Supplemental Digital Content is available in the text
Keywords: antibiotic resistance, Burkholderia pseudomallei, clinical isolates, molecular analysis
Abstract
Burkholderia pseudomallei is the causative agent of meliodosis, and the cases in China are gradually increasing. The present retrospective study aimed to surveil the molecular epidemiological characteristics and antibiotic resistance of B pseudomallei isolates. B pseudomallei strains were isolated and verified from meliodosis patients with relevant epidemiological information from 2004 to 2016 in Hainan, China. Pulsed-field gel electrophoresis based on Spe I digestion was carried out, and antimicrobial resistance of B pseudomallei strains was observed against 9 frequently-used antimicrobials. A total of 164 B pseudomallei isolates were successfully divided into 60 pulsed-field gel electrophoresis (PFGE) patterns, including 33 clusters and 27 single types, at an 85% similarity level. The isolates also exhibited a high level of ceftazidime resistance rate (12.8%, 21/164). B pseudomallei strains were mainly heterogenous with no predominant type, but there were some clonal populations, dominate clusters prevalent and the resistance rates of cephems antimicrobial increased significantly between 2004 and 2016 along with the number of melioidosis cases collected in Hainan (cefoperazone-sulbactam [SCF], rs = 0.96, P = .04; ceftazidime [CAZ], rs = 0.98, P = .01). In conclusion, this study will help to enhance our understanding of molecular characteristics and antibiotic resistance of B pseudomallei.
1. Introduction
Melioidosis is a serious and fatal zoonosis, caused by the Gram negative bacterium of Burkholderia pseudomallei. It contributes to a high mortality rate that can be up to 40% even with appropriate treatment.[1,2]B pseudomallei has been listed as a Tier-1 (top tier) select agent by the US Centers for Disease Control and Prevention (CDC) because of its potential as a bioweapon.[3] The endemic areas are mainly distributed within latitudes 20°N and 20°S. Now more and more cases of melioidosis are reported in China, including the South China Sea and the Taiwan Strait, such as Hainan,[4] Hongkong,[5] Taiwan,[6] and Guangxi.[7]
A mathematical modeling study of 2015 has predicted global annual burden of melioidosis to be 165,000 cases with 89,000 deaths, Hainan, in southern China, is also involved.[2] However, melioidosis is hard to be cured. Previous studies show that B pseudomallei isolates from persistent infections are reported to resistant to antibiotics, such as penicillin, first- and second-generation cephalosporins, macrolides, rifamycins, colistin, and aminoglycosides.[8,9] In our previous research, B pseudomallei could evade inherent immunity by 3 novel miRNAs of MIR4458, MIR4667-5p, and MIR4668-5p targeted regulation of ATG10.[10] Therefore, monitoring pathogenic bacteria of B pseudomallei may be a useful way to understand the epidemic characteristics, antimicrobial resistance characteristics, and provide a basis for the control of the disease.
Molecular typing methods are powerful tools for epidemiological investigation of B pseudomallei,[11,12] which can provide insight about bacterial diversity and distribution. A variety of molecular tools, such as pulsed-field gel electrophoresis (PFGE), multi-locus sequence typing (MLST), and ribotyping have been used to infer epidemiology and genetic relatedness between B pseudomallei isolates in tropical countries, particularly in Northern Australia, Malaysian, and Thailand.[11–13] Among molecular typing methods, PFGE is the gold standard method among the molecular typing methods with a standardized PulseNet protocol.[14] PFGE with a high degree of resolution is necessary to detect subtle differences in what most likely are genetically related strains. It is suitable for characterizing strains that cause localized outbreaks. PFGE is also an important strategy to make it possible to compare bacterial DNA fingerprints from different epidemic areas and provide laboratory evidence.
Aiding the data of active surveillance, outbreak investigation and source tracking for melioidosis is crucial to current and future prevention approaches. In Hainan, southern China, epidemiological data on genome fingerprinting of local strains are still limited. Therefore, the objective of this retrospective study is to molecularly characterize the B pseudomallei isolated from patients in Hainan with PFGE to find out if there exist predominant genotypes and to determine the antibiotic susceptibility profile by minimum inhibitory concentration (MIC) methods.
