Abstract
Bacterial cell division is mediated by the divisome which is organized by the Z ring, a cytoskeletal element formed by the polymerization of the tubulin homologue FtsZ. Despite billions of years of bacterial evolution, the Z ring is nearly universal among bacteria that have a cell wall and divide by binary fission. Recent studies have revealed the mechanism of cooperative assembly of FtsZ and that the Z ring consists of patches of FtsZ filaments tethered to the membrane that treadmill to distribute the septal biosynthetic machinery. Here, we summarize these advances and discuss questions raised by these new findings.
Keywords: Z ring, FtsZ, treadmilling, tubulin
The Z ring
Cell division in most bacteria is mediated by a multiprotein complex called the divisome [1–5]. At the heart of this complex is a cytoskeletal element called the Z ring [6] which consists of membrane anchored filaments of FtsZ, the ancestral homolog of eukaryotic tubulin [7]. Formation of a Z ring is the first step in bacterial cytokinesis and it not only serves as a scaffold to recruit all other division proteins to form the divisome, but also guides the synthesis of septal peptidoglycan (PG) resulting in cell constriction. In addition, the Z ring may provide the force necessary to invaginate the cytoplasmic membrane, which is reinforced by the growth of septal PG [8, 9]. Despite billions of years of bacterial evolution and a variety of cell shapes and sizes, the Z ring is used by most prokaryotic cells for division, so understanding how the Z ring forms and how it organizes and regulates cell division is of extreme importance.
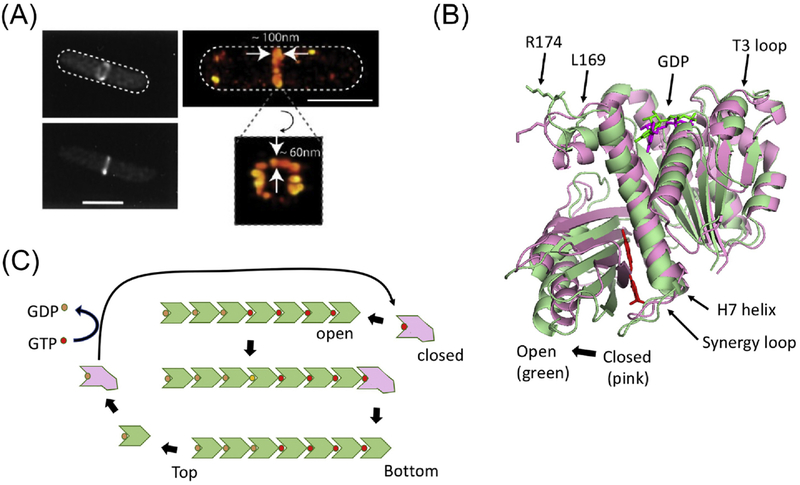
Despite tremendous efforts to determine the structure of the Z ring, we do not have a definitive architecture. As a result, in vitro efforts are hampered by the lack of a definitive goal for reconstitution and the propensity of FtsZ filaments to form irregular contacts resulting in bundling [10]. By conventional fluorescence microscopy the Z ring appears to be a continuous ring-like structure, but super resolution microcopy revealed that the Z ring is actually discontinuous and consists of small clusters of FtsZ assemblies of unknown structure [11–14] (Fig. 1A). In Escherichia coli, each cluster is estimated to contain 683 ± 439 FtsZ molecules, with an average length of ~120 nm and a width of ~100 nm [14, 15]. These clusters move circumferentially at the division site in both directions with a velocity of about ~30 nm/sec. A cluster persists for about 15 seconds (travelling about 30% of the circumference) before it disassembles and new clusters appear [16].
Figure 1. Cooperative assembly of FtsZ.
(A) Conventional and super resolution images of the Z ring (from ref. 14). (B) The two conformations of FtsZ. The open (green – PDB:3VOB) and closed (pink-PDB:2RHL) forms of FtsZ (from Staphylococcus aureus) are superimposed. The PC190723 inhibitor (red) and is present in the structure of the open form. Residues corresponding to R174 and L169 are indicated as are other important features. (C) A model for the cooperative assembly of FtsZ. In this model FtsZ in the closed form adds to the bottom end (the kinetic plus end) of a filament and switches conformation to the open form. Subsequent GTP hydrolysis leads to weakening of longitudinal bonds and loss of subunits from the minus end. Such subunits revert to the closed form and undergo nucleotide exchange before adding to the growing end.
Formation of a Z ring proceeds through two steps. First, FtsZ monomers polymerize into single-stranded filaments that are tethered to the membrane. Second, these dynamic membrane anchored FtsZ filaments are subjected to spatial regulation (Box 1), crosslinked by accessory proteins and linked to the Ter signal so that a Z ring is formed at midcell [5, 17]. Recent studies reveal that FtsZ filaments treadmill, the basis for this behavior and how these filaments recruit the remaining divisome proteins and direct septal PG synthesis [15, 18–21]. These findings provide important insights into our understanding of the Z ring but at the same time raise many questions. In this review, we examine what is known about the Z ring in the model organism E. coli.
Box 1. Spatial regulation of the Z ring: using the GTPase to explore space.
The GTPase activity associated with FtsZ filaments powers treadmilling and allows the filaments to explore intracellular space for conditions that are favorable for assembly into a Z ring. In eukaryotic cells microtubules extending from centrosomes also explore intracellular space by growing and shrinking (dynamic instability) until contact is made with a kinetochore to provide stability leading to formation of a spindle [28]. For the Z ring the spatial inputs include a positive Ter signal (linked to the terminus of the chromosome) and two negative signals, one linked to the shape of the cell (Min system) and another linked to the position of the nucleoid (SlmA). None of these regulators are essential, as they have partially overlapping roles. Loss of Min has the most dramatic effect resulting in the formation of minicells, whereas loss of the Ter signal leads to increased frequency of spiral septa which don’t always go to completion. Loss of SlmA has little effect during balanced growth but is important to delay Z ring formation when DNA segregation is impeded and avoid guillotining of the DNA.
