Abstract
The aim of this study is to investigate the expression of pancreatic progenitor cell differentiation and proliferation factor (PPDPF) and its relationship with clinicopathological factors in hepatocellular carcinoma (HCC).
A total of 135 patients diagnosed with HCC who underwent curative surgery were enrolled in this study. The expression of PPDPF was examined by real time-polymerase chain reaction (RT-PCR), western blot, and immunohistochemistry. The prognostic value for each sample was explored.
Both RT-PCR and western blot revealed PPDPF expression was upregulated in HCC. Higher PPDPF expression was also observed in HCC (54.07%) detected by immunohistochemistry (IHC), which was significantly associated with tumors size (P = .003), Edmondson-Steiner Grading (P = .021), recurrence (P = .010), and Diolame complete (P = .023). Patients with higher PPDPF expression had increased cancer progression and poorer prognosis than those with lower expression (P = .043). Multivariate analysis indicated PPDPF as an independent prognostic factor (P = .014).
Aberrance PPDPF expression might be a useful predictor and could serve as a potential therapeutic target for HCC.
Keywords: hepatocellular carcinoma, pancreatic progenitor cell differentiation and proliferation factor, prognosis
1. Introduction
Hepatocellular carcinoma (HCC) is considered as the most common cause of cancer-related mortality in worldwide.[1] HCC incidence is highest in East Asia and Africa due to the hepatitis B virus (HBV) infection and rapid increases in prevalence have occurred in Western countries caused by hepatitis C virus (HCV) infection and alcohol abuse.[2,3] Despite great progress of the complicated treatment acquired in the decade, HCC patients still have a poor prognosis, and the average survival time since initial diagnosis is dismal.[4] Tumor recurrence and metastasis after liver resection seem unavoidable, which are major contributor to the unfavorable prognosis.[5] Therefore, therapeutic intervention is needed for targeting new clinically applicable molecules for an early treatment of HCC.
Pancreatic progenitor cell differentiation and proliferation factor (PPDPF) locates in chromosome 20. The role of PPDPF is rarely explored, and the main molecular function is reported as a key regulator of exocrine pancreas development controlled by retinoic acid and ptf1a; PPDPF could promote the exocrine and inhibit the endocrine development through regulating the expression of cell cycle genes: cyclinG1, cyclinD1, p21Cip, p27Kip.[6]
In the National Center for Biotechnology Information (NCBI) database, the human PPDPF ortholog is expressed in relatively high levels in multiple tissues including the pancreas, colon, stomach, prostate, and kidney. Interestingly, the EST expression profile also indicates that higher level of PPDPF ortholog has been detected in several cancers including pancreatic cancer, liver cancer, and kidney cancer. In addition to structural mutations, many growth factor receptors and their ligands are overexpressed in liver cancers.[7] Since PPDPF can promote cell proliferation, it is worth studying the role of this gene in liver cancer.
The PPDPF consists with a putative signaling molecule containing 2 SH2 and 2 SH3 domains as well as other conserved domains,[6] suggesting that PPDPF may function in response to signals from adjacent mesoderm tissues. Multiple intercellular signals including transforming growth factor beta, Hedgehog, and Notch are critical for the tumor development.[8] It is not clear which signal or signals PPDPF responds to in order to promote the carcinogenesis.
In the present study, we evaluate the expression of PPDPF and investigate its relationship with clinicopathological characteristics and patients’ prognosis. This study will provide insight into the function of PPDPF genes and liver cancer progression.
2. Materials and methods
2.1. Patients and samples
A total of 135 HCC tissue specimens and 91 adjacent normal liver tissue cases were obtained by surgical resection samples from HCC patients admitted at Affiliated Hospital of Jiangsu University from 2008 to 2015. Both kinds of tissues were confirmed by pathological examination, formalin-fixed, and paraffin embedded. Clinicopathological features were collected from the hospital electronic database. Overall survival (OS) was followed up from the date of surgery until the death date or the last follow-up. This study was approved by the Review Board of Hospital Ethics Committee (Approval Number: JDFY20160527007) and the informed consent from each participant was obtained for every specimen examined.
