Abstract
Recent evidence suggests that numerous long non-coding RNAs (lncRNAs) are dysregulated in cancer, and have critical roles in tumour development and progression. The present study investigated the ghrelin receptor antisense lncRNA growth hormone secretagogue receptor opposite strand (GHSROS) in breast cancer. Reverse transcription-quantitative polymerase chain reaction revealed that GHSROS expression was significantly upregulated in breast tumour tissues compared with normal breast tissue. Induced overexpression of GHSROS in the MDA-MB-231 breast cancer cell line significantly increased cell migration in vitro, without affecting cell proliferation, a finding similar to our previous study on lung cancer cell lines. Microarray analysis revealed a significant repression of a small cluster of major histocompatibility class II genes and enrichment of immune response pathways; this phenomenon may allow tumour cells to better evade the immune system. Ectopic overexpression of GHSROS in the MDA-MB-231 cell line significantly increased orthotopic xenograft growth in mice, suggesting that in vitro culture does not fully capture the function of this lncRNA. This study demonstrated that GHSROS may serve a relevant role in breast cancer. Further studies are warranted to explore the function and therapeutic potential of this lncRNA in breast cancer progression.
Keywords: long non-coding RNA, ghrelin receptor gene, antisense transcript, breast cancer, cell migration, tumour growth
Introduction
The human genome produces coding and non-coding RNA (ncRNA) transcripts; however, ncRNAs represent the predominant RNA species (1). Long ncRNAs (lncRNAs) are >200 nucleotides in length, have low protein-coding potential, and often exhibit spatial- and temporal-specific expression patterns (2). They are important regulators of gene expression, acting through a range of different mechanisms, in cis or trans (3,4). Given their broad role in a diverse range of biological processes, it is appreciated that they serve major roles in physiology and disease (5). Notably, a large number of lncRNAs have been reported to mediate cancer-associated processes (3). However, the role and expression patterns of the majority of lncRNAs in cancer remain largely unknown.
Breast cancer is the most commonly diagnosed cancer and a leading cause of cancer-associated mortality in women (6). Given the significant incidence of breast cancer in the population, there is a need to explore the therapeutic potential of novel molecular targets, particularly for triple-negative breast cancer, which is diagnosed in 15% of patients with breast cancer (7). Due to a lack of targeted therapies, these patients require more aggressive treatment regimens (7). Historically, molecular classification and therapeutic targeting of breast cancer-associated genes has focused on protein-coding genes, which represent <1% of the genome (8). It is now appreciated that many lncRNAs are feasible biomarkers and targets for molecular therapies (9-12). A key example includes HOX transcript antisense intergenic RNA (HOTAIR), which is upregulated in primary and metastatic breast tumours (13-15). Overexpression of HOTAIR in breast cancer activates an oestrogen receptor (ER)-associated transcriptional program, to enhance cancer growth and tamoxifen resistance in breast cancer (16). Similarly, the highly conserved and abundant lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is upregulated in a broad range of tumour types, including metastatic breast cancer, and stimulates cell proliferation and migration (17). Furthermore, targeting MALAT1 in a mouse model of mammary carcinoma using modified antisense oligonucleotides (ASOs) significantly reduces breast cancer metastasis and slows primary tumour growth (18). Taken together, these studies highlight the value of studying the expression and function of lncRNAs in breast cancer.
The lncRNA growth hormone secretagogue receptor (GHSR) opposite strand (GHSROS) (19) is a single-exon gene antisense to the intronic region of GHSR; GHSR is also known as the ghrelin receptor gene (19). Our previous study demonstrated that GHSROS expression is elevated in non-small cell lung cancer and that its induced overexpression increases migration in lung adenocarcinoma cell lines (19). However, to the best of our knowledge, the expression pattern and functional role of GHSROS in breast cancer remains unknown. The present study analysed the expression of GHSROS in breast tissues and derived cell lines, and determined the effects of GHSROS overexpression in vitro (MDA-MB-231 and MCF10A cell lines) and in vivo (MDA-MB-231 tumour xenografts in mice).
Materials and methods
Cell culture
Cell lines were obtained from the American Type Culture Collection (ATCC). The MDA-MB-231 (HTB-26), MDA-MB-468 (HTB-132) and MDA-MB-453 (HTB-131) breast cancer cell lines were maintained in Dulbecco's modified Eagle's medium:Nutrient Mixture F-12 medium (DMEM/F12) supplemented with 10% foetal bovine serum (FBS), 100 U/ml penicillin G and 100 µg/ml streptomycin (all from Thermo Fisher Scientific, Inc., Waltham, MA, USA). The MCF10A breast-derived non-tumourigenic cell line (CRL-10317) was maintained in DMEM/F12 supplemented with 5% heat-inactivated horse serum, 50 U/ml penicillin G, 100 µg/ml streptomycin, 10 µg/ml bovine insulin (all from Thermo Fisher Scientific, Inc.), 0.5 µg/ml hydrocortisone, 200 µg/ml cholera toxin (both from Sigma Aldrich; Merck KGaA) and 20 ng/ml epidermal growth factor (Thermo Fisher Scientific, Inc.). The T-47D (HTB-133) and MCF-7 (HTB-22) cell lines were maintained in RPMI-1640 medium supplemented with 10% FBS, 100 U/ml penicillin G, 100 µg/ml streptomycin and 10 µg/ml bovine insulin (Thermo Fisher Scientific, Inc.). The HMEC human mammary epithelial cell-derived, non-malignant cell line (PCS-600-010) was grown in HuMEC Ready Medium and Supplement kit (Thermo Fisher Scientific, Inc.). All cell lines were maintained at 37°C in a humidified incubator (Panasonic) containing 5% CO2; cells were grown in tissue culture flasks (Corning, Inc.), and passaged at 2- to 3-day intervals upon reaching 70% confluence using TrypLE Select (Thermo Fisher Scientific, Inc.). Cell morphology and viability were monitored by microscopic observation and regular Mycoplasma testing was performed (Universal Mycoplasma Detection kit; ATCC). Approval for cell line use was granted by the Queensland University of Technology (QUT) Human Research Ethics Committee (Brisbane, Australia).
Production of GHSROS-overexpressing cell lines
For gain-of-function studies, full-length GHSROS was generated as previously described (19). The full-length GHSROS transcript, amplified from the A549 (CCL-185; ATCC) lung adenocarcinoma cell line, was cloned into the pTargeT mammalian expression vector (Promega Corporation). MDA-MB-231 and MCF10A cell lines were transfected with 1 µg GHSROS-pTargeT plasmid DNA or vector alone (empty vector) using Lipofectamine LTX (Invitrogen; Thermo Fisher Scientific, Inc.) as per the manufacturer's instructions. Cells (2×105/well) were seeded in a 6-well plate 24 h prior to transfection. Following incubation at room temperature for 5 min, cells were transfected for 24 h at 37°C in Lipofectamine LTX and further selected with geneticin (G418; Invitrogen; Thermo Fisher Scientific, Inc.) at concentrations of 500 µg/ml for MCF10A and 600 µg/ml for MDA-MB-231 cells. Transfected cells were grown in the presence of G418 for ≥2 weeks prior to the performance of functional analyses. For in vivo xenograft experiments, MDA-MB-231 cells stably overexpressing luciferase pGL4.51[luc2/CMV/Neo] (MDA-MB-231luc) were obtained (provided by Dr Eloïse Dray, QUT). pReceiver-Lv105 lentiviral vectors (containing full-length GHSROS or no insert) pre-packaged in lentiviral particles were purchased from GeneCopoeia, Inc. Briefly, to optimise transduction, a titration of 0.1-10 µl viral particles was performed in the PC3 prostate cancer cell line (CRL-1435; ATCC) and the lowest, most viable dose was used. MDA-MB-231 cells were seeded at 50-60% confluence and were transduced with GHSROS or empty vector control lentiviral constructs at a multiplicity of infection of 1 in the presence of 8 µg/ml polybrene (Sigma-Aldrich; Merck KGaA) according to the manufacturer's protocol. After 48 h at 37°C, transduced cells were selected with 1 µg/ml puromycin (Invitrogen; Thermo Fisher Scientific, Inc.) and expanded. GHSROS expression was confirmed ~3 weeks after selection by reverse transcription-quantitative polymerase chain reaction (RT-qPCR), then every 2-3 weeks and prior to every functional experiment.
RNA extraction and RT-qPCR
Cell lines were centrifuged at 133 × g for 5 min and total RNA was extracted from cell pellets using an RNeasy Plus Mini kit and a genomic DNA (gDNA) eliminator spin column (Qiagen GmbH). Total RNA was quantified using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc., Wilmington, DE, USA), diluted 1:5 in RNase-free water, and frozen at -80°C until further use. To remove contaminating gDNA, 1 µg RNA was treated with DNase prior to cDNA synthesis with Superscript III (Invitrogen; Thermo Fisher Scientific, Inc.); RT was performed according to the manufacturer's protocol. RT-qPCR was performed with SYBR-Green PCR Master Mix (Qiagen GmbH) using primers listed in Table I on an AB7500 FAST sequence detection thermal cycler (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) or a ViiA Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). PCR cycles were performed under the following conditions: 10 min at 95°C; 40 cycles of 15 sec at 95°C and 1 min at 60°C, followed by melting curve analysis. All experiments were performed in triplicate. A negative control (water instead of template) was used for each primer set. Baseline and quantification cycle (Cq) values were obtained using ABI 7500 Prism and the relative expression levels of mRNA were calculated using the comparative 2−ΔΔCq method (20). Expression was normalised to the housekeeping gene β-actin (ACTB). Statistical analyses were performed using GraphPad Prism v.6.01 software (GraphPad Software, Inc.).
