Abstract
Osteosarcoma (OS) is the most common primary bone malignancy, mainly affecting children and adolescents. Currently, surgical resection combined with adjuvant chemotherapy has been standardized for OS treatment. Despite great advances in chemotherapy for OS, its clinical prognosis remains far from satisfactory; this is due to chemoresistance, which has become a major obstacle to improving OS treatment. Autophagy, a catabolic process through which cells eliminate and recycle their own damaged proteins and organelles to provide energy, can be activated by chemotherapeutic drugs. Accumulating evidence has indicated that autophagy plays the dual role in the regulation of OS chemoresistance by either promoting drug resistance or increasing drug sensitivity. The aim of the present review was to demonstrate thatautophagy has both a cytoprotective and an autophagic cell death function in OS chemoresistance. In addition, methods to detect autophagy, autophagy inducers and inhibitors, as well as autophagy-mediated metastasis, immunotherapy and clinical prognosis are also discussed.
Keywords: osteosarcoma, chemoresistance, cytoprotection, autophagy, autophagic cell death
1. Introduction
Osteosarcoma (OS) is the most frequent primary malignant bone tumor that predominantly occurs in children and adolescents, and accounts for ~15% of all bone malignancies (1,2). Its predilection sites include distal femur (43%), proximal tibia (23%) and humerus (10%). Since chemotherapy was introduced in the 1970s, the 5-year survival rate for OS has markedly improved from <20 to 70% (3). Doxorubicin, cisplatin and methotrexate are the most commonly used chemotherapy drugs in the treatment of OS (4). Despite great advances in chemotherapy for OS, survival rates have reached a plateau and remained unsatisfactory during the past three decades (5). Drug resistance is one of the main reasons contributing to this (6); 35-45% of OS patients are not sensitive to chemotherapy drugs, with their 5-year survival rate at only 5-20% (7,8). Chemoresistance often leads to treatment failure and poor prognosis. It has become a major obstacle to improving OS treatment. Therefore, elucidating the underlying molecular mechanisms implicated in OS chemoresistance is urgently required.
Autophagy, initially discovered by Ohsumi in 1992 (who received the 2016 Nobel Prize in Physiology or Medicine for his outstanding contributions to the field), is a catabolic process via which cells eliminate and recycle their own damaged proteins and organelles to provide energy (9). There are three types of autophagy, including microautophagy, macroautophagy and chaperone-mediated autophagy (10). The difference between these autophagic processes is the substrates delivered to the lysosomes (11). Microautophagy refers to the direct engulfment of cytoplasmic material by lysosomes for degradation. It can be activated by signaling molecules on the surface of damaged organelles, such as mitochondria or peroxisomes, leading to the fusion of lysosomes with these organelles (12). Macroautophagy is the process during which damaged organelles are first enclosed in double-membrane vesicles (also known as autophagosomes) and then fused with lysosomes to become autophagolysosomes (12). Chaperone-mediated autophagy is selective for specific substrate proteins containing a pentapeptide amino acid sequence, which can be recognized by molecular chaperone and then carried into lysosomes for degradation (10,12). This review focused on macroautophagy (hereafter referred to as autophagy).
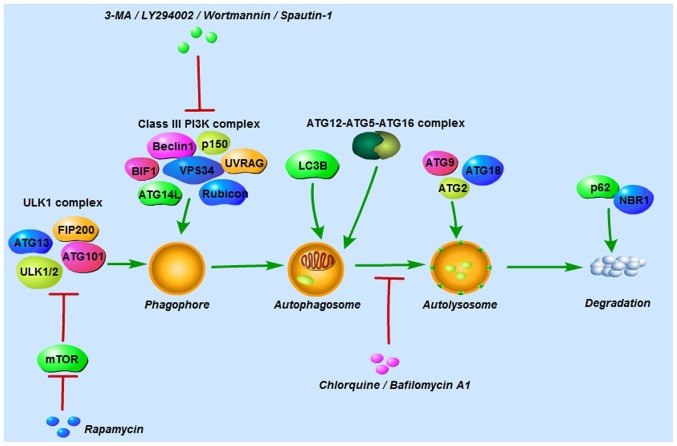
The autophagic process can be mainly divided into 4 steps: i) Formation of the phagophore to wrap the damaged material; ii) elongation and closure of the phagophore followed by autophagosome generation; iii) fusion of autophagosomes and lysosomes to form autolysosomes; and iv) content degradation and recycling (9). Autophagy can be triggered under stressful conditions, such as starvation, hypoxia and cytotoxicity, to maintain cell survival by providing energy (11). To date, >30 autophagy-related proteins (ATGs) have been found to participate in autophagy regulation (9). Autophagy is initiated by the UNC-51-like kinase (ULK1) complex comprising ULK1/2, ATG13, ATG101 and focal adhesion kinase family interacting protein of 200 kDa (FIP200), and the class III phosphoinositide 3-kinase (PI3K) complex consisting of Beclin-1, vacuolar protein sorting 34, p150, ultraviolet irradiation resistance-associated gene, BAX-interacting factor-1, ATG14-like protein and Run domain Beclin-1-interacting and cysteine-rich domain-containing protein (9,10,13). The ULK1 complex is negatively regulated by mammalian target of rapamycin (mTOR) in nutrient-rich conditions; conversely, in nutrient-deprived conditions, mTOR is inhibited and the ULK1 complex is then activated to induce autophagy (9). Autophagosome formation is controlled by the ATG12 and LC3 conjugation systems. In the first system, ATG12 and ATG5 are conjugated in the presence of ATG7 and ATG10. ATG12-ATG5 conjugation then binds to ATG16 to form the ATG12-ATG5-ATG16 complex (9). In the second system, the protease ATG4 cleaves microtubule-associated protein 1-light chain 3 (LC3; also known as ATG8) to LC3-I, which is then conjugated with phosphatidylethanolamine and converted into LC3-II (11). In this process, LC3-II is translocated from the cytoplasm to the autophagosome membrane. For that reason, LC3-II is considered an important marker for autophagosomes. ATG2, ATG9 and ATG18 are involved in autolysosome formation, and p62 and neighbor of BRCA1 protein in degradation and recycling regulation (Fig. 1) (10).
Figure 1.
Autophagy-related proteins, and autophagy inducers and inhibitors involved in the autophagic process. Autophagy is initiated by the ULK1 complex and the class III PI3K complex. The former is composed of ULK1/2, ATG13, ATG101 and FIP200, and the latter of Beclin-1, VPS34, p150, UVRAG, BIF1, ATG14L and rubicon. Autophagosome formation is controlled by the ATG12 and LC3 conjugation systems. ATG2, ATG9 and ATG18 are involved in autolyso-some formation, and p62 and NBR1 in the regulation of degradation and recycling. Rapamycin activates autophagy by inhibiting mTOR, a negative regulator of the ULK1 complex. 3-MA, LY294002, wortmannin and spautin-1 suppress early autophagy by inhibiting the class III PI3K complex. Chloroquine and bafilomycin A1 inhibit late autophagy by blocking the fusion of autophagosomes and lysosomes. ULK1, UNC-51-like kinase; ATG, autophagy-related protein; FIP200, focal adhesion kinase family interacting protein of 200; VPS34, vacuolar protein sorting 34; UVRAG, ultraviolet irradiation resistance-associated gene; BIF1, BAX-interacting factor-1; ATG14L, ATG14-like protein; rubicon, Run domain Beclin-1-interacting and cysteine-rich domain-containing protein; mTOR, mammalian target of rapamycin; 3-MA, 3-methyladenine; NBR1, neighbor of BRCA1 protein.
