Abstract
Cancer stem cells (CSCs) are postulated to play significant role in the pathogenesis, progression as well as drug resistance of breast cancer. Nucleostemin (NS) is thought to be a key molecule for stemness, and the clinical impact of NS immunoreactivity in breast cancer can indicate its actual role and future therapeutic potentials.
The current study is an observational study with an attempt to evaluate the correlation between NS expression (protein and gene expression levels) and different clinicopathological attributes of invasive breast cancer. For that reason, we investigated NS immunohistochemistry expression on commercial tissue microarray (TMA) of 102 patients and 51 archival specimens from patients admitted to Saqr Hospital, Ras Al Khaimah and diagnosed in Al Baraha Hospital, Dubai, UAE. In addition, the association between NS (GNL3) gene expression and different prognostic parameters as well as patient outcome was also evaluated using 2 large publicly available databases.
Interestingly, we found NS expression to be associated with less differentiated and more advance stage. In addition, NS expression was significantly higher in larger size (P = .001) and LN-positive tumors (P = .007). Notably, NS expression was significantly correlated to P53 positive (P = .037) status. Furthermore, NS was found to be more expressed in the highly aggressive breast cancer subtypes including human epidermal growth factor receptor 2 (HER-2) and triple negative breast cancer (TNBC) subtypes. Moreover, our results also showed that high GNL3 gene expression to be associated with poor patient outcome and higher chances of tumor recurrence.
Our results highlight NS expression as a marker of aggressive phenotype and poor outcome and indicate its possible use as a potential target for CSC-associated breast cancer management.
Keywords: breast cancer, G Protein Nucleolar 3, patient outcome, prognosis, stem cells markers
1. Introduction
Despite all the advances in the oncology field, breast cancer remains one of the leading causes of death in women.[1,2] Moreover, disease recurrence and drug resistance are still major challenges for breast cancer survivors.[3] Hence, identification of new markers that can predict patients with high risk of development of tumor recurrence and drug resistance might help not only in designing more aggressive therapeutic protocols to treat such patients but also in the discovery of novel and more precise therapeutic options.
One of the proposed mechanisms to explain the tumor recurrence is the presence of cancer stem cells (CSCs).[2] Despite the fact that they represent a small proportion of the tumor cells, CSCs are believed to be responsible for tumor initiation and to play a major role in tumor recurrence, metastasis, and drug resistance.[4–6] This might be due to their unique features like self-renewal and pluripotency.[7,8] Thus, investigating markers associated with breast CSCs might provide an important tool to identify patients with higher risk of more aggressive disease behavior.
Nucleostemin (NS) is GTP-binding protein that was shown to be involved in different processes including ribosome synthesis, pre-rRNA processing as well as genome protection.[9–11] The main site of NS accumulation is the nucleoli. Following the binding of NS with GTP it moves to the nucleoplasm, where it interacts with group of proteins like p53 to execute different functions including cell cycle progression, apoptosis, proliferation as well as self-renewal.[12,13] Indeed, NS was found to be up-regulated in breast cancer cell lines and was later proved to play an essential role in stem cells maintenance through regulation of their self-renewal capacity.[14] In addition, it was previously reported that NS gene is markedly expressed in the nucleoli of undifferentiated cells including stem cells compared to more differentiated cells.[15] Moreover, numerous studies confirmed its expression in a wide spectrum of cancers including colon cancer,[16] lung cancer,[17] brain tumors,[18] breast cancer,[12] and hepatocellular carcinoma.[19]
We report here for the first time, the prognostic value of NS expression in a multiethnic cohort of breast cancer patients from United Arab Emirates (UAE) compared to a cohort of commercial tissue microarray (TMA) of breast cancer patients.
Our results revealed an important association between NS protein, as well as, gene expression levels with well-known, poor clinicopathological parameters including larger tumor size, advanced tumor stage, high grade as well as more aggressive breast cancer molecular subtypes. Finally, our results also showed that higher NS gene (GNL3) expression was associated with higher possibility of tumor recurrence. Together, these findings highlight the promising use of NS as a marker of tumor progression and poor patient outcome.
2. Materials and methods
2.1. Patients’ cohorts
A total of 431 breast lesions (excluding mastitis and breast abscess) were admitted to the Surgery Department, Saqr Hospital, Ras Al Khiamah, UAE in the period from 1998 to 2011. Ninety-five cases were diagnosed as primary invasive breast carcinoma in the Pathology Department, Al Baraha Hospital, Dubai, UAE, out of which, only 51 cases with available complete records and paraffin blocks with sufficient tissue were included in the study.
The other cohort was a commercial TMA purchased from Pantomics (BRC1021, Richmond, USA) and consisted of 102 cores. It included 5 normal/benign cores, 6 in situ carcinomas and 91 cores of invasive breast carcinoma from different histological and molecular subtypes.
The majority of the patients of the UAE cohort were Asians 21/51 (41.2%), while 12 (23.5%) patients were UAE nationals and 16 (31.4%) were from other Arab countries. For the UAE cohort, the patients involved in the study have given their permission to be involved in this study. The study protocol was approved by the Research and Ethics Committee of RAK Medical and Health Sciences University—UAE. Moreover, for the commercial TMA, the providing company assured that the patients involved in this cohort gave a written informed consent and tissues were collected with high ethical standards
2.2. Immunohistochemistry
The tissue sections were deparaffinized, rehydrated using a series of methanol concentrations, then immersed in hydrogen peroxide solution for 30 minutes to ensure endogenous peroxidase activity blockage. This was followed by heat-induced antigen retrieval with citrate buffer at pH 6.0 for 30 minutes. The slides then incubated with rabbit anti-NS polyclonal antibody (bs-6846R) at 4°C overnight, followed by HRP conjugation. The reaction was visualized with diaminobenzidine and the slides were then counterstained with Mayer's hematoxylin and examined manually.
2.3. Immunohistochemistry scoring
All the cases were evaluated for immunostaining in a blind manner from there clinic-pathological parameters by 3 independent pathologists. All the information related to patient's identification remains anonymous in adherence to the ethical and legal guidelines.