Table I.
Primer sequences used in the present study.
| Primer | Gene name | Primer sequence (5′-3′) |
|---|---|---|
| GHSROS | Growth hormone secretagogue receptor opposite strand | F: ACATTCAGCAAATCCAGTTAATGACA R: CGACTGGAGCACGAGGACACTTGA |
| GHSROS-RT linker | Growth hormone secretagogue receptor opposite strand | CGACTGGAGCACGAGGACACTGACAACAGAATTCACTACTTCCCCAAA |
| ACTB | β-actin (housekeeping gene) | F: ACTCTTCCAGCCTTCCTTCCT R: CAGTGATCTCCTTCTGCATCCT |
| HLA-DRB3 | MHC class II, DR β3 | QIAGEN QuantiTect Primer Assay |
| HLA-DRA | MHC class II, DR α | QIAGEN QuantiTect Primer Assay |
| HLA-DPB1 | MHC, class II, DP β1 | QIAGEN QuantiTect Primer Assay |
| HLA-DPA1 | MHC class II, DP α1 | QIAGEN QuantiTect Primer Assay |
| TBX3 | T-box 3 | QIAGEN QuantiTect Primer Assay |
| HTR1F | 5-hydroxytryptamine receptor 1F | QIAGEN QuantiTect Primer Assay |
| TENM1 | Teneurin transmembrane protein 1 | QIAGEN QuantiTect Primer Assay |
F, forward primer; R, reverse primer; RT, reverse transcription; MHC, major histocompatibility complex.
GHSROS expression in human tissue specimens
The expression of GHSROS in human breast tissue was quantified by RT-qPCR (as aforementioned) using cDNA panels of breast tumour and normal breast tissue samples. Briefly, TissueScan Cancer Survey Tissue qPCR panels BCRT101, BCRT102, BCRT103 and BCRT104 were arrayed onto a single 384-well reaction plate by OriGene Technologies, Inc. Data are expressed as mean fold-change using the comparative 2−ΔΔCq method (compared to non-malignant control tissue on the same plate and normalised to ACTB).
Cell proliferation assays
Proliferation assays were performed using an xCELLigence real-time cell analysis (RTCA) DP instrument (ACEA Biosciences, Inc.). This system employs sensor impedance technology to quantify the status of the cell using a unit-less parameter termed the cell index (CI) (21). The CI represents the status of the cell based on the measured relative alterations in electrical impedance that occur in the presence and absence of cells in the wells [generated by the software, according to the formula CI = (Zi-Z0)/15 Ω, where Zi is the impedance at an individual point of time during the experiment and Z0 is the impedance at the start of the experiment] (21,22). Impedance is measured at three different frequencies (10, 25 and 50 kHz). Briefly, 5×103 cells were trypsinised, seeded into a 96-well plate (E-plate) and grown for 48 h at 37°C in 150 µl growth media. The CI was measured every 15 min and all experiments were performed in triplicate, with at least three independent repeats.
Cell migration assays
Migration assays were performed using an xCELLigence RTCA DP instrument (ACEA Biosciences, Inc.), and cell invasion and migration plates (CIM-Plate®; ACEA Biosciences, Inc.). Briefly, 5×104 cells/well in 150 µl serum-free media were seeded on the upper electronically integrated Boyden chamber above a membrane with a pore size of 8-µm. The lower chamber contained 160 µl media supplemented with 10% FBS as a chemoattractant. The CI was measured every 15 min for 18 h (as an indication of the rate of cell migration onto the underside of the top chamber). All experiments were performed in triplicate, with at least three independent repeats.
Oligonucleotide microarray analysis
Oligonucleotide micro-array analysis was performed using RNA extracted from MDA-MB-231 cell lines overexpressing GHSROS or vector controls. The MDA-MB-231 breast cancer cell line (three biological replicates) was transfected independently three times with GHSROS-pTargeT or empty pTargeT vector as aforementioned, and RNA was extracted and its purity analysed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.). Total RNA (500 ng) was processed and hybridised to Affymetrix Human Gene Arrays 1.0 by the Ramaciotti Centre for Gene Function Analysis (Sydney, Australia). The array (n=3, GHSROS-pTargeT and empty control) was quantile normalised and log2-transformed using the R statistical programming language v3.12 (www.r-project.org). Gene annotations were obtained from the National Center for Biotechnology Information (Platform GPL6244; downloaded September 2017; www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL6244). Differential expression was determined using R package 'limma' v3.33.12 (23). Differentially expressed genes were defined as an absolute fold-change ≥1.5 and P≤0.05.
To identify associations between differentially expressed genes, STRING (v10.5) functional analysis (24) was implemented. STRING integrates predicted and experimentally confirmed relationships between proteins that are likely to contribute to a common biological purpose. Using default parameters, differentially expressed genes were mapped into the STRING user interface and interactions were partitioned into distinct clusters using k-means analysis (25). Interaction networks were exported into Inkscape (v0.91; inkscape.org). To test for gene enrichment, differentially expressed genes were analysed using the Kyoto Encylopedia of Genes and Genomes (KEGG) (26) pathway database within the STRING functional analysis tool. STRING categorises KEGG pathway terms and calculates an 'enrichment score' or EASE score (a modified Fisher's exact test-derived P-value).
Orthotopic mammary fat pad in vivo xenografts in mice
Experiments were approved by the University of Queensland (Brisbane, Australia) and QUT animal ethics committees (TRI/QUT/328/16). Mice were housed under specific pathogen-free conditions in individually ventilated cages (Tecniplast S.p.A.) at room temperature (20-23°C), 40-60% relative humidity and under a 12-h light/dark cycle. Food and water were provided ad libitum. MDA-MB-231-GHSROS or MDA-MB-231-Vector cell lines were injected at a 1:1 ratio with growth factor-reduced Matrigel (Corning, Inc.) (n=8-10/cell line) directly into the right inguinal mammary fat pad of 3-week old female NOD.Cg-Prkdc SCID IL-2rgtm1WjL/SzJ (NSG) mice (weight, 20-25 g; generated by the Jackson Laboratory; provided by Animal Resource Centre). Tumour growth was measured twice weekly with digital calipers (ProSciTech Pty Ltd.) and tumour volume was calculated using a formula for the volume of an ellipse: V = π/6(d1 × d2)3/2, where d1 and d2 represent perpendicular tumour measurements (27). In addition, tumour size and growth were monitored weekly by bioluminescent imaging (28). Briefly, mice were injected intraperitoneally with 150 mg/kg firefly luciferase substrate D-Luciferin (PerkinElmer, Inc.) diluted in PBS (Thermo Fisher Scientific, Inc.) 10 min prior to imaging. Following anaesthesia with isoflurane, bioluminescent imaging was performed using an IVIS Spectrum in vivo imaging system (PerkinElmer, Inc.) (28). Images were analysed using the associated Living Image Software (PerkinElmer, Inc.; version 4.7.1). Briefly, total flux in photons/sec (p/s) was used as a surrogate for primary tumour size and determined within a defined region of interest, individually, for each mouse (28). Animals were sacrificed once tumour volume reached 1,000 mm3, or earlier according to other ethical endpoints.
Statistical analyses
Data are presented as the mean ± standard error of the mean of at least two independent experiments, and were evaluated by Student's t-test, one-way or two-way ANOVA with Dunnett's or Bonferroni's post hoc test, Mann-Whitney-Wilcoxon test or Kruskal-Wallis test with Dunn's post hoc test for multiple groups. P≤0.05 was considered to indicate a statistically significant difference. Statistical analyses were performed using GraphPad Prism v.6.01 (GraphPad Software, Inc.).
Results
GHSROS expression is elevated in breast cancer tissue and breast cancer-derived cell lines
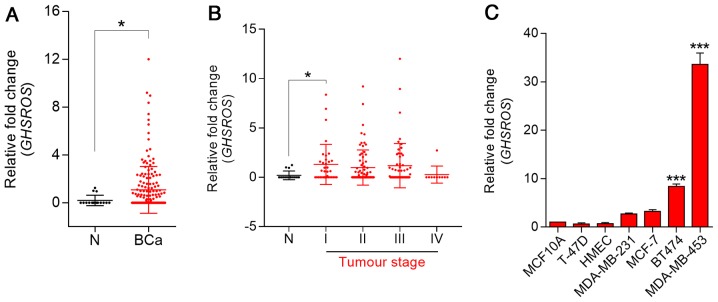
To investigate the role of GHSROS in breast cancer, RT-qPCR was performed on cDNA array panels of normal breast and breast cancer tissues. GHSROS expression could be detected in three out of 16 normal breast tissue samples (Fig. 1A). In contrast, 47% (83/176) of tumours expressed GHSROS at significantly higher levels than the normal breast tissues examined (P=0.030). In addition, GHSROS was significantly elevated in stage I (P=0.0077) breast cancer samples (Fig. 1B; Table II). GHSROS expression was not associated with a range of other clinical parameters and features, including age, hormone receptor status [ER, progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2)] and metastasis (Table II). Similar to the normal tissue specimens, very low levels of GHSROS expression were observed in the MCF10A and HMEC non-malignant cell lines compared with most breast cancer cell lines (Fig. 1C). GHSROS was expressed at higher levels in triple-negative (ER−, PR− and HER2−) MDA-MB-231 (2.71±0.27 fold-change, P=0.3601) and MDA-MB-453 (33.60±3.33 fold-change, P=0.0001) cell lines, and in the ER−/PR+/HER2+ BT474 (8.36±0.94 fold-change, P=0.0001) and ER+/PR+/HER2+ MCF7 (3.28±0.65 fold-change, P=0.0652) cell lines compared with the MCF10A cell line (Fig. 1C).
