Abstract
This study aimed to investigate the correlation of long noncoding RNA (lncRNA) ZNFX1 antisense RNA (ZFAS1) with disease risk, severity, inflammation markers, and prognosis in sepsis patients.
A total of 202 sepsis patients were consecutively enrolled, and 200 healthy volunteers were also recruited as healthy controls (HCs). Plasma samples of all patients and HCs were collected. LncRNA ZFAS1 expression was determined by quantitative polymerase chain reaction assay and inflammatory cytokines levels were detected by enzyme-linked immunosorbent assay.
The median value of lncRNA ZFAS1 expression in sepsis patients was (0.639 [0.325–1.071]), which was lower compared to HCs (1.957 [0.876–3.245], P < .001], and receiver operating characteristics (ROC) curve revealed that lncRNA ZFAS1 expression had a good diagnostic value for sepsis with area under curve (AUC) of 0.814 (95% confidence interval [CI]: 0.771–0.857). Spearman test disclosed that lncRNA ZFAS1 expression was negatively correlated with Acute Physiology and Chronic Health Evaluation (APACHE) II score (r = −0.505, P < .001), and it was negatively associated with levels of C-creative protein (r = −0.241, P = .001), tumor necrosis factor-α (r = −0.253, P < .001), and interleukin (IL)-6 (r = −0.177, P = .012) while positively associated with IL-10 level (r = 0.173, P = .014). Also, lncRNA ZFAS1 expression was lower in survivor group compared to nonsurvivor group (P < .001), and it presented with a good predictive value on distinguishing nonsurvivors from survivors in sepsis patients with AUC of 0.628 (95% CI: 0.538–0.717).
Circulating lncRNA ZFAS1 expression negatively correlates with disease risk, severity, and inflammatory markers levels, and might predict worse survival in sepsis patients.
Keywords: inflammation markers, long noncoding RNA ZNFX1 antisense RNA, prognosis, risk, sepsis
1. Introduction
Sepsis is potentially morbid organ dysfunction caused by dysregulated host response to infection, and it is characterized by early activation of inflammatory responses, immune suppression as well as coagulation.[1] According to global statistics, more than 30 million people suffer from sepsis each year, and the incidence rates of hospital-treated sepsis and severe sepsis are 437 and 270 per 100,000 person-year, respectively.[2] Although the applications of antibiotics, vasoactive agents, and renal dialysis are effective and time sensitive in the management of sepsis, the mortality rate of sepsis still ranges from 29% to 70%, leading to millions of deaths worldwide every year.[3,4] Therefore, exploring novel and accurate biomarkers is necessary to increase the sensitivity of diagnosis, monitor disease progression, and improve prognosis in sepsis patients.
Long noncoding RNAs (lncRNAs) are defined as RNA transcripts whose length is longer than 200 nucleotides and do not code proteins.[5] Accumulating reports have disclosed that lncRNAs are involved in inflammatory processes in several diseases such as carcinomas, neurogenically diseases, and immune diseases via involving in gene transcription regulation, epigenetic regulation, and translation.[6–8] LncRNA ZFAS1, located on chromosome 20, has been observed to participate in the development and progression of diseases with inflammatory disorder including rheumatoid arthritis (RA) and acute myocardial infarction (AMI), while the role of lncRNA ZFAS1 remains unclear in sepsis, a typical disease with inflammatory disorder.[8–10] Therefore, this study aimed to investigate the correlation of lncRNA ZFAS1 with disease risk, severity, inflammation markers, and prognosis in sepsis patients.
2. Materials and methods
2.1. Participants
A total of 202 sepsis patients admitted to Huangshi Central Hospital, Affiliated Hospital of Hubei Polytechnic University from January 2015 to December 2017 were consecutively enrolled in this study. The inclusion criteria were: Diagnosed as sepsis according to the American College of Chest Physicians/Society of Critical Care Medicine consensus definition: confirmed infection + systemic inflammatory response syndrome (SIRS). SIRS was defined by 2 or more of the following conditions: temperature >38°C or <36°C; heart rate >90 beats per minute; respiratory rate >20 breaths per minute or PaCO2 < 32 mm Hg; white blood cell count >12,000 cu mm, <4000 cu mm, or >10% immature forms[11]; Age above 18 years. Exclusion criteria were: History of solid tumors, hematologic malignancies; History of severe chronic diseases of the heart, liver, kidney, or lung; History of severe systematic immune diseases; Received immunosuppressive drugs within 3 months before admission. In the meanwhile, 200 healthy volunteers were recruited in this study from Huangshi Central Hospital, Affiliated Hospital of Hubei Polytechnic University as well and severed as healthy controls (HCs) whose age and gender was matched to sepsis patients.