2.2. RNA preparation and quantitative real-time PCR
The complementary DNA (cDNA) was prepared using the kit purchased from Promega after the total RNA was extracted using Trizol (Invitrogen, CA). Primers for the genes tested in the present experiments were designed using software PRIMER5 (version 5.0 for windows, CA). The mRNA level of PPDPF in the HCC tissues and paired non-cancerous tissues was examined by quantitative real-time PCR using SYBR Green Realtime PCR Master Mix (TOYOBO, Kita-ku, Osaka, Japan) following the instructions of the manufacturer. Sequences of quantitative real-time PCR primers are listed as follows:
18S Forward primer: 5′-TAAATCAGTTATGGTT CCTT-3′
18S Reverse primer: 5′-CGACTACCATCGAAAG TTGA-3′
PPDPF Forward primer: 5′-CGGTCTTCTCTGCAAATGGGC-3′
PPDPF Reverse primer: 5′-TGGCTGGTGGGATCTGGG-3′
2.3. Western blotting
Human polyclonal anti-rabbit PPDPF antibody was purchased from Proteintech (IL) and antibody to actin was purchased from Sigma (St. Louis, MO). All other chemicals were obtained either from Sigma (St. Louis, MO) or Fisher Scientific (Houston, TX).
After being determined the protein concentration, the cellular protein was subjected to dodecyl sulfate, sodium salt–polyacrylamide gel electrophoresis (SDS–PAGE), and the gel-fractionated proteins were transferred to nitrocellulose membranes (Bio-Rad, CA) and reacted with appropriate antibodies. Signals were detected with ECL kit (Pierce, Rockford, IL) by using one dimensional Image analysis.
2.4. Immunohistochemistry
Formalin-fixed paraffin-embedded tissue sections (4 μm thick) were stained using the immunoperoxidase method with avidin-biotin complex as described previously.[9] The monoclonal anti-PPDPF antibody was from Proteintech (IL). Sections were incubated with the anti-PPDPF monoclonal antibody at 1:100 dilutions overnight at 4 °C. The absence of nonspecific staining was confirmed by the control staining omitting the primary antibody. Labeling for marker was carried out using the Envision Plus Detection Kit (DAKO, Carpinteria, CA) followed the manufacturer's protocol. Nuclei were counterstained with hematoxylin. Immunohistochemical results were judged by 3 pathologists who were unaware of the clinical data. The staining results referred to PPDPF high or low expression were judged by German semiquantitative scoring system which considers both the stain intensity and the area extent as previous described.[10]
2.5. Statistical analysis
For comparison of PPDPF mRNA in 2 different groups, Student t test was used. Statistical analysis of correlation between PPDPF expression and clinical parameters was performed using chi-squared test. The overall survival (OS) was estimated using the Kaplan–Meier method from the date of surgery. Univariate analysis of the clinicopathological variables was performed using the log-rank test. Multivariate regression was performed using the Cox proportional hazards model. All analyses were performed using SPSS statistical software (version 16.0 for windows; SPSS Inc., Chicago, IL). All P-values were derived from 2-tailed tests and P < .05 was considered statistically significant.
3. Results
3.1. Expression of PPDPF in HCC
To determine the significance of PPDPF in HCC, we first analyzed microarray data sets in the TCGA database (http://xena.ucsc.edu/; data ID: TCGA. LIHC sample Map/HiSeqV2_exon). As showed in Fig. 1A, we found PPDPF mRNA levels were significantly increased in HCC samples as compared with normal liver tissue (P = .036).
Figure 1.
