Abstract
Plants possess the ability to recognize microbe-associated molecular patterns (MAMPs) and PAMPs through the PRRs, and initiate pattern-triggered immunity. MAMPs are derived from cell-envelope components, secreted materials and cytosolic proteins from bacteria, oomycetes or fungi, and some MAMPs play a similar function in the innate immunity in mammals. Chitin is a representative fungal MAMP and triggers defense signaling in a wide range of plant species. The chitin receptors CEBiP and CERK1 on the plasma membrane have LysM (lysin motif) in their ectodomains. These molecules play an important role for the defense responses in rice and Arabidopsis, strictly recognizing the size and acetylated form of chitin oligosaccharides. However, related LysM receptors also play major roles for the signaling in root nodule and arbuscular mycorrhizal symbiosis. This review summarizes current knowledge on the molecular mechanisms of the defense and symbiosis signaling mediated by LysM receptors, including the activation steps of chitin-induced defense signaling downstream of LysM receptors.
Keywords: Plant immunity, chitin, LysM receptor, CEBiP, CERK1, signal transduction, rice, Arabidopsis, symbiosis
Introduction
Global food security is one of the most important problems not only at present, but also in the future, considering the inadequate food supply of > 800 million people today and the rapidly increasing world population, which is expected to exceed 9.7 billion in 2050.1 The fact that at least 10% of global food production is lost by plant disease is thus a serious problem for the future.1 To overcome such a threat of plant disease, it is important to understand the immune system of plants. Plants possess the ability to recognize microbe-associated molecular patterns (MAMPs) and PAMPs through the PRRs located on the plasma membrane and initiate pattern-triggered immunity (PTI) with various defense responses. This PTI is first line of plant defense against the invasion of pathogens and contributes to the basal resistance of plants. PTI is considered to play a similar role in the innate immunity of mammals.2
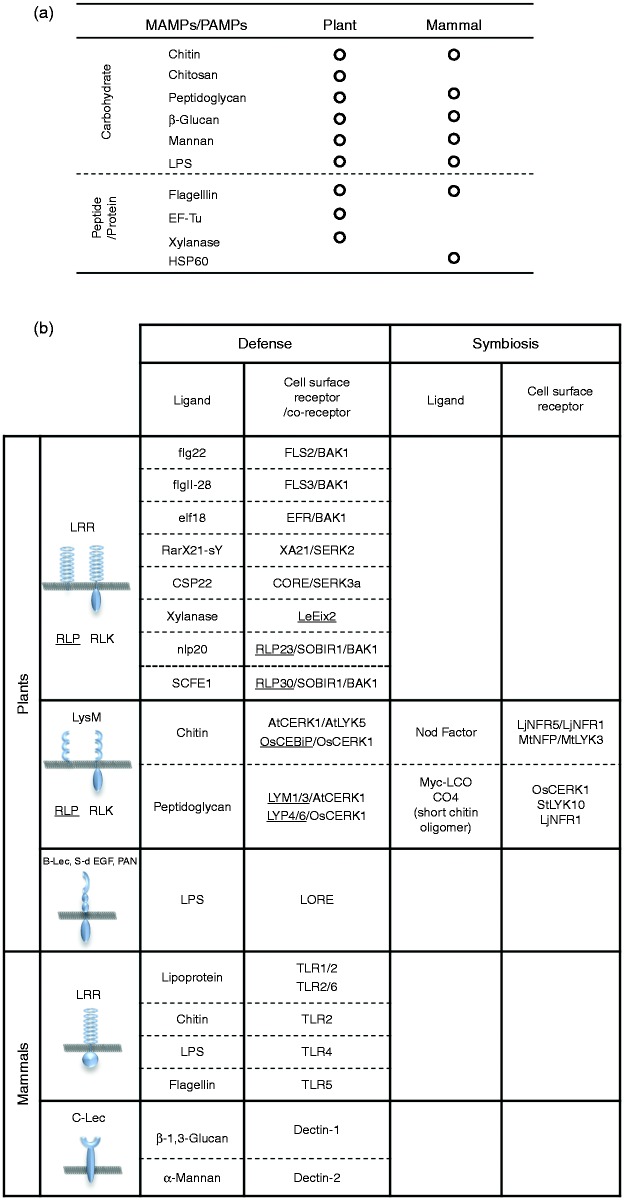
MAMPs are conserved molecules in a range of microorganisms, not present in higher plants and animals; they are often essential for the life of the microorganism they come from, suggesting that they cannot easily be abandoned or mutated. The properties of MAMPs have also made them useful for plants and animals to distinguish between self and non-self. MAMPs recognized by plants have been isolated from the cell envelopes, secreted materials and cytosolic protein of bacteria, fungi and oomycetes. Representative MAMPs include flagellin, translation elongation factor Tu, LPS, peptidoglycan (PGN), chitin, chitosan, lipopeptides and β-glucan.2,3 Interestingly, most of these MAMPs are also known to be recognized by the innate immune system of mammals (Figure 1a).