2.2. Ethics statement and approval
The Ethics Committee of Huangshi Central Hospital, Affiliated Hospital of Hubei Polytechnic University approved the protocol of this study. This study was conducted under the Statement of Helsinki. All the participants or their family members signed the informed consents.
2.3. Plasma sample collection
Blood sample was obtained from sepsis patients before any treatment was performed, and from HCs after their enrollment. After collection of blood sample, plasma was isolated and stored at −80°C for further detection.
2.4. LncRNA ZFAS1 expression determination
Quantitative polymerase chain reaction assay was performed to determine lncRNA ZFAS1 expression in plasma from both sepsis patients and HCs. In brief, total RNA was extracted using TRIzol reagent (Invitrogen, Waltham, Massachusetts), and was transcribed reversely by PrimerScript real-time reagent kit (TaKaRa, kusatsu, Japan), then SYBR Premix Ex TaqTM II (TaKaRa) was applied for the quantitation of lncRNA ZFAS1 expression. Quibit (Invitrogen) was used to examine the quality of plasma RNA after extraction. GAPDH was used as internal references, and it also served as internal control for plasma RNA quality. LncRNA ZFAS1 expression was calculated by 2-ΔΔct method (2{[Mean ct of lncZFAS1–mean ct of GAPDH]–[mean ct of control–mean ct of GAPDH]}).[12] The calculated result was the relative quantitative expression value of lncRNA ZFAS1 compared with the internal reference. The primer of lncRNA ZFAS1 was: 5′–CTATTGTCCTGCCCGTTAGAG-3,′ reverse 5′-GTCAGGAGATCGAAGGTTGTAG-3′. The primer of GAPDH was: GAGTCCACTGGCGTCTTCAC; reverse: ATCTTGAGGCTGTTGTCATACTTCT.
2.5. Inflammatory cytokine level measurement
Enzyme-linked immunosorbent assay (ELISA) was conducted to determine the expressions of tumor necrosis factor-α (TNF-α), interleukin (IL)-1β, IL-6, IL-10, and IL-17 in plasma from sepsis patients using commercial ELISA kits (R&D, Minneapolis, Minnesota). The detailed procedure of ELISA was: prepare plasma samples by 2-fold dilution into calibrator diluent RD5T; equilibrate reagents to room temperature and prepare samples, reagents, standards, and controls following instruction of the kit; remove excess microplate strips from the plate frame; add 50 μL of assay diluent RD1W and 50 μL of sample into each well; aspirate each well and wash, repeat for 4× and invert the plate then blot it against clean paper towels; add 100 μL of mouse IL-10 conjugate into each well then cover with new strip and incubate for 2 hours on shaker under room temperature; repeat step 5 then add 100 μL of substrate solution into each well and incubate for 30 minutes on benchtop under room temperature; add 100 μL of stop solution into each well and gently mix; determine the optical density of each well within 30 minutes and calculate the results according to the standard curve; Standard substances of TNF-α, IL-1β, IL-6, IL-10, and IL-17 (R&D) was used as positive control, which was the quality control of our measurements.
2.6. Data collection
Information on age, gender, and body mass index (BMI) of sepsis patients and HCs were recorded. In addition, laboratory indexes including serum creatinine (Scr) level, albumin level, white blood cell (WBC) count, and C-creative protein (CRP) level were collected in sepsis patients, and Acute Physiology and Chronic Health Evaluation (APACHE) II scores were assessed.