The negative regulatory systems provide spatial inputs and share a common mode of action. The Min system uses a well characterized oscillatory mechanism to prevent formation of the Z ring away from midcell and SlmA prevents the Z ring from forming over the origin proximal region of the nucleoid. Each inhibitor is positioned in the cell by binding to a surface (SlmA to the DNA and MinC/MinD to the membrane). Binding to the surface activates the inhibitor leading to binding to the CCTP of FtsZ and in a second step disrupting the filaments [45].
The link between the Z ring and the terminus of replication (Ter) has emerged recently. It consists of ZapA, which binds to FtsZ filaments, MatP, which organizes the Ter region of the chromosome into a macrodomain, and ZapB, which links the two [95]. The Ter signal has an important role in maintaining Z ring geometry, ensuring completion of the septum and may turn out to be very widespread among bacteria. Consistent with this ZapA is very conserved among bacteria [75]. Although, ZapB and MatP are only found in bacteria closely related to E. coli, a coiled coil protein in Caulobacter crescentus appears to be more broadly distributed and a functional homologue of ZapB [96]. Also, MatP is among a set of proteins (including SeqA, HolD, MukBEF) that are only present in bacteria that contain the Dam methylase (enteric bacteria). It is likely that a functional homologue exists in more distantly related bacteria [97].
Cooperative assembly of FtsZ
FtsZ is a filament forming GTPase in the tubulin superfamily [22–24]. It consists of an N-terminal polymerization domain connected to a highly conserved C-terminal peptide (CCTP or CTC) of ~8 amino acids by an intrinsically disordered linker region of variable length (50 amino acids in E. coli) [25, 26]. Each domain is critical for FtsZ’s function in cell division, but polymerization only requires the globular domain which has the tubulin fold. Like tubulin, FtsZ undergoes GTP-dependent assembly, however, unlike tubulin, which usually assembles into microtubules that each contain 13 protofilaments, FtsZ assembles into single-stranded filaments of about 100-200 nm in length in buffers that mimic in vivo conditions [10]. Polymerization occurs in a head to tail manner with the GTP bound end of one subunit contacting the opposite end of the adjacent subunit. This longitudinal interaction generates the catalytic site for the GTPase resulting in the hydrolysis of GTP and a weakening of the longitudinal interaction and subsequent disassembly [27].
Cooperative assembly of cytoskeletal proteins includes: nucleation, elongation and steady state. The rate limiting step is nucleation because it is unfavorable compared to elongation. For actin and microtubules in eukaryotic cells, spontaneous nucleation is rare and these proteins rely on nucleation factors to get around this kinetic barrier [28, 29]. However, purified FtsZ (>1 μM) readily assembles into polymers upon the addition of GTP, suggesting that nucleation is a negligible kinetic barrier [10]. Currently, it is not clear whether there are nucleation factors for FtsZ inside cells, but if there are, they are not critical. Spontaneous nucleation of multi-stranded polymers like actin and microtubules depends upon multiple subunits forming an energetically unfavorable seed which is elongated by an incoming subunit making contacts with several subunits in the seed. How a linear single-stranded polymer like FtsZ could assemble cooperatively was unclear, but it was proposed it could if FtsZ existed in two conformations with a conformational change accompanying assembly [30–33]. This has been confirmed as FtsZ has been crystallized in two conformations: one in filaments (open) and another (closed) in the monomer [21] (Fig. 1B).
At steady state, FtsZ filaments treadmill in which subunits are added to one end of the filament (the plus end) and dissociate from the opposite end (the minus end) [34] (Fig. 1C). A transient delay in the GTPase after assembly and a delay in release of the subunits after GTP is hydrolyzed results in filaments with an average length of 40-50 subunits [10]. The treadmilling behavior of FtsZ filaments is at the heart of the regulation of bacterial cell division as dynamic filaments are responsive to spatial regulators and are required to distribute PG synthesis at the septum [15, 18, 35, 36].
Since FtsZ filaments treadmill, the question becomes what is the polarity? If we define the GTP binding domain of an FtsZ molecule as the top surface and the opposite end as the bottom surface (Fig. 1C), an FtsZ filament could grow from either end. It was reasoned that mutations at the top and bottom surfaces of FtsZ should have different effects on assembly and therefore cell division [37]. Testing a number of top and bottom mutants revealed only bottom end mutants displayed toxicity, implying that FtsZ filaments grow from the bottom end. However, none displayed substoichiometric toxicity and most still polymerized. Reinvestigation of this question using top and bottom surface mutants defective in longitudinal interactions made after the structure of FtsZ in a filament was known revealed that a bottom surface mutant was extremely toxic in vivo and antagonized polymerization of wild type FtsZ in vitro, whereas a top surface mutant had no effect [19]. These findings demonstrated that FtsZ subunits are added to the bottom end of a filament (plus end). In a proposed model (Box 2), FtsZ with GTP bound exists in two conformations with the closed form the dominant form. Above the critical concentration enough FtsZ is in the open form to nucleate assembly. A subunit in the closed form adds to the bottom (growing) end of the filament and the energy of binding induces the newly added subunit to switch to the open form, generating a new high affinity end (Fig. 1B). As subunits are added, GTP hydrolysis leads to the weakening of longitudinal interactions and the dissociation of subunits from the opposite end. This result was somewhat surprising since microtubules grow with the opposite polarity with new subunits added to the top end [28]. It also indicated that inhibitors, like SulA (produced in response to DNA damage in E. coli) and MciZ (produced during sporulation in Bacillus subtilis), cap filaments resulting in disassembly [19, 38]. SulA had previously been thought to inhibit assembly by sequestration [31, 39].
Box 2: Structural basis for cooperative assembly of FtsZ.
Cooperative assembly of FtsZ involves a polymerization associated conformational switch, which was validated by recent structures of FtsZ in the monomer form and in a filament. The first evidence for the polymerization associated conformational switch came from the structure of FtsZ (Staphylococcus aureus) with the inhibitor PC190723, a compound that forces FtsZ into the open or filament form and dramatically reduces the critical concentration for assembly [93, 94]. In the structure, PC190723 is inserted into the cleft between the N-terminal and C-terminal subdomains forcing a 27° rotation around the H7 helix that connects the two domains (Fig. 1B). The H7 helix also shifts downward about one helical pitch to accommodate this rotation. The H6-H7 loop moves away from the nucleotide binding pockets and the guanine ring rotates. This structure represents the open form of FtsZ whereas previously reported structures are in the closed form. Structures of various S. aureus mutants in the monomer and filament confirmed that the structure with PC190723 represents the open form of FtsZ [21].