Figure 1.
MAMPs/PAMPs and their receptors/co-receptors of plants and mammals.2–4,12,13,26,57,58 (a) MAMPs/PAMPs recognized by innate immune systems in plants and mammals. (b) Motifs/domains in the surface receptors and corresponding ligands for defense/symbiosis signaling in plants and mammals. S-d EGF: S-domain protein/epidermal growth factor; PAN: plasminogen-apple-nematode; C-Lec: C-type lectin.
The structure of major cell-surface PRRs/co-receptors for MAMPs in plants and mammals are summarized in Figure 1b. The plant cell-surface PRRs can be classified into three major groups with their motifs/domains in the extracellular region of receptors. The first group of PRRs includes those molecules carrying leucine-rich repeat (LRR) domain in the extracellular region and consists of corresponding receptor-like proteins (RLP) and receptor-like kinases (RLK). Most PRRs of this group seem to recognize peptide/protein MAMP ligands. In mammals, TLRs are also well characterized LRR receptors in innate immunity (Figure 1b).
Interestingly, although bacterial flagellin is recognized by LRR-type receptors in both plants and mammals, the molecular machinery involved in the activation of downstream signaling is significantly different. Arabidopsis FLS2/BAK1 complex bound to flg22, a fragment of flagellin, directly activates downstream molecules through their kinase activities. However, TLRs do not have intrinsic enzymatic activity and the receptor complex requires the recruitment of a specific set of adaptor proteins and cytoplasmic kinases.4,5 Furthermore, the recognition site for flagellin is different between human and plants.6 In the latter case, it is known that even different plant species, rice and Arabidopsis recognize different epitopes.6,7
Lysin motif (LysM) is a widely distributed protein motif in prokaryotes and eukaryotes and also known to bind to peptidoglycan and chitin.8 Plant LysM-type PRRs contain two groups of molecules: GPI-anchored RLPs (LysM-RLPs) and LysM-RLKs (Figure 1b). LysM-PRRs were identified in the plasma membrane and plasmodesmata and serve for the defense signaling evoked by carbohydrate MAMPs (Figure 2a). LysM-containing proteins have also been reported to be involved in the innate immunity of some invertebrates, red swamp crayfish (Procambarus clarkia) and kuruma shrimp (Marsupenaeus japonicas), although the defense mechanism is still unclear.9,10
Figure 2.
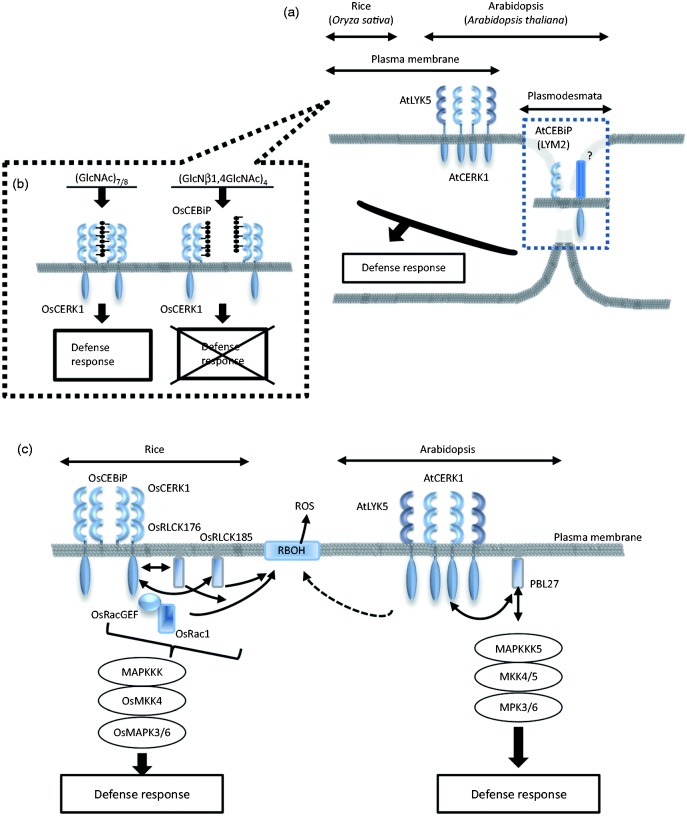
LysM receptor-mediated immune signaling in rice and Arabidopsis. (a) LysM receptors on the cell surface of rice and Arabidopsis. (b) Activation model of OsCEBiP-OsCERK1 complex by chitin heptamer or octamer [(GlcNAc)7/8]. [β-GlcN-(1→4)-GlcNAc]4 does not induce OsCEBiP dimerization and complex formation with OsCERK1. (c) Models of chitin signaling in rice and Arabidopsis.
Interestingly, in plants, structurally related LysM receptors are also required for the symbioses signaling for root nodule (RN) and arbuscular mycorrhizal (AM) formation (Figure 1b).