2.7. Statistics
Statistical analysis was performed using SPSS 21.0 software (SPSS Inc, NewYork City, State of New York) and GraphPad Prism 5.0.1 (GraphPad Software, NewYork City, State of New York). Data were mainly displayed as mean ± standard deviation, median (quartile 25th–75th) or count (percentage). Wilcoxon rank sum test was used for comparison of skew distributed continuous data including comparison of lncRNA ZFAS1 expression between HCs and sepsis patients, as well as comparison of lncRNA ZFAS1 expression between survivors and nonsurvivors. As for comparison of normally distributed continuous data including age and BMI between sepsis patients and HCs, t test was used. Chi-squared test was used to determine the difference in ratio of gender between sepsis patients and HCs. Correlation of lncRNA ZFAS1 with APACHE II score and inflammatory markers was analyzed by Spearman test. In addition, ROC curve was performed to assess the value of lncRNA ZFAS1 in distinguishing sepsis patients from HCs, and in predicting survival in sepsis patients. P < .05 was considered significant.
3. Results
3.1. Baseline characteristics
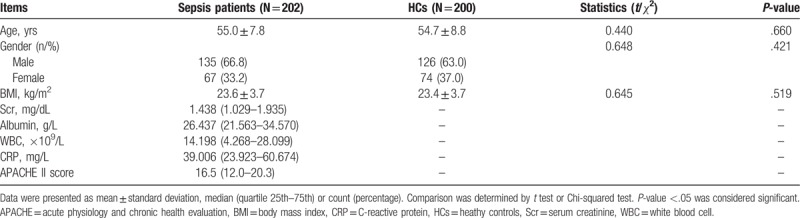
There was no difference in age (P = .365), gender (P = .421), and BMI (P = .519) between sepsis group and HCs group (Table 1). The mean ages in sepsis group and HC group were 55.0 ± 7.8 years and 54.7 ± 8.8 years, respectively. There were 135 males and 67 females in sepsis group, 126 males and 74 females in HC group. The mean values of BMI for sepsis group and HC group were 23.6 ± 3.7 (kg/m2) and 23.4 ± 3.7 (kg/m2), respectively. In sepsis group, the median concentrations of Scr, albumin, WBC, and CRP were 1.438 (1.029–1.935) (mg/dL), 26.437 (21.563–34.570) (g/L), 14.198 (4.268–28.099) (×109/L), and 39.006 (23.923–60.674) (mg/L) respectively, and the median APACHE II score was 16.5 (12.0–20.3).
Table 1.
Baseline characteristics of participants.
3.2. Blood culture information of sepsis patients
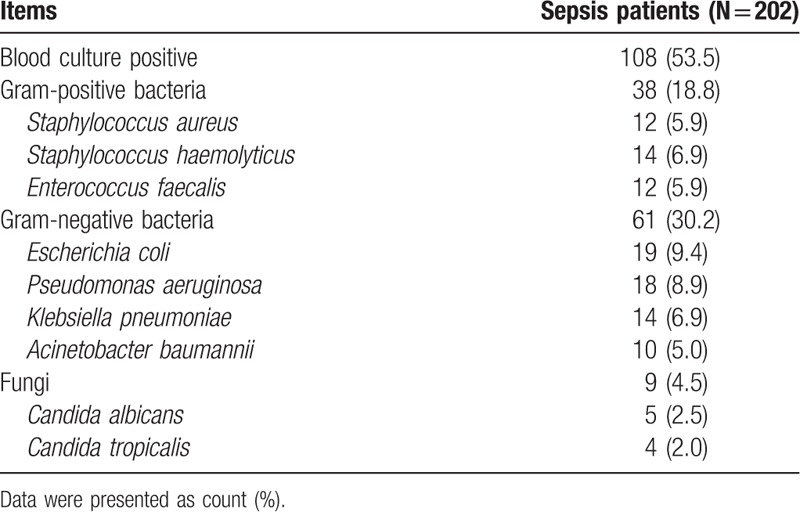
There were 108 patients (53.5%) with blood culture positive, and the numbers of patients with confirmed gram-positive bacteria, gram-negative bacteria, and fungi infection were 38 (18.8%), 61 (30.2%), and 9 (4.5%), respectively (Table 2). The detailed blood culture information is shown in Table 2.
Table 2.
Blood culture status of sepsis patients.