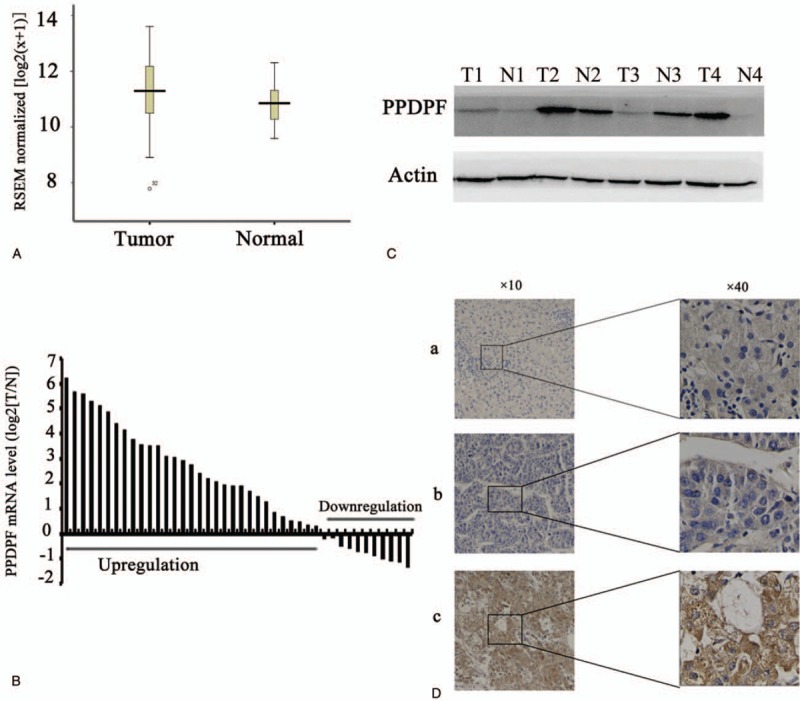
PPDPF expression is upregulated in HCC. (A) Analysis from the TCGA database (http://xena.ucsc.edu/) shows that mRNA expression levels of PPDPF is significantly higher in HCC compared with normal liver tissues. (B) Quantitative real-time PCR results of the relative expression level of PPDPF in 42 pairs of HCC and normal liver tissue samples. All of the reactions were performed in triplicate and results represent the mean ± SD (P < .05). The expression of PPDPF is normalized to beta-actin. (C) Expression of PPDPF protein in 4 randomly selected paired HCC samples was analyzed using western blot. N, normal; T, primary HCC. (D) Immunohistochemistry (IHC) results of PPDPF in human HCC specimens and adjacent normal tissues. a: expression of PPDPF in the paired adjacent tissues; b: low expression of PPDPF in HCC tissue; c: High PPDPF expression in HCC tissue. HCC = hepatocellular carcinoma, PCR = polymerase chain reaction, PPDPF = pancreatic progenitor cell differentiation and proliferation factor.
We further assessed the mRNA expression of PPDPF on 42 pair human HCC specimens and their matched normal tissues by real-time RT-PCR test. As shown in Fig. 1B, upregulation of the PPDPF gene occurred in 31 of 42 (73.8%) HCC compared with the paired normal liver tissues. Elevated levels of PPDPF protein were also found in 4 randomly picked pair HCC samples with different expression levels of PPDPF mRNA, as shown by western blot analysis (Fig. 1C).
In addition, in order to further confirm the result demonstrated above, we employed HCC tissues slides with follow-up data from 135 HCC specimens and 91 paired adjacent non-cancerous tissues. The IHC staining demonstrated that PPDPF expression was dramatically enhanced in 54.07% of the HCC samples (73/135) when compared with the adjacent non-tumorous specimens (22/91, 24.17%) (Table 1) (Fig. 1D).
Table 1.
High expression of PPDPF in HCC.
