Abstract
Angiogenesis has an important role in tumour cell growth and metastasis. Anisomycin has been shown to inhibit tumour cell growth. However, whether anisomycin can inhibit angiogenesis of tumours has not been reported. The present study demonstrated that there was a positive correlation between tumour angiogenesis and the number of CD44+/CD133+ serous human ovarian cancer stem cells (HuOCSCs). Subsequently, it was confirmed that anisomycin significantly inhibited the proliferation, invasion, tumorigenic ability and tumour angiogenesis of HuOCSCs. Gene expression profiling by cDNA microarrays revealed that the expression levels of vascular endothelial cell markers, platelet-derived growth factors, Notch pathway components and 27 tumour angiogenesis-related genes were significantly decreased in the anisomycin-treated group compared with the control group. Further experiments demonstrated that the expression levels of endogenous long non-coding RNA (lncRNA) maternally expressed 3 (Meg3) were significantly decreased in anisomycin-treated HuOCSCs, whereas the expression levels of microRNA (miR)-421 were significantly increased. The results of luciferase reporter assays indicated that, when miR-421 was overexpressed in cells, the luciferase activities of wild-type platelet derived growth factor receptor α (PDGFRA) 3′ untranslated region and Meg3 reporter plasmids were significantly decreased. Overexpression of miR-421 in HuOCSCs significantly enhanced the anisomycin-mediated inhibition of HuOCSC proliferation. Taken together, the present results demonstrated that anisomycin inhibited the activation downstream of the Notch1 pathway by attenuating the molecular sponge effect of the lncRNA-Meg3/miR-421/PDGFRA axis, ultimately inhibiting angiogenesis, proliferation and invasion in ovarian cancer cells.
Keywords: ovarian cancer, angiogenesis, anisomycin, long non-coding RNA maternally expressed 3, microRNA-421, platelet derived growth factor receptor α
Introduction
Angiogenesis is crucial for the growth and metastasis, and therefore the prognosis, of malignant solid tumours (1-4). Angiogenesis is necessary for the malignant progression of solid tumours after penetrating the epithelial basement membrane (1-4). Generally, there is a positive correlation between the rate and extent of angiogenesis and the unfavourable prognosis of tumours (1-4). Newly formed blood vessels in tumours are characterized by unique structures, such as incomplete vessel walls that lack vascular smooth muscles and are composed of only porous endothelial cells and a flaky basement membrane (1-4). A growing number of studies have noted that the structural features of new blood vessels provide favourable conditions for the distant metastasis of malignant tumour cells (1-5). Therefore, the number of new blood vessels in malignant solid tumours is an important independent prognostic factor (1-4,6).
Anisomycin [also known as 3,4-Pyrrolidinediol,2- [(4-methoxyphenyl)methyl]-,3-acetate,(2R,3S,4S)] is the first confirmed anti-protozoal antibiotic extracted from Streptomyces griseolus (7,8). Anisomycin inhibits the formation of peptide bonds in most cells by binding to the 60S ribosomal subunit and, thereby, inhibiting protein synthesis (7,8). Previous studies have shown that anisomycin can promote the production of amyloid β (Aβ) 1-42 causing neurotoxicity, as well as inhibit the proliferation of certain cancer cells. Yu et al (9) found that anisomycin inhibited the proliferation of Jurkat T cells by promoting the expression of p53, p21 and p27, thus stopping the cells from entering the S and G2/M phases. Seo et al (10) reported that anisomycin could induce apoptosis in renal tumour cells by downregulating Bcl-2, c-FLIPL and Mcl-1. Liu et al (11) also demonstrated that anisomycin induced the apoptosis of cancer cells in glucocorticoid-resistant acute lymphoblastic leukaemia by facilitating the phosphorylation of the mitogen-activated protein kinase p38 and the activation of JNK. Therefore, anisomycin may be a promising chemotherapeutic drug.
Long non-coding RNAs (lncRNAs) are transcripts of >200 nt in length that are produced by RNA polymerase II and matured by modifications, including alternative splicing, 5′-end capping, and 3′-end polyadenylation (7,12-15). LncRNA lacks the complete open reading frame required for translation, does not encode proteins and has a poorly conserved primary structure (7,12-15). An increasing number of studies have found that lncRNA extensively regulates eukaryotic cell function and the development of diseases (7,12-15). LncRNA can regulate the activity of genes at the transcriptional level, but can also regulate the translation and silencing of mRNAs at the post-transcriptional level (7,12-15). At the transcriptional level, lncRNA modulates gene transcription by mimicking DNA elements, competitively binding transcription factors or forming scaffold structures, and recruiting multiple trans-acting factors (7,12-15). At the post-transcriptional level, lncRNA acts as a 'molecular sponge' that binds to a target microRNA (miRNA) and blocks its activity, abolishing its RNA interfering effect on the target gene (7,12-15). Multiple studies have indicated that lncRNAs regulate tumour cell proliferation, metastasis and angiogenesis at multiple levels (16-20).
Based on the evidence mentioned above, the present study explored the effects of anisomycin on CD44+/CD133+ human ovarian cancer stem cells (HuOCSCs), and specifically on their in vitro and in vivo proliferation, invasion, angiogen-esis and in vivo tumorigenicity. The results demonstrated that anisomycin could significantly inhibit the growth and angiogenesis of ovarian cancer cells in vitro and in vivo. Next, the present study examined the epigenetic mechanism of the anisomycin-mediated inhibition of angiogenesis in ovarian cancer cells. Anisomycin was demonstrated to attenuate the molecular sponge effect of lncRNA-Meg3, releasing miR-421, which then silenced the expression of the target gene platelet derived growth factor receptor α (PDGFRA), ultimately inhibiting angiogenesis.
Materials and methods
Isolation and culture of primary HuOCSCs
The experiment was performed according to previously described methods (21). Briefly, surgically isolated tissues from 4 ovarian cancer patients (Table I) were minced, digested with 0.25% trypsin (Gibco; Thermo Fisher Scientific, Inc.), and centrifuged at 4°C, 450 × g for 5 min. The cell precipitates were collected and incubated with mouse anti-human CD133-FITC antibodies (1:100; cat. no. 11-1339-42; eBioscience; Thermo Fisher Scientific, Inc.) and rabbit anti-human CD44-APC antibodies (1:100; cat. no. 17-0441-82; eBioscience; Thermo Fisher Scientific, Inc.) at 4°C for 30 min. Next, CD44+/CD133+ OCSCs were sorted from the sample by fluorescence-activated cell sorting. After sorting, single cells were plated at 1,000 cells/ml in DMEM/F12 (Gibco; Thermo Fisher Scientific, Inc.), supplemented with 10 ng/ml basic fibroblast growth factor, 10 ng/ml epidermal growth factor, 5 µg/ml insulin and 0.5% BSA (all from Sigma-Aldrich; Merck KGaA, and cultured at 37°C with 5% CO2. Cells of the fourth passage were used to perform subsequent experiments. The study involving human tissues was approved by the Ethics Review Committee of Shanghai Geriatric Institute of Chinese Medicine of Research in Human Production, authorized by Shanghai Municipal Government; written informed consent was provided by all patients, in accordance with The Declaration of Helsinki.
Table I.
Characteristics of the patient cohort.
| Characteristic | Patients (n=4) |
|---|---|
| Age median (range) | (39-62) |
| ≤40 | 1 |
| 40-60 | 2 |
| ≥60 | 1 |
| Surgical staging | |
| I a, b, c | 1 |
| II a, b, c | 2 |
| III a, b, c | 1 |
| IV | - |
| Histopathology | |
| Serous | 4 |
| Endometrioid | - |
| Mucinous | - |
| Clear cells | - |
| Others (mixed epithelial, undifferentiated) | - |
| Tumor grade | |
| 1 | 1 |
| 2 | 2 |
| 3 or clear cell | 1 |
| Unknown | - |
| Treatments | |
| Primary surgery | 4 |
| Radical surgery | - |
| Secondary surgery | - |
| Platinum-based chemotherapy | 1 |
| Radiotherapy | - |
Treatment of CD44+/CD133+ HuOCSCs with anisomycin
Anisomycin (purity >98%) and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich (Merck KGaA). According to the IC50 value determined by MTT assay, anisomycin was applied at a final concentration of 31.8 mM in all the assays. As a control, the same volume of DMSO was applied to the cells.
Histopathology by haematoxylin and eosin (H&E) staining
Briefly, all fresh tissues were soaked in 4% paraformaldehyde (Sigma-Aldrich; Merck KGaA) at room temperature for 30 min. The tissues were dehydrated using an ethanol gradient and embedded in paraffin. Then, the tissues were sectioned (thickness, 6 µm) and soaked in xylene for dewaxing. The tissue sections were stained with haematoxylin and eosin (Sigma-Aldrich; Merck KGaA) and were finally cleared with xylene and mounted with neutral resin (Sigma-Aldrich; Merck KGaA). Five random fields of each tissue section from a total 4 slides were observed (magnification, ×200) with a light microscope (Olympus CX23; Olympus Corporation).
Immunohistochemistry (IHC) analysis
Briefly, all fresh tissues were soaked in 4% paraformaldehyde (Sigma-Aldrich; Merck KGaA) at room temperature for 30 min. The tissues were dehydrated using an ethanol gradient and embedded in paraffin. Then, the tissues were sectioned (thickness, 6 µm) and soaked in xylene for dewaxing. Next, sections were rinsed with 3% phosphate buffer (Sigma-Aldrich; Merck KGaA). Mouse anti-human CD34 and CD31 primary antibodies (1:200; cat nos. 3569 and 3528, respectively; Cell Signaling Technology, Inc.) were added for 60 min at room temperature, and then anti-mouse horseradish peroxidase-conjugated secondary antibody (1:200; cat no. 7076; Cell Signaling Technology, Inc.) was added for 60 min at room temperature. Finally, ABC chromo-genic reagent (VECTASTAIN Elite ABC kit; cat no. PK-6100; Vector Laboratories, Inc.) was used for the color reaction. PBS (pH 7.4) was used as a negative control in the place of the primary antibody. Five random fields of each tissue section were observed (magnification, ×200) and analyzed using IPP software (Image-Pro Plus Version 6.0; Media Cybernetics Co. Ltd.). ImageJ 1.42q (National Institutes of Health) was used to analyze the area of the positive cells on IHC staining.
miR-421 overexpression by RNA mimics transfection
miR-421 and miR-421 mutant (miR-mut; a miRNA mimics negative control) oligonucleotide RNAs were purchased from Shanghai GenePharma Co., Ltd. HuOCSCs were transfected with 0.2 mg miR-421 mimics or miR-mut using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The cells were used for subsequent experiments at 24 h post-transfection.
Cell proliferation assay
Briefly, a total 1,000 cells from each group in 100 µl medium were seeded in a 96-well plate. After 24 h, 10 µl of MTT solution (Sigma-Aldrich; Merck KGaA) was added to each group of cells for incubation at 37°C for 3 h. The medium was discarded, 150 µl of DMSO was added to each well, and the plate was shaken for 15 sec to mix well. The culture plate was placed in a microplate reader to record the absorbance value at 450 nm. The formula for calculating the cell proliferation inhibition rate (%) was: [1-(OD value of experimental group of cells-blank)/(OD value of control group of cells-blank)] ×100.