LOS-specific reduced elicitation (LORE), which was identified as an RLK essential for LPS signaling in Arabidopsis, represents another example of plant PRRs. LORE contains a bulb-type lectin (B-Lec) domain in the extracellular region, but it is not known whether LORE itself binds LPS.11 In mammals, Dectin1 and Dectin2, cell-surface receptors that bind carbohydrate ligands, β-(1→3)-linked glucan and α-mannan, respectively, also have a lectin domain (C-type) in the extracelluar region.12 However, LPS-induced immune responses in mammals is mediated by the sequential interaction of lipid binding protein, CD14 and TLR4/MD2 complex.5 Although knowledge of the plant receptors for LPS is still very limited, these results suggest that the mechanism of perception of MAMPs by PRRs is significantly different between plants and mammals (Figure 1b).
This review focuses on the recognition mechanism of chitin by plant LysM receptors and the activation of defense signaling downstream of these receptors. Current knowledge on the relationships between the defense and symbiosis signaling mediated by LysM receptors are also discussed.
Chitin recognition in rice and Arabidopsis
Chitin [β-(1→4)-linked polycondensate of N-acetylglucosamine] is a main cell-wall component of fungi and its fragments act as a MAMP, eliciting defense signaling in a wide range of plants.13 The chitin recognition system in rice comprises two types of LysM proteins: OsCEBiP and OsCERK1. OsCEBiP was identified as a plasma membrane protein that binds chitin oligosaccharides with a high affinity. Although OsCEBiP was expected to play a critical role in chitin signaling, it does not have a cytoplasmic domain.14 However, OsCERK1, an RLK essential for chitin signaling, does not bind to chitin, despite the presence of the LysM-containing ectodomain.13,15 These results suggest that OsCEBiP binds chitin oligosaccharides with the extracellular region and forms a complex with OsCERK1 to induce immune signaling. Concerning to the structural requirements of ligands, the N-acetyl groups of chitin oligosaccharides are essential to induce defense responses and chitosan oligosaccharides, which do not have N-acetyl groups and cannot induce any defense response in rice cells.13 Moreover, the length of chitin oligosaccharide is also important to induce the defense responses and the oligosaccharides larger than a hexamer show a very high activity.
Hayafune et al. analyzed chitin and OsCEBiP interaction in more detail.16 Deletion and domain-swapping experiments indicated that the middle of three LysMs of OsCEBiP is essential for the binding of chitin oligosaccharides. Epitope mapping by NMR spectroscopy indicated the preferential binding of longer oligosaccharides and the importance of N-acetyl groups for binding. Moreover, docking simulation and the analysis of dimerization using a recombinant protein and chitin-related oligosaccharides provided the basis for a unique sandwich-type chitin-binding model, in which OsCEBiP dimerizes through the binding of one chitin oligosaccharide from the opposite sides by two OsCEBiP molecules (Figure 2b). Recently, the crystal structure of OsCEBiP-chitin tetrasaccharide complex has been reported.15 In this paper, the authors also describe the importance of the middle LysM of OsCEBiP for chitin binding and propose a dimerization model, in which two OsCEBiP molecules also share a single chitin oligosaccharide and ‘slide’ along the oligosaccharide.
In Arabidopsis, AtCERK1 was identified as an essential LysM-RLK to activate chitin signaling.17 Several groups have indicated that AtCERK1 binds chitin directly and that AtCERK1 acts as an ‘all-in one’ receptor.13,18,19 It is known that three orthologs of OsCEBiP exist in the Arabidopsis genome. Two of them, LYM1 and LYM3, bind PGN and induce defense responses in the presence of AtCERK1, indicating that AtCERK1 mediates both chitin and PGN signaling, probably depending on the nature of the receptor complex (Figure 1b).20 Another homolog, AtCEBiP/LYM2, binds chitin oligosaccharides as similar to rice OsCEBiP but is not required for chitin signaling mediated by AtCERK1.19 However, AtCEBiP/LYM2 was shown to contribute to disease resistance, casting a question on its function in plant immunity.21,22 Faulkner et al. reported that AtCEBiP/AtLYM2 is localized in the plasmodesmata and contributes to chitin-induced changes of plasmodesmata flux, which might contribute to disease resistance (Figure 2a).21 Concerning the chitin receptor complex in Arabidopsis, Cao et al. later showed that another LysM-RLK, AtLYK5, binds chitin oligosaccharides with a higher affinity than AtCERK1.23 They proposed that AtLYK5 are the major chitin binding proteins in Arabidopsis and form a signaling complex with AtCERK1. Further studies would clarify the validity of these chitin receptor models and also provide further information on the chitin perception in the plasmodesmata. In addition to the LysM proteins described, two LRR-RLKs (LIK1 and IOS1) were reported to interact with AtCERK1 (as a CERK1-interacting RLKs), though the biological significance of such interactions remains to be clarified.24,25 However, chitosan is also known to induce defense responses in Arabidopsis through AtCERK1-independent pathway; the PRR(s) for chitosan signaling is(are) still unknown.26
