Abstract
Compelling evidence demonstrates the crucial role of the commensal microbiota in host physiology and the detrimental effects of its perturbations following antibiotic treatment. However, the effects of commensal microbiota on intestinal mucosa antimicrobial molecules have not been elucidated systematically. Here, we investigate the impacts of antibiotic-induced depletion and subsequent restoration of the intestinal microbiota on the murine antimicrobial molecules in intestinal mucosa. Our results demonstrate that depletion of commensal microbiota leads to intestinal mucosa atrophy and reduction of antimicrobial molecules, including lysozyme, regenerating islet-derived protein 3 gamma (RegIIIγ), and cryptdin 5 mRNA, whereas subsequent reconstitution of intestinal microbiota by fecal microbiota transplantation (FMT) rescues mucosa morphology and antimicrobials. Importantly, our study shows that down-regulation of aryl hydrocarbon receptor (AhR), interleukin-22 (IL-22), and phosphorylated Stat3 (p-Stat3) is associated with decreased antimicrobials, which might mediate the antibiotic-associated intestinal mucosa injury. Last, exogenous activation of the AhR/IL-22/Stat3 signaling pathway with the AhR agonist 6-formylindolo(3,2-b)carbazole (Ficz) rescued antimicrobial molecule levels markedly after antibiotic treatment to levels similar to those following reconstitution of intestinal microbiota by FMT. Together, our results demonstrate that the AhR/IL-22/Stat3 signaling pathway is involved in the modulation of intestinal mucosa antimicrobial molecules by commensal microbiota and suggest this pathway as a promising target in the treatment of antibiotic-associated gut barrier damage.
Keywords: Commensal microbiota, intestinal mucosa, antimicrobial molecules, aryl hydrocarbon receptor, IL-22
Introduction
The human gastrointestinal tract harbors a great number of microorganisms, including bacteria, viruses, fungi, protozoa, and helminths, which are collectively referred to as the commensal microbiota. In recent years, the commensal microbiota has been established as an indispensable factor in host physiology processes such as food digestion, vitamin synthesis, and fat metabolism.1 Moreover, an important aspect underlining the indispensability of gut microbiota is their contribution to the development, maturation, and regulation of the immune system, including systemic immunity of the host and local immunity of the gut.2 Studies in isolator-raised germ-free (GF) mice revealed fundamental impairments in mucosal immunity, including decreased IgA levels, atrophic intestinal epithelia, and reduced lamina propria lymphocytes.3 However, the effects of commensal microbiota on other important immune compartments such as intestinal-epithelium-derived antimicrobial molecules remain unclear.
The intestinal epithelium is regarded as the physical and immune barrier of the gut mucosa. Intestinal enterocytes consists of columnar cells, goblet cells, Paneth cells, and enteroendocrine cells, which can produce and secrete a series of antimicrobial molecules for epithelial protection, including lysozyme, regenerating islet-derived protein 3 gamma (RegIIIγ), cryptdins, and secretory phospholipase A2.4 Cryptdins create pores in bacterial cell membranes, thus affecting the growth of Gram-negative and Gram-positive bacteria.5 RegIIIγ binds peptidoglycans in the membrane and exerts bactericidal activity against Gram-positive bacteria.6 Lysozyme hydrolyzes peptidoglycan in bacterial cells and is required to prevent excessive inflammatory responses.7 The creation and release of these products from enterocytes are promoted by IL-22 via the IL-22Rα1/IL-10Rβ/Stat3 pathway.8 These products limit excessive growth of the gut microbiota by disrupting the integrity of the bacterial cell membrane or wall to regulate the balance of intestinal homeostasis. Production and secretion reduction of these antimicrobials result in bacterial translocation from the intestinal tract to the circulation, leading to gut-derived infection, sepsis, or even multiple organ failure.9 In addition, Paneth-cell-derived antimicrobials can be damaged cellular induction of autophagy and overexpression of LC3.10 The intestinal epithelium and its antimicrobial products are necessary for the maintenance of intestinal homeostasis, but it remains unclear whether the production of these antimicrobial molecules is dependent on gut microbiota.
Aryl hydrocarbon receptor (AhR), a basic helix–loop–helix protein, is a member of the Per-AhR-nuclear translocator-Sim superfamily of proteins. AhR is a cytosolic transcription factor that is expressed ubiquitously in vertebrate cells and can be activated by a large variety of natural and synthetic ligands, including environmental, dietary, and endogenous aromatic compounds.11 In its inactive state, AhR resides in the cytosol bound to several cochaperones such as heat shock protein 90. Upon ligand binding, chaperones are released and AhR translocates to the nucleus, where it binds to its dimerization partner, AhR-nuclear translocator, thus initiating the transcription of a variety of target genes.12 Recently, AhR has gained attention because it represents an important link between the environment and immunity. Recent studies showed that activation of AhR by the microbiota within the gastrointestinal tract is essential for promoting local IL-22 production. Specifically, tryptophan catabolites generated via metabolism by the microbiota are involved in mucosal immune responses via AhR modulation.13 However, whether the expression of AhR depends on the commensal microbiota and its role in maintaining antimicrobial molecules is unknown.