2. Material and methods
2.1. Case collection and bacterial strains Identification
The cases were obtained from the people's Hospital of Sanya City and the people's Hospital of Haikou City in Hainan from December 2004 to December 2016, including people from other parts of China who have traveled to Hainan, such as Fujian Province, Hunan Province, and so on. B pseudomallei strains were isolated from patients’ blood, sputum, pus, and urine (Table S1). Our treatment of B pseudomallei infection included 2 phases which were based on a series of clinical trials.[15,16] First, treatment with agents to which B pseudomallei was susceptible required 2 to 4 weeks of parenteral therapy (e.g., with ceftazidime, imipenem, or meropenem) as acute phase therapy. Second, oral “eradication” therapy (e.g., with trimethoprim-sulfamethoxazole, amoxicillin-clavulanate, or combination therapy) was followed for 3 to 6 months at the aim of preventing relapse. The outcome was recorded when patients of melioidosis left the hospital.
B pseudomallei were acquired from culture-confirmed clinical specimens at 37 °C for 2 to 3 days and identified by VITEK-2 identification system (BioMérieux, Marcy-I’Etoile, France), with an excellent specificity level of identification (>99%). The B pseudomallei isolates were further confirmed by 16S ribosomal DNA gene sequencing as Dance et al[17] described before. The verified B pseudomallei was then stored in LB media containing 15% glycerol at –80 °C until they were used.
2.2. Pulsed-field gel electrophoresis (PFGE)
PFGE was performed to determine genomic DNA fingerprinting profiles of B pseudomallei isolates according to the procedures developed by the CDC.[18] Briefly, the isolates were inoculated into 5 mL LB medium overnight at 37 °C. One milliliter LB cultures were centrifuged at 6000 rmp for 5 minutes, the supernatants were discarded and the pellets were resuspended in cell suspension buffer (100 mM Tris–HCl, 100 mM EDTA pH 8.0) and adjusted to absorbance values of approximately 1.4 to 1.8 measured at a wavelength of 600 nm with Bio-Rad iMark Microplate Absorbance Reader (Bio-Rad). Ten microliter Proteinase K (20 mg/mL) was added into 200 μL of the bacterial suspension, and an equal volume of melted 1% SeaKem Gold agarose (FMC Bio-Products, Rockville, ME) containing 1% sodium dodecyl sulfate prepared in Tris EDTA buffer (TE; 10 mM Tris, 1 mM EDTA pH 8.0).
The mixture was dispensed into a sample mold (Bio-Rad, Hercules, CA). After solidification, the plugs were transferred to a tube containing 1 mL lysis buffer (50 mM Tris–HCl, 50 mM EDTA pH 8.0, 1% sarcosyl) and 0.1 mg of proteinase K per mL. Cells were lysed 2 hours in a water bath at 54 °C. After lysis, the plugs were washed twice with deionized distilled water and 4 times with TE buffer (10 mM Tris–HCl, 1 mM EDTA pH 8.0) for 15 minutes per wash at 54 °C. Agarose-embedded DNA was digested with 20 U of Spe I (TaKaRa, Dalian, China) 5 hours in a water bath at 37 °C. The plugs were then placed in a 1% SeaKem Gold agarose gel. Restriction fragments were separated by electrophoresis in 0.5 M Tris-borate-EDTA buffer at 14 °C for 20 hours by using a Chef Mapper (Bio-Rad) with pulse times of 3 to 50 seconds. The gel was stained with Goldview, and DNA bands were visualized with a UV transilluminator.
2.3. Antimicrobial susceptibility testing
Antimicrobial susceptibility testing for the B pseudomallei isolates was determined via the BD Phoenix-100 Automated Microbiology System (Becton, Dickinson and Company, Sparks, MD). The minimum inhibitory concentrations (MICs) were interpreted by the standards of Clinical and Laboratory Standards Institute (CLSI) supplement M100: 2017.[19] Susceptibility to 9 commonly used antimicrobials was evaluated. The antimicrobial agents were as follows: imipenem (IPM), meropenem (MEM), ceftazidime (CAZ), amoxicillin-clavulanate (AMC), piperacillin-tazobactam (TZP), trimethoprim-sulfamethoxazole (SXT), levofloxacin (LEV), tetracycline (TET) and Cefoperazone-sulbactam (SCF). Escherichia coli ATCC 25922, Enterococcus faecalis ATCC 29212, Staphylococcus aureus ATCC 29213, and Pseudomonas aeruginosa ATCC 27853 were used as quality-control organisms in antimicrobial MIC determinations to ensure that the dilution range for each antimicrobial agent was properly controlled.