2. Methods for detecting autophagy
As autophagy is a dynamic multi-stage process, it is necessary to identify whether autophagy occurs in stressful conditions induced by chemotherapeutic agents, such as starvation, hypoxia, and cytotoxicity, and which steps of autophagy, if any, are affected. Given that LC3-II is the only protein marker for autophagosomes, one of the key characteristics of autophagy, LC3-II detection has been widely used in autophagy-related research. However, it may yield opposite results only by analyzing the LC3-II expression. For example, an increased LC3-II expression can either represent increased autophagosome formation or reduction of autophagosome degradation (14). More and more methods of monitoring autophagy are being identified. Transmission electron microscopy (TEM), first used to detect autophagy in the 1950s, is now considered the golden standard for autophagy detection, as it is the only tool to morphologically observe the ultrastructure of autophagosomes in the nm range (14). Two autophagic vacuoles, initial autophagic vacuole (AVi) and degradative autophagic vacuole (AVd), can be observed in a TEM image. The defining structure of AVi, also referred to as autophagosomes, is that intact organelles are sequestrated by a special double-membrane structure (14). And the defining structure of AVd, also referred to as autolysosomes, is that degraded organelles are sequestrated by only one limiting membrane (14). However, the limitation of TEM is that it can only statically observe a certain stage of autophagy rather than the entire process. Therefore, greater attention has been paid to autophagic flux monitoring. The utilization of tandem monomeric red fluorescent protein (mRFP)-green fluorescent protein (GFP)-LC3 via confocal microscopy is one of the most utilized approaches in autophagic flux monitoring (14). This method is based on the principle that GFP signal is quenched in the acid environment of lysosomes, whereas RFP is stable. To be specific, when autophagosomes have not yet been fused with lysosomes, GFP and RFP fluorescence are colocalized in autophagosomes displaying yellow dots. When autophagosomes fuse with lysosomes to form autolysosomes, only RFP fluorescence is localized in autolysosomes (14). In order to improve understanding of autophagy, experts in autophagy from all over the world published the 3rd edition of the guidelines for monitoring autophagy in 2016 (14). In addition to the methods mentioned above, several other assays were introduced in this edition. They strongly recommend that multiple assays should be used to monitor autophagy instead of one (14).
3. Autophagy inducers and inhibitors
Autophagy inducers and inhibitors are indispensable in the regulation of autophagy. The most commonly used inducers are rapamycin and its analogs, including temsirolimus, everolimus and deforolimus, which activate autophagy by inhibiting mTOR, a negative regulator of autophagy (11). As autophagy can be blocked at different stages, a large number of inhibitors have been used in different mechanisms. At an early stage, 3-methyladenine (3-MA), LY294002 and wortmannin can suppress autophagy by inhibiting class III PI3K (15). Another novel PI3K inhibitor, spautin-1 has been shown to degrade the class III PI3K complex via Beclin-1 (15). It was demonstrated by Schott et al (16) that pre-treatment with spautin-1 enhanced the canine OS cell inhibition induced by doxorubicin. At a later stage, chloroquine and its derivatives (such as hydroxychloroquine), which were originally used as anti-malarial drugs, are capable of preventing lysosomal acidification and blocking the fusion of autophagosomes and lysosomes (10). Bafilomycin A1, an inhibitor of vacuolar-type H+-ATPase, also prevents lysosome acidification (Fig. 1) (15).
4. Dual role of autophagy in OS chemoresistance
As autophagy can be triggered by chemotherapy drugs, a growing number of studies have focused on the association between autophagy and chemoresistance in tumor cells (11,16). Of note, autophagy has been shown to play a dual role in cancer; either tumor-promoting or tumor-suppressing. On the one hand, autophagy helps tumor cells survive in the presence of chemotherapy drugs by eliminating its own damaged organelles and proteins (17). On the other hand, excessive autophagy ultimately leads to cell death (17). This double-edged sword effect of autophagy was observed by O'Farrill and Gordon (11), who found that autophagy inhibition resulted in increased sensitivity of LM7 metastatic human OS cells to gemcitabine, but decreased sensitivity in K7M3 metastatic murine OS cells. Consistent with the above findings, Hollomon et al (18) revealed that autophagy inhibition via ATG5 knockdown reduced camptothecin-induced cell death in DLM8 metastatic murine OS cells but increased it in K7M3 cells. These contradictory outcomes largely depend on the stage and type of tumor (10).
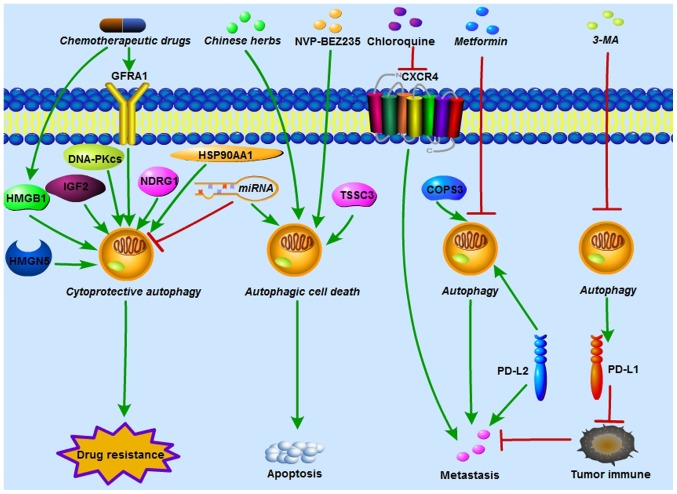
In OS, accumulating evidence has indicated that autophagy plays a crucial role in chemoresistance, either by promoting drug resistance or increasing drug sensitivity. Various oncogenic and tumor-suppressing genes have been confirmed to regulate OS chemoresistance via autophagy activation or inhibition. In autophagy-related OS chemoresistance, autophagy can act as either a cytoprotective process or autophagic cell death (Fig. 2).
Figure 2.
Autophagy regulates OS chemoresistance, metastasis and tumor immunity. HMGB1, GFRA1, HMGN5, IGF2, DNA-PKcs, NDRG1 and HSP90AA1 induced by chemotherapeutic drugs activate cytoprotective autophagy and contribute to chemoresistance in OS. In addition, miRNAs increase OS chemosensitivity by either inhibiting cytoprotective autophagy or inducing autophagic cell death. NVP-BEZ235 (a PI3K/mTOR inhibitor), TSSC3 and certain Chinese herbs enhance chemosensitivity in OS by increasing apoptosis which is dependent of autophagic cell death. COPS3 knockdown and metformin reduce autophagy-mediated metastasis in OS. Polymeric chloroquine decreased CXCR4-mediated OS metastasis, and this effect was autophagy-independent. PD-L1 suppression by 3-MA and PD-L2 knockdown enhanced immunological response and inhibited OS metastasis. HMGB1, High mobility group box 1; GFRA1, GDNF receptor α1; HMGN5, high-mobility group nucleosome-binding domain 5; IGF2, insulin growth factor 2; DNA-PKcs, DNA-dependent protein kinase catalytic subunit; miRNA, microRNA; NDRG1, N-myc downstream-regulated gene 1; HSP90AA1, heat shock protein 90AA1; OS, osteosarcoma; TSSC3, tumor-suppressing STF cDNA 3; COPS3, COP9 signalosome subunit 3; CXCR4, chemokine receptor 4; PD-L, programmed death ligand; 3-MA, 3-methyladenine.
Autophagy acts as a cytoprotective process contributing to OS chemoresistance
Directly targeting autophagy with either ATG silencing or autophagy modulators is a commonly used method to determine autophagy-mediated OS chemoresistance. Silencing of ATG14, also termed Beclin-1-associated autophagy-related key regulator, increased cisplatin-induced apoptosis in SaOS-2 cells (19). Beclin-1 inhibition enhanced the sensitivity of both MG63 and cisplatin-resistant MG63 cells to cisplatin in vitro and in vivo (20). Autophagy inhibition with chloroquine triggered apoptotic cell death in SaOS-2 cells which were resistant to cisplatin (21). Inhibition of autophagy via either ATG7 small interfering (si)RNA or 3-MA enhanced doxorubicin cytotoxicity in U2OS and SaOS-2 cells (22). It was reported by Zhou et al (23) that celecoxib, a selective cyclo-oxygenase-2 inhibitor, exerted an antitumor effect on 143B and U2OS cells. ATG5 silencing, and autophagy inhibitors chloroquine or SAR405 further enhanced cell proliferation inhibition and celecoxib-induced apoptosis. Guo et al (24) observed that rapamycin, an autophagy inducer, decreased paclitaxel-induced apoptosis in MG63. On the contrary, pretreatment with 3-MA, an autophagy inhibitor, increased MG63 apoptosis induced by paclitaxel. It was first revealed by Liu et al (25) that apatinib, a highly selective inhibitor of vascular endothelial growth factor receptor-2, induced OS cells apoptosis and autophagy. In addition, autophagy inhibition via 3-MA markedly enhanced apatinib-induced apoptosis in KHOS cells.