AhR is activated by catabolites derived from gut microbiota and then promotes local intestinal IL-22 production to help maintain the levels of mucosal antimicrobials. In this study, we hypothesized that the gut microbiota can modulate the production and secretion of antimicrobial molecules and the AhR/IL-22/Stat3 signaling pathway might be involved in this process.
Methods
Animals
Animal protocols were approved by the animal care and use committee of Jinling Hospital, Nanjing, China. Male 6-wk-old C57BL/6 J mice were used and all animals were bred, raised, and housed in the facilities of the Model Animal Research Center of Nanjing University under specific-pathogen-free conditions. Animals were kept under ambient temperature (20–26℃) and 40–70% humidity conditions with a 12-h light/dark cycle. All mice had free access to a standard laboratory diet and water before the study and were unrestricted in activity.
Generation of antibiotic treatment mice and reconstitution of intestinal commensal microbiota by fecal transplantation
Eighteen 6-wk-old male mice were divided randomly into 3 groups (n = 6 for each group): the distilled water treatment group (Normal), the antibiotic treatment group (Abx), and the fecal microbiota transplantation group (FMT). To deplete the intestinal microbiota virtually, Abx and FMT mice were transferred to sterile cages and subjected to antibiotic treatment for 4 wk. An antibiotic mixture (4Abx) consisting of ampicillin (1 g/l), neomycin (1 g/l), metronidazole (1 g/l), and vancomycin (500 mg/l) was administered in distilled drinking water.14 Successful eradication of intestinal microbiota was confirmed by quantitative polymerase chain reaction (qPCR) of fecal samples. The Normal group received distilled water as a blank control. All mice received 60Co-irradiation-treated sterile food.
After antibiotic treatment for 4 wk, the antibiotic mixture was withdrawn and replaced with sterile drinking water. The Abx group received sterile drinking water continuously. After sterile drinking water treatment for 3 d, mice in the FMT group received FMT for 4 wk. Fresh fecal samples were collected from Normal group mice, pooled, and dissolved in 10 ml of sterile PBS (Life Technologies, Carlsbad, CA, USA) and the supernatant was applied perorally by gavage (0.25 ml/mouse) twice per wk for 4 wk to reconstitute a complex intestinal microbiota. All mice received 60Co-irradiation-treated sterile food and were then sacrificed at the same time.
Administration of AhR agonist
The AhR agonist 6-formylindolo(3,2-b)carbazole (Ficz; Enzo Life Sciences, Lausen, Switzerland) was resuspended in DMSO (Sigma-Aldrich, St. Louis, MO, USA) and administered intraperitoneally. After antibiotic treatment, the antibiotic mixture was withdrawn and replaced with sterile drinking water. Another six Abx mice received sterile food and Ficz injection intraperitoneally (1 µg/mouse) once per wk as described by Lamas et al.15 for 4 wk (same as FMT duration) and were sacrificed together with the other groups.
Sampling procedures
Mice were weighed and anesthetized by intraperitoneal injection of ketamine (100 mg/kg body mass). Whole small intestines (from the Treitz ligament to terminal ileum) were removed under sterile conditions. Small intestinal stool samples were harvested from the terminal ileum, homogenized mechanically, and used for DNA extraction and bacterial quantification. Three segments of the terminal ileum, approximately 1.0 cm in length, were collected and used for morphological analysis, protein extraction, and RNA analysis. The segment used for histology was immediately fixed in 5% formalin, whereas the left two segments were immediately snap frozen in liquid nitrogen and stored at –80℃ for molecular analysis. The same anatomical part of distal ileum was used for the same type of analysis.
qPCR for fecal bacterial quantification and cryptdin 5 and LC3 mRNA levels in ileal tissues
Small intestinal stool samples were harvested from the terminal ileum, homogenized mechanically, and used for DNA extraction with the E.Z.N.A. Stool DNA kit (OMEGA Bio, Norcross, GA, USA). DNA was then subjected to qPCR using a SYBR Green PCR kit (Alkali Scientific, Pompano Beach, FL, USA) and the StepOne Real-time PCR system (Applied Biosystems, Foster City, CA, USA). The fold changes in the 16 S rDNA gene were calculated and normalized to the β-actin (Actb) genome.16
Total RNA from distal ileal tissue samples was extracted using RNAiso Plus (Takara Bio, Dalian, China) according to the manufacturer’s protocol. The purity and concentration of RNA was determined by measuring the optical density at 260 and 280 nm. One microgram of RNA was used as the template for reverse transcription with random primers and reverse transcriptase used in a cDNA synthesis reaction according to the manufacturer’s instructions (RR047A; Takara Bio). cDNA was then subjected to qPCR as described above. The mRNA expression levels of the cryptdin gene were calculated and normalized against those of GAPDH.