For NS, cases were considered positive if 10% or more tumor cells exhibited nucleolus/nucleoplasm staining. As regards ER, progesterone receptor (PR), human epidermal growth factor receptor 2 (HER-2), and p53, the assessment of the UAE cohort had been done as previously described.[20] Moreover, the cases were classified into molecular subtypes according to immunohistochemistry surrogate of molecular breast cancer subtypes that was previously used.[21]
2.4. Data mining
Two large, publicly available databases were used in our report; the first is the gene expression-based outcome (GOBO) database, which includes data of 1881 breast cancer patients. This tool enabled us to investigate the association between NS (GNL3) gene expression and various prognostic parameters including grade and molecular subtypes, in addition to its correlation with patient outcome represented as relapse-free survival (RFS). The median (middle quantile) was used for dividing the cases into high or low expression groups as previously described.[22]
We also used Curtis dataset (around 2000 patient) of the ONCOMINE database to investigate the association between NS (GNL3) gene expression and tumor progression presented as tumor stage and also its association with p53 status.
3. Statistical analysis
The association between NS expression and different clinic-pathological parameters were estimated using chi-square test (χ2) test. P <.05 was considered to be statistically significant.
4. Results
4.1. NS expression in 2 different patients’ cohorts
Initially, we investigated the expression levels of NS in 2 different patients’ cohorts, the UAE cohort (51 cases) and the commercial TMA cohort (102 cases). Our results revealed that NS is expressed in 43.13% of the UAE cohort compared to the 78.35% in the commercial TMA cohort. This difference might be attributed to the fact that patients from UAE cohort had mostly early stage disease compared to more advance disease in the TMA cohort (Tables 1 and 2).
Table 1.
Association between Immunohistochemical expression of NS and the clinicopathological parameters in the UAE cohort.
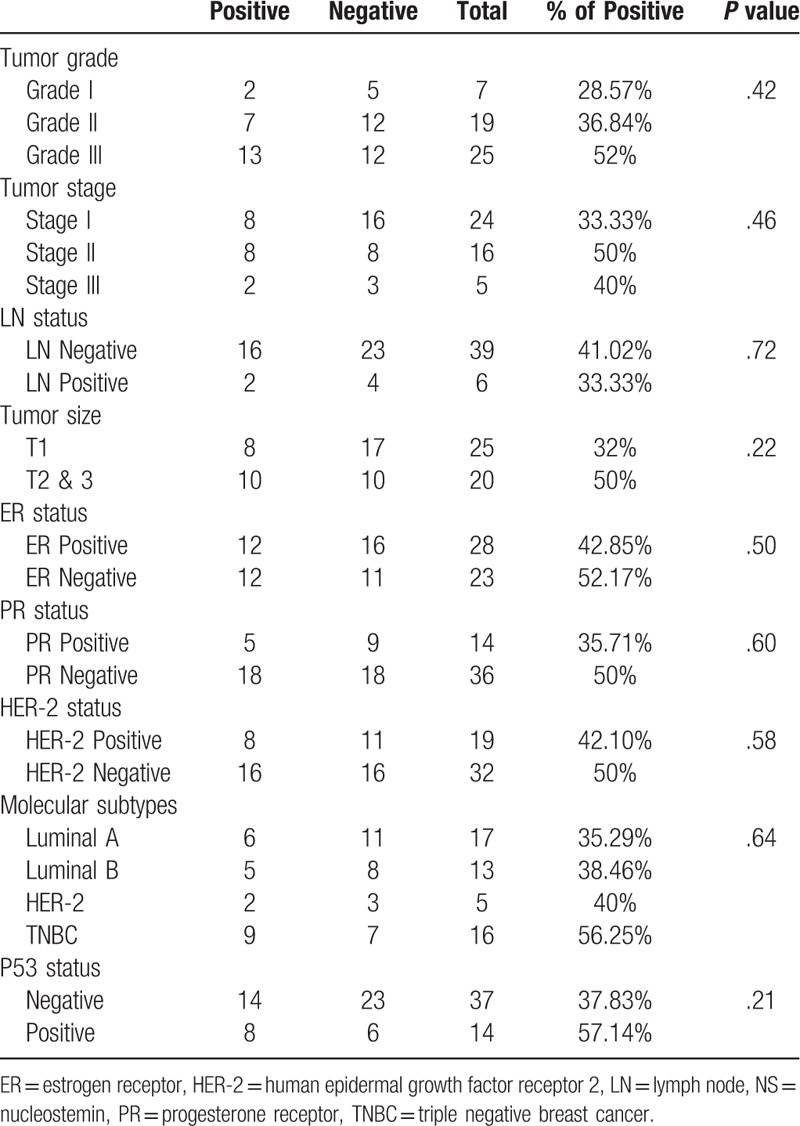
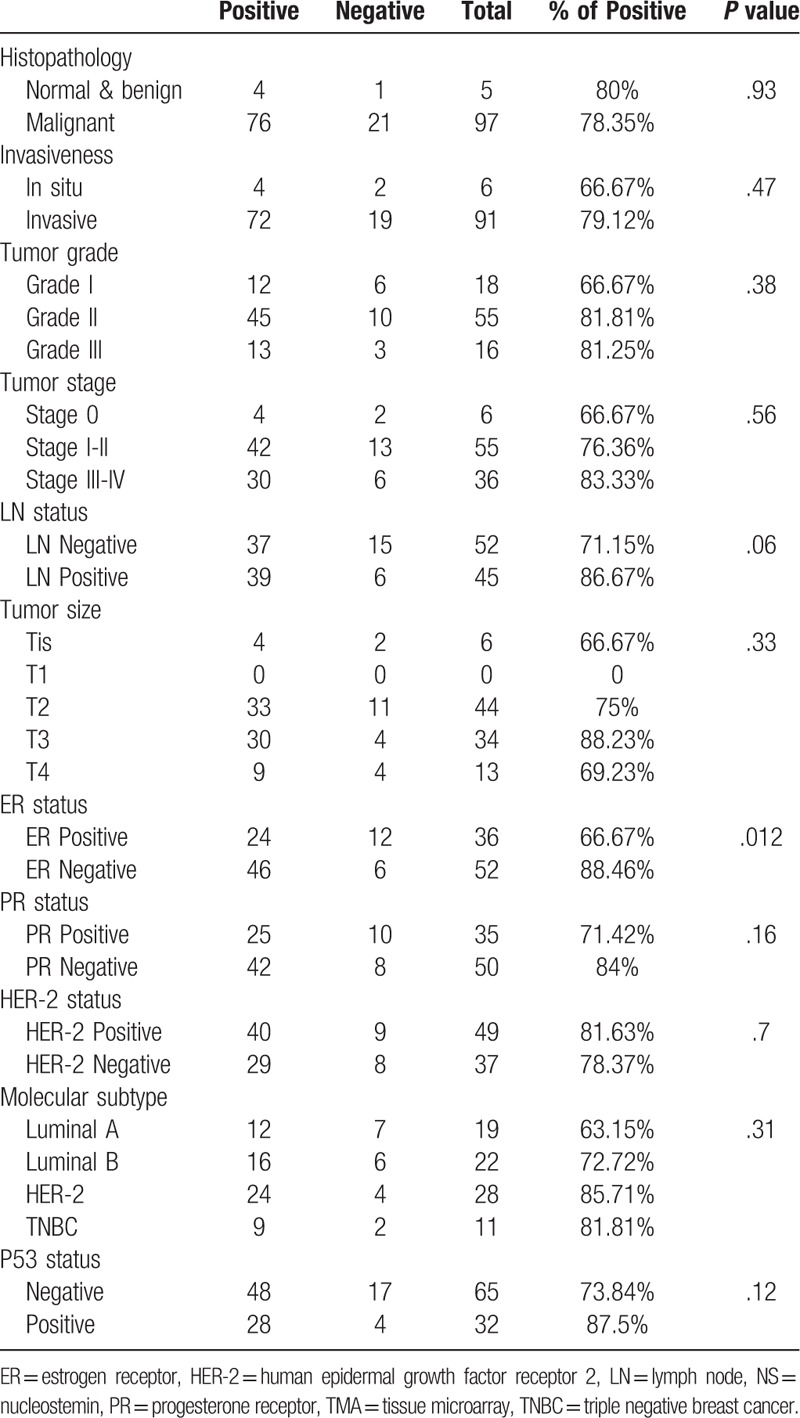
Table 2.
Association between Immunohistochemical expression of NS and the clinicopathological parameters in the commercial TMA.
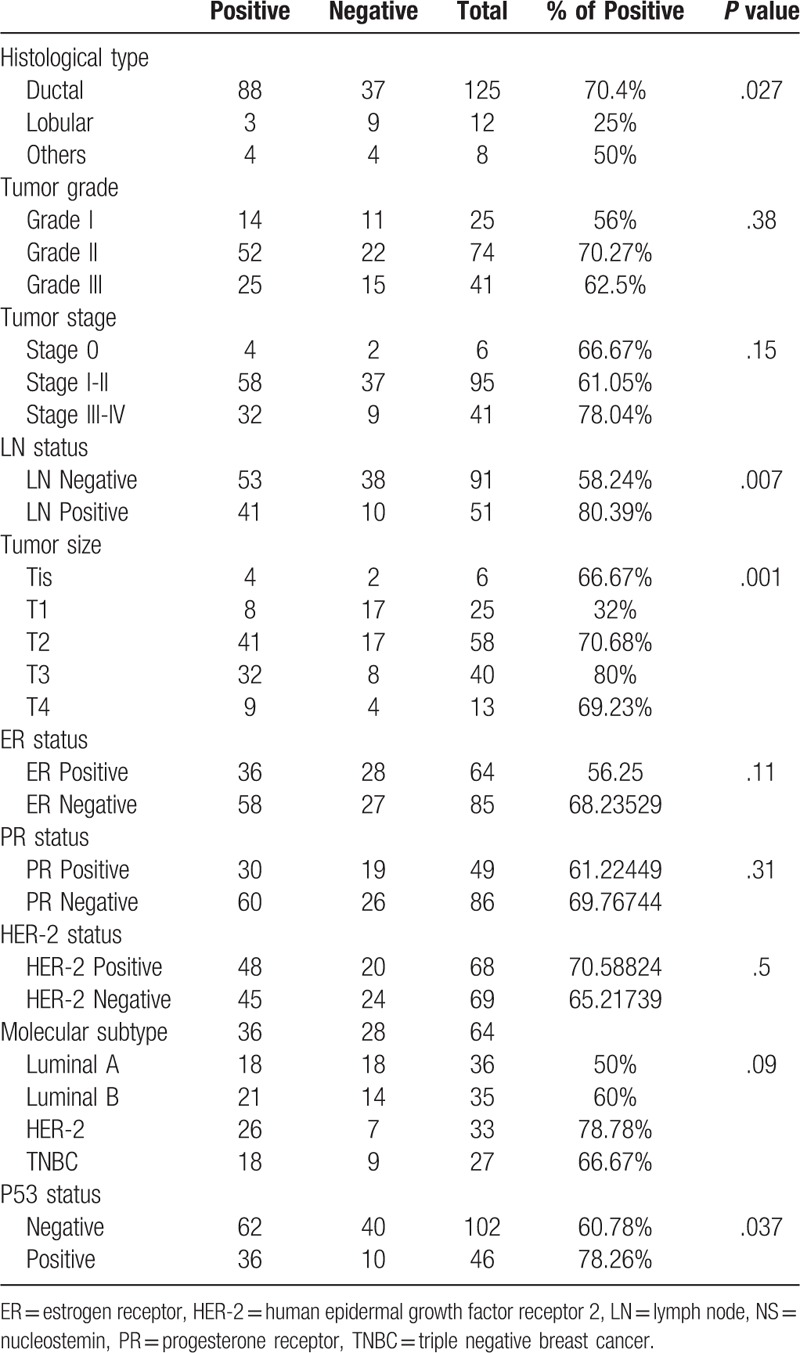
Interestingly, examining the NS status of the entire cohort according to the different histopathological subtypes, revealed that NS was predominantly expressed in ductal carcinomas (70.4%), while lobular carcinomas showed significantly lower expression of NS with only (25%) of cases exhibiting positive NS expression (P = .027) (Table 3).