Activation of chitin signaling through the autophosphorylation of CERK1
Autophosphorylation of CERK1 immediately after chitin perception is a critical event for the activation of downstream responses, but the function of each phosphorylation site is not well understood (Figure 2c). Petutschnig et al. previously identified three in vivo phosphorylation sites (S268, S274 and T519) of AtCERK1 after chitin treatment, although their function in chitin signaling was not evaluated.18 Recently, Suzuki et al. identified 41 Ser/The/Tyr and 15 Ser/Thr residues as in vitro and in vivo phosphorylation sites of AtCERK1, respectively.27 Evaluation of the function of these phosphorylation sites by site-directed mutagenesis indicated that the phosphorylation of three residues (T479, T573 and Y428) are essential for chitin signaling. Mutation of T479 by site-directed mutagenesis resulted in the loss of chitin responses. Also, an in vitro kinase assay showed that the T479A mutant almost completely lost auto/transphosphorylation activity, indicating that T479 contributes to chitin signaling through the regulation of the kinase activity. Threonine residues corresponding to T479 in the P + 1 loop of the activation segment, such as T455 of BAK1,28 are conserved in many RLKs and their phosphorylation seems to play an important role for the activation of these kinases, probably by connecting the activation segment and catalytic loop. Mutation of T573 in the C-terminal region was also shown to decrease the chitin responses and in vitro auto/transphosphorylation. As the C-terminal region of BRI1 was reported to negatively regulate their phosphorylation activity,29 it is possible that the phosphorylation of T573 has a similar function for the regulation of AtCERK1 kinase activity. Although the Y428F mutant completely lost chitin responsiveness, this mutation did not affect the in vitro kinase activity, indicating Y428 regulates chitin signaling independently of the regulation of kinase activity. Interestingly, the corresponding tyrosine residue, Y836 in EFR, a non-RD type RLK with very low kinase activity, was also shown to be critical for the elf18-induced response but is not involved in the regulation of kinase activity.30 The fact that the corresponding tyrosine residues are also conserved in other RLKs indicates that the phosphorylation of these residues plays an important role in the corresponding signaling pathway, too, though the phosphorylation status, as well as their possible function in these molecules, are not clear.
Signal transduction downstream of CERK1
Several signaling components immediately downstream of CERK1 have been identified. OsRLCK185 and OsRLCK176, which belong to RLCK subfamily VII, were shown to interact with OsCERK1 and contribute to chitin and PGN signaling in rice (Figure 2c).31,32 OsRLCK185/176-RNAi lines were shown to decrease the defense responses triggered by these MAMPs. These RLCKs might have overlapping functions downstream of OsCERK1. Recently, OsRLCK57, OSRLCK107 and OsRLCK118 were also found to contribute to chitin and PGN signaling in rice.33 In Arabidopsis, PBL27, an Arabidopsis ortholog of OsRLCK185, selectively regulates the defense responses downstream of AtCERK1, such as the activation of MAPK cascade, defense gene expression and callose deposition, but does not contribute to flg22 signaling.34 PBL27 was more preferably phosphorylated than BIK1 by CERK1, indicating the importance of the substrate specificity of RLKs for the determination of downstream components. Recently, PBL27 was shown to directly phosphorylate MAPKKK535 and activate the downstream MAPK cascade, MKK4/5 and MPK3/6, showing that PBL27 directly links the cell-surface RLK and MAPK cascade for triggering defense responses. In rice, OsRLCK185 was also reported to activate the downstream MAPK cascade through the phosphorylation of OsMAPKKK18 and OsMAPKKK24, indicating that the similar downstream signaling of CERK1 exists in Arabidopsis and rice.36,37 BIK1 is also a member of RLCK subfamily VII and regulated by FLS2/BAK1 complex.38,39 While Zhang et al. reported that BIK1 is involved in the regulation of AtCERK1-mediated chitin responses,39 Lu et al. reported that BIK1 phosphorylation was induced by flg22 but not by chitin.38 In addition, it was shown that OsRacGEF, a Rac/ROP family protein interacts with the kinase domain of OsCERK1 and is activated through the phosphorylation by OsCERK1.40 The phosphorylated OsRacGEF then activates a small GTPase, OsRac1, which regulates chitin-induced defense responses.
