.3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Magri L, Zhang F, Marsh NO, Albrecht S, Huynh JL, … Casaccia P (2015). Chromatin landscape defined by repressive histone methylation during oligodendrocyte differentiation. Journal of Neuroscience, 35(1), 352–365. 10.1523/JNEUROSCI.2606-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Moyon S, Hernandez M, & Casaccia P (2016). Epigenetic control of oligodendrocyte development: Adding new players to old keepers. Current Opinion in Neurobiology, 39, 133–138. 10.1016/j.conb.2016.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, & Lammerding J (2011). The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. Journal of Biological Chemistry, 286(30), 26743–26753. 10.1074/jbc.M111.233700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenço T, & Grãos M (2016). Modulation of oligodendrocyte differentiation by mechanotransduction. Frontiers in Cellular Neuroscience, 10, 277 10.3389/FNCEL.2016.00277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenço T, Paes de Faria J, Bippes CA, Maia J, Lopes-da-Silva JA, Relvas JB, & Grãos M (2016). Modulation of oligodendrocyte differentiation and maturation by combined biochemical and mechanical cues. Scientific Reports, 6, 21563 10.1038/srep21563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low BC, Pan CQ, Shivashankar GV, Bershadsky A, Sudol M, & Sheetz M (2014). YAP/TAZ as mechanosensors and mechanotransducers in regulating organ size and tumor growth. FEBS Letters, 588(16), 2663–2670. 10.1016/j.febslet.2014.04.012 [DOI] [PubMed] [Google Scholar]
- Magri L, Gacias M, Wu M, Swiss VA, Janssen WG, & Casaccia P (2014). C-Myc-dependent transcriptional regulation of cell cycle and nucleosomal histones during oligodendrocyte differentiation. Neuroscience, 276, 72–86. 10.1016/j.neuroscience.2014.01.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhija E, Jagielska A, Zhu L, Bost AC, Ong W, Chew SY, … Van Vliet KJ (2018). Mechanical strain alters cellular nuclear dynamics at early stages of oligodendrocyte differentiation. Frontiers in Cellular Neuroscience, 12, 59 10.3389/fncel.2018.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Husstege M, Muggironi M, Liu A, & Casaccia-Bonnefil P (2002). Histone deacetylase activity is necessary for oligodendrocyte lineage progression. Journal of Neuroscience, 22(23), 10333–10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins RP, Finan JD, Farshid G, & Lee DA (2012). Mechanical regulation of nuclear structure and function. Annual Review of Biomedical Engineering, 14(1), 431–455. 10.1146/annurev-bioeng-071910-124638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeendarbary E, Weber IP, Sheridan GK, Koser DE, Soleman S, Haenzi B, … Franze K (2017). The soft mechanical signature of glial scars in the central nervous system. Nature Communications, 8, 14787 10.1038/ncomms14787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, & Leblond CP (1970). Electron microscopic identification of three classes of oligodendrocytes and a preliminary study of their proliferative activity in the corpus callosum of young rats. Journal of Comparative Neurology, 139(1), 1–29. 10.1002/cne.901390102 [DOI] [PubMed] [Google Scholar]
- Mosqueira D, Pagliari S, Uto K, Ebara M, Romanazzo S, Escobedo-Lucea C, … Forte G (2014). Hippo pathway effectors control cardiac progenitor cell fate by acting as dynamic sensors of substrate mechanics and nanostructure. ACS Nano, 8(3), 2033–2047. 10.1021/nn4058984 [DOI] [PubMed] [Google Scholar]
