Abstract
HNH is one of two endonuclease domains of the clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein Cas9 that perform site-specific cleavage of double-stranded DNA. We engineered a novel construct of this critical nuclease from Streptococcus pyogenes Cas9 that not only maintains the wild-type amino acid sequence and fold, but displays enhanced thermostability when compared to the full-length Cas9 enzyme. Here, we report backbone and side chain assignments of the HNH nuclease as a foundational step toward the characterization of protein dynamics and allostery in CRISPR-Cas9.
Keywords: CRISPR, Cas9, HNH nuclease, NMR assignments
Biological context
The CRISPR-associated protein 9 (Cas9) system provides an exciting tool for precision editing (Charpentier and Doudna 2013; Charpentier and Marraffini 2014; Wang et al. 2016) and regulation (Dominguez et al. 2016) of genomic DNA with possible applications to the treatment of heritable human diseases (Maeder and Gersbach 2016; Strong and Musunuru 2017; Xiong et al. 2016) and cancer (Martinez-Lage et al. 2018; Tian et al. 2019). The Cas9 endonuclease system is comprised of a large (1368 residue) multidomain and multifunctional polypeptide chain and either two RNA chains (Jiang and Doudna 2017), tracrRNA and crRNA (Deltcheva et al. 2011; Jinek et al. 2012), or a combined RNA oligonucleotide, referred to as single-guide RNA. The Cas9 polypeptide chain houses two endonuclease domains, HNH and RuvC; a large alpha helical recognition domain (REC), and a Protospacer Adjacent Motif (PAM) interacting (PI) domain (Jiang and Doudna 2017). The power of Cas9 lies in the elegant simplicity of its targeting mechanism. Cas9 binds to a target 20-nucleotide DNA sequence by creating a DNA–RNA hybrid with the crRNA. However, Cas endonucleases are inactive without the recognition of a unique PAM sequence directly downstream of the DNA–RNA duplex (Jiang and Doudna 2017). The allosteric network that transmits the signal of PAM recognition to the endonucleases is not well understood, but to better control the Cas9 machinery for use in the treatment of human disease, knowledge of this allosteric network is vital. Molecular dynamics simulations have suggested the presence of an allosteric pathway connecting the PI domain to the alpha helical REC lobe and HNH domain (Oakes et al. 2016; Palermo et al. 2017). Signaling between the HNH and RuvC nucleases is required for concerted cleavage of double-stranded DNA, but the protein motions responsible for propagating this information have not been identified. NMR is a valuable tool in this endeavor, and the utility of NMR in studies of dynamic allostery is well documented (Lisi and Loria 2016a, b).
At over 160 kDa, the full-length Cas9 protein presents an extremely difficult target to study by solution NMR, especially considering its low thermostability that precludes the use of elevated temperatures to enhance spectral quality of large proteins (Arbogast et al. 2015). Thus, we have engineered a construct of the critical HNH nuclease (15.4 kDa), and report NMR resonance assignments for its amide backbone (HN, NH, Cα, Cβ, CO) and aliphatic side chains. These assignments, along with those of the Met-Ile-Leu-Val-Ala-Thr methyl groups, provide a basis for the study of the structure and dynamic network within HNH, which will be expanded on a “per domain” basis to map the entire Cas9 allosteric pathway by NMR.
Methods and experiments
Sample preparation
A codon-optimized sequence for the HNH domain (residues 775–909) from S. pyogenes Cas9 was inserted into the pET28a vector with an N-terminal His6-tag and a TEV protease cleavage site. The plasmid was transformed into BL21(DE3) cells (Novagen). Labeled samples were grown in M9 minimal media supplemented with 15N ammonium chloride (1.0 g/L) and 13C glucose (2.0 g/L; Cambridge Isotope Labs). Cells were grown at 37 °C until reaching an OD600 of 0.8–0.9 and were subsequently induced with 0.5 mM IPTG. HNH was expressed for 18 h at 20 °C. Cells were collected by centrifugation at 5500 rpm for 25 min and resuspended in a buffer of 20 mM HEPES, 300 mM KCl and 5 mM imidazole at pH 8.0. The resuspended cells were lysed by sonication and the lysate was centrifuged at 15,000 rpm for 45 min to remove cellular debris. The His6-tagged HNH domain was purified from the supernatant by Ni–NTA affinity chromatography and eluted with the same buffer containing 220 mM imidazole. The eluent was dialyzed against a buffer of 20 mM HEPES and 80 mM KCl at pH 7.4. The N-terminal His6-tag was cleaved following incubation of HNH with TEV protease at room temperature for 4 h. The tag and TEV protease were removed by Ni–NTA chromatography and HNH was dialyzed against a buffer of 20 mM HEPES, 80 mM KCl, 1 mM EDTA, 1 mM DTT, and 7% 2H2O at pH 7.4. Even with no changes to its amino acid sequence, the HNH domain has a thermal unfolding temperature that is nearly 20 °C higher than that of full-length Cas9, and is stable at concentrations of 1.2–1.7 mM for NMR experiments.