In addition to directly modulating autophagy as mentioned above, several upstream target genes and signaling pathways have been demonstrated to regulate autophagy-mediated OS chemoresistance (Table I).
Table I.
Autophagy acts as a cytoprotective process contributing to OS chemoresistance.
| First author, year | Target gene/signaling pathway | Autophagy | Alteration | OS cell lines | Chemotherapeutic agents | Resistance | Reference |
|---|---|---|---|---|---|---|---|
| Huang, 2012 and 2012 | HMGB1 | Beclin-1-PI3KC3 | |||||
| ULK1-Matg13-FIP200 | ↑ | MG-63, SaOS-2, U-2OS | DOX, CDDP and MTX | ↑ | (26,27) | ||
| Kim, 2017 and 2018 | GFRA1 | Beclin-1, LC3-II | ↑ | MG-63 | CDDP | ↑ | (29,30) |
| Meng, 2016 | miR-140-5p | LC3-II | ↑ | SaOS-2 | DOX, CDDP | ↑ | (33) |
| Wei, 2017 | miR-140-5p | Beclin-1, ATG5, LC3-II | ↓ | HOS, U-2OS MG-63 | DOX, CDDP and MTX | ↓ | (34) |
| Chen, 2014 | miR-155 | LC3-II, ATG5 | ↑ | SaOS-2 | DOX, CDDP | ↑ | (35) |
| Xu, 2016 | miR-30a | Beclin-1, LC3-II | ↓ | MG-63/Dox-resistant cells | DOX, CDDP and MTX | ↓ | (31) |
| Chen, 2017 | miR-410 | LC3-II, ATG16L1 | ↓ | U-2OS, MG-63 | DOX, CDDP and RAPA | ↓ | (32) |
| Guo, 2014 | miR-22 | LC3-II, ATG7 | ↓ | MG-63 | DOX, CDDP | ↓ | (37) |
| Li, 2014 | miR-22 | LC3-II, ATG7 | ↓ | U-2OS, MG-63 | DOX, CDDP | ↓ | (38) |
| Wang, 2019 | miR-22 | Beclin-1, LC3-II, ATG5 | ↓ | MG-63 | CDDP | ↓ | (39) |
| Chang, 2014 | miR-101 | LC3-II, ATG5 | ↓ | U-2OS | DOX | ↓ | (40) |
| Zhou, 2015 | miR-143 | LC3-II, ATG2B | ↓ | SaOS-2/Dox-resistant cells, U-2OS/Dox-resistant cells | DOX | ↓ | (41) |
| Li, 2016 | miR-199a-5p | Beclin-1, LC3-II | ↓ | MG-63 | CDDP | ↓ | (42) |
| Yang, 2014 | HMGN5 | Beclin-1, LC3-II | ↑ | U-2OS, MG-63 | DOX, CDDP and MTX | ↑ | (47) |
| Shimizu, 2014 | IGF2 | LC3-II, ATG7 | ↑ | SaOS-2, U-2OS | DOX, CDDP | ↑ | (48) |
| Zhen, 2016 | DNA-PKcs | Beclin-1, LC3-II | ↑ | U-2OS, MG-63 | Salinomycin | ↑ | (49) |
| Wang, 2017 | NDRG1 | LC3-II | ↑ | MG-63.2 | CA-4 | ↑ | (50) |
| Xiao, 2018 | HSP90AA1 | LC3-II | ↑ | MG-63 | DOX, CDDP | ↑ | (51) |
| Tao, 2017 | Wnt/β-catenin | Beclin-1 | ↓ | MG-63 | Gemcitabine | ↓ | (52) |
| Mukherjee, 2017 | JNK | LC3-II, ATG5, ATG12 | ↓ | HOS | CDDP | ↓ | (53) |
| Zhang, 2017 | JNK | ATG5 | ↑ | MG-63 | Curcumin | ↑ | (54) |
| Guan, 2016 | Caveolin-1, PI3K-Akt-JNK | Beclin-1, LC3-II, ATG5, ATG7 | ↓ | SaOS-2/Taxol-resistant cells, U-2OS/Taxol-resistant cells | Taxol | ↓ | (55) |
OS, osteosarcoma; miR, microRNA; HMGB1, high mobility group box 1; GFRA1, glial cell line-derived neurotrophic factor receptor α1; HMGN5, high-mobility group nucleosome-binding domain 5; IGF2, insulin growth factor 2; DNA-PKcs, DNA-dependent protein kinase catalytic subunit; NDRG1, N-myc downstream-regulated gene 1; HSP90AA1, heat shock protein 90AA1; JNK, Jun N-terminal kinase; PI3KC3, class III phosphoinositide 3-kinase catalytic subunit 3; ULK1, UNC-51-like kinase; ATG, autophagy-related protein; LC3, microtubule-associated protein 1-light chain 3; CDDP, cisplatin; DOX, doxorubicin; MTX, methotrexate; RAPA, rapamycin; CA-4, combretastatin A-4.
High mobility group box 1 (HMGB1)
HMGB1, a chromatin-binding nuclear protein with 215 amino acid residues, is composed of three different domains: An A box, B box and C-terminal acidic tail (26,27). It can localize in the nucleus, cytoplasm and cell surface, and it can be released extracellularly. Different forms of HMGB1 exhibit different functions. For example, nuclear HMGB1 regulates DNA replication, recombination, transcription and repair, and sustains genomic stability (28). Cytoplasmic HMGB1 contributes to cell motility and autophagy. Cell surface HMGB1 is associated with neurite outgrowth and platelet activation (28). Extracellular HMGB1 is implicated in cancer cell activation, inflammation progression and apoptosis of monocyte-lineage and immune cells (28). When it comes to HMGB1-mediated autophagy in OS, it was first reported by Huang et al (26,27) that HMGB1 overexpres-sion induced autophagy by regulating Beclin-1-PI3K catalytic subunit 3 and ULK1-mATG13-FIP200 complex formation, and increased the drug resistance of MG-63, SaOS-2 and U-2OS cells to doxorubicin, cisplatin and methotrexate. Conversely, the suppression of HMGB1 by short hairpin (sh) RNA inhibited autophagy and enhanced sensitivity to these chemotherapeutic agents.
Glial cell line-derived neurotrophic factor (GDNF) receptor α1 (GFRA1)
The GDNF family, consisting of GDNF, neurturin, artemin and persephin, plays a crucial role in the development and maintenance of the nervous system (29,30). GFRA1 is the receptor of GDNF, and the binding of GFRA1 with GDNF promotes neuronal cell survival and differentiation (29,30). Of note, it was found by Kim et al (29,30) that GFRA1-mediated autophagy was also implicated in OS cisplatin resistance. They demonstrated that GFRA1 was significantly upregulated in the presence of cisplatin, but not doxorubicin and methotrexate in two OS cell lines (MG-63 and U-2OS). In addition, GFRA1 induced autophagy in MG-63 cells by activating SRC-AMPK signaling following cisplatin treatment.
microRNAs (miRNAs/miRs)
miRNAs are a class of small non-coding RNAs (18-25 nucleotides) that can negatively regulate gene expression by binding to the 3′-untranslated region (3′-UTR) of their target mRNAs, and modulate mRNA and protein expression at the post-transcriptional level (31). The dysregulation of miRNAs has been identified in the carcinogenesis of various malignancies, including OS (32). Recently, they have emerged as key regulators of OS chemosensitivity or chemotherapy resistance by targeting autophagy; notably, certain miRNAs have been shown to lead to contradictory outcomes due to their dual role in OS chemo-resistance. For example, miR-140-5p functioned as a tumor promoter and was clearly upregulated in SaOS-2 and MG-63 cells following doxorubicin and cisplatin treatment (33). miR-140-5p overexpression induced autophagy, as confirmed by increased GFP-LC3 puncta and LC3-II, and decreased p62, which contributed to OS chemoresistance. Conversely, it was revealed by another study (34) that miR-140-5p serves as a tumor suppressor and is downregulated in 40 clinical OS tissues and three OS cell lines (HOS, U-2OS and MG63). Overexpression of miR-140-5p increased the sensitivity of OS cells to doxorubicin, cisplatin and methotrexate by inhibiting autophagy, as detected by TEM, confocal microscopy and western blotting. In addition, certain miRNAs contribute to OS chemoresistance not only via autophagy activation, but also by inhibiting autophagy. One study demonstrated that miR-155 promoted OS chemoresistance by inducing autophagy (35). Conversely, miR-155 inhibited autophagy by regulating the PTEN-PI3K/AKT/mTOR pathway, and enhanced resistance of MG63 cells to doxorubicin in another study (36).