2.4. Data analysis
PFGE results were analyzed using BioNumerics 7.6.1 (Applied Maths, Belgium). A dendrogram of PFGE patterns was constructed using unweighted pair-group method (UPGMA) with a position tolerance of 1.5% and optimization of 1%. Strains showing Dice coefficient of ≥0.85 are grouped into the same PFGE type and subtypes. The evolution of resistance rate and PFGE type over time in Hainan was studied by Spearman rank correlation coefficient (rs) using SPSS (SPSS, version 20). P value was calculated with Pearson chi-square test and Fisher exact test. Statistical level of significance was set at P < .05.
2.5. Ethical standards
This study was approved by the Human Research Ethics Committee of the Third Military Medical University, which is a member of the Chongqing City Ethics Committees of China. All clinical cases were anonymized without personal information and therefore, written informed consent was not required or obtained from the patients involved.
3. Result
3.1. Clinical characteristics
One hundred and sixty-four strains of B pseudomallei with a clear clinical background were collected (Table S1). Of the 164 cases identified, 138 (84.1%) were men, the mean age was 49.1 years (range, 11–82 years old) and 58.54% was farmers. All collected regions except Dingan, Baisha, and Baoting were located along the coast, and the population density of coastal areas was higher than that of inland areas (Fig. 1). Of these, 102 (62.2%) were isolated from blood, 32 (19.51%) were from sputum, 27 (16.46%) were from pus, and 3 (1.83%) were from urine samples. The effect was not entirely satisfactory, only 34.75% (57/164) of the patients were cured, and 65.24% (107/164) were not able to respond to treatment or just improved, including 7 death cases, when they left the hospital.
Figure 1.
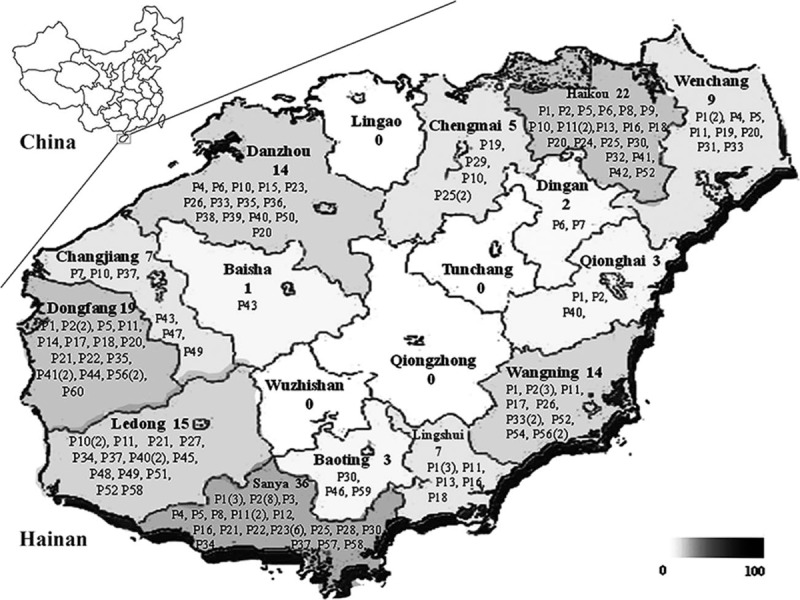
Map of Hainan Island and distribution of the 60 PFGE types. The map showed the major regions in the island. The PFGE types of 164 clinical isolates were labeled in each region in which melioidosis patients came from. The number of the strains is shown in the brackets. PFGE = pulsed-field gel electrophoresis.
3.2. Molecular typing
A total of 164 clinical isolates were identified as B pseudomallei and all strains were successfully divided by Spe I-digested PFGE. B pseudomallei C006 (BPC006, NC_018527.1and NC_018529.1), the first genome sequence of a B pseudomallei isolate in China, was chose as a standard strain.[20] PFGE analysis resulted in the characterization of 60 types of PFGE fingerprints with Dice coefficient 85%, as shown in Fig. 2. A total of 27 out of 60 types were single types with similarity coefficient ranged from 59.7% to 84.6%. Another 33 clusters were shared by 137 unrelated clinical strains of B pseudomallei with similarity coefficient ranging from 85.3% to 100%. Remarkably, there were 4 isolates (100%) in cluster P40 shared the same band type. Herein P1 and P2 were identified as dominate clusters in which each of the cluster had 3 subtypes and contained 12 (7.32%) and 18 (10.98%) cases, respectively. Specially, the PFGE type of P2 increased significantly between 2004 and 2016 with melioidosis cases (rs = 0.96, P = .04, Fig. 3C). The other clusters occurring in ≥5 cases were P11 (9, 5.49%), P10 (8, 4.88%), P23 (7, 4.27%), and P33 (5, 3.05%), as shown in Fig. 3A.
Figure 2.