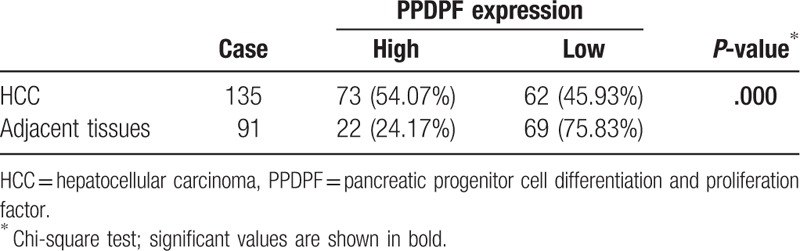
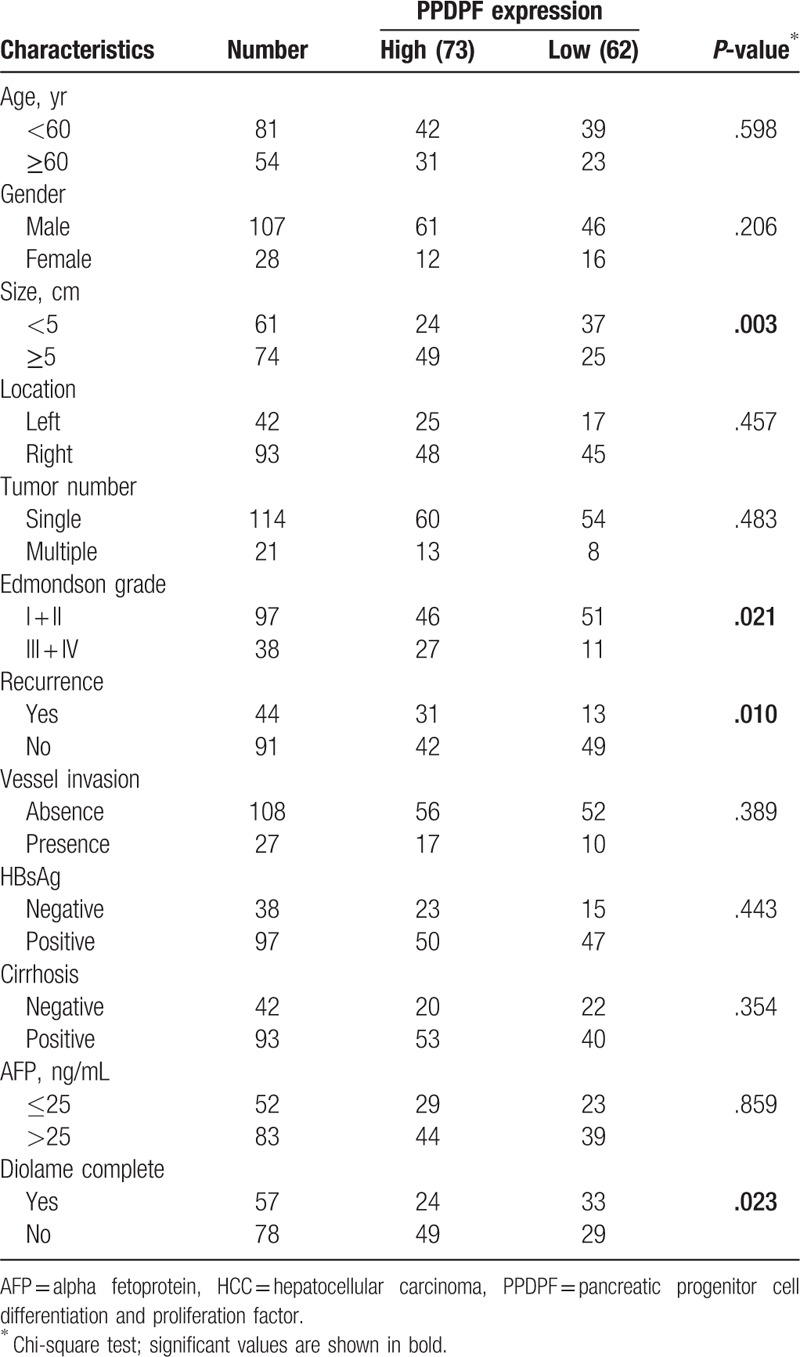
Furthermore, analysis revealed that PPDPF overexpression was significantly correlated with tumors size (P = .003), Edmondson-Steiner Grading (P = .021), recurrence (P = .010), and Diolame complete (P = .023). However, no statistical connections were found between PPDPF expression and other clinicopathological parameters, such as age, sex, HBsAg level, α-fetoprotein (AFP) level, tumor number, cirrhosis, or vascular invasion (Table 2).