The majority of miRNAs that function as tumor suppressors increase OS chemosensitivity by negatively regulating autophagy. Xu et al (31) found that miR-30a was down-regulated, while ATGs Beclin-1 and LC3-II were increased in doxorubicin-resistant MG-63 cells. Furthermore, miR-30a overexpression enhanced OS chemosensitivity by suppressing Beclin-1-mediated autophagy, which could be partly reversed by rapamycin, an autophagy activator. Chen et al (32) observed that miR-410 sensitized U-2OS and MG-63 cells to doxorubicin and cisplatin via ATG16L1 inhibition. Certain studies have shown that miR-22 increases OS chemosensitivity by inhibiting HMGB1-mediated autophagy (37,38). Consistent with these findings, miR-22 can also sensitize MG-63 cells to cisplatin via metadherin-mediated autophagy (39). miR-101 blocks doxorubicin-induced autophagy and enhance U-2OS cell chemosensitivity (40). miR-143 was found to reverse chemoresistance in SaOS-2 and U-2OS doxorubicin-resistant cells through the inhibition of autophagy (41). miR-199a-5p was reported to reduce the resistance of MG-63 cells to cispl-atin by inhibiting autophagy, as indicated by the decreased expression of LC3-II and Beclin-1 (42). Long non-coding RNA (LncRNA) small nucleolar RNA host gene 15 was found to increase proliferation, invasion, migration and autophagy in MG-63 cells by negatively regulating miR-141 (43). LncRNA CTA was reported to reduce doxorubicin resistance in SaOS-2, MG-63 and doxorubicin-resistant MG-63 cells by suppressing miR-210 and autophagy (44).
In contrast with the aforementioned studies, Yu et al (45) revealed that miR-100 and Beclin-1 expression levels were markedly reduced in cisplatin-resistant MG-63 cells, compared with their sensitive counterparts, and miR-100 upregulation enhanced cisplatin-induced apoptosis via mTOR inhibition and autophagy activation. Similar to their findings, it was confirmed by Wu et al (46) that miR-145-3p overexpression promoted apoptosis and autophagy in U-2OS and MG-63 cells by negatively regulating histone deacetylase 4.
Certain other genes are also implicated in OS chemoresistance via autophagy. High-mobility group nucleosome-binding domain 5 was required for OS chemoresistance by upregulating autophagy (47). Insulin growth factor 2 was shown to maintain OS cell survival in the presence of chemotherapeutic drugs by activating autophagy. Blocking autophagy with chloroquine or bafilomycin A restored chemosensitivity (48). Zhen et al (49) demonstrated that DNA-dependent protein kinase catalytic subunit (DNA-PKcs) was involved in autophagy-mediated sali-nomycin resistance in OS cells. The knockdown of DNA-PKcs by its inhibitors, shRNA and miR-101, reduced salinomycin resistance by inhibiting autophagy. Wang et al (50) found that N-myc downstream-regulated gene 1 (NDRG1) was associated with OS chemoresistance. Combretastatin A-4 (CA-4), a tubulin-depoly-merizing agent with antitumor effects, activated cytoprotective autophagy in OS. A synergistic cytotoxic effect was observed when CA-4 was combined with chloroquine. Furthermore, NDRG1 inhibition by siRNA enhanced the sensitivity of OS cells to CA-4 by suppressing autophagosome-lysosome fusion (50). Heat shock protein 90AA1 (HSP90AA1) was confirmed to regulate OS drug resistance via autophagy (51). The overexpression of HSP90AA1 promoted autophagy and led to increased resistance. This pro-survival effect of HSP90AA1 could be reversed by 3-MA. Conversely, the suppression of HSP90AA1 enhanced chemosensitivity by inhibiting autophagy (51).
Signaling pathways
Accumulating evidence has indicated that the regulation of autophagy-related signaling pathways is implicated in OS chemoresistance. Wnt/β-catenin signaling pathway activation enhanced sensitivity of MG-63 cells to gemcitabine by attenuating Beclin-1-mediated autophagy (52). Mukherjee et al (53) observed that a negative feedback loop between the Jun N-terminal kinase (JNK) pathway and autophagy, and the inhibition of both, led to maximal cisplatin sensitivity in HOS cells. Similar to the results of the present study, it was revealed by Zhang et al (54) that the inhibition of autophagy with 3-MA enhanced the apoptosis of MG63 cells induced by curcumin, a chemotherapeutic drug derived from the rhizome of the East Indian plant Curcuma longa. It was further confirmed that this cell apoptosis promoted by 3-MA was dependent on the JNK pathway. In order to investigate the association between caveolin-1 and taxol resistance, Guan et al (55) established taxol-resistant SaOS-2 and U-2OS cells by gradually increasing taxol concentration for 6 months. Reduced caveolin-1 expression and enhanced autophagy activity were identified in taxol-resistant cells compared with their parental cells. In addition, caveolin-1 overexpression reduced taxol resistance by attenuating PI3K-Akt-JNK-dependent autophagy.
Autophagy acts as autophagic cell death reversing OS chemo-resistance
For a long time, autophagy has been considered to have a crucial pro-survival effect on OS chemoresistance, as it can maintain tumor cell growth in response to chemotherapeutic drugs by eliminating and recycling its own damaged proteins and organelles to provide energy (23,25). However, an increasing number of studies have focused on the other primary outcome of autophagy: Autophagic cell death characterized by excessive autophagy, one of the three main forms of programmed cell death (PCD) (56,57). The other two forms of PCD are apoptosis and programmed necrosis (56,57). An intricate cross-talk between apoptosis and autophagy is most widely discussed in OS chemoresistance-related studies (56,57). Autophagic cell death, different from cytoprotective autophagy, can be increased by autophagy activation or decreased by autophagy inhibition.
Autophagy induced by rapamycin inhibits the proliferation of SaOS-2 and U-2OS in vitro and tumor growth in mice xenograft models in vivo (58). Tumor-suppressing STF cDNA 3-induced autophagy was found to be indispensable for the suppression of OS tumorigenesis and metastasis in vitro and in vivo (59). NVP-BEZ235, a PI3K/mTOR inhibitor, increases cisplatin-induced apoptosis in U-2OS and SaOS-2 cells by turning cytoprotective autophagy into pro-death autophagy (60). Voacamine, a bisindolic alkaloid extracted from Peschiera fuchsiaefolia, enhances the chemosensitivity of doxorubicin-resistant U-2OS cells by inducing autophagic cell death rather than apoptosis (61).
Recently, several Chinese herbs have been reported to exert their antitumor effects on OS via autophagic cell death. For example, Huang et al (62) indicated that honokiol, extracted from Magnolia trees, inhibited HOS and U-2OS cell proliferation by inducing both apoptosis and autophagy. They further discovered that the honokiol-induced cell death was largely dependent on autophagic cell death, as shown by the results that honokiol-induced cell death was more clearly reversed by 3-MA compared with Z-VAD-FMK, a widely used caspase inhibitor. Autophagy induced by tanshinone IIA, isolated from the herb Salvia miltiorrhiza, was reported to be cytotoxic to 143B cells (63). Brazilin, purified from Biancaea sappan wood, induces autophagic cell death in MG-63 cells (64). Liu et al (65) suggested that andrographolide reduced MG-63 and U-2OS cell viability by inducing autophagy, but not apoptosis. In addition, the inhibition of autophagy via 3-MA and Beclin-1 silencing could rescue the cytotoxic effects of andrographolide, indicating that autophagic cell death contributed to the tumor-suppressing effect of andrographolide. Furthermore, marrubenol, escin and chamaejasmine can also inhibit OS by inducing autophagic cell death (66-69). Surprisingly, different active ingredients from the same herb can induce opposing autophagy functions in the same OS cell line by activating the same pathway. Cytoprotective autophagy and autophagic cell death were induced by curcumin and curcumol, respectively, in MG-63 cells via the JNK pathway (Table II) (54,70).