Annexin V/propidium iodide (PI) staining and flow cytometry
The experiment was performed according to the instruction manual of the Annexin V-FITC Apoptosis Detection kit (Beyotime Institute of Biotechnology). Briefly, adherent cells were digested using trypsin. The cells were washed with PBS once, centrifuged, and gently resuspended in 195 µl of Annexin V-FITC binding solution. Next, 5 µl of Annexin V-FITC was added, and the sample was gently mixed. Finally, 10 µl of PI staining solution was added, and the sample was gently mixed and incubated at 20°C in the dark for 30 min. The cells were then detected using a flow cytometer (Cytomics FC 500; Beckman Coulter, Inc.) and analysed using FlowJo software (Version 7.6.1; FlowJo, LLC).
Transwell invasion assay
The upper chambers of the tran-swell filters (8.0-µm pore, 6.5-mm polycarbonate; Corning, Inc.) were pre-coated with 50 µl Matrigel (Corning, Inc.). Cells (2×105) were resuspended in 200 µl serum-free DMEM/F12 and seeded on the upper chambers of the Transwell filters. A total of 600 µl DMEM/F12 containing 10% FBS was added to the bottom chambers. Cells were allowed to invade for 24 h at 37°C in a humidified incubator with 5% CO2. Cells invaded to the lower surface of the filter were fixed in 4% paraformaldehyde at room temperature for 30 min, and stained with DAPI (Beyotime Institute of Biotechnology). Invaded cells were then counted using a fluorescent microscope (Olympus CX23; Olympus Corporation). Five random optical fields were counted for each Transwell filter.
Capillary tubule formation assay
All steps were performed according to a previously described method (22). In brief, human umbilical vein epithelial cells (HUVECs) were divided into three experimental groups: the non-treated group, the anisomycin-treated group (anisomycin was applied at a final concentration of 31.8 mM; dilution for 1:1,000) and the vehicle-treated group (the same volume of DMSO was applied to the cells). Each group cells was plated on Matrigel-coated 6-chamber slides (2×105 cells/chamber), in the presence or absence of treatments as aforementioned. After 6 h of incubation, the cells were photographed using a light microscope (Olympus CX23; Olympus Corporation). To quantitate the data, the number of branch points in four non-overlapping fields was counted.
In vivo xenograft experiments
Female BALB/c nude mice (n=4; weight, ~20 g; age, 8 weeks) were obtained from the Shanghai Research Center for Model Organisms (Shanghai, China). This study was approved (permit no. SRCMR20160018) by the Animal Ethics Committee of Shanghai Research Center for Model Organisms, and the experimental protocols were in compliance with the Experimental Animal Regulations of the Ministry of Science and Technology National Science and Technology Commission (Beijing, China). All mice were maintained in colonies of 1 per cage for 7 days, and housed in a temperature-controlled room under a standard light-dark cycle, with free access to food and water. Two types of cells HuOCSCs were inoculated into the back of all mice: DMSO-pretreated cells (total 1×105 cells in 100 µl per mouse) were inoculated into the scapula and anisomycin-pretreated cells (total 1×105 cells in 100 µl per mouse) were inoculated into the lower back. Each experimental group comprised four mice. After 35 weeks, the mice were sacrificed and the tumors were surgically excised. Tumors were weighed, and the volume was calculated as follows: Tumor volume (mm3) = [longest axis (mm) x shortest axis (mm)2]/2.
For the histological analysis of the xenograft tumors, the fresh tumor tissues were soaked in 4% paraformaldehyde (Sigma-Aldrich; Merck KGaA) at room temperature for 30 min. The tumor tissues were dehydrated using an ethanol gradient and embedded in paraffin. Then, the paraffin blocks were sectioned (thickness, 6 µm) and soaked in xylene for dewaxing. The tumor tissue sections were stained with hematoxylin and eosin (Sigma-Aldrich; Merck KGaA) and were finally cleared with xylene and mounted with neutral resin (Sigma-Aldrich; Merck KGaA). Three random fields of each tumor tissue section from a total 4 slides were observed (magnification, ×200) using a light microscope (Olympus CX23; Olympus Corporation). In addition, immunofluorescence analysis was performed. The tumor tissue sections were rinsed with 3% phosphate buffer, and mouse anti-CD31 (clone P2B1; cat. no. ab24590; Abcam), rabbit anti-CD146 (clone EPR3208; cat. no. ab75769; Abcam), mouse anti-coagulation factor VIII (FVIII; clone GMA-012; ab78852; Abcam) and rabbit anti-fms related tyrosine kinase 1 (Flt1; clone Y103; cat. no. ab32152; Abcam) were added at 1:200 dilution for 60 min at room temperature. Then, the tissue sections were incubated with goat anti-mouse (Alexa Fluor® 555 pre-adsorbed; 1:200; ab150118; Abcam) or goat anti-rabbit secondary antibody (Alexa Fluor® 488 pre-adsorbed; 1:200; ab150077; Abcam) and 5 µg/ml DAPI at room temperature for 60 min. The tumor tissue sections were then washed thoroughly with TBST [25 mM Tris/HCl (pH 8.0), 125 mM NaCl and 0.05% Tween-20] and observed under a fluorescence microscope (DMI3000; Leica Microsystems, Inc.). PBS (pH 7.4) was used as a negative control in the place of the primary antibodies. Three random fields of each tissue section were observed (magnification, ×200) and analyzed using IPP software (Image-Pro Plus Version 6.0; Media Cybernetics Co. Ltd.). ImageJ 1.42q (National Institutes of Health) was used to analyze the area of the positive cells on the immunofluorescence staining.
Extraction of total RNA and quantitative PCR (qPCR)
Total RNA from each group of cells was extracted using TRIzol reagent (Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. Total RNA was treated with DNase I (Sigma-Aldrich; Merck KGaA), quantified and reverse transcribed into cDNA using the ReverTra Ace-α First Strand cDNA Synthesis kit (Toyobo Life Science). qPCR was performed with a RealPlex4 real-time PCR detection system from Eppendorf Co. Ltd. SYBR Green Real-Time PCR Master Mix (Toyobo Life Science) was used as the fluorescent dye. qPCR was performed as follows: 95°C for 5 min, then 40 cycles of denaturation at 95°C for 15 sec, annealing at 58°C for 30 sec and extension at 72°C for 42 sec, followed by a final extension step at 72°C for 10 min. The relative gene expression levels were calculated using the 2−ΔΔCq method (ΔCq=Cq of gene -Cq of 18s rRNA; ΔΔCq=ΔCq of experimental group-ΔCq of control group) (23). The mRNA expression levels were normalised to the expression levels of 18s rRNA. The primers for amplification of each gene were as follows: Notch1, forward 5′-CGC TGA CGG AGT ACA AGTG-3′ and reverse 5′-GTA GGA GCC GAC CTC GTT G-3′; d-like canonical Notch ligand 1 (DLL1), forward 5′-TGT GAC GAG TGT ATC CGC TAT-3′ and reverse 5′-GTG TGC AGT AGT TCA GGT CCT-3′; hes family bHLH transcription factor 1 (HES1), forward 5′-CCT GTC ATC CCC GTC TAC AC-3′ and reverse 5′-CAC ATG GAG TCC GCC GTA A-3′; recombination signal binding protein for immunoglobulin κ J region (RBPJ), forward 5′-AAC AAA TGG AAC GCG ATG GTT-3′ and reverse 5′-GGC TGT GCA ATA GTT CTT TCC TT-3′; PDGFRA, forward 5′-TTG AAG GCA GGC ACA TTT ACA-3′ and reverse 5′-GCG ACA AGG TAT AAT GGC AGA AT-3′; platelet derived growth factor receptor β (PDGFRB), forward 5′-TGA TGC CGA GGA ACT ATT CAT CT-3′ and reverse 5′-TTT CTT CTC GTG CAG TGT CAC-3′; miR-421, forward 5′-ATC AAC AGA CAT TAA TTG GGC GC-3′ and reverse 5′-GCT GTC AAC GAT ACG CTAC CTA-3′; miR-145, forward 5′-GTC CAG TTT TCC CAG GAA TCC CT-3′ and reverse 5′-GCT GTC AAC GAT ACG CTA CCT A-3′; lncRNA-Meg3, forward 5′-GCC CAT CTA CAC CTC AC-3′ and reverse 5′-ATC CTT TGC CAT CCT G-3′; and 18S rRNA, forward 5′-CAG CCA CCC GAG ATT GAG CA-3′ and reverse 5′-TAG TAG CGA CGG GCG GTG TG-3′.
Western blot analysis
Briefly, cells were lysed in 2X loading lysis buffer [50 mM Tris-HCl (pH 6.8), 2% sodium dodecyl sulfate, 10% β-mercaptoethanol, 10% glycerol and 0.002% bromophenol blue]. The concentration of total protein was then determined by Enhanced BCA Protein Assay kit (Beyotime Institute of Biotechnology). Total proteins (25 µg/µl) from each group were then separated by 12% SDS-PAGE and transferred onto Hybrid-polyvinylidene fluoride membranes (EMD Millipore). The membranes were blocked with 5% (w/v) non-fat dried milk in TBST and washed four times with TBST at room temperature (15 min per wash) prior to incubation with the primary antibodies (all final dilution 1:1,000) in 5% non-fat dried milk in TBST) at 37°C for 1 h. The primary antibodies were: Mouse anti-CD31 (clone P2B1; cat. no. ab24590; Abcam), rabbit anti-CD146 (clone EPR3208; cat. no. ab75769; Abcam), mouse anti-FVIII (clone GMA-012; ab78852; Abcam), rabbit anti-vascular endothelial growth factor receptor 1 (VEGFR1; clone Y103; cat. no. ab32152; Abcam), rabbit anti-PDGFRA (clone D1E1E; cat. no. 3174; Cell Signalling Technology, Inc.), rabbit anti-PDGFRB (clone 28E1; cat. no. 3169, Cell Signalling Technology, Inc.), rabbit anti-argonaute 2 (Ago2; clone C34C6; cat no. 2897; Cell Signalling Technology, Inc.), Notch Activated Targets Antibody Sampler kit (cat. no. 68309; Cell Signalling Technology, Inc.), and rabbit anti-GAPDH antibody (clone EPR16891; cat. no. ab181602; Abcam). After washing, the membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse secondary antibody (1:1,000; cat. nos. ab97051 and ab6789; Abcam) at 37°C for 1 h. After washing four times with TBST at room temperature (15 min per wash), the immunoreactive protein bands were visualized by enhanced chemiluminescence (ECL) using an ECL kit (PerkinElmer, Inc.).