3.3. LncRNA ZFAS1 expression in sepsis patients and HCs
The median value of lncRNA ZFAS1 in sepsis patients was 0.639 (0.325–1.071), which was lower than that in HCs group (1.957 [0.876–3.245], P < .001) (Fig. 1A). Furthermore, ROC curve revealed that lncRNA ZFAS1 had a good diagnostic value for sepsis with area under curve (AUC) of 0.814 (95% confidence interval [CI]: 0.771–0.857). Sensitivity and specificity values were 92.1% and 63.5%, respectively, at the best cut-off point where the largest sum of sensitivity and specificity occurred (Fig. 1B).
Figure 1.
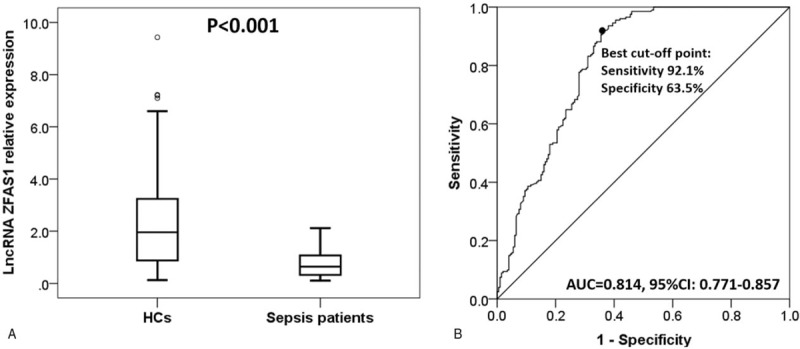
Long noncoding RNA ZNFX1 antisense RNA (lncRNA ZFAS1) expression in sepsis patients and healthy controls (HCs). LncRNA ZFAS1 expression was notably lower in sepsis patients compared to HCs (A). LncRNA ZFAS1 exhibited a good diagnostic value for sepsis (B). The difference of lncRNA ZFAS1 expression between 2 groups was determined by Wilcoxon rank sum test. Receiver operating characteristic curve was used to assess the diagnostic value of lncRNA ZFAS1 for sepsis. P < .05 was considered significant.
3.4. Correlation of lncRNA ZFAS1 expression with bacteria and fungi types
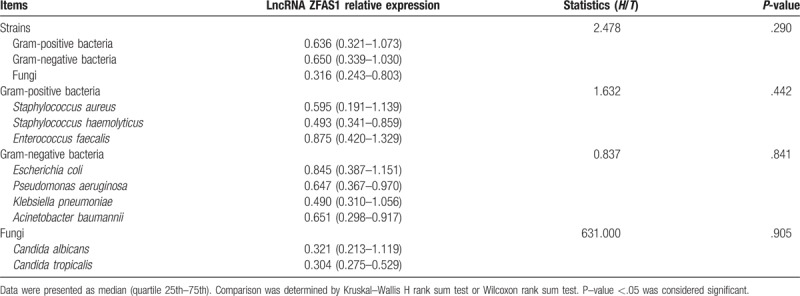
Comparison of lncRNA ZFAS1 expression among patients with different bacteria and fungi types was determined by Kruskal–Wallis H rank sum test or Wilcoxon rank sum test, which disclosed that there was no correlation of lncRNA ZFAS1 with bacteria or fungi types (all P > .05) (Table 3).
Table 3.
LncRNA ZFAS1 relative expression among sepsis patients with different pathogenic bacteria.
3.5. Correlation between lncRNA ZFAS1 expression and APACHE II score in sepsis patients
Spearman test was performed to evaluate the correlation between lncRNA ZFAS1 expression and APACHE II score, which disclosed that lncRNA ZFAS1 expression was negatively correlated with APACHE II score in sepsis patients (r = −0.505, P < .001) (Fig. 2).
Figure 2.