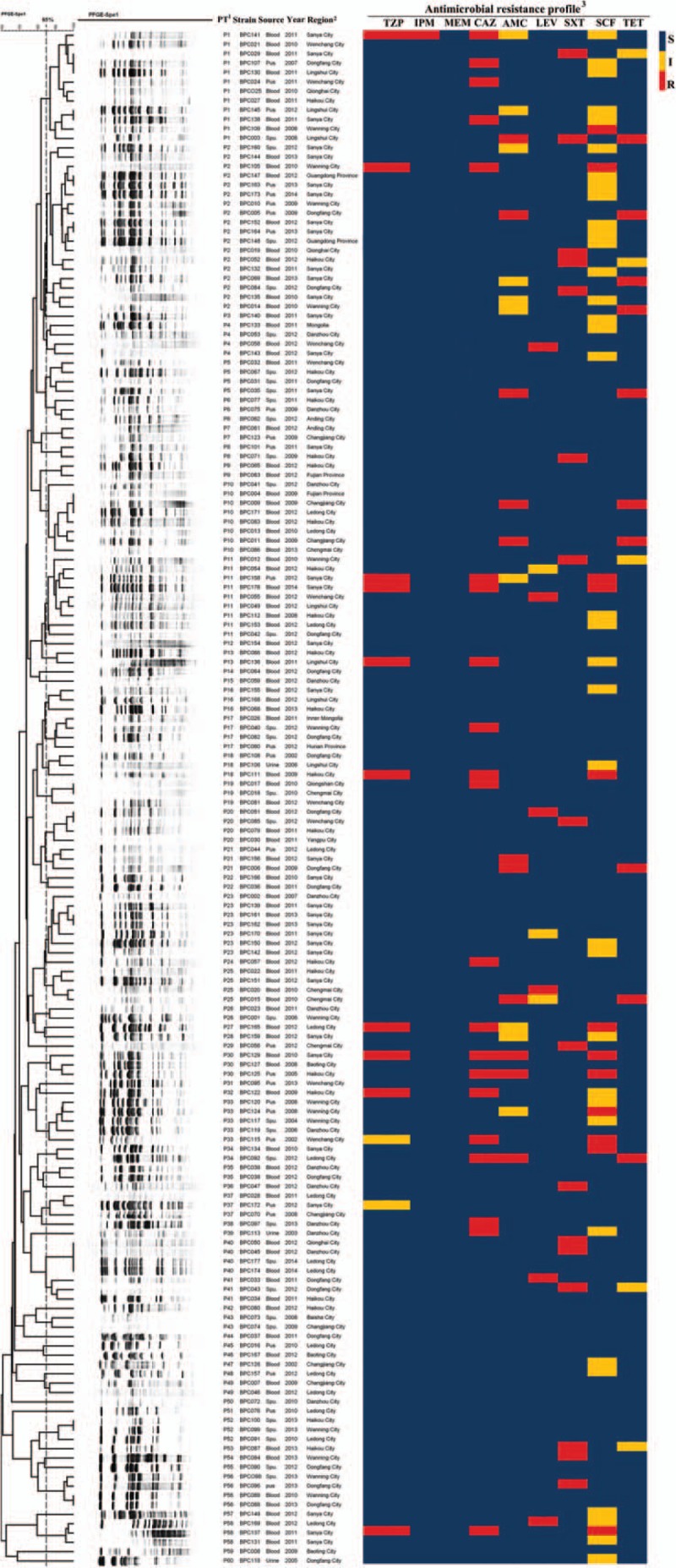
Dendrogram of PFGE patterns for 164 B pseudomallei isolates detected in Hainan, China, 2004 to 2016. 1. PT, PFGE type. Molecular typing by PFGE at 85% similarity. 2. The regions are mainly from cities within Hainan Province. People from other parts of China who have traveled to Hainan are also included. 3. MICs test of B pseudomallei. Nine commonly used antimicrobials were tested, imipenem (IPM), meropenem (MEM), ceftazidime (CAZ), amoxicillin-clavulanate (AMC), piperacillin-tazobactam (TZP), trimethoprim-sulfamethoxazole (SXT), levofloxacin (LEV), tetracycline (TET), and cefoperazone-sulbactam (SCF). Blue: sensitive (S); yellow: intermediate (I); red: resistant (R). MIC = minimum inhibitory concentration, PFGE = pulsed-field gel electrophoresis.
Figure 3.