Primers were designed and synthesized by Invitrogen (Carlsbad, CA, USA). Primer sequences were as follows: 16S rDNA, forward 5′-AGAGTTTGATCCTGGCTCAG-3′ and reverse 5′-TGCTGCCTCCCGTAGGAGT-3′; Actb, forward 5′-ATGACCCAGATCATGTTTGA-3′ and reverse 5′-TACGACCAGAGGCATACAG-3′. cryptdin 5, forward 5′-GCTCCTGCTCAACAATTCTCC-3′ and reverse 5′-CAGCTGCAGCAGAATACGAA-3′. LC3, forward 5′- AAGATCCCAGTGATTATAGAGCGA-3′ and reverse 5′-ATTGCTGTCCCGAATGTCTC-3′. GAPDH, forward 5′-AGGCCGGTGCTGAGTATGTC-3′ and reverse 5′-TGCCTGCTTCACCACCTTCT-3′. Data were calculated using the 2−ΔΔCt method.
Hematoxylin and eosin staining of ileal tissue samples
After dehydration and wax embedding, tissue samples were cut into 5 -µm-thick slices and stained with hematoxylin and eosin (H&E) following standard protocols. Ileal villus length and crypt depth were measured in coded sections (with six intact villus crypt units per section; two sections were calculated to give an average for each mouse) using a calibrated eyepiece by two investigators blinded to the sample identities. Data were analyzed with a computer-supported image analysis system (NIS-Elements AR 3.0 software; Nikon, Tokyo, Japan).
Immunohistochemistry (IHC) of lysozyme in ileal tissue samples
The wax blocks were cut into 5 -µm-thick slices and deparaffinized. After Ag retrieval and blocking with 5% BSA in PBS with 1% Tween 20 (Sigma-Aldrich, St. Louis, MO, USA) for 1 h, ileal tissue slices were incubated with an anti-lysozyme Ab (Abcam, Cambridge, UK) overnight (8–12 h) at 4℃. Subsequently, the sections were processed using a 3,3′-diaminobenzidine detection kit (ZSGB-Bio, Beijing, China) according to the manufacturer’s instructions. Next, the sections were counterstained with H&E and the coverslips were fixed with 50% neutral resins. For quantification of the number of Paneth cells, sections of all mice were stained and cells were counted in eight crypts in representative microscopic fields for each mouse. Results were normalized and are expressed as a percentage.
Western blot analysis of lysozyme, RegIIIγ, Stat3, phosphorylated Stat3, and AhR
Ileal tissue samples were homogenized in lysis buffer containing 20 mM Tris-HCl, pH 7.4, 2.5 mM EDTA, 200 mM NaCl, 200 mM NaCl, and 1% NP-40 using a hand-held tissue homogenizer (Kinematica, Luzern, Switzerland). Samples were centrifuged at 12,000 g for 20 min at 4℃. The protein concentration of supernatants was determined using a bicinchoninic acid protein assay kit (KeyGEN, Nanjing, China). Equal amounts of total protein extracted from ileal tissue samples were heated at 95℃ for 10 min and then separated by electrophoresis on 12% Bis-Tris protein gels (Invitrogen). Proteins were transferred onto polyvinylidene fluoride membranes (Millipore, MA, USA), washed with PBS containing Tween 20, blocked with 5% BSA (Sigma-Aldrich) for 1 h, and then incubated with primary Abs (anti-lysozyme, 1:5000, ab108508, Abcam; anti-RegIIIγ, 1:1000, ab198216, Abcam; anti-AhR, 1:500, ab84833, Abcam; anti-Stat3, 1:1000, No. 12640, Cell Signaling Technology, Danvers, MA, USA; anti-pStat3, 1:1000, No. 9131, Cell Signaling Technology) overnight at 4℃. β-Actin (1:5000, A1978, Sigma-Aldrich) and GAPDH (1:5000, G8795, Sigma-Aldrich) were used as internal controls. Subsequently, the membranes were washed and incubated with their corresponding secondary Abs for 1 h at 25℃ with constant agitation. After being washed with PBS 3 times for 5 min each, the membranes were incubated with electrochemiluminescence solution for 5 min and the bands of target proteins were detected using Kodak film. Grayscale analysis of the bands was performed using ImageJ software (NIH, Bethesda, MD, USA). Data are presented as the expression ratios of the target protein relative to internal controls.
Immunofluorescence of AhR in ileal tissue samples
Ileal tissues were fixed in 4% paraformaldehyde overnight, dehydrated, and embedded in paraffin. Tissue slices (5 µm in thickness) were mounted on positively charged glass and dewaxed. Ag retrieval was achieved by incubation in 0.01 M sodium citrate buffer, pH 6.0, for 20 min in a boiling steamer. Slides were then blocked with normal goat serum blocking reagent (Invitrogen) for 30 min and subjected to sequential incubation with anti-AhR (1:50, ab84833, Abcam) at 4℃ overnight and fluorophore-conjugated secondary Abs at 24℃ for 2 h and nuclei were visualized with 4[prime],6-diamidino-2-phenylindole (1:2000, ab104139, Abcam). Between each incubation step, the slides were washed with PBS three times for 5 min each. Sections were mounted using fluorescent mounting medium. Confocal images were obtained with a Zeiss (Oberkochen, Germany) LSM 710 imaging system.
ELISA of IL-22 in ileal tissue samples
Distal ileal tissue was homogenized as described above and samples of equal concentration were centrifuged at 10,000 g for 15 min at 4℃. The supernatant was extracted and recentrifuged. The concentration of IL-22 in the supernatant was measured using an ELISA kit (DLdevelop, Wuxi, China) following the manufacturer’s instructions.