Effect of anisomycin on vascular development of zebrafish embryos
In a 6-well plate, a total of 30 24-h post-fertilization zebrafish embryos of the fli1a-EGFP/casper strain (Shanghai Research Centre for Model Organisms, Shanghai, China) were added to each well. DMSO (0.1%) was added to the E3 embryonic culture medium (0.58 g/l NaCl, 0.27 g/l KCl, 0.97 g/l CaCl2·2H2O, 0.16 g/l MgCl2·6H2O, 1% methylene blue; pH 7.2; all from Sigma-Aldrich; Merck KGaA) in the control group, and 31.8 mM anisomycin was used in the experimental group. After 8 h of treatment, the medium was replaced with fresh embryonic culture medium, and the phenotypic changes of zebrafish embryos were photographed under a Nikon SMZ 1500 stereomicroscope (Nikon Corporation; magnification, ×40). The numbers of fully-developed intersegmental blood vessels (ISV) and dorsal longitudinal anastomotic vessels (DLAV) in each embryo were counted. The inhibition rate of angiogenesis was calculated using the following formula (24): % inhibition of angiogenesis = [1-(average number of ISV in the embryoid body in the experimental group/average numbers of ISV and DLAV in the embryoid body in the control group)] ×100.
cDNA microarray analysis
Total RNA samples from each group of cells were labeled using Agilent's Low RNA Input Fluorescent Linear Amplification kit (Agilent Technologies, Inc.), according to the manufacturer's protocol. Cy3-dCTP or Cy5-dCTP was incorporated during reverse transcription of 5 µg total RNA into cDNA. Different fluorescently labeled cDNA probes were mixed in 30 µl hybridization buffer (3X SSC, 0.2% SDS, 5X Denhardt's solution and 25% formamide) and applied to the microarray (CapitalBio human mRNA microarray V2.0; CapitalBio Technology, Inc.) at 42°Cfor 16 h. After hybridization, the slide was washed with 0.2% SDS/2X SSC at 42°C for 5 min, and then washed with 0.2X SSC at room temperature for 5 min. The fluorescent images of the hybridized microarray were scanned with an Agilent Whole Human Genome 4×44 microarray scanner system (Agilent Technologies, Inc.). Images and quantitative data of the gene expression levels were analyzed using the Agilent's Feature Extraction (FE) software, version 9.5 (Agilent Technologies, Inc.). Raw data were normalized with a robust multi-array average algorithm and analyzed using GeneSpring software (version 13.1; Agilent Technologies, Inc.). Differentially expressed genes (DEGs) were then identified via the fold-change (Sample group/Control group) in expression, as well as the P-value calculated from a t-test. The threshold set for upregulated and downregulated genes was: Log10 (Sample group/Control group) >3 was considered significantly upregulated expression; and log10 (Sample group/Control group) <-2 was considered significantly downregulated expression. Hierarchical clustering was then performed to demonstrate the expression profile of the DEGs among samples using MultiExperiment Viewer (version 4.9.0; http://mev.tm4.org/). Gene Ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG; https://www.genome.jp/kegg/) pathway analyses of the identified DEGs were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) database. Finally, protein-protein interaction networks were constructed using the Search Tool for the Retrieval of Interacting Genes (STRING version 11.0; https://string-db.org/) database and visualized using Cytoscape software (version 3.4.0; https://cytoscape.org/).
Luciferase reporter assay
All steps of the luciferase reporter assay were performed as previously described (21). NIH-3T3 cells were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China), and cultured with DMEM medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10 % FBS (Gibco; Thermo Fisher Scientific, Inc.). NIH-3T3 cells were seeded at 3×104/well in 48-well plates and co-transfected with 400 ng of miR-421 mimics or miR-134-mut control, 20 ng of pGL3 cm-PDGFRA-3UTR-WT or pGL3 cm-PDGFRA-3UTR-Mut (NovoBio, Ltd.), pGL3 cm-Meg3-WT (NovoBio, Ltd.) or pGL3 cm-Meg3-Mut (NovoBio, Ltd.), and pRL-TK (Promega Corporation) using Lipofectamine 2000 reagent (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. At 48 h post-transfection, the Firefly and Renilla luciferase activities were measured using the dual-luciferase reporter assay system (Promega Corporation).
RNA immunoprecipitation (RIP)-PCR
RIP experiments were performed using the Magna RIP RNA-Binding Protein Immunoprecipitation kit (EMD Millipore). All the steps of RIP were performed as previously described (12). Briefly, cells from all groups were lysed (500 µl per plate) in a modified cell lysis buffer used for western blotting and immunoprecipitation (20 mM Tris pH7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, sodium pyrophosphate, β-glycerophosphate, Na3VO4 and leupeptin; Beyotime institute of Biotechnology). After lysis, each sample was centrifuged at 4°C, 1,200 × g for 10 min to clear the insoluble debris and the supernatant was pre-incubated with 20 µg protein A agarose beads (Beyotime institute of Biotechnology) by rocking for 30 min at 4°C, followed by centrifugation (4°C, 450 × g for 2 min) and transfer to a new 1.5 ml tube. The mouse anti-human Ago2 monoclonal antibody (clone C34C6; cat no. 2897; Cell Signaling Technology, Inc.; 1:100) was added for 90 min, before the re-addition of 20 µg of protein A agarose beads to capture the immune complexes. The agarose beads were then washed three times with ice-cold homogenization buffer. Then, the co-precipitated RNAs were isolated by resuspending the beads in TRIzol RNA extraction reagent (Thermo Fisher Scientific, Inc.), and extracted using the RNeasy Mini kit (Qiagen GmbH). Total RNAs were subjected to reverse transcription using a ReverTra Ace-α First Strand cDNA Synthesis kit (Toyobo Life Science). PCR amplification was performed for 31 cycles as follows: Denaturation at 95°C for 30 sec, annealing at 65°C for 30 sec and extension at 72°C for 42 sec, using Easy-Load™ PCR Master Mix (cat. no. D7251-1 ml; Beyotime Institute of Biotechnology). The amplification products were visualized by 1.2% agarose gel electrophoresis. The primers for amplification of each gene were as follows: miR-421, forward 5′-ATC AAC AGA CAT TAA TTG GGC GC-3′ and reverse 5′-GCT GTC AAC GAT ACG CTA CCT A-3′; and lncRNA-Meg3, forward 5′-GCC CAT CTA CAC CTC AC-3′ and reverse 5′-ATC CTT TGC CAT CCT G-3′.
Statistical analysis
Each experiment was performed as least three times, and data are presented as the mean ± standard error where applicable. Differences were evaluated using one-way ANOVA followed by Dunnett's test. Statistical analysis was performed by GraphPad Prism Version 5.00 (GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
Angiogenesis is positively correlated to the number of CD44+/CD133+ cells in ovarian cancer tissues
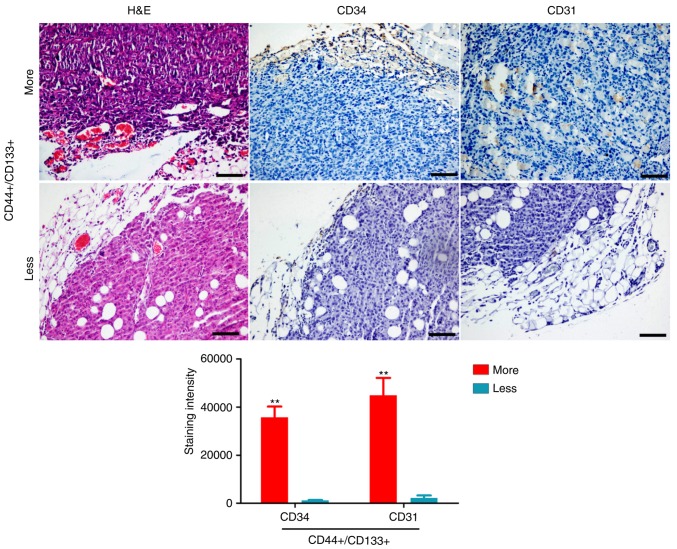
First, the association between HuOCSCs and tumour angiogenesis was investigated. Flow cytometry analysis revealed a large difference in the number of CD44+/CD133+ HuOCSCs in serous ovarian cancer tissues from four different patients (Fig. 1). Subsequently, H&E staining revealed that there were more new blood vessels in tissues from P2 patient with increased numbers of CD44+/CD133+ cells than in tissues from P4 patient with less CD44+/CD133+ cells (Fig. 2). Immunohistochemical staining confirmed that the expression of CD31 and CD34 was significantly higher in tissues from P2 patient with more CD44+/CD133+ cells than in tissues from P4 patient with less CD44+/CD133+ cells (Fig. 2). Therefore, these results indicated that higher content of CD44+/CD133+ cells in ovarian cancer tissues may be associated with increased angiogenesis.
Figure 1.
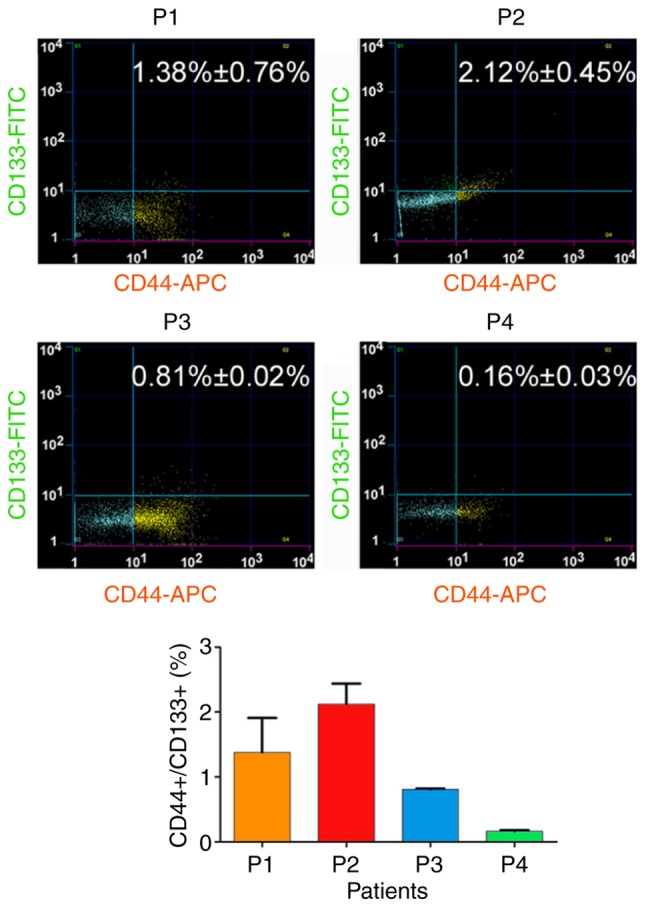
Proportion of CD44+/CD133+ cancer stem cells in ovarian cancer tissues of each patient by flow cytometry.
Figure 2.
New blood vessels in ovarian cancer tissues. Representative images and quantification from H&E and immunohistochemical staining for the blood vessel markers CD34 and CD31 (magnification, ×100). **P<0.01.