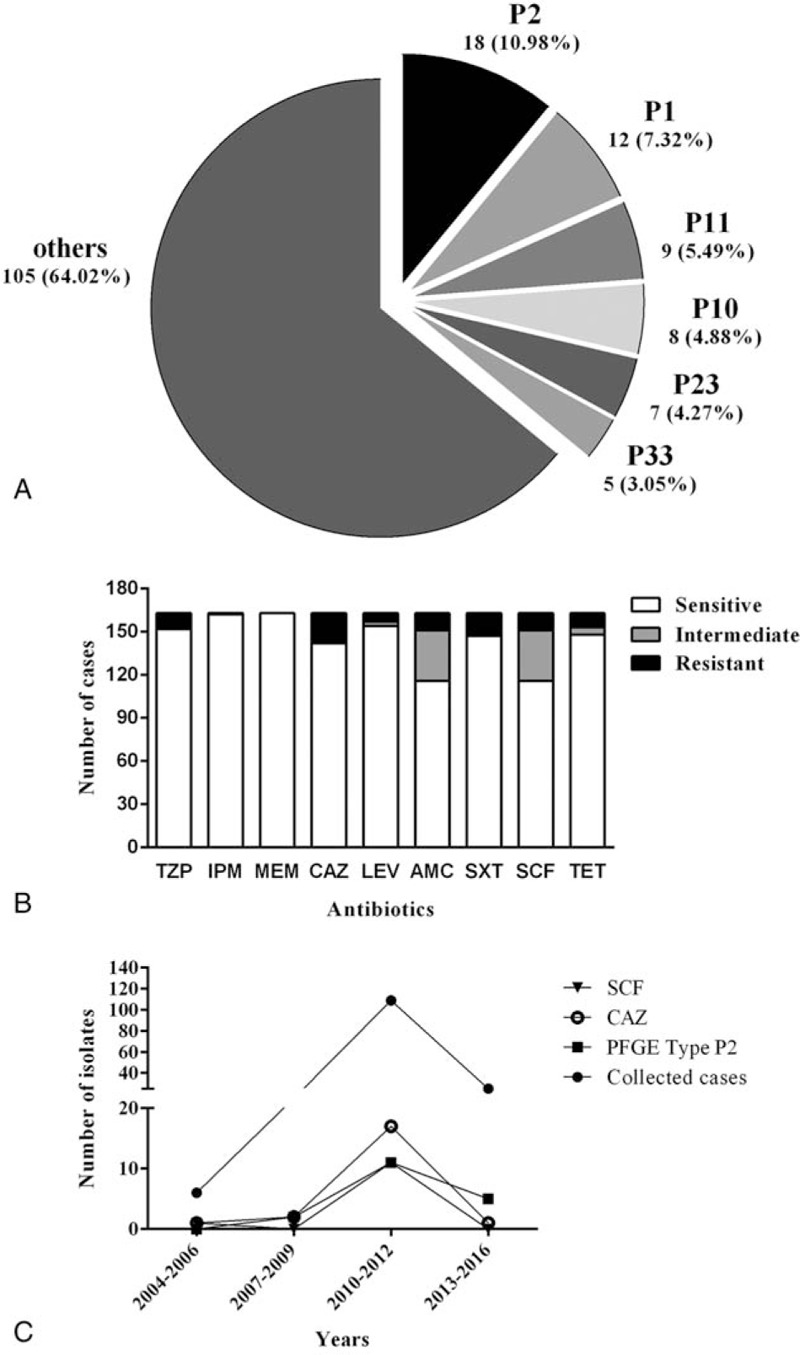
PFGE type P2 and the resistance rates of cephems antimicrobial increased significantly. A. PFGE profiles of the 164 B pseudomallei isolates from Hainan, China. B. The number of cases in antimicrobial susceptibility test. C. PFGE type of P2 and cephems antimicrobial increased significantly between 2004 and 2016 with melioidosis cases collected in Hainan. P2, rs = 0.96, P = .04; SCF, rs = 0.96, P = .04; CAZ, rs = 0.98, P = .01, by Spearman rank correlation coefficient (rs). CAZ = ceftazidime, PFGE = pulsed-field gel electrophoresis, SCF = cefoperazone-sulbactam.