Figure 1.
GHSROS is expressed at low levels in normal breast tissue and at higher levels in breast cancer (BCa). (A) GHSROS expression in clinical BCa (n=176) and N breast samples (n=16), as determined by RT-qPCR. *P≤0.05, Mann-Whitney-Wilcoxon test. (B) GHSROS expression stratified by clinical stage of BCa, as determined by RT-qPCR. N (n=16), stage I (n=35), stage II (n=74), stage III (n=57) and stage IV (n=10). *P≤0.05, Kruskal-Wallis with Dunn's post hoc test. (C) GHSROS expression in the MCF10A and HMLE normal-breast derived cell lines, and the MDA-MB-231, MCF-7, T-47D, BT474 and MDA-MB-453 BCa cell lines compared to the MCF-10A cell line, as determined by RT-qPCR. ***P≤0.001 vs. MCF-10A, one-way ANOVA with Dunnett's post hoc test. All experiments were performed independently three times (n=3) with three replicates per experiment (n=3). Data are presented as the mean ± standard error of the mean, and relative expression levels were determined using the comparative 2−ΔΔCq method. Samples were normalised using β-actin. BCa, breast cancer; GHSROS, growth hormone secretagogue receptor opposite strand; N, normal; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Table II.
GHSROS expression, as determined by reverse transcription-quantitative polymerase chain reaction, and clinicopathological parameters in breast cancer and normal breast clinical specimens.
| Clinicopathological parameters | Total number (n) | Sample n demonstrating expression | Sample % demonstrating expression | P-value |
|---|---|---|---|---|
| N/T | 16/176 | 3/83 | 12.5/47.2 | 0.0300a |
| Clinical stage | ||||
| N | 16 | 2 | 12.5 | NA |
| I | 35 | 20 | 57.1 | 0.0406a |
| II | 74 | 33 | 44.6 | 0.2364 |
| III | 57 | 27 | 47.4 | 0.1359 |
| IV | 10 | 1 | 10 | >0.9999 |
| Clinical features | ||||
| ER+/ER− | 74/59 | 32/33 | 43.2/55.9 | 0.3829 |
| PR+/PR− | 67/57 | 27/29 | 40.3/50.9 | 0.3402 |
| HER2+/HER2− | 42/93 | 18/44 | 42.9/47.3 | 0.6185 |
| ER−, PR−, HER2−/Other | 33/152 | 17/66 | 51.5/43.4 | 0.7110 |
| M/Non-M | 17/159 | 5/76 | 29.4/47.8 | 0.4236 |
GHSROS expression in tumour tissues stratified by clinical stage and clinical features was compared to normal breast tissues. All tumour tissue was compared against normal breast tissue. Clinical features were assessed within tumour tissues. P-values were calculated using the Kruskal-Wallis with Dunn's post hoc test for multiple comparisons. ER, oestrogen receptor; HER2, human epidermal growth factor receptor 2; M, metastatic tumour; non-M, non-metastasised tumour (primary breast tumour confined to site); N, normal breast tissue; NA, non-applicable; PR, progesterone receptor; T, tumour tissue.
P≤0.05 compared with N. For some samples, clinical information was missing; therefore, these samples were excluded from that analysis. The mean age at diagnosis was 55.8 years.
Ectopic overexpression of GHSROS in breast-derived cell lines promotes in vitro migration, but not in vitro proliferation
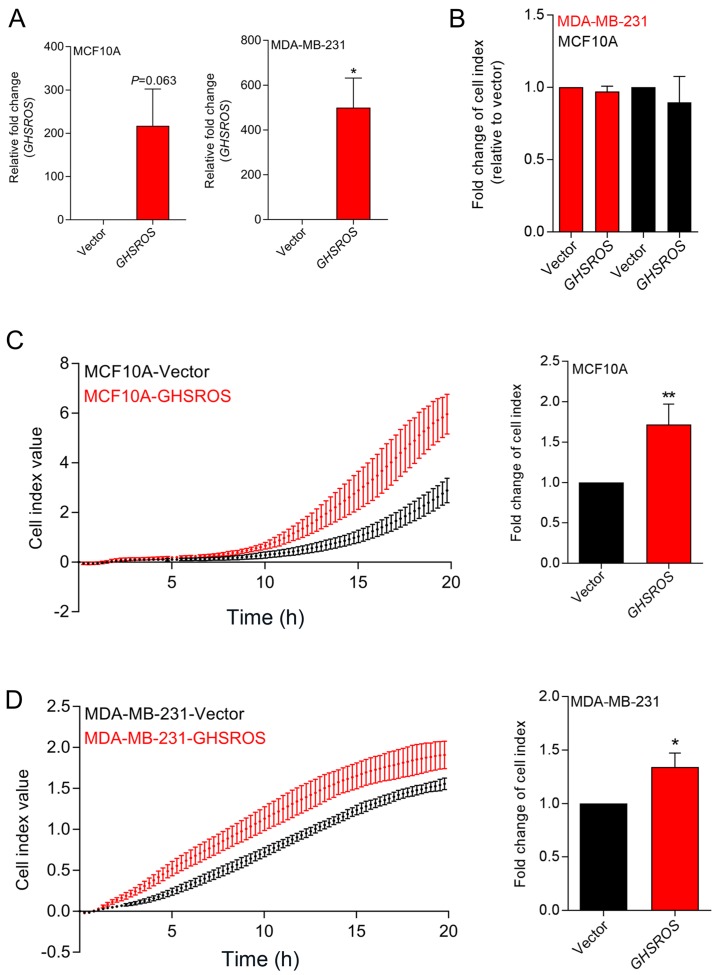
In order to investigate GHSROS function, MCF10A normal breast-derived cells and MDA-MB-231 breast cancer cells stably overexpressing full-length GHSROS (MCF10A-GHSROS and MDA-MB-231-GHSROS), and corresponding vector control cell lines (MCF10A-Vector and MDA-MB-231-Vector), were generated (Fig. 2A). The MDA-MB-231 cell line was chosen for overexpression studies as it has relatively low endogenous levels of GHSROS. Using real-time cell analysis (xCELLigence system), it was observed that GHSROS overexpression did not significantly increase the in vitro proliferation rate of cultured MCF10A (0.96±0.04 fold-change, P=0.18) and MDA-MD-231 cell lines (0.89±0.81 fold-change, P=0.29) over 72 h (Fig. 2B). Conversely, overexpression of GHSROS significantly increased the rate of migration of MCF10A (1.71±0.43 fold-change, P=0.040; Fig. 2C) and MDA-MB-231 (1.34±0.26 fold-change, P=0.045; Fig. 2D) cell lines over an 18-h period.
Figure 2.
GHSROS promotes cell migration, but not cell proliferation, in MCF10A and MDA-MB-231 breast cancer cell lines in vitro. (A) Relative expression of GHSROS in MDA-MD-231-GHSROS, MDA-MB-231-Vector, MCF10A-GHSROS and MCF10A-Vector cell lines. Expression was normalised to the housekeeping gene β-actin using the comparative 2−ΔΔCq method of quantification. Results are relative to each vector control. (B) Proliferation was not significantly increased in the MCF10A or MDA-MB-231 cell lines overexpressing GHSROS compared to cells expressing vector alone when assessed using an xCELLigence real-time cell analyser for 72 h. (C) GHSROS overexpression increased MCF10A migration across a porous membrane (pore size, 8 µM). Left panel, representative plot of raw cell index impedance measurements from 0 to 20 h after cell seeding. Right panel, GHSROS overexpression increased cell migration at 18 h. (D) GHSROS overexpression increased MDA-MB-231 migration across a porous membrane. Left panel, representative plot of raw cell index impedance measurements from 0 to 20 h after overexpression of GHSROS. Right panel, GHSROS overexpression increased cell migration 18 h after passaging. Data are presented as the mean ± standard error of the mean (n=3). *P≤0.05, **P≤0.01 vs. vector control; Student's t-test. GHSROS, growth hormone secretagogue receptor opposite strand.