- Moyon S, Huynh JL, Dutta D, Zhang F, Ma D, Yoo S, … Casaccia P (2016). Functional characterization of DNA methylation in the oligodendrocyte lineage. Cell Reports, 15(4), 748–760. 10.1016/j.celrep.2016.03.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz S, Sánchez P, Schmitt S, Snaidero N, Mitkovski M, Velte C, … Simons M (2015). Actin filament turnover drives leading edge growth during myelin sheath formation in the central nervous system. Developmental Cell, 34(2), 139–151. 10.1016/j.devcel.2015.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrdlik A, Kukalev A, Louvet E, Farrants A-KO, Caputo L, & Percipalle P (2008). The histone acetyltransferase PCAF associates with actin and hnRNP U for RNA polymerase II transcription. Molecular and Cellular Biology, 28(20), 6342–6357. 10.1128/MCB.00766-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgushi M, Minaguchi M, & Sasai Y (2015). Rho-signaling-directed YAP/TAZ activity underlies the long-term survival and expansion of human embryonic stem cells. Cell Stem Cell, 17, 448–461. 10.1016/j.stem.2015.07.009 [DOI] [PubMed] [Google Scholar]
- Philimonenko VV, Zhao J, Iben S, Dingová H, Kyselá K, Kahle M, … Grummt I (2004). Nuclear actin and myosin I are required for RNA polymerase I transcription. Nature Cell Biology, 6(12), 1165–1172. 10.1038/ncb1190 [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, … Young RA (2005). Genome-wide map of nucleosome acetylation and methylation in yeast. Cell, 122(4), 517–527. 10.1016/j.cell.2005.06.026 [DOI] [PubMed] [Google Scholar]
- Rosenberg SS, Kelland EE, Tokar E, De la Torre AR, & Chan JR (2008). The geometric and spatial constraints of the microenvironment induce oligodendrocyte differentiation. Proceedings of the National Academy of Sciences of the United States of America, 105(38), 14662–14667. 10.1073/pnas.0805640105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruscielewicz T, Nam J, Damanakis E, John GR, Raine CS, & Melendez-Vasquez CV (2014). Accelerated repair of demyelinated CNS lesions in the absence of non-muscle myosin IIB. Glia, 62(4), 580–591. 10.1002/glia.22627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha K, Keung AJ, Irwin EF, Li Y, Little L, Schaffer DV, & Healy KE (2008). Substrate modulus directs neural stem cell behavior. Biophysical Journal, 95(9), 4426–4438. 10.1529/biophysj.108.132217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serebryannyy LA, Cruz CM, & de Lanerolle P (2016). A role for nuclear actin in HDAC 1 and 2 regulation. Scientific Reports, 6, 28460 10.1038/srep28460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Li J, & Casaccia-Bonnefil P (2005). Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. Journal of Cell Biology, 169(4), 577–589. 10.1083/jcb.200412101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Liu A, Li J, Wolubah C, & Casaccia-Bonnefil P (2008). Epigenetic memory loss in aging oligodendrocytes in the corpus callosum. Neurobiology of Aging, 29(3), 452–463. 10.1016/j.neurobiolaging.2006.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher F, Boddeke E, Olah M, & Copray S (2012). Dynamic changes in Ezh2 gene occupancy underlie its involvement in neural stem cell self-renewal and differentiation towards oligodendrocytes. PLoS One, 7(7), e40399 10.1371/journal.pone.0040399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher F, Rößler R, Brouwer N, Balasubramaniyan V, Boddeke E, & Copray S (2008). Differentiation of neural stem cells into oligodendrocytes: Involvement of the Polycomb group protein Ezh2. Stem Cells, 26(11), 2875–2883. 10.1634/stemcells.2008-0121 [DOI] [PubMed] [Google Scholar]
- Shimizu T, Osanai Y, Tanaka KF, Abe M, Natsume R, Sakimura K, & Ikenaka K (2016). YAP functions as a mechanotransducer in oligodendrocyte morphogenesis and maturation. Glia, 65, 360–374. 10.1002/glia.23096 [DOI] [PubMed] [Google Scholar]
- Stüven T, Hartmann E, & Görlich D (2003). Exportin 6: A novel nuclear export receptor that is specific for profilin·actin complexes. The EMBO Journal, 22(21), 5928–5940. 10.1093/emboj/cdg565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streitberger KJ, Sack I, Krefting D, Pfuller C, Braun J, Paul F, & Wuerfel J (2012). Brain viscoelasticity alteration in chronic-preogressive multiple sclerosis. PLoS One, 7(1), e29888 10.1016/j.psyneuen.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiss VA, Ngyuen T, Dugas J, Ibrahim A, Barres B, Androulakis IP, & Casaccia P (2011). Identification of a gene regulatory network necessary for the initiation of oligodendrocyte differentiation. PLoS One, 6(4), e18088 10.1371/journal.pone.0018088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, … Harte PJ (2009). CBP-mediated acetylation of histone H3 lysine 27 antagonizes drosophila polycomb silencing. Development, 136(18), 3131–3141. 10.1242/dev.037127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P, & Reinberg D (2007). Facultative heterochromatin: Is there a distinctive molecular signature? Molecular Cell, 28(1), 1–13. 10.1016/j.molcel.2007.09.011 [DOI] [PubMed] [Google Scholar]