Table 3.
Immunohistochemical expression of NS in the studied clinicopathological parameters in both cohorts.
4.2. NS expression is associated with poorly differentiated breast cancer
Keeping in mind the chief role of NS in maintaining stemness and the strong association between stemness and loss of differentiation, we examined the association between NS protein expression and tumor grade. Indeed, in UAE cohort, poorly differentiated (grade III) tumors showed around double (52%) the expression of NS compared to the well differentiated (grade I) tumors (28.57%) (Table 1)(Fig. 1A).
Figure 1.
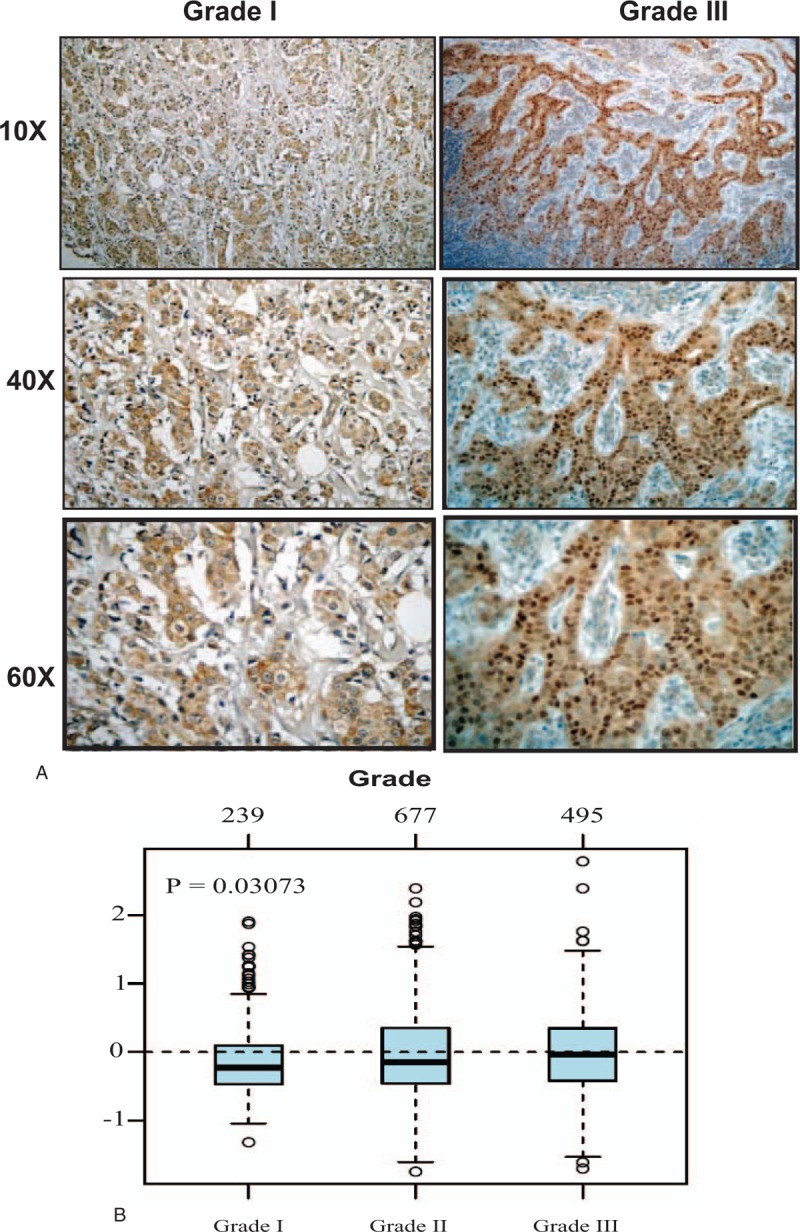
The association between NS expression and tumor grade. (A) Representative pictures of immunohistochemical expression of NS in breast cancer samples of different grades. (B) GNL3 gene expression and its association with tumor grade in 1881 breast cancer samples obtained from GOBO database. GOBO = gene expression based outcome, NS = nucleostemin.
The same trend was also observed in the larger commercial TMA, with the highest NS expression detected in the moderately differentiated and poorly differentiated (grade III) (81.81% and 81.25% respectively) tumors compared to the well differentiated (grade I) tumors (66.67%) (Table 2).
To further analyze this point, we next evaluated the association between NS gene (GNL3) and tumor grade in more than 1881 breast cancer samples using GOBO database. Indeed, GNL3 gene expression showed a significant association (P = .03) with poorly differentiated (grade III) tumors compared to well-differentiated tumors (Fig. 1B).
4.3. NS expression is associated with more advanced stage and larger tumor size
Next, we studied the association between NS and other well established clinicopathological parameters including tumor size, lymph node (LN) involvement as well as tumor stage.
Our results observed more frequent NS positivity with increasing tumor size in both cohorts (Tables 1 and 2). Noticeably, the UAE cohort displayed more NS expression in the LN negative tumors compared to the LN positive tumors (41% and 33.33% respectively) (Table 1), whereas, the larger commercial TMA showed higher NS immunoreactivity in the LN positive tumors (86.67%) compared to the LN negative tumors (71.15%) (P = .06) (Table 2).
Overall, assessing the relation between NS expression and tumor stage, which is regarded as a major indicator of disease progression, revealed that, stage II & III tumors of the UAE cohort showed more frequent NS expression (50% and 40% respectively) compared to stage I tumors (33.33%) (Table 1). The same trend was also observed in the commercial TMA with the highest NS expression detected in the more advanced stage III & IV tumors (83.33%) and least expressed in the early stage 0 cases (66.67%) (Table 2).
Interestingly, when we combined cases from both cohorts (Table 3), we found a significant association between NS immunoreactivity and larger tumor size (P = .001) and positive LN status (P = .007), although this association was not significant when examined in each cohort separately. In addition, NS was also found to be more expressed in the more advanced stages III and IV (78.04%) compared to early stages I and II (61.05%) and in situ (stage 0) (66.67%) tumors of the entire cohort, however, this trend did not reach statistical significance (Table 3) (Fig. 2A).