Anisomycin inhibits the activity of CD44+/CD133+HuOCSCs in vitro
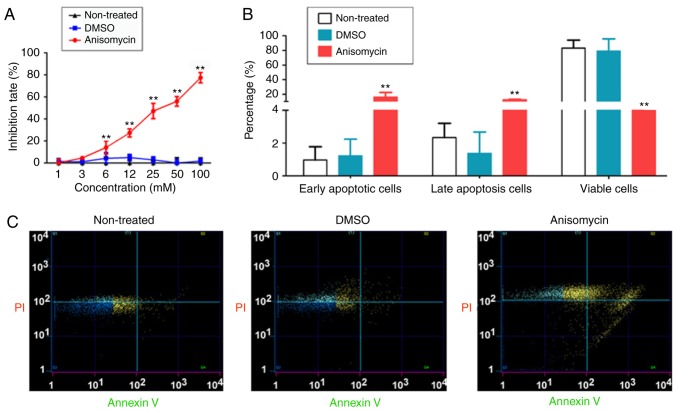
The effect of different concentrations (1-100 mM) of anisomycin was next analysed on the proliferation of CD44+/CD133+ HuOCSCs in vitro. The MTT assay results demonstrated that after 36 h of cell treatment, the inhibition rate of cell proliferation increased significantly with the increase of anisomycin concentration (Fig. 3A). The results showed that anisomycin caused inhibition of cell proliferation in a dose-dependent manner (IC50=31.8 mM). Flow cytometry analysis of Annexin V/PI double staining indicated that treatment of HuOCSCs with anisomycin for 36 h significantly increased the proportion of apoptotic cells in HuOCSCs, and significantly decreased the proportion of viable cells (Fig. 3B and C). In addition, Transwell invasion assays demonstrated that treatment of HuOCSCs with anisomycin for 36 h significantly weakened their invasive ability in Matrigel (Fig. 4A).
Figure 3.
Anisomycin inhibits proliferation of CD44+/CD133+ HuOCSCs. (A) Proliferation of HuOCSCs, treated as indicated, was evaluated with the MTT assay. (B) Quantification and (C) representative plots from Annexin V/PI staining and flow cytometry analysis of HuOCSCs, treated as indicated. **P<0.01 vs. non-treated cells (n=4). HuOCSCs, human ovarian cancer stem cells.
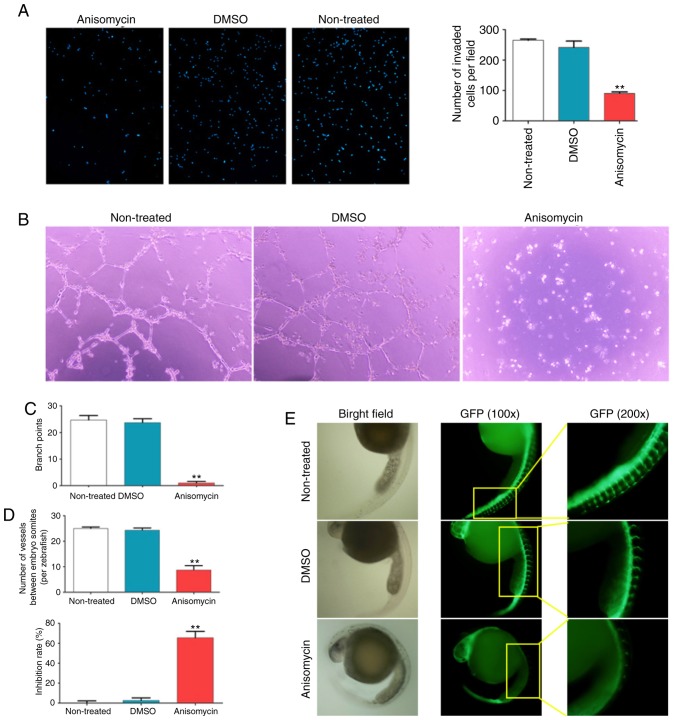
Figure 4.
Anisomycin inhibits HuOCSC invasion and angiogenesis in vitro. (A) A Transwell invasion assay was used to investigate the invasive properties of CD44+/CD133+ HuOCSCs following anisomycin treatment (n=4). Magnification, ×100. (B) Representative images and (C) quantification of the ability of HUVECs to form 3-dimensional tubular structures in Matrigel following anisomycin treatment (n=4). Magnification, ×100. (D) Quantification and (E) representative images of the growth and development of intersegmental vessels and dorsal longitudinal anastomotic vessels in zebrafish embryos following anisomycin treatment (n=30). Magnification, ×40. **P<0.01 vs. non-treated. HuOCSCs, human ovarian cancer stem cells; GFP, green fluorescent protein.
Next, the effects of anisomycin on angiogenesis were investigated. The results of an in vitro HUVEC capillary tubule formation assay showed that 36 h of treatment with anisomycin at the IC50 dose significantly inhibited the ability of HUVECs to form 3-dimensional tubular structures in Matrigel (Fig. 4B and C). Furthermore, zebrafish embryos of the fli1a-EGFP/casper strain were incubated with anisomycin at the IC50 dose for 24 h and the results demonstrated that anisomycin significantly inhibited the growth and development of ISV and DLAV (Fig. 4D and E); the inhibition rate of vascular development in the anisomycin-treated group was significantly higher compared with the DMSO-treated and non-treated groups (Fig. 4D). These results indicated that anisomycin significantly inhibited the proliferation and invasion of HuOCSCs, as well as inhibited angiogenesis of HUVECs.
Anisomycin inhibits the activity of CD44+/CD133+ HuOCSCs in vivo
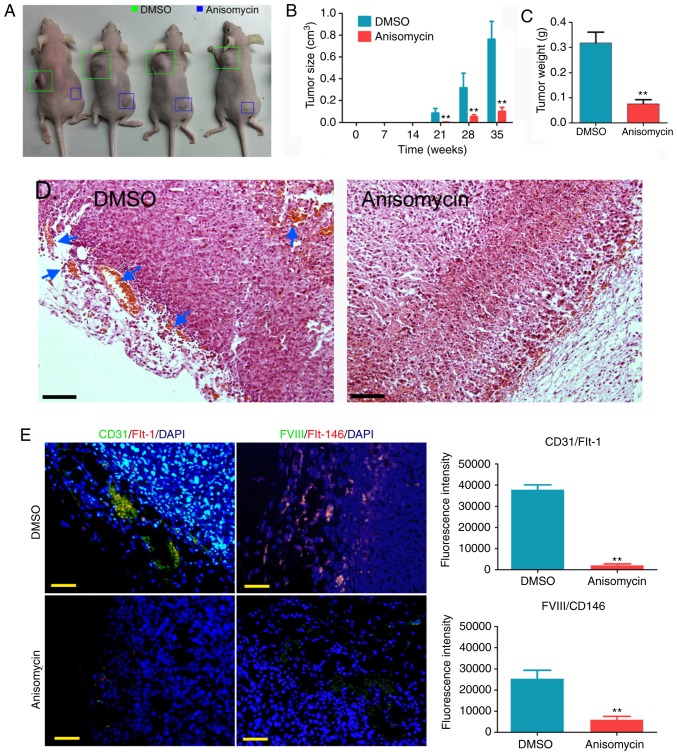
First, CD44+/CD133+ HuOCSCs were divided into two groups, one group was treated with anisomycin at IC50 dose, and the other group was treated with an equal volume of DMSO. After the two groups of cells were treated for 36 h as aforementioned, an equal number of cells was used to subcutaneously inoculate mice at the shoulder or hip. The two groups of nude mice were kept under the same conditions for 35 weeks. It was observed that tumours grew on both groups of nude mice at week 21 post-inoculation, but the tumours formed by DMSO-treated cells were markedly larger than those formed by anisomycin-treated cells (Fig. 5A). Over time, the volume of tumours formed by DMSO-treated HuOCSCs increased significantly, while the volume of tumours formed by anisomycin-treated HuOCSCs increased slowly; a statistically significant difference in tumour volume was observed between the two groups (Fig. 5B). At week 35, all nude mice were sacrificed, and the weights of tumours in each group were measured. The results demonstrated that the weight of the tumours formed by anisomycin-treated HuOCSCs was significantly decreased compared with that formed by DMSO-treated cells (Fig. 5C). Subsequently, H&E staining of the xenograft tumours revealed minimal neovascularization in the tumour tissues formed by anisomycin-treated HuOCSCs, whereas marked neovascularization was observed in the tumour tissues formed by DMSO-treated cells (Fig. 5D). In addition, immunofluorescence staining revealed that the density of CD31/Flt-1 and FVIII/CD146 double-positive cells in the tumour tissues formed by anisomycin-treated HuOCSCs was significantly decreased compared with that in the tumour tissues formed by DMSO-treated cells (Fig. 5E). Therefore, these results indicated that anisomycin significantly inhibited the proliferation and angiogenic ability of CD44+/CD133+ HuOCSCs in vivo.
Figure 5.
Anisomycin inhibits the in vivo activity of CD44+/CD133+ HuOCSCs. (A) Tumours formed by anisomycin-pre-treated and DMSO-pretreated HuOCSCs in nude mice. (B) Volume of tumours throughout the experiment. (C) Final tumour weights at 35 weeks. (D) H&E staining histopathology analysis of the xenografted tumours. Blood vessels are indicated by blue arrows. Magnification, ×200. (E) Immunofluorescence staining of the density of CD31/Flt-1 and FVIII/CD146 double-positive cells in the xenografted tumours. Magnification, ×200. **P<0.01 vs. DMSO group (n=4). HuOCSCs, human ovarian cancer stem cells; Flt-1, fms related tyrosine kinase 1; FVIII, coagulation factor VIII.
Anisomycin inhibits the expression of multiple angiogenesis-related genes
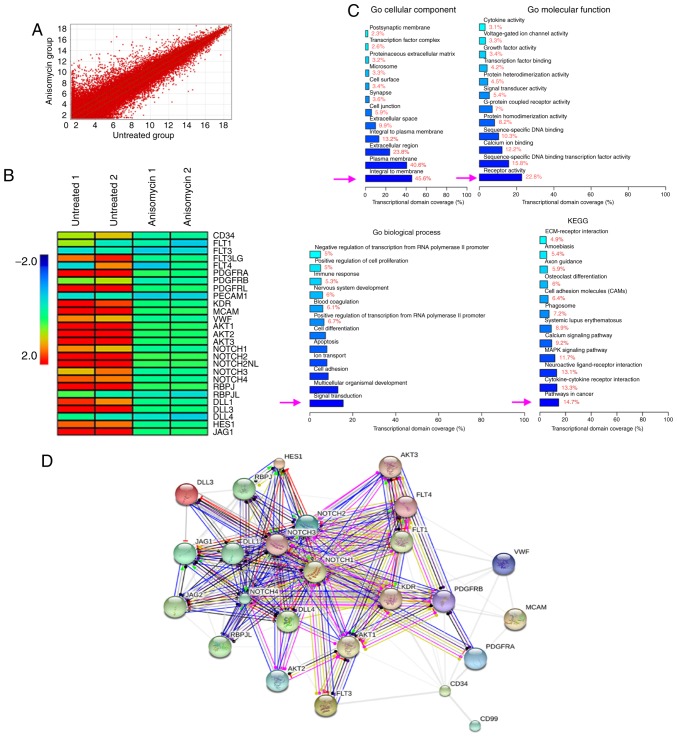
Using cDNA microarrays, changes in the expression profiles of a total of 14,740 genes were identified between the anisomycin-treated and the untreated groups (Fig. 6A). Among them, the mRNA expression levels of 1,797 genes changed significantly following treatment with anisomycin in CD44+/CD133+ HuOCSCs. The results indicated that the expression of vascular endothelial cell markers, platelet-derived growth factors (PDGFs), Notch pathway components, and 27 tumour angiogenesis-related genes, all showed significant downregulation (Fig. 6B; Table II). GO analysis revealed that in terms of cellular components, the most significant differences in gene expression were observed for genes included in the 'integral to membrane' section (Fig. 6C). In terms of molecular function, the most significant differences in gene expression were observed for genes included in the 'receptor activity' section (Fig. 6C). In terms of biological process, the most significant differences in gene expression were observed for genes included in the 'signal transduction' section (Fig. 6C). In addition, with regards to KEGG analysis, the most significant differences in gene expression between the anisomycin-treated and untreated groups were observed for genes included in the 'pathways in cancer' and 'cytokine-cytokine receptor interaction' sections (Fig. 6C). Further analysis with the STRING bioinformatics tool (https://string-db.org/) identified a network of interactions among angiogenesis-related proteins. The software predictions indicated that the vascular endothelial cell marker, PDGFs, Notch pathway components, and the proteins encoded by 27 angiogenesis-related genes were intrinsically interrelated, with multiple predicted interactions between them (Fig. 6D). These data indicated that anisomycin significantly inhibited the expression of multiple angiogenesis-related genes in ovarian cancer stem cells.