GTP is critical for FtsZ assembly, but how GTP binding contributes to polymerization is not clear. However, it is clear that GTP binding does not directly induce the conformational changes to the open form because structures of the FtsZ monomer bound with GDP or GTP are in the closed form. The GTP γ-phosphate stabilizes the T3 loop of FtsZ and interacts extensively with the synergy loop of the adjacent subunit in the filament. This provides the longitudinal interactions necessary for subunit addition which cannot be achieved with GDP. GTP bound monomers likely exist in an equilibrium between the two forms with the closed form being predominant. Below the critical concentration, too few FtsZ-GTP molecules are in the open form to generate productive assembly, however, above the critical concentration, sufficient molecules exist in the open form to form a nucleus triggering cooperative assembly [19]. A time delay in GTP hydrolysis (or phosphate release) determines the length of the filament. It is also possible that other factors may affect subunit addition or release which affect filament length.
Membrane tethering of FtsZ filaments-flexibility and avidity
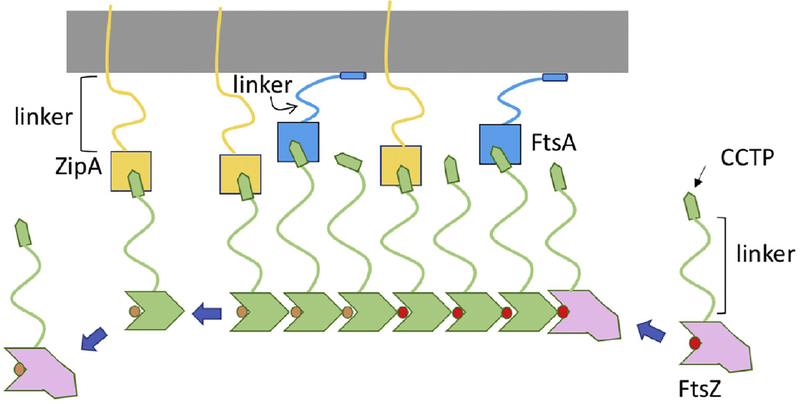
In E. coli the core components of the Z ring are FtsZ and its membrane anchors ZipA and FtsA [7]. As FtsZ filaments form they attach to the cytoplasmic membrane through association with the membrane anchors mediated by the CCTP of FtsZ (Fig. 2). The CCTP is at the tip of a 50 amino acid intrinsically disordered segment (linker) that connects it to the globular tubulin domain [25, 40]. ZipA also has a 100 amino acid intrinsically disordered segment connecting its transmembrane region to a globular domain that binds FtsZ and FtsA has a 25 amino acid intrinsically disordered segment that connects its globular actin domain that binds FtsZ to a membrane binding amphipathic helix [41–43]. Unstructured regions (intrinsically disordered) are common among proteins involved in endocytosis in eukaryotic cells and it is thought that such regions allow flexibility in recruiting binding partners to the membrane [44]. Having the CCTP at the tip of a long intrinsically disordered region of FtsZ provides flexibility needed for it to interact with so many different partners (FtsA, ZipA, MinC/MinD, ZapD, SlmA and possibly more) [25, 41, 45–48]. Furthermore, these interactions preferentially occur with filamentous FtsZ and the quantity of membrane anchors is such that all CCTPs in the Z ring are probably occupied (~6,700 FtsZ [with 30% of FtsZ in the ring], 2,000 ZipA and 1,000 FtsA per cell) [49]. The long flexible region of ZipA may enhance its ability to capture FtsZ filaments whereas the short flexible region in FtsA may aid it in connecting to the other division proteins [50].
Figure 2. Linking FtsZ filaments to the membrane.
FtsZ filaments bind to membrane anchors (FtsA and ZipA). This binding involves the CCTP (conserved C-terminal peptide) of FtsZ binding to FtsA and ZipA. The CCTP is an example of a MoRF (molecular recognition feature) more common in eukaryotic proteins. Such motifs display weak affinity and this is true for the CCTP binding to FtsA or ZipA, however, an FtsZ filament displays strong binding due to avidity (many CCTPs binding many membrane anchors). The intrinsically disordered regions (linker) in FtsZ, ZipA and FtsA increase flexibility allowing interaction with their partners. The long linker in ZipA may help it snare FtsZ filaments while the long linker in FtsZ with the CCTP at the tip may function like a grappling hook.
Reconstitution experiments showed that either FtsA or ZipA can tether FtsZ filaments to the membrane consistent with either being able to support Z ring formation in vivo [7, 34, 51, 52]. Furthermore, FtsZ filaments form bundles that gradually form ring-like chiral structures with a diameter of ~1 μm. Since the bundles in the rings display motion due to treadmilling, the filaments within the bundle likely have the same orientation and their dynamics coordinated. Also, replacing the CCTP of FtsZ with a membrane targeting sequence (MTS) leads to chiral vortices on membranes, indicating that FtsA and ZipA are not required for this behavior [53]. In fact, membrane targeting may not be necessary as FtsZ forms ring-like structures in the presence of crowding agents or when expressed in yeast cells [54, 55], indicating that formation of ring-like structures is an intrinsic property of FtsZ. However, the membrane anchors have additional functions during division besides tethering FtsZ to the membrane.
The CCTP of FtsZ is highly conserved in the bacterial domain, although different proteins are used as membrane anchors [56, 57]. The CCTP is an example of a MoRF in a prokaryotic protein which are more prevalent in eukaryotic proteins [58]. Consistent with it being a MoRF the structure of the CCTP bound to various partners varies indicating it is conformationally flexible with the bound structure dictated by the partner [43, 46, 48, 59]. In vitro reconstitution experiments show that FtsZ monomers (or the CCTP peptide) bind ZipA and FtsA with the typical weak affinity of a MoRF (20-50 μM), whereas oligomers bind with high affinity (in the nM range) [43, 60]. This differential affinity is not only seen with ZipA, but also with other proteins that bind the CCTP, such as the spatial regulator SlmA [60]. This differential affinity ensures that FtsZ polymers are tethered to the membrane but once a subunit of FtsZ dissociates from a polymer it will also dissociate from ZipA or FtsA while FtsA and ZipA can bind to additional CCTPs within a filament. This differential affinity for polymers versus monomers is likely critical for FtsZ filaments to treadmill on the membrane and also allows negative spatial regulators to target filaments.