Symbiotic signaling triggered by LysM receptors
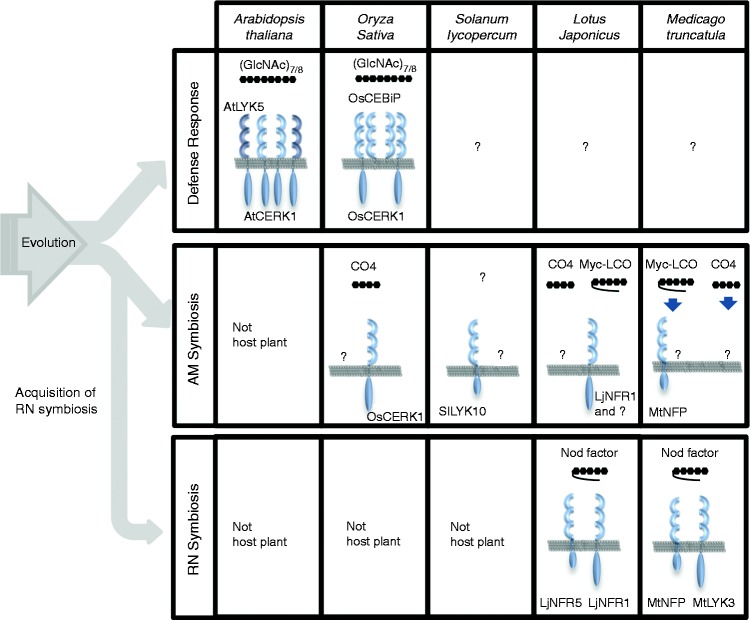
LysM-RLKs are known to play an essential role in inducing symbiosis, besides their function in innate immunity. LysM receptor pairs, such as NFR1 and NFR5 in Lotus japonicus and LYK3 and Nod Factor Perception (NFP) in Medicago truncatula are required for the formation of nitrogen-fixing nodules on the host roots (Figure 1b). Perception of the signaling molecules secreted by rhizobia, so called Nod factors (NF), by these receptor complexes is essential for inducing early responses of the root nodule (RN) symbiosis.41 NFs are lipo-chitooligosaccharides (LCOs), which are structurally related to chitin oligosaccharides for triggering PTI. NF receptor LjNFR1/ MtLYK3 is the closest homolog of chitin receptor AtCERK1 in Arabidopsis. These facts suggest that the initiation processes of immunity and symbiosis have some similarity, probably reflecting the common evolutional origin (Figure 3).
Figure 3.
Hypothetical scheme of the evolution of defense and symbiosis systems in plant.
Concerning the origin of the RN symbiosis, it has been widely accepted that RN symbiosis was established on the basis of AM symbiosis.42 AM symbiosis is the most widespread symbiosis present in terrestrial plants with the exception of Brassicaceae and some other species. Interestingly, CERK1 homolog genes in Brassicaceae, including Arabidopsis AtCERK1, do not have the conserved three-amino acid motif (YAQ or YAR) in the kinase domain, which is important for RN formation.43 In the case of L. japonicus, ljnfr1 but not ljnfr5 knockout mutants slightly diminish AM colonization on the roots.44 These facts suggest that AtCERK1 homolog genes are involved in AM symbiosis in non-legume plants, except Brassicaceae, and could be the origin of the NF receptor in legume. In fact, recent studies revealed that OsCERK1 is involved in AM symbiosis in addition to PTI. Although a significant reduction of AM symbiosis was observed in oscerk1 knockout mutant and RNAi lines, knockout mutants of oscebip, the chitin-binding receptor in rice, did not show any mycorrhizal phenotypes.44,45 This finding suggests that perception of longer-chain chitin oligosaccharides, such as heptamers or octamers, are not necessary for AM symbiosis.45
Signal molecules for AM symbiosis have also been reported. Maillet et al. showed that a mixture of sulfated and non-sulfated LCOs (Myc-LCOs) enhanced AM infection and root branching in several plant families, including M. truncatula.46 Ca2+ spiking, which is known as a unique event in RN and AM symbiosis, is also observed in the root of M. truncatula and L. japonicus treated with a synthesized Myc-LCO.46,47 However, short chain chitin oligomers such as tetramers (CO4) also induced the Ca2+ spiking in the root of M. truncatula and L. japonicus.47,48 Genre et al. showed that the CO4-induced Ca2+ spiking is independent of NFP,48 and Sun et al. also reported a similar observation, although the response for Myc-LCO is dependent on NFP in M. truncatula.47 In the case of rice, Ca2+ spiking was induced in the root treated with CO4, whereas no Ca2+ spiking or the expression of AM marker genes was observed in the Myc-LCO treated rice root.44,47 Recently, experiments using the transgenic rice lines expressing a nuclear Ca2+-sensing chameleon probe have revealed that, by comparison with WT plants, the oscerk1 mutant is defective in response to crude germinated AM fungal spore exudates and CO4.49 These results indicate that short chitin oligomers such as CO4 are a likely candidate of the signal molecule for AM symbiosis in these plants.
Although OsCERK1 has been shown to regulate both symbiosis and immunity in rice, it is still unclear how a single RLK (OsCERK1) triggers the proper process, symbiosis or immune response. In the case of immunity, OsCEBiP mediates chitin binding, as shown in the model described by Hayafune et al.16 Similarly, an unknown receptor might play a key role in binding ‘Myc factor’ and initiate symbiosis. Although several reports showed that NjNFR5/MtNFP knockdown mutant exhibited the reduction of AM colonization, for example in Parasponia and tomato,50,51 no such effects have been reported for OsNFR5/OsRLK2, the single NjNFR5/MtNFP homolog gene in rice, and GSE or CO4 was able to elicit Ca2+ spiking in osnfr5 mutants.52 It is not clear at present whether the presence of the redundant receptor resulted in this phenotype or another receptor mediates this process in rice. Whether the identification of AM receptors in model plants is one of the most important target for understanding the symbiosis remains to be elucidated. Part of the reason for this difficulty may be functional redundancy between the members of the very large family of LysM-RLKs present, especially in legume genomes. In the case of L. japonicus, Rasmussen et al. have shown that Lys11, an AM-inducible NFR5/NFP paralog gene, is not essential for the initiation of AM symbiosis, though this gene can complement the function of NFR5 for RN symbiosis.53 Further biochemical, as well as molecular genetic, studies of LysM-RLKs/RLPs could lead to the identification of the ligand–receptor pairs for AM symbiosis.