Table II.
Autophagy acts as autophagic cell death, reversing OS chemoresistance.
| First author, year | Autophagy inducers/Chinese herbs | Autophagy | Alteration | OS cell lines | Chemotherapeutic agents | Sensitivity/autophagic cell death | Reference |
|---|---|---|---|---|---|---|---|
| Zhao, 2015 | Rapamycin | LC3-II | ↑ | SaOS-2, U-2OS | / | ↑ | (58) |
| Zhao, 2018 | TSSC3 | ATG5, LC3-II | ↑ | SaOS-2 | / | ↑ | (59) |
| Huang, 2018 | NVP-BEZ235 | LC3-II | ↑ | U-2OS, SaOS-2 | CDDP | ↑ | (60) |
| Meschini, 2007 | Voacamine | Autophagosomes, | ↑ | U-2OS-R | DOX | ↑ | (61) |
| Huang, 2018 | Honokiol | ATG5, LC3-II | ↑ | HOS, U-2OS | / | ↑ | (62) |
| Yen, 2018 | Tanshinone IIA | LC3-II | ↑ | 143B | / | ↑ | (63) |
| Kang, 2018 | Brazilin | LC3-II, ATG5, ATG7, ATG10, ULK1 | ↑ | MG-63 | / | ↑ | (64) |
| Liu, 2017 | Andrographolide | ATG5, Beclin-1, LC3-II | ↑ | MG-63, U-2OS | / | ↑ | (65) |
| Zhang, 2018 | Marrubenol | Beclin-1, LC3-II | ↑ | SaOS-2 | / | ↑ | (66) |
| Liu, 2017; Zhu, 2017 | Escin | Beclin-1, LC3-II, ATG5, ATG12 | ↑ | U-2OS, HOS, SaOS-2 | / | ↑ | (67,68) |
| Yang, 2019 | Chamaejasmine | Beclin-1, LC3-II, ATG7 | ↑ | MG-63 | / | ↑ | (69) |
| Zhang, 2017 | Curcumol | LC3-II | ↑ | MG-63 | / | ↑ | (70) |
OS, osteosarcoma; TSSC3, tumor-suppressing STF cDNA 3; ATG, autophagy-related protein; LC3, microtubule-associated protein 1-light chain 3; CDDP, cisplatin; DOX, doxorubicin.
5. Autophagy and metastasis
Metastasis (particularly lung metastasis), detected in 13-27% of patients with OS at diagnosis and 40% at progressive stage, is one of the main reasons contributing to unfavorable prognosis (71). It is estimated that ~30-40% of patients with OS show poor response to chemotherapy due to metastasis (72). It has been revealed by certain studies that autophagy is also implicated in OS metastasis. Zhang et al (73) reported that COP9 signalosome subunit 3 knockdown reduced OS metastasis by inhibiting Beclin-1. In addition, both 3-MA and Beclin-1 silencing induced anti-metastasis effects. It was reported by Bao et al (72) that metformin, mainly used in the treatment of type II diabetes, inhibited OS metastasis via miR-570-3p-mediated suppression of lung cancer metastasis-related protein 1 and ATG12. miR-506-3p reversed epithelial-to-mesenchymal transition, which is closely associated with cancer metastasis, by suppressing autophagy in OS cells (74). It has already been reported that chemokine receptor 4 (CXCR4) is crucial for the regulation of OS metastasis; in our previous study, it was found that CXCR4 inhibition with AMD3100 significantly reduces OS survival and metastasis (75). Yu et al (76) discovered that polymeric chloroquine decreased CXCR4-mediated U-2OS cell metastasis by promoting the internalization of surface CXCR4 receptors, which made CXCR4 inaccessible for binding with its ligand, chemokine 12. However, no change in LC3 expression was observed when cells were treated with polymeric chloroquine, indicating that this anti-metastasis effect was independent of autophagy. Whether CXCR4 influences autophagy in the regulation of OS chemoresistance and metastasis remains largely unknown; this will be the focus of future studies (Fig. 2).
6. Autophagy and immunotherapy
Recently, immunotherapy has emerged as a novel therapeutic method for OS; due to their immune function, T-cells can help kill cancer cells. The binding of programmed death ligand-1 (PD-L1) to PCD protein-1 (PD-1) attenuates the anti-tumor effects of T-cells, ultimately leading to tumor immune escape, chemoresistance and metastasis (77,78). Yu et al (77) indicated that PD-L1 suppression via photodynamic therapy combined with the autophagy inhibitor 3-MA enhanced the immune response, and inhibited OS growth and metastasis in vitro and in vivo. Similarly, Ren et al (78) revealed the pro-metastatic function of PD-L2, another ligand of PD-1, in OS. In addition, PD-L2 knockdown was found to decrease OS migration and invasion by inhibiting Beclin-1 expression (Fig. 2).
7. Autophagy as a prognostic marker in OS
It is noteworthy that certain clinical studies have explored whether autophagy could be used to predict treatment response and survival rate in OS. Livingston et al (79) detected LC3B and HSP27 expression in 394 tumor samples, including pre-treatment, post-treatment and metastatic samples from 260 OS patients via immunohistochemistry. It was revealed that the percentage of LC3B-positive samples in the pre-treatment, post-treatment and metastatic groups were 34, 50 and 67%, respectively. Furthermore, patients with positive LC3B and negative HSP27 expression exhibited the highest 10-year survival rate (75%), whereas those with negative LC3B and positive HSP27 expression the worst (25%), indicating that LC3B and HSP27 were associated with favorable and poor outcomes in OS, respectively. Lu et al (80) demonstrated that p62 was detected in 54/70 OS samples (77.1%), and that its overexpression was associated with tumor size, metastasis, clinical stage and poor prognosis. Conversely, Ma et al (81) discovered that the 5-year survival rate of patients with OS with low p62 expression was lower than that of patients with high p62 expression, suggesting that decreased p62 expression was associated with higher metastasis and chemotherapy resistance rates in OS.
8. Conclusion
Chemoresistance is one of the most important factors contributing to treatment failure and poor prognosis in OS. Autophagy, a catabolic process via which cells eliminate and recycle their own damaged proteins and organelles to provide energy, can be activated by chemotherapeutic drugs. Accumulating evidence indicates that autophagy serves a dual role in the regulation of OS chemoresistance, by either exerting cytoprotection or causing autophagic cell death. Therefore, both the elimination of cytoprotective autophagy and the stimulation of autophagic cell death could enhance OS chemosensitivity. In addition, autophagy is also implicated in OS metastasis, immunotherapy and clinical prognosis. It is anticipated that targeting autophagy may be a promising therapeutic strategy for OS.
Acknowledgments
Not applicable.