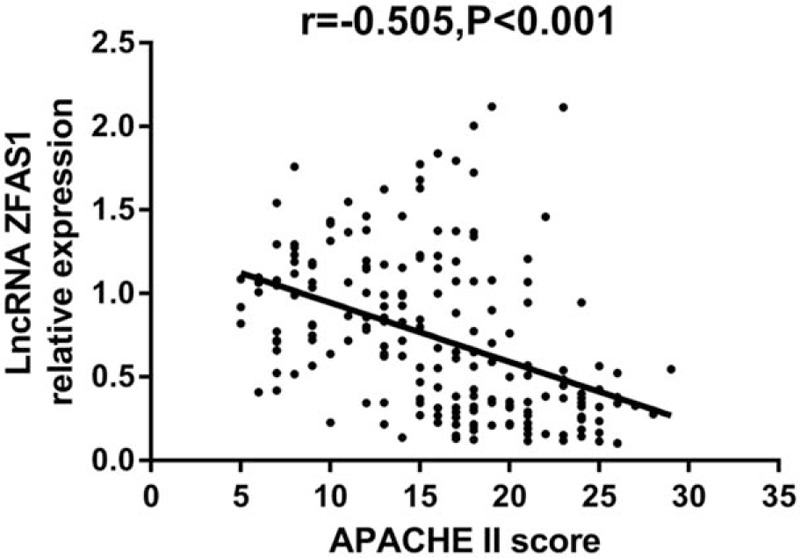
Correlation between long noncoding RNA ZNFX1 antisense RNA (lncRNA ZFAS1) expression and acute physiology and chronic health evaluation (APACHE) II score. LncRNA ZFAS1 expression was negatively correlated with disease severity. Correlation between expression of lncRNA ZFAS1 and APACHE II score was determined by Spearman test and P < .05 was considered significant.
3.6. Correlation of lncRNA ZFAS1 expression with inflammation markers in sepsis patients
LncRNA ZFAS1 expression was negatively associated with level of CRP (r = −0.241, P = .001) (Fig. 3A), TNF-α (r = −0.253, P < .001) (Fig. 3B), and IL-6 (r = −0.177, P = .012) (Fig. 3D), while positive correlation was observed between lncRNA ZFAS1 expression and IL-10 in sepsis patients (r = 0.173, P = .014) (Fig. 3E). There was no correlation between lncRNA ZFAS1 expression and IL-1β (r = −0.086, P = .222) (Fig. 3C) or IL-17 (r = −0.093, P = .189) (Fig. 3F).
Figure 3.
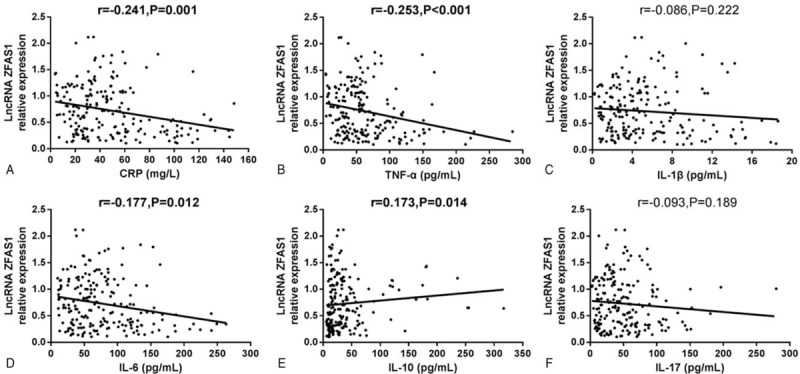
Correlation of long noncoding RNA ZNFX1 antisense RNA (lncRNA ZFAS1) expression with inflammation markers in sepsis patients. LncRNA ZFAS1 expression was negatively correlated with C-creative protein (CRP) (A), tumor necrosis factor-α (TNF-α) (B), and interleukin (IL)-6 (D). LncRNA ZFAS1 expression was positively correlated with IL-10 (E). No correlation between lncRNA ZFAS1 expression and IL-1β (C) or IL-17 (F) was observed. Correlations between lncRNA ZFAS1 expression and inflammation markers were individually determined by Spearman correlation tests and P < .05 was considered significant.