Table 2.
Correlation between PPDPF expression and clinicopathological features of HCC patients.
3.2. Prognostic value of PPDPF expression in HCC
Notably, HCC patients with PPDPF-positive tumor exhibited shorter overall survival than did patients with PPDPF negative tumors (P = .043). HCC patients with higher PPDPF expression exhibited shorter median OS time (19 months) compared with patients who had lower levels of PPDPF expression (31.0 months) (Fig. 2). In addition, HCC patients with higher PPDPF expression levels had shorter overall survival than did those with lower PPDPF expression levels (1-, 3-, and 5-year OS: 54.1%, 33.2%, and 19.7% vs 72.1%, 50.6%, and 35.6%, respectively).
Figure 2.
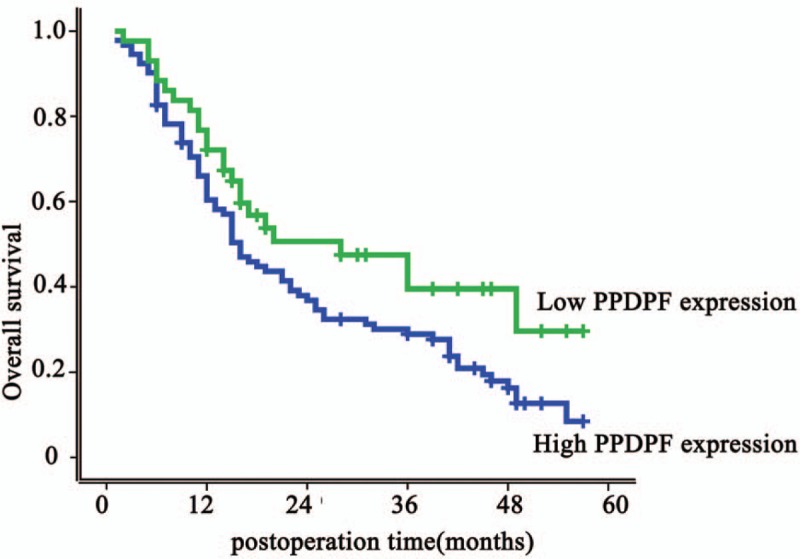
Kaplan–Meier overall survival curves of patients with HCC according to pancreatic progenitor cell differentiation and proliferation factor (PPDPF) expression. Patients with high expression of PPDPF had a significantly poor survival rate than patients with low PPDPF expression (P = .043). HCC = hepatocellular carcinoma.