Figure 2.
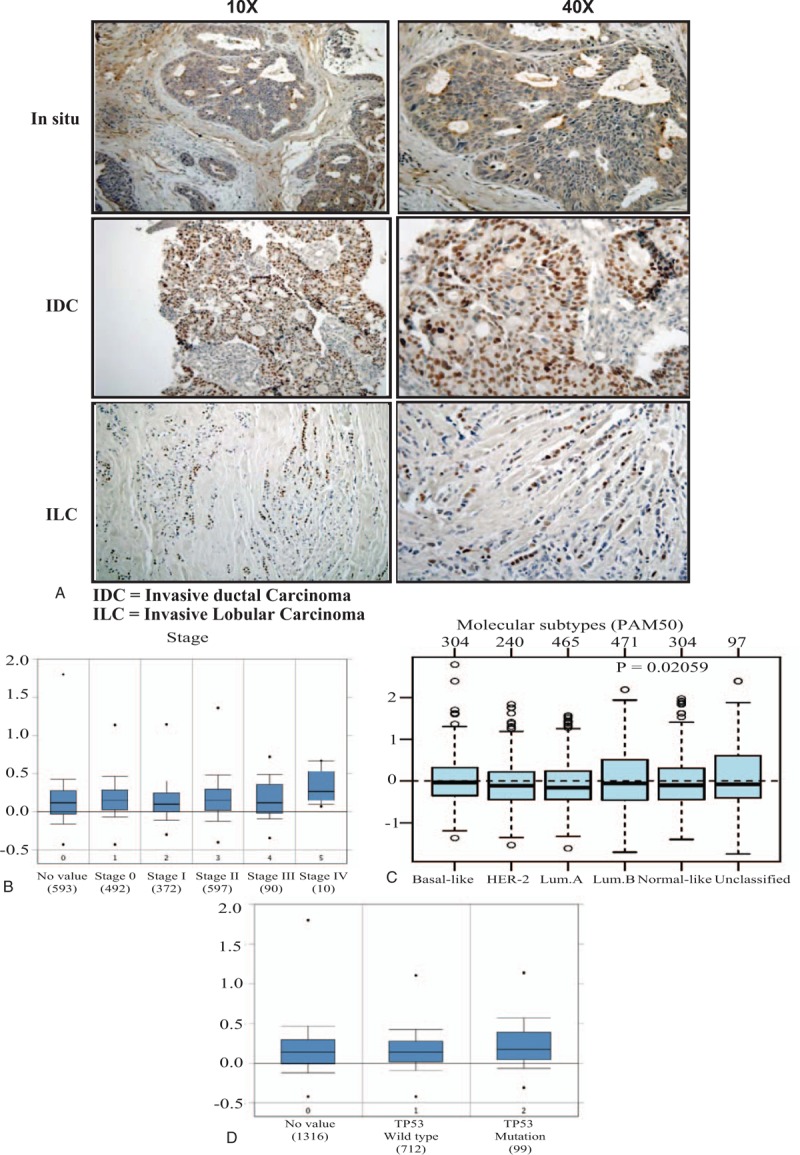
The association between NS expression and tumor stage, molecular subtype and P53 status. (A) Representative pictures of immunohistochemical expression of NS in breast cancer samples from early in situ carcinoma and more advanced invasive ductal and lobular carcinoma. (B) GNL3 gene expression and its association with tumor stage in more than 2000 breast cancer samples obtained from Curtis dataset of ONCOMINE database. (C) GNL3 gene expression and its association with breast cancer molecular subtypes classified according to PAM50 in 1881 breast cancer samples obtained from GOBO database. (D) GNL3 gene expression and its association with P53 status in more than 2000 breast cancer samples obtained from Curtis dataset of ONCOMINE database. GOBO = gene expression based outcome, NS = nucleostemin.
To further explore this point, we next evaluated GNL3 gene expression in more than 2000 breast cancer samples obtained from Curtis dataset of ONCOMINE database (Fig. 2B). As expected, GNL3 expression was highest in the more advanced stage IV tumors in comparison to the less advanced cases (median = 0.3335) and least in the early stage I disease (median = 0.108).
These findings highlight that NS expression is associated with breast cancer progression and advanced disease.
4.4. NS expression is associated with more aggressive breast cancer subtypes
Considering that breast cancer represents a heterogeneous group of diseases with distinct histological, molecular as well as clinical subgroups, we attempted to investigate the expression levels of NS in association with classical markers like ER, PR, and HER-2 as well as its expression in different breast cancer molecular subtypes.
Interestingly, while NS showed no significant association with the PR and HER-2 status, it showed an important association with estrogen receptor (ER) negative status reaching to significant levels in the commercial TMA (P = .012). This association with ER-negative tumor was further supported though our finding that NS expression showed the highest expression in the ER-negative and highly aggressive triple negative breast cancer (TNBC) tumors (56.25%) in the UAE cohort, with lowest expression in the least aggressive, ER-positive, luminal A tumors (35.29%) (Table 1). Furthermore, comparable results were obtained using the larger commercial TMA with the highest NS expression detected in the HER-2 (85.71%) followed by TNBC tumors (81.81%) (Both ER-negative tumors) and the least expression in luminal A tumors (63.15%) (Table 2).
Combined analyses of both cohorts revealed an obvious trend of association between NS expression and the 2 more aggressive (ER-negative) breast cancer subtypes (HER-2 and TNBC) (P = .09) (Table 3).
To further clarify the role of NS in different breast cancer subtypes, we next evaluated the expression levels of GNL3 gene expression in different molecular subtypes (PAM 50 classification) using GOBO database. Indeed, GNL3 expression was shown to be significantly higher in TNBC tumors as well as luminal B tumors with the least expression in the luminal A tumors (P = .02) (Fig. 2 C). This highlights the strong association between NS and its gene with more aggressive subtypes.