3.3. Determination of MIC's to antibiotics
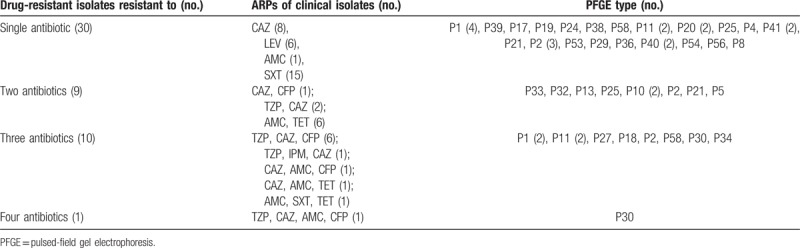
Antimicrobial susceptibility testing was conducted for the 164 B pseudomallei isolates, as shown in Figs. 2 and 3B. The detailed information on the resistance rates to all of the tested antimicrobials was listed in Table 1. The highest resistance rate was observed for CAZ, which reached 12.8% (n = 21), followed by resistance to SXT (n = 16; 9.8%), SCF and AMC (n = 12; 7.4%, uniformly), and 5 other antimicrobials with resistance isolates under 10 (6.1%). There was almost no resistance noted to carbapenems (IPM, MEM), except BPC141, the only one isolates was found resistant to IPM. Multidrug-resistant isolates which were resistant to >1 drug were also found, as shown in the Table 2. Notability, the resistant strains in P1, the dominate clusters, showed the highest proportion which reached 58.3% (7/12), followed by strains in cluster P11 (44.4%, 4/9). Specially, the resistance rates of cephems antimicrobial increased significantly between 2004 and 2016 along with the number of melioidosis cases collected in Hainan (SCF, rs = 0.96, P = .04; CAZ, rs = 0.98, P = .01), as shown in Fig. 3C (Table S2).
Table 1.
Antimicrobial resistance rates of the 164 B pseudomallei isolates.
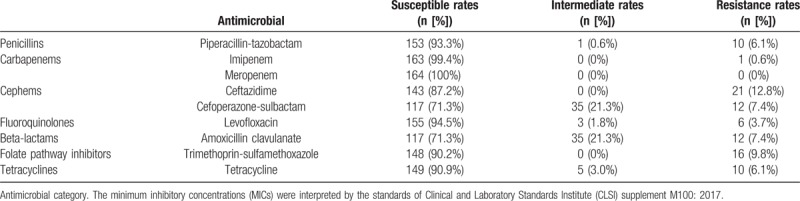
Table 2.
Antimicrobial resistance profiles (ARPs) of clinical isolates.
4. Discussion
Melioidosis has emerged as an important cause of morbidity and mortality. Hainan Province has long been an endemic focus of melioidosis,[21–23] our team previously investigated 102 B pseudomallei strains by MLST, which were also included in this retrospective study showed the phylogenetic relationships of dominant STs in Hainan was significant linked with B pseudomallei isolates from Thailand.[22] However, the isolates in previous research were only 102 up to 2014 and the used typing technology was mainly about sequencing and comparison of internal fragments of 7 housekeeping genes in the genome. The epidemiological data on the complete genome fingerprinting and antimicrobial susceptibility profile were still limited. The present study was based on PFGE-Spe I and MICs test to investigate the outbreak situation and drug sensitivity of B pseudomallei, it reported a 13 years retrospective study of 164 melioidosis cases in the province Hainan of China. According to the clinical cases collection, the population density of melioidosis in coastal areas was higher than that in inland areas as the phenomenon found in Wang research.[23] But it still needed further investigation (e.g., Environmental strains of both coastal and inland areas across Hainan should be collected to better understand the true ecological distribution of B pseudomallei) to clarify if coastal areas were more conducive to B pseudomallei growth and persistence than inland regions.
One hundred and thirty-seven clinical isolates were shown to share 33 PFGE fingerprint types with similarity coefficient >0.85, suggesting that there was a close genetic relatedness among those strains even though they were isolated from different patients who were from different locations. But no particular PFGE type in a concentrated outbreak was found. Generally, PFGE based clustering tree branched out extensively, including 27 single types and 33 clusters, which suggested the isolates of B pseudomallei were heterogeneous in this retrospective study. The result in Hainan province of China was also similar to the genome fingerprinting of B pseudomallei isolates from Malaysian and Thailand patients.[11,12] However, it was noteworthy that there were different isolates, especially cluster P40 containing 4 isolates, shared a same band type, which indicated that there were some clonal populations of B pseudomallei were prevalent in Hainan, China.