Figure 6.
Anisomycin inhibits the expression of multiple angiogenesis-related genes. (A) The results of cDNA microarrays revealed 1,797 genes with signifi-cant differences in expression following anisomycin treatment. (B) Heatmap analysis of the expression levels of vascular endothelial cell markers, PDGFs, Notch pathway components and 27 tumour angiogenesis-related genes, that were significantly downregulated in the anisomycin-treated group. (C) GO and KEGG enrichment analysis results. (D) Interaction network for the angiogenesis-related proteins identified by the microarray analysis. PDGF, platelet-derived growth factor; GO, Gene Ontology; KEGG, Kyoto Encyclopaedia of Genes and Genomes.
Table II.
Results of angiogenesis-related genes from the cDNA microarray analysis.
| Genbank accession no. | Gene symbol | Fold change (Anisomycin/Untreated) | Official full name | Genomic coordinates |
|---|---|---|---|---|
| NM_001025109 | CD34 | 0.113 | CD34 molecule | chr1:208062091-208062032 |
| NM_001160031 | FLT1 | 0.181 | fms-related tyrosine kinase 1 | chr13:28979986-28979927 |
| NM_004119 | FLT3 | 0.543 | fms-related tyrosine kinase 3 | chr13:28578051-28577993 |
| NM_001459 | FLT3LG | 0.032 | fms-related tyrosine kinase 3 ligand | chr19:49979743-49979802 |
| AB209637 | FLT4 | 0.066 | fms-related tyrosine kinase 4 | chr5:180046720-180046661 |
| NM_006206 | PDGFRA | 0.002 | platelet-derived growth factor receptora | chr4:55163989-55164048 |
| NM_002609 | PDGFRB | 0.156 | platelet-derived growth factor receptorb | chr5:149493523-149493464 |
| NM_006207 | PDGFRL | 0.006 | platelet-derived growth factor receptor-like | chr8:17500476-17500535 |
| NM_000442 | PECAM1 | 0.520 | platelet/endothelial cell adhesion molecule 1 | chr17:62400603-62400544 |
| NM_002253 | KDR | 0.016 | kinase insert domain receptor | chr4:55945456-55945397 |
| NM_006500 | MCAM | 0.002 | melanoma cell adhesion molecule | chr11:119180061-119180002 |
| NM_000552 | VWF | 0.065 | von Willebrand factor | chr12:6058242-6058183 |
| NM_005163 | AKT1 | 0.003 | AKT serine/threonine kinase 1 | chr14:105236034-105235975 |
| NM_001626 | AKT2 | 0.004 | AKT serine/threonine kinase 2 | chr19:40738762-40738703 |
| NM_005465 | AKT3 | 0.003 | AKT serine/threonine kinase 3 | chr1:243665260-243665201 |
| NM_017617 | NOTCH1 | 0.067 | notch receptor 1 | chr9:139389823-139389764 |
| NM_024408 | NOTCH2 | 0.001 | notch receptor 2 | chr1:120454917-120454858 |
| NM_203458 | NOTCH2NL | 0.001 | notch 2 N-terminal like | chr1:145281999-145282058 |
| NM_000435 | NOTCH3 | 0.059 | notch receptor 3 | chr19:15270548-15270489 |
| NM_004557 | NOTCH4 | 0.043 | notch receptor 4 | chr6:32162683-32162624 |
| NM_203284 | RBPJ | 0.001 | recombination signal binding protein for immunoglobulin κ J region | chr4:26432686-26432745 |
| NM_014276 | RBPJL | 0.300 | recombination signal binding protein for immunoglobulin κ J region-like | chr20:43942760-43943087 |
| NM_005618 | DLL1 | 0.037 | δ-like canonical Notch ligand 1 | chr6:170591701-170591642 |
| NM_203486 | DLL3 | 0.013 | δ-like canonical Notch ligand 3 | chr19:39998477-39998536 |
| NM_019074 | DLL4 | 0.394 | δ-like canonical Notch ligand 4 | chr15:41229655-41229714 |
| NM_005524 | HES1 | 0.044 | hes family bHLH transcription factor 1 | chr3:193854276-193854463 |
| NM_000214 | JAG1 | 0.007 | jagged canonical Notch ligand 1 | chr20:10619120-10619061 |
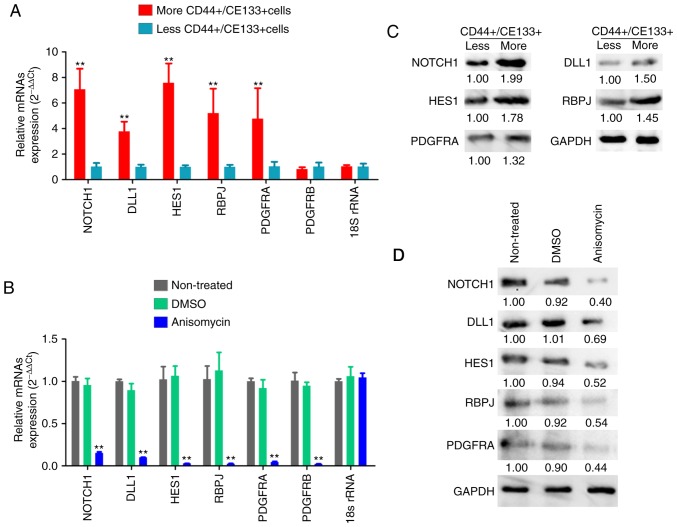
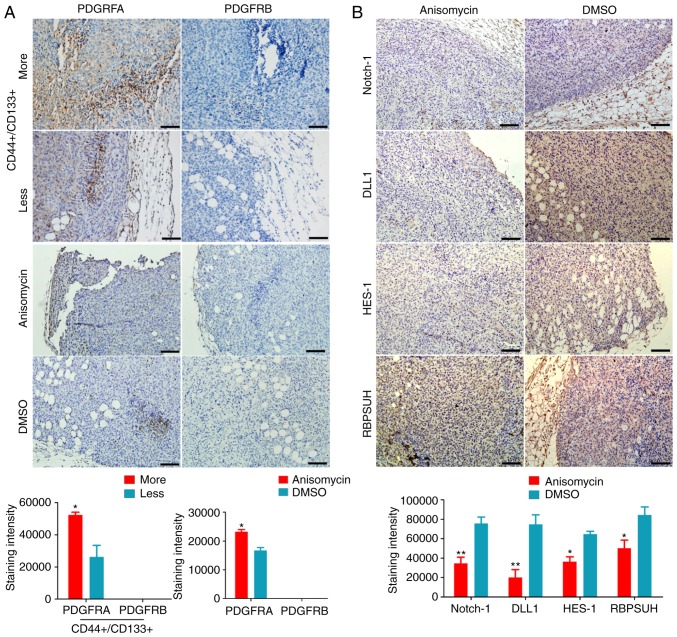
Subsequently, the expression levels of the Notch pathway components and PDGF receptors were examined by qPCR and western blotting. qPCR results demonstrated that the mRNA expression levels of key factors of the Notch pathway (Notch-1, Dll1, Hes1 and RBPJ) and PDGFRA were significantly increased in ovarian cancer tissues from P2 patient with more CD44+/CD133+ HuOCSCs compared with those from P4 patient with less CD44+/CD133+ HuOCSCs (Fig. 7A). The results from the western blot analysis were similar those of qPCR (Fig. 7C). However, after CD44+/CD133+ HuOCSCs were treated with anisomycin, the results of qPCR and western blot analyses revealed that the expression levels of key factors of the Notch pathway (Notch-1, Dll1, Hes1 and RBPJ) and PDGFRA and PDGFRB in the anisomycin-treated cells were significantly decreased compared with those in the DMSO-treated and non-treated groups (Fig. 7B and D). In addition, immunohistochemical staining indicated that, firstly, PDGFRA was highly expressed in ovarian cancer tissues with more CD44+/CD133+HuOCSCs compared with tissues with less CD44+/CD133+ HuOCSCs (Fig. 8A); PDGFRB was mostly undetectable in both groups (Fig. 8A). Secondly, in the xenograft tumours of nude mice, the expression levels of key factors of the Notch pathway and PDGFRA were significantly increased in the tumours formed by anisomycin-treated HuOCSCs compared with those in tumours formed by DMSO-treated cells (Fig. 8B). The present results suggested that anisomycin significantly inhibited the activation of the Notch pathway and PDGFRs in HuOCSCs.
Figure 7.
Anisomycin affects the expression of PDGFRs and Notch pathway components in ovarian cancer stem cells. (A) mRNA expression levels of key factors of the Notch pathway, PDGFRA and PDGFRB in ovarian cancer tissues with more and less CD44+/CD133+ HuOCSCs. **P<0.01. (B) mRNA expression levels of key factors of the Notch pathway, PDGFRA and PDGFRB in CD44+/CD133+ HuOCSCs following anisomycin treatment. **P<0.01 vs. non-treated (n=4). (C) Western blotting results from the experimental groups of panel A. (D) Western blotting results from the experimental groups of panel B. PDGFR, platelet-derived growth factor receptor; HuOCSCs, human ovarian cancer stem cells; DLL1, δ-like canonical Notch ligand 1; HES1, hes family bHLH transcription factor 1; RBPJ, recombination signal binding protein for immunoglobulin κ J region.
Figure 8.
Expression levels of PDGFRs and key factors of the Notch pathway by immunohistochemical staining. (A) The expression levels of PDGFRs in ovarian cancer tissues from patients. Magnification, ×200. *P<0.05. (B) The expression levels of key Notch pathway factors in the xenograft tumours derived from anisomycin-treated or DMSO-treated ovarian cancer stem cells. Magnification, ×200. *P<0.05 and **P<0.01 (n=4). PDGFR, platelet-derived growth factor receptor; DLL1, δ-like canonical Notch ligand 1; HES1, hes family bHLH transcription factor 1; RBPJ, recombination signal binding protein for immunoglobulin κ J region.