4.5. NS expression is associated with P53 positive expression
Previous studies suggested that NS plays an important role in the tumor suppressor p53 inactivation through its interaction with MDM2 leading to cell cycle progression and cell survival.[23,24] Indeed, p53 is known for its short half-life, however, when it is mutated, usually this leads to its stability and accumulation in cells.
As anticipated, UAE and TMA cohorts demonstrated higher NS expression among p53 positive tumors (57.14% and 87.5% respectively) (Tables 1 and 2). Furthermore, when the analysis was done for the entire study cohort, we found a significant association between NS expression and p53 status (P = .037) (Table 3).
This was further confirmed using Curtis breast cancer database, which also revealed that GNL3 gene expression was higher in the cases associated with mutated p53 compared to wild type p53 (Fig. 2 D). This proves that in breast cancer, higher NS expression is associated with p53 mutation and inactivation, which might explain its role in cell cycle progression and cell survival.
4.6. GNL3 gene expression to be associated with poor patients’ outcome represented as shorter distance metastasis survival
For better evaluation of the prognostic role of NS in breast cancer, we next appraised the association between GNL3 gene expression levels and patients’ outcome with relapse-free survival (RFS) as an end point in 1881 breast cancer patient's samples using GOBO database.
Indeed, the results disclosed that high GNL3 gene expression levels are significantly associated with poor patient outcome presented as shorter relapse free survival (RFS) (P = .00034) (Fig. 3A).
Figure 3.
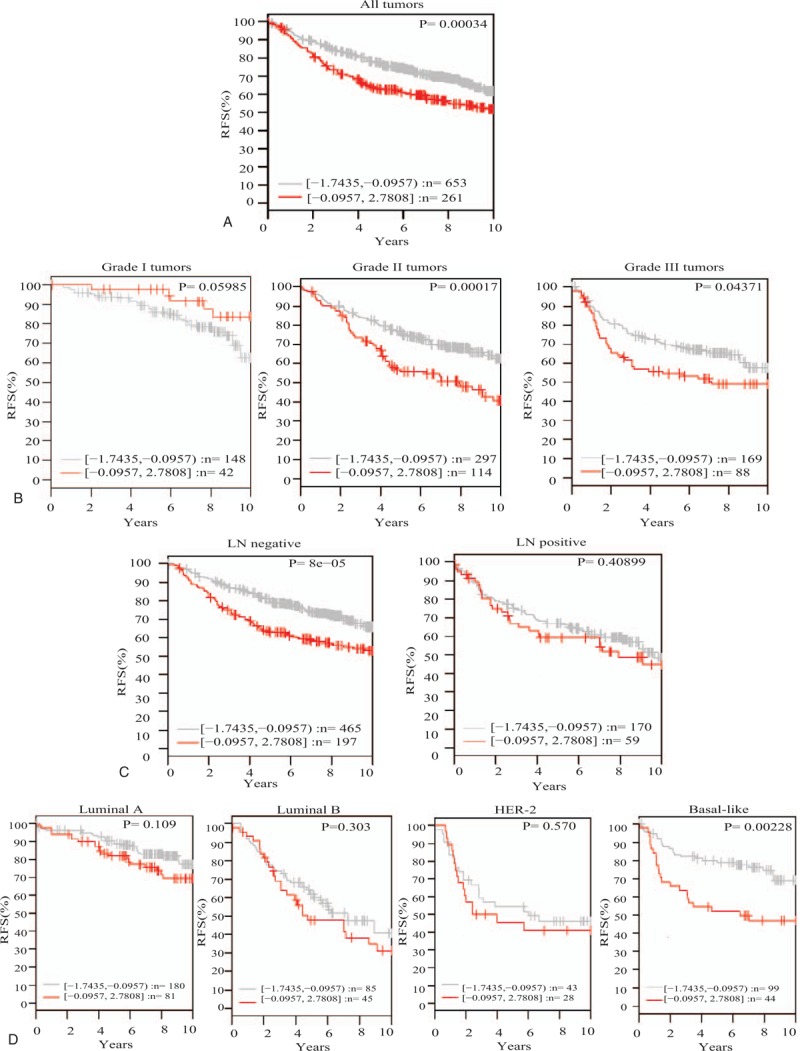
GNL3 gene expression and its association with patient outcome represented as relapse free survival in 1881 breast cancer samples obtained from GOBO database. (A) GNL3 gene expression and its association with patient outcome in the whole patients sample. (B) GNL3 gene expression and its association with patient outcome in different tumor grades. (C) GNL3 gene expression and its association with patient outcome in LN negative and positive samples. (D) GNL3 gene expression and its association with patient outcome in different breast cancer subtypes. GOBO = gene expression based outcome, LN = lymph node, NS = nucleostemin.
Moreover, classification of those patients according to their tumors’ differentiation levels, revealed that while, GNL3 gene expression levels had no effect on patients’ outcome in the well-differentiated tumors, it showed a significant association with poor patients’ outcome in the less-differentiated moderate and poorly differentiated tumors (P = .00017, P = .04371 respectively) (Fig. 3B).
In addition, GNL3 gene expression levels showed a significant association with patient outcome in the early LN negative tumors with no effect on the outcome of LN positive tumors. This indicates that NS might play an important role in early breast cancer tumorogenesis and disease progression (Fig. 3C).
Finally, and owing to the essential and emerging role of breast cancer molecular subtypes in determining patients’ outcome and therapeutic options for breast cancer, next we reviewed the association between GNL3 gene expression levels and patients’ outcome in different breast cancer subtypes. Interestingly, GNL3 gene expression showed no significant association with patients’ outcome in luminal A, B, and HER-2, though it showed significant association with poor patients’ outcome in the highly aggressive, stem cells- enriched, basal-like breast cancer tumors (Fig. 3D).
5. Discussion
Breast CSCs are believed to play a crucial role in breast cancer aggressiveness, in addition to their association with tumor recurrence and drug resistance.[3] Consequently, members of pathways involved in breast cancer stemness might provide ideal candidates to predict patients with higher risk of poor outcome. Earlier research proved that NS is implicated in regulating stem cell maintenance and proliferation in different types of cancer cells.[25] However, its role in the prognosis and prediction of patient outcome in breast cancer patient still needs to be further elucidated.