Statistical analysis
Data are expressed as the mean ± SD. Mean variability among all groups was compared by one-way analysis of variance with Tukey’s post hoc test for multiple comparisons. Statistical calculations were performed using SPSS version 19.0 software (SPPS, Chicago, IL, USA). Two-sided P values ≤ 0.05 were considered significant.
Results
Macroscopic and microscopic sequelae following depletion and reconstitution of intestinal microbiota
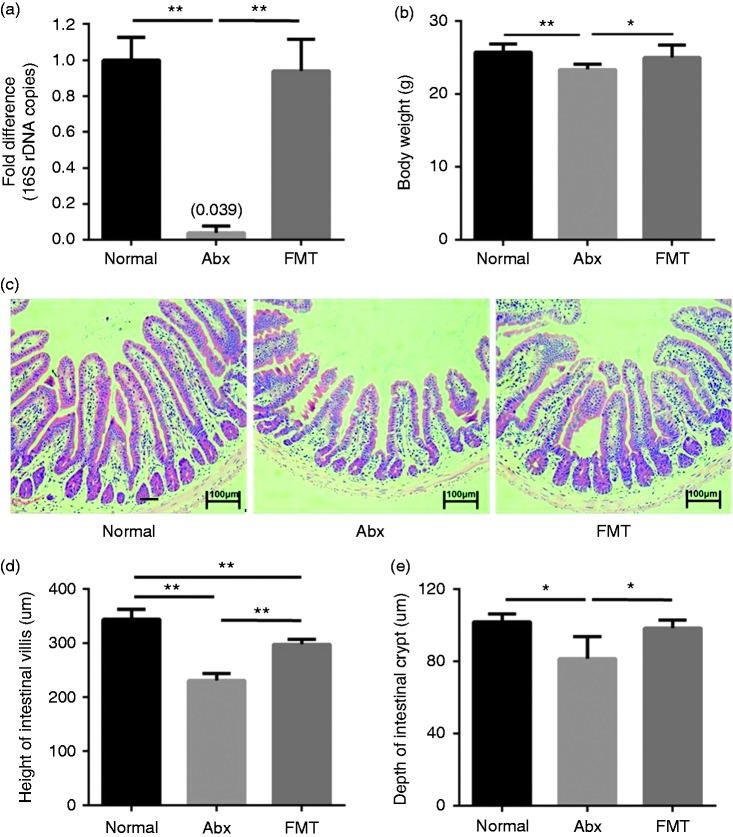
To confirm the successful depletion and reconstitution of the intestinal microbiota, we used qPCR to determine the fold change in 16 S rDNA, which is found in all bacterial chromosomes. More than 95% of the intestinal microbiota was depleted by broad-spectrum antibiotics and was reconstituted by FMT (Figure 1a). Body mass significantly decreased following antibiotic treatment (P < 0.01) and returned to normal levels after FMT (Figure 1b). Robust intestinal villi are essential for the maintenance of the gut barrier. Histological assessment of the ileum samples by H&E staining revealed severe blunting of the villi following antibiotic treatment and showed that FMT improved mucosal morphology (Figure 1c). Villus height was significantly lower following antibiotic treatment, but was rescued markedly by FMT, although the height remained significantly lower than that in the normal group (Figure 1d). In addition, crypt depth decreased in the Abx group, but returned to normal levels following FMT (Figure 1e).
Figure 1.
Macroscopic and microscopic sequelae following depletion and reconstitution of intestinal microbiota (n = 6). (a) Fold changes of bacterial 16 S rDNA levels in fecal samples. Antibiotic treatment decreased the number of small intestinal microbes efficiently. (b–e), Body mass (b), representative distal ileum H&E-stained histology image (c), villus height (d), and crypt depth (e) in conventional mice (Normal group), antibiotic treatment mice (Abx group), and subsequent FMT mice (FMT group). Scale bars represent 100 µm. *P < 0.05, **P < 0.01.