The bundling dilemma
Since the Z ring is composed of patches of FtsZ that presumably contain multiple FtsZ filaments, a question is whether formation of these structures depends on lateral interactions between FtsZ filaments. A term used widely in the literature for FtsZ lateral interactions is bundling, however, this term is often used to describe the effect on FtsZ filaments caused by proteins, such as ZapA, ZapC and ZapD, that can crosslink filaments in vitro. For clarity, we use bundling to describe FtsZ lateral interactions only and use crosslinking for the effect mediated by Zap proteins (Fig. 3).
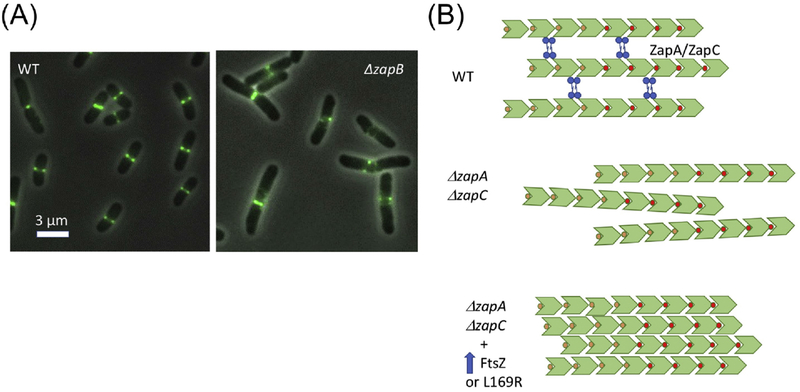
Figure 3. Bundling vs cross-linking of FtsZ filaments.
(A) Images of FtsZ-GFP in wild type (WT) and a mutant lacking the Ter signal (ΔzapB). (B) FtsZ filaments are present in patches in the Z ring. The filaments are presumed to be aligned in parallel based upon the directional movement seen in bundles and patches. In WT cells the primary interaction between filaments may be due to crosslinking proteins (ZapA, ZapC, and ZapD). In the absence of two of these (ZapA and ZapC) cells are elongated and cells contain spiral Z rings (similar to images in (A)). Overexpression of FtsZ or the presence of FtsZ-L169R may lead to increased bundling that compensates for the loss of Zap proteins.
FtsZ filaments readily form bundles or sheets in the presence of divalent cation metals [10, 61]. These bundles, however, do not contain repetitive subunit interactions. The dynamic bundles seen with the assembly of FtsZ-MTS on planar membranes is presumably similar. Although the C-terminal linker region and the CCTP can affect FtsZ filament bundling in vitro, whether they do so in vivo is not clear [62–64].
Genetic evidence for a role for bundling of FtsZ filaments in cell division rests on the behavior of various mutants. Some FtsZ mutants are affected in bundling in vitro and are also affected in Z-ring function. One such mutant R174D (Fig. 1B) polymerizes, but does not form bundles in the presence of calcium (a potent bundler) and bundles less than WT in other buffers suggesting that it may be defective in lateral interactions [65, 66]. The R174D mutant is incorporated into the Z ring and is dominant-negative but it is not clear why. However, the toxicity of R174D can be overcome by coexpression of another mutant, L169R, which is more prone to bundle [66, 67]. In vivo, L169R assembles into ring-like and helical structures at the division site that persist at the newly formed cell poles, consistent with stronger lateral interactions. Intriguingly, R174 was recently shown to be ADP-ribosylated by a Type VI secretion effector from Serratia proteamaculans [68]. This modification prevents FtsZ polymerization and results in a cell division block. The region around R174 is also a binding site for a toxin (from a chromosomal encoded toxin/antitoxin pair) that targets FtsZ as well as the actin homologue MreB involved in the elongasome [69, 70].
Other FtsZ mutants thought to be deficient for lateral interaction are K121L and D304L [71]. The affected residues are located at the lateral face of the FtsZ filament, but distinct from L169-R174. These two mutants were implicated in lateral interaction by photo-induced cross-linking. Furthermore, it is assumed the filaments are antiparallel which appears inconsistent with treadmilling patches. When expressed in wild type cells, the mutants localized to the Z ring presumably because they can co-polymerize with wild type FtsZ. However, these two mutants polymerized in vitro and displayed a similar degree of bundling as the wild type. Neither K121L nor D304L was functional in cell division, but it is not clear whether they are defective in Z ring formation or in a step after Z ring formation.
Despite evidence for FtsZ bundling, direct evidence for FtsZ lateral interaction is still missing. Furthermore, analysis of FtsZ filaments at the septum by cryo-tomography indicates that the space between individual filaments is too large for lateral interactions [72, 73]. This observation suggests that either the bundling of FtsZ filaments observed in vitro is not physiologically relevant or the cryo-tomography does not image all filaments. Thus, filament bundling in vivo is still a question and it may be that it occurs with some FtsZ mutants or when FtsZ is over expressed.