NMR experiments
NMR data for backbone assignment were collected on a Varian Inova 600 MHz spectrometer at 25 °C equipped with pulsed field gradients and a triple resonance probe. The backbone assignments were completed using the following TROSY triple resonance experiments (Pervushin et al. 1997): 1H–15N HSQC, HNCA, HN(CO)CA, HN(CA)CB, HN(COCA)CB, HN(CA)CO, and HNCO (Salzmann et al. 1998). NMR data for side chain assignment were collected on a Bruker Avance III 600 MHz spectrometer at 25 °C equipped with pulsed field gradients and a triple resonance cryo-probe. Side chain assignments were completed using the following experiments: 1H–13C ctHSQC, 15N-edited TOCSY (Marion et al. 1989), (H)CCH-TOCSY, HC(C) H-TOCSY (Bax et al. 1990), and HA(CACO)NH (Feng et al. 1996). All spectra were processed in NMRPipe (Delaglio et al. 1995) and analyzed in Sparky (Lee et al. 2015). Three-dimensional correlations and assignments were made in CARA (Keller 2005).
Assignment and data deposition
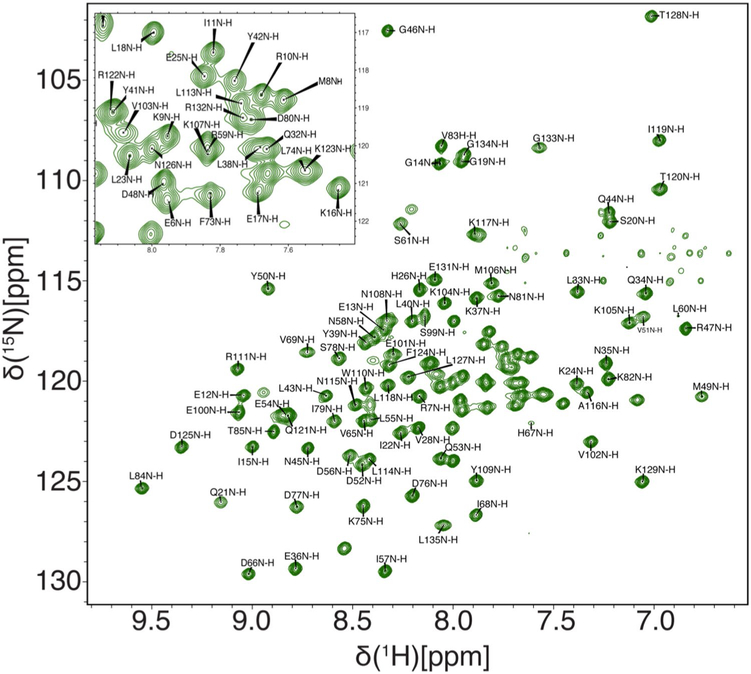
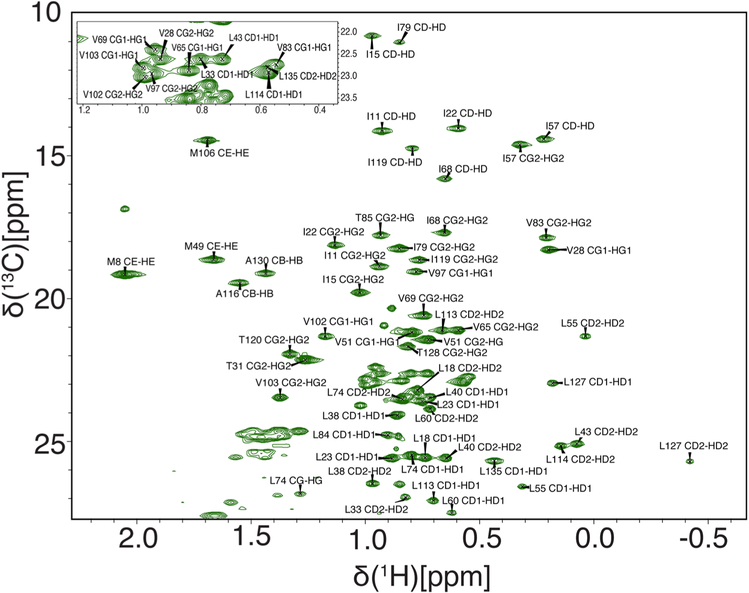
The assigned 1H–15N TROSY HSQC spectrum for HNH is shown in Fig. 1. 80.7% of the backbone 1H–15N resonances have been assigned including 80.7% of Cα, 77.0% of Cβ, and 77.0% of CO. Since future work is geared toward the creation of larger Cas9 constructs, we also assigned the aliphatic side chains and Met-Ile-Leu-Val-Ala-Thr methyl resonances, which are useful probes in NMR spectra of proteins > 40 kDa (Ollerenshaw et al. 2003). The assigned methyl region of the 1H–13C ctHSQC spectrum for HNH is shown in Fig. 2. 84.8% of the aliphatic sidechain 1H–13C resonances have been assigned including 96.8% of Met-Ile-Leu-Val-Ala-Thr methyl resonances. A list of the 1H, 13C, and 15N chemical shifts has been deposited into the BioMagResBank under accession number 27949.