Impact of broad-spectrum antibiotic treatment and subsequent FMT on intestinal mucosal antimicrobials
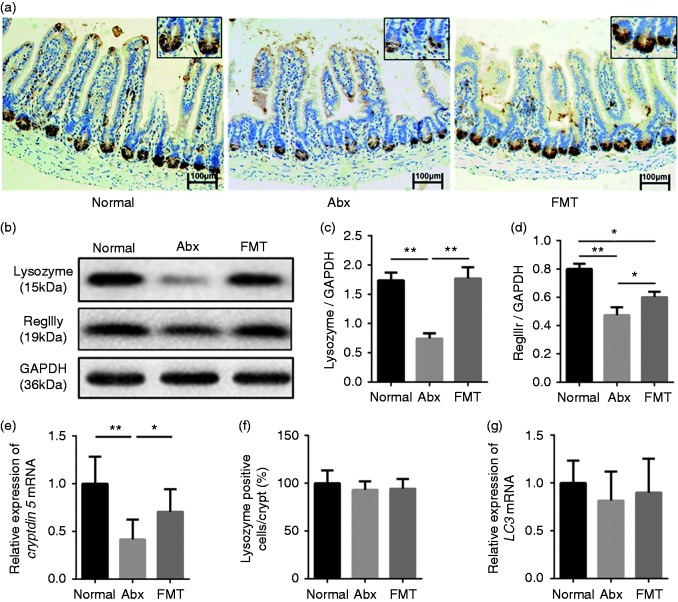
Cryptdins, lysozyme, and RegIIIγ are the major antimicrobial molecules produced and released by the intestinal epithelium. IHC showed that lysozyme was localized at the base of the ileal crypts and that its distribution was much sparser following antibiotic treatment. Reconstitution of the intestinal microbiota restored the distribution density of lysozyme in ileal crypts (Figure 2a). In addition, Western blot analysis showed that the protein levels of lysozyme were decreased significantly in the Abx group compared with the Normal group and subsequent FMT improved the expression of lysozyme protein significantly, which is consistent with the IHC results (Figure 2b and c). In mice, two forms of lysozyme are expressed, one specific for Paneth cells (lysozyme P) in the intestine and the other expressed in neutrophils and macrophages (lysozyme M). The Ab used was generated against total lysozyme (both types). However, because no difference in polymorphonuclear neutrophil or macrophage presence was detected, as illustrated by the IHC data on lysozyme, the results obtained by Western blot analysis reflected changes in Paneth-cell-derived lysozyme. In addition, antibiotic treatment diminished protein levels of RegIIIγ, which could be rescued partially by FMT (Figure 2b and d). Next, the gene expression of cryptdin 5, which is expressed exclusively by Paneth cells, was investigated. Strikingly, the mRNA levels of cryptdin 5 showed the same tendency as lysozyme and RegIIIγ protein levels (Figure 2e). Quantification of Paneth cells revealed equal numbers in the three groups, excluding the possibility that the observed decrease in lysozyme expression and cryptdin 5 mRNA levels in Paneth cells were caused by reduced cell numbers (Figure 2f). To exclude the effects of starvation and the occurrence of autophagy on antimicrobial proteins, the mRNA levels of LC3 were detected and showed no significant difference among the three groups (Figure 2g). Therefore, depletion of intestinal microbiota down-regulated the levels of mucosal antimicrobials, whereas reconstitution of intestinal microbiota by FMT enabled partial rescue of antimicrobial molecules, restoring bactericidal function of intestinal mucosa.
Figure 2.
Impact of broad-spectrum antibiotic treatment and subsequent FMT on intestinal mucosa antimicrobial molecules (n = 6). (a) Immunohistochemistry showing lysozyme expression in ileal tissue from different groups. Scale bars represent 100 µm. (b–d) Western blot analysis of lysozyme and RegIIIγ expression in ileal tissue and normalization to GAPDH levels. (e) Relative expression of cryptdin 5 mRNA. (f) Lysozyme-positive cells in crypts. (g) Relative expression of LC3 mRNA. *P < 0.05, **P < 0.01.
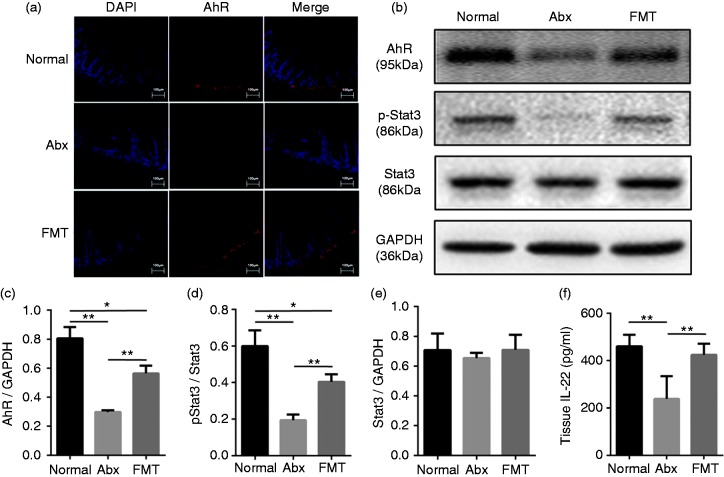
Suppression of the AhR/IL-22/Stat3 pathway is involved in antimicrobial molecule decrease following depletion of intestinal microbiota
AhR is an important sensor of catabolites derived from gut microbiota and promotes local intestinal IL-22 production. Previous studies have shown that IL-22 plays a prominent role in the production and secretion of antimicrobial molecules by activating IL-22 R and Stat3 phosphorylation. Therefore, we hypothesized that the AhR/IL-22/Stat3 pathway was involved in modulating antimicrobial molecules following antibiotic treatment and FMT. As expected, immunofluorescence showed that AhR was down-regulated significantly following antibiotic treatment and restored partially by subsequent FMT (Figure 3a). The protein levels of AhR as measured by Western blot analysis showed the same changes (Figure 3b and c). IL-22 levels in the Abx group were significantly lower than those in the Normal group and subsequent FMT rescued this change to normal levels (Figure 3f). Moreover, although the expression of Stat3 was independent of intestinal microbiota (Figure 3b and e), the p-Stat3/Stat3 ratio in distal ileal tissue was significantly lower following antibiotic treatment but was rescued partially by FMT (Figure 3b and d). Together, these data suggest that the AhR/IL-22/Stat3 signaling pathway is associated with the modulation of intestinal mucosa antimicrobial molecules by commensal microbiota.