Cross-linking FtsZ filaments into a Z ring
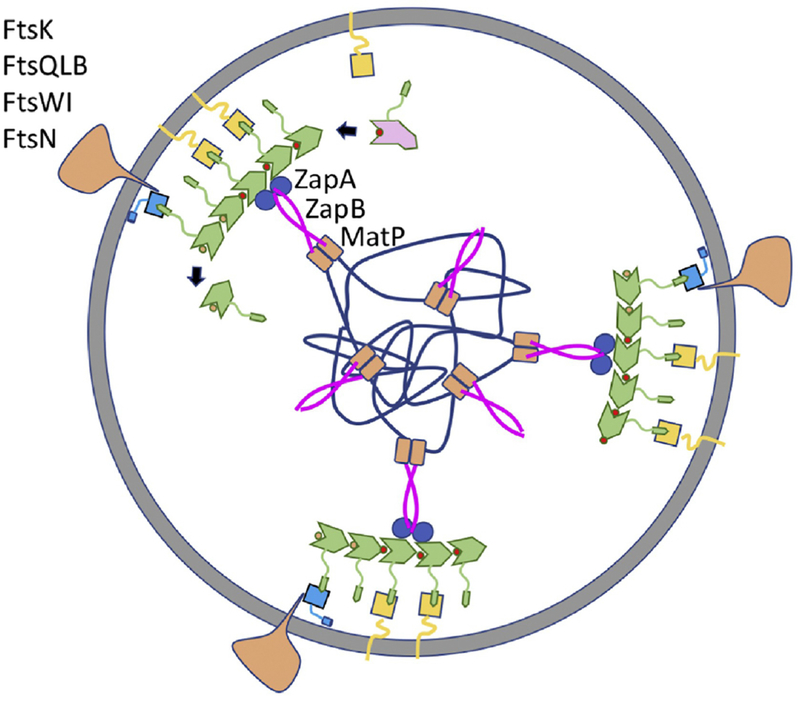
While it is uncertain whether FtsZ filaments bundle in vivo, many proteins can crosslink FtsZ filaments in vitro. These FtsZ associated proteins (Zaps) include ZapA, ZapC and ZapD. Importantly, deletion of all three results in increased cell filamentation and poor viability [74]. Among these ZapA is very widely conserved whereas the others appear to be restricted to enteric bacteria [75]. Although ZapC is a monomer, it contains two FtsZ binding sites so it can cross-link FtsZ filaments [76]. ZapD binds to the CCTP and forms dimers so it can also cross-link FtsZ filaments [48]. ZapA exists as dimers and tetramers so it too can crosslink filaments and does so in vitro, however, there is some question if it does so in vivo [77]. It is unique among the three in being part of a higher order structure (Ter signal, Fig. 4) that promotes cooperative and rapid ring assemblies in vivo with a very sharp appearance [17, 78]. The Ter signal forms in the absence of FtsZ and precedes formation of the Z ring [79]. ZapA is linked to MatP, which condenses the terminus region of the chromosome [80], through ZapB. Without ZapA or ZapB, FtsZ filaments often form a loose spiral structure instead of a Z ring (Fig. 3A) [77]. This deficiency is further exacerbated by the loss of ZapC or ZapD [74]. The hampered Z ring assembly in the absence of these proteins indicates crosslinking by Zap proteins is critical to form sharp Z rings in vivo and ensure that the Z ring does not spiral away which can result in aborted divisions [77].
Figure 4. Cell wall synthesis distributed by treadmilling FtsZ filaments linked to Ter.
FtsZ filaments attached to the membrane assemble into the Z ring (FtsZ, FtsA and ZipA colored as in Fig. 2), which consists of patches of FtsZ filaments that move circumferentially around the septum in both directions. Assembly at midcell is aided by the presence of the Ter signal consisting of ZapA bound to FtsZ linked to the MatP macrodomain by ZapB. The septal peptidoglycan synthetic machinery (depicted as a single large complex) consists of enzymes (FtsW-transglcosylase and Ftsl-transpeptidase) and proteins involved in their recruitment (FtsK, FtsQLB) and regulation (FtsQLB, FtsN).
Although attempts were made to distinguish FtsZ filament bundling from cross-linking, the line may not be so clear. One intriguing observation is that overexpression of FtsZ can largely suppress the division defect of a ΔzapA ΔzapC mutant, suggesting that increasing the number of FtsZ filaments may potentiate bundling and compensate for the loss of cross-linking [52]. Consistent with this, increasing the FtsZ level results in Z rings that have the same width but contain more FtsZ [77]. Furthermore, the division defect of the ΔzapA ΔzapC mutant can also be suppressed by the L169R mutant, which bundles more readily in vitro [52, 67]. Perhaps, bundling of FtsZ filaments is not very efficient in vivo so this task is aided by Zap proteins. However, when the level of FtsZ is increased or certain mutants are present, bundling occurs and compensates for the loss of cross-linking (Fig. 3).
Linking the Z ring with cell wall synthetic enzymes
The Z ring was postulated to guide peptidoglycan synthesis upon its discovery, but how was not clear [2, 6]. This idea was further reinforced with the observation that altered FtsZ structures lead to altered septal geometry [81]. Recent analysis of the dynamics of the Z ring and septal PG synthetic enzymes (FtsW and Ftsl [82]) demonstrated that FtsZ treadmills in vivo and that these enzymes follow the treadmilling patches of FtsZ filaments at the division site [15, 18]. Also, incorporation of septal PG appears to follow the FtsZ patches. These observations suggest that treadmilling FtsZ filaments distribute the incorporation of septal PG by linking up with the septal PG synthetic enzymes. But what is the linkage? Although FtsZ has been reported to interact with downstream division proteins in some bacterial species, this job is probably mediated by the membrane anchors in most bacteria. In E. coli the membrane anchor FtsA is the key player (ZipA can be readily bypassed) to bring the PG synthases to the Z ring [83]. FtsA recruits downstream division proteins (FtsK and FtsQLB) that recruit FtsW and Ftsl (Fig. 4). Mutations that bypass ZipA (ftsA*), impair FtsA’s self-interaction but enhance the interaction with the downstream protein FtsN [84, 85]. This led to a model in which polymeric FtsA is unable to interact with downstream proteins because the site for interaction with downstream proteins is buried in the self-interaction interface. Thus, polymerization of FtsA and its ability to link downstream proteins to the Z ring are mutually exclusive. ZipA may contribute to activating FtsA for recruitment by competing for the CCTP of FtsZ or possibly by interacting directly with FtsA [84, 86].
The pseudo-ABC transporter FtsEX is also important for linking the Z ring with downstream division proteins [87]. FtsEX localizes to the Z ring as it forms and has multiple functions in cell division including promoting assembly of the divisome, septal PG synthesis and cell separation [88, 89]. However, FtsEX can be bypassed by many conditions, indicating that it is a regulator of division and not a core component. FtsA interacts with FtsX but when this interaction is disrupted, downstream proteins are not recruited although both FtsA and FtsEX are at the Z ring [20]. This finding along with the observation that FtsA* mutants can bypass FtsEX, suggests that FtsEX regulates assembly of the divisome by antagonizing FtsA’s oligomerization status. Another protein that can link the Z ring with the cell wall synthetic enzymes is FtsN, the trigger that starts septal PG synthesis and is normally the last essential protein recruited to the Z ring [90, 91]. It interacts directly with FtsA [92], but its ability to recruit only comes into play when conditions exist that enhance the FtsA-FtsN interaction and allow the bypass of ZipA or FtsEX [85].