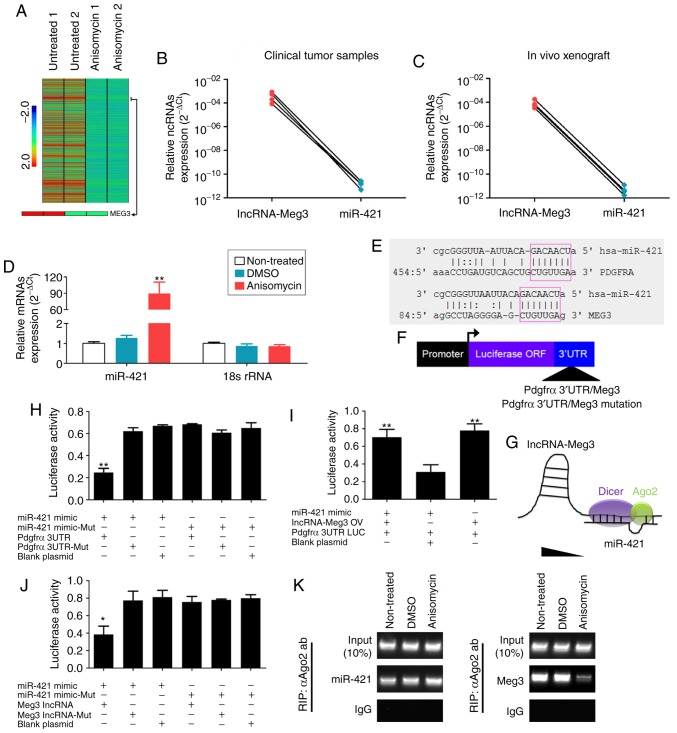
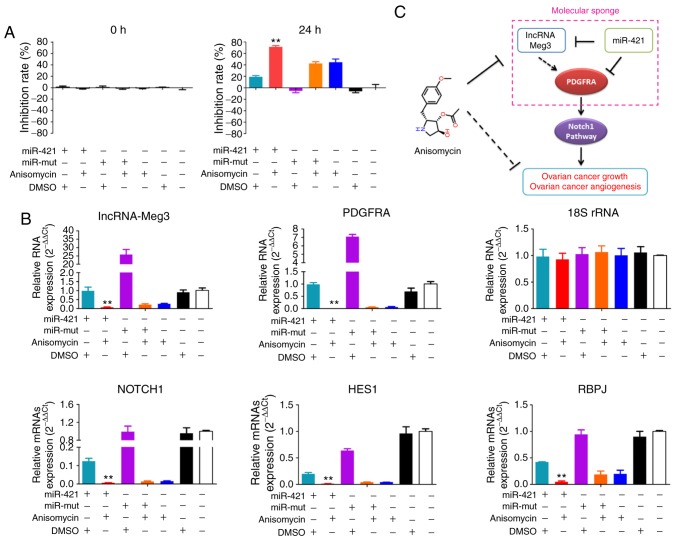
Anisomycin attenuates the molecular sponge effect in the lncRNA-Meg3/miR-421/PDGFRA axis
Through in-depth analysis of the cDNA microarray data, it was revealed that the expression levels of lncRNA-Meg3 in the anisomycin-treated group were significantly lower compared with that in the control group (Fig. 9A). Furthermore, qPCR results suggested an opposite trend in expression between lncRNA-Meg3 and miR-421 expression levels in both tumour samples of patients and subcutaneous xenograft tumour tissues of nude mice (Fig. 9B and C). In addition, after HuOCSCs were treated with anisomycin, their endogenous miR-421 expression levels were significantly higher compared with that of control cells (Fig. 9D).
Figure 9.
Anisomycin attenuates the molecular sponge effect in the lncRNA-Meg3/miR-421/PDGFRA axis. (A) Heatmap showing that the expression of lncRNA-Meg3 in the anisomycin-treated group was significantly lower than that of the control group. (B) Expression levels in clinical tumour samples and (C) in xenograft tumours suggest an inverse trend in expression for lncRNA-Meg3 and miR-421. (D) Expression levels of miR-421 in HuOCSCs following anisomycin treatment. **P<0.01 vs. non-treated (n=4). (E) Schematic of the complementary sites of mature miR-421 and the 3'UTR of lncRNA-Meg3 and PDGFRA mRNA. (F) Schematic of the structure of luciferase reporter plasmids. (G) The molecular sponge effect of lncRNA-Meg3/miR-421/PDGFRA. (H-J) Results of the luciferase reporter assays. **P<0.01 vs. blank plasmid (n=3). (K) RIP results revealed that the binding between Meg3 and Ago2 decreased significantly following treatment of HuOCSCs with anisomycin. lncRNA, long non-coding RNA; Meg3, maternally expressed 3; miR, microRNA; PDGFRA, platelet derived growth factor receptor α; HuOCSCs, human ovarian cancer stem cells; RIP, RNA immunoprecipitation; Ago2, argonaute 2; ORF, open reading frame; UTR, untranslated region; mut, mutant; IgG, immunoglobulin G.
Next, the effect of miR-421 overexpression was investigated in HuOCSCs. First, RT-qPCR results confirmed that miR-421 was successfully overexpressed following mimics transfection, compared with a mutant control or untransfected cells (Fig. S1). Following exogenous miR-421 overexpression in HuOCSCs, RT-qPCR results indicated that both expression levels of lncRNA-Meg3 and PDGFRA were significantly reduced compared with control-transfected cells (Fig. S1). According to bioinformatics analysis using TargetScanHuman (Version 7.0; http://www.targetscan.org), the mature miR-421 complementarily pairs with 7 bases at a specific site (97-104 bp) of lncRNA-Meg3, suggesting that lncRNA-Meg3 may be one of the targets of miR-421 (Fig. 9E). At the same time, it was also identified that the mature miR-421 complementarily pairs with 7 bases at a specific site (471-477 bp) of the 3′UTR of the PDGFRA gene, suggesting that PDGFRA may also be one of the targets of miR-421 (Fig. 9E). To examine the potential regulatory effect of miR-421 on lncRNA-Meg3 and PDGFRA, luciferase reporter plasmids were constructed for these two genes (Fig. 9F). The results of the luciferase reporter assay demonstrated that when miR-421 was overexpressed in cells, the luciferase activity of the PDGFRA 3′UTR reporter was significantly decreased, while the other plasmid combinations did not affect luciferase activity (Fig. 9H). In the aforementioned system, the luciferase activity did not decrease significantly after overexpression of lncRNA-Meg3 (Fig. 9I). However, when miR-421 was overexpressed in cells, expression of the luciferase gene carrying wild-type Meg3 was significantly decreased, while the other plasmid combinations did not affect luciferase activity (Fig. 9J). Finally, the results of RIP assay indicated that binding between Meg3 and Ago2 decreased significantly following treatment of HuOCSCs with anisomycin; however, there was no significant change in the binding level between miR-421 and Ago2 (Fig. 9G and K). The present results suggested that lncRNA-Meg3 and PDGFRA may be potential targets of miR-421; anisomycin downregulated the expression of endogenous lncRNA-Meg3, leading to redundant miR-421, which subsequently inhibited PDGFRA expression.
Overexpression of miR-421 significantly enhances the inhibitory effect of anisomycin on HuOCSCs by suppressing the activity of the Notch-1 pathway
MTT assay results showed that treating miR-421-overexpressing HuOCSCs with anisomycin for 24 h resulted in a significant inhibition of cell proliferation; the inhibition rate of proliferation was significantly higher compared with that of HuOCSCs overex-pressing miR-421 alone or HuOCSCs treated with anisomycin alone (Fig. 10A). qPCR results demonstrated that, following treatment of miR-421-overexpressing HuOCSCs with aniso-mycin for 24 h, the mRNA expression levels of endogenous lncRNA-Meg3, PDGFRA, Notch1, HES1 and RBPJ were significantly decreased; the expression levels were significantly lower than those of HuOCSCs overexpressing miR-421 alone or HuOCSCs treated with anisomycin alone (Fig. 10B). These results suggested that overexpression of miR-421 significantly enhanced the inhibitory effect of anisomycin on HuOCSCs by suppressing the activity of the Notch-1 pathway.
Figure 10.
miR-421 enhances the inhibitory effect of anisomycin on HuOCSCs. (A) MTT results showed that overexpression of miR-421 and treatment with anisomycin resulted in more severe proliferation inhibition of HuOCSCs, than either treatment alone. **P<0.01 vs. DMSO group (n=4). (B) Quantitative PCR results showed that overexpression of miR-421 and treatment with anisomycin decreased the expression of lncRNA-Meg3, PDGFRA, Notch1, HES1 and RBPJ. **P<0.01 vs. DMSO group (n=4). (C) Schematic of the proposed molecular mechanism of anisomycin inhibition of the activity and angiogenesis of ovarian cancer cells, by attenuating the molecular sponge effect in the lncRNA-Meg3/miR-421/PDGFRA axis. miR, microRNA; HuOCSCs, human ovarian cancer stem cells; lncRNA, long non-coding RNA; Meg3, maternally expressed 3; PDGFRA, platelet derived growth factor receptor α; HES1, hes family bHLH transcription factor 1; RBPJ, recombination signal binding protein for immunoglobulin κ J region; mut, mutant.
Discussion
Cancer stem cells exhibit a high proliferative and invasive activity and resistance to various chemotherapeutic drugs, which are important causes of tumour metastasis and recurrence (13,25-28). The discovery of cancer stem cells put forward new demands on tumour chemotherapy. Since cancer stem cells are resistant to most traditional chemotherapeutic drugs, such as cisplatin and paclitaxel, it is necessary to develop more effective drugs for tumour chemotherapy (21,29-31). Anisomycin has been confirmed to have significant inhibitory effects on a variety of solid tumours and is a promising chemotherapeutic drug candidate (7-11). Our previous report has revealed that anisomycin inhibits the proliferation and invasion of ovarian cancer stem cells by enhancing the activity of lncRNA-BACE1-AS and increasing the release of endogenous Aβ42 (7). The present study suggested that anisomycin may have the potential to inhibit cancer stem cells. Based on the existing reports (7-11), it was speculated that the inhibitory effect of anisomycin on cancer stem cells may depend on multi-target regulation. Therefore, it is meaningful to explore its in-depth mechanism. The present study first compared the difference in gene expression profiles between anisomycin-treated ovarian cancer stem cells and control cells using cDNA microarrays. Considering that the effect of anisomycin on tumour angiogen-esis had not been reported previously, signalling pathways and molecules related to tumour angiogenesis were selected as the main research focus. The cDNA microarray results suggested that anisomycin treatment significantly inhibited the expression of Notch pathway components and of vascular endothelial cell markers. This result strongly suggested the molecular biology basis for the inhibitory effect of anisomycin on tumour angiogenesis. In order to confirm this hypothesis in vitro, MTT, Transwell and 3-dimensional angiogenesis assays were performed in vitro; in vivo, an angiogenesis assay in transgenic zebrafish embryos and xenograft tumour experiments in nude mice were performed. The results of these experiments demonstrated that anisomycin could effectively inhibit the angiogenic ability of ovarian cancer stem cells in vitro and in vivo, thus confirming the hypothesis that anisomycin can effectively inhibit angiogenesis in ovarian cancer.