Despite the importance of AM symbiosis in the ecosystem, its initiation process is not still fully understood. One of the reasons is that Arabidopsis, the best characterized model plant, cannot establish the AM symbiosis. In contrast, rice can establish AM symbiosis, while the innate immune system has been studied extensively because of its importance as a crop. This means rice could be a good model with which to understand the mechanism of how LysM receptors regulate these opposite responses, symbiosis and immunity. These studies will lead us to further understanding of the mechanism how plants distinguish friend and foe.
Conclusion
The understanding of the plant immune system against fungi is important because 80% of serious plant diseases are caused by fungi (Introduction to Fungi: http://www.apsnet.org/). In fact, 10–30% of the annual rice harvest is lost by the rice blast fungus, which is a devastating pathogen.54
Chitin, a representative fungal MAMP, is a major cell wall component of fungi and induces defense signaling through plant LysM receptors. During the past decade, our knowledge on the molecular mechanism of chitin recognition by the corresponding receptors and the activation of downstream signaling has been developed. However, the details of the receptor complex formation triggered by chitin oligosaccharides and the activation of downstream signaling remain for future studies. It is also not clear why rice and Arabidopsis adopted partly different chitin signaling systems during evolution.
Although not discussed in this short review, it has also been revealed that some fungal pathogens secrete LysM effectors that directly bind to chitin or chitin oligosaccharides released from cell walls by plant chitinases to escape from the chitin-triggered immunity, providing an excellent example of host–pathogen co-evolution.13,55
Moreover, plant LysM receptors are also essential molecules for the signaling in RN and AM symbiosis. Interestingly, it was shown that that OsCERK1 has a dual function in defense and symbiosis signaling in rice.45 However, how the single RLK regulates these opposite signaling remains to be clarified. The knowledge of rice could be a clue to understanding the more complicated system in legumes in order to distinguish between pathogen, AM fungi and rhizobia. Understanding of the molecular mechanism of plant immunity, as well as symbiosis mediated by LysM receptors, would contribute to solve the global problem of food supply by providing the basis of ideas for the development of disease-resistant crops and the manipulation of symbiosis with useful microbes in agriculture.
Finally, although chitin also activates mammalian innate immunity, its receptor is still obscure and seems very different from those of plants.5,56 Whereas cell-surface LysM receptors in plants also play an essential role for PGN-induced immunity, in mammals, PGN seems to be detected by cytosolic receptors,5 indicating that the perception of PGN is also very different from plants. It is interesting to understand how and why such different machineries for the detection of common MAMPs have been developed during the evolution of immune systems in plants and mammals.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Grants-in-Aid for Scientific Research on Innovative Areas (15H01240 to HK), Grants-in-Aid for Young Scientists (No. 26850028 to YD), JSPS Overseas Research Fellowships (to KM) and MEXT-Supported Program for the Strategic Research Foundation at Private Universities 2014-2018 (S1411023) to HK from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
References
- 1.Strange RN, Scott PR. Plant disease: a threat to global food security. Annu Rev Phytopathol 2005; 43: 83–116. [DOI] [PubMed] [Google Scholar]
- 2.Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 2009; 60: 379–406. [DOI] [PubMed] [Google Scholar]
- 3.Boutrot F, Zipfel C. Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu Rev Phytopathol 2017; 55: 257–286. [DOI] [PubMed] [Google Scholar]
- 4.Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol 2015; 33: 257–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kieser KJ, Kagan JC. Multi-receptor detection of individual bacterial products by the innate immune system. Nat Rev Immunol 2017; 17: 376–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zipfel C, Felix G. Plants and animals: a different taste for microbes? Curr Opin Plant Biol 2005; 8: 353–360. [DOI] [PubMed] [Google Scholar]
- 7.Katsuragi Y, Takai R, Furukawa T, et al. CD2-1, the C-terminal region of flagellin, modulates the induction of immune responses in rice. Mol Plant Microbe Interact 2015; 28: 648–658. [DOI] [PubMed] [Google Scholar]
- 8.Buist G, Steen A, Kok J, Kuipers OP. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol 2008; 68: 838–847. [DOI] [PubMed] [Google Scholar]