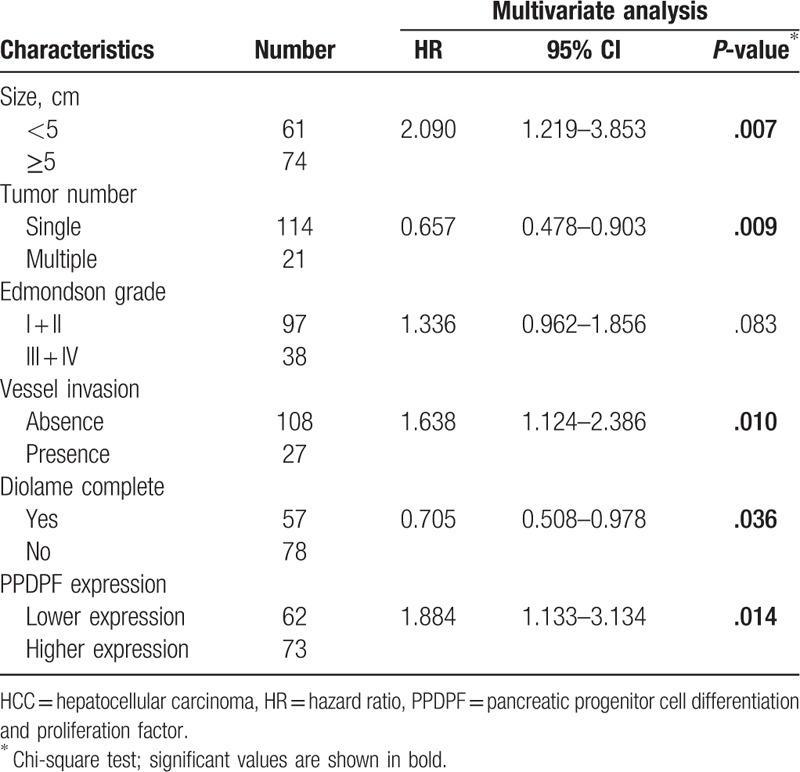
In addition to PPDPF expression, univariate analysis demonstrated that tumor size, tumor number, Edmondson grade, vessel invasion, diolame complete were also significantly associated with OS, while others were not (Table 3). Moreover, multivariate analysis revealed that higher expression of PPDPF, tumor size, tumor number, vessel invasion, diolame complete were independent risk factors associated with decreased survival (Table 4). Collectively, the clinical data indicated that PPDPF is correlated with poor prognosis in HCC patients.
Table 3.
Univariate analysis of risk factors associated with overall survival of HCC patients.
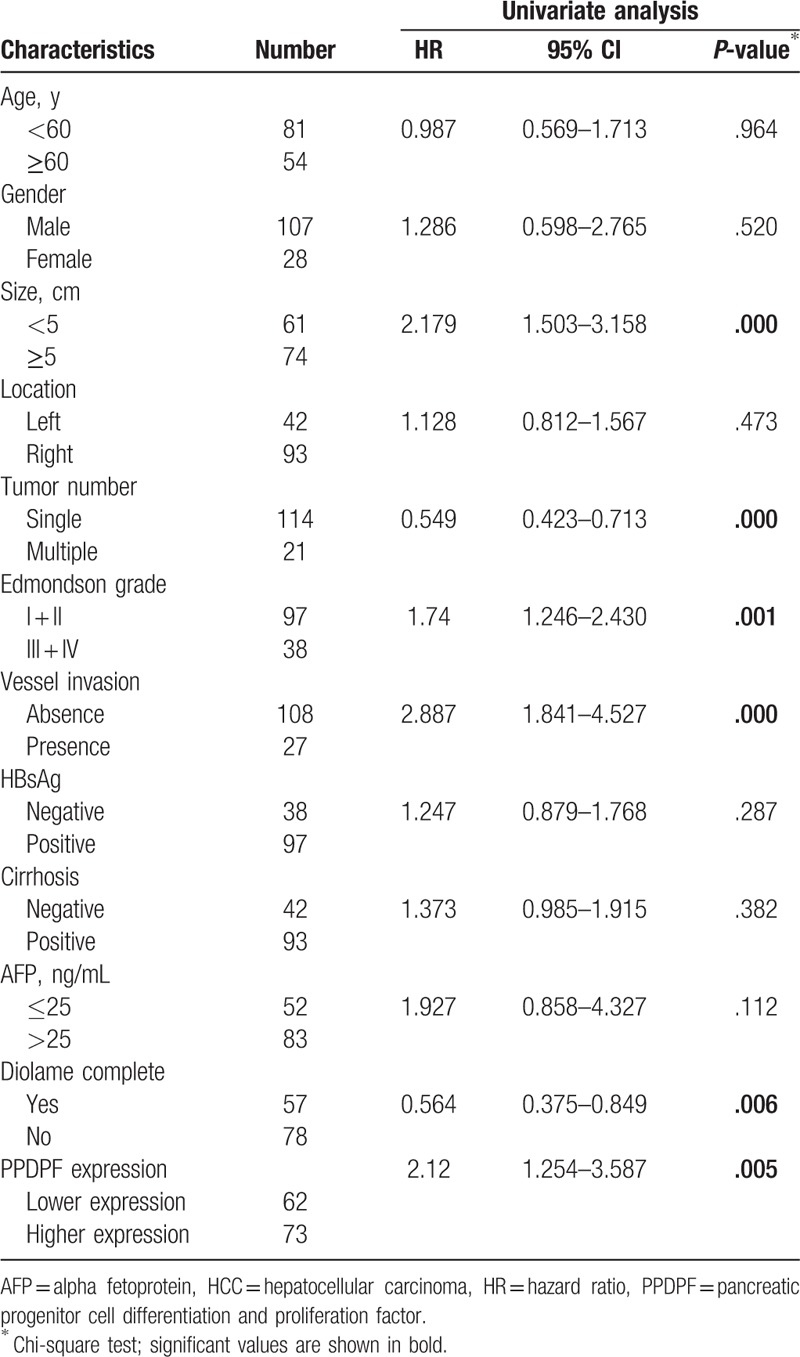
Table 4.
Multivariate analysis of risk factors associated with overall survival of HCC patients.
4. Discussion
To the best of our knowledge, this study represents the first observation of expression of PPDPF and its prognostic role in patients with liver cancer. NCBI database had reported that PPDPF was highly expressed in a variety of human cancers, including liver, colon, and prostate. Coinciding with this result, we demonstrated that both PPDPF mRNA and protein expression were dramatically increased in HCC tissues. Furthermore we obtained increased expression of PPDPF in HCC tissues by IHC, compared with normal adjacent tissues (54.07% vs 24.17%).
Besides, the increased PPDPF expression was correlated with large tumor size, Edmondson-Steiner Grading, Diolame complete and recurrence, poor prognosis, and overall survival rates. Furthermore, we demonstrated that patients whose liver cancer cells were positive for PPDPF had a mean survival of 19.17 ± 1.92 months compared with 31.25 ± 3.4 months for those whose cells not expressed PPDPF. The results imply that higher PPDPF expression in liver cancer may be considered an indicator of poor prognosis in patients who underwent radical resection with curative surgery.