Figure 3.
Inhibition of the AhR/IL-22/Stat3 pathway is involved in antimicrobial molecule decrease following depletion of intestinal microbiota (n = 6). (a) Immunofluorescence showing AhR expression in ileal tissue from different groups. Scale bars represent 100 µm. (b–e) Western blot analysis of AhR, Stat3, and p-Stat3 expression in ileal tissue and ratio of pStat3/Stat3. (f) ELISA analysis of IL-22 levels in ileal tissue from different groups. *P < 0.05, **P < 0.01.
Stimulation of AhR activates the IL-22/Stat3 pathway and restores antimicrobial molecules following antibiotic treatment
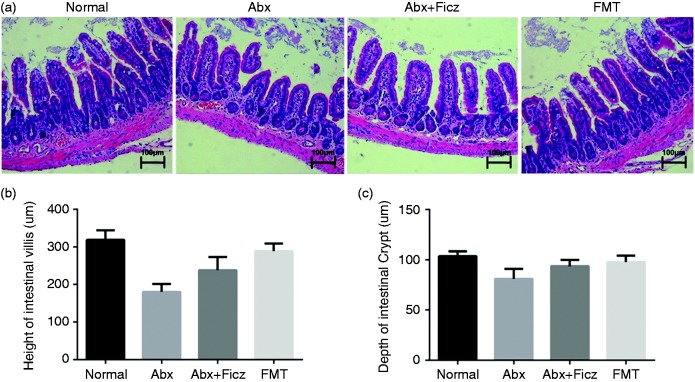
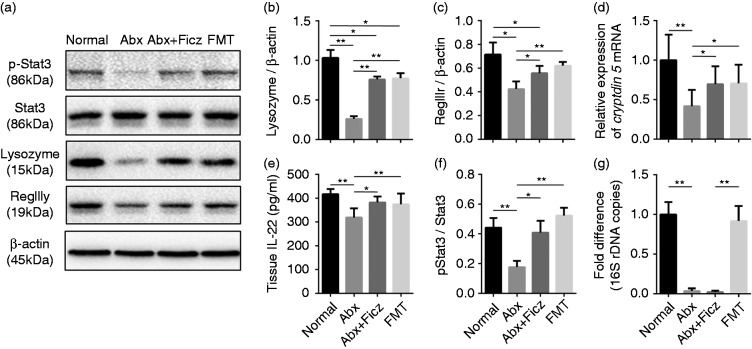
We found that down-regulation of the AhR/IL-22/Stat3 signaling pathway is associated with the reduction of antimicrobial molecules following antibiotic treatment, which could be rescued partially by subsequent FMT. To further assess the role of the AhR/IL-22/Stat3 pathway in the regulation of antimicrobial molecules by intestinal microbiota, we administered Ficz, an AhR agonist, to antibiotic-treated mice. After a 4-wk treatment with AhR agonist, mucosal atrophy (Figure 4) and reduction of lysozyme, RegIIIγ, and cryptdin 5 mRNA were improved in the Abx mice treated with Ficz compared with their untreated counterparts, almost reaching the levels observed in the FMT group (Figure 5a to d). In addition, administration of Ficz increased IL-22 levels and the ratio of p-Stat3/Stat3 in Abx mice significantly, reaching the levels in the Normal and FMT groups (Figure 5e and f), indicating that activation of AhR improved antimicrobial molecule levels through the IL-22/Stat3 pathway. To exclude that the restoration of the mucosal morphology, antimicrobial component expression, and stat3 phosphorylation was due to agonist treatment rather than being a result of growth and expansion of the remaining microbiota during the 4-wk Ficz treatment period, bacterial quantification was assessed in all mice before they were sacrificed. As expected, the fecal bacterial load in Ficz-treated mice was still extremely low and showed no significant difference compared with their untreated counterparts (Figure 5g). Together, these data demonstrate that commensal microbiota modulates intestinal mucosa antimicrobial molecules, a process in which the AhR/IL-22/Stat3 signaling pathway is involved.
Figure 4.
Ficz administration improved mucosal morphology following antibiotic treatment (n = 6). (a) Representative distal ileum H&E-stained histology image from different groups. Scale bars represent 100 µm. Also shown are villus height (b) and crypt depth (c) in the different groups. *P < 0.05, **P < 0.01.
Figure 5.
Ficz administration activated the AhR/IL-22/Stat3 pathway and restored antimicrobial molecules following antibiotic treatment (n = 6). (a–c) Western blot analysis of lysozyme and RegIIIγ expression in ileal tissue normalized to β-actin. (d) Relative expression of cryptdin 5 mRNA, (e) IL-22 levels, and (f) p-Stat3/Stat3 ratio in ileal tissue from different groups. (g) Fold changes of bacterial 16S rDNA levels in fecal samples. *P < 0.05, **P < 0.01.