Fig. 1.
1H–15N TROSY HSQC spectrum of the HNH domain from S. pyogenes Cas9 collected at 25 °C and 600 MHz. The inset shows assigned residues within the crowded center of the spectrum
Fig. 2.
1H–13C ctHSQC spectrum of HNH collected at 25 °C and 600 MHz. The spectrum depicts the methyl resonances of Met-Ile-Leu-Val-Ala-Thr residues, with the crowded central region of the spectrum shown as an inset
Acknowledgements
This work was supported by start-up funds from Brown University and funds from the COBRE Center of Computational Biology of Human Disease (P20GM109035) to GPL. HBB is supported by a NIH/NIGMS training grant in Pharmacological Sciences (2T32GM077995).
Footnotes
Conflict of interest The authors declare no conflict of interest.
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Arbogast LW, Brinson RG, Marino JP (2015) Mapping monoclonal antibody structure by 2D 13C NMR at natural abundance. Anal Chem 87:3556–3561 [DOI] [PubMed] [Google Scholar]
- Bax A, Clore GM, Gronenborn AM (1990) H-1-H-1 correlation via isotropic mixing of C-13 magnetization, a new 3-dimensional approach for assigning H-1 and C-13 spectra of C-13-enriched proteins. J Magn Reson 88:425–431 [Google Scholar]
- Charpentier E, Doudna JA (2013) Biotechnology: rewriting a genome. Nature 495:50–51 [DOI] [PubMed] [Google Scholar]
- Charpentier E, Marraffini LA (2014) Harnessing CRISPR-Cas9 immunity for genetic engineering. Curr Opin Microbiol 19:114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293 [DOI] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E (2011) CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471:602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez AA, Lim WA, Qi LS (2016) Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol 17:5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Rios CB, Montelione GT (1996) Phase labeling of C-H and C-C spin-system topologies: application in PFG-HACANH and PFG-HACA(CO)NH triple-resonance experiments for determining backbone resonance assignments in proteins. J Biomol NMR 8:98–104 [DOI] [PubMed] [Google Scholar]
- Jiang F, Doudna JA (2017) CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys 46:505–529 [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R (2005) Optimizing the process of nuclear magnetic resonance spectrum analysis and computer aided resonance assignment. ETH, Zurich [Google Scholar]
- Lee W, Tonelli M, Markley JL (2015) NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31:1325–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisi GP, Loria JP (2016a) Solution NMR spectroscopy for the study of enzyme allostery. Chem Rev 116:6323–6369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisi GP, Loria JP (2016b) Using NMR spectroscopy to elucidate the role of molecular motions in enzyme function. Prog Nucl Magn Reson Spectrosc 92–93:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Gersbach CA (2016) Genome-editing technologies for gene and cell therapy. Mol Ther 24:430–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion D, Driscoll PC, Kay LE, Wingfield PT, Bax A, Gronenborn AM, Clore GM (1989) Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H-15N Hartmann-Hahn-multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: application to interleukin 1 beta. Biochemistry 28:6150–6156 [DOI] [PubMed] [Google Scholar]
- Martinez-Lage M, Puig-Serra P, Menendez P, Torres-Ruiz R, Rodriguez-Perales S (2018) CRISPR/Cas9 for cancer therapy: hopes and challenges. Biomedicines 6:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes BL, Nadler DC, Flamholz A, Fellmann C, Staahl BT, Doudna JA, Savage DF (2016) Profiling of engineering hotspots identifies an allosteric CRISPR-Cas9 switch. Nat Biotechnol 34:646–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollerenshaw JE, Tugarinov V, Kay LE (2003) Methyl TROSY: explanation and experimental verification. Magn Reson Chem 41:843–852 [Google Scholar]
- Palermo G, Ricci CG, Fernando A, Basak R, Jinek M, Rivalta I, Batista VS, McCammon JA (2017) Protospacer adjacent motif-induced allostery activates CRISPR-Cas9. J Am Chem Soc 139:16028–16031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervushin K, Riek R, Wider G, Wuthrich K (1997) Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA 94:12366–12371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K (1998) TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci USA 95:13585–13590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong A, Musunuru K (2017) Genome editing in cardiovascular diseases. Nat Rev Cardiol 14:11–20 [DOI] [PubMed] [Google Scholar]
- Tian X, Gu T, Patel S, Bode AM, Lee MH, Dong Z (2019) CRISPR/Cas9—an evolving biological tool kit for cancer biology and oncology. NPJ Precis Oncol 3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, La Russa M, Qi LS (2016) CRISPR/Cas9 in genome editing and beyond. Annu Rev Biochem 85:227–264 [DOI] [PubMed] [Google Scholar]
- Xiong X, Chen M, Lim WA, Zhao D, Qi LS (2016) CRISPR/Cas9 for human genome engineering and disease research. Annu Rev Genomics Hum Genet 17:131–154 [DOI] [PMC free article] [PubMed] [Google Scholar]