In this context, we report in the present study, the prognostic value of NS expression in a multiethnic cohort of breast cancer patients from UAE compared to a cohort of commercial TMA of breast cancer. Interestingly, our study showed a variable NS expression among the 2 cohorts. Indeed, UAE cohort showed lower NS expression compared to the commercial TMA, which could be attributed to the difference of clinicopathological features of both cohorts. Particularly, the UAE cohort included predominantly stage I and II tumors, whereas the commercial TMA cohort showed more diversity in the tumors’ stage with around one-third being of the more advanced stage III and IV.
Importantly, we should mention that our report has certain limitations. One of the main limitations we observe is the limited sample size of the UAE patient's cohort, in addition to the limited clinicopathological as well as follow up information available for those patients. This resulted in insignificant association between NS expression and all of the clinic-pathological parameters in the UAE cohort. This limitation associated with sample size is not restricted to our report, but it is also observed in large group of histopathological studies. This is attributed to the fact that many of these reports are retrospective and based upon availability of cases and was not based on sample size calculations essential for primary research question evaluation.[26] However, and to overcome this limitation, we investigate the NS expression in additional cohort (commercial TMA purchased from Pantomics), which contains another 102 cases. Moreover, we thoroughly reviewed and analyzed 2 publicly available databases to explore the correlation of NS expression and the well-known prognostic parameters in around 2000 breast cancer samples. The results from both databases endorsed our results and provided compelling evidence that NS expression in breast cancer samples is associated with loss of tumor differentiation, advanced tumor stage and tumor progression.
The association between high NS expression and more aggressive phenotype was observed in other reports. Indeed, Yoshida et al, 2014[27] reported NS, to be a marker of advanced malignant phenotype in oral squamous cell carcinoma. In addition, and consistent with earlier studies,[27,28] our results also revealed that NS expression is increased with tumor progression presented as larger tumor size, LN metastasis, and advanced tumor stage. However, Kobayashi et al 2014 [12] showed that NS had no significant association with tumor size, LN involvement as well as distant metastasis.
Another noteworthy finding of the present study was the detection of higher NS immunorecativity in the more aggressive molecular subtypes including HER-2 and TNBC, the latter being usually associated with worse prognosis and characterized by high-grade tumors[29] and enriched with stemness and epithelial to mesenchymal transition-related genes.[30] In support of our results, Lin et al 2010 concluded that in mammary tumors, basal cell type expressed more NS than the luminal cell type. Furthermore, NS-enriched mammary tumor cells displayed stronger in vitro and in vivo tumorigenic activities.[31]
Remarkably, our results revealed a significant association between positive NS expression and positive p53 expression, which usually denotes p53 mutation and inactivation. This is in agreement with Kobayashi et al 2014[12] who showed a similar association and demonstrated that tumors characterized by higher levels of co-expression of NS and p53 are associated with worse prognosis presented as disease-free survival (DFS).
In his review, Tsai 2014, discussed the possible interplay between NS and p53 and stated that in cells with wild-type p53, NS depletion turns on p53 and prompt cell cycle arrest, however, this might be completely or partially reversed upon p53 knockdown.[32–34] Furthermore, the fact that NS interacts with MDM2 to modulate the activity of p53 was proved by many authors.[23,24] On the other hand, some authors showed accelerated death of NS-knockout MEF cells upon loss of p53[35] inferring that p53 is not essential for NS function in cell proliferation maintenance.
Finally, we analyzed data of 1881 breast cancer patients to probe the association between NS gene (GNL3) expression levels and patients’ outcome presented as relapse-free survival (RFS), which is an important indicator of tumor recurrence. The results confirmed a significant association between GNL3 expression and poor patients’ outcome presented as shorted RFS. This was more evident in the more aggressive grade II and III as well as basal-like breast cancer subtype. This goes with other reports that also showed NS expression to be a marker of tumor recurrence[28] and poor patient outcome.[12]
In conclusion, our results disclosed that NS protein, as well as gene expression levels, can be used as markers of tumor progression, aggressive histological phenotype as well as higher risk of tumor recurrence in breast cancer, implying that NS may be a potential target for CSC-associated breast cancer management.
Author contributions
Conceptualization: Manal M Sami, Ibrahim Hachim.
Data curation: Manal M Sami, Mahmood Y Hachim, Ibrahim Hachim, Ahmed H Elbarkouky, Vanessa M. López -Ozuna.
Formal analysis: Manal M Sami, Mahmood Y Hachim, Ibrahim Hachim, Ahmed H Elbarkouky, Vanessa M. López -Ozuna.
Investigation: Manal M Sami, Mahmood Y Hachim, Ahmed H Elbarkouky, Vanessa M. López -Ozuna.
Methodology: Manal M Sami, Mahmood Y Hachim.
Project administration: Manal M Sami, Ibrahim Hachim.
Resources: Manal M Sami.
Validation: Manal M Sami, Ibrahim Hachim.
Writing – original draft: Manal M Sami, Mahmood Y Hachim, Ibrahim Hachim.
Writing – review & editing: Manal M Sami, Ibrahim Hachim, Vanessa M. López -Ozuna.
Footnotes
Abbreviations: CSC = cancer stem cell, ER = estrogen receptor, GOBO = gene expression based outcome, HER-2 = human epidermal growth factor receptor 2, LN = lymph node, NS = nucleostemin, PR = progesterone receptor, TMA = tissue microarray, TNBC = triple negative breast cancer.
This study was supported by Research Grant from RAK College of Medical Sciences, RAKMHSU, UAE.
The authors declare no conflict of interest.