Discussion
With increasing evidence demonstrating the indispensability of commensal bacteria in host physiology, complex mutualistic host–microbiota relationships, particularly microbiota-induced changes to intestinal immune homeostasis, have gained increased interest in recent years. Although GF mice have advanced our understanding of the impact of microbiota on the immune system greatly, they cannot be used to determine the role of microbiota-induced changes in host and gut immunity later in life, such as environmental changes and drug use.17 Previous studies have established profound immunological changes such as reduced numbers of IgA-producing plasma cells and lamina propria CD4+ T cells and smaller germinal centers in mesenteric lymph nodes in GF mice.18 Furthermore, increased evidence regarding pathologies related to antibiotic therapy, including antibiotic-associated gut barrier dysfunction, gut-derived infections, and diarrhea, has revealed the impact of antibiotic-induced perturbations of the intestinal microbiota in host physiology.19 Intestinal-epithelium-derived antimicrobial molecules are important components of intestinal mucosal innate immunity and play vital roles in regulating intestinal microbiota and maintaining homeostasis. Conversely, the effects of commensal microbiota on antimicrobial molecules have not yet been investigated fully.
In the present study, we used a mouse model suitable for investigating the interactions among microbiota, antibiotics, and intestinal mucosal immunity in conventionally raised and developed mice.20 Following broad-spectrum antibiotic treatment, mice were virtually lacking the intestinal microbiota, as shown by qPCR. We detected decreased antimicrobial molecules following broad-spectrum antibiotic treatment, indicating that the gut microbiota is necessary for maintaining the production of these antimicrobials. Our results agree with those of Zhang et al., who demonstrated that symbiotic bacteria can protect lysozyme from degradation in lysosome and direct selective cargo sorting through the Nod2–LRRK2–Rab2a complex in Paneth cells.21 Another previous study showed that diminished antimicrobial protein production and aberrant Paneth cell morphological features occurred during starvation.10 Paneth cells of starved mice showed increased induction of autophagy, which may have caused the deviant granules observed in these cells. Upon experimental starvation in which up to 25% of body mass was lost, there were profound effects on the intestine and certainly on the expression of Paneth cell antimicrobials.10 To exclude that the effects of microbiota depletion is not caused by mechanisms related to starvation and occurrence of autophagy, expression of LC3 was detected and showed no significant difference, indicating that other mechanisms must be involved.
To understand fully the influences of commensal microbiota on these antimicrobial molecules, we reconstructed the intestinal microbiota using FMT. Reintroduction of complex microbiota into the host via FMT is a well-known method of therapy dating back to the Chinese Dong Jin dynasty and is a therapeutic option for the treatment of refractory Clostridium difficile infection-induced acute necrotizing enterocolitis.22,23 Remarkably, FMT recovered mucosal morphology and antimicrobial molecule levels effectively, which further suggested that commensal microbiota are indispensable for maintaining intestinal mucosal immunity.
AhR has been widely reported to be involved in the regulation of intestinal physiology. Multiple studies have suggested that the AhR pathway is an effective targeting strategy for diseases such as multiple sclerosis, inflammatory bowel diseases, and cancer.24 AhR-mediated IL-22 production from adaptive and innate cells normally confers protection from bacterial infections and wound healing.25 Here, we found that antibiotic treatment led to reduced AhR levels and decreased IL-22 and phosphorylation Stat3 levels in ileal tissues, which are downstream targets of AhR and play critical roles in promoting antimicrobial molecules. Strikingly, these measurements were restored significantly by FMT, indicating that AhR is important in the modulation of intestinal antimicrobial molecules. Therefore, we administered Ficz to activate AhR exogenously in Abx mice and found significantly increased IL-22 and phosphorylation Stat3 levels, which contributed to up-regulated antimicrobial molecule levels. Our data agree with previous findings demonstrating the importance of microbiota-driven signaling for the expansion of AhR activity and IL-22 levels in the gut.15 Disordered gut microbiota failed to metabolize tryptophan into metabolites that act as AhR ligands, leading to decreased AhR activity and IL-22 levels, ultimately delaying the recovery from dextran sulfate sodium-induced colitis.15 It is fascinating that the microbiota had not recovered during the administration of Ficz as late as 4 wk after stopping antibiotic treatment. The reason that the 5% of remaining microbes failed to expand may be that the effects of antibiotics last several weeks even after they are withdrawn. Together, our data demonstrate that commensal microbiota probably modulates antimicrobial molecules via the AhR/IL-22/Stat3 signaling pathway.
Although these in vivo results are promising, there are some limitations to our study. First, the exogenous agonist activated the AhR/IL-22/Stat3 signaling pathway and rescued antimicrobials markedly after antibiotic treatment, whereas these measurements failed to return to normal levels, indicating that other factors and pathways might participate in the dys-regulation of intestinal antimicrobials following antibiotic treatment. Whether the commensal microbiota affects these factors in terms of antimicrobial molecule modulation requires further analysis. Second, the induction of antimicrobials that occurs via the microbiota-induced AhR relies on the effect of a single agonist tested and additional agonist would strengthen this specific connection. Third, commensal microbiota modulates various intestinal immune cells and other cytokines, such as CD4+ and CD8+ T lymphocytes, dendritic cells, regulatory T cells, IFN-γ, IL-17, and IL-10, all of which may be involved in the production of antimicrobial molecules.26 Therefore, in vitro experiments are anticipated to further confirm the role of the AhR//IL-22/Stat3 signaling pathway in the maintenance of antimicrobial molecules.