GHSROS regulates genes associated with cancer and the immune response
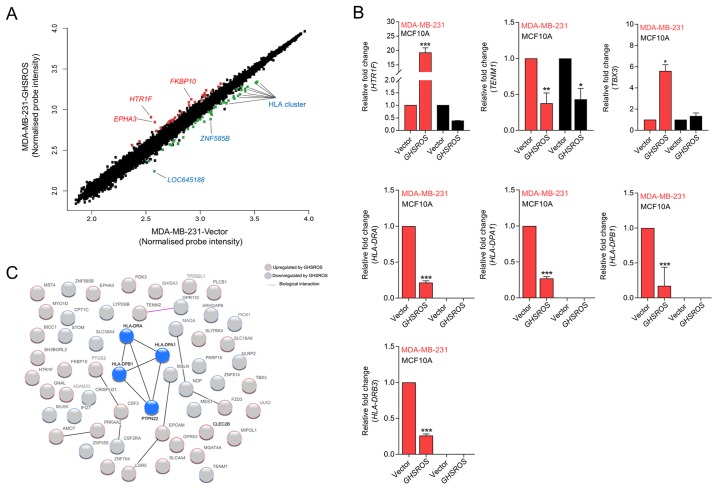
To gain further insight into the function of GHSROS in breast cancer, gene expression microarrays were performed on RNA isolated from GHSROS-overexpressing MDA-MB-231 cells and vector control cells. A total of 76 genes were differentially regulated by >1.5-fold in MDA-MB-231 cells overexpressing GHSROS (36 upregulated and 40 downregulated) (Fig. 3A and Table III).
Figure 3.
GHSROS significantly differentially regulates 76 genes in the MDA-MB-231 breast cancer cell line. (A) Scatter plot visualization of induced (red) or repressed (green) genes identified by microarray. The threshold was set at as log2 1.5 fold-change and Q≤0.05 (Benjamini Hochberg-adjusted P-value). EPHA3, HTR1F, FKBP10, LOC645188, ZNF585B and HLA genes were affected by GHSROS. (B) Expression levels of TBX3, HTR1F, TENM1, HLA-DRA, HLA-DRB3, HLA-DPA1 and HLA-DPB1 were measured in cultured MDA-MB-231-GHSROS, MDA-MB-231-Vector, MCF10A-Vector and MCF10A-GHSROS cells by reverse transcription-quantitative polymerase chain reaction. Expression was normalised to the housekeeping gene β-actin. Results are relative to the respective vector control. Data are presented as the mean ± standard error of the mean (n=2); ***P≤0.001 vs. vector control, two-way ANOVA with Bonferroni's post hoc analysis. (C) STRING network consisting of 56 proteins encoded by genes differentially expressed in MDA-MB-231-GHSROS cells. Nodes represent differentially expressed genes. Genes induced (red border) or repressed (blue border) by GHSROS are indicated. Lines between protein nodes indicate biological associations inferred or experimentally demonstrated. Differentially expressed HLA genes are represented by blue nodes in the centre of the cluster. EPHA3, EPH receptor A3; FKBP10, FK506 binding protein 10; GHSROS, growth hormone secretagogue receptor opposite strand; HLA-DRA, MHC, class II, DR α; HLA-DRB3, MHC, class II, DR β3; HLA-DPA1, MHC, class II, DP α1; HLA-DPB1, MHC, class II, DP β1; HTR1F, 5-hydroxytryptamine receptor 1F; MHC, major histocompatibility complex; TBX3, T-box 3; TENM1, teneurin transmembrane protein 1; ZNF585B, zinc finger protein 585B.
Table III.
Differentially expressed genes in MDA-MB-231-GHSROS cells compared with in empty vector control cells.
| Gene symbol | Gene name | Fold-change | P-value | Affymetrix probe |
|---|---|---|---|---|
| HTR1F | 5-Hydroxytryptamine receptor 1F | 3.12 | 0.0040 | 8081067 |
| EPHA3 | EPH receptor A3 | 2.27 | 0.0190 | 8081081 |
| FKBP10 | FK506 binding protein 10 | 2.23 | 0.0032 | 8007154 |
| SHISA3 | Shisa family member 3 | 2.13 | 0.0046 | 8094870 |
| MYO1D | Myosin ID | 2.07 | 0.0111 | 8014115 |
| STK26 | Serine/threonine protein kinase 26 | 1.91 | 0.0008 | 8169949 |
| BICC1 | BicC family RNA binding protein 1 | 1.81 | 0.0021 | 7927681 |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 | 1.79 | 0.0106 | 7922976 |
| EPCAM | Epithelial cell adhesion molecule | 1.77 | 0.0089 | 8098439 |
| SH3BGRL2 | SH3 domain binding glutamate rich protein like 2 | 1.70 | 0.0008 | 8120833 |
| FZD3 | Frizzled class receptor 3 | 1.68 | 0.0021 | 8145611 |
| PRKAA2 | Protein kinase AMP-activated catalytic subunit α2 | 1.63 | 0.0118 | 7901720 |
| SLC16A6 | Solute carrier family 16 member 6 | 1.63 | 0.0045 | 8017843 |
| SLC4A4 | Solute carrier family 4 member 4 | 1.63 | 0.0056 | 8095585 |
| GPR63 | G protein-coupled receptor 63 | 1.62 | 0.0092 | 8128316 |
| MIPOL1 | Mirror-image polydactyly 1 | 1.62 | 0.0013 | 7973985 |
| LGR5 | Leucine rich repeat containing G protein-coupled receptor 5 | 1.58 | 0.0184 | 7957140 |
| PLCB1 | Phospholipase C β1 | 1.58 | 0.0222 | 8060854 |
| ADAM23 | ADAM metallopeptidase domain 23 | 1.56 | 0.0083 | 8047788 |
| SLITRK4 | SLIT and NTRK like family member 4 | 1.52 | 0.0070 | 8175574 |
| GNAL | G protein subunit α L | 1.52 | 0.0006 | 8020164 |
| AMOT | Angiomotin | 1.52 | 0.0128 | 8174576 |
| MGAT4A | mannosyl (α-1,3-)-glycoprotein β-1,4-N-acetylglucosaminyltransferase, isozyme A | 1.52 | 0.0418 | 8054135 |
| ULK2 | Unc-51 like autophagy activating kinase 2 | 1.50 | 0.0008 | 8013399 |
| OR2A9P///OR2A20P | Olfactory receptor family 2 subfamily A member 9 pseudogene///olfactory receptor family 2 subfamily A member 20 pseudogene | 1.50 | 0.0373 | 8136983 |
| ZNF704 | Zinc finger protein 704 | 1.50 | 0.0032 | 8151496 |
| TBX3 | T-box 3 | 1.49 | 0.0041 | 7966690 |
| SUPT20HL1/// | SPT20 homolog, SAGA complex component-like 1///SPT20 | 1.48 | 0.0245 | 8166509 |
| SUPT20HL2 | homolog, SAGA complex component-like 2 | |||
| SUPT20HL1/// | SPT20 homolog, SAGA complex component-like 1///SPT20 | 1.48 | 0.0245 | 8171844 |
| SUPT20HL2 | homolog, SAGA complex component-like 2 | |||
| TENM2 | Teneurin transmembrane protein 2 | 1.47 | 0.0391 | 8109752 |
| PDK3 | Pyruvate dehydrogenase kinase 3 | 1.47 | 0.0220 | 8166511 |
| OR2A9P///OR2A20P | Olfactory receptor family 2 subfamily A member 9 pseudogene///olfactory receptor family 2 subfamily A member 20 pseudogene | 1.47 | 0.0334 | 8143629 |
| TPD52L1 | Tumour protein D52-like 1 | 1.46 | 0.0024 | 8121838 |
| TMEM56-RWDD3/// | TMEM56-RWDD3 readthrough///transmembrane | 1.46 | 0.0363 | 7903162 |
| TMEM56 | protein 56 | |||
| CLEC2B | C-type lectin domain family 2 member B | 1.45 | 0.0498 | 7961083 |
| CSF3 | Colony stimulating factor 3 | 1.45 | 0.0052 | 8006999 |
| PICK1 | Protein interacting with PRKCA 1 | −1.47 | 0.0386 | 8072989 |
| MAOA | Monoamine oxidase A | −1.48 | 0.0217 | 8166925 |
| CRISPLD1 | Cysteine rich secretory protein LCCL domain containing 1 | −1.49 | 0.0009 | 8146967 |
| MSLN | Mesothelin | −1.50 | 0.0059 | 7992071 |
| ZNF558 | Zinc finger protein 558 | −1.53 | 0.0070 | 8033667 |
| PTPN22 | Protein tyrosine phosphatase, non-receptor type 22 | −1.54 | 0.0004 | 7918657 |
| NDP | NDP, norrin cystine knot growth factor | −1.54 | 0.0004 | 8172220 |
| IFI27 | Interferon α inducible protein 27 | −1.56 | 0.0225 | 7976443 |
| ZNF814 | Zinc finger protein 814 | −1.56 | 0.0033 | 8039692 |
| PARP15 | poly(ADP-ribose) polymerase family member 15 | −1.56 | 0.0147 | 8082086 |
| CPT1C | Carnitine palmitoyltransferase 1C | −1.56 | 0.0267 | 8030448 |
| RNU5D-1 | RNA, U5D small nuclear 1 | −1.57 | 0.0412 | 7915592 |
| ARHGAP6 | Rho GTPase activating protein 6 | −1.58 | 0.0042 | 8171313 |
| MEST | Mesoderm specific transcript | −1.63 | 0.0026 | 8136248 |
| STOM | stomatin | −1.67 | 0.0110 | 8163896 |
| NLRP2 | NLR family pyrin domain containing 2 | −1.72 | 0.0001 | 8031398 |
| MUSK | Muscle associated receptor tyrosine kinase | −1.79 | 0.0389 | 8157173 |
| CSF2RA | Colony stimulating factor 2 receptor α subunit | −1.80 | 0.0030 | 8165735 |
| CSF2RA | Colony stimulating factor 2 receptor α subunit | −1.80 | 0.0030 | 8176306 |
| LYPD6B | LY6/PLAUR domain containing 6B | −1.97 | 0.0188 | 8045664 |
| TENM1 | Teneurin transmembrane protein 1 | −1.97 | 0.0238 | 8174937 |
| TGIF2LY/// | TGFB induced factor homeobox 2 like, Y-linked///TGFB | −1.97 | 0.0002 | 8176397 |
| TGIF2LX | induced factor homeobox 2 like, X-linked | |||
| HLA-DPB1 | Major histocompatibility complex, class II, DP β1 | −1.99 | 0.0485 | 8118594 |