3.7. Predicting value of lncRNA ZFAS1 expression for survival in sepsis patients
There were 62 nonsurvivors and 140 survivors in sepsis patients, and lncRNA ZFAS1 expression was 0.532 (0.204–1.068) in nonsurvivor group, which was decreased compared to survivor group (0.714 [0.374–1.079]) (P < .001) (Fig. 4A). In addition, the predictive value of lncRNA ZFAS1 on prognosis in sepsis patients was further assessed, and we found that lncRNA ZFAS1 presented with a good potential to distinguish nonsurvivors from survivors in sepsis patients with AUC of 0.628 (95% CI: 0.538–0.717). Sensitivity and specificity values were 92.1% and 35.5%, respectively, at the best cut-off point (Fig. 4B).
Figure 4.
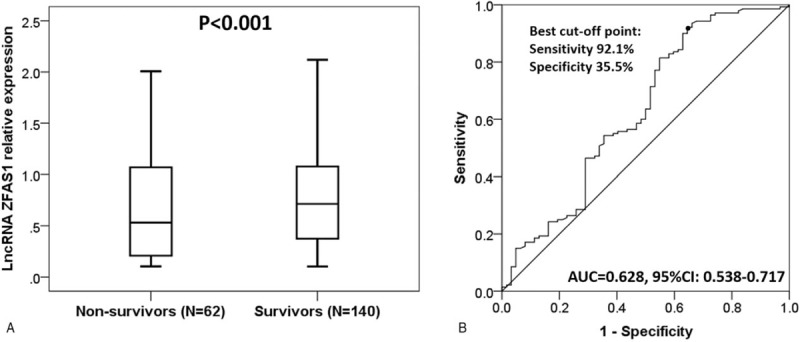
Predicting value of long noncoding RNA ZNFX1 antisense RNA (lncRNA ZFAS1) expression for survival in sepsis patients. LncRNA ZFAS1 expression in nonsurvivor group was lower than that in survivor group (A). LncRNA ZFAS1 had good potential to distinguish survivors and nonsurvivors in sepsis patients (B). Wilcoxon rank sum test was performed to determine the difference in lncRNA ZFAS1 expression between nonsurvivor and survivor groups. Receiver operating characteristic curve was used to assess the ability of lncRNA ZFAS1 in distinguishing sepsis nonsurvivors from survivors. P < .05 was considered significant.
4. Discussion
Our study revealed that: LncRNA ZFAS1 expression was lower in sepsis patients compared to HCs, and it presented with a good value in predicting sepsis susceptibility; LncRNA ZFAS1 expression was negatively correlated with APACHE II score and inflammatory markers levels; LncRNA ZFAS1 expression was lower in nonsurvivor group compared to survivor group, and it had the potential to distinguish nonsurvivors from survivors in sepsis patients.
According to previous studies, some lncRNAs devote to inflammatory disorders through interaction with multiple genes and signaling pathways. For example, a nuclear factor kappa B (NF-κB)-linked lncRNA is reported to suppress inflammation by preventing the overexpression of NF-κB, thereby reducing cell metastasis in breast cancer.[13] Another interesting animal study revealed that downregulation of lncRNA-EPS enhances inflammation by interacting with heterogeneous nuclear ribonucleoprotein to restrain immune response genes in mice macrophages.[14] These studies imply that some lncRNAs may serve as anti-inflammatory genes in diseases with inflammatory disorders via altering inflammation-related genes and corresponding signaling pathways.
LncRNA ZFAS1, a novelly identified lncRNA in sepsis patients, is commonly expressed in human bone marrow, ovary, and thyroid. LncRNA ZFAS1 has been extensively reported to promote cell proliferation, migration, and invasion in various carcinomas such as gastric cancer and osteosarcoma,[5,15] while only 2 studies reported investigate the function of lncRNA ZFAS1 in inflammatory diseases, which disclose that lncRNA ZFAS1 stimulates cells apoptosis via regulating level of miRNA-150 and CRP in AMI, and it regulates migration and invasion of fibroblast-like synoviocytes by targeting the expression of miRNA-27a in RA.[8,10] Considering sepsis as a highly inflammatory disease and known from the previous study that lncRNA ZFAS1 involved in inflammation processes, we hypothesized that lncRNA ZFAS1 expression might also be dysregulated in sepsis patients and it might have diagnostic potential for sepsis. In this present study, we observed that plasma lncRNA ZFAS1 expression was lower in sepsis patients compared to HCs, and it had good diagnostic value for sepsis, which testified our hypothesis. Also, lncRNA ZFAS1 expression was negatively associated with disease severity that was assessed by APACHE II score. The possible explanations were: Firstly, lncRNA ZFAS1 correlated with increased systemic inflammation. Thus it could probably raise the susceptibility of sepsis and enhance the disease severity through regulating inflammation. This was further validated in our subsequent experiments. Secondly, lncRNA ZFAS1 might regulate several genes or signaling pathways to affect cellular activities and cause apoptosis of cells including nephrocytes, hepatocytes, and myocardial cells, thereby resulting in severe organ dysfunction in sepsis patients.