Abbreviations
- OS
osteosarcoma
- ATGs
autophagy-related proteins
- ULK1
UNC-51-like kinase
- FIP200
focal adhesion kinase family interacting protein of 200 kDa
- mTOR
mammalian target of rapamycin
- LC3
light chain 3
- TEM
transmission electron microscopy
- AVi
initial autophagic vacuole
- AVd
degradative autophagic vacuole
- 3-MA
3-methyladenine
- PI3K
class III phosphoinositide 3-kinase
- HMGB1
high mobility group box 1
- GDNF
glial cell line-derived neurotrophic factor
- GFRA1
GDNF receptor α1
- miRNAs/miRs
microRNAs
- DNA-PKcs
DNA-dependent protein kinase catalytic subunit
- NDRG1
N-myc downstream-regulated gene 1
- CA-4
combretastatin A-4
- HSP90AA1
heat shock proteins 90AA1
- JNK
Jun N-terminal kinase
- PCD
programmed cell death
- CXCR4
chemokine receptor 4
- PD-L1
programmed death ligand-1
- PD-1
programmed cell death protein-1
- HSP27
heat shock protein 27
Funding
No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
YXL was involved in designing the study, literature review and drafting of the manuscript. YXL was also responsible for designing the figures. HYY, JYL, YRC and FL participated in acquisition and analysis of data, and discussion of the manuscript. ZMH and SSH designed the study and revised the manuscript critically. All of the authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Morrow JJ, Bayles I, Funnell APW, Miller TE, Saiakhova A, Lizardo MM, Bartels CF, Kapteijn MY, Hung S, Mendoza A, et al. Positively selected enhancer elements endow osteosarcoma cells with metastatic competence. Nat Med. 2018;24:176–185. doi: 10.1038/nm.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Isakoff MS, Bielack SS, Meltzer P, Gorlick R. Osteosarcoma: Current treatment and a collaborative pathway to success. J Clin Oncol. 2015;33:3029–3035. doi: 10.1200/JCO.2014.59.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li S, Sun W, Wang H, Zuo D, Hua Y, Cai Z. Research progress on the multidrug resistance mechanisms of osteosarcoma chemotherapy and reversal. Tumour Biol. 2015;36:1329–1338. doi: 10.1007/s13277-015-3181-0. [DOI] [PubMed] [Google Scholar]
- 5.Xu J, Wang H, Hu Y, Zhang YS, Wen L, Yin F, Wang Z, Zhang Y, Li S, Miao Y, et al. Inhibition of CaMKIIα activity enhances antitumor effect of fullerene C60 nanocrystals by suppression of autophagic degradation. Adv Sci (Weinh) 2019;6:1801233. doi: 10.1002/advs.201801233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen R, Wang G, Zheng Y, Hua Y, Cai Z. Drug resistance-related microRNAs in osteosarcoma: Translating basic evidence into therapeutic strategies. J Cell Mol Med. 2019;23:2280–2292. doi: 10.1111/jcmm.14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Duan G, Feng S. MicroRNA-301a modulates doxorubicin resistance in osteosarcoma cells by targeting AMP-activated protein kinase alpha 1. Biochem Biophys Res Commun. 2015;459:367–373. doi: 10.1016/j.bbrc.2015.02.101. [DOI] [PubMed] [Google Scholar]
- 8.Luetke A, Meyers PA, Lewis I, Juergens H. Osteosarcoma treatment - where do we stand? A state of the art review. Cancer Treat Rev. 2014;40:523–532. doi: 10.1016/j.ctrv.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Camuzard O, Santucci-Darmanin S, Carle GF, Pierrefite-Carle V. Role of autophagy in osteosarcoma. J Bone Oncol. 2019;16:100235. doi: 10.1016/j.jbo.2019.100235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar A, Singh UK, Chaudhary A. Targeting autophagy to overcome drug resistance in cancer therapy. Future Med Chem. 2015;7:1535–1542. doi: 10.4155/fmc.15.88. [DOI] [PubMed] [Google Scholar]
- 11.O'Farrill JS, Gordon N. Autophagy in osteosarcoma. Adv Exp Med Biol. 2014;804:147–160. doi: 10.1007/978-3-319-04843-7_8. [DOI] [PubMed] [Google Scholar]
- 12.D'Arcy MS. Cell death: A review of the major forms of apoptosis, necrosis and autophagy. Cell Biol Int. 2019;43:582–592. doi: 10.1002/cbin.11137. [DOI] [PubMed] [Google Scholar]
- 13.Nagelkerke A, Sweep FC, Geurts-Moespot A, Bussink J, Span PN. Therapeutic targeting of autophagy in cancer. Part I: Molecular pathways controlling autophagy. Semin Cancer Biol. 2015;31:89–98. doi: 10.1016/j.semcancer.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagelkerke A, Bussink J, Geurts-Moespot A, Sweep FC, Span PN. Therapeutic targeting of autophagy in cancer. Part II: Pharmacological modulation of treatment-induced autophagy. Semin Cancer Biol. 2015;31:99–105. doi: 10.1016/j.semcancer.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 16.Schott CR, Ludwig L, Mutsaers AJ, Foster RA, Wood GA. The autophagy inhibitor spautin-1, either alone or combined with doxorubicin, decreases cell survival and colony formation in canine appendicular osteosarcoma cells. PLoS One. 2018;13:e0206427. doi: 10.1371/journal.pone.0206427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He H, Ni J, Huang J. Molecular mechanisms of chemoresistance in osteosarcoma (Review) Oncol Lett. 2014;7:1352–1362. doi: 10.3892/ol.2014.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hollomon MG, Gordon N, Santiago-O'Farrill JM, Kleinerman ES. Knockdown of autophagy-related protein 5, ATG5, decreases oxidative stress and has an opposing effect on camptothecin-induced cytotoxicity in osteosarcoma cells. BMC Cancer. 2013;13:500. doi: 10.1186/1471-2407-13-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Z, Tao L, Shen C, Liu B, Yang Z, Tao H. Silencing of Barkor/ATG14 sensitizes osteosarcoma cells to cisplatin induced apoptosis. Int J Mol Med. 2014;33:271–276. doi: 10.3892/ijmm.2013.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu W, Li W, Zhou Y, Zhang C. Inhibition of beclin1 affects the chemotherapeutic sensitivity of osteosarcoma. Int J Clin Exp Pathol. 2014;7:7114–7122. [PMC free article] [PubMed] [Google Scholar]
- 21.Shen C, Wang W, Tao L, Liu B, Yang Z, Tao H. Chloroquine blocks the autophagic process in cisplatin-resistant osteosarcoma cells by regulating the expression of p62/SQSTM1. Int J Mol Med. 2013;32:448–456. doi: 10.3892/ijmm.2013.1399. [DOI] [PubMed] [Google Scholar]
- 22.Xu ZM, Li CB, Liu QL, Li P, Yang H. Ginsenoside Rg1 prevents doxorubicin-induced cardiotoxicity through the inhibition of autophagy and endoplasmic reticulum stress in mice. Int J Mol Sci. 2018;19:19. doi: 10.3390/ijms19113658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou P, Li Y, Li B, Zhang M, Xu C, Liu F, Bian L, Liu Y, Yao Y, Li D. Autophagy inhibition enhances celecoxib-induced apoptosis in osteosarcoma. Cell Cycle. 2018;17:997–1006. doi: 10.1080/15384101.2018.1467677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Y, Huang C, Li G, Chen T, Li J, Huang Z. Paxilitaxel induces apoptosis accompanied by protective autophagy in osteosarcoma cells through hypoxia-inducible factor-1α pathway. Mol Med Rep. 2015;12:3681–3687. doi: 10.3892/mmr.2015.3860. [DOI] [PubMed] [Google Scholar]
- 25.Liu K, Ren T, Huang Y, Sun K, Bao X, Wang S, Zheng B, Guo W. Apatinib promotes autophagy and apoptosis through VEGFR2/STAT3/BCL-2 signaling in osteosarcoma. Cell Death Dis. 2017;8:e3015. doi: 10.1038/cddis.2017.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang J, Ni J, Liu K, Yu Y, Xie M, Kang R, Vernon P, Cao L, Tang D. HMGB1 promotes drug resistance in osteosarcoma. Cancer Res. 2012;72:230–238. doi: 10.1158/0008-5472.CAN-11-2001. [DOI] [PubMed] [Google Scholar]