| TGIF2LY/// | TGFB induced factor homeobox 2 like, Y-linked///TGFB | −1.99 | 0.0002 | 8168646 |
| TGIF2LX | induced factor homeobox 2 like, X-linked | |||
| ADGRF1 | Adhesion G protein-coupled receptor F1 | −2.00 | 0.0038 | 8126820 |
| SLC38A4 | Solute carrier family 38 member 4 | −2.04 | 0.0081 | 7962559 |
| HLA-DPB1 | Major histocompatibility complex, class II, DP β1 | −2.07 | 0.0403 | 8178220 |
| SPANXC///SPANXD | SPANX family member C///SPANX family member D | −2.10 | 0.0027 | 8175558 |
| HLA-DPA1 | Major histocompatibility complex, class II, DP α1 | −2.22 | 0.0466 | 8125556 |
| HLA-DPA1 | Major histocompatibility complex, class II, DP α1 | −2.22 | 0.0466 | 8178891 |
| HLA-DPA1 | Major histocompatibility complex, class II, DP α1 | −2.22 | 0.0470 | 8180100 |
| HLA-DPB1 | Major histocompatibility complex, class II, DP β1 | −2.31 | 0.0391 | 8179519 |
| ZNF585B | zinc finger protein 585B | −2.31 | 0.0001 | 8036389 |
| HLA-DRA/// | Major histocompatibility complex, class II, DR α///major | −2.35 | 0.0065 | 8118548 |
| HLA-DQA1 | histocompatibility complex, class II, DQ α1 | |||
| HLA-DRA/// | Major histocompatibility complex, class II, DR α///major | −2.36 | 0.0058 | 8179481 |
| HLA-DQA1 | histocompatibility complex, class II, DQ α1 | |||
| LOC645188 | Uncharacterised LOC645188 | −2.39 | 0.0003 | 8170257 |
| HLA-DRA/// | Major histocompatibility complex, class II, DR α///major | −2.55 | 0.0072 | 8178193 |
| HLA-DQA1 | histocompatibility complex, class II, DQ α1 | |||
| LOC105369230/// | HLA class II histocompatibility antigen, DRB1-7β chain///major | −2.62 | 0.0054 | 8180003 |
| HLA-DRB6/// | histocompatibility complex, class II, DR β6 (pseudogene)///major | |||
| HLA-DRB5/// | histocompatibility complex, class II, DR β5///major | |||
| HLA-DRB4/// | histocompatibility complex, class II, DR β4///major | |||
| HLA-DRB3/// | histocompatibility complex, class II, DR β4///major | |||
| HLA-DRB1/// | histocompatibility complex, class II, DR β3///major | |||
| LOC105369230/// | histocompatibility complex, class II, DR β1///HLA class II | |||
| HLA-DRB5/// | histocompatibility antigen, DRB1-7β chain///major | |||
| HLA-DRB4/// | histocompatibility complex, class II, DR β5///major | |||
| HLA-DRB3/// | histocompatibility complex, class II, DR β4///major | |||
| HLA-DRB1/// | histocompatibility complex, class II, DR β3///major | |||
| HLA-DQB1 | histocompatibility complex, class II, DR β1///major histocompatibility complex, class II, DQ β1 | |||
| LOC105369230/// | HLA class II histocompatibility antigen, DRB1-7 β chain///major | −2.66 | 0.0063 | 8178811 |
| HLA-DRB6/// | histocompatibility complex, class II, DR β6 (pseudogene)///major | |||
| HLA-DRB5/// | histocompatibility complex, class II, DR β5///major | |||
| HLA-DRB4/// | histocompatibility complex, class II, DR β4///major | |||
| HLA-DRB3/// | histocompatibility complex, class II, DR β3///major | |||
| HLA-DRB1/// | histocompatibility complex, class II, DR β1///HLA class II | |||
| LOC105369230/// | histocompatibility antigen, DRB1-7β chain///major | |||
| HLA-DRB5/// | histocompatibility complex, class II, DR β5///major | |||
| HLA-DRB4/// | histocompatibility complex, class II, DR β4///major | |||
| HLA-DRB3/// | histocompatibility complex, class II, DR β3///major | |||
| HLA-DRB1/// | histocompatibility complex, class II, DR β1///major | |||
| HLA-DQB1 | histocompatibility complex, class II, DQ β1 | |||
| LOC105369230/// | HLA class II histocompatibility antigen, DRB1-7β chain///major | −2.71 | 0.0068 | 8178802 |
| HLA-DRB6/// | histocompatibility complex, class II, DR β6 (pseudogene)///major | |||
| HLA-DRB5/// | histocompatibility complex, class II, DR β5///major | |||
| HLA-DRB4/// | histocompatibility complex, class II, DR β4///major | |||
| HLA-DRB3/// | histocompatibility complex, class II, DR β3///major | |||
| HLA-DRB1/// | histocompatibility complex, class II, DR β1///HLA class II | |||
| LOC105369230/// | histocompatibility antigen, DRB1-7β chain///major histocompatibility | |||
| HLA-DRB5/// | complex, class II, DR β5///major histocompatibility complex, | |||
| HLA-DRB4/// | class II, DR β4///major histocompatibility complex, class II, | |||
| HLA-DRB3/// | DR β3///major histocompatibility complex, class II, DR β1///major | |||
| HLA-DRB1///HLA-DQB1 | histocompatibility complex, class II, DQ β1 |
Bold text represents higher expression in MDA-MB-231-GHSROS cells; unbold text represents lower expression in MDA-MB-231-GHSROS cells. P-values were obtained using the R package 'limma' (moderated t-test).
The most upregulated genes in the microarray were 5-hydroxy-tryptamine (5-HT) receptor 1F (HTR1F, 3.1 fold-change, P=0.0040) and EPH receptor A3 (EPHA3, 2.3 fold-change, P=0.0190) (Table III). Significant upregulation of HTR1F was verified by RT-qPCR in the MDA-MB-231-GHSROS cell line (19.2±1.173 fold-change, P=0.0002; Fig. 3B). Conversely, HTR1F expression was reduced in MCF10A-GHSROS cells (-2.68±0.002 fold-change, P>0.999; Fig. 3B), although this finding was not statistically significant. Notably, a number of established oncogenes were differentially expressed in the MDA-MB-231-GHSROS microarray data. These included teneurin transmembrane protein 1 (TENM1), which was downregulated in MDA-MB-231-GHSROS cells (-2.00 fold-change, P=0.0238. These data were validated by RT-qPCR in MDA-MB-231-GHSROS cells (-2.65±0.286 fold-change, P=0.0048), and TENM1 was similarly downregulated in MCF10A-GHSROS cells (-2.31±0.262 fold-change, P=0.020; Fig. 3B). The T-box transcription factor gene T-box 3 (TBX3) is associated with breast cancer cell migration and growth (29) and was upregulated (1.5 fold-change, P=0.0041; Table III) in the MDA-MB-231-GHSROS microarray. Upregulation of TBX3 was confirmed by RT-qPCR in MDA-MB-231-GHSROS cells (5.60±0.82 fold-change, P=0.025; Fig. 3B); however, TBX3 expression was not altered in MCF10A-GHSROS cells (Fig. 3B).
In the present dataset, it was observed that a single probe matched to several class II major histocompatibility complex (MHC) genes. A number of MHC class II genes were significantly repressed by GHSROS [including MHC, class II, DR β3 (HLA-DRB3), -2.7 fold-change, P=0.0068; MHC, class II, DR β1, -2.7 fold-change, P=0.0063; MHC, class II, DR α (HLA-DRA), -2.4 fold-change, P=0.0058; MHC, class II, DP β1 (HLA-DPB1), -2.3 fold-change, P=0.039; Table III]. MHC gene loci are complex; their exons are highly similar and person-to-person variation in exon sequences can confound microarray probe hybridisation (30,31). To firmly establish which MHC-II genes were differentially expressed upon forced GHSROS overexpression, selected genes were validated by RT-qPCR. Using RT-qPCR, it was confirmed that GHSROS overexpression in MDA-MB-231 cells induced downregulation of the MHC-II genes HLA-DRA (-4.62±0.024 fold-change, P<0.0001), HLA-DPB1 (-4.53±0.30 fold-change, P<0.0001), MHC, class II, DP α1 HLA-DPA1 (-3.69±0.02 fold-change, P<0.0001) and HLA-DRB3 (-3.84±0.02 fold-change, P<0.0001) (Fig. 3B). These transcripts were not detected in the MCF10A-GHSROS or MCF10A-Vector cell lines, which is consistent with the fact that normal breast epithelial tissue is typically MHC-II negative (32).