In the retrospective study, B pseudomallei showed high resistance to cephems antimicrobial (20%, CAZ, n = 21; SCF, n = 12). The founding was completely at odds with those from other melioidosis-endemic areas, for example, Northeast Thailand (resistance rates to CAZ = 8/4030, 2011).[24] It may cause by misuse and overuse of antibiotics.[25] Meanwhile, the high resistance rate to ceftazidime may also link to increased expression of serine β-lactamases (classes A and D) or deletion of penicillin binding protein 3.[26,27] What's more, SXT, as the oral antibiotic drugs on the second phase for curing infection, the resistance rate was 9.8% (16/164), which was lower than the rate reported from Thailand (13% by E-test),[28] the widespread co-trimoxazole resistance in clinical B pseudomallei isolates was likely to result in BpeEF-OprC efflux pump expression.[29] In our retrospective study, B pseudomallei prevalent in Hainan were still susceptible to IPM and MEM, excepted one isolate.
5. Conclusion
We used SpeI-digested PFGE and MICs test approach to investigate the outbreak situation and drug sensitivity of B pseudomallei, it reported a 13 years retrospective study of 164 melioidosis cases in the province Hainan of China. This study will help to enhance our understanding of molecular characteristics and antibiotic resistance of B pseudomallei.
6. Limitations
However, there were limitations in our conclusion. It was well known that some methods over-estimate resistance rates of B pseudomallei, the resistance rates require one more detection methods, such as standard microdilution MICs and E-test. Meanwhile the high rate might be jammed by the little scale of the included cases in Hainan. The increased expression of β-lactamase and BpeEF-OprC efflux pump, or deletion of penicillin binding protein 3 in the resistance strains need further experimental verification. To some extent, this work may shed light on molecular characteristics and antibiotic resistance of B pseudomallei and contributes to the prevention and control of B pseudomallei in the important endemic area of China.
Author contributions
Data curation: Chen Jiangao, Qian Li.
Formal analysis: Lu Xiaoxue.
Funding acquisition: Mao Xuhu.
Methodology: Rao Chenglong.
Project administration: Qian Li.
Resources: Tang Mengling, Chen Hai.
Software: Rao Chenglong.
Validation: Cao Liusu, Deng Ling.
Writing – original draft: Rao Chenglong.
Writing – review & editing: Hu Zhiqiang.
Supplementary Material
Footnotes
Abbreviations: AMC = amoxicillin-clavulanate; CAZ = ceftazidime; CDC = Centers for Disease Control and Prevention; CLSI = Clinical and Laboratory Standards Institute; IPM = imipenem; LEV = levofloxacin; MEM = meropenem; MIC = minimum inhibitory concentration; MLST = multilocus sequence typing; PCR = polymerase chain reaction; PFGE = pulsed-field gel electrophoresis; SCF = cefoperazone-sulbactam; SXT = trimethoprim-sulfamethoxazole; TET = tetracycline; TZP = piperacillin-tazobactam.
RC and HZ have contributed equally to this work.
This study was supported by grants from National Natural Science Foundation of China (NSFC, No. 81772141) and Third Military Medical University Science Foundation of Outstanding Youth (2017YQRC-07).
The authors report no conflicts of interest.
Supplemental Digital Content is available for this article.
References
- [1].Wiersinga WJ, Currie BJ, Peacock SJ. Melioidosis. N Engl J Med 2012;367:1035–44. [DOI] [PubMed] [Google Scholar]
- [2].Limmathurotsakul D, Golding N, Dance DA, et al. Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nat Microbiol 2016;1:15008. [DOI] [PubMed] [Google Scholar]
- [3].Wiersinga WJ, van der Poll T, White NJ, et al. Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei. Nat Rev Microbiol 2006;4:272–82. [DOI] [PubMed] [Google Scholar]
- [4].Yang S. Melioidosis research in China. Acta Trop 2000;77:157–65. [DOI] [PubMed] [Google Scholar]
- [5].Currie BJ, Dance DA, Cheng AC. The global distribution of Burkholderia pseudomallei and melioidosis: an update. Trans R Soc Trop Med Hyg 2008;102suppl:S1–4. [DOI] [PubMed] [Google Scholar]
- [6].Ko WC, Cheung BM, Tang HJ, et al. Melioidosis outbreak after typhoon, southern Taiwan. Emerg Infect Dis 2007;13:896–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ma G, Zheng D, Cai Q, et al. Prevalence of Burkholderia pseudomallei in Guangxi, China. Epidemiol Infect 2010;138:37–9. [DOI] [PubMed] [Google Scholar]