References
- [1].Lu J, Steeg PS, Price JE, et al. Breast cancer metastasis: challenges and opportunities. Cancer Res 2009;69:4951–3. [DOI] [PubMed] [Google Scholar]
- [2].Finicelli M, Benedetti G, Squillaro T, et al. Expression of stemness genes in primary breast cancer tissues: the role of SOX2 as a prognostic marker for detection of early recurrence. Oncotarget 2014;5:9678–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Al-Ejeh F, Smart CE, Morrison BJ, et al. Breast cancer stem cells: treatment resistance and therapeutic opportunities. Carcinogenesis 2011;32:650–8. [DOI] [PubMed] [Google Scholar]
- [4].Ahmad A. Pathways to breast cancer recurrence. ISRN Oncol 2013;2013:290568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Anderson EC, Hessman C, Levin TG, et al. The role of colorectal cancer stem cells in metastatic disease and therapeutic response. Cancers (Basel) 2011;3:319–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer 2005;5:275–84. [DOI] [PubMed] [Google Scholar]
- [7].Badve S, Nakshatri H. Breast-cancer stem cells-beyond semantics. Lancet Oncol 2012;13:e43–8. [DOI] [PubMed] [Google Scholar]
- [8].Ali HR, Dawson SJ, Blows FM, et al. Cancer stem cell markers in breast cancer: pathological, clinical and prognostic significance. Breast Cancer Res 2011;13:R118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhu Q, Yasumoto H, Tsai RY. Nucleostemin delays cellular senescence and negatively regulates TRF1 protein stability. Mol Cell Biol 2006;26:9279–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Meng L, Hsu JK, Zhu Q, et al. Nucleostemin inhibits TRF1 dimerization and shortens its dynamic association with the telomere. J Cell Sci 2011;124:3706–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lin T, Meng L, Lin TC, et al. Nucleostemin and GNL3L exercise distinct functions in genome protection and ribosome synthesis, respectively. J Cell Sci 2014;127:2302–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kobayashi T, Masutomi K, Tamura K, et al. Nucleostemin expression in invasive breast cancer. BMC Canc 2014;14:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bernardi R, Pandolfi PP. The nucleolus: at the stem of immortality. Nat Med 2003;9:24–5. [DOI] [PubMed] [Google Scholar]
- [14].Tsai RY. Turning a new page on nucleostemin and self-renewal. J Cell Sci 2014;127:3885–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yoshida R, Fujimoto T, Kudoh S, et al. Nucleostemin affects the proliferation but not differentiation of oral squamous cell carcinoma cells. Cancer Sci 2011;102:1418–23. [DOI] [PubMed] [Google Scholar]
- [16].O’Brien CA, Pollett A, Gallinger S, et al. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007;445:106–10. [DOI] [PubMed] [Google Scholar]
- [17].Eramo A, Lotti F, Sette G, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ 2008;15:504–14. [DOI] [PubMed] [Google Scholar]
- [18].Tamase A, Muraguchi T, Naka K, et al. Identification of tumor-initiating cells in a highly aggressive brain tumor using promoter activity of nucleostemin. Proc Natl Acad Sci U S A 2009;106:17163–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yuan F, Cheng Q, Li G, et al. Nucleostemin knockdown sensitizes hepatocellular carcinoma cells to ultraviolet and serum starvation-induced apoptosis. PLoS One 2015;10:e0141678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sami MM, Hachim IY, Elbarkouky AH. Breast cancer profile in Ras Al Khaimah, United Arab [Emirates–a histopathological and immunohistochemical study. Hamdan Med J 2014;7:1–24. [Google Scholar]
- [21].Hachim IY, Hachim MY, Lopez VM, et al. Prolactin receptor expression is an independent favorable prognostic marker in human breast cancer. Appl Immunohistochem Mol Morphol 2016;24:238–45. [DOI] [PubMed] [Google Scholar]
- [22].Ringner M, Fredlund E, Häkkinen J, et al. GOBO: gene expression-based outcome for breast cancer online. PLoS One 2011;6:e17911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dai MS, Sun XX, Lu H. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol Cell Biol 2008;28:4365–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Meng L, Lin T, Tsai RY. Nucleoplasmic mobilization of nucleostemin stabilizes MDM2 and promotes G2-M progression and cell survival. J Cell Sci 2008;121:4037–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tsai RY, McKay RD. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev 2002;16:2991–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Knijn N, Simmer F, Nagtegaa ID. Recommendations for reporting histopathology studies: a proposal. Virchows Arch 2015;466:611–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yoshida R, Nakayama H, Nagata M, et al. Overexpression of nucleostemin contributes to an advanced malignant phenotype and a poor prognosis in oral squamous cell carcinoma. Br J Cancer 2014;111:2308–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Nakajima TE, Yoshida H, Okamoto N, et al. Nucleostemin and TWIST as predictive markers for recurrence after neoadjuvant chemotherapy for esophageal carcinoma. Cancer Sci 2012;103:233–8. [DOI] [PubMed] [Google Scholar]
- [29].Carey L, Winer E, Viale G, et al. Triple-negative breast cancer: disease entity or title of convenience. Nat Rev Clin Oncol 2010;7:683–92. [DOI] [PubMed] [Google Scholar]
- [30].Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist 2010;15suppl 5:39–48. [DOI] [PubMed] [Google Scholar]
- [31].Lin T, Meng L, Li Y, et al. Tumor-initiating function of nucleostemin-enriched mammary tumor cells. Cancer Res 2010;70:9444–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ma H, Pederson T. Depletion of the nucleolar protein nucleostemin causes G1 cell cycle arrest via the p53 pathway. Mol Biol Cell 2007;18:2630–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Paridaen JT, JansonF E, Utami KH, et al. The nucleolar GTP-binding proteins Gnl2 and nucleostemin are required for retinal neurogenesis in developing zebrafish. Dev Biol 2011;355:286–301. [DOI] [PubMed] [Google Scholar]
- [34].Yamashita M, Nitta F, Nagamatsu G, et al. Nucleostemin is indispensable for the maintenance and genetic stability of hematopoietic stem cells. Biochem Biophys Res Commun 2013;441:196–201. [DOI] [PubMed] [Google Scholar]
- [35].Meng L, Lin T, Peng G, et al. Nucleostemin deletion reveals an essential mechanism that maintains the genomic stability of stem and progenitor cells. Proc Natl Acad Sci U S A 2013;110:11415–20. [DOI] [PMC free article] [PubMed] [Google Scholar]