In another model, the polymerization status of FtsA is proposed to control the recruitment process by regulating the bundling of FtsZ protofilaments, although a mechanism is unclear [66]. FtsA was observed to form mini-rings on flat membranes consisting of 12 FtsA molecules that negatively affect the bundling of FtsZ filaments in vitro, whereas FtsA* which was unable to form mini-rings, enhanced the bundling of FtsZ filaments. Because ZipA was thought to cause FtsZ bundling in vitro and FtsA* bypasses ZipA, FtsZ bundling was proposed to be critical for the recruitment process. Since FtsA mini-rings antagonize the bundling of FtsZ filaments recruitment of downstream proteins is blocked. As the cell cycle proceeds, FtsA becomes less oligomeric such that FtsZ protofilaments bundle, resulting in the recruitment of downstream proteins and activation of septal PG synthesis. However, mini-rings of FtsA have not been seen in vivo and it is not clear how bundling of FtsZ filaments results in recruitment of downstream proteins. Recently, it was found that ZipA did not cause FtsZ bundling in vitro, arguing against the idea that enhanced bundling of FtsZ filaments accounts for the bypass of ZipA by FtsA* [52]. Nevertheless, these authors confirmed that FtsA* is less oligomeric and ZipA interacts with FtsA directly and negatively regulates its polymerization, supporting the first model.
Concluding Remarks
The Z ring is a universal feature of bacterial cytokinesis that have a cell wall and divide by binary fission. The more we focus on it the more details emerge and questions are raised, especially the organization of dynamic patches of FtsZ and how they distribute the enzymes that make septal PG. Nonetheless, in the past few years many things have been learned including that FtsZ treadmills and distributes septal PG machinery, the basis for cooperative assembly of FtsZ and how kinetic polarity is achieved. An important feature in Z ring construction is the use of proteins with regions of intrinsic disorder that allow flexibility in accommodating binding partners. Also, the use of a MoRF by FtsZ to bind so many of its partners allows FtsZ polymers to bind with high affinity whereas monomers bind with weak affinity. This feature is likely to be important for FtsZ dynamics and for FtsA and associated divisome proteins to track treadmilling filaments. The capacity of FtsZ to bundle in vitro may play a role in vivo whereas a role for crosslinking proteins is more established. Past studies on spatial regulation focused on the fascinating negative regulators of the Z ring which has overshadowed the positive regulation extending from the Ter region. Although not essential, at least in E. coli, it is possible that this feature is widespread in the bacterial world.
Outstanding Questions.
How is the width of Z rings maintained at ~100 nm, even if FtsZ levels are increased?
What is the organization of FtsZ filaments within patches in the ring?
Why do FtsZ filaments align in the same orientation and how is the treadmilling behavior of FtsZ filaments in bundles (in vitro) and in patches (in vivo) coordinated?
If lateral interaction of FtsZ filament exists in vivo, are the reported residues critical?
Can crosslinking of FtsZ filaments be replaced by bundling, and if so, what is the basis?
Is the Ter linkage widespread among bacteria and what is the structure?
How are divisome proteins other that FtsN linked to the Z ring via FtsA?
Highlights.
The Z ring is discontinuous and consists of patches of FtsZ assemblies that treadmill circumferentially at the division site.
The mechanism of FtsZ treadmilling was revealed by determination of the conformational changes that accompany assembly and the kinetic polarity of FtsZ filaments.
FtsZ employs an intrinsically disordered linker and a short linear motif (SLiM) to achieve flexibility and avidity in tethering FtsZ filaments to the membrane.
Evidence exists that crosslinking and bundling of FtsZ filaments contribute to assembly of the Z ring but how filaments are organized in patches awaits further investigation.
Treadmilling FtsZ filaments are coupled to the septal PG machinery through membrane anchors.
Acknowledgements
We thank Dr. Sebastien Pichoff for critical review of the manuscript. Work in the laboratory of Joe Lutkenhaus is funded by grant R01GM29764 from NIH. We apologize to those authors we could not cite due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Nanninga N (1991) Cell division and peptidoglycan assembly in Escherichia coli. Mol Microbiol 5 (4), 791–5. [DOI] [PubMed] [Google Scholar]
- 2.Lutkenhaus J (1993) FtsZ ring in bacterial cytokinesis. Mol Microbiol 9 (3), 403–9. [DOI] [PubMed] [Google Scholar]
- 3.Haeusser DP and Margolin W (2016) Splitsville: structural and functional insights into the dynamic bacterial Z ring. Nat Rev Microbiol 14 (5), 305–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Du S and Lutkenhaus J (2017) Assembly and activation of the Escherichia coli divisome. Mol Microbiol 105 (2), 177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lutkenhaus J et al. (2012) Bacterial cytokinesis: From Z ring to divisome. Cytoskeleton (Hoboken) 69 (10), 778–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bi EF and Lutkenhaus J (1991) FtsZ ring structure associated with division in Escherichia coli. Nature 354 (6349), 161–4. [DOI] [PubMed] [Google Scholar]
- 7.Pichoff S and Lutkenhaus J (2002) Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J 21 (4), 685–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osawa M et al. (2009) Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J 28 (22), 3476–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coltharp C et al. (2016) Defining the rate-limiting processes of bacterial cytokinesis. Proc Natl Acad Sci U S A 113 (8), E1044–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erickson HP et al. (2010) FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev 74 (4), 504–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strauss MP et al. (2012) 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis. PLoS Biol 10 (9), e1001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma X et al. (1996) Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci U S A 93 (23), 12998–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holden SJ et al. (2014) High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. Proc Natl Acad Sci U S A 111 (12), 4566–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyu Z et al. (2016) Influence of FtsZ GTPase activity and concentration on nanoscale Z-ring structure in vivo revealed by three-dimensional Superresolution imaging. Biopolymers 105 (10), 725–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang X et al. (2017) GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355 (6326), 744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez AJ et al. (2019) Movement dynamics of divisome proteins and PBP2x:FtsW in cells of Streptococcus pneumoniae. Proc Natl Acad Sci U S A 116 (8), 3211–3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buss J et al. (2015) A multi-layered protein network stabilizes the Escherichia coli FtsZ-ring and modulates constriction dynamics. PLoS Genet 11 (4), e1005128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bisson-Filho AW et al. (2017) Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355 (6326), 739–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du S et al. (2018) FtsZ filaments have the opposite kinetic polarity of microtubules. Proc Natl Acad Sci U S A 115 (42), 10768–10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du S et al. (2016) FtsEX acts on FtsA to regulate divisome assembly and activity. Proc Natl Acad Sci U S A 113 (34), E5052–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagstaff JM et al. (2017) A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments. MBio 8 (3) e00254–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukherjee A and Lutkenhaus J (1994) Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol 176 (9), 2754–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nogales E et al. (1998) Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol 5 (6), 451–8. [DOI] [PubMed] [Google Scholar]