- [8].Khosravi Y, Vellasamy KM, Mariappan V, et al. Antimicrobial susceptibility and genetic characterisation of Burkholderia pseudomallei isolated from Malaysian patients. ScientificWorldJournal 2014;2014:132971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lazar Adler NR, Govan B, Cullinane M, et al. The molecular and cellular basis of pathogenesis in melioidosis: how does Burkholderia pseudomallei cause disease? FEMS Microbiol Rev 2009;33:1079–99. [DOI] [PubMed] [Google Scholar]
- [10].Li Q, Fang Y, Zhu P, et al. Burkholderia pseudomallei survival in lung epithelial cells benefits from miRNA-mediated suppression of ATG10. Autophagy 2015;11:1293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Azura MN, Norazah A, Kamel AG, et al. DNA fingerprinting of septicemic and localized Burkholderia pseudomallei isolates from Malaysian patients. Southeast Asian J Trop Med Public Health 2011;42:114–21. [PubMed] [Google Scholar]
- [12].Koonpaew S, Ubol MN, Sirisinha S, et al. Genome fingerprinting by pulsed-field gel electrophoresis of isolates of Burkholderia pseudomallei from patients with melioidosis in Thailand. Acta Trop 2000;74:187–91. [DOI] [PubMed] [Google Scholar]
- [13].Cheng AC, Ward L, Godoy D, et al. Genetic diversity of Burkholderia pseudomallei isolates in Australia. J Clin Microbiol 2008;46:249–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Talon D, Cailleaux V, Thouverez M, et al. Discriminatory power and usefulness of pulsed-field gel electrophoresis in epidemiological studies of Pseudomonas aeruginosa. J Hosp Infect 1996;32:135–45. [DOI] [PubMed] [Google Scholar]
- [15].Jenney AW, Lum G, Fisher DA, et al. Antibiotic susceptibility of Burkholderia pseudomallei from tropical northern Australia and implications for therapy of melioidosis. Int J Antimicrob Agents 2001;17:109–13. [DOI] [PubMed] [Google Scholar]
- [16].Dance D. Treatment and prophylaxis of melioidosis. Int J Antimicrob Agents 2014;43:310–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dance DAB, Sarovich D, Price EP, et al. Human infection with Burkholderia thailandensis, China, 2013. Emerg Infect Dis 2018;24:953–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ribot EM, Fair MA, Gautom R, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 2006;3:59–67. [DOI] [PubMed] [Google Scholar]
- [19].CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 27th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2017. [Google Scholar]
- [20].Fang Y, Huang Y, Li Q, et al. First genome sequence of a Burkholderia pseudomallei Isolate in China, strain BPC006, obtained from a melioidosis patient in Hainan. J Bacteriol 2012;194:6604–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Fang Y, Chen H, Li YL, et al. Melioidosis in Hainan, China: a restrospective study. Trans R Soc Trop Med Hyg 2015;109:636–42. [DOI] [PubMed] [Google Scholar]
- [22].Fang Y, Zhu P, Li Q, et al. Multilocus sequence typing of 102 Burkholderia pseudomallei strains isolated from China. Epidemiol Infect 2016;144:1917–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang XM, Zheng X, Wu H, et al. Multilocus sequence typing of clinical isolates of Burkholderia pseudomallei collected in Hainan, a tropical island of Southern China. Am J Trop Med Hyg 2016;95:760–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wuthiekanun V, Amornchai P, Saiprom N, et al. Survey of antimicrobial resistance in clinical Burkholderia pseudomallei isolates over two decades in Northeast Thailand. Antimicrob Agents Chemother 2011;55:5388–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Laxminarayan R, Duse A, Wattal C, et al. Antibiotic resistance-the need for global solutions. Lancet Infect Dis 2013;13:1057–98. [DOI] [PubMed] [Google Scholar]
- [26].Papp-Wallace KM, Becka SA, Taracila MA, et al. Exposing a beta-Lactamase “Twist”: the Mechanistic Basis for the High Level of Ceftazidime Resistance in the C69F Variant of the Burkholderia pseudomallei PenI beta-Lactamase. Antimicrob Agents Chemother 2016;60:777–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Godfrey AJ, Wong S, Dance DA, et al. Pseudomonas pseudomallei resistance to beta-lactam antibiotics due to alterations in the chromosomally encoded beta-lactamase. Antimicrob Agents Chemother 1991;35:1635–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wuthiekanun V, Cheng AC, Chierakul W, et al. Trimethoprim/sulfamethoxazole resistance in clinical isolates of Burkholderia pseudomallei. J Antimicrob Chemother 2005;55:1029–31. [DOI] [PubMed] [Google Scholar]
- [29].Podnecky NL, Wuthiekanun V, Peacock SJ, et al. The BpeEF-OprC efflux pump is responsible for widespread trimethoprim resistance in clinical and environmental Burkholderia pseudomallei isolates. Antimicrob Agents Chemother 2013;57:4381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