In our present study, the number of patients recruited was inadequate and there is a possibility of bias. Due to the insidious nature of HCC, most patients diagnosed first time are in advanced tumor stage; patients with early stage are very rare. This may have had an underlying bias on the present study. In future studies, it is necessary to include more patients with early-stage. The majority of our HCC cases accompany with HBV infection. If possible, we would include more patients with HCC without HBV and examine the PPDPF expression pattern. However, in the multivariable Cox proportional hazards models, the analyses strongly demonstrated that PPDPF expression may be a useful biomarker of predicting outcome of patients with HCC.
The mechanism by which PPDPF expression portends a worse prognosis is not fully known. As previous describe, the PPDPF consist of SH2, SH3 domains, and CLK2 kinase binding site. The former SH domains are considered associating with proteins of signaling pathways involving in the cytoskeleton, the Ras associated protein, and many others.[11] The CLK2 kinase has shown to interact with serine/arginine-rich (SR) proteins of the spliceosomal complex, which is responsible for the SR proteins to control RNA splicing.[12] This protein kinase is involved in the regulation of several cellular processes and may serve as a link between cell cycle progression, apoptosis, and telomere length regulation.[13] In the development of exocrine of pancreas, PPDPF could inhibit the cell cycle arrest through mediating cell cycle inhibitor genes p21Cip, p27Kip, and cyclin G1, it is likely that the high expression of PPDPF might associated with dysfunction of the cell cycle.[6] Given the pivotal roles of PPDPF in the regulation of cell division, abnormal expression of PPDPF may perturb its normal biological functions, which might ultimately induce cancer progression. According to previous study and our results, we can hypothesize that the higher expression of PPDPF might promote tumorigenesis and progression in HCC. Further studies are required to prove our hypothesis.