- 27.Huang J, Liu K, Yu Y, Xie M, Kang R, Vernon P, Cao L, Tang D, Ni J. Targeting HMGB1-mediated autophagy as a novel therapeutic strategy for osteosarcoma. Autophagy. 2012;8:275–277. doi: 10.4161/auto.8.2.18940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pistoia V, Pezzolo A. Involvement of HMGB1 in resistance to tumor vessel-targeted, monoclonal antibody-based immunotherapy. J Immunol Res. 2016;2016:3142365. doi: 10.1155/2016/3142365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim M, Jung JY, Choi S, Lee H, Morales LD, Koh JT, Kim SH, Choi YD, Choi C, Slaga TJ, et al. GFRA1 promotes cisplatin-induced chemoresistance in osteosarcoma by inducing autophagy. Autophagy. 2017;13:149–168. doi: 10.1080/15548627.2016.1239676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim M, Kim DJ. GFRA1: A novel molecular target for the prevention of osteosarcoma chemoresistance. Int J Mol Sci. 2018;19:19. doi: 10.3390/ijms19041078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu R, Liu S, Chen H, Lao L. MicroRNA-30a downregulation contributes to chemoresistance of osteosarcoma cells through activating Beclin-1-mediated autophagy. Oncol Rep. 2016;35:1757–1763. doi: 10.3892/or.2015.4497. [DOI] [PubMed] [Google Scholar]
- 32.Chen R, Li X, He B, Hu W. MicroRNA-410 regulates autophagy-related gene ATG16L1 expression and enhances chemosensitivity via autophagy inhibition in osteosarcoma. Mol Med Rep. 2017;15:1326–1334. doi: 10.3892/mmr.2017.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei R, Cao G, Deng Z, Su J, Cai L. miR-140-5p attenuates chemotherapeutic drug-induced cell death by regulating autophagy through inositol 1,4,5-trisphosphate kinase 2 (IP3k2) in human osteosarcoma cells. Biosci Rep. 2016;36:36. doi: 10.1042/BSR20160238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meng Y, Gao R, Ma J, Zhao J, Xu E, Wang C, Zhou X. MicroRNA-140-5p regulates osteosarcoma chemoresistance by targeting HMGN5 and autophagy. Sci Rep. 2017;7:416. doi: 10.1038/s41598-017-00405-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L, Jiang K, Jiang H, Wei P. miR-155 mediates drug resistance in osteosarcoma cells via inducing autophagy. Exp Ther Med. 2014;8:527–532. doi: 10.3892/etm.2014.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L, Tang B, Han H, Mao D, Chen J, Zeng Y, Xiong M. miR-155 affects osteosarcoma MG-63 cell autophagy induced by adriamycin through regulating PTEN-PI3K/AKT/mTOR signaling pathway. Cancer Biother Radiopharm. 2018;33:32–38. doi: 10.1089/cbr.2017.2306. [DOI] [PubMed] [Google Scholar]
- 37.Guo S, Bai R, Liu W, Zhao A, Zhao Z, Wang Y, Wang Y, Zhao W, Wang W. miR-22 inhibits osteosarcoma cell proliferation and migration by targeting HMGB1 and inhibiting HMGB1-mediated autophagy. Tumour Biol. 2014;35:7025–7034. doi: 10.1007/s13277-014-1965-2. [DOI] [PubMed] [Google Scholar]
- 38.Li X, Wang S, Chen Y, Liu G, Yang X. miR-22 targets the 3′UTR of HMGB1 and inhibits the HMGB1-associated autophagy in osteosarcoma cells during chemotherapy. Tumour Biol. 2014;35:6021–6028. doi: 10.1007/s13277-014-1797-0. [DOI] [PubMed] [Google Scholar]
- 39.Wang P, Zhao ZQ, Guo SB, Yang TY, Chang ZQ, Li DH, Zhao W, Wang YX, Sun C, Wang Y, et al. Roles of microRNA-22 in suppressing proliferation and promoting sensitivity of osteo-sarcoma cells via metadherin-mediated autophagy. Orthop Surg. 2019;11:285–293. doi: 10.1111/os.12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang Z, Huo L, Li K, Wu Y, Hu Z. Blocked autophagy by miR-101 enhances osteosarcoma cell chemosensitivity in vitro. ScientificWorldJournal. 2014;2014:794756. doi: 10.1155/2014/794756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou J, Wu S, Chen Y, Zhao J, Zhang K, Wang J, Chen S. microRNA-143 is associated with the survival of ALDH1+CD133+ osteosarcoma cells and the chemoresistance of osteosarcoma. Exp Biol Med (Maywood) 2015;240:867–875. doi: 10.1177/1535370214563893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Jiang W, Hu Y, Da Z, Zeng C, Tu M, Deng Z, Xiao W. MicroRNA-199a-5p inhibits cisplatin-induced drug resistance via inhibition of autophagy in osteosarcoma cells. Oncol Lett. 2016;12:4203–4208. doi: 10.3892/ol.2016.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu K, Hou Y, Liu Y, Zheng J. LncRNA SNHG15 contributes to proliferation, invasion and autophagy in osteosarcoma cells by sponging miR-141. J Biomed Sci. 2017;24:46. doi: 10.1186/s12929-017-0353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z, Liu Z, Wu S. Long non-coding RNA CTA sensitizes osteosarcoma cells to doxorubicin through inhibition of autophagy. Oncotarget. 2017;8:31465–31477. doi: 10.18632/oncotarget.16356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu Z, Li N, Jiang K, Zhang N, Yao LL. MiR-100 up-regulation enhanced cell autophagy and apoptosis induced by cisplatin in osteosarcoma by targeting mTOR. Eur Rev Med Pharmacol Sci. 2018;22:5867–5873. doi: 10.26355/eurrev_201809_15913. [DOI] [PubMed] [Google Scholar]
- 46.Wu G, Yu W, Zhang M, Yin R, Wu Y, Liu Q. MicroRNA-145-3p suppresses proliferation and promotes apotosis and autophagy of osteosarcoma cell by targeting HDAC4. Artif Cells Nanomed Biotechnol. 2018;46(Suppl 2):579–586. doi: 10.1080/21691401.2018.1464459. [DOI] [PubMed] [Google Scholar]
- 47.Yang C, Gao R, Wang J, Yuan W, Wang C, Zhou X. High-mobility group nucleosome-binding domain 5 increases drug resistance in osteosarcoma through upregulating autophagy. Tumour Biol. 2014;35:6357–6363. doi: 10.1007/s13277-014-1833-0. [DOI] [PubMed] [Google Scholar]
- 48.Shimizu T, Sugihara E, Yamaguchi-Iwai S, Tamaki S, Koyama Y, Kamel W, Ueki A, Ishikawa T, Chiyoda T, Osuka S, et al. IGF2 preserves osteosarcoma cell survival by creating an autophagic state of dormancy that protects cells against chemotherapeutic stress. Cancer Res. 2014;74:6531–6541. doi: 10.1158/0008-5472.CAN-14-0914. [DOI] [PubMed] [Google Scholar]
- 49.Zhen YF, Li ST, Zhu YR, Wang XD, Zhou XZ, Zhu LQ. Identification of DNA-PKcs as a primary resistance factor of salinomycin in osteosarcoma cells. Oncotarget. 2016;7:79417–79427. doi: 10.18632/oncotarget.12712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H, Li W, Xu J, Zhang T, Zuo D, Zhou Z, Lin B, Wang G, Wang Z, Sun W, et al. NDRG1 inhibition sensitizes osteosarcoma cells to combretastatin A-4 through targeting autophagy. Cell Death Dis. 2017;8:e3048. doi: 10.1038/cddis.2017.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao X, Wang W, Li Y, Yang D, Li X, Shen C, Liu Y, Ke X, Guo S, Guo Z. HSP90AA1-mediated autophagy promotes drug resistance in osteosarcoma. J Exp Clin Cancer Res. 2018;37:201. doi: 10.1186/s13046-018-0880-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tao H, Chen F, Liu H, Hu Y, Wang Y, Li H. Wnt/β-catenin signaling pathway activation reverses gemcitabine resistance by attenuating Beclin1-mediated autophagy in the MG63 human osteosarcoma cell line. Mol Med Rep. 2017;16:1701–1706. doi: 10.3892/mmr.2017.6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mukherjee S, Dash S, Lohitesh K, Chowdhury R. The dynamic role of autophagy and MAPK signaling in determining cell fate under cisplatin stress in osteosarcoma cells. PLoS One. 2017;12:e0179203. doi: 10.1371/journal.pone.0179203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, Chen P, Hong H, Wang L, Zhou Y, Lang Y. JNK pathway mediates curcumin-induced apoptosis and autophagy in osteosarcoma MG63 cells. Exp Ther Med. 2017;14:593–599. doi: 10.3892/etm.2017.4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guan J, Yuan Z, He J, Wu Z, Liu B, Lin X, Mo L, Mo H. Overexpression of caveolin-1 reduces Taxol resistance in human osteosarcoma cells by attenuating PI3K-Akt-JNK dependent autophagy. Exp Ther Med. 2016;12:2815–2822. doi: 10.3892/etm.2016.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ouyang L, Shi Z, Zhao S, Wang FT, Zhou TT, Liu B, Bao JK. Programmed cell death pathways in cancer: A review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012;45:487–498. doi: 10.1111/j.1365-2184.2012.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li J, Yang Z, Li Y, Xia J, Li D, Li H, Ren M, Liao Y, Yu S, Chen Y, et al. Cell apoptosis, autophagy and necroptosis in osteosarcoma treatment. Oncotarget. 2016;7:44763–44778. doi: 10.18632/oncotarget.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao S, Lu N, Chai Y, Yu X. Rapamycin inhibits tumor growth of human osteosarcomas. J BUON. 2015;20:588–594. [PubMed] [Google Scholar]