In conclusion, our study demonstrates that the commensal microbiota is of indispensable importance for maintaining normal antimicrobial molecule levels, in which the AhR//IL-22/Stat3 signaling pathway is involved. Although antibiotic treatment is unavoidable under specific clinical conditions, it is crucial to consider the effects of this therapy on intestinal mucosal immunity. Targeting the AhR/IL-22/Stat3 pathway might be a promising approach for treating antibiotic-associated gut barrier damage.
Acknowledgments
We would like to thank Dr. Xiang Gao at the Model Animal Research Center of Nanjing University for the Stat3 and p-Stat3 Abs.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported by the National Natural Science Foundation of China (grant numbers 81470797 and 81770531); the Science Foundation of Outstanding Youth in Jiangsu Province (grant number BK20170009); and the Research Special Fund for Public Welfare Industry of Health from National Health and Family Planning Commission of China (grant number 201502022).
References
- 1.Blander JM, Longman RS, Iliev ID, Sonnenberg GF, Artis D. Regulation of inflammation by microbiota interactions with the host. Nat Immunol 2017; 18: 851–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 2009; 9: 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fiebiger U, Bereswill S, Heimesaat MM. Dissecting the interplay between intestinal microbiota and host immunity in health and disease: lessons learned from germfree and gnotobiotic animal models. Eur J Microbiol Immunol (Bp) 2016; 6: 253–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol 2011; 9: 356–368. [DOI] [PubMed] [Google Scholar]
- 5.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature 2002; 415: 389–395. [DOI] [PubMed] [Google Scholar]
- 6.Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J Exp Med 2007; 204: 1891–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganz T. Antimicrobial polypeptides. J Leukoc Biol 2004; 75: 34–38. [DOI] [PubMed] [Google Scholar]
- 8.Wolk K, Kunz S, Witte E, Friedrich M, Asadullah K, Sabat R. IL-22 increases the innate immunity of tissues. Immunity 2004; 21: 241–254. [DOI] [PubMed] [Google Scholar]
- 9.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A 2008; 105: 20858–20863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodin CM, Lenaerts K, Grootjans J, et al. Starvation compromises Paneth cells. Am J Pathol 2011; 179: 2885–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu YZ, Hogenesch JB, Bradfield CA. The PAS superfamily: sensors of environmental and developmental signals. Annu Rev Pharmacol Toxicol 2000; 40: 519–561. [DOI] [PubMed] [Google Scholar]
- 12.Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol 2003; 43: 309–334. [DOI] [PubMed] [Google Scholar]
- 13.Zelante T, Iannitti RG, Cunha C, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013; 39: 372–385. [DOI] [PubMed] [Google Scholar]
- 14.Ochi T, Feng Y, Kitamoto S, et al. Diet-dependent, microbiota-independent regulation of IL-10-producing lamina propria macrophages in the small intestine. Sci Rep 2016; 6: 27634–27634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamas B, Richard ML, Leducq V, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med 2016; 22: 598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iliev ID, Funari VA, Taylor KD, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 2012; 336: 1314–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grover M, Kashyap PC. Germ-free mice as a model to study effect of gut microbiota on host physiology. Neurogastroenterol Motil 2014; 26: 745–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macpherson AJ, Martinic MM, Harris N. The functions of mucosal T cells in containing the indigenous commensal flora of the intestine. Cell Mol Life Sci 2002; 59: 2088–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becattini S, Taur Y, Pamer EG. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol Med 2016; 22: 458–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ekmekciu I, von Klitzing E, Fiebiger U, et al. Immune responses to broad-spectrum antibiotic treatment and fecal microbiota transplantation in mice. Front Immunol 2017; 8: 397–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Q, Pan Y, Yan R, et al. Commensal bacteria direct selective cargo sorting to promote symbiosis. Nat Immunol 2015; 16: 918–926. [DOI] [PubMed] [Google Scholar]
- 22.Zhang F, Luo W, Shi Y, Fan Z, Ji G. Should we standardize the 1,700-year-old fecal microbiota transplantation? Am J Gastroenterol 2012; 107 1755; author reply: 1755–1756. [DOI] [PubMed] [Google Scholar]
- 23.Fischer M, Kao D, Kelly C, et al. Fecal microbiota transplantation is safe and efficacious for recurrent or refractory Clostridium difficile infection in patients with inflammatory bowel disease. Inflamm Bowel Dis 2016; 22: 2402–2409. [DOI] [PubMed] [Google Scholar]
- 24.Cella M, Colonna M. Aryl hydrocarbon receptor: Linking environment to immunity. Semin Immunol 2015; 27: 310–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirchberger S, Royston DJ, Boulard O, et al. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J Exp Med 2013; 210: 917–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature 2016; 535: 65–74. [DOI] [PubMed] [Google Scholar]