Subsequently, the present study aimed to investigate how anisomycin inhibited the expression of the Notch-1 pathway components. After screening the data from the cDNA micro-array analysis, the present study further focused on members of the PDGFR family. In the anisomycin-treated group, the expression of the PDGFRs was significantly decreased. PDGF is a basic protein stored in the α particles in platelets and is a low molecular weight mitogen (32-34). PDGF can stimulate multiple cell types that are arrested in the G0/G1 phase, including fibroblasts, glial cells and smooth muscle cells, to enter the proliferative cycle (33-35). PDGFRs are receptors for the PDGF family, located on the surface of the cell membrane and they belong to the family of tyrosine kinase receptors, with molecular weights of 170-180 kDa. There are two subtypes of PDGFR, namely, PDGFRA and PDGFRB (32-34). PDGFR is a peptide chain consisting of a domain specifically recognized by PDGF at the extracellular N-terminus, an intermediate hydrophobic domain of a single-stranded transmembrane sequence, and a domain with tyrosine protein kinase activity at the intracellular C-terminus (32-34). PDGF binds to PDGFR on the cell membrane to form a dimeric complex, activates autophosphorylation of the tyrosine residues of the intracellular domain, or promotes phosphorylation of tyrosine residues that activate specific target proteins, thereby transmitting signals into the cell, activating downstream signalling pathways, and directly inducing cell proliferation, differentiation, growth and development (35,36). Although both PDGFRA and PDGFRB bind to PDGF, their affinity differs greatly (32-34). The α subunit has a high affinity for both the a chain and b chain of PDGF; the β subunit has a high affinity only for the b chain (32-34). Therefore, the α subunit can bind to PDGF-aa, PDGF-ab, and PDGF-bb, while the β subunit can only bind to PDGF-bb and PDGF-ab (32-34). It has been reported that, in tumour cells, PDGF binds to PDGFR to form a dimer, which induces tumour cell proliferation, invasion, metastasis and angiogenesis, by activating the downstream Notch-1 pathway, making it a very important signalling activator in tumour angiogenesis events (32-34,37-39). Using bioinformatics analysis, the present study systematically predicted the correlation among Notch-1 signalling molecules, angiogenesis markers and PDGFR family members, and mapped the protein interaction network. The present results confirmed that there was a close relationship among the three components.
Next, the present study investigated whether there was a direct relationship between the inactivation of PDGFR and anisomycin. Bioinformatics analysis predicted that the PDGFRA gene may be one of the potential target genes of miR-421. The expression levels of miR-421 were significantly increased in ovarian cancer stem cells treated with anisomycin, while PDGFRA was significantly decreased in anisomycin-treated cells, due to the RNA interfering effect induced by miR-421. Furthermore, the expression levels of endogenous lncRNA-Meg3 was significantly decreased in ovarian cancer stem cells following treatment with anisomycin. Then, the present study investigated the relationship between lncRNA-Meg3, PDGFRA and miR-421. Bioinformatics analysis predicted that lncRNA-Meg3 may also be a potential target gene of miR-421. Therefore, miR-421 could silence both lncRNA-Meg3 and PDGFRA. This regulatory network would be in agreement with the lncRNA-miRNA 'molecular sponge' effect (12,13,15,27).
An increasing number of studies have found that there is some interaction between lncRNAs and miRNAs, and lncRNAs can exert their regulatory effects on the miRNA network through the competitive endogenous RNA (ceRNA) effect (13,30). The ceRNA theory is based on classical miRNA regulation mechanisms. miRNAs inhibit translation of target genes primarily by reducing mRNA stability. In a cell, a gene is often regulated by multiple miRNAs, and a miRNA can also regulate the expression of multiple genes. The ceRNA theory proposes that lncRNAs within cells carry miRNA binding sites of different types and amounts, and RNAs carrying the same miRNA binding sites are called ceRNAs. ceRNAs can compete with one another for binding to the same miRNA, reducing the concentration of free miRNAs, thereby reducing the inhibition efficiency of the miRNA to target genes to some extent (40-44). Functional studies have shown that lncRNAs can act as ceRNAs (13,17,40,44,45). Each ceRNA in a cell may contain a large number of different miRNA binding sites that can compete for binding to multiple miRNAs (40-44). At the same time, since each miRNA regulates a variety of different target genes, each ceRNA can achieve a 'cross-talk' with a variety of gene regulation processes through the regulatory network of miRNAs (40-44). In this process, lncRNAs can act like 'sponges' to adsorb different miRNAs and participate in the regulatory network of miRNAs. This complex function has been termed as the 'molecular sponge effect' of lncRNAs (40-44).
There have been many reports on the functions of lncRNA-Meg3, PDGFRA and miR-421 in tumours, suggesting that these three molecules have very important regulatory effects on tumour proliferation and invasion (35,37,45-52). However, there have been no reports discussing the three in combination. The present study found an intrinsic association between lncRNA-Meg3, PDGFRA and miR-421. A high expression of lncRNA-Meg3 is present in ovarian cancer cells, which binds to and consumes endogenous miRNA-421 and blocks its inhibitory effect on target genes. Thus, PDGFRA can be stably expressed. However, after ovarian cancer stem cells are treated with anisomycin, the expression of lncRNA-Meg3 is inhibited by anisomycin, attenuating its molecular sponge effect on miRNA-421; a large amount of miR-421 can then specifically bind to the target gene PDGFRA and inhibit its expression (Fig. 10C).
In summary, the present study has elucidated a novel mechanism underlying the anisomycin-mediated inhibition of angiogenesis in ovarian cancer, by targeting the molecular sponge effect in the lncRNA-Meg3/miR-421/PDGFRA axis.
Supplementary Data
Acknowledgments
We are very grateful to Professor Chuan Chen of the Shanghai Geriatric Institute of Chinese Medicine for his guidance on this study.
Funding
This work was supported by the Shanghai Natural Science Foundation (grant no. 16ZR1434000), the Development Fund for Shanghai Talents (grant no. 2017054), the Fund for Xinglin Talents of Shanghai University of Traditional Chinese Medicine (grant no. 201707081) and the National Natural Science Foundation of China (grant no. 81973899).
Availability of data and materials
The datasets used or analysed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
WY, ZN and SY performed the majority of the experiments in the study. HP, YH and YX contributed to the analysis of experimental data. TL contributed to the study design, manuscript writing and provided experimental funding support.
Ethics approval and consent to participate
The study involving human tissues was approved by the Ethics Review Committee of Shanghai Geriatric Institute of Chinese Medicine of Research in Human Production, authorized by Shanghai Municipal Government; written informed consent was provided by all patients, in accordance with The Declaration of Helsinki. The study involving animals was approved (permit no. SRCMR20160018) by the Animal Ethics Committee of Shanghai Research Centre for Model Organisms, and the experimental protocols were in compliance with the Experimental Animal Regulations of the Ministry of Science and Technology National Science and Technology Commission (Beijing, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jain RK, Carmeliet P. SnapShot: Tumor angiogenesis. Cell. 2012;149:1408–1408. e1401. doi: 10.1016/j.cell.2012.05.025. [DOI] [PubMed] [Google Scholar]
- 2.Rivera LB, Bergers G. CANCER. Tumor angiogenesis, from foe to friend. Science. 2015;349:694–695. doi: 10.1126/science.aad0862. [DOI] [PubMed] [Google Scholar]
- 3.Melero-Martin JM, Dudley AC. Concise review: Vascular stem cells and tumor angiogenesis. Stem Cells. 2011;29:163–168. doi: 10.1002/stem.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuczynski EA, Vermeulen PB, Pezzella F, Kerbel RS, Reynolds AR. Vessel co-option in cancer. Nat Rev Clin Oncol. 2019;16:469–493. doi: 10.1038/s41571-019-0181-9. [DOI] [PubMed] [Google Scholar]
- 5.Ho IA, Toh HC, Ng WH, Teo YL, Guo CM, Hui KM, Lam PY. Human bone marrow-derived mesenchymal stem cells suppress human glioma growth through inhibition of angiogenesis. Stem Cells. 2013;31:146–155. doi: 10.1002/stem.1247. [DOI] [PubMed] [Google Scholar]
- 6.Apte RS, Chen DS, Ferrara N. VEGF in signaling and disease: Beyond discovery and development. Cell. 2019;176:1248–1264. doi: 10.1016/j.cell.2019.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Q, Liu X, Xu L, Wang Y, Wang S, Li Q, Huang Y, Liu T. Long non-coding RNA BACE1-AS is a novel target for anisomycin-mediated suppression of ovarian cancer stem cell proliferation and invasion. Oncol Rep. 2016;35:1916–1924. doi: 10.3892/or.2016.4571. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Hu J, Song H, Wu T. Antibiotic anisomycin selectively targets leukemia cell lines and patient samples through suppressing Wnt/beta-catenin signaling. Biochem Biophys Res Commun. 2018;505:858–864. doi: 10.1016/j.bbrc.2018.09.183. [DOI] [PubMed] [Google Scholar]
- 9.Yu C, Xing F, Tang Z, Bronner C, Lu X, Di J, Zeng S, Liu J. Anisomycin suppresses Jurkat T cell growth by the cell cycle-regulating proteins. Pharmacol Rep. 2013;65:435–444. doi: 10.1016/S1734-1140(13)71019-3. [DOI] [PubMed] [Google Scholar]
- 10.Seo BR, Min KJ, Kim S, Park JW, Park WK, Lee TJ, Kwon TK. Anisomycin treatment enhances TRAIL-mediated apoptosis in renal carcinoma cells through the down-regulation of Bcl-2, c-FLIP(L) and Mcl-1. Biochimie. 2013;95:858–865. doi: 10.1016/j.biochi.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Ge J, Li Q, Gu L, Guo X, Ma ZG, Zhu YP. Anisomycin induces apoptosis of glucocorticoid resistant acute lymphoblastic leukemia CEM-C1 cells via activation of mitogen-activated protein kinases p38 and JNK. Neoplasma. 2013;60:101–110. doi: 10.4149/neo_2013_014. [DOI] [PubMed] [Google Scholar]