In conclusion, our observations have provided evidence for the roles of PPDPF in HCC; high expression of PPDPF is related to unfavorable prognosis in HCC. PPDPF may be considered as a promising therapeutic target in control HCC.
Author contributions
Data curation: Xi Li.
Formal analysis: Xiaoyan Ma.
Methodology: Jiangxin Zhang.
Supervision: Xuqing Wang.
Writing – original draft: Zhengfa Mao.
Writing – review & editing: Xin Fan.
Footnotes
Abbreviations: HBV = hepatitis B virus, HCC = hepatocellular carcinoma, HCV = hepatitis C virus, IHC = immunohistochemistry, PPDPF = pancreatic progenitor cell differentiation and proliferation factor, RT-PCR = real time-polymerase chain reaction.
This work was sponsored by National Natural Science foundation of China, Grant numbers: 81502372; Natural Science Foundation of Jiangsu Province (Youth Fund), Grant numbers: BK20130475; Jiangsu Provincial Medical Youth Talent (QNRC2016838, QNRC2016839); Zhenjiang Science and Technology Pillar Program, Grant numbers: SH2014035; The Foundation for Young Scientists of affiliated Hospital of Jiangsu University (Grant numbers: JDFYRC2013009, JDFYRC2016002).
The authors report no conflicts of interest.
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- [2].GBD 2016 Disease and Injury Incidence and Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017;390:1211–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115–32. [DOI] [PubMed] [Google Scholar]
- [4].Cho YY, Lee JH, Chang Y, et al. Comparison of overall survival between antiviral-induced viral suppression and inactive phase chronic hepatitis B patients. J Viral Hepat 2018;25:1161–71. [DOI] [PubMed] [Google Scholar]
- [5].Matsushima H, Takami Y, Ryu T, et al. Prognosis of hepatocellular carcinoma patients who achieved long-term recurrence-free survival after curative therapy: impact of the Albi Grade. J Gastrointest Surg 2018;22:1230–8. [DOI] [PubMed] [Google Scholar]
- [6].Jiang Z, Song J, Qi F, et al. Exdpf is a key regulator of exocrine pancreas development controlled by retinoic acid and ptf1a in zebrafish. PLoS Biol 2008;6:e293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Marquardt JU. The role of transforming growth factor-beta in human hepatocarcinogenesis: mechanistic and therapeutic implications from an integrative multiomics approach. Gastroenterology 2018;154:17–20. [DOI] [PubMed] [Google Scholar]
- [8].Cesi G, Walbrecq G, Margue C, et al. Transferring intercellular signals and traits between cancer cells: extracellular vesicles as “homing pigeons”. Cell Commun Signal 2016;14:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sun ZJ, Wang Y, Cai Z, et al. Involvement of Cyr61 in growth, migration, and metastasis of prostate cancer cells. Br J Cancer 2008;99:1656–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Deng YZ, Yao F, Li JJ, et al. RACK1 suppresses gastric tumorigenesis by stabilizing the beta-catenin destruction complex. Gastroenterology 2012;142:812.e15–23.e15. [DOI] [PubMed] [Google Scholar]
- [11].Knight JM, Mandal P, Morlacchi P, et al. Small molecule targeting of the STAT5/6 Src homology 2 (SH2) domains to inhibit allergic airway disease. J Biol Chem 2018;293:10026–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Petsalaki E, Zachos G. Clks 1, 2 and 4 prevent chromatin breakage by regulating the Aurora B-dependent abscission checkpoint. Nat Commun 2016;7:11451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhao S, Chen D, Geng Q, et al. The highly conserved LAMMER/CLK2 protein kinases prevent germ cell overproliferation in Drosophila. Dev Biol 2013;376:163–70. [DOI] [PubMed] [Google Scholar]