- 24.de Boer PA et al. (1992) Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J Bacteriol 174 (1), 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang KH et al. (2013) FtsZ ring stability: of bundles, tubules, crosslinks, and curves. J Bacteriol 195 (9), 1859–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliva MA et al. (2004) Structural insights into FtsZ protofilament formation. Nat Struct Mol Biol 11 (12), 1243–50. [DOI] [PubMed] [Google Scholar]
- 27.Mukherjee A and Lutkenhaus J (1998) Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J 17 (2), 462–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desai A and Mitchison TJ (1997) Microtubule polymerization dynamics. Annu Rev Cell Dev Biol 13, 83–117. [DOI] [PubMed] [Google Scholar]
- 29.Chesarone MA and Goode BL (2009) Actin nucleation and elongation factors: mechanisms and interplay. Curr Opin Cell Biol 21 (1), 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miraldi ER et al. (2008) Allosteric models for cooperative polymerization of linear polymers. Biophys J 95 (5), 2470–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dajkovic A et al. (2008) Investigation of regulation of FtsZ assembly by SulA and development of a model for FtsZ polymerization. J Bacteriol 190 (7), 2513–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huecas S et al. (2008) Energetics and geometry of FtsZ polymers: nucleated self-assembly of single protofilaments. Biophys J 94 (5), 1796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michie KA and Lowe J (2006) Dynamic filaments of the bacterial cytoskeleton. Annu Rev Biochem 75, 467–92. [DOI] [PubMed] [Google Scholar]
- 34.Loose M and Mitchison TJ (2014) The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat Cell Biol 16 (1), 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lutkenhaus J (2007) Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem 76, 539–62. [DOI] [PubMed] [Google Scholar]
- 36.Arumugam S et al. (2014) MinCDE exploits the dynamic nature of FtsZ filaments for its spatial regulation. Proc Natl Acad Sci U S A 111 (13), E1192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Redick SD et al. (2005) Mutants of FtsZ targeting the protofilament interface: effects on cell division and GTPase activity. J Bacteriol 187 (8), 2727–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bisson-Filho AW et al. (2015) FtsZ filament capping by MciZ, a developmental regulator of bacterial division. Proc Natl Acad Sci U S A 112 (17), E2130–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y et al. (2012) SulA inhibits assembly of FtsZ by a simple sequestration mechanism. Biochemistry 51 (14), 3100–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gardner KA et al. (2013) The C-terminal linker of Escherichia coli FtsZ functions as an intrinsically disordered peptide. Mol Microbiol 89 (2), 264–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hale CA and de Boer PA (1997) Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88 (2), 175–85. [DOI] [PubMed] [Google Scholar]
- 42.Pichoff S and Lutkenhaus J (2007) Identification of a region of FtsA required for interaction with FtsZ. Mol Microbiol 64 (4), 1129–38. [DOI] [PubMed] [Google Scholar]
- 43.Szwedziak P et al. (2012) FtsA forms actin-like protofilaments. EMBO J 31 (10), 2249–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dafforn TR and Smith CJ (2004) Natively unfolded domains in endocytosis: hooks, lines and linkers. EMBO Rep 5 (11), 1046–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du S and Lutkenhaus J (2014) SlmA Antagonism of FtsZ Assembly Employs a Two-pronged Mechanism like MinCD. PLoS Genet 10 (7), e1004460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schumacher MA and Zeng W (2016) Structures of the nucleoid occlusion protein SlmA bound to DNA and the C-terminal domain of the cytoskeletal protein FtsZ. Proc Natl Acad Sci U S A 113 (18), 4988–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen B and Lutkenhaus J (2009) The conserved C-terminal tail of FtsZ is required for the septal localization and division inhibitory activity of MinC(C)/MinD. Mol Microbiol 72 (2), 410–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schumacher MA et al. (2017) Structure of the Z Ring-associated Protein, ZapD, Bound to the C-terminal Domain of the Tubulin-like Protein, FtsZ, Suggests Mechanism of Z Ring Stabilization through FtsZ Cross-linking. J Biol Chem 292 (9), 3740–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li GW et al. (2014) Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157 (3), 624–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohashi T et al. (2002) Structural evidence that the P/Q domain of ZipA is an unstructured, flexible tether between the membrane and the C-terminal FtsZ-binding domain. J Bacteriol 184 (15), 4313–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conti J et al. (2018) FtsA reshapes membrane architecture and remodels the Z-ring in Escherichia coli. Mol Microbiol 107 (4), 558–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krupka M et al. (2018) Escherichia coli ZipA Organizes FtsZ Polymers into Dynamic Ring-Like Protofilament Structures. MBio 9 (3) e01008–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramirez-Diaz DA et al. (2018) Treadmilling analysis reveals new insights into dynamic FtsZ ring architecture. PLoS Biol 16 (5), e2004845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Srinivasan R et al. (2008) The bacterial cell division protein FtsZ assembles into cytoplasmic rings in fission yeast. Genes Dev 22 (13), 1741–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Popp D et al. (2009) FtsZ condensates: an in vitro electron microscopy study. Biopolymers 91 (5), 340–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duman R et al. (2013) Structural and genetic analyses reveal the protein SepF as a new membrane anchor for the Z ring. Proc Natl Acad Sci U S A 110 (48), E4601–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krol E et al. (2012) Bacillus subtilis SepF binds to the C-terminus of FtsZ. PLoS One 7 (8), e43293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan J et al. (2016) Molecular recognition features (MoRFs) in three domains of life. Mol Biosyst 12 (3), 697–710. [DOI] [PubMed] [Google Scholar]