- 12.Liu T, Zhang H, Zheng J, Lin J, Huang Y, Chen J, Yu Z, Guo L, Pan W, Xiong Y, Chen C. SPION-mediated miR-141 promotes the differentiation of HuAESCs into dopaminergic neuron-like cells via suppressing lncRNA-HOTAIR. J Cell Mol Med. 2018;22:2299–2310. doi: 10.1111/jcmm.13512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu T, Chi H, Chen J, Chen C, Huang Y, Xi H, Xue J, Si Y. Curcumin suppresses proliferation and in vitro invasion of human prostate cancer stem cells by ceRNA effect of miR-145 and lncRNA-ROR. Gene. 2017;631:29–38. doi: 10.1016/j.gene.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Sarfi M, Abbastabar M, Khalili E. Long noncoding RNAs biomarker-based cancer assessment. J Cell Physiol. 2019;234:16971–16986. doi: 10.1002/jcp.28417. [DOI] [PubMed] [Google Scholar]
- 15.Liu SJ, Lim DA. Modulating the expression of long non-coding RNAs for functional studies. EMBO Rep. 2018;19:e46955. doi: 10.15252/embr.201846955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu AD, Sun YY, Ma QJ, Xu F. lncRNA-ATB promotes viability, migration, and angiogenesis in human micro-vascular endothelial cells by sponging microRNA-195. J Cell Biochem. 2019;120:14360–14371. doi: 10.1002/jcb.28692. [DOI] [PubMed] [Google Scholar]
- 17.Zhao J, Li L, Han ZY, Wang ZX, Qin LX. Long noncoding RNAs, emerging and versatile regulators of tumor-induced angiogenesis. Am J Cancer Res. 2019;9:1367–1381. [PMC free article] [PubMed] [Google Scholar]
- 18.Sheng SR, Wu JS, Tang YL, Liang XH. Long noncoding RNAs: Emerging regulators of tumor angiogenesis. Future Oncol. 2017;13:1551–1562. doi: 10.2217/fon-2017-0149. [DOI] [PubMed] [Google Scholar]
- 19.Sun X, Huang T, Zhang C, Zhang S, Wang Y, Zhang Q, Liu Z. Long non-coding RNA LINC00968 reduces cell proliferation and migration and angiogenesis in breast cancer through up-regulation of PROX1 by reducing hsa-miR-423-5p. Cell Cycle. 2019;18:1908–1924. doi: 10.1080/15384101.2019.1632641. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Cheng Y, Dai X, Yang T, Zhang N, Liu Z, Jiang Y. Low long noncoding RNA growth arrest-specific transcript 5 expression in the exosomes of lung cancer cells promotes tumor angiogenesis. J Oncol. 2019;2019:2476175. doi: 10.1155/2019/2476175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng W, Liu T, Wan X, Gao Y, Wang H. MicroRNA-199a targets CD44 to suppress the tumorigenicity and multidrug resistance of ovarian cancer-initiating cells. FEBS J. 2012;279:2047–2059. doi: 10.1111/j.1742-4658.2012.08589.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Zheng J, Lin J, Chen J, Yu Z, Chen C, Liu T. miR-758 mediates oxLDL-dependent vascular endothelial cell damage by suppressing the succinate receptor SUCNR1. Gene. 2018;663:1–8. doi: 10.1016/j.gene.2018.04.029. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez-Hernandez A, Brea-Calvo G, Fernandez-Ayala DJ, Cordero M, Navas P, Sanchez-Alcazar JA. Nuclear caspase-3 and caspase-7 activation, and poly(ADP-ribose) polymerase cleavage are early events in camptothecin-induced apoptosis. Apoptosis. 2006;11:131–139. doi: 10.1007/s10495-005-3276-y. [DOI] [PubMed] [Google Scholar]
- 25.Si Y, Liu J, Shen H, Zhang C, Wu Y, Huang Y, Gong Z, Xue J, Liu T. Fisetin decreases TET1 activity and CCNY/CDK16 promoter 5hmC levels to inhibit the proliferation and invasion of renal cancer stem cell. J Cell Mol Med. 2019;23:1095–1105. doi: 10.1111/jcmm.14010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Zheng J, Shen H, Huang Y, Liu T, Xi H, Chen C. Curcumin suppresses in vitro proliferation and invasion of human prostate cancer stem cells by modulating DLK1-DIO3 imprinted gene cluster MicroRNAs. Genet Test Mol Biomarkers. 2018;22:43–50. doi: 10.1089/gtmb.2017.0179. [DOI] [PubMed] [Google Scholar]
- 27.Fang K, Liu P, Dong S, Guo Y, Cui X, Zhu X, Li X, Jiang L, Liu T, Wu Y. Magnetofection based on superparamagnetic iron oxide nanoparticle-mediated low lncRNA HOTAIR expression decreases the proliferation and invasion of glioma stem cells. Int J Oncol. 2016;49:509–518. doi: 10.3892/ijo.2016.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu T, Xu F, Du X, Lai D, Liu T, Zhao Y, Huang Q, Jiang L, Huang W, Cheng W, Liu Z. Establishment and characterization of multi-drug resistant, prostate carcinoma-initiating stem-like cells from human prostate cancer cell lines 22RV1. Mol Cell Biochem. 2010;340:265–273. doi: 10.1007/s11010-010-0426-5. [DOI] [PubMed] [Google Scholar]
- 29.Shao Y, Zhang L, Cui L, Lou W, Wang D, Lu W, Jin D, Liu T. LIN28B suppresses microRNA let-7b expression to promote CD44+/LIN28B+ human pancreatic cancer stem cell proliferation and invasion. Am J Cancer Res. 2015;5:2643–2659. [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Y, Liu T, Huang Y. MicroRNA-134 suppresses endometrial cancer stem cells by targeting POGLUT1 and Notch pathway proteins. FEBS Lett. 2015;589:207–214. doi: 10.1016/j.febslet.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 31.Qin W, Xiong Y, Chen J, Huang Y, Liu T. DC-CIK cells derived from ovarian cancer patient menstrual blood activate the TNFR1ASK1-AIP1 pathway to kill autologous ovarian cancer stem cells. J Cell Mol Med. 2018;22:3364–3376. doi: 10.1111/jcmm.13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams LT. Signal transduction by the platelet-derived growth factor receptor. Science. 1989;243:1564–1570. doi: 10.1126/science.2538922. [DOI] [PubMed] [Google Scholar]
- 33.Hoch RV, Soriano P. Roles of PDGF in animal development. Development. 2003;130:4769–4784. doi: 10.1242/dev.00721. [DOI] [PubMed] [Google Scholar]
- 34.Heldin CH. Targeting the PDGF signaling pathway in tumor treatment. Cell Commun Signal. 2013;11:97. doi: 10.1186/1478-811X-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H, Gu X, Liu Y, Wang J, Wirt SE, Bottino R, Schorle H, Sage J Kim SK. PDGF signalling controls age-dependent proliferation in pancreatic β-cells. Nature. 2011;478:349–355. doi: 10.1038/nature10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi MH, Lee IK, Kim GW, Kim BU, Han YH, Yu DY, Park HS, Kim KY, Lee JS, Choi C, et al. Regulation of PDGF signalling and vascular remodelling by peroxiredoxin II. Nature. 2005;435:347–353. doi: 10.1038/nature03587. [DOI] [PubMed] [Google Scholar]
- 37.Tam WL, Lu H, Buikhuisen J, Soh BS, Lim E, Reinhardt F, Wu ZJ, Krall JA, Bierie B, Guo W, et al. Protein kinase C α is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell. 2013;24:347–364. doi: 10.1016/j.ccr.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wieland E, Rodriguez-Vita J, Liebler SS, Mogler C, Moll I, Herberich SE, Espinet E, Herpel E, Menuchin A, Chang-Claude J, et al. Endothelial notch1 activity facilitates metastasis. Cancer Cell. 2017;31:355–367. doi: 10.1016/j.ccell.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 39.Ramasamy SK, Kusumbe AP, Wang L, Adams RH. Endothelial Notch activity promotes angiogenesis and osteo-genesis in bone. Nature. 2014;507:376–380. doi: 10.1038/nature13146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang M, Mao C, Ouyang L, Liu Y, Lai W, Liu N, Shi Y, Chen L, Xiao D, Yu F, et al. Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ. 2019 doi: 10.1038/s41418-019-0394-6. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luan W, Zhang X, Ruan H, Wang J, Bu X. Long noncoding RNA OIP5-AS1 acts as a competing endogenous RNA to promote glutamine catabolism and malignant melanoma growth by sponging miR-217. J Cell Physiol. 2019 doi: 10.1002/jcp.28335. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 42.Xu TP, Ma P, Wang WY, Shuai Y, Wang YF, Yu T, Xia R, Shu YQ. KLF5 and MYC modulated LINC00346 contributes to gastric cancer progression through acting as a competing endogeous RNA and indicates poor outcome. Cell Death Differ. 2019 doi: 10.1038/s41418-018-0236-y. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong H, Hu J, Zou K, Ye M, Chen Y, Wu C, Chen X, Han M. Activation of LncRNA TINCR by H3K27 acetylation promotes trastuzumab resistance and epithelial-mesenchymal transition by targeting MicroRNA-125b in breast cancer. Mol Cancer. 2019;18:3. doi: 10.1186/s12943-018-0931-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Wu XS, Wang F, Li HF, Hu YP, Jiang L, Zhang F, Li ML, Wang XA, Jin YP, Zhang YJ, et al. LncRNA-PAGBC acts as a microRNA sponge and promotes gallbladder tumorigenesis. EMBO Rep. 2017;18:1837–1853. doi: 10.15252/embr.201744147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Wu N, Wang J, Li Z. LncRNA MEG3 inhibits cell proliferation and induces apoptosis in laryngeal cancer via miR-23a/APAF-1 axis. J Cell Mol Med. 2019 doi: 10.1111/jcmm.14549. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joglekar-Javadekar M, Van Laere S, Bourne M, Moalwi M, Finetti P, Vermeulen PB, Birnbaum D, Dirix LY, Ueno N, Carter M, et al. Characterization and targeting of platelet-derived growth factor receptor alpha (PDGFRA) in inflammatory breast cancer (IBC) Neoplasia. 2017;19:564–573. doi: 10.1016/j.neo.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ong HS, Gokavarapu S, Tian Z, Li J, Xu Q, Cao W, Zhang CP. PDGFRA mRNA is overexpressed in oral cancer patients as compared to normal subjects with a significant trend of overexpression among tobacco users. J Oral Pathol Med. 2017;46:591–597. doi: 10.1111/jop.12571. [DOI] [PubMed] [Google Scholar]
- 48.Gao S, Li E, Gao H. Long non-coding RNA MEG3 attends to morphine-mediated autophagy of HT22 cells through modulating ERK pathway. Pharm Biol. 2019;57:536–542. doi: 10.1080/13880209.2019.1651343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Binabaj MM, Bahreyni A, Khazaei M, Avan A, Hassanian SM. The prognostic value of long noncoding RNA MEG3 expression in the survival of patients with cancer: A meta-analysis-response. J Cell Biochem. 2019 doi: 10.1002/jcb.28532. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 50.Meng Q, Li S, Liu Y, Zhang S, Jin J, Zhang Y, Guo C, Liu B, Sun Y. Circular RNA circSCAF11 accelerates the glioma tumorigenesis through the miR-421/SP1/VEGFA axis. Mol Ther Nucleic Acids. 2019;17:669–677. doi: 10.1016/j.omtn.2019.06.022. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y, Han X, Li Q, Wang C, Lou Z, Wang X. Long noncoding RNA HOXD-AS1 induces epithelial-mesenchymal transition in breast cancer by acting as a competing endogenous RNA of miR-421. J Cell Biochem. 2019;120:10633–10642. doi: 10.1002/jcb.28353. [DOI] [PubMed] [Google Scholar]
- 52.Yin Y, Xu L, Chang Y, Zeng T, Chen X, Wang A, Groth J, Foo WC, Liang C, Hu H, Huang J. N-Myc promotes therapeutic resistance development of neuroendocrine prostate cancer by differentially regulating miR-421/ATM pathway. Mol Cancer. 2019;18:11. doi: 10.1186/s12943-019-0941-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used or analysed during the current study are available from the corresponding author on reasonable request.