- 9.Shi XZ, Feng XW, Sun JJ, et al. Involvement of a LysM and putative peptidoglycan-binding domain-containing protein in the antibacterial immune response of kuruma shrimp Marsupenaeus japonicus. Fish Shellfish Immunol 2016; 54: 489–498. [DOI] [PubMed] [Google Scholar]
- 10.Shi XZ, Zhou J, Lan JF, et al. A lysin motif (LysM)-containing protein functions in antibacterial responses of red swamp crayfish, Procambarus clarkii. Dev Comp Immunol 2013; 40: 311–319. [DOI] [PubMed] [Google Scholar]
- 11.Ranf S, Gisch N, Schaffer M, et al. A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat Immunol 2015; 16: 426–433. [DOI] [PubMed] [Google Scholar]
- 12.Plato A, Hardison SE, Brown GD. Pattern recognition receptors in antifungal immunity. Semin Immunopathol 2015; 37: 97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shinya T, Nakagawa T, Kaku H, Shibuya N. Chitin-mediated plant-fungal interactions: catching, hiding and handshaking. Curr Opin Plant Biol 2015; 26: 64–71. [DOI] [PubMed] [Google Scholar]
- 14.Kaku H, Nishizawa Y, Ishii-Minami N, et al. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci U S A 2006; 103: 11086–11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S, Wang J, Han Z, et al. Molecular mechanism for fungal cell wall recognition by rice chitin receptor OsCEBiP. Structure 2016; 24: 1192–1200. [DOI] [PubMed] [Google Scholar]
- 16.Hayafune M, Berisio R, Marchetti R, et al. Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc Natl Acad Sci U S A 2014; 111: E404–E413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miya A, Albert P, Shinya T, et al. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci U S A 2007; 104: 19613–19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petutschnig EK, Jones AM, Serazetdinova L, et al. The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J Biol Chem 2010; 285: 28902–28911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shinya T, Motoyama N, Ikeda A, et al. Functional characterization of CEBiP and CERK1 homologs in arabidopsis and rice reveals the presence of different chitin receptor systems in plants. Plant Cell Physiol 2012; 53: 1696–1706. [DOI] [PubMed] [Google Scholar]
- 20.Willmann R, Lajunen HM, Erbs G, et al. Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc Natl Acad Sci U S A 2011; 108: 19824–19829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faulkner C, Petutschnig E, Benitez-Alfonso Y, et al. LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc Natl Acad Sci USA 2013; 110: 9166–9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narusaka Y, Shinya T, Narusaka M, et al. Presence of LYM2 dependent but CERK1 independent disease resistance in Arabidopsis. Plant Signal Behav 2013; 8: e25345–e25345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao Y, Liang Y, Tanaka K, et al. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. Elife 2014; 3: e03766–e03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le MH, Cao YR, Zhang XC, Stacey G. LIK1, a CERK1-interacting kinase, regulates plant immune responses in Arabidopsis. PLOS ONE 2014; 9: e102245–e102245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeh YH, Panzeri D, Kadota Y, et al. The Arabidopsis malectin-like/LRR-RLK IOS1 is critical for BAK1-dependent and BAK1-independent pattern-triggered immunity. Plant Cell 2016; 28: 1701–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malerba M, Cerana R. Chitosan effects on plant systems. Int J Mol Sci 2016; 17: E996–E996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki M, Shibuya M, Shimada H, et al. Autophosphorylation of specific threonine and tyrosine residues in Arabidopsis CERK1 is essential for the activation of chitin-induced immune signaling. Plant Cell Physiol 2016; 57: 2312–2322. [DOI] [PubMed] [Google Scholar]
- 28.Yan L, Ma Y, Liu D, et al. Structural basis for the impact of phosphorylation on the activation of plant receptor-like kinase BAK1. Cell Res 2012; 22: 1304–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Li X, Meisenhelder J, et al. Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev Cell 2005; 8: 855–865. [DOI] [PubMed] [Google Scholar]
- 30.Macho AP, Schwessinger B, Ntoukakis V, et al. A bacterial tyrosine phosphatase inhibits plant pattern recognition receptor activation. Science 2014; 343: 1509–1512. [DOI] [PubMed] [Google Scholar]
- 31.Ao Y, Li ZQ, Feng DR, et al. OsCERK1 and OsRLCK176 play important roles in peptidoglycan and chitin signaling in rice innate immunity. Plant J 2014; 80: 1072–1084. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi K, Yamada K, Ishikawa K, et al. A receptor-like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe 2013; 13: 347–357. [DOI] [PubMed] [Google Scholar]