As to the prognostic value of lncRNA ZFAS1 in sepsis patients, we found that lncRNA ZFAS1 expression was positively associated with survival. This might result from that: Low expression of lncRNA ZFAS1 might promote inflammation and increase disease severity in sepsis patients, leading to worsened prognosis; LncRNA ZFAS1 might affect the sensitivity of drugs such as antibiotics and vasoactive agents, thereby altering the treatment efficacy and impacting prognosis in sepsis patients. However, the mechanisms of lncRNA ZFAS1 affecting drug sensitivity need to be verified in future studies.
Sepsis is an endpoint of a complexed, infection-induced process, and the hyperinflammatory response is the critical pathological manifestation. Chemotaxis of inflammatory cells and release of cytokines trigger the cascade of inflammation mediators such as TNF-α, IL-6, IL-1, and IL-8, which further activated granulocytes to injure endothelial cell and facilitate platelet adhesion.[8,10] The inflammation response also increases the release of oxygen radicals and lipid metabolites, thereby eventually lead to cascade effects, which damages histocytes and impairs organ function.[8,10] In the present study, we discovered that lncRNA ZFAS1 was negatively associated with CRP, TNF-α, and IL-6 in sepsis patients, and this might imply the mechanism of lncRNA ZFAS1 in sepsis. Moreover, lncRNA ZFAS1 has been shown to sponge microRNA-27a and microRNA-150, which participate in inflammation response by regulating CRP and NF-κB in inflammatory diseases.[16,17] Although there is currently no experimental evidence about the function of lncRNA ZFAS1 in sepsis, the close correlation of lncRNA ZFAS1 with inflammatory response helps illuminate the value of lncRNA ZFAS1 in sepsis.
Pathogenic blood culture is currently the common standard diagnostic approach for sepsis and selection of antibiotics, while it takes a long time for culturing and the positive rate is <50%.[18] Under the condition of negative blood culture results, critical illness rating system such as APACHE II and SAPS are used for prognosis of patients. However, they cannot independently predict infection in sepsis. From our result, lncRNA ZFAS1 expression was negatively correlated with disease risk and severity, which suggested that it could be used as an assistant biomarker to predict the risk of sepsis during blood testing routine. The advantage of lncRNA ZFAS1 as a biomarker for diagnosis of sepsis is that it only needs a small amount of blood sample and is quick. However, using biomarkers (e.g., PCT, a critical biomarker for sepsis infection) in the diagnosis of sepsis is not deterministic, and predictive value of lncRNA ZFAS1 on sepsis still lacks experimental support.[19]
There were several limitations to our study. Firstly, the sample size was relatively small, thereby leading to decreased statistic power. Secondly, the detailed molecular mechanism of lncRNA ZFAS1 in sepsis was not investigated, especially the role in regulating inflammation as well as its effect on drug sensitivity. Thirdly, this study was single-centered, which might be subject to selection bias due to region restriction. Thus, further study with a larger sample size from multicenter is greatly needed. Moreover, the negative correlation of lncRNA ZFAS1 with inflammatory markers did not necessarily suggest that low lncRNA ZFAS1 level could predict advanced sepsis severity. Instead, it was possible that the inflammatory reaction in sepsis reduced the expression of lncRNA ZFAS1. Therefore, the correlation of lncRNA ZFAS1 with disease severity needed further validation.
5. Conclusion
In conclusion, circulating lncRNA ZFAS1 expression negatively correlates with disease risk, severity, and inflammatory markers levels, and might predict worse survival in sepsis patients.