Differentially expressed genes identified by microarray analysis were interrogated for biological interactions using the STRING tool (24). Within the functional protein-protein interaction (PPI) network a small, distinct interaction was detected between the MHC-II gene set (HLA-DRA, HLA-DPA1 and HLA-DPB1) and protein tyrosine phosphatase, non-receptor type 22 (lymphoid) (Fig. 3C; expected interactions, 7; observed interactions, 15; PPI enrichment P=0.0099, hypergeometric test). KEGG pathway analysis of 56 genes analysed in STRING demonstrated that the 40 genes downregulated in MDA-MB-231-GHSROS cells were enriched for pathways typically associated with the expression of MHC-II genes (Fig. 3C; Table IV), including 'antigen processing and presentation' [Benjamini-Hochberg corrected P-value, Benjamini-Hochberg false discovery rate (BH-FDR)=0.0020], 'asthma' (BH-FDR=0.0011) and 'graft-versus-host disease' (BH-FDR=0.0011) pathways (Table IV).
Table IV.
Enriched KEGG pathway terms for 40 genes downregulated in MDA-MB-231-GHSROS cells.
| KEGG ID | Pathway description | Gene count | BH-FDR | Genes |
|---|---|---|---|---|
| 5310 | Asthma | 3 | 0.00107a | HLA-DPA1, HLA-DPB1, HLA-DRA |
| 5330 | Allograft rejection | 3 | 0.00107a | HLA-DPA1, HLA-DPB1, HLA-DRA |
| 5332 | Graft-versus-host disease | 3 | 0.00107a | HLA-DPA1, HLA-DPB1, HLA-DRA |
| 4940 | Type I diabetes mellitus | 3 | 0.00111a | HLA-DPA1, HLA-DPB1, HLA-DRA |
| 4672 | Intestinal immune network for IgA production | 3 | 0.00119a | HLA-DPA1, HLA-DPB1, HLA-DRA |
| 5150 | Staphylococcus aureus infection | 3 | 0.00132a | HLA-DPA1, HLA-DPB1, HLA-DRA |
| 5320 | Autoimmune thyroid disease | 3 | 0.00132a | HLA-DPA1, HLA-DPB1, HLA-DRA |
| 5416 | Viral myocarditis | 3 | 0.00146a | HLA-DPA1, HLA-DPB1, HLA-DRA |
| 5321 | Inflammatory bowel disease (IBD) | 3 | 0.00185a | HLA-DPA1, HLA-DPB1, HLA-DRA |
| 4612 | Antigen processing and presentation | 3 | 0.00201a | HLA-DPA1, HLA-DPB1, HLA-DRA |
| 5140 | Leishmaniasis | 3 | 0.00209a | HLA-DPA1, HLA-DPB1, HLA-DRA |
| 5323 | Rheumatoid arthritis | 3 | 0.00356a | HLA-DPA1, HLA-DPB1, HLA-DRA |
| 5322 | Systemic lupus erythematosus | 3 | 0.00443a | HLA-DPA1, HLA-DPB1, HLA-DRA |
| 5145 | Toxoplasmosis | 3 | 0.0069a | HLA-DPA1, HLA-DPB1, HLA-DRA |
| 4514 | Cell adhesion molecules | 3 | 0.0114a | HLA-DPA1, HLA-DPB1, HLA-DRA |
| 4145 | Phagosome | 3 | 0.0121a | HLA-DPA1, HLA-DPB1, HLA-DRA |
| 5164 | Influenza A | 3 | 0.0179a | HLA-DPA1, HLA-DPB1, HLA-DRA |
| 5152 | Tuberculosis | 3 | 0.0181a | HLA-DPA1, HLA-DPB1, HLA-DRA |
| 5168 | Herpes simplex infection | 3 | 0.0183a | HLA-DPA1, HLA-DPB1, HLA-DRA |
| 5169 | Epstein-Barr virus infection | 3 | 0.0221a | HLA-DPA1, HLA-DPB1, HLA-DRA |
| 5166 | HTLV-I infection | 3 | 0.0465a | HLA-DPA1, HLA-DPB1, HLA-DRA |
KEGG pathway passed the 5% BH-FDR-corrected P-value. BH-FDR, Benjamini-Hochberg false discovery rate; HLA-DPA1, MHC class II, DP α1; HLA-DPB1, MHC class II, DP β1; HLA-DRA, MHC class II, DR α; KEGG, Kyoto Encyclopedia of Genes and Genomes; MHC, major histocompatibility complex.
GHSROS increases orthotopic breast xenograft growth
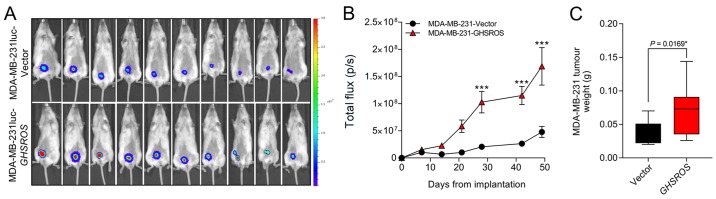
In order to investigate the effect of GHSROS on tumour growth in vivo an orthotopic xenograft model was used (33). MDA-MB-231luc-GHSROS and MDA-MB-231luc-Vector cells were injected into the mammary fat pad of NSG mice (Fig. 4A). Compared to vector controls, mammary fat pad xenograft tumour volumes [measured by total flux (p/s)] were significantly increased in MDA-MB-231luc-GHSROS mice at day 28 post-implantation (Mann-Whitney, P=0.0002), day 42 post-implantation (P≤0.0001) and at the experimental (ethical) endpoint (day 49, P≤0.0001) (Fig. 4B). No gross metastases were observed in either experimental group. Upon excision, the MDA-MB-231luc-GHSROS tumours weighed significantly more post mortem (Mann-Whitney-Wilcoxon test, P=0.017; Fig. 4C).
Figure 4.
GHSROS promotes orthotopic MDA-MB-231 xenograft tumour growth in vivo. (A) NOD.Cg-Prkdc SCID IL-2rgtm1WjL/SzJ mice were injected in the mammary fat pad with MDA-MB-231luc cell lines stably overexpressing GHSROS (n=10) or empty vector (n=10). Representative IVIS images showing total flux (bioluminescence) at day 49 (endpoint) demonstrating that tumours were larger in mice with GHSROS-overexpressing tumours. (B) Time course for MDA-MB-231luc-GHSROS (n=10) and luc-Vector control (n=10) mammary fat pad xenograft tumour bioluminescence. Tumour bioluminescent imaging [total flux (p/s)] was measured using the IVIS Spectrum in vivo imaging system. ***P≤0.001 vs. vector control, two-way ANOVA with Bonferroni's post hoc analysis. (C) Tumour weights of MDA-MB-231 xenografts (GHSROS overexpressing, n=10; vector, n=10) at the end point. *P<0.05 vs. vector control, Mann-Whitney-Wilcoxon test. Data are presented as the mean ± standard error of the mean. GHSROS, growth hormone secretagogue receptor opposite strand; p/s, photons/sec.
Discussion
Our previous study demonstrated that GHSROS, a lncRNA derived from a gene antisense to the ghrelin receptor gene, is expressed by non-small cell lung tumours and increases lung cancer cell migration in vitro (19). Similarly, in this study it was demonstrated that GHSROS was expressed in breast cancer and promoted in vitro cell line migration. GHSROS also promoted MDA-MB-231 breast cancer cell xenograft tumour growth and altered the expression of cancer-associated and immune genes. While lncRNAs are rarely highly conserved, nor abundantly expressed, they can serve integral regulatory roles in a large array of cellular processes, including pathological conditions such as tumourigenesis (34,35). In a previous study, genome-wide RNA-seq profiling of 22 paired tumour and non-malignant tissues from ER+ breast cancer samples demonstrated that the expression of natural antisense transcript lncRNAs are increased in tumour samples (36), suggesting that they may have important regulatory roles.
A number of lncRNAs differentially expressed in breast cancer function as critical mediators of breast cancer tumourigenesis (35,37,38). For example, the lncRNAs H19 (39), MALAT1 (40), HOTAIR (41) and taurine upregulated 1 (42) have a higher gene expression in breast cancer tissues and derived cell lines (41,43). lncRNAs have also been proposed as potential breast cancer biomarkers. MALAT1 is an abundant and highly conserved lncRNA that can be detected in the serum of patients with breast cancer and at much higher levels than in patients with benign breast disease (40). Although the present study did not detect a direct relationship between GHSROS expression and clinical parameters (including ER, PR and HER2 status), further studies interrogating a larger patient cohort are warranted.
GHSROS increased in vitro cell migration, but not proliferation, of the MDA-MB-231 breast cancer cell line and the non-tumourigenic, normal-breast derived MCF10A cell line. Our previous study observed similar effects in non-small-cell lung carcinoma cell lines overexpressing GHSROS (19). MDA-MB-231 breast cancer cells metastasise from the mammary fat pads of NSG mice, with gross tumours observed in multiple tissues, including the lungs, lymph nodes and liver, 53 days after implantation (44). This model is, therefore, useful for investigating the effect of gene expression on metastasis. In the current study, mice bearing GHSROS-overexpressing xenografts were euthanised 49 days after injection as they had reached the ethical endpoint. However, gross metastases were not observed and therefore, it was not possible to determine if GHSROS affected metastasis in these animals.