- 59.Zhao GS, Gao ZR, Zhang Q, Tang XF, Lv YF, Zhang ZS, Zhang Y, Tan QL, Peng DB, Jiang DM, et al. TSSC3 promotes autophagy via inactivating the Src-mediated PI3K/Akt/mTOR pathway to suppress tumorigenesis and metastasis in osteosarcoma, and predicts a favorable prognosis. J Exp Clin Cancer Res. 2018;37:188. doi: 10.1186/s13046-018-0856-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang JC, Cui ZF, Chen SM, Yang LJ, Lian HK, Liu B, Su ZH, Liu JS, Wang M, Hu ZB, et al. NVP-BEZ235 synergizes cisplatin sensitivity in osteosarcoma. Oncotarget. 2017;9:10483–10496. doi: 10.18632/oncotarget.23711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meschini S, Condello M, Marra M, Formisano G, Federici E, Arancia G. Autophagy-mediated chemosensitizing effect of the plant alkaloid voacamine on multidrug resistant cells. Toxicol In Vitro. 2007;21:197–203. doi: 10.1016/j.tiv.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 62.Huang K, Chen Y, Zhang R, Wu Y, Ma Y, Fang X, Shen S. Honokiol induces apoptosis and autophagy via the ROS/ERK1/2 signaling pathway in human osteosarcoma cells in vitro and in vivo. Cell Death Dis. 2018;9:157. doi: 10.1038/s41419-017-0166-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yen JH, Huang ST, Huang HS, Fong YC, Wu YY, Chiang JH, Su YC. HGK-sestrin 2 signaling-mediated autophagy contributes to antitumor efficacy of Tanshinone IIA in human osteosarcoma cells. Cell Death Dis. 2018;9:1003. doi: 10.1038/s41419-018-1016-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kang Y, He P, Wang H, Ye Y, Li X, Xie P, Wu B. Brazilin induces FOXO3A-dependent autophagic cell death by disturbing calcium homeostasis in osteosarcoma cells. Cancer Chemother Pharmacol. 2018;82:479–491. doi: 10.1007/s00280-018-3633-5. [DOI] [PubMed] [Google Scholar]
- 65.Liu Y, Zhang Y, Zou J, Yan L, Yu X, Lu P, Wu X, Li Q, Gu R, Zhu D. Andrographolide induces autophagic cell death and inhibits invasion and metastasis of human osteosarcoma cells in an autophagy-dependent manner. Cell Physiol Biochem. 2017;44:1396–1410. doi: 10.1159/000485536. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Y, Ma R, Cheng S, Gu G. Marrubenol inhibits osteosarcoma cancer cell growth by inducing autophagic cell death and inhibiting cancer cell migration and invasion. J BUON. 2018;23:729–734. [PubMed] [Google Scholar]
- 67.Liu ZR, Sun LZ, Jia TH, Jia DF. β-Aescin shows potent antiproliferative activity in osteosarcoma cells by inducing autophagy, ROS generation and mitochondrial membrane potential loss. J BUON. 2017;22:1582–1586. [PubMed] [Google Scholar]
- 68.Zhu J, Yu W, Liu B, Wang Y, Shao J, Wang J, Xia K, Liang C, Fang W, Zhou C, et al. Escin induces caspase-dependent apoptosis and autophagy through the ROS/p38 MAPK signalling pathway in human osteosarcoma cells in vitro and in vivo. Cell Death Dis. 2017;8:e3113. doi: 10.1038/cddis.2017.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang D, Zhang H, Wu J, Ma R, Li Z, Wang K, Yang F. The role of chamaejasmine in cellular apoptosis and autophagy in MG-63 cells. Biosci Rep. 2019;39:39. doi: 10.1042/BSR20181707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang C, Wang LM. Inhibition of autophagy attenuated curcumol-induced apoptosis in MG-63 human osteosarcoma cells via Janus kinase signaling pathway. Oncol Lett. 2017;14:6387–6394. doi: 10.3892/ol.2017.7010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71.Liao YX, Zhou CH, Zeng H, Zuo DQ, Wang ZY, Yin F, Hua YQ, Cai ZD. The role of the CXCL12-CXCR4/CXCR7 axis in the progression and metastasis of bone sarcomas (Review) Int J Mol Med. 2013;32:1239–1246. doi: 10.3892/ijmm.2013.1521. [DOI] [PubMed] [Google Scholar]
- 72.Bao X, Zhao L, Guan H, Li F. Inhibition of LCMR1 and ATG12 by demethylation-activated miR-570-3p is involved in the anti-metastasis effects of metformin on human osteosarcoma. Cell Death Dis. 2018;9:611. doi: 10.1038/s41419-018-0620-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang F, Yan T, Guo W, Sun K, Wang S, Bao X, Liu K, Zheng B, Zhang H, Ren T. Novel oncogene COPS3 interacts with Beclin1 and Raf-1 to regulate metastasis of osteosarcoma through autophagy. J Exp Clin Cancer Res. 2018;37:135. doi: 10.1186/s13046-018-0791-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang D, Bao F, Teng Y, Li Q, Li J. MicroRNA-506-3p initiates mesenchymal-to-epithelial transition and suppresses autophagy in osteosarcoma cells by directly targeting SPHK1. Biosci Biotechnol Biochem. 2019;83:836–844. doi: 10.1080/09168451.2019.1569496. [DOI] [PubMed] [Google Scholar]
- 75.Liao YX, Fu ZZ, Zhou CH, Shan LC, Wang ZY, Yin F, Zheng LP, Hua YQ, Cai ZD. AMD3100 reduces CXCR4-mediated survival and metastasis of osteosarcoma by inhibiting JNK and Akt, but not p38 or Erk1/2, pathways in in vitro and mouse experiments. Oncol Rep. 2015;34:33–42. doi: 10.3892/or.2015.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu F, Li J, Xie Y, Sleightholm RL, Oupicky D. Polymeric chloroquine as an inhibitor of cancer cell migration and experimental lung metastasis. J Control Release. 2016;244:347–356. doi: 10.1016/j.jconrel.2016.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu W, Wang Y, Zhu J, Jin L, Liu B, Xia K, Wang J, Gao J, Liang C, Tao H. Autophagy inhibitor enhance ZnPc/BSA nanoparticle induced photodynamic therapy by suppressing PD-L1 expression in osteosarcoma immunotherapy. Biomaterials. 2019;192:128–139. doi: 10.1016/j.biomaterials.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 78.Ren T, Zheng B, Huang Y, Wang S, Bao X, Liu K, Guo W. Osteosarcoma cell intrinsic PD-L2 signals promote invasion and metastasis via the RhoA-ROCK-LIMK2 and autophagy pathways. Cell Death Dis. 2019;10:261. doi: 10.1038/s41419-019-1497-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Livingston JA, Wang WL, Tsai JW, Lazar AJ, Leung CH, Lin H, Advani S, Daw N, Santiago-O'Farrill J, Hollomon M, et al. Analysis of HSP27 and the autophagy marker LC3B+ puncta following preoperative chemotherapy identifies high-risk osteo-sarcoma patients. Mol Cancer Ther. 2018;17:1315–1323. doi: 10.1158/1535-7163.MCT-17-0901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lu Y, Wang Q, Zhou Y, Sun L, Hu B, Xue H, Li M, Zhang K, Ren C, Duan N, et al. Overexpression of p62 is associated with poor prognosis and aggressive phenotypes in osteosarcoma. Oncol Lett. 2018;15:9889–9895. doi: 10.3892/ol.2018.8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ma H, Li X, Wang J, Hornicek FJ, Garbutt CC, Chang X, Duan Z. Expression and clinical implication of autophagy-associated protein p62 in osteosarcoma. Oncology. 2018;95:52–60. doi: 10.1159/000487437. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.