Author contributions
Conceptualization: Bibo Shao.
Data curation: Yahuan Xu.
Formal analysis: Yahuan Xu, Bibo Shao.
Investigation: Yahuan Xu.
Methodology: Yahuan Xu.
Resources: Bibo Shao.
Supervision: Bibo Shao.
Writing – original draft: Yahuan Xu.
Writing – review & editing: Bibo Shao.
Footnotes
Abbreviations: AMI = acute myocardial infarction, AUC = area under curve, BMI = body mass index, IRG = immune response genes, NF-κB = nuclear factor kappa B, RA = rheumatoid arthritis, Scr = serum creatinine, WBC = white blood cell.
The authors have no funding and conflicts of interest to disclose.
References
- [1].Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016;315:801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fleischmann C, Scherag A, Adhikari NK, et al. Assessment of global incidence and mortality of hospital-treated sepsis. Current estimates and limitations. Am J Respir Crit Care Med 2016;193:259–72. [DOI] [PubMed] [Google Scholar]
- [3].Stevenson EK, Rubenstein AR, Radin GT, et al. Two decades of mortality trends among patients with severe sepsis: a comparative meta-analysis∗. Crit Care Med 2014;42:625–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Dellinger RP, Levy MM, Rhodes A, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock, 2012. Intensive Care Med 2013;39:165–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Li N, Sun ZH, Fang M, et al. Long non-coding RNA ZFAS1 sponges miR-486 to promote osteosarcoma cells progression and metastasis in vitro and vivo. Oncotarget 2017;8:104160–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Leucci E, Vendramin R, Spinazzi M, et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature 2016;531:518–22. [DOI] [PubMed] [Google Scholar]
- [7].Faghihi MA, Modarresi F, Khalil AM, et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med 2008;14:723–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wu T, Wu D, Wu Q, et al. Knockdown of long non-coding rna-zfas1 protects cardiomyocytes against acute myocardial infarction via anti-apoptosis by regulating miR-150/CRP. J Cell Biochem 2017;118:3281–9. [DOI] [PubMed] [Google Scholar]
- [9].Zhang Y, Jiao L, Sun L, et al. LncRNA ZFAS1 as a SERCA2a inhibitor to cause intracellular Ca(2+) overload and contractile dysfunction in a mouse model of myocardial infarction. Circ Res 2018;122:1354–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ye Y, Gao X, Yang N. LncRNA ZFAS1 promotes cell migration and invasion of fibroblast-like synoviocytes by suppression of miR-27a in rheumatoid arthritis. Hum Cell 2018;31:14–21. [DOI] [PubMed] [Google Scholar]
- [11].Bone RC, Balk RA, Cerra FB, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest 1992;101:1644–55. [DOI] [PubMed] [Google Scholar]
- [12].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- [13].Liu B, Sun L, Liu Q, et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 2015;27:370–81. [DOI] [PubMed] [Google Scholar]
- [14].Atianand MK, Hu W, Satpathy AT, et al. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell 2016;165:1672–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Xu W, He L, Li Y, et al. Silencing of lncRNA ZFAS1 inhibits malignancies by blocking Wnt/beta-catenin signaling in gastric cancer cells. Biosci Biotechnol Biochem 2018;82:456–65. [DOI] [PubMed] [Google Scholar]
- [16].Laszlo I, Trasy D, Molnar Z, et al. Sepsis: from pathophysiology to individualized patient care. J Immunol Res 2015;2015:510436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rittirsch D, Flierl MA, Ward PA. Harmful molecular mechanisms in sepsis. Nat Rev Immunol 2008;8:776–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Scheer CS, Fuchs C, Grundling M, et al. Impact of antibiotic administration on blood culture positivity at the beginning of sepsis: a prospective clinical cohort study. Clin Microbiol Infect 2018;doi: 10.1016/j.cmi.2018.05.016. [DOI] [PubMed] [Google Scholar]
- [19].Almansa R, Martin S, Martin-Fernandez M, et al. Combined quantification of procalcitonin and HLA-DR improves sepsis detection in surgical patients. Sci Rep 2018;8:11999. [DOI] [PMC free article] [PubMed] [Google Scholar]