Although no changes in proliferation were observed in the present two-dimensional cell models, tumour size was significantly greater in orthotopic MDA-MB-231-GHSROS xenograft tumours. In vitro models of cell proliferation do not replicate numerous aspects of cancer progression (33), including the tumour microenvironment and growth factors, which are not present in an in vitro system (45).
Microarray analysis of MDA-MB-231-GHSROS cells cultured in vitro was conducted to reveal potential GHSROS-regulated genes. The most highly upregulated gene, HTR1F, belongs to a subgroup of 5-HT/serotonin receptors and is significantly associated with breast cancer recurrence (46). This gene was downregulated in GHSROS-overexpressing MCF10A cells, which also exhibited increased in vitro migration, suggesting that HTR1F is unlikely to play a major role in GHSROS-mediated cell migration. Upregulation of the 5-HT signalling pathways in metastatic breast cancer is pro-oncogenic, stimulating pro-proliferative, invasive and anti-apoptotic pathways (46). Conversely, normal physiological levels of the ligand, 5-HT, induce growth inhibition and apoptosis in breast cancer cell lines, presumably by increased expression of its receptors (47). TBX3 is a transcription factor that is considered a key driver of breast cancer progression, which was elevated in GHSROS-overexpressing breast cancer cells. TBX3 is elevated in metastatic breast cancer, is correlated with reduced metastasis-free survival, and potently promotes cell survival and tumour growth in early-stage breast cancer cell models (29,48,49). Additionally, TBX3 overexpression stimulates cell migration in normal breast and breast cancer cells (49). Therefore, it was hypothesised that TBX3 may represent a key mediator of the effects of GHSROS in breast cancer.
Transcripts encoding a number of subunits of MHC-II were repressed in MDA-MB-231 cells overexpressing GHSROS. MHC-II genes encode cell surface proteins primarily involved in antigen presentation and adaptive immunity (30,50,51). Reduced tumour expression of the MHC-II complex increases breast tumour aggressiveness and results in poor overall survival (50,52). Conversely, increased MHC-II expression is associated with a positive prognosis in triple-negative breast cancer (50). Given that in vivo xenograft models using human cell lines require immunocompromised or syngeneic mice, the role of GHSROS in antitumour immunity is currently challenging to investigate and could not be assessed in the present xenograft model. Humanised mice that support human cell lines and patient-derived xenografts (53,54) will be critical in assessing if GHSROS overexpression indeed facilitates immune system evasion. The present study hypothesised that GHSROS, by downregulating critical components of the acquired immune system, may promote breast tumour cell survival.
There are several limitations to the present study. Firstly, gene expression was not examined between tumours and corresponding adjacent, normal tissue. Future studies will aim to detect the expression of GHSROS in larger, well-characterised clinical cohorts. Secondly, overexpression of GHSROS should be complemented by knockdown experiments, using, for example, short hairpin RNA or modified ASOs. We recently generated ASOs targeting GHSROS, revealing that these compounds reciprocally regulate GHSROS-mediated expression and function of prostate cancer cell lines (Thomas et al, unpublished data). We aim to assess these ASOs in a future breast cancer study. Finally, 3D invasion assays, complementing the 2D in vitro assays employed in this study, may provide critical further insights into the function of GHSROS. 2D assays, particularly invasion assays (22), are not fully representative of gene function and may lead to discordant results, as observed when comparing 2D in vitro proliferation assay data and in vivo xenograft tumour growth in this study. 3D invasion assays will be employed in a future study to further determine the function of GHSROS.
In conclusion, the present study examined the expression and function of the lncRNA GHSROS in breast cancer, suggesting a potential role for GHSROS in breast cancer cell migration and tumour growth. These data expand on recent findings regarding GHSROS in lung cancer (19) and provide a rationale for further investigations into this lncRNA in cancer.
Acknowledgments
The authors would like to acknowledge the scientific and technical assistance of the Translational Research Institute Preclinical Imaging Facility.
Funding
This work was supported by the National Health and Medical Research Council Australia (grant nos. 1002255 and 1059021; to LKC, ACH, RNV and IS), the Cancer Council Queensland (grant no. 1098565; to ACH, RNV, LKC and IS), the Australian Research Council (grant no. DP140100249; to ACH and LKC), a QUT Vice-Chancellor's Senior Research Fellowship (to IS), the Movember Foundation and the Prostate Cancer Foundation of Australia through a Movember Revolutionary Team Award, the Australian Government Department of Health, and the Australian Prostate Cancer Research Centre, Queensland (LKC and CCN), Queensland University of Technology. The Translational Research Institute is supported by a grant from the Australian Government.
Availability of data and materials
The datasets generated and/or analysed during the current study are available in the Gene Expression Omnibus repository, [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE35001]
Authors' contributions
PBT, IS, PLJ and LKC conceived and designed the study, and interpreted the data. PBT, AS, MM, IS, LKC and PLJ performed laboratory experiments. IS and PBT performed computational biology analyses. PBT, LKC, PLJ and IS wrote the article. All authors (PBT, IS, PLJ, MDG, MM, GJC, AS, ETS, CW, EJW, CCN, ACH, RML, RNV and LKC) contributed to the conception and design of the study, interpretation of the data and writing of the manuscript. LKC approved the final version of the manuscript.
Ethics approval and consent to participate
Experiments were approved by the University of Queensland and QUT animal ethics committees (TRI/QUT/328/16). Approval for cell line use was granted by the QUT Human Research Ethics Committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 2.Deveson IW, Hardwick SA, Mercer TR, Mattick JS. The Dimensions, dynamics, and relevance of the mammalian noncoding transcriptome. Trends Genet. 2017;33:464–478. doi: 10.1016/j.tig.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Hu X, Sood AK, Dang CV, Zhang L. The role of long noncoding RNAs in cancer: The dark matter matters. Curr Opin Genet Dev. 2018;48:8–15. doi: 10.1016/j.gde.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 5.Sun M, Kraus WL. From discovery to function: The expanding roles of long non-coding RNAs in physiology and disease. Endocr Rev. 2015 Jan 7; doi: 10.1210/er.2014-1034. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 7.Australian Institute of Health and Welfare Cancer in Australia: Actual incidence data from 1982 to 2013 and mortality data from 1982 to 2014 with projections to 2017. Asia Pac J Clin Oncol. 2018;14:5–15. doi: 10.1111/ajco.12761. [DOI] [PubMed] [Google Scholar]
- 8.ENCODE Project Consortium The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 9.Kumar M, DeVaux RS, Herschkowitz JI. Molecular and cellular changes in breast cancer and new roles of lncRNAs in breast cancer initiation and progression. Prog Mol Biol Transl Sci. 2016;144:563–586. doi: 10.1016/bs.pmbts.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 10.Amorim M, Salta S, Henrique R, Jerónimo C. Decoding the usefulness of non-coding RNAs as breast cancer markers. J Transl Med. 2016;14:265. doi: 10.1186/s12967-016-1025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerk S, Schwarzenbacher D, Adiprasito JB, Stotz M, Hutterer GC, Gerger A, Ling H, Calin GA, Pichler M. Current status of long non-coding RNAs in human breast cancer. Int J Mol Sci. 2016;17:17. doi: 10.3390/ijms17091485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soudyab M, Iranpour M, Ghafouri-Fard S. The role of long non-coding RNAs in breast cancer. Arch Iran Med. 2016;19:508–517. [PubMed] [Google Scholar]
- 13.Bhan A, Hussain I, Ansari KI, Kasiri S, Bashyal A, Mandal SS. Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. J Mol Biol. 2013;425:3707–3722. doi: 10.1016/j.jmb.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He X, Bao W, Li X, Chen Z, Che Q, Wang H, Wan XP. The long non-coding RNA HOTAIR is upregulated in endometrial carcinoma and correlates with poor prognosis. Int J Mol Med. 2014;33:325–332. doi: 10.3892/ijmm.2013.1570. [DOI] [PubMed] [Google Scholar]
- 15.Bhan A, Mandal SS. Estradiol-induced transcriptional regulation of long non-coding RNA, HOTAIR. Methods Mol Biol. 2016;1366:395–412. doi: 10.1007/978-1-4939-3127-9_31. [DOI] [PubMed] [Google Scholar]