- Urbanski MM, Kingsbury L, Moussouros D, Kassim I, Mehjabeen S, Paknejad N, & Melendez-Vasquez CV (2016). Myelinating glia differentiation is regulated by extracellular matrix elasticity. Scientific Reports, 6, 33751 10.1038/srep33751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev A, Kaneko KJ, Shu H, Zhao Y, & Depamphilis ML (2001). TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/yes-associated protein localized in the cytoplasm. Genes & Development, 15(10), 1229–1241. 10.1101/gad.888601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Rusielewicz T, Tewari A, Leitman EM, Einheber S, & Melendez-Vasquez CV (2012). Myosin II is a negative regulator of oligodendrocyte morphological differentiation. Journal of Neuroscience Research, 90(8), 1547–1556. 10.1002/jnr.23036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Tewari A, Einheber S, Salzer JL, & Melendez-Vasquez CV (2008). Myosin II has distinct functions in PNS and CNS myelin sheath formation. Journal of Cell Biology, 182(6), 1171–1184. 10.1083/jcb.200802091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, & Crabtree GR (1996). Diversity and specialization of mammalian SWI / SNF complexes. Genes & Development, 4(17), 2117–2130. 10.1101/gad.10.17.2117 [DOI] [PubMed] [Google Scholar]
- Wolffe AP, & Pruss D (1996). Targeting chromatin disruption: Transcription regulators that acetylate histones. Cell, 84, 817–819. 10.1016/S0092-8674(00)81059-4 [DOI] [PubMed] [Google Scholar]
- Wu M, Hernandez M, Shen S, Sabo JK, Kelkar D, Wang J, … Cassacia P (2012). Differential modulation of the oligodendrocyte transcriptome by sonic hedgehog and bone morphogenetic protein 4 via opposing effects on histone acetylation. The Journal of Neuroscience, 32(19), 6651–6664. 10.1523/JNEUROSCI.4876-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuerfel J, Paul F, Beierbach B, Hamhaber U, Klatt D, Papzoglou S, … Sack I (2010). MR-elastography reveals degradation of tissue integrity in multiple sclerosis. NeuroImage, 49(3), 2520–2525. 10.1016/j.neuroimage.2009.06.018 [DOI] [PubMed] [Google Scholar]
- Xie X, Almuzzaini B, Drou N, Kremb S, Yousif A, Ostlund Farrants AK, … Percipalle P (2018). β-Actin-dependent global chromatin organization and gene expression programs control cellular identity. The FASEB Journal, 32(3), 1296–1314. 10.1096/fj.201700753R [DOI] [PubMed] [Google Scholar]
- Ye F, Chen Y, Hoang T, Montgomery RL, Zhao X, Bu H, … Lu QR (2009). HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nature Neuroscience, 12(7), 829–838. 10.1038/nn.2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X, … Lu QR (2013). Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell, 152(1–2), 248–261. 10.1016/j.cell.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo a., & Crabtree GR (1998). Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell, 95(5), 625–636. 10.1016/S0092-8674(00)81633-5 [DOI] [PubMed] [Google Scholar]
- Zhong W, Tian K, Zheng X, Li L, Zhang W, Wang S, & Qin J (2013). Mesenchymal stem cell and chondrocyte fates in a multishear microdevice are regulated by yes-associated protein. Stem Cells and Development, 22(14), 2083–2093. 10.1089/scd.2012.0685 [DOI] [PubMed] [Google Scholar]
- Zuchero JB, Fu MM, Sloan SA, Ibrahim A, Olson A, Zaremba A, … Barres BA (2015). CNS myelin wrapping is driven by actin disassembly. Developmental Cell, 34(2), 152–167. 10.1016/j.devcel.2015.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]