- 33.Li Z, Ao Y, Feng D, et al. OsRLCK 57, OsRLCK107 and OsRLCK118 positively regulate chitin- and PGN-induced immunity in rice. Rice (N Y) 2017; 10: 6–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shinya T, Yamaguchi K, Desaki Y, et al. Selective regulation of the chitin-induced defense response by the Arabidopsis receptor-like cytoplasmic kinase PBL27. Plant J 2014; 79: 56–66. [DOI] [PubMed] [Google Scholar]
- 35.Yamada K, Yamaguchi K, Shirakawa T, et al. The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J 2016; 35: 2468–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang C, Wang G, Zhang C, et al. OsCERK1-mediated chitin perception and immune signaling requires receptor-like cytoplasmic kinase 185 to activate an MAPK cascade in rice. Mol Plant 2017; 10: 619–633. [DOI] [PubMed] [Google Scholar]
- 37.Yamada K, Yamaguchi K, Yoshimura S, et al. Conservation of chitin-induced MAPK signaling pathways in rice and Arabidopsis. Plant Cell Physiol 2017; 58: 993–1002. [DOI] [PubMed] [Google Scholar]
- 38.Lu D, Wu S, Gao X, et al. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci U S A 2010; 107: 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Li W, Xiang T, et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 2010; 7: 290–301. [DOI] [PubMed] [Google Scholar]
- 40.Akamatsu A, Wong HL, Fujiwara M, et al. An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe 2013; 13: 465–476. [DOI] [PubMed] [Google Scholar]
- 41.Oldroyd GED. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 2013; 11: 252–263. [DOI] [PubMed] [Google Scholar]
- 42.Parniske M. Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol 2008; 6: 763–775. [DOI] [PubMed] [Google Scholar]
- 43.Nakagawa T, Kaku H, Shimoda Y, et al. From defense to symbiosis: limited alterations in the kinase domain of LysM receptor-like kinases are crucial for evolution of legume-Rhizobium symbiosis. Plant J 2011; 65: 169–180. [DOI] [PubMed] [Google Scholar]
- 44.Zhang XW, Dong WT, Sun JH, et al. The receptor kinase CERK1 has dual functions in symbiosis and immunity signalling. Plant J 2015; 81: 258–267. [DOI] [PubMed] [Google Scholar]
- 45.Miyata K, Kozaki T, Kouzai Y, et al. The bifunctional plant receptor, OsCERK1, regulates both chitin-triggered immunity and arbuscular mycorrhizal symbiosis in rice. Plant Cell Physiol 2014; 55: 1864–1872. [DOI] [PubMed] [Google Scholar]
- 46.Maillet F, Poinsot V, Andre O, et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 2011; 469: 58–63. [DOI] [PubMed] [Google Scholar]
- 47.Sun JH, Miller JB, Granqvist E, et al. Activation of symbiosis signaling by arbuscular mycorrhizal fungi in legumes and rice. Plant Cell 2015; 27: 823–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Genre A, Chabaud M, Balzergue C, et al. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol 2013; 198: 179–189. [DOI] [PubMed] [Google Scholar]
- 49.Carotenuto G, Chabaud M, Miyata K, et al. The rice LysM receptor-like kinase OsCERK1 is required for the perception of short-chain chitin oligomers in arbuscular mycorrhizal signaling. New Phytol 2017; 214: 1440–1446. [DOI] [PubMed] [Google Scholar]
- 50.Buendia L, Wang TM, Girardin A, Lefebvre B. The LysM receptor-like kinase SlLYK10 regulates the arbuscular mycorrhizal symbiosis in tomato. New Phytol 2016; 210: 184–195. [DOI] [PubMed] [Google Scholar]
- 51.den Camp RO, Streng A, De Mita S, et al. LysM-type mycorrhizal receptor recruited for Rhizobium symbiosis in nonlegume Parasponia. Science 2011; 331: 909–912. [DOI] [PubMed] [Google Scholar]
- 52.Miyata K, Hayafune M, Kobae Y, et al. Evaluation of the role of the LysM receptor-like kinase, OsNFR5/OsRLK2 for AM symbiosis in rice. Plant Cell Physiol 2016; 57: 2283–2290. [DOI] [PubMed] [Google Scholar]
- 53.Rasmussen SR, Fuchtbauer W, Novero M, et al. Intraradical colonization by arbuscular mycorrhizal fungi triggers induction of a lipochitooligosaccharide receptor. Sci Rep 2016; 6: 29733–29733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Talbot NJ. On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu Rev Microbiol 2003; 57: 177–202. [DOI] [PubMed] [Google Scholar]
- 55.Sanchez-Vallet A, Mesters JR, Thomma BPHJ. The battle for chitin recognition in plant-microbe interactions. FEMS Microbiol Rev 2015; 39: 171–183. [DOI] [PubMed] [Google Scholar]
- 56.Elieh Ali Komi D, Sharma L, Dela Cruz CS. Chitin and its effects on inflammatory and immune responses. Clin Rev Allergy Immunol 2017. Mar 1 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hind SR, Strickler SR, Boyle PC, et al. Tomato receptor FLAGELLIN-SENSING 3 binds flgII-28 and activates the plant immune system. Nat Plants 2016; 2: 16128–16128. [DOI] [PubMed] [Google Scholar]
- 58.Wang L, Albert M, Einig E, et al. The pattern-recognition receptor CORE of Solanaceae detects bacterial cold-shock protein. Nat Plants 2016; 2: 16185–16185. [DOI] [PubMed] [Google Scholar]