- 16.Xue X, Yang YA, Zhang A, Fong KW, Kim J, Song B, Li S, Zhao JC, Yu J. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2016;35:2746–2755. doi: 10.1038/onc.2015.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aiello A, Bacci L, Re A, Ripoli C, Pierconti F, Pinto F, Masetti R, Grassi C, Gaetano C, Bassi PF, et al. MALAT1 and HOTAIR long non-coding RNAs play opposite role in estrogen-mediated transcriptional regulation in prostate cancer cells. Sci Rep. 2016;6:38414. doi: 10.1038/srep38414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, Kim Y, MacLeod AR, Krainer AR, Norton L, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30:34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whiteside EJ, Seim I, Pauli JP, O'Keeffe AJ, Thomas PB, Carter SL, Walpole CM, Fung JN, Josh P, Herington AC, et al. Identification of a long non-coding RNA gene, growth hormone secretagogue receptor opposite strand, which stimulates cell migration in non-small cell lung cancer cell lines. Int J Oncol. 2013;43:566–574. doi: 10.3892/ijo.2013.1969. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Ke N, Wang X, Xu X, Abassi YA. The xCELLigence system for real-time and label-free monitoring of cell viability. Methods Mol Biol. 2011;740:33–43. doi: 10.1007/978-1-61779-108-6_6. [DOI] [PubMed] [Google Scholar]
- 22.Limame R, Wouters A, Pauwels B, Fransen E, Peeters M, Lardon F, De Wever O, Pauwels P. Comparative analysis of dynamic cell viability, migration and invasion assessments by novel real-time technology and classic endpoint assays. PLoS One. 2012;7:e46536. doi: 10.1371/journal.pone.0046536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartigan JA, Wong MA. Algorithm AS 136: A k-means clustering algorithm. J R Stat Soc Ser C Appl Stat. 1979;28:100–108. [Google Scholar]
- 26.Du J, Yuan Z, Ma Z, Song J, Xie X, Chen Y. KEGG-PATH: Kyoto encyclopedia of genes and genomes-based pathway analysis using a path analysis model. Mol Biosyst. 2014;10:2441–2447. doi: 10.1039/C4MB00287C. [DOI] [PubMed] [Google Scholar]
- 27.Russell PJ, Raghavan D, Gregory P, Philips J, Wills EJ, Jelbart M, Wass J, Zbroja RA, Vincent PC. Bladder cancer xenografts: A model of tumor cell heterogeneity. Cancer Res. 1986;46:2035–2040. [PubMed] [Google Scholar]
- 28.Lim E, Modi KD, Kim J. In vivo bioluminescent imaging of mammary tumors using IVIS spectrum. J Vis Exp. 2009 Apr 29; doi: 10.3791/1210. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amir S, Simion C, Umeh-Garcia M, Krig S, Moss T, Carraway KL, III, Sweeney C. Regulation of the T-box transcription factor Tbx3 by the tumour suppressor microRNA-206 in breast cancer. Br J Cancer. 2016;114:1125–1134. doi: 10.1038/bjc.2016.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trowsdale J, Knight JC. Major histocompatibility complex genomics and human disease. Annu Rev Genomics Hum Genet. 2013;14:301–323. doi: 10.1146/annurev-genom-091212-153455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boegel S, Löwer M, Schäfer M, Bukur T, de Graaf J, Boisguérin V, Türeci O, Diken M, Castle JC, Sahin U. HLA typing from RNA-Seq sequence reads. Genome Med. 2012;4:102. doi: 10.1186/gm403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabibzadeh SS, Sivarajah A, Carpenter D, Ohlsson-Wilhelm BM, Satyaswaroop PG. Modulation of HLA-DR expression in epithelial cells by interleukin 1 and estradiol-17 beta. J Clin Endocrinol Metab. 1990;71:740–747. doi: 10.1210/jcem-71-3-740. [DOI] [PubMed] [Google Scholar]
- 33.Kocatürk B, Versteeg HH. Orthotopic injection of breast cancer cells into the mammary fat pad of mice to study tumor growth. J Vis Exp. 2015 Feb 8; doi: 10.3791/51967. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lanzós A, Carlevaro-Fita J, Mularoni L, Reverter F, Palumbo E, Guigó R, Johnson R. Discovery of cancer driver long noncoding RNAs across 1112 tumour genomes: New candidates and distinguishing features. Sci Rep. 2017;7:41544. doi: 10.1038/srep41544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu MT, Hu JW, Yin R, Xu L. Long noncoding RNA: An emerging paradigm of cancer research. Tumour Biol. 2013;34:613–620. doi: 10.1007/s13277-013-0658-6. [DOI] [PubMed] [Google Scholar]
- 36.Wenric S, ElGuendi S, Caberg JH, Bezzaou W, Fasquelle C, Charloteaux B, Karim L, Hennuy B, Frères P, Collignon J, et al. Transcriptome-wide analysis of natural antisense transcripts shows their potential role in breast cancer. Sci Rep. 2017;7:17452. doi: 10.1038/s41598-017-17811-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nie L, Wu HJ, Hsu JM, Chang SS, Labaff AM, Li CW, Wang Y, Hsu JL, Hung MC. Long non-coding RNAs: Versatile master regulators of gene expression and crucial players in cancer. Am J Transl Res. 2012;4:127–150. [PMC free article] [PubMed] [Google Scholar]
- 38.Pan YF, Feng L, Zhang XQ, Song LJ, Liang HX, Li ZQ, Tao FB. Role of long non-coding RNAs in gene regulation and oncogenesis. Chin Med J (Engl) 2011;124:2378–2383. [PubMed] [Google Scholar]
- 39.Collette J, Le Bourhis X, Adriaenssens E. Regulation of human breast cancer by the long non-coding RNA H19. Int J Mol Sci. 2017;18:18. doi: 10.3390/ijms18112319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miao Y, Fan R, Chen L, Qian H. Clinical significance of long non-coding RNA MALAT1 expression in tissue and serum of breast cancer. Ann Clin Lab Sci. 2016;46:418–424. [PubMed] [Google Scholar]
- 41.Avazpour N, Hajjari M, Tahmasebi Birgani M. HOTAIR: A promising long non-coding RNA with potential role in breast invasive carcinoma. Front Genet. 2017;8:170. doi: 10.3389/fgene.2017.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiu HS, Somvanshi S, Patel E, Chen TW, Singh VP, Zorman B, Patil SL, Pan Y, Chatterjee SS, Sood AK, et al. Cancer Genome Atlas Research Network Pan-Cancer Analysis of lncRNA regulation supports their targeting of cancer genes in each tumor context. Cell Rep. 2018;23:297–312.e12. doi: 10.1016/j.celrep.2018.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iorns E, Drews-Elger K, Ward TM, Dean S, Clarke J, Berry D, El Ashry D, Lippman M. A new mouse model for the study of human breast cancer metastasis. PLoS One. 2012;7:e47995. doi: 10.1371/journal.pone.0047995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagini S. Breast Cancer: Current molecular therapeutic targets and new players. Anticancer Agents Med Chem. 2017;17:152–163. doi: 10.2174/1871520616666160502122724. [DOI] [PubMed] [Google Scholar]
- 45.Cavo M, Fato M, Peñuela L, Beltrame F, Raiteri R, Scaglione S. Microenvironment complexity and matrix stiffness regulate breast cancer cell activity in a 3D in vitro model. Sci Rep. 2016;6:35367. doi: 10.1038/srep35367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kopparapu PK, Tinzl M, Anagnostaki L, Persson JL, Dizeyi N. Expression and localization of serotonin receptors in human breast cancer. Anticancer Res. 2013;33:363–370. [PubMed] [Google Scholar]
- 47.Pai VP, Marshall AM, Hernandez LL, Buckley AR, Horseman ND. Altered serotonin physiology in human breast cancers favors paradoxical growth and cell survival. Breast Cancer Res. 2009;11:R81. doi: 10.1186/bcr2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Weinberg MS, Zerbini L, Prince S. The oncogenic TBX3 is a downstream target and mediator of the TGF-β1 signaling pathway. Mol Biol Cell. 2013;24:3569–3576. doi: 10.1091/mbc.e13-05-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peres J, Davis E, Mowla S, Bennett DC, Li JA, Wansleben S, Prince S. The highly homologous T-Box transcription factors, TBX2 and TBX3, have distinct roles in the oncogenic process. Genes Cancer. 2010;1:272–282. doi: 10.1177/1947601910365160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Forero A, Li Y, Chen D, Grizzle WE, Updike KL, Merz ND, Downs-Kelly E, Burwell TC, Vaklavas C, Buchsbaum DJ, et al. Expression of the MHC Class II pathway in triple-negative breast cancer tumor cells is associated with a good prognosis and infiltrating lymphocytes. Cancer Immunol Res. 2016;4:390–399. doi: 10.1158/2326-6066.CIR-15-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doonan BP, Haque A. HLA Class II antigen presentation in prostate cancer cells: A novel approach to prostate tumor immu-notherapy. Open Cancer Immunol J. 2010;3:1–7. doi: 10.2174/1876401001003010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thibodeau J, Bourgeois-Daigneault MC, Lapointe R. Targeting the MHC Class II antigen presentation pathway in cancer immunotherapy. OncoImmunology. 2012;1:908–916. doi: 10.4161/onci.21205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walsh NC, Kenney LL, Jangalwe S, Aryee KE, Greiner DL, Brehm MA, Shultz LD. Humanized mouse models of clinical disease. Annu Rev Pathol. 2017;12:187–215. doi: 10.1146/annurev-pathol-052016-100332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao Y, Shuen TWH, Toh TB, Chan XY, Liu M, Tan SY, Fan Y, Yang H, Lyer SG, Bonney GK, et al. Development of a new patient-derived xenograft humanised mouse model to study human-specific tumour microenvironment and immunotherapy. Gut. 2018;67:1845–1854. doi: 10.1136/gutjnl-2017-315201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the Gene Expression Omnibus repository, [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE35001]