- 59.Mosyak L et al. (2000) The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J 19 (13), 3179–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Du S et al. (2015) Oligomerization of FtsZ converts the FtsZ tail motif (conserved carboxy-terminal peptide) into a multivalent ligand with high avidity for partners ZipA and SlmA. Mol Microbiol 95 (2), 173–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mukherjee A and Lutkenhaus J (1999) Analysis of FtsZ assembly by light scattering and determination of the role of divalent metal cations. J Bacteriol 181 (3), 823–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huecas S et al. (2017) Self-Organization of FtsZ Polymers in Solution Reveals Spacer Role of the Disordered C-Terminal Tail. Biophys J 113 (8), 1831–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sundararajan K and Goley ED (2017) The intrinsically disordered C-terminal linker of FtsZ regulates protofilament dynamics and superstructure in vitro. J Biol Chem 292 (50), 20509–20527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buske PJ and Levin PA (2012) Extreme C terminus of bacterial cytoskeletal protein FtsZ plays fundamental role in assembly independent of modulatory proteins. J Biol Chem 287 (14), 10945–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koppelman CM et al. (2004) R174 of Escherichia coli FtsZ is involved in membrane interaction and protofilament bundling, and is essential for cell division. Mol Microbiol 51 (3), 645–57. [DOI] [PubMed] [Google Scholar]
- 66.Schoenemann KM et al. (2018) Gain-of-function variants of FtsA form diverse oligomeric structures on lipids and enhance FtsZ protofilament bundling. Mol Microbiol 109 (5), 676–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haeusser DP et al. (2015) A mutation in Escherichia coli ftsZ bypasses the requirement for the essential division gene zipA and confers resistance to FtsZ assembly inhibitors by stabilizing protofilament bundling. Mol Microbiol 97 (5), 988–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ting SY et al. (2018) Bifunctional Immunity Proteins Protect Bacteria against FtsZ-Targeting ADP-Ribosylating Toxins. Cell 175 (5), 1380–1392 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan Q et al. (2011) YeeV is an Escherichia coli toxin that inhibits cell division by targeting the cytoskeleton proteins, FtsZ and MreB. Mol Microbiol 79 (1), 109–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heller DM et al. (2017) CbtA toxin of Escherichia coli inhibits cell division and cell elongation via direct and independent interactions with FtsZ and MreB. PLoS Genet 13 (9), e1007007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guan F et al. (2018) Lateral interactions between protofilaments of the bacterial tubulin homolog FtsZ are essential for cell division. Elife 7, e35578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Szwedziak P et al. (2014) Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. Elife 3, e04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Z et al. (2007) The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J 26 (22), 4694–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Durand-Heredia J et al. (2012) Identification of ZapD as a cell division factor that promotes the assembly of FtsZ in Escherichia coli. J Bacteriol 194 (12), 3189–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gueiros-Filho FJ and Losick R (2002) A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev 16 (19), 2544–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schumacher MA et al. (2016) Structural and Functional Analyses Reveal Insights into the Molecular Properties of the Escherichia coli Z Ring Stabilizing Protein, ZapC. J Biol Chem 291 (5), 2485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Buss J et al. (2013) In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol Microbiol 89 (6), 1099–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bailey MW et al. (2014) Evidence for divisome localization mechanisms independent of the Min system and SlmA in Escherichia coli. PLoS Genet 10 (8), e1004504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buss JA et al. (2017) ZapA and ZapB form an FtsZ-independent structure at midcell. Mol Microbiol 104 (4), 652–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mercier R et al. (2008) The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell 135 (3), 475–85. [DOI] [PubMed] [Google Scholar]
- 81.Addinall SG and Lutkenhaus J (1996) FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol Microbiol 22 (2), 231–7. [DOI] [PubMed] [Google Scholar]
- 82.Taguchi A et al. (2019) FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat Microbiol. 4(4):587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Geissler B et al. (2003) A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc Natl Acad Sci U S A 100 (7), 4197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pichoff S et al. (2012) FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA’s self-interaction competes with its ability to recruit downstream division proteins. Mol Microbiol 83 (1), 151–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pichoff S et al. (2018) Disruption of divisome assembly rescued by FtsN-FtsA interaction in Escherichia coli. Proc Natl Acad Sci U S A. 115(29):E6855–E6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vega DE and Margolin W (2019) Direct Interaction between the Two Z Ring Membrane Anchors FtsA and ZipA. J Bacteriol 201 (4) e00579–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schmidt KL et al. (2004) A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J Bacteriol 186 (3), 785–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang DC et al. (2011) An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc Natl Acad Sci U S A 108 (45), E1052–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arends SJ et al. (2009) ATP-binding site lesions in FtsE impair cell division. J Bacteriol 191 (12), 3772–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu B et al. (2015) Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Mol Microbiol 95 (6), 945–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gerding MA et al. (2009) Self-enhanced accumulation of FtsN at Division Sites and Roles for Other Proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J Bacteriol 191 (24), 7383–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Busiek KK et al. (2012) The early divisome protein FtsA interacts directly through its 1c subdomain with the cytoplasmic domain of the late divisome protein FtsN. J Bacteriol 194 (8), 1989–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Matsui T et al. (2012) Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus. Acta Crystallogr D Biol Crystallogr 68 (Pt 9), 1175–88. [DOI] [PubMed] [Google Scholar]
- 94.Elsen NL et al. (2012) Mechanism of action of the cell-division inhibitor PC190723: modulation of FtsZ assembly cooperativity. J Am Chem Soc 134 (30), 12342–5. [DOI] [PubMed] [Google Scholar]
- 95.Mannik J and Bailey MW (2015) Spatial coordination between chromosomes and cell division proteins in Escherichia coli. Front Microbiol 6, 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Woldemeskel SA et al. (2017) A conserved coiled-coil protein pair focuses the cytokinetic Z-ring in Caulobacter crescentus. Mol Microbiol 105 (5), 721–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brezellec P et al. (2006) DomainSieve: a protein domain-based screen that led to the identification of dam-associated genes with potential link to DNA maintenance. Bioinformatics 22 (16), 1935–41. [DOI] [PubMed] [